Abstract
Microbial symbionts can affect host phenotypes and, thereby, ecosystem functioning. The microbiome is increasingly being recognized as an important player in the tripartite interaction between parasitic flatworms, snail intermediate hosts, and the snail microbiome. In order to better understand these interactions, transplant experiments are needed, which rely on the development of a reliable and reproducible protocol to obtain microbiome-disturbed snails. Here, we report on the first successful snail bacteriome transplants, which indicate that Biomphalaria glabrata can accrue novel bacterial assemblies depending on the available environmental bacteria obtained from donor snails. Moreover, the phylogenetic relatedness of the donor host significantly affected recipients’ survival probability, corroborating the phylosymbiosis pattern in freshwater snails. The transplant technique described here, complemented by field-based studies, could facilitate future research endeavors to investigate the role of specific bacteria or bacterial communities in parasitic flatworm resistance of B. glabrata and might ultimately pave the way for microbiome-mediated control of snail-borne diseases.
Keywords: freshwater snail, microbiome, parasite, Schistosomiasis, transplant, tripartite interaction
The development of a bacteriome transplant protocol could prove to be a key element in understanding the tripartite interaction between freshwater snails, their microbiome, and flatworm parasites.
Introduction
Evidence is mounting that environmental factors affect the community of host-associated microorganisms and their gene functions also known as microbiomes (see for example the review of Stock et al. 2021 and references therein). For instance, microbiome metabolism can shift due to global warming, ecological interactions in the microbiome can alter due to antibiotic exposure, and skin microbiota can become dysbiotic due to eutrophication (Callens et al. 2018, Fontaine and Kohl 2020, Krotman et al. 2020). In turn, microbiomes can play an important role in ecosystem functioning by modifying host phenotypes (Moreira et al. 2009, Koch and Schmid-Hempel 2012, Ridaura et al. 2013, Macke et al. 2017). The microbiome is known to affect the host's physiology, behavior, and disease resistance (Greyson-Gaito et al. 2020).
The latter is pertinent for the transmission potential of diseases (Ford and King 2016) and changes in the microbiome of intermediate hosts may thus indirectly affect ecosystem health (Flandroy et al. 2018). Intermediate hosts, parasites, and microbes interact to determine infection outcome and, subsequently, the potential for disease transmission to definitive hosts (Brinker et al. 2019). There are precedents with mosquito-borne diseases, such as malaria, dengue, and chikungunya, where the release of microbiome-altered vectors reduced disease prevalence, both at a local and at a regional scale (Moreira et al. 2009, Hoffmann et al. 2011, Pinto et al. 2021). It is hypothesized that microbiomes might affect the outcome of parasitic flatworm infections in snails by competing for resources, producing antimicrobials or stimulating the host's immune response (Huot et al. 2020, Portet et al. 2021, Stock et al. 2021, Le Clec'h et al. 2022).
Such findings have been stimulating schistosomiasis research to elucidate the tripartite interaction between the freshwater snail host, its bacteriome (the bacterial microbiome) and its parasitic flatworm community. Schistosomiasis is a parasitic disease caused by schistosome flatworms (Digenea, Platyhelminthes). It burdens millions of people and animals globally due to inefficient control measures (Sokolow et al. 2018). Different snail-parasite strain combinations exhibit various reciprocity to infection, of which, the underlying pattern is often ill-understood (Sène et al. 2004, Portet et al. 2021). One intermediate host, the Brazilian snail Biomphalaria glabrata, serves as a model system for studying schistosomiasis, placing it at the forefront of research focused on the snail-bacteriome-flatworm interaction (Duval et al. 2015, Huot et al. 2020, Portet et al. 2021, Le Clec'h et al. 2022). A decades-long gap exists with earlier publications describing and manipulating the bacteria of B. glabrata (Chernin 1957, 1960). These works, however, provide the foundation for current efforts on host-bacteriome interactions, more in particular for bacteriome transplants.
Transplant experiments are manipulations whereby a number of bacterial strains or entire bacterial communities are actively or passively transplanted from a host or substrate to another host (Greyson-Gaito et al. 2020). These experiments help us to decipher the host-bacteria interaction and showed, for example, that the host-associated bacterial community determines bumblebee resistance to parasites (Koch and Schmid-Hempel 2012), water flea tolerance to toxic cyanobacteria (Macke et al. 2017) and mice's tendencies to become obese (Ridaura et al. 2013). Such transplant experiments, however, depend on the development of a reliable and reproducible protocol to obtain bacteriome-disturbed recipients. The current study describes the optimization of the protocol designed by Chernin in 1957, its thorough validation with molecular tools, and the first successful transplants of bacterial communities across a taxonomic range of freshwater snails.
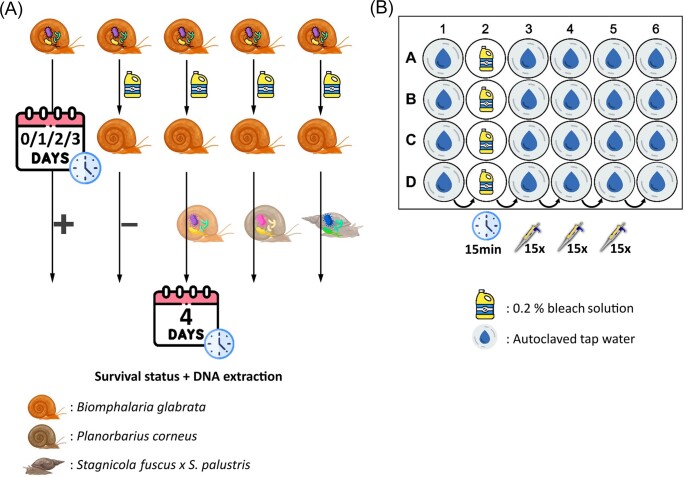
A phylosymbiosis pattern between freshwater snails and their bacteriome has been reported (Huot et al. 2020). Therefore, we hypothesize that exposure to a donor inoculum from a phylogenetically more distantly related snail would reduce the survivability of bacteriome-disturbed snails as opposed to exposure to a donor inoculum from a conspecific or a phylogenetically more closely related snail. To test this and determine the most suitable conditions for bacterial transplants in the study system the experimental design (Fig. 1A) aimed to test 1) whether bacteriome-disturbed snails would acquire a donor bacteriome, 2) what the optimal timing for exposure is, and 3) how the donor-recipient phylogenetic relationship affects the transplant outcome.
Figure 1.
