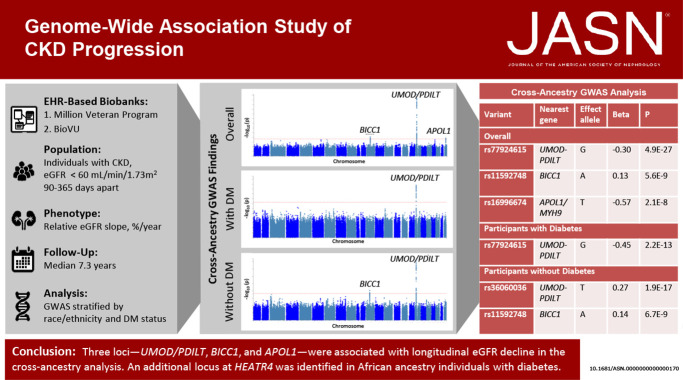
Visual Abstract
Keywords: CKD, human genetics, progression of chronic renal failure, Genome-Wide Association Study
Abstract
Significance Statement
Rapid progression of CKD is associated with poor clinical outcomes. Most previous studies looking for genetic factors associated with low eGFR have used cross-sectional data. The authors conducted a meta-analysis of genome-wide association studies of eGFR decline among 116,870 participants with CKD, focusing on longitudinal data. They identified three loci (two of them novel) associated with longitudinal eGFR decline. In addition to the known UMOD/PDILT locus, variants within BICC1 were associated with significant differences in longitudinal eGFR slope. Variants within HEATR4 also were associated with differences in eGFR decline, but only among Black/African American individuals without diabetes. These findings help characterize molecular mechanisms of eGFR decline in CKD and may inform new therapeutic approaches for progressive kidney disease.
Background
Rapid progression of CKD is associated with poor clinical outcomes. Despite extensive study of the genetics of cross-sectional eGFR, only a few loci associated with eGFR decline over time have been identified.
Methods
We performed a meta-analysis of genome-wide association studies of eGFR decline among 116,870 participants with CKD—defined by two outpatient eGFR measurements of <60 ml/min per 1.73 m2, obtained 90–365 days apart—from the Million Veteran Program and Vanderbilt University Medical Center's DNA biobank. The primary outcome was the annualized relative slope in outpatient eGFR. Analyses were stratified by ethnicity and diabetes status and meta-analyzed thereafter.
Results
In cross-ancestry meta-analysis, the strongest association was rs77924615, near UMOD/PDILT; each copy of the G allele was associated with a 0.30%/yr faster eGFR decline (P = 4.9×10−27). We also observed an association within BICC1 (rs11592748), where every additional minor allele was associated with a 0.13%/yr slower eGFR decline (P = 5.6×10−9). Among participants without diabetes, the strongest association was the UMOD/PDILT variant rs36060036, associated with a 0.27%/yr faster eGFR decline per copy of the C allele (P = 1.9×10−17). Among Black participants, a significantly faster eGFR decline was associated with variant rs16996674 near APOL1 (R2=0.29 with the G1 high-risk genotype); among Black participants with diabetes, lead variant rs11624911 near HEATR4 also was associated with a significantly faster eGFR decline. We also nominally replicated loci with known associations with eGFR decline, near PRKAG2, FGF5, and C15ORF54.
Conclusions
Three loci were significantly associated with longitudinal eGFR change at genome-wide significance. These findings help characterize molecular mechanisms of eGFR decline and may contribute to the development of new therapeutic approaches for progressive CKD.
Introduction
The rate of decline in kidney function varies widely among individuals with CKD and is incompletely explained by known risk factors. More rapid CKD progression is associated with worse clinical outcomes, including higher risks of ESKD, cardiovascular events, and mortality, independent of cross-sectional eGFR.1,2
Previous genome-wide association studies (GWASs) have evidenced associations of common variants of several genes with eGFR and CKD from a single time point.3–10 Among these, UMOD is a well-established causal gene for the development of CKD, and GATM and PRKAG2 have been repeatedly associated with eGFR.5 Such variants are informative for identifying and understanding mechanisms of predisposition to disease, but not necessarily for influencing disease progression. Because progression of CKD may depend on factors unrelated to the primary disorder (e.g., glomerular hypertension, inflammation, fibrosis), unique loci may exist for renal function decline beyond those identified for a one-time measure of eGFR.11,12 One GWAS of longitudinal change in eGFR has been conducted exclusively among individuals with CKD.13 However, limited statistical power, owing to a small sample size, may have hampered the discovery of novel hits.
