Abstract
Objective:
We report the development and validation of a combined DNA/RNA next-generation sequencing (NGS) platform to improve the evaluation of pancreatic cysts.
Background and Aims:
Despite a multidisciplinary approach, pancreatic cyst classification, such as a cystic precursor neoplasm, and the detection of high-grade dysplasia and early adenocarcinoma (advanced neoplasia) can be challenging. NGS of preoperative pancreatic cyst fluid improves the clinical evaluation of pancreatic cysts, but the recent identification of novel genomic alterations necessitates the creation of a comprehensive panel and the development of a genomic classifier to integrate the complex molecular results.
Methods:
An updated and unique 74-gene DNA/RNA-targeted NGS panel (PancreaSeq Genomic Classifier) was created to evaluate 5 classes of genomic alterations to include gene mutations (e.g., KRAS, GNAS, etc.), gene fusions and gene expression. Further, CEA mRNA (CEACAM5) was integrated into the assay using RT-qPCR. Separate multi-institutional cohorts for training (n=108) and validation (n=77) were tested, and diagnostic performance was compared to clinical, imaging, cytopathologic, and guideline data.
Results:
Upon creation of a genomic classifier system, PancreaSeq GC yielded a 95% sensitivity and 100% specificity for a cystic precursor neoplasm, and the sensitivity and specificity for advanced neoplasia were 82% and 100%, respectively. Associated symptoms, cyst size, duct dilatation, a mural nodule, increasing cyst size, and malignant cytopathology had lower sensitivities (41–59%) and lower specificities (56–96%) for advanced neoplasia. This test also increased the sensitivity of current pancreatic cyst guidelines (IAP/Fukuoka and AGA) by >10% and maintained their inherent specificity.
Conclusions:
PancreaSeq GC was not only accurate in predicting pancreatic cyst type and advanced neoplasia but also improved the sensitivity of current pancreatic cyst guidelines.
Keywords: intraductal oncocytic papillary neoplasm, intraductal papillary mucinous neoplasm, mucinous cystic neoplasm, pancreatic ductal adenocarcinoma, pancreatic neuroendocrine tumor, serous cystadenoma, pseudocyst
Pancreatic cysts are a diverse group of lesions with protean clinical, imaging, and pathologic features. Broadly, cysts can be subdivided into non-neoplastic and neoplastic cysts.1 Non-neoplastic cysts include pseudocysts, retention cysts, and others that can arise due to chronic pancreatitis. The remaining cysts are neoplastic and consist of cystic neoplasms without malignant potential, noninvasive cystic precursors to invasive pancreatic ductal adenocarcinoma (PDAC), malignant cystic neoplasms, and cystic degeneration of solid malignancies. Serous cystadenomas (SCAs) are neoplastic cysts and, while they can grow to be large, these neoplasms have no malignant potential. In contrast, IPMNs, MCNs, and intraductal oncocytic papillary neoplasms (IOPNs) are noninvasive cystic neoplasms that may progress to invasive PDAC, and hence referred to as “cystic precursor neoplasms”.2 Due to their histologic features, IOPNs are universally regarded to harbor at least high-grade dysplasia. Cystic pancreatic neuroendocrine tumors (cPanNETs) are malignant cystic neoplasms, whereas solid pancreatic malignancies can also present as cystic lesions (eg, PDAC).
Considering the wide array of entities, it is not surprising that the frequent detection of a cyst by imaging represents a clinical conundrum. In the preoperative setting, classifying a cyst can be difficult. Many neoplastic cysts, such as IPMNs and SCAs, and even non-neoplastic cysts, can clinically mimic one another. This issue is more challenging when attempting to identify high-grade dysplasia and early PDAC (advanced neoplasia) arising in an IPMN/MCN. As a result, consensus- and evidence-based guidelines have been developed to improve the evaluation of pancreatic cysts.3–6 However, current guidelines, such as those by the American Gastroenterology Association (AGA) and the International Association of Pancreatology (IAP/Fukuoka), demonstrate suboptimal sensitivity and specificity for detecting advanced neoplasia.7,8
To address these limitations, targeted DNA-based next-generation sequencing (NGS) of pancreatic cyst fluid was developed and validated in the retrospective and prospective clinical settings.9–13 Studies have shown that mutations in KRAS/GNAS and VHL achieve a high specificity for IPMNs/MCNs and SCAs, respectively.14–16 Furthermore, genomic alterations in “high-risk” genes, such as TP53, SMAD4, PIK3CA, and PTEN, are associated with advanced neoplasia.9–12,17 There are, however, numerous shortcomings associated with the sensitivity of this targeted DNA-based approach. For instance, class 2 and class 3 BRAF mutations alterations have been described as a unique feature of IPMNs. Similarly, recurrent gene fusions (GFs), such as those involving BRAF and PRKACA/B were discovered to be frequently associated with IPMNs and IOPNs, respectively.2,18,19 To our knowledge, none of the aforementioned genomic alterations including the assessment of GF are evaluable with current, clinically available gene panels for pancreatic cyst fluid testing.18 Moreover, the estimation of mutant allele frequencies (AFs) for individual genes and identification of copy number alterations (CNAs) should be of consideration when determining advanced neoplasia.10,18,20 Hence, an updated version of DNA-based NGS panels, such as PancreaSeq, is required.10,18 The aims of this study were to: (1) develop a DNA/RNA-based NGS panel (PancreaSeq Genomic Classifier) for the evaluation of endoscopic ultrasound-fine needle aspiration (EUS-FNA) cyst fluid, (2) create a genomic classifier to simplify the data analysis and reporting of cystic precursor neoplasms and associated advanced neoplasia, and (3) validate PancreaSeq GC to determine its diagnostic performance in comparison to clinical and imaging parameters for the assessment of pancreatic cysts.
MATERIALS AND METHODS
Study Population
Study approval was obtained from the University of Pittsburgh Institutional Review Board (IRB# STUDY19070069). EUS-FNA pancreatic cyst fluid specimens for training and validation were taken from the specimen archives of the Molecular and Genomic Pathology (MGP) laboratory at the University of Pittsburgh Medical Center (UPMC) between 2012 and 2019. Since 2006, pancreatic cyst fluid specimens have been submitted to the UPMC MGP laboratory from multiple institutions for routine clinical care in the assessment of a pancreatic cyst (Fig. 1).2,7,10,14,15 Cross-referencing surgical resection pathology follow-up that was submitted from participating institutions, pancreatic cyst fluid specimens with sufficient nucleic acids were identified and in total consisted of 185 specimens with confirmed diagnostic pathology (Supplementary Table 1, Supplemental Digital Content 1, http://links.lww.com/SLA/A622). The specimens were further subdivided into training (n=108) and validation (n=77) cohorts (see Supplementary Data, Supplemental Digital Content 1, http://links.lww.com/SLA/A622 for statistical considerations). Patient data included demographics, clinical presentation, EUS findings, CEA levels if available, cytopathologic diagnoses, and diagnostic surgical diagnoses.21 IPMNs, MCNs, and IOPNs were defined as cystic precursor neoplasms, whereas an IPMN, MCN, and IOPN with high-grade dysplasia and/or an associated invasive adenocarcinoma were categorized as advanced neoplasia.
FIGURE 1.
