Unlike pan-FGFR inhibitors, RLY-4008 was designed to be selective for FGFR2 and induces clinical responses in FGFR2-altered solid tumors without clinically significant FGFR1-mediated hyperphosphatemia and FGFR4-mediated diarrhea.
Abstract
Oncogenic activation of fibroblast growth factor receptor 2 (FGFR2) drives multiple cancers and represents a broad therapeutic opportunity, yet selective targeting of FGFR2 has not been achieved. Although the clinical efficacy of pan-FGFR inhibitors (pan-FGFRi) validates FGFR2 driver status in FGFR2 fusion–positive intrahepatic cholangiocarcinoma, their benefit is limited by incomplete target coverage due to FGFR1- and FGFR4-mediated toxicities (hyperphosphatemia and diarrhea, respectively) and the emergence of FGFR2 resistance mutations. RLY-4008 is a highly selective, irreversible FGFR2 inhibitor designed to overcome these limitations. In vitro, RLY-4008 demonstrates >250- and >5,000-fold selectivity over FGFR1 and FGFR4, respectively, and targets primary alterations and resistance mutations. In vivo, RLY-4008 induces regression in multiple xenograft models—including models with FGFR2 resistance mutations that drive clinical progression on current pan-FGFRi—while sparing FGFR1 and FGFR4. In early clinical testing, RLY-4008 induced responses without clinically significant off-isoform FGFR toxicities, confirming the broad therapeutic potential of selective FGFR2 targeting.
Significance:
Patients with FGFR2-driven cancers derive limited benefit from pan-FGFRi due to multiple FGFR1–4-mediated toxicities and acquired FGFR2 resistance mutations. RLY-4008 is a highly selective FGFR2 inhibitor that targets primary alterations and resistance mutations and induces tumor regression while sparing other FGFRs, suggesting it may have broad therapeutic potential.
See related commentary by Tripathi et al., p. 1964.
This article is featured in Selected Articles from This Issue, p. 1949
INTRODUCTION
FGFR2 is a member of the fibroblast growth factor receptor (FGFR) family and plays a key role in cell survival and proliferation (1, 2). FGFR2 genetic alterations have been reported in many solid tumors, most commonly in intrahepatic cholangiocarcinoma (iCCA; 10%–16%; refs. 3, 4), endometrial cancer (7.5%–11%), and gastric/gastroesophageal junction cancer (3.7%–7.9%; refs. 5, 6). Oncogenic activation of FGFR2 can occur via gene amplification, activating mutation, or chromosomal rearrangement (1). FGFR2 fusions are the most common FGFR2 alteration in iCCA, occurring in 10% to 15% of cases (7), but are also found in a variety of other tumor types (5, 6). These oncogenic fusions typically consist of FGFR2 exons 1 to 17—encoding the intact extracellular and kinase domains—fused to a 3′ partner gene that permits protein dimerization, resulting in a functional protein that is constitutively active (7, 8). It was recently demonstrated that the dimerization domain of a fusion partner is not a requirement for oncogenicity; rather the truncation of FGFR2 after exon 17 (producing the previously identified FGFR2-C3 isoform; refs. 9, 10) is an oncogenic alteration itself, in which loss of the C-terminus is the key determinant of oncogenicity (11). These findings suggest that a broad population of patients with FGFR2 alterations could benefit from FGFR2-targeted therapy.
Clinical proof of concept of FGFR2 as a therapeutic target is illustrated by the approval of pan-FGFR inhibitors (pan-FGFRi; non–isoform-selective inhibitors of FGFRs 1–4) for the treatment of patients with cholangiocarcinoma bearing FGFR2 fusions/rearrangements (12–14) and for patients with urothelial carcinoma bearing FGFR2 or FGFR3 genetic alterations (15). However, the high degree of homology (in sequence and in structure) between the kinase domains of FGFRs has presented a challenge for traditional structure-based drug design; therefore, these inhibitors are not selective. Despite achieving response rates of 23% to 42% in cholangiocarcinoma (16–19), their clinical benefit has been limited by their incomplete target coverage due to side effects that prevent optimal dosing, and the emergence of FGFR2 resistance mutations (16–23). One of the most common adverse effects of pan-FGFRi is hyperphosphatemia, which was reported in 55% to 91% of patients [all Common Terminology Criteria for Adverse Events (CTCAE) grades] in phase II clinical trials and requires management via dietary modification, phosphate binders, and dose reduction or interruption (16–19). Hyperphosphatemia is attributed to the inhibition of FGFR1, which then increases phosphate reabsorption in the kidney (24). In addition, diarrhea is reported in 15% to 36% of patients (all CTCAE grades), owing to inhibition of FGFR4 (16–19, 25). Selective inhibition of FGFR2 is expected to provide superior target coverage, leading to substantially improved response rates.
Increased activity against on-target resistance mutations is also expected to improve durability, as disease progression during treatment of FGFR2-altered cholangiocarcinoma with pan-FGFRi is often driven by acquired resistance mutations. Common acquired FGFR2 resistance mutations include kinase domain mutations at the gatekeeper residue (FGFR2V564F/L/I) as well as FGFR2N549K/D/H and FGFR2K659N/M mutations that promote ligand-independent kinase activation (1, 21–23, 26, 27). In a report of 46 patients with advanced FGFR2 fusion/rearrangement–positive cholangiocarcinoma treated with pan-FGFRi, monoclonal (n = 1 mutation; 24%) or polyclonal (n >1 mutation; 26%) FGFR2 kinase domain mutations were detected in 50% of patients at progression (23). Rapid emergence of on-target polyclonal resistance occurs at the approved doses of pan-FGFRi, as described in the literature (21, 22) and as evidenced by patients who enrolled on the ReFocus phase I/II trial (28). Of 25 FGFR2 fusion/rearrangement–positive cholangiocarcinoma patients evaluable by circulating tumor DNA (ctDNA), one of six (17%) FGFRi-naive patients, five of 12 (42%) patients with one previous line of pan-FGFRi therapy, and six of seven (86%) patients with two or more previous lines of pan-FGFRi therapy at their approved doses had FGFR2 kinase domain resistance mutations at baseline (29). Preclinical evidence suggests that bypass resistance, through the switch of dependence from FGFR2 to other receptor tyrosine kinases such as EGFR, may be responsible for resistance in patients lacking on-target mutations (30).
To overcome these limitations, we leveraged differences in the conformational dynamics between FGFR2 and other FGFRs to identify novel opportunities for optimal and selective FGFR2 inhibition. Here we describe RLY-4008, the first highly selective, irreversible, small-molecule FGFR2 inhibitor specifically designed to overcome the limitations of pan-FGFRi via targeting of oncogenic FGFR2 alterations and resistance mutations. We present preclinical characterization in biochemical assays, cell-based assays, and in vivo cancer models that validates RLY-4008's unique mechanism of action. To augment these data, we present three case studies from the ongoing phase I/II, first-in-human study of RLY-4008 (ReFocus) in which RLY-4008 has shown high response rates in patients with FGFRi-naive cholangiocarcinoma harboring an FGFR2 fusion or rearrangement, as well as responses in patients with other FGFR2-altered tumors and activity against common FGFR2 resistance mutations (31). These case studies demonstrate that RLY-4008 induces a durable radiographic response in pan-FGFRi–naive and pretreated patients without clinically significant off-isoform toxicity (hyperphosphatemia, diarrhea), suggesting its unique preclinical profile translates to the clinical setting.
RESULTS
RLY-4008 Is a Potent and Selective Irreversible Inhibitor of FGFR2
Despite significant investment in traditional structure-based drug design, selective targeting of FGFR2 has remained elusive. The kinase domain of FGFR2 is 95%, 96%, and 88% similar to the kinase domains of FGFR1, FGFR3, and FGFR4, respectively, and is 87% identical to both FGFR1 and FGFR3, and 77% identical to FGFR4 (Fig. 1A). This high sequence and structural similarity among FGFRs includes a cysteine residue (Cys491) at the tip of the phosphate-binding loop (P-loop) that has been successfully targeted by the irreversible pan-FGFRi futibatinib (32). We leveraged differences in conformational dynamics between FGFR2 and other FGFRs observed through long timescale molecular dynamics simulations to enable the design of RLY-4008, an FGFR2-selective inhibitor that covalently binds to Cys491 (Fig. 1B). In FGFR1, the P-loop has a wide range of motion and displays rapid dynamics, whereas the FGFR2 P-loop is less dynamic. The reversible binding of RLY-4008 exploits these differences to promote a rigid and extended P-loop in FGFR1 that disfavors covalent bond formation while minimally affecting the conformation of the P-loop in FGFR2, enabling efficient covalent bond formation and leading to FGFR2 selectivity (Supplementary Video). Notably, all residues that bind RLY-4008 are conserved between FGFR2 and FGFR1; 32/34 and 31/34 binding site residues are conserved between FGFR2 and FGFR3 and FGFR4, respectively (Fig. 1A).
Figure 1.
RLY-4008 is a potent and selective irreversible inhibitor of FGFR2. A, Sequence alignment of the kinase domains of FGFR1–4 indicates a high degree of similarity among paralogs. RLY-4008 binding site residues are boxed; residues shown in pink identify amino acid differences between FGFR2 and paralogs within this region. Numbering refers to the FGFR2 IIIc isoform. B, Chemical structure of RLY-4008, N-(4-(4-amino-5-(3-fluoro-4-((4-methylpyrimidin-2-yl)oxy)phenyl)-7-methyl-7H-pyrrolo[2,3-d]pyrimidin-6-yl)phenyl)methacrylamide. C, Crystal structure of RLY-4008 in complex with FGFR2 (PDB: 8STG). Protein is shown in green, and inhibitor carbons are shown in magenta. The Cys491 sulfur is shown in gold and as a covalent adduct with RLY-4008. D and E, Rate of covalent labeling of FGFR2 (red) and FGFR1 (blue) by RLY-4008 (D) and futibatinib (E) as measured by intact mass over time. Triplicate biological replicates are reported. F, RLY-4008 concentration-dependent modification rate against FGFR2 (red) and FGFR1 (blue). RLY-4008 against FGFR2: kinact = 6.45 × 10−2 per second; KI = 1.87 μmol/L; kinact/KI = 3.45 × 10−2 per second/(μmol/L). RLY-4008 against FGFR1: kinact = 2.33 × 10−3 per second; KI = 6.14 μmol/L; kinact/KI = 3.79 × 10−4 per second/(μmol/L). G, Fold change in biochemical IC50 values of the indicated inhibitors between FGFR2 and FGFR1, FGFR3, and FGFR4. Average fold change of three independent experiments each containing two biological replicates is reported. Error bars indicate SD. H, TREEspot depicting selectivity of RLY-4008 screened against 468 kinases via KINOMEscan (DiscoverX, Eurofins). At the test concentration of 500 nmol/L, three kinases showed greater than 75% inhibition: FGFR2 (94.1%), MEK5 (92.4%), and MKNK2 (89%). Image generated using TREEspot Software Tool and reprinted with permission from KINOMEscan, a division of DiscoveRx Corporation, ©DiscoverX Corporation 2010.
The covalent binding and inhibition of FGFR2 by RLY-4008 were characterized by both biophysical and biochemical techniques. A crystal structure of the RLY-4008 and FGFR2 inhibitor:kinase complex showed binding of RLY-4008 in the ATP-binding pocket, with the acrylamide warhead forming a covalent bond with Cys491 [Fig. 1C; Protein Data Bank (PDB) ID: 8STG]. The pyrrolo-pyrimidine core of the compound occupies a similar binding pose as the adenine of ATP and is stabilized by hydrogen bond interactions with the hinge residues (aa 565–567). Notably, RLY-4008 stabilizes FGFR2 in a conformation that has not been observed in published FGFR2 crystal structures. To compare structures, we superimposed all ligand-bound and ligand-unbound FGFR2 kinase domain structures in the PDB and found consistency in the N-lobe orientation with respect to the C-lobe. In contrast, the N-lobe of RLY-4008–bound FGFR2 is rotated in a novel conformation (Supplementary Fig. S1A). Compared with the structure of FGFR2 bound to an ATP analogue, the N-lobe of RLY-4008–bound FGFR2 is rotated by ∼18 degrees and the C-helix center of mass is rotated out by ∼4.5 Å (PDB ID: 2PVF). The reacted warhead holds Cys491 in an unfavorable torsion angle, pulling the P-loop into a down conformation and into the inhibited state. Structural changes in the back pocket propagate through the hydrophobic spine to accommodate RLY-4008 (Supplementary Fig. S1B). RLY-4008 engages an ensemble of P-loop conformations in FGFR2 that optimally position Cys491 relative to the warhead, facilitating covalent bond formation. In contrast, molecular dynamics simulations of the reversible binding complex between RLY-4008 and FGFR1 reveal that RLY-4008 induces a rigid, extended P-loop conformation in FGFR1 that does not align the Cys for covalent bond formation, resulting in markedly weaker inhibition.
Mass spectrometry was performed to confirm that RLY-4008 covalently modifies the sulfhydryl group of Cys491 on FGFR2. Incubation of recombinant FGFR2 with an excess of RLY-4008 resulted in an intact mass shift consistent with the formation of a covalent RLY-4008:FGFR2 complex (Supplementary Fig. S1C). After approximately 1 hour at room temperature, FGFR2 was completely labeled with a single molecule of RLY-4008; no evidence of multiple labeling events was detected. To confirm the targeting of Cys491 by RLY-4008, recombinant unlabeled FGFR2 and covalently complexed samples were analyzed by liquid chromatography/mass spectrometry (LC/MS; Supplementary Fig. S1D). A peptide unique to the covalently modified sample was confirmed to have a mass consistent with RLY-4008 adduct formation, and a combination of exact mass and tandem mass spectrometry indicated that RLY-4008 was bound to Cys491.
To compare the rate of covalent labeling of FGFR2 by RLY-4008 to that of FGFR1, 100 μmol/L RLY-4008 was incubated with recombinant FGFR2 or FGFR1 at room temperature and covalent adduct formation was detected by intact mass shift over time. Given the assumption that 100 μmol/L is significantly greater than the compound's reversible equilibrium binding constant, these conditions allowed for the determination of the rate of covalent modification (kinact, where t1/2 = 0.693/kinact). Although FGFR2 was labeled rapidly (t1/2 = 8 s), labeling of FGFR1 occurred much more slowly (t1/2 = 351 s; Fig. 1D), consistent with the observation from molecular dynamics simulations that RLY-4008 induces a conformation in FGFR1 that is not favorable for covalent bond formation. In contrast, the irreversible pan-FGFRi, futibatinib, covalently labeled both FGFR1 and FGFR2 at a similar rate (FGFR1 t1/2 = 8 s; FGFR2 t1/2 = 13 s) under identical experimental conditions (Fig. 1E), consistent with its reported covalent inhibition of all FGFRs (33).
For an irreversible inhibitor such as RLY-4008, potency is expressed by the ratio of the inactivation rate constant (kinact) to the binding constant (KI) (kinact/KI) as FGFR2 is covalently modified by RLY-4008 over time. In vitro mass spectrometry studies were performed to determine the kinact/KI ratio of modification of FGFR1 and FGFR2 by RLY-4008 (Fig. 1F). Varying concentrations of RLY-4008 were incubated with a fixed concentration of wild-type (WT) FGFR2 or FGFR1 protein, and the amount of unmodified protein remaining over time was detected by LC/MS. These data were used to determine the rate of covalent adduct formation at each concentration of RLY-4008. By analyzing that rate as a function of RLY-4008 concentration, the reversible inhibitor dissociation constant was obtained. The kinact/KI ratio of modification of FGFR1 and FGFR2 by RLY-4008 is 3.79 × 10−4 per s/μmol/L and 3.45 × 10−2 per s/μmol/L, respectively, indicating that RLY-4008 covalently modifies FGFR2 at a rate >90 times faster than FGFR1 (Supplementary Table S1).
This mechanism of action of RLY-4008 affords not only selectivity for FGFR2 over FGFR1 but also a high degree of selectivity for FGFR2 over all FGFR family members. RLY-4008 demonstrates potent inhibition of FGFR2 enzyme activity in biochemical assays (IC50 = 3 nmol/L) but markedly weaker inhibition of other FGFRs (Supplementary Table S2). RLY-4008 has >250-fold selectivity over FGFR1, and >80- and >5,000-fold selectivity over FGFR3 and FGFR4, respectively (Fig. 1G). In contrast, pan-FGFRi demonstrate little to no selectivity (Fig. 1G; Supplementary Table S2). The covalent adduct formation between RLY-4008 and FGFR2 affords significant potency, as a noncovalent analogue with a saturated warhead demonstrates weak inhibition of FGFR2 (IC50 = 1,344 nmol/L; Supplementary Fig. S1E).
Evaluation of RLY-4008 activity showed a high degree of selectivity against 468 kinases and disease-relevant kinase mutants using the KINOMEscan Profiling Service scanMAX (Eurofins; Fig. 1H). At the test concentration of 500 nmol/L (which resulted in 94.1% inhibition of FGFR2), only two additional kinases showed greater than 75% inhibition: MEK5 (92.4%) and MKNK2 (89%). Neither kinase has a cysteine residue in the same position on the P-loop as the FGFRs and is thus not expected to be inhibited irreversibly by RLY-4008. Therefore, FGFR2 is the only kinase expected to be irreversibly inhibited by RLY-4008, resulting in previously unachieved selectivity.
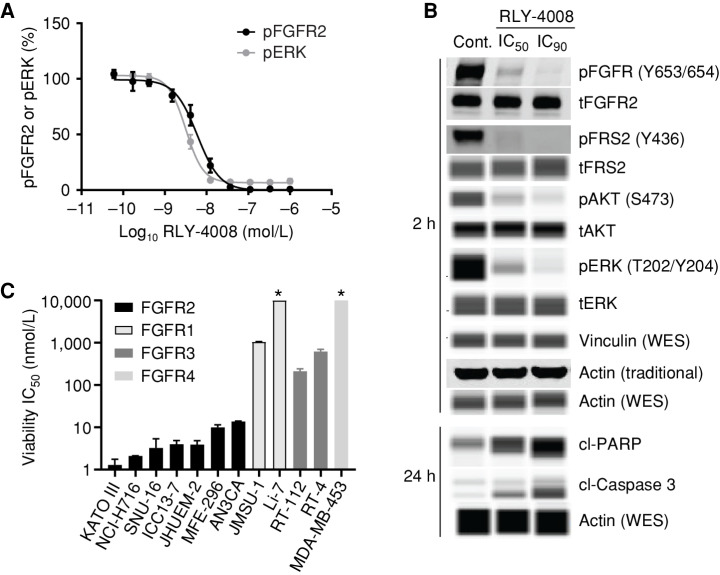
RLY-4008 Inhibits FGFR2-Mediated Signaling and Selectively Inhibits Proliferation in FGFR2-Driven Cancer Cell Lines
To demonstrate the effects of RLY-4008 on cells, we initially used the FGFR2-amplified gastric cancer cell line SNU-16. FGFR2 inhibition was determined by measuring FGFR2 phosphorylation (pFGFR2; Tyr653/654) and phosphorylation of its downstream effector, ERK1/2 (pERK; Thr202/Tyr204), following 2 hours of treatment (Fig. 2A). The cellular potency (expressed as IC50) of RLY-4008 is 6 nmol/L (pFGFR) and 3 nmol/L (pERK), consistent with the observed biochemical potency on FGFR2. RLY-4008 also demonstrated a dose-dependent reduction of phosphorylation of FGFR2 signaling pathway nodes, including fibroblast growth factor receptor substrate 2 (FRS2), AKT, and ERK (Fig. 2B). Following a 24-hour treatment, RLY-4008 induced dose-dependent cleavage of caspase-3 and poly (ADP-ribose) polymerase (PARP)—early markers of apoptosis (Fig. 2B). Notably, RLY-4008 does not inhibit FGFR2 phosphorylation in the FGFR2 fusion–positive (FGFR2–OPTN) iCCA cell line ICC13-7 in which an FGFR2–OPTN fusion protein containing the Cys491Ser mutation is overexpressed (ICC13-7–FGFR2C491S; Supplementary Fig. S1F), confirming the on-target nature of RLY-4008 cellular activity.
Figure 2.
RLY-4008 inhibits FGFR2-mediated signaling and proliferation in cells. A, Inhibition of FGFR2-mediated signaling in SNU-16 cells. Cells were incubated with RLY-4008 for 2 hours prior to lysis and analysis via pFGFR2 (Y653/654) and pERK (T202/Y204) HTRF (PerkinElmer). B, Inhibition of FGFR2-mediated signaling and induction of apoptosis in SNU-16 cells. Immunoblots of cell lysates generated from cells treated with DMSO (Control), IC50, or IC90 concentrations of RLY-4008 (as determined by pFGFR2 HTRF) for the indicated times. Samples were analyzed via traditional Western or via WES (Protein Simple). Loading controls were actin (traditional) and actin and vinculin (WES). cl, cleaved; p, phospho; t, total. C, Viability IC50 values for RLY-4008 in FGFR2-, FGFR1-, FGFR3-, and FGFR4-dependent cancer cell lines (Supplementary Table S3). Cells were treated for 96 hours and cellular viability was assayed using CellTiter-Glo (Promega). Average IC50 of two independent experiments each containing two biological replicates is reported. Error bars indicate SD. * Indicates that IC50 was not reached in this cell line (maximum RLY-4008 concentration = 10 μmol/L).
To determine the selectivity of RLY-4008 in cells, we evaluated its effects on the proliferation of FGFR2-dependent (KATO III, NCI-H716, SNU-16, ICC13-7, JHUEM-2, MFE-296, AN3CA), FGFR1-dependent (JMSU-1, Li-7), FGFR3-dependent (RT-112, RT-4), and FGFR4-dependent (MDA-MB-453) cell lines (Fig. 2C; Supplementary Table S3). RLY-4008 inhibited cellular proliferation with IC50 <14 nmol/L in FGFR2-dependent cell lines including those derived from FGFR2-amplified gastric carcinoma (KATO III, SNU-16) and colorectal adenocarcinoma (NCI-H716), FGFR2 fusion–positive iCCA (ICC13–7), and FGFR2-mutant endometrial adenocarcinoma (MFE-296, FGFR2N549K and AN3CA, FGFR2K310R;N549K) and breast carcinoma (JHUEM-2, FGFR2C383R). In contrast to pan-FGFRi, RLY-4008 did not have strong inhibitory activity in FGFR1-, FGFR3-, or FGFR4-dependent cell lines. The potency (expressed as IC50) of RLY-4008 on these cell lines was 212 nmol/L to >10 mmol/L, demonstrating a high degree of selectivity for RLY-4008 on FGFR2 relative to other FGFR family members in cellular assays (Supplementary Table S3). RLY-4008 did not inhibit the proliferation of ICC13-7–FGFR2C491S, confirming that the antiproliferative activity of RLY-4008 is on-target (Supplementary Fig. S1G).
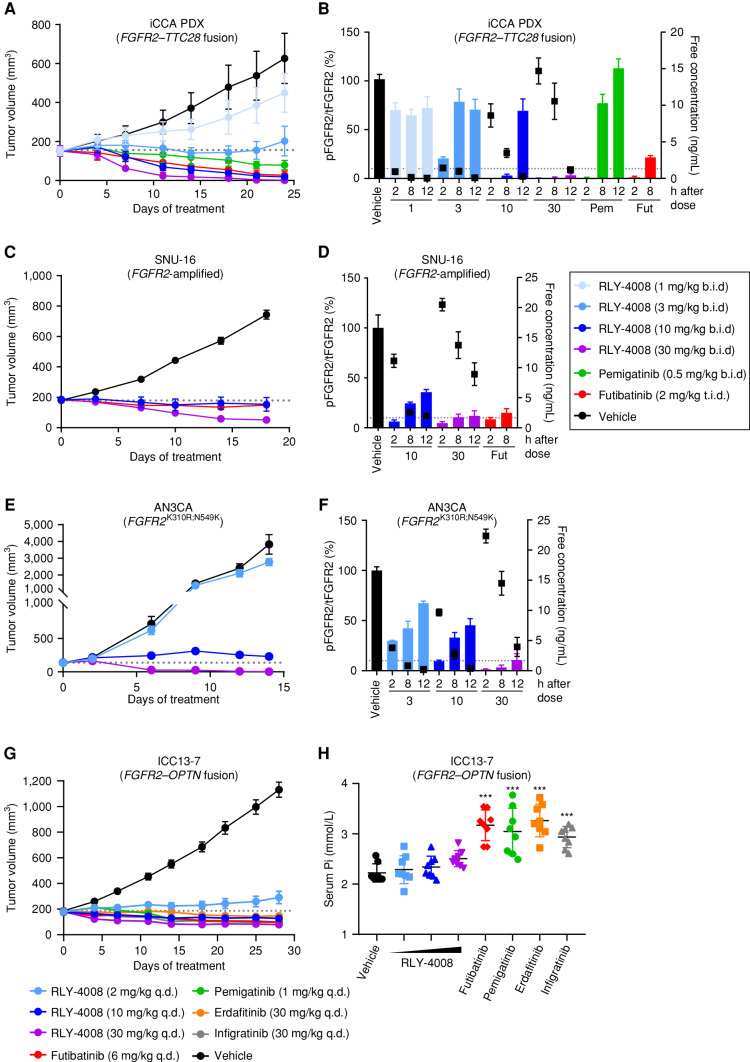
RLY-4008 Demonstrates Antitumor Activity in FGFR2-Altered Cancer Xenograft Models and Spares FGFR1 In Vivo
Given the observation that RLY-4008 inhibits the proliferation of multiple FGFR2-altered cancer cell lines in vitro, we evaluated the effects of RLY-4008 in subcutaneous xenograft mouse models harboring different FGFR2 alterations. These models included an FGFR2 fusion–positive (FGFR2–TTC28) iCCA patient-derived xenograft (Fig. 3A and B) and two cell line–derived xenografts, SNU-16 (FGFR2-amplified gastric carcinoma; Fig. 3C and D) and AN3CA (FGFR2-mutant endometrial adenocarcinoma; FGFR2K310R;N549K; Fig. 3E and F). RLY-4008, administered orally, twice daily from 1 to 30 mg/kg, exhibited dose-dependent antitumor activity and induced tumor regression in all models (Fig. 3A, C, and E). RLY-4008 was well tolerated at all doses, showing no adverse effects on body weight (Supplementary Fig. S2A). Pemigatinib and futibatinib, dosed to achieve exposures similar to those achieved at their recommended human doses of 13.5 mg and 20 mg, respectively (Supplementary Table S4), were less efficacious than 30 mg/kg of RLY-4008.
Figure 3.
Treatment with RLY-4008 leads to dose-dependent inhibition of FGFR2 and tumor regression in multiple FGFR2-altered tumor models and spares FGFR1 in vivo. A–F, Refer to boxed legend. A, C, E, and G, Dotted line indicates tumor volume prior to initiation of treatment. B, D, and F, Dotted line indicates 10% pFGFR2/tFGFR2 (90% inhibition of pFGFR2). A, Antitumor activity of RLY-4008 compared with pemigatinib and futibatinib in an FGFR2–TTC28 iCCA patient-derived xenograft (PDX) model (n = 6/group). Data are mean ± SEM. B, Dose-dependent inhibition of FGFR2 in FGFR2–TTC28 tumors. Animals were sacrificed, and tumors were harvested at the indicated time points after the final dose on the third day of dosing. Tumor lysates were analyzed via pFGFR2 (Y653/654) ELISA and tFGFR2 HTRF; pFGFR2 normalized to tFGFR2 is reported (n = 3/group). Free plasma concentration of RLY-4008 is reported. Data are mean ± SEM. Fut, futibatinib; Pem, pemigatinib. C, Antitumor activity of RLY-4008 compared with futibatinib in the SNU-16 gastric cancer xenograft model (n = 7/group). Data are mean ± SEM. D, Dose-dependent inhibition of FGFR2 in SNU-16 tumors. Animals were sacrificed and tumors were harvested at the indicated time points after the final dose on the fourth day of dosing. Tumor lysates were analyzed via pFGFR2 (Y653/654) and tFGFR2 HTRF; pFGFR2 normalized to tFGFR2 is reported (n = 3/group). Free plasma concentration of RLY-4008 is reported. Data are mean ± SEM. E, Antitumor activity of RLY-4008 in the AN3CA endometrial cancer xenograft model (n = 8/group). Data are mean ± SEM. F, Dose-dependent inhibition of FGFR2 in AN3CA tumors. Animals were sacrificed and tumors were harvested at the indicated time points after the final dose on the third day of dosing. Tumor lysates and plasma were analyzed and reported as in B (n = 3/group). G, Antitumor activity of RLY-4008 and pan-FGFRi futibatinib, pemigatinib, erdafitinib, and infigratinib in an FGFR2–OPTN iCCA cell line–derived xenograft model, ICC13–7 (n = 8/group). Data are mean ± SEM. H, RLY-4008 spares FGFR1 in vivo. Two hours after the final dose of the study shown in G, blood was collected from all animals for serum phosphate analysis (n = 8/group). Data are mean ± SEM. ***, P < 0.0001, one-way ANOVA. b.i.d., twice daily; q.d., once daily; t.i.d., three times daily.
Pharmacokinetic and pharmacodynamic (PK/PD) analyses of plasma exposure and target engagement in tumor samples demonstrated dose-dependent inhibition of FGFR2 in all models (Fig. 3B, D, and F). In each model, at a dose of 30 mg/kg twice daily, RLY-4008 led to ≥90% pFGFR2 inhibition that was sustained throughout the 12-hour dosing interval and resulted in tumor regression. These data demonstrate that sustained ∼90% inhibition of pFGFR2 by RLY-4008 is achievable at exposures that are well tolerated. In the FGFR2 fusion–positive iCCA model (Fig. 3A and B), <90% sustained pFGFR2 inhibition also led to tumor regression, suggesting FGFR2 fusion–positive tumors have a greater dependency on FGFR2 signaling and are thus more sensitive to inhibition of FGFR2. Studies in additional FGFR2 fusion–positive non-iCCA patient-derived xenograft models confirm this finding (Supplementary Fig. S2B–S2E).
To evaluate the effects of RLY-4008 and pan-FGFRi on FGFR1 activity at efficacious doses in vivo, we measured tumor volume and serum phosphate levels in the FGFR2 fusion–positive (FGFR2–OPTN) iCCA xenograft model, ICC13-7. This model—implanted in nonobese diabetic/severe combined immunodeficiency (NOD SCID) mice—does not tolerate twice-daily dosing; thus all compounds were dosed once daily. Futibatinib, pemigatinib, erdafitinib, and infigratinib were dosed to achieve exposures similar to those achieved at their approved human doses of 20 mg, 13.5 mg, 9 mg, and 125 mg, respectively (Supplementary Table S4). All compounds caused tumor regression (Fig. 3G), whereas only pan-FGFRi demonstrated a statistically significant increase in serum phosphate, consistent with clinical observations (32%–47% over vehicle; P < 0.0001, one-way ANOVA; Fig. 3H). RLY-4008 led to tumor regression at doses of 10 and 30 mg/kg once daily without significantly altering serum phosphate levels (P > 0.2, one-way ANOVA), demonstrating that RLY-4008 is the only molecule tested that spares FGFR1 at efficacious exposures in vivo (Fig. 3H).
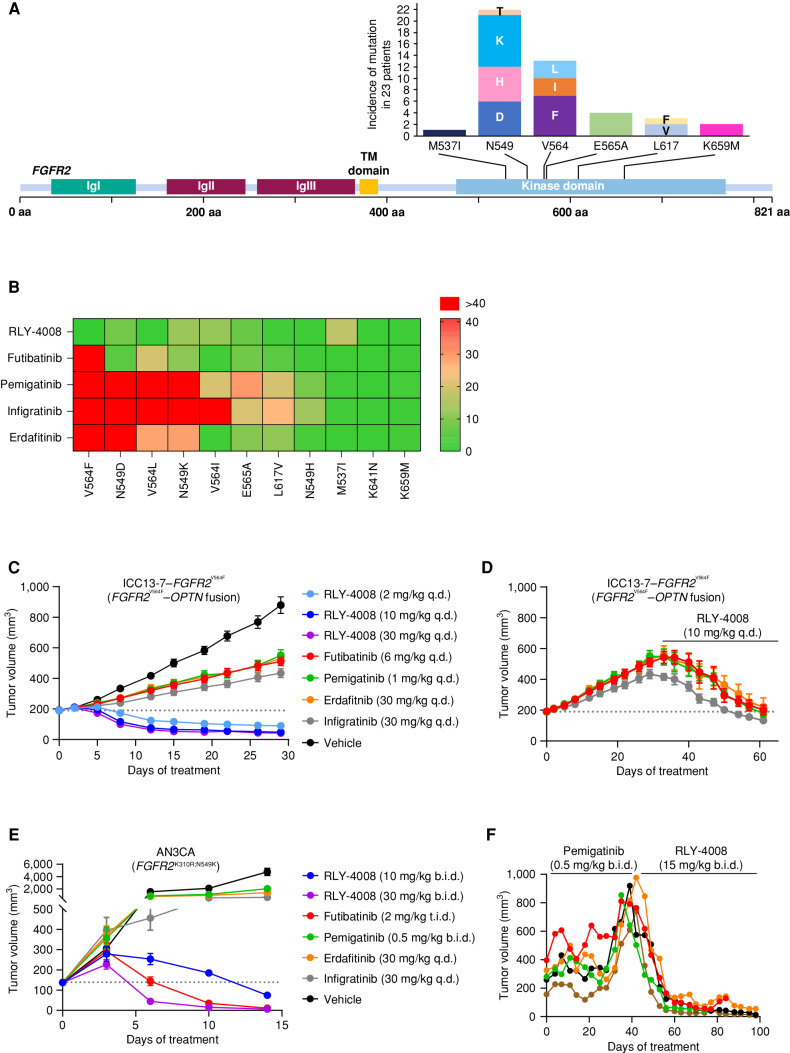
RLY-4008 Retains Potency on FGFR2 Resistance Mutations
The emergence of on-target resistance mutations limits the clinical efficacy of current pan-FGFRi (Fig. 4A; refs. 23, 34). RLY-4008 is designed to target primary FGFR2 alterations and acquired resistance mutations. We tested this by generating a panel of 293T cell lines expressing WT or mutant FGFR2 and measured the level of pFGFR2 following 2 hours of compound treatment (Fig. 4B; Supplementary Table S5). Two of the most common FGFR2 kinase domain mutations detected in FGFR2 fusion–positive iCCA at progression on pan-FGFRi are the FGFR2V564F “gatekeeper” mutation and the FGFR2N549K mutation, which relieves the “molecular brake” (21–23, 27). Our data align with these clinical findings, showing that current pan-FGFRi do not retain potency on FGFR2V564F, resulting in a >55-fold shift in IC50 between FGFR2WT and this mutation. In contrast, RLY-4008 is more potent on FGFR2V564F than FGFR2WT. For the FGFR2N549K mutation, RLY-4008 and futibatinib demonstrate an ∼10-fold shift in potency relative to FGFR2WT, whereas other pan-FGFRi demonstrate shifts of ∼30 to >100-fold. Consistent with this, in cellular proliferation assays, RLY-4008 demonstrates a 3- to 5-fold IC50 shift between cells expressing an FGFR2WT kinase domain and those harboring the FGFR2N549K mutation (Supplementary Table S3). Overall, RLY-4008 exhibits broad coverage of FGFR2 resistance mutations.
Figure 4.
RLY-4008 is active on mutations associated with acquired resistance to pan-FGFRi. A, Acquired resistance mutations in the FGFR2 kinase domain are commonly found in patients with FGFR2 fusion– or rearrangement–positive iCCA treated with pan-FGFRi. The graph indicates the number of times the indicated mutant allele was detected in tissue or ctDNA in 23 patients (out of 46) who developed FGFR2 kinase domain mutations at progression on pan-FGFRi. Figure art adapted from Varghese et al. and patient data are from Goyal et al. (23, 34). B, Heat map displaying the fold change in potency (IC50) for the indicated inhibitors against the indicated FGFR2 mutant as compared with FGFR2 WT. Numbering of mutant residues refers to the FGFR2 IIIc isoform to remain consistent with the usage of this nomenclature. Following 2 hours of incubation with the compound, FGFR2 inhibition was determined via pFGFR2 (Y653/654) HTRF assay, and IC50 values against FGFR2 WT and FGFR2 mutants were calculated. The average fold change of three independent experiments each containing two biological replicates was used to derive a heat map in GraphPad Prism. Fold change of one indicates equivalent potency on FGFR2 WT and the indicated FGFR2 mutant. C–E, Dotted line indicates tumor volume prior to initiation of treatment. C, Antitumor activity of RLY-4008 compared with futibatinib, pemigatinib, erdafitinib, and infigratinib in the ICC13-7–FGFR2V564F xenograft model (n = 8/group). Only RLY-4008 leads to tumor regression. Data are mean ± SEM. D, Following 28 days of treatment on the indicated inhibitors, animals on pan-FGFRi in the study shown in C were changed to treatment with RLY-4008 10 mg/kg once daily. Tumor regression was observed in all animals receiving RLY-4008. Data are mean ± SEM. E, Antitumor activity of RLY-4008 compared with futibatinib, pemigatinib, erdafitinib, and infigratinib in the AN3CA (FGFR2K310R;N549K) endometrial cancer xenograft model (n = 7/group). Only RLY-4008 and futibatinib treatment lead to tumor regression. Data are mean ± SEM. F, RLY-4008 overcomes acquired resistance to pemigatinib in vivo. Antitumor activity of pemigatinib followed by RLY-4008 in an FGFR2–TTC28 iCCA patient-derived xenograft model. Animals were dosed with pemigatinib for 40 days, followed by treatment with RLY-4008 from days 42–98. Each line represents one animal. b.i.d., twice daily; q.d., once daily; t.i.d., three times daily.
To assess the in vivo activity of RLY-4008 on FGFR2V564F, we generated an FGFR2 fusion–positive iCCA cell line with this mutation via CRISPR-mediated knock-in (ICC13-7–FGFR2V564F). ICC13-7–FGFR2V564F xenograft tumors had similar growth kinetics to ICC13-7 tumors (Figs. 3G and 4C). RLY-4008 and pan-FGFRi were administered at the same dose and once-daily schedule as in the ICC13-7 model. Strikingly, RLY-4008 caused rapid regression of ICC13-7–FGFR2V564F tumors at a dose of 2 mg/kg once daily (Fig. 4C), lower than that required to drive regression of ICC13-7 tumors (Fig. 3G). These data are consistent with the increased potency of RLY-4008 on FGFR2V564F compared with FGFR2WT in cellular assays (Fig. 4B). In contrast, pan-FGFRi were ineffective (Fig. 4C), consistent with the finding that they do not retain potency on FGFR2V564F (Fig. 4B). Notably, in ICC13-7–FGFR2V564F tumors that progressed on pan-FGFRi, RLY-4008 (10 mg/kg once daily) induced rapid regression and restored body weight (Fig. 4D; Supplementary Fig. S2F).
To assess the in vivo activity of RLY-4008 and pan-FGFRi on FGFR2N549K, we used the AN3CA (FGFR2-mutant endometrial adenocarcinoma; FGFR2K310R;N549K) xenograft model. RLY-4008 induced tumor regression at a dose of 10 mg/kg twice daily and resulted in complete tumor regression after 14 days of dosing with 30 mg/kg twice daily (Fig. 4E). Futibatinib was the only pan-FGFRi to demonstrate antitumor activity in this model. These results are consistent with the ∼10-fold shift in potency between FGFR2WT and FGFR2N549K observed for RLY-4008 and futibatinib (Fig. 4B), indicating that this relatively modest cellular shift does not prevent RLY-4008 from achieving levels of target inhibition required for complete tumor regression in vivo. As anticipated from the cellular assays, pemigatinib, erdafitinib, and infigratinib demonstrated minimal antitumor activity in this model.
Because the lack of tolerability of pan-FGFRi and the emergence of resistance limits their efficacy, we sought to model this in vivo and to determine whether RLY-4008 is efficacious following the emergence of resistance to pan-FGFRi. We treated an FGFR2 fusion–positive (FGFR2–TTC28) iCCA patient-derived xenograft model with pemigatinib (0.5 mg/kg twice daily, to achieve exposures similar to those achieved at the approved human dose of 13.5 mg once daily; Supplementary Table S4) and monitored tolerability and efficacy. Following 6 weeks of treatment, five animals whose tumors progressed on pemigatinib were changed to the regimen of 15 mg/kg RLY-4008 twice daily. Two additional animals that progressed on pemigatinib were added to this treatment group after ∼10 weeks of treatment. RLY-4008 caused an immediate and rapid regression of all seven tumors (Fig. 4F; Supplementary Fig. S2G). Together, these data demonstrate the unique profile of RLY-4008—unprecedented selectivity for FGFR2 and potent targeting of both primary alterations and acquired resistance mutations—suggesting that RLY-4008 may overcome the key limitations of currently available pan-FGFRi.
RLY-4008 Demonstrates Selective FGFR2 Targeting and Meaningful Clinical Activity in Patients Refractory to Chemotherapy: Clearance of FGFR2-Mutant Clones and Induction of Durable Radiographic Response without Clinically Relevant Hyperphosphatemia or Diarrhea
Clinical activity of RLY-4008 is illustrated by patient cases from an ongoing phase I/II trial in patients with solid tumors (ReFocus; NCT04526106; ref. 31).
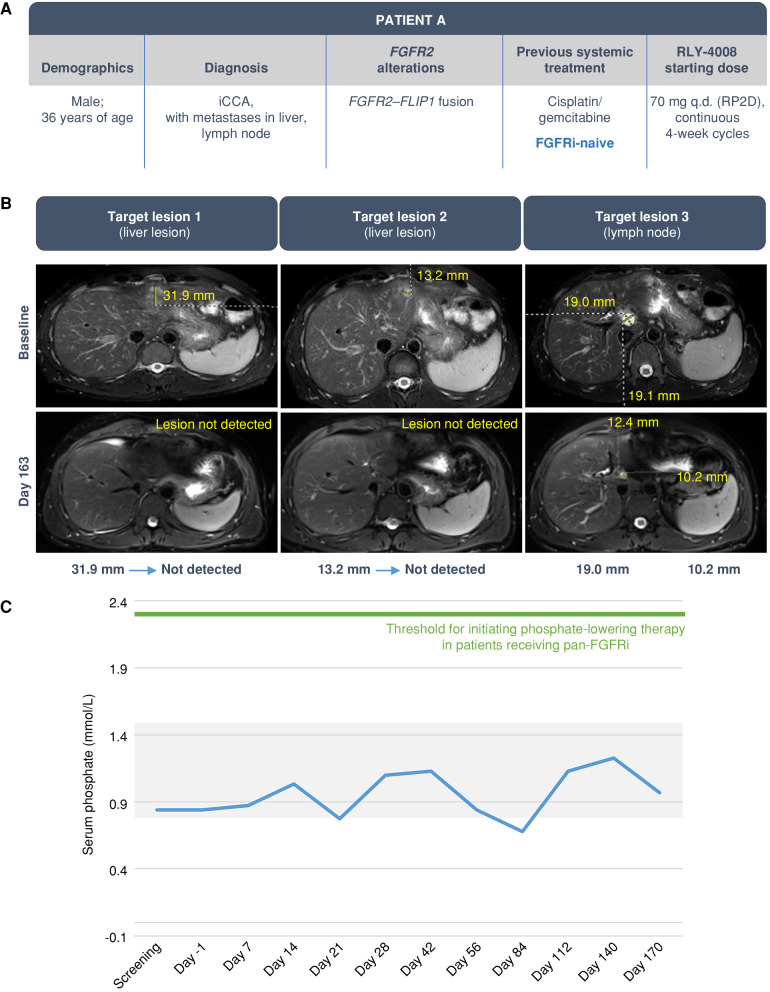
Patient A, a 36-year-old male (Fig. 5) with advanced, unresectable iCCA bearing an FGFR2–FLIP1 fusion who was previously treated with cisplatin/gemcitabine, was enrolled in the ReFocus phase I dose escalation and treated with RLY-4008 at 70 mg once daily (Fig. 5A). After 163 days of therapy, near complete tumor regression was observed, with an 84% reduction in the sum of target lesions and a confirmed partial response (PR) per RECIST version 1.1 (Fig. 5B). Given the marked tumor reduction, the patient underwent surgical resection of his liver tumors with curative intent on day 179 and completed 6 months of adjuvant treatment with RLY-4008. There was no elevation of serum phosphate levels above the normal range throughout treatment with RLY-4008 (Fig. 5C) and no diarrhea, confirming selective inhibition of FGFR2. RLY-4008 was generally well tolerated with low-grade, reversible, and manageable FGFR2 on-target toxicities including onycholysis, dry eye, and stomatitis; no dose interruption or reduction was necessary.
Figure 5.
Clinical response in an FGFRi-naive iCCA patient with liver and lymph node metastases. The patient was treated with RLY-4008 at 70 mg once daily, the RP2D. A, Summary of key patient and disease characteristics. B, CT scans of liver and lymph node metastases at baseline (top) and after 163 days of RLY-4008 treatment (bottom) show profound tumor regression. C, Serum phosphate over the course of treatment with RLY-4008. The shaded area represents the normal range for serum phosphate (0.8–1.5 mmol/L). q.d., once daily; RP2D, recommended phase II dose.
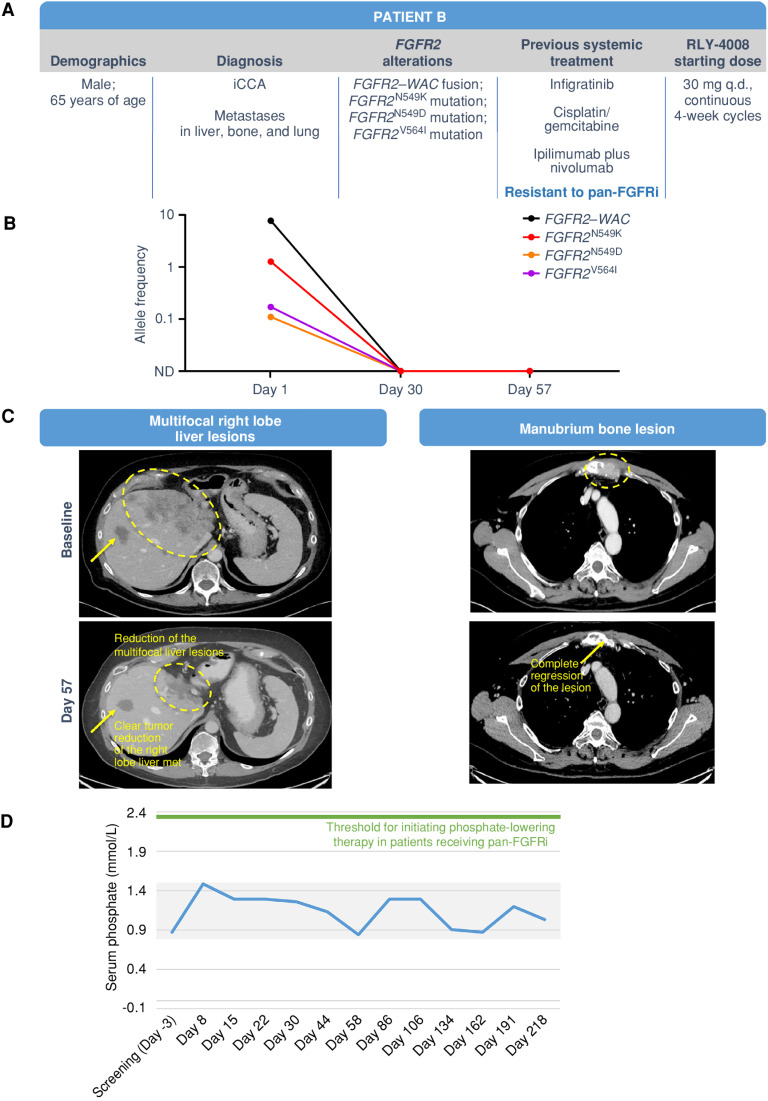
Patient B (Fig. 6) was a 65-year-old male with metastatic iCCA bearing an FGFR2–WAC fusion and the FGFRi resistance mutations FGFR2N549K, FGFR2N549D, and FGFR2V564I. He was enrolled in the ReFocus phase I dose escalation and treated with RLY-4008, initially at 30 mg once daily and subsequently at 50 and 70 mg once daily (per protocol, which permitted intrapatient dose escalation based on tolerability). This patient had received three lines of prior systemic therapy, including a pan-FGFRi (infigratinib), gemcitabine/cisplatin, and immunotherapy (ipilimumab plus nivolumab; Fig. 6A). Clinically, the patient had a dramatic reduction in pain at all tumor sites and an increase in appetite within a couple of weeks of initiating RLY-4008. By day 30, ctDNA analysis demonstrated complete clearance of FGFR2N549K, FGFR2N549D, and FGFR2V564I clones (Fig. 6B), consistent with the preclinical profile of RLY-4008 and confirming clinical activity against FGFRi resistance mutations. A marked radiographic PR per RECIST version 1.1 was achieved by day 57 (change in the sum of target lesions from baseline: −72%; Fig. 6C), which improved further during subsequent cycles, reaching a change of −84% by day 162. No clinically meaningful serum phosphate elevation or diarrhea occurred during treatment with RLY-4008, confirming selective FGFR2 inhibition (Fig. 6D). The patient developed reversible and manageable FGFR2 on-target toxicities including low-grade onychauxis and paronychia, reversible low-grade retinal disorders (subretinal fluid, cystoid macular edema), and grade 3 hand–foot syndrome that improved to grade 1 with dose modification. The patient remained on RLY-4008 and was responding to treatment until disease progression on day 218.
Figure 6.
Clinical response in a patient with pan-FGFRi–resistant iCCA with liver, bone, and lung metastases. The patient was treated with RLY-4008 starting at 30 mg once daily. A, Summary of key patient and disease characteristics. B, ctDNA analysis demonstrated complete clearance of FGFR2N549K, FGFR2N549D, and FGFR2V564 L clones by day 30. C, Left: CT scans of right lobe liver metastasis (arrow) and multifocal liver lesions (circled) at baseline (top) and on day 57 of RLY-4008 treatment (bottom) show a rapid, marked reduction in tumor volume. Right: CT scans of manubrium bone lesion at baseline (top) and on day 57 of RLY-4008 treatment (bottom) show complete regression of lesion. D, Serum phosphate during treatment with RLY-4008. Shaded area represents the normal range for serum phosphate (0.8–1.5 mmol/L). ND, not detectable; q.d., once daily.
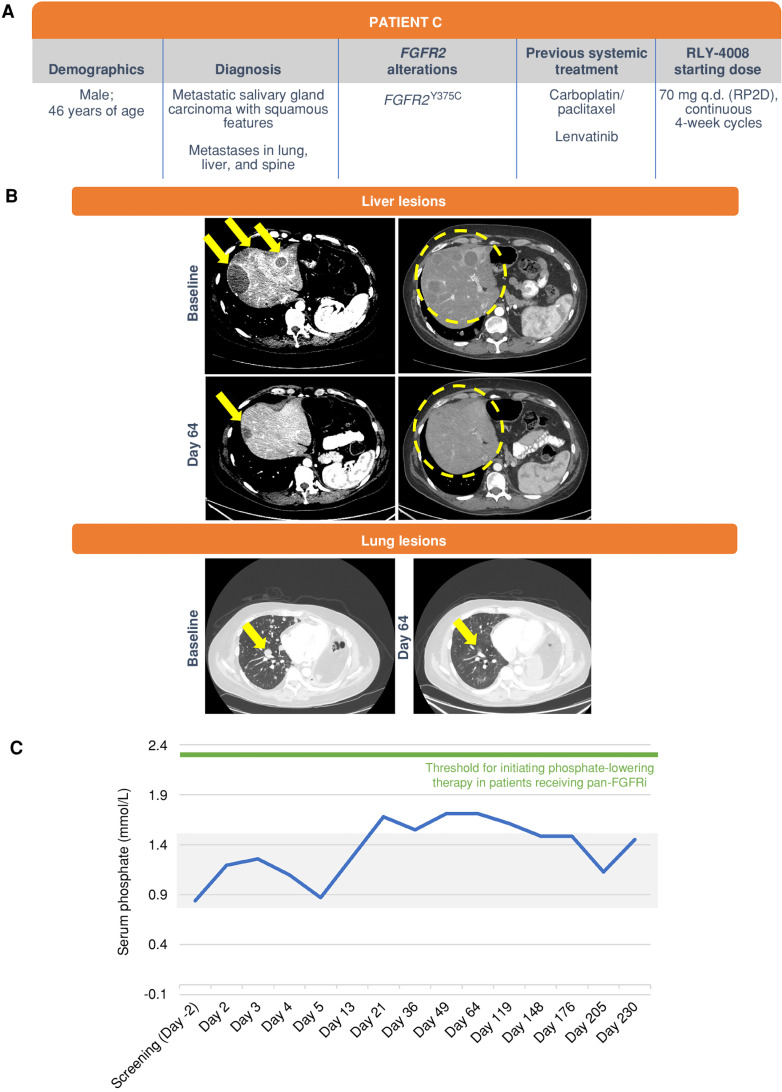
Patient C (Fig. 7), a 46-year-old male with metastatic carcinoma of the right parotid salivary gland bearing an FGFR2Y375C mutation, was enrolled and treated with RLY-4008 starting at 70 mg once daily (Fig. 7A). He had received two prior lines of systemic therapy including carboplatin/paclitaxel and lenvatinib. At day 64 (Fig. 7B), he achieved a PR with a 62% reduction in the sum of target lesions, near complete resolution of liver metastases, and regression of lung and adrenal metastases. ctDNA analysis demonstrated complete clearance of FGFR2Y375C clones. Clinically, the patient had decreased pain and could be weaned off oxygen. Deepening of the response to −67% was observed on day 119. RLY-4008 was well tolerated overall, with reversible and manageable low-grade FGFR2 on-target toxicities, including stomatitis, onycholysis, and paronychia. Slight elevation of serum phosphate was observed (grade 1), which self-resolved without intervention (Fig. 7C). No diarrhea was reported. The patient remained on RLY-4008 and was responding to treatment until disease progression on day 230.
Figure 7.
Clinical response in a patient with metastatic salivary gland carcinoma with squamous features and metastases in the lung, liver, and spine. The patient was treated with RLY-4008 at 70 mg once daily, the RP2D. A, Summary of key patient and disease characteristics. B, Liver lesions, left: CT scans of liver metastases on the dome at baseline (top; arrows) and day 64 of RLY-4008 treatment (bottom; arrow indicates remaining lesion with marked reduction in volume) show dramatic regression of lesions. Liver lesions, right: CT scans of diffuse metastatic lesions within the liver parenchyma at baseline and on day 64 of RLY-4008 treatment show a near complete regression of such lesions (circles). Lung lesions: CT scan of lung metastases at baseline and on day 64 of RLY-4008 treatment show overall decrease in lung metastases and near resolution of a lesion (arrow). C, Serum phosphate over the course of treatment with RLY-4008. The shaded area represents the normal range for serum phosphate (0.8–1.5 mmol/L). q.d., once daily; RP2D, recommended phase II dose.
In summary, the responses shown in these treatment-refractory patients are consistent with the unique, highly selective preclinical profile of RLY-4008. In patient B, whose tumor exhibited FGFR2 resistance mutations after treatment with infigratinib, clearance of resistant clones and the associated response provide clinical proof of concept of the broad mutational coverage of RLY-4008.
DISCUSSION
Although pan-FGFRi have improved clinical outcomes for patients with FGFR2-driven tumors, their clinical benefit is limited by the development of FGFR2 resistance mutations and side effects that include FGFR1-mediated hyperphosphatemia and FGFR4-mediated diarrhea (16–19, 22, 23, 26). Designed based on the differences in conformational dynamics between FGFR2 and other FGFRs, RLY-4008 is the first FGFR2-selective, small-molecule inhibitor to enter the clinic. RLY-4008 is >250-fold selective over FGFR1, and >80- and >5,000-fold selective over FGFR3 and FGFR4, respectively. In vivo, RLY-4008 induces tumor regression in FGFR2 fusion–positive, FGFR2-amplified and FGFR2-mutant xenograft models. In addition, RLY-4008 has potent in vitro and in vivo activity against FGFR2 mutations that drive clinical resistance to available pan-FGFRi. These include FGFR2N549K, which relieves the “molecular brake” and causes constitutive and robust FGFR2 signaling, and the common gatekeeper mutation FGFR2V564F (1, 21–23, 27). In fact, RLY-4008 is more potent on FGFR2V564F than on FGFR2WT, as observed in cell line and xenograft experiments. Further, xenograft tumors bearing the FGFR2V564F mutation that progressed on pan-FGFRi showed a striking and rapid regression when treated with RLY-4008. Finally, RLY-4008 spares FGFR1 in vivo, indicating the potential to avoid FGFR1-mediated toxicities in the clinic. Taken together, the preclinical profile of RLY-4008 highlights its potential for the treatment of FGFR2-driven cancers. A first-in-human study in patients with unresectable or metastatic solid tumors with FGFR2 alterations is ongoing, and promising early clinical data consistent with the preclinical profile of RLY-4008 have been presented (29, 31).
The broad clinical potential of RLY-4008 is highlighted by the patient cases discussed above, which show promising efficacy in pan-FGFRi–naive and pretreated cholangiocarcinoma as well as in solid tumors beyond cholangiocarcinoma. In the pan-FGFRi–naive setting, patient A had a near-complete tumor regression with RLY-4008 and subsequently underwent surgery with curative intent followed by adjuvant therapy. This level of tumor regression is unusual in unresectable cholangiocarcinoma and suggests future study of multimodal approaches with the combination of RLY-4008, surgical resection, and subsequent adjuvant treatment may be warranted. In the pan-FGFRi–resistant setting, patient B had a confirmed PR to RLY-4008 following progression on infigratinib. This patient had polyclonal FGFR2 resistance with mutations FGFR2N549K, FGFR2N549D, and FGFR2V564I. Treatment with RLY-4008 overcame this resistance, achieving complete clearance of these mutations by day 30. This patient had confirmed radiographic response and disease stabilization for greater than 7 months, which is significant, as patients with cholangiocarcinoma who have failed chemotherapy and pan-FGFRi therapy have no effective treatment options and poor outcomes (expected median progression-free survival is ∼2–4 months; ref. 35). To our knowledge, this is the first patient with FGFR2N549K-positive iCCA to achieve a confirmed PR with FGFR-targeted therapy. RLY-4008 also demonstrated clinical activity with durable response in FGFR2N549K-positive breast cancer, suggesting further testing in FGFR2-mutant tumors, which is ongoing in ReFocus. Beyond FGFR2 fusion–positive iCCA, a third patient diagnosed with metastatic carcinoma of the salivary gland bearing an FGFR2Y375C mutation achieved a confirmed PR, highlighting the tumor-agnostic potential of selective FGFR2 inhibition. Consistent with the preclinical profile of RLY-4008, these cases highlight the efficacy, broad mutational coverage, and tumor-agnostic potential of RLY-4008. RLY-4008 was well tolerated, with reports of mainly low-grade, FGFR2-mediated adverse events. Serum phosphate levels remained mostly within the normal range without any intervention throughout treatment and diarrhea was not reported, consistent with preclinical data demonstrating that RLY-4008 spares FGFR1 and FGFR4.
Development of FGFR2 resistance mutations limits the efficacy of pan-FGFRi. Notably, both futibatinib and RLY-4008 retain activity on the FGFR2N549K mutation—each with a ∼10-fold decrease in potency from WT FGFR2—and both inhibitors are efficacious in the AN3CA (FGFR2K310R;N549K) xenograft model, unlike other FGFRi (Fig. 4E). Despite the observed efficacy, it is notable that the FGFR2N549K mutation was detected in multiple patients who progressed on futibatinib in one study (23). This may be attributable to side effects of futibatinib that limit its maximum daily dose or require treatment interruptions in more than half of patients (19). We anticipate that consistent FGFR2-selective inhibition will better maintain target coverage of resistance mutations. Similarly, FGFR2V564F/L/I mutations were detected in multiple patients who progressed on futibatinib and in patients who progressed on treatment with a reversible pan-FGFRi (23). In contrast to futibatinib, pemigatinib, infigratinib, and erdafitinib, RLY-4008 retains preclinical activity on FGFR2V564L and has increased potency on FGFR2V564F (Fig. 4B); although pan-FGFRi were not efficacious in FGFR2V564F-expressing xenografts, RLY-4008 induced rapid tumor regression. The clinical importance of broad mutational coverage has yet to be fully determined. However, evidence from patients with FGFRi-resistant iCCA suggests that tumor heterogeneity is widespread, highlighting the importance of an FGFR2-selective inhibitor with broad mutational coverage (34, 36).
The initial safety profile of RLY-4008 provides insight into the unique toxicity in healthy tissue specific to selective FGFR2 inhibition. Although RLY-4008 avoids the hyperphosphatemia (FGFR1) and diarrhea (FGFR4) observed with pan-FGFRi, on-target FGFR2 effects are observed including oral (stomatitis), skin (palmar–plantar erythrodysesthesia), and nail toxicities (29). These events are generally low-grade, readily monitorable, and reversible; however, they may require supportive care measures and dose modifications. “On-target, off-tumor” adverse events therefore represent a potential limitation of selective FGFR2 inhibition. To address this, future drug development efforts will require alternative approaches to enable selective targeting of oncogenic FGFR2 alterations while sparing FGFR2 in healthy tissue.
Although pan-FGFRi improve treatment outcomes for patients with FGFR2-driven tumors, selective targeting of FGFR2 has remained elusive. RLY-4008 was designed based on the differential protein dynamics between FGFR2 and other FGFRs; this approach has delivered a degree of selectivity not previously achieved for FGFR2. RLY-4008 is the first highly selective, small-molecule FGFR2 inhibitor to enter the clinic; a phase I/II study in FGFR2-driven solid tumors is ongoing (ReFocus; NCT04526106; ref. 28). It is our hope that this novel, motion-based approach to drug discovery—which can also be applied to other targets in oncology and beyond—will deliver sustained clinical benefit for patients.
METHODS
Throughout the article, amino acids are numbered according to the FGFR2-IIIc isoform. Use of the FGFR2-IIIb isoform in specific experiments is indicated here.
Cloning and Protein Purification for Crystallization
The FGFR2 kinase domain (aa P458-E768) was cloned into a modified pET15-b vector with an N-terminal hexahistadine tag (His6) followed by the TEV sequence (MGSSHHHHHHENLYFQS). BL21 (DE3) Escherichia coli cells were cotransformed with this plasmid and a plasmid encoding YopH, and grown in Luria-Bertani (LB) media supplemented with 100 μg/mL ampicillin and 50 μg/mL kanamycin at 37°C. At OD600 = 0.4, the temperature was lowered to 18°C. At OD600 = 0.6, isopropyl-β-D-thiogalactopyranoside was added to a final concentration of 750 mmol/L. After 18 hours, cells were harvested in wash buffer (600 mmol/L NaCl, 50 mmol/L HEPES, pH 8.0, 1 mmol/L TCEP, 10 mmol/L imidazole, 10% glycerol) supplemented with 0.1% TritonX-100, 1 mmol/L PMSF, and protease inhibitor tablet (Pierce Universal Nuclease). Cells were lysed by sonication and cleared by centrifugation. Tagged FGFR2 kinase domain protein was captured on TALON resin preequilibrated in wash buffer and eluted in 25 mmol/L NaCl, 50 mmol/L HEPES, pH 8.0, 1 mmol/L TCEP, 400 mmol/L imidazole, and 10% glycerol. Eluted protein was incubated overnight at 4°C with TEV protease and RLY-4008 in a 1:1.5 (protein:compound) molar ratio to digest the tag and form the FGFR2–RLY-4008 covalent adduct. Cleavage of the protein was confirmed by SDS-PAGE. Protein was treated with lambda phosphatase followed by anion exchange chromatography using a MonoQ column. Purified protein was supplemented with additional RLY-4008 (1:1 molar ratio) and concentrated before gel filtering with a Superdex–S200 Hi Load 16/60 column equilibrated with 30 mmol/L HEPES, pH 7.5, 150 mmol/L NaCl, and 1 mmol/L TCEP. Purified protein was concentrated to approximately 6.5 to 8 mg/mL.
Synthesis and Characterization of RLY-4008
The route of synthesis and characterization of RLY-4008 is reported in patent WO 2022/109577 A1 (Structure I-1; ref. 37).
Protein Crystallization and Structure Determination
Crystals of the FGFR2–RLY-4008 covalent adduct were generated by the hanging-drop vapor diffusion method [18°C, equilibrated against 1 μL well solution (0.1 M Tris, pH 8, PEG 8K 18%–20% w/v)]. After 48 hours, the rods were harvested in well solution supplemented with 27% glycerol. Crystals grew in the space group P212121 a = 62.51; b = 91.34; c = 129.62. X-ray diffraction experiments were carried out at the Swiss Light Source at beamline X06SA, and reflections were recorded at wavelength 1.000032 Å. Reflections were integrated using XDS (38) and reduced using an autoPROC applying anisotropic cutoff of the data (global phasing). Phases for the data were initially calculated by molecular replace in Phaser (ref. 39; RRID:SCR_014219) using coordinates of the C-lobe as a search model (aa residues 568–765 of PDB ID: 2PVF). The search produced a solution with two chains in the asymmetric unit and an interpretable electron density map. The N-lobe of each model was docked and placed by rigid body refinement, which produced initial phases. Subsequent rounds of model building in Coot (ref. 40; RRID:SCR_014222) and refinement in Phenix (ref. 41; RRID:SCR_014224) were performed. Composite omit maps were calculated to guide model adjustment. Ligand restraints for RLY-4008 were generated in the eLBOW module of Phenix. Regions of the structure were left unmodeled where electron density was uninterpretable.
Purification of FGFR1 and FGFR2 WT Kinase Domains
WT FGFR1 kinase domain (aa A458-E765) was purified from E. coli using Talon FF, Resource Q, HisTrap HP, and Superdex 200 columns at Wuxi Biortus Biosciences Co. Ltd. Protein was treated with CIP prior to Resource Q column, and tag was removed via protease digestion prior to HisTrap HP column. Purified protein was stored in 20 mmol/L Tris pH 8.0, 20 mmol/L NaCl, 2 mmol/L TCEP. WT FGFR2 kinase domain (aa P458-E768) was purified by an identical process and purified protein was stored in 30 mmol/L HEPES pH 8.0, 150 mmol/L NaCl, 1 mmol/L TCEP. Theoretical molecular weights for WT FGFR1 and FGFR2 kinase domain are 35434.91 Da and 36143.85 Da, respectively. Molecular weights were confirmed experimentally using SYNAPT G2-Si LC/MS (FGFR1 = 35432 Da and FGFR2 = 36141 Da).
LC/MS Sample Preparation and Data Analysis
FGFR1 or FGFR2 kinase domain was diluted in reaction buffer (25 mmol/L HEPES, pH 7.4, 10 mmol/L MgCl2, 20 mmol/L NaCl, 1 mmol/L DTT) at 0.1 mg/mL and mixed at a 1:1 ratio with reaction buffer containing 2× RLY-4008 (of indicated concentration; 200 μmol/L for RLY-4008 and futibatinib in rate of covalent bond formation experiments). At indicated time points, 10 μL reaction was quenched with 2 μL 5% formic acid; samples were then diluted to 40 μL. Using SYNAPT G2-Si LC/MS, ∼0.2 μg of material was injected per sample. Primary LC/MS data were processed using BioPharmaLynx 1.3.5 (Waters Technologies Corporation) and deconvoluted in batch using identical settings. Peak intensities corresponding to nonmodified FGFR1 or FGFR2 kinase domain mass and mass plus RLY-4008 were recorded.
Using peak intensity data of unmodified and modified protein, the percent unmodified protein remaining was calculated. Percent unmodified protein over time followed an exponential decay pattern and was analyzed using GraphPad Prism to obtain the apparent rate of decay (kobs). kobs at each RLY-4008 concentration was plotted against compound concentration to determine how efficiently FGFR1 and FGFR2 kinase domains were modified by RLY-4008. Modification efficiency is defined by kinact/KI, parameters obtained from this plot: kinact is the maximum kobs achievable at infinite compound concentration; KI is the compound concentration needed to achieve 50% of the maximum kobs. For the rate of covalent bond formation experiments with RLY-4008 and futibatinib, compound concentration was significantly higher than the compound's respective IC50 value. Thus, it was assumed that the concentration tested (100 μmol/L final) was significantly higher than the reversible equilibrium constant of the initial encounter complex; therefore, the apparent rate of decay is equivalent to the rate of covalent adduct formation (kinact).
Intact Mass Analysis
Intact mass analysis was performed by ESI qTOF MS (Waters, Synapt G2Si qTOF MS). FGFR2 protein samples were desalted prior to MS using a Waters Acquity H class UPLC system. Samples were loaded onto a protein trap (Waters BEH C4) and desalted with a linear gradient of water and acetonitrile with 0.1% formic acid over 5 minutes. The instrument was monitored from 600 to 2,000 m/z. The protein spectra were deconvoluted by MaxEnt software. The mass accuracy of the measurements was approximately 0.005%.
LC/MS Peptide Mapping
Peptide mapping by LC/MS was performed on trypsin-digested FGFR2 using a Waters Acquity H class UPLC coupled to a mass spectrometer (Synapt XS qTOF MS, Waters). Briefly, 50 μg of protein was diluted to approximately 0.2 mg/mL (25 μL), denatured with 45 μL of 8 M guanidine HCl, and reduced with 5 μL of 100 mmol/L DTT at 25°C for 60 minutes. The sample was diluted with 75 μL 50 mmol/L Na HEPES, pH 7.5, and digested with 10 μL of 0.2 mg/mL trypsin at 37°C for 22 hours. Digested protein (50 μL) was injected into a 1 × 100 mm BEH C18 1.7 μmol/L column, and peptides were separated under reversed-phase conditions using a linear gradient of water and acetonitrile with 0.1% formic acid.
FGFR1 and FGFR2 Kinase Activity Assay
RLY-4008 was preincubated with FGFR1 or FGFR2 kinase (Carna Biosciences) and substrate peptide (FLPeptide30, PerkinElmer) in 50 mmol/L HEPES, pH 7.5, 10 mmol/L MgCl2, 1 mmol/L EGTA, 0.01% Brij-35, 2 mmol/L DTT, and 0.05% BSA for 30 minutes at room temperature. Compound volume was 1% final assay volume. ATP was added to a total concentration of 100 μmol/L and incubated for 90 minutes, and reactions were quenched using 100 mmol/L HEPES, pH 7.5, 35.5 mmol/L EDTA, 0.015% Brij-35, 0.0002% coating reagent #3 (PerkinElmer), and 5% DMSO. The substrate conversion ratio was calculated for each RLY-4008 concentration and was then used to calculate IC50 values.
Cell Culture and Cell Line Engineering
KATO III (HTB-103, RRID:CVCL_0371), NCI-H716 (CCL-251, RRID:CVCL_1581), SNU-16 (CRL-5974, RRID:CVCL_0076), AN3CA (HTB-111, RRID:CVCL_0028), RT-4 (HTB-2, RRID:CVCL_0036), MDA-MB-453 (HTB-131, RRID:CVCL_0418), and HEK-293 (CRL-1573, RRID:CVCL_0045) were purchased from ATCC in 2017 and 2018; JMSU-1 (ACC505, RRID:CVCL_2081), MFE-296 (ACC419, RRID:CVCL_1406), and RT-112 (ACC418, RRID:CVCL_1670) were purchased from DSMZ between August 2018 and November 2018; Li-7 (RCB1941, RRID:CVCL_3840) and JHUEM-2 (RCB1551, RRID:CVCL_4656) were purchased from RIKEN BRC in November 2018. ICC13-7 (RRID:CVCL_A1VD) was a gift from Nabeel Bardeesy (Massachusetts General Hospital). Cell lines were authenticated by short tandem repeat DNA profiling by the cell line bank from which they were obtained or by IDEXX BioAnalytics (ICC13-7). They were all provided after testing Mycoplasma-free and were routinely tested while in culture. All cell lines were cultured for less than 1 month after thaw and used within 10 passages from receipt. Cell lines were cultured at 37°C in 5% CO2 humidified air in media recommended by the vendor.
FGFR2IIIb WT and mutant open reading frames were cloned into a pLenti-P2A-Puro (OriGene) expression vector. Lentiviral particles were produced in Lenti-X 293T cells (Takara Bio) using a Lenti-vpac packaging kit (OriGene) per the manufacturer's protocol. Lentivirus-infected cells were selected for stable expression using 2 μg/mL puromycin. The V564F mutation was knocked into the endogenous FGFR2–OPTN fusion in the ICC13-7 cell line using CRISPR-mediated homology-directed repair. Briefly, ICC13-7 cells were cotransfected with pLentiCRISPRv2 (GenScript) plasmid containing single-guide RNA (sgRNA; adjacent to V564) and a single-stranded oligo donor cassette (containing V564F mutation and silent PAM mutation). The cells expressing sgRNA and Cas9 were enriched with 4 μg/mL puromycin treatment for 3 days. Following puromycin selection, single-cell clones were selected and sequenced to confirm the presence of the desired V564F mutation along with the silent PAM mutation. The sgRNA sequence was as follows: 5′-CTAAAGGCAACCTCCGAGAA. The single-stranded oligodeoxynucleotide sequence was as follows: 5′-TCTCTTTAGGGAGCTTCTCTTCTTCCTCAACAGGGCCTCTCTATGTCATATTTGAGTATGCTTCTAAAGGCAACCTCCGAGAATACCTCCGAGCCCGGAGGC.
Cell Lysates and Immunoblotting
SNU-16 cells were treated with DMSO or RLY-4008 for 2 hours or 24 hours at 37°C with 5% CO2. Cells were washed with cold PBS and lysed with cold RIPA buffer supplemented with cOmplete and PhosSTOP tablets (Roche) for 30 minutes on ice. Following 4°C centrifugation for 10 minutes at 14,000 rpm, supernatant concentrations were determined via BCA assay. Lysates were mixed with 6× Laemmli's buffer for traditional SDS-PAGE/immunoblot analysis or with ProteinSimple WES MasterMix for WES (ProteinSimple) analysis. The following antibodies were from Cell Signaling Technology: pFGFR (Y653/654; #3471, RRID:AB_331072), FGFR2 (#23328, RRID:AB_2798862), pFRS2-α (Y436; #3861, RRID:AB_2231950), pS6 (S235/236; #4858, RRID:AB_916156), S6 (#2217, RRID:AB_331355), pAKT (S473; #4058, RRID:AB_331168), AKT (#9272, RRID:AB_329827), pERK (T202/204; #9101, RRID:AB_331646), ERK (#9102, RRID:AB_330744), cleaved PARP (#5625, RRID:AB_10699459), cleaved Caspase-3 (#9664, RRID:AB_2070042), vinculin (#13901, RRID:AB_2728768), actin (#3700, RRID:AB_2242334), and actin (#4970, RRID:AB_2223172). FRS2 (#MAB4069, RRID:AB_2106236) was from R&D Systems.
Cell Proliferation
Adherent cells were seeded in 100 μL media into a black, 96-well, clear-bottom plate, including a day 0 untreated plate to be read after 24 hours. After 24 hours of incubation at 37°C and 5% CO2, cells were treated with DMSO or test compound in an additional 100 μL for 96 hours at 37°C and 5% CO2. Suspension cells were seeded in a volume of 100 μL in black, 96-well, clear-bottom plates containing DMSO or test compound and incubated at 37°C and 5% CO2 for 96 hours, including a day 0 untreated plate to be read after 24 hours. Following incubation, plates and CellTiter-Glo 2.0 (Promega) were equilibrated to room temperature for 30 minutes. For adherent cells, 100 μL media were removed from each well followed by the addition of 100 μL CellTiter-Glo 2.0 to all wells. Plates were placed on a shaker (protected from light) at room temperature for 30 minutes and read on an EnVision plate reader. Data were normalized by subtracting day 0 values from all treated sample measurements followed by normalization to DMSO controls and conversion to percent viability. A sigmoidal four-parameter curve (4PL; GraphPad Prism, RRID:SCR_002798) was used to determine the IC50.
In Vivo Studies
All procedures relating to animal handling, care, and treatment were approved by the Institutional Animal Care and Use Committee of the companies performing the studies and followed the guidance of the Association for Assessment and Accreditation of Laboratory Animal Care. RLY-4008 and erdafitinib were formulated in 0.5% methylcellulose/2% TPGS and dosed orally as suspensions. Infigratinib was formulated in 70% PEG300/30% D5W and futibatinib was formulated in 0.5% methylcellulose/2% TPGS or 20% HP-β-CD at pH 3.0; they were dosed orally as suspensions. Pemigatinib was formulated in 10% DMA/90% 20% HP-β-CD at pH 3.5 and dosed orally as a solution. All compound doses are expressed as mg/kg free base.
The SNU-16, ICC13-7, and ICC13-7–FGFR2V564F xenograft studies were conducted at Pharmaron, Inc. For SNU-16, female BALB/c nude mice were inoculated subcutaneously on the right flank with 1 × 107 cells in 0.1 mL RPMI 1640:BD Matrigel mixture (1:1). For ICC13-7 and ICC13-7–FGFR2V564F xenografts, female NOD SCID mice were inoculated in the same manner. The AN3CA xenograft study was conducted at Shanghai ChemPartner Co., Ltd. Female BALB/c nude mice were inoculated subcutaneously on the right flank with 5 × 106 cells in 0.2 mL EMEM:BD Matrigel mixture (1:1). The patient-derived xenograft studies were conducted at Crown Bioscience in female BALB/c nude mice. Treatment was initiated when the average tumor volume was 100 to 200 mm3 for efficacy studies and 250 to 350 mm3 for PK/PD studies. Body weight was measured daily, and tumors were measured twice weekly in two dimensions using a caliper. Tumor volume was expressed in mm3 using the formula: V = 0.5 a × b2, where a and b are the long and short diameters of the tumor, respectively.
In Vitro and In Vivo PD Analysis
For cellular PD assays, SNU-16 cells and 293T cells stably expressing FGFR2 WT and mutants were plated in 384 well plates (SNU-16: 60,000 cells/well; 293T: 25,000 cells/well). SNU-16 cells were immediately incubated with RLY-4008 for 2 hours at 37°C and 5% CO2; 293T cells were incubated overnight at 37°C and 5% CO2 prior to compound addition. Phospho-FGFR2 (Tyr653/654) Cellular HTRF (PerkinElmer; 63ADK054PEH) or Phospho-ERK (Thr202/Tyr204) Cellular HTRF (PerkinElmer; 64AERPEH) assays were performed per the manufacturer's instructions. Data were fit to a sigmoidal four-parameter curve (4PL; GraphPad Prism, RRID:SCR_002798) to determine IC50.
For tumor PD by HTRF assay, tumors were lysed in lysis buffer #2 (component of PerkinElmer 63ADK054PEH/63ADK057PEH kits) supplemented with cOmplete protease inhibitor tablet (Roche) and homogenized in a TissueLyser (Qiagen). pFGFR2 and tFGFR2 (PerkinElmer; 63ADK057PEH) HTRF assays were carried out per manufacturer's protocol. For tumor PD by pFGFR2 ELISA, 96-well ELISA plates were coated with 50 μL tFGFR2 antibody (Cell Signaling Technology; #23328, RRID:AB_2798862) in PBS at 0.5 μg/mL and incubated overnight at room temperature. Plates were washed 4× with 400 μL/well wash buffer (0.05% Tween-20 in PBS) and blocked with 150 μL/well blocking buffer (3% BSA + 0.05% Tween-20 in PBS, filtered) at room temperature for 2 hours. Plates were washed as above followed by loading 50 μL/well of 2 mg/mL sample in sample diluent (1% BSA + 0.05% Tween-20 in PBS). Plates were incubated overnight at 4°C and washed as above. Anti-phosphotyrosine 4G10-biotin antibody (Millipore #16-103, RRID:AB_310777) was diluted to 0.005 mg/mL in sample diluent, and 50 μL was added to each well. Following incubation for 2 hours at room temperature, plates were washed as above. Streptavidin-HRP (Thermo Scientific) was diluted 1:200 in sample diluent, and 50 μL/well was added. Plates were incubated for 25 minutes at room temperature protected from light on a shaker (300 rpm). Plates were washed, and 50 μL/well 1:1 Pico SuperSignal Chemiluminescent substrate (Thermo Scientific) was added and incubated for 5 minutes before plates were read on the EnVision Plate Reader (US Luminescence).
ReFocus Phase I/II Study
ReFocus is a global, open-label, phase I/II, first-in-human study (NCT04526106) with anticipated enrollment of ∼490 patients. Key objectives of phase I are to define the maximum tolerated dose, safety profile, PK, and preliminary antitumor activity of RLY-4008 in patients with unresectable or metastatic solid tumors with FGFR2 alterations. The key objectives of phase II are to define the overall response rate and duration of response per RECIST version 1.1 for advanced, FGFR2-altered solid tumors and cholangiocarcinoma. Additional phase II objectives are to assess the safety, PK, and PD of RLY-4008 at the recommended phase II dose (RP2D) and to explore potential mechanisms of resistance. The study was initiated in September 2020, and the phase I dose escalation (N = 116) has now been completed. Phase II is ongoing with solid tumor agnostic and cholangiocarcinoma cohorts treated at the RP2D determined in phase I. Presented data herein are preliminary; data cutoff was January 30, 2023.
The study was conducted in accordance with the Declaration of Helsinki and was reviewed and approved by the Institutional Review Board of each clinical site. Written informed consent was obtained from all patients before study entry. Patients eligible for study participation were ≥18 years old and had Eastern Cooperative Oncology Group performance status 0 to 1; had no significant corneal or retinal disorder or uncontrolled central nervous system metastases; had no known tumor genetic alteration for which there is an approved targeted therapy, apart from FGFR2; and had adequate cardiac function. Additional enrollment criteria are provided in Supplementary Appendix S1. RLY-4008 was administered orally, once daily, in 4-week cycles. Adverse events were graded per CTCAE. Response was evaluated per RECIST version 1.1. Levels of ctDNA in plasma were assessed by next-generation sequencing using 74-gene Guardant360 CDx (Guardant Health).
Data Sharing Statement
Further information and requests for access to data and/or reagents should be made directly to datasharingrequest@relaytx.com.
Supplementary Material
Potency (kinact/KI) of RLY-4008 against FGFR1 and FGFR2
Biochemical inhibition of FGFR by RLY-4008 and pan-FGFRi
Antiproliferative effect of RLY-4008 and pan-FGFRi in human tumor cell lines
In vivo exposures of pan-FGFRi in mouse studies
Fold change in potency against FGFR2 acquired resistance mutations
Supplementary Figure 1. RLY-4008 is an irreversible inhibitor of FGFR2.
Supplementary Figure 2. Treatment with RLY-4008 is well-tolerated and leads to dose-dependent inhibition of FGFR2 and tumor regression in multiple FGFR2-altered tumor models.
ReFocus entry criteria
three references cited in supplementary data
In this schematic movie, apo FGFR1 (left panel) demonstrates rapid P-loop dynamics, while apo FGFR2 (right panel) displays less P-loop motion.
Supplementary video legend describing schematic video of P-loop dynamics
Acknowledgments
The authors thank Dr. Nabeel Bardeesy (Massachusetts General Hospital) for sharing the ICC13-7 cell line to enable this work. The authors also thank the patients, their families, and caregivers for their support of this research. This study was sponsored by Relay Therapeutics, Inc. Medical writing assistance was provided by Christine Elsner from BOLDSCIENCE Inc. and was funded by Relay Therapeutics, Inc. V. Subbiah is an Andrew Sabin Family Foundation Fellow at The University of Texas MD Anderson Cancer Center. V. Subbiah acknowledges the support of the Jacquelyn A. Brady Fund. V. Subbiah is supported by HHS/NIH/NCI grants R01CA242845 and R01CA273168. The MD Anderson Cancer Center Department of Investigational Cancer Therapeutics is supported by the Cancer Prevention & Research Institute of Texas (no. RP1100584), the Sheikh Khalifa Bin Zayed Al Nahyan Institute for Personalized Cancer Therapy (no. 1U01 CA180964), an HHS/NIH/National Center for Advancing Translational Sciences grant (no. UL1 TR000371), and the MD Anderson Cancer Center Support Grant (no. P30 CA016672). A.M. Schram acknowledges an ASCO Conquer Cancer Foundation Career Development Award, NCI P30CA008748 Cancer Clinical Investigator Team Leadership Award, and the Memorial Sloan Kettering Cancer Center Support Grant (P30 CA008748).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
V. Subbiah reports grants from Relay Therapeutics, Inc. during the conduct of the study; research funding/grant from Relay Therapeutics, Inc. for clinical trial research support; support for clinical trials from AbbVie, Agensys, Inc., Alfasigma, Altum, Amgen, Bayer, BERG Health, Blueprint Medicines Corporation, Boston Biomedical, Inc., Boston Pharmaceuticals, Celgene Corporation, D3 Bio, Inc., Dragonfly Therapeutics, Inc., Exelixis, Fujifilm, GSK, Idera Pharmaceuticals, Inc., Incyte Corporation, Inhibrx, Loxo Oncology, MedImmune, MultiVir, Inc., NanoCarrier, Co., the National Comprehensive Cancer Network, the NCI-Cancer Therapy Evaluation Program, Northwest Biotherapeutics, Novartis, PharmaMar, Pfizer, Relay Therapeutics, Inc., Roche/Genentech, Takeda, Turning Point Therapeutics, The University of Texas MD Anderson Cancer Center, and Vegenics Pty Ltd.; travel support from the American Society of Clinical Oncology, the European Society for Medical Oncology, Helsinn Healthcare, Incyte Corporation, Novartis, and PharmaMar; consultancy/advisory board participation for Helsinn Healthcare, Jazz Pharmaceuticals, Incyte Corporation, Loxo Oncology/Eli Lilly, MedImmune, Novartis, QED Therapeutics, Relay Therapeutics, Inc., Daiichi Sankyo, and R-Pharm US; and other relationship with PeerView and Medscape. V. Sahai reports personal fees from AstraZeneca, Autem, Taiho, Delcath Systems, Kinnate, Amplity, GSK, Helsinn, and Histosonics, grants, personal fees, and nonfinancial support from Cornerstone, grants and personal fees from Ipsen, Incyte, and Servier, grants from Actuate Therapeutics, Bristol Myers Squibb, Boehringer Ingelheim, Celgene, Clovis, Exelixis, Fibrogen, MedImmune, the NCI, PanCAN, Relay Therapeutics, Inc. Repare, Syros, and Transthera, and nonfinancial support from BeiGene outside the submitted work. D.T. Moustakas reports a patent for WO2020231990 A1 (2020-11-19) pending. H. Schönherr reports a patent for WO2020231990 A1 (2020-11-19) pending and a patent for WO2022109577 A1 (2022-05-27) pending. A.M. Taylor reports a patent for WO2020231990 pending to Relay Therapeutics, Inc. B.M. Hudson reports a patent for WO2020231990 A1 (2020-11-19) pending. D. Pal reports other support from Relay Therapeutics, Inc. outside the submitted work. P. Ayaz reports a patent for US20230104574A1 pending, a patent for TW202108141A pending, a patent for EP3968999A1 pending, a patent for CA3137458A1 pending, a patent for AU2020274083A1 pending, and a patent for WO2020231990A1 pending. D.A. Sharon reports patents for US20230104574A1, TW202108141A, EP3968999A1, CA3137458A1, AU2020274083A1, and WO2020231990A1 pending. L. Goyal reports personal fees from Alentis Therapeutics AG, Black Diamond, Basilea, Blueprint Medicines, Genentech, Exelixis, Eisai/H3Biomedicine, Incyte Corporation, Kinnate, Merck, QED Therapeutics, Relay Therapeutics, Inc., Sirtex Medical Ltd., The Servier Group, Surface Oncology, Taiho Oncology, Tyra Biosciences, and Transthera Bio and participation on data safety monitoring boards for AstraZeneca during the conduct of the study. A.M. Schram reports personal fees and other support from Relay Therapeutics, Inc. during the conduct of the study, as well as other support from Revolution Medicines outside the submitted work. S. Kamath reports personal fees from Seagen, Exelixis, Guardant Health, Foundation Medicine, and Tempus outside the submitted work. C. Sherwin reports personal fees from Relay Therapeutics, Inc. outside the submitted work. O. Schmidt-Kittler reports personal fees from Relay Therapeutics, Inc. during the conduct of the study. K. Jen reports other support from Relay Therapeutics, Inc. outside the submitted work. F. Ricard reports personal fees and other support from Relay Therapeutics, Inc. outside the submitted work. B.B. Wolf reports personal fees and other support from Relay Therapeutics, Inc. outside the submitted work. D.E. Shaw is the sole beneficial owner and Chief Scientist of D. E. Shaw Research, which has a multitarget collaboration and licensing agreement with Relay Therapeutics, Inc. By way of his ownership of D. E. Shaw Research, D.E. Shaw also has an indirect equity interest in Relay Therapeutics, Inc. D.A. Bergstrom is an employee of and shareholder in Relay Therapeutics, Inc. J. Watters reports personal fees from Relay Therapeutics, Inc. during the conduct of the study, as well as personal fees from RADD Pharmaceuticals outside the submitted work. J.B. Casaletto is an employee of and shareholder in Relay Therapeutics, Inc. No disclosures were reported by the other authors.
Authors’ Contributions
V. Subbiah: Conceptualization, resources, data curation, supervision, validation, investigation, visualization, writing–original draft, writing–review and editing. V. Sahai: Resources, investigation, writing–review and editing. D. Maglic: Conceptualization, resources, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. K. Bruderek: Resources, data curation, formal analysis, validation, investigation, visualization, methodology, writing–review and editing. B.B. Toure: Conceptualization, resources, data curation, formal analysis, supervision, investigation, methodology. S. Zhao: Conceptualization, resources, data curation, formal analysis, validation, investigation, visualization, methodology, writing–review and editing. R. Valverde: Conceptualization, data curation, software, supervision, methodology. P.J. O'Hearn: Data curation, formal analysis, investigation, methodology, writing–review and editing. D.T. Moustakas: Conceptualization, resources, data curation, formal analysis, visualization, methodology, writing–review and editing. H. Schönherr: Conceptualization, resources, data curation, formal analysis, investigation, methodology, writing–review and editing. N. Gerami-Moayed: Resources, data curation, formal analysis, investigation, visualization, methodology, writing–review and editing. A.M. Taylor: Conceptualization, resources, data curation, formal analysis, investigation, methodology, writing–review and editing. B.M. Hudson: Conceptualization, resources, data curation, formal analysis, visualization, writing–review and editing. D.J. Houde: Resources, data curation, software, formal analysis, investigation, methodology, writing–review and editing. D. Pal: Data curation, formal analysis, validation, visualization. L. Foster: Data curation, methodology, writing–review and editing. H. Gunaydin: Conceptualization, investigation, methodology, writing–review and editing. P. Ayaz: Investigation, writing–review and editing. D.A. Sharon: Investigation, writing–review and editing. L. Goyal: Resources, validation, investigation, visualization, methodology, writing–review and editing. A.M. Schram: Resources, validation, investigation, writing–review and editing. S. Kamath: Resources, validation, investigation, writing–review and editing. C.A. Sherwin: Conceptualization, resources, supervision, methodology, project administration, writing–review and editing. O. Schmidt-Kittler: Resources, formal analysis, writing–review and editing. K.Y. Jen: Resources, data curation, investigation, writing–review and editing. F. Ricard: Resources, data curation, investigation, writing–review and editing. B.B. Wolf: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. D.E. Shaw: Supervision, writing–review and editing. D.A. Bergstrom: Conceptualization, resources, supervision, writing–review and editing. J. Watters: Conceptualization, resources, data curation, formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. J.B. Casaletto: Conceptualization, resources, data curation, formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing.