A: Experimental setup. Snails were sterilized on the same day as, and one day, two days and three days prior to, receiving a donor inoculum. Bacteriome-disturbed Biomphalaria glabrata individuals (n=8) of three maternal lines (n=3) were exposed to three donor inocula isolated from: B. glabrata individuals (same species but different maternal line), Planorbarius corneus individuals (family: Planorbidae), and Stagnicola fuscus x S. palustris individuals (superorder: Hygrophila). Additionally, for each factorial combination of time (n=4) and maternal line (n=3), positive (untreated samples) and negative (bleach-treated samples without donor inoculum exposure) controls were included (four technical replicates). Survival status of the egg/juvenile was noted on the day of the donor inoculum exposure, three days post donor inoculum exposure and four days post donor inoculum exposure when the specimens were sacrificed for DNA extraction. Fig. 1B: The 24-well cell culture plate setup used during the sterilization protocol. The wells of the first column receive one egg each, prior to bleach exposure. The egg from A1 is transferred to the well A2 which contains a 0.2% bleach solution and remains there for 15 min. After the 15 min bleach exposure, the egg was washed in autoclaved tap water in A3, A4 and A5 by pipetting up and down 15 times in each well before being moved to the incubation well A6. This process was done simultaneously for all four rows (A, B, C, and D) for a total of four eggs per plate. The incubation time in column six depended on the assigned condition of that sample in accordance with Fig. 1A (0, 1, 2, or 3 days) before being exposed to a donor bacteriome. The survival status of each specimen was noted, and irrespective of whether the specimen had hatched, still in the egg, or died, was exposed to the donor inoculum in their incubation well in column six.
Material and methods
Cultivation of biomphalaria glabrata
All individuals used in this study originated from Biomphalaria glabrata colonies in our laboratory. Biomphalaria glabrata (Brazil strain) has been a stock population in the Host-Pathogen-Environment Interactions lab at the University of Perpignan Via Domitia in France since its collection from Recife, Brazil in 1975. This strain has been previously studied through molecular markers and appeared to exhibit a low genetic diversity, probably due to the many cycling events in the laboratory (Dheilly et al. 2015). We received 30 specimens and egg clutches of this B. glabrata Brazil strain in June 2020, started maternal lines from individual egg clutches in October 2020, and conducted the transplant experiment described here in April 2022.
Biomphalaria glabrata Brazil strain individuals were kept in 2 l and 5 l aquaria, depending on the cohort size, on a 12/12 day/night cycle at 25°C (clear white light was generated by OSRAM FH 28 W/840 HE). Each aquarium had a constant air supply provided through aeration stones. Fresh organic salad (pesticide-free) was provided three times per week at libitum. Aquaria were cleaned on a weekly basis with soap and mechanical rubbing, followed by an industrial washing machine program at 55°C and drying in the oven. Aeration stones were cleaned by boiling in water for 10 min, overnight incubation in diluted bleach (1 : 3) and, finally, dried by connecting to the air line.
Donor bacteriome
Bacteriome-disturbed B. glabrata individuals (see below ‘sterilization experiment') were exposed to three donor bacteriomes. The donor bacteriome hosts were selected according to a taxonomical gradient of relatedness to B. glabrata: same species but different maternal line (B. glabrata), same subfamily yet different tribe (Planorbarius corneus; collected from an artificial pond with GPS-coordinates: 50.999559, 4.955650), and the same superorder yet different family (Stagnicola fuscus x S. palustris; collected from an artificial pond with GPS-coordinates: 50.915337, 4.686979). All three donor snail species were molecularly identified (see suppl. Data: ‘Donor snail host identification’, Figs S1 and S2). The partial COI sequences of B. glabrata, P. corneus and S. fuscus x S. palustris, and the ITS2 rDNA sequence of S. fuscus x S. palustris were uploaded to GenBank (OQ954055-OQ954057, OQ955479).
Sterilization experiment
Prior to donor bacteriome exposure, germ-free or microbiome-disturbed recipients had to be obtained. This was done as follows. At the start of each day, egg clutches of each maternal line were collected and assessed for developmental status under a stereo microscope (Olympus SZX10). Late-development clutches were selected (Fig. S3), whereby, surrounding eggs were sliced open and debris was removed from around the egg of interest. Egg clusters were dissected for the three maternal lines. ±38 eggs were dissected to ensure that sufficient specimens were available for further manipulations. The eggs were kept separately per maternal line in autoclaved tap water (see next sentence for details) in 6-well plates to transport to the sterile biosafety cabinet. Dechlorinated tap water was autoclaved per litre, and is further referred to as ‘autoclaved tap water’. This water was aliquoted in tubes of 50 ml and was stored next to the snail aquaria at 25 °C. The UV light was turned on 15 min before turning on the circulation in the biosafety cabinet. The working surface and required utensils were disinfected with 70% ethanol. Bleach was diluted with autoclaved tap water to obtain a working solution of 0.2% Sodium hypochlorite or 0.042% active chlorine. Snail eggs would not fit through normal 200 µl tips. Therefore, 200 µl tips were trimmed with sterilized scissors to have a larger opening. All plates required for that day were filled according to Fig. 1B. Briefly, the second column of the 24-well plates were filled with 1 ml of the 0.2% bleach solution. All other wells were filled with 1 ml of autoclaved tap water. Each first-column well received one dissected egg. Subsequently, eggs were exposed to 0.2% bleach for 15 min. The timer started when the first egg entered the bleach solution. The following eggs for that plate were added at 30 sec intervals. Once all four eggs were in the bleach solution, we would swirl the plate gently for 10 s. This process was repeated for a total of six plates. Next, the first egg, now exposed to bleach for 15 min, was placed in the next well containing autoclaved tap water. We pipetted up and down 15 times in each well before transferring the egg to the next well to remove any remaining bleach. This manipulation was repeated two times until the egg entered the final well where it was allowed to hatch. This was repeated for all four eggs of that plate. Finally, the lid-tray interface was closed with parafilm.
Transplant experiment
To test if (i) treated snails would acquire a given donor bacteriome, (ii) what the optimal timing for exposure would be, and (iii) how the phylogenetic relationship between the recipient and donor host species affects the transplant outcome, we designed the following experimental setup (Fig. 1A). Snails were sterilized three days, two days, one day prior to and on the same day as receiving a donor inoculum. On the day of exposure donor inocula from B. glabrata and P. corneus were obtained by separately crushing three individual snails with their shell with a sterile pestle and topping the volume with autoclaved tap water until 1 ml. 300 µl of the suspension of each individual snail specimen was pooled in a 2 ml tube per donor type and topped till 1.4 ml with autoclaved tap water. Diluting the donor inocula with autoclaved tap water was required for pipet manipulations. The same protocol was used for S. fuscus x S. palustris, however, here only two snails were available. From the donor inocula 20 µl was each time added to the desired well. This was done for three separate maternal lines of lab-reared B. glabrata each time for four technical replicates (four individual snails were exposed to the same conditions). Each factorial combination (time (n=4) * maternal line (n=3)) received a donor inoculum from, either, a fourth maternal line of lab-reared B. glabrata, a wild-caught P. corneus or a wild-caught S. fuscus x S. palustris. Additionally, for each factorial combination of time and maternal line a positive (dissected without bleach exposure) and a negative (dissected with bleach exposure but no donor inoculum) control were included. Survival status of the egg/juvenile was noted at the day of the donor inoculum exposure, three days post donor inoculum exposure and four days post donor inoculum exposure when the specimens were sacrificed and the experiment ended. Snails were pooled by up to three specimens per factorial combination (time (n=4) * maternal line (n=3) * donor bacteriome (n=3) *survival status (n=3)) in order to obtain sufficient DNA for subsequent analyses. All negative controls and the positive controls of day 0 and day three were tested for the presence of bacteria in the growth assay in combination with microscopic investigation and qPCR. These and all other samples were included for 16S metabarcoding.