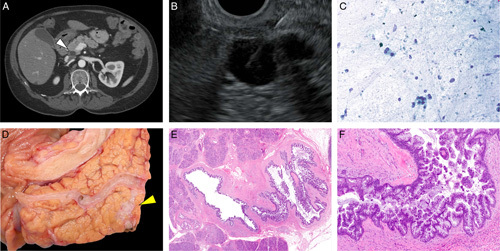
A pancreatic cyst that was evaluated by PancreaSeq GC. A and B, An incidentally identified 1.2 cm cyst (white arrowhead, pancreatic cyst) that communicated with the pancreatic duct and clinically suspicious for a branch-duct IPMN without concerning imaging features. C, Cytopathology of EUS-FNA pancreatic cyst fluid detected atypical ductal cells in a background of debris. Upon surgical resection, D, the macroscopic (yellow arrowhead, pancreatic cyst) and E, microscopic findings were consistent with an IPMN involving the side-branch. F, In addition, high-grade dysplasia was identified within the IPMN; however, no definitive invasive adenocarcinoma was seen. PancreaSeq GC has a cystic precursor score of 6 and a risk score of 5 (Supplementary Table 1, Supplemental Digital Content 1, http://links.lww.com/SLA/A622, Case 28).
PancreaSeq GC and Data Analysis
Nucleic acids were isolated from EUS-FNA pancreatic cyst fluid specimens that were collected into a DNA/RNA preservation solution. DNA and mRNA were isolated using the MagNA Pure Compact instrument (Roche). Extracted DNA and RNA were quantitated on the Glomax Discover using the QuantiFluor ONE dsDNA System and the QuantiFluor RNA system, respectively (Promega). NGS libraries of genes summarized within the Supplementary Data, Supplemental Digital Content 1, http://links.lww.com/SLA/A622 were created from 10 ng of DNA and 10 ng of RNA using the Ion AmpliSeq Library kit PLUS and Ion Xpress Barcode Adapters as previously described.22,23 These 74 genes include those implicated in pancreatic cysts and associated neoplasms and include “hot-spot” mutations, such as KRAS, GNAS, etc., copy number alterations/loss of heterozygosity, GFs, gene expression alterations (GEAs; KRT7, KRT20, and PGK1) and CHGA RNA expression. The libraries were normalized for template preparationon the Ion Chef and sequenced on an Ion S5 System according to the manufacturer’s instructions (Thermo Fisher Scientific). The Torrent Suite Software v5.12 (Thermo Fisher Scientific) and an in-house developed software Variant Explorer v2 were used for data analysis and interpretation. In parallel, one-step quantitative reverse transcription PCR (RT-qPCR) for CEACAM5 was performed.
CEACAM5 mRNA Expression by RT-qPCR
Primers and probes for the CEACAM5 gene and the GUSB housekeeping control gene were designed and validated in-house to measure mRNA expression of these genes. One-step quantitative reverse transcription PCR (RT-qPCR) was performed using TaqMan One-step RT to Ct Master Mix kit and run on the ABI7500 real-time PCR instrument according to manufacturer’s instructions (Applied Biosystems). Relative quantity of CEACAM5 in Gene Expression Units (GEU) was calculated according to the 2−ΔΔCt method.24 Additional details are provided within the Supplementary Data, Supplemental Digital Content 1, http://links.lww.com/SLA/A622 section.
PancreaSeq GC Data Analysis
For the prediction of a cystic precursor neoplasm, each detected genomic alteration was annotated to receive a value (0–3) based on the strength of its association with a specific cystic precursor neoplasm. A Genomic Classifier (GC) score for cystic precursor neoplasms was calculated as a sum of the following individual values: GC cystic precursor neoplasm score = x KRAS,GNAS,RNF43,BRAF,PRKACA&PRKACB + x CEACAM5 + x KRT7/20, x = weighted value, 0 to 3. In parallel, CHGA expression is performed for each sample and evaluated for the presence of a neuroendocrine tumor in samples that did not meet a cutoff for a cystic precursor neoplasm. A CHGA value of >50% was deemed as positive result. For the prediction of advanced neoplasia arising from a cystic precursor neoplasm, a similar GC score was developed with each identified genomic alteration receiving a value (0–3) based on its reported association with high-grade dysplasia and PDAC.2,7,10,14,15,19,25–28 A risk score for advanced neoplasia for PancreaSeq GC was calculated as follows: GC risk score = xSNV/indel + xAF + xCNA/LOH + xGF + xGEA); x = weighted value, 0 to 3; SNV/indel [single nucleotide variant/small insertions and deletions, AF, CNA/LOH (loss of heterozygosity), GFs, and GEAs are indicators of genomic alteration type]. Weighted values and scoring were calculated using bioinformatic algorithms using the aforementioned in-house developed Variant Explorer v2 software. Additional details are provided within the Supplementary Data, Supplemental Digital Content 1, http://links.lww.com/SLA/A622 section.
Statistical Analysis
Empirical sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) with 95% Wilson confidence intervals were calculated for each diagnostic parameter. The true positive fraction (sensitivity) and the false positive fraction (1-specificity) were plotted against the false positive rate in receiver-operative characteristic curves. The area under the curve (AUC) was computed by the trapezoidal method by the method of DeLong.29 All statistical analyses were performed using the SPSS Statistical software, V.28 (IBM), and statistical significance was defined as a P value of <0.05.
RESULTS
Establishing a Genomic Classifier for Cystic Precursor Neoplasms
The PancreaSeq GC was trained with 108 preoperative EUS-FNA pancreatic cyst fluid specimens that correspond to 72 cystic precursor neoplasms and 36 other neoplastic and non-neoplastic pancreatic cysts (Fig. 2). All 108 specimens were sufficient for targeted DNA/RNA-based next-generation sequencing and identified genomic alterations consisted of: KRAS (44%), GNAS (32%), TP53 (19%), SMAD4 (14%), RNF43 (9%), BRAF (6%), PIK3CA (6%), VHL (6%), MEN1 (4%), CTNNB1 (3%), PTEN (2%), and PRKACA/B (2%). Genomic alterations in the MAPK genes (KRAS and BRAF) and GNAS had a sensitivity and a specificity of 81% and 100% for a cystic precursor neoplasm (Table 1). In the absence of MAPK/GNAS mutations, VHL alterations were identified in 2 of 3 (66%) serous cystadenomas. Further, MEN1 alterations that did not co-occur with mutations in MAPK/GNAS were present in 3 of 10 (30%) cPanNETs. Gene expression analysis revealed elevated CHGA mRNA in all 10 (100%) cPanNETs and one pseudocyst.
FIGURE 2.
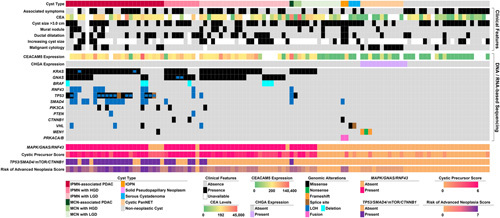
An oncoplot of 108 pancreatic cyst fluid specimens that were used for PancreaSeq GC training and includes cyst type, associated clinical features, and DNA/RNA-based sequencing results. At the bottom is a summary of genomic alterations involving MAPK gene, GNAS, and/or RNF43 alterations; cystic precursor score; summary genomic alterations involving TP53, SMAD4, mTOR genes, and CTNNB1; and risk score.
TABLE 1.
