Abstract
A rickettsial isolate (isolate MOAa) belonging to the spotted fever group (SFG) was obtained from the lone star tick Amblyomma americanum. We used PCR to characterize the genes for the rickettsial outer membrane proteins rOmpA and rOmpB. We sequenced the PCR products (domains I of both the rompA gene and the rompB gene) of MOAa and WB-8-2, another rickettsial isolate from A. americanum. To place MOAa and WB-8-2 and two other nonpathogenic isolates (Rickettsia rickettsii Hlp2 and Rickettsia montana M5/6) with respect to their putative sister species, we included them in a phylogenetic analysis of 9 Rickettsia species and 10 Rickettsia strains. Our phylogenetic results implied three evolutionary lineages of SFG rickettsiae and that WB-8-2 and MOAa were most closely related to R. montana. New World isolates were not the most closely related to each other (they did not form a clade). Rather, our results supported four independent origins (introductions) of rickettsiae into North America from different Old World regions. The results of our phylogenetic analysis did not support the hypothesis of a stable coevolution of rickettsiae and their tick hosts. Finally, we examined the rompA gene of a nonpathogenic rickettsial symbiont isolated from the tick Ixodes scapularis. In a phylogenetic analysis, the symbiont was placed as the sister to R. montana and its isolates. The relationship of this symbiont to R. montana raised questions as to the potential origin of pathogenic SFG rickettsiae from nonpathogenic tick symbionts, or vice versa.
Rickettsiae are obligate intracellular, gram-negative bacteria which infect arthropods and vertebrates, causing typhus and spotted fevers in humans. The typhus group is typically louse-borne, while the larger and more diverse spotted fever group (SFG) is usually associated with ixodid ticks (13). The tick hosts of SFG rickettsiae serve as both pathogen reservoirs and vectors. The SFG rickettsiae are further differentiated from the typhus group by the presence of 190- and 120-kDa outer membrane proteins (rickettsial outer membrane proteins rOmpA and rOmpB, respectively) which elicit protective immunity in vertebrate hosts (4, 12, 21, 22). The rOmp proteins may play important roles in determining the host range and pathogenicities of SFG rickettsiae (1–4, 41).
Studies of rickettsiae have focused on determining how they move between arthropod and vertebrate hosts and other biological aspects of their disease epidemiology. These studies have traditionally been difficult, because rickettsiae are obligate intracellular parasites with fastidious growth requirements (67). Identification of different species, strains, and isolates required laborious culturing techniques and diagnostic tests. Recently, the application of molecular biology-based techniques, such as PCR, has improved investigators’ ability to identify rickettsiae (16, 56) and has contributed to the better characterization of nonpathogenic strains and studies of the evolutionary relationships (phylogeny) among them (57–61, 63).
Pathogenic SFG rickettsiae are distributed worldwide, sharing geographic and tick host ranges with closely related nonpathogenic symbionts (13). Pathogenic and nonpathogenic rickettsiae must be distinguished for accurate census results. During a survey of lone star ticks (Amblyomma americanum) collected in Missouri (June 1992), we isolated a new rickettsia (hereafter referred to as isolate MOAa). A. americanum, which is not a major vector of virulent SFG rickettsiae (23), is known to harbor a nonpathogenic SFG rickettsia, isolate WB-8-2 (11). Female black-legged ticks, Ixodes scapularis, also harbor within their ovarian tissues (37) prokaryotes that resemble SFG rickettsiae, but, like A. americanum, I. scapularis ticks do not appear to be vectors of pathogenic SFG rickettsiae (9). We used PCR to amplify the genomic coding sequences of the rompA and rompB genes (19, 56) in order to identify MOAa as an SFG rickettsia and to determine the relation of the I. scapularis symbiont to pathogenic and nonpathogenic SFG rickettsiae. The amplification products of MOAa were compared with those from three strains of nonpathogenic SFG rickettsiae: WB-8-2 (11), Rickettsia montana M5/6 (7, 20) and Rickettsia rickettsii Hlp2 (20, 50). We established that MOAa and the I. scapularis symbiont are SFG rickettsiae and sequenced the rompA PCR products to characterize further their relationship with other SFG rickettsiae. Here we present evidence that MOAa and the I. scapularis symbiont are most closely related to R. montana M5/6 on the basis of a phylogenetic analysis of the first 459 bp of the rompA-coding sequence. This analysis included rickettsial species and strains for which rompA sequences were available.
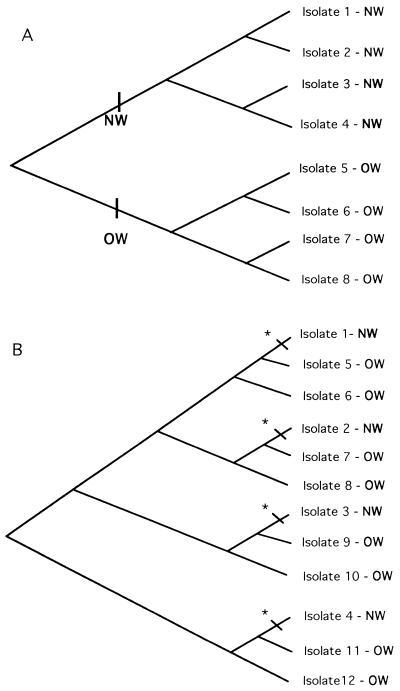
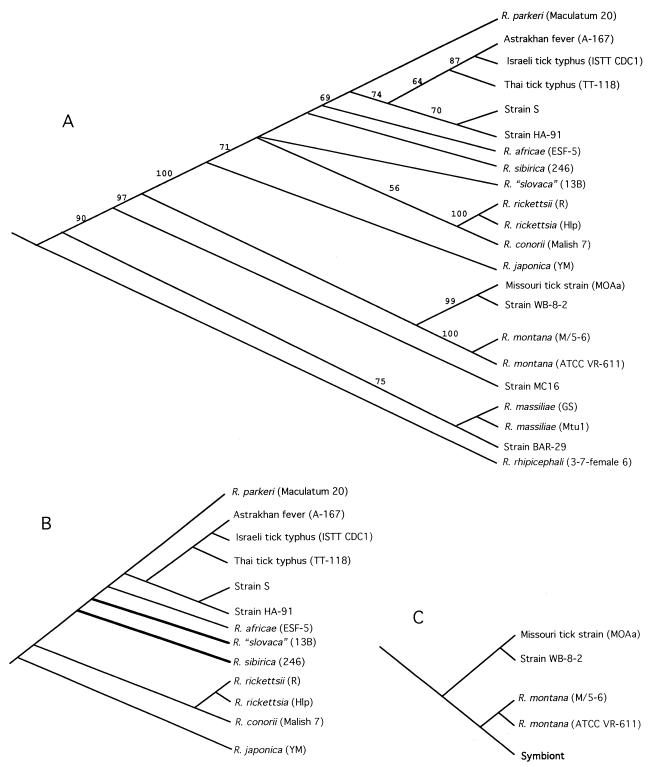
Phylogenies provide important frameworks for the testing of a hypothesis (8). An evolutionary tree can be used to examine geographic distribution patterns (biogeography) and the coevolution of organisms (14, 25, 45). For example, if New World (NW) rickettsial isolates are most closely related to one another (i.e., if they form a clade) and if Old World (OW; broadly defined as being of Austral-Asian-European-African origin) isolates form a clade, then we predict that we will find the pattern presented in Fig. 1A. This pattern implies a single Gondwanaland origin for NW isolates. On the other hand, if we find the pattern presented in Fig. 1B, in which NW isolates are not the closest relatives but, rather, appear to evolve from different OW isolates, then we conclude that there have been independent origins of NW isolates. Using comparative biology-based methods, we can also test whether rickettsial isolates and their arthropod hosts have a strict coevolutionary relationship or whether rickettsial host shifts occur.
FIG. 1.
Alternative hypotheses for the origin of rickettsiae in the NW. (A) NW rickettsiae originate once in the NW. Hence, all NW isolates are more closely related to one another (form a clade) than to any OW isolates. (B) NW rickettsiae originate multiple times in the NW. Hence, individual NW isolates are more closely related to OW isolates than to other NW isolates. Asterisks indicate transitions from OW to NW.
Our phylogeny has implications for the origin of NW strains, the evolution of tick host associations, and the evolution of pathogenicity. Our trees imply that at least four independent introductions of SFG rickettsiae into the NW have occurred from different regions of the OW. These introductions most likely followed mammalian immigration patterns. Furthermore, our results do not support the hypothesis of strict coevolution between tick vectors and their associated rickettsial pathogens. Rather, rickettsiae appear to switch readily to different tick hosts over evolutionary time. Lastly, our phylogeny suggests that nonpathogenic strains and tick rickettsial symbionts may play an important role in the evolution of the SFG rickettsiae. We suggest that pathogenic forms of SFG rickettsiae may have evolved from tick symbionts through changes in surface antigen gene proteins.
MATERIALS AND METHODS
Tick cell culture.
Tick cells, lines RAE25 (34), IDE2 (44), and DALBE3 (54), derived from embryonated eggs of Rhipicephalus appendiculatus, I. scapularis, and Dermacentor albipictus ticks, respectively, were used. Cultures were grown to near confluency in culture flasks (25 cm2) with 5 ml of L-15B medium (pH 7) supplemented with 5% fetal bovine serum, 10% tryptose phosphate broth, and 10 μg of a bovine lipoprotein concentrate per ml (43). No antibiotics were used.
Rickettsiae.
