Abstract
Eukaryotes have historically been studied as parasites, but recent evidence suggests they may be indicators of a healthy gut ecosystem. Here, we describe the eukaryome along the gastrointestinal tract of children aged 2–5 years and test for associations with clinical factors such as anaemia, intestinal inflammation, chronic undernutrition, and age. Children were enrolled from December 2016 to May 2018 in Bangui, Central African Republic and Antananarivo, Madagascar. We analyzed a total of 1104 samples representing 212 gastric, 187 duodenal, and 705 fecal samples using a metabarcoding approach targeting the full ITS2 region for fungi, and the V4 hypervariable region of the 18S rRNA gene for the overall eukaryome. Roughly, half of all fecal samples showed microeukaryotic reads. We find high intersubject variability, only a handful of taxa that are likely residents of the gastrointestinal tract, and frequent co-occurrence of eukaryotes within an individual. We also find that the eukaryome differs between the stomach, duodenum, and feces and is strongly influenced by country of origin. Our data show trends towards higher levels of Fusarium equiseti, a mycotoxin producing fungus, and lower levels of the protist Blastocystis in stunted children compared to nonstunted controls. Overall, the eukaryome is poorly correlated with clinical variables. Our study is of one of the largest cohorts analyzing the human intestinal eukaryome to date and the first to compare the eukaryome across different compartments of the gastrointestinal tract. Our results highlight the importance of studying populations across the world to uncover common features of the eukaryome in health.
Keywords: human intestinal eukaryome; biogeography; fungi, protists; Sub-Saharan Africa; microbiota
This study uses amplicon sequencing to unravel the intestinal eukaryome of almost 1000 African children and shows that it is influenced by geographic location, gut biogeography, and nutritional status.
Introduction
The human gastrointestinal (GI) microbiome is a complex community comprised of bacteria, archaea, viruses, and eukaryotes (fungi, protists, and helminths). Many studies have mapped the bacterial microbiome across life stages, geographic locations, and health states and demonstrated its importance in health and disease (Yatsunenko et al. 2012, Luan et al. 2015, Castanys-Muñoz et al. 2016, Dominguez-Bello et al. 2019). Despite the clear contributions of some host-associated eukaryotes to human health, eukaryotes have been less often characterized than the bacterial members of the gut microbiome, particularly in large-scale studies (Parfrey et al. 2014, Stensvold and van der Giezen 2018, Mann et al. 2020). Hurdles include the lower biomass of eukaryotes in the gut (eukaryotes constitute < 0.1% of the total biomass of the microbiota; Qin et al. 2010), the smaller community studying eukaryotes, particularly outside of parasitology, technical difficulties arising from less well-curated databases (del Campo et al. 2020), and difficulty separating gut residents from eukaryotes introduced via diet (Suhr and Hallen-Adams 2015, Mann et al. 2020).
Studies using 18S rRNA targeted amplicon sequencing showed that protists and fungi, especially members of the Saccharomycetales, are the dominant eukaryotes in the human GI tract (Parfrey et al. 2014, Scanlan et al. 2018). Protists are a common part of the gut community (Stensvold et al. 2011a, b, 2020, Forsell et al. 2012, Roser et al. 2013, Parfrey et al. 2014, Krogsgaard et al. 2015, 2018, Jokelainen et al. 2017, Stensvold and van der Giezen 2018, Stensvold 2019, Mann et al. 2020) and are regularly detected in the feces of infants and toddlers (Jokelainen et al. 2017). In the past, studies describing the human eukaryome using specific Polymerase Chain Reaction (PCR) primers reported that the protist genera Blastocystis and Dientamoeba (Scanlan and Marchesi 2008, Velasco et al. 2011, Roser et al. 2013, El Safadi et al. 2014, Turkeltaub et al. 2015, Andersen and Stensvold 2016, Jokelainen et al. 2017) are widespread in healthy humans. Amoebae, especially Entamoeba coli, have been also described as commensal members of the human gut microbiome (ten Hove et al. 2007, Bruijnesteijn van Coppenraet et al. 2009, Stensvold et al. 2011b, Krogsgaard et al. 2018). Further, the presence of Blastocystis has been associated with sanitation levels, water source, and contact with animals or other infected humans (El Safadi et al. 2014, Beghini et al. 2017, Scanlan et al. 2018) and is generally reported at higher prevalence in low- and middle-income countries. Fungi are also commonly found in the intestinal microbiota of many different mammals, including humans (El Mouzan et al. 2017, Hallen-Adams and Suhr 2017, Nash et al. 2017, Auchtung et al. 2018, Lavrinienko et al. 2021, Boutin et al. 2021, Sun et al. 2021). In humans, fungi colonize the infant intestinal tract shortly after birth (Schei et al. 2017). Thus, there is consistent detection of eukaryotes in the intestinal microbiota of healthy subjects.
Recent cross-kingdom analyses show that fungi (Huseyin et al. 2017) and microeukaryotes (Stensvold 2019) are active participants in the GI ecosystem and influence health and disease through interactions with each other, other microbes, and the host. Microeukaryotes including Blastocystis, Giardia, Entamoeba (Verweij et al. 2004, ten Hove et al. 2007, Velasco et al. 2011), and a variety of fungi (Luan et al. 2015, Limon et al. 2019, Richard and Sokol 2019) have been linked to GI disease, antibiotic-associated diarrhea, and chemotherapy-induced enteric disorders (Stensvold and van der Giezen 2018), inflammatory bowel disease (IBD; El Mouzan et al. 2017, Sovran et al. 2018, Limon et al. 2019, Richard and Sokol 2019), and asthma (Arrieta et al. 2018, Goldman et al. 2018, Boutin et al. 2021) [reviewed in Huseyin et al. (2017)]. Further, the human mycobiome is changed in obesity (Mar Rodriguez et al. 2015) and fungal–bacterial interactions are perturbed in patients suffering of IBD (Sovran et al. 2018). Other microeukaryotes, such as Giardia intestinalis and Entamoeba histolytica, are pathogens that directly cause substantial morbidity and mortality (Roberts et al. 2013). Last, helminths, which are often considered pathogens, are well known to have an immunoregulatory function (Walsh et al. 2009, Broadhurst et al. 2012, Zaiss et al. 2015, Finlay et al. 2016, Gause and Maizels 2016, Giacomin et al. 2016, Ramanan et al. 2016) and the presence of several helminths has been associated with changes in the bacterial community (Walk et al. 2010, Cantacessi et al. 2014, McKenney et al. 2015, Zaiss et al. 2015, Giacomin et al. 2016). There is thus a clear link between alterations in the fungal and microeukaryotic community of the intestinal tract and disease.
In contrast to these observations, a growing number of studies show that protists such as E. coli and Blastocystis are common in healthy people (El Safadi et al. 2014, Krogsgaard et al. 2015, Andersen and Stensvold 2016, Beghini et al. 2017, Nieves-Ramirez et al. 2018), suggesting that they might also be important to maintain proper gut homeostasis (Audebert et al. 2016, Andersen and Stensvold 2016, Beghini et al. 2017) and are, thus indicators of a healthy gut ecosystem (Stensvold and van der Giezen 2018). Indeed, some of them, as an example Blastocystis (Krogsgaard et al. 2015) or Dientamoeba are even more common in healthy individuals than in comparison groups with immune mediated disease such as IBD (Andersen and Stensvold 2016, Beghini et al. 2017) and irritable bowel syndrome [reviewed in Stensvold and van der Giezen (2018)]. Further, colonization with helminths were virtually universal in human populations before the adoption of modern, highly sanitized lifestyles (Goncalves et al. 2003) and their drastic decrease in urban industrial populations has been discussed as a possible contributor to the concomitant rise of autoimmune disease (Rook 2012). Another recent observation is that coinfection with different eukaryotic pathogens can lead to reduced virulence in some conditions [reviewed in Venter et al. (2022)]. In this light, reduced diversity of helminths and other microeukaryotes (Parfrey et al. 2014) in industrialized countries may be altering the gut ecosystem directly through loss of species and indirectly by the associated loss of interactions.
Thus, the role of microeukaryotes in the intestinal tract seems to be complex and to depend on the particular GI ecosystem they inhabit.
There is clear evidence of geographic differences in the bacterial microbiome, mainly mediated through diet (De Filippo et al. 2010). Further, globalization and urbanization have been shown to be associated with a major loss in microbial diversity compared to a more traditional lifestyle (Clemente et al. 2015, Obregon-Tito et al. 2015, Smits et al. 2017, Jha et al. 2018, Pasolli et al. 2019). For the eukaryome, to date, most studies have been performed on a single geographic location. A small study in South Africa has shown that the mycobiome is affected by urbanization (Kabwe et al. 2020). A larger study in China, spanning several ethnicities and geographic regions showed very high variability across different geographic regions (Sun et al. 2021), mainly reflecting dietary habits and the urbanization gradient. For microeukaryotes, there are some geographic differences in the subtypes of Blastocystis (Alfellani et al. 2013). These studies clearly show the need to investigate the overall eukaryome make-up across different geographic locations.
So far, most studies assessing the microbiome focus on fecal samples as an overall read-out of the microbial intestinal community. There is, however, clear evidence for bacterial community changes along the GI tract (Vonaesch et al. 2018); there are likely similar changes in the eukaryotic community. The different segments of the GI tract have different roles and physiology, with nutrient absorption taking place mainly in the small intestine and fecal samples representing an overall read-out of the lower GI tract microbiota.
In this study, we aimed to characterize and compare the intestinal and fecal eukaryome in children aged 2–5 years living in two African countries: Madagascar and the Central African Republic (CAR). We then investigate the relationship between the eukaryome composition and clinical factors such as stunted growth, iron deficiency, and intestinal inflammation.
Methods
Study cohort, sample collection, metadata, and biobanking
This study was carried out in the context of the AFRIBIOTA project, a case-control study for stunting in children aged 2–5 years in two different study sites, Bangui, CAR and Antananarivo, Madagascar (Vonaesch et al. 2018). Recruitment took place between December 2016 and March 2018. In the context of this analysis, only children recruited in the community were included. Metadata including age, nutritional status, iron levels, hemoglobin, and socio-economic factors were collected using a standardized questionnaire. Complete blood count, C-reactive protein (CRP), and ferritin levels were measured at the Clinical Biology Center (CBC) of the Institut Pasteur de Madagascar and the Laboratoire d'Analyse Médicale at the Institut Pasteur de Bangui within 4 h after blood collection. Ferritin levels were corrected for systemic inflammation as described in (Thurnham et al. 2010). Hemoglobin values were adjusted for altitude as described in Centers for Disease Control (CDC; 1989) and Sullivan et al. (2008), and anemia was defined as less than 110 g/l according to WHO criteria (Onis 2006, OMS 2011). All participants received oral and written information about the study and legal representatives of the children provided written consent to participate in the study. The study protocol for AFRIBIOTA has been approved by the Institutional Review Board of the Institut Pasteur (2016–06/IRB) and the National Ethical Review Boards of Madagascar (55/MSANP/CE, 19 May 2015) and CAR (173/UB/FACSS/CSCVPER/16). Detailed inclusion and exclusion criteria and recruitment procedures are described elsewhere (Vonaesch et al. 2018); importantly, children with severe acute disease were excluded, as were children that had recently taken antibiotics (Vonaesch et al. 2018). Based on median height of the WHO reference population (Onis 2006, World Health Organization 2007), the children were classified in three groups: severe stunting (height-for-age z-score ≤ −3SD), moderate stunting (height-for-age z-score between −3SD and −2SD), and not stunted (height-for-age z-score ≥ −2SD). Caregivers were instructed to collect feces in the morning before coming to the hospital. Gastric and duodenal samples were collected using a pediatric nasogastric tube (Vygon, France), and were only collected for stunted children (ethical constraints). The procedure was put in place for studying the bacterial community of the small intestine, which was suspected, and later shown, to be implicated in the pathophysiology of stunting (Vonaesch et al. 2018, 2022). We, thus had the unique opportunity to analyze the intestinal eukaryome from these samples. Once the gastric, duodenal, or fecal samples were collected, they were aliquoted, frozen at −20°C and transferred the same day to a −80°C freezer (Bangui), or directly snap-frozen in liquid nitrogen and then transferred to a −80°C freezer (Antananarivo). DNA extraction was performed on site in Antananarivo and Bangui and extracted DNA was shipped on dry ice. Biobanking and sample distribution was performed by the Unité de Bactériologie Expérimentale, Institut Pasteur de Madagascar, the Laboratoire d'Analyse Médicale, Institut Pasteur de Bangui, and the Clinical Investigation and Access to BioResources Platform (ICAReB) at the Institut Pasteur, Paris. The STORMS checklist for this study is included as File S1 (Supporting Information).
DNA extraction and sequencing
Gastric, duodenal, and fecal samples were extracted by commercial kits (QiaAmp cador® Pathogen Mini or cador® Pathogen 96 QIAcube® HT Kit, Qiagen, which are kits using the same chemistry for manual or automatic extraction) following the manufacturer’s recommendations with an additional bead-beating step to increase mechanical disruption as described in Vonaesch et al. (2018). DNA extraction was compared between the two sites using bacterial ZymoBiomics community standards (Zymobiomics, D6300) and DNA contamination was assessed using parallel processed negative controls (molecular grade water). Samples were stored at −80°C until sequencing. Extracted DNA samples were shipped on dry ice to a commercial provider where library generation and sequencing was performed (Microbiome Insights, Canada).