Diagnostic Performance of PancreaSeq GC Testing and Other Modalities Based on a Training Cohort of 108 Surgically Confirmed Pancreatic Cysts
| Parameter | Sensitivity (95% CI) | Specificity (95% CI) | PPV (95% CI) | NPV (95% CI) |
|---|---|---|---|---|
| IPMN | ||||
| MAPK/GNAS mutations | 90% (0.78–0.96) | 86% (0.73–0.94) | 88% (0.76–0.95) | 88% (0.75–0.95) |
| Elevated CEA (n=83)* | 71% (0.55–0.84) | 68% (0.52–0.81) | 70% (0.54–0.82) | 70% (0.53–0.83) |
| Elevated CEACAM5 (n=94)* | 92% (0.80–0.97) | 84% (0.69–0.93) | 87% (0.74–0.94) | 90% (0.76–0.97) |
| Cystic precursor score (≥3) | 98% (0.93–1.00) | 82% (0.69–0.91) | 86% (0.75–0.93) | 98% (0.86–1.00) |
| IPMN with advanced neoplasia | ||||
| MAPK/GNAS & TP53, SMAD4, CTNNB1 and/or mTOR gene mutations | 74% (0.55–0.87) | 95% (0.86–0.98) | 86% (0.67–0.97) | 89% (0.79–0.94) |
| Associated clinical symptoms (n=106)* | 50% (0.32–0.68) | 50% (0.38–0.62) | 30% (0.19–0.45) | 70% (0.56–0.81) |
| Index cyst size >3.0 cm (n=107)* | 61% (0.42–0.77) | 37% (0.26–0.49) | 30% (0.20–0.42) | 68% (0.51–0.81) |
| Main pancreatic duct dilatation (n=106)* | 50% (0.32–0.68) | 73% (0.61–0.82) | 44% (0.28–0.62) | 77% (0.65–0.86) |
| Presence of a mural nodule (n=106)* | 16% (0.06–0.34) | 84% (0.73–0.91) | 29% (0.11–0.56) | 70% (0.59–0.79) |
| Increasing index cyst size (n = 45)* | 33% (0.13–0.61) | 37% (0.21–0.56) | 21% (0.08–0.43) | 52% (0.30–0.74) |
| Malignant cytopathology (n=106)† | 24% (0.12–0.43) | 99% (0.92–1.00) | 89% (0.51–0.99) | 74% (0.64–0.82) |
| Risk of advanced neoplasia score (≥4) | 82% (0.65–0.93) | 92% (0.83–0.97) | 82% (0.65–0.93) | 92% (0.83–0.97) |
| IPMN, MCN, and IOPN | ||||
| MAPK/GNAS mutations | 81% (0.69–0.89) | 100% (0.88–1.00) | 100% (0.92–1.00) | 72% (0.58–0.83) |
| Elevated CEA (n=83)* | 69% (0.54–0.80) | 79% (0.60–0.91) | 86% (0.71–0.94) | 58% (0.41–0.73) |
| Elevated CEACAM5 (n=94)* | 80% (0.68–0.89) | 97% (0.80–1.00) | 98% (0.89–1.00) | 68% (0.52–0.81) |
| Cystic precursor score (≥3) | 90% (0.80–0.96) | 100% (0.88–1.00) | 100% (0.93–1.00) | 84% (0.69–0.93) |
| IPMN, MCN, and IOPN with advanced neoplasia | ||||
| MAPK/GNAS & TP53, SMAD4, CTNNB1 and/or mTOR gene mutations | 71% (0.54–0.84) | 97% (0.89–1.00) | 93% (0.76–0.99) | 86% (0.76–0.93) |
| Associated clinical symptoms (n=106)* | 47% (0.31–0.64) | 49% (0.37–0.61) | 32% (0.20–0.47) | 64% (0.50–0.77) |
| Index cyst size >3.0 cm (n=107)* | 65% (0.47–0.79) | 39% (0.27–0.51) | 36% (0.25–0.49) | 68% (0.51–0.81) |
| Main pancreatic duct dilatation (n=106)* | 50% (0.33–0.67) | 74% (0.62–0.84) | 50% (0.33–0.67) | 74% (0.62–0.84) |
| Presence of a mural nodule (n=106)* | 19% (0.08–0.37) | 86% (0.75–0.93) | 41% (0.19–0.67) | 67% (0.57–0.77) |
| Increasing index cyst size (n=45)* | 41% (0.19–0.67) | 39% (0.22–0.59) | 29% (0.13–0.51) | 52% (0.30–0.74) |
| Malignant cytopathology (n=106)† | 22% (0.10–0.39) | 99% (0.91–1.00) | 89% (0.51–0.99) | 70% (0.60–0.79) |
| Risk of advanced neoplasia score (≥4) | 84% (0.69–0.93) | 97% (0.89–1.00) | 94% (0.79–-0.99) | 92% (0.83–0.97) |
CEA indicates carcinoembryonic antigen; IOPN, intraductal oncocytic papillary neoplasm; IPMN, intraductal papillary mucinous neoplasm; LOH, loss of heterozygosity; MCN, mucinous cystic neoplasm; NPV, negative predictive value; PPV, positive predictive value.
n designates the number of patients with data available for analysis.
Malignant cytopathology was defined as at least suspicious for adenocarcinoma.
CEA indicates carcinoembryonic antigen; IOPN, intraductal oncocytic papillary neoplasm; IPMN, intraductal papillary mucinous neoplasm; LOH, loss of heterozygosity; MCN, mucinous cystic neoplasm; NPV, negative predictive value; PPV, positive predictive value.
RT-qPCR for CEACAM5 was able to be performed for 94 (87%) cases and ranged from 0 to 140,400 gene expression units (GEU). A cutoff of >200 GEU was calculated to separate cystic precursor neoplasms from other cystic lesions of the pancreas with the highest sensitivity and highest specificity (Supplementary Figure 1, Supplemental Digital Content 1, http://links.lww.com/SLA/A622). The sensitivity and specificity of CEACAM5 for a cystic precursor neoplasm were 80% and 97%, respectively, and yielded an AUC of 0.924. Conventional CEA testing data was available for 83 patients and, at a cutoff of ≥192 ng/mL, CEA had a sensitivity of 69% and a specificity of 79% for a cystic precursor neoplasm, and yielded an AUC of 0.818.
Integrating the presence versus absence of specific genomic alterations, gene expression (eg, CHGA for exclusion), and elevated CEACAM5, a cystic precursor score was developed to separate IPMNs, MCNs, and IOPNs from other pancreatic cysts. This scoring system (0 to 6) had a discriminating cutoff of 3 as a marker of a cystic precursor neoplasm that attained the highest sensitivity and highest specificity with an AUC of 0.946. Using this cutoff, the sensitivity, specificity, PPV, and NPV of PancreaSeq GC for an IPMN, MCN, or IOPN was 90%, 100%, 100%, and 84%, respectively (Supplementary Figure 2A, Supplemental Digital Content 1, http://links.lww.com/SLA/A622).
Establishing a Genomic Classifier for Risk of Advanced Neoplasia
Among the 72 cystic precursor neoplasms within the training cohort, 38 cases harbored advanced neoplasia. In combination with MAPK/GNAS mutations, genomic alterations in TP53, SMAD4, CTNNB1, and/or mTOR genes (PIK3CA, PTEN, and AKT1) had a sensitivity of 71% and a specificity of 97% for advanced neoplasia. Integrating mutant AF, LOH/CNA, gene fusions, and gene expression alterations, a risk score for advanced neoplasia was created (0 to 7) with a discriminating cutoff of 4 for advanced neoplasia that attained the highest sensitivity and highest specificity with an AUC of 0.955. Based on this cutoff, the sensitivity, specificity, PPV, and NPV of PancreaSeq GC for advanced neoplasia were 84%, 97%, 84%, and 92%, respectively (Supplementary Figure 2B, Supplemental Digital Content 1, http://links.lww.com/SLA/A622). In comparison, clinical symptoms, cyst size of >3.0 cm, main pancreatic duct dilatation, a mural nodule on EUS, and increasing cyst size were all associated with lower sensitivities and lower specificities for advanced neoplasia. Similarly, a cytopathologic diagnosis of at least suspicious for adenocarcinoma had a lower sensitivity, but the specificity was higher at 99%.
