Abstract
Breda virus (BRV), a member of the genus Torovirus, is an established etiological agent of disease in cattle. BRV isolates have been detected in the stools of neonatal calves with diarrhea in both Iowa and Ohio and in several areas of Europe. However, this virus has been reported only once in Canada. Therefore, a study was performed to determine the extent to which bovine torovirus is present in calves with diarrhea from farms in southern Ontario. A total of 118 fecal samples from symptomatic calves and 43 control specimens from asymptomatic calves were examined by electron microscopy (EM) and reverse transcription-PCR (RT-PCR) for the presence of torovirus. Torovirus RNA was detected in 43 of the 118 diarrheic samples (36.4%) by RT-PCR with primers designed in the conserved 3′ end of the torovirus genome. By EM, torovirus particles were observed in 37 of the 118 specimens (31.4%). All but one of these samples were also positive by RT-PCR. The incidence of torovirus in the asymptomatic control specimens by RT-PCR was only 11.6%. To establish the identity of the particles observed in the diarrheic specimens, five of the amplicons from samples positive by both RT-PCR and EM were cloned and sequenced. Nucleotide sequence analysis revealed that the bovine torovirus found in southern Ontario manifests between 96 and 97% sequence identity to the BRV type 1 strain found in Iowa. This study shows that bovine torovirus is a common virus in the fecal specimens of calves with diarrhea from farms in southern Ontario and thus may be an important pathogen of cattle.
The etiology of infectious diarrhea in calves has been attributed to rotavirus, coronavirus, calicivirus, parvovirus, and astrovirus (14–16, 22), agents of defined morphology that can be readily detected by electron microscopy (EM). Breda virus (BRV), a member of the genus Torovirus, was first associated with enteritis of calves in 1982 (25). This agent has been relatively infrequently reported because it is more difficult to recognize by EM and because, unlike the torovirus prototype, Berne virus (BEV), it cannot as yet be grown in cell culture, which has precluded the development of routine immunospecific diagnostic tests. However, the partial sequencing of the 3′ end of the BRV genome (11) has allowed for the application of reverse transcription-PCR (RT-PCR) for the diagnosis of bovine torovirus infections.
Prospective studies in The Netherlands, in which viruses were examined in symptomatic and asymptomatic calves by enzyme-linked immunosorbent assay (ELISA), demonstrated that torovirus was present in 6.4% of calves with diarrhea compared with only 1.7% of asymptomatic controls. In contrast, rotavirus was found in 37.4% of symptomatic animals and 13.9% of controls in this study (12). Epidemiologic studies have shown that bovine torovirus is widespread in The Netherlands (10), Germany and Switzerland (19), the United Kingdom (2), and the United States (17, 23, 26), with approximately 90% of dairy cattle being seropositive. In one study from Belgium, bovine toroviruses were found to play a role in respiratory, digestive, and reproductive disorders of cattle (18). In a recent study from Saskatchewan, Canada, BRV-like particles were detected in 42 of 221 fecal or intestinal specimens of symptomatic calves (7).
The aim of the present study was to determine the incidence of torovirus excretion in calves with diarrhea from farms in southern Ontario by RT-PCR and EM and to compare this with the excretion of other enteric pathogens including bovine rotaviruses and coronaviruses.
MATERIALS AND METHODS
Specimens.
A total of 118 stool specimens from calves with diarrhea and 43 specimens from asymptomatic calves were obtained from the Animal Health Laboratory (AHL) in Guelph, Ontario, Canada. These specimens were submitted to the AHL by veterinarians in the region of southern Ontario between April 1995 and March 1997. The majority of the calves were between 2 and 60 days old at the time of specimen collection. Since testing for torovirus was performed retrospectively, the diarrheic specimens had all previously been tested at the AHL for bovine coronavirus by an in-house ELISA system (1, 3), for bovine type A rotavirus by latex agglutination (Microgen Bioproducts Ltd., Camberley, United Kingdom), and for bovine viral diarrhea virus (BVDV) by isolation in cell culture (4). Furthermore, all specimens were coded, examined for the presence of viruses by negative-contrast EM, and tested for bovine torovirus by RT-PCR in a blinded fashion.