For ITS2 sequencing, the primers ITS4 (5′-CCTCCGCTTATTGATATGC-3′) and fITS7 (5′-CCGTGARTCATCGAATCTTTG-3′) were used (Ihrmark et al. 2012). Primer choice was based on Nash et al. (2017). PCR conditions were identical to the ones described in Gweon et al. (2015). 18S primers TAReuk454FWD1 (5′-CCAGCASCYGCGGTAATTCC-3′) and TAReukREV3 (5′-ACTTTCGTTCTTGATYRA-3′) were chosen to preferentially amplify protists (Stoeck et al. 2010, Maritz et al. 2017). In brief, library synthesis and amplification were performed using Phusion High-Fidelity PCR Master Mix (Thermo Scientific, catalogue #F-531S) in a 20 μl reaction volume, and a two-step PCR amplification strategy according to the following protocol: 98°C for 30 s, 10 cycles of 98°C for 10 s, 53°C for 30 s, 72°C for 30 s; and then 25 cycles of 98°C for 10 s, 48°C for 30 s, 72°C for 30 s, and ending at 72°C for 10 min. Sequencing was performed on an Illumina MiSeq using the 300-bp paired-end kit (v.3). Only high quality forward reads (R1) were used for the analysis.
For a subset of the data, a second library using the same 18S rRNA V4 amplicon primers (TAReuk454FWD1 and TAReukREV3) and a peptide nucleic acid primer to block amplification of mammalian sequences (5′-TCTTAATCATGGCCTCAGTT-3′) (Mann et al. 2020) was performed at the University of British Columbia, Vancouver, Canada using identical PCR conditions. The mammalian blocking primer was designed to reduce the amplification of human reads and thus increase sequencing depth for nonhuman eukaryotic reads (Mann et al. 2020). Sequencing of this second data set was performed at Dalhousie University on a MiSeq Illumina sequencer using 300+300 bp paired-end V3 chemistry as described in Kozich et al. (2013).
Bioinformatic analysis
18S rRNA V4 datasets
An average of 48 893 (minimum: 1; maximum: 369 607; total: 42 879 181) reads were generated for the 18S dataset.
Demultiplexed reads were obtained from the two sequencing facilities and were processed into amplicon sequence variants (ASVs) using the DADA2 pipeline (Callahan et al. 2016) with a minimum sequence length of 150 and maximum expected error of 8. Overall, for the 18S rRNA dataset using no blocking primer, we obtained 694 978 clean sequences after running the Dada2 pipeline and filtering out any nonmicroeukaryotic reads such as vertebrates or plants. On average, we had 2632 sequences per sequencing-positive sample (i.e. with any sequences amplified; median: 1638; minimum: 0; maximum: 21 327). Taxonomy for the 18S dataset was assigned in a multistep process. First, taxonomy was assigned using DADA2 and with the integrated tool SINA (Pruesse et al. 2012) and against the SILVA database (version 128) after which any ASV that could not be assigned a taxonomy was compared to the PR2 database (version 4.11.1) (Guillou et al. 2013). ASVs present in only one sample and at a relative abundance of less than 0.1% of the total dataset were removed. Samples with fewer than 5000 reads were removed from downstream analyses.
An average of 30 203 (minimum: 289; maximum: 267 972; total: 9 393 270) reads were generated for the 18S dataset using a blocking primer. Overall, for the 18S rRNA dataset using the blocking primer, we obtained 3 415 973 clean sequences after running the Dada2 pipeline and filtering out any nonmicroeukaryotic reads such as vertebrates or plants. On average, we had 6222 sequences per sequencing-positive sample (i.e. with any sequences amplified; median: 983; minimum: 2; maximum: 84 623). ASVs for this dataset were generated in an identical manner to the full 18S dataset and samples with fewer reads than 500 were removed from downstream analyses. 18S sample processing files can be found at https://github.com/Parfreylab/afribiota.
Taxonomy was refined by cross-referencing with the other databases and by placement in phylogenetic trees for key taxa as described below. Suspected taxonomic misannotations were checked using BLASTn and the NCBI NT database (McGinnis and Madden 2004, Ye et al. 2006, Johnson et al. 2008) and the taxonomic string assignment for Entamoeba and Saccharomycetales, which are erroneous in the Silva database, were corrected manually. All corrections to the original taxonomy file are listed in Table S1 (Supporting Information). Code for the bioinformatic analysis is available at https://github.com/Parfreylab/afribiota.
ITS2 dataset
An average of 57 170 (minimum: 24, maximum: 339 136) sequences per sample were generated for the ITS2 dataset. ASVs were generated using the DADA2 pipeline with a minimum sequence length of 50 and maximum expected error set to 6 and 8 for the forward and reverse reads, respectively. After dereplication, ASVs comprised of fewer than 50 reads or those with less than 0.1% overall relative abundance in the full dataset were removed. On average, we obtained 40 576 clean sequences per sample for the ITS2 dataset after running the Dada2 pipeline (minimum: 0; maximum: 217 506). Samples with fewer than 5000 reads were removed from downstream analyses. Taxonomy was assigned using the UNITE database (version 8.0) (Koljalg et al. 2005). Fungal ASVs that could not be assigned beyond the kingdom level using the UNITE database (36.7% of the total dataset) were run through BLAST against the NCBI NT database but no further taxonomic information could be resolved and the sequences were thus just assigned at Kingdom level. All code for sample processing is available at https://github.com/parfreylab/afribiota. Rarefaction curves for all datasets are given in Figure S1 (Supporting Information). All raw sequencing data is deposited in ENA, accession number PRJEB57073.
Annotation of taxa as environmental, gut-associated, or potentially gut-associated taxa
Taxa were individually assessed and screened for human-association using expert knowledge and Google search. Taxa were classified as gut-associated if the taxon has been associated with human infection or repeatedly described to be part of the eukaryotic microbiome in peer-reviewed scientific journals. Taxa were classified as possibly gut-associated if the taxon has been associated with human infection, but in very few reports or cases. Taxa were classified as environmental if they are known plant parasites, are known to inhabit aquatic environments or soil, or are known to be associated with nonhuman eukaryotes such as insects. If there was no hit for the taxon in the Google search or if the taxonomic level did not allow distinguishing the taxon enough to make a claim about the most likely origin or association, taxa were classified as from unknown origin.
Construction of phylogenetic trees
Taxonomy was refined by phylogenetic analysis for key eukaryotes by constructing backbone trees and then placing ASVs within these trees. A total of five backbone trees were generated, one for Blastocystis, Entamoeba, Trichostomatia, Diplomonada, and Trichomonada. For each backbone tree, we retrieved available sequences from the SILVA (v.132), PR2 (v.4.12.0) and NCBI databases. Sequence accession numbers used for the backbone trees are presented in Table S2 (Supporting Information). We also used out-group taxa selected based on the literature to root the trees. Sequences were first sorted and clustered at 99% similarity using usearch (v.8.1.1831_i86linux32) or vsearch (v.2.15.1) (Rognes et al. 2016). Sequences were then aligned using mafft (v.6.814b or v.7.475) (Katoh et al. 2009) and trimmed using trimAl (v.1.2rev59 or v.1.4.1) (Capella-Gutierrez et al. 2009), and the alignment inspected using aliview (v.1.26) (Larsson 2014). A maximum-likelihood phylogenetic tree was then constructed using a GTR model and 100 bootstrap replicates with RAxML (v.8) 95 (Stamatakis 2015, Kozlov et al. 2019). ASVs retrieved from the Afribiota dataset were then placed into the resulting backbone tree using SINA (v.1.7.1) (Pruesse et al. 2012). The annotation of 102 ASVs was then updated based on the backbone trees (Table S1, Supporting Information). Blastocystis ASVs were further curated based on the alignment and tree. Two ASVs fall outside of known subtypes (DALASV146 and DALASV723); DALASV146 matches at 100% similarity with accession OM057456.1, which is sampled from Hoolock Gibbon. Subtypes ST1, ST2, and ST3 were represented by many ASVs and were further collapsed into subclusters if sequences were >97% similar and formed a clade in the phylogenetic tree (Table S1 and Figure S4, Supporting Information). Subclusters did not show one to one correspondence with Blastocystis allele designations because the sequence fragment here is shorter than the fragment used for allele designation and missing informative sites; for this reason we refer to subclusters by the ASV# of the ASV with highest relative abundance rather than allele. The most similar allele for each ASV within a subcluster was determined using the Blastocystis Public databases for molecular typing and microbial genome diversity (Stensvold et al. 2012) and reported along with ASV in Table S1 (Supporting Information). Conda environment and scripts used to generate the Entamoeba tree are available at https://github.com/aemann01/afribiota and the ones to generate the Blastocystis, Trichostomatia, Diplomonada, and Trichomonada trees at https://github.com/parfreylab/afribiota.
Measurement of fecal markers of inflammation
Fecal calprotectin concentration was assayed in duplicate by a ‘sandwich’ type enzyme-linked immunosorbent assay, which uses a polyclonal antibody system (Calprest; Eurospital, Italy). The assay was performed according to the manufacturer’s instructions yielding a measurement range of 15–5000 µg/g. Fecal α1-antitrypsin was measured using an immune-nephelemetric method adapted on the BN ProSpec system (Siemens, Germany). Briefly, stool samples were diluted 1:5 in 0.15 M NaCl then shaken vigorously by the mean of a vortex until complete homogenization. The homogenate was centrifuged at 10 000 g for 15 min at 4°C and the supernatant was used for analysis, which was performed at two different dilutions (1:5 and 1:500 final dilutions) to avoid any prozone phenomena. Using this method, the range of measurement was 0.01–20 mg/g (Rodriguez-Otero et al. 2012).
Identification of microeukaryotes by microscopy
The identification of parasites was performed on a subset of subjects, which provided enough stool samples according to methods reported previously (Habib et al. 2021). In short, fecal samples were examined microscopically using the Merthiolate–Iodine–Formaldehyde (MIF) and Kato-Katz (KK) techniques for helminths and the direct smear method (mounting without colouring) for protozoans according to standard techniques. The analyses were performed at the Medical Center and the Experimental Bacteriology Unit of the Institut Pasteur de Madagascar and the Institut Pasteur de Bangui and validated by a medical doctor for diagnostic purposes.
Biostatistical analysis
The final, filtered dataset with >5000 sequences per sample comprised the following: for the 18S rRNA dataset without human blocking primers 33 gastric samples, 53 duodenal samples, and 464 fecal samples, for the 18S rRNA with human blocking primer 23 duodenal samples, 241 fecal samples and for the ITS2 dataset 158 gastric samples, 145 duodenal samples, and 315 fecal samples. Biostatistical analyses were performed using R, version 3.4.1 and the R packages Phyloseq (version 1.22.3) (McMurdie and Holmes 2013), microbiome (version 1.0.2) (Callahan et al. 2016), vegan (version 2.4–6) (Dixon 2009), DESeq2 (version 1.18.1) 103 (Anders and Huber 2010, Anders et al. 2013), and ggplot2 (version 3.3.0) (Journal of Statistical Software, 2010). For the 18S dataset, taxa without annotation beyond the kingdom level were excluded from the final analysis because many were misannotated bacterial sequences. Alpha diversity was quantified using the combined Shannon index using rarefied data including singletons. All other analyses were performed on datasets with singletons filtered out. Beta diversity was quantified using the Bray–Curtis dissimilarity index for data of relative abundance and the Jaccard index for presence–absence datasets (Bray and Curtis 1957). The Jaccard index was preferred over the Sorensen index to have equal weight for all taxa regardless of their prevalence across the dataset. Differences of diversity tests between samples were performed using nonparametric multivariate analysis of variance (PERMANOVA) with the function ‘adonis’ in the R package vegan (Dixon 2009) or using an iterative, logistic regression. P-values were corrected for multiple comparisons using the Benjamini–Hochberg procedure. Multivariate analyses of differentially abundant taxa as well as the presence of given taxa were performed on combined samples from both countries as well as on data from each country independently. Multivariate models were corrected for gender, age (in months), and country of origin and stratified on sample type, then on country of origin. For fecal samples, the multivariate models were also corrected for calprotectin levels as a measure of intestinal inflammation. Metadata, ASV tables and taxonomy tables can be found in the Appendix (Supporting Information) and the code is available at https://github.com/parfreylab/afribiota.
Results
Description of study population and the presence of eukaryotic reads
We analyzed a total of 1104 biological samples from a cohort of children from Madagascar and CAR with two primer sets to investigate the fungal (ITS2 primers) and microeukaryotic (18S rRNA gene primers) components of the gut microbiome. The sample filtering workflow is summarized in Figure S3 (Supporting Information). The overall characteristics of the children included in the study are given in Table S3 (Supporting Information) and the distribution of the main clinical variables in Figure S2 (Supporting Information).
Host, dietary, and environmental taxa were frequently amplified with 18S primers, as is common for studies of the mammalian gut (Mann et al. 2020). Removing sequences corresponding to plants or vertebrates from the 18S dataset left 464 fecal samples and 53 duodenal samples that had microeukaryotic reads (66% and 28%, respectively) (Figure S3A, Supporting Information). We also amplified a subset of 312 samples with 18S primers plus a mammalian blocking primer; 241 fecal samples (92%), and 23 duodenal samples (72%) were above the threshold of 500 sequences set by the background (negative control; Figure S3B, Supporting Information). After filtering out plant and vertebrate reads, the microeukaryotic reads were very low in the duodenal and gastric samples. They were a median of 99 reads in the duodenal samples and nine reads in the gastric samples using no blocking primer compared to a median read count of 988 for fecal samples and 49 reads in the duodenal samples and 2886 reads in fecal samples in the dataset using a blocking primer. Thus, for the two 18S datasets, the statistical analyses were therefore only performed for the fecal samples. Amplification of ITS and 18S was uneven across samples, so we asked whether samples with reads above threshold levels were correlated with country of origin or clinical variables. There were significantly more microeukaryotic positive samples in Madagascar compared to Bangui (18S dataset with a blocking primer: P < .001 and without a blocking primer: P = .014). Further, there was a weak trend to more microeukaryotic positive samples in older children compared to younger children (18S dataset with a blocking primer P = .38, 18S dataset without a blocking primer: P = .026). Anemia in the fecal 18S dataset using a human blocking primer was the only clinical variable significantly associated with the presence of microeukaryotic reads. The dataset generated with a blocking primer yielded higher diversity and more eukaryotes likely to be gut residents (Figure S5, Supporting Information). We, thus focus in our analysis on this dataset and present results without the blocking primer in the supplement.