DNA extraction
Snails were crushed with their shell with a sterile pestle in 100 µl of autoclaved tap water by up to three specimens per factorial combination in order to obtain sufficient DNA for subsequent analyses. The complete homogenate was used for recipients of donor bacteriomes. For all negative controls and the positive controls at day zero and three, 90 µl of this homogenate was used for DNA extraction (5 µl was used for microscopic investigation and 5 µl for the growth assay). DNA of all samples was extracted using the E.Z.N.A.® Mollusc DNA Kit (OMEGA Bio-tek, Norcross, GA, USA) according to the manufacturer's protocol and eluted through two elution steps of 50 μl, totalling to 100 μl of DNA extract. DNA concentrations were determined using the Qubit dsDNA high-sensitivity assay (Thermo Fisher Scientific) for all DNA extracts. 3 ng of template DNA (max. 10 µl) was used for qPCR. This indicated that DNA concentrations were low for some samples (mean = 0.16 ng / µl, sd = 0.14 ng / µl). Therefore, the DNA for all samples was concentrated with the Eppendorf™ Concentrator Plus for 30 min at 30 °C, halving the total volume. Subsequently, Qubit measurements were repeated on all samples (mean = 0.35 ng / µl, sd = 0.33 ng / µl). This protocol increased the total amount of DNA per PCR reaction from 0.3 ng to 0.6 ng.
Screening for germ-free samples
We screened for germ-free samples through a growth assay, microscopic investigation and qPCR. For the growth assay, 5 µl of the homogenate was added to 4 ml of R2A medium. The R2A medium was selected as it is a medium capable of growing a broad range of bacteria (Aditi et al., in prep.). The R2A medium consisted of 0.5 g/l BactoTM Proteose peptone (Gibco), 0.5 g/l Casamino acids (Gibco), 0.5 g/l Yeast extract (OXOID), 0.5 g/l Dextrose [D-(+)-Glucose] C6H1206–M 180, 16 g/mol (Carl Roth), 0.5 g/l Soluble starch (Merck), 0.3 g/l Dipotassium hydrogen phosphate (K2HPO4) (Merck), 0.024 g/l Magnesium sulphate (MgSO4.7H20) (Duchefa), 0.3 g/l Sodium pyruvate 100 mM (100x) (Gibco) and topped till one litre with distilled water. The resulting medium was autoclaved and stored at room temperature. The sterile 14 ml tube (Greiner Bio-one) was vortexed briefly and incubated with constant mechanical mixing at 25 °C for 72 h. Prior to OD measurements at 600 nm, the R2A medium was pipetted up and down several times to ensure adequate mixing and homogeneous OD values per sample. Measurements were done on 1 ml of medium in the Genesys 10S UV-Vis Spectrophotometer (Thermo Fisher Scientific) using the Macro-cuvette, 1 ml (Greiner Bio-one). R2A medium incubated with 5 µl of autoclaved tap water was used as a negative to obtain 0 OD values. Samples were considered germ-free if the OD measurement was 0.
To microscopically assess for bacterial presence,, the LIVE/DEAD® BacLightTM Bacterial Viability Kit L13152 (Thermo Fisher Scientific) was used. Each dye was dissolved in 2.5 ml of autoclaved tap water and 10 µl of a (2/3 Propidium Iodide and 1/3 SYTO9) mix to 5 µl of sample. Then it was incubated in the dark for 15 min. Subsequently, 6 µl of the sample was placed under a cover glass and the slide was visually inspected under a fluorescence microscope (Olympus BX51, U-MWB2 Blue fluorescence filter, metal halide lamp). Twenty fields of view were examined for bacterial presence at random, unless a piece of tissue/shell/matrix was seen in the field of view, then this received special attention as bacteria were notably more likely to grow on such substrates, thus increasing our chances of finding bacteria in supposedly bacteriome-disturbed samples. If no bacteria were detected in these 20 fields of view, the sample was classified as germ-free.
qPCR reactions targeting bacterial 16 s rRNA were run on the LightCycler® 480 (Roche). Each reaction consisted of 10 µl of LightCycler® 480 SYBR Green I Master (Life Science), 1 µl 16S-Fw (5′-AGACACGGTCCAGACTCCTAC-3′) at 10 µM, 1 µl 16S-Rv (5′-CTTGCACCCTCCGTATTACCG-3′) at 10 µM, 3 ng of template DNA (n µl) and 8-n µl of sterile Milli-Q water to a final reaction volume of 20 µl, according to the protocol developed by Callens et al. (2018). The temperature cycle was as follows, 5 min of initial denaturation at 95°C, followed by 45 cycles of 10 sec at 95°C, 20 sec at 60°C and 5 sec at 72°C. Each sample was done in triplicate. Standards were obtained by extracting DNA from a pure Eisheria coli culture, measuring the DNA concentration and comparing this weight to the weight of a single E. coli genome. Standards of the desired concentration were mixed with 4 µl of salmon sperm in a total volume of 5 µl to prevent plasmid loss caused by nonspecific binding to surfaces. The melting and amplification curves were calculated through the LightCycler® 480 software (v. 1.5.1) and exported as reports to pdf and excel. The excel data set was then further analysed in R (v. 4.2.2, 64-bit) using RStudio (v. 2022.12.0). Samples were considered germ-free if the Cp value was not significantly different from negative controls in post-hoc pairwise comparisons calculated through the contrast and lsmeans functions with a Scheffe correction for multiple comparisons (emmeans package, v. 1.8.6, Lenth 2023).