R. rickettsii Hlp2, R. montana M5/6, and isolate WB-8-2 (52) were kindly provided by Robert Heinzen (Rocky Mountain Laboratories, National Institutes of Health). We grew rickettsial isolate MOAa from an adult female A. americanum tick collected in June 1992 in Bolling County, Mo., by Dorothy Feir (St. Louis University). Ticks collected in the field were surface disinfected (5 min each in 0.1% aqueous benzalkonium chloride–70% ethanol and three rinses in sterile water). Internal organs from individual ticks were aseptically removed and were transferred to a 96-well tissue culture plate containing complete BSK medium (100 μl per well) (6). One week later the contents of seven wells (with the contents of seven ticks) that had remained free of bacterial and fungal contamination were pooled and transferred to a 25-cm2 culture of RAE25 tick cells (see below). One month later, with weekly feedings with tick cell culture medium (44), the culture was found by Giemsa staining to be heavily infected with rickettsia-like organisms. The MOAa organisms were highly cytopathogenic and were propagated by a weekly or a biweekly transfer of 1 to 5% of a culture (in which they had infected and lysed nearly 100% of their host cells) to a fresh, established tick cell culture. MOAa reacted positively with a monoclonal antibody (monoclonal antibody 13-2) (provided by Robert Heinzen, Rocky Mountain Laboratories, National Institutes of Health) specific to an epitope of the 120-kDa heat-sensitive rOmpB protein of SFG rickettsiae (2) by the indirect immunofluorescence method (26, 42).
An SFG rickettsial symbiont associated with the ovarian tissues of I. scapularis (47) was also analyzed. Ticks in the fifth laboratory generation were used. The colony was started with adult females collected in 1988 from hunter-killed white-tailed deer in Minnesota and Wisconsin.
Preparation of rickettsiae for PCR.
Lines IDE2 and DALBE3 (grown in 25-cm2 flasks) were infected by adding 0.1 ml of a previously infected culture to an established, confluent cell layer. The medium was identical to the tick cell maintenance medium. Infected cultures were incubated at 34°C for 7 to 10 days. Rickettsiae were harvested from cultures in which more than 90% of the cells were infected, as judged from Giemsa-stained cell spreads. The cells were centrifuged at 100 × g for 10 min at room temperature. The cell pellet was resuspended in 2 ml of fresh culture medium, and the cells were mechanically ruptured by passing the suspension 10 times through a 27-gauge needle. Large debris was removed by centrifugation at 275 × g for 10 min. The supernatant was diluted to 5 ml in L-15B and was filtered sequentially through 5- and 0.8-μm-pore-size filters. The rickettsiae in the filtrate were concentrated by centrifugation at 13,500 × g for 15 min at 4°C and lysed in 100 μl of PCR buffer with nonionic detergents and proteinase K (27). The lysates were stored at −20°C.
Ovarian tissues were dissected from flat adult female I. scapularis ticks, and symbiont template DNA was prepared for PCR as described by O’Neill et al. (48).
Primers for PCR and internal probes.
Primers were synthesized at the University of Minnesota Microchemical Facility. Lyophilized primers were dissolved in TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]) and were used at a final concentration of 0.1 to 0.5 μM. We used the primer pair Rr190.70p and Rr190.602n (56) for the rompA gene of R. rickettsii and those of Gage et al. (19) for the rompB gene. In keeping with the nomenclature of Regnery et al. (56), we refer to the rompB gene-specific primers as Rr120.501p and Rr120.1000n, respectively. For internal probes, 20-mer oligonucleotides were synthesized and dissolved in TE as described above. For the R. rickettsii rompB gene, we used the oligonucleotide Rr120.741 (19). For rompA, nucleotides 341 to 360, primer Rr190.341 (4), were used.
PCR conditions.
For the rompB gene fragment, the DNA in 1 μl of lysate was amplified in 100 μl of reaction buffer, as specified by the manufacturer (GIBCO BRL, Gaithersburg, Md.). Samples were heated to 95°C for 5 min and were then amplified by using the program specified by Gage et al. (19) for 35 cycles, with a final extension step of 10 min at 72°C. For the rompA gene fragment, 3 μl of rickettsial lysates was amplified in 100 μl of buffer containing primers at a concentration of 0.1 μM each. The DNA was denatured for 5 min at 95°C and was then amplified for 35 cycles (45 s at 94°C, 30 s at 55°C, and 90 s at 72°C), with a final extension step of 10 min at 72°C.
Electrophoretic analysis of PCR products.
Ninety-microliter aliquots of the PCR mixtures were successively extracted with equal volumes of phenol and chloroform, followed by ethanol precipitation overnight at −20°C. Precipitates were resuspended in 90 μl of TE buffer, and 10-μl aliquots were digested with AluI (rompA) or DraI (rompB) under standard conditions (Stratagene, La Jolla, Calif.). Equal aliquots (equivalent to 10% of the original PCR mixtures) of undigested and digested samples were electrophoresed at 5 V/cm on 1% agarose gels in 1× Tris-acetate buffer (TAE; pH 8.0). The gels were stained with ethidium bromide, and DNA was visualized by UV illumination.
Southern blot analysis.
PCR samples that had undergone agarose gel electrophoresis were transferred to a BA-S85 membrane (Schleicher & Schuell, Keene, N.H.) by standard procedures (38). The blotted membranes were prehybridized for 24 h at room temperature in sealed bags containing 0.25 ml of hybridization solution (3× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 4 mg of heparin per ml, 2 mg of sodium pyrophosphate per ml, 0.2% sodium dodecyl sulfate) (14) per cm2 of membrane. After prehybridization, radiolabelled internal rompA or rompB probes (see below) were added to a final concentration of 105 cpm/ml, and hybridization was continued for 16 h at room temperature. The membranes were washed twice in 2× SSC–0.1% sodium dodecyl sulfate at room temperature for 5 min, followed by two washes at 40°C for 5 min. The membranes were air dried and autoradiographed with Kodak X-OMAT AR film at −70°C with an intensifying screen.
Preparation of radiolabelled probes.
One-nanomole aliquots of the 20-mer rompA and rompB oligonucleotides were digested with calf intestinal alkaline phosphatase under standard conditions (Stratagene) and 5′ end labelled with [32P]ATP (Amersham, Arlington Heights, Ill.) and polynucleotide kinase (New England Biolabs, Beverly, Mass.). Labelled probes were separated from unincorporated radionucleotide with Chromaspin 10 columns (Clonetech, Palo Alto, Calif.). The specific activities of the probes were 9 × 105 cpm (Cerenkov)/μg for rompB and 2.2 × 106 cpm/μg for rompA.
DNA sequencing.
The 459-bp rompA and 376-bp rompB PCR amplification products were purified by electrophoresis on 1% agarose gels followed by band capture with an NA-45 nitrocellulose membrane (Schleicher & Schuell). Gel-purified amplification products were treated with exonuclease I and shrimp alkaline phosphatase, extracted with phenol and chloroform, and ethanol precipitated. Precipitates were resuspended in TE buffer. Manual sequencing was performed via the dideoxy chain termination method (62) with [35S]dATP (Amersham) and the Sequenase PCR product sequencing kit (United States Biochemicals, Cleveland, Ohio). The sequencing reaction mixtures were electrophoresed on 6% polyacrylamide gels and autoradiographed with Kodak X-OMAT AR film.
The PCR products of the I. scapularis ovarian symbiont were cloned into the T vector of pBluescript II (TA cloning) (39). Three clones of the insert were sequenced in both directions with the T3 and T7 primers and the ABI dye primer cycle sequencing kit (Applied Biosystems) and an ABI 373A autosequencer.
Phylogenetic analyses.
To place MOAa and two other isolates (isolates Hlp2 and WB-8-2) with respect to their putative sister species, we obtained rompA sequences from GenBank for nine other Rickettsia species and eight strains (Table 1). The sequences of the rompA genes were easily aligned by locating conserved amino acid codons. After alignment, individual codon sites shared by two or more taxa (species and isolates) were treated as individual characters. Ambiguous sequence information was treated as missing data. All codon positions were equally weighted, and trees were first rooted with Rickettsia rhipicephali on the basis of the 16S rRNA results of Stothard and Fuerst (63). Alternate outgroups were tested to ascertain tree topology robustness, as discussed in the Results. Similarly, the rompB sequence for R. rickettsii R (22) was obtained from GenBank, and we report partial sequences for the strains R. rickettsii Hlp2, R. montana M5/6, MOAa, and WB-8-2.