In the fungal ITS2 dataset 95% of the gastric samples sequenced (158 samples), 77% of the duodenal samples sequenced (145 samples), and 45% of the fecal samples sequenced (315 samples) had ITS2 (fungal) reads above the threshold of 5000 reads set by the negative control (Figure S3C, Supporting Information). Gastric and duodenal samples had a higher prevalence of fungi-positive samples compared to feces (P < .0001). The prevalence of samples with fungal reads in Madagascar (43%) was significantly higher compared to Bangui (33%; P = .03) and there were also more fungal reads per sample in Madagascar compared to Bangui (P = .03). The presence of fungal reads was not significantly associated with any of the clinical parameters measured in the duodenum or feces.
Thus, our data shows that most of the human samples contain eukaryotic reads and that the presence of an intestinal eukaryome is not strongly influenced by clinical parameters but is influenced by the children's country of residence and age.
The fecal eukaryome of African children is dominated by Blastocystis and fungi
Characterizing the eukaryome reveals a handful of fungi and microeukaryotes that are prevalent across populations, while the overall eukaryome is low diversity and the distribution of most taxa patchy across individuals and populations (Figure 1A and B; Figure S8A, Supporting Information). After filtering for low abundance and low-prevalence taxa we recovered 219 ASVs in the ITS dataset of which 99 (45%) are unassigned at species level and 48 (22%) at phylum level.
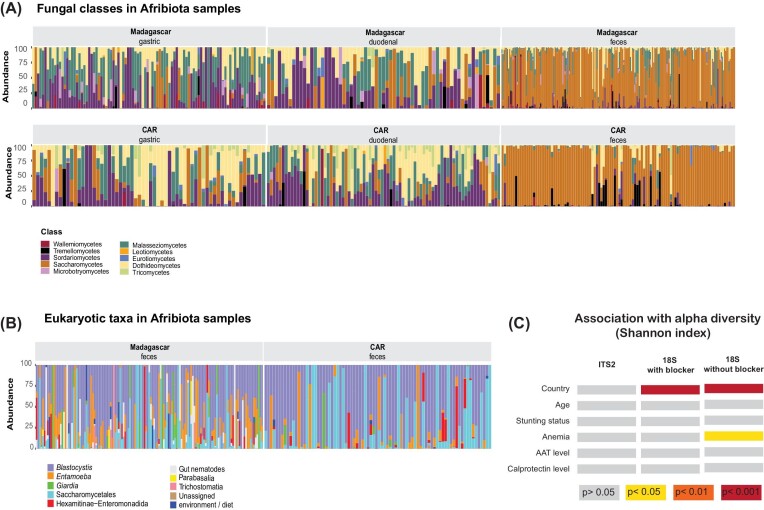
Figure 1.
Composition of the Afribiota samples as revealed by ITS2 sequencing targeting fungi (A), the broader 18S primers using a mammalian blocker (B) as well as association of the overall diversity with different clinical outcomes (C). Significant association with the Shannon index as a measure of alpha diversity were assessed using a Wilcoxon rank-sum test. The colour code illustrating the degree of significance of the association is given on the bottom of the figure.
After filtering for low-prevalence taxa (present in less than 50 sequences and less than 0.01% of the samples) and removing plants and vertebrates (18S rRNA dataset), we recovered 57 ASVs in the 18S rRNA dataset without using a mammalian blocking primer, and 127 ASVs in the reduced 18S rRNA dataset using a mammalian blocking primer.
As expected, for the ITS2 dataset, most of the taxa belong to the phylum Ascomycota or Basidiomycota. Overall, we identified two different fungal classes, 23 different orders, 41 different families, 60 different genera, and 78 different species (Tables S5–S10, Supporting Information). We identified a suite of eukaryotes spanning a broad taxonomic range that include many protists and helminths that are familiar from parasitology textbooks (Roberts et al. 2013), such as Entamoeba, Giardia, Trichuris, and Ascaris (Figure 3). The list of taxa detected is very likely an incomplete catalog of the microeukaryotic diversity in this cohort due to rather low sequencing depth and because many microeukaryotes are found at low frequency. We assessed whether using a mammalian blocking primer consistently increased the recovered diversity across microeukaryotic taxa by analyzing a subset of 100 fecal samples that were amplified with and without the mammalian blocking primer and compared taxon prevalence (Figure S5, Supporting Information). The mammalian blocking primer yielded higher sensitivity and diversity for protist genera, but depressed detection and prevalence of fungal and helminth genera (Figure S5, Supporting Information). We detected higher diversity of ASVs overall using the mammalian blocking primer (Shannon Index, P < .001; Figure S5, Supporting Information). This was mediated by both higher richness (Chao1, P < .001) and higher evenness of the community structure (Inverse Simpson, P < .001). Thus, the rest of the analysis is focused on the dataset using mammalian blocking primer as it is most representative of the diversity in the gut eukaryome and particularly for protists. Results on the slightly larger dataset not using mammalian blocking primers are presented in the supplementary data (Figures S6, S8–S10, S13, Tables S8, S9, and S19, Supporting Information).
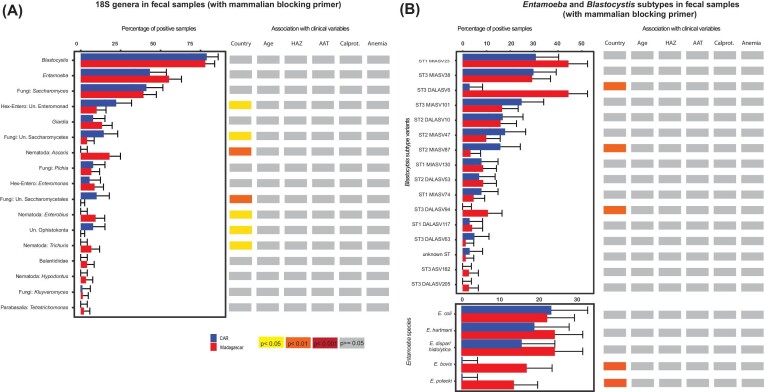
Figure 3.
Differences in the fecal microeukaryome in relation to geographic location and clinical variables. (A) All genera as detected by 18S sequencing using a mammalian blocking primer and (B) split in different Blastocystis clusters or Entamoeba species based on phylogenetic trees. Samples were considered to be positive for a given genus if they had at least a single sequence relating to this genus. Groups were compared using the Pearson chi-squared test and Benjamini–Hochberg correction for multiple testing. The colour code illustrating the degree of significance of the association is given on the bottom of the figure.
We found two genera to be present in at least 50% of samples in the 18S dataset: Blastocystis and Entamoeba. Using mammalian blocking primer allowing for preferential detection of protists, we detected Blastocystis to be present in 75% of all fecal samples analyzed. Stratified by country, 50% of all samples from Madagascar amplified Blastocystis and Entamoeba and 50% of all samples from CAR Blastocystis.
In the dataset using a blocking primer, there were no consistent trends to co-occurrence or coexclusion of any of the eukaryotes with each other at lower taxonomic level (Figures S6A–C, Supporting Information). However, there were clear negative correlations between fungi and protozoa/helminths at higher taxonomic level (Figures S6D–G, Supporting Information). The same trends were observed in the dataset not using a mammalian blocker (data not shown). Several co-occurrences and coexclusions were observed in the fungal ITS2 dataset, yet often with weak associations (Figure S7, Supporting Information). There was also extensive coexistence of different Blastocystis subtypes within a single individual (Table S4, Supporting Information).
In summary, our data shows that the eukaryome of African children shows a high intersubject variability and is dominated by the protists Blastocystis and Entamoeba and different fungi.
The mycobiome of African children is varied and dominated by members of Saccharomyces
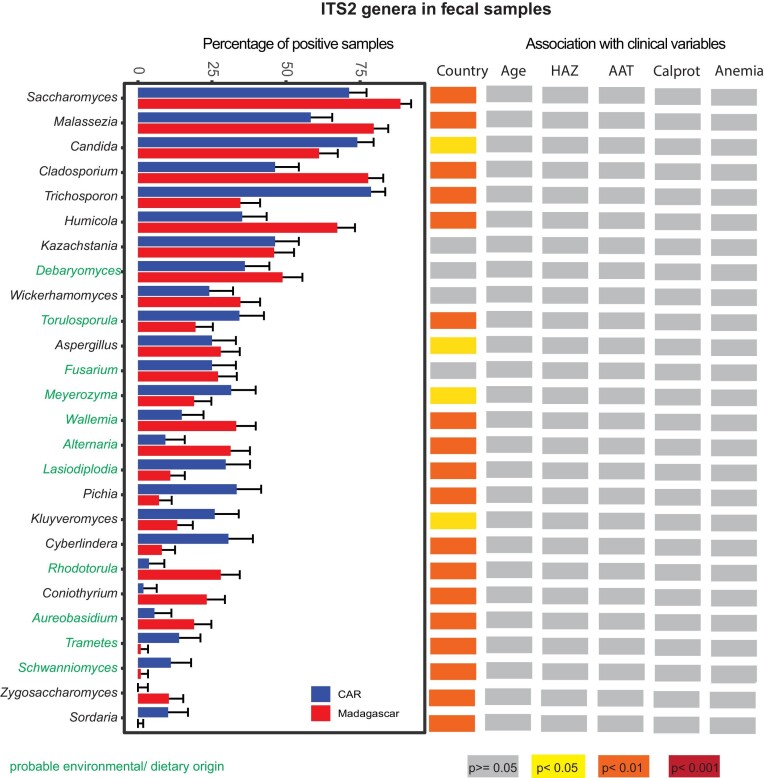
The fecal mycobiome was largely dominated by Ascomycota (Figure 2) and more precisely by members of the group Saccharomycetales (average rel. abundance 74%; Tables S5 and S6, Supporting Information). There were very few fungi conserved on the lower taxonomic level. We detected five fungal genera to be present in at least 50% of all fecal samples and with a relative abundance of at least 0.001%: Humicola, Malassezia, Cladosporium, Candida, and Saccharomyces and two fungal species: Humicola grisea and Malassezia restricta.
Figure 2.
Differences in the fecal mycobiome in relation to geographic location and different clinical outcomes. Samples were considered to be positive for a given genus if they had at least a single reads relating to this genus. Genera indicated in green are of probable environmental origin. Groups were compared using the Pearson chi-squared test and Benjamini–Hochberg correction for multiple testing. The colour code illustrating the degree of significance of the association is given on the bottom of the figure.
Thus, our data highlights a dominance of Ascomycota in the GI tract of African children with members of the Saccharomycetales as the main constituents of the fecal mycobiome.
The fecal eukaryome of children is strongly influenced by the country of residence
Overall, our data reveals that the fecal eukaryome of African children is influenced by country of residence (Figure 1C–4; Figures S8B, S9B–D, S10, Supporting Information). For all amplicon datasets, country of origin significantly contributes to the overall beta diversity when assessing both relative abundance (Bray–Curtis dissimilarity index; Figure 4D) and presence–absence (Jaccard index; Figures S9B–D, Supporting Information). Further, the prevalence of several taxa was significantly different between the two countries in a bivariate analysis (Figure 2 and 3; Figure S7B, Supporting Information). The relative abundance of several of these taxa remained significantly associated with the country of origin in a multivariate model correcting for sequencing run, total fungal reads and intestinal inflammation (Figure S10, Supporting Information).
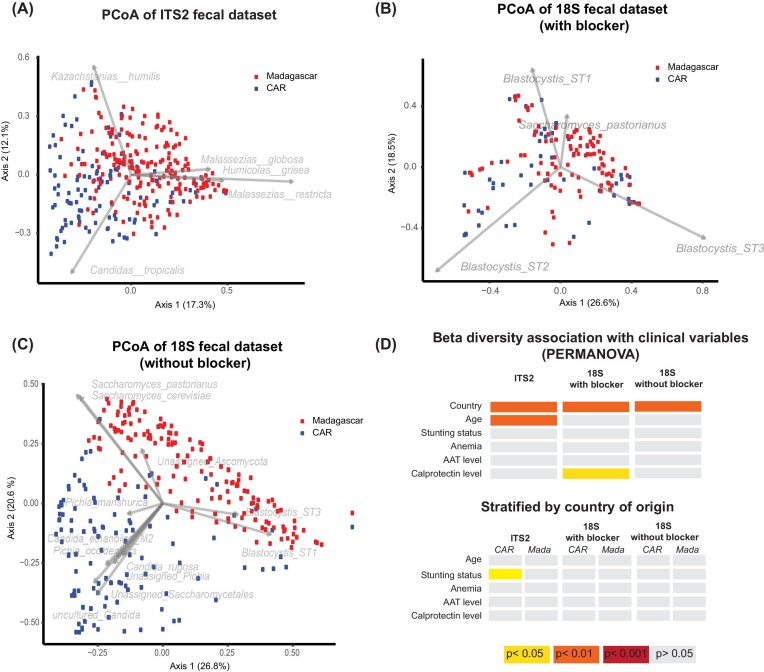
Figure 4.
Differences in the microeukaryome (18S dataset) based on country of origin. PCoA plot based on the normalized Bray–Curtis dissimilarity index (log10) of the fecal dataset iteratively rarefied to 1000 microeukaryotic reads (A) with the ITS2 primers targeting fungi (CAR: n = 100, Madagascar: n = 202), (B) with a mammalian blocker using 18S primers (CAR: n = 54, Madagascar: n = 99), (C) without a mammalian blocker using 18S primers (CAR: n = 110, Madagascar: n = 154). Samples from CAR are coloured in blue, samples from Madagascar in red. (D) Association of different clinical factors with Beta Diversity using a Permanova analysis on dispersion. The colour code is given in the figure.
