Abstract
The vacuolating cytotoxin of Helicobacter pylori (VacA) is known to cause cell damage to mammalian cells and is suspected to give rise to gastric epithelial lesions that might lead to peptic ulcer disease. As shown recently, the gene encoding VacA exhibits genetic variation, with three different families of signal sequences (s1a, s1b, and s2) and two families of midregion sequences (m1 and m2). In order to investigate the relationship between the presence of specific vacA genotypes and peptic ulceration, the vacA genotypes of 158 clinical isolates of H. pylori were determined. The study group consisted of 106 patients with duodenal ulceration; 52 patients with nonulcer dyspepsia (NUD) were used as controls. H. pylori of genotype s1 was isolated from 96% of the patients with ulcerations, whereas genotype s2 was only present in 4%, indicating a strong correlation between the vacA genotype and peptic ulceration (P < 0.001). In contrast, 31% of the patients from the NUD control group were infected with strains of vacA genotype s2. Particular midregion genotypes (m1 and m2) were not associated with clinical manifestations. The midregions from 18% of the isolates could not be classified by the proposed scheme. DNA sequencing revealed high homology between the untypeable midregions and that of genotype m1, with multiple base pair exchanges, some affecting the primer annealing site. Compared to those of m1 and m2 alleles, the divergent midregions from untypeable strains showed clustering, indicating the presence of a further subfamily of sequences in the midregion of vacA in German isolates, for which we propose the term “m1a.” A new specific primer that we designed for typing m1a isolates might be useful in other studies.
Helicobacter pylori is a gram-negative spiral bacterium that colonizes the gastric mucosa and is able to persist over decades if the infection is not treated. The chronic infection often occurs without symptoms, but some individuals develop severe features of upper gastrointestinal diseases such as peptic ulcer disease (PUD), gastric adenocarcinoma, and mucosa-associated lymphoid tissue (MALT) lymphoma (1, 13). The vacuolating cytotoxin is one of the putative virulence factors of H. pylori that might lead to ulcerations. It induces massive formation of acidic vacuoles in the cytoplasm of gastric epithelial cells in vivo (14) as well as in vitro in primary epithelial cells (9) or in permanent cell lines. It has also been demonstrated that oral administration of purified VacA to mice causes injury of the gastric mucosa (14). The gene coding for the cytotoxin exhibits a mosaic of different alleles, which can be separately detected by PCR (2, 3, 6). For the N-terminal region of vacA, coding for the signal peptide, three different families of sequences, termed s1a, s1b, and s2, can be differentiated. The midregion of vacA is represented by two different families of alleles, termed m1 and m2 (2).
The aim of this study was (i) to perform vacA typing of H. pylori isolated from German patients in order to test the applicability of this method to isolates from a population not yet examined, (ii) to evaluate the association of vacA genotypes with peptic ulceration and nonulcer dyspepsia (NUD), and (iii) to assess the association of vacA genotypes with the presence of the pathogenicity marker cagA.
MATERIALS AND METHODS
Patients.
Independent clinical isolates of H. pylori were obtained from 158 adults who underwent gastroduodenoscopy. Of these, 106 patients (mean age, 51 years) presenting with duodenal ulcerations (PUD) with a minimum ulcer size of 5 mm have been enrolled in a multicenter study (7) including 28 centers all over Germany. Biopsies of a control group consisting of 52 patients (mean age, 47 years) with NUD were taken by two gastroenterologists. Strains from patients with gastric ulceration, gastric cancer, or MALT lymphoma or who had taken antimicrobial agents 4 weeks prior to endoscopy were not included in the study.
Isolation and culture conditions.
During endoscopy one antrum and one corpus mucosal biopsy were obtained from each patient. Each biopsy specimen was placed in a transport medium (Portagerm Pylori; Biomerieux) and sent to the laboratory within 24 h. The specimens were ground with a pellet pestle, spread on solid-agar plates, and incubated under microaerobic conditions at 37°C for 2 to 5 days. Yeast extract-cysteine blood agar base (Difco), supplemented with 0.0005% hemin, 0.007% potassium hydroxide, 0.001% vitamin K, 10% horse serum, 10% washed human erythrocytes, vancomycin (10 mg/liter), trimethoprim (5 mg/liter), and nystatin (1 mg/liter) after autoclaving, was used as the growth medium. Bacteria were identified as H. pylori by standard criteria (12). The primary cultures grown from antrum and corpus biopsy specimens were transferred to Microbank cryovials (Mast Diagnostica) and stored frozen at −70°C. These are referred to as stock cultures.
PCR-based typing.
The DNA sequences of the primer oligonucleotides used for PCR and sequencing and the sizes of the corresponding PCR products are listed in Table 1. For PCR analysis, bacteria from frozen stock cultures were grown for 48 h in brucella broth supplemented with 10% fetal calf serum at 37°C with shaking under microaerobic conditions. The cells were harvested by centrifugation and resuspended in distilled water. A 1-μl aliquot from a 100-fold dilution of this suspension was analyzed in a PCR mixture (50 ml) containing 1 U of Taq DNA polymerase and 25 pmol of each primer in a buffer system described previously (4). Amplification was done in 30 cycles consisting of 94, 50, and 72°C (for 1 minute each). The PCR products were electrophoretically separated on a 2.5% agarose gel and stained with ethidium bromide.
TABLE 1.
Primers used in the study
| Gene and DNA region amplified | Primer | Nucleotide sequence | Product size (bp)a | Source or reference |
|---|---|---|---|---|
| vacA | 2 | |||
| s1 | VA1-F | ATGGAAATACAACAAACACAC | ||
| VA1-R | CTGCTTGAATGCGCCAAAC | 259 | ||
| s2 | VA1-F | ATGGAAATACAACAAACACAC | ||
| VA1-R | CTGCTTGAATGCGCCAAAC | 286 | ||
| s1a | SS1-Fb | GTCAGCATCACACCGCAAC | 190 | |
| s1b | SS3-Fb | AGCGCCATACCGCAAGAG | 187 | |
| s2 | SS2-Fb | GCTAACACGCCAAATGATCC | 199 | |
| m1 | VA3-F | GGTCAAAATGCGGTCATGG | ||
| VA3-R | CCATTGGTACCTGTAGAAAC | 290 | ||
| m2 | VA4-F | GGAGCCCCAGGAAACATTG | ||
| VA4-R | CATAACTAGCGCCTTGCAC | 352 | ||
| vacA | This study | |||
| mx (m1a) | VA3.1-Rc | CTGTTAGTGCCCGCAGAAAC | 290 | |
| mx (m1a) | VA3.2-Rc | CTAATGCTGTTAGTGCCCGC | 290 | |
| Midregion | VA6-F | TCAATATCAACAAGCTC | ||
| VA5-R | CCGCATGCTTTAATGTC | 785 | ||
| cagA | 5 | |||
| Midregion | CAG-Ld | TGCTAAATTAGACAACTTGAGCGA | ||
| CAG-Rd | AATAATCAACAAACATCACGCCAT | 289 |
Size of PCR products is given for individual strains and can vary among strains.
Used in combination with primer VA1-R.
Used in combination with primer VA3-F; the oligonucleotide sequence is located at positions 2821 to 2840 of the vacA DNA sequence, EMBL database accession no. Z26883 (15).
Primers for detection of cagA were kindly provided by Antonello Covacci.
DNA sequence analysis of the vacA midregion.
To determine the nucleotide sequence of the vacA midregion, a 785-bp fragment was amplified by using the two primers VA6-F and VA5-R, which bind to conserved regions flanking the midregion of the vacA gene (Table 1). The PCR products were purified by using the QIAquik PCR purification kit from Qiagen and sequenced with the PRISM ready reaction dye cycle sequencing kit (Applied Biosystems), including fluorescence-labeled dideoxynucleotides. The fluorescent reaction products were separated on a denaturing polyacrylamide gel and analyzed with an Applied Biosystems 373A automated DNA-sequencing machine. The DNA sequence was analyzed with the HUSAR software package provided by the German Cancer Research Center in Heidelberg. The detailed alignments were done with GAP, CLUSTAL, or BESTFIT software. The dendrogram was constructed with pc/gene software, version 6.8, from IntelliGenetics Inc.
Detection of cagA.
The presence of the cagA gene was ascertained by PCR with primer oligonucleotides CAG-L and CAG-R (Table 1) under the cycling conditions described above.
Nucleotide sequence accession numbers.
The vacA midregion of strains 003, 013, 026, 031, and 049 have been assigned EMBL database accession no. Y14740, Y14741, Y14742, Y14743, and Y14744, respectively.
RESULTS
Distribution of vacA alleles in patient groups.
A total of 158 strains were investigated to determine their vacA genotypes. Of these, 106 were isolated from patients who suffered from duodenal ulceration (PUD) and 52 were isolated from patients with NUD. All three alleles of the vacA signal sequence could be detected and typed in all isolates as either s1a, s1b, or s2. Allele s1a was found in the PUD group with higher frequency (90%) than in the NUD group (58%). The allele s2, known to be associated with less-cytotoxic activity and with the absence of the pathogenicity-related gene cagA, was found in the NUD group at a much higher frequency (31%) than in the PUD group (4%). Both associations revealed statistical significance (P < 0.001 by the χ2 test). The midregion subtypes earlier defined as m1 and m2 (2, 3) were detectable in only 81.5% of the isolates, whereas in 18.5% no PCR product was obtained by using the primers specific for m1 and m2. The midregion subtype present in these strains was given the preliminary designation mx, which could be identified as a distinct subtype of m1 by further criteria (see below). With respect to this classification, the m1 and m2 alleles were equally distributed over the PUD and NUD groups. The data are summarized in Table 2.
TABLE 2.
Distribution of vacA alleles in patient groups
| vacA allele | No. (%) of isolates
|
|
|---|---|---|
| Study group PUD (n = 105) | Control group NUD (n = 52) | |
| s1a | 95 (90) | 30 (58) |
| s1b | 6 (6) | 6 (11) |
| s2 | 4 (4) | 16 (31) |
| m1 | 30 (29) | 19 (36) |
| mx (m1a) | 23 (22) | 6 (12) |
| m2 | 52 (49) | 27 (52) |
Different vacA genotypes in strains from one patient.
For each patient, the isolate from the gastric antrum and the isolate from the gastric corpus were analyzed separately. We had only one case in which H. pylori isolates with more than one vacA allele were obtained from a single patient. Strains 091A (isolated from the antrum) and 091C (isolated from the corpus) showed genotypes s1a m2 and s2 m2, respectively. This implied that the patient was infected with more than one strain. The results from this patient were not included in Table 2.
Further characterization of the midregion subtype mx.
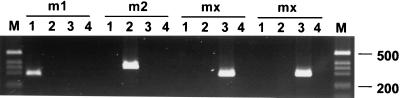
The 29 strains which gave no PCR product with the primers for typing m1 or m2 alleles were further investigated by DNA sequence analysis and by PCR with specifically designed primer oligonucleotides. For this purpose, the DNA sequences of 785-bp fragments including the divergent vacA midregions from five independent randomly selected untypeable isolates were determined. The analysis revealed six single-base-pair transitions within the binding site for the m1 antisense primer, VA3-R, whereas the binding site m1 sense primer, VA3-F, was completely conserved (Table 3). Binding sites for primers specific for the m2 allele were absent. In each of the five clinical isolates, which originated from patients living in different areas of Germany, the nucleotide exchanges were identical, with the exception that in isolate 026 an additional C-to-T transition was present (Table 3). In order to further investigate whether this variation was responsible for the insufficient amplification and to classify further mx strains, two new primers, VA3.1-R and VA3.2-R, were designed. The nucleotide sequence of primer VA3.1-R was identical with that of the altered binding site found in mx strains, and the primer VA3.2-R carried the sequence 5 bp upstream from the original annealing site (Tables 1 and 3). To determine whether these primers were suitable for investigation of the mx subtype, individual m1, m2, and mx strains were analyzed by PCR with primers VA3.1-R and VA3.2-R in combination with primer VA3-F (Fig. 1). This analysis revealed that primer VA3.1-R gave a strong amplification product with mx strains whereas primer VA3.2-R gave only weak amplification of the mx midregion sequence. Neither primer amplified the midregions of m1 or m2 strains, indicating that primer VA3.1-R, especially, could be useful to specifically detect the mx subtype. This was further confirmed by analysis of all 29 mx strains which could be successfully classified by this method (data not shown).
TABLE 3.
Mutations in the primer annealing site for VA3-R in mx (m1a) strains
| Source | Sequence
|
|
|---|---|---|
| VA3-F | VA3-R | |
| Primera | GGTCAAAATGCGGTCATGG | CCATTGGTACCTGTAGAAAC |
| Isolateb | ||
| 003 | GGTCAAAATGCGGTCATGG | CTGTTAGTGCCCGCAGAAAC |
| 013 | GGTCAAAATGCGGTCATGG | CTGTTAGTGCCCGCAGAAAC |
| 026 | GGTCAAAATGCGGTCATGG | CTGTTAGTGTCCGCAGAAAC |
| 031 | GGTCAAAATGCGGTCATGG | CTGTTAGTGCCCGCAGAAAC |
| 049 | GGTCAAAATGCGGTCATGG | CTGTTAGTGCCCGCAGAAAC |
Oligonucleotide sequences specific for detection of vacA genotype m1.
DNA sequences complementary to the primer annealing sites in the primarily untypeable isolates (m1a) 003 to 049. Nucleotides which differ are in boldface.
FIG. 1.
PCR analysis of the vacA midregion of defined m1, m2, and mx strains. Isolates 005C (m1), 004C (m2), 003C (mx), and 013C (mx) were analyzed with the four primer combinations which specifically detect the vacA subtypes m1 (VA3-F and -R) (lanes 1), m2 (VA4-F and -R) (lanes 2), and mx (VA3.1-R and VA3-F and VA3.2-R and VA3-F) (lanes 3 and 4, respectively). PCR products were separated on a 2.5% agarose gel and stained with ethidium bromide. The size of defined marker DNA fragments (1-kb DNA ladder; BRL) is indicated in base pairs to the right.
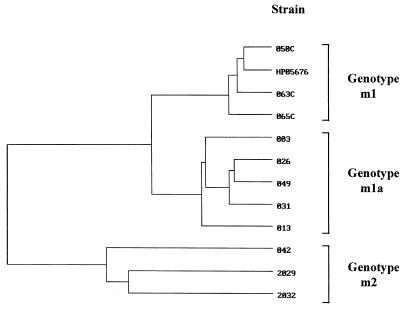
To further assess the homology among the mx, m1, and m2 alleles on the basis of the DNA sequence, the 785-bp fragments from five strains were aligned with the corresponding vacA sequences from databases. This analysis revealed that the mx region exhibits a significantly closer homology with m1 strains (88%) than with m2 strains (74%). The fact that the homology was significantly higher within mx strains (95 to 98%) indicated that the mx group represents a cluster resembling a new subtype of m1, which could be present only in European isolates (Table 4 and Fig. 2). In this context it is interesting to note that the vacA sequence published earlier by Schmitt and Haas (15) is also of the mx type.
TABLE 4.
Nucleotide sequence identity observed in alignments of 785-bp fragments from the vacA divergent midregions and homologous regions in EMBL database sequences
| Strain and isolate | % Identity with:
|
|||
|---|---|---|---|---|
| Hp05676a (m1) | HpCyttoxb (m1) | Hp07145c (m1) | Hp05677d (m2) | |
| mx (m1a) | ||||
| 003 | 89 | 95 | 91 | 74 |
| 013 | 88 | 96 | 90 | 74 |
| 026 | 89 | 98 | 91 | 75 |
| 031 | 88 | 97 | 90 | 75 |
| 049 | 89 | 98 | 92 | 74 |
| Range | 88–89 | 95–98 | 90–92 | 74–75 |
| m1 | ||||
| 050 | 99 | 87 | 95 | 74 |
| 057 | 94 | 94 | 94 | 74 |
| 063 | 98 | 87 | 94 | 73 |
| 092 | 96 | 89 | 95 | 73 |
| 065 | 97 | 86 | 94 | 72 |
| 2137 | 99 | 87 | 95 | 72 |
| Range | 94–99 | 86–94 | 94–95 | 72–74 |
| m2 | ||||
| 2029 | 89 | 81 | 80 | 97 |
| 2032 | 78 | 74 | 77 | 96 |
| 042 | 78 | 79 | 79 | 93 |
| Range | 78–89 | 74–81 | 77–80 | 93–97 |
FIG. 2.
Dendrogram showing the relationships among vacA midregion sequences of subtypes m1, m2, and m1a. The midregion sequences from 11 German H. pylori isolates were aligned. The DNA regions analyzed correspond to nucleotides 2629 to 3078 in the vacA gene HP05676 deposited in the GenBank and EMBL databases, which served as a reference (6).
Relationship between vacA genotype and CagA status.
The presence of the pathogenicity marker gene cagA was highly correlated with the vacA signal sequence type s1a (P < 0.001 by the χ2 test). This link between cagA and allele s1a was not exclusive, as indicated by the detection of six cagA-negative isolates exhibiting the vacA genotype s1a and, conversely, three cagA-positive isolates exhibiting the vacA genotype s2.
DISCUSSION
The present study revealed that the vacA allele s1a of H. pylori is strongly associated with PUD. On the other hand, the vacA allele s2 was predominantly found in isolates from patients with NUD and was very rare in strains isolated from ulcer patients. Therefore, the vacA allele s2 is not suspected to contribute to PUD.
It was reported in previous studies (2, 3, 6) that the observed cytotoxic activity was higher in s1a m1 than in s1a m2 strains and that cytotoxicity seemed to be completely absent in s2 m2 strains. It was proposed, as the most likely explanation for the high prevalence of the s1a genotype in the ulcer group, that the elevated toxicity of s1a m1 strains might contribute to the development of ulcerations. However, this explanation is not consistent with the findings of the present study, which indicate that, in contrast to the genotype of the signal sequence, the genotype of the midregion showed no correlation with the outcome of disease. If it is true that the ulcerative effect is exclusively dependent on cytotoxic activity, one would expect a higher frequency of m1 genotypes in the ulcer group and correspondingly more m2 genotypes in the NUD group. One explanation for these contradictory observations could be that the toxin assay with HeLa cells might not accurately reflect the in vivo effect occurring in the duodenal epithelial cells. It is supposed that the cytotoxin enters the cell by receptor-mediated endocytosis (8), and if this is true, the presence of specific receptor molecules on the surface of the target cell should influence the efficacy of the cytotoxin. Another explanation might be that factors other than VacA influence the development of ulcers. As shown before, and also demonstrated in this study, the vacA genotype s1a is strongly associated with the presence of the pathogenicity marker cagA (2, 3), whereas strains with vacA genotype s2 mostly lacked the cagA gene. This may imply that vacA genotype s1a is a marker for elevated virulence. Interestingly, the vacA midregion allele m2 was correlated with a negative cagA status (P < 0.001 by the χ2 test) (Table 5).
TABLE 5.
Relationship between cagA status and vacA genotype of 158 H. pylori clinical isolates
| vacA allele | No. of isolates with cagA status:
|
|
|---|---|---|
| Positive | Negative | |
| s1aa | 117 | 6 |
| s1b | 10 | 2 |
| s2b | 3 | 19 |
| m1 | 51 | 1 |
| mx (m1a) | 28 | 2 |
| m2c | 54 | 21 |
Signal sequence allele s1a correlates with the presence of cagA (P < 0.001 by χ2 test).
Signal sequence allele s2 correlates with absence of cagA (P < 0.001 by χ2 test).
vacA midregion genotype m2 correlates with absence of cagA (P < 0.001 by χ2 test).
It was one of the objectives of this study to evaluate whether the vacA genotype might be a predictor of the duodenal ulcer risk of an infected person. However, our conclusions are limited by the fact that the NUD control group, in contrast to the PUD group, was heterogeneous and included patients with erosive gastritis as well as persons who might have suffered from ulcers before or might suffer from ulcers in the future. This potential selection bias could in part explain the relatively high prevalence of genotype s1a in the NUD group.
Nearly one-fifth of the H. pylori strains which were isolated harbored a vacA genotype that could not be classified by the proposed scheme (2, 3) and which we gave the preliminary name mx. The genotypes of all 29 of these isolates have been confirmed by allele-specific PCR, and 5 were additionally analyzed by DNA sequencing. Because the mx type investigated in this study highly resembled the midregion type m1, we would propose the term “m1a,” analogous to the terms s1a and -b used for the signal sequence subtypes (2, 3). The results of the present study also provided evidence that all m1a isolates are of clonal origin and represent a distinct midregion genotype. To see such clear-cut differences from the results of vacA typing of strains of North American origin was surprising (2). To date, there is one other published study about H. pylori isolates from Japan, which reported that 84 of 87 clinical isolates were untypeable with respect to the midregion of the vacA gene (11). It would be of interest to evaluate in further studies if the m1a genotype is restricted exclusively to Western Europe or if it is also found in other geographic regions. We presume that geographical clustering of vacA genotypes in different populations exists. The primer we designed for detecting genotype m1a might be a useful tool in such studies.
ACKNOWLEDGMENTS
We thank Antonello Covacci for providing primers for detection of cagA. We are grateful to Henriette Ries and Stefanie Pietsch for excellent technical assistance.
REFERENCES
- 1.Asaka M, Kimura T, Kato M, Kudo M, Miki K, Ogoshi K, Kato T, Tatsuga M, Graham D D Y. Possible role of Helicobacter infection in early gastric cancer development. Cancer. 1994;73:2691–2694. doi: 10.1002/1097-0142(19940601)73:11<2691::aid-cncr2820731107>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 2.Atherton J C, Cao P, Peek R M, Tummuru M K R, Blaser M J, Cover T L. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. J Biol Chem. 1995;270:17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 3.Atherton J C, Peek M R, Tham K T, Cover T L, Blaser M J. Clinical and pathological importance of heterogeneity in vacA, the cytotoxin gene of Helicobacter pylori. Gastroenterology. 1997;112:92–95. doi: 10.1016/s0016-5085(97)70223-3. [DOI] [PubMed] [Google Scholar]
- 4.Bereswill S, Pahl A, Bellemann P, Zeller W, Geider K. Sensitive and species-specific detection of Erwinia amylovora by polymerase chain reaction analysis. Appl Environ Microbiol. 1992;58:3522–3526. doi: 10.1128/aem.58.11.3522-3526.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, Massone A, Papini E, Xiang Z, Figura N, Rappuoli R. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci USA. 1993;50:5791–5795. doi: 10.1073/pnas.90.12.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cover T L, Tummuru M K R, Cao P, Thompson S A, Blaser M J. Divergence of genetic sequences for vacuolating cytotoxin among Helicobacter pylori strains. J Biol Chem. 1994;269:10566–10573. [PubMed] [Google Scholar]
- 7.Dammann, H. G., U. R. Fölsch, E. G. Hahn, D. H. von Kleist, H. U. Klör, T. Kirchner, and M. Kist. 1997. Seven vs 14 day treatment with pantoprazole, clarithromycin and metronidazole for cure of H. pylori infection in duodenal ulcer patients. Gut 41(Suppl. 1):95. [DOI] [PubMed]
- 8.Garner J A, Cover T L. Binding and internalization of the Helicobacter pylori vacuolating cytotoxin by epithelial cells. Infect Immun. 1996;64:4197–4203. doi: 10.1128/iai.64.10.4197-4203.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris P R, Cover T L, Crowe D R, Orenstein J M, Graham M F, Blaser M J, Smith P D. Helicobacter pylori cytotoxin induces vacuolation of primary human mucosal epithelial cells. Infect Immun. 1996;64:4867–4871. doi: 10.1128/iai.64.11.4867-4871.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hazell S L, Andrews R H, Mitchell H M, Daskalopoulous G. Genetic relationship among isolates of Helicobacter pylori: evidence for the existence of a Helicobacter pylori species-complex. FEMS Microbiol Lett. 1997;150:27–32. doi: 10.1111/j.1574-6968.1997.tb10345.x. [DOI] [PubMed] [Google Scholar]
- 11.Ito Y, Azuma T, Ito S, Miyaji H, Hirai M, Yamazaki Y, Sato F, Kato T, Kohli Y, Kuriyama M. Analysis and typing of the vacA gene from cagA-positive strains of Helicobacter pylori isolated in Japan. J Clin Microbiol. 1997;35:1710–1714. doi: 10.1128/jcm.35.7.1710-1714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kist M. Diagnostische Verfahrensrichtlinien der DGHM: Isolierung und Identifizierung von Bakterien der Gattungen Campylobacter und Helicobacter. Zentralbl Bakteriol. 1991;276:124–139. [PubMed] [Google Scholar]
- 13.McColl K E. Helicobacter pylori: clinical aspects. J Infect. 1997;34:7–13. doi: 10.1016/s0163-4453(97)80003-5. [DOI] [PubMed] [Google Scholar]
- 14.Phadnis S H, Ilver D, Janzon L, Normark S, Westblom T U. Pathological significance and molecular characterization of the vacuolating cytotoxin gene of Helicobacter pylori. Infect Immun. 1994;62:1557–1565. doi: 10.1128/iai.62.5.1557-1565.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmitt W, Haas R. Genetic analysis of the Helicobacter pylori vacuolating cytotoxin: structural similarities with IgA protease type exported protein. Mol Microbiol. 1994;12:307–319. doi: 10.1111/j.1365-2958.1994.tb01019.x. [DOI] [PubMed] [Google Scholar]
- 16.Telford J L, Ghiara P, Dell’Ocro M, Comanducci M, Burroni D, Bugnoli M, Tecce M F, Censini S, Covacci A, Xiang Z, Papini E, Montecucco C, Parente L, Rappuoli R. Gene structure of the Helicobacter pylori cytotoxin and evidence of its key role in gastric disease. J Exp Med. 1994;179:1653–1658. doi: 10.1084/jem.179.5.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tummuru M K R, Cover T L, Blaser M J. Mutation of the cytotoxin-associated cagA gene does not affect the vacuolating cytotoxin activity of Helicobacter pylori. Infect Immun. 1994;62:2609–2613. doi: 10.1128/iai.62.6.2609-2613.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]