Sex-biased T-cell exhaustion in glioblastoma is, in part, regulated by T cell–intrinsic factors, such as the X chromosome inactivation escape gene Kdm6a, which highlights the potential for a precision therapy approach based on patient biologic sex.
Abstract
Sex differences in glioblastoma (GBM) incidence and outcome are well recognized, and emerging evidence suggests that these extend to genetic/epigenetic and cellular differences, including immune responses. However, the mechanisms driving immunologic sex differences are not fully understood. Here, we demonstrate that T cells play a critical role in driving GBM sex differences. Male mice exhibited accelerated tumor growth, with decreased frequency and increased exhaustion of CD8+ T cells in the tumor. Furthermore, a higher frequency of progenitor exhausted T cells was found in males, with improved responsiveness to anti–PD-1 treatment. Moreover, increased T-cell exhaustion was observed in male GBM patients. Bone marrow chimera and adoptive transfer models indicated that T cell–mediated tumor control was predominantly regulated in a cell-intrinsic manner, partially mediated by the X chromosome inactivation escape gene Kdm6a. These findings demonstrate that sex-biased predetermined behavior of T cells is critical for inducing sex differences in GBM progression and immunotherapy response.
Significance:
Immunotherapies in patients with GBM have been unsuccessful due to a variety of factors, including the highly immunosuppressive tumor microenvironment in GBM. This study demonstrates that sex-biased T-cell behaviors are predominantly intrinsically regulated, further suggesting sex-specific approaches can be leveraged to potentially improve the therapeutic efficacy of immunotherapy in GBM.
See related commentary by Alspach, p. 1966.
This article is featured in Selected Articles from This Issue, p. 1949
INTRODUCTION
Glioblastoma (GBM) is the most common primary malignant brain tumor, and patients with GBM experience poor prognosis despite aggressive current standard-of-care therapies, including surgical resection, radiotherapy, and chemotherapy with temozolomide (1). One reason that GBM is difficult to treat is its highly immunosuppressive tumor microenvironment (TME). GBM tumors are infiltrated with suppressive myeloid populations, including macrophages, myeloid-derived suppressor cells (MDSC), and microglia (2). A reduction in T cells in the circulation further contributes to poor anti-GBM immunity, with sequestration of naive T cells in the bone marrow and involution of primary and secondary lymphoid organs suggested as mechanisms of T-cell reduction (3, 4). Clinical trials in GBM using immune-checkpoint inhibitors (ICI) such as anti–PD-1 and anti-CTLA4 monoclonal antibodies (mAb) have generally not shown significant improvement in overall survival, even when used in combination therapies with existing treatment options (5–7), with the exception of a single trial that demonstrated a modest but statistically significant survival benefit when ICI was used in the neoadjuvant setting (8). Given the unique TME and anatomic immune privilege in GBM, there is a pressing need to understand how to reinvigorate immune responses in GBM.
Adding to the complexity of GBM treatment is a sex difference in disease outcome, with male patients exhibiting a 1.6-fold higher incidence and poorer prognosis after treatment compared with female patients (9). Although tumor-intrinsic factors underlying sex differences have been identified in GBM (10, 11), sex differences in antitumor immunity may also contribute. In general, females exhibit stronger immune responses than males, as mostly shown in autoimmune and infectious diseases, and the differences are attributed to sex hormones and/or sex chromosomes (12). Overall, male-biased prevalence of cancers in nonreproductive organs has been reported (13), yet the underlying mechanisms remain to be elucidated.
In many tumors, T-cell function is disrupted, and addressing this has been the focus of many immunotherapies. T-cell exhaustion refers to a dysfunctional state of T cells that is characterized by high expression of inhibitory receptors, poor effector function, and decreased proliferative potential and is mediated by epigenetic remodeling (14). Chronic antigen stimulation in infection and cancers induces T-cell exhaustion, resulting in impaired control of disease. Exhausted T cells are comprised of two distinct subsets: stem-like/progenitor exhausted T cells (PEX) with self-renewal properties, which then transition to the irreversible stage of terminally exhausted T cells (TEX). Progenitor exhausted T cells can temporarily differentiate into “effector-like” T cells in response to anti–PD-1 blockade that effectively inhibits tumor growth (15, 16). Male-biased T-cell exhaustion was observed in a variety of cancers, including melanoma and bladder cancer (17, 18), demonstrating that sex differences in T-cell exhaustion led to divergent disease outcomes in males and females. While GBM is highly enriched with exhausted T cells (19), it remains unknown the extent to which sex-biased T-cell exhaustion mediates the sex differences in GBM survival.
We previously demonstrated sex-specific behaviors of MDSC subsets in GBM (20). In this study, we hypothesized that the sex differences in response to GBM extend beyond the myeloid lineage and, here, report sex differences in T-cell exhaustion that underlie differential ICI response in GBM.
RESULTS
T Cells Are a Critical Driver of Sex Differences in GBM
We have previously shown sex differences in survival using the syngeneic mouse glioma models SB28 and GL261 (20), with male mice experiencing a worse outcome than female mice. To investigate the role of immune cell populations in this sex-based survival difference, we utilized immunocompromised mouse strains with different degrees of immunodeficiency. Intracranial tumor implantation revealed that the sex-biased survival difference observed in wild-type mice was not present in immune-deficient NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice (Fig. 1A; Supplementary Fig. S1A). We also did not observe any difference in survival of NSG mice challenged with a lower number of tumor cells (Supplementary Fig. S1B), indicating the abolished sex difference was not due to accelerated tumor growth in immune-deficient models. To further specify the immune cell population responsible for the survival difference observed in immunocompetent B6 mice, we used RAG1−/− mice that lack only mature T and B lymphocytes, whereas innate immune cells remain intact. We observed no sex difference in survival outcomes in tumor-bearing RAG1−/− mice (Fig. 1A; Supplementary Fig. S1A), suggesting a role for lymphocytes. The male-biased aggressive tumor growth was not due to elevated female immune responses against male-specific antigens expressed by the tumor cells (SB28 and GL261), as we confirmed that neither tumor cell line contains a Y chromosome (21) or expresses Y chromosome–encoded genes, particularly the H-Y minor histocompatibility antigen encoding gene (smcy; Supplementary Fig. S2A and S2B).
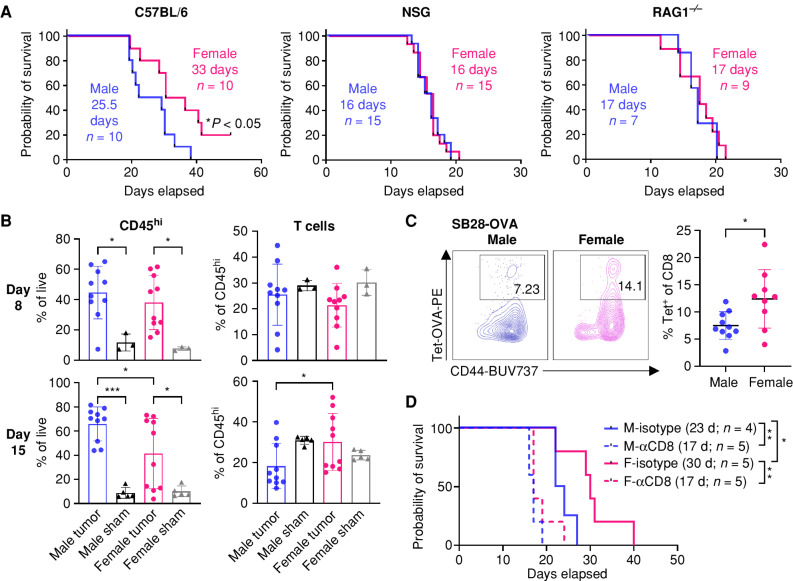
Figure 1.
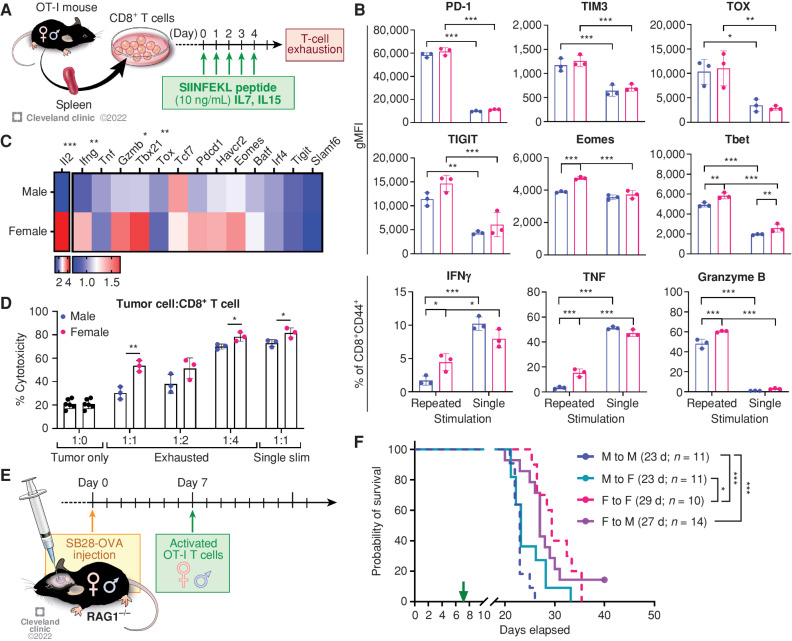
T cells drive sex differences in GBM survival. A, Survival analysis was performed after intracranial injection of the mouse GBM cell line SB28 in immunocompetent B6 mice (15,000 cells/injection) and immune-deficient NSG (10,000 cells/injection) and RAG1−/− mice (15,000 cells/injection). Median survival days and number of animals are indicated in the graph. Data combined from two to three independent experiments. Statistical significance was determined by log-rank test, considering P < 0.05 to be significant. B, Frequency of CD45hi immune cells and CD3+ T cells from the tumor-bearing left hemisphere of SB28-injected mice or the left hemisphere of the sham-injected group on days 8 and 15. Data are shown as mean ± SD from two independent experiments. n = 10 for SB28-bearing mice and n = 3–5 for sham-injected mice. One-way ANOVA with the Tukey multiple comparisons test was performed to determine statistical significance (*, P < 0.05; ***, P < 0.001). C, Frequency of OVA-specific CD8+ T cells was measured using tetramer (Tet) antibody from the tumor-bearing left hemisphere of SB28-OVA (25,000 cells/mouse)–injected B6 wild-type mice on day 14 after tumor implantation. Data are shown as mean ± SD from two independent experiments. n = 9–10/group. *, P < 0.05 was determined by an unpaired t test. D, Kaplan–Meier curves depicting survival of SB28-bearing male and female mice treated with anti-CD8–depleting antibody. Log-rank test was performed to determine statistical significance (*, P < 0.05; **, P < 0.01). d, days; F, female; M, male.