In the 18S dataset without a mammalian blocker, prevalence of Ascaris, Trichuris, Entamoeba, and Blastocystis was slightly higher in Madagascar while Candida, Pichia, two different groups of unassigned Saccharomycetales, and one unknown enteromonad were more prevalent in CAR (Figure S8B, Supporting Information). When correcting for sequencing run, total microeukaryotic reads, and intestinal inflammation (alpha-antitrypsin levels and calprotectine levels), the two genera of unassigned Saccharomycetales, Pichia, Entamoeba and Candida remained significantly associated with the country of origin of the full dataset of fecal samples. In the reduced dataset using a mammalian blocking primer, we further found Entamoeba polecki to be significantly more prevalent in Madagascar compared to CAR (FDR = 0.007) in a multivariate model correcting for anemia status, intestinal inflammation, age and sequencing depth. Differences in alpha diversity according to the country of origin were visible in the 18S dataset but not in the ITS2 dataset (Figure 1C). The differences in microeukaryotes across taxa was further confirmed on a subset of samples and microeukaryotes using microscopy-based approaches (Table S19, Supporting Information).
Thus, our data show a clear influence of the country of origin on the eukaryotic community of fecal samples.
High genetic diversity of Blastocystis and frequent mixed colonization
In the dataset sequenced using a mammalian blocking primer, 78% of all fecal samples had at least one subtype of Blastocystis detected in their feces (189/241). The most prevalent subtype detected was ST3 (53% prevalence; 128/241), closely followed by ST1 (46%; 111/241) and ST2 (33%; 79/241; Tables S6–S9, Supporting Information). Of the Blastocystis positive samples, 49% had only one Blastocystis subtype detected by 18S amplicon sequencing and 51% displayed mixed colonization (Table S4, Supporting Information). Overall, there was a slightly higher prevalence of Blastocystis ST3 in Madagascar compared to CAR (FDR = 0.05). Blastocystis subtypes were further divided into subclusters using phylogenetic analysis (Figure 3B; Table S1, Supporting Information). Several of the subclusters showed a different distribution according to the country of origin of the children: There was a higher prevalence of subclusters ST3-DALASV6 and ST3-DALASV94 in feces from Malagasy children compared to CAR children. We observed further a lower prevalence of subcluster ST2-MIASV87 in Madagascar compared to CAR (Figure 3B).
Together, these results indicate that children frequently have mixed colonization of distinct Blastocystis subtypes and genetic variability is high within Blastocystis subtypes. Some genetic variants (subclusters) show country-specific differences in prevalence.
Different species of Entamoeba are part of the intestinal eukaryome
All sequences from Entamoeba were placed in a phylogenetic tree (Figure S4, Supporting Information) to refine the classification at species level. We detected five Entamoeba species: E. coli, E. dispar/histolytica (the ASVs detected there are all more similar to the nonpathogenic E. dispar, though it is not possible to confidently distinguish these species by amplicon sequencing), E. polecki, E. bovis, and E. hartmanni. Entamoeba polecki, previously known as Entamoeba chattoni, was detected in 8% of samples (20/242). We detected E. dispar/histolytica in 21% of all samples (50/242), E. coli in 22% (54/242), E. hartmanni in 22% (53/242), E. polecki in 8% (20/242), and E. bovis in 10% (25/242). Roughly, one-fourth of the Entamoeba positive samples had more than one Entamoeba species present. Further, for both, Blastocystis and Entamoeba, we saw a negative correlation with the relative abundance of fungi and a positive correlation in between Blastocystis and Entamoeba (Figure S6, Supporting Information).
In conclusion, our data shows the presence of different species of Entamoeba in fecal samples with pronounced country-specific differences in the prevalence of specific subtaxa. Most samples show only a single Entamoeba species at the time.
Stunting is associated with altered abundance of certain members of the eukaryome
We next assessed if different clinical factors, including anemia, stunting status, and environmental enteric disease (measured through the inflammatory markers fecal calprotectin and alpha-1-antitrypsin) are associated with significant changes in the eukaryome composition in the GI tract (Figure 2 and 3; Figure S7B, Supporting Information). Overall, clinical factors contributed only marginally to the beta diversity (Figire 4D). Further, there was little influence of these clinical parameters on alpha diversity (Figure 1C), except for anemia and calprotectin levels, which contributed marginally to the alpha diversity of the 18S dataset using mammalian blockers. In a bivariate model, prevalence of Ascaris, Trichuris, and Saccharomycetales was associated with anemia (Figure S8, Supporting Information). However, no specific taxa were consistently associated with anemia, alpha-1-antitrypsin, or calprotectin levels in a multivariate analysis assessing for relative abundance of the reads in the 18S nor the ITS2 datasets (data not shown).
We then tested for associations between taxa prevalence and relative abundance and stunted growth in the ITS and 18S datasets (Figure S13, Supporting Information). In the ITS dataset the relative abundance of one taxon— Fusarium equiseti—was associated with stunted growth in CAR in a bivariate analysis assessing stunted growth as a categorical variable (corrected P = .03) and in a multivariate analysis (DeSeq2) that corrected for age, sequencing run and total fungal reads (P = .001). Fusarium equiseti was also found with higher prevalence in stunted children compared to nonstunted controls in a multivariate logistic regression model (P = .03) correcting for country of origin, calprotectin levels, sequencing run, total fungal reads, and gender. Together, these data suggest that there might be more F. equiseti in stunted children compared to nonstunted controls.
In the 18S dataset using the mammalian blocking primers, there was a trend of reduced Blastocystis relative abundance in stunted children compared to nonstunted controls. This was observed in the overall dataset (including both study sites) performing a bivariate correlation analysis (FDR = 0.07). Stratifying the analysis by country, this trend was observed in bivariate analysis using stunting either as a categorical variable in Antananarivo (FDR = 0.08), as a continuous variable (FDR = 0.016), but not in CAR. The association with Blastocystis was not significant in a multivariate model correcting for intestinal inflammation, anemia, age, and gender, which are known modulators of the microbiota. The same trends were observed in the 18S dataset not using a mammalian blocking primer. Further, in both 18S datasets, several members of the Saccharomycetales were consistently associated with stunting in Bangui, CAR.
Thus, our data indicates that there are consistent trends for lower Blastocystis and Saccharomycetales levels and higher F. equiseti levels in stunted children in both study sites. The associations are independent of intestinal inflammation and/or anemia.
The human eukaryome differs along the GI tract
We examined the mycobiome and eukaryome along the GI tract. Data for the eukaryome of the upper GI tract are sparse; most of the microeukaryotic reads in the gastric and duodenal samples belong to host, dietary, and environmental sources, while gut residents, such as Entamoeba and Blastocystis, were sporadically observed. After filtering out vertebrate, arthropod, and plant reads (which are likely of dietary origin) there were only few gastric and duodenal samples with microeukaryotic reads with and without the mammalian blocking primer (Figure S3A and B, Supporting Information).
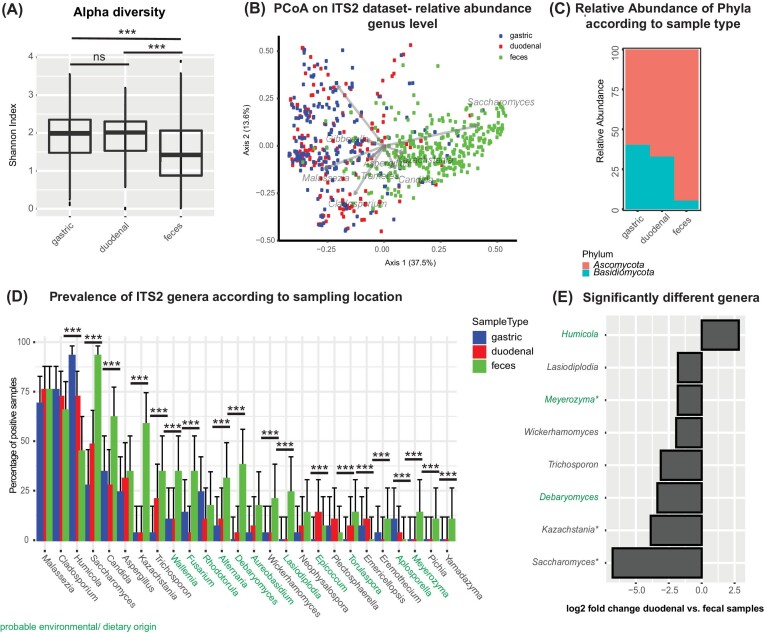
The mycobiome (assessed through ITS2 sequencing) of the upper and lower GI tract differed significantly in its alpha- and beta-diversity. Fecal samples had a higher number of observed taxa, but lower evenness and thus a lower overall diversity as measured by the Shannon index (Figure 5A; P < .001). Further, the gastric and duodenal samples showed a similar composition (Figure 5C), and both differed significantly from the fecal samples when assessing relative abundance (Bray–Curtis index, Figure 5B; P = .006) and presence/absence (Jaccard index; Figure S9A, Supporting Information; P = .002). The ratio of Basidiomycota/Ascomycota was significantly higher in gastric and duodenal samples compared to fecal samples (Figure 5C; P = .003). Several members of Saccharomycetales were found in higher levels in the feces compared to the upper GI tract, including Saccharomyces, Kasachstania, Debaryomyces, Wickerhamomyces, and Meyerozyma, while Humicola is more abundant in the upper GI samples (Figure 5D). Further, these fungal genera remained significantly different in between fecal and duodenal samples in a multivariate analysis correcting for inflammatory status, sequencing run and total fungal read count in the combined dataset (Figure 5E), and when the ITS dataset was stratified by country of origin (Figure S11, Supporting Information). The same trends were also observed in a reduced sample set including only subjects with samples for all three GI tract locations (Figure S12, Supporting Information).
Figure 5.
Differences in the mycobiome along the GI tract. (A) Alpha diversity in the different compartments as measured by the Shannon index. (B) PCoA plot based on the normalized Bray–Curtis dissimilarity index (log10) of the dataset iteratively rarefied to 5000 fungal sequences (gastric: N = 148, duodenal: N = 132, and feces: N=299). (C) Relative abundance of the different phyla according to sample type. (D) Differences in the mycobiome in relation to sampling location along the GI tract. Samples were considered to be positive for a given genus if they had at least a single sequence relating to this genus. Groups were compared using the Pearson chi-squared test and Benjamini–Hochberg correction for multiple testing. *P < .05; **P < .01; ***P < .005; comparison without an indication are nonsignificant. (E) Fungal genera showing significant differences in their relative abundance between duodenal and fecal samples in a DeSeq2 model correcting for sequencing depth.
Our data thus clearly shows that the eukaryome differs in between different sites of the GI tract.
Discussion
Eukaryotes are important members of the GI tract, contributing to the consumption of nutrients and vitamin production (Schei et al. 2017) and directly modulating both the bacterial microbiota composition (Morton et al. 2015, Zaiss et al. 2015, Audebert et al. 2016, Nieves-Ramirez et al. 2018, Stensvold and van der Giezen 2018) and the immune system (Underhill and Iliev 2014, Zaiss et al. 2015, McFarlane et al. 2017, Richard and Sokol 2019). While the field of parasitology is well-established, the diversity and ecology of human intestinal eukaryome and variability across individuals remains poorly studied. We describe the human eukaryome and mycobiome in a large cohort of children living in Africa without diarrhea or acute GI symptoms. We find a diverse community of eukaryotes, which does not seem to be strongly influenced by clinical factors. The results here are consistent with the emerging viewpoint that many eukaryotes present in the human GI tract may be minimally harmful, commensal or of potential benefit (Parfrey et al. 2014, Andersen and Stensvold 2016, Stensvold and van der Giezen 2018). Pathogenic protists such as E. histolytica and Cryptosporidium sp. are major sources of morbidity and mortality among children in resource poor settings (Turkeltaub et al. 2015), and have rightly garnered much research attention. These taxa are rare here, likely because this study excluded children with acute GI disease (Vonaesch et al. 2018). Further, many carriers of E. histolytica and G. intestinalis do not show (severe) symptoms and their virulence is determined by a complex interplay between parasite, host, and microbiota composition (Marie and Petri 2014). Last, it has also been shown that the eukaryotic microbiota composition is associated with the bacterial microbiota composition (Morton et al. 2015) and that the bacterial community has in turn an influence on Entamoeba virulence in the human GI tract. Indeed, there is increased awareness that not only bacteria, but the whole microbial community inhabiting the GI tract, including protists, helminths, fungi, archaea, and viruses play an important role in the overall ecosystem as well as in cross-talk with the host and regulation of virulence [reviewed in Hirt (2019), Rowan-Nash et al. (2019), and Ungaro et al. (2019)]. Integrated studies across all domains of life would, therefore, be increasingly needed to understand the real implication of the human microbiome in health and disease.
To our knowledge, this is the first description of the eukaryotic community comparing different GI compartments and the first sequencing-based study to assess the eukaryotes in the human stomach and small intestine. Few gastric and duodenal samples yielded microeukaryotic reads in the 18S datasets, so we cannot compare the community across the GI tract. However, similar taxa were observed in the upper GI tract and in feces with Blastocystis most prevalent. Our data reveals that there is a clear compartmentalization in the communities found along the GI tract for fungi. Fungal composition was very similar between the gastric and duodenal samples, and both are distinct from the fecal composition, similar to the bacterial community (Vonaesch et al. 2018, 2022). While this might be expected, seen the vastly different environments encountered at each site, this has so far never been reported. Earlier research described few acid-tolerant fungi like Candida and Phialemonium (von Rosenvinge et al. 2013) [reviewed in Hallen-Adams and Suhr (2017)] in the stomach and culture-based approaches identified Candida in the small intestine (Heyworth and Brown 1975, Minoli et al. 1981). In our study, we assessed the fungal composition of 196 duodenal samples from two different countries and detect a vast diversity of strains, most of which are probably transients. Overall, the two most frequently detected taxa were H. grisea (70% in Madagascar, 84% in CAR) and M. restricta (52% in Madagascar, 71% in CAR). Humicola grisea is likely an environmental transient rather than a true colonizer of the gut: It is because it is a well-known thermophilic, soil-dwelling fungus and was not part of the eukaryotic community described in earlier studies assessing the feces in urban industrialized countries (Suhr and Hallen-Adams 2015, Hallen-Adams and Suhr 2017). Malassezia and Cladosporium, the two other most prevalent taxa in the upper GI tract, which are also frequently found in the feces here and in urban industrialized countries (Nash et al. 2017, Auchtung et al. 2018), show similar prevalence in all three compartments, suggesting that they could be true colonizers but further experimental work is necessary (Hallen-Adams and Suhr 2017). Malassezia restricta is also known to be a natural member of the human skin microbiota (Vijaya Chandra et al. 2020). Malassezia has also been associated with IBD in a recent patient study and its direct implication has been validated in subsequent experiments in mice (Limon et al. 2019).