Validating the PancreaSeq Genomic Classifier
The genomic classifiers for cystic precursor neoplasms and risk of advanced neoplasia were validated on an independent set of 77 preoperative EUS-FNA pancreatic cyst fluid specimens (Fig. 3). A PancreaSeq GC score of ≥3 for cystic precursor neoplasms had a sensitivity, specificity, PPV, and NPV for an IPMN, MCN, or IOPN was 95%, 100%, 100%, and 86%, respectively (Table 2 and Supplementary Figure 3A, Supplemental Digital Content 1, http://links.lww.com/SLA/A622). Alone, conventional CEA testing of ≥192 ng/mL demonstrated a high specificity of 100%, but a low sensitivity of 65% for predicting a cystic precursor neoplasm. Based on a PancreaSeq GC risk score of ≥4, the sensitivity, specificity, PPV, and NPV for advanced neoplasia were 82%, 100%, 100%, and 91%, respectively (Supplementary Figure 3B, Supplemental Digital Content 1, http://links.lww.com/SLA/A622). Analogous to the training cohort, clinical symptoms, cyst size of >3.0 cm, main pancreatic duct dilatation, a mural nodule on EUS, and increasing cyst size were all associated with lower sensitivities and lower specificities for advanced neoplasia. A cytopathologic diagnosis of at least suspicious for adenocarcinoma also had a lower sensitivity and a lower specificity than PancreaSeq GC. The diagnostic performance of PancreaSeq GC and other modalities for the combined training and validation cohorts are summarized in Supplementary Table 2, Supplemental Digital Content 1, http://links.lww.com/SLA/A622 and was superior in comparison to DNA-based testing alone as summarized in Supplementary Table 3, Supplemental Digital Content 1, http://links.lww.com/SLA/A622. Of note, although all adenocarcinomas associated with a cystic precursor neoplasm were all early-stage carcinomas (pT1aN0 or pT1bN0), a separate analysis was performed to evaluate the diagnostic performance of PancreaSeq GC for high-grade dysplasia is a cystic precursor neoplasm and summarized in Supplementary Table 4, Supplemental Digital Content 1, http://links.lww.com/SLA/A622.
FIGURE 3.

An oncoplot of 77 pancreatic cyst fluid specimens that were used for PancreaSeq GC validation and includes cyst type, associated clinical features, and DNA/RNA-based sequencing results. At the bottom are the corresponding cystic precursor and risk scores.
TABLE 2.
Diagnostic Performance of PancreaSeq GC Testing and Other Modalities Based on Validation Cohort of 77 Surgically Confirmed Pancreatic Cysts
| Parameter | Sensitivity (95% CI) | Specificity (95% CI) | PPV (95% CI) | NPV (95% CI) |
|---|---|---|---|---|
| IPMN | ||||
| MAPK/GNAS mutations | 85% (0.72–0.93) | 91% (0.71–0.99) | 96% (0.85–0.99) | 72% (0.53–0.87) |
| Elevated CEA (n=49)* | 66% (0.48–0.80) | 79% (0.49–0.94) | 89% (0.69–0.97) | 48% (0.27–0.69) |
| Elevated CEACAM5 (n=74)* | 96% (0.85–0.99) | 83% (0.61–0.94) | 93% (0.81–0.98) | 91% (0.68–0.98) |
| Cystic precursor score (≥3) | 95% (0.84–0.99) | 95% (0.74–1.00) | 98% (0.89–1.00) | 87% (0.65–0.97) |
| IPMN with advanced neoplasia | ||||
| MAPK/GNAS & TP53, SMAD4, CTNNB1 and/or mTOR gene mutations | 80% (0.59–0.92) | 98% (0.88–1.00) | 95% (0.74–1.00) | 91% (0.80–0.97) |
| Associated clinical symptoms (n=73)* | 40% (0.22–0.61) | 75% (0.60–0.86) | 46% (0.25–0.67) | 71% (0.56–0.82) |
| Index cyst size >3.0 cm (n=75)* | 52% (0.32–0.72) | 54% (0.40–0.68) | 36% (0.21–0.54) | 69% (0.52–0.83) |
| Main pancreatic duct dilatation (n=76)* | 64% (0.43–0.81) | 77% (0.62–0.87) | 57% (0.37–0.75) | 81% (0.67–0.91) |
| Presence of a mural nodule (n=77)* | 48% (0.28–0.68) | 79% (0.65–0.89) | 52% (0.31–0.73) | 76% (0.62–0.86) |
| Increasing index cyst size (n=37)* | 50% (0.22–0.78) | 64% (0.43–0.81) | 40% (0.18–0.67) | 73% (0.50–0.88) |
| Malignant cytopathology (n=77)† | 52% (0.32–0.72) | 94% (0.83–0.99) | 81% (0.54–0.95) | 80% (0.68–0.89) |
| Risk of advanced neoplasia score (≥4) | 80% (0.59–0.92) | 96% (0.86–0.99) | 91% (0.69–0.98) | 91% (0.79–0.97) |
| IPMN, MCN, and IOPN | ||||
| MAPK/GNAS mutations | 81% (0.69–0.90) | 100% (0.78–1.00) | 100% (0.91–1.00) | 62% (0.42–0.79) |
| Elevated CEA (n=49)* | 65% (0.48–0.80) | 100% (0.63–1.00) | 100% (0.84–1.00) | 39% (0.21–0.61) |
| Elevated CEACAM5 (n=74)* | 91% (0.80–0.97) | 89% (0.64–0.98) | 96% (0.86–0.99) | 76% (0.53–0.91) |
| Cystic precursor score (≥3) | 95% (0.85–0.99) | 100% (0.78–1.00) | 100% (0.92–1.00) | 86% (0.63–0.96) |
| IPMN, MCN, and IOPN with advanced neoplasia | ||||
| MAPK/GNAS & TP53, SMAD4, CTNNB1 and/or mTOR gene mutations | 78% (0.57–0.91) | 100% (0.91–1.00) | 100% (0.81–1.00) | 89% (0.78–0.96) |
| Associated clinical symptoms (n=73)* | 41% (0.23–0.61) | 76% (0.61–0.87) | 50% (0.29–0.71) | 69% (0.54–0.81) |
| Index cyst size >3.0 cm (n=75)* | 52% (0.32–0.71) | 56% (0.41–0.70) | 40% (0.24–0.58) | 68% (0.51–0.81) |
| Main pancreatic duct dilatation (n=76)* | 59% (0.39–0.77) | 76% (0.61–0.86) | 57% (0.37–0.75) | 77% (0.62–0.88) |
| Presence of a mural nodule (n=77)* | 48% (0.29–0.68) | 80% (0.66–0.90) | 57% (0.35–0.76) | 74% (0.60–0.85) |
| Increasing index cyst size (n=37)* | 50% (0.24v0.76) | 65% (0.43–0.83) | 47% (0.22–0.73) | 68% (0.45–0.85) |
| Malignant cytopathology (n=77)† | 52% (0.32–0.71) | 96% (0.85–0.99) | 88% (0.60–0.98) | 79% (0.66–0.88) |
| Risk of advanced neoplasia score (≥4) | 82% (0.61–0.93) | 100% (0.91–1.00) | 100% (0.82–1.00) | 91% (0.79–0.97) |
n designates the number of patients with data available for analysis.