16S rRNA metabarcoding
To characterize the bacteriome of our samples, a 16S rRNA metabarcoding approach was used. The PCR protocol targeting the V3 and V4 hypervariable regions of the 16S rRNA gene (Klindworth et al. 2013) was adapted from (Huot et al. 2020). PCR reactions were conducted in triplicate for all samples in a 25 µl total volume. 12.5 µl of PCRBIO HS VeriFiTM Mix (PCR BiosystemsTM), 0.75 µl of the primer 341F (5′-CCTACGGGNGGCWGCAG-3′) at 20 µM, 0.75 µl of the primer 805R (5′-GACTACHVGGGTATCTAATCC-3′) at 20 µM and a variable volume of DNA (0.6 ng of DNA per sample) and water (11 µl—the volume of DNA) to complete the volume to 25 µl and obtain a total DNA input of 0.6 ng. An initial denaturation at 98°C was done for 5 min, followed by 25 cycles of denaturation at 98 °C for 45 s, annealing at 60°C for 15 sec and extension at 72°C for 10 sec and, finally, an elongation step at 72°C for 7 min. The resulting triplicate PCR products were pooled per sample and subsequently purified with magnetic beads (CleanNGS, GC Biotech) following the manufacturer's protocol. The purified PCR products were sent to the Genomics core at UZ Leuven for sequencing (Illumina Miseq v3). First a quality control was performed through a fragment analyzer, and the DNA concentration was measured through Qubit. Then a second PCR reaction attached the Illumina adapters and indexes to the DNA fragments through the Genomics core tails. This PCR was done in a total volume of 20 µl and consisted of 9 µl PCR1 product (concentration 0.5-5 ng/ µl), 0.5 µl of the forward P7 primer (5 µM), 0.5 µl of the Reverse P5 primer (5 µM) and 10 µl of Phusion high fidelity PCR master mix (Thermo Fisher Scientific). PCR conditions were the following. First an initial denaturation step at 94°C for 30 s, 15 cycles of a denaturation step at 94°C for 10 s, an annealing step at 51°C for 30 sec and an elongation step at 72°C for 30 sec and, finally, an elongation step at 72°C for 1 min. After this PCR a second quality control was done with the fragment analyzer to make sure the adapters have joined properly (i.e. fragments increased by +-60 bp). Qubit analysis was performed once more and a final equimolar library was created. Finally, 14 pM was loaded with 37% PhiX spike-in on the Illumina MiSeq v3 sequencer (2 × 300 bp with the 600-cycle kit).
Mock communities
In order to optimize and quantify any biases during our pipeline, two types of mock communities were included in the 16S metabarcoding run. First, the 10 Strain Even Mix Genomic Material MSA-1000™ (ATCC) is a DNA-based mock community that allowed us to exclude any potential extraction bias. It consists of 10% Bacillus pacificus (ATCC 10987), 10% Bifidobacterium adolescentis (ATCC 15703), 10% Clostridium beijerinckii (ATCC 35702), 10% Deinococcus radiodurans (ATCC BAA-816), 10% Enterococcus faecalis (ATCC 47077), 10% Escherichia coli (ATCC 700926), 10% Lactobacillus gasseri (ATCC 33323), 10% Cereibacter sphaeroides (ATCC 17029), 10% Staphylococcus epidermidis (ATCC 12228) and 10% Streptococcus mutans (ATCC 700610). Second, the 10 Strain Even Mix cell Material MSA-1000™ (ATCC) is a cell-based mock community that allowed us to assess any potential extraction bias by comparing the results to the DNA-based mock community (2 × 107 whole cells/vial ± 1 log). It consists of freeze-dried cell material of the same 10 strains and ratios as the MSA-1000™ mock community. The mock communities also allow us to determine the suitable threshold below which spurious sequences should be removed as discussed by Reitmeier et al. (2021).
Analysis of sequencing data
The raw reads of our 16S rRNA metabarcoding approach were processed based on Janssens et al. (2022) with the QIIME2 pipeline v2022.2 (Bolyen et al. 2019). First, the paired-end demultiplexed sequence reads were imported and their quality scores were assessed through MultiQC (Ewels et al. 2016). The run resulted in 4 708 922 raw sequences (min 22 608, max 142 259), 123 622 allocated to mock communities. Using the denoise function of DADA2 within QIIME2, both forward and reverse reads were trimmed for 15 bp and truncated at 280 bp for the forward read and 240 bp for the reverse read, while demanding a 30 bp overlap between both reads. Furthermore, the paired-end sequences were filtered, denoised and dereplicated, chimeras were filtered and an amplicon sequence variants (ASVs) table was created using this denoise function. As a result, on average 59.4% of the reads were maintained per sample (sd= 9.7%). A total of 3 161 ASVs across all samples was obtained with an average length of 425.82 bp (sd= 12.27). The Silva 138 SSU Ref NR 99 database (Quast et al. 2013) was used to assign taxonomy to the ASVs. It contained the extracted sequences of the V3-V4 region using the forward primer (5′-CCTACGGGNGGCWGCAG-3′), reverse primer (5′-GACTACHVGGGTATCTAATCC-3′) and truncation length (465 bp). The R pipeline of Callahan et al. (2016) was modified for this study and ran with R using RStudio on a Windows machine (Windows 11). Briefly, the resulting feature table, taxonomy, metadata and phylogenetic tree were imported in R through the phyloseq package (v. 1.42.0, McMurdie and Holmes 2013). The R package Decontam (v. 1.18.0, Davis et al. 2018) was used to look for contaminating DNA using the prevalence method and a 0.1 threshold. The PCR negative (n = 1) and extraction negative (n = 1) controls were used as negative samples to detect contamination. The mock community dataset did not have contamination in the 21 detected ASVs according to the Decontam package. The transplant experiment had 64 contaminating ASVs out of the total 3 156 ASVs removed from the dataset. Further manual cleaning and filtering of the dataset was performed in R based on Janssens et al. (2022), whereby chloroplast and mitochondrial ASVs were removed from the dataset and ASV taxonomic information was corrected. The Vegan package (v. 2.6–4, Oksanen et al. 2013) was used to construct a rarefaction species richness curve (Heck Jr., van Belle and Simberloff 1975). Samples were rarefied to the sampling depth of the sample with the fewest reads (i.e. 15 321) using the ‘rarefy_even_depth’ function of the phyloseq package (rngseed = 711), while ensuring that the plateau phase was reached for all samples. Consequently, 208 ASVs were removed from the dataset. Subsequently, low abundance taxa (cumulative relative read abundance < 0.5%, 2 046 ASVs; see end of this section for determining this threshold) were removed through the ‘prune_taxa’ function of the phyloseq package. Subsequent analyses excluded snail specimens that were still in the egg at the end of the experiment or died prior to donor inoculum exposure. Alpha diversity measures included the Shannon (phyloseq package, function ‘estimate_richness’) and the Faith's phylogenetic diversity (picante package, v. 1.8.2 (Kembel et al. 2010); function ‘pd’). A generalized linear model with gamma distribution was used to model the effect of treatment on alpha diversity (stats package, v4.2.2). Significance was tested with an ANOVA type III test (car package, v3.1–2, Fox and Weisberg 2019). Outliers, influential observations and normality were assessed using the outliertest (car package), cooks.distance (stats package), and shapiro.test (stats package) functions, respectively. A Tukey pairwise post-hoc comparison was conducted through the glth function (multcomp package, v 1.4–25; Hothorn et al. 2008). Beta diversity measures included the Bray Curtis and Jaccard index, visualized through NMDS (phyloseq package, function ‘ordinate’) and RDA (vegan package, function ‘capscale’). An analysis of variance model was used to model the effect of treatment on beta diversity (stats package). Significance was tested with an ANOVA type III test (car package). For model assumptions, we tested whether the ratio of maximum variance to minimum variance did not exceed five, and outliers, influential observations and normality were assessed as mentioned above. The micrUBIfuns (v. 0.1.0., Sousa 2020) function was modified to study the within group beta diversity for several diversity measures. Core bacteriome analyses were conducted with the microbiome package (v. 1.20.0, Lahti and Shetty 2012) using different detection (0.01–0.001) and prevalence (0.85–0.95, value determined to allow the taxon to be absent from a maximum of one sample) values. For the analyses of the core bacteriome we adhered as much as possible to the guidelines of Neu et al. (2021) and conducted the analyses on non-rarefied, rarefied and rarefied with relative abundance thresholds of 0.25% and 0.5%. Kaplan-Meier survival curves were created (survival package, v. 3.4–0; function ‘survfit’) and significance tested (p- and χ2-values, survival package, function ‘survdiff’, Therneau 2020) based on a subset of the dataset that excluded snail specimens still in the egg at the end of the experiment and specimens that died prior to inoculum exposure. Finally, to assess a suitable threshold below which spurious sequences should be removed (Reitmeier et al. 2021), a maximum likelihood (ML) tree was calculated through the IQ-tree web servers (http://iqtree.cibiv.univie.ac.at), and nodal support was assessed through 1 000 bootstrap replicates (Trifinopoulos et al. 2016). The resulting tree was midpoint rooted in Figtree (version 1.4.4) and tip labels were edited. The resulting tree was exported in PDF format to Adobe Acrobat Pro DC (version 2022.003.20310), in which we adjusted the position of ill-placed nodal support values and italicized species names. Subsequently, a cumulative relative read abundance threshold of 0.5% was selected based on Fig. S4 and Table S1, limiting our dataset as much as possible to the expected ASVs.