In line with previous results from urban industrialized countries (Stensvold et al. 2011b, Parfrey et al. 2014) our data reveals that the fecal eukaryome of African children is dominated by fungi and protists, especially Blastocystis and Entamoeba. The patterns that there are few taxa in any given individual and high variability across the population are perhaps surprising when evaluated in the framework of the bacterial microbiome. Indeed, the fecal prokaryome harbours overall more taxa and a much higher proportion are widely shared across individuals. However, these patterns have been previously observed in humans and other mammals (Parfrey et al. 2014, Bachmann et al. 2015). The communities observed in this cohort are more diverse than in industrialized countries both in terms of the broader taxonomic groups represented (e.g. Parabasalids, Blastocystis) and greater diversity within taxonomic groups. At the individual level this means there are more so called mixed colonizations, e.g. multiple subtypes and strains of Blastocystis and Entamoeba were detected in ∼50% of individuals that harbour these taxa. We did not detect Chilomastix, Dientamoeba, nor Cryptosporidium in our 18S datasets, despite the fact they are widespread in the human fecal eukaryome (Barratt et al. 2011, Roser et al. 2013, Turkeltaub et al. 2015, Jokelainen et al. 2017, Greigert et al. 2018). There is evidence that D. fragilis is more prevalent in westernized compared to traditional communities (Barratt et al. 2011), which could explain the absence of D. fragilis in our study. Another hypothesis is that low sequencing depth was insufficient to detect D. fragilis and Chillomastix, which are low prevalence in the microscopy-based analysis. Last, we identified three primer mismatches of the primers for Parabasalids (Dientamoeba) and for Fornicata (including Chilomastix), which could explain the missing reads. Cryptosporidium is mainly associated with diarrhoea (Checkley et al. 1998, Costa et al. 2011, Mekonnen et al. 2019), so likely not detected in our cohort due to the choice of children included in the study. In line with this hypothesis, Cryptosporidium was also not detected through microscopy.
Last, recent evidence suggests that there are age-dependent changes not only in the bacterial microbiota community but also in the eukaryotes. Indeed, two studies in the USA and Ireland showed Blastocystis to be less prevalent in children compared to adults (Scanlan et al. 2016, 2018). This contrasts with the high prevalence of protist in the cohort of children described here. This difference might be due to a very high exposure to contaminated drinking water in these areas (Habib et al. 2021, Vonaesch et al. 2021), favouring the colonization by these microorganisms in exposed children. This would be in line with a previous study in children from Mexico, showing a high prevalence of Blastocystis already in early life (Partida-Rodriguez et al. 2021). More work is needed to understand the early life dynamics of eukaryotes within the human GI tract.
The fecal mycobiome of African children resembles the mycobiome reported in westernized countries [reviewed in Richard and Sokol (2019)]: in most studies, Candida (particularly Candida albicans), Saccharomyces (particularly S. cerevisiae), Penicillium, Aspergillus, Cryptococcus, Malassezia (particularly M. restricta), Cladosporium, Galactomyces, Debaryomyces, and Trichosporon, were detected in decreasing prevalence. In addition to these species, we frequently detected Humicola, Kazachstania, and Wickerhamomyces. Humicola grisea is an environmental fungus (Wang et al. 2019). Kazachstania humilis, also called Candida humilis and K. exigua are normally found in fermented food (Garcia-Ortega et al. 2022). Wickerhamomyces anomalus also called Saccharomyces anomalus or Pichia anomala is often associated with spoilage or processing of food (Masneuf-Pomarede et al. 2015) and has also been found in the GI tract of insects (Cappelli et al. 2020). Very few eukaryotic taxa were present in the feces of at least 50% of children, including H. grisea and M. restricta in the ITS2 dataset. Additional studies on describing the mycobiome of the food and environment in conjunction with the gut mycobiome, including ideally also source-tracking and/or repeated sampling would be needed to determine where gut fungi originate, and which are true colonizers of the gut (Lavrinienko et al. 2021). Here and in other studies fungal diversity within a sample (alpha diversity) is relatively low, Ascomycota and Basidiomycota are the dominant phyla, and many of the same genera are found (Hoffmann et al. 2013, Chehoud et al. 2015, Richard et al. 2015, Nash et al. 2017, Richard and Sokol 2019). The Ascomycota/Basidiomycota ratio was previously associated with gut health and has been shown to be lowered in adults (Sokol et al. 2017) as well as a children (Chehoud et al. 2015) suffering of IBD. However, in our study set, there was no association between this ratio and either stunting, anemia, or intestinal inflammation. However, there was a clear difference in the ratio by country of origin with higher Basidiomycota levels in Madagascar compared to CAR.
Which of the eukaryotes truly colonize the human gut and which are only transient is a fundamental but unanswered question and one, i.e. increasingly debated for fungi (Hallen-Adams and Suhr 2017, Fiers et al. 2019). Recent evidence suggests that fungal colonization of the intestinal tract of healthy individuals is minimal (Auchtung et al. 2018), and raises the possibility that a small minority of the fungi detected in the gut mycobiome are residents of the gut (exemplified by Candida albicans; Fiers et al. 2019). More than two-thirds of all species reported in two previous studies were found only in a single sample, suggesting that they come from environmental sources (Suhr and Hallen-Adams 2015, Hallen-Adams and Suhr 2017). We hypothesized that true residents should be likely shared across several individuals, while food contaminants might be distributed less consistently. We thus tried to address this point by filtering the taxa according to their prevalence. Of note that however even commonly detected gut fungi, such as Debaryomyces hansenii or Penicillium, do not grow at 37°C and might thus not be true residents of the gastrointestinal tract (Auchtung et al. 2018). Other common fungi may be transients and originate from food (e.g. Saccharomyces cerevisiae) are plant pathogens (such as Fusarium, Alternaria, and Botrytis), fungal communities from other parts of the body (Malassezia), or from the environment (Aspergillus) [reviewed in Auchtung et al. (2018)]. It is impossible to determine without experimental evidence if fungi are transient or resident members of the microbiota (Fiers et al. 2019). Similarly, several of the microeukaryotes detected, such as the rotifer Rotaria, are common inhabitants of freshwater and therefore likely environmental contaminants. Even within known gut taxa we detect strains that are likely transient in humans and true residents of other mammals, such as E. bovis. These results reiterate the importance of critically examining the eukaryotic community rather than assuming all sequences obtained are members of the eukaryome.
Transient fungi might contribute to intestinal disturbances, especially if they produce mycotoxins (Smith et al. 2012); F. equiseti, Penicillium, Fusarium, and Aspergillus are among the mycotoxin producers detected here (Goswami et al. 2008, Munkvold 2017). Fusarium equiseti is a plant pathogen of cereals, field weeds, durian, goji berries, among others and is found in both, tropical and temperature regions (Goswami et al. 2008, Munkvold 2017). Further, colonization with F. equiseti has been associated with a vegetarian diet (Hallen-Adams and Suhr 2017). Interestingly, we found a consistent trend towards higher levels of F. equiseti in stunted children compared to healthy controls. This association might be due to a diet low in meat and high in plant matter consumed by stunted children (Vonaesch et al. 2021) and favouring the colonization by F. equiseti (Hallen-Adams and Suhr 2017). It is also tempting to speculate that this fungus is contributing to the pathophysiology underlying stunted growth, however, mycotoxin profiling as well as experimental data are needed to establish a causal relationship between these toxins and intestinal disturbances.
The diversity of protists and nematodes detected here resembles earlier reports from humans living in Sub-Saharan Africa with high prevalence of Blastocystis, Entamoeba, Trichomonads, and yeasts (Parfrey et al. 2014, Greigert et al. 2018). The catalogue of gut eukaryotes observed here is comparable to other parts of the world, albeit with higher diversity than typically found in Europe (Forsell et al. 2012, Scanlan et al. 2018, Lhotska et al. 2020). Nematodes (especially Ascaris and Trichuris trichiura) were mainly found in Madagascar and only rarely in the CAR, possibly related to deworming medicine or lifestyle choices. However, local hotspots of nematode colonization were previously reported from several places in Africa and are not associated with deworming campaigns (Moser et al. 2017, Schulz et al. 2018).
Diversity within common gut protists is high: diverse Entamoeba species colonize the intestines of humans and nonhuman primates (Stensvold et al. 2011b, Elsheikha et al. 2018, Stensvold 2019, Dos Santos Zanetti et al. 2021), and in this cohort we frequently detected E. polecki, E. dispar/E.histolytica, E. hartmanni, and E. coli. Entamoeba bovis is most often associated with ungulates and is a potential transient here. Entamoeba histolytica, a true pathogen, is detected less frequently and difficult to distinguish from E. dispar by 18S sequencing (a specific qPCR is needed to make the molecular distinction). The diversity of Entamoeba detected here is high, and half of individuals positive for Entamoeba harbor multiple species and/or subtypes.
We find a trend towards greater Entamoeba presence in nonstunted compared to stunted children. Two studies assessing a possible correlation between infection with specific pathogens and stunted growth are currently ongoing within the Afribiota project.
Blastocystis is the most common protist within this cohort and widespread in humans and other mammals. Overall, Blastocystis ST1, ST2, and ST3 are predominant here, as in other human populations (Stensvold et al. 2020, Forsell et al. 2012), and ST4, which is common in Europe is absent. The diversity of Blastocystis is similar between countries , though the prevalence of several strains (subclusters) differs between countries . In contrast to two previous studies in Europe (Flemish Gut Project and Twins UK) (Tito et al. 2019), more than half of the children included in our study showed concomitant presence of several Blastocystis subtypes in their feces. Our results suggest that different Blastocystis subtypes and Entamoeba species often coexist. This confirms a recent study in Cameroon in which it was shown that some subtypes of Blastocytis and Entamoeba species show a positive correlation in their occurrence (Even et al. 2021).
One surprising finding that emerges from our study is the low correlation between the eukaryome and clinical variables. In previous studies, particular fungal taxa were associated with increased inflammation in the context of IBD and/or colorectal cancer (Ye et al. 2006). In industrialized countries lower prevalence of Blastocystis has been reported in patients with inflammatory diseases including colorectal cancer or Crohn’s disease compared to control individuals (Beghini et al. 2017, Tito et al. 2019). Indeed, these microaerobic protists thrive in low oxygen levels, while higher oxygen levels are a hallmark of gut inflammation, and Blastocystis could, therefore, be a marker of a ‘healthy gut environment’, meaning a gut environment that remains largely anaerobic (Audebert et al. 2016, Stensvold et al. 2020). Entamoeba is also reported to be inversely correlated with inflammatory diseases (Morton et al. 2015). Here we see trends of lower prevalence for E. coli in stunted children compared to nonstunted controls, though the association was not significant after correcting for multiple testing. While these results might be confounded by the fact that we had only limited sequencing depth and a very high variability of taxa for fungi, likely decreasing statistical power, our results suggest the eukaryotic community, at least in our two study sites, is not dramatically reorganized by the clinical variables measured. More research is needed to better elucidate the role of eukaryotes to the pathophysiology associated with a dysbiotic microbiota.
Overall, our results from two sites in Sub-Saharan Africa show little correlation between stunting and the eukaryome, or individual eukaryote prevalence. This could suggest that eukaryotes are less directly involved in the pathophysiology of chronic childhood undernutrition, as suggested also in a targeted analysis of the parasites in the Madagascar study site of Afribiota (Habib et al. 2021). Our results could also be a reflection that eukaryotes are less influenced by the altered gut environment compared to bacteria (Vonaesch et al. 2018). Further studies are, however, needed to corroborate this point and exclude any technical bias or site-specific effects. Earlier reports showing a direct role of helminths in undernutrition led to the recommendation of WHO for systematic antihelminthic treatments. However, our observation is consistent with a recent meta-analysis on 80 studies showing that helminths are not directly involved in undernutrition (Raj et al. 2022). In line with this observation, a recent meta-analysis found very little impact of antihelminthic treatments on stunting or development (Taylor-Robinson et al. 2019). However, the cross-sectional design of our study does not allow to capture events that initiate pathophysiology of stunting and therefore, cannot rule out a role for eukaryotes in leading to long term undernutrition. Further, as many children harbor several eukaryotes at the same time, it is possible that effects of individual taxa are shielded because, e.g. different eukaryotes might have opposite effects on the immune system that cancel each other out.
This work provides a foundation for future studies assessing the eukaryome in disease contexts and longitudinal studies on the establishment and role of the human eukaryome. Future studies should also assess for interactions between prokaryotes and eukaryotes in the gut microbiome to reveal cross-kingdom community structures and dynamics potentially influencing gut homeostasis and disease and assess how these interactions are shaped by diet and subsistence, as previously shown to play a role in a study on nonhuman primates (Sharma et al. 2022) as well as in several studies in humans (Morton et al. 2015, Rowan-Nash et al. 2019).
One strength of our study lies in the fact that we combine the analysis of the 18S rRNA gene for the overall eukaryome with and without an additional primer blocking amplification of human DNA and the internally transcribed spacer (ITS) gene for targeted analyses of the mycobiome and that we confirm presence of given eukaryotes on a subset of samples using microscopy. We show that the use of a mammalian blocking primer altered the observed community structure by detecting higher diversity within a sample, especially of protists and microeukaroytes detected from a higher proportion of samples. This shows a limitation of amplicon-based profiling calls for standardized primers and protocols to allow for comparisons between different studies and locations. Combining the ITS and 18S rRNA gene approaches we were able to show that fungal and microeukaryotic diversity in the gut of African children are only marginally correlated with clinical factors, yet strongly shaped by geographic location, most likely through diet and other environmental exposures.