Malignant cytopathology was defined as at least suspicious for adenocarcinoma.
CEA indicates carcinoembryonic antigen; IOPN, intraductal oncocytic papillary neoplasm; IPMN, intraductal papillary mucinous neoplasm; LOH, loss of heterozygosity; MCN, mucinous cystic neoplasm; NPV, negative predictive value; PPV, positive predictive value.
Evaluation of cPanNETs and Non-neoplastic Pancreatic Cysts
In total, our study also included 24 cPanNET and 24 non-neoplastic cysts. PancreaSeq GC (elevated CHGA) had 100% sensitivity, 99% specificity, 96% PPV, and 100% NPV for a diagnosis of a cPanNETs. A cytopathologic diagnosis of at least suspicious for a neuroendocrine neoplasm had a lower sensitivity of 83%, but 100% specificity for a cPanNET. The diagnostic performance of PancreaSeq GC was also evaluated to determine its ability to distinguish a non-neoplastic pancreatic cyst from a neoplastic pancreatic cyst. A negative PancreaSeq GC that denotes an absence of gene mutations, gene fusions, CNAs, and GEAs had a sensitivity, specificity, PPV, and NPV of 95%, 95%, 69%, and 99%, respectively, for a non-neoplastic pancreatic cyst.
Comparison and Integration of PancreaSeq GC to Current Pancreatic Cyst Guidelines
The clinicopathologic features of all patients with this study are summarized in Supplementary Table 5, Supplemental Digital Content 1, http://links.lww.com/SLA/A622 with respect to PancreaSeq GC scores. Considering current pancreatic cyst guidelines have primarily focused on detecting advanced neoplasia in IPMNs, a subanalysis of only 107 resected IPMNs (training and validation cohorts) revealed PancreaSeq GC attained a sensitivity and a specificity of 81% and 98%, respectively, for advanced neoplasia (Table 3). A comparison of criteria for surgical management from the AGA guidelines and the IAP/Fukuoka guidelines showed lower sensitivities (64% and 77%) and lower specificities (66% and 40%). Incorporating PancreaSeq GC as another criterion to the AGA guidelines and IAP/Fukuoka guidelines increased the sensitivity to 91% and 95%, respectively.
TABLE 3.
Comparison of PancreaSeq GC Testing and Pancreatic Cyst Guideline Recommendations
| Parameter | Sensitivity (95% CI) | Specificity (95% CI) | PPV (95% CI) | NPV (95% CI) |
|---|---|---|---|---|
| IPMN with advanced neoplasia among IPMNs only* | ||||
| AGA guidelines | 64% (0.49–0.75) | 66% (0.51–0.78) | 68% (0.54–0.80) | 61% (0.47–0.74) |
| IAP/Fukuoka guidelines | 77% (0.64–0.87) | 40% (0.27–0.55) | 60% (0.47–0.71) | 61% (0.42–0.77) |
| PancreaSeq GC† | 81% (0.68–0.90) | 98% (0.88–1.00) | 98% (0.87–1.00) | 82% (0.69–0.90) |
| AGA guidelines OR PancreaSeq GC† | 91% (0.80–0.97) | 66% (0.51–0.78) | 75% (0.63–0.85) | 87% (0.71–0.95) |
| IAP/Fukuoka guidelines OR PancreaSeq GC† | 95% (0.85–0.99) | 40% (0.27–0.55) | 64% (0.53–0.74) | 87% (0.65–0.97) |
| AGA guidelines AND PancreaSeq GC† | 53% (0.39–0.66) | 98% (0.88–1.00) | 97% (0.82–1.00) | 65% (0.53–0.75) |
| IAP/Fukuoka guidelines AND PancreaSeq GC† | 63% (0.49–0.75) | 98% (0.88–1.00) | 97% (0.84–1.00) | 70% (0.58–0.80) |
| IPMN with advanced neoplasia among all pancreatic cysts‡ | ||||
| AGA guidelines | 63% (0.49–0.75) | 77% (0.69–0.84) | 56% (0.43–0.68) | 82% (0.73–0.88) |
| IAP/Fukuoka guidelines | 77% (0.64–0.87) | 59% (0.50–0.68) | 47% (0.37–0.57) | 85% (0.75–0.91) |
| PancreaSeq GC† | 81% (0.68–0.90) | 94% (0.88–0.98) | 87% (0.74–0.94) | 91% (0.85–0.95) |
| AGA guidelines OR PancreaSeq GC† | 91% (0.80–0.97) | 75% (0.66–0.82) | 63% (0.51–0.73) | 95% (0.88–0.98) |
| IAP/Fukuoka guidelines OR PancreaSeq GC† | 95% (0.85–0.99) | 57% (0.48–0.66) | 51% (0.41–0.60) | 96% (0.88–0.99) |
| AGA guidelines AND PancreaSeq GC† | 53% (0.39–0.66) | 97% (0.91–0.99) | 88% (0.72–0.96) | 82% (0.74–0.87) |
| IAP/Fukuoka guidelines AND PancreaSeq GC† | 63% (0.49–0.75) | 97% (0.91–0.99) | 90% (0.75–0.97) | 85% (0.78–0.90) |
| IPMN, MCN, and IOPN with advanced neoplasia among all pancreatic cysts‡ | ||||
| AGA guidelines | 62% (0.49–0.74) | 79% (0.70–0.85) | 61% (0.48–0.73) | 79% (0.71–0.86) |
| IAP/Fukuoka guidelines | 75% (0.62–0.84) | 60% (0.50–0.69) | 50% (0.40–0.60) | 81% (0.71–0.89) |
| PancreaSeq GC† | 83% (0.71–0.91) | 99% (0.95–1.00) | 98% (0.89–1.00) | 91% (0.85–0.95) |
| AGA guidelines OR PancreaSeq GC† | 92% (0.82–0.97) | 79% (0.70–0.86) | 70% (0.59–0.79) | 95% (0.88–0.98) |
| IAP/Fukuoka guidelines OR PancreaSeq GC† | 95% (0.86–0.99) | 60% (0.50–0.69) | 56% (0.46–0.66) | 96% (0.88–0.99) |
| AGA guidelines AND PancreaSeq GC† | 52% (0.40–0.65) | 99% (0.95–1.00) | 97% (0.83–1.00) | 80% (0.72–0.86) |
| IAP/Fukuoka guidelines AND PancreaSeq GC† | 62% (0.49–0.74) | 99% (0.95–1.00) | 98% (0.85–1.00) | 83% (0.75–0.89) |
Based on 107 surgically confirmed IPMNs (both training and validation study cohorts) with correlative clinical, imaging, and cytopathologic findings.
Positive PancreaSeq GC testing was defined based on a risk of advanced neoplasia score of ≥4.
Based on 180 surgically confirmed pancreatic cysts (both training and validation study cohorts) with correlative clinical, imaging, and cytopathologic findings.
CEA indicates carcinoembryonic antigen; IOPN, intraductal oncocytic papillary neoplasm; IPMN, intraductal papillary mucinous neoplasm; LOH, loss of heterozygosity; MCN, mucinous cystic neoplasm; NPV, negative predictive value; PPV, positive predictive value.