Results and discussion
The germ-free status was assigned to five out of a total of twelve bleach-treated samples (42%; Fig. S5, Table S2) and was affected by the maternal line of the recipient (Fig. S6b, χ2 (2, n = 35) = 44, P<0.001). The timespan between sterilization and DNA extraction did not affect the germ-free status of a sample (Fig. S6a, χ2 (3, n = 35) = 2.2, P=0.53). The true germ-free rate of individual snail specimens is expected to have been higher than the detected 42% because the tested samples all comprised of three individual snails. Hence, one contaminated snail is sufficient to label none of the three specimens as germ-free. Importantly, the detection of germ-free snail specimens indicates that vertical transmission within B. glabrata eggs is, at the minimum, not omnipresent. This does not exclude vertical transmission on the egg surface or within the gelatinous matrix of the egg clutch.
Sterilization on the same day as exposure to a donor inoculum induced a high mortality rate (Fig. S6a, χ2 (3, n = 280) = 107, P < 0.001). We speculate that the snail specimens might need some time (minimally 24 h) to recover from bleach exposure. Perhaps, opportunistic pathogens are at play here, whereby otherwise symbiotic bacteria turn pathogenic when the opportunity arises (Brown et al. 2012). However, since the experimental setup required all specimens to be dissected and bleach-exposed per time factor, perhaps this pattern is due to a batch effect. Despite the utmost care taken during the experiment we cannot, at this point, rule out such an experimental artifact. Nevertheless, excluding the samples that were sterilized at the same day as exposure to a donor inoculum, resulted in the same pattern in survival probability across the different inoculum types for the remaining samples.
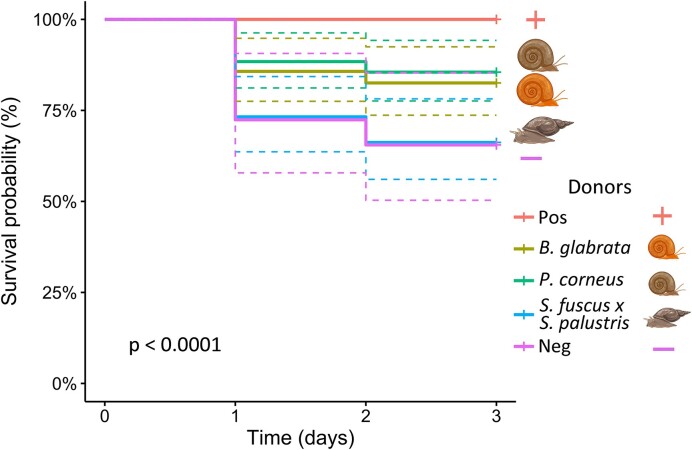
The donor inoculum affected overall survival chance of juvenile snails (Fig. 2, χ2 (4, n = 280) = 26, P<0.001). Moreover, bacteriome-disturbed snails that received a P. corneus donor inoculum survived better than snails without inoculum or with a S. fuscus x S. palustris donor inoculum (χ2 (2, n = 60) = 6.79, P = 0.02). A similar trend, although not significant, is notable for snails that received a B. glabrata donor bacteriome (χ2 (2, n = 55) = 5, P = 0.07). The combination of all these results corroborate our hypothesis and the phylosymbiosis pattern reported by Huot et al. (2020), more in particular that exposure to a donor inoculum from a phylogenetically more distantly related snail reduces the fitness of bacteriome-disturbed snails as opposed to exposure to a donor inoculum from a conspecific or a phylogenetically more closely related snail. Interestingly, survivorship was not significantly different when bacteriome-disturbed snails were exposed to a donor inoculum originating from a conspecific or a species from the same family (Planorbidae). A pattern possibly explained by the wild origin of the latter species as species kept in laboratory conditions tend to rapidly loose microbial diversity (Voulgari-Kokota et al. 2022, Baldassarre et al. 2023), counteracting the phylogenetic distance between the host species. However, since the alpha diversity measures between both donor types was not significantly different this seems unlikely (Fig. S8, P = 1). Mollusks have been reported to form novel microbial assemblies following colonization events in novel regions. Since this is often reported for invasive species (Bankers et al. 2021, Chiarello et al. 2022), it could indicate that this group has a readily adaptable bacteriome if needed.
Figure 2.
Kaplan–Meier survival curve across all days per donor type, excluding specimens dead at the start of the experiment or unhatched at the end of the experiment. ‘Pos’ refers to positive controls (untreated samples), and ‘Neg’ refers to negative controls (bleach-treated samples without donor inoculum exposure). The donor inocula were isolated from Biomphalaria glabrata, Planorbarius corneus, and Stagnicola fuscus x S. palustris individuals. The difference in survival probability between the different donor treatments was highly significant (P<0.0001).
The recipients of donor inocula across all three treatments shared ASVs belonging to the family Pseudomonaceae, Comamonadaceae and Flavobacteriaceae when considering a prevalence of minimally 95% (maximum missing in one sample) and a relative read abundance of minimally 1%. These core members made up 49% of the reads across all samples (sd = 12%), providing a set of potentially ecologically relevant taxa for future research. However, the persistence of this core bacteriome through time remains uncertain. ASVs belonging to the family Pirellulaceae were part of the core bacteriome of bleach-treated snails and occurred in all but one sample. This core member had a highly variable abundance across all negative samples (32%, sd = 28%). It also did not occur in the PCR negative, indicating that it is not due to post-experiment contamination.