Identifying individuals with a higher genetic risk of rapid kidney function decline will likely improve prediction of ESKD risk, advance prognostication of CKD outcomes, and provide a pool of genetic markers that may lead to actionable protein targets and disease mechanisms.
To investigate the role of common genetic variants in longitudinal eGFR change among individuals with CKD, we conducted a GWAS of the rate of eGFR decline among 109,570 participants from the Million Veteran Program (MVP) and 7300 participants within Vanderbilt University Medical Center's DNA biobank (BioVU).
Methods
Participants
The MVP is a large health care system observational cohort study and biobank supported by the Department of Veterans Affairs (VA) of participants recruited from the patient populations of 63 VA medical facilities. This study was approved by the VA central and site-specific institutional review boards, and all participants provided informed consent. Study population and recruitment methods have been previously described.14 In brief, MVP recruitment commenced in 2011 and is ongoing, with over 775,000 participants enrolled to date. Each participating veteran completed baseline and lifestyle surveys and gave permission to access the VA electronic health record data and obtain a blood sample for genetic analysis.
The BioVU is Vanderbilt University Medical Center's DNA repository. The BioVU resource and its ethical, privacy and other protection, has been described.15,16 In brief, BioVU accrues DNA samples during routine clinical care from patients at the Vanderbilt University Medical Center (VUMC) who have not opted out of participation, using blood that would otherwise be discarded after clinical testing. The samples are deidentified and considered nonhuman subjects research. Samples and genetic data within BioVU are linked to a deidentified and research-enabled version of VUMC's electronic medical record, with detailed longitudinal clinical data dating back to the 1990s.
For the purposes of this study, we restricted analyses to participants with CKD, defined by the presence of two outpatient eGFR measurements <60 ml/min per 1.73 m2 within a minimum of 90 days and a maximum of 365 days apart. We excluded individuals who were on dialysis or had received a kidney transplantation before cohort entry. We excluded serum creatinine values <0.4 and >20 mg/dl because they likely represented laboratory or data entry errors. We excluded participants with >24 outpatient eGFR measurements per year and those with <2 years of follow-up time within the VA or VUMC's electronic health record. In addition, analyses were restricted to individuals with a minimum of four longitudinal eGFR measurements to optimize estimation of the slope. Participants were censored at ESKD, defined as chronic dialysis initiation or kidney transplantation.
Genotyping and Imputation
MVP genotyping was performed using an Affymetrix Axiom biobank array, the MVP 1.0 Genotyping Array. Quality control pipelines included the exclusion of duplicate samples, those with discordant reported and genotyped sex, and samples with more heterozygosity than expected. One participant randomly selected from each pair of related individuals (closer than halfway between second and third-degree relatives or closer) was excluded. After using EAGLE v2 to prephase each chromosome, genotypes from the 1000 Genomes Project Phase 3 were imputed into MVP participants using Minimac3 software.17–19 Ethnicity was assigned using a harmonized ancestry and ethnicity variable on the basis of an algorithm that integrates genetically inferred ancestry on the basis of the top 30 principal components with self-identified ethnicity. Details have been described elsewhere.20 On the basis of the harmonized ancestry and ethnicity variable, MVP participants with genotype data are assigned to one of the following four nonoverlapping groups: non-Hispanic White, non-Hispanic Black, non-Hispanic Asian, and Hispanic participants.
In BioVU, GWAS-level genotyping was performed using a custom Illumina MEGA-Ex chip, which includes >2 million common and rare variants before imputation. We obtained genotyped data in PLINK format from the Vanderbilt core after the following quality control steps: excluding either samples or variants with ≥5% missingness, mismatched identifiers as detected by identity by descent checks, and nonconcordance between reported sex and genetically determined sex. Overlapping variants with 1000 Genomes demonstrated ≥99.98% variant call concordance using HapMap sample aliquots. After filtering out suboptimal markers and individuals with call rate <95%, variants across the genome were phased and imputed to the 1000 Genomes panel using the IMPUTE2 program.