TABLE 1.
rOmpA sequencesa of SFG rickettsia species and strains used in our analyses
| Rickettsial group and strain | Strain designation | GenBank accession no. | Geographic origin | Vector tick |
|---|---|---|---|---|
| Validated SFG species | ||||
| R. africae | ESF-5 | U43790 | Ethiopia | Amblyomma variegatum |
| R. conorii | Malish 7 | U01028 | South Africa | Unknown |
| R. japonica | YM | U43795 | Japan | Dermacentor taiwanensis |
| R. massiliae | Mtu1 | U43799 | France | Dermacentor turanicus |
| R. massiliae | GS | U43793 | Greece | Rhipicephalus sanguineus |
| R. montana | ATCC VR-611 | U43801 | Montana | Dermacentor andersoni and Dermacentor variabilis |
| R. montana | M5/6 | AF045223 | Montana | D. andersoni and D. variabilis (?) |
| R. parkeri | Maculatum 20 | U43802 | Mississippi | Amblyomma maculatum |
| R. rhipicephali | 3-7-female 6 | U43803 | Mississippi | R. sanguineus, D. andersoni, and D. variabilis |
| R. rickettsii | R | M31227 | Montana | D. andersoni |
| R. rickettsii | Hlp2 | AF045220 | Montana | Haemaphysalis leporispalustris |
| R. sibirica | 246 | U43807 | Siberia | Dermacentor nutalli |
| Unclassified strains or unrecognized species | ||||
| Astrakhan fever | A-167 | U43791 | Astrakhan, Russia | Rhipicephalus pumilio |
| Strain HA-91 | HA-91 | U4379 | Mongolia | Haemaphysalis asiaticum |
| Israeli tick typhus | ISTT CDC1 | U43797 | Israel | R. sanguineus |
| Strain MC16 | MC16 | U43800 | Morocco | Hyalomma marginatum |
| Missouri tick | MOAa | AF045221 | Missouri | Amblyomma americanum |
| “R. slovaca” | 13-B | U43808 | Slovakia | Dermacentor marginalis |
| Strain S | S | U4380 | Armenia | R. sanguineus |
| Strain BAR-29 | BAR-29 | U43792 | Spain | Rhipicephalus species (?) |
| Thai tick typhus | TT-118 | U43809 | Thailand | Ixodes or Rhipicephalus spp. |
| WB-8-2 | WB-8-2 | AF045222 | United States | A. americanum |
| I. scapularis symbiont | ISS | AB002268 | Minnesota | Ixodes scapularis |
The sequences are from nucleotides 70 to 602 of domain I.
We used three phylogenetic approaches: the maximum-parsimony (MP), neighbor-joining (NJ), and maximum-likelihood (ML) approaches. These three methods have different underlying assumptions, strengths, and weaknesses (65). If the same clades are obtained, regardless of the type of analysis, then these results are robust to the different underlying assumptions (5). There is no consensus on which tree indices (i.e., consensus indices, bootstrapping indices, or decay indices) best measure tree robustness, because each will fail to show significance under certain conditions (65). Thus, at this time, our best estimate of whether a particular topology accurately reflects phylogenetic history is the concordant results of these three methods (5).
For MP analyses, sequence data were analyzed by using the heuristic search algorithms implemented with PAUP, version 3.1.1 (MULPARS, TBR) (64). Taxa were randomly shuffled (the random addition repetition option is 50 in PAUP) to avoid island topologies (36). Bootstrap analyses (18) were performed to examine the relative support for the branches. A phylogenetic signal was assessed by using random trees and skewness tests (28, 30). For NJ analyses, the two-parameter model of Kimura et al. (33) was used and was implemented with MEGA. An ML analysis was performed by using Phylip, version 3.5c (18), the empirical base frequencies, and a transition/transversion ratio of 2.0. We examined whether the trees obtained by MP analysis were significantly worse than the trees obtained by ML analysis using the test of Kishino and Hasegawa (32) implemented with Phylip and specified user trees.
We tested alternative hypotheses for the origin of NW rickettsias, arthropod host patterns, and an alternate hypothesis of rickettsial relationships using two approaches. We investigated if constraining the MP analysis to recover certain groups increased the tree length significantly. From a random tree analysis, we generated standard deviations (SDs). We then compared the tree length for the unconstrained and constrained MP analyses and tested whether the alternative hypotheses had significantly greater log likelihood (by the test of Kishino and Hasegawa [32]; Phylip).
The rompA sequence of the symbiont of I. scapularis was added, and the analyses described above were repeated to determine which cluster of SFG species it was most likely related to and to ascertain if the inclusion of this symbiont would influence the tree structure.
Nucleotide sequence accession numbers.
GenBank accession numbers for the rompA sequences of R. rickettsii Hlp2, isolate MOAa, isolate WB-8-2, R. montana M5/6, and the I. scapularis symbiont used in our phylogenetic analyses are AF045220, AF045221, AF45222, AF045223, and AB002268, respectively. The GenBank accession numbers for the rompB sequences of R. rickettsii Hlp2, R. montana M5/6, isolate WB-8-2, and isolate MOAa are AF045224, AF045225, AF45226, and AF045227, respectively.
RESULTS
Characterization of isolates.
The biological properties of the MOAa isolate, including intracellular growth in cultured tick cells and ultrastructural morphology, as well as its geographic origin, suggested that it was an SFG Rickettsia (42). We confirmed this hypothesis with a DNA detection and typing system (19, 20, 56), based on PCR amplification of DNA sequences specific to the 190- and 120-kDa outer membrane proteins (rOmpA and rOmpB, respectively) of SFG rickettsiae. The partial rompA sequence of MOAa was found to be closely related to that of WB-8-2, an SFG rickettsia frequently detected in natural populations of A. americanum (11, 23).
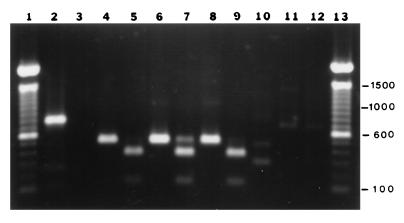
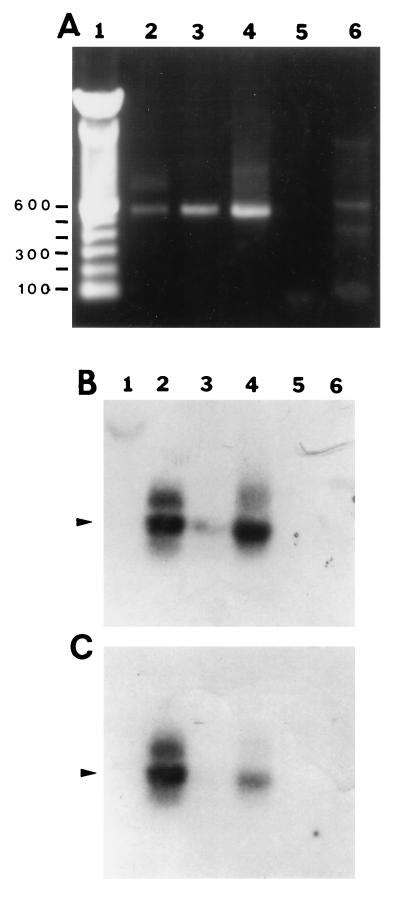
We amplified the predicted 500-bp rompB PCR product from template DNA isolated from R. rickettsii Hlp2, R. montana M5/6, and isolate MOAa (Fig. 2, lanes 4, 6, and 8, respectively). Furthermore, DraI restriction endonuclease digestion of the PCR products yielded the predicted 375- and 125-bp fragments (lanes 5, 7, and 9, respectively). Nonspecific PCR amplification products obtained with template DNA derived from tick cell lines RAE25, IDE2, and IDE8 were resolved from the rickettsial amplification products by electrophoresis (lanes 10, 11, and 12, respectively). In Southern blot experiments with a probe complementary to nucleotides 741 to 760 of the R. rickettsii rompB gene, strong hybridization signals were observed with the PCR amplification products from R. rickettsii, R. montana, and MOAa when the blots were washed at 40°C or lower (Fig. 3A and B, lanes 2, 3, and 5, respectively). When we washed the blots under higher-stringency conditions (50°C), as recommended by Gage et al. (19), only R. rickettsii gave a detectable signal (data not shown). Hybridization signals were not observed in lanes containing aliquots of rompB PCR mixtures with template DNA from tick cell lines ISE6 and IDE2 or Borrelia burgdorferi (Fig. 3A and B, lanes 4, 7, and 6, respectively).
FIG. 2.
Detection and verification of specificity of a 500-bp PCR amplification product from the rompB gene. Ten-microliter aliquots of 100-μl PCR mixtures were visualized with ethidium bromide on a 1% agarose gel. Lanes 1 and 13, 100-bp marker ladder (BRL); lane 2, human control DNA amplification product in the Gibco PCR kit; lane 3, B. burgdorferi control reaction; lanes 4 and 5, R. rickettsii Hlp2 amplification product and DraI restriction endonuclease digest, respectively; lanes 6 and 7, R. montana M5/6 amplification product and DraI digest, respectively; lanes 8 and 9, MOAa amplification product and DraI digest, respectively; lanes 10, 11, and 12, amplification products from tick cell lines RAE25, IDE2, and IDE8, respectively. Numbers on the right are in base pairs.
FIG. 3.
Verification of specificity of the rompB PCR amplification product by Southern blotting. Oligonucleotide Rr120.742 was used as an internal probe. (A) Three-microliter (rickettsia or spirochete) or 10-μl (tick cell line) aliquots of PCR mixtures were electrophoresed on a 1% agarose gel with Tris-acetate buffer and stained with ethidium bromide. Lanes 1 and 8, 100-bp marker; lane 2, R. rickettsii Hlp2; lane 3, R. montana M5/6; lane 4, ISE6 cell line; lane 5, MOAa; lane 6, B. burgdorferi; lane 7, tick cell line IDE2. (B) Southern blot of the gel shown in panel A. Numbers on the right are in base pairs.
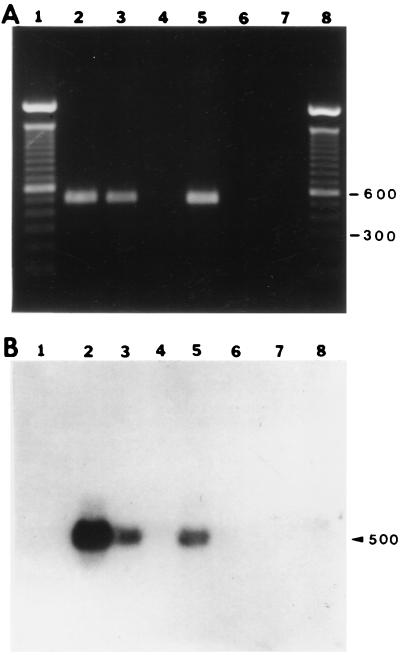
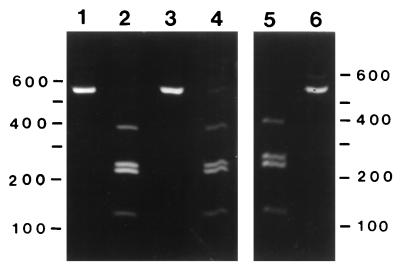
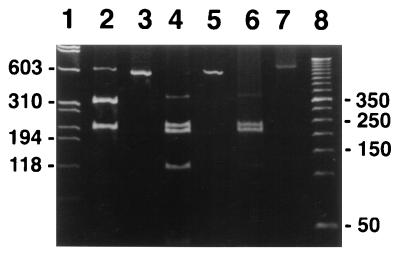
We amplified the predicted rompA PCR product of approximately 530 bp from template DNA isolated from MOAa, WB-8-2, and R. rickettsii Hlp2 (Fig. 4, lanes 1, 3, and 6, respectively) and from R. montana M5/6 (Fig. 5A, lane 3). The specificities of the amplification products for MOAa, WB-8-2, and R. rickettsii Hlp2 were confirmed by AluI digestions, which yielded the predicted three fragments: fragments of approximately 120, 210, and 220 bp (Fig. 4, lanes 2, 4, and 5, respectively). A faint band, probably an incompletely digested fragment, of approximately 400 bp was also observed. The R. montana M5/6 PCR product had one AluI digestion site, and fragments of approximately 300 and 200 bp were obtained (data not shown). The specificities of the MOAa rompA PCR amplification products were further confirmed by Southern blot analysis with an internal probe complementary to nucleotides 341 to 361 of the rompA gene. Hybridization signals corresponding to the PCR amplification products from R. rickettsii Hlp2, R. montana M5/6, and MOAa (Fig. 5A, lanes 2, 3, and 4, respectively) were observed on blots washed at 22°C (Fig. 5B, lanes 2, 3, and 4, respectively), although the signal from R. montana M5/6 was weak. Hybridization signals were not observed from rompA PCR mixtures with template DNA from a spirochete isolate or the tick cell line IDE8 (Fig. 5A and B, lanes 5 and 6, respectively). We noted that blots washed at 40°C or higher resulted in the loss of the signal from R. montana M5/6 (Fig. 5B, lane 3) and greatly weakened the signal from isolate MOAa (lane 4), while the signal from R. rickettsii Hlp2 (lane 2) remained comparatively strong.
FIG. 4.
Detection and verification of specificity of PCR amplification product from the rickettsial rompA gene. Ten-microliter aliquots of 100-μl PCR mixtures were visualized with ethidium bromide on an 8% acrylamide gel. Lanes 1 and 2, MOAa amplification product and AluI digest, respectively; lanes 3 and 4, WB-8-2 amplification product and AluI digest, respectively; lanes 5 and 6, R. rickettsii Hlp2 AluI digest and amplification product, respectively. Note that the two brightest bands are the predicted 208- and 212-bp fragments; the 400-bp band is an incompletely digested fragment. Numbers on the right and left are in base pairs.
FIG. 5.
Verification of the specificity of the rompA PCR amplification product by Southern blotting. Oligonucleotide Rr190.341 was used as an internal probe. (A) Five-microliter aliquots of PCR mixtures were electrophoresed on a 1% agarose gel with Tris-acetate buffer and stained with ethidium bromide. Lane 1, 100-bp ladder; lane 2, R. rickettsii Hlp2; lane 3, R. montana M5/6; lane 4, MOAa; lane 5, B. burgdorferi; lane 6, tick cell line IDE8. Numbers on the left are in base pairs. (B and C) Southern blot of the gel shown in panel A and subjected to final washes at 22 and 40°C, respectively. Arrowheads indicate positions of amplification products.
We also amplified an approximately 530-bp rompA PCR product from the template DNA of the I. scapularis ovarian symbiont (Fig. 6, lane 3). An AluI digest of the symbiont yielded two fragments, one of approximately 300 bp and another of approximately 200 bp (Fig. 6, lane 2).
FIG. 6.
Detection of a 530-bp amplification product from the rompA gene. Three-microliter aliquots of 100-μl PCR mixtures and 10 μl of AluI digestion products were visualized with ethidium bromide on an 8% acrylamide gel in 1× TBE (Tris-borate-EDTA) buffer. Lane 1, HaeIII digest of φX174 DNA (molecular weight marker); lanes 2 and 3, I. scapularis symbiont AluI digest and amplification product, respectively; lanes 4 and 5, R. rickettsii Hlp2 AluI digest and amplification product, respectively; lanes 6 and 7, MOAa AluI digest and amplification product, respectively; lane 8, 50-bp molecular size marker. Numbers on the right and left are in base pairs.
Characterization of rompA and rompB fragments.
Nucleotide sequence analysis of the PCR products confirmed the AluI restriction results and allowed us to deduce the amino acid sequence of the rompA gene in R. rickettsii Hlp2, R. montana M5/6, isolate MOAa, isolate WB-8-2, and the I. scapularis symbiont. We confirmed that the rompA genes of R. rickettsii Hlp2, WB-8-2, and MOAa had two AluI restriction sites. In contrast, the rompA genes of the I. scapularis symbiont and R. montana M5/6 were each cut at only one site, and they were different sites.
For both rompA and rompB, the PCR products corresponded to sequences encoding their respective domains I (4, 22). Our rompA fragment begins at amino acid residue 8 and ends at amino acid residue 170 (4). Our rompB product begins at residue 115 (nucleotide 568) and ends at residue 238 (nucleotide 940) (22). For the aligned 459 bp of rompA, 38 of 153 sites were variable for the first codon and 43 and 38 sites were variable for the second and third codons, respectively. The elevated variation in the second codon positions compared to the variation in the third codon is observed in proteins involved in immune antibody-antigen systems (55). The first codon position exhibited an A bias (>35%) and a G bias (>39%). The third codon position had a pronounced T bias (>53%). However, across all codon sites, the average nucleotide composition percentages were not strikingly different (A = 27%, T = 31%, C = 19%, and G = 23%).
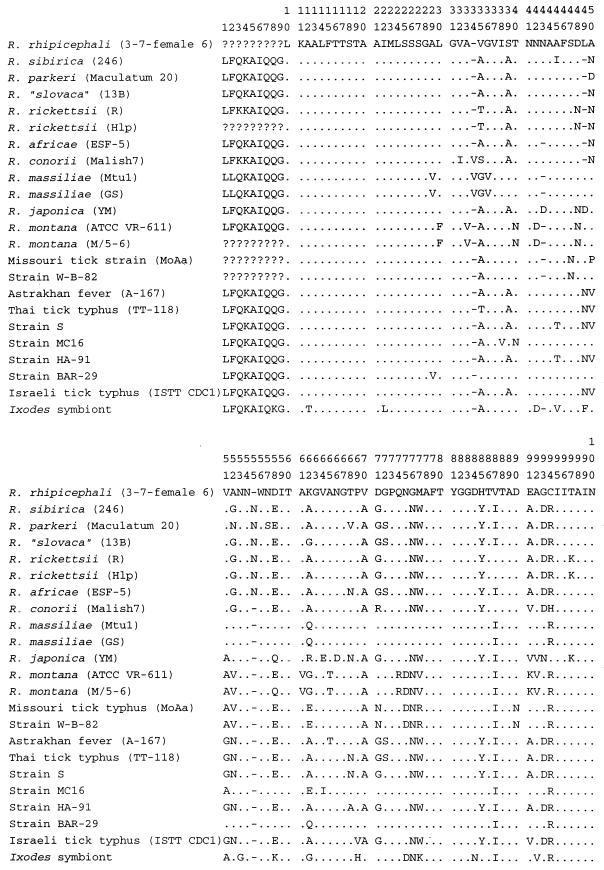
The deduced amino acid sequences for rompA are given in Fig. 7. In the rompA fragment, codons for the following amino acids were most common: alanine (16%), glycine (12%), asparagine (12%), threonine (12%), and valine (8%). R. rhipicephali was the only isolate with a cysteine residue. Within the first 55 amino acid residues of rompA, four insertion or deletion events occur (residues 34, 43, 49, and 55; Fig. 7). Of the amino acid substitutions, 13 of 19 (68%) are nonsynonymous and 6 of 19 (32%) are synonymous. When nonsynonymous substitutions occurred, 8 of 13 (62%) changed the hydrophilicity-hydrophobicity profile.
FIG. 7.
Deduced amino acid sequence alignment of the Rickettsia rOmpA proteins. Sequences were aligned with the R. rickettsii sequence beginning at amino acid 8 (4) (A, alanine; C, cystine; D, aspartic acid; E, glutamic acid; F, phenylalanine; G, glycine; H, histidine; I, isoleucine; K, lysine; L, leucine; M, methionine; N, asparagine; P, proline; Q, glutamine; R, arginine; S, serine; T, threonine; V, valine; Y, tyrosine; dots, identity; dashes, deletion; ?, unknown).