EM.
Fecal specimens were diluted with an equal volume of 1% (wt/vol) ammonium acetate and clarified by centrifugation at 9,000 × g for 15 min at 4°C. The supernatant was transferred to a new tube and centrifuged at 12,000 × g for 15 min at 4°C. Each sample was applied to a 400-mesh grid precoated with polyvinyl formal and carbon. The grids were stained for 1 min with 2% phosphotungstic acid (pH 7.0) and examined by negative-contrast EM with a Philips EM 300 microscope, at a magnification of ×50,000 (13).
RNA extraction.
Fecal specimens were diluted in an equal volume (wt/vol) of phosphate-buffered saline and clarified by centrifugation at 9,000 × g for 15 min at 4°C. The supernatant was transferred to a new tube and centrifuged at 12,000 × g for 15 min at 4°C. In a separate room, with dedicated micropipetters and aerosol-resistant tips, viral RNA was extracted from 100 μl of the partially purified supernatant with TRIzol reagent (Gibco BRL, Burlington, Ontario, Canada) according to the manufacturer’s protocol. Each RNA pellet was resuspended in 10 μl of DNase-free, RNase-free double-distilled water (ddH2O) (5 prime-3 prime Inc., Boulder, Colo.) and stored at −80°C.
RT-PCR.
All specimens were tested for the presence of torovirus RNA by RT-PCR. For each run of specimens assayed, a negative control (ddH2O) and a control RNA extracted from a stool specimen shown to contain only bovine rotavirus by EM were included. RNA extracted from a BRV type 1 (BRV-1) (Iowa strain)-positive stool specimen (obtained from G. Woode, Texas A&M University) was tested as a positive control in every third RT-PCR assay due to the limited amount of control stool available. Oligonucleotide primers (General Synthesis and Diagnostics, Toronto, Ontario, Canada) were designed from the 3′ end of the BEV genome (DDBJ accession no. D00563). The sense primer (5′TAATGGCACTGAAGACTC3′) and the antisense primer (5′ACATAACATCTTACATGG3′) bracketed a genome fragment of 219 bases, which included the 3′ end of the N protein coding region and most of the 3′ noncoding region upstream of the poly(A) tail.
The RT reaction and PCR mixtures were set up in an isolated room, with dedicated micropipetters and aerosol-resistant tips. Reactions were then performed in another room designated for PCR amplification. For the RT reaction, a 10-μl RNA aliquot was incubated for 5 min at 65°C and added to 10 μl of the RT mixture containing 5 mM MgCl2; 10 mM Tris-HCl (pH 8.3); 50 mM KCl; 1.25 mM (each) dATP, dCTP, dGTP, and dTTP (Promega, Madison, Wis.); 2.6 μM random hexamer primers; 20 U of RNase Guard (Gibco BRL); and 50 U of Moloney murine leukemia virus reverse transcriptase (Gibco BRL). The reaction mixture was overlaid with sterile mineral oil and incubated at room temperature for 10 min. The RT reaction was performed in a Perkin-Elmer (Mississauga, Ontario, Canada) 480 thermal cycler at 42°C for 30 min and 99°C for 5 min, and then the reaction mixture was held at 5°C for 5 min.
For the PCR, 10 μl of the RT reaction was added to the PCR mixture containing 1 mM MgCl2, 8 mM Tris-HCl (pH 8.3), 40 mM KCl, 2.5 U of Amplitaq DNA polymerase (Perkin-Elmer Cetus and Applied Biosystems Inc.), and 50 pmol of each primer. The total volume of the PCR was 50 μl. The reaction mixture was overlaid with sterile mineral oil and amplified in a Perkin-Elmer 480 thermal cycler with an initial denaturation at 94°C for 2 min, followed by 35 cycles consisting of denaturation at 95°C for 40 s, annealing at 50°C for 1 min, and extension at 72°C for 1 min 30 s. Reaction mixtures were then incubated at 72°C for 10 min and held at 4°C. Products were analyzed by electrophoresis through a 1.2% agarose gel containing ethidium bromide and viewed under a UV transilluminator.
