Abstract
NADPH-cytochrome P450 reductase delivers electrons required by heme oxygenase, squalene monooxygenase, fatty acid desaturase, and 48 human cytochrome P450 enzymes. While conformational changes supporting reductase intramolecular electron transfer are well defined, intermolecular interactions with these targets are poorly understood, in part because of their transient association. Herein the reductase FMN domain responsible for interacting with targets was fused to the N-terminus of three drug-metabolizing and two steroidogenic cytochrome P450 enzymes to increase the probability of interaction. These artificial fusion enzymes were profiled for their ability to bind their respective substrates and inhibitors and to perform catalysis supported by cumene hydroperoxide. Comparisons with the isolated P450 enzymes revealed that even the oxidized FMN domain causes substantial and diverse effects on P450 function. The FMN domain could increase, decrease, or not affect total ligand binding and/or dissociation constants depending on both P450 enzyme and ligand. As examples, FMN domain fusion has no effect on inhibitor ketoconazole binding to CYP17A1 but substantially altered CYP21A2 binding of the same compound. FMN domain fusion to CYP21A2 resulted in differential effects dependent on whether the ligand was 17α-hydroxyprogesterone versus ketoconazole. Similar enzyme-specific effects were observed on steady-state kinetics. These observations are most consistent with FMN domain interacting with the proximal P450 surface to allosterically impact P450 ligand binding and metabolism separate from electron delivery. The variety of effects on different P450 enzymes and on the same P450 with different ligands suggests intricate and differential allosteric communication between the P450 active site and its proximal reductase-binding surface.
Keywords: cytochrome P450, redox partner, allostery, ligand binding, NADPH-cytochrome P450 reductase
Cytochrome P450 enzymes are a class of heme-containing monooxygenases critical for diverse functions in the human body. Most well known for their roles in xenobiotic metabolism, many human P450 enzymes are also critical in various endogenous biosynthetic pathways, including the biosynthesis of steroid hormones, bile acids, eicosanoids, and vitamins.
The dozen or so human xenobiotic P450 enzymes are responsible for the metabolism of over 70% of drugs on the market (1), with CYP1, CYP2, and CYP3 families largely responsible for this activity. Indeed, CYP3A4 and CYP2D6 alone contribute to over 50% of CYP-related drug metabolism (2). Other drug-metabolizing P450 enzymes are somewhat more selective but still important for selected classes of drugs or individual drugs. For example, the CYP2 family enzyme CYP2A6 is only responsible for 3% of CYP-related drug metabolism but is critically important for the metabolism and clearance of nicotine and its metabolite cotinine (3).
In human steroidogenesis six different P450 enzymes convert cholesterol into mineralocorticoids such as aldosterone controlling blood pressure, into glucocorticoids such as cortisol that modulate stress and immune responses, and into androgenic and estrogenic sex steroids that control reproductive development and fertility. Within this pathway CYP17A1 and CYP21A2 occur at particularly critical junctions. In the absence of CYP17A1 activity, the formation of mineralocorticoids occurs but not that of glucocorticoids or sex steroids. The CYP21A2 enzyme is similarly a committed step in the generation of mineralocorticoids and glucocorticoids over the formation of androgens and estrogens.
Whether involved in drug metabolism or endogenous pathways like steroidogenesis, cytochrome P450 enzymes perform substrate oxidation using their heme prosthetic group to activate molecular oxygen. To accomplish such catalysis almost all human P450 enzymes require two electrons delivered from a redox partner protein. For 48 of the 57 human P450 enzymes, this redox partner protein is NADPH-cytochrome P450 reductase. This reductase abstracts a hydride from NADPH, accepts both electrons via its FAD-containing domain, then transfers them one at a time to its FMN-containing domain, which can then pass electrons to the P450 heme (4).
In order to accomplish this, the reductase FMN-containing domain must release from its FAD-containing domain to interact with the P450. Thus, there are large-scale conformational dynamics involved between domains. The reductase FAD-to-FMN interdomain electron transfer occurs in a “closed” conformation, for which there is a crystal structure indicating that the distance between the two flavins is 3.5 Å (5). However, conformational changes in a hinge region between the two reductase flavin-containing domains are thought to allow the two flavin domains to move apart from each other to permit the FMN domain to bind the proximal P450 surface. Several lines of evidence support this concept. First, restricting conformational change by engineering a disulfide bond between the two flavin domains markedly decreases P450 reduction (6). Second, mutations in the linker affect electron transfer supporting P450 catalysis (7). Third, modification of the linker between the two flavin domains revealed a set of “open” conformations, in which the FMN domain would be available for interaction with a P450 enzyme (8). Finally, these domain motions are consistent with NMR and X-ray scattering experiments (9). The FMN domain of reductase must undergo these domain movements twice in order to transfer both electrons required for P450 catalysis (4). Unfortunately, relatively little is known about the details of this transient interaction between the reductase FMN-containing domain and any of the 48 relevant human P450 enzymes.
Both clinical and experimental data suggest that the interactions between reductase and individual P450 enzymes may vary. Clinically, some of the strongest evidence has been seen in patients with Antley–Bixler syndrome. This rare genetic disorder results in severe skeletal and developmental malformations (10, 11). Early studies attributed the condition to mutations in the fibroblast growth factor receptor 2 causing the observed skeletal malformations (12). However, subsequent investigation of patients with Antley–Bixler syndrome with developmental malformations consisting of ambiguous or malformed genitalia revealed abnormal steroid profiles (13). The steroidogenic P450 enzymes CYP17A1 and CYP21A2 had decreased activities, but sequencing revealed no mutations in these enzymes (14, 15, 16, 17). Instead it was discovered that mutations in the NADPH-cytochrome P450 reductase were the cause. Since this reductase is also required by drug-metabolizing P450 enzymes, one would expect similar deficiencies in their catalytic functions, but this was not uniformly the case. Instead, reductase mutations can have quite varied effects on different cytochrome P450 enzymes (18). For example, the patient mutation Q153R in the reductase FMN domain decreased both CYP17A1 hydroxylase and lyase activity by ∼70% (17) but had less consistent effects on the activities of drug-metabolizing P450 enzymes. While the catalysis of CYP3A4, CYP2D6, and CYP1A2 was moderately increased (∼120%, ∼130%, and ∼140% of wildtype activity, respectively), the activity of CYP2C19 was actually increased dramatically, to ∼280% of wildtype activity (18, 19, 20, 21). Another A115V mutation supported CYP17A1 and CYP3A4 catalysis at ∼70 to 85% of normal, but decimated CYP1A2 and CYP2C19 catalysis (17, 19, 21). In general, activities can increase, decrease, or remain unchanged depending on the individual P450 enzyme (18). Further investigation of different reductase mutants confirmed the enzyme-specific effect and also revealed a substrate-dependent effect (20, 22). Not only are the catalytic activities of specific P450 enzymes differentially affected but the binding affinities and thermodynamic properties of the different reductase–P450 interactions also vary among P450 enzymes (23).
Altogether the above data suggest that the interaction between reductase and P450 is likely to be different for individual human P450 enzymes. Unfortunately, this interaction is rather poorly understood for any human P450 enzyme. Kinetic studies suggest that the FMN domain of reductase only transiently binds to P450 enzymes rather than forming a stable complex (24, 25). Perhaps because this interaction is necessarily transient, no structures are known of a mammalian P450–reductase complex. While a recent cryo-EM structure of the soluble, bacterial, natural P450/reductase fusion protein CYP102A1 was recently solved supporting the conformational changes of flavin domains relative to each other and the P450 enzyme (26), this protein functions as a homodimer and has limited relevance to membrane-bound mammalian P450 enzymes with their redox partner.
While there are no structures of mammalian P450 enzymes with reductase, there are now structures of individual human P450 enzymes (27) and of reductase (5), including open conformations where the FMN domain would be available to interact with a P450 enzyme (8). Thus attempts have been made to dock the two proteins together (28, 29, 30). These docking studies informed selective mutagenesis experiments that implicated the proximal side of the P450, particularly the C-helix, in interaction with the FMN domain of reductase (30, 31, 32). This is further supported by biophysical techniques such as chemical cross-linking and hydrogen–deuterium exchange (33, 34, 35). Lastly, evidence suggests that these P450–reductase interactions are largely mediated by electrostatic interactions (31, 32, 36) and/or hydrophobics (37, 38).
Similar mitochondrial P450 systems have been studied recently and provide evidence of redox partners affecting P450 function. Mitochondrial P450 enzymes instead use soluble adrenodoxin and membrane-bound adrenodoxin reductase for electron delivery, with adrenodoxin acting as an electron shuttle between the P450 and adrenodoxin reductase, functionally similar to the reductase FMN domain. For these mitochondrial P450 enzymes, evidence suggests that adrenodoxin aids not only in this electron delivery but also in allosteric modulation of these P450 enzymes. Adrenodoxin binding has been shown to modulate ligand binding to CYP11B1, CYP11B2, and CYP24A1 and catalytic turnover of both CYP11B enzymes (39, 40, 41). Furthermore, these results are supported by studies that suggest adrenodoxin binding modulates positioning of substrate in the CYP11A1 active site required for catalysis (42). In contrast, adrenodoxin binding does not appear to have an allosteric effect on CYP27C1 (43). Taken together, these results suggest that the adrenodoxin redox partner not only can allosterically modulate P450 activity but can also do so in an isoform-specific manner. From this, one could imagine that microsomal P450 enzymes may also experience allosteric modulation from their redox partner protein NADPH cytochrome P450 reductase.
Herein the aim is to investigate the effects of the reductase FMN domain on the function of individual microsomal cytochrome P450 enzymes. Because the reductase FMN domain shuttles between its own FAD domain and the P450, and the reductase–P450 interaction is transient, the strategy pursued herein is to promote the FMN domain–P450 interaction by generating artificial fusion proteins containing individual P450 enzymes linked to the FMN-containing domain of reductase. Such fusion enzymes were generated for three human drug-metabolizing P450 enzymes, CYP2A6, CYP2D6, and CYP3A4, and for two human steroidogenic P450 enzymes, CYP17A1 and CYP21A2. Comparison with the corresponding individual P450 enzymes revealed various effects of the reductase FMN domain on ligand binding and catalysis. These experiments were performed without reduction of the FMN domain so that allosteric modulation of the P450 function could be distinguished from the effects of electron transfer.
Results
Rationale, generation, and characterization of recombinant fusion P450 enzymes
There exist natural fusions of reductase with its redox target, such as in the case of nitric oxide synthase (44) and the bacterial CYP102/BM3 (45). There are also multiple examples of artificial fusions of various mammalian catalytically active P450 enzymes with rat reductase (46, 47, 48, 49). Herein stabilization of the interaction of the human reductase electron-delivering FMN domain with human P450 enzymes was used to facilitate studying their interactions. To (1) avoid competition between the FMN domain interacting with either the P450 or with its own FAD domain and to (2) sequester the P450 in close local proximity of the reductase FMN domain, P450 enzymes and the reductase FMN domain were designed as a single polypeptide with an intervening linker for flexibility. The linker was designed using analysis of naturally occurring linkers (50), a structural analysis of the distance required to allow the N-terminal FMN domain to reach the proximal P450 surface, and successful design of such linkers for studying P450–adrenodoxin interactions (39). A seven-amino-acid linker AKKTSS has successfully been used to create a similar artificial fusion protein consisting of the N-terminal redox partner adrenodoxin and C-terminal CYP11B2 to generate a crystallographic structure of this protein–protein complex, although the linker is disordered in the electron density (40). Finally, the linker employed herein was also used to artificially link adrenodoxin and CYP11B2, and this construct also demonstrated strong allosteric effects of adrenodoxin on CYP11B2 ligand interactions (39).