The rompA sequence from the I. scapularis symbiont is similar to that of the SFG rickettsias in both nucleotide and amino acid compositions. This isolate is slightly richer in histidine than the other rompA proteins (4 of 151 [2.7%] versus 2 of 151 [1.4%]). Although the overall sequence and amino acid residue compositions are similar, the arrangement of the amino acids is strikingly different. Twenty unique substitutions for this symbiont occur along the length of our aligned sequence (Fig. 7). Many of these amino acid substitutions are caused by single nucleotide substitutions at either the first (8 of 20) or the second (6 of 20) codon position.
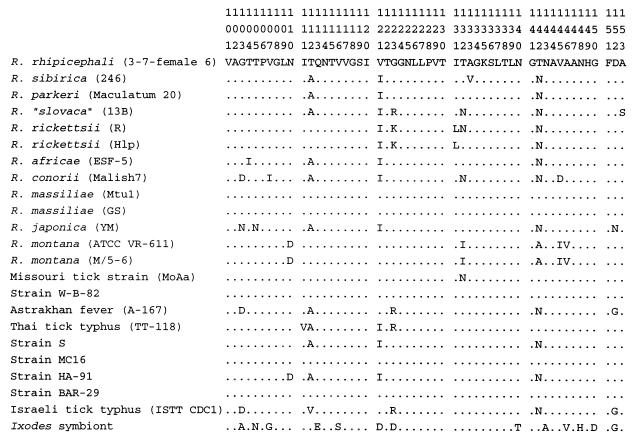
The nucleotide and amino acid base compositions for the rompB fragment are similar to those for the rompA fragment. For the aligned 376 bp of rompB, 12 first codon sites, 10 second codon sites, and 10 third codon sites were variable for the 125 sites of each codon. Nonpolar amino acid residues are slightly more common than polar amino acid residues: alanine (15%), glycine (17.5%), isoleucine (8.5%), leucine (10.5%), asparagine (8%), and threonine (12.5%). Of the 125 translated amino acids, 19 are variable, and at position 94, valine is inserted in two strains, WB-8-2 and MOAa (Fig. 8). As with rompA, 68% (13 of 19) of the amino acid substitutions are nonsynonymous and 62% (8 of 13) of these change the hydrophilic-hydrophobic nature of the residue.
FIG. 8.
Deduced amino acid sequence alignment of the rickettsial rOmpB proteins. Sequences were aligned with the R. rickettsii sequence (22) beginning at amino acid 114 (symbols and letters are as defined in the legend to Fig. 7).
Phylogenetic results: rickettsial relationships.
When the 497 bp of rompA are analyzed by the MP method, two trees result (length [L] = 167, consistency index [CI] = 70, retention index [RI] = 85). SFG divides into three clades, all with high bootstrap support (90, 97, and 100%, respectively). The largest clade (the Rickettsia japonica clade) comprises seven recognized SFG species and five strains that subdivide into two smaller clades, the Astrakhan fever rickettsia clade and the R. rickettsii clade (Fig. 9A). “Rickettsia slovaca” is placed either as the sister to the R. rickettsii clade or basal to the Astrakhan fever rickettsia and R. rickettsii clades (Fig. 9A). The second clade comprises the two R. montana isolates (isolates ATCC VR-611 and M5/6) and isolates WB-8-2 and MOAa. Rickettsia massiliae places basally and is associated with the strains MC16 and BAR-29. The same network is recovered regardless of which outgroup is used (data not shown), and random ordering of the taxa did not produce alternate topologies. The topology obtained by the MP method is much shorter than that which would be obtained randomly. Bootstrap values are low within the Astrakhan fever rickettsia clade and the R. massiliae clade, but given the low character support for these branches (Fig. 9A), this conservative test will fail to be significant (29). The tree obtained by the NJ method recovers the three clades obtained by MP analysis but differs from the tree obtained by MP analysis in that “R. slovaca” is placed basally to the Rickettsia africae clade and R. africae and Rickettsia sibirica are placed as sister taxa (data not shown). Importantly, the tree obtained by the ML method also agrees with the tree obtained by the MP method except for the placements of “R. slovaca” and R. sibirica (Fig. 9B). Furthermore, the trees obtained by the MP method are not significantly worse than the tree obtained by the ML method (P < 0.05).
FIG. 9.
Relationships of the SFG rickettsiae and placement of the Ixodes symbiont using the rompA fragment. (A) Strict consensus of MP trees (L = 112, CI = 89, RI = 94). The trees obtained by the MP method are nearly 7 SDs shorter than the shortest random tree (mean random tree length = 354, SD = 17, g1 = −0.64, g2 = 0.22). Numbers indicate bootstrap support above 50%. (B) R. japonica clade of the tree obtained by the ML method with differences highlighted with boldface lines. (C) R. montana clade showing placement of Ixodes symbiont.
We constrained our MP analysis to recover one taxon pair (Rickettsia conorii and R. sibirica) proposed by Stothard and Fuerst (63). With this tree constraint, two trees result (L = 175, CI = 66, and RI = 82). A strict consensus of the constraint trees recovers the three clades found in the tree obtained by the MP method, but with different relationships for the R. japonica clade (data not shown). We also removed all taxa except those in common with this study. Two trees were obtained, but neither recovered the topology of Stothard and Fuerst (63). Finally, a tree with this taxon pair constraint is significantly worse than the tree obtained by the ML method (P < 0.05).
The rompB data could be analyzed only for R. rickettsii and the four isolates due to a lack of data for the other species. The unrooted network recovered the pairs (R. rickettsii plus isolate Hlp2 and isolates M5/6 and WB-8-2 plus isolate MOAa) found in the larger rompA analysis. Because of the limited number of taxa, no further analyses were performed.
Relationship of the Ixodes symbiont.
The inclusion of the symbiont in our analysis affected only the placements of R. sibirica and “R. slovaca,” taxa that were unstable in the original analyses. The symbiont places as the sister to R. montana M5/6 on a long branch (37 autapomorphies) in all six trees obtained by the MP method (L = 209, CI = 69, RI = 84; Fig. 9C). The trees obtained by the MP method are nearly 7 SDs shorter than the shortest tree obtained randomly (L = 342, mean random tree L = 406, SD = 17.7, g1 = −0.52, g2 = 0.16). The tree obtained by the ML method also places the symbiont as the sister to R. montana plus the M5/6 isolate (data not shown). As in our original analyses, placement of “R. slovaca” and R. sibirica is problematic; however, these isolates always place within the largest clade rooted with R. japonica (consensus not shown). From these results and the previous analyses without the symbiont, we conclude that the relationships within the R. japonica clade are not stable.
DISCUSSION
MOAa isolate.
The MOAa isolate, when transferred to tick cell culture, displayed the intracellular growth typical of Rickettsia species. Furthermore, a monoclonal antibody (monoclonal antibody 13-2) that is specific for R. rickettsii rOmpB bound to MOAa, suggesting that the isolate was a member of the SFG rickettsiae (42).
The results of the PCR-based diagnostic assays specific for the SFG rompA and rompB genes (19, 20, 56) support the status of MOAa as a member of the SFG rickettsiae. PCR mixtures with the rompA and rompB primers yielded the expected products from MOAa and previously uncharacterized isolate WB-8-2. Furthermore, predicted restriction fragment patterns were obtained with AluI and DraI. Southern analysis of the rompA and rompB products with oligonucleotide probes indicated a difference in gene products, suggesting sequence divergence in the respective areas of the gene(s). Sequence divergence was confirmed by sequencing of the PCR products. Nevertheless, MOAa and WB-8-2 are members of the SFG rickettsiae and are closely related.
I. scapularis symbiont.
The ultrastructural morphology of the I. scapularis symbiont is similar to that of the SFG rickettsiae (37). However, unlike the other nonpathogenic rickettsiae studied in the project, we were unable to culture the symbiont in tick cell cultures. This result may reflect its restriction to the ovarian tissues and its apparent inability to cause a generalized infection in the tick. Nevertheless, our rompA PCR data support the status of the symbiont as a member of the SFG rickettsiae. Our sequence analysis demonstrated the symbiont’s relationship with R. montana M5/6, WB-8-2, and MOAa and delineated it from R. rickettsii Hlp2.
Phylogenetic utility of rOmpA and rOmpB.
Both rOmpA and rOmpB are outer membrane proteins and play an important role in rickettsial interactions in both vertebrate and invertebrate hosts. Like vertebrate major histocompatibility complex genes, the genes for these proteins are likely to evolve more rapidly than nonimmune system genes (e.g., 16S rRNA genes) because they are subject to selective pressures exerted by host immune systems. Domains I of both rompA and rompB exhibit nucleotide variation capable of resolving evolutionary relationships among isolates and species of SFG. We focused on domain I of rompA because of the specificity of the nucleotide sequences in that region for SFG rickettsiae. Domain II, which contains the tandem repeats (21), is more conserved among the three species sequenced to date, and repeats may not be homologues due to rearrangements (12). Thus, the use of domain II in phylogenetic studies is more difficult due to gene paralogy problems (65) and low variability. Nevertheless, the structure of the repeat units of rompA has been associated with rickettsial pathogenicity and invasion (1, 4). Because we lacked repeat unit sequence data, our analysis does not allow us to draw conclusions on the role of the rOmpA protein in these nonpathogenic strains. The occurrence of rOmpA in seemingly nonpathogenic rickettsiae indicates that this protein not only is important for mammalian pathogenicity but may also play a role in the survival of symbiotic rickettsiae in the tick host. Although taxon sampling was more limited, the quality and quantity of nucleotide variation in rompB are comparable to those in rompA.
A novel result of our phylogenetic analysis is the placement of isolates WB-8-2 and MOAa. All of our analyses place MOAa as being most closely related to R. montana and WB-8-2 (Fig. 9). In modern systematic practice (45, 66) evolutionary trees are reflected in the taxonomy (nomenclature) when evidence is compelling. To reflect our phylogenetic results, we propose that both strain WB-8-2 and strain MOAa be treated nomenclaturally as R. montana isolates.
Phylogeny of SFG rickettsiae.
We found that both the rompA and the rompB genes have great promise for being of use in investigating the evolutionary relationships of the strains and species in the SFG group. Phylogenies have been proposed for SFG rickettsiae by using the 16S rRNA gene (59, 63), the citrate synthase gene (60), restriction fragment length polymorphisms of rompA (21), and pulsed-field gel electrophoresis of the SFG genome (58, 61). However, none of these studies include all known rickettsial species. Furthermore, some analyses are phenetic (58, 61) and other approaches are phylogenetic (63), which further limits comparability among them. Our data do not support the relationships among the SFG rickettsiae proposed by some (21, 58, 61, 63). The lack of congruence is most likely the result of taxon sampling in prior studies, because one of the studies used the same gene, rompA (21). Our results are congruent with the most recent study in which the citrate synthase gene, gltA, was used (60).
Our results confirm that three evolutionary lineages of SFG exist (Fig. 9) (60). This interpretation is supported by the high bootstrap values obtained by our analyses (90, 97, and 100%, respectively). Our R. japonica clade is more robustly supported than it was in the earlier study (100 versus 78%), but the other two clades have comparable support (100 and 97%, respectively) (60). As in our study, the study of Roux et al. (60) does not have significant bootstrap support within the R. japonica clade. More characters (greater gene variation) are needed to provide a robust estimate of these relationships. The two studies disagree on rooting. In our study, the R. massiliae isolate is basal, whereas in the other study the R. montana isolate is basal. Again, discordance is most likely due to differences in taxon sampling and rooting. Complete overlap of taxon sampling is not possible because many taxa either lack rompA or have not yet been sequenced. The rooting issue is discussed below (see the section Biogeographic origins of NW rickettsiae).
An understanding of phylogenetic relationships provides a predictive framework for investigating the evolution of SFG rickettsias by comparative biology-based methods (8, 18). The pattern of relationships has implications for the origin of SFG rickettsiae in the NW and the evolution of their tick host associations. Furthermore, the placement of the Ixodes symbiont with the R. montana clade provides an interesting insight into the possible origins of SFG rickettsiae. Nonpathogenic SFG rickettsiae appear to be widespread among diverse tick genera, and some possess outer membrane proteins also found in pathogenic species. The association with ticks and the presence of outer membrane proteins suggest a mechanism by which pathogenic strains may continuously evolve from symbiotic rickettsiae through changes in outer membrane proteins. Ticks harboring these rickettsiae could be viewed as evolutionary reservoirs for the next generation of emerging pathogens.
Infections of individual ticks with multiple microbial pathogens have been documented for I. scapularis (49, 53). However, evidence as to whether or not pathogenic and nonpathogenic rickettsiae can coexist in the same tick is equivocal. Rocky Mountain wood ticks, Dermacentor andersoni, previously infected with the nonpathogenic organism R. montana could not be superinfected with R. rickettsii (9). Moreover, dual infections with different SFG rickettsial species have not been detected during epidemiological surveys (20, 51). In contrast, the nonpathogenic rickettsia Rickettsia peacockii (formerly, the East Side agent) and virulent R. rickettsii can coexist within the same tick (10), but the former interferes with the ability of D. andersoni to transmit the latter (46). The rompA gene sequence of R. peacockii is most closely related to those clustered within our largest SFG clade which includes the R. rickettsii and Astrakhan fever rickettsia clades, suggesting a close relationship between this nonpathogenic organism and virulent R. rickettsii. Considering that rOmpA is believed to play a role in rickettsial invasion of and movement through mammalian host cells (26), it is of interest that the R. peacockii rOmpA protein is either truncated or not expressed (46), suggesting a possible molecular basis for its lack of mammalian infectivity. Although at present it is not known whether all populations of I. scapularis carry the rickettsial symbiont described here, infection with this organism might be a factor in the inability of this tick species to transmit virulent R. rickettsii. Interference seems to be strongest between closely related species, as in the case of R. rickettsii and R. peacockii (46). Neither the mechanism nor the molecular basis responsible for the rickettsial interference phenomenon in ticks has been elucidated.
Biogeographic origins of NW rickettsiae.
Analysis of the biogeographic origins of the NW rickettsiae was based exclusively on North American isolates because of a lack of sufficient information on South American isolates. We assume an OW origin for the genus Rickettsia, because the typhus group and other, unplaced rickettsiae originate in the OW and place basally with respect to our root of Rickettsia rhipicephali (63). Roux et al. (60) suggested an NW origin for rickettsias, but their outgroup, Bartonella henselae, is a very distant outgroup (see Fig. 1 of Roux et al. [60]). In cases in which distant outgroups are used, root placement will be nearly random and is not reliable (65). With this one exception, the patterns that we describe below are the same as those found in the study of Roux et al. (60).
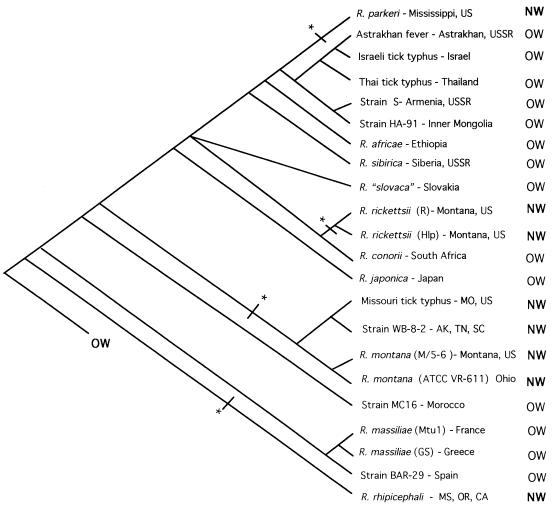
A comparison of our tree (Fig. 10) with the two alternative patterns in Fig. 1 demonstrates that our results support the hypothesis of multiple NW introductions (Fig. 1B) and not the hypothesis of a single origin (Fig. 1A). Our tree obtained by the MP method predicts that SFG rickettsiae have independently been introduced into the NW as many as four times (Fig. 9). The presumed direction is from OW to NW, because the immediate outgroups are OW, and comparative methods use the most parsimonious reconstruction of ancestral states (8, 25). Within the Astrakhan fever rickettsia-R. rickettsii clade, R. parkeri (NW) originated separately from R. rickettsii R (NW) and its nonpathogenic strain, Hlp2 (NW). R. montana (NW) and three associated strains arose independently of these and R. rhipicephali (NW).
FIG. 10.
Origin of NW species and biogeography of SFG rickettsiae. Geographic data were obtained from GenBank and Roux et al. (57). The immediate ancestor was assumed to be an OW isolate (59, 63). NW, NW isolates; Old World isolates. Asterisks indicate transition from OW to NW.
We asked if trees that gave either a single origin of North American species or two or three origins were significantly different from our trees obtained by the MP and ML methods by using the test of Kishino and Hasegawa (32). When we constrained all NW rickettsias to originate once, the resultant three trees were significantly longer than the trees obtained by the MP method (+48 steps; SD = 17) and significantly worse than the tree obtained by the ML method (P < 0.05). Similarly, if we test the likelihood of two independent origins of North American rickettsias, the trees are significantly less likely than the tree obtained by the ML method (P < 0.05). Even three independent origins are significantly worse than the four of the tree obtained by the ML method (P < 0.05).
Assuming that NW SFG rickettsiae arose from four independent origins, we note that Rickettsia parkeri is associated with strains geographically found in the former USSR, Israel, and Asia (Fig. 10). This result is also supported by recent work that showed an association of R. parkeri and Astrakhan fever rickettsia (60). R. rickettsii is associated with R. conorii from South Africa, but we did not include in our analysis other African R. conorii strains, which might allow identification of the sister strain or species of R. rickettsii. Our data are consistent with an African origin of R. rickettsii. The remaining NW species, R. montana and R. rhipicephali, are associated with Mediterranean rickettsial species and strains. This biogeographic pattern suggests that the introduction of SFG is related to mammalian immigration influxes from the OW.
Evolution of arthropod hosts and rickettsias.
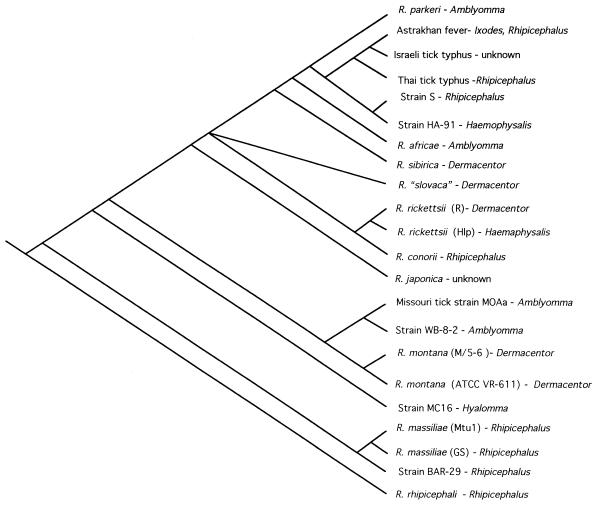
Our trees obtained by the MP and ML methods imply frequent arthropod host shifts by SFG rickettsias. The tick genus Rhipicephalus is associated with all the basal, NW, and Mediterranean rickettsiae (Fig. 11). Subsequent shifts are then made to ticks in the genera Hyalomma, Dermacentor, Amblyomma, Haemaphysalis, and Ixodes. We constrained our analyses to produce single origins of arthropod vectors; that is, all rickettsiae with a particular tick host originate as a clade. There were four constraint groups: Dermacentor rickettsiae, Amblyomma rickettsiae, Haemaphysalis rickettsiae, and Rhipicephalus rickettsiae. The remaining two rickettsiae whose hosts are unknown were unconstrained. The resultant 85 trees were 246 steps longer than the trees obtained by the MP method (CI = 44, RI = 56; >4 SDs). The log-likelihood test also discounts a single origin of tick hosts (P < 0.05). Thus, a hypothesis of classic coevolution (15) between ticks and rickettsiae is not supported. Our data are most consistent with the hypothesis that SFG rickettsiae were initially associated with Rhipicephalus ticks and subsequently radiated into other arthropod hosts multiple times. Elucidating the exact pattern of arthropod host switching will depend upon further studies of hosts and the addition of other Rickettsia species and strains in a larger analysis (24).