In the prospective clinical setting, identifying IPMNs can be challenging. Therefore, the AGA guidelines, the IAP/Fukuoka guidelines, and PancreaSeq GC were evaluated for their ability to identify IPMNs, MCNs, and IOPNs with advanced neoplasia among the entire study cohort. The sensitivity and specificity of the AGA guidelines were 62% and 79%, respectively, whereas the IAP/Fukuoka guidelines yielded a sensitivity of 75% and a specificity of 60%. PancreaSeq GC alone had an 83% sensitivity and 99% specificity. Combining PancreaSeq GC to the AGA guidelines and the IAP/Fukuoka guidelines as another criterion increased the sensitivities to 92% and 95%, respectively, whereas the specificities were 79% and 60%, respectively.
DISCUSSION
The heterogeneous nature of pancreatic cysts necessitates a multi-biomarker approach to the evaluation of pancreatic cyst fluid. PancreaSeq GC was designed to improve upon current molecular assays by incorporating a 74-gene panel to evaluate the intricacies and the nuances of not only neoplastic cysts (eg, mutant AF, gene fusions, CHGA, etc.), but also non-neoplastic pancreatic cysts.2,10,19 Considering the complexities of this novel panel, a genomic classifier was created for the detection of cystic precursor neoplasms and their risk of advanced neoplasia. Using a two-phase biomarker approach, PancreaSeq GC with a cystic precursor score of ≥3 achieved 95% sensitivity, 100% specificity, 100% PPV, and 86% NPV for either an IPMN, MCN, or IOPN. In addition, the sensitivity, specificity, PPV, and NPV for advanced neoplasia using a PancreaSeq GC risk score of ≥4 were 82%, 100%, 100%, and 91%, respectively.
The discrimination between cystic precursor neoplasms and other pancreatic cysts is challenging but imperative to stratifying patients for appropriate surveillance protocols and management options. This issue becomes more critical when dispelling both patient and physician anxiety with regard to the risk of an underlying PDAC, such as in the case of an SCA mimicking an IPMN. Not surprisingly, the classification of pancreatic cyst type requires a multimodal assessment of clinical features, imaging findings, and pancreatic cyst fluid analysis. Pancreatic cyst fluid testing for elevated CEA is one of the most accurate assays to the diagnosis of a cystic precursor neoplasm, such as an IPMN.30 Herein, an elevated CEA within the training and validation cohorts showed sensitivities of 69% and 65%, respectively, and specificities of 79% and 100%, respectively. Similarly, a recent meta-analysis of 31 studies by Faias et al found elevated CEA to be associated with a pooled sensitivity of 67% and a pooled specificity of 80% for IPMNs and MCNs.31 However, the authors noted significant inconsistencies in sensitivity and specificity for CEA across studies. They attributed the lack of reproducible performance due to the wide range of CEA analyzers in clinical practice. Thus, to standardize CEA testing and provide an integrative molecular analysis with the understanding that aspirated pancreatic cyst fluid is limited in volume, PancreaSeq GC quantitatively measures CEACAM5, the corresponding mRNA for CEA, was superior to CEA, and had a sensitivity and specificity of 91% and 89%, respectively. Furthermore, the combination of CEACAM5 along with other alterations that correspond to a PancreaSeq GC cystic precursor score of ≥3 improved the sensitivity to 95% and specificity to 100%. Importantly, the NPV of PancreaSeq GC was 86%, whereas CEA testing was only 39%.
Upon establishing a diagnosis of a cystic precursor neoplasm, equally problematic is determining the presence versus absence of advance neoplasia. Several clinicopathologic features suggestive of advanced neoplasia have been identified: large cyst size (>3.0 cm), symptoms related to duct obstruction, main pancreatic duct dilatation, a mural nodule, and high-grade epithelial atypia by cytopathology.3–6 Individually, however, these parameters demonstrate suboptimal diagnostic performance in either sensitivity, specificity, or both.10,32,33 Recently, numerous studies have reported the association of genomic alterations in TP53, SMAD4, CTNNB1, and the mTOR genes (“high-risk” genes) with advanced neoplasia.7,10–12 But the presence of high-risk genomic alterations in the absence of MAPK/GNAS mutations can be found in SCAs and cPanNETs.10,34 These findings therefore underscore the importance of defining pancreatic cyst type when evaluating for advanced neoplasia. Furthermore, we previously reported the importance of mutant AFs in the detection of advanced neoplasia arising from a cystic precursor neoplasm.10 In rare instances, the mutant AF for MAPK/GNAS alterations can be >55% through either deletion of the wild-type allele or copy number gain of the mutant allele. This phenomenon is known as mutant allele-specific imbalance or LOH and corresponds to the identification of advanced neoplasia. Another point of consideration is the mutant AF for TP53 and PIK3CA. In combination with MAPK/GNAS mutations, TP53 and PIK3CA alterations have not only been described in advanced neoplasia, but also low-grade dysplasia. However, in the setting of low-grade dysplasia, the mutant AF for TP53 and PIK3CA is at a low level as compared to MAPK/GNAS mutations.
Considering the intricacies of interpreting concurrent genomic alterations (eg, MAPK/GNAS with TP53) and mutant AFs, the creation of a genomic classifier that integrates various molecular permutations associated with advanced neoplasia became necessary. Within the validation cohort, a PancreaSeq GC risk score of ≥4 was superior in diagnostic performance as compared to clinicopathologic features. Moreover, PancreaSeq GC improved the sensitivity of the IAP/Fukuoka and AGA guidelines. Nonetheless, the IAP/Fukuoka and the AGA guidelines were designed with the intent of evaluating IPMNs alone. Within a subanalysis of the 107 surgically confirmed IPMNs within this study, PancreaSeq GC continued to attain higher sensitivity and higher specificity than either guideline. Furthermore, the addition of PancreaSeq GC as a criterion improved the sensitivities of each guideline by >10%. Additional advantages of PancreaSeq GC are its ability to identify cPanNETs and non-neoplastic cysts. In fact, PancreaSeq GC attained a higher sensitivity for cPanNETs of 100% as compared to cytopathologic evaluation of 83%. Similar to pancreatic cysts, PanNETs are also increasing in incidence due to the frequent use of abdominal imaging, and, thus, additional biomarkers are required to aid in stratifying risk of distant metastasis.18,35–38 PancreaSeq GC may represent the foundation for which to build prognostic biomarkers for PanNET patients.
There are several limitations to this study that should be acknowledged. It is retrospective in design and because a surgical diagnosis is required to assess test performance, the study suffers from a surgical selection bias. However, all EUS-FNA pancreatic cyst fluid specimens were collected in real-time and as part of clinical care. We also recognize the presence of a testing selection bias as specimens used within this study were previously deemed satisfactory for molecular analysis. The effect of a testing selection bias is likely to be minimal considering our previously reported failure rate of 2% to 7%.7,10,15 It is also important to note that the sensitivity of MAPK/GNAS mutations with high-risk gene alterations for advanced neoplasia was lower within the training and validation cohorts than previously reported.10 Nucleic acid degradation may explain this discrepancy as the specimens used were approximately 3 to 10 years old. Prospective testing is therefore required to determine the true diagnostic performance of PancreaSeq GC and is currently underway. However, additional studies will be required to ascertain the optimal approach for PancreaSeq GC testing and how PancreaSeq GC should be incorporated into current and future pancreatic cyst guidelines.
In summary, PancreaSeq GC is a DNA/RNA-based targeted NGS panel for the evaluation of EUS-FNA pancreatic cyst fluid to aid in the classification of pancreatic cyst type, such as cystic precursor neoplasms, and the detection of advanced neoplasia. Considering the large breadth of DNA and RNA alterations associated with neoplastic and non-neoplastic pancreatic cysts, a genomic classifier to integrate the molecular results of PancreaSeq GC was required. Among 185 EUS-FNA pancreatic cyst fluid specimens from multiple institutions, PancreaSeq GC showed superior performance in the identification of cystic precursor neoplasms and advanced neoplasia as compared to standard clinical, imaging, cytopathologic, and ancillary fluid parameters. Moreover, the addition of PancreaSeq GC improved the diagnostic sensitivity of current pancreatic cyst guidelines, such as the IAP/Fukuoka and AGA guidelines, while maintaining their inherent diagnostic specificities.
Supplementary Material
Footnotes
M.N.N., A.I.W., and A.D.S.: study concept and design. M.N.N., A.I.W., D.M.S., M.A.M., M.G., Y.T.L., R.E.B., A.M.O.L., K.M., W.G.P., P.R.P., P.M.P., N.K., J.D., J.J.E., A.D., S.R.M., M.B.W., V.K., B.A.B., W.M., S.T., K.J.F., E.A., Y.B., S.R., J.N., W.S., A.K., K.F., J.C., R.D., H.S., .S.S., A.S., C.G., T.S., T.T., H.D.V., A.T., L.M.S., K.S., P.D.B., R.H.H., A.P., A.Z., K.K.L., M.O., H.Z., R.M., J.H., Y.E.N., and A.D.S: acquisition of clinical and pathological data. M.N.N., A.I.W., D.M.S., L.M.S., Y.E.N., and A.D.S: analysis and interpretation of data. M.N.N., A.I.W., D.M.S., Y.E.N., and A.D.S: drafting of the manuscript.
This study was supported in part by the Department of Defense (W81XWH-21-1-0709), National Cancer Institute (NIH/NCI 1U01CA200466 and 5R37CA263622), the Pittsburgh Liver Research Center at the University of Pittsburgh (NIH/NIDDK P30DK120531), the University of Pittsburgh Medical Center, and the National Pancreas Foundation, Western Pennsylvania Chapter.
A.D.S. has received an honorarium from Foundation Medicine Inc. M.N.N. and Y.E.N. own intellectual property related to the PancreaSeq technology and receive royalties from University of Pittsburgh. R.H.H. has the potential to receive royalty payments from Thrive Earlier Detection for the GNAS invention in an arrangement reviewed and approved by the Johns Hopkins University in accordance with its conflict-of-interest policies. The remaining authors report no conflicts of interest.
Supplemental Digital Content is available for this article. Direct URL citations are provided in the HTML and PDF versions of this article on the journal's website, www.annalsofsurgery.com.
Contributor Information
Maria Grupillo, Email: maria.grupillo@chp.edu.
Yi-Tak Lai, Email: laiym@upmc.edu.
Randall E. Brand, Email: brandre@upmc.edu.
Anne Marie O’Broin-Lennon, Email: amlennon@jhmi.edu.
Kevin McGrath, Email: mcgrkm@upmc.edu.
Walter G. Park, Email: wgpark@stanford.edu.
Patrick R. Pfau, Email: prp@medicine.wisc.edu.
Patricio M. Polanco, Email: patricio.polanco@utsouthwestern.edu.
Nisa Kubiliun, Email: nisa.kubiliun@utsouthwestern.edu.
John DeWitt, Email: jodewitt@iu.edu.
Jeffrey J. Easler, Email: jjeasler@iu.edu.
Aamir Dam, Email: dam42nam@protonmail.com.
Shaffer R. Mok, Email: shaffer.mok@moffitt.org.
Michael B. Wallace, Email: Wallace.Michael@mayo.edu.
Vivek Kumbhari, Email: vkumbhari@gmail.com.
Brian A. Boone, Email: brian.boone@hsc.wvu.edu.
Wallis Marsh, Email: Wallis.Marsh@hsc.wvu.edu.
Shyam Thakkar, Email: shyam.thakkar@hsc.wvu.edu.
Kimberly J. Fairley, Email: kfairley329s@protonmail.com.
Elham Afghani, Email: eafghan1@jhmi.edu.
Yasser Bhat, Email: BhatY@sutterhealth.org.
Sanjay Ramrakhiani, Email: Sanhys392@protonmail.com.
John Nasr, Email: nasr9482s@protonmail.com.
Wasseem Skef, Email: Wskef89@protonmail.com.
Nikhil R. Thiruvengadam, Email: nthiruvengadam@llu.edu.
Asif Khalid, Email: khalax@upmc.edu.
Kenneth Fasanella, Email: fasanellake@upmc.edu.
Jennifer Chennat, Email: chennatjs@upmc.edu.
Rohit Das, Email: dasr@upmc.edu.
Harkirat Singh, Email: singhh3@upmc.edu.
Savreet Sarkaria, Email: SarkariaS123@protonmail.com.
Adam Slivka, Email: slivkaa@upmc.edu.
Charles Gabbert, Email: gabbertcc2@upmc.edu.
Tarek Sawas, Email: Tarek.Sawas@UTSouthwestern.edu.
Thomas Tielleman, Email: thomas.tielleman@utsouthwestern.edu.
Hendrikus Dutch Vanderveldt, Email: Dutch.Vanderveldt@UTSouthwestern.edu.
Anna Tavakkoli, Email: anna.tavakkoli@utsouthwestern.edu.
Katelyn Smith, Email: smithkm13@upmc.edu.
Phoenix D. Bell, Email: bellpd@upmc.edu.
Ralph H. Hruban, Email: rhruban@jhmi.edu.
Alessandro Paniccia, Email: panicciaa2@upmc.edu.
Amer Zureikat, Email: zureikatah@upmc.edu.
Kenneth K. Lee, Email: leek@upmc.edu.
Melanie Ongchin, Email: ongchinmc3@upmc.edu.
Herbert Zeh, Email: Herbert.Zeh@UTSouthwestern.edu.
Rebecca Minter, Email: minter@surgery.wisc.edu.
Jin He, Email: jhe11@jhmi.edu.
Aatur D. Singhi, Email: ardy360@gmail.com.