Cloning and DNA sequencing.
Selected PCR products were purified by the Wizard PCR Preps DNA purification system (Promega), cloned into a pCR-Script Amp SK+ cloning vector, and transformed into Epicurian coli XL1-blue MRF′ Kan supercompetent cells (pCR-Script Amp SK+ cloning kit; Stratagene, La Jolla, Calif.) as per the manufacturer’s recommendations. Clones were screened by PCR with the same primers as described above. Plasmids containing inserts were purified from broth cultures with the Wizard Miniprep DNA purification system (Promega) and sequenced with the fmol DNA sequencing system (Promega) according to the manufacturer’s recommendations. Sequence data were analyzed with the computer program GCG, version 8 (Genetics Computer Group, Inc., Madison, Wis.). As an additional precaution against plasmid contamination of PCRs, all cloning and sequencing assays were undertaken only after the RT-PCR experiments on stool samples had been completed.
Statistical analysis.
The chi-square test was used to establish statistical significance.
RESULTS
EM.
Of the 118 diarrheic specimens that were examined, 37 (31.4%) were found to be positive for bovine torovirus by negative-contrast EM. In contrast, torovirus particles were detected in only 2 of the 43 asymptomatic control specimens examined (Table 1).
TABLE 1.
EM and RT-PCR results for the detection of torovirus in stool specimens from diarrheic and asymptomatic calves in southern Ontarioa
| RT-PCR result | No. (%) of calves
|
|||||
|---|---|---|---|---|---|---|
| Symptomatic
|
Asymptomatic
|
|||||
| EM+ | EM− | Total | EM+ | EM− | Total | |
| Positive | 36 (30.5) | 7 (5.9) | 43 (36.4)b | 2 (4.7) | 3 (6.9) | 5 (11.6) |
| Negative | 1 (0.9) | 74 (62.7) | 75 (63.4) | 0 | 38 (88.4) | 38 (88.4) |
| Total | 37 (31.4) | 81 (68.6) | 118 | 2 (4.7) | 41 (95.3) | 43 |
Specimens were examined by EM for the presence (EM+) or absence (EM−) of bovine torovirus particles, coded, and tested by RT-PCR. The RT-PCR was performed with primers designed from the 3′ end of the BEV genome. Positive specimens gave amplicons of 219 bases in length.
P = 0.0023 (chi-square test).
The torovirus particles detected by EM were morphologically similar to the BRV previously reported in the stool specimens of diarrheic calves in Iowa and Ohio (26). These particles measured between 100 and 120 nm at their largest diameter, and they exhibited torus-, crescent-, and rod-shaped conformations. The particles were clearly enveloped and bore a fringe of peplomers, each measuring approximately 10 nm in length (Fig. 1).
FIG. 1.
Electron micrograph of a calf fecal specimen showing torus- and crescent-shaped bovine torovirus particles. Bar, 100 nm.
RT-PCR.
By RT-PCR analysis, a DNA fragment of 219 bases was detected in 43 of the 118 (36.4%) specimens from symptomatic calves. Of these 43 samples, 36 were shown to contain torovirus particles by EM. Only one diarrheic specimen that was negative by RT-PCR was found to contain torovirus by EM (Table 1). Repeat testing was performed on all discordant samples to affirm the initial RT-PCR results.