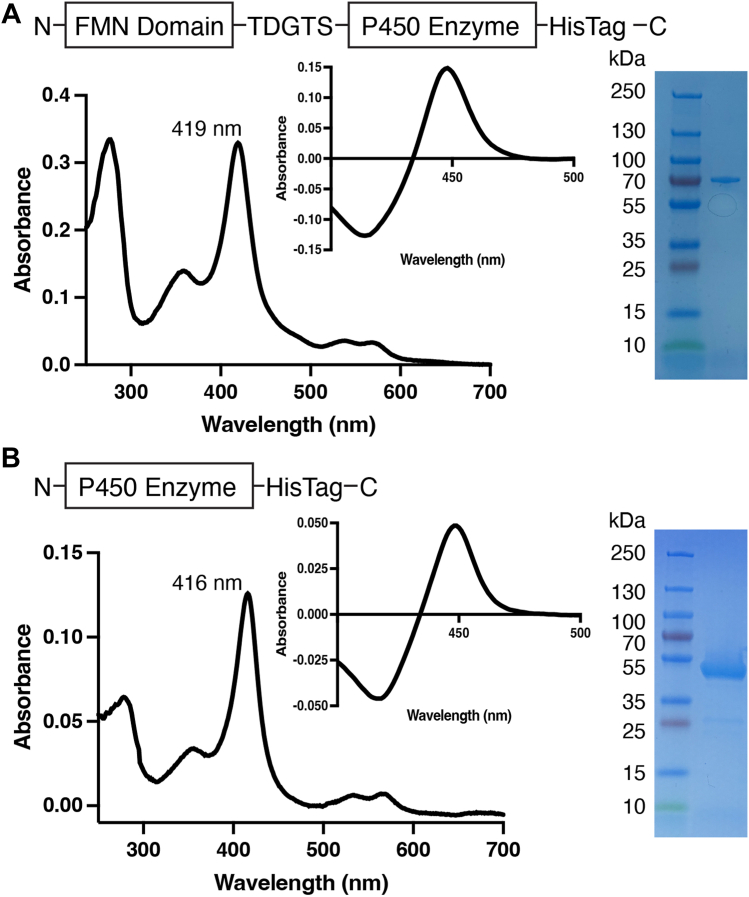
The fusion enzymes used in these studies consist of the human NADPH-cytochrome P450 reductase FMN-containing domain (FMND) at the N terminus, followed by a short five-amino-acid linker (TDGTS), then the P450 catalytic domain, and a final C-terminal His-tag (Figs. 1A, S1 and S2). Herein these artificial fusion proteins are denoted as FMND/P450—for example, FMND/CYP2A6 for the CYP2A6 fusion. These fusion proteins were expressed in Escherichia coli and highly purified, and the integrity of the fusion proteins was assessed by SDS-PAGE, with all five yielding the expected 75 to 77 kDa molecular weight and only minor contaminants in some cases (e.g., Figs. 1A and S1). The folding of the reductase FMN domain and flavin incorporation is difficult to assess spectroscopically. While the extinction coefficient for FMN is much lower and absorbance is overwhelmed by the absorbance of the P450 heme, the reductase FMN domain has successfully been expressed and purified independently and is quite stable (51). The integrity of the P450 domain was analyzed by UV-visible absorbance, as well as the reduced carbon monoxide difference spectra. Each fusion displayed a typical Soret peak with maximal absorbance at ∼418 nm (e.g., Fig. 1A), similar to the isolated P450 enzymes (e.g., Fig. 1B) and consistent with isolation of the water-bound heme iron. The reduced carbon monoxide difference spectra for both isolated P450 enzymes and the FMND/P450 fusion proteins primarily had maximal absorbance at 450 nm, consistent with proper incorporation of the heme prosthetic group (e.g., Fig. 1, A and B insets), although steroidogenic proteins had some P420 species present, particularly for the FMND/CYP17A1 fusion. Spectra and gels for all proteins are shown in Fig. S3. In summary, all fusion and nonfusion P450 enzymes appear to be pure and to have correctly incorporated heme.
Figure 1.
Generation and characterization ofrepresentative FMND/P450 fusion enzyme compared withthe corresponding isolated P450 enzyme.A, fusion enzymes consisted of the reductase FMN domain plus a five-amino-acid linker plus the catalytic P450 domain with a histidine tag (top). This resulted in purified protein with a water-bound Soret peak in the absolute spectrum (main image) and a typical reduced-carbon monoxide difference spectrum (inset). The purified protein runs on an SDS-PAGE gel at the expected molecular weight of 75 to 77 kDa (right). B, the individual P450 enzymes that the fusions were compared with consisted of only the catalytic P450 domain with a histidine tag (top). This resulted in purified protein with a water-bound Soret peak in the absolute spectrum (main image) and a typical reduced-carbon monoxide difference spectrum (inset). The purified protein runs on an SDS-PAGE gel at the expected molecular weight of 54 to 56 kDa (right). The specific examples shown are for (A) FMND/CYP2A6 and (B) CYP2A6. Fig. S3 shows these data for all proteins, including repetition of FMND/CYP2A6 and CYP2A6 to facilitate comparisons.
Effects of reductase FMN domain on ligand binding mode
Each protein pair consisting of an isolated P450 protein and the corresponding FMND/P450 fusion protein was evaluated for the ability to bind both a native substrate and an inhibitor. Ligands were chosen that are known to be effective substrates or inhibitors for the respective P450 enzyme and have low background absorbance to enable assessment of binding using a UV-visible binding assay. For the drug-metabolizing enzymes CYP3A4, CYP2D6, and CYP2A6, the substrates were hydrocortisone, thioridazine, and coumarin, while the inhibitors were clotrimazole, prinomastat, and pilocarpine, respectively. For the steroidogenic CYP17A1 and CYP21A2 all major native steroid substrates were evaluated (progesterone, pregnenolone and their 17α-hydroxy versions for CYP17A1 and progesterone and 17 α-hydroxyprogesterone for CYP21A2), along with the shared inhibitor ketoconazole.
Cytochrome P450 ligand binding at the catalytic heme can be monitored through shifts in the maximal absorbance of the heme Soret peak, which is usually best observed as difference spectra. While some compounds can bind to P450 enzymes without causing a spectral shift, particularly if they bind far from the active site, the presence of a spectral shift upon addition of a small molecule is generally consistent with its binding in the active site.
P450 enzyme substrates typically bind and displace the heme-coordinated water molecule, resulting in a Soret peak shift to a shorter wavelength, resulting in a type I difference spectra (52) with a peak at ∼386 to 390 nm and trough at ∼419 to 424 nm. The drug-metabolizing CYP3A4, CYP2D6, and CYP2A6 bound their respective hydrocortisone, thioridazine, and coumarin substrates, and all demonstrated this expected type I spectral shift, as did the corresponding fusion proteins (e.g., Figs. 2A and S4). The same type I shift was observed for steroidogenic CYP21A2 binding its substrates progesterone and 17α-hydroxyprogesterone and for CYP17A1 binding its substrates progesterone, 17α-hydroxyprogesterone, pregnenolone, and 17α-hydroxypregnenolone, whether or not the FMN domain was fused (Fig. S5).
Figure 2.
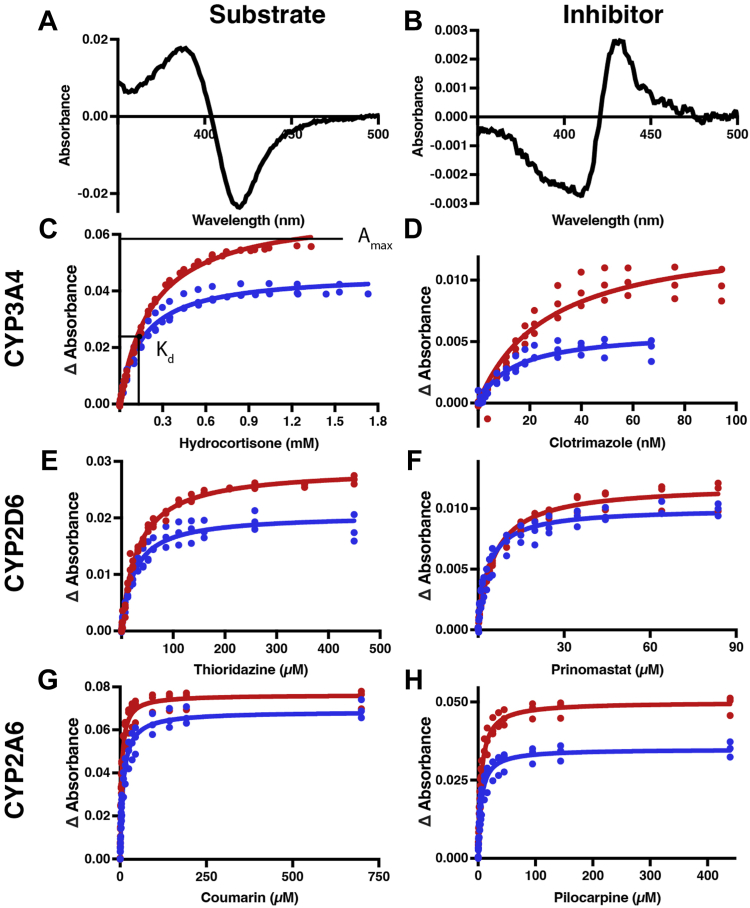
Binding of substrates and inhibitors to drug-metabolizing P450 enzymes (red) compared with the corresponding FMND/P450 fusion proteins (blue). Representative difference spectra for titrations of FMND/CYP3A4 binding its substrate hydrocortisone with a type I binding mode (A) and inhibitor clotrimazole with a type II binding mode (B). C–H, plotting the changes in absorbance (peak–trough) from such difference spectra versus the corresponding ligand concentration and fitting to a single-site binding equation allows comparison of the maximal absorbance (ΔAmax) and dissociation constant (Kd) for the isolated P450 (red) and fusion (blue) proteins, as visualized in C. Ligand binding titrations are shown for CYP3A4 substrate hydrocortisone (C) and inhibitor clotrimazole (D), for CYP2D6 substrate thioridazine (E) and inhibitor prinomastat (F), and for CYP2A6 substrate coumarin (G) and inhibitor pilocarpine (H). Each titration was completed in triplicate on different days with all data points shown. Fitted values and 95% confidence intervals are shown in Table 1.
Inhibitors that bind in the P450 active site and directly coordinate to the heme iron result in a shift of the Soret peak to longer wavelengths. Such type II (52) difference spectra typically have a peak at ∼427 to 435 nm and trough at ∼407 to 414 nm and are common for P450 inhibitors with a lone pair-containing nitrogen, such as in the azole- or pyridine-containing inhibitors used herein. The drug-metabolizing CYP3A4, CYP2D6, and CYP2A6 bound their respective clotrimazole, prinomastat, and pilocarpine inhibitors with the expected type II shifts, as did the corresponding FMND/P450 fusion enzymes (e.g., Figs. 2B and S4). Whether or not the FMN domain was fused, the same type II shifts were also observed for steroidogenic CYP17A1 and CYP21A2 binding the common inhibitor ketoconazole (Fig. S5). Thus, the presence of the FMN domain did not change the binding mode for any of these substrates or inhibitors.
Effects of reductase FMN domain on ligand binding by drug-metabolizing P450 enzymes
Both type I and type II spectral shifts can be readily quantitated by measuring the difference between the peak and trough (Δ absorbance) in the difference spectra (Fig. S4). Plotting these absorbance changes throughout titration with a ligand (e.g., Fig. 2C) allows the determination of both maximal saturation (reported as ΔAmax) and the ligand affinity or dissociation constant (Kd) (e.g., Fig. 2C and Table 1). The ratio of ΔAmax/Kd is a measure of binding efficiency analogous to kcat/Km for catalysis. Experiments for fusion and nonfusion forms of P450 enzymes were performed identically; thus, the resulting differences in Kd or ΔAmax are logically due to the presence of the FMN domain in the fusion protein. Herein all titrations were performed with the FMN domain in the oxidized state because the goal was to determine the effects of protein–protein interactions separate from any effects due to electron transfer. The effects of FMN domain fusion were variable across the different drug-metabolizing enzymes (Fig. 2, C–H and Table 1). Almost all of the isolated P450 enzymes and their corresponding fusion enzymes have binding parameters with nonoverlapping 95% confidence intervals. This is consistent with statistically significant effects. For the major drug-metabolizing cytochrome P450 3A4, fusion of the FMND reduced total binding of its hydrocortisone substrate by 33% but simultaneously decreased the Kd by a similar amount (30%). As a result the binding efficiency (ΔAmax/Kd) for this substrate remained essentially unchanged. For the inhibitor clotrimazole the FMND/CYP3A4 fusion results in a 58% reduction in final saturation and a 49% reduction in the Kd value compared with CYP3A4. As a result the binding efficiency for this inhibitor similarly remained little changed.
Table 1.
Ligand binding data for drug-metabolizing P450 enzymes ± fusion with the reductase FMN-containing domain
 |
The 95% confidence intervals are provided in gray.
a Substrates for CYP3A4, CYP2D6, and CYP2A6 were hydrocortisone, thioridazine, and coumarin, respectively. Kd units are μM except for hydrocortisone, which is mM.
b Inhibitors for CYP3A4, CYP2D6, and CYP2A6 were clotrimazole, prinomastat, and pilocarpine, respectively. Kd units are μM except for clotrimazole, which is nM.