Our study has a few limitations: as a single sample was taken from each child and as we included only children aged 2–5 years. Therefore, the study does not allow assessing for dynamic changes nor does it allow to make any assumptions about early life succession. Further, storage protocols slightly differed across the two sites and DNA was extracted with the same kit but in two different geographic locations and by different experimenters. Thus, we might have overestimated geographic differences. The overall impact of geography on the microeukaryome was, however, confirmed on a subset of samples using microscopy. Further, seen that we only included two study sites in our project, the results about the overall diversity of eukaryotes might be specific to our study context and might not apply to other LIMC settings. Further, since the implementation of our study, new amplicon-based sequencing methods have been developed, allowing for the detection of a broader range of eukaryotes within the human microbiome (Popovic et al. 2018). Last, PCR based approaches used for the taxonomic profiling of a given microbiota are at best semiquantitative in nature and typically do not integrate various taxonomic groups. This hurdle can somehow be overcome by metatranscriptomic approaches, which can provide a more global, unbiased and quantitative approach, as exemplified in a recent publication on idiopathic chronic diarrhea in macaques (Westreich et al. 2019). Thus, as any sequencing-based study, the data presented here is likely not representative of the true diversity of eukaryotes found in the feces of these children.
Nevertheless, by comparing the eukaryome of almost 1000 children of whom roughly half showed fecal read counts for 18S rRNA gene and/or the ITS2 region in two different geographic locations of Sub-Saharan Africa, our data contributes valuable insights about the human eukaryome and sets the stage for more targeted analyses of eukaryome dynamics and of the role of the eukraoyme in health and disease.
In conclusion, our study clearly shows that African children harbour a specific eukaryome, which is compartmentalized along different sites of the GI tract and is strongly influenced by country of residence.
Group authorship Afribiota investigators
AFRIBIOTA Investigators (Group authorship in alphabetical order):
Laurence Barbot-Trystram, Hôpital Pitié-Salpêtrière, Paris, France.
Robert Barouki, Hôpital Necker- Enfants maladies, Paris, France.
Alexandra Bastaraud, Institut Pasteur de Madagascar, Antananarivo, Madagascar.
Jean-Marc Collard, Institut Pasteur de Madagascar, Antananarivo, Madagascar.
Maria Doria, Institut Pasteur, Paris, France.
Darragh Duffy, Institut Pasteur, Paris, France.
B. Brett Finlay, University of British Columbia, Vancouver, Canada.
Serge Ghislain Djorie, Institut Pasteur de Bangui, Bangui, Central African Republic.
Tamara Giles-Vernick, Institut Pasteur, Paris, France.
Milena Hasan, Institut Pasteur, Paris, France.
Bolmbaye Privat Godje, Complexe Pédiatrique de Bangui, Bangui, Central African Republic.
Jean-Chrysostome Gody, Complexe Pédiatrique de Bangui, Bangui, Central African Republic.
Francis Allen Hunald, Service de Chirurgie pédiatrique, Centre Hospitalier Universitaire Joseph Ravoahangy Andrianavalona (CHU-JRA), Antananarivo, Madagascar.
Nathalie Kapel, Hôpital Pitié-Salpêtrière, Paris, France.
Jean-Pierre Lombart, Institut Pasteur de Bangui, Bangui, Central African Republic.
Alexandre Manirakiza, Institut Pasteur de Bangui, Bangui, Central African Republic.
Synthia Nazita Nigatoloum, Complexe Pédiatrique de Bangui, Bangui, Central African Republic.
Laura Wegener Parfrey, University of British Columbia, Vancouver, Canada.
Lisette Raharimalala, Centre de Santé Materno-Infantile, Tsaralalana, Antananarivo, Madagascar.
Maheninasy Rakotondrainipiana, Institut Pasteur de Madagascar, Antananarivo, Madagascar.
Rindra Randremanana, Institut Pasteur de Madagascar, Antananarivo, Madagascar.
Harifetra Mamy Richard Randriamizao, Centre Hospitalier Universitaire Joseph Ravoahangy Andrianavalona (CHU-JRA), Antananarivo, Madagascar.
Frédérique Randrianirina, Institut Pasteur de Madagascar, Antananarivo, Madagascar.
Annick Robinson, Centre Hospitalier Universitaire Mère Enfant de Tsaralalana, Antananarivo, Madagascar.
Pierre-Alain Rubbo, Institut Pasteur de Bangui, Bangui, République Centrafricaine.
Philippe Sansonetti, Institut Pasteur, Paris, France.
Laura Schaeffer, Institut Pasteur, Paris, France.
Ionela Gouandjika-Vassilache, Institut Pasteur de Bangui, Bangui, République Centrafricaine.
Pascale Vonaesch, Institut Pasteur, Paris, France.
Sonia Sandrine Vondo, Complexe Pédiatrique de Bangui, Bangui, Central African Republic.
Inès Vigan-Womas, Institut Pasteur de Madagascar, Antananarivo, Madagascar.
Supplementary Material
Acknowledgements
The authors wish to thank all children and their families that participated in the AFRIBIOTA project. Further, they wish to thank the AFRIBIOTA Consortium, the participating hospitals in Bangui and Antananarivo as well as the Institut Pasteur, the Institut Pasteur de Madagascar and de Bangui, and members of the scientific advisory board for their continuous support. Further, they wish to thank the Centre de Recherche Translationelle and the Direction Internationale of the Institut Pasteur, and especially Paméla Palvadeau, Jane Lynda Deuve, Cécile Artaud, Nathalie Jolly, Sophie Jarijon, Mamy Ratsialonina, and Jean-François Damaras for precious help in setting-up and steering the AFRIBIOTA project. We would also like to thank Jean-Marc Collard, Pierre-Alain Rubbo, Dieu-Merci Welekoi-Yapondo, Lova Andrianonimiadana, Laurence Arowas, and Marie-Noelle Ungeheuer for managing the biobank, Rachelle S. Loo for help with the sequencing, and Lova Andrianonimiadana, Jean-Robert Mbecko Hugues Sanke, and Jean-Marc Collard for extracting DNA. We also thank Magali Chabe, Pauline Scanlan, Heather Hallen-Adams, and two anonymous reviewers for constructive feedback. This project was funded by the Total Foundation, Institut Pasteur, Pasteur Foundation Switzerland and a donation by the Odyssey Re-Insurance Company. P.V. was supported by an Early Postdoctoral Fellowship (P2EZP3_152159), an Advanced Postdoctoral Fellowship (P300PA_177876) as well as a Return Grant (P3P3PA_177877) from the Swiss National Science Foundation, a Roux-Cantarini Fellowship (2016), and a L’Oréal-UNESCO for Women in Science France Fellowship (2017) and the Forschungsfonds of the University of Basel. P.J.S. is a HHMI Senior Foreign Scholar and CIFAR scholar in the human microbiota consortium. Work in LWP’s group is funded by the Human Frontier Science Program RGY0078/2015 and Canada Research Chair in Protist Ecology and work in P.V.’s group is funded by the NCCR Microbiomes NCCR Microbiomes, a National Centre of Competence in Research, funded by the Swiss National Science Foundation (grant number 180575).
Notes
The Afribiota Investigators are listed in the acknowledgements.
Reviewer Name(s): Magali Chabé, Heather Hallen-Adams, Pauline Scanlan
Contributor Information
Pascale Vonaesch, Unité de Pathogénie Microbienne Moléculaire, Institut Pasteur, 25-28 Rue du Dr Roux, 75015 Paris, France.
Vincent Billy, Departments of Botany and Zoology, and Biodiversity Research Centre, University of British Columbia, 3200-6270 University Boulevard, V6T1Z4 Vancouver, Canada.
Allison E Mann, Departments of Botany and Zoology, and Biodiversity Research Centre, University of British Columbia, 3200-6270 University Boulevard, V6T1Z4 Vancouver, Canada.
Evan Morien, Departments of Botany and Zoology, and Biodiversity Research Centre, University of British Columbia, 3200-6270 University Boulevard, V6T1Z4 Vancouver, Canada.
Azimdine Habib, Unité de Bactériologie Expérimentale, Institut Pasteur de Madagascar, BP1274 Ambatofotsikely Avaradoha 101 Antananarivo, Madagascar.
Jean-Marc Collard, Unité de Bactériologie Expérimentale, Institut Pasteur de Madagascar, BP1274 Ambatofotsikely Avaradoha 101 Antananarivo, Madagascar.
Michel Dédé, Laboratoire d’Analyse médicale, Institut Pasteur de Bangui, Avenue De Independence Bangui, 923 Central African Republic.
Nathalie Kapel, Laboratoire de Coprologie Fonctionnelle, Assistance Publique- Hôpitaux de Paris, Hôpital Pitié-Salpêtrière, 47-83 Bd de l’Hôpital, 75013 Paris, France.
Philippe J Sansonetti, Unité de Pathogénie Microbienne Moléculaire, Institut Pasteur, 25-28 Rue du Dr Roux, 75015 Paris, France.
Laura Wegener Parfrey, Departments of Botany and Zoology, and Biodiversity Research Centre, University of British Columbia, 3200-6270 University Boulevard, V6T1Z4 Vancouver, Canada.
for the Afribiota Investigators:
Laurence Barbot-Trystram, Robert Barouki, Alexandra Bastaraud, Jean-Marc Collard, Maria Doria, Darragh Duffy, B Brett Finlay, Serge Ghislain Djorie, Tamara Giles-Vernick, Milena Hasan, Bolmbaye Privat Godje, Jean-Chrysostome Gody, Francis Allen Hunald, Nathalie Kapel, Jean-Pierre Lombart, Alexandre Manirakiza, Synthia Nazita Nigatoloum, Laura Wegener Parfrey, Lisette Raharimalala, Maheninasy Rakotondrainipiana, Rindra Randremanana, Harifetra Mamy Richard Randriamizao, Frédérique Randrianirina, Annick Robinson, Pierre-Alain Rubbo, Philippe Sansonetti, Laura Schaeffer, Ionela Gouandjika-Vassilache, Pascale Vonaesch, Sonia Sandrine Vondo, and Inès Vigan-Womas
Authors’ contributions
P.V., P.J.S., and L.W.P. designed the study protocol. P.V. performed the bioinformatic and biostatistical analyses and V.B. and A.M. constructed the phylogenetic trees under the supervision of L.W.P. N.K. and A.N. performed the measurements of fecal calprotectin and alpha-1-antitrypsin. P.V. assured scientific, logistic, and reglementary coherence of Afribiota and coordinated the study. P.V., L.W.P. and P.J.S. acquired funding. P.V. and L.W.P. wrote the manuscript. All authors critically read and approved the final manuscript.
Conflict of interest statement
None declared.