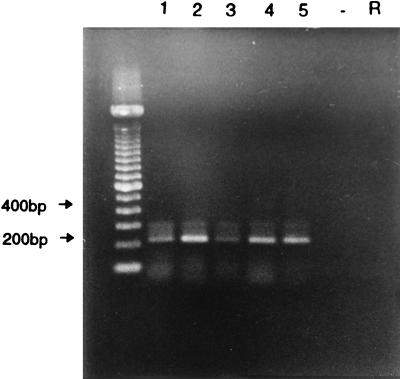
Among the 43 asymptomatic control samples tested, only 5 (11.6%) were positive for torovirus by RT-PCR, and 2 of these were also positive by EM. All 38 specimens that were negative by RT-PCR were also negative by EM (Table 1). Therefore, there was a significantly greater number of torovirus-positive specimens in the symptomatic calf population than in the asymptomatic calves (P = 0.0023). None of the negative control and rotavirus control samples gave a positive result by RT-PCR. Figure 2 shows representative amplification products after agarose gel electrophoresis.
FIG. 2.
Gel electrophoresis of RT-PCR products from five torovirus-positive fecal specimens. Lanes marked − and R represent a negative control (ddH2O) and a bovine rotavirus sample, respectively. A 100-bp ladder was used as the molecular size marker.
Other viruses.
Of the 118 specimens from diarrheic calves, 29 were positive for viruses other than bovine torovirus, including coronavirus, rotavirus, BVDV, and small round-structured viruses (SRSVs) as seen by EM. Of the 43 specimens that were positive for torovirus by RT-PCR, 10 were also positive for another virus. Of these, five had torovirus and rotavirus, two were positive for torovirus and coronavirus, one had torovirus and BVDV, and two were found to contain a mixed infection of torovirus, rotavirus, and coronavirus (Table 2).
TABLE 2.
Summary of viruses present in the stools of symptomatic and asymptomatic calves from Ontario farmsa
| Virus present | No. (%) of calves
|
|
|---|---|---|
| Symptomatic (n = 118) | Asymptomatic (n = 43) | |
| Torovirus alone | 33 (27.9)b | 5 (11.6) |
| Torovirus plus rotavirusc | 5 (4.2) | 0 |
| Torovirus plus coronavirus | 2 (1.7) | 0 |
| Torovirus plus BVDV | 1 (0.9) | 0 |
| Torovirus plus SRSV | 0 | 0 |
| Torovirus, rotavirus, plus coronavirus | 2 (1.7) | 0 |
| Rotavirus alonec | 3 (2.5) | 2 (4.7) |
| Coronavirus alone | 5 (4.2) | 1 (2.3) |
| BVDV alone | 3 (2.5) | 0 |
| SRSV alone | 1 (0.9) | 0 |
| Rotavirus plus coronavirus | 6 (5.2) | 0 |
| Rotavirus plus BVDV | 1 (0.9) | 0 |
| No virus present | 56 (47.4) | 35 (81.4) |
The presence of torovirus was determined by RT-PCR, that of rotavirus was determined by latex agglutination and EM, that of coronavirus was determined by ELISA and EM, that of BVDV was determined by cell culture, and that of SRSV was determined by EM.
P = 0.016.
The number of other viruses present in torovirus-positive diarrheic specimens was compared to the number of other viruses present in torovirus-negative diarrheic specimens by chi-square analysis (P = 0.8).
In addition, 19 of the 75 diarrheic specimens that were negative for torovirus by RT-PCR were positive for other viruses. Of these, three had rotavirus, five had coronavirus, three were positive for BVDV, one contained SRSV, and seven had mixed infections of either rotavirus and coronavirus or rotavirus and BVDV (Table 2). There was no significant difference between torovirus-positive diarrheic specimens and torovirus-negative diarrheic specimens for the presence of other viruses in the stools (P = 0.8).
Only three of the asymptomatic control samples that were negative for torovirus by both EM and RT-PCR contained other viruses, including two specimens with rotavirus and one specimen with coronavirus as seen by EM.
Cloning and sequencing of torovirus-positive RT-PCR products.