In both cohorts, we restricted analyses to genetic variants with minor allele frequency (MAF) >0.01. Principal component analysis was performed using common genetic variants (MAF>0.05) using EIGENSOFT.21 For the purposes of this study, analyses were restricted to non-Hispanic White and non-Hispanic Black participants.
Phenotype
The primary study phenotype was the relative annualized change in kidney function, defined by the exponentiated slope of log-transformed eGFR minus one and interpreted as percentage change in eGFR per year. This phenotype was chosen on the basis of ongoing work demonstrating minimal dependence of relative eGFR change on baseline eGFR and straightforward interpretation of regression coefficients. Estimated GFR was calculated using the Chronic Kidney Disease Epidemiology Collaboration serum creatinine equation.22 Only outpatient eGFR measurements were included in the slope estimation, with additional restriction to one creatinine per day, given the following rules: (1) If a participant had multiple outpatient creatinine measurements on a given day and the difference between the trough and peak of creatinine measurements was ≤0.3 ml/dl, the average of creatinine measurements for that day was retained for eGFR and slope estimation, and (2) if a participant had multiple outpatient creatinine measurements on a given day and the difference between the trough and peak of creatinine measurements was >0.3 ml/dl, measurements from that day were excluded from the slope estimation, to exclude measurements reflecting acute fluctuations in kidney function.
To generate the relative eGFR slopes for GWASs, we performed linear mixed model (LMM) analysis of the repeated log-transformed eGFR measurements stratified by ethnicity and diabetes status at baseline using the R package lme4. In each analysis, we included baseline age, sex, and time (in years) as fixed effects and a random intercept and random slope in the model. The random intercept term accounts for unobserved individual-level factors that contribute to differences in eGFR levels that are not explained by age, sex, or time. The inclusion of random intercepts in the LMM analysis allows for estimation of the fixed effects of genetic variants on changes in eGFR over time while accounting for individual-level differences in eGFR at age and time of zero. This can lead to more accurate and efficient estimation of the fixed effects of the genetic variants and improve the overall fit of the model. We used the inverse-normal transformed best linear unbiased predictor (BLUP) of the random slopes obtained from the LMM as our trait of interest in the GWAS.
In secondary analyses, we also examined the absolute change in kidney function, defined as the annualized slope of eGFR in ml/min per 1.73 m2 per year. The absolute eGFR slopes were generated in a similar manner as the relative eGFR slopes described above, without log transformation before the LMM analysis.
Statistical Analysis
Cohort entry (baseline) was defined as the date of the second CKD-confirmatory eGFR measurement. Diabetes was defined by the use of any antidiabetic medications or presence of at least two outpatient International Classification of Diseases codes for diabetes (ICD9 250.*) on separate dates within 365 days before baseline. Hypertension was defined by the presence of a hypertension code, prescription of an antihypertensive drug, or having two systolic BP measurements >140 mm Hg and/or two diastolic BP measurements >90 mm Hg. Body mass index was estimated using the closest weight to the baseline and height mode, as weight in kilograms divided by height in meters squared. Urine albumin-to-creatinine ratio closest to baseline was assessed in mg/g and calculated as urinary albumin (mg/L)/urinary creatinine (mg/dl)×100. Microalbuminuria was defined as urine albumin-to-creatinine ratio >30 mg/g.
GWAS analyses were performed in PLINK version 2a using linear regression of the inverse normally transformed BLUPs of eGFR random slopes as the dependent variable and genotypes (allelic dosage) as predictors, under an additive genetic model. Covariates included age at baseline, sex, and the first ten principal components of ancestry. To retain the interpretation of the genetic effect estimates in the original scale of eGFR slopes (before the inverse-normal transformation), we multiplied the genetic effect estimates and standard errors by the standard deviation of the untransformed BLUPs of eGFR random slopes.