References
- 1. Babina IS, Turner NC. Advances and challenges in targeting FGFR signalling in cancer. Nat Rev Cancer 2017;17:318–32. [DOI] [PubMed] [Google Scholar]
- 2. Katoh M. Fibroblast growth factor receptors as treatment targets in clinical oncology. Nat Rev Clin Oncol 2019;16:105–22. [DOI] [PubMed] [Google Scholar]
- 3. Jain A, Borad MJ, Kelley RK, Wang Y, Abdel-Wahab R, Meric-Bernstam F, et al. Cholangiocarcinoma with FGFR genetic aberrations: a unique clinical phenotype. JCO Precis Oncol 2018;2:1–12. [DOI] [PubMed] [Google Scholar]
- 4. Kendre G, Murugesan K, Brummer T, Segatto O, Saborowski A, Vogel A. Charting co-mutation patterns associated with actionable drivers in intrahepatic cholangiocarcinoma. J Hepatol 2023;78:614–26. [DOI] [PubMed] [Google Scholar]
- 5. Helsten T, Elkin S, Arthur E, Tomson BN, Carter J, Kurzrock R. The FGFR landscape in cancer: analysis of 4,853 tumors by next-generation sequencing. Clin Cancer Res 2016;22:259–67. [DOI] [PubMed] [Google Scholar]
- 6. Gu W, Yang J, Wang Y, Xu J, Wang X, Du F, et al. Comprehensive identification of FGFR1–4 alterations in 5 557 Chinese patients with solid tumors by next-generation sequencing. Am J Cancer Res 2021;11:3893–906. [PMC free article] [PubMed] [Google Scholar]
- 7. Goyal L, Kongpetch S, Crolley VE, Bridgewater J. Targeting FGFR inhibition in cholangiocarcinoma. Cancer Treat Rev 2021;95:102170. [DOI] [PubMed] [Google Scholar]
- 8. Wu YM, Su F, Kalyana-Sundaram S, Khazanov N, Ateeq B, Cao X, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov 2013;3:636–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cha JY, Maddileti S, Mitin N, Harden TK, Der CJ. Aberrant receptor internalization and enhanced FRS2-dependent signaling contribute to the transforming activity of the fibroblast growth factor receptor 2 IIIb C3 isoform. J Biol Chem 2009;284:6227–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pearson A, Smyth E, Babina IS, Herrera-Abreu MT, Tarazona N, Peckitt C, et al. High-level clonal FGFR amplification and response to FGFR inhibition in a translational clinical trial. Cancer Discov 2016;6:838–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zingg D, Bhin J, Yemelyanenko J, Kas SM, Rolfs F, Lutz C, et al. Truncated FGFR2 is a clinically actionable oncogene in multiple cancers. Nature 2022;608:609–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. TRUSELTIQ® (infigratinib). Highlights of prescribing information. [cited 2022 Oct 26]. Available from: https://www.accessdata.fda.gov/drugsatfda_docs/label/2021/214622s000lbl.pdf.
- 13. PEMAZYRE® (pemigatinib). Highlights of prescribing information. [cited 2022 Oct 26]. Available from: https://www.accessdata.fda.gov/drugsatfda_docs/label/2022/213736s002lbl.pdf.
- 14. LYTGOBI® (futibatinib). Highlights of prescribing information. [cited 2022 Oct 26]. Available from: https://www.accessdata.fda.gov/drugsatfda_docs/label/2022/214801s000lbl.pdf.
- 15. BALVERSA® (erdafitinib) Highlights of prescribing information. [cited 2022 Oct 26]. Available from: https://www.accessdata.fda.gov/drugsatfda_docs/label/2019/212018s000lbl.pdf.
- 16. Javle M, Roychowdhury S, Kelley RK, Sadeghi S, Macarulla T, Weiss KH, et al. Infigratinib (BGJ398) in previously treated patients with advanced or metastatic cholangiocarcinoma with FGFR2 fusions or rearrangements: mature results from a multicentre, open-label, single-arm, phase 2 study. Lancet Gastroenterol Hepatol 2021;6:803–15. [DOI] [PubMed] [Google Scholar]
- 17. Javle M, Lowery M, Shroff RT, Weiss KH, Springfeld C, Borad MJ, et al. Phase II study of BGJ398 in patients with FGFR-altered advanced cholangiocarcinoma. J Clin Oncol 2018;36:276–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Abou-Alfa GK, Sahai V, Hollebecque A, Vaccaro G, Melisi D, Al-Rajabi R, et al. Pemigatinib for previously treated, locally advanced or metastatic cholangiocarcinoma: a multicentre, open-label, phase 2 study. Lancet Oncol 2020;21:671–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goyal L, Meric-Bernstam F, Hollebecque A, Valle JW, Morizane C, Karasic TB, et al. Futibatinib for FGFR2-rearranged intrahepatic cholangiocarcinoma. N Engl J Med 2023;388:228–39. [DOI] [PubMed] [Google Scholar]
- 20. Lacouture ME, Sibaud V, Anadkat MJ, Kaffenberger B, Leventhal J, Guindon K, et al. Dermatologic adverse events associated with selective fibroblast growth factor receptor inhibitors: overview, prevention, and management guidelines. Oncologist 2021;26:e316–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Silverman IM, Hollebecque A, Friboulet L, Owens S, Newton RC, Zhen H, et al. Clinicogenomic analysis of FGFR2-rearranged cholangiocarcinoma identifies correlates of response and mechanisms of resistance to pemigatinib. Cancer Discov 2021;11:326–39. [DOI] [PubMed] [Google Scholar]
- 22. Goyal L, Saha SK, Liu LY, Siravegna G, Leshchiner I, Ahronian LG, et al. Polyclonal secondary FGFR2 mutations drive acquired resistance to FGFR inhibition in patients with FGFR2 fusion-positive cholangiocarcinoma. Cancer Discov 2017;7:252–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goyal L, Baiev I, Zhang K, Harris W, Karasic TB, Damjanov N, et al. Landscape of acquired resistance to selective FGFR inhibitors in FGFR2 fusion or rearrangement+ cholangiocarcinoma. In: Proceedings of the 32nd EORTC/AACR/NCI Virtual Symposium; 2020Oct 24–25 (Virtual meeting). Abstract nr 49.
- 24. Gattineni J, Alphonse P, Zhang Q, Mathews N, Bates CM, Baum M. Regulation of renal phosphate transport by FGF23 is mediated by FGFR1 and FGFR4. Am J Physiol Renal Physiol 2014;306:F351–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kommalapati A, Tella SH, Borad M, Javle M, Mahipal A. FGFR inhibitors in oncology: insight on the management of toxicities in clinical practice. Cancers 2021;13:2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Goyal L, Shi L, Liu LY, Fece de la Cruz F, Lennerz JK, Raghavan S, et al. TAS-120 overcomes resistance to ATP-competitive FGFR inhibitors in patients with FGFR2 fusion-positive intrahepatic cholangiocarcinoma. Cancer Discov 2019;9:1064–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen H, Ma J, Li W, Eliseenkova AV, Xu C, Neubert TA, et al. A molecular brake in the kinase hinge region regulates the activity of receptor tyrosine kinases. Mol Cell 2007;27:717–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schram AM, Kamath SD, El-Khoueiry AB, Borad MJ, Mody K, Mahipal A, et al. First-in-human study of highly selective FGFR2 inhibitor, RLY-4008, in patients with intrahepatic cholangiocarcinoma and other advanced solid tumors. J Clin Oncol 39, 2021(suppl 15; abstr TPS4165). [Google Scholar]
- 29. Goyal L, Borad MJ, Subbiah V, Mahipal A, Kamath S, Mody K, et al. First results of RLY-4008, a potent and highly selective FGFR2 inhibitor in a first-in-human study in patients with FGFR2-altered cholangiocarcinoma and multiple solid tumors [abstract]. In: Proceedings of the AACR-NCI-EORTC Virtual International Conference on Molecular Targets and Cancer Therapeutics; 2021 Oct 7–10. Philadelphia (PA): AACR; Mol Cancer Ther 2021;20(12 Suppl):Abstract nr P02-02 .
- 30. Wu Q, Zhen Y, Shi L, Vu P, Greninger P, Adil R, et al. EGFR inhibition potentiates FGFR inhibitor therapy and overcomes resistance in FGFR2 fusion-positive cholangiocarcinoma. Cancer Discov 2022;12:1378–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hollebecque A, Borad MJ, Goyal L, Schram A, Park JO, Cassier PA, et al. Efficacy of RLY-4008, a highly selective FGFR2 inhibitor in patients (pts) with an FGFR2-fusion or rearrangement (f/r), FGFR inhibitor (FGFRi)-naïve cholangiocarcinoma (CCA): ReFocus trial. Ann Oncol 2022;33 Suppl 7:S1381. Abstract nr LBA12. Available from: 10.1016/j.annonc.2022.08.006. [DOI] [Google Scholar]
- 32. Kalyukina M, Yosaatmadja Y, Middleditch MJ, Patterson AV, Smaill JB, Squire CJ. TAS-120 cancer target binding: defining reactivity and revealing the first fibroblast growth factor receptor 1 (FGFR1) irreversible structure. ChemMedChem 2019;14:494–500. [DOI] [PubMed] [Google Scholar]
- 33. Sootome H, Fujita H, Ito K, Ochiiwa H, Fujioka Y, Ito K, et al. Futibatinib is a novel irreversible FGFR 1-4 inhibitor that shows selective antitumor activity against FGFR-deregulated tumors. Cancer Res 2020;80:4986–97. [DOI] [PubMed] [Google Scholar]
- 34. Varghese AM, Patel J, Janjigian YY, Meng F, Selcuklu SD, Iyer G, et al. Noninvasive detection of polyclonal acquired resistance to FGFR inhibition in patients with cholangiocarcinoma harboring FGFR2 alterations. JCO Precis Oncol 2021;5:PO.20.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gile JJ, Wookey V, Zemla TJ, Shi Q, Jin Z, Alberts SR, et al. Outcomes following FGFR inhibitor therapy in patients with cholangiocarcinoma. Target Oncol 2022;17:529–38. [DOI] [PubMed] [Google Scholar]
- 36. Krook MA, Bonneville R, Chen H-Z, Reeser JW, Wing MR, Martin DM, et al. Tumor heterogeneity and acquired drug resistance in FGFR2-fusion-positive cholangiocarcinoma through rapid research autopsy. Cold Spring Harb Mol Case Stud 2019;5:a004002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lescarbeau A, Beamish-Cook J, Prentice Z, Kendall T, Dumas A, Isaak E, et al., inventors; Relay Therapeutics, Inc. and D. E. Shaw Research, LLC, assignees. FGFR inhibitors and methods of making and using the same. World Intellectual Property Organization, International Bureau WO 2022/109577 Al. 2022 May 27.
- 38. Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr 2010;66:125–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr 2007;40:658–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 2010;66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liebschner D, Afonine PV, Baker ML, Bunkóczi G, Chen VB, Croll TI, et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol 2019;75:861–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Potency (kinact/KI) of RLY-4008 against FGFR1 and FGFR2
Biochemical inhibition of FGFR by RLY-4008 and pan-FGFRi
Antiproliferative effect of RLY-4008 and pan-FGFRi in human tumor cell lines
In vivo exposures of pan-FGFRi in mouse studies
Fold change in potency against FGFR2 acquired resistance mutations
Supplementary Figure 1. RLY-4008 is an irreversible inhibitor of FGFR2.
Supplementary Figure 2. Treatment with RLY-4008 is well-tolerated and leads to dose-dependent inhibition of FGFR2 and tumor regression in multiple FGFR2-altered tumor models.
ReFocus entry criteria
three references cited in supplementary data
In this schematic movie, apo FGFR1 (left panel) demonstrates rapid P-loop dynamics, while apo FGFR2 (right panel) displays less P-loop motion.
Supplementary video legend describing schematic video of P-loop dynamics



![Figure 1. RLY-4008 is a potent and selective irreversible inhibitor of FGFR2. A, Sequence alignment of the kinase domains of FGFR1–4 indicates a high degree of similarity among paralogs. RLY-4008 binding site residues are boxed; residues shown in pink identify amino acid differences between FGFR2 and paralogs within this region. Numbering refers to the FGFR2 IIIc isoform. B, Chemical structure of RLY-4008, N-(4-(4-amino-5-(3-fluoro-4-((4-methylpyrimidin-2-yl)oxy)phenyl)-7-methyl-7H-pyrrolo[2,3-d]pyrimidin-6-yl)phenyl)methacrylamide. C, Crystal structure of RLY-4008 in complex with FGFR2 (PDB: 8STG). Protein is shown in green, and inhibitor carbons are shown in magenta. The Cys491 sulfur is shown in gold and as a covalent adduct with RLY-4008. D and E, Rate of covalent labeling of FGFR2 (red) and FGFR1 (blue) by RLY-4008 (D) and futibatinib (E) as measured by intact mass over time. Triplicate biological replicates are reported. F, RLY-4008 concentration-dependent modification rate against FGFR2 (red) and FGFR1 (blue). RLY-4008 against FGFR2: kinact = 6.45 × 10−2 per second; KI = 1.87 μmol/L; kinact/KI = 3.45 × 10−2 per second/(μmol/L). RLY-4008 against FGFR1: kinact = 2.33 × 10−3 per second; KI = 6.14 μmol/L; kinact/KI = 3.79 × 10−4 per second/(μmol/L). G, Fold change in biochemical IC50 values of the indicated inhibitors between FGFR2 and FGFR1, FGFR3, and FGFR4. Average fold change of three independent experiments each containing two biological replicates is reported. Error bars indicate SD. H, TREEspot depicting selectivity of RLY-4008 screened against 468 kinases via KINOMEscan (DiscoverX, Eurofins). At the test concentration of 500 nmol/L, three kinases showed greater than 75% inhibition: FGFR2 (94.1%), MEK5 (92.4%), and MKNK2 (89%). Image generated using TREEspot Software Tool and reprinted with permission from KINOMEscan, a division of DiscoveRx Corporation, ©DiscoverX Corporation 2010.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/d9ea/10481131/d7e8a83dc76e/2012fig1.jpg)