FIG. 11.
Evolution of tick hosts and their SFG rickettsiae (tick host data were obtained from Roux et al. [57]).
Origin of SFG rickettsiae.
The trees obtained by both the MP and the ML methods place the Ixodes symbiont as a member of the R. montana clade (Fig. 9C). It is premature, however, to conclude that we have definitively established the symbiont’s relationship. A classic problem with long-branch taxa (many autapomorphies) is that they are attracted to other long branches by chance (17, 65). The addition of other SFG taxa or symbionts could affect this symbiont’s placement. We conclude that further testing with additional gene regions is required.
The occurrence of rompA in the I. scapularis symbiont raises the intriguing possibility that nonpathogenic mutualists have converted into pathogenic rickettsiae (or vice versa). With so few rompA sequences for nonpathogenic rickettsiae, it is not possible to determine the direction of this invasion, nor is it possible to determine whether there have been single or multiple conversions of mutualists into pathogens. Little is known about rickettsial genetic exchange mechanisms, but lateral gene transfer within the tick is speculated to play a role in the generation of new strains of the spirochete B. burgdorferi (31, 35, 40). Resolution of these questions requires that more nonpathogenic tick symbionts be surveyed for rOmpA proteins. Further study of these rickettsial symbionts of ticks will provide important insights into the evolution of pathogenic Rickettsia strains and may answer the vexing question of the origin of rickettsias themselves.
ACKNOWLEDGMENTS
We thank Robert Zink for comments on the manuscript.
This work was supported by NIH grant AR37909 (to T. J. Kurtti) and NSF grant DEB-9306755 (to S. J. Weller).
Footnotes
Paper no. 981170002 of the Minnesota Agricultural Experiment Station.
REFERENCES
- 1.Anacker R L, List R H, Mann R E, Wiedbrauk D L. Antigenic heterogeneity in high- and low-virulence strains of Rickettsia rickettsii revealed by monoclonal antibodies. Infect Immun. 1986;51:653–660. doi: 10.1128/iai.51.2.653-660.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anacker R L, Mann R E, Gonzales C. Reactivity of monoclonal antibodies to Rickettsia rickettsii with spotted fever and typhus group rickettsiae. J Clin Microbiol. 1987;25:167–171. doi: 10.1128/jcm.25.1.167-171.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anacker R L, Philip R N, Williams J C, List R H, Mann R E. Biochemical and immunochemical analysis of Rickettsia rickettsii strains of various degrees of virulence. Infect Immun. 1984;44:559–564. doi: 10.1128/iai.44.3.559-564.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson B E, McDonald G A, Jones D C, Regnery R L. A protective protein antigen of Rickettsia rickettsii has tandemly repeated, near-identical sequences. Infect Immun. 1990;58:2760–2769. doi: 10.1128/iai.58.9.2760-2769.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avise J C, Nelson W S. Reply. Mol Phylogenet Evol. 1995;4:353–356. [Google Scholar]
- 6.Barbour A G. Isolation and cultivation of Lyme disease spirochetes. Yale J Biol Med. 1984;57:521–525. [PMC free article] [PubMed] [Google Scholar]
- 7.Bell E J, Kohls G M, Stoenner H G, Lackman D B. Nonpathogenic rickettsias related to the spotted fever group isolated from ticks, Dermacentor variabilis and Dermacentor andersoni from eastern Montana. J Immunol. 1963;90:770–781. [PubMed] [Google Scholar]
- 8.Brooks D R, Mclennan D A. Phylogeny, ecology and behavior. Chicago, Ill: University of Chicago Press; 1991. [Google Scholar]
- 9.Burgdorfer W. Ecological and epidemiological considerations of Rocky Mountain spotted fever and scrub typhus. In: Walker D H, editor. Biology of rickettsial diseases. Boca Raton, Fla: CRC Press, Inc.; 1988. pp. 33–50. [Google Scholar]
- 10.Burgdorfer W, Hayes S F, Mavros A J. Nonpathogenic rickettsiae in Dermacentor andersoni: a limiting factor for the distribution of Rickettsia rickettsii. In: Burgdorfer W, Anacker R L, editors. Rickettsiae and rickettsial diseases. New York, N.Y: Academic Press, Inc.; 1981. pp. 585–594. [Google Scholar]
- 11.Burgdorfer W, Hayes S F, Thomas L A, Lancaster J L. A new spotted fever group rickettsia from the lone star tick, Amblyomma americanum. In: Burgdorfer W, Anacker R L, editors. Rickettsiae and rickettsial diseases. New York, N.Y: Academic Press, Inc.; 1981. pp. 595–602. [Google Scholar]
- 12.Crocquet-Valdes P A, Weiss K, Walker D H. Sequence analysis of the 190-kDa antigen-encoding gene of Rickettsia conorii (Malish 7 strain) Gene. 1994;140:115–119. doi: 10.1016/0378-1119(94)90740-4. [DOI] [PubMed] [Google Scholar]
- 13.Dasch G A, Weiss E. The genera Rickettsia, Rochalimaea, Ehrlichia, Cowdria, and Neorickettsia. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. Vol. 3. New York, N.Y: Springer-Verlag; 1992. pp. 2407–2470. [Google Scholar]
- 14.Eggleston P E, Raven P H, editors. Phylogenetics and ecology. London, United Kingdom: Academic Press; 1994. [Google Scholar]
- 15.Ehrlich P R, Raven P H. Butterflies and plants: a study in coevolution. Evolution. 1964;18:586–608. [Google Scholar]
- 16.Eremeeva M, Yu X, Raoult D. Differentiation among spotted fever group rickettsiae species by analysis of restriction fragment length polymorphism of PCR-amplified DNA. J Clin Microbiol. 1994;32:803–810. doi: 10.1128/jcm.32.3.803-810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felsenstein J. Cases in which parsimony and compatibility methods will be positively misleading. Syst Zool. 1978;27:783–791. [Google Scholar]
- 18.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 19.Gage K L, Gilmore R D, Karstens R H, Schwan T G. Detection of Rickettsia rickettsii in saliva, hemolymph and triturated tissues of infected Dermacentor andersoni ticks by polymerase chain reaction. Mol Cell Probes. 1992;6:333–341. doi: 10.1016/0890-8508(92)90010-u. [DOI] [PubMed] [Google Scholar]
- 20.Gage K L, Schrumpf M E, Karstens R H, Burgdorfer W, Schwan T G. DNA typing of rickettsiae in naturally infected ticks using a polymerase chain reaction/restriction fragment length polymorphism system. Am J Trop Med Hyg. 1994;50:247–260. doi: 10.4269/ajtmh.1994.50.247. [DOI] [PubMed] [Google Scholar]
- 21.Gilmore R D, Hackstadt T. The DNA polymorphism in the conserved 190 kD antigen gene repeat among spotted fever group rickettsiae. Biochim Biophys Acta. 1991;1097:77–80. doi: 10.1016/0925-4439(91)90027-7. [DOI] [PubMed] [Google Scholar]
- 22.Gilmore R D, Joste N, McDonald G A. Cloning, expression and sequence analysis of the gene encoding the 120 kD surface-exposed protein of Rickettsia rickettsii. Mol Microbiol. 1989;3:1579–1586. doi: 10.1111/j.1365-2958.1989.tb00143.x. [DOI] [PubMed] [Google Scholar]
- 23.Goddard J, Norment B R. Spotted fever group rickettsiae in the lone star tick, Amblyomma americanum (Acari: Ixodidae) J Med Entomol. 1986;23:465–472. doi: 10.1093/jmedent/23.5.465. [DOI] [PubMed] [Google Scholar]
- 24.Hafner M S, Nadler S A. Phylogenetic trees support the coevolution of parasites and their hosts. Nature (London) 1988;332:258–259. doi: 10.1038/332258a0. [DOI] [PubMed] [Google Scholar]
- 25.Harvey P H, Pagel M D. The comparative method in evolutionary biology. Oxford, United Kingdom: Oxford University Press; 1991. [Google Scholar]
- 26.Heinzen R A, Hayes S F, Peacock M G, Hackstadt T. Directional actin polymerization associated with spotted fever group rickettsia infection of Vero cells. Infect Immun. 1993;61:1926–1935. doi: 10.1128/iai.61.5.1926-1935.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higuchi H. Simple and rapid preparation of samples for PCR. In: Ehrlich H A, editor. PCR technology, principles and applications for DNA amplification. New York, N.Y: Stockton Press; 1989. pp. 31–38. [Google Scholar]
- 28.Hillis D M. Discriminating between phylogenetic signal and random noise in DNA sequences. In: Miyamoto M M, Cracraft J, editors. Phylogenetic analysis of DNA sequences. New York, N.Y: Oxford University Press; 1991. pp. 278–294. [Google Scholar]
- 29.Hillis D M, Bull J J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol. 1993;42:182–192. [Google Scholar]
- 30.Hillis D M, Huelsenbeck J P. Signal, noise, and reliability in molecular phylogenetic analyses. J Hered. 1992;83:189–195. doi: 10.1093/oxfordjournals.jhered.a111190. [DOI] [PubMed] [Google Scholar]
- 31.Jauris-Heipke S, Liegl G, Preac-Mursic V, Rössler D, Schwab E, Soutschek E, Will G, Wilske B. Molecular analysis of genes encoding outer surface protein C (OspC) of Borrelia burgdorferii sensu lato: relationship to ospA genotype and evidence of lateral gene exchange of ospC. J Clin Microbiol. 1995;33:1860–1866. doi: 10.1128/jcm.33.7.1860-1866.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kishino H, Hasegawa M. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in Hominoidea. J Mol Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- 33.Kumar S, Tamura K, Nei M. MEGA (molecular evolutionary genetic analysis), version 1.02. Privately distributed by authors. University Park: Pennsylvania State University; 1994. [Google Scholar]
- 34.Kurtti T J, Munderloh U G, Samish M. Effect of medium supplements on tick cells in culture. J Parasitol. 1982;68:930–935. [PubMed] [Google Scholar]
- 35.Livey I, Gibbs C P, Schuster R, Dorner F. Evidence for lateral transfer and recombination in OspC variation in Lyme disease Borrelia. Mol Microbiol. 1995;18:257–269. doi: 10.1111/j.1365-2958.1995.mmi_18020257.x. [DOI] [PubMed] [Google Scholar]
- 36.Maddison W P, Donoghue M J, Maddison D R. Outgroup analysis and parsimony. Syst Zool. 1984;33:83–103. [Google Scholar]
- 37.Magnarelli L A, Andreadis T G, Stafford I K C, Holland C J. Rickettsiae and Borrelia burgdorferi in Ixodid ticks. J Clin Microbiol. 1991;29:2798–2804. doi: 10.1128/jcm.29.12.2798-2804.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 39.Marchuk D, Drumm M, Saulino A, Collins F S. Construction of T-vectors, a rapid and general system for direct cloning of unmodified PCR products. Nucleic Acids Res. 1991;19:1154. doi: 10.1093/nar/19.5.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Margolis N, Hogan D, Tilly K, Rosa P A. Plasmid location of Borrelia purine biosynthesis gene homologs. J Bacteriol. 1994;176:6427–6432. doi: 10.1128/jb.176.21.6427-6432.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moulder J W. Comparative biology of intracellular parasitism. Microbiol Rev. 1985;49:298–337. doi: 10.1128/mr.49.3.298-337.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Munderloh, U. G., S. F. Hayes, J. Cummings, and T. J. Kurtti. Microscopic analysis of spotted fever rickettsia movement through tick cells. Microsc. Microanal., in press.
- 43.Munderloh U G, Kurtti T J. Formulation of medium for tick cell culture. Exp Appl Acarol. 1989;7:219–229. doi: 10.1007/BF01194061. [DOI] [PubMed] [Google Scholar]
- 44.Munderloh U G, Liu Y, Wang M, Chen C, Kurtti T J. Establishment, maintenance and description of cell lines from the tick Ixodes scapularis. J Parasitol. 1994;80:533–543. [PubMed] [Google Scholar]
- 45.Nelson G, Platnick N. Systematics and biogeography. Cladistics and vicariance. New York, N.Y: Columbia University Press; 1981. [Google Scholar]
- 46.Niebylski M L, Schrumpf M E, Burgdorfer W, Fischer E R, Gage K L, Schwan T G. Rickettsia peacockii sp. nov., a new species infecting wood ticks, Dermacentor andersoni, in western Montana. Int J Syst Bacteriol. 1997;47:446–452. doi: 10.1099/00207713-47-2-446. [DOI] [PubMed] [Google Scholar]
- 47.Noda H, Munderloh U G, Kurtti T J. Endosymbionts of ticks and their relationship to Wolbachia and tick-borne pathogens of man and animals. Appl Environ Microbiol. 1997;63:3926–3932. doi: 10.1128/aem.63.10.3926-3932.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O’Neill S L, Giordano R, Colbert A M E, Karr T L, Robertson H M. 16S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc Natl Acad Sci USA. 1992;89:2699–2702. doi: 10.1073/pnas.89.7.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pancholi P, Kolbert C P, Mitchell P D, Reed K D, Dumler J S, Bakken J S, Telford III S R, Persing D H. Ixodes dammini as a potential vector of human granulocytic ehrlichiosis. J Infect Dis. 1995;172:1007–1212. doi: 10.1093/infdis/172.4.1007. [DOI] [PubMed] [Google Scholar]
- 50.Parker R R, Pickens E G, Lackman D B, Bell E J, Thraikill F B. Isolation and characterization of Rocky Mountain spotted fever from the rabbit tick Haemaphysalis leporispalustris Packard. Public Health Rep. 1951;66:455–463. [PMC free article] [PubMed] [Google Scholar]
- 51.Philip R N, Casper E A. Serotypes of spotted fever group rickettsiae isolated from Dermacentor andersoni (Stiles) ticks in western Montana. Am J Trop Med Hyg. 1981;30:230–238. doi: 10.4269/ajtmh.1981.30.230. [DOI] [PubMed] [Google Scholar]
- 52.Philip R N, Casper E A, Burgdorfer W, Gerloff R K, Hughes L E, Bell E J. Serologic typing of rickettsiae of the spotted fever group by microimmunofluorescence. J Immunol. 1978;121:1961–1968. [PubMed] [Google Scholar]
- 53.Piesman J, Hicks T C, Sinsky R J, Obiri G. Simultaneous transmission of Borrelia burgdorferi and Babesia microti by individual nymphal Ixodes dammini. J Clin Microbiol. 1987;25:2012–2013. doi: 10.1128/jcm.25.10.2012-2013.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Policastro P, Munderloh U G, Fischer E R, Hackstadt T. Rickettsia rickettsii growth and temperature-inducible protein expression in embryonic tick cell lines. J Med Microbiol. 1997;46:839–845. doi: 10.1099/00222615-46-10-839. [DOI] [PubMed] [Google Scholar]
- 55.Potts W K, Wakeland E K. Evolution of diversity at the major histocompatibility complex. Trends Ecol Evol. 1990;5:181–187. doi: 10.1016/0169-5347(90)90207-T. [DOI] [PubMed] [Google Scholar]
- 56.Regnery R L, Spruill C L, Plikaytis B D. Genotypic identification of rickettsiae and estimation of intraspecies divergence for portions of two rickettsial genes. J Bacteriol. 1991;173:1576–1589. doi: 10.1128/jb.173.5.1576-1589.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roux V, Fournier P-E, Raoult D. Differentiation of spotted fever group rickettsiae by sequencing and analysis of restriction fragment length polymorphism of PCR-amplified DNA of the gene encoding the protein rOmpA. J Clin Microbiol. 1996;34:2058–2065. doi: 10.1128/jcm.34.9.2058-2065.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roux V, Raoult D. Genotypic identification and phylogenetic analysis of the spotted fever group rickettsiae using pulsed-field gel electrophoresis. J Bacteriol. 1993;175:4895–4904. doi: 10.1128/jb.175.15.4895-4904.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roux V, Raoult D. Phylogenetic analysis of the genus Rickettsia by 16S rDNA sequencing. Res Microbiol. 1995;146:385–396. doi: 10.1016/0923-2508(96)80284-1. [DOI] [PubMed] [Google Scholar]
- 60.Roux V, Rydkina E, Eremeeva M, Raoult D. Citrate synthase gene comparison, a new tool for phylogenetic analysis, and its application for the rickettsiae. Int J Syst Bacteriol. 1997;47:252–261. doi: 10.1099/00207713-47-2-252. [DOI] [PubMed] [Google Scholar]
- 61.Roux V, Drancourt M, Raoult D. Determination of genome sizes of rickettsia spp. within the spotted fever group, using pulsed-field electrophoresis. J Bacteriol. 1992;174:7455–7457. doi: 10.1128/jb.174.22.7455-7457.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stothard D R, Fuerst P A. Evolutionary analysis of the spotted fever and typhus groups of rickettsia using 16S rRNA sequences. Syst Appl Microbiol. 1995;18:52–61. [Google Scholar]
- 64.Swofford D L. PAUP—phylogenetic analysis using parsimony (MAC version 3.0g). Champaign: Illinois Natural History Survey; 1989. [Google Scholar]
- 65.Swofford D L, Olsen G J, Wadell P J, Hillis D M. Phylogenetic inference. In: Hillis D M, Moritz C, Mable B K, editors. Molecular systematics. Sunderland, Mass: Sinauer; 1996. [Google Scholar]
- 66.Wiley E O. Phylogenetic systematics. The principles and practice of phylogenetic systematics. New York, N.Y: John Wiley & Sons, Inc.; 1981. [Google Scholar]
- 67.Winkler H H. Rickettsia species (as organisms) Annu Rev Microbiol. 1990;44:131–153. doi: 10.1146/annurev.mi.44.100190.001023. [DOI] [PubMed] [Google Scholar]