REFERENCES
- 1.Ayoub F, Davis AM, Chapman CG. Pancreatic cysts-an overview and summary of society guidelines, 2021. JAMA. 2021;325:391–392. [DOI] [PubMed] [Google Scholar]
- 2.Singhi AD, Wood LD, Parks E, et al. Recurrent rearrangements in PRKACA and PRKACB in intraductal oncocytic papillary neoplasms of the pancreas and bile duct. Gastroenterology. 2020;158:573–582 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanaka M, Fernandez-Del Castillo C, Kamisawa T, et al. Revisions of international consensus Fukuoka guidelines for the management of IPMN of the pancreas. Pancreatology. 2017;17:738–753. [DOI] [PubMed] [Google Scholar]
- 4.European Study Group on Cystic Tumours of the Pancreas. European evidence-based guidelines on pancreatic cystic neoplasms. Gut. 2018;67:789–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vege SS, Ziring B, Jain R, et al. American gastroenterological association institute guideline on the diagnosis and management of asymptomatic neoplastic pancreatic cysts. Gastroenterology. 2015;148:819–822; quize12–13. [DOI] [PubMed] [Google Scholar]
- 6.Elta GH, Enestvedt BK, Sauer BG, et al. ACG Clinical Guideline: Diagnosis and Management of Pancreatic Cysts. Am J Gastroenterol. 2018;113:464–479. [DOI] [PubMed] [Google Scholar]
- 7.Singhi AD, Zeh HJ, Brand RE, et al. American Gastroenterological Association guidelines are inaccurate in detecting pancreatic cysts with advanced neoplasia: a clinicopathologic study of 225 patients with supporting molecular data. Gastrointest Endosc. 2016;83:1107–1117 e2. [DOI] [PubMed] [Google Scholar]
- 8.Dbouk M, Brewer Gutierrez OI, Lennon AM, et al. Guidelines on management of pancreatic cysts detected in high-risk individuals: an evaluation of the 2017 Fukuoka guidelines and the 2020 International Cancer of the Pancreas Screening (CAPS) consortium statements. Pancreatology. 2021;21:613–621. [DOI] [PubMed] [Google Scholar]
- 9.Rosenbaum MW, Jones M, Dudley JC, et al. Next-generation sequencing adds value to the preoperative diagnosis of pancreatic cysts. Cancer Cytopathol. 2017;125:41–47. [DOI] [PubMed] [Google Scholar]
- 10.Singhi AD, McGrath K, Brand RE, et al. Preoperative next-generation sequencing of pancreatic cyst fluid is highly accurate in cyst classification and detection of advanced neoplasia. Gut. 2018;67:2131–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Springer S, Masica DL, Dal Molin M, et al. A multimodality test to guide the management of patients with a pancreatic cyst. Sci Transl Med. 2019;11:eaav4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Springer S, Wang Y, Dal Molin M, et al. A combination of molecular markers and clinical features improve the classification of pancreatic cysts. Gastroenterology. 2015;149:1501–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rift CV, Melchior LC, Kovacevic B, et al. Targeted next generation sequencing of endoscopic ultrasound-guided through-the-needle-biopsies from pancreatic cystic lesions. Gastrointest Endosc. 2023;97:50–58. [DOI] [PubMed] [Google Scholar]
- 14.Singhi AD, Nikiforova MN, Fasanella KE, et al. Preoperative GNAS and KRAS testing in the diagnosis of pancreatic mucinous cysts. Clin Cancer Res. 2014;20:4381–4389. [DOI] [PubMed] [Google Scholar]
- 15.Nikiforova MN, Khalid A, Fasanella KE, et al. Integration of KRAS testing in the diagnosis of pancreatic cystic lesions: a clinical experience of 618 pancreatic cysts. Mod Pathol. 2013;26:1478–1487. [DOI] [PubMed] [Google Scholar]
- 16.Schmitz D, Kazdal D, Allgauer M, et al. KRAS/GNAS-testing by highly sensitive deep targeted next generation sequencing improves the endoscopic ultrasound-guided workup of suspected mucinous neoplasms of the pancreas. Genes Chromosomes Cancer. 2021;60:489–497. [DOI] [PubMed] [Google Scholar]
- 17.Jones M, Zheng Z, Wang J, et al. Impact of next-generation sequencing on the clinical diagnosis of pancreatic cysts. Gastrointest Endosc. 2016;83:140–148. [DOI] [PubMed] [Google Scholar]
- 18.Paniccia A, Polanco PM, Boone BA, et al. Prospective, multi-institutional, real-time next-generation sequencing of pancreatic cyst fluid reveals diverse genomic alterations that improve the clinical management of pancreatic cysts. Gastroenterology. 2023;164:117–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JW, Hruban RH, Brosens LAA, et al. RNA sequencing identifies frequent mitogen-activated protein kinase-associated fusion genes in intraductal tubulopapillary neoplasms of the pancreas. Gastroenterology. 2023;164:1310–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krasinskas AM, Moser AJ, Saka B, et al. KRAS mutant allele-specific imbalance is associated with worse prognosis in pancreatic cancer and progression to undifferentiated carcinoma of the pancreas. Mod Pathol. 2013;26:1346–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lokuhetty D, White V, Watanabe R, et al. Digestive System Tumours: WHO Classification of Tumours. International Agency for Research on Cancer; 2019. [Google Scholar]
- 22.Nikiforova MN, Wald AI, Roy S, et al. Targeted next-generation sequencing panel (ThyroSeq) for detection of mutations in thyroid cancer. J Clin Endocrinol Metab. 2013;98:E1852–E1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beadling C, Wald AI, Warrick A, et al. A multiplexed amplicon approach for detecting gene fusions by next-generation sequencing. J Mol Diagn. 2016;18:165–175. [DOI] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. [DOI] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Research Network. Electronic address aadhe, Cancer Genome Atlas Research N. Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell. 2017;32:185–203 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tate JG, Bamford S, Jubb HC, et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019;47:D941–D947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singhi AD, George B, Greenbowe JR, et al. Real-time targeted genome profile analysis of pancreatic ductal adenocarcinomas identifies genetic alterations that might be targeted with existing drugs or used as biomarkers. Gastroenterology. 2019;156:2242–2253 e4. [DOI] [PubMed] [Google Scholar]
- 29.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 30.Brugge WR, Lewandrowski K, Lee-Lewandrowski E, et al. Diagnosis of pancreatic cystic neoplasms: a report of the cooperative pancreatic cyst study. Gastroenterology. 2004;126:1330–1336. [DOI] [PubMed] [Google Scholar]
- 31.Faias S, Cravo M, Chaves P, et al. Comparative analysis of glucose and carcinoembryonic antigen in the diagnosis of pancreatic mucinous cysts: a systematic review and meta-analysis. Gastrointest Endosc. 2021;94:235–247. [DOI] [PubMed] [Google Scholar]
- 32.Scheiman JM, Hwang JH, Moayyedi P. American Gastroenterological Association technical review on the diagnosis and management of asymptomatic neoplastic pancreatic cysts. Gastroenterology. 2015;148:824–48 e22. [DOI] [PubMed] [Google Scholar]
- 33.Roldan J, Harrison JM, Qadan M, et al. Evolving trends in pancreatic cystic tumors: a 3-decade single-center experience with 1290 resections. Ann Surg. 2023;277:491–497. [DOI] [PubMed] [Google Scholar]
- 34.Kim SA, Kim MS, Kim MS, et al. Pleomorphic solid pseudopapillary neoplasm of the pancreas: degenerative change rather than high-grade malignant potential. Hum Pathol. 2014;45:166–174. [DOI] [PubMed] [Google Scholar]
- 35.Hackeng WM, Brosens LAA, Kim JY, et al. Non-functional pancreatic neuroendocrine tumours: ATRX/DAXX and alternative lengthening of telomeres (ALT) are prognostically independent from ARX/PDX1 expression and tumour size. Gut. 2022;71:961–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singhi AD, Liu TC, Roncaioli JL, et al. Alternative lengthening of telomeres and loss of DAXX/ATRX expression predicts metastatic disease and poor survival in patients with pancreatic neuroendocrine tumors. Clin Cancer Res. 2017;23:600–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pea A, Yu J, Marchionni L, et al. Genetic analysis of small well-differentiated pancreatic neuroendocrine tumors identifies subgroups with differing risks of liver metastases. Ann Surg. 2020;271:566–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roy S, LaFramboise WA, Liu TC, et al. Loss of chromatin-remodeling proteins and/or cdkn2a associates with metastasis of pancreatic neuroendocrine tumors and reduced patient survival times. Gastroenterology. 2018;154:2060–2063 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]