Bleach-treated samples, negative controls and the S. fuscus x S. palustris donor inoculum had a lower alpha diversity compared to the other samples (Fig. S8, P < 0.01). Samples that were inoculated with B. glabrata or P. corneus were not significantly different from their respective donor inocula. In contrast, samples that were exposed to the S. fuscus x S. palustris donor inoculum were significantly different from the donor. This discrepancy is presumed to be a consequence of a sequencing bias of the S. fuscus x S. palustris donor inoculum as quality checks prior to sequencing indicated failed amplification but the sample was sequenced nevertheless.
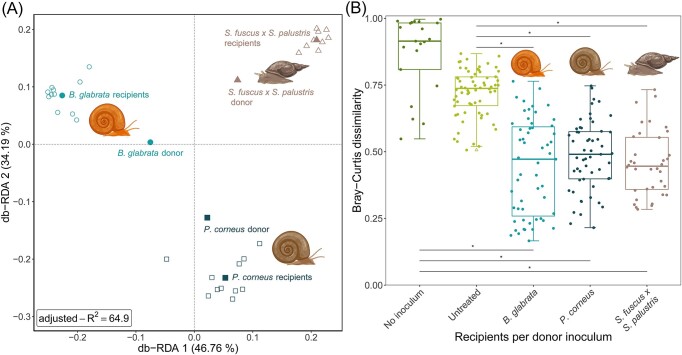
The type of donor inoculum affected the bacterial community composition of the recipients (Fig. 3 and Fig. S9, P < 0.005). Contrary to expectations, Fig. 3A shows each treatment type to cluster most closely to its donor bacteriome yet more distant from the centroid. The previously reported phylosymbiosis pattern would suggest that a snail species assimilates more similar communities from different microbial pools, hence recipient samples were expected to be closer to the centroid. This can potentially be explained by (i) the ‘Anna Karenina principle’, whereby dysbiotic communities tend to be more dissimilar than healthy communities, (ii) alternative stable states of bacterial communities or (iii) the checkerboard pattern, whereby certain taxa tend to exclude each other from the same habitat (Levy and Borenstein 2014, Zaneveld et al. 2017). Our data seems to indicate that the former hypothesis is least probable, because individuals that received the same bacterial inoculum display lower dissimilarities compared to controls (Fig. 3b; Fig. S9) (Zaneveld et al. 2017). As it is unclear at this point whether the different bacteriomes have different metabolic patterns, one cannot ascertain between alternative stable states or the checkerboard pattern. To discern between the three previously listed hypotheses, future experiments should assess the stability, variability and metabolic function of the bacterial community, following a transplant event, after a small perturbation and over time (Zaneveld et al. 2017). The latter should also reveal how long the bacterial transplants persist over time. Notably, our data set might have been skewed against rare taxa due the 40 PCR cycles used to create our sequencing library and the threshold value used to remove spurious sequences. Nevertheless, the inclusion of such a threshold value is desirable as it improves reproducibility of bacteriome analyses (Reitmeier et al. 2021).
Figure 3.
Beta diversity measures across the different treatments. A RDA plot showing the beta diversity of the samples that received a donor inoculum (Bray-Curtis dissimilarity). The model contained only the donor type and had a R² value of 64.9%. B The within-group Bray–Curtis dissimilarity (n=233). ‘No inoculum’ samples (negative controls) were dissected and exposed to bleach but received no donor inoculum. Untreated samples (positive controls) were dissected but were not exposed to bleach and did not receive a donor inoculum. Outliers are indicated by triangles. The lower and upper hinges correspond to the first and third quartiles (the 25th and 75th percentiles). The whiskers extend from the hinge to the largest value no further than 1.5 times the interquartile range from the hinge. ‘*’ indicates P<0.0001.
Future studies can apply our transplant protocol to decipher how snail genotypes interact with microbiota to drive resistance to flatworm parasite infection similar to the experimental setup of Macke et al. (2017) for Daphnia genotype x microbiome interactions with respect to toxic cyanobacteria tolerance. The described technique has been designed on an oviparous freshwater snail system and might, therefore, not necessarily be applicable to all other snail species. For example, dissecting and sterilizing eggs of ovoviviparous snails will come with a new set of challenges. Nevertheless, the technique and patterns reported here provide a foundation for future research to build upon. For example, it could help with two interesting hypotheses with respect to the ovoviviparous freshwater snail Potamopyrgus antipodarum (Takacs-Vesbach et al. 2016). First, the data of Takacs-Vesbach et al. (2016) suggests a link between the bacteriome and the reproductive mode of P. antipodarum. Second, the authors bring forward that snail genotype-bacteriome interactions are determining snail susceptibility to Microphallus infections. Following optimization of our protocol, it could provide the foundation for a reciprocal transplant experiment, testing a causal link between these factors.
Another interesting pattern is the correlation between bacterial diversity and the Guadeloupe resistance complex (CRC) genomic region in B. glabrata (Allan et al. 2018). Our transplant experiment could be applied in a reciprocal transplant experiment followed by an exposure experiment to Schistosoma mansoni to detect a direct link between the CRC genomic region, bacteriome and infected resistance of B. glabrata to schistosome parasites.
The microbiome can determine the transmission potential of vectors (Ford and King 2016). Previous works hypothesize this might also be the case for the tripartite interaction between parasitic flatworms, snails and their microbiome (Huot et al. 2020, Portet et al. 2021, Stock et al. 2021, Le Clec'h et al. 2022). The outcome of the tripartite interaction may have profound effects on ecosystem health as parasitic flatworm biomass is similar to that of birds or fishes in estuaries (Kuris et al. 2008), but is also highly relevant for human and animal health (Flandroy et al. 2018). The microbiome will prove an invaluable factor in better understanding the epidemiology of parasitic flatworms, especially given the wide spread of invasive and occasionally extremely permissive snail species (Lounnas et al. 2017, Carolus et al. 2019, Bankers et al. 2021), or the inconclusive results on snail-parasite interactions in endemic areas (Picquet et al. 1996, Southgate et al. 2000, Sène et al. 2004, Webster et al. 2012).
This study reveals the potential for bacterial community transplant experiments in freshwater snails. Although such controlled laboratory experiments will undoubtedly provide vital insights into the tripartite interaction between snails, bacteria and parasitic flatworms, extrapolation of patterns and protocols to field settings might prove complicated (Greyson-Gaito et al. 2020). However, correlative field-based studies could fill these caveats and, likewise, transplant experiments could help discern correlation from causation in a controlled setting (Stock et al. 2021). Correlative studies, based on museum collections for example (Chalifour et al. 2022), could help in identifying bacterial strains involved in snail resistance. The potential of these strains should then be further examined in transplant experiments in controlled lab environments and mesocosm studies prior to manipulations in the field. We advocate in doing these contained experiments ahead of unconfined biological manipulations as the outcome can be hard to predict. Nevertheless, an improved understanding of the tripartite interaction between hosts, bacteria and pathogens will prove paramount to fully grasp disease epidemiology and field-based correlative studies might help guide experimental efforts.