References
- Alfellani MA, Stensvold CR, Vidal-Lapiedra Aet al. Variable geographic distribution of Blastocystis subtypes and its potential implications. Acta Trop. 2013;126:11–18. 10.1016/j.actatropica.2012.12.011. [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, McCarthy DJ, Chen Yet al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc. 2013;8:1765–86. 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- Andersen LO, Stensvold CR. Blastocystis in health and disease: are we moving from a clinical to a public health perspective?. J Clin Microbiol. 2016;54:524–8. 10.1128/JCM.02520-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrieta MC, Arévalo A, Stiemsma Let al. Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting. J Allergy Clin Immunol. 2018;142:424–34. 10.1016/j.jaci.2017.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auchtung TA, Fofanova TY, Stewart CJet al. Investigating colonization of the healthy adult gastrointestinal tract by fungi. mSphere. 2018;3:3–18.: 10.1128/mSphere.00092-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audebert C, Even G, Cian Aet al. Colonization with the enteric protozoa Blastocystis is associated with increased diversity of human gut bacterial microbiota. Sci Rep. 2016;6:25255. 10.1038/srep25255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann V, et al. Bile salts modulate the mucin-activated type VI secretion system of pandemic Vibrio cholerae. PLoS NeglTrop Dis. 2015;9:e0004031. 10.1371/journal.pntd.0004031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt JL, Harkness J, Marriott Det al. A review of Dientamoeba fragilis carriage in humans: several reasons why this organism should be considered in the diagnosis of gastrointestinal illness. Gut Microbes. 2011;2:3–12. 10.4161/gmic.2.1.14755. [DOI] [PubMed] [Google Scholar]
- Beghini F, Pasolli E, Truong TDet al. Large-scale comparative metagenomics of Blastocystis, a common member of the human gut microbiome. ISME J. 2017;11:2848–63. 10.1038/ismej.2017.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutin RCT, Sbihi H, McLaughlin RJet al. Composition and associations of the infant gut fungal microbiota with environmental factors and childhood allergic outcomes. mBio. 2021;12:e0339620. 10.1128/mBio.03396-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray JR, Curtis JT. An ordination of the upland forest communities of Southern Wisconsin. Ecol Monogr. 1957;27:325–49. 10.2307/1942268. [DOI] [Google Scholar]
- Broadhurst MJ, Ardeshir A, Kanwar Bet al. Therapeutic helminth infection of macaques with idiopathic chronic diarrhea alters the inflammatory signature and mucosal microbiota of the colon. PLoS Pathog. 2012;8:e1003000. 10.1371/journal.ppat.1003000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruijnesteijn van Coppenraet LE, Wallinga JA, Ruijs GJet al. Parasitological diagnosis combining an internally controlled real-time PCR assay for the detection of four protozoa in stool samples with a testing algorithm for microscopy. Clin Microbiol Infect. 2009;15:869–74. 10.1111/j.1469-0691.2009.02894.x. [DOI] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJet al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan BJ, Sankaran K, Fukuyama JAet al. Bioconductor workflow for microbiome data analysis: from raw reads to community analyses. F1000Res. 2016;5:1492. 10.12688/f1000research.8986.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantacessi C, Giacomin P, Croese Jet al. Impact of experimental hookworm infection on the human gut microbiota. J Infect Dis. 2014;210:1431–4. 10.1093/infdis/jiu256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–3. 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappelli A, Favia G, Ricci I. Wickerhamomyces anomalus in mosquitoes: a promising yeast-based tool for the “Symbiotic Control” of mosquito-borne diseases. Front Microbiol. 2020;11:621605. 10.3389/fmicb.2020.621605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanys-Muñoz E, Martin MJ, Vazquez E. Building a beneficial microbiome from birth. Adv Nutr. 2016;7:323–30. 10.3945/an.115.010694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control (CDC) . CDC criteria for anemia in children and childbearing-aged women. Morb Mortal Wkly Rep. 1989;38:400–4. [PubMed] [Google Scholar]
- Checkley W, et al. Effects of Cryptosporidium parvum infection in Peruvian children: growth faltering and subsequent catch-up growth. Am J Epidemiol. 1998;148:497–506. [DOI] [PubMed] [Google Scholar]
- Chehoud C, Albenberg LG, Judge Cet al. Fungal signature in the gut microbiota of pediatric patients with inflammatory bowel disease. Inflamm Bowel Dis. 2015;21:1948–56. 10.1097/MIB.0000000000000454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente JC, Pehrsson EC, Blaser MJet al. The microbiome of uncontacted Amerindians. Sci Adv. 2015;1:e1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa LB, JohnBull EA, Reeves JTet al. Cryptosporidium-malnutrition interactions: mucosal disruption, cytokines, and TLR signaling in a weaned murine model. J Parasitol. 2011;97:1113–20. 10.1645/GE-2848.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippo C, Cavalieri D, Di Paola Met al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Nat Acad Sci USA. 2010;107:14691–6. 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Campo J, Bass D, Keeling PJ. The eukaryome: diversity and role of microeukaryotic organisms associated with animal hosts. Funct Ecol. 2020;34:2045–54. 10.1111/1365-2435.13490. [DOI] [Google Scholar]
- Dixon P. VEGAN, a package of R functions for community ecology. J Veg Sci. 2009;14:927–30. 10.1111/j.1654-1103.2003.tb02228.x. [DOI] [Google Scholar]
- Dominguez-Bello MG, Godoy-Vitorino F, Knight Ret al. Role of the microbiome in human development. Gut. 2019;68:1108–14. 10.1136/gutjnl-2018-317503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos Zanetti A, Malheiros AF, de Matos TAet al. Diversity, geographical distribution, and prevalence of Entamoeba spp. in Brazil: a systematic review and meta-analysis. Parasite. 2021;28:17. 10.1051/parasite/2021028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Mouzan M, Wang F, Al Mofarreh Met al. Fungal microbiota profile in newly diagnosed treatment-naive children with Crohn’s disease. J Crohns Colitis. 2017;11:586–92. 10.1093/ecco-jcc/jjw197. [DOI] [PubMed] [Google Scholar]
- El Safadi D, Gaayeb L, Meloni Det al. Children of Senegal River Basin show the highest prevalence of Blastocystis sp. ever observed worldwide. BMC Infect Dis. 2014;14:164. 10.1186/1471-2334-14-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsheikha HM, Regan CS, Clark CG. Novel Entamoeba findings in nonhuman primates. Trends Parasitol. 2018;34:283–94. 10.1016/j.pt.2017.12.008. [DOI] [PubMed] [Google Scholar]
- Even G, Lokmer A, Rodrigues Jet al. Changes in the human gut microbiota associated with colonization by Blastocystis sp. and Entamoeba spp. in non-industrialized populations. Front Cell Infect Microbiol. 2021;11:533528. 10.3389/fcimb.2021.533528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers WD, Gao IH, Iliev ID. Gut mycobiota under scrutiny: fungal symbionts or environmental transients?. Curr Opin Microbiol. 2019;50:79–86. 10.1016/j.mib.2019.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay CM, Stefanska AM, Walsh KPet al. Helminth products protect against autoimmunity via innate type 2 cytokines IL-5 and IL-33, which promote eosinophilia. J Immunol. 2016;196:703–14. 10.4049/jimmunol.1501820. [DOI] [PubMed] [Google Scholar]
- Forsell J, Granlund M, Stensvold CRet al. Subtype analysis of Blastocystis isolates in Swedish patients. Eur J Clin Microbiol Infect Dis. 2012;31:1689–96. 10.1007/s10096-011-1416-6. [DOI] [PubMed] [Google Scholar]
- Garcia-Ortega LF, Colón-González M, Sedeño Iet al. Draft genome sequence of a Kazachstania humilis strain isolated from agave fermentation. Microbiol Resour Announc. 2022;11:e0115421. 10.1128/mra.01154-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gause WC, Maizels RM. Macrobiota—helminths as active participants and partners of the microbiota in host intestinal homeostasis. Curr Opin Microbiol. 2016;32:14–8. 10.1016/j.mib.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomin P, Agha Z, Loukas A. Helminths and intestinal flora team up to improve gut health. Trends Parasitol. 2016;32:664–6. 10.1016/j.pt.2016.05.006. [DOI] [PubMed] [Google Scholar]
- Goldman DL, Chen Z, Shankar Vet al. Lower airway microbiota and mycobiota in children with severe asthma. J Allergy Clin Immunol. 2018;141:808–11. 10.1016/j.jaci.2017.09.018. [DOI] [PubMed] [Google Scholar]
- Goncalves ML, Araujo A, Ferreira LF. Human intestinal parasites in the past: new findings and a review. Mem Inst Oswaldo Cruz. 2003;98:103–18. 10.1590/s0074-02762003000900016. [DOI] [PubMed] [Google Scholar]
- Goswami RS, Dong YH, Punja ZK. Host range and mycotoxin production by Fusarium equiseti isolates originating from ginseng fields. Can J Plant Pathol. 2008;30:155–60. 10.1080/07060660809507506. [DOI] [Google Scholar]
- Greigert V, Abou-Bacar A, Brunet Jet al. Human intestinal parasites in Mahajanga, Madagascar: the kingdom of the protozoa. PLoS ONE. 2018;13:e0204576. 10.1371/journal.pone.0204576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillou L, Bachar D, Audic Set al. The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 2013;41:D597–604. 10.1093/nar/gks1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gweon HS, Oliver A, Taylor Jet al. PIPITS: an automated pipeline for analyses of fungal internal transcribed spacer sequences from the Illumina sequencing platform. Methods Ecol Evol. 2015;6:973–80. 10.1111/2041-210X.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib A, Andrianonimiadana L, Rakotondrainipiana Met al. High prevalence of intestinal parasite infestations among stunted and control children aged 2 to 5 years old in two neighborhoods of Antananarivo, Madagascar. PLoS Negl Trop Dis. 2021;15:e0009333. 10.1371/journal.pntd.0009333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallen-Adams HE, Suhr MJ. Fungi in the healthy human gastrointestinal tract. Virulence. 2017;8:352–8. 10.1080/21505594.2016.1247140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyworth B, Brown J. Jejunal microflora in malnourished Gambian children. Arch Dis Child. 1975;50:27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt RP. Mucosal microbial parasites/symbionts in health and disease: an integrative overview. Parasitology. 2019;146:1109–15. 10.1017/S0031182019000647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann C, Dollive S, Grunberg Set al. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS ONE. 2013;8:e66019. 10.1371/journal.pone.0066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huseyin CE, O'Toole PW, Cotter PDet al. Forgotten fungi-the gut mycobiome in human health and disease. FEMS Microbiol Rev. 2017;41:479–511. 10.1093/femsre/fuw047. [DOI] [PubMed] [Google Scholar]
- Ihrmark K, Bödeker IT, Cruz-Martinez Ket al. New primers to amplify the fungal ITS2 region–evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol Ecol. 2012;82:666–77. 10.1111/j.1574-6941.2012.01437.x. [DOI] [PubMed] [Google Scholar]
- Jha AR, Davenport ER, Gautam Yet al. Gut microbiome transition across a lifestyle gradient in Himalaya. PLoS Biol. 2018;16:e2005396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M, Zaretskaya I, Raytselis Yet al. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36:W5–9. 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokelainen P, Hebbelstrup Jensen B, Andreassen BUet al. Dientamoeba fragilis, a commensal in children in Danish day care centers. J Clin Microbiol. 2017;55:1707–13. 10.1128/JCM.00037-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journal of Statistical Software . ggplot2: Elegant Graphics for Data Analysis. researchgate.net. Berlin: Springer, 2010. [Google Scholar]
- Kabwe MH, Vikram S, Mulaudzi Ket al. The gut mycobiota of rural and urban individuals is shaped by geography. BMC Microbiol. 2020;20:257. 10.1186/s12866-020-01907-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Asimenos G, Toh H. Multiple alignment of DNA sequences with MAFFT. Methods Mol Biol. 2009;537:39–64. 10.1007/978-1-59745-251-9_3. [DOI] [PubMed] [Google Scholar]
- Koljalg U, Larsson KH, Abarenkov Ket al. UNITE: a database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytol. 2005;166:1063–8. 10.1111/j.1469-8137.2005.01376.x. [DOI] [PubMed] [Google Scholar]
- Kozich JJ, Westcott SL, Baxter NTet al. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–20. 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov AM, Darriba D, Flouri Tet al. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35:4453–5. 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogsgaard LR, Andersen LO, Johannesen TBet al. Characteristics of the bacterial microbiome in association with common intestinal parasites in irritable bowel syndrome. Clin Transl Gastroenterol. 2018;9:161. 10.1038/s41424-018-0027-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogsgaard LR, Engsbro AL, Stensvold CRet al. The prevalence of intestinal parasites is not greater among individuals with irritable bowel syndrome: a population-based case-control study. Clin Gastroenterol Hepatol. 2015;13:507–13. 10.1016/j.cgh.2014.07.065. [DOI] [PubMed] [Google Scholar]
- Larsson A. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics. 2014;30:3276–8. 10.1093/bioinformatics/btu531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavrinienko A, Scholier T, Bates STet al. Defining gut mycobiota for wild animals: a need for caution in assigning authentic resident fungal taxa. Anim Microbiome. 2021;3:75. 10.1186/s42523-021-00134-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lhotska Z, Jirků M, Hložková Oet al. A study on the prevalence and subtype diversity of the intestinal protist Blastocystis sp. in a gut-healthy human population in the Czech Republic. Front Cell Infect Microbiol. 2020;10:544335. 10.3389/fcimb.2020.544335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limon JJ, Tang J, Li Det al. Malassezia is associated with Crohn’s disease and exacerbates Colitis in mouse models. Cell Host Microbe. 2019;25:377–88. 10.1016/j.chom.2019.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan C, Xie L, Yang Xet al. Dysbiosis of fungal microbiota in the intestinal mucosa of patients with colorectal adenomas. Sci Rep. 5:7980:2015. 10.1038/srep07980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann AE, Mazel F, Lemay MAet al. Biodiversity of protists and nematodes in the wild nonhuman primate gut. ISME J. 2020;14:609–22. 10.1038/s41396-019-0551-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mar Rodriguez M, Pérez D, Javier Chaves Fet al. Obesity changes the human gut mycobiome. Sci Rep. 2015;5:14600. 10.1038/srep14600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie C, Petri WA Jr. Regulation of virulence of Entamoeba histolytica. Annu Rev Microbiol. 2014;68:493–520. 10.1146/annurev-micro-091313-103550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maritz JM, Rogers KH, Rock TMet al. An 18S rRNA workflow for characterizing protists in sewage, with a focus on Zoonotic Trichomonads. Microb Ecol. 2017;74:923–36. 10.1007/s00248-017-0996-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masneuf-Pomarede I, Bely M, Marullo Pet al. The genetics of non-conventional wine yeasts: current knowledge and future challenges. Front Microbiol. 2015;6:1563. 10.3389/fmicb.2015.01563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane AJ, McSorley HJ, Davidson DJet al. Enteric helminth-induced type I interferon signaling protects against pulmonary virus infection through interaction with the microbiota. J Allergy Clin Immunol. 2017;140:1068–78. 10.1016/j.jaci.2017.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32:W20–25. 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney EA, Williamson L, Yoder ADet al. Alteration of the rat cecal microbiome during colonization with the helminth Hymenolepis diminuta. Gut Microbes. 2015;6:182–93. 10.1080/19490976.2015.1047128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e61217. 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekonnen GK, Mengistie B, Sahilu Get al. Etiologies of diarrhea and drug susceptibility patterns of bacterial isolates among under-five year children in refugee camps in Gambella Region, Ethiopia: a case control study. BMC Infect Dis. 2019;19:1008. 10.1186/s12879-019-4599-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoli G, Terruzzi V, Butti GCet al. Candidiasis of the duodenum and jejunum. Gastroenterology. 1981;81:825. [PubMed] [Google Scholar]