Analyses were stratified by ethnicity and diabetes status at baseline. Genomic control parameters were estimated for each cohort and appropriate genomic control correction was applied to input statistics before performing meta-analysis to correct for residual cryptic relatedness or population stratification. Fixed-effect variance-weighted meta-analysis of analyses in Black and White participants and participants with diabetes and nondiabetic participants was conducted within each cohort, and study-specific estimates and standard errors were combined using METAL.23 Variants with imputation quality <0.4 were excluded from all analyses. An effective minor allele count more than 30 was used to filter out variants, which was calculated as 2×MAF(1-MAF)×Sample Size per phenotype×info score. A total of 11,619,996 autosomal genetic variants were included in the cross-ancestry diabetic strata meta-analysis, and a total of 11,652,122 autosomal variants were included in the cross-ancestry nondiabetic strata analysis. Quantile-quantile plots, Manhattan plots, and regional association plots were produced using R, LocusZoom,24 and the functional annotation and mapping software tool FUMA, respectively.25 Proportion of variance explained (PVE) was estimated using the formula PVE=t2/(n−2+t2), where t=β/standard error of the mean. Linkage disequilibrium score regression was used to estimate the single nucleotide polymorphism (SNP)-based heritability of eGFR slope among White participants with diabetes and White nondiabetic participants.
Conditional analyses were performed using Genome-wide Complex Traits Analysis to test whether multiple independent risk alleles existed at any of the genome-wide significant loci, using an approximate linkage disequilibrium structure from an external reference sample, forward stepwise selection procedure, and summary-level statistics from the transethnic meta-analysis. The reference samples were constructed from 5000 randomly selected BioVU participants of European ancestry and 5000 randomly selected BioVU participants of African ancestry. We set the collinearity restriction to 0.90 and defined genome-wide significance as P < 5×10−8. We defined loci as 500 kb up and down the lead variants found in the Genome-wide Complex Traits Analysis-COJO analysis.
For primary replication, we selected 12 candidate genetic variants previously reported to be associated with kidney function decline from a recently published report.26 We additionally examined associations from the genome-wide significant variants from our analyses within the summary statistics from Gorski et al.26 for independent replication of our findings. Statistical significance for replication analyses was set at Bonferroni-corrected α=0.05/12=0.0036. To evaluate the overall correlation between the ability of longitudinal and cross-sectional data to identify eGFR associations, we compared the magnitude of the β coefficients and statistical significance of all tested genetic variants with P < 0.01 between our analyses and those from a recently published meta-analysis of cross-sectional creatinine-based eGFR from the Chronic Kidney Diseases Genetics (n=765,348) Consortium and the UK Biobank (n=436,581).11,27
We also performed a haplotype association analysis among non-Hispanic Black participants, to test the effect of known risk alleles in APOL1 on the relative slope of eGFR. We defined the APOL1 haplotype as the combination of G1 and G2 haplotypes, where G1 consists of missense mutations at rs73885319 and rs60919145 and G2 consists of rs71785313. BioVU used a proxy genetic variant, rs12106505, for G2 because rs71785313 was not genotyped or imputed in the BioVU cohort. Black participants with two G1 alleles or one G1 and one G2 alleles were classified as APOL1 high risk and those with only one G1 or G2 allele or no G1 or G2 alleles were defined as APOL1 low risk.
Results
Among participants with CKD and diabetes in the MVP and BioVU, mean (SD) eGFR at baseline was 52 (±7) ml/min per 1.73 m2 and 49 (±17) ml/min per 1.73 m2, respectively, and median (interquartile range [IQR]) relative kidney function decline was −1.3 (−4.2, 0.9)%/yr and −2.38 (−10.8, +3.9)%/yr, respectively (Table 1). Overall median (IQR) follow-up time among participants with diabetes was 6.5 years (4.2, 10.5). Among those with CKD without diabetes in the MVP and BioVU, mean (SD) eGFR at baseline was 52 (±8) ml/min per 1.73 m2 and 51.4 (±18) ml/min per 1.73 m2, respectively, and median (IQR) relative kidney function decline was −0.40 (−2.1, 1.1)%/yr and −0.74 (−6.40, +4.41)%/yr, respectively (Table 1). Median (IQR) follow-up time among participants without diabetes was 7.9 years (4.5, 11.2). Participants with faster longitudinal decline in eGFR were more likely to be older, have diabetes, and have microalbuminuria.
Table 1.