Supplementary Material
Acknowledgements
Our major gratitude goes out to Benjamin Gourbal and the ‘Université de Perpignan Via Domitia’ for providing the lab-reared Biomphalaria glabrata. Without these samples our research would not have been possible. Our gratitude also goes out to Maxim Vinarski and Katrin Schniebs for providing valuable insights in the identification of the Stagnicola fuscus x S. palustris snail. We also appreciate the constructive comments provided by the editor and reviewers on our manuscript. We also want to thank Eve Toulza and Bart Lievens for their constructive feedback on the study and methodology design during the PhD committee meetings. Furthermore, our gratitude goes out to Shira Houwenhuyse, Manon Coone, Cyril Hammoud, Amruta Rajarajan and Martijn Callens for providing the foundations of our pipeline. Finally, we want to thank Willem Stock for his critical remarks on our dataset and all who was involved in maintaining the snail culture. The computational resources and services used in this work were provided by the VSC (Flemish Supercomputer Center), funded by the Research Foundation—Flanders (FWO) and the Flemish Government.
Contributor Information
Ruben Schols, Department of Biology, Royal Museum for Central Africa, 3080 Tervuren, Belgium; Laboratory of Aquatic Biology, KU Leuven, Campus Kortrijk, 8500 Kortrijk, Belgium.
Isabel Vanoverberghe, Laboratory of Aquatic Biology, KU Leuven, Campus Kortrijk, 8500 Kortrijk, Belgium.
Tine Huyse, Department of Biology, Royal Museum for Central Africa, 3080 Tervuren, Belgium.
Ellen Decaestecker, Laboratory of Aquatic Biology, KU Leuven, Campus Kortrijk, 8500 Kortrijk, Belgium.
Author contributions
Ruben Schols (Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Visualization, Writing – original draft, Writing – review & editing), Isabel Vanoverberghe (Conceptualization, Methodology, Resources, Writing – original draft, Writing – review & editing), Tine Huyse (Funding acquisition, Project administration, Resources, Supervision, Validation, Writing – review & editing), and Ellen Decaestecker (Conceptualthisization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing – review & editing).
Conflict of interest
The authors declare no competing interests.
Funding
This work was supported by BRAIN-be 2.0 under the MicroResist project [Grant number B2/191/P1/MicroResist]. RS received a RBZS travel grant to fund his stay in Kortrijk, Belgium.
Data Availability
The metabarcoding data set generated and analysed during the current study is freely available on the European Nucleotide Service (ENA), under the bioproject PRJEB60803. The COI and ITS2 sequences used for the identification of the donor snail species were uploaded to GenBank and can be retrieved under accession numbers OQ954055-OQ954057, OQ955479.
References
- Allan ERO, Tennessen JA, Sharpton TJet al. Allelic Variation in a Single Genomic Region Alters the Microbiome of the Snail Biomphalaria glabrata. J Hered. 2018;109:604–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldassarre L, Reitzel AM, Fraune S. Genotype–environment interactions determine microbiota plasticity in the sea anemone Nematostella vectensis. PLOS Biol. 2023;21:e3001726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankers L, Dahan D, Neiman Met al. Invasive freshwater snails form novel microbial relationships. Evol Appl. 2021;14:770–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E, Rideout JR, Dillon MRet al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinker P, Fontaine MC, Beukeboom LWet al. Host, Symbionts, and the Microbiome: the Missing Tripartite Interaction. Trends Microbiol. 2019;27:480–8. [DOI] [PubMed] [Google Scholar]
- Brown SP, Cornforth DM, Mideo N. Evolution of virulence in opportunistic pathogens: generalism, plasticity, and control. Trends Microbiol. 2012;20:336–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan BJ, Sankaran K, Fukuyama JAet al. Bioconductor Workflow for Microbiome Data Analysis: from raw reads to community analyses. F1000Res. 2016;5:1492. 10.12688/f1000research.8986.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callens M, Watanabe H, Kato Yet al. Microbiota inoculum composition affects holobiont assembly and host growth in Daphnia. Microbiome. 2018;6:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carolus H, Muzarabani KC, Hammoud Cet al. A cascade of biological invasions and parasite spillback in man-made Lake Kariba. Sci Total Environ. 2019;659:1283–92. [DOI] [PubMed] [Google Scholar]
- Chalifour BN, Elder LE, Li J. Gut microbiome of century-old snail specimens stable across time in preservation. Microbiome. 2022;10:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernin E. a Method of Securing Bacteriologically Sterile Snails (Australorbis glabratus). Exp Biol Med. 1957;96:204–10. [DOI] [PubMed] [Google Scholar]
- Chernin E. Infection of Australorbis glabratus with Schistosoma mansoni under Bacteriologically Sterile Conditions. Exp Biol Med. 1960;105:292–6. [DOI] [PubMed] [Google Scholar]
- Chiarello M, Bucholz JR, McCauley Met al. Environment and Co-occurring Native Mussel Species, but Not Host Genetics, Impact the Microbiome of a Freshwater Invasive Species (Corbicula fluminea). Front Microbiol. 2022;13:800061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis NM, Proctor DM, Holmes SPet al. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. 2018;6:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dheilly NM, Duval D, Mouahid Get al. A family of variable immunoglobulin and lectin domain containing molecules in the snail Biomphalaria glabrata. Develop Comparat Immunol. 2015;48:234–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval D, Galinier R, Mouahid Get al. A Novel Bacterial Pathogen of Biomphalaria glabrata: a Potential Weapon for Schistosomiasis Control?. PLoS Negl Trop Dis. 2015;9:e0003489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels P, Magnusson M, Lundin Set al. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flandroy L, Poutahidis T, Berg Get al. The impact of human activities and lifestyles on the interlinked microbiota and health of humans and of ecosystems. Sci Total Environ. 2018;627:1018–38. [DOI] [PubMed] [Google Scholar]
- Fontaine SS, Kohl KD. Gut microbiota of invasive bullfrog tadpoles responds more rapidly to temperature than a noninvasive congener. Mol Ecol. 2020;29:2449–62. [DOI] [PubMed] [Google Scholar]
- Ford SA, King KC. Harnessing the power of defensive microbes: evolutionary implications in nature and disease control. PLoS Pathog. 2016;12:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J, Weisberg S. An R Companion to Applied Regression. Third. Thousand Oaks, CA: Sage, 2019. [Google Scholar]
- Greyson-Gaito CJ, Bartley TJ, Cottenie Ket al. Into the wild: microbiome transplant studies need broader ecological reality. Proc R Soc B. 2020;287:20192834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck KL Jr, van Belle G, Simberloff D. Explicit Calculation of the Rarefaction Diversity Measurement and the Determination of Sufficient Sample Size. Ecology. 1975;56:1459–61. [Google Scholar]
- Hoffmann AA, Montgomery BL, Popovici Jet al. Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature. 2011;476:454–7. [DOI] [PubMed] [Google Scholar]
- Hothorn T, Bretz F, Westfall P. Simultaneous inference in general parametric models. Biom J. 2008;50:346–63. [DOI] [PubMed] [Google Scholar]
- Huot C, Clerissi C, Gourbal Bet al. Schistosomiasis vector snails and their microbiota display a Phylosymbiosis pattern. Front Microbiol. 2020;10:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens L, Van de Maele M, Delnat Vet al. Evolution of pesticide tolerance and associated changes in the microbiome in the water flea Daphnia magna. Ecotoxicol Environ Saf. 2022;240:113697. [DOI] [PubMed] [Google Scholar]
- Kembel SW, Cowan PD, Helmus MRet al. Picante: r tools for integrating phylogenies and ecology. Bioinformatics. 2010;26:1463–4. [DOI] [PubMed] [Google Scholar]
- Klindworth A, Pruesse E, Schweer Tet al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch H, Schmid-Hempel P. Gut microbiota instead of host genotype drive the specificity in the interaction of a natural host-parasite system. Ecol Lett. 2012;15:1095–103. [DOI] [PubMed] [Google Scholar]
- Krotman Y, Yergaliyev TM, Alexander Shani Ret al. Dissecting the factors shaping fish skin microbiomes in a heterogeneous inland water system. Microbiome. 2020;8:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuris AM, Hechinger RF, Shaw JCet al. Ecosystem energetic implications of parasite and free-living biomass in three estuaries. Nature. 2008;454:515–8. [DOI] [PubMed] [Google Scholar]
- Lahti L, Shetty S. microbiome R package. 2012.