To understand immunologic differences between male and female hosts, we profiled tumor-infiltrating immune cells using flow cytometry at two different time points (Fig. 1B; Supplementary Fig. S3A and S3B). On day 8 after tumor implantation, when T cells are expected to be fully primed and activated, there was increased CD45hi immune cell infiltration with tumor compared with the sham group (Fig. 1B). Males and females showed comparable levels of total immune cells as well as T cells at this early time point. On day 15 after tumor implantation, when some mice start to show neurologic symptoms as a result of an advanced tumor, we found that CD45hi immune cells were significantly higher in males compared with females. Further analysis revealed that CD45hi immune cells in female tumors were more enriched in T cells (Fig. 1B), whereas male tumors had a higher ratio of macrophages (Supplementary Fig. S3B). We did not observe any major differences in other immune cell subsets on either day 8 or day 15 (Supplementary Fig. S3B).
To test whether sex differences exist in tumor-specific T-cell responses, we measured the ovalbumin (OVA)-specific CD8+ T-cell population using a tetramer antibody after implantation of OVA-expressing tumor cells (SB28-OVA) into male and female wild-type B6 mice, in which the sex difference in survival was observed (Supplementary Fig. S4A). Similar to polyclonal T cells (Fig. 1B), OVA-specific CD8+ T cells were significantly higher in female mice (Fig. 1C).
To further investigate the extent to which the sex difference in survival was specifically driven by CD8+ T cells, we depleted CD8+ T cells using anti-CD8–depleting antibody prior to tumor implantation in wild-type B6 mice. The survival advantage of female mice was completely abrogated when CD8+ T cells were depleted in both models, SB28 (Fig. 1D) and GL261 (Supplementary Fig. S4B), suggesting a crucial role for CD8+ T cells in driving the sex differences in survival observed in mouse GBM models.
Male T Cells Become Exhausted More Quickly than Female T Cells, with a Higher Frequency of Progenitor Exhausted T-cell Subsets
We next performed extensive immune cell profiling focused on T cells to understand how T cells underlie the sex differences in GBM progression. Higher frequencies of CD8+ and CD4+ T-cell populations were observed in CD45hi immune cells infiltrated into female tumors compared with male tumors on day 15 after tumor implantation, whereas the frequency of Foxp3+ cells was comparable, indicating that the differences are likely from effector T cells (Fig. 2A). Next, we asked whether female T cells were primed and activated earlier, thereby more effectively attenuating tumor growth. To address this possibility, we evaluated activated T-cell phenotypes in the draining lymph nodes at an earlier time point (day 8) after tumor implantation. No differences in phenotype or functionality (Supplementary Fig. S5A) or in tumor-specific tetramer+ T cells (Supplementary Fig. S5B) were observed between male and female T cells in draining lymph nodes, indicating that differential kinetics of T-cell activation was likely not the reason for the sex differences in T-cell infiltration into tumors.
Figure 2.
More male CD8+ T cells are functionally exhausted and skewed toward a progenitor exhausted T-cell phenotype. Tumor-infiltrating T cells were analyzed on day 15 after implantation of SB28 tumor cells. A, Frequency of T-cell subsets in CD45hi immune cells. Data are combined from two independent experiments. n = 11–12 for the SB28-injected group and n = 4 for the sham-injected group. B, Inhibitory receptor expression in CD8+ and CD4+Foxp3− effector T cells (Teff). Data are combined from two independent experiments. n = 10–12 for the SB28-injected group and n = 4 for the sham-injected group. C, Intracellular cytokine expression in CD8+ and CD4+Foxp3− effector T cells was measured after ex vivo stimulation. Data are combined from two independent experiments. n = 7–10 for the SB28-injected group and n = 3 for the sham-injected group. D, Exhausted T-cell subsets in CD8+ T cells: TEX (CD8+CD44+PD-1+TCF1−TIM3+), PEX (CD8+CD44+PD-1+TCF1+TIM3−), and EFF (CD8+CD44+TCF1−TIM3−). Data are combined from two independent experiments. n = 9–10 for the SB28-injected group and n = 4 for the sham-injected group. E, TOX expression in each CD8+ T-cell subset. gMFI, geometric mean fluorescence intensity. Intracellular expression of granzyme B (F) and IFNγ+TNF+ (G) in each CD8+ T-cell subset after ex vivo stimulation. Data are combined from two independent experiments. n = 5–7 for SB28-injected group. Two-way ANOVA with the Tukey multiple comparisons test (A–C and E–G) or unpaired Student t test (D) was performed (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Further analysis revealed that male CD8+ T cells express a higher frequency of inhibitory receptors, including PD-1, CTLA4, and LAG3, but not TIM3, compared with female CD8+ T cells (Fig. 2B). Additionally, male CD8+ T cells showed decreased levels of intracellular cytokine expression compared with female CD8+ T cells following ex vivo stimulation (Fig. 2C). Notably, loss of TNF occurs at an earlier stage of exhaustion, whereas IFNγ production is maintained after exhaustion (16, 22). CD4+Foxp3− effector T cells showed differences such as expression of CTLA4 and LAG3, but these differences were not as prominent as in CD8+ T cells (Fig. 2B). The differences in inhibitory receptor and cytokine expression were not observed at the early time point (day 8; Supplementary Fig. S5C–S5E), consistent with the findings in Fig. 1B. We also confirmed these findings using another GBM model, GL261, which showed no clear difference in T-cell frequency but differences in inhibitory receptor expression as well as TNF expression (Supplementary Fig. S6A–S6C). These phenotypic differences were observed only in T cell–infiltrating tumors, as no significant changes in inhibitory receptor and cytokine expression were found in T cells from blood and bone marrow (Supplementary Fig. S7A–S7D).
The increased expression of inhibitory receptors and decreased cytokine expression in male CD8+ T cells prompted us to hypothesize that male and female CD8+ T cells exhibit differential exhaustion status. It is well established that exhausted CD8+ T cells are comprised of two subsets, stem-like/progenitor exhausted CD8+ T cells (PEX; CD8+CD44+PD-1+TCF1+TIM3−) and terminally exhausted CD8+ T cells (TEX; CD8+CD44+PD-1+TCF1−TIM3+; ref. 16). Therefore, we evaluated the frequency of exhausted T-cell subsets and effector T cells (EFF; CD8+CD44+TCF1−TIM3−) in tumor-infiltrating CD8+ T cells (Supplementary Fig. S8A). Strikingly, male CD8+ T cells contained a significantly higher ratio of PEX compared with female CD8+ T cells, whereas female T cells had elevated frequencies of EFF (Fig. 2D). A higher frequency of the PEX subset in males was also observed in the GL261 model (Supplementary Fig. S8B). No difference was observed in the TEX population between male and female CD8+ T cells (Fig. 2D), as well as at the early time point (Supplementary Fig. S8C). TOX, a defined T-cell exhaustion marker (16), was highly expressed in the TEX subset, whereas its expression was significantly higher in male TEX compared with their female counterparts (Fig. 2E). Intracellular expression of granzyme B confirmed the gating strategy for exhausted T-cell subsets, as TEX showed the highest level of granzyme B, whereas PEX showed minimal expression as described previously (ref. 16; Fig. 2F). As exhausted CD8+ T cells have reduced capacity to produce multiple cytokines (16), we evaluated the proportion of cells simultaneously producing IFNγ and TNF. Interestingly, all three subsets of female CD8+ T cells were more polyfunctional, suggesting that the fundamental sex difference in T-cell functionality exists independent of exhaustion status (Fig. 2G). Collectively, these data indicate that male and female T cells undergo exhaustion at different rates, with higher PEX in males and higher effector cytokine production in female CD8+ T cells, which may result in survival differences.
Males Benefit from Anti–PD-1 Therapy More than Females
PEX are better able to control tumor growth and respond to anti–PD-1 treatment (15, 16). Thus, we hypothesized that males will respond better than females to anti–PD-1 mAb treatment due to the high frequency of progenitor cells in male tumors. To test this possibility, we treated male and female mice bearing SB28 tumors with anti–PD-1 mAb or isotype antibody (Fig. 3A). In accordance with our prediction, anti–PD-1 mAb treatment significantly extended the survival of male mice compared with the isotype-treated group (Fig. 3B). However, the treatment effect was mild in females, as previously reported (23).
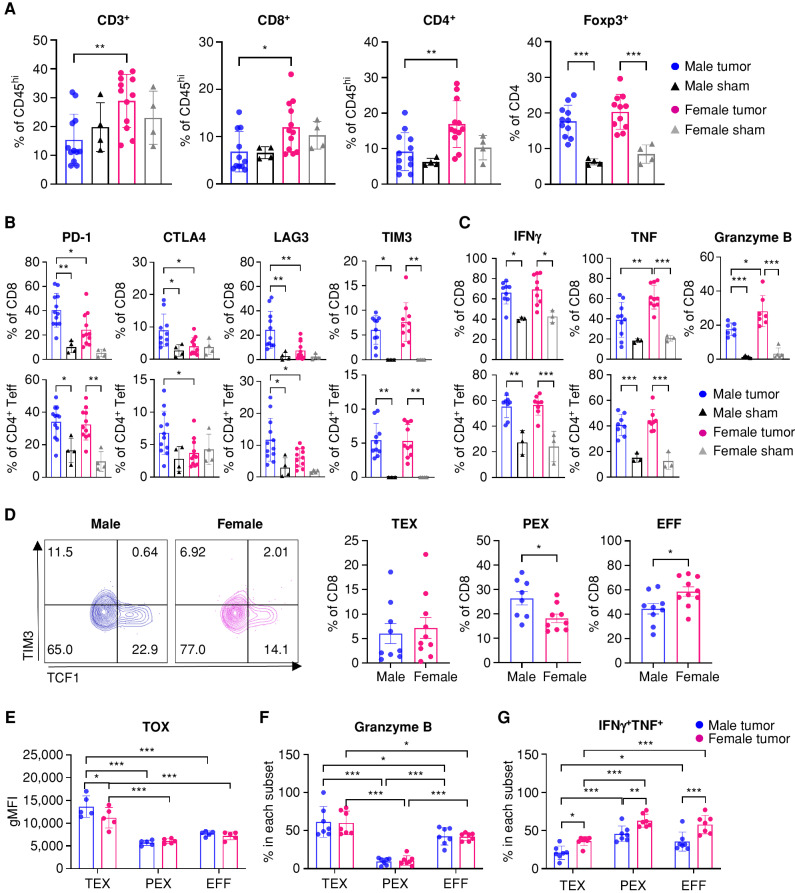
Figure 3.