To confirm that the 219-base product obtained by RT-PCR was indeed bovine torovirus and to determine the degree of heterogeneity among samples, amplicons obtained from five different specimens were cloned and sequenced. Clones were screened by PCR with the same primers, and at least two clones per sample were found to contain the 219-base fragment. These clones were isolated and sequenced, and their nucleotide sequences were compared to the 3′ regions of the BRV-1 and BEV genomes (Fig. 3). The nucleotide sequences of each of the five torovirus-positive isolates were found to be between 96 and 97% identical to BRV-1 and between 95 and 96% identical to BEV in this area. The nucleotide substitutions were interspersed throughout the 219-base fragment. None of these substitutions caused changes in the predicted amino acid sequence of the BRV-1 nucleocapsid (N) protein, and substitutions at nucleotide positions 23 and 25 caused only one amino acid change, from glutamine to lysine, in the predicted amino acid sequence of the BEV N protein.
FIG. 3.
Alignment of the nucleotide sequence from the 3′ end of the BRV-1 (Iowa strain) genome with those of five bovine torovirus-positive samples from diarrheic calves in southern Ontario (a to e) and the BEV genome. Nucleotides identical to the consensus sequence are shown as dots.
DISCUSSION
Although bovine torovirus has been established as a widespread agent of diarrhea in calves, its prevalence has been investigated to only a limited extent, largely because it cannot be grown in cell culture. This inability to isolate the virus has precluded the large-scale preparation of reference antisera and antigens for the development of commercial immunospecific diagnostic tests such as ELISA and latex agglutination, which are currently used to diagnose coronavirus and type A rotavirus infections (3, 8, 9). Consequently, the etiology of a substantial proportion of viral diarrhea in calves due to BRV may not be diagnosed, which impacts on the establishment of containment measures and the potential incentive towards the development of vaccines.
The RT-PCR was successfully applied to the diagnosis of torovirus in fecal specimens of calves in this study. While molecular approaches such as hybridization with cDNA probes have been previously reported (11), our findings show that RT-PCR is a useful method for routine diagnosis of torovirus. The assay compared favorably with direct EM and was more sensitive in that it detected tonovirus in 6 additional specimens beyond the 37 detected by EM alone. It is unlikely that these specimens represent false positives because of the exhaustive precautions taken to control for contamination of the RT-PCR assay. Only one specimen that was positive by EM was negative by RT-PCR. This may have been due to nonspecific inhibitors present in the stool, as has been shown previously for other gastroenteritis viruses (20). Due to the limited amounts of BRV-1 control stool available, the presence of inhibitors could not be verified by spiking an aliquot of the discordant sample with a known amount of BRV-1 control specimen and repeating the RT-PCR assay.
In this study, bovine torovirus was present significantly more frequently (36.4%) in calves with diarrhea than in the asymptomatic controls (11.6%; P = 0.0023). Thus, bovine torovirus is associated with the symptoms of enteritis in these symptomatic cases. The excretion of torovirus, as well as other viruses including rotavirus and coronavirus, in asymptomatic animals has been previously reported (6), and our findings are consistent with these observations.
In a number of cases, other viruses were also present along with torovirus. That is, 10 of 43 diarrheic specimens that were positive for bovine torovirus by RT-PCR were also shown to contain other viruses. Nevertheless, the incidence of torovirus alone in the diarrheic specimens (27.9%) was still significantly greater than the presence of bovine torovirus in asymptomatic calves (11.6%; P = 0.016). However, it remains impossible to determine whether bovine torovirus was the primary cause of diarrhea in the symptomatic calves who had concomitant infections with other viral agents of enteritis. This phenomenon of mixed infection was also observed among the diarrheic specimens that were negative by EM and RT-PCR which contained other viruses and has previously been reported in other studies of animals and humans (14, 15, 21, 24).
This study has added to our understanding of the epidemiology of torovirus enteritis by demonstrating that, in keeping with previous observations from North America (7, 17, 23, 26) and Europe (2, 10, 18, 19), it is also prevalent in the southern Ontario cattle population. Moreover, bovine torovirus was shown to be a common pathogen in the stools of symptomatic calves, exceeding the prevalence of bovine rotavirus and bovine coronavirus in this study. This was also the case in the Saskatchewan study, in which 19% of symptomatic specimens were found to contain BRV-like particles, whereas only 9.5% were positive for rotavirus as detected by EM (7).