For CYP2D6 (Table 1), fusion of the FMN domain had similar effects on binding of the substrate thioridazine: a 28% reduction in total binding at saturation but a 22% increase in affinity such that the binding efficiency was quite similar to the isolated CYP2D6. However, FMND/CYP2D6 binding of the inhibitor pilocarpine had a 17% reduction in total binding but caused a larger 37% increase in the affinity compared with CYP2D6 alone. As a result, the ΔAmax/Kd binding efficiency was increased 32% by fusion of the FMN domain. Thus for CYP3A4 and CYP2D6, the presence of the reductase FMN domain typically decreased total ligand binding but increased affinities for both substrates and inhibitors.
Distinct results were observed for the drug-metabolizing CYP2A6 enzyme. Fusion of the FMN domain resulted in relatively little change in total ligand binding for its substrate coumarin but a more than 2-fold increase in the Kd value. Evaluation of inhibitor pilocarpine binding conversely demonstrated no change in the affinity and a 30% decrease in total binding. Thus for CYP2A6 the effects of the FMN domain on ligand binding appear to be different from those for CYP3A4 and CYP2D6 and more distinct for these two ligands.
Effects of reductase FMN domain on ligand binding by steroidogenic P450 enzymes
Having observed the diversity of effects of the FMN domain on the quite promiscuous drug-metabolizing P450 enzymes, the question arose what the effects might be on more substrate-selective cytochrome P450 enzymes. Human P450 enzymes CYP17A1 and CYP21A2 are much more selective for steroidal substrates. CYP17A1 has four major, very structurally similar substrates, and CYP21A2 has two such substrates, all of which were examined alongside ketoconazole, a nonsteroidal inhibitor of both enzymes.
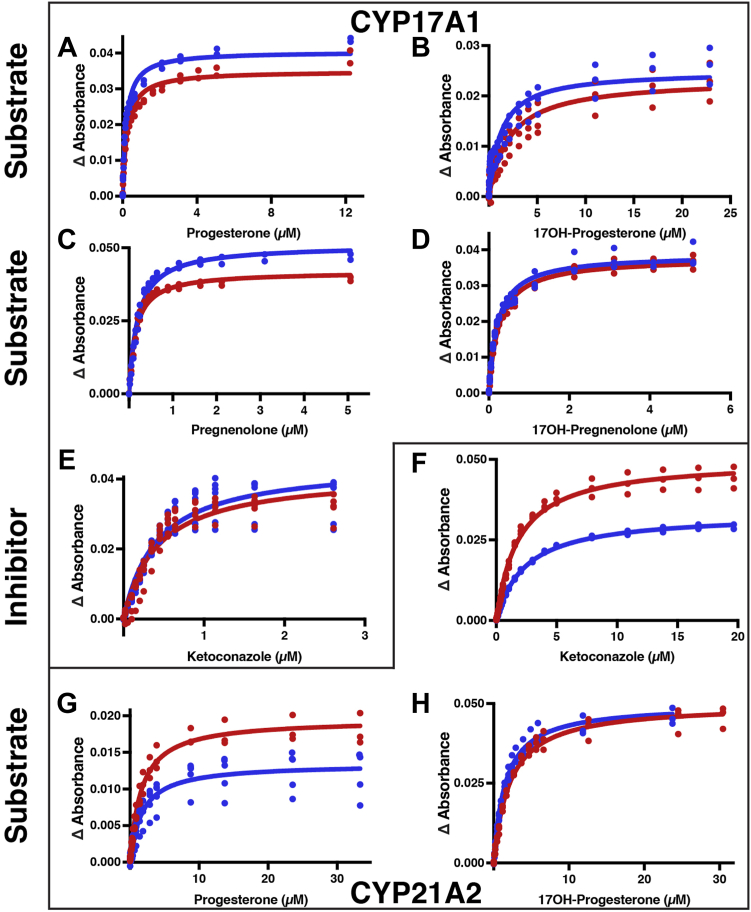
Titrations of the steroidogenic P450 enzymes CYP17A1 and CYP21A2 with their native substrates and with the type II inhibitor ketoconazole are shown in Fig. S5, with the corresponding binding curves shown in Figure 3, and the fitted values for total absorbance change (ΔAmax) and affinity (Kd) summarized in Table 2. A variety of effects are observed. Fusion of the FMN domain increased overall binding efficiency of CYP17A1 for progesterone (38%) and 17α-hydroxyprogesterone (86%). This occurred as a result of a 15% increase in CYP17A1 total saturation by progesterone and a 17% increase in affinity, whereas there was a smaller increase in saturation by 17α-hydroxyprogesterone but a 43% increase in affinity (Fig. 3, A and B and Table 2). Both parameters were increased for pregnenolone (Fig. 3C and Table 2) such that the binding efficiency for pregnenolone was largely unchanged. Fusion of the FMN domain to CYP17A1 had little effect on saturation by 17α-hydroxypregnenolone but a 12% decrease in Kd (Fig. 3D) such that the overall binding efficiency was only 16% higher (Table 2). However, no significant changes in either saturation or affinity were observed for inhibitor ketoconazole (Fig. 3E and Table 2).
Figure 3.
Binding of substrates and inhibitors to the steroidogenic CYP17A1 and CYP21A2 enzymes (red) compared with the corresponding fusion proteins (blue). The changes in difference spectra absorbance (peak–trough) versus ligand concentration over the course of a titration are depicted for the individual P450 enzyme (red) compared with the corresponding FMND/CYP fusion (blue) proteins. Ligand binding titrations are shown for CYP17A1 binding its major native substrates progesterone (A), 17α-hydroxyprogesterone (B), pregnenolone (C), and 17α-hydroxy pregnenolone (D), as well as the inhibitor ketoconazole (E). Titrations are also shown for the CYP21A2 inhibitor ketoconazole (F) and its major native substrates progesterone (G) and 17α-hydroxyprogesterone (H). Each titration was completed in duplicate or more with all data points shown. Fitted values and 95% confidence intervals are shown in Table 2.
Table 2.
Ligand binding data for steroidogenic P450 enzymes ± fusion with the reductase FMN-containing domain
 |
The 95% confidence intervals are provided in gray.
a Kd units are μM.
Fusion of the FMN domain to CYP21A2 similarly had clear ligand-specific results. Fusion resulted in a 31% decrease in total absorbance change due to progesterone binding without substantially altering the affinity, while for the 17α-hydroxyprogesterone substrate, the opposite was true. The Kd value is decreased by ∼27% while the total absorbance change was unmodified. Fusion of the FMN domain has effects on both parameters for binding of the ketoconazole inhibitor. For this ligand ∼30% decreases are observed for both the maximal absorbance and the affinity, yielding an overall almost 50% reduction in binding efficiency.
Thus fusion of the reductase FMN domain had very distinct effects, even when the substrates are very similar (e.g., CYP21A2 with progesterone and 17α-hydroxyprogesterone) and for the same ligand with different P450 isoforms (e.g., ketoconazole binding to CYP17A1 versus CYP21A2).
P450 metabolism using artificial reductant cumene hydroperoxide
The cytochrome P450 catalytic cycle is initiated with substrate entering the P450 active site, typically displacing water from the heme iron, which in turn increases its redox potential favoring acceptance of an electron from cytochrome P450 reductase. Molecular oxygen can then bind, followed by the transfer of the second electron to form a peroxoanion intermediate. Initial protonation forms a hydroperoxide intermediate while a second protonation results in the cleavage of the oxygen–oxygen bond to form the catalytic oxyferryl intermediate.
The artificial fusion enzymes used in the current experiments lack the full-length reductase enzyme, having only the reductase FMN domain fused to the N-terminal to the P450 enzyme. As the FAD domain is required to bind NADPH and donate electrons to the FMN domain for P450 delivery, these artificial fusion enzymes are not catalytically active on their own with NADPH. While addition of free FAD domain can allow for delivery of electrons to the FMN domain and ultimately to the P450 (5, 8, 51), the FAD domain would also compete with the P450 for binding of the reductase FMN domain. Thus the strategy employed herein uses cumene hydroperoxide to drive metabolism. This artificial reductant uses the “peroxide shunt” to directly form the hydroperoxo intermediate, which subsequently only requires a single protonation event to yield the catalytic oxyferryl active species. This approach is commonly used to investigate P450 mechanism and P450–redox partner interactions (53, 54, 55, 56). Although P450 catalysis supported by artificial peroxide reductants is often substantially slower (57, 58), this provides a method to investigate the effect of the FMN domain on steps of the P450 catalytic cycle subsequent to substrate binding that does not relate to electron transfer from the redox partner. Product formation was measured. Michaelis–Menten curves were determined for each cytochrome P450 in both fused and nonfused forms (Fig. 4), and the resulting Km and kcat values were determined (Table 3).
Figure 4.
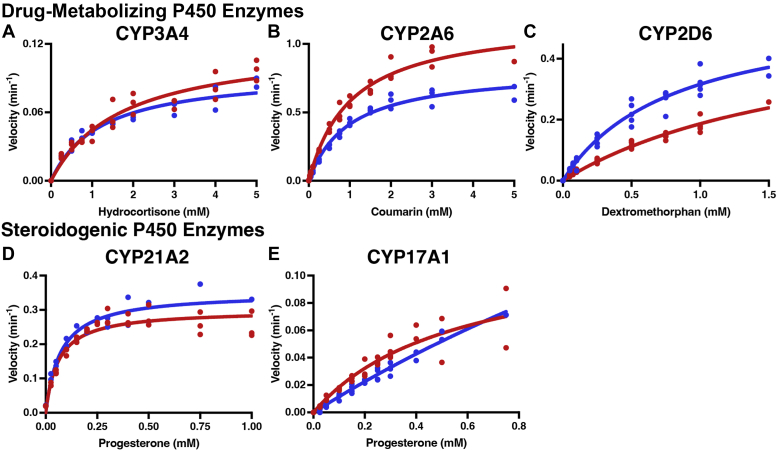
Substrate metabolism for drug-metabolizing (CYP3A4, CYP2D6, CYP2A6) and steroidogenic (CYP17A1 and CYP21A2) P450 enzymes (red) compared with the corresponding FMND/P450 fusion protein (blue). Metabolism of the respective substrates was supported by cumene hydroperoxide. The effects of FMN domain fusion are shown for CYP3A4 hydrocortisone metabolism to 6β-hydroxycortisol (A), CYP2A6 coumarin metabolism to 7-hydroxycoumarin (B), CYP2D6 metabolism of dextromethorphan to dextrorphan (C), CYP21A2 progesterone 21-hydroxylation (D), and CYP17A2 progesterone 17α-hydroxylation (E). Each reaction series was completed in duplicate or more on different days with all data points shown. Fitted values and 95% confidence intervals are shown in Table 3.
Table 3.
Steady-state kinetic parameters for P450 enzymes ± fusion with the reductase FMN-containing domain
 |
The CYP17A1 activity did not reach saturation, and thus the kcat and Km numbers are unreliable and not reported here.
The 95% confidence intervals are provided in gray.
a Km units are mM except for progesterone, which is μM.
Again a variety of results were observed across the drug-metabolizing and steroidogenic P450 enzymes. Addition of the FMN domain to CYP3A4 decreased kcat by ∼21% and Km by ∼30%, such that the overall kcat/Km efficiency was little changed. Conversely, fusion of the reductase FMN domain to CYP21A2 resulted in ∼17 to 19% increases in both kcat and Km, thus similarly leaving the catalytic efficiency unchanged. However, FMND/CYP2A6 fusion resulted in a 31% decrease in the kcat with little effect on Km, and for CYP2D6 a 58% decrease in Km was observed without a significant change in kcat. Thus for these latter two enzymes, fusion of the FMN domain decreased and increased catalytic efficiency, respectively, although the CYP2D6 kinetic parameters were less well defined. Unfortunately, progesterone 17α-hydroxylation by CYP17A1 was so poorly supported by cumene hydroperoxide that saturation could not be reached (Fig. 4E) and comparisons between the CYP17A1 and FMND/CYP17A1 are poorly defined, although it does appear that the trend was for the FMN domain to increase kcat as well as Km.