Baseline characteristics by relative slope tertile and diabetes
| Participant Characteristic | Million Veteran Program (n=109,570) | BioVU (n=7,300) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Participants with Diabetes (n=44,782) | Participants without Diabetes (n=64,788) | Participants with Diabetes (n=1,642) | Participants without Diabetes (n=5,658) | |||||||||
| Tertile of relative eGFR decline, %/yr | < −3.0 | −3.0 to 0.1 | >0.1 | < −1.4 | −1.4 to 0.5 | >0.5 | < −7.3 | (−7.3, +1.5) | >1.5 | < −3.9 | −3.9, 2.0 | >2.0 |
| N | 14,928 | 14,927 | 14,927 | 21,596 | 21,596 | 21,596 | 555 | 533 | 554 | 1,886 | 1,886 | 1,886 |
| Age, yr, mean (SD) | 67 (9) | 68 (8) | 67 (8) | 69 (10) | 69 (9) | 67 (10) | 65 (12) | 66 (11) | 66 (11) | 64 (17) | 63 (17) | 59 (22) |
| Female sex, n (%) | 477 (3) | 538 (4) | 716 (5) | 828 (4) | 1,184 (5) | 1,527 (7) | 271 (49) | 278 (52) | 259 (47) | 935 (50) | 1,025 (54) | 959 (51) |
| Non-Hispanic Black race, n (%) | 3,712 (25) | 2,273 (15) | 2,735 (18) | 3,404 (16) | 2,264 (10) | 3,055 (14) | 89 (16) | 68 (19) | 83 (15) | 254 (14) | 151 (8) | 231 (12) |
| eGFR, ml/min per 1.73 m2; mean (SD) | 51 (8) | 52 (7) | 52 (7) | 50 (8) | 53 (6) | 52 (7) | 48 (19) | 54 (16) | 46 (17) | 51 (19) | 56 (16) | 47 (17) |
| Follow-up time, yr; median (IQR) | 6 (4–10) | 8 (5–12) | 6 (4–10) | 9 (5–12) | 9 (5–12) | 7 (4–11) | 2 (1–6) | 5 (3–7) | 2 (1–4) | 2 (1–5) | 6 (4–9) | 2 (1–5) |
| Systolic BP, mm Hg; mean (SD) | 135 (19) | 131 (17) | 129 (17) | 134 (18) | 131 (16) | 129 (17) | 136 (23) | 134 (20) | 132 (22) | 133 (23) | 131 (46) | 129 (21) |
| Body mass index, kg/m2; mean (SD) | 32 (6) | 32 (6) | 33 (6) | 29 (5) | 29 (5) | 30 (5) | 32 (7) | 32 (7) | 33 (7) | 29 (7) | 28 (7) | 28 (7) |
| Microalbuminuria; n (%) | 3,917 (54) | 2,372 (31) | 2,067 (26) | 715 (36) | 334 (17) | 382 (18) | 130 (54) | 124 (35) | 86 (35) | 216 (41) | 142 (21) | 136 (28) |
IQR, interquartile range.
Cross-Ancestry Overall GWAS Results
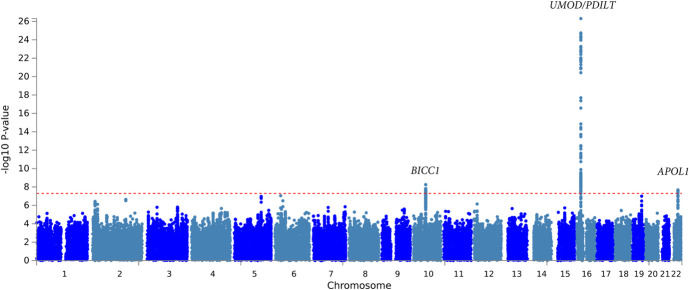
The overall meta-analysis of Black and White individuals with and without diabetes identified variants in three independent regions, which exceeded the threshold of genome-wide significance for association with eGFR decline (Figure 1, Table 2, Supplemental Figure 1A, Supplemental Table 2, Supplemental Table 3). The strongest association was with rs77924615, intronic in the UMOD/PDILT region (P = 4.9×10−27; Table 2, Supplemental Figure 2, Supplemental Table 1). Conditional analysis of variants within the UMOD/PDILT region (with adjustment for the sentinel variant) did not reveal any additional independent genome-wide significant loci within the gene region. After adjustment for variation at rs77924615, the next most significant locus within the UMOD/PDILT region was rs36060036 (P = 1.04×10−4). The rs77924615 variant showed similar magnitudes of effect among Black participants (−0.20% faster eGFR decline per additional G allele), but was not statistically significantly associated with eGFR decline, either overall or within subgroups with diabetes (Table 2 and Supplemental Figure 3). Overall correlation between longitudinal and cross-sectional data eGFR associations was ρ=0.39 (Supplemental Figure 4).