Sequence analysis of a subset of the torovirus-positive RT-PCR amplicons confirmed that the particles detected in the fecal specimens of calves in southern Ontario are related to the torovirus prototype, BEV, and to BRV-1, since their sequences demonstrated between 95 and 97% sequence identity in the 3′ noncoding region of the genomes of these viruses (11). There was also a small yet defined amount of genomic heterogeneity among the bovine torovirus clones despite the conserved nature of the 3′ end of the torovirus genome. This could be interpreted as evidence that genetic variation may exist among toroviruses isolated from different outbreaks. A more complete analysis of the sequences of specific viral genes, especially the peplomer and hemagglutinin-esterase genes, the latter of which has recently been described for BRV (5), is needed to address this hypothesis.
ACKNOWLEDGMENT
This research was supported by a grant from the Medical Research Council of Canada.
REFERENCES
- 1.Athanassious R, Marsolais G, Assaf R, Dea S, Descouteaux J P, Dulude S, Monpetit C. Detection of bovine coronavirus and type A rotavirus in neonatal calf diarrhea and winter dysentery of cattle in Quebec: evaluation of three diagnostic methods. Can Vet J. 1994;35:163–169. [PMC free article] [PubMed] [Google Scholar]
- 2.Brown D W G, Hall C, Green J, Lamouliatte F, Flewett T H. Detection of Breda virus antigen and antibody in humans and animals by enzyme immunoassay. J Clin Microbiol. 1987;25:637–640. doi: 10.1128/jcm.25.4.637-640.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carman P S, Hazlett M J. Bovine coronavirus infection in Ontario, 1990–1991. Can Vet J. 1992;33:812–814. [PMC free article] [PubMed] [Google Scholar]
- 4.Carman S, van Dreumel T, Ridpath J, Hazlet M, Alves D, Dubovi E, Tremblay R, Bolin S, Godkin A, Anderson N. Severe acute bovine viral diarrhea in Ontario, 1993–1995. J Vet Diagn Invest. 1998;10:27–35. doi: 10.1177/104063879801000106. [DOI] [PubMed] [Google Scholar]
- 5.Cornelissen L A H M, Wierda C M H, van der Meer F J, Herrewegh A A P M, Horzinek M C, Egberink H F, de Groot R J. Hemagglutinin esterase, a novel structural protein of torovirus. J Virol. 1997;71:5277–5286. doi: 10.1128/jvi.71.7.5277-5286.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crouch C F, Acres S D. Prevalence of rotavirus and coronavirus antigens in the feces of normal cows. Can J Comp Med. 1984;48:340–342. [PMC free article] [PubMed] [Google Scholar]
- 7.Durham P J K, Hassard L E, Norman G R, Yemen R L. Viruses and virus-like particles detected during examination of feces from calves and piglets with diarrhea. Can Vet J. 1989;30:876–881. [PMC free article] [PubMed] [Google Scholar]
- 8.Ellens D J, de Leeuw P W. ELISA for diagnosis of rotavirus infections in calves. J Clin Microbiol. 1977;6:530–532. doi: 10.1128/jcm.6.5.530-532.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellens D J, van Balken J A M, de Leeuw P W. Proceedings of the International Symposium on Neonatal Diarrhea, Saskatoon, Canada. 1978. Diagnosis of bovine coronavirus infections with hemadsorption-elution-hemagglutination assay (HEHA) and with enzyme-linked immunosorbent assay (ELISA) pp. 321–330. [Google Scholar]