Discussion
Rationale for comparative evaluation
Artificial FMN domain/P450 fusion proteins were designed for three drug-metabolizing P450 enzymes, CYP3A4, CYP2D6, and CYP2A6, and for the steroidogenic P450 enzymes CYP17A1 and CYP21A2. While one cannot rule out nonspecific contributions of the short five-amino-acid linker or comment on what the effects of changes in linker length or composition might be, there are several relevant pieces of information that suggest the TDGTS linker is reasonable. This same linker has been previously used to artificially link mitochondrial CYP11B2 (C-terminal domain) with its native redox partner adrenodoxin (N-terminal domain), and this construct displayed allosteric effects on CYP11B2 ligand binding similar to titrations with high concentrations of free adrenodoxin. The linker certainly restrains the two domains in the same vicinity and likely therefore increases transient interactions and also has the potential to stabilize this interaction. A similar-length AKKTSS linker between adrenodoxin and CYP11A1 (59) and CYP11B2 (40) facilitated the determination of structures of these complexes with the adrenodoxin located as expected on the proximal face of these P450 enzymes.
Thus, the artificial FMND/P450 fusion enzymes were compared with the corresponding P450 enzyme to determine the effects of the FMN domain presence on ligand binding and catalysis. Since experiments for fusion and nonfusion forms were performed identically, differences in binding or catalytic parameters can be attributed to the presence of the FMN domain. Experiments were all performed with the FMN domain in its oxidized state to separate any effects of electron transfer from protein–protein effects in P450 enzyme function.
Ligand binding characterization of drug-metabolizing P450 enzymes
P450 enzymes and their corresponding versions fused with the reductase FMN domain were functionally characterized by their ability to bind ligands, both a canonical substrate and an inhibitor. Monitoring the amount of shift in the Soret peak is typically taken to reflect the amount of ligand binding in the active site immediately adjacent to the heme. Although there are small molecules that can bind in the active site and are metabolized that do not cause a spectral shift, substrates used herein cause spectral shifts, and the same types of spectral shifts, for both the isolated P450 enzymes and the corresponding FMND/P450 fusion protein. The resulting binding curves can be used to quantitate the binding affinity or dissociation constant (Kd) and overall change in absorbance at ligand saturation (ΔAmax).
The results herein revealed that the effects of reductase FMN domain fusion to P450 ligand binding affinity are highly variable with respect to both the P450 and the ligand. For almost all of the drug-metabolizing enzymes there are significant effects observed on both ΔAmax and Kd values for both substrates and inhibitors (Table 1). The only exceptions are the Kd for CYP2A6 binding an inhibitor, which is unchanged, and potentially the Kd for binding of the CYP2D6 substrate and CYP3A4 substrate and inhibitor, for which the 95% confidence intervals slightly overlap for the fused and isolated proteins. However, the trend does appear to be that these Kd values, as well as the Kd value for CYP2D6 with inhibitor, generally decrease due to fusion of the FMN domain. Thus the presence of the FMN domain increases affinity for these ligands. However, CYP2A6 demonstrates the opposite effect, with fusion of the FMN domain resulting in a 2-fold increase in the Kd value for substrate binding and no difference in Kd value for inhibitor binding. Thus for CYP2A6 fusion of the FMN domain has no effect on affinity for the inhibitor but decreases affinity for the substrate, illustrating the ligand dependence of this effect.
For the drug-metabolizing enzymes, the presence of the FMN domain consistently decreased the overall absorbance change/response for all ligands, from a 10% decrease in ΔAmax for CYP2A6 binding substrate to 58% decrease for CYP3A4 binding inhibitor (Table 1). While affinity is relatively self-explanatory, this magnitude of the absorbance change (ΔAmax) likely reflects changes in the population of P450 enzymes competent to bind ligand in the active site near or interacting with the heme iron to initiate a spectral change. In several cases this decreased response was offset by increased affinity (Kd) described above such that the overall binding efficiency (ΔAmax/Kd) was unchanged (CYP3A4 with both substrate and inhibitor and CYP2D6 with substrate). In other instances, the net effect was increased binding efficiency (CYP2D6 inhibitor binding) or decreased binding efficiency (CYP2A6 inhibitor and substrate binding).
Thus the ability of each of the major drug-metabolizing cytochrome P450 enzymes to bind ligands was substantially altered by the presence of the reductase FMN domain, generally decreasing the total population of enzyme binding ligand, but increasing, decreasing, or having no effect on the affinity for those ligands. The variability of this effect is quite remarkable.
Effect of the reductase FMN domain on steroidogenic cytochrome P450 ligand binding
While drug-metabolizing P450 enzymes can bind and metabolize many structurally diverse ligands, fewer ligands are compatible with the active sites of P450 enzymes that act in endogenous metabolic pathways such as steroidogenesis. This appears to be related to the active site lability, with evidence suggesting that drug-metabolizing P450 enzymes active sites are much more flexible than those of P450 enzymes acting on fewer, more structurally conserved substrates (27).
To determine if the P450- and ligand-dependent effects of FMN domain fusion carried over to more selective P450 enzymes, two steroidogenic cytochrome P450 enzymes were employed. CYP17A1 and CYP21A2 have in common two major substrates, progesterone and 17α-hydroxyprogesterone, while CYP17A1 has two additional closely related pregnenolone and 17α-hydroxypregnenolone substrates. Both are also inhibited by the type II ligand ketoconazole. Thus these enzymes permit a closer inspection of both ligand- and P450-dependent effects.
Overview of the effects of FMN domain fusion on the population of steroidogenic P450 proteins binding ligand (ΔAmax) yields some new information. Like the decreases observed in overall ligand binding for the drug-metabolizing enzymes (Fig. 2 and Table 1), this same trend was observed in several instances with steroidogenic P450 enzymes (30% decrease in CYP21A2 binding progesterone and inhibitor). However, FMN domain fusion had little effect on ΔAmax for others (CYP17A1 binding inhibitor, 17α-hydroxyprogesterone, and 17α-hydroxypregnenolone and CYP21A2 binding 17α-hydroxyprogesterone) and actually increased maximal ligand binding by 15 to 22% for some (CYP17A1 with progesterone and pregnenolone).
The effects of FMN domain fusion on ligand affinity for steroidogenic P450 enzymes are as diverse as those observed for drug-metabolizing P450 enzymes. FMN domain fusion had relatively little effect on the binding of some ligands; the confidence intervals overlapped for the CYP17A1 and FMND/CYP17A1 Kd values with all four substrates and the inhibitor ketoconazole (Table 2). However, for CYP21A2 addition of the FMN domain decreased the dissociation constant for 17α-hydroxyprogesterone by 27% but increased it by 31% for the inhibitor ketoconazole (Table 2). Thus, the FMN domain also has P450-specific and ligand-specific effects on steroidogenic cytochrome P450 enzymes.
Two comparisons are particularly notable in underscoring the differential effects of the FMN domain. First, comparison of the effects of FMN domain fusion on CYP21A2 binding of 17α-hydroxyprogesterone versus ketoconazole (Fig. 3H versus Fig. 3F) illustrates the ligand dependency of the FMN domain effect. Second, comparison of the effects on CYP17A1 and CYP21A2 with the common inhibitor ketoconazole are notable (Fig. 3E versus Fig. 3F). While essentially no effect is observed for either ΔAmax or Kd for CYP17A1, for CYP21A2 concomitant decreases in ΔAmax and increases in the dissociation constant Kd result in an overall ∼50% decrease in binding efficiency.
Effects of FMN domain fusion on substrate metabolism
In the present experiments the FMN domain is oxidized to separate effects of the protein–protein interaction from electron delivery and thus cannot reduce the P450 heme. As a result, an artificial reductant was employed to support catalysis. This artificial reductant was cumene hydroperoxide, which provides both the electrons and oxygen atoms directly to the cytochrome P450 heme active site (57, 58). Thus, as in the ligand binding experiments, the FMN domain is oxidized so the observed effects are expected to be related to the FMND–P450 interaction.
Comparison between fusion and nonfusion forms of these P450 enzymes again demonstrate variable effects on catalysis by different P450 enzymes. Fusion of the FMN domain results in Kcat values variably decreased 21 to 31% (CYP3A4 and CYP2A6), unchanged (CYP2D6), or increased by 17% (CYP21A2). Similarly, Km values are increased by 19% (CYP21A2), unaffected (CYP2A6), or decreased by 30 to 58% (CYP2D6 and CYP3A4). The combination of these effects is to increase catalytic efficiency (kcat/Km) as in the case of CYP2D6 and CYP3A4, decrease catalytic efficiency as occurs with CYP2A6, or compensate to yield unchanged efficiency as occurs with CYP21A2.
Conclusions
Fusion of the reductase FMN domain to a P450 enzyme could theoretically have three different impacts on P450 function. First, the FMN domain could extend freely from the N terminus of the P450, not associating with the P450 catalytic domain. In this instance, one would expect that both substrate binding and peroxide-supported catalysis would remain unchanged in comparison with the isolated P450 enzyme. The results herein clearly demonstrate that fusion of the reductase FMN domain to five different human cytochrome P450 enzymes causes substantial modifications to their ligand binding and catalytic capabilities, suggesting this is not likely the case. Second, the fused FMN domain could orient in such a way as to impede substrate binding or catalysis. Indeed for the drug-metabolizing P450 enzymes the FMN domain fusion consistently decreased maximal binding, but this idea is at odds with frequent observation of increased ligand binding affinity, increased overall binding for some ligands of the steroidogenic P450 enzymes evaluated, and substantially increased catalytic efficiency for CYP2D6. Third, localization of the FMN domain via fusion to the P450 could promote interaction between the FMN-bound surface of the FMN domain and the proximal cytochrome P450 surface. This is most consistent with the current results if the FMN domain binds to the proximal P450 surface and allosterically impacts substrate binding and metabolism quite apart from its ability to deliver electrons. This type of interaction has recently been observed when the redox partner adrenodoxin binds to this corresponding proximal surface of mitochondrial P450 enzymes (39, 40).
The variety of effects that the FMN domain has on different P450 enzymes and on the same P450 with different ligands suggests intricate and differential allosteric communication between the P450 active site and the proximal surface 15 to 20 Å away where the FMN domain binds. This concept is consistent with several other lines of evidence. First, the same reductase FMN domain interacts with and delivers electrons to support the catalysis of 48 different human microsomal cytochrome P450 enzymes, which are not identical on their proximal, FMN domain-binding surface, as well as heme oxygenase, squalene monooxygenase, and fatty acid desaturases. Second, some patient-identified reductase mutations have differential effects on the catalysis by individual cytochrome P450 enzymes (17, 18, 19, 20, 21). In addition, screening a mutant reductase library revealed different sets of mutations that promoted catalysis of CYP1A2, CYP2A6, or CYP3A4 while being neutral or detrimental to catalysis by the other P450 enzymes (60). Such studies have indicated that FMN domain mutants have substrate-specific effects on catalysis by CYP1A2 (22) and have even been reported to affect the regiospecificity of CYP1A2 caffeine metabolism (61). These FMN domain mutants are not thought to impact the structure of the FMN domain but are close to sites thought to be involved in P450 binding. Rather they have been suggested to differentially affect binding individual P450 enzymes in a substrate-specific way, further corroborating close communication between the P450 FMN domain-binding surface and the P450 active site and/or substrate entry.
While the importance of conformational changes essential to NADPH-cytochrome P450 function has been discussed previously, there is also substantial evidence that cytochrome P450 enzymes are also dynamic. Many P450 structures have been determined revealing a closed state with a buried, isolated active site, but obviously channels must open to permit substrate entry and product egress. Other structures have trapped some P450 enzymes with various openings to the surface, some of which are sufficient for the passage of small molecules. Thus conformational change is usually required for substrate binding. It is also possible that redox partner binding on the proximal side of the P450 could allosterically modulate P450 dynamics related to ligand access and egress. This idea is consistent with the studies herein and other reports. For example, when the redox partner protein cytochrome b5 is present, CYP17A1 conformational changes far from the binding site in areas associated with substrate binding are observed by solution NMR (62). NMR has also revealed substantial variations when the same cytochrome b5 protein interacts with different drug-metabolizing and steroidogenic human P450 enzymes (63). The same seems to be likely for the single reductase protein interacting with multiple human P450 enzymes and other enzymes. Another literature report indicates reductase perturbs CYP3A4 conformations as observed by hydrogen–deuterium exchange (34). Methods like NMR and hydrogen–deuterium exchange are advantageous to probe these protein–protein interactions because they require little or no modification of the system being used. However, other methods employing more extensive labeling and cross-linking are largely also consistent with reductase and cytochrome b5 binding on the proximal P450 surface, while silent on the impacts on P450 function.
For some time there has been an emerging understanding of the formation of protein–protein complexes (64). It is thought that generation of the complex occurs first through formation of an encounter complex consisting of two proteins generally oriented with the productive surfaces facing each other, often guided by electrostatic interactions (the 3D search) but with a variety of relative orientations and potentially still solvated. Encounter complexes can either resolve or evolve to a more specific complex as short-range forces “lock in” a specific complex (a 2D search). When it comes to redox proteins, it is thought that formation of the encounter complex can be enough for intermolecular electron tunneling to occur (65). Thus encounter complexes might be sufficient for the reductase FMN domain to support catalysis of the many P450 and other enzymes such as heme oxygenase (66) and could be evolutionarily favorable considering their diverse binding surfaces. However, it is less likely that formation of an encounter complex alone would result in the type of communication observed herein with the active site or the ligand-dependent effects. This suggests more specific interactions at the protein–protein interface, resulting in distinct conformational changes across different P450 enzymes and P450–ligand complexes.
Further understanding of the allosteric effects of reductase binding and definition of differential interactions between human P450 enzymes and the reductase FMN domain could potentially be used to design P450-specific small molecule disruptors as an orthogonal route to modulating specific P450 catalysis to treat disease or selectively impair xenobiotic metabolism.
Experimental procedures
Protein expression
CYP3A4
Both construct and expression of CYP3A4 catalytic domain omitting the N-terminal transmembrane helix were described (67) (Fig. S1).
CYP2D6
The construct yielding CYP2D6 catalytic domain was described (63) (Fig. S1). Expression was based on a previous method (68) with modifications. E. coli strain DH5α cells containing the pGro7 plasmid (Takara Bio) were transformed with the CYP2D6 expression plasmid. All media contained 100 μg/ml carbenicillin and 20 μg/ml chloramphenicol to select for the CYP2D6 and pGro7 plasmids, respectively. A single colony from the transformation was grown in 5 ml of lysogeny broth (LB) for 7 h at 37 °C with shaking at 250 rpm. Subsequently, 200 μl of this culture was used to inoculate 200 ml of LB, which was grown for 16 h at 37 °C with shaking at 250 rpm. Expression was performed using 1 l terrific broth (TB) in three 2.8-l Fernbach flasks without baffling. Each flask was inoculated with 10 ml of the starter culture and grown at 37 °C with shaking at 215 rpm until the absorbance (A600) reached 0.3, at which time 1 mM of heme precursor δ-aminolevulinic acid and 4 mg/ml of chaperone inducer L-arabinose were added to the cultures and the temperature was reduced to 28 °C. At A600 0.6 to 0.8, 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to initiate CYP2D6 expression, and shaking was reduced to 190 rpm. Cultures were allowed to grow for 24 h before harvesting and freezing the cell pellets at −80 °C until use.
CYP2A6
The CYP2A6 catalytic domain construct was described (69, 70) (Fig. S1), and expression and purification protocols were based on a method previously described (69) with modifications. E. coli strain DH5α cells containing the pGro7 plasmid (Takara Bio) were transformed with the CYP2A6 expression plasmid. All media contained 100 μg/ml carbenicillin and 20 μg/ml chloramphenicol to select for the CYP2A6 and pGro7 plasmids, respectively. A single colony from the transformation was grown in 5 ml of LB for 7 h at 37 °C with shaking at 250 rpm. Subsequently, 200 μl of this culture was used to inoculate 200 ml of LB, which was grown for 16 h at 37 °C with shaking at 250 rpm. Expression was performed using 14 1-l flasks with 250 ml of TB containing 2 mg/ml of chaperone inducer L-arabinose. Each flask was inoculated with 15 ml of the starter culture and grown at 37 °C with shaking at 200 rpm. At A600 of 0.6 to 0.8, 1 mM of heme precursor δ-aminolevulinic acid and 1 mM IPTG were added to initiate CYP2A6 expression. Temperature was reduced to 25 °C, and shaking was reduced to 190 rpm. Cultures were allowed to grow for 72 h before harvesting and freezing the cell pellets at −80 °C until use.
CYP17A1
Construct (71) and expression of the catalytic domain were described (72) (Fig. S1).
CYP21A2
Construct (73) and expression of the catalytic domain were described (72) (Fig. S1).
Fusion proteins
Constructs of fusion proteins were generated and inserted into the pCWori+ plasmid in between the Ndel and HindIII restriction sites (GenScript). All constructs yielded a fusion protein with an N-terminal human FMN domain (residues 62–244) followed by a five-amino-acid linker region corresponding to the amino acids TDGTS followed by a C-terminal human P450 identical to the constructs mentioned previously for the nonfusion P450 enzymes and including a C-terminal histidine tag (Fig. S1).
Plasmids for fusion proteins were transformed into DH5α E. coli and were selected for using 100 μg/ml carbenicillin in all media. Single colonies from transformation were inoculated into 50 ml LB and grown for 6 h 37 °C with shaking at 250 rpm. This culture was subsequently used to inoculate 200 ml LB and then grown 16 h at 37 °C with shaking at 250 rpm. Alternatively, single colonies were directly inoculated into the 200-ml culture. Expression was performed using 1 l TB in three 2.8-l Fernbach flasks without baffling. Each flask was inoculated with 10 ml of the starter culture and grown at 37 °C with shaking at 220 rpm until the A600 reached 0.3, at which time 1 mM of heme precursor δ-aminolevulinic acid was added. At A600 of 0.6 to 0.9, 1 mM IPTG was added to initiate expression, and temperature and shaking were reduced to 26 °C and 190 rpm, respectively. Cultures were allowed to grow for 48 to 72 h before harvesting and freezing the cell pellets at −80 °C until use.
Protein purification
CYP3A4
CYP3A4 was purified as described (63).
CYP2D6
CYP2D6 was purified as described (63).
CYP2A6
Harvested cells were thawed and spheroplasts were isolated as described (69, 74), but with slight modifications to omit 2-mercaptoethanol and increase the lysozyme concentration to 0.2 mg/ml. The cell pellet was then resuspended in buffer (500 mM potassium phosphate pH 7.4, 300 mM sodium chloride, 20% glycerol, 1 mM phenylmethylsulfonyl fluoride) corresponding to 10% of culture volume using a Dounce homogenizer. Cells were lysed using a French Press at 10,000 psi or by sonicating (Fisher Ultrasonic dismembrator D100, Horn: Fisher 15-338-269) six times on ice at 30-s intervals at 10% amplitude with rest on ice in between pulses. The sample was then incubated with 4.8 mM Cymal-5 detergent (Anatrace C325S) for 1 h at 4 °C to extract CYP2A6 from bacterial membranes. The sample was clarified by 1-h ultracentrifugation at ∼119,000g at 4 °C. The supernatant was loaded onto a 30-ml nickel-nitrilotriacetic acid (Ni-NTA) Agarose (Qiagen) column preequilibrated with buffer (100 mM potassium phosphate pH 7.4, 200 mM sodium chloride, 20% glycerol, 4.8 mM Cymal-5). The column was washed with 6 column volumes (CVs) of buffer, followed by 6 CVs of the same buffer containing 8 mM histidine. The protein was washed further with a linear gradient elution over 4 CVs to elution buffer (10 mM potassium phosphate pH 7.4, 100 mM sodium chloride, 20% glycerol, 4.8 mM Cymal-5, 2 mM EDTA, 80 mM histidine) and then eluted with additional 2 CVs of elution buffer.
Fractions to pool for further purification were selected based on their absorbance at 418 nm (heme). This was then diluted 5-fold into ion exchange buffer (5 mM potassium phosphate pH 7.4, 20% glycerol, 1 mM EDTA) with 4.8 mM Cymal-5. This was loaded onto two preequilibrated 5-ml carboxymethyl columns (Cytiva) connected in series and washed with 10 CVs of ion exchange buffer. This was further washed with a 5-CV linear gradient up to the ion exchange elution buffer (50 mM potassium phosphate pH 7.4, 500 mM sodium chloride, 20% glycerol, 1 mM EDTA) and was then further washed with elution buffer until complete protein elution. Fractions were again pooled based on heme absorbance, concentrated, and loaded onto a 200-ml Superdex 200 (Cytiva) size exclusion chromatography (SEC) column preequilibrated with ion exchange elution buffer.
CYP21A2
Protein was purified as described (72).
CYP17A1
Protein was purified as described (72).
FMND/CYP3A4
Harvested cells were thawed and resuspended in approximately 50 ml of buffer per liter of E. coli culture. The buffer contained 500 mM potassium phosphate pH 7.4, 500 mM sodium chloride, 20% glycerol, 0.67 mg/ml magnesium chloride, 0.67 mg/ml DNAase, 1.33 mg/ml lysozyme, and 200 μl HALT protease inhibitors. This resuspension was homogenized using a Dounce homogenizer prior to two passes of microfluidization (Avestin) at 10,000 to 15,000 psi. The sample was incubated with 14 mM 3-((3-cholamidopropyl) dimethylammonio)-1-propanesulfonate (CHAPS) and 0.3 mg/ml DNAase for 1 h at 4 °C to extract FMND/CYP3A4 from bacterial membranes. The sample was clarified by 1-h ultracentrifugation at ∼119,000g at 4 °C. The supernatant was loaded onto a 30-ml Ni-NTA Agarose (Qiagen) column preequilibrated with buffer (100 mM potassium phosphate pH 7.4, 500 mM sodium chloride, 20% glycerol, 14 mM CHAPS). The column was washed with 3 CVs of this buffer, followed by 5 CVs of the same buffer containing 10 mM histidine. The protein was eluted with a similar buffer containing high histidine concentrations (10 mM potassium phosphate pH 7.4, 200 mM sodium chloride, 80 mM histidine, 10 mM CHAPS, 20% glycerol).
Fractions to pool for further purification were selected based on their absorbance at 418 nm (heme). This was then diluted 5-fold into ion exchange buffer (5 mM potassium phosphate pH 7.4, 200 mM sodium chloride, 20% glycerol) containing 2 mM CHAPS. This was loaded onto a preequilibrated 5-ml quaternary ammonium (Q) column (Cytiva) and washed with 3 CVs of ion exchange buffer. The column was further washed with a 20-CV linear gradient to the ion exchange elution buffer (50 mM potassium phosphate pH 7.4, 500 mM sodium chloride, 20% glycerol) and was then further washed with elution buffer until complete protein elution. Fractions were again pooled based on heme absorbance, concentrated, and loaded onto an Superdex 200 increase (10/300) (Cytiva) SEC column preequilibrated with ion exchange elution buffer.
FMND/CYP2D6
Harvested cells were thawed and resuspended in approximately 80 ml of buffer per liter E. coli culture. The buffer contained 500 mM potassium phosphate pH 7.4, 500 mM sodium chloride, 20% glycerol, and two SigmaFast protease inhibitor tablets. This resuspension was homogenized using a Dounce homogenizer. The sample was then divided into 50-ml aliquots and sonicated (Fisher Ultrasonic dismembrator D100, Horn: Fisher 15-338-269) six times on ice at 30-s intervals at 10% amplitude with rest on ice in between pulses. Lysozyme was added to 0.3 mg/ml and samples were sonicated in the same way two more times. The lysate was incubated with 14 mM CHAPS and 0.3 mg/ml DNAase for 1 h at 4 °C to extract FMND/CYP2D6 from bacterial membranes. The sample was clarified by 1-h ultracentrifugation at ∼119,000g at 4 °C. The supernatant was loaded onto a 30-ml Ni-NTA Agarose (Qiagen) column preequilibrated with buffer (100 mM potassium phosphate pH 7.4, 500 mM sodium chloride, 20% glycerol, 14 mM CHAPS). The column was washed with 3 CVs of buffer, followed by a linear gradient over 5 CVs up to the same buffer containing 10 mM histidine. Then after a 2-CV hold at 10 mM histidine, the protein was eluted with the same buffer containing 80 mM histidine.
Fractions to pool for further purification were selected based on their absorbance at 418 nm (heme). This was then diluted 5-fold into ion exchange buffer (5 mM potassium phosphate pH 7.4, 100 mM sodium chloride, 20% glycerol) containing 2 mM CHAPS. This was loaded onto a preequilibrated 5-ml Q column (Cytiva) and washed with 5 CVs of ion exchange buffer. The column was further washed with a 20-CV linear gradient up to the ion exchange elution buffer (50 mM potassium phosphate pH 7.4, 500 mM sodium chloride, 20% glycerol) and was then further washed with elution buffer until complete protein elution. Fractions were again pooled based on heme absorbance and were concentrated and loaded onto an Superdex 200 increase (10/300) (Cytiva) SEC column preequilibrated with ion exchange elution buffer.
FMND/CYP2A6
Harvested cells were thawed and resuspended in 50 to 80 ml of buffer per liter E. coli culture. The buffer contained 500 mM potassium phosphate pH 7.4, 500 mM sodium chloride, 20% glycerol, two SigmaFast protease inhibitor tablets, and 0.3 mg/ml lysozyme. This resuspension was homogenized using a Dounce homogenizer. The sample was then lysed by microfluidization (Avestin) at 10,000 to 15,000 psi or by sonication. For sonication, the sample was divided into 50-ml aliquots and sonicated (Fisher Ultrasonic dismembrator D100, Horn: Fisher 15-338-269) six times on ice at 30-s intervals at 10% amplitude with rest on ice in between pulses. The lysate was incubated with 4.8 mM Cymal-5 and 0.3 mg/ml DNAase for 1 h at 4 °C to extract protein from membranes. The sample was clarified by 1-h ultracentrifugation at ∼119,000g at 4 °C. The supernatant was loaded onto a 30-ml Ni-NTA Agarose (Qiagen) column preequilibrated with buffer (100 mM potassium phosphate pH 7.4, 500 mM sodium chloride, 20% glycerol, 4.8 mM Cymal-5). The column was washed with 3 CVs of buffer, followed by a linear gradient over 5 CVs up to the same buffer containing 20 mM histidine. Then after a 2-CV hold at 20 mM histidine, the protein was eluted with the same buffer containing 80 mM histidine.
Fractions to pool for further purification were selected based on their absorbance at 418 nm (heme). This was then diluted 5-fold into ion exchange buffer (5 mM potassium phosphate pH 7.4, 100 mM sodium chloride, 20% glycerol) containing 4.8 mM Cymal-5. This was loaded onto a preequilibrated 5-ml Q column (Cytiva) or two 5-ml diethylaminoethyl (Cytiva) columns connected in series and washed with 5 CVs of ion exchange buffer. This was further washed with a 20-CV linear gradient to the ion exchange elution buffer (50 mM potassium phosphate pH 7.4, 500 mM sodium chloride, 20% glycerol) and was then further washed with elution buffer until complete protein elution. Fractions were again pooled based on heme, concentrated, and loaded onto an Superdex 200 increase (10/300) (Cytiva) SEC column preequilibrated with ion exchange elution buffer.
FMND/CYP17A1
Harvested cells were thawed and resuspended in 100 ml of buffer per liter E. coli culture. The buffer contained 500 mM potassium phosphate pH 7.4, 500 mM sodium chloride, 20% glycerol, and two SigmaFast protease inhibitor tablets. In some cases, 0.3 mg/ml lysozyme, and 0.1 mg/ml DNAase and magnesium chloride were added. The resuspension was homogenized using a Dounce homogenizer. The sample was then lysed using either sonication or microfluidization. For sonication, the sample was divided into 50-ml aliquots and sonicated (Fisher Ultrasonic dismembrator D100, Horn: Fisher 15-338-269) six times on ice at 30-s intervals at 10% amplitude with rest on ice in between pulses. For microfluidization, the sample was run through a microfluidizer (Avestin) for two passes at 10,000 to 15,000 psi. FMND/CYP17A1 was then extracted from bacterial membranes with a 1-h incubation with 1% Emulgen 913 (Desert Biologicals) at 4 °C, and the sample was clarified by 1-h ultracentrifugation at ∼119,000g at 4 °C. The supernatant was loaded onto a 30-ml Ni-NTA Agarose (Qiagen) column preequilibrated with buffer (100 mM potassium phosphate pH 7.4, 500 mM sodium chloride, 0.2% Emulgen 913, 20% glycerol). The column was washed with 2 CVs of buffer, followed by a 2-CV linear gradient wash to the same buffer containing 10 mM histidine. Then after a 4-CV hold at 10 mM histidine, the protein is eluted in a high histidine buffer (10 mM potassium phosphate pH 7.4, 200 mM sodium chloride, 0.2% Emulgen 913, 20% glycerol, 80 mM histidine).
Fractions to pool for further purification were selected based on their absorbance at 418 nm (heme). This solution was then diluted 5-fold into ion exchange buffer (5 mM potassium phosphate pH 7.4, 100 mM sodium chloride, 20% glycerol) containing 0.2% Emulgen 913. This was loaded onto a preequilibrated 5-ml Q column (Cytiva) and washed with 5 CVs of ion exchange buffer. This was further washed with a 20-CV linear gradient to 60% of the ion exchange elution buffer (50 mM potassium phosphate pH 7.4, 500 mM sodium chloride, 20% glycerol) and was then washed with 100% elution buffer until complete protein elution. Fractions were again pooled based on heme absorbance, concentrated, and loaded onto an Superdex 200 increase (10/300) (Cytiva) SEC column preequilibrated with ion exchange elution buffer.
FMND/CYP21A2
Harvested cells were thawed and resuspended in 50 to 100 ml of buffer per liter E. coli culture. The buffer contained 500 mM potassium phosphate pH 7.4, 500 mM sodium chloride, 20% glycerol, 0.3 mg/ml lysozyme, 0.3 mg/ml DNAase, 0.3 mg/ml magnesium chloride, and two SigmaFast protease inhibitor tablets. The resuspension was homogenized using a Dounce homogenizer prior to two passes of microfluidization (Avestin) at 10,000 to 15,000 psi. FMND/CYP21A2 was then extracted from bacterial membranes with a 1-h incubation with 1% Emulgen 913 (Desert Biologicals) at 4 °C, and the sample was clarified by 1-h ultracentrifugation at ∼119,000g at 4 °C. The supernatant was loaded onto a 30-ml Ni-NTA Agarose (Qiagen) column preequilibrated with buffer (100 mM potassium phosphate pH 7.4, 500 mM sodium chloride, 20% glycerol, 0.2% Emulgen 913). The column was then washed with 5 CVs of buffer, followed by a 5-CV linear gradient wash to the same buffer containing 10 mM histidine. Then after a 2- to 5-CV hold at 10 mM histidine, the protein was eluted in a high histidine buffer (10 mM potassium phosphate pH 7.4, 200 mM sodium chloride, 20% glycerol, 0.2% Emulgen 913, 80 mM histidine). Fractions to pool for further purification were selected based on their absorbance at 418 nm (heme). This was then diluted 5-fold into ion exchange buffer (5 mM potassium phosphate pH 7.4, 100 mM sodium chloride, 20% glycerol) containing 0.2% Emulgen 913. This was loaded onto a preequilibrated 5-ml Q column (Cytiva) or two 5-ml diethylaminoethyl columns and washed with 5 CVs of ion exchange buffer. The Q column (Cytiva) or diethylaminoethyl (Cytiva) columns were then washed with a 20-CV (Q column) or 40-CV (diethylaminoethyl column) linear gradient to the ion exchange elution buffer (50 mM potassium phosphate pH 7.4, 500 mM sodium chloride, 20% glycerol). The fractions were again pooled based on heme, concentrated, and loaded onto an Superdex 200 increase (10/300) (Cytiva) SEC column with ion exchange elution buffer.
P450 quality assessment
The quality and quantity of P450 proteins were analyzed by UV-visible spectrum, SDS-PAGE, and reduced carbon monoxide difference spectrum. Then proteins were flash frozen in liquid nitrogen in the SEC buffer and stored until use in assays.
Ligand binding assays
Ligand binding assays were performed to measure spectral absorbance changes resulting from ligand binding to P450 heme to determine the dissociation constant (Kd) and the maximal absorbance at saturation (Amax) for substrates and inhibitors. Assay conditions for each P450 were identical to those of its respective fusion FMND/P450 enzyme. Most experiments were performed by preparing 1 μM P450 in 100 mM potassium phosphate pH 7.4, 20% glycerol and transferring 1 ml of sample to each of two quartz cuvettes with path lengths of 1 cm. P450 concentration was quantitated based on Soret absorbance as described (75).
The UV-visible spectrophotometer was used to baseline the reference and sample cuvettes between 300 and 500 nm. Ligand stocks dissolved in solvent were added into the sample cuvette, while an equal volume of solvent was added into a reference cuvette. For most experiments, cuvettes were incubated for 8 min at room temperature and the difference spectra were collected over 300 to 500 nm. The difference between the apex wavelength for the peak and the trough of the difference spectrum was plotted against the ligand concentration, and these data were fitted to the one site specific binding equation in GraphPad Prism.
Some differences in experimental conditions occurred for specific P450/ligand assays. The stability of FMND/CYP3A4 required the addition of 100 mM sodium chloride in the buffer, and the tight binding of clotrimazole required a lower protein concentration of 0.05 μM, which dictated the use of 5-cm-pathlength quartz cuvettes with 5 ml of sample in each. The stability of FMND/CYP21A2 required the reduction of the incubation time to 1 min and the addition of 200 mM sodium chloride and 0.2% Emulgen 913 for binding assays with progesterone and ketoconazole.
Lastly, the stability of CYP2D6 with thioridazine also required the reduction of incubation time to 1 min. In addition, the spectral properties of thioridazine required the use of tandem cuvettes in order to control for the inherent absorbance on this ligand. The tandem cuvettes have two chambers of 1-cm pathlength each. In each cuvette, one chamber holds protein as described above and the other chamber holds buffer. In the sample cuvette, ligand is added to the chamber containing protein, while an equal volume of dimethyl sulfoxide is added to the chamber with buffer. In the reference cuvette, the ligand is instead added to the chamber without protein and dimethyl sulfoxide is added to the chamber with protein. The experiment is otherwise carried out as described previously.
Metabolism assays
General
Catalytic assays were performed to measure product production from each P450 and FMND/P450 from its respective substrate. Assay conditions for each P450 were identical to those of its respective fusion FMND/P450 enzyme. P450 was incubated in 300 μl of potassium phosphate buffer (CYP2D6, CYP3A4, and CYP17A1: 50 mM potassium phosphate pH 7.4, 50 mM sodium chloride; CYP2A6: 50 mM potassium phosphate pH 7.4, 50 mM sodium chloride, 1% glycerol; CYP21A2: 100 mM potassium phosphate pH 7.4, 20% glycerol). The amounts of P450 or fusion FMND/P450 were 200 pmol (CYP2A6 and CYP17A1) and 400 pmol (CYP2D6, CYP3A4, and CYP21A2). P450 and FMND/P450 were quantitated based on their reduced carbon monoxide difference spectra measuring absorbance at 450 nm with the extinction coefficient 91 mM−1 cm−1 (76).
Substrate was added with varied concentration ranges: (CYP2A6 coumarin, 0–3 mM; CYP2D6 dextromethorphan, 0–1.5 mM; CYP3A4 hydrocortisone, 0–5 mM; CYP17A1 progesterone, 0–0.75 mM; CYP21A2 progesterone 0–1 mM). Samples were preincubated at 37 °C for 3 min (CYP2A6, CYP3A4, CYP17A1, and CYP21A2) or 5 min (CYP2D6). Reactions were initiated by addition of an artificial reductant, cumene hydroperoxide (CYP2D6, 0.05 mM; CYP3A4, 0.1 mM; CYP2A6, 0.2 mM; CYP17A1 and CYP21A2, 1 mM) and proceeded for 40 min (CYP2A6 and CYP2D6) or 1 h (CYP3A4, CYP17A1, and CYP21A2). Reactions were then terminated by addition of 150 μl of 40% trichloroacetic acid. Internal standards appropriate for each reaction were spiked into the samples, and then samples were centrifuged at 5000g for 10 min prior to injection onto a Luna C18 column (5 μm, 250 × 4.60 mM, Phenomenex).
For all catalytic assays, analytes were quantitated using a calibration curve of authentic standards prepared in the same manner as the samples. For samples and standards, quantitation was performed using the ratio of product peak over the internal standard peak. Data were fit to the Michaelis–Menten equation using GraphPad Prism.
CYP2A6 coumarin metabolism assay
For CYP2A6 enzymes, the substrate coumarin was separated from its 7-hydroxycoumarin product and 4-hydroxycoumarin internal standard using 50% 20 mM potassium phosphate pH 2.8 and 50% methanol for 15 min at 37 °C. Analytes were detected by absorbance at 330 nm.
CYP2D6 dextromethorphan metabolism assay
For CYP2D6 enzymes, the substrate dextromethorphan was separated from its dextrorphan product and 10-ketodextromethorphan internal standard using aqueous phase (10 mM potassium phosphate pH 3.5) and organic phase (27.8% acetonitrile, 44.4% methanol, 27.8% water). The column was run at 37 °C using 30% organic phase for 6 min followed by a linear gradient over 14 min to 80% organic phase. The column was held at 80% organic phase for 4 min. Between samples, the column was washed with 100% organic phase for 6 min and then reequilibrated at 30% organic phase for 5 min. Analytes were detected by absorbance at 280 nm.
CYP3A4 hydrocortisone metabolism assay
For CYP3A4 enzymes, the substrate hydrocortisone was separated from its 6β-hydroxycortisol product from its estradiol internal standard using aqueous phase (water) and organic phase (acetonitrile). The column was run at 40 °C using 15% organic phase for 3 min, followed by a linear gradient over 22 min to 70% organic phase. Between samples, the column was washed with 100% organic phase for 5 min and then reequilibrated at 15% organic phase for 5 min. Analytes were detected by absorbance at 240 nm.
CYP17A1 and CYP21A2 progesterone metabolism assay
For CYP17A1 and CYP21A2 enzymes, 17α-hydroxyprogesterone served as the internal standard for 21-hydroxyprogesterone quantitation and 21-hydroxyprogesterone was used as the internal standard for 17α-hydroxyprogesterone quantitation. The substrate progesterone was separated from 17α-hydroxyprogesterone and 21-hydroxyprogesterone using aqueous phase (0.2% acetic acid in water) and organic phase (acetonitrile). The column was run at 40 °C using 60% organic phase for 6 min followed by a linear gradient over 9 min to 80% organic phase. The column was then held at 80% organic phase for 5 min. Between samples, the column was washed with 100% organic phase for 5 min and then reequilibrated at 15% organic phase for 5 min. Analytes were detected by absorbance at 240 nm.
Data availability
All of the data are included in this article or the supplemental information.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Lily (Heeseo) Kim and Hyun Gi Yun provided technical assistance in repeating assays.
Author contributions
S. D. B.-H. and E. E. S. conceptualization; S. D. B.-H. methodology; S. D. B.-H. validation; S. D. B.-H. formal analysis; S. D. B.-H. investigation; S. D. B.-H. resources; S. D. B.-H. and E. E. S. writing – original draft; S. D. B.-H. and E. E. S. writing – review & editing; S. D. B.-H. visualization; E. E. S. supervision; E. E. S. project administration; E. E. S. funding acquisition.
Funding and additional information
This work was supported by NIH T32 GM007767 (supporting S. D. B.-H.) and R37 GM076343 (to E. E. S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Reviewed by members of the JBC Editorial Board. Edited by Joseph Jez
Supporting information
References
- 1.Guengerich F.P. Cytochrome P450 and chemical toxicology. Chem. Res. Toxicol. 2008;21:70–83. doi: 10.1021/tx700079z. [DOI] [PubMed] [Google Scholar]
- 2.Zhao M., Ma J., Li M., Zhang Y., Jiang B., Zhao X., et al. Cytochrome P450 enzymes and drug metabolism in humans. Int. J. Mol. Sci. 2021;22:12808–12824. doi: 10.3390/ijms222312808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson K.M., Su D., Zhang D.L. Cytochrome P450. 2021. Characteristics of major drug metabolizing cytochrome P450 enzymes; pp. 27–54. [Google Scholar]
- 4.Iyanagi T., Xia C., Kim J.J. NADPH-cytochrome P450 oxidoreductase: prototypic member of the diflavin reductase family. Arch. Biochem. Biophys. 2012;528:72–89. doi: 10.1016/j.abb.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang M., Roberts D.L., Paschke R., Shea T.M., Masters B.S., Kim J.J. Three-dimensional structure of NADPH-cytochrome P450 reductase: prototype for FMN- and FAD-containing enzymes. Proc. Natl. Acad. Sci. U. S. A. 1997;94:8411–8416. doi: 10.1073/pnas.94.16.8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xia C., Hamdane D., Shen A.L., Choi V., Kasper C.B., Pearl N.M., et al. Conformational changes of NADPH-cytochrome P450 oxidoreductase are essential for catalysis and cofactor binding. J. Biol. Chem. 2011;286:16246–16260. doi: 10.1074/jbc.M111.230532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campelo D., Esteves F., Brito Palma B., Costa Gomes B., Rueff J., Lautier T., et al. Probing the role of the hinge segment of cytochrome P450 oxidoreductase in the interaction with cytochrome P450. Int. J. Mol. Sci. 2018;19:3914. doi: 10.3390/ijms19123914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamdane D., Xia C., Im S.C., Zhang H., Kim J.J., Waskell L. Structure and function of an NADPH-cytochrome P450 oxidoreductase in an open conformation capable of reducing cytochrome P450. J. Biol. Chem. 2009;284:11374–11384. doi: 10.1074/jbc.M807868200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis J., Gutierrez A., Barsukov I.L., Huang W.C., Grossmann J.G., Roberts G.C. Domain motion in cytochrome P450 reductase: conformational equilibria revealed by NMR and small-angle x-ray scattering. J. Biol. Chem. 2009;284:36628–36637. doi: 10.1074/jbc.M109.054304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poddevin F., Delobel B., Courreges P., Bayart M. Antley-Bixler syndrome: case report and review of the literature. Genet. Couns. 1995;6:241–246. [PubMed] [Google Scholar]
- 11.Antley R., Bixler D. Trapezoidocephaly, midfacial hypoplasia and cartilage abnormalities with multiple synostoses and skeletal fractures. Birth Defects Orig. Artic. Ser. 1975;11:397–401. [PubMed] [Google Scholar]
- 12.Chun K., Siegel-Bartelt J., Chitayat D., Phillips J., Ray P.N. FGFR2 mutation associated with clinical manifestations consistent with Antley-Bixler syndrome. Am. J. Med. Genet. 1998;77:219–224. doi: 10.1002/(sici)1096-8628(19980518)77:3<219::aid-ajmg6>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 13.Reardon W., Smith A., Honour J.W., Hindmarsh P., Das D., Rumsby G., et al. Evidence for digenic inheritance in some cases of Antley-Bixler syndrome? J. Med. Genet. 2000;37:26–32. doi: 10.1136/jmg.37.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arlt W., Walker E.A., Draper N., Ivison H.E., Ride J.P., Hammer F., et al. Congenital adrenal hyperplasia caused by mutant P450 oxidoreductase and human androgen synthesis: analytical study. Lancet. 2004;363:2128–2135. doi: 10.1016/S0140-6736(04)16503-3. [DOI] [PubMed] [Google Scholar]
- 15.Flück C.E., Tajima T., Pandey A.V., Arlt W., Okuhara K., Verge C.F., et al. Mutant P450 oxidoreductase causes disordered steroidogenesis with and without Antley-Bixler syndrome. Nat. Genet. 2004;36:228–230. doi: 10.1038/ng1300. [DOI] [PubMed] [Google Scholar]
- 16.Miller W.L., Agrawal V., Sandee D., Tee M.K., Huang N., Choi J.H., et al. Consequences of POR mutations and polymorphisms. Mol. Cell. Endocrinol. 2011;336:174–179. doi: 10.1016/j.mce.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang N., Pandey A.V., Agrawal V., Reardon W., Lapunzina P.D., Mowat D., et al. Diversity and function of mutations in p450 oxidoreductase in patients with Antley-Bixler syndrome and disordered steroidogenesis. Am. J. Hum. Genet. 2005;76:729–749. doi: 10.1086/429417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pandey A.V., Flück C.E. NADPH P450 oxidoreductase: structure, function, and pathology of diseases. Pharmacol. Ther. 2013;138:229–254. doi: 10.1016/j.pharmthera.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 19.Agrawal V., Huang N., Miller W.L. Pharmacogenetics of P450 oxidoreductase: effect of sequence variants on activities of CYP1A2 and CYP2C19. Pharmacogenet. Genomics. 2008;18:569–576. doi: 10.1097/FPC.0b013e32830054ac. [DOI] [PubMed] [Google Scholar]
- 20.Sandee D., Morrissey K., Agrawal V., Tam H.K., Kramer M.A., Tracy T.S., et al. Effects of genetic variants of human P450 oxidoreductase on catalysis by CYP2D6 in vitro. Pharmacogenet. Genomics. 2010;20:677–686. doi: 10.1097/FPC.0b013e32833f4f9b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flück C.E., Mullis P.E., Pandey A.V. Reduction in hepatic drug metabolizing CYP3A4 activities caused by P450 oxidoreductase mutations identified in patients with disordered steroid metabolism. Biochem. Biophys. Res. Commun. 2010;401:149–153. doi: 10.1016/j.bbrc.2010.09.035. [DOI] [PubMed] [Google Scholar]
- 22.Esteves F., Urban P., Rueff J., Truan G., Kranendonk M. Interaction modes of microsomal cytochrome P450s with its reductase and the role of substrate binding. Int. J. Mol. Sci. 2020;21:6669. doi: 10.3390/ijms21186669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yablokov E.O., Sushko T.A., Ershov P.V., Florinskaya A.V., Gnedenko O.V., Shkel T.V., et al. A large-scale comparative analysis of affinity, thermodynamics and functional characteristics of interactions of twelve cytochrome P450 isoforms and their redox partners. Biochimie. 2019;162:156–166. doi: 10.1016/j.biochi.2019.04.020. [DOI] [PubMed] [Google Scholar]
- 24.Miwa G.T., Lu A.Y. The association of cytochrome P-450 and NADPH-cytochrome P-450 reductase in phospholipid membranes. Arch. Biochem. Biophys. 1984;234:161–166. doi: 10.1016/0003-9861(84)90337-0. [DOI] [PubMed] [Google Scholar]
- 25.Taniguchi H., Imai Y., Iyanagi T., Sato R. Interaction between NADPH-cytochrome P-450 reductase and cytochrome P-450 in the membrane of phosphatidylcholine vesicles. Biochim. Biophys. Acta. 1979;550:341–356. doi: 10.1016/0005-2736(79)90220-7. [DOI] [PubMed] [Google Scholar]
- 26.Su M., Chakraborty S., Osawa Y., Zhang H. Cryo-EM reveals the architecture of the dimeric cytochrome P450 CYP102A1 enzyme and conformational changes required for redox partner recognition. J. Biol. Chem. 2020;295:1637–1645. doi: 10.1074/jbc.RA119.011305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scott E.E., Godamudunage M.P. In: Raven E., Ikeda-Saito M., editors. The Royal Society of Chemistry; Cambridge, UK: 2018. Structures of Human Cytochrome P450 Enzymes: Variations on a Theme in Dioxygen-dependent Heme Enzymes; pp. 249–273. Chapter 11. [Google Scholar]
- 28.Sündermann A., Oostenbrink C. Molecular dynamics simulations give insight into the conformational change, complex formation, and electron transfer pathway for cytochrome P450 reductase. Protein Sci. 2013;22:1183–1195. doi: 10.1002/pro.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allorge D., Bréant D., Harlow J., Chowdry J., Lo-Guidice J.M., Chevalier D., et al. Functional analysis of CYP2D6.31 variant: homology modeling suggests possible disruption of redox partner interaction by Arg440His substitution. Proteins. 2005;59:339–346. doi: 10.1002/prot.20399. [DOI] [PubMed] [Google Scholar]
- 30.Bridges A., Gruenke L., Chang Y.T., Vakser I.A., Loew G., Waskell L. Identification of the binding site on cytochrome P450 2B4 for cytochrome b5 and cytochrome P450 reductase. J. Biol. Chem. 1998;273:17036–17049. doi: 10.1074/jbc.273.27.17036. [DOI] [PubMed] [Google Scholar]
- 31.Jang H.H., Jamakhandi A.P., Sullivan S.Z., Yun C.H., Hollenberg P.F., Miller G.P. Beta sheet 2-alpha helix C loop of cytochrome P450 reductase serves as a docking site for redox partners. Biochim. Biophys. Acta. 2010;1804:1285–1293. doi: 10.1016/j.bbapap.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen A.L., Kasper C.B. Role of acidic residues in the interaction of NADPH-cytochrome P450 oxidoreductase with cytochrome P450 and cytochrome c. J. Biol. Chem. 1995;270:27475–27480. doi: 10.1074/jbc.270.46.27475. [DOI] [PubMed] [Google Scholar]
- 33.Bumpus N.N., Hollenberg P.F. Cross-linking of human cytochrome P450 2B6 to NADPH-cytochrome P450 reductase: Identification of a potential site of interaction. J. Inorg. Biochem. 2010;104:485–488. doi: 10.1016/j.jinorgbio.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ducharme J., Sevrioukova I.F., Thibodeaux C.J., Auclair K. Structural dynamics of cytochrome P450 3A4 in the presence of substrates and cytochrome P450 reductase. Biochemistry. 2021;60:2259–2271. doi: 10.1021/acs.biochem.1c00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nisimoto Y. Localization of cytochrome c-binding domain on NADPH-cytochrome P-450 reductase. J. Biol. Chem. 1986;261:14232–14239. [PubMed] [Google Scholar]
- 36.Nadler S.G., Strobel H.W. Role of electrostatic interactions in the reaction of NADPH-cytochrome P-450 reductase with cytochromes P-450. Arch. Biochem. Biophys. 1988;261:418–429. doi: 10.1016/0003-9861(88)90358-x. [DOI] [PubMed] [Google Scholar]
- 37.Kenaan C., Zhang H., Shea E.V., Hollenberg P.F. Uncovering the role of hydrophobic residues in cytochrome P450-cytochrome P450 reductase interactions. Biochemistry. 2011;50:3957–3967. doi: 10.1021/bi1020748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voznesensky A.I., Schenkman J.B. The cytochrome P450 2B4-NADPH cytochrome P450 reductase electron transfer complex is not formed by charge-pairing. J. Biol. Chem. 1992;267:14669–14676. [PubMed] [Google Scholar]
- 39.Loomis C.L., Brixius-Anderko S., Scott E.E. Redox partner adrenodoxin alters cytochrome P450 11B1 ligand binding and inhibition. J. Inorg. Biochem. 2022;235 doi: 10.1016/j.jinorgbio.2022.111934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brixius-Anderko S., Scott E.E. Structural and functional insights into aldosterone synthase interaction with its redox partner protein adrenodoxin. J. Biol. Chem. 2021;296 doi: 10.1016/j.jbc.2021.100794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hartfield K.A., Stout C.D., Annalora A.J. The novel purification and biochemical characterization of a reversible CYP24A1:adrenodoxin complex. J. Steroid Biochem. 2013;136:47–53. doi: 10.1016/j.jsbmb.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 42.Mast N., Annalora A.J., Lodowski D.T., Palczewski K., Stout C.D., Pikuleva I.A. Structural basis for three-step sequential catalysis by the cholesterol side chain cleavage enzyme CYP11A1. J. Biol. Chem. 2011;286:5607–5613. doi: 10.1074/jbc.M110.188433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Glass S.M., Webb S.N., Guengerich F.P. Binding of cytochrome P450 27C1, a retinoid desaturase, to its accessory protein adrenodoxin. Arch. Biochem. Biophys. 2021;714 doi: 10.1016/j.abb.2021.109076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geller D.A., Lowenstein C.J., Shapiro R.A., Nussler A.K., Di Silvio M., Wang S.C., et al. Molecular cloning and expression of inducible nitric oxide synthase from human hepatocytes. Proc. Natl. Acad. Sci. U. S. A. 1993;90:3491–3495. doi: 10.1073/pnas.90.8.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruettinger R.T., Wen L.P., Fulco A.J. Coding nucleotide, 5' regulatory, and deduced amino acid sequences of P-450BM-3, a single peptide cytochrome P-450:NADPH-P-450 reductase from Bacillus megaterium. J. Biol. Chem. 1989;264:10987–10995. [PubMed] [Google Scholar]
- 46.Fisher C.W., Shet M.S., Caudle D.L., Martin-Wixtrom C.A., Estabrook R.W. High-level expression in Escherichia coli of enzymatically active fusion proteins containing the domains of mammalian cytochromes P450 and NADPH-P450 reductase flavoprotein. Proc. Natl. Acad. Sci. U. S. A. 1992;89:10817–10821. doi: 10.1073/pnas.89.22.10817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shet M.S., Fisher C.W., Arlotto M.P., Shackleton C.H., Holmans P.L., Martin-Wixtrom C.A., et al. Purification and enzymatic properties of a recombinant fusion protein expressed in Escherichia coli containing the domains of bovine P450 17A and rat NADPH-P450 reductase. Arch. Biochem. Biophys. 1994;311:402–417. doi: 10.1006/abbi.1994.1255. [DOI] [PubMed] [Google Scholar]
- 48.Shet M.S., Fisher C.W., Holmans P.L., Estabrook R.W. Human cytochrome P450 3A4: enzymatic properties of a purified recombinant fusion protein containing NADPH-P450 reductase. Proc. Natl. Acad. Sci. U. S. A. 1993;90:11748–11752. doi: 10.1073/pnas.90.24.11748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Faulkner K.M., Shet M.S., Fisher C.W., Estabrook R.W. Electrocatalytically driven omega-hydroxylation of fatty acids using cytochrome P450 4A1. Proc. Natl. Acad. Sci. U. S. A. 1995;92:7705–7709. doi: 10.1073/pnas.92.17.7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Argos P. An investigation of oligopeptides linking domains in protein tertiary structures and possible candidates for general gene fusion. J. Mol. Biol. 1990;211:943–958. doi: 10.1016/0022-2836(90)90085-Z. [DOI] [PubMed] [Google Scholar]
- 51.Zhao Q., Modi S., Smith G., Paine M., McDonagh P.D., Wolf C.R., et al. Crystal structure of the FMN-binding domain of human cytochrome P450 reductase at 1.93 A resolution. Protein Sci. 1999;8:298–306. doi: 10.1110/ps.8.2.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schenkman J.B., Remmer H., Estabrook R.W. Spectral studies of drug interaction with hepatic microsomal cytochrome. Mol. Pharmacol. 1967;3:113–123. [PubMed] [Google Scholar]
- 53.Guengerich F.P., Johnson W.W. Kinetics of ferric cytochrome P450 reduction by NADPH-cytochrome P450 reductase: rapid reduction in the absence of substrate and variations among cytochrome P450 systems. Biochemistry. 1997;36:14741–14750. doi: 10.1021/bi9719399. [DOI] [PubMed] [Google Scholar]
- 54.Keizers P.H., Schraven L.H., de Graaf C., Hidestrand M., Ingelman-Sundberg M., van Dijk B.R., et al. Role of the conserved threonine 309 in mechanism of oxidation by cytochrome P450 2D6. Biochem. Biophys. Res. Commun. 2005;338:1065–1074. doi: 10.1016/j.bbrc.2005.10.066. [DOI] [PubMed] [Google Scholar]
- 55.Lin H.L., Kenaan C., Zhang H., Hollenberg P.F. Reaction of human cytochrome P450 3A4 with peroxynitrite: nitrotyrosine formation on the proximal side impairs its interaction with NADPH-cytochrome P450 reductase. Chem. Res. Toxicol. 2012;25:2642–2653. doi: 10.1021/tx3002753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoshimoto F.K., Gonzalez E., Auchus R.J., Guengerich F.P. Mechanism of 17alpha,20-lyase and new hydroxylation reactions of human cytochrome P450 17A1: 18O labeling and oxygen surrogate evidence for a role of a perferryl oxygen. J. Biol. Chem. 2016;291:17143–17164. doi: 10.1074/jbc.M116.732966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Strohmaier S.J., De Voss J.J., Jurva U., Andersson S., Gillam E.M.J. Oxygen surrogate systems for supporting human drug-metabolizing cytochrome P450 enzymes. Drug Metab. Dispos. 2020;48:432–437. doi: 10.1124/dmd.120.090555. [DOI] [PubMed] [Google Scholar]
- 58.Chefson A., Zhao J., Auclair K. Replacement of natural cofactors by selected hydrogen peroxide donors or organic peroxides results in improved activity for CYP3A4 and CYP2D6. Chembiochem. 2006;7:916–919. doi: 10.1002/cbic.200600006. [DOI] [PubMed] [Google Scholar]
- 59.Strushkevich N., MacKenzie F., Cherkesova T., Grabovec I., Usanov S., Park H.W. Structural basis for pregnenolone biosynthesis by the mitochondrial monooxygenase system. Proc. Natl. Acad. Sci. U. S. A. 2011;108:10139–10143. doi: 10.1073/pnas.1019441108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Esteves F., Campelo D., Gomes B.C., Urban P., Bozonnet S., Lautier T., et al. The role of the FMN-domain of human cytochrome P450 oxidoreductase in its promiscuous interactions with structurally diverse redox partners. Front Pharmacol. 2020;11:299. doi: 10.3389/fphar.2020.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Esteves F., Almeida C.M.M., Silva S., Saldanha I., Urban P., Rueff J., et al. Single mutations in cytochrome P450 oxidoreductase can alter the specificity of human cytochrome P450 1A2-mediated caffeine metabolism. Biomolecules. 2023;13:1083. doi: 10.3390/biom13071083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Estrada D.F., Skinner A.L., Laurence J.S., Scott E.E. Human cytochrome P450 17A1 conformational selection: modulation by ligand and cytochrome b5. J. Biol. Chem. 2014;289:14310–14320. doi: 10.1074/jbc.M114.560144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bart A.G., Scott E.E. Structural and functional effects of cytochrome b5 interactions with human cytochrome P450 enzymes. J. Biol. Chem. 2017;292:20818–20833. doi: 10.1074/jbc.RA117.000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ubbink M. The courtship of proteins: understanding the encounter complex. FEBS Lett. 2009;583:1060–1066. doi: 10.1016/j.febslet.2009.02.046. [DOI] [PubMed] [Google Scholar]
- 65.Volkov A.N. Structure and function of transient encounters of redox proteins. Acc. Chem. Res. 2015;48:3036–3043. doi: 10.1021/acs.accounts.5b00343. [DOI] [PubMed] [Google Scholar]
- 66.Spencer A.L., Bagai I., Becker D.F., Zuiderweg E.R., Ragsdale S.W. Protein/protein interactions in the mammalian heme degradation pathway: heme oxygenase-2, cytochrome P450 reductase, and biliverdin reductase. J. Biol. Chem. 2014;289:29836–29858. doi: 10.1074/jbc.M114.582783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Godamudunage M.P., Grech A.M., Scott E.E. Comparison of antifungal azole interactions with adult cytochrome P450 3A4 versus neonatal cytochrome P450 3A7. Drug Metab. Dispos. 2018;46:1329–1337. doi: 10.1124/dmd.118.082032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang A., Savas U., Hsu M.H., Stout C.D., Johnson E.F. Crystal structure of human cytochrome P450 2D6 with prinomastat bound. J. Biol. Chem. 2012;287:10834–10843. doi: 10.1074/jbc.M111.307918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smith B.D., Sanders J.L., Porubsky P.R., Lushington G.H., Stout C.D., Scott E.E. Structure of the human lung cytochrome P450 2A13. J. Biol. Chem. 2007;282:17306–17313. doi: 10.1074/jbc.M702361200. [DOI] [PubMed] [Google Scholar]
- 70.DeVore N.M., Smith B.D., Urban M.J., Scott E.E. Key residues controlling phenacetin metabolism by human cytochrome P450 2A enzymes. Drug Metab. Dispos. 2008;36:2582–2590. doi: 10.1124/dmd.108.023770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.DeVore N.M., Scott E.E. Structures of cytochrome P450 17A1 with prostate cancer drugs abiraterone and TOK-001. Nature. 2012;482:116–119. doi: 10.1038/nature10743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Burris-Hiday S.D., Loomis C.L., Richard A.M., Scott E.E. Generation of human steroidogenic cytochrome P450 enzymes for structural and functional characterization. Meth. Enzymol. 2023 doi: 10.1016/j.jbc.2023.105112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fehl C., Vogt C.D., Yadav R., Li K., Scott E.E., Aube J. Structure-based design of inhibitors with improved selectivity for steroidogenic cytochrome P450 17A1 over cytochrome P450 21A2. J. Med. Chem. 2018;61:4946–4960. doi: 10.1021/acs.jmedchem.8b00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wester M.R., Stout C.D., Johnson E.F. Purification and crystallization of N-terminally truncated forms of microsomal cytochrome P450 2C5. Methods Enzymol. 2002;357:73–79. doi: 10.1016/s0076-6879(02)57667-9. [DOI] [PubMed] [Google Scholar]
- 75.Peterson J.A. Camphor binding by Pseudomonas putida cytochrome P-450. Arch. Biochem. Biophys. 1971;144:678–693. doi: 10.1016/0003-9861(72)90518-8. [DOI] [PubMed] [Google Scholar]
- 76.Omura T., Sato R. Fractional solubilization of haemoproteins and partial purification of carbon monoxide-binding cytochrome from liver microsomes. Biochim. Biophys. Acta. 1963;71:224–226. doi: 10.1016/0006-3002(63)91015-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data are included in this article or the supplemental information.