Figure 1.
Manhattan plot of the strength of association of genetic variants with eGFR decline (%/yr) in cross-ancestry analyses among individuals with CKD. The y axis represents −log10 P-values for a linear mixed model of genetic variant dosage on repeated log-transformed eGFR measurements, adjusted for age, sex, and first ten principal components of ancestry, stratified by diabetes at baseline and ethnicity, and then meta-analyzed for overall cross-ancestry results. The x axis indicates the chromosomal position of each SNP. A dotted red line marks the P = 1×10−8 threshold.
Table 2.
Associations of independent genetic variants with relative slope of eGFR
| Variant Characteristics | Cross-Ancestry Analysis | White Participants | Black Participants | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifier | Nearest Gene | Chr | Position | EA | OA | EAF | β | SEM | P | EAF | β | SEM | P | EAF | β | SEM | P | ||||
| Overall | |||||||||||||||||||||
| rs77924615 | UMOD-PDILT | 16 | 20392332 | G | A | 0.80 | −0.30 | 0.03 | 4.9×10−27 | 0.78 | −0.30 | 0.03 | 7.2×10−27 | 0.81 | −0.20 | 0.20 | 0.32 | ||||
| rs11592748 | BICC1 | 10 | 60284915 | A | G | 0.57 | 0.13 | 0.02 | 5.6×10−9 | 0.55 | 0.13 | 0.02 | 2.4×10−8 | 0.24 | 0.21 | 0.10 | 0.07 | ||||
| rs16996674 | APOL1/MYH9 | 22 | 36726652 | T | C | 0.26 | −0.57 | 0.10 | 2.1×10−8 | 0.00 | — | — | — | 0.26 | −0.57 | 0.10 | 2.1×10−8 | ||||
| Participants with diabetes | |||||||||||||||||||||
| rs77924615 | UMOD-PDILT | 16 | 20392332 | G | A | 0.80 | −0.45 | 0.06 | 2.2×10−13 | 0.80 | −0.46 | 0.06 | 1.5×10−13 | 0.94 | −0.10 | 0.40 | 0.81 | ||||
| rs11624911 | HEATR4 | 14 | 74026568 | A | C | 0.18 | 0.02 | 0.06 | 0.75 | 0.20 | −0.02 | 0.06 | 0.69 | 0.03 | 0.29 | 0.05 | 1.3×10−8 | ||||
| Participants without diabetes | |||||||||||||||||||||
| rs36060036 | UMOD-PDILT | 16 | 20361950 | T | C | 0.16 | 0.27 | 0.03 | 1.9×10−17 | 0.16 | 0.27 | 0.03 | 2.4×10−18 | 0.07 | 0.32 | 0.21 | 0.12 | ||||
| rs11592748 | BICC1 | 10 | 60284915 | A | G | 0.57 | 0.14 | 0.02 | 6.7×10−9 | 0.56 | 0.14 | 0.03 | 3.0×10−8 | 0.78 | 0.25 | 0.13 | 0.06 | ||||
β from a linear mixed model of genetic variant dosage on repeated log-transformed eGFR measurements, adjusted for age, sex, and first ten principal components of ancestry, stratified by ethnicity and diabetes, and then meta-analyzed for overall cross-ancestry results. EA, effect allele; OA, other allele; EAF, effect allele frequency; β, regression estimate, difference in % decline/yr per additional copy of the effect allele; SEM, standard error of the mean.
We additionally identified an intronic variant in BICC1 (rs11592748), which was significantly associated with eGFR slope (Figure 1, Supplemental Figure 5). Every additional A allele at the locus was associated with a 0.13% slower slope in eGFR (P = 5.6×10−9). Variation at the third genome-wide significant polymorphism, intronic within the APOL1/MYH9 region (rs16996674) and present only among Black participants, was associated with a 0.57% faster eGFR slope (P = 2.1×10−8) (Figure 1, Supplemental Figure 6).