Males are more responsive to anti–PD-1 therapy. A, Schematics depicting treatment regimen for anti–PD-1 and immune profiling. B, Kaplan–Meier curves depicting survival of male and female SB28-bearing mice treated with anti–PD-1 or isotype antibodies (10 mg/kg) starting from day 7 after intracranial tumor implantation. Combined results from three independent experiments with log-rank test (**, P < 0.01; ***, P < 0.001). Median survival length and number of animals are indicated. d, days; F, female; M, male. C–F, Immunophenotyping was performed on tumor-infiltrating immune cells on day 18, 2 days after the last treatment. Data are combined from two independent experiments. n = 9–10 for the anti–PD-1 treatment group and n = 7–8 for the isotype antibody-treated group. C, Percentage of CD8+ T cells in CD45hi cells. D, Proliferation marker Ki-67 expression in CD8+ T cells. Data are shown as mean ± SD of n = 5/group from one of two independently repeated experiments. E, Frequency of exhausted T-cell subsets in CD8+ T cells: TEX, PEX, and EFF. F, Percentages of intracellular CD8+ T cells expressing IFNγ, TNF, and granzyme B. Two-way ANOVA with the Tukey multiple comparisons test was performed (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
To further interrogate the survival differences between males and females in response to anti–PD-1 mAb treatment, we analyzed tumor-infiltrating T cells using flow cytometry 2 days after the last treatment. The frequency of tumor-infiltrating CD8+ T cells was increased in both males and females with anti–PD-1 mAb treatment (Fig. 3C), whereas other immune cells in the tumor showed a similar trend regardless of sex (Supplementary Fig. S9A). Expression of PD-1, CTLA4, LAG3, and TIM3 was not altered (Supplementary Fig. S9B). Interestingly, we observed a significant increase of Ki-67+ CD8+ T cells (Fig. 3D) and CD4+ effector T cells (Supplementary Fig. S9C) in males, suggesting that male T cells became more proliferative upon anti–PD-1 mAb treatment. PD-1 blockade also led to a significant decrease in the TEX and PEX subsets in males, whereas the decrease was moderate in females (Fig. 3E). Cytokine expression was elevated in both male and female CD8+ T cells (Fig. 3F), but not in CD4+ effector T cells (Supplementary Fig. S9D), with anti–PD-1 mAb treatment. We also evaluated immunologic changes on day 14 (after 3 doses of anti–PD-1 mAb) and found a significant reduction in the PEX subset and an increase in the EFF subset with PD-1 treatment in both sexes (Supplementary Fig. S9E). Additionally, decreased expression of TOX and TIGIT was found in female CD8+ T cells, whereas a transcription factor related to T-cell effector function, Eomes, but not Tbet, was increased in both sexes (Supplementary Fig. S9F).
To test whether anti–PD-1 treatment directly affects T cells already infiltrated into the tumor and not by recruiting newly primed T cells from lymph nodes, we treated male mice with FTY720. FTY720 treatment sequesters T cells in lymph nodes, thereby blocking further T-cell infiltration into the tumor (24). Combining anti–PD-1 antibody with FTY720 did not abrogate the survival benefit in male mice compared with PD-1 blockade alone (Supplementary Fig. S9G), indicating PD-1 blockade was able to reinvigorate the effector function of T cells in the tumor.
Taken together, these results indicate that anti–PD-1 blockade was more effective on male T cells, with significant changes in reducing exhausted T-cell subsets and increasing proliferation compared with females, which may lead to survival benefit upon treatment in males.
Immune Cell–Intrinsic Regulation of Sex Differences in GBM
Next, we asked whether the sex differences in T-cell phenotypes and GBM survival were driven by a hematopoietic immune cell–intrinsic or cell–extrinsic manner. To test this, we generated sex-matched or mismatched bone marrow chimeras by transferring T cell–depleted bone marrow cells into lethally irradiated recipient mice (Fig. 4A). We depleted preexisting T cells in the bone marrow to prevent induction of graft-versus-host disease in the female-to-male (F to M) group as well as to newly generate T cells from the donor hematopoietic stem cells and fully matured in the recipients’ thymuses (Supplementary Fig. S10A). The immune components of the recipients were successfully reconstituted by the donor cells after 6 weeks (CD45.1+ cells >95%), and no significant difference in the frequency of circulating immune cells was observed at a steady state (Supplementary Fig. S10B).
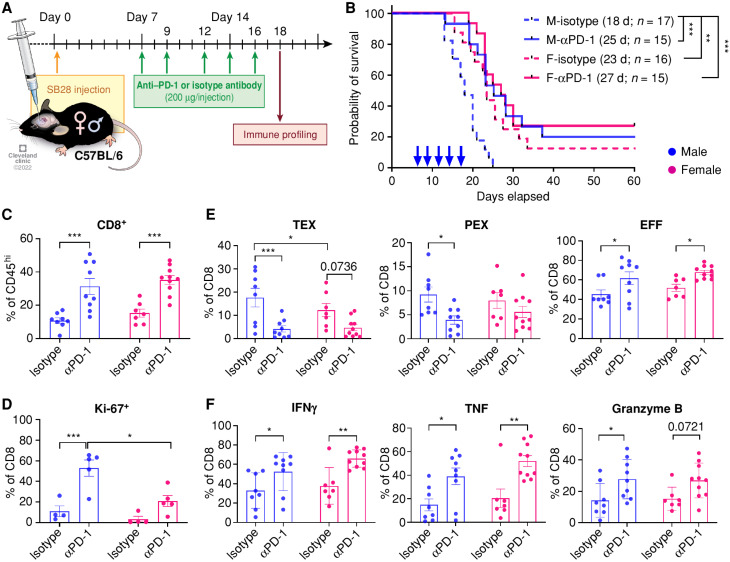
Figure 4.
Immune cell–intrinsic and cell–extrinsic effect in GBM survival. A, A schematic of the generation of bone marrow chimera models. Immune profiling was performed from the tumor-bearing hemisphere on day 14 after tumor implantation (SB28, 10,000 cells/injection). i.c., intracranial. B, Frequency of T-cell subsets in CD45hi cells. Data are combined from four independent experiments. n = 8–18 per group. One-way ANOVA with the Tukey multiple comparisons test (*, P < 0.05; **, P < 0.01). C, Percentage of exhausted T-cell subsets in CD8+ T cells. Data are combined from three independent experiments. n = 4–11 per group. One-way ANOVA with Tukey multiple comparisons test (*, P < 0.05; **, P < 0.01). D, Kaplan–Meier curves depicting survival of bone marrow chimeras after intracranial injection of SB28 cells. Data are combined from three independent experiments. n = 23–25 per group. Statistical significance was determined by the log-rank test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). d, days. E, Schematic showing the generation of mixed bone marrow chimera models. Immune profiling was performed from the tumor-bearing hemisphere on day 14 after tumor implantation (SB28, 10,000 cells/injection). F, Frequency of the PEX subset in male (CD45.1+) or female (CD90.1+) CD8+ T cells. Dotted line indicates cells from the same recipient. G, Ki-67 expression in male (CD45.1+) or female (CD90.1+) CD8+ T cells. Data are combined from two independent experiments. n = 6–7 per group. A paired t test was performed (*, P < 0.05; **, P < 0.01).
We first analyzed tumor-infiltrating lymphocytes in the bone marrow chimera mice 14 days after tumor implantation to assess the environmental effect on T cells. We found a higher frequency of CD3+ T cells in the female-to-female group (F to F; female control) compared with the male-to-male group (M to M; male control) in both CD8+ T cells and CD4+ T cells, but not in Foxp3+ cells (Fig. 4B), confirming that T cells in the bone marrow chimera recapitulate their behavior observed in the B6 wild-type mouse model (Fig. 2A). Interestingly, the frequency of infiltrating T cells in the F to M group was significantly increased compared with the M to M group but comparable to the F to F group (Fig. 4B). Meanwhile, the male-to-female (M to F) group showed mixed results, as the frequency of CD4+ T cells was significantly lower than that of the F to F group; in contrast, CD8+ T cells did not behave in the same way (Fig. 4B). Although there was no difference in inhibitory receptor expression (Supplementary Fig. S10C), we found that the PEX subset was largely increased in the M to M group, with a significantly decreased EFF subset (Fig. 4C). No difference was observed in the TEX subset. Importantly, neither the M to F nor the F to M group showed an increase in the PEX subset, suggesting critical roles for a combined hematopoietic cell–intrinsic and cell–extrinsic effect on the higher frequency of PEX in male mice shown in Fig. 2D.
Next, we performed survival analysis on the bone marrow chimera mice after the implantation of SB28 cells to assess immune cell–intrinsic and cell–extrinsic effects on tumor growth. The control groups, M to M and F to F, replicated the survival differences (Fig. 4D), as observed in intact B6 mice (Fig. 1A). Reconstitution of female recipients with male immune cells (M to F) significantly shortened the survival of female mice similar to the M to M group, as we have previously shown (20), and the F to M group showed extended survival comparable to the F to F group. This survival result was consistent with T-cell frequency in the tumor (Fig. 4B) but not correlated with T-cell subsets (Fig. 4C), implying that more complex mechanisms beyond T-cell exhaustion govern tumor progression. Overall, these data support that the sex of immune cell origin is a critical factor that contributes to tumor progression.
We further questioned whether male and female T cells react differently under the same environmental conditions, including other immune cells. To answer this question, we generated a mixed bone marrow chimera by transferring male and female T cell–depleted bone marrow cells mixed at a 1:1 ratio into male or female recipients (Fig. 4E). After reconstitution, we did not observe any differences between male and female CD8+ T cells at steady state (Supplementary Fig. S11A). Survival analysis after intracranial implantation of SB28 cells revealed no difference between male and female recipients, corroborating our findings of a hematopoietic stem cell–intrinsic mechanism that drives sex differences (Supplementary Fig. S11B). Next, we analyzed the tumor-infiltrating T cells on day 14 after tumor implantation and found no significant difference in CD8+ T-cell infiltration into the tumor between male and female recipient mice (Supplementary Fig. S11C). Strikingly, there was a significant increase in the PEX subset in male (CD45.1+) CD8+ T cells compared with female (CD90.1+) T cells in the same recipient mouse regardless of recipient sex (Fig. 4F). As PEX cells have a high capacity for self-renewal (15), we measured the expression of Ki-67, a cell proliferation marker (Fig. 4G). As expected, male (CD45.1+) T cells showed significantly higher levels of Ki-67 than female (CD90.1+) T cells in both sexes of recipients. Meanwhile, we did not observe any differences in the TEX subset and other exhaustion markers (PD-1 and TOX; Supplementary Fig. S11D and S11E). These data further support the importance of a combined hematopoietic cell–intrinsic and cell–extrinsic effect on the higher frequency of PEX in males.