- Le Clec'h W, Nordmeyer S, Anderson TJCet al. Snails, microbiomes, and schistosomes: a three-way interaction?. Trends Parasitol. 2022;38:353–5. [DOI] [PubMed] [Google Scholar]
- Lenth RV. emmeans: Estimated Marginal Means, aka Least-Squares Means. 2023.
- Levy R, Borenstein E. Metagenomic systems biology and metabolic modeling of the human microbiome. Gut Microbes. 2014;5:265–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lounnas M, Correa AC, Vázquez AAet al. Self-fertilization, long-distance flash invasion and biogeography shape the population structure of Pseudosuccinea columella at the worldwide scale. Mol Ecol. 2017;26:887–903. [DOI] [PubMed] [Google Scholar]
- Macke E, Callens M, De Meester Let al. Host-genotype dependent gut microbiota drives zooplankton tolerance to toxic cyanobacteria. Nat Commun. 2017;8:1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, Holmes S. phyloseq: an R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS One. 2013;8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira LA, Iturbe-Ormaetxe I, Jeffery JAet al. A Wolbachia Symbiont in Aedes aegypti Limits Infection with Dengue, Chikungunya, and Plasmodium. Cell. 2009;139:1268–78. [DOI] [PubMed] [Google Scholar]
- Neu AT, Allen EE, Roy K. Defining and quantifying the core microbiome: challenges and prospects. Proc Natl Acad Sci USA. 2021;118:e2104429118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt Ret al. Community ecology package. R Packag Version. 2013;2:321–6. [Google Scholar]
- Picquet M, Ernould JC, Vercruysse Jet al. The epidemiology of human schistosomiasis in the Senegal river basin. Trans R Soc Trop Med Hyg. 1996;90:340–6. [DOI] [PubMed] [Google Scholar]
- Pinto SB, Riback TIS, Sylvestre Get al. Effectiveness of Wolbachia-infected mosquito deployments in reducing the incidence of dengue and other Aedes-borne diseases in Niterói, Brazil: a quasi-experimental study. PLoS Negl Trop Dis. 2021;15:e0009556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portet A, Toulza E, Lokmer Aet al. Experimental infection of the Biomphalaria glabrata vector snail by schistosoma mansoni parasites drives snail microbiota dysbiosis. Microorganisms. 2021;9. 10.3390/microorganisms9051084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz Pet al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitmeier S, Hitch TCA, Treichel Net al. Handling of spurious sequences affects the outcome of high-throughput 16S rRNA gene amplicon profiling. ISME COMMUN. 2021;1:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FEet al. Gut Microbiota from Twins Discordant for Obesity Modulate Metabolism in Mice. Science. 2013;341:1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sène M, Southgate VR, Vercruysse J. Bulinus truncatus, intermediate host of Schistosoma haematobium in the Senegal River Basin (SRB). Bull La Soc Pathol Exot. 2004;97:29–32. [PubMed] [Google Scholar]
- Sokolow SH, Wood CL, Jones IJet al. To Reduce the Global Burden of Human Schistosomiasis, Use ‘Old Fashioned’ Snail Control. Trends Parasitol. 2018;34:23–40. 10.1016/j.pt.2017.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa A. Set of functions for microbiome analyses. 2020.
- Southgate VR, de Clercq D, Sène Met al. Observations on the compatibility between Bulinusspp. and Schistosoma haematobium in the Senegal River basin. Ann Trop Med Parasitol. 2000;94:157–64. [DOI] [PubMed] [Google Scholar]
- Stock W, Callens M, Houwenhuyse Set al. Human impact on symbioses between aquatic organisms and microbes. Aquat Microb Ecol. 2021;87:113–38. [Google Scholar]
- Takacs-Vesbach C, King K, Van Horn Det al. Distinct Bacterial Microbiomes in Sexual and Asexual Potamopyrgus antipodarum, a New Zealand Freshwater Snail. PLoS One. 2016;11:e0161050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therneau TM. a Package for Survival Analysis in R. 2020.
- Trifinopoulos J, Nguyen L-T, von Haeseler Aet al. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016;44:W232–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voulgari-Kokota A, Beukeboom LW, Wertheim Bet al. Houseflies harbor less diverse microbiota under laboratory conditions but maintain a consistent set of host-associated bacteria. Sci Rep. 2022;12:11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster BL, Emery AM, Webster JPet al. Genetic Diversity within Schistosoma haematobium: DNA Barcoding Reveals Two Distinct Groups. PLoS Negl Trop Dis. 2012;6:e1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaneveld JR, McMinds R, Vega Thurber R. Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat Microbiol. 2017;2:17121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The metabarcoding data set generated and analysed during the current study is freely available on the European Nucleotide Service (ENA), under the bioproject PRJEB60803. The COI and ITS2 sequences used for the identification of the donor snail species were uploaded to GenBank and can be retrieved under accession numbers OQ954055-OQ954057, OQ955479.