Abstract
Six strains of a hitherto-undescribed gram-positive, catalase-negative, facultatively anaerobic coccus from human sources were characterized by phenotypic and molecular taxonomic methods. Comparative 16S rRNA gene sequencing studies demonstrated that the unknown strains are genealogically homogeneous and constitute a new subline within the genus Gemella. The unknown bacterium was readily distinguished from Gemella haemolysans, the type species of the genus Gemella, and from Gemella morbillorum by biochemical tests and electrophoretic analysis of whole-cell proteins. On the basis of phylogenetic and phenotypic evidence, it is proposed that the unknown bacterium from clinical specimens be classified as Gemella bergeriae sp. nov. The type strain of G. bergeriae is CCUG 37817 (= strain 617-93).
Members of the genus Gemella (1) consist of catalase-negative, facultatively anaerobic, gram-positive cocci which occur in pairs (often with adjacent sides flattened), tetrads, or short chains and which possess DNA with a low G+C content (2). Two species, Gemella haemolysans and Gemella morbillorum, are currently recognized (2). G. haemolysans, the type species of the genus, was originally classified as a member of the gram-negative genus Neisseria (19), whereas G. morbillorum was initially classified as Diplococcus morbillorum (18). The latter species was subsequently assigned to the genera Peptostreptococcus and Streptococcus before its placement in the genus Gemella (13). Despite their somewhat checkered taxonomic histories, G. haemolysans and G. morbillorum closely resemble each other phenotypically (3, 8, 9), and 16S rRNA gene sequence analysis has shown that they represent two genealogically highly related species (20).
Both G. haemolysans and G. morbillorum are residents of the mucous membranes of humans and some other animals (2). In healthy people, G. haemolysans has been found in the oral cavity and upper respiratory tract whereas G. morbillorum is in addition found as a component of the normal human intestinal flora. Gemellae, like some other commensal bacteria of the human microbiota, are opportunistic pathogens, causing severe localized and generalized infection, particularly in immunocompromised patients (5–7, 12, 14, 15, 17). In this article, we report the phenotypic and phylogenetic characterization of some catalase-negative, gram-positive, Gemella-like isolates from human clinical specimens. On the basis of the phylogenetic findings and the phenotypic distinctiveness of the unknown isolates from humans, a new species, Gemella bergeriae sp. nov., is described.
MATERIALS AND METHODS
Strains.
Five bacterial isolates (46-86, 617-93, 767-96, 1239-85, and 1625-92) and a single strain (CCUG 31456) originating from human clinical specimens were referred to the Centers for Disease Control and Prevention (CDC; Atlanta, Ga.) and to the Culture Collection of the University of Göteborg (CCUG; Göteborg, Sweden), respectively, for identification. Strains 46-86 (CCUG 37968) and 617-93 (CCUG 37817) were isolated from blood cultures of persons with subacute bacterial endocarditis. Strains 767-96 (CCUG 37818), 1239-85 (CCUG 37967), and 1625-92 (CCUG 37969) were also isolated from blood cultures of hospitalized patients, but clinical information on these patients was not provided with the cultures. The sixth strain, CCUG 31456, was isolated from blood cultures of a patient with endocarditis from Stockholm, Sweden.
Biochemical tests.
All strains were cultured on Columbia agar (Difco, Detroit, Mich.) supplemented with 5% horse blood at 37°C in air plus 5% CO2. The strains were biochemically characterized by using the API Rapid ID32 Strep and API ZYM systems according to the manufacturer’s instructions (API bioMérieux, Marcy l’Etoile, France).
PAGE analysis of whole-cell proteins.
Polyacrylamide gel electrophoresis (PAGE) analysis of whole-cell proteins was performed as described by Pot et al. (16). For densitometric analysis normalization and interpretation of protein patterns, the GelCompar GCW 3.0 software package (Applied Maths, Kortrijk, Belgium) was used.
DNA base composition.
The moles percent G+C content of DNA was determined by thermal denaturation as described by Garvie (11).
16S rRNA gene sequence analysis.
A large fragment of the 16S rRNA gene (corresponding to positions 30 to 1521 of the Escherichia coli 16S rRNA gene) was amplified by PCR with conserved primers close to the 3′ and 5′ ends of the gene. The PCR products were purified by using a Prep-A-Gene kit (Bio-Rad, Hercules, Calif.) according to the manufacturer’s instructions and directly sequenced by using a Taq Dye-Deoxy terminator cycle sequencing kit (Applied Biosystems, Foster City, Calif.) and an automatic DNA sequencer (model 373A; Applied Biosystems). The closest known relatives of the new isolates were determined by performing database searches. These sequences and those of other known related strains were retrieved from the GenBank database or Ribosomal Database Project library and aligned with the newly determined sequences by using the program PILEUP (4). The resulting multiple-sequence alignment was corrected manually, and a distance matrix was calculated by using the programs PRETTY (10) and DNADIST (using the Kimura-2 correction parameter [10]). A phylogenetic tree was constructed by the neighbor-joining method with the program NEIGHBOR (10). The stability of the groupings was estimated by bootstrap analysis (500 replications) using the programs DNABOOT, DNADIST, NEIGHBOR, and CONSENCE (10).
Nucleotide sequence accession number.
The 16S rRNA gene sequence of strain 617-93 (= CCUG 37817T) has been deposited in GenBank under accession no. Y13365.
RESULTS AND DISCUSSION
The six clinical isolates were ovoid in shape and formed single cells, pairs, or short chains. All of the strains were gram-positive, catalase-negative, oxidase-negative facultative anaerobes which produced pinhead colonies on blood agar. The strains did not grow in broth containing 6.5% NaCl, at 10 or 45°C. The strains resembled gemellae in their appearance and their relative nonreactivity in the biochemical tests employed. All of the isolates produced acid from glucose and were similar to each other in not producing acid from d-arabitol, l-arabinose, cyclodextrin, glycogen, lactose, melibiose, melezitose, methyl-β-d-glucopyranoside, pullulan, ribose, sorbitol, sucrose, trehalose, or d-xylose. Three of the six strains showed weak acid production from mannitol; acid production from maltose was either negative or weak. In commercial API systems, all six isolates produced phosphoamidase, pyrrolidonyl arylamidase, pyrazinamidase, and ester lipase C8; variable activity was shown for esterase C4 (five of six strains positive), and one of six strains displayed acid phosphatase activity. None of the strains produced arginine dihydrolase, alkaline phosphatase, alanine-phenylalanine-proline arylamidase, N-acetyl-β-glucosaminidase, chymotrypsin, α-fucosidase, α-galactosidase, β-galactosidase, β-galacuronidase, β-glucuronidase, glycyl-tryptophan arylamidase, lipase C14, β-mannosidase, trypsin, urease, or valine arylamidase. In conventional disk tests for pyrrolidonyl arylamidase and leucine aminopeptidase, all of the strains tested were positive, although strain 617-93 was weakly positive in both tests. All of the strains failed to hydrolyze esculin, gelatin, and hippurate; they did not reduce nitrate and were Voges-Proskauer and indole negative. The close phenotypic affinity among the clinical isolates was confirmed by PAGE analysis of whole-cell proteins, in which the six strains formed a robust and tight cluster. The group embracing the unknown clinical isolates was quite separate from all other gram-positive, catalase-negative reference organisms examined, including G. haemolysans and G. morbillorum (Fig. 1).
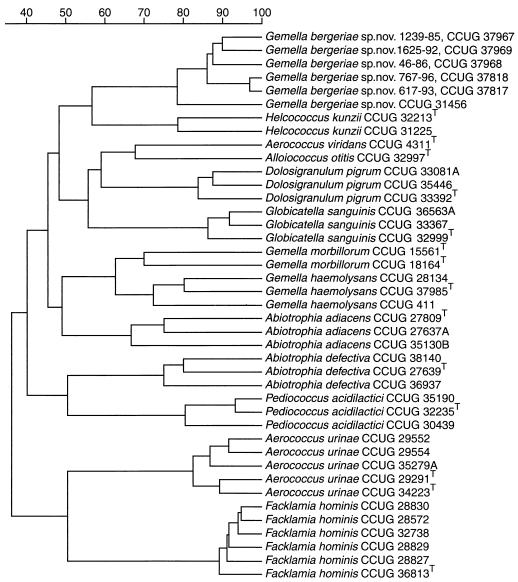
FIG. 1.
Similarity dendogram based on whole-cell protein patterns of G. bergeriae sp. nov. and related species. Levels of correlation are expressed as percentages of similarity for convenience.
To establish the phylogenetic affinities of the clinical isolates, their partial 16S rRNA gene sequences were examined. The sequence of a large fragment of >1,400 nucleotides was determined for each strain. No nucleotide differences were detected among the six strains (i.e., 100% sequence similarity was exhibited), thereby demonstrating their genealogical homogeneity. Sequence searches of the GenBank and Ribosomal Database Project libraries revealed that the unknown organism was phylogenetically most closely associated with species of the genus Gemella. A tree depicting the phylogenetic affinity of the unknown coccus with closely related taxa is shown in Fig. 2. The unknown coccus formed a distinct line branching at the base of the Gemella clade (comprising G. haemolysans and G. morbillorum). Bootstrap resampling showed the grouping of the unknown clinical bacterium with the genus Gemella to be robust (bootstrap value, 100%). Earlier studies have shown that G. haemolysans and G. morbillorum, although genotypically separate species (approximately 20% DNA-DNA relatedness), are nevertheless genealogically highly related, exhibiting only 1.6% sequence divergence (corresponding to 24 base differences) in their 16S rRNAs (20). In the present study, the 16S rRNA of the unknown coccus displayed 5.6 and 5.4% sequence divergence from those of G. haemolysans and G. morbillorum, respectively. These somewhat higher values, together with the deeper branching position in the tree, demonstrate that the unidentified bacterium is phylogenetically more distant from G. haemolysans and G. morbillorum than these two species are from each other. The high overall 16S rRNA gene sequence similarity (approximately 95%) and phenotypic resemblance of the unknown bacterium to G. haemolysans and G. morbillorum are, however, clearly consistent with its assignment to this genus.
FIG. 2.
Unrooted tree showing the phylogenetic relationships of G. bergeriae sp. nov. and some other low-G+C-content gram-positive bacteria. The tree, constructed by the neighbor-joining method, was based on a comparison of approximately 1,320 nucleotides. Bootstrap values, each expressed as a percentage of 500 replications, are given at branching points. Asterisks indicate sequences from the Institute of Food Research database. The specific epithet of Alloiococcus otitis is now A. otitidis.
The two currently recognized Gemella species are considered commensal with humans, but each has been shown to be a causative agent of endocarditis and other infections (2, 5–7, 12, 14, 15, 17). For example, G. haemolysans has also been reported in clinical cases of septicemia and meningitis (14, 15) and an infection of a total knee arthroplasty (6). All of the strains in this study originated from human clinical material. Strain 617-93 (= CCUG 37817T) was sent to the CDC from P. Coudron, Veterans Affairs Hospital, Richmond, Va. This strain was isolated from four of eight blood culture bottles that had been inoculated with blood from a 42-year-old male patient with a history of malaise, night sweats, and fever. The patient had mitral valve prolapse and periodontitis. A clinical diagnosis of subacute endocarditis was made; the patient remained hospitalized for 3 months and eventually underwent mitral valve replacement surgery. One other patient also had a diagnosis of subacute bacterial endocarditis (isolate 48-86). This culture was submitted to the CDC from the Virginia State Health Department. The remaining three cultures examined (767-96, 1239-85, and 1625-92) at the CDC were isolated from blood cultures of patients, but no clinical diagnoses were provided. They were submitted from the state health department laboratories of Arizona, Florida, and Maryland. All five patients in the United States survived their infections. In view of the fact that the patient from which strain 617-93 was isolated had severe periodontitis, it is likely that the source of this strain was the oral cavity. Strain CCUG 31456 also originated from the blood of a patient with endocarditis (in Sweden). No further details on the case were available. It appears that the described unknown coccus, like the other gemellae, represents another opportunistic human pathogen. Due to limitations in tests used in routine diagnostic laboratories, it is not inconceivable that the hitherto-unknown coccus may have been misidentified as G. haemolysans and/or G. morbillorum in the past. Characteristics that we consider useful for distinguishing the new bacterium from G. haemolysans and G. morbillorum in the diagnostic laboratory are summarized in Table 1. On the basis of both phenotypic and phylogenetic evidence, we believe that the unknown bacterium from clinical sources should be classified as a new species of the genus Gemella, for which the name G. bergeriae is proposed.
TABLE 1.
Characteristics useful for differentiating G. bergeriae sp. nov., G. haemolysans, and G. morbilloruma
| Test | G. bergeriae sp. nov. | G. haemo- lysans | G. morbil- lorum |
|---|---|---|---|
| Production of acid from: | |||
| Maltose | − | + | + |
| Sucrose | − | V | + |
| Production of: | |||
| Alkaline phosphatase | − | V | − |
| Alanyl-phenylalanyl-proline arylamidase | − | V | V |
| Acid phosphatase | Vb | + | −c |
| Phosphoamidase | + | −c | − |
+, positive; −, negative; V, variable.
One of six strains was weakly positive.
Some strains may display weak activity.
Description of Gemella bergeriae sp. nov.
Gemella bergeriae (ber.ger.iae L. n., named after Ulrich Berger in recognition of his contributions to the microbiology of gemellae). Cells are gram-positive, non-spore-forming cocci that occur singly, in pairs, or in short chains. Cocci are sometimes elongated. Colonies on blood agar plates after 48 h are small, circular, entire, low convex, translucent to opaque, and smooth. Nonpigmented. Some strains (three of six) are hemolytic on horse blood agar. Growth does not occur in broth containing 6.5% NaCl, at 10 or 45°C. Facultatively anaerobic. Catalase and oxidase negative. Gas is not produced from Mann, Rogosa, or Sharpe broth. Acid is produced from glucose. Some strains produce acid (weak reaction) from maltose and mannitol. Acid is not produced from d-arabitol, l-arabinose, cyclodextrin, glycogen, lactose, melibiose, melezitose, methyl-β-d-glucopyranoside, pullulan, ribose, sorbitol, sucrose, trehalose, or d-xylose. Using the API system, phosphoamidase, pyrrolidonyl arylamidase, pyrazinamidase, and ester lipase C8 are detected; esterase C4 and phosphatase acid activity is detected in some strains. Leucine aminopeptidase is positive, as is pyrrolidonyl arylamidase, by disk test methodology, but tests incorporated into dehydrated commercial identification systems (API) using these substrates give variable results. Arginine dihydrolase, alkaline phosphatase, alanine-phenylalanine-proline arylamidase, N-acetyl-β-glucosaminidase, chymotrypsin, α-fucosidase, α-galactosidase, β-galactosidase, β-galacuronidase, β-glucuronidase, glycyl-tryptophan arylamidase, lipase C14, β-mannosidase, trypsin, urease, and valine arylamidase activities are not detected. Esculin, gelatin, and hippurate are not hydrolyzed. Voges-Proskauer and indole tests are negative. Nitrate is not reduced. The G+C content of the DNA is 32.5 mol%. The type strain of Gemella bergeriae is strain 617-93 (= CCUG 37817). The type strain produces acid from mannitol and is pyroglutamic acid arylamidase negative. Isolated from clinical specimens. Habitat unknown.
ACKNOWLEDGMENTS
This work was supported in part by a grant from the European Union (BI02-CT94-3098).
The protein profiling conducted by Eva Åkervall is gratefully acknowledged.
REFERENCES
- 1.Berger U. A proposed new genus of gram-negative cocci: Gemella. Int Bull Bacteriol Nomencl Taxon. 1961;11:17–19. [Google Scholar]
- 2.Berger U. The genus Gemella. In: Balows A, Trüper H G, Dworkin M, Harder W, Schliefer K-H, editors. The prokaryotes. 2nd ed. New York, N.Y: Springer-Verlag; 1992. pp. 1643–1653. [Google Scholar]
- 3.Berger U, Pervanidis A. Differentiation of Gemella haemolysans (Thjötta and Böe 1938) Berger 1960 from Streptococcus morbillorum (Prévot 1933) Holdeman and Moore 1974. Zentbl Bakteriol Parasitenkd Infektionskr Hyg Abt Orig Reihe A. 1986;261:311–321. doi: 10.1016/s0176-6724(86)80048-7. [DOI] [PubMed] [Google Scholar]
- 4.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Durack D T, Kaplan E L, Bisno A L. Apparent failures of endocarditis prophylaxis. Analysis of 52 cases submitted to a national registry. JAMA. 1983;250:2318–2322. [PubMed] [Google Scholar]
- 6.Eggelmeier F, Petit P, Dijkmans B A C. Total knee arthroplasty infection due to Gemella haemolysans. Br J Rheumatol. 1992;31:67–69. doi: 10.1093/rheumatology/31.1.67. [DOI] [PubMed] [Google Scholar]
- 7.Etienne J, Reverdy M E, Gruer L D, Delorme V, Fleurette J. Evaluation of the API 20 STREP system for species identification of streptococci associated with infective endocarditis. Eur Heart J. 1984;5:25–27. doi: 10.1093/eurheartj/5.suppl_c.25. [DOI] [PubMed] [Google Scholar]
- 8.Facklam R R, Smith P B. The gram-positive cocci. Hum Pathol. 1976;7:187–194. doi: 10.1016/s0046-8177(76)80022-6. [DOI] [PubMed] [Google Scholar]
- 9.Facklam R R. The major differences in the American and British Streptococcus taxonomy schemes with special reference to Streptococcus milleri. Eur J Clin Microbiol. 1984;3:91–93. [Google Scholar]
- 10.Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 11.Garvie E I. Streptococcus raffinolactis Orla-Jensen and Hansen, a group N streptococcus found in raw milk. Int J Syst Bacteriol. 1978;28:190–193. [Google Scholar]
- 12.Kaufhold A, Franzen D, Lütticken R. Endocarditis caused by Gemella haemolysans. Infection. 1989;17:385–387. doi: 10.1007/BF01645552. [DOI] [PubMed] [Google Scholar]
- 13.Kilpper-Bälz R, Schleifer K H. Transfer of Streptococcus morbillorum to the genus Gemella as Gemella morbillorum comb. nov. Int J Syst Bacteriol. 1988;38:442–443. [Google Scholar]
- 14.Mitchell R G, Teddy P J. Meningitis due to Gemella haemolysans after radiofrequency trigeminal rhizotomy. J Clin Pathol. 1985;38:558–560. doi: 10.1136/jcp.38.5.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petit Y J, Layre J C, Lamaury I, Perez C, Jonquet O, Janbon F. Purulent meningitis caused by Gemella haemolysans. Presse Med. 1993;22:444. [PubMed] [Google Scholar]
- 16.Pot B, Vandamme P, Kersters K. Analysis of electrophoretic whole-organism protein fingerprints. In: Goodfellow M, O’Donnell A G, editors. Modern microbial methods. Chemical methods in prokaryotic systematics. Chichester, United Kingdom: John Wiley and Sons Ltd.; 1994. pp. 493–521. [Google Scholar]
- 17.Pradeep R, Ali M, Encarnacion C F. Retropharyngeal abscess due to Gemella morbillorum. Clin Infect Dis. 1997;24:284–285. doi: 10.1093/clinids/24.2.284. [DOI] [PubMed] [Google Scholar]
- 18.Prévot A R. Etudes de systematique bactérienne. I. Lois generales. II. Cocci anaerobies. Ann Soc Nat Zool Biol Anim. 1933;15:23–26. [Google Scholar]
- 19.Thjötta T, Böe J. Neisseria haemolysans. A haemolytic species of Neisseria Trevisan. Acta Pathol Microbiol Scand Suppl. 1938;37:727–731. [Google Scholar]
- 20.Whitney A M, O’Connor S P. Phylogenetic relationship of Gemella morbillorum to Gemella haemolysans. Int J Syst Bacteriol. 1993;43:832–838. doi: 10.1099/00207713-43-4-832. [DOI] [PubMed] [Google Scholar]