Conditional analyses of the BICC1 and APOL1 regions did not identify any secondary association signals, indicating no additional independently associated SNPs after conditioning on the region's lead SNP. In aggregate, the loci (rs77924615, rs11592748 and rs16996674) explained 0.20% of the variance in eGFR slope, and associations replicated at P < 0.003 in the analysis by Gorski et al. (Supplemental Table 4).26 The amount of variance in relative eGFR slope explained by the joint effect of all genetic variants (SNP heritability, h2 SNP) was 2.4% among participants without diabetes and 3.0% among those with diabetes.
GWAS Stratified by Diabetes
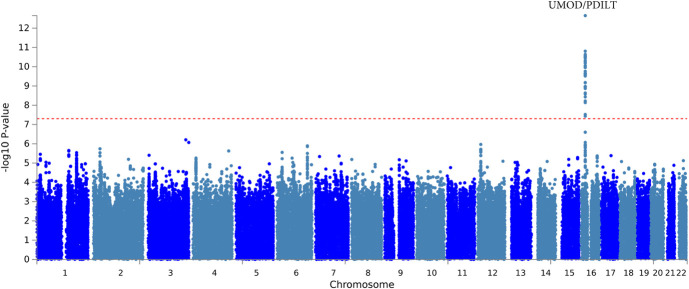
Cross-ancestry meta-analysis of participants with diabetes (n=46,424) uncovered 27 variants from one gene region, UMOD/PDILT, associated with decline in kidney function at genome-wide significance (Figure 2, Supplemental Figure 7). The genetic variant with the strongest independent association, rs77924615, is intronic in PDILT; every additional minor allele of G was associated with a 0.45%/yr faster decline in eGFR (P = 2.24×10−13; Table 2, Supplemental Table 5). Among White participants with diabetes, rs77924615 was also the strongest independent association (P = 1.54×10−13; Table 2, Supplemental Table 6). This variant was not associated with decline in kidney function among Black participants with diabetes (P = 0.81; Table 2). Among Black participants with diabetes, the top independently associated locus was at rs11624911, upstream of HEATR4, for which every additional copy of the A allele was associated with a 2.94%/yr slower decline in eGFR (P = 1.30×10−8; Supplemental Figure 8, Supplemental Table 7). Results were similar for eGFR slope characterized on the absolute scale in ml/min per 1.73 m2 per year (Supplemental Table 8, Supplemental Table 9, Supplemental Table 10).
Figure 2.
Manhattan plot of the strength of association of genetic variants with eGFR decline (%/yr) in cross-ancestry analyses among individuals with CKD and without diabetes. The y axis represents −log10 P-values for a linear mixed model of genetic variant dosage on repeated log-transformed eGFR measurements among individuals without diabetes, adjusted for age, sex, and first ten principal components of ancestry, stratified by ethnicity, and then meta-analyzed for overall cross-ancestry results. The x axis indicates the chromosomal position of each SNP. A dotted red line marks the P = 1×10−8 threshold.
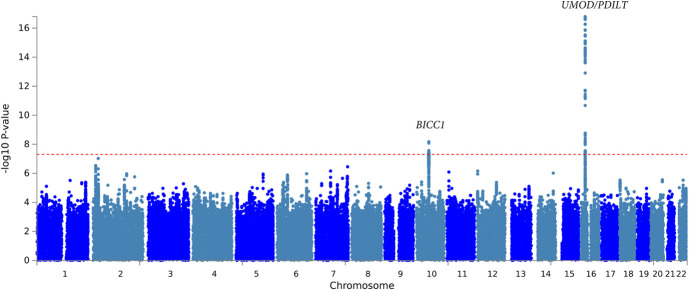
Among participants without diabetes, the UMOD/PDILT region again housed the top independent variant, rs36060036, associated with a 0.27%/yr faster decline in eGFR per copy of the T allele, in cross-ancestry analysis (P = 1.90×10−17; Figure 3, Table 2, Supplemental Figure 9, Supplemental Table 11). In addition, each copy of the minor allele of rs11592748, intronic in BICC1, was associated with a 0.14%/yr slower decline in eGFR in the cross-ancestry analysis of individuals without diabetes (P = 6.73×10−9; