Abstract
The protective and absorptive functions of the intestinal epithelium rely on differentiated enterocytes in the villi. The differentiation of enterocytes is orchestrated by sub‐epithelial mesenchymal cells producing distinct ligands along the villus axis, in particular Bmps and Tgfβ. Here, we show that individual Bmp ligands and Tgfβ drive distinct enterocytic programs specific to villus zonation. Bmp4 is expressed from the centre to the upper part of the villus and activates preferentially genes connected to lipid uptake and metabolism. In contrast, Bmp2 is produced by villus tip mesenchymal cells and it influences the adhesive properties of villus tip epithelial cells and the expression of immunomodulators. Additionally, Tgfβ induces epithelial gene expression programs similar to those triggered by Bmp2. Bmp2‐driven villus tip program is activated by a canonical Bmp receptor type I/Smad‐dependent mechanism. Finally, we establish an organoid cultivation system that enriches villus tip enterocytes and thereby better mimics the cellular composition of the intestinal epithelium. Our data suggest that not only a Bmp gradient but also the activity of individual Bmp drives specific enterocytic programs.
Keywords: enterocytes, epithelial differentiation, intestinal mesenchymal cells, small intestine, Tgfβ / Bmp signalling
Subject Categories: Development, Signal Transduction
While moving upwards, the intestinal villi enterocytes acquire different cellular fates, which are promoted by a gradient of Bmp ligands. In addition to this gradient, individual Bmps have distinct activities in the differentiation of enterocytes.
Introduction
The small intestinal epithelium consists of diverse types of differentiated cells that are important for a wide range of functions including absorption of nutrients and protection of the body from the intestinal content. The differentiated cells arise in the crypt from the intestinal epithelial stem cells (IESCs), specifically from their progeny, the transit‐amplifying cells (Spit et al, 2018; Gehart & Clevers, 2019). The differentiated cells exit the crypt, migrate upwards along the villus, and are eventually shed into the intestinal lumen from the villus tip. It is thought that various stromal signals along the crypt–villus axis orchestrate the differentiation process. The details of this complex process have only just begun to be elucidated. Since dysregulation underpins many intestinal diseases, it is imperative to define the output of the various signals provided along the crypt–villus axis and their role during intestinal homeostasis.
Enterocytes are the most abundant differentiated intestinal epithelial cells with a lifetime of approximately 4 days (Schwanhäusser et al, 2011; Harnik et al, 2021). Besides absorbing nutrients, enterocytes constitute a physical and biochemical barrier, produce antimicrobial peptides and can act as immunomodulators (Moor et al, 2018a; Harnik et al, 2021). Enterocytes display a zonated gene expression pattern along the crypt–villus axis (Moor et al, 2018a). Enterocytes at the bottom of the villus express antimicrobial programs, while in the centre and top of the villus they sequentially activate genes connected to the absorption of amino acids, carbohydrates and lipids. In addition, as they migrate towards the villus tip, enterocytes upregulate genes related to cell adhesion and immunomodulation (Moor et al, 2018a; Harnik et al, 2021).
The zonated gene expression signatures, as well as the fate of enterocytes, are most likely governed by sub‐epithelial mesenchymal cells producing morphogen gradients. In the crypt, Pdgfralow mesenchymal cells secrete Wnt ligands that support the renewal of IESCs and intestinal homeostasis (Degirmenci et al, 2018; Greicius et al, 2018; Brügger et al, 2020; McCarthy et al, 2020a). Besides Wnts, they also produce inhibitors of bone morphogenic proteins (Bmp) such as gremlin possibly creating an insulating zone around the stem cells and proliferating progenitors, separating IESCs from differentiated cells. In the villus, Pdgfrahigh mesenchymal cells express various Bmp ligands (further referred to as Bmps) (Brügger et al, 2020; McCarthy et al, 2020a).
Bmps belong to the transforming growth factor β (Tgfβ) superfamily of secreted morphogens. They can form homo‐ or heterodimers that signal via canonical Smad‐dependent pathway or induce various non‐canonical signalling routes (Wang et al, 2014; Salazar et al, 2016). In the intestine, the canonical Bmp pathway directly restricts the stemness of IESCs and thus counteracts Wnt signalling (Qi et al, 2017). Additionally, Bmps trigger the differentiation of epithelial cells, in particular enterocytes, along the crypt–villus axis. Bmp2 and Bmp4 are the two most abundant Bmp ligands in the intestine. The combination of Bmp2 and Bmp4 activates genes responsible for lipid uptake (Beumer et al, 2022). Of note, Bmp2 and Bmp4 do not form heterodimers inter se, thus a different mode of action of these two ligands is plausible (Wang et al, 2014). However, the individual roles of Bmp2 versus Bmp4 remain unknown. The transcriptional switch from carbohydrate to lipid metabolism (initiated either by Bmp2 or Bmp4) occurs near the centre of the villus and the resulting subtype of enterocytes are found also in the upper part of the villus, but not at the villus tip (Beumer et al, 2022). Additionally in the enterocytes, Bmps activate c‐Maf, a master regulator of lipid and amino acid uptake (Cosovanu et al, 2022; González‐Loyola et al, 2022). Hence, Bmps trigger the differentiation of epithelial cells, in particular enterocytes, manifested as sequentially activated and zonated gene expression along crypt–villus axis.
Although it is well‐described that along the villus the features of the enterocytes gradually change, we still do not understand how exactly this process is regulated. Is it simply the vaguely described Bmp gradient that is responsible for this process? Why are then various Bmps expressed in the villus, if just one Bmp can form the gradient and guide the entire process? Are the individual intestinal Bmps redundant or interchangeable? Do they act distinctly? How do they contribute to unique features of villus tip enterocytes? To address these questions, we set out to study the role of distinct members of the Tgfβ superfamily, particularly Bmps and Tgfβ, in regulating the differentiation of intestinal cell types and the functional zonation of the villus using in vitro organoids models and in vivo mouse models. We find Bmp2 and Tgfβ induce distinct villus tip programs, while Bmp4 regulates the villus centre genes. Furthermore, we establish a new organoid culture system, enriched in villus tip enterocytes.
Results
Villus mesenchymal cells induce differentiation of intestinal epithelial cells via Bmp
Villus mesenchymal cells corresponding to Pdgfrahigh cells in the small intestine are expected to control differentiation of epithelial cells (Bahar Halpern et al, 2020; Brügger et al, 2020; McCarthy et al, 2020a). To understand how they influence the fate of the epithelial cells and to explore the role of factors secreted by them, we generated an immortalized small intestinal Pdgfrahigh cell line.
Despite the long‐term in vitro cultivation and immortalization procedure (4–6 weeks), the immortalized villus mesenchymal cells (ivMCs) retained key features typical for in situ to Pdgfrahigh cells of the small intestine such as the expression of Bmps and the absence of canonical Wnts (Fig EV1A and B). Hence, ivMCs cell line can serve as a proxy for Pdgfrahigh cells of the small intestine. In co‐cultivation experiments, ivMCs induced differentiation of freshly isolated intestinal crypts into secretory lineage such as goblet cells (marked by Muc2) and enterocytes (Krt20, Alpi, and Sim—positive); the cell morphology also changed (Fig 1A and B). Importantly, ivMCs promoted the terminal differentiation of enterocytes determined by the expression of villus tip genes (Fig 1B). The induction of key villus tip marker gene Laminin3 (Lama3) could be partially abrogated by treatment with the Bmp type I receptor inhibitor LDN‐193189, pointing to the possible role of Bmps in terminal enterocytic differentiation (Fig EV1C and D). This fact together with recently published data (Beumer et al, 2022; Cosovanu et al, 2022; González‐Loyola et al, 2022) directed our attention to study the role of Bmps in more detail.
Figure EV1. Characterization of ivMCs cells.
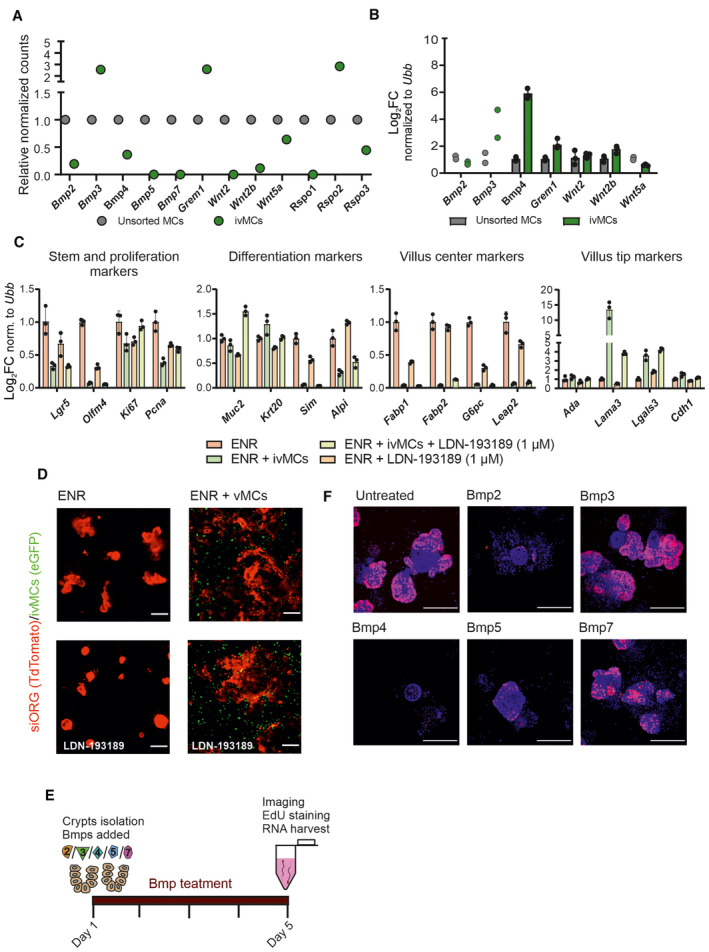
- Immortalized cultured ivMCs still express Bmp ligands as determined by RNAseq. Dots indicate relative normalized counts, unsorted mesenchymal cells (MCs) were freshly isolated and short‐time cultured mesenchymal cells were from PdgfraH2BeGFP animals. Time in culture: 9 weeks after the isolation from PdgfraH2BeGFP animals, 7 weeks after the transduction with immortalization retroviral particles and 3 weeks after sort/establishment of ivMCs.
- qRT–PCR of indicated Bmps and Wnts in ivMCs after 6 weeks in the culture (total culture time since primary MSc isolation—12 weeks). (Expression levels normalized to ubiquitin B (Ubb), unsorted freshly isolated MSCs parallel set as 1, each dot represents one technical replicate of one representative experiment and error bars show SD). If n = 2 technical replicates, error bars and mean are not shown.
- Blocking Bmp type I receptor by LDN‐193189 inhibitor (1 μM) partially revert enterocytic differentiation induced by ivMCs as determined by qRT–PCR. (Expression levels normalized to Ubiquitin B (Ubb), ENR parallel set as 1, each dot represents one technical replicate of one representative experiment and error bars show SD).
- Co‐cultivation of intestinal organoids (siORG, marked by TdTomato) with immortalized villus mesenchymal cells (ivMCs, expressing nuclear eGFP) in the presence of Bmp type I receptor inhibitor LDN‐193189 inhibitor (1 μM). ENR – organoids in cultivation medium (no vMCs), ENR + ivMCs organoids in cultivation medium with ivMCs. (Native fluorescent microscopy, scale bar, 200 μm).
- Scheme of 96 h Bmp treatment of freshly isolated crypts.
- Proliferation of freshly isolated intestinal crypts cultivated with indicated Bmps for 96 h determined by immunostaining for EdU incorporation (red) and DAPI (blue) counterstains nuclei. (Immunocytochemistry, scale bar, 200 μm).
Figure 1. Bmp ligands secreted by villus Mesenchymal Cells are sufficient to boost intestinal epithelial differentiation.

- Co‐cultivation of intestinal organoids (siORG, marked by TdTomato) with immortalized villus mesenchymal cells (ivMCs, expressing nuclear eGFP); timepoint: day 6. Upper panel shows the scheme of the experimental setup. ENR – organoids in cultivation medium (no ivMCs) and ENR + ivMCs organoids in cultivation medium with ivMCs (Native fluorescent microscopy. Scale bar, 200 μm).
- ivMCs induce differentiation of co‐cultured intestinal organoids as determined by qRT–PCR for stem cell (Lgr5, Olfm4) and proliferation (Ki67, Pcna) and differentiation markers (Muc2‐goblet cells, Krt20‐differentiated epithelial cells, Sim and Alpi‐enterocytes). Enhanced terminal enterocytic differentiation was checked by villus centre markers (Fabp1, Fabp2, G6pc and Leap2) and villus tip markers (Ada, Lama3, Lgals3 and Cdh1) (Expression levels normalized to Ubiquitin B (Ubb), ENR parallel set as 1, error bars show SD and each dot represents one technical replicate of one representative experiment). If n = 2 technical replicates, error bars and mean are not shown.
- Expression of individual Tgfβ and Bmp ligands in Pdgfra mesenchymal cells of small intestine (SI). (Dot plot, size of the dot represents the percentage of the cells expressing the transcript and colour intensity indicates the expression level. Only ligands determined to be expressed are shown. Used dataset: GSM3747599 (McCarthy et al, 2020a; Data Ref: McCarthy et al, 2020b))
- The impact of individual Bmp ligands on the growth of freshly established intestinal organoids. (After 96 h of culturing in medium containing 500 ng/ml of indicated Bmp ligand. Brightfield images. Scale bar, 200 μm)
- Proliferation of freshly established intestinal organoids cultivated with indicated Bmp for 96 h determined by quantification of EdU incorporation (% of EdU‐positive cells). Each dot represents one cultured organoid. Average value for each treatment is indicated by line; error bars show SD.
- Relative expression of differentiation (Muc2‐goblet cells, Krt20‐differentiated epithelial cells, Sim and Alpi‐enterocytes) and stem (Lgr5 and Olfm4) markers in intestinal crypts upon 96 h cultivation with indicated Bmp (500 ng/ml). (qRT–PCR, expression levels normalized to Ubiquitin B (Ubb), error bars show SD, untreated parallel set as 1 and each dot represents one technical replicate of one representative experiment).
Source data are available online for this figure.
Re‐analysis of published scRNAseq dataset revealed that sub‐epithelial mesenchymal cells marked by Pdgfra in the small intestine display a complex expression pattern of distinct Tgfβ superfamily members (Fig 1C and Appendix Fig S1). The striking complexity and diversity of Bmps secreted by sub‐epithelial cells lead us to ask if individual Bmps have similar effects on epithelial cells, or if a particular Bmp has its specific role in the regulation of intestinal differentiation.
To address this question, we initially assessed the impact of individual Bmps on gene expression, morphology and cellular composition of organoids. Freshly isolated intestinal crypts were treated by selected recombinant Bmp ligands (Fig EV1E). After 96 h (i.e. time corresponding to the average life of enterocytes in the villus), Bmp‐treated organoids showed distinct changes in shape and survival (Fig 1D), proliferation rate (Figs 1E and EV1F) and gene expression (Fig 1F). The induced changes resembled those observed after ivMCs co‐cultivation with Bmp2 and Bmp4 showing the strongest effect. Based on expressed markers, Bmp2, Bmp4 and Bmp7 promoted differentiation to goblet cells (Muc2) and enterocytes (Krt20, Sim and Alpi). Additionally, the IESC pool was exhausted and proliferation was reduced, mirroring enhanced differentiation at the cost of IESCs (Fig 1F).
Individual Bmps activate distinct enterocytic differentiation programs
During their movement upwards, the enterocytes quickly change their position and could be exposed to particular levels of distinct Bmp ligands only for a short time (Harnik et al, 2021). To model short‐term Bmp exposure and to delineate Bmp‐driven differentiation programs, we treated intestinal crypts (72 after the isolation) with individual recombinant Bmps for 24 h followed by RNA sequencing (Fig EV2A). Short‐term treatment had less impact on the growth and viability of the organoids than 96 h of treatment (Figs EV2B and 1D).
Figure EV2. Bmp2 induces villus tip programs in enterocytes, whereas Bmp4 activates lipid metabolism taking place in the centre of the villus.
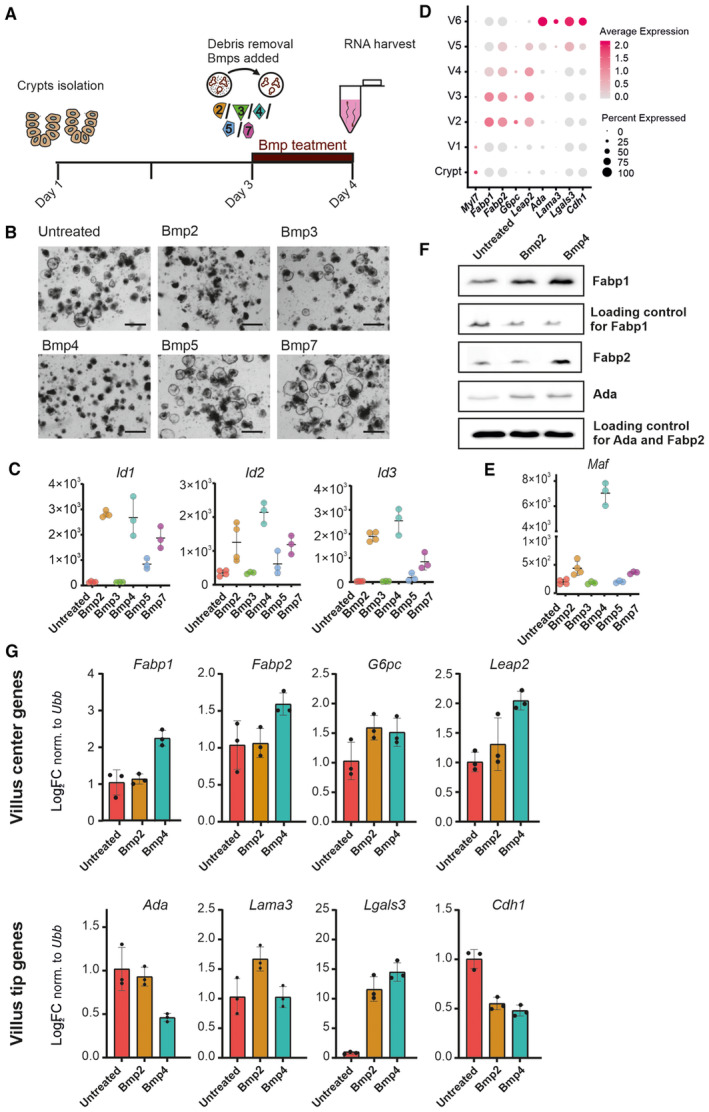
- Scheme of 24 h Bmp treatment of freshly isolated crypts including debris removal step prior to the treatment.
- Brightfield images of freshly isolated crypts treated with 24 h with indicated Bmp (Scale bar, 200 μm).
- Expression of generic Bmp target genes Id1, Id2 and Id3 upon 24 h Bmp treatment. (Graphs show normalized counts determined by RNAseq, n = at least 3 for each treatment and error bars denote SD).
- The expression of indicated villus centre genes and villus tip genes throughout the villus. Dot plot: size of the dot represents the percentage of the cells expressing the transcript and colour indicates the expression level. Used datasets: GSM2644349 and GSM2644350 (Yan et al, 2017a; Data ref: Yan et al, 2017b) reanalysed by Moor et al (2018a), Data ref: Moor et al (2018b).
- The expression of c‐Maf, the master regulator of lipid metabolism and uptake, is initiated preferentially by Bmp4. (Graph shows normalized counts determined by RNAseq, n = at least 3 for each treatment, timepoint: 24 h, error bars denote SD).
- Fabp1 and Fabp2 proteins connected to lipid uptake are expressed upon Bmp4 treatment. Immunoblot of freshly established organoids treated for 48 h as indicated by Bmp2 (500 ng/ml) or Bmp4 (500 ng/ml). Ada is a protein involved in purine metabolism that is expressed at the villus tip. All proteins are expressed after Bmp treatment, but Fapb1 and Fabp2 are specifically regulated by Bmp4.
- qRT–PCR for villus centre genes and villus tip genes in freshly isolated crypts cultivated 24 h in Bmp2 (500 ng/ml) and Bmp4 (500 ng/ml). (Expression levels normalized to ubiquitin B (Ubb), untreated parallel set as 1, each dot represents one technical replicate of one representative experiment, and error bars show SD).
Principal component analysis of differentially expressed genes based on at least three independent experiments revealed that individual Bmps have distinct effects (Fig 2A). Bmp2‐ and Bmp4‐treated samples grouped separately, suggesting a specific downstream transcriptional regulation for each of them. Bmp5‐ and Bmp7‐treated samples grouped together between Bmp2 and untreated samples, pointing to similar but milder effect as compared to Bmp2. Bmp3 overlapped with untreated samples and did not dramatically affect the expression of stem cells or differentiation markers, indicating that Bmp3 had no effect on the organoids. It could be connected to its specific role of acting as a Bmp antagonist rather than the inducer of Bmp‐activated programs (Bragdon et al, 2011; Wang et al, 2014) (Fig 2A). The missing activation capability of Bmp3 was recently shown also for human intestinal organoids (Beumer et al, 2022). Hierarchical clustering, based on the top 10 differentially expressed genes for each treatment, confirmed that Bmp2 and Bmp4 treatments have different genetic outputs (Fig 2B). This differential output is not related to differences in the specific activity of the recombinant Bmps, since both Bmp2 and Bmp4 activated the generic Bmp targets Id1 and Id3 to a similar extent (Fig EV2C). As in the case of long‐term (96 h) treatment also, a short exposure to Bmps (24 h) reduced the stemness and proliferation activity of the intestinal organoids, especially in the case of Bmp2 and Bmp4. On the other hand, short Bmp treatment promoted enterocytic differentiation marked by Krt20 and AldlB (Appendix Fig S2A and B). Interestingly, two enterocytic markers Sim and Alpi were induced by Bmp4 only (Appendix Fig S2A), already suggesting the differences in the impact of Bmp2 versus Bmp4.
Figure 2. Individual Bmps drive distinct differentiation programs in enterocytes.
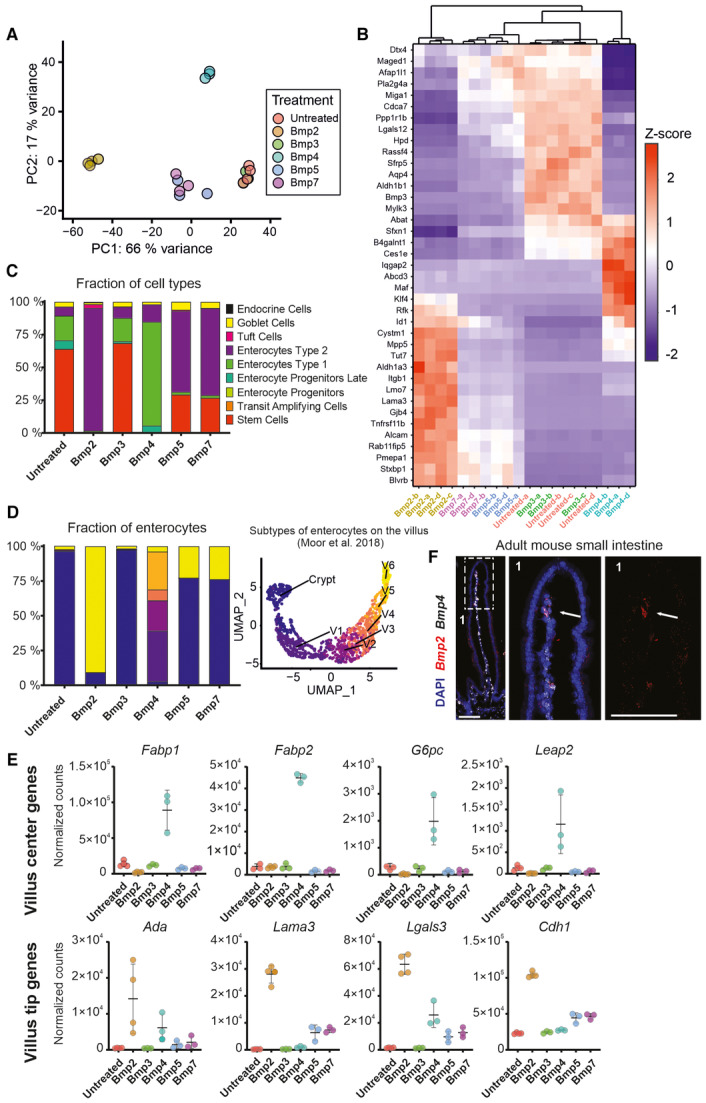
- Principal component analysis (PCA) of the transcriptomes of intestinal organoids treated with indicated Bmp (500 ng/ml) for 24 h. PC1 and PC2 explain 66% and 17% of the variance respectively. (RNAseq samples, n = at least 3 for each Bmp treatment and control).
- Heatmap showing changes in expression of indicated genes after 24 h of indicated Bmp (500 ng/ml) treatment in the intestinal organoids. Hierarchical clustering of Bmp treatments shows differences in gene regulation by Bmp2 versus Bmp4. (n = at least 3 per each treatment and/or control, top 10 differentially expressed genes per each treatment were selected and are shown. Z‐score was calculated using standard formula z = (x – μ) / σ).
- Ratio of intestinal epithelial cell types in organoids treated with indicated Bmp (500 ng/ml) for 24 h. Bmp2 and Bmp4 induce distinct enterocytic programs. (Bar graph depicting RNAseq dataset deconvoluted by CellAnneal software to GSE92332 (Haber et al, 2017a; Data ref: Haber et al, 2017b) organoids scRNAseq dataset as reference).
- Ratio of enterocyte subtypes in the intestinal organoids treated with indicated Bmp (500 ng/ml) for 24 h (left graph). V1 to V6 denote enterocytes according to crypt–villus axis (C = crypt, V1 = close to crypt and V6 = villus tip) as determined by Moor et al, 2018a, and shown on UMAP analysis of 10.5281/zenodo.3403670 dataset (right graph). Colours in the bar graph correspond to enterocyte sub‐type from UMAP. Bar graph depicting RNAseq dataset deconvoluted by CellAnneal software to GSM2644349 and GSM2644350 (Yan et al, 2017a; Data ref: Yan et al, 2017b) scRNAseq reference dataset reanalysed by Moor et al (2018a), Data ref: Moor et al (2018b).
- Whereas Bmp4 triggers expression of villus centre genes connected to lipid metabolism, Bmp2 preferentially activates the expression of villus tip genes. (Graphs show normalized counts determined by RNAseq, n = at least 3 for each treatment, timepoint: 24 h, error bars denote SD).
- Bmp2 is expressed by non‐epithelial cells at the villus tip, whereas Bmp4 is produced along the whole villus. (Single‐molecule RNA hybridization, Bmp2‐red, Bmp4‐white, DAPI‐blue counterstains nuclei, 1 indicate inset shown on the right as magnified and arrows point to Bmp2 signal located at the villus tip mesenchyme, scale bar, 100 μm).
Source data are available online for this figure.
To identify which cellular programs are governed by individual Bmps, we deconvoluted our RNAseq data using Cellanneal package (preprint: Buchauer & Itzkovitz, 2021). We used a single‐cell RNA‐sequencing (scRNAseq) dataset generated from untreated wild‐type small intestinal organoids as a reference (Haber et al, 2017a; Data ref: Haber et al, 2017b). The deconvolution allowed us to determine cellular composition of the RNAseq samples from organoids upon Bmp treatments. All the epithelial cell types (including Enterocyte Type 1 and Enterocyte Type 2) were already annotated in the reference scRNAseq dataset (Haber et al, 2017a; Data ref: Haber et al, 2017b). In general, the short‐term Bmp treatment induced enterocyte differentiation, most likely at the expense of IESCs and transit‐amplifying cells (Fig 2C). Interestingly, Bmp2 treatment increased the Enterocyte Type 2 signature, corresponding to terminally differentiated enterocytes, whereas Bmp4 promoted differentiation into Enterocyte Type 1, characterized by the expression of metabolic genes. Bmp5 and Bmp7 acted like Bmp2, but they did not reduce the number of IECs so drastically. In addition to triggering an Enterocyte Type 2 program, they stimulated differentiation into goblet cells (Fig 2C). We further focused our comparison mainly on Bmp2 and Bmp4, since they had the greatest impact on the organoids and showed a similar activation of generic targets, while depicting remarkable differences in inducing distinct cell populations of enterocytes.
Moor and colleagues assigned enterocytes identities to the enterocytes based on their position along the crypt–villus axis (Crypt, V1–V6) (Moor et al, 2018a) (Fig EV2D). Taking advantage of this work, we also deconvoluted our RNAseq using the Moor dataset (Moor et al, 2018a; Data ref: Moor et al, 2018b). Based on this framework, Bmp2 induced differentiation into villus tip enterocytes (V6), whereas Bmp4 triggered differentiation into villus base and centre enterocytes (V1‐V4) (Fig 2D). As Bmp2 and Bmp4 induce the enterocytic differentiation, they both affected the expression of antimicrobial genes (encoding antimicrobial peptides) expressed at the bottom of the villus (Moor et al, 2018a; Harnik et al, 2021) in repressive manner (Appendix Fig S3A). Many of the Bmp4‐induced genes encode for important regulators of lipid metabolism and absorption, such as fatty‐acid‐binding proteins Fabp1 and Fabp2 (Figs 2E, 3C, and EV2F), as well as a master regulator of amino acid and lipid uptake Maf (Cosovanu et al, 2022; González‐Loyola et al, 2022) (Fig EV2E). In the same line, the genes‐encoding proteins involved in the formation of chylomicrons (i.e. ultra‐low‐density lipoprotein particles) were upregulated by Bmp4 (Appendix Fig S3B). In contrast to Bmp4, Bmp2 preferentially stimulated the expression of villus tip genes (V6 fraction) associated with adhesion such as Laminin3 (Lama3), E‐cadherin (Cdh1), Galectin3 (Lgals3) or modulation of immune system Adenosine deaminase (Ada) (Fig 2E). The differential activation of Fabp1, Leap3 and Lama3 was confirmed by real‐time qPCR (Fig EV2G).
Figure 3. Bmp2 treatment induces the expression of villus tip proteins, whereas Bmp4 treatment leads to upregulation of Fabp2 connected to lipid metabolism in the intestinal organoids.

- Bmp2 treatment induces the expression of the villus tip markers Nt5e (Cd73 protein). (Graph shows normalized counts determined by RNAseq, n = at least 3 for each treatment, timepoint: 24 h, error bars denote SD).
- Scheme of the experimental pipeline used for immunostaining of Bmp‐treated organoids. (Scale bar, 100 μm).
- Expression of Cd73 (Nt5e gene) (green) and Fabp2 (red) upon the Bmp2 and Bmp4 treatment in the intestinal organoids. Immunocytochemistry, DAPI (blue) counterstains nuclei, timepoint: 48 h. (Scale bar, 100 μm).
- Quantification of Cd73 (Nt5e gene) and Fabp2 immunostainings determined as the ratio of specific signal to DAPI. (Each dot represents one organoid, timepoint: 48 h, n = at least 20 organoids, graphs show: min., first quartile, median, third quartile, max., P‐values counted using t‐test).
- Relative Ada enzymatic activity in intestinal organoids after Bmp2 and Bmp4 treatment. (Each dot represents individual measurement, timepoint: 48 h, SD).
Source data are available online for this figure.
Taken together, these observations suggest that Bmp2 activates villus tip programs and Bmp4, a broader central villus program. To explore this further, we looked at the expression pattern of the Bmp2 and Bmp4. mRNA in situ hybridization (RNAscope) revealed that Bmp4 was expressed in non‐epithelial cells from the centre to the top of the villus. In contrast, but consistent with its proposed action at the villus tip, Bmp2 is produced by a few cells at the tip of the villus (Fig 2F).
Besides enhanced expression of adhesive molecules, the villus tip enterocytes specifically express proteins acting as immunomodulators such as Ada and Cd73 (encoded by Nt5e gene) involved in catabolism of purines (Moor et al, 2018a). The active transcription of these proteins at the villus tip correlates with the protein expression (Moor et al, 2018a; Harnik et al, 2021). Bmp2 treatment specifically induced the transcription of Ada and Nt5e (Cd73) (Figs 2E and 3B). In contrast to Bmp4, Bmp2 increased the presence of Cd73 (Nt5e)‐positive cells in the intestinal organoids (Figs 3A,C, and D, and EV3). The enzymatic activity of Ada converting adenosine to inosine is enhanced upon Bmp2 treatment, suggesting increased level of the active protein (Fig 3E).
Figure EV3. Bmp2 treatment leads to enhanced expression of Cd73 protein after 24 h.
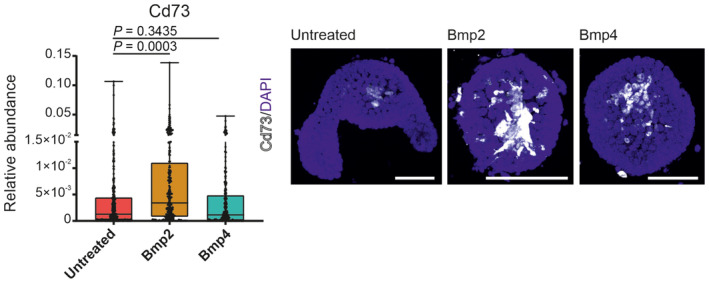
Quantification of Cd73 (gene Nt5e) immunostainings determined as the ratio of specific signal to DAPI. (Each dot represents one organoid, n = at least 20 organoids, t‐test, graphs show: min., first quartile, median, third quartile and max). Expression of Cd73 (Nt5e gene) (white) upon the Bmp2 and Bmp4 treatment in the intestinal organoids. Immunocytochemistry, DAPI (blue) counterstains nuclei. (Scale bar, 100 μm).
Altogether, these observations support a positional effect in which subset of villus mesenchymal cells control the villus tip program in enterocytes by Bmp2.
Tgfβ promotes villus tip differentiation of enterocytes similar to Bmp2
Besides Bmps, Pdgfrahigh villus mesenchymal cells express Tgfβ (Fig 1C). The RNAseq also revealed that Bmp2 and Bmp5/7 (forming heterodimers with Bmp2) treatments activate the expression of Tgfβ1 in the intestinal organoids (Fig EV4A). Interestingly, in advanced colon cancer, Tgfβ secreted by surrounding mesenchymal cells initiates a Wnt independency in colon cancer cells (Han et al, 2020). To assess the role of Tgfβ on healthy small intestinal epithelium, we treated intestinal organoids with Tgfβ ligand using the same setup as for Bmps. The impact of Tgfβ on the growth and morphology of the organoids was analogous to that of Bmps (Figs 4A and B, and EV4B and C). Tgfβ repressed stemness and proliferation, while induced epithelial differentiation (Appendix Fig S4A and B). Tgfβ seems to activate a program rather similar to that triggered Bmp2 (respectively, Bmp2/5/7) (Figs 4B and C, and EV4C–E, Appendix Fig S4B). Additionally, we saw that Tgfβ treatment reduced the expression of villus centre genes (Figs 4C and EV4F and H). We hypothesize this was because Tgfβ promoted the differentiation of villus tip enterocytes (Enterocytes Type 2), reducing the number of villus centre enterocytes and consequently the overall levels in these RNAseq experiments (Fig EV4D). Activation of villus tip genes and reduced villus centre signature upon Tgfβ treatment were apparent also at protein level (Appendix Fig S4B). Of note is the increased fraction of tuft cells (Fig EV4E), observed also in the case of Bmp2 treatment (Fig 2C).
Figure EV4. Tgfβ represses villus centre genes, but it activates villus tip genes.

- Tgfb1 expression is induced by Bmp2 treatment in freshly established intestinal organoids. (Graph shows normalized counts determined by RNAseq, n = at least 3 for each treatment, timepoint: 24 h, error bars denote SD).
- Brightfield images of freshly isolated crypts treated for 96 h with Tgfβ. (Scale bar, 200 μm).
- Principal component analysis (PCA) of the transcriptomes of intestinal crypts treated with Tgfβ for 24 h. PC1 and PC2 explain 96 and 2% of the variance respectively. (RNAseq samples, n = 3 for each treatment and control).
- Expression of generic Bmp target genes Id1, Id2 and Id3 in freshly established intestinal organoids cultivated for 24 h with Tgfβ. (Graph shows normalized counts determined by RNAseq, n = at least 3 for each biological/independent treatment, timepoint: 24 h, error bars denote SD).
- Proportional changes in indicated epithelial cell types in the freshly established intestinal organoids treated with Tgfβ (500 ng/ml) for 24 h. (Bar graph depicting deconvoluted RNAseq dataset compared by CellAnneal software to GSE92332 organoids scRNAseq dataset).
- qRT–PCR for villus centre genes and villus tip genes in freshly established intestinal organoids cultivated 24 h with Tgfβ (500 ng/ml). (Expression levels normalized to ubiquitin B (Ubb), n = 3, untreated parallel set as 1, error bars show SD).
- Venn diagrams of intersecting upregulated or downregulated genes |log2FC| ≥ 2, P < 0.05 comparing Tgfβ and Bmp2 treatment.
- The expression of Fabp1 and Fabp2 proteins connected to lipid uptake is decreased in freshly established organoids treated for 48 h by Tgfβ (500 ng/ml). (Immunoblot).
- Expression of Tgfβ/Bmp family members in freshly established intestinal organoids cultivated with Tgf‐β (500 ng/ml). (Graph shows normalized counts determined by RNAseq, n = 3 for each treatment, timepoint: 24 h error bars denote SD).
- Expression of Tgfβ/Bmp family members in freshly established intestinal organoids cultivated with indicated Bmps (500 ng/ml). (Graph shows normalized counts determined by RNAseq, n = 3 for each treatment, timepoint: 24 h error bars denote SD).
Figure 4. Tgfβ promotes differentiation program similar to Bmp2 and different from Bmp4.

- Effect of Tgfβ treatment (after 24 h, 500 ng/ml) on the morphology and growth of freshly established intestinal organoids. (Brightfield, scale bar, 200 μm).
- Principal component analysis (PCA) of the transcriptomes of intestinal organoids treated with Tgfβ integrated into independent Bmps treatment experiment. PC1 and PC2 explain 69% and 17% of the variance respectively. (RNAseq samples, n = 3 for each treatment and control, Tgfβ, timepoint: 24 h).
- Tgfβ induces the expression of villus tip genes and represses villus centre genes. (Graphs show normalized counts determined by RNAseq, n = 3 for each treatment, timepoint: 24 h, error bars denote SD).
- Relevant upregulated and downregulated gene ontology (GO) terms and KEGG pathways based on differentially expressed genes with |log2FC| ≥ 2, P < 0.05, as calculated using Cytoscape platform (STRING App).
- Venn diagrams of intersecting upregulated or downregulated genes |log2FC| ≥ 2; P < 0.05 indicate high overlap between genes regulated by Tgfβ and Bmp2, but low overlap in genes simultaneously regulated by Tgfβ and Bmp4.
Source data are available online for this figure.
The dichotomy of programs regulated by Bmp family members led us to further characterize the impact of Bmp2 and Tgfβ on one hand and Bmp4 on the other, in terms of affected biological processes and altered signalling pathways. As shown before, Bmp4 preferentially upregulated genes connected to metabolism (lipid metabolism) and nutrient absorption, whereas Bmp2/Tgfβ altered cell adhesion, cell migration and locomotion (Fig 4D). Almost 80% of genes upregulated by Tgfβ were also activated by Bmp2, pointing to their overlapping activity (Figs 4E and EV4G). On the contrary, the overlap in upregulated genes between Tgfβ and Bmp4 was lower (Fig 4E). Hence, GO (Gene Ontology) term analysis and KEGG (Kyoto Encyclopaedia of Genes and Genomes) signalling pathway analysis confirmed that whereas Bmp4 governs differentiation of villus centre enterocytes by triggering genes important for lipid metabolism and uptake, Bmp2 and Tgfβ alter the cellular adhesion activating villus tip programs. Based on our RNAseq uncovered, Tgfβ treatment may induce its own expression in the organoids, suggesting a positive feedback loop (Fig EV4I and Datasets EV1 and EV2). We also checked, how individual Bmps affected their epithelial expression (Fig EV4J).
Villus tip differentiation driven by Bmp2 depends on canonical BmpRI/Smad4 pathway
Bmp ligands can act via the canonical mechanism that starts upon the binding of Bmp to heterotetrameric complexes formed by Type I and Type II receptors. Interaction of ligand with the receptor complex initiates a cascade of phosphorylation events, ultimately resulting in the phosphorylation of receptor‐regulated Smad (R‐Smad1/5/8). Phosphorylated R‐Smad associates with core Smad4. Consequently, the Smad complex translocates to the nucleus where it triggers the expression of target genes. However, Bmps can also induce target gene expression via various non‐canonical, Smad‐independent ways (Wang et al, 2014; Salazar et al, 2016). To dissect whether Bmp2 activates villus tip enterocytic programs via the canonical pathway, we administered Bmp receptor Type I inhibitor LDN‐193189 to wild‐type mice (Fig 5A). Short‐term repression of Bmp receptor complex (4 days) altered villus tip differentiation: reduced expression of villus tip marker gene Ada (Figs 5B–D and EV5A) and Lama3 (Figs 5D and EV5A) was observed, pointing to the role of Bmps in regulation of their expression and thus in the induction of villus tip fate. The expression of villus centre gene Fabp2 was mildly repressed by blocking Bmp receptor Type I, confirming the previous findings (Beumer et al, 2022). Of note is the fact that the epithelial morphology and proliferation stayed preserved after LDN‐193189 treatment, showing intact villi and proliferating crypts (Fig EV5B). As villus tip genes showed stronger repression in comparison to villus centre genes, it is possible that the Bmp2‐driven villus tip program is more sensitive to the pathway perturbations. On the other hand, zonated gene expression within the centre of the villus (controlled preferentially by Bmp4) might be more resilient. An alternative explanation is that LDN‐193189 was not able to penetrate as effectively the central villus region as to villus tip. The requirement of Bmp receptor complex to trigger villus tip gene expression/program corroborates the results from the organoid‐ivMCs co‐cultivation model (Fig EV1C).
Figure 5. The inhibition of Bmp type I receptor blocks the differentiation of intestinal enterocytes.

- Scheme of Bmp type I receptor inhibitor LDN‐193189‐4HCl administration.
- Four days of treatment with LDN‐193189‐4HCl (50 mg/kg) reduces the expression of Ada (green) protein at the villus tips. (Immunohistochemistry, DAPI (blue) counterstains nuclei and E‐cadherin (white) marks epithelial cells, scale bar, 100 μm).
- Quantification of Ada signal range at the villus tip as visualized in (B). (Each dot represents one villus, n = at least 20 villi, graphs show: min., first quartile, median, third quartile, max., P‐values counted using t‐test).
- Changes in the expression of villus tip and villus centre genes after LDN‐193189‐4HCl (150 mg/kg) treatment determined by qRT–PCR normalized to β‐actin (β‐act). (Each dot represents one technical replicate of one representative experiment, error bars show SD).
Source data are available online for this figure.
Figure EV5. Blocking Bmp receptor type I represses the expression of villus tip genes, Bmp2‐induced differentiation is mediated through Smad4, and long‐term cultivation of intestinal organoids results in reduced differentiation state, but enhances stem cell/proliferating state.
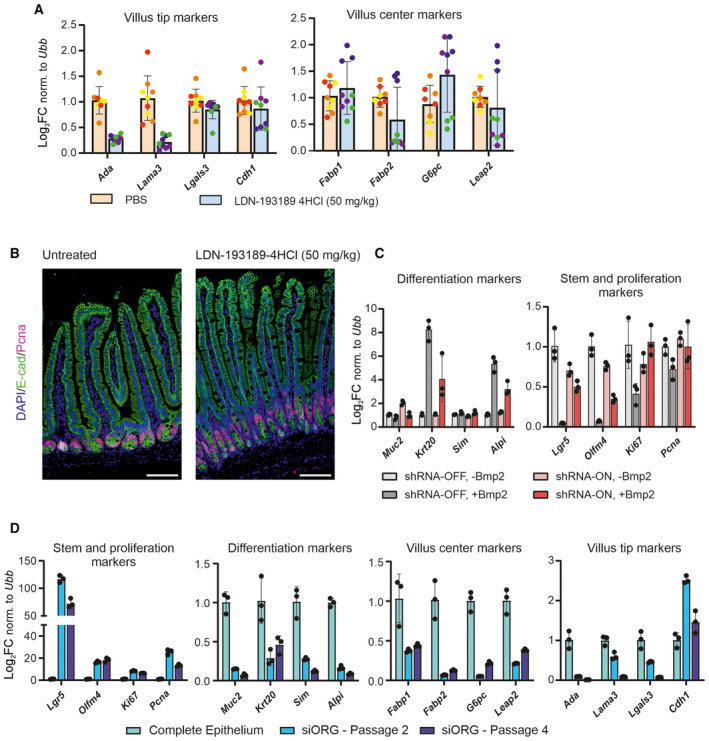
- Four‐day treatment with LDN‐193189‐4HCl (50 mg/kg) inhibitor does not significantly affect the expression of the villus centre genes but it decreases the expression of villus tip genes. (qRT–PCR, expression levels normalized to ubiquitin B (Ubb), n = 3 animals in each condition, error bars show SD and untreated parallel set as 1).
- The epithelial morphology and active proliferation after the 4 days of treatment with LDN‐193189‐4HCl (50 mg/kg) inhibitor. Immunofluorescence, E‐cadherin (green) marks epithelial cells, Pcna (red) stains proliferating cells and DAPI (blue) counterstains nuclei. (Scale bar, 200 μm).
- Smad4 knockdown (shRNA‐ON) alleviates Bmp2‐induced differentiation marked by relative expression of differentiation markers (Muc2, Krt20, Sim and Alpi) and restores the expression of stem cell and proliferation genes (Lgr5, Olfm4, Ki67 and Pcna). (qRT–PCR, expression levels normalized to ubiquitin B (Ubb), n = 3, error bars show SD, untreated parallel set as 1 and treatment as in Fig 6A–C).
- qRT–PCR for indicated genes in complete intestinal epithelium and intestinal organoids cultivated in standard medium after passage 2 (circa 2 weeks after the isolation), respectively, passage 4 (circa 4 weeks after the isolation). (Expression levels normalized to Ubiquitin B (Ubb), n = 3, untreated parallel set as 1, error bars show SD).
To further determine whether Bmp2‐induced enterocytic differentiation requires canonical Smad‐dependent route, we generated organoids with a doxycyclin‐inducible shRNA against Smad4 (shRNASmad4). Despite not all organoids expressed shRNASmad4 as marked by coupled TdTomato, the doxycycline induction reduced the total expression of Smad4 (Fig 6A and B). When the organoids with reduced Smad4 were treated with Bmp2 for 24 h, the activation of villus tip genes was lower than in wild‐type organoids (Fig 6C). The Smad4 depletion also restored proliferation and alleviated IESCs exhaustion caused by Bmp2 treatment (Fig EV5C). Importantly, in contrast to freshly isolated crypts used for the previous organoid experiments, shRNASmad4 organoids spent long time in the culture, were transduced and passaged many times. Nevertheless, they still responded to Bmp2 treatment by activating villus tip program dependent on core Smad4.
Figure 6. Villus tip differentiation induced by Bmp2 depends on Smad4 and can be achieved in long‐term organoid culture through continual low doses of Bmp2.
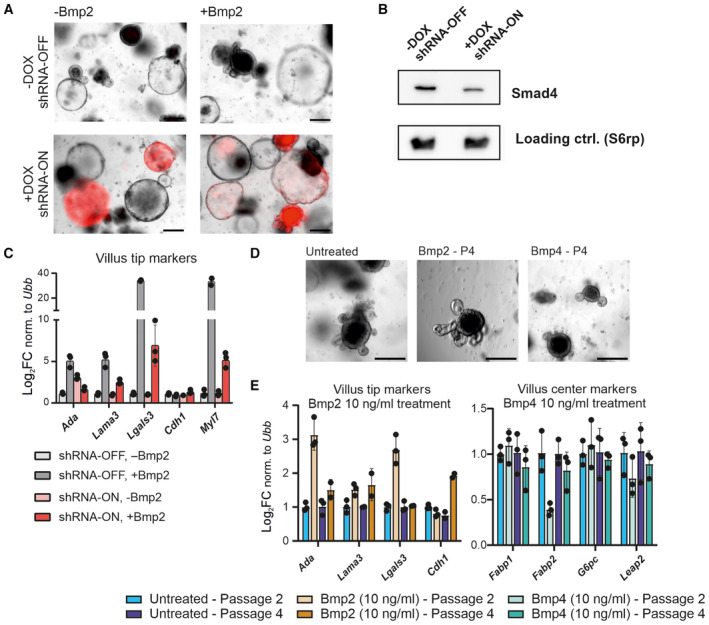
- Induction of shRNA against Smad4 upon doxycycline (+Dox) treatment (3 days), followed by Bmp2 treatment for additional 24 h. shRNA activation is visualized by the expression of coupled TdTomato (red), scale bar, 200 μm.
- The shRNA against Smad4 (+Dox / shRNA‐ON) leads to reduced levels of Smad4. (Immunoblot, after 4 days ± Dox).
- Smad4 knock‐down (shRNA‐ON) alleviates Bmp2‐induced differentiation marked by relative expression of villus tip genes. (qRT–PCR, expression levels normalized to Ubiquitin B (Ubb), each dot represents one technical replicate of one representative experiment, error bars show SD, untreated parallel set as 1, treatment as in A).
- Intestinal organoids cultivated for four passages (circa 4 weeks) in the presence of Bmp2 or Bmp4 (10 ng/ml) are viable. (Brightfield images, scale bar, 200 μm).
- Long‐term (4 weeks) organoid cultivation in the presence of Bmp2 (10 ng/ml) enriches the expression of villus tip genes. (qRT–PCR, expression levels normalized to Ubiquitin B (Ubb), each dot represents one technical replicate of one representative experiment, error bars show SD, untreated parallel set as 1, P2—collected 4 days after second passage and P4—collected 4 days after fourth passage).
Source data are available online for this figure.
Mimicking the conditions at the villus tip via long‐term cultivation in the presence of Bmp2
Long‐term cultivated intestinal organoids do not completely recapitulate intestinal epithelium: they rarely contain villus tip cells and most enterocytes have a villus bottom identity (Grün et al, 2015; Haber et al, 2017a; Fujii et al, 2018). To enrich villus tip cells and make organoids more similar to real intestinal epithelium, at least in certain aspects, we tried to establish long‐term organoid culture using medium containing half concentrations of Bmp2 (250 ng/ml). Bmp2‐treated organoids could survive up to two passages. Using even lower Bmp2 concentration (10 ng /ml), the organoids could be propagated in Bmp2‐containing medium for minimally 4 weeks (at least 4 passages). Addition of Bmp2 to standard cultivation medium enriched for villus tip signatures around passage 2 (Fig 6D and E), again underscoring the role of Bmp2 as an inducer of villus tip expression in the enterocytes. Hence, we established a system suitable to study various aspects of villus tip biology in the future. Although this system is still far from in vivo situation, it can serve as a tool to study enterocytic differentiation, as under the standard cultivation conditions organoids lost some villus tip markers (the expression of other is strongly reduced) already around passage 2, whereas are enriched for stem cells and proliferating cells (Fig EV5D).
Bmp2 potentially cooperates with Wnt5a to promote the villus tip differentiation of enterocytes
A specific subset of villus mesenchymal cells—Lgr5‐positive mesenchymal cells—are localized in the villus tip. These cells express Bmp2, Bmp4 and Wnt5a (Bahar Halpern et al, 2020). Bahar Halpern and colleagues showed that the elimination of these cells led to loss of villus tip identity in enterocytes (Bahar Halpern et al, 2020). In our experiments with a short‐term inhibition of Bmp receptor type (Fig 5B–D), the villus tip identity was also lost. These data suggest that Bmp2 could be the main factor controlling this identity. Our experiments also suggest a synergistic action of Bmp2 and Wnt5a, and partial role of Wnt5a in activating villus tip genes (Fig 7A–C). Further research is needed to confirm this putative synergy and to elucidate the exact mechanism.
Figure 7. Wnt5a and Bmp2 jointly induce enterocytic differentiation.

- qRT–PCR for villus tip genes and villus centre genes in freshly established organoids cultivated 24 or 96 h with Bmp2 (500 ng/ml), Wnt5a (500 ng/ml) or both Bmp2 + Wnt5a (each 500 ng/ml). Whereas Bmp2 induces villus tip genes on its own, it can act synergistically with Wnt5a. For the short‐term cultivation, the effect is specific for villus tip genes (notice the different scale in left graphs). (Expression levels normalized to Hmbs, untreated parallel set as 1, each dot represents one technical replicate of one representative experiment and error bars show SD). If n = 2 technical replicates, error bars and mean are not shown.
- Expression of E‐cadherin (red) and Bmp2, Bmp4, Wnt5a and Bmp2 + Wnt5a treatment in the intestinal organoids. Immunocytochemistry, DAPI (blue) counterstains nuclei. (Immunocytochemistry, timepoint: 24 h, 500 ng/ml each protein, scale bar, 200 μm).
- Quantification of E‐cadherin immunostainings determined as the ratio of specific signal to DAPI. (Each dot represents one organoid, n = at least 20 organoids, t‐test, graphs show: min., first quartile, median, third quartile= and max).
Source data are available online for this figure.
Discussion
Proper spatial and temporal control of cellular differentiation is an ultimate requirement for normal intestinal functions. The balance between continual supply of IESCs and differentiation of epithelial progenitors depends on the surrounding non‐epithelial cells which create an adequate microenvironment. Whereas the role of an intestinal stem cells niche supporting the maintenance of IESCs has been clearly documented in recent years (Degirmenci et al, 2018; Greicius et al, 2018; McCarthy et al, 2020a), the existence and function of a “differentiation niche” remain enigmatic. However, the existence of such a niche is flagged by the presence of villus‐localized Pdgfrahigh mesenchymal cells expressing mixture of Bmps. Bmp signalling acts as a negative regulator of crypt formation, restricting the stemness (Haramis et al, 2004; Qi et al, 2017). The epithelial differentiation, however, is not a default state that is automatically triggered in the absence of the stem cell programs. The lack of IESCs‐promoting signals is not sufficient to achieve a successful differentiation. Indeed, Bmps are needed to actively promote epithelial differentiation along the crypt–villus axis. Bmps produced by sub‐epithelial villus mesenchymal cells contribute to the formation of sequential and zonated gene expression in enterocytes (Beumer et al, 2022). The Bmp gradient along the villus is classically thought to trigger the sequential gene expression and thus the differentiation (Spit et al, 2018; Gehart & Clevers, 2019). But why are there so many distinct Bmps expressed by villus mesenchymal cells if the gradient could simply be formed by a single ligand? What if the identity of the individual Bmps also matters? By highlighting distinct roles of different Bmp ligands, our results demonstrate the importance of their individual identity in the differentiation process.
Two highly expressed Bmps are Bmp2 and Bmp4. A combination of Bmp2 and Bmp4 was shown to push the differentiation of enterocytes by activating genes involved in lipid metabolism and uptake (Beumer et al, 2022). Two studies highlighted c‐Maf as a key regulator of this metabolic differentiation (Cosovanu et al, 2022; González‐Loyola et al, 2022). One study showed that Bmp2 induces expression of c‐Maf, while the other described Bmp4 as the trigger (both in vitro) (Cosovanu et al, 2022; González‐Loyola et al, 2022). We also found that both Bmp2 and Bmp4 can induce Maf but that Bmp4 was much more potent (Fig EV2E). Crucially, however, our transcriptome analysis suggests that Bmp2 and Bmp4 trigger distinct programs. Whereas Bmp4 governs the lipid metabolism and uptake, Bmp2 regulates the cellular locomotion, adhesion and expression of immunomodulators (Figs 2E and 3B–E). The activation of distinct programs can be connected to expression pattern of Bmps. Whereas Bmp4 is expressed along the villus, Bmp2 is produced in few cells at the very tip of the villus (Fig 2F). The pattern of expression corresponds to enterocytic zonation—enterocytes in the middle to the top of the villus express metabolic genes—, whereas villus tip enterocytes are characterized by changed adhesion and production of immunomodulators (Moor et al, 2018a; Harnik et al, 2021) (Fig 2D and E).
The role of Bmp2 in priming villus tip identity goes in line with the observation that rare Lgr5‐positive cells localized at the villus tip promote the terminal differentiation of enterocytes (Bahar Halpern et al, 2020). Besides Bmp2 and Bmp4, Lgr5‐positive cells also secrete Wnt5a. Our pilot experiments suggest the potential of Wnt5a to induce villus tip features in parallel to Bmp2. Importantly, there might be a synergy between Bmp2 and Wnt5a in this process (Fig 7A–C). The fact, that Wnt5a could not induce the expression of villus centre genes (lipid metabolism and uptake) (Beumer et al, 2022) may point to specific synergism between Bmp2 and Wnt5a since Bmp2 is not expressed at the centre of the villus. Nevertheless, as mentioned, further research (beyond this work) is required to confirm or disprove this phenomenon.
Villus tip epithelial identity is triggered by Bmp2 in canonical, Smad‐dependent way (Fig 6A–C). The enterocytes quickly move upwards along the villus. Thus, time dictates how long the distinct Bmp signalling can act. Whereas the short‐term exposure drives preferentially enterocytic programs (Figs 2C and D, 3B–E, and EV2A), the longer exposure additionally drives the differentiation into goblet cells (Fig 1F), as also shown recently in an independent study (Beumer et al, 2022).
Bmp5 and Bmp7 are the other ligands expressed by Pdgfrahigh villus mesenchymal cells. They triggered a program similar to that elicited by Bmp2 (Figs 2D and E, and EV2A). This goes in line with what is known about Bmps homo‐ or heterodimerization. Bmp5 heterodimerizes just with Bmp2, whereas Bmp7 can form heterodimers with both Bmp2 and Bmp4. On the other hand, Bmp2 does not heterodimerize with Bmp4 (Bragdon et al, 2011; Wang et al, 2014; Salazar et al, 2016). This may indirectly support the distinct roles of Bmp2 vs. Bmp4. Regulation of adhesive properties can also be modulated by Tgfβ, acting similarly to Bmp2 on healthy organoids. Of note, also Tgfβ elicited a differentiation program similar to that of Bmp2 (Figs 3C and E, and EV4F and G). Tgfβ seems to act mainly as immunoregulator in the healthy intestine (Bauché & Marie, 2017; Stolfi et al, 2020), but older reports indicate it can affect the intestinal epithelium, consistent with our observations. Tgfβ enhances the cellular adhesion of intestinal epithelial cell lines grown as monolayer in vitro (Howe et al, 2005). In the colon, Tgfβ promotes the regeneration of the intestinal epithelium after mechanical damage (Miyoshi et al, 2012). The mechanisms at least partially depend on affecting the focal adhesion that limits the proliferation, finally facilitating the reappearance of new colonic crypts (Miyoshi et al, 2012). Hence, Tgfβ may act as an alternative regulator of cellular adhesion, activating the gene signature present in villus tip cells.
Another important aspect of our work is the experimental design and establishment of an appropriate organoid culture system. Since intestinal organoids cultivated long‐term under the standard conditions do not recapitulate the composition of differentiated enterocytes (Grün et al, 2015; Haber et al, 2017a; Beumer et al, 2022), we used freshly isolated crypts to keep the responsiveness to differentiating stimuli and respectively established a longer‐term cultivation system. As the lower concentration of Bmp2 used in long‐term experimental set‐up showed a similar trend as originally used higher concentration, it points to ligand specificity of particular Bmps rather than to the concentration threshold. This may be in line with findings indicating (in other systems) that not only the concentration but also the different combination of Bmp ligands drive various cellular responses due to distinct context‐dependent perception by receptor complexes (Antebi et al, 2017; Klumpe et al, 2022). In fact, the distribution of Bmp/Tgfβ receptors and their contribution to processing distinct Bmp stimuli should be studied further.
Taking together, we showed that individual Bmp ligands secreted by sub‐epithelial mesenchymal cells can induce distinct differentiation programs, especially in enterocytes. Whereas Bmp4 activates lipid metabolism and uptake in the centre of the villus, Bmp2 modulates properties of villus tip enterocytes, governing their terminal differentiation. The specific role of Bmp2 can be connected to its expression at the villus tip. Hence, the division of labour in enterocytes may at least partially mirror the specific action and expression of particular Bmps.
A limitation of our study is that the key experiments are based on freshly established intestinal organoids, while the situation in vivo may be more complex. However, the in vivo villus tip‐specific expression of Bmp2 correlates with the gene signature of villus tip enterocytes that is achieved also by Bmp2 treatment in organoids. Another qualifier that needs to be noted is that the study focuses on proximal small intestine. Finally, the work focuses to describe the impact of individual roles of Bmps on the gene expression of the epithelial cells. Hence, the mechanism of how distinct expression of Bmps is controlled is not part of this work.
Materials and Methods
Reagents and Tools table
| Reagent/Resource | Reference or Source | Identifier or Catalog Number |
|---|---|---|
| Experimental Models | ||
| M. musculus strain: C57BL/6 (wild type) | Mice were provided from general mouse stock at IMG mouse facility | |
| M. musculus strain: B6.129S4‐Pdgfratm11(EGFP)Sor/J (Pdgfrα‐eGFP) | The Jackson Laboratory (www.jax.org) | 007669 |
| M. musculus strain: ROSA‐nonSTOP‐TdTomato | This strain was generated for this project as a result of a one‐time crossing of ROSA‐TdTomato strain and Actin‐Cre strain. The first parental ROSA‐TdTomato strain is available at The Jackson Laboratory (www.jax.org) under the number 007908 and the second parental strain can be found under MGI ID 5285392. | |
| M. musculus strain: Lgr5‐EGFP‐IRES‐creERT2 | The Jackson Laboratory (www.jax.org) | 008875 |
| Cell line: Cultrex HA‐R‐Spondin1‐Fc 293T Cells | R&DSystems | 3710‐001‐01 |
| Cell line: mNoggin‐Fc 293T Cells | Provided by Wim de Lau from Clevers group at The Hubrecht institute in the Netherlands. | |
| Cell line: Platinum‐E | Cell Biolabs Inc. | RV‐101 |
| Recombinant DNA | ||
| p1322 HPV‐16 E6, viral insert | Addgene | 8642 |
| pMXs‐Puro Retroviral Expression Vector | Cell Biolabs Inc. | RTV‐012 |
| pTRIPZ | Dharmacon | No number available |
| Antibodies | ||
| Ada (sheep, polyclonal) | R&DSystems | AF7048 |
| Akt (rabbit, monoclonal) | Cell Signaling | 4691 |
| AldlB (rabbit, monoclonal) | Abcam | ab75751 |
| Cd73 (mouse, monoclonal) | Biolegend | 127210 |
| E‐cadherin (mouse, monoclonal) | BD Transduction | 610181 |
| Fapb1 (goat, polyclonal) | R&DSystems | AF1565 |
| Fabp2 (goat, polyclonal) | R&DSystems | AF1486 |
| Ki67 (rabbit, polyclonal) | Abcam | ab15580 |
| Muc2 (mouse, monoclonal) | Santa Cruz | sc‐15334 |
| Pcna (rabbit, polyclonal) | Abcam | ab18197 |
| S6rp (rabbit, monoclonal) | Cell Signalling | 2217 |
| Oligonucleotides and other sequence‐based reagents | ||
| Set of qRT‐PCR primers | This study | Table EV2 |
| Smad4 shRNA oligo pair | This study | Table EV1 |
| Chemicals, Enzymes and other reagents | ||
| Bmp2 | Peprotech | 120‐02C |
| Bmp3 | Peprotech | 120‐24B |
| Bmp4 | Peprotech | 315‐27 |
| Bmp5 | Peprotech | 120‐39 |
| Bmp7 | Peprotech | 120‐03P |
| Tgfβ | Peprotech | 100‐21C |
| Egf | Invitrogen | PMG8043 |
| Noggin | Sigma | SRP3227 |
| R‐spo1 | Peprotech | 315‐32 |
| Corning® Matrigel® Growth Factor Reduced | Corning | 356231 |
| Advanced DMEM/F12 | Invitrogen | 12634‐010 |
| GlutaMax 100× | Invitrogen | 35050‐068 |
| Hepes 1M | Invitrogen | 15630‐056 |
| Penicillin/Streptomycin 10K U/ml 10K μg/ml 100× | Invitrogen | 15140‐122 |
| N2 supplment 100× | Invitrogen | 17502‐048 |
| B27 supplement 50× | Invitrogen | 17504‐044 |
| N‐Acetylcysteine | Sigma‐Aldrich | A9165‐5G |
| Primocin | Invivogen | ant‐pm‐2 |
| Corning® Cell Recovery Solution | Merck | CLS354253 |
| MesenCultTM Expansion Kit (Mouse) | Stemcell Technologies | 05513 |
| TrypLE™ Express Enzyme (1×) | Gibco | 12604013 |
| Gentle Cell Dissociation Reagent | Stemcell Technologies | 100‐0485 |
| Collagenase D | Merck | 11088866001 |
| Dispase | Corning | CLS354235 |
| FBS | Gibco | A52567 |
| DMEM | Home‐made | |
| Opti‐MEM | Gibco | 31985070 |
| Lipofectamine 2000 reagent | invitrogen | 11668019 |
| Lipofectamine 3000 reagent | Invitrogen | L3000015 |
| DMSO | SERVA | 20385 |
| Lenti‐X Concentrator | Clontech Laboratories, Inc. | PT4421‐2 |
| Retro‐X Concentrator | Clontech Laboratories, Inc. | PT5063‐2 |
| LDN‐193189 | Medchemexpress | HY‐12071 |
| LDN‐193189 – 4HCl | Medchemexpress | HY‐12071A |
| Software | ||
| R 4.1.0 | https://www.r‐project.org | |
| RStudio 2022.02.3 | https://www.rstudio.com | |
| heatmaply 1.3.0 |
https://github.com/talgalili/heatmaply (Galili et al, 2017) |
|
| dplyr 1.0.7 |
(Wickham et al, 2023) |
|
| ggplot2 3.3.5 | https://ggplot2.tidyverse.org (Wickham, 2016) | |
| ggVennDiagram 1.2.2 | ||
| Seurat 4.0.6 |
(Hao et al, 2021) |
|
| Fiji (ImageJ) 1.53q |
https://imagej.net/software/fiji/ (Schindelin et al, 2012) |
|
| GraphPad Prism 8.4.3 (686) | https://www.graphpad.com/scientific‐software/prism/ | |
| CorelDRAW 2020 22.1.1.523 | https://www.coreldraw.com | |
| Cytoscape 3. 9. 1 | ||
| stringApp 1.7.1 | ||
| Cellanneal |
https://github.com/LiBuchauer/cellanneal (preprint: Buchauer & Itzkovitz, 2021) |
|
| FlowJo 10.7.1 | https://www.flowjo.com | |
| Other | ||
| Probe – Mm‐Bmp2 | ACD Bio | 406661 |
| Probe – Mm‐Bmp4‐C3 | ACD Bio | 401301‐C3 |
| Positive Control Probe_M | ACD Bio | 320881 |
| Negative Control Probe | ACD Bio | 320871 |
| Fluorescent Mplex Reagent Kit | ACD Bio | 320850 |
| RNeasy Mini Kit | QIAGEN | 74106 |
| RevertAid First Strand cDNA Synthesis Kit | ThermoFisher SCIENTIFIC | K1622 |
| LightCycler 480 SYBR Green I Master | Roche | 04887352001 |
| EdU Cell Proliferation Kit | Baseclick | BCK‐EdU647 |
| Adenosine Deaminase Activity Assay Kit (Fluorometric) | Sigma‐Aldrich | EPI020 |
Methods and Protocols
Mice
Wild‐type (C57BL/6) mice were obtained from our own breeding colony housed at the Institute of Molecular Genetics, Prague, Czech Republic. PdgfraH2BeGFP (Hamilton et al, 2003) was purchased from Jackson Laboratories, USA (Stock number: 007669). TdTomato‐positive crypts were isolated from Actin‐Cre (Gt(ROSA)26Sor(ACTB‐Cre,‐EGFP)) crossed to Ai14 reporter strain (B6;129S6‐Gt(ROSA)26Sortm14(CAG‐tdTomato)Hze/J). Mice were 10–15 weeks old at the time of treatments and crypt isolations, both sexes were used, taken randomly from few cages. No specific blinding was applied.
To block Bmp type I receptors, wild‐type animals were intraperitoneally injected with LDN‐193189‐4HCl (Medchemexpress) dissolved in PBS (50 or 150 mg/kg in 100 μl volume, one injection per day) or vehicle (PBS) for 4 consecutive days. The mice were sacrificed 24 h after the last injection.
Housing of mice and in vivo experiments were performed in compliance with the European Communities Council Directive of 24 November 1986 (86/609/EEC) and national and institutional guidelines. Animal care and experimental procedures were approved by the Animal Care Committee of the Institute of Molecular Genetics (no. 2482/2021). For animal experiments, at least three animals were used in each experimental group (per individual treatment). All presented images (images of organoids and histological staining) were chosen as representative images for each condition.
Mesenchymal cells isolation and immortalization
Mesenchymal cells from duodenum of PdgfraH2BeGFP mouse were isolated as described previously (Degirmenci et al, 2018; Brügger et al, 2020). The duodenal tissue was harvested, opened longitudinally and rinsed. The tissue was gently rocked for 30 min at room temperature in Gentle Cell Dissociation Reagent (Stemcell Technologies) to remove epithelial cells. After the detachment of epithelium, remaining tissue pieces were digested for 35 min in DMEM supplemented with 1 mg/ml collagenase D and 0.3 mg/ml dispase in thermomixer at 37°C with gentle shaking (60 rpm in horizontally oriented 50 ml Falcon tube). Afterwards, cells were filtered through 70 μm cell strainer and seeded. Twenty‐four hours after seeding, non‐adherent cells were washed out. The cells were cultivated in MesenCult medium. The medium was changed three times a week. The mesenchymal cells were immortalized by transduction of (HPV16)‐E6 gene using retroviral particles. For the retroviral production PlatinumE cells were transfected with pMXs‐EF1‐Puro (Cell Biolabs) retroviral construct containing (HPV16)‐E6 insert re‐cloned from p1322HPV‐16‐E6 vector. Lipofectamine 3000 and OptiMEM medium were used for transfection according to manufacturer's protocol. Transfection medium was changed for cultivation medium 6 h after transfection. Viral particles were produced for 72 h. Retroviral particles were concentrated overnight at 4°C by RetroX (Clontech) concentrator and used for infection of mesenchymal cells by spinoculation (30°C, 450 g, 1 h). Seventy‐two hours after the spinoculation, the medium with viral particles was changed and cells were selected with puromycin (1 μg/ml) and further expanded. The immortalized cells were sorted based on internal eGFP fluorescence intensity to eGFPlow eGFPhigh and further separately expanded. Occasionally, the cells were retreated with puromycin. The cells were passaged with dissociation by TripleLE Express solution (Gibco).
Crypt isolation and culture
Crypts from proximal small intestine were isolated and cultured as described (Sato et al, 2009, 2011). The dissected intestine was flushed out and opened longitudinally. Villi were shaved off by cover glass. After repeated washing in ice‐cold PBS, the fragments were gently rocked for 30 min at 4°C in 5 mM EDTA in PBS. Crypts were released by gentle hand shaking for 30 s into ice‐cold PBS, filtered through 70 μm cell strainer and seeded into Matrigel (Corning) domes on 24‐well plates (3 domes/25 μl per well) and cultivated in complete organoid medium supplemented with mEGF (50 ng/ml), mNoggin (100 ng/ml) and 10% of Rspondin1‐conditioned medium prepared using Cultrex HA‐R‐Spondin1‐Fc 293T cell line (R&D systems) according to cell line datasheet—ENR medium. Individual recombinant Bmp proteins (Peprotech) or Tgfβ (Peprotech) (500 ng/ml) were added to ENR medium at the time of seeding and crypts were cultivated with Bmps or Tgfβ for 96 h (with one medium change after 48 h) for initial experiments shown in Figs 1, EV1, and EV4B. For EdU incorporation assay, the medium was changed daily.
For RNAseq and subsequent confirmative quantitative real‐time PCR (qRT–PCR) experiments, the crypts were initially seeded in ENR medium (without any Bmp/Tgfβ). After 48 h, the debris was removed: old medium was removed and Matrigel domes containing organoids were broken with 1 ml plastic tip with cut‐off tip and transferred into 15 ml tube containing 10 ml of ice‐cold PBS. The crypts were incubated 10 min on ice, sedimented and then upper 8 ml of PBS containing cell debris was sucked out and replaced by fresh PBS. Organoids were centrifugated (300 g, 4°C and 5 min) and put into fresh Matrigel domes. Recombinant Bmps or Tgfβ (500 ng/ml) were added for 24 h.
When crypts were cultivated for western blotting, the debris removal step was omitted, and organoids were kept in recombinant Bmps for 48 h without medium change.
For the long‐term villus tip, enriched culture ENRB medium was used, composed of ENR supplemented with Bmp2 or Bmp4 (10 or 250 ng/ml). The organoids were passaged approximately once a week.
EdU staining and image quantification
Duodenal crypts were isolated as mentioned above and seeded on glass‐bottomed eight‐well chamber (iBidi) in 10 μl Matrigel drops for 5 days in ENR medium containing individual Bmps (500 ng/ml). After 96 h, they were incubated in medium containing 10 μM EdU for 45 min and stained with EdU Cell Proliferation Kit (Baseclick) according to manufacturer's protocol. Afterwards, the organoids were counterstained with DAPI. Z‐stack confocal images of individual organoids were taken using Dragonfly 503 spinning disc microscope (Andor). EdU and DAPI signal volumes were equally computed using Fiji software. Finally (EdU overlapping with DAPI)/DAPI ratio (i.e. percentage of proliferating cells) was calculated. For automated quantification, we used our macro written by Light Microscopy core facility at Institute of Molecular Genetics of the Czech Academy of Sciences.
Co‐cultivation of vMCs with intestinal crypts
Intestinal villus mesenchymal cells (ivMCs) corresponding to immortalized eGFPhigh cells were seeded on 96‐well plate 2 days before the addition of organoid fragments. TdTomato‐positive crypts were isolated and cultivated as described above in advance to co‐cultivation start. For the co‐cultivation experiment, organoids were mechanically dissociated by pipetting. Organoid chunks were mixed in 10% Matrigel dissolved in ENR organoid medium. Twenty microlitre of crypt/Matrigel/ERN medium suspension containing fragments from roughly 10–20 organoid fragments were added to ivMCs (70–80% confluent at the time of mixing) or empty wells. The plate was incubated 10 min at 37°C and afterwards gently covered with 100 μl complete ENR organoid medium optionally containing 1 μM LDN‐193189 (Medchemexpress). The co‐cultures were analysed after 3 days of cultivation.
Organoid immunostaining, imaging and image analysis
Organoids were generated from isolated crypts of the murine small intestine as done previously (Yang et al, 2021). Organoids were kept in IntestiCult Organoid Growth Medium (Stemcell Technologies) with 100 μg/ml penicillin–streptomycin for amplification and maintenance. Mechanically split organoids were plated in a 96‐well plate and treated with 500 ng/ml Bmp2 or 500 ng/ml Bmp4 in ENR medium for 24 or 48 h until the fixation. The procedure for immunostaining was performed as described before (Yang et al, 2021). Primary antibody (1:200) of CD73 (BioLegend, RRID: AB_11218786), Fabp2 (Sigma, MABS1694), AldlB (Abcam, ab75751) and secondary antibodies (Thermo Fisher Scientific, 1:2,000) were diluted in blocking buffer and applied as indicated in Yang et al (2021). Cell nuclei were stained with 20 μg/ml DAPI (Invitrogen) in PBS for 10 min at room temperature.
High‐throughput imaging was done with an automated spinning disk microscope from Yokogawa (CellVoyager 7000S), with an enhanced CSU‐W1 spinning disk (Microlens‐ enhanced dual Nipkow disk confocal scanner), a 40× (NA = 0.95) Olympus objective and a Neo sCMOS camera (Andor, 2,560 × 2,160 pixels). For imaging, an intelligent imaging approach was used in the Yokogawa CV7000 (Search First module of Wako software 2.0) as described before (Yang et al, 2021). Z‐planes spanning a range up to 80 μm and 2 μm z‐steps were acquired.
Organoid segmentation in maximum intensity projections (MIPs) was adapted from before (Yang et al, 2021). For each acquired confocal z‐stack field, MIPs were generated. All MIP fields of a well were stitched together to obtain MIP well overviews for each channel. The high‐resolution well overviews were used for organoid segmentation and feature extraction. Organoid segmentation based on DAPI channel was generated by a full convolutional neural network (FCN) as in Yang et al (2021). Artefacts of segmentations were identified by setting size‐threshold and visual inspection, annotated in Fiji (Version 2.9.0) and removed by a customized python script. From the segmented MIPs, we calculated the size of each individual organoid from label masks. Using the individual organoid masks from the MIP, we cropped for each organoid the signals of all recorded channels. Uniformed threshold for intensity was applied to all samples to generate the segmentation of positive regions. Then, the ratio of each individual organoid was calculated by the size of positive region dividing the size of organoid label mask (see Fig 3B). Graphs were created using Graph Pad Prism 8.4.3. Quantification of relative protein abundance within organoids was done on at least 15 organoids within each treatment. Each organoid is considered one biological replicate.
Measurement of Ada enzymatic activity
After 48 h of Bmps treatment (the same setup as for western blot), the organoids were harvested into ice‐cold PBS and centrifuged at 4°C, 300 g and 5 min. The supernatant was removed, and the pelleted organoids were analysed for Ada activity using Adenosine Deaminase Activity Assay Kit (Fluorometric, Sigma‐Aldrich, EPI020). The pelleted organoids were pre‐processed according to the kit manufacturer's protocol. The fluorescence measurement was performed using EnVision 2105 (PerkinElmer) instrument. The Ada activity was related to the untreated samples prior to the plotting using Graph Pad Prism 8.4.3. Relative Ada activity was measured in two independent experiments with two technical replicates.
Smad4 shRNA organoids line preparation and experiment
Lentiviral construct‐containing doxycycline‐inducible shRNA was generated by inserting oligos targeting Smad4 (see Table EV1 for sequences) into pTripz (Dharmacon) vector. Viral particles were prepared according to pTRIPZ technical manual in HEK 293 FT cell line using Lipofectamine 2000 (Thermo Fisher) for the transfection of lentiviral vectors. After 72 h, lentiviral suspension was collected and concentrated overnight using Lenti‐X concentrator (Clontech). Small intestinal organoids were pre‐treated with WntSur 0.5 μM, Nicotinamide 10 mM and Y‐27632 10 μM for 96 h before the lentiviral infection. The organoids were disintegrated into single‐cell suspension using Triple LE Express reagent. The cells were spinoculated with concentrated viral suspension (32°C, 600 g and 1 h) and kept at 37°C for another 5 h. Afterwards, they were embedded into Matrigel and kept in ENR containing WntSur 0.5 μM, Nicotinamide 10 mM and Y‐27632 10 μM. Furthermore, the transduced organoids were selected using puromycin 2 μg/ml.
Before sorting, organoids were pre‐treated with doxycycline 1 μg/ml. On the day of sort, organoids were disintegrated to single cell suspension using Triple LE Express reagent. Cells with the highest RFP signal were sorted into ENR‐containing Wnt‐Surrogate (WntSur) 0.5 μM, Nicotinamide 10 mM, Y‐27632 10 μM and 10% Matrigel, centrifugated and embedded into Matrigel and medium containing WntSur 0.5 μM, Nicotinamide 10 mM and Y‐27632 10 μM. The sorting step was repeated twice. Finally, WntSur 0.5 μM, Nicotinamide 10 mM and Y‐27632 10 μM were gradually removed. The organoids were occasionally reselected with 2 μg/ml puromycin.
For the experiment, the organoids were passaged and treated ± doxycycline (Dox) (1 μg/ml) for 3 days. Afterwards, Bmp2 500 ng/ml (± Dox) or vehicle (± Dox) were added and organoids were cultured for an additional 24 h. Then, they were photographed and harvested.
Note: Despite repeated selection and sorting, not all organoids expressed shRNASmad4 as marked by coupled TdTomato upon Dox treatment.
Histology, immunohistochemistry and microscopy
Dissected duodenum was flushed out by ice‐cold PBS, then cut into 1 cm pieces and fixed in 4% PFA overnight at 4°C. Afterwards, tissues were repeatedly washed with PBS, dehydrated, embedded in paraffin and cut into 5 μm sections. Deparaffinized tissue sections were subjected to antigen retrieval in 2.4 mM sodium citrate and 1.6 mM citric acid, pH 6, for 25 min in a steamer. For Ada staining, the frozen sections were prepared as described previously (Fazilaty et al, 2021).
Sections were washed with PBST (0.1% Tween‐20 in PBS) and blocked for 30 min at RT in blocking buffer (5% BSA and 5% heat‐inactivated normal goat serum in PBST). Following overnight incubation at 4°C with primary antibody (1:100, in blocking buffer), sections were washed in PBST and incubated with secondary antibody (1:400, in blocking buffer) for 1 h at room temperature. The antibodies are indicated in the Reagents and Tools Table. Nuclei were stained with DAPI (1:1,000) in blocking solution for 5 min at RT. Sections were imaged on DM6000 (Leica) or SP8 laser scanning confocal microscope (Leica). The pictures were equally processed by ImageJ (Fiji) software. All presented images (images of organoids and histological staining) were chosen as representative images for each condition. The Ada signal range on histological images was measured on sections obtained from three animals within each experimental group.
Quantitative real‐time PCR
For cultured crypts/organoids, Matrigel was washed out by ice‐cold PBS and organoids pelleted. In the case of LDN‐193189‐4HCl treatment in vivo, the complete intestinal epithelium of proximal small intestine from animals was isolated as described (Gracz et al, 2012). Total RNA from crypts or intestinal epithelium was extracted using RNeasy Micro Kit (Qiagen) according to manufacturer's protocol including DNAseI treatment. cDNA was synthetized with RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher). qRT–PCR was performed in three replicates consisting of pooled independent samples. Fold change values were counted using ΔΔCT method. Graphs were created using Graph Pad Prism 8.4.3. The primer sequences are indicated in Table EV2.
In situ mRNA hybridization
mRNAs were detected and visualized using RNAscope method (Advanced Cell Diagnostics, Germany) on small intestine tissue sections according to manufacturer's protocol (RNAscope Fluorescent Multiplex Assay). Probe sets for Bmp2 and Bmp4 were designed by Advanced Cell Diagnostics. Images were taken by Leica SP8 laser scanning confocal microscope (Leica). Pictures were processed equally using the ImageJ (FIJI) software.
Western blotting
Cultivated crypts were harvested and treated with recovery solution for 30 min on ice to remove residual Matrigel. Afterwards, they were pelleted and lysed in buffer containing: 50 mM HEPES/KOH pH 7.4, 1% Triton X‐100, 50 mM NaF, 5 mM Na2H2P2O7, 400 mM NaCl, 40 mM β‐glycerol phosphate, 12.5 mM EGTA pH 8, 1 mM Na3VO4, 1 mM benzamidin, 0.5 mM PMSF, 1.5 mM MgCl2, leupeptin 0.04 μl/ml, pepstatin 0.04 μl/ml and antipain 0.04 μl/ml. Total concentration of proteins was determined using standard Bradford assay.
Proteins were separated on 15% acrylamide‐Tris‐BIS‐0.1% SDS gel, and 6.16% acrylamide‐Tris‐BIS gel was used as stacking. Afterwards, proteins were transferred to nitrocellulose membrane using semi‐dry Blotter BioRad (2.5 mA/cm2, 10 W, 15 V, 55 min). All antibodies were diluted according to manufacturer's recommendations. Antibodies were visualized with chemiluminescent substrate SuperSignal West Pico PLUS. Used antibodies are indicated in the Reagents and Tools Table.
RNA sequencing
For control samples (untreated samples, at least n = 3 independent experiments, i.e. biological replicates, each consisted of pooled technical replicates) and experimental samples (individual Bmp or Tgfβ treatment for 24 h, for each Bmp, n = 3 independent experiments, each consisted of pooled technical replicates), total RNA was isolated with the same procedure as for qRT–PCR. Sequencing libraries were prepared from total RNA using the KAPA mRNA Hyperprep Kit, followed by size distribution analysis in the Agilent 2100 Bioanalyzer using the High Sensitivity DNA Kit (Agilent). Libraries were sequenced in two runs of the Illumina NextSeq 500 instrument (Illumina, USA) using a 75 nt single‐end configuration. The complete processing of isolated RNA was done by Genomics and Bioinformatics core facility at the Institute of Molecular Genetics of the Czech Academy of Sciences.
Computational RNA‐sequencing analysis
The nf‐core/rnaseq bioinformatics pipeline version 3.5 (Ewels et al, 2020) was used for subsequent read processing. Individual steps included removal of sequencing adapters and low‐quality reads with Trim Galore! (www.bioinformatics.babraham.ac.uk/projects/trim_galore) and quantification of gene expression with Salmon (Patro et al, 2017) using GRCm39 reference (Howe et al, 2021). Estimated expression per gene served as input for differential expression analysis using the DESeq2 R Bioconductor package (Love et al, 2014). Genes exhibiting a minimum absolute log2 fold change of 2 (|log2 FC| ≥ 2) and statistical significance (adjusted P‐value < 0.05) between the compared sample groups were considered differentially expressed. The initial mapping and analysis of sequencing data were done by Genomics and Bioinformatics core facility at the Institute of Molecular Genetics of the Czech Academy of Sciences.
Heatmap
For each treatment, top 10 differentially expressed genes were selected. Z‐score was calculated using standard formula z = (x – μ) / σ. The heatmap was generated with heatmaply package in R.
Deconvolution of RNAseq datasets
The percentage value charts were created by using CellAnneal software with default deconvolution values kept. Normalized RNA counts from RNAseq experiments were used. Pseudobulked datasets GSE92332 from Haber et al (2017a), Data ref: Haber et al (2017b) and Moor et al (2018a), Data ref: Moor et al (2018b) were used as reference datasets for different visualizations. The datasets were pre‐processed using standard Seurat workflow. Biological replicates were averaged. The code used for pre‐processing raw scRNA‐seq count matrices is available. Graphs were created in Graph Pad Prism 8.4.3.
Author contributions
Linda Berková: Data curation; software; formal analysis; validation; investigation; visualization; methodology; writing – original draft; writing – review and editing. Hassan Fazilaty: Investigation; visualization; methodology; writing – review and editing. Qiutan Yang: Validation; investigation; visualization; methodology. Jan Kubovčiak: Formal analysis; investigation. Monika Stastna: Investigation. Dusan Hrckulak: Investigation. Martina Vojtechova: Investigation. Tosca Dalessi: Investigation. Michael David Brügger: Investigation. George Hausmann: Writing – review and editing. Prisca Liberali: Resources; supervision; methodology. Vladimir Korinek: Resources. Konrad Basler: Resources; supervision. Tomas Valenta: Conceptualization; resources; data curation; formal analysis; supervision; funding acquisition; investigation; methodology; writing – original draft; project administration; writing – review and editing.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Appendix S1
Expanded View Figures PDF
Table EV1
Table EV2
Dataset EV1
Dataset EV2
PDF+
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Acknowledgements
O. Stepanek kindly shared Actin‐Cre animals with us. For the technical help with RNAseq, we thank Genomic and Bioinformatic Facility of IMG, Prague, directed by M. Kolar. This work was supported by Czech Science Foundation grant 21‐26025S and The project National Institute for Cancer Research (Programme EXCELES, ID Project No. LX22NPO5102)—Funded by the European Union—Next Generation EU. The lab of Konrad Basler was supported by the Swiss National Science Foundation and the Kanton of Zürich.
EMBO reports (2023) 24: e56454
Contributor Information
Konrad Basler, Email: konrad.basler@mls.uzh.ch.
Tomas Valenta, Email: tomas.valenta@mls.uzh.ch.
Data availability
The sequencing data generated in this study are deposited in GEO with the accession number: GSE216699 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE216699). The used codes are available from GitHub: https://github.com/LindtaB/Bmps
References
- Antebi YE, Linton JM, Klumpe H, Bintu B, Gong M, Su C, McCardell R, Elowitz MB (2017) Combinatorial signal perception in the BMP pathway. Cell 170: 1184–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahar Halpern K, Massalha H, Zwick RK, Moor AE, Castillo‐Azofeifa D, Rozenberg M, Farack L, Egozi A, Miller DR, Averbukh I et al (2020) Lgr5+ telocytes are a signaling source at the intestinal villus tip. Nat Commun 11: 1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauché D, Marie JC (2017) Transforming growth factor β: a master regulator of the gut microbiota and immune cell interactions. Clin Trans Immunol 6: e136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer J, Puschhof J, Yengej FY, Zhao L, Martinez‐Silgado A, Blotenburg M, Begthel H, Boot C, van Oudenaarden A, Chen YG et al (2022) BMP gradient along the intestinal villus axis controls zonated enterocyte and goblet cell states. Cell Rep 38: 110438 [DOI] [PubMed] [Google Scholar]
- Bragdon B, Moseychuk O, Saldanha S, King D, Julian J, Nohe A (2011) Bone morphogenetic proteins: a critical review. Cell Signal 23: 609–620 [DOI] [PubMed] [Google Scholar]
- Brügger MD, Valenta T, Fazilaty H, Hausmann G, Basler K (2020) Distinct populations of crypt‐associated fibroblasts act as signaling hubs to control colon homeostasis. PLoS Biol 18: e3001032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchauer L, Itzkovitz S (2021) Cellanneal: a user‐friendly deconvolution software for omics data. bioRxiv 10.48550/ARXIV.2110.08209 [PREPRINT] [DOI] [Google Scholar]
- Cosovanu C, Resch P, Jordan S, Lehmann A, Ralser M, Farztdinov V, Spranger J, Mülleder M, Brachs S, Neumann C (2022) Intestinal epithelial c‐Maf expression determines enterocyte differentiation and nutrient uptake in mice. J Exp Med 219: e20220233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degirmenci B, Valenta T, Dimitrieva S, Hausmann G, Basler K (2018) GLI1‐expressing mesenchymal cells form the essential Wnt‐secreting niche for colon stem cells. Nature 558: 449–453 [DOI] [PubMed] [Google Scholar]
- Doncheva NT, Morris JH, Gorodkin J, Jensen LJ (2019) Cytoscape String App: network analysis and visualization of proteomics data. J Proteome Res 18: 623–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels PA, Peltzer A, Fillinger S, Patel H, Alneberg J, Wilm A, Garcia MU, Di Tommaso P, Nahnsen S (2020) The nf‐core framework for community‐curated bioinformatics pipelines. Nat Biotechnol 38: 276–278 [DOI] [PubMed] [Google Scholar]
- Fazilaty H, Brügger MD, Valenta T, Szczerba BM, Berkova L, Doumpas N, Hausmann G, Scharl M, Basler K (2021) Tracing colonic embryonic transcriptional profiles and their reactivation upon intestinal damage. Cell Rep 36: 109484 [DOI] [PubMed] [Google Scholar]
- Fujii M, Matano M, Toshimitsu K, Takano A, Mikami Y, Nishikori S, Sugimoto S, Sato T (2018) Human intestinal organoids maintain self‐renewal capacity and cellular diversity in niche‐inspired culture condition. Cell Stem Cell 23: 787–793 [DOI] [PubMed] [Google Scholar]
- Galili T, O'Callaghan A, Sidi J, Sievert C (2017) Heatmaply: an R package for creating interactive cluster heatmaps for online publishing. Bioinformatics 34: 1600–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao CH, Yu G, Cai P (2021) ggVennDiagram: An intuitive, easy‐to‐use, and highly customizable R package to generate Venn diagram. Front Genet 12: 1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehart H, Clevers H (2019) Tales from the crypt: new insights into intestinal stem cells. Nat Rev Gastroenterol Hepatol 16: 19–34 [DOI] [PubMed] [Google Scholar]
- González‐Loyola A, Bernier‐Latmani J, Roci I, Wyss T, Langer J, Durot S, Munoz O, Prat‐Luri B, Delorenzi M, Lutolf MP et al (2022) c‐MAF coordinates enterocyte zonation and nutrient uptake transcriptional programs. J Exp Med 219: e20212418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracz AD, Puthoff BJ, Magness ST (2012) Identification, isolation, and culture of intestinal epithelial stem cells from murine intestine. In Somatic stem cells, Singh SR (ed), pp 89–107. Totowa, NJ: Humana Press; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius G, Kabiri Z, Sigmundsson K, Liang C, Bunte R, Singh MK, Virshup DM (2018) PDGFRα+ pericryptal stromal cells are the critical source of Wnts and RSPO3 for murine intestinal stem cells in vivo . Proc Natl Acad Sci U S A 115: E3173–E3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grün D, Lyubimova A, Kester L, Wiebrands K, Basak O, Sasaki N, Clevers H, van Oudenaarden A (2015) Single‐cell messenger RNA sequencing reveals rare intestinal cell types. Nature 525: 251–255 [DOI] [PubMed] [Google Scholar]
- Haber AL, Biton M, Rogel N, Herbst RH, Shekhar K, Smillie C, Burgin G, Delorey TM, Howitt MR, Katz Y et al (2017a) A single‐cell survey of the small intestinal epithelium. Nature 551: 333–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber AL, Biton M, Rogel N, Herbst RH, Shekhar K, Smillie C, Burgin G, Delorey TM, Howitt MR, Katz Y et al (2017b) Gene Expression Omnibus GSE92332. (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE92332). [DATASET]
- Hamilton TG, Klinghoffer RA, Corrin PD, Soriano P (2003) Evolutionary divergence of platelet‐derived growth factor alpha receptor signaling mechanisms. Mol Cell Biol 23: 4013–4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T, Goswami S, Hu Y, Tang F, Zafra MP, Murphy C, Cao Z, Poirier JT, Khurana E, Elemento O et al (2020) Lineage reversion drives WNT independence in intestinal cancer. Cancer Discov 10: 1590–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Hao S, Andersen‐Nissen E, Mauck WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M et al (2021) Integrated analysis of multimodal single‐cell data. Cell 184: 3573–3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramis APG, Begthel H, van den Born M, van Es J, Jonkheer S, Offerhaus GJA, Clevers H (2004) De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science 303: 1684–1686 [DOI] [PubMed] [Google Scholar]
- Harnik Y, Buchauer L, Ben‐Moshe S, Averbukh I, Levin Y, Savidor A, Eilam R, Moor AE, Itzkovitz S (2021) Spatial discordances between mRNAs and proteins in the intestinal epithelium. Nat Metab 3: 1680–1693 [DOI] [PubMed] [Google Scholar]
- Howe KL, Reardon C, Wang A, Nazli A, McKay DM (2005) Transforming growth factor‐β regulation of epithelial tight junction proteins enhances barrier function and blocks enterohemorrhagic Escherichia coli O157:H7‐induced increased permeability. Am J Pathol 167: 1587–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe KL, Achuthan P, Allen J, Allen J, Alvarez‐Jarreta J, Amode MR, Armean IM, Azov AG, Bennett R, Bhai J et al (2021) Ensembl 2021. Nucleic Acids Res 49: D884–D891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpe HE, Langley MA, Linton JM, Su CJ, Antebi YE, Elowitz MB (2022) The context‐dependent, combinatorial logic of BMP signaling. Cell Syst 13: 388–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol 15: 550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy N, Manieri E, Storm EE, Saadatpour A, Luoma AM, Kapoor VN, Madha S, Gaynor LT, Cox C, Keerthivasan S et al (2020a) Distinct mesenchymal cell populations generate the essential intestinal BMP signaling gradient. Cell Stem Cell 3: 391–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy N, Manieri E, Storm EE, Saadatpour A, Luoma AM, Kapoor VN, Madha S, Gaynor LT, Cox C, Keerthivasan S et al (2020b) Gene Expression Omnibus GSM3747599. (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM3747599). [DATASET] [DOI] [PMC free article] [PubMed]
- Miyoshi H, Ajima R, Luo CT, Yamaguchi TP, Stappenbeck TS (2012) Wnt5a potentiates TGF‐β signaling to promote colonic crypt regeneration after tissue injury. Science 338: 108–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moor AE, Harnik Y, Ben‐Moshe S, Massasa EE, Rozenberg M, Eilam R, Bahar Halpern K, Itzkovitz S (2018a) Spatial reconstruction of single enterocytes uncovers broad zonation along the intestinal villus Axis. Cell 175: 1156–1167 [DOI] [PubMed] [Google Scholar]
- Moor AE, Harnik Y, Ben‐Moshe S, Massasa EE, Rozenberg M, Eilam R, Bahar Halpern K, Itzkovitz S (2018b) Zenodo 3403670. (https://zenodo.org/record/3403670) [DATASET] [DOI] [PubMed]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C (2017) Salmon provides fast and bias‐aware quantification of transcript expression. Nat Methods 14: 417–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z, Li Y, Zhao B, Xu C, Liu Y, Li H, Zhang B, Wang X, Yang X, Xie W et al (2017) BMP restricts stemness of intestinal Lgr5+ stem cells by directly suppressing their signature genes. Nat Commun 8: 13824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar VS, Gamer LW, Rosen V (2016) BMP signalling in skeletal development, disease and repair. Nat Rev Endocrinol 12: 203–221 [DOI] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ et al (2009) Single Lgr5 stem cells build crypt‐villus structures in vitro without a mesenchymal niche. Nature 459: 262–265 [DOI] [PubMed] [Google Scholar]
- Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, Clevers H (2011) Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469: 415–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda‐Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B et al (2012) Fiji:an open‐source platform for biological‐image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M (2011) Global quantification of mammalian gene expression control. Nature 473: 337–342 [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spit M, Koo BK, Maurice MM (2018) Tales from the crypt: intestinal niche signals in tissue renewal, plasticity and cancer. Open Biol 8: 180120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolfi C, Troncone E, Marafini I, Monteleone G (2020) Role of TGF‐Beta and Smad7 in gut inflammation, fibrosis and cancer. Biomolecules 11: 17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RN, Green J, Wang Z, Deng Y, Qiao M, Peabody M, Zhang Q, Ye J, Yan Z, Denduluri S et al (2014) Bone morphogenetic protein (BMP) signaling in development and human diseases. Genes Dis 1: 87–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H (2016) ggplot2: elegant graphics for data analysis. New York, NY: Springer‐Verlag New York; [Google Scholar]
- Wickham H, François R, Henry L, Müller K, Vaughan D (2023) Dplyr: a grammar of data manipulation. (https://dplyr.tidyverse.org, https://github.com/tidyverse/dplyr)
- Yan KS, Gevaert O, Zheng GXY, Anchang B, Probert CS, Larkin KA, Davies PS, Cheng ZF, Kaddis JS, Han A et al (2017a) Intestinal enteroendocrine lineage cells possess homeostatic and injury‐inducible stem cell activity. Cell Stem Cell 21: 78–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan KS, Gevaert O, Zheng GXY, Anchang B, Probert CS, Larkin KA, Davies PS, Cheng ZF, Kaddis JS, Han A et al (2017b) Gene Expression Omnibus GSE99457 (GSM2644349 and GSM 2644350). (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE99457) [DATASET]
- Yang Q, Xue SL, Chan CJ, Rempfler M, Vischi D, Maurer‐Gutierrez F, Hiiragi T, Hannezo E, Liberali P (2021) Cell fate coordinates mechano‐osmotic forces in intestinal crypt formation. Nat Cell Biol 23: 733–744 [DOI] [PMC free article] [PubMed] [Google Scholar]