- 10.Koopmans M, van den Boom U, Woode G N, Horzinek M C. Seroepidemiology of Breda virus in cattle using ELISA. Vet Microbiol. 1989;19:233–243. doi: 10.1016/0378-1135(89)90069-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koopmans M, Snijder E J, Horzinek M C. cDNA probes for the diagnosis of bovine torovirus (Breda virus) infection. J Clin Microbiol. 1991;29:493–497. doi: 10.1128/jcm.29.3.493-497.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koopmans M, van Wuijckhuise-Sjouke L, Schukken Y H, Cremers H, Horzinek M C. Association of diarrhea in cattle with torovirus infections on farms. Am J Vet Res. 1991;11:1769–1773. [PubMed] [Google Scholar]
- 13.Middleton P J, Szymanski M, Petric M. Viruses associated with acute gastroenteritis in young children. Am J Dis Child. 1977;131:733–737. doi: 10.1001/archpedi.1977.02120200015004. [DOI] [PubMed] [Google Scholar]
- 14.Moerman A, van Zijderveld F G, de Leeuw P W. Neonatal calf diarrhea. Annu Rep Cent Vet Inst (Lelystad) 1986;1986:36–41. [Google Scholar]
- 15.Snodgrass D R, Terzolo H R, Sherwood D. Aetiology of diarrhea in young calves. Vet Rec. 1986;119:31–34. doi: 10.1136/vr.119.2.31. [DOI] [PubMed] [Google Scholar]
- 16.Storz J, Bates R C. Parvovirus infections in calves. J Am Vet Med Assoc. 1973;163:884–886. [Google Scholar]
- 17.Van Kruiningen H J, Castellano V P, Koopmans M, Harris L L. A serologic investigation for coronavirus and Breda virus antibody in winter dysentery of dairy cattle in the northeastern United States. J Vet Diagn Invest. 1992;4:450–452. doi: 10.1177/104063879200400415. [DOI] [PubMed] [Google Scholar]
- 18.Vanopdenbosch E, Wellemans G, Oudewater J, Petroff K. Prevalence of torovirus infections in Belgian cattle and their role in respiratory, digestive, and reproductive disorders. Vlaams Diergeneeskd Tijdschr. 1985;61:187–191. [Google Scholar]
- 19.Weiss M, Steck F, Kaderli R, Horzinek M C. Antibodies to Berne virus in horses and other animals. Vet Microbiol. 1984;9:523–531. doi: 10.1016/0378-1135(84)90014-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilde J, Eiden J, Yolken R. Removal of inhibitory substances from human fecal specimens for detection of group A rotaviruses by reverse transcriptase and polymerase chain reactions. J Clin Microbiol. 1990;28:1300–1307. doi: 10.1128/jcm.28.6.1300-1307.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woode G N. 12th World Congress on Cattle Diseases, The Netherlands. Vol. 1. 1982. Etiology of enteric viral infections of calves: pathological and clinical aspects; pp. 201–208. [Google Scholar]
- 22.Woode G N, Bridger J C. Isolation of small viruses resembling astroviruses and caliciviruses from acute enteritis of calves. J Med Microbiol. 1978;11:441–452. doi: 10.1099/00222615-11-4-441. [DOI] [PubMed] [Google Scholar]
- 23.Woode G N, Mohammed K A, Saif L J. Diagnostic methods for the newly discovered “Breda” group of calf enteritis inducing viruses. Proc Annu Meet Am Assoc Vet Lab Diagn. 1983;3:533–538. [Google Scholar]
- 24.Woode G N, Pholenz J F, Gourley N E K. Astrovirus and Breda virus infection of dome cell epithelium of bovine ileum. J Clin Microbiol. 1984;19:623–630. doi: 10.1128/jcm.19.5.623-630.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woode G N, Reed D E, Runnels P L, Herrig M A, Hill H T. Studies with an unclassified virus isolated from diarrheal calves. Vet Microbiol. 1982;7:221–240. doi: 10.1016/0378-1135(82)90036-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woode G N, Saif L J, Quesada M, Winand N J, Pholenz J F, Gourley N E K. Comparative studies on three isolates of Breda virus calves. Am J Vet Res. 1985;46:1003–1010. [PubMed] [Google Scholar]