Collectively, these findings suggest that the sex difference in antitumor immunity is not only regulated in a hematopoietic immune cell–intrinsic manner but is also subject to environmental influences.
T cell–Intrinsic Regulation of Sex Differences
Our findings in our in vivo models suggested that male and female T cells are activated to develop to different functional stages during tumor progression (Fig. 2). Thus, we hypothesized that male and female T cells undergo distinct types of cellular reprogramming in the highly suppressive TME. To test this hypothesis, we induced T-cell exhaustion in vitro by repeated stimulation of OT-I cells with SIINFEKL peptides (Fig. 5A). Compared with the cells stimulated only once (single stim), both male and female T cells showed increased expression of the exhaustion markers PD-1, TIM3, TOX, and TIGIT upon repeated stimulation (Fig. 5B). However, intracellular cytokine levels measured by flow cytometry after polyclonal stimulation (phorbol 12-myristate 13-acetate/ionomycin) showed that female exhausted T cells retained their functionality, with higher expression of IFNγ, TNF, and granzyme B, in addition to the transcription factors Tbet and Eomes (Fig. 5B). qPCR analysis confirmed that female exhausted T cells exhibited significantly higher expression of genes encoding antitumor effector cytokines (Ifng, Gzmb, and Il2) and a transcription factor related to effector function (Tbx21; Fig. 5C). Meanwhile, transcript levels of markers associated with exhaustion status were comparable (Pdcd1, Havcr2, Tox, Batf, Irf4, and Tigit) between male and female cells (Fig. 5C). To compare the cytotoxic functionality of male and female exhausted T cells, we cocultured in vitro exhausted T cells with SB28-OVA tumor cells at different ratios and measured the viability of tumor cells after 24 hours. Female exhausted T cells showed significantly higher cytotoxicity compared with their male counterparts at ratios of 1:1 and 1:4 (Fig. 5D). These data suggest that male and female T cells may undergo distinct cell-intrinsic regulation of their functional state during exhaustion.
Figure 5.
Cell-intrinsic regulation of sex differences in T-cell function. A, Schematics depicting in vitro generation of exhausted T cells. B, Exhaustion markers and cytokine expression were measured on day 5 by flow cytometry after polyclonal stimulation with a stimulation cocktail for 4 hours. Data are shown as mean ± SD (n = 3) and are representative of three independent experiments. Two-way ANOVA with Tukey multiple comparisons test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). gMFI, geometric mean fluorescence intensity. C, qPCR analysis on exhausted T cells. Relative expression levels normalized to exhausted T cells from one male are shown (n = 4). An unpaired t test was performed. D, Male and female in vitro exhausted T cells were cultured with SB28-OVA cells for 24 hours, and viability of tumor cells was measured by flow cytometry. Multiple t test (*, P < 0.05; **, P < 0.01). E, A schematic depicting the adoptive transfer model. F, Kaplan–Meier curves depicting survival of male and female RAG1−/− mice bearing SB28-OVA tumor cells after adoptive transfer of OT-I cells. Data shown are combined from three independent experiments and n = 10–14 each group. Statistical significance was determined by the log-rank test (*, P < 0.05; ***, P < 0.001). d, days.
Next, we evaluated the intrinsic role of T cells in driving the observed sex differences in GBM survival. To avoid female T cell–mediated rejection of males, we utilized an adoptive transfer model using the OVA-OT-I system in RAG1−/− mice (Fig. 5E). Transfer of female T cells delivered a survival advantage to recipients, as female OT-I cells significantly extended survival of male recipients compared with the M to M group (Fig. 5F). In contrast, male OT-I cells shortened the survival of female recipients (Fig. 5F). These results indicate that the ability of T cells to control tumor growth is predominantly determined by the sex of the originating host, not the recipient's environment.
Male-Biased T-cell Exhaustion in GBM Patients and Human T Cells
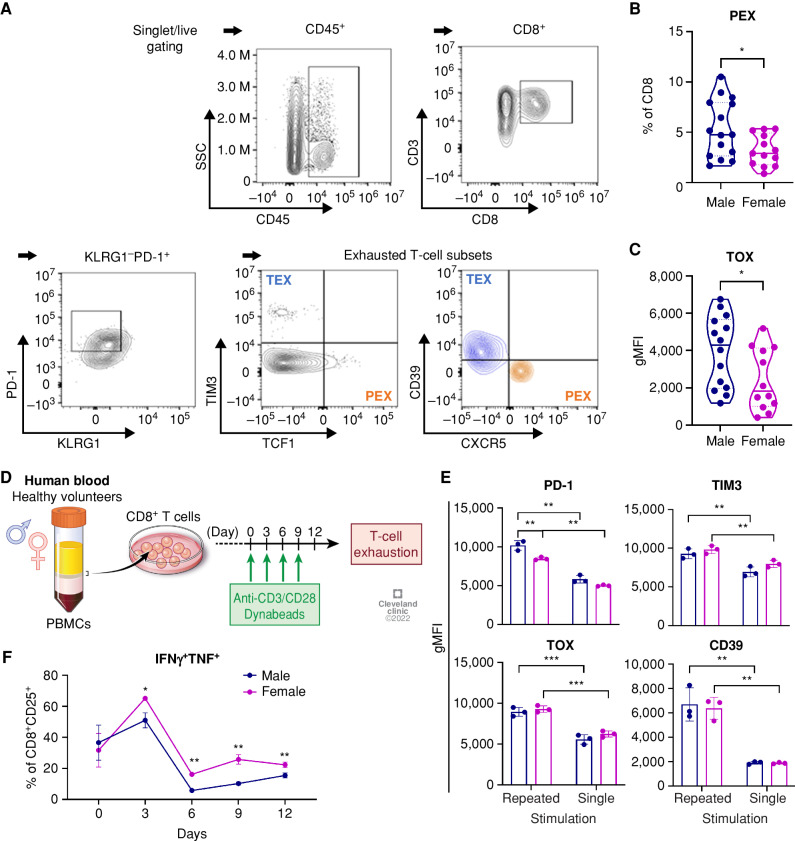
To investigate whether sex differences in T cells are recapitulated in human GBM patients, we analyzed exhausted CD8+ T-cell subsets from GBM patient tumors using flow cytometry. KLRG1 and PD-1 expression was used to exclude the short-lived effector T-cell population, and exhausted T-cell subsets were determined based on the expression of TCF1, TIM3, and CXCR5 (ref. 25; Fig. 6A). There was no significant difference in the percentage of CD8+ T cells and the PD-1+KLRG1− population from male and female tumors (Supplementary Fig. S12A). Meanwhile, an increased frequency of PEX (CD8+KLRG1−PD-1+CXCR5+TCF1+TIM3−) was found in male compared with female tumor samples (Fig. 6B), whereas no difference was observed in the subsets of TEX (CD8+KLRG1−PD-1+CXCR5−TCF1−TIM3+; Supplementary Fig. S12B). Additionally, increased expression of the exhaustion marker TOX was found in CD8+ T cells from male tumors (Fig. 6C; Supplementary Fig. S12C), whereas the expression levels of other markers were comparable (Supplementary Fig. S12D). These results indicate that T cells from patients with GBM exhibit a male-biased exhaustion status in line with our mouse model.
Figure 6.
Sex differences in exhausted T cells in GBM patients. A, A gating strategy for exhausted T-cell subsets from GBM patient tumors. B and C, Frequency of PEX (CD8+KLRG1−PD-1+CXCR5−TCF1+TIM3−; B) and TOX expression (C) in CD8+ T cells from tumors of male (n = 18) and female (n = 14) patients with isocitrate dehydrogenase (IDH) wild-type GBM tumors. Unpaired t test (*, P < 0.05). gMFI, geometric mean fluorescence intensity. D,In vitro induction of exhaustion in human CD8+ T cells. E, Exhaustion marker expression in CD8+ T cells on day 12 after stimulation. Two-way ANOVA with Tukey multiple comparisons test (**, P < 0.01; ***, P < 0.001). gMFI, geometric mean fluorescence intensity. F, Intracellular expression of IFNγ+TNF+ in CD8+ T cells during repeated stimulation. Multiple unpaired t test was performed (*, P < 0.05; **, P < 0.01). Data are shown as mean ± SD (n = 3) and are representative of two independent experiments.
Additionally, we reanalyzed a single-cell RNA sequencing (scRNA-seq) dataset on immune cells from patients with GBM previously published by Ravi and colleagues (26) and evaluated sex differences in CD8+ T cells (Supplementary Fig. S13A and S13B). Consistent with our mouse study and flow analysis of GBM patient tumors, male CD8+ T cells expressed higher levels of TCF7 and TOX, whereas female CD8+ T cells expressed higher levels of effector cytokines such as IFNG, TNF, and GZMK (Supplementary Fig. S13C and S13D).
To further address sex differences in T cells under exhaustion conditions, we performed an in vitro exhaustion assay by repeatedly stimulating human CD8+ T cells isolated from peripheral blood mononuclear cells (PBMC) of healthy volunteers (Fig. 6D). Both male and female T cells exhibited elevated expression levels of exhaustion markers (PD-1, TIM3, TOX, and CD39) after repeated stimulation for 12 days compared with their singly stimulated counterparts (Fig. 6E). Intracellular cytokine expression analysis revealed that the ability of T cells to produce dual cytokines dramatically decreased after the second stimulation (day 6), whereas female CD8+ T cells consistently expressed higher levels of IFNγ and TNF compared with male cells (Fig. 6F). Interestingly, qPCR analysis showed significantly higher expression of a set of transcription factors related to T-cell exhaustion (IRF4, TOX, TCF1, EOMES, and MYC) in female T cells on day 6, but not on day 12, which suggests differential transcriptional regulation during exhaustion in male and female T cells (Supplementary Fig. S14A and S14B).