- Morton ER, Lynch J, Froment Aet al. Variation in rural African gut microbiota is strongly correlated with colonization by Entamoeba and subsistence. PLos Genet. 2015;11:e1005658. 10.1371/journal.pgen.1005658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser W, Schindler C, Keiser J. Efficacy of recommended drugs against soil transmitted helminths: systematic review and network meta-analysis. BMJ. 2017;358:j4307. 10.1136/bmj.j4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkvold GP. Fusarium species and their associated mycotoxins. Methods Mol Biol. 2017;1542:51–106. 10.1007/978-1-4939-6707-0_4. [DOI] [PubMed] [Google Scholar]
- Nash AK, Auchtung TA, Wong MCet al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome. 2017;5:153. 10.1186/s40168-017-0373-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieves-Ramirez ME, Partida-Rodríguez O, Laforest-Lapointe Iet al. Asymptomatic intestinal colonization with Protist Blastocystis is strongly associated with distinct microbiome ecological patterns. mSystems. 2018;3:e00007–18. 10.1128/mSystems.00007-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obregon-Tito AJ, Tito R, Metcalf Jet al. Subsistence strategies in traditional societies distinguish gut microbiomes. Nat Commun. 2015;6:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OMS . Concentrations en hémoglobine permettant de diagnostiquer l'anémie et d'en évaluer la sévérité. Geneva: World Health Organization, 2011. [Google Scholar]
- Onis M. WHO child growth standards based on length/height, weight and age. Acta Paediatr. 2006;95:76–85. 10.1111/j.1651-2227.2006.tb02378.x. [DOI] [PubMed] [Google Scholar]
- Parfrey LW, Walters WA, Lauber CLet al. Communities of microbial eukaryotes in the mammalian gut within the context of environmental eukaryotic diversity. Front Microbiol. 2014;5:298. 10.3389/fmicb.2014.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partida-Rodriguez O, Nieves-Ramirez M, Laforest-Lapointe Iet al. Exposure to parasitic protists and Helminths changes the intestinal community structure of bacterial communities in a cohort of mother-child binomials from a semirural setting in Mexico. mSphere. 2021;6:e0008321. 10.1128/mSphere.00083-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasolli E, Asnicar F, Manara Set al. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell. 2019;176:649–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic A, Bourdon C, Wang PWet al. Design and application of a novel two-amplicon approach for defining eukaryotic microbiota. Microbiome. 2018;6:228. 10.1186/s40168-018-0612-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Peplies J, Glockner FO. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–9. 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes Jet al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj E, Calvo-Urbano B, Heffernan Cet al. Systematic review to evaluate a potential association between helminth infection and physical stunting in children. Parasit Vectors. 2022;15:135. 10.1186/s13071-022-05235-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan D, Bowcutt R, Lee SCet al. Helminth infection promotes colonization resistance via type 2 immunity. Science. 2016;352:608–12. 10.1126/science.aaf3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard ML, Lamas B, Liguori Get al. Gut fungal microbiota: the Yin and Yang of inflammatory bowel disease. Inflamm Bowel Dis. 2015;21:656–65. 10.1097/MIB.0000000000000261. [DOI] [PubMed] [Google Scholar]
- Richard ML, Sokol H. The gut mycobiota: insights into analysis, environmental interactions and role in gastrointestinal diseases. Nat Rev Gastroenterol Hepatol. 2019;16:331–45. 10.1038/s41575-019-0121-2. [DOI] [PubMed] [Google Scholar]
- Roberts LS, Janovy JJ, Nadler S, Schmidt GD, Larry S. Foundations of Parasitology. 9th edn.New York: McGraw-Hill Publishing, 2013. [Google Scholar]
- Rodriguez-Otero P, Porcher R, Peffault de Latour Ret al. Fecal calprotectin and alpha-1 antitrypsin predict severity and response to corticosteroids in gastrointestinal graft-versus-host disease. Blood. 2012;119:5909–17. 10.1182/blood-2011-12-397968. [DOI] [PubMed] [Google Scholar]
- Rognes T, Flouri T, Nichols Bet al. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook GA. Hygiene hypothesis and autoimmune diseases. Clin Rev Allergy Immunol. 2012;42:5–15. 10.1007/s12016-011-8285-8. [DOI] [PubMed] [Google Scholar]
- Roser D, Simonsen J, Nielsen HVet al. Dientamoeba fragilis in Denmark: epidemiological experience derived from four years of routine real-time PCR. Eur J Clin Microbiol Infect Dis. 2013;32:1303–10. 10.1007/s10096-013-1880-2. [DOI] [PubMed] [Google Scholar]
- Rowan-Nash AD, Korry BJ, Mylonakis Eet al. Cross-domain and viral interactions in the microbiome. Microbiol Mol Biol Rev. 2019;83;e00044. 10.1128/MMBR.00044-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan PD, Hill CJ, Ross RPet al. The intestinal protist Blastocystis is not a common member of the healthy infant gut microbiota in a Westernized country (Ireland). Parasitology. 2018;145:1274–8. 10.1017/S0031182018000033. [DOI] [PubMed] [Google Scholar]
- Scanlan PD, Knight R, Song SJet al. Prevalence and genetic diversity of Blastocystis in family units living in the United States. Infect Genet Evol. 2016;45:95–7. 10.1016/j.meegid.2016.08.018. [DOI] [PubMed] [Google Scholar]
- Scanlan PD, Marchesi JR. Micro-eukaryotic diversity of the human distal gut microbiota: qualitative assessment using culture-dependent and -independent analysis of faeces. ISME J. 2008;2:1183–93. 10.1038/ismej.2008.76. [DOI] [PubMed] [Google Scholar]
- Schei K, Avershina E, Øien Tet al. Early gut mycobiota and mother-offspring transfer. Microbiome. 2017;5:107. 10.1186/s40168-017-0319-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz JD, Moser W, Hurlimann Eet al. Preventive chemotherapy in the fight against soil-transmitted Helminthiasis: achievements and limitations. Trends Parasitol. 2018;34:590–602. 10.1016/j.pt.2018.04.008. [DOI] [PubMed] [Google Scholar]
- Sharma AK, Davison S, Pafco Bet al. The primate gut mycobiome-bacteriome interface is impacted by environmental and subsistence factors. NPJ Biofilms Microbiomes. 2022;8:12. 10.1038/s41522-022-00274-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LE, Stoltzfus RJ, Prendergast A. Food chain mycotoxin exposure, gut health, and impaired growth: a conceptual framework. Adv Nutr. 2012;3:526–31. 10.3945/an.112.002188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits SA, Leach J, Sonnenburg EDet al. Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science. 2017;357:802–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Leducq V, Aschard Het al. Fungal microbiota dysbiosis in IBD. Gut. 2017;66:1039–48. 10.1136/gutjnl-2015-310746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sovran B, Planchais J, Jegou Set al. Enterobacteriaceae are essential for the modulation of colitis severity by fungi. Microbiome. 2018;6:152. 10.1186/s40168-018-0538-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. Using RAxML to infer phylogenies. Curr Protoc Bioinformatics. 2015;51:6 14 11–14. 10.1002/0471250953.bi0614s51. [DOI] [PubMed] [Google Scholar]
- Stensvold CR, Alfellani M, Clark CG. Levels of genetic diversity vary dramatically between Blastocystis subtypes. Infect Genet Evol. 2012;12:263–73. 10.1016/j.meegid.2011.11.002. [DOI] [PubMed] [Google Scholar]
- Stensvold CR, Lebbad M, Verweij JJ. The impact of genetic diversity in protozoa on molecular diagnostics. Trends Parasitol. 2011a;27:53–58. 10.1016/j.pt.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Stensvold CR, Lebbad M, Victory ELet al. Increased sampling reveals novel lineages of Entamoeba: consequences of genetic diversity and host specificity for taxonomy and molecular detection. Protist. 2011b;162:525–41. 10.1016/j.protis.2010.11.002. [DOI] [PubMed] [Google Scholar]
- Stensvold CR, Tan KSW, Clark CGB. Blastocytosis. Trends Parasitol. 2020;36:315–6. 10.1016/j.pt.2019.12.008. [DOI] [PubMed] [Google Scholar]
- Stensvold CR, van der Giezen M. Associations between gut microbiota and common luminal intestinal parasites. Trends Parasitol. 2018;34:369–77. 10.1016/j.pt.2018.02.004. [DOI] [PubMed] [Google Scholar]
- Stensvold CR. Pinning down the role of common luminal intestinal parasitic protists in human health and disease—status and challenges. Parasitology. 2019;146:695–701. 10.1017/S0031182019000039. [DOI] [PubMed] [Google Scholar]
- Stoeck T, Bass D, Nebel Met al. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol Ecol. 2010;19:21–31. 10.1111/j.1365-294X.2009.04480.x. [DOI] [PubMed] [Google Scholar]
- Suhr MJ, Hallen-Adams HE. The human gut mycobiome: pitfalls and potentials–a mycologist’s perspective. Mycologia. 2015;107:1057–73. 10.3852/15-147. [DOI] [PubMed] [Google Scholar]
- Sullivan KM, Mei Z, Grummer-Strawn Let al. Haemoglobin adjustments to define anaemia. Trop Med Int Health. 2008;13:1267–71. 10.1111/j.1365-3156.2008.02143.x. [DOI] [PubMed] [Google Scholar]
- Sun Y, Zuo T, Cheung CPet al. Population-level configurations of gut mycobiome across 6 ethnicities in urban and rural China. Gastroenterology. 2021;160:272–86 e211. 10.1053/j.gastro.2020.09.014. [DOI] [PubMed] [Google Scholar]
- Taylor-Robinson DC, Maayan N, Donegan Set al. Public health deworming programmes for soil-transmitted helminths in children living in endemic areas. Cochrane Database Syst Rev. 2019;9:CD000371. 10.1002/14651858.CD000371.pub7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Hove R, Schuurman T, Kooistra Met al. Detection of diarrhoea-causing protozoa in general practice patients in the Netherlands by multiplex real-time PCR. Clin Microbiol Infect. 2007;13:1001–7. 10.1111/j.1469-0691.2007.01788.x. [DOI] [PubMed] [Google Scholar]
- Thurnham DI, McCabe LD, Haldar Set al. Adjusting plasma ferritin concentrations to remove the effects of subclinical inflammation in the assessment of iron deficiency: a meta-analysis. Am J Clin Nutr. 2010;92:546–55. 10.3945/ajcn.2010.29284. [DOI] [PubMed] [Google Scholar]
- Tito RY, Chaffron S, Caenepeel Cet al. Population-level analysis of Blastocystis subtype prevalence and variation in the human gut microbiota. Gut. 2019;68:1180–9. 10.1136/gutjnl-2018-316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkeltaub JA, McCarty TR 3rd, Hotez PJ. The intestinal protozoa: emerging impact on global health and development. Curr Opin Gastroenterol. 2015;31:38–44. 10.1097/MOG.0000000000000135. [DOI] [PubMed] [Google Scholar]
- Underhill DM, Iliev ID. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol. 2014;14:405–16. 10.1038/nri3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungaro F, Massimino L, D'Alessio Set al. The gut virome in inflammatory bowel disease pathogenesis: from metagenomics to novel therapeutic approaches. Unit Eur Gastroenterol J. 2019;7:999–1007. 10.1177/2050640619876787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco J, González F, Díaz Tet al. Profiles of enteropathogens in asymptomatic children from indigenous communities of Mérida, Venezuela. J Infect Dev Ctries. 2011;5:278–85. [DOI] [PubMed] [Google Scholar]
- Venter F, Matthews KR, Silvester E. Parasite co-infection: an ecological, molecular and experimental perspective. Proc Biol Sci. 2022;289:20212155. 10.1098/rspb.2021.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij JJ, Blangé RA, Templeton Ket al. Simultaneous detection of Entamoeba histolytica, Giardia lamblia, and Cryptosporidium parvum in fecal samples by using multiplex real-time PCR. J Clin Microbiol. 2004;42:1220–3. 10.1128/JCM.42.3.1220-1223.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijaya Chandra SH, Srinivas R, Dawson TL Jret al. Cutaneous Malassezia: commensal, pathogen, or protector?. Front Cell Infect Microbiol. 2020;10:614446. 10.3389/fcimb.2020.614446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Rosenvinge EC, Song Y, White JRet al. Immune status, antibiotic medication and pH are associated with changes in the stomach fluid microbiota. ISME J. 2013;7:1354–66. 10.1038/ismej.2013.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonaesch P, Araújo JR, Gody JCet al. Stunted children display ectopic small intestinal colonization by oral bacteria, which cause lipid malabsorption in experimental models. Proc Natl Acad Sci USA. 2022;119:e2209589119. 10.1073/pnas.2209589119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonaesch P, Djorie SG, Kandou KJEet al. Factors associated with stunted growth in children under five years in Antananarivo, Madagascar and Bangui, Central African Republic. Matern Child Health J. 2021;25:1626–37. 10.1007/s10995-021-03201-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonaesch P, Morien E, Andrianonimiadana Let al. Stunted childhood growth is associated with decompartmentalization of the gastrointestinal tract and overgrowth of oropharyngeal taxa. Proc Nat Acad Sci USA. 2018;115:E8489–98. 10.1073/pnas.1806573115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonaesch P, Randremanana R, Gody JCet al. Identifying the etiology and pathophysiology underlying stunting and environmental enteropathy: study protocol of the AFRIBIOTA project. BMC Pediatr. 2018;18:236. 10.1186/s12887-018-1189-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walk ST, Blum AM, Ewing SA-Set al. Alteration of the murine gut microbiota during infection with the parasitic helminth Heligmosomoides polygyrus. Inflamm Bowel Dis. 2010;16:1841–9. 10.1002/ibd.21299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh KP, Brady MT, Finlay CMet al. Infection with a helminth parasite attenuates autoimmunity through TGF-beta-mediated suppression of Th17 and Th1 responses. J Immunol. 2009;183:1577–86. 10.4049/jimmunol.0803803. [DOI] [PubMed] [Google Scholar]
- Wang XW, Yang FY, Meijer Met al. Redefining Humicola sensu stricto and related genera in the Chaetomiaceae. Stud Mycol. 2019;93:65–153. 10.1016/j.simyco.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westreich ST, Ardeshir A, Alkan Zet al. Fecal metatranscriptomics of macaques with idiopathic chronic diarrhea reveals altered mucin degradation and fucose utilization. Microbiome. 2019;7:41. 10.1186/s40168-019-0664-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization WHO child growth standards: head circumference-for-age, arm circumference-for-age, triceps skinfold-for-age and subscapular skinfold-for-age: methods and development. Geneva: World Health Organization, 2007. [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJet al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–7. 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, McGinnis S, Madden TL. BLAST: improvements for better sequence analysis. Nucleic Acids Res. 2006;34:W6–9. 10.1093/nar/gkl164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaiss MM, Rapin A, Lebon Let al. The intestinal microbiota contributes to the ability of helminths to modulate allergic inflammation. Immunity. 2015;43:998–1010. 10.1016/j.immuni.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.