X. Inactivation Escape Gene Kdm6a Contributes to the Sex Differences in T-cell Exhaustion
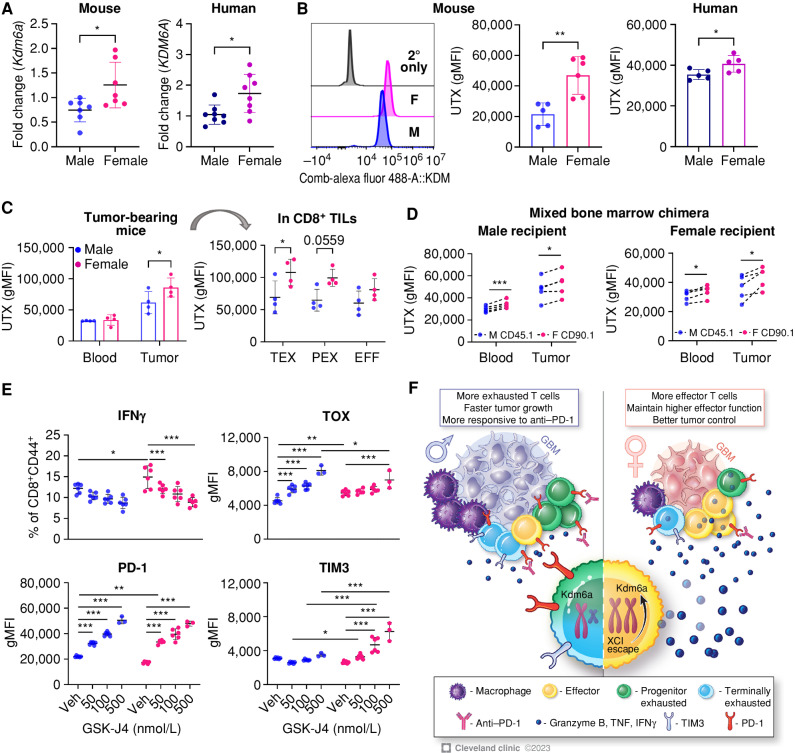
Some X chromosome–encoded genes can escape inactivation, thereby leading to elevated gene expression in female cells compared with male cells. These genes, termed X chromosome inactivation (XCI) escape genes, have been reported to contribute to sex differences mainly in autoimmune diseases (27). Therefore, we tested whether XCI escape genes are responsible for the sex-biased phenotype of exhausted T cells. First, we measured the mRNA level of XCI escape genes and found Kdm6a expression was significantly higher in female in vitro exhausted T cells than male cells (Fig. 7A; Supplementary Fig. S15A). Interestingly, the difference was not observed in unstimulated T cells (Supplementary Fig. S15B), suggesting that XCI escape may be induced by stimulation. Expression of UTX (the protein encoded by Kdm6a) as measured by flow cytometry was significantly higher in female T cells in murine and human in vitro exhausted T cells (Fig. 7B). We also found a trend of increased KDM6A expression in female CD8+ T cells from a GBM patient tumor scRNA-seq dataset (Supplementary Fig. S15C). In tumor-bearing mice, the sex difference in UTX expression was found only in tumor-infiltrating CD8+ T cells, not in blood, and the difference was more prominent in the TEX population (Fig. 7C). To test whether UTX expression is intrinsically regulated, we measured UTX expression in T cells from mixed bone marrow chimera models after tumor implantation (Figs. 4E and 7D). In both male and female recipients, female (CD90.1+) T cells showed a significantly higher expression of UTX than male (CD45.1+) T cells in tumor and blood, indicating female T cell–intrinsic regulation of UTX expression (Fig. 7D). Based on this, we questioned whether UTX (Kdm6a) was related to T-cell exhaustion and sex-biased differences in T cells. Thus, we inhibited UTX function in in vitro exhausted T cells using a small-molecule inhibitor, GSK-J4 (28). UTX blockade reduced cytokine production (IFNγ) and increased exhaustion markers (PD-1, TIM3, and TOX), abolishing the sex differences between male and female exhausted T cells (Fig. 7E). As UTX is a histone demethylase that targets histone H3 lysine 27 trimethylation (H3K27me3), we observed that GSK-J4 treatment indeed increased H3K27me3 (Supplementary Fig. S16A). In contrast, blockade of the KDM5 family did not induce the same effect as the UTX blocker (Supplementary Fig. S16B). Additionally, UTX blockade in human in vitro exhausted T cells also abrogated the sex-biased exhausted T-cell phenotypes (Supplementary Fig. S16C). These data indicate that UTX (Kdm6a) plays critical roles in regulating T-cell exhaustion and that higher expression of UTX in female T cells contributes to sex-biased T-cell exhaustion in a T cell–intrinsic manner. Taken together, our findings indicate that male T cells are more prone to exhaustion, which leads to accelerated tumor growth in males but potentially provides them with a larger benefit from anti–PD-1 mAb therapy, whereas female T cells tend to maintain higher functionality and protect the host from tumor progression (Fig. 7F).
Figure 7.
Higher UTX (Kdm6a) expression in female exhausted T cells is associated with sex-biased T-cell exhaustion status. A, mRNA expression level of Kdm6a in murine and human in vitro exhausted T cells measured by qPCR analysis. Relative expression levels normalized to exhausted T cells from one male are shown. Unpaired t test (*, P < 0.05). B, Expression of UTX (encoded by Kdm6a) in murine and human in vitro exhausted T cells measured by flow cytometry. A representative histogram of UTX expression in murine cells is shown. Data are combined from two independent experiments. Unpaired t test (*, P < 0.05; **, P < 0.01). gMFI, geometric mean fluorescence intensity. C, UTX expression in CD8+ T cells isolated from SB28-bearing male and female B6 mice on day 14 after tumor implantation. Two-way ANOVA with the Tukey multiple comparisons test (*, P < 0.05). TIL, tumor-infiltrating lymphocyte. D, UTX expression in CD8+ T cells from SB28-bearing mixed bone marrow chimera mice on day 14 after tumor implantation. Paired t test (*, P < 0.05; ***, P < 0.001). E, Male and female OT-I cells were treated with GSK-J4 at varying doses during in vitro induction of exhaustion (days 2–5), and expression level of IFNγ and exhaustion markers was measured by flow cytometry on day 5. Data are shown as mean ± SD (n = 6) and are representative of two independent experiments. Two-way ANOVA with the Tukey multiple comparisons test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Veh, vehicle. F, Proposed model of sex-biased T-cell phenotype and functionality mediated by the XCI gene Kdm6a in patients with GBM.
DISCUSSION
Sex differences are emerging as a major contributor to cancer progression and therapeutic response through distinct genetic, epigenetic, and immunologic mechanisms (29). We previously reported a sex difference in MDSC localization in GBM whereby males had increased monocytic MDSCs in the TME, whereas females had increased granulocytic MDSCs in the periphery. This difference was leveraged for the development of sex-specific therapies that were validated in preclinical models (20). Importantly, whereas functional and targetable sex differences present within certain myeloid populations, we demonstrate here that GBM-infiltrating T cells actually mediate sex difference in overall survival. Using preclinical models and human patient validation, we demonstrate increased T-cell exhaustion in males compared with females, with males displaying an enhanced response to single-agent ICI treatment. Mechanistically, the T cell–dependent survival difference was predominantly due to hematopoietic immune cell–intrinsic differences along with the impact of the environment including sex hormones.
Our observation that males are more responsive to single-agent ICI treatment, based on sex differences in T-cell exhaustion and inhibitor receptor expression, supports clinical trial data in many cancers whereby males show an enhanced therapeutic response and females develop more adverse events (30–32). The latter observation is likely due to enhanced immune activation status in women. Interestingly, in patients with lung cancer, females showed an enhanced response to PD-1/PD-L1 when combined with chemotherapy compared with males (33), suggesting that additional stimuli are required to activate an antitumor immune response in females. For GBM, this will likely be needed, as ICI monotherapies have not shown strong clinical benefit and current immunotherapy strategies are now focused on combination therapies (34). Furthermore, immediate priorities currently include sex-specific assessment of combination strategies in preclinical models and using sex as stratification criteria for early-stage clinical trials.
Our preclinical data also demonstrate that male T cells are more exhausted than female T cells, which has also recently been seen in several malignancies including melanoma, colon cancer, and bladder cancer models (17, 18). These observations were also validated in human GBM patients and are consistent with reports in patients with melanoma (35) but opposite to what has been observed in patients with lung cancer (36). Although both of these tumors are more responsive to ICIs than GBM (37, 38) and have an increased mutational burden compared with GBM (39), it is unclear why there are differences and why GBM appears to more closely phenocopy melanoma. This is likely due to a combination of differences in driver mutations, standard-of-care therapies, and anatomic locations. For GBM in particular, the specialized neural–immune microenvironment is likely to provide unique stimuli (40), and sex differences in other neural cell types may also affect T-cell function. Future studies focusing on the interaction between brain-specific mechanisms and T cells are likely to help clarify the molecular underpinnings of sex differences in T-cell function and may reveal sex-biased mechanisms that could be leveraged for next-generation therapies.
Androgen in particular has recently received attention for its role in regulating T-cell exhaustion, with contradictory molecular mechanisms (17, 18). These groups have shown that blockade of androgen receptor signaling restored CD8+ T-cell function, with increased responsiveness in anti–PD-1/PD-L1 treatment in males, in line with a previous report on castration-resistant prostate cancer (41). Our in vitro data, however, along with our adoptive transfer studies and bone marrow chimera studies, suggest a predominant cell-intrinsic underpinning of male T-cell exhaustion. We reached these conclusions based on the maintained difference in T-cell exhaustion in males versus females ex vivo in the absence of the influence of sex hormones. Yet this does not rule out the possibility that the initial impact of sex hormones on stem cell stage or before isolation has been maintained through epigenetic memory, which needs further investigation. These cell-intrinsic differences could be potentially derived from sex chromosomes via expression of genes escaping XCI or micro-RNAs highly enriched in X chromosomes (27). Indeed, we found differential expression of Kdm6a (UTX) between male and female exhausted T cells, and its pharmacologic blockade abolished the sex differences we observed in exhausted T cells. Differential expression of epigenetic regulators encoded on sex chromosomes has been reported in microglia (42) and natural killer cells (43), leading to sex-biased inflammatory responses. Given that sex differences in GBM are observed throughout all age groups, whereas sex hormone levels vary (44), delineating the effects of sex hormones and sex chromosomes in sexually dimorphic GBM immunity requires further investigation.
Sex differences in T-cell responses in GBM are likely derived from a combination of sex hormone–derived influences and cell intrinsically derived influences. The molecular drivers of these and how the two intersect should be the focus of future studies to better enable us to understand immunologic sex differences and tailor therapies accordingly. Additionally, an aspect outside of the scope of this study but still of pressing interest is the extent to which sex differences in antigen-presenting cells are present and affect T-cell behavior. A meta-analysis of GBM clinical trials using autologous dendritic cells showed that female patients had a more robust survival advantage compared with male patients, providing a rationale to understand underlying mechanisms (45). Taken together, our study identifies T cells as a critical component driving sex differences in GBM progression and a male-biased T-cell exhaustion state that could potentially interrogate sex-specific immunotherapy responses in patients with cancer. Our study provides insight into the immunologic mechanisms underlying sex differences in GBM and further emphasizes a need for sex-specific treatment strategies.
METHODS
Cell Lines
The syngeneic mouse GBM cell lines SB28 and SB28-OVA were kindly gifted by Dr. Hideho Okada (University of California San Francisco). GL261 cells were obtained from the Developmental Therapeutic Program, NCI. All cell lines were treated with 1:100 MycoRemoval Agent (MP Biomedicals) upon thawing and routinely tested for Mycoplasma spp. (Lonza). All cell lines were maintained in complete RPMI 1640 (Media Preparation Core, Cleveland Clinic) supplemented with 10% FBS (Thermo Fisher Scientific), 1% penicillin/streptomycin (Media Preparation Core), and GlutaMAX (Gibco). Cells were maintained in humidified incubators held at 37°C and 5% CO2 and not grown for more than 15 passages.
Mice
All animal procedures were performed in accordance with the guidelines and protocols approved by the Institutional Animal Care and Use Committee (IACUC) at the Cleveland Clinic. C57BL/6 (RRID:IMSR_JAX:000664), B6 CD45.1 (B6.SJL-PtprcaPepcb/BoyJ; RRID:IMSR_JAX:002014), B6 CD90.1 (B6.PL-Thy1a/CyJ; RRID:IMSR_JAX:000406), RAG1−/− (B6.129S7-Rag1tm1Mom/J; RRID:IMSR_JAX:002216), and OT-I TCR transgenic [C57BL/6-Tg(TcraTcrb)1100Mjb/J; RRID:IMSR_JAX:003831] mice were purchased from The Jackson Laboratory as required. NSG mice were obtained from the Biological Research Unit (BRU) at Lerner Research Institute, Cleveland Clinic. All animals were housed in a specific pathogen–free facility of the BRU, with a light–dark period of 12 hours each.
For tumor implantation, 5- to 6-week-old mice were anesthetized, fit to the stereotaxic apparatus, and intracranially injected with 10,000 to 25,000 tumor cells in 5 μL RPMI-null media into the left hemisphere approximately 0.5 mm rostral and 1.8 mm lateral to the bregma with 3.5 mm depth from the scalp. In some experiments, 5 μL null media were injected into age- and sex-matched animals for sham controls. Animals were monitored over time for the presentation of neurologic and behavioral symptoms associated with the presence of a brain tumor.
To deplete CD8+ T cells, B6 mice were intraperitoneally injected with 200 μg of anti-CD8 antibody (Bio X Cell, cat. #0061) or isotype antibody (Bio X Cell, cat. #BE0090) on day −1 and 0 of tumor implantation and then 100 μg once every 5 days until humane endpoint.
In some experiments, mice were treated with anti–PD-1 (Bio X Cell, cat. #BE0273; RRID: AB_2687796) or isotype antibody (Bio X Cell, cat. #BE0089, RRID:AB_1107769) intraperitoneally starting 7 days after tumor implantation, and injections were repeated every 2 to 3 days five times. In some experiments, FTY720 (Sigma, cat. #SML-0700) was administered in the drinking water at 3 μg/mL, starting from day 6 and lasting until day 21. FTY720-containing water was replaced every 3 to 4 days.
In the adoptive transfer model, RAG1−/− mice received an intracranial injection of SB28-OVA tumor cells (15,000 cells/mouse). OT-I splenocytes were activated in vitro with 2 μg of SIINFEKL peptides (Sigma) in the presence of recombinant human IL2 (50 U/mL, PeproTech) for 3 days. Activated CD8+ T cells (5 × 106 cells/mouse) were transferred intravenously into SB28-OVA tumor–bearing mice on day 7 after tumor implantation.
To generate bone marrow chimeras, 5-week-old B6.CD45.2 male and female mice were irradiated with 12 Gy in total, given in two fractions 3 to 4 hours apart. Bone marrow cells were obtained from tibias and femurs of 5-week-old B6.CD45.1 mice, and existing T cells were depleted using Thy1.2 (Bio X Cell, cat. # BE0066, RRID:AB_1107682) and rabbit complement. A total of 5 × 106 T cell–depleted bone marrow cells were injected retro-orbitally into the irradiated recipients. For mixed bone marrow chimeras, bone marrow cells were collected from 5-week-old B6.CD45.1 male mice and age-matched B6.CD90.1 female mice, and T cells were depleted as described above. Then male and female cells were mixed at a 1:1 ratio, and a total of 6 × 106 T cell–depleted bone marrow cells were retro-orbitally injected into the lethally irradiated recipient mice. Animals were given Sulfatrim in drinking water for 2 weeks to prevent infection. After 6 to 8 weeks, animals were subjected to tumor implantation.
Immunophenotyping by Flow Cytometry
At the indicated time points, a single-cell suspension was prepared from the tumor-bearing left hemisphere by enzymatic digestion using collagenase IV (Sigma) and DNase I (Sigma), followed by straining with a 40-μm filter. Cells were stained with LIVE/DEAD Fixable Stains (Thermo Fisher) on ice for 15 minutes. After washing with PBS, cells were resuspended in an Fc receptor blocker (Miltenyi Biotec) diluted in PBS/2% BSA and incubated on ice for 10 minutes. For surface staining, fluorochrome-conjugated antibodies were diluted in Brilliant buffer (BD) at 1:100 to 1:250, and cells were incubated on ice for 30 minutes. After washing with PBS/2% BSA buffer, cells were then fixed with Foxp3/Transcription Factor Fixation Buffer (eBioscience) overnight. For tetramer staining, cells were incubated with tetramer antibody diluted in PBS/2% BSA at 1:1,000 dilution after the FcR blocker step on ice for 30 minutes, followed by surface staining. For intracellular staining, antibodies were diluted in Foxp3/Transcription Factor Permeabilization Buffer (perm buffer) at 1:250 to 1:500, and cells were incubated at room temperature for 45 minutes. For intracellular cytokine detection, cells were stimulated using Cell Stimulation Cocktail plus protein transport inhibitor (eBioscience) in complete RPMI for 4 hours. After stimulation, cells were subjected to the staining procedures described above. For UTX and H3K27me3 staining, cells were intracellularly stained with primary antibody (anti-UTX, anti-H3K27me3) at 1:500 diluted in perm buffer at room temperature for 45 minutes. Following two washing steps with perm buffer, a secondary antibody conjugated with fluorochrome was diluted in perm buffer at 1:1,000, and cells were incubated at room temperature for 45 minutes. Following washing steps, stained cells were acquired with a BD LSRFortessa (BD) or Aurora (Cytek) and analyzed using FlowJo software (v10, BD Biosciences).
Reagents
For immunophenotyping in mouse models, the following fluorophore-conjugated antibodies were used: CD11b (M1/70, cat. #563553), CD11c (HL3, cat. #612796), Ly6G (1A8, cat. #560603), CD3 (145–2C11, cat. #564379), and CD44 (IM7, cat. #612799) from BD Biosciences. CTLA4 (UC10–4B9, cat. #106312), PD-1 (29F.1A12, cat. #135241), B220 (RA3–6B2, cat. #103237), Ki-67 (11Fb, cat. #151215), TIM3 (RMT3–23, cat. #119727), I-A/I-E (M5/114.15.2, cat. #107606), CD45 (30-F11, cat. #103132), LAG3 (C9B7W, cat. #125224), NK1.1 (PK136, cat. #108716), CD4 (GK1.5, cat. #100422), CD8 (6206.7, cat. #100712), Ly6C (HK1.4, cat. #128024), CD68 (FA-11, cat. #137024), granzyme B (QA18A28, cat. #396413), TNF (MP6-XT22, cat. #506329), IFNγ (XMG1.2, cat. #505846), and TIGIT (1G9, cat. #142108) were obtained from BioLegend. Anti-Foxp3 (FJK-16s, cat. #12-5773) antibody was obtained from eBioscience. Anti-TOX (TXRX10, cat. #12-6502) antibody was purchased from Invitrogen, and anti-TCF1 (C63D9, cat. #6709S) antibody was obtained from Cell Signaling Technology. Antibodies against PD-1 (RMP1-30, cat. #752354), Eomes (X4-83, cat. #567171), Tbet (O4-46, cat. #563320), and Ly108 (13G3, cat. #748564) were obtained from BD Biosciences. For the tetramer assay, anti-CD8 antibody (KT15, cat. #ab22504) was obtained from Abcam, and PE-conjugated H-2K(b) SIINFEKL tetramer was provided by the NIH Tetramer Core Facility.
For analysis of GBM patient samples, the following fluorophore-conjugated antibodies were used: CD45 (HI30, cat. #563791), CD3 (SP34-2, cat. #557757), CD4 (SK3, cat. #612749), PD-1 (EH12.2, cat. #564104), and CTLA4 (BN13, 561717) from BD Biosciences. CD8 (RPA-T8, cat. #301038), TIGIT (A15153G, cat. #372718), KLRG1 (2F1/KLRG1, cat. #138415), TIM3 (F38-2E2, cat. #345030), and TBET (4B10, cat. #644817) were purchased from BioLegend. Anti-CD39 (A1, cat. #67-0399) antibody was obtained from eBioscience. Anti-TOX and anti-TCF1 antibodies were obtained as described above.
For analysis of UTX, anti-KDM6A antibody (rabbit polyclonal, cat. #PA5-31828) and anti-rabbit goat IgG-AlexaFluor488 antibody (cat. #A-11008) were obtained from Thermo Fisher Scientific. Anti-H3K27me3 antibody (C36B11, cat. #9733S) was purchased from Cell Signaling Technology. Anti–histone H3 antibody (cat. #819407) was obtained from BioLegend. The KDM inhibitors GSK-J4 (cat. #4594) and JQKD-82 (cat. #76765) were obtained from Tocris.
Tumor-Infiltrating Immune Cell Isolation
At the indicated time points, mice were euthanized via CO2 asphyxiation, followed by cervical dislocation as approved by the IACUC. The tumor-bearing left hemisphere of the brain was collected and dissociated into a single-cell suspension. In brief, tissue was minced and incubated with collagenase D (1 mg/mL; Roche) and DNase I (0.1 mg/mL; Roche) at 37°C for up to 45 minutes. Digested tissue was filtered through a 70-μm cell strainer, and lymphocytes were enriched by gradient centrifugation using 30% Percoll solution (Sigma). The enriched immune cells were subjected to flow cytometric analysis as described above.
In Vitro Generation of Exhausted T Cells
To induce exhaustion of mouse T cells, CD8+ T cells were isolated from splenocytes of OT-I mice using a magnetic bead isolation kit (STEMCELL Technologies) and cultured following a previously published protocol (46). In brief, 10 ng of SIINFEKL peptides were added into the culture every day until day 5 (repeated stimulation), or for single stimulation, peptides were added only once on day 0, and cells were washed on day 2 and rested until day 5.
To induce exhaustion of human T cells, blood was obtained from healthy volunteers upon written informed consent following the Cleveland Clinic Institutional Review Board (IRB) protocol (IRB07-918). Fresh PBMCs were isolated using a Ficoll density gradient, and CD8+ T cells were subsequently isolated using the STEMCELL human CD8+ isolation kit (STEMCELL Technologies) following the manufacturer's instructions. Induction of T-cell exhaustion was performed following the previously published method with modification (47). Briefly, cells were cultured in complete RPMI containing recombinant human IL2 (30 U/mL, PeproTech) and stimulated with anti-CD3/anti-CD28 Dynabeads (Invitrogen) at a bead:cell ratio of 1:10. Every 3 days, cells were harvested, washed, and cultured with fresh beads for up to 12 days. For single-stimulated controls, cells were harvested on day 3 and cultured without further stimulation for up to 12 days.
Real-time Quantitative PCR
Total RNA was isolated using an RNeasy Mini Kit (Qiagen), and cDNA was synthesized using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). qPCR reactions were performed using Fast SYBR Green Master Mix (Thermo Fisher Scientific) on an Applied Biosystems StepOnePlus Real-Time PCR system. The threshold cycle (Ct) value for each gene was normalized to the expression levels of Gapdh (mouse) or ACTIN (human), and relative expression was calculated by normalizing to the delta Ct value of one male T-cell datapoint. Primer sequences were obtained from PrimerBank (48) or previously published articles and are listed in Supplementary Table S1 (mouse) and Supplementary Table S2 (human).
In Vitro Inhibition of UTX (GSK-J4 Treatment)
To test the effect of UTX blockade during T-cell exhaustion, GSK-J4 was added to the culture medium from day 2 (mouse) or day 3 (human) of stimulation during in vitro exhaustion assays. GSK-J4 was dissolved and diluted in dimethylsulfoxide (DMSO), and the same amount of DMSO was added as a vehicle control in each experiment.
GBM Patient Samples
Cryopreserved single-cell suspension samples were collected by the Rosa Ella Burkhardt Brain Tumor Bank after obtaining written informed consent from the patients. All studies were conducted in accordance with ethical guidelines and approved by the Cleveland Clinic IRB (IRB2559). Samples from GBM patients diagnosed as isocitrate dehydrogenase (IDH) mutations were excluded from our study. Cells were thawed in a 37°C water bath and washed twice with warm complete RPMI. Cells were stained with LIVE/DEAD Fixable Stains for 15 minutes on ice and washed, followed by incubation with an Fc receptor blocker (Miltenyi Biotec) for 15 minutes on ice. Surface marker staining was performed for 30 minutes on ice with the following antibodies: CD45, CD3, CD4, CD8, CD44, PD-1, TIM3, CD39, KLRG1, and TIGIT. Cells were then fixed with Foxp3/Transcription Factor Fixation Buffer (Invitrogen) overnight at 4°C, and intracellular staining was performed in permeabilization buffer for the following markers: TBET, TCF1, CTLA4, and TOX. Stained samples were acquired by Cytek Aurora and analyzed by FlowJo software.
Analysis of Previously Published scRNA-seq Data
Previously published scRNA-seq data were acquired from Ravi and colleagues (26) and analyzed using Seurat v4.0 (49). Ravi and colleagues normalized the gene expression values by dividing each cell by the total number of transcripts, multiplied by 10,000, and then did a natural log transformation. They then removed batch effects and scaled the data using a regression model with sample batch information and percentage of ribosomal and mitochondrial gene expression. The cell type of each individual cell was used as determined by Ravi and colleagues. Briefly, the authors determined cell type through three different methods: expression of signature markers of immune cells; an automated annotation tool, SingelR; and gene set variation analysis. The results from all three methods were combined, and cells were assigned to clusters using the highest enrichment in all models.
Statistical Analysis
GraphPad Prism (Version 9, GraphPad Software Inc., RRID:SCR_002798) software was used for data presentation and statistical analysis. Unpaired or paired t test or one-way/two-way analysis of variance (ANOVA) was used with a multiple comparisons test, as indicated in the figure legends. Survival analysis was performed by the log-rank test. P < 0.05 was considered statistically significant (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Data Availability Statement
All data generated in this study are available upon request from the corresponding author, Dr. Justin D. Lathia (lathiaj@ccf.org).
Supplementary Material
Supplementary Fig. S1. Immune cell-dependent survival differences between males and females in murine glioblastoma models. Supplementary Fig. S2. Mouse syngeneic GBM cell lines do not contain a Y chromosome. Supplementary Fig. S3. Frequencies of tumor-infiltrating immune cells in SB28 model. Supplementary Fig. S4. Sex difference in survival in SB28-OVA model and GL261 model. Supplementary Fig. S5. No sex difference in T cells was observed at an earlier time point. Supplementary Fig. S6. Male T cells are more exhausted in the GL261 model. Supplementary Fig. S7. Phenotype of T cells in the periphery does not replicate the sex differences shown in tumor-infiltrating T cells. Supplementary Fig. S8. Frequencies of exhausted T cell subsets. Supplementary Fig. S9. PD1 blockade enhanced immune responses Supplementary Fig. S10. No significant difference in the immune cell composition of bone marrow chimera mice before tumor implantation. Supplementary Fig. S11. Male and female CD8+ T cells in mixed bone marrow chimera model. Supplementary Fig. S12. Phenotyping tumor-infiltrating CD8+ T cell from GBM patient tumors. Supplementary Fig. S13. scRNA-seq analysis of tumor-infiltrating CD8+ T cells from GBM patients. Supplementary Fig. S14. mRNA expression level of transcription factors in human in vitro exhausted T cells. Supplementary Fig. S15. Expression level of X chromosome inactivation (XCI) genes in exhausted T cells. Supplementary Fig. S16. Inhibition of UTX abrogates sex differences in T cell exhaustion.
Table S1. qPCR primers used for mouse study Table S2. qPCR primers used for human study
Acknowledgments
We thank the members of the Lathia laboratory for insightful discussions. We are grateful to Drs. Jill Barnholtz-Sloan (NCI), Josh Rubin (Washing University in St. Louis), Jim Connor (Penn State College of Medicine), and Mike Berens (TGEN) for their useful suggestions. We thank Drs. Jingqin (Rosy) Luo and Tamara Abou-Antoun (Washing University in St. Louis) for their technical advice. We greatly appreciate the editorial assistance of Dr. Erin Mulkearns-Hubert (Cleveland Clinic) and illustrative work of Ms. Amanda Mendelsohn from the Center for Medical Art and Photography at the Cleveland Clinic. We acknowledge the technical help from Cleveland Clinic Flow Cytometry Core. We also thank the NIH Tetramer Core Facility (contract number 75N93020D00005) for providing H-2K SIINFEKL tetramers. This work is supported by NIH grants R35 NS127083 (J.D. Lathia), P01 CA245705 (J.D. Lathia), K99 CA248611 (D. Bayik), R35 NS105068 (H. Okada), R01 DK128143 (W.A. Goodman), F30 CA250254 (A. Lauko), T32 GM007250 (A. Lauko), R01 HL158801 (S.J. Cameron), T32 5T32AI007024-40 (D.C. Watson), TL1 5TL1TR002549-03 (D.C. Watson), K99 CA277242 (D.C. Watson), and F31 CA264849 (K.E. Kay). This work was also supported the American Brain Tumor Association (J.D. Lathia), Case Comprehensive Cancer Center (J.D. Lathia), Cleveland Clinic/Lerner Research Institute (J.D. Lathia), and the American Society of Transplantation Research Network (M. Nicosia).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
D. Bayik reports grants from the NCI during the conduct of the study. A. Lauko reports grants from the NIH during the conduct of the study. M.S. Ahluwalia reports grants from AstraZeneca, Bristol Myers Squibb, Bayer, Seagen, and Novartis, and personal fees from Insightec, GSK, Xoft, Nuvation, Cellularity, SDP Oncology, Apollomics, Prelude, Janssen Oncology, Tocagen, Voyager Therapeutics, Viewray, Caris Lifesciences, Pyramid Biosciences, Modifi Biosciences, Bugworks, Bayer, and Sumitomo Pharma Oncology outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
J. Lee: Conceptualization, formal analysis, investigation, methodology, writing–original draft. M. Nicosia: Conceptualization, methodology, writing–review and editing. E.S. Hong: Formal analysis, visualization. D.J. Silver: Investigation. C. Li: Investigation. D. Bayik: Investigation. D.C. Watson: Investigation. A. Lauko: Investigation. K.E. Kay: Investigation. S.Z. Wang: Investigation. S. Johnson: Resources. M. McGraw: Resources. M.M. Grabowski: Resources. D.D. Kish: Resources. A.B. Desai: Resources, writing–review and editing. W.A. Goodman: Resources, writing–review and editing. S.J. Cameron: Resources, writing–review and editing. H. Okada: Resources. A. Valujskikh: Writing–review and editing. R.L. Fairchild: Writing–review and editing. M.S. Ahluwalia: Resources. J.D. Lathia: Conceptualization, supervision, funding acquisition, writing–original draft.
References
- 1. Stupp R, Taillibert S, Kanner A, Read W, Steinberg D, Lhermitte B, et al. Effect of Tumor-treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma: a randomized clinical trial. JAMA 2017;318:2306–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Quail DF, Joyce JA. The microenvironmental landscape of brain tumors. Cancer Cell 2017;31:326–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chongsathidkiet P, Jackson C, Koyama S, Loebel F, Cui X, Farber SH, et al. Sequestration of T cells in bone marrow in the setting of glioblastoma and other intracranial tumors. Nat Med 2018;24:1459–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ayasoufi K, Pfaller CK, Evgin L, Khadka RH, Tritz ZP, Goddery EN, et al. Brain cancer induces systemic immunosuppression through release of non-steroid soluble mediators. Brain 2020;143:3629–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reardon DA, Brandes AA, Omuro A, Mulholland P, Lim M, Wick A, et al. Effect of nivolumab vs bevacizumab in patients with recurrent glioblastoma: the CheckMate 143 phase 3 randomized clinical trial. JAMA Oncol 2020;6:1003–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Omuro A, Reardon DA, Sampson JH, Baehring J, Sahebjam S, Cloughesy TF, et al. Nivolumab plus radiotherapy with or without temozolomide in newly diagnosed glioblastoma: results from exploratory phase I cohorts of CheckMate 143. Neurooncol Adv 2022;4:vdac025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Medikonda R, Dunn G, Rahman M, Fecci P, Lim M. A review of glioblastoma immunotherapy. J Neurooncol 2021;151:41–53. [DOI] [PubMed] [Google Scholar]
- 8. Cloughesy TF, Mochizuki AY, Orpilla JR, Hugo W, Lee AH, Davidson TB, et al. Neoadjuvant anti–PD-1 immunotherapy promotes a survival benefit with intratumoral and systemic immune responses in recurrent glioblastoma. Nat Med 2019;25:477–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ostrom QT, Rubin JB, Lathia JD, Berens ME, Barnholtz-Sloan JS. Females have the survival advantage in glioblastoma. Neuro Oncol 2018;20:576–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ippolito JE, Yim AK, Luo J, Chinnaiyan P, Rubin JB. Sexual dimorphism in glioma glycolysis underlies sex differences in survival. JCI Insight 2017;2:e92142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang W, Warrington NM, Taylor SJ, Whitmire P, Carrasco E, Singleton KW, et al. Sex differences in GBM revealed by analysis of patient imaging, transcriptome, and survival data. Sci Transl Med 2019;11:eaao5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Klein SL, Flanagan KL. Sex differences in immune responses. Nat Rev Immunol 2016;16:626–38. [DOI] [PubMed] [Google Scholar]
- 13. Rubin JB, Lagas JS, Broestl L, Sponagel J, Rockwell N, Rhee G, et al. Sex differences in cancer mechanisms. Biol Sex Differ 2020;11:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Belk JA, Daniel B, Satpathy AT. Epigenetic regulation of T cell exhaustion. Nat Immunol 2022;23:848–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Siddiqui I, Schaeuble K, Chennupati V, Fuertes Marraco SA, Calderon-Copete S, Pais Ferreira D, et al. Intratumoral Tcf1(+)PD-1(+)CD8(+) T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity 2019;50:195–211. [DOI] [PubMed] [Google Scholar]
- 16. Miller BC, Sen DR, Al Abosy R, Bi K, Virkud YV, LaFleur MW, et al. Subsets of exhausted CD8(+) T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol 2019;20:326–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang C, Jin J, Yang Y, Sun H, Wu L, Shen M, et al. Androgen receptor-mediated CD8(+) T cell stemness programs drive sex differences in antitumor immunity. Immunity 2022;55:1268–83. [DOI] [PubMed] [Google Scholar]
- 18. Kwon H, Schafer JM, Song NJ, Kaneko S, Li A, Xiao T, et al. Androgen conspires with the CD8(+) T cell exhaustion program and contributes to sex bias in cancer. Sci Immunol 2022;7:eabq2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Woroniecka K, Chongsathidkiet P, Rhodin K, Kemeny H, Dechant C, Farber SH, et al. T-cell exhaustion signatures vary with tumor type and are severe in glioblastoma. Clin Cancer Res 2018;24:4175–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bayik D, Zhou Y, Park C, Hong C, Vail D, Silver DJ, et al. Myeloid-derived suppressor cell subsets drive glioblastoma growth in a sex-specific manner. Cancer Discov 2020;10:1210–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xu J, Peng X, Chen Y, Zhang Y, Ma Q, Liang L, et al. Free-living human cells reconfigure their chromosomes in the evolution back to uni-cellularity. Elife 2017;6:e28070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wherry EJ, Blattman JN, Murali-Krishna K, van der Most R, Ahmed R. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J Virol 2003;77:4911–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Genoud V, Marinari E, Nikolaev SI, Castle JC, Bukur V, Dietrich PY, et al. Responsiveness to anti–PD-1 and anti-CTLA-4 immune checkpoint blockade in SB28 and GL261 mouse glioma models. Oncoimmunology 2018;7:e1501137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chiba K. FTY720, a new class of immunomodulator, inhibits lymphocyte egress from secondary lymphoid tissues and thymus by agonistic activity at sphingosine 1-phosphate receptors. Pharmacol Ther 2005;108:308–19. [DOI] [PubMed] [Google Scholar]
- 25. Dolina JS, Van Braeckel-Budimir N, Thomas GD, Salek-Ardakani S. CD8(+) T cell exhaustion in cancer. Front Immunol 2021;12:715234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ravi VM, Neidert N, Will P, Joseph K, Maier JP, Kückelhaus J, et al. T-cell dysfunction in the glioblastoma microenvironment is mediated by myeloid cells releasing interleukin-10. Nat Commun 2022;13:925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Libert C, Dejager L, Pinheiro I. The X chromosome in immune functions: when a chromosome makes the difference. Nat Rev Immunol 2010;10:594–604. [DOI] [PubMed] [Google Scholar]
- 28. Kruidenier L, Chung CW, Cheng Z, Liddle J, Che K, Joberty G, et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature 2012;488:404–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Haupt S, Caramia F, Klein SL, Rubin JB, Haupt Y. Sex disparities matter in cancer development and therapy. Nat Rev Cancer 2021;21:393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wallis CJD, Butaney M, Satkunasivam R, Freedland SJ, Patel SP, Hamid O, et al. Association of patient sex with efficacy of immune checkpoint inhibitors and overall survival in advanced cancers: a systematic review and meta-analysis. JAMA Oncol 2019;5:529–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Unger JM, Vaidya R, Albain KS, LeBlanc M, Minasian LM, Gotay CC, et al. Sex differences in risk of severe adverse events in patients receiving immunotherapy, targeted therapy, or chemotherapy in cancer clinical trials. J Clin Oncol 2022;40:1474–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Conforti F, Pala L, Pagan E, Corti C, Bagnardi V, Queirolo P, et al. Sex-based differences in response to anti–PD-1 or PD-L1 treatment in patients with non-small-cell lung cancer expressing high PD-L1 levels. A systematic review and meta-analysis of randomized clinical trials. ESMO Open 2021;6:100251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Conforti F, Pala L, Bagnardi V, Viale G, De Pas T, Pagan E, et al. Sex-based heterogeneity in response to lung cancer immunotherapy: a systematic review and meta-analysis. J Natl Cancer Inst 2019;111:772–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lim M, Xia Y, Bettegowda C, Weller M. Current state of immunotherapy for glioblastoma. Nat Rev Clin Oncol 2018;15:422–42. [DOI] [PubMed] [Google Scholar]
- 35. Loo K, Tsai KK, Mahuron K, Liu J, Pauli ML, Sandoval PM, et al. Partially exhausted tumor-infiltrating lymphocytes predict response to combination immunotherapy. JCI Insight 2017;2:e93433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Conforti F, Pala L, Pagan E, Bagnardi V, De Pas T, Queirolo P, et al. Sex-based dimorphism of anticancer immune response and molecular mechanisms of immune evasion. Clin Cancer Res 2021;27:4311–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010;363:711–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Herbst RS, Baas P, Kim DW, Felip E, Pérez-Gracia JL, Han JY, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 2016;387:1540–50. [DOI] [PubMed] [Google Scholar]
- 39. Schaettler MO, Richters MM, Wang AZ, Skidmore ZL, Fisk B, Miller KE, et al. Characterization of the genomic and immunologic diversity of malignant brain tumors through multisector analysis. Cancer Discov 2022;12:154–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhao F, Li B, Yang W, Ge T, Cui R. Brain-immune interaction mechanisms: Implications for cognitive dysfunction in psychiatric disorders. Cell Prolif 2022;55:e13295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Guan X, Polesso F, Wang C, Sehrawat A, Hawkins RM, Murray SE, et al. Androgen receptor activity in T cells limits checkpoint blockade efficacy. Nature 2022;606:791–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Qi S, Al Mamun A, Ngwa C, Romana S, Ritzel R, Arnold AP, et al. X chromosome escapee genes are involved in ischemic sexual dimorphism through epigenetic modification of inflammatory signals. J Neuroinflammation 2021;18:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cheng MI, Li JH, Riggan L, Chen B, Tafti RY, Chin S, et al. The X-linked epigenetic regulator UTX controls NK cell-intrinsic sex differences. Nat Immunol 2023;24:780–91. [DOI] [PubMed] [Google Scholar]
- 44. Ostrom QT, Kinnersley B, Wrensch MR, Eckel-Passow JE, Armstrong G, Rice T, et al. Sex-specific glioma genome-wide association study identifies new risk locus at 3p21.31 in females, and finds sex-differences in risk at 8q24.21. Sci Rep 2018;8:7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shireman JM, Ammanuel S, Eickhoff JC, Dey M. Sexual dimorphism of the immune system predicts clinical outcomes in glioblastoma immunotherapy: a systematic review and meta-analysis. Neurooncol Adv 2022;4:vdac082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhao M, Kiernan CH, Stairiker CJ, Hope JL, Leon LG, van Meurs M, et al. Rapid in vitro generation of bona fide exhausted CD8+ T cells is accompanied by Tcf7 promotor methylation. PLoS Pathog 2020;16:e1008555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dunsford LS, Thoirs RH, Rathbone E, Patakas A. A human in vitro T cell exhaustion model for assessing immuno-oncology therapies. In: Tan SL, editor. Immuno-oncology: cellular and translational approaches. New York: Springer US; 2020. p. 89–101. [Google Scholar]
- 48. Spandidos A, Wang X, Wang H, Seed B. PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res 2010;38:D792–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, et al. Integrated analysis of multimodal single-cell data. Cell 2021;184:3573–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. S1. Immune cell-dependent survival differences between males and females in murine glioblastoma models. Supplementary Fig. S2. Mouse syngeneic GBM cell lines do not contain a Y chromosome. Supplementary Fig. S3. Frequencies of tumor-infiltrating immune cells in SB28 model. Supplementary Fig. S4. Sex difference in survival in SB28-OVA model and GL261 model. Supplementary Fig. S5. No sex difference in T cells was observed at an earlier time point. Supplementary Fig. S6. Male T cells are more exhausted in the GL261 model. Supplementary Fig. S7. Phenotype of T cells in the periphery does not replicate the sex differences shown in tumor-infiltrating T cells. Supplementary Fig. S8. Frequencies of exhausted T cell subsets. Supplementary Fig. S9. PD1 blockade enhanced immune responses Supplementary Fig. S10. No significant difference in the immune cell composition of bone marrow chimera mice before tumor implantation. Supplementary Fig. S11. Male and female CD8+ T cells in mixed bone marrow chimera model. Supplementary Fig. S12. Phenotyping tumor-infiltrating CD8+ T cell from GBM patient tumors. Supplementary Fig. S13. scRNA-seq analysis of tumor-infiltrating CD8+ T cells from GBM patients. Supplementary Fig. S14. mRNA expression level of transcription factors in human in vitro exhausted T cells. Supplementary Fig. S15. Expression level of X chromosome inactivation (XCI) genes in exhausted T cells. Supplementary Fig. S16. Inhibition of UTX abrogates sex differences in T cell exhaustion.
Table S1. qPCR primers used for mouse study Table S2. qPCR primers used for human study
Data Availability Statement
All data generated in this study are available upon request from the corresponding author, Dr. Justin D. Lathia (lathiaj@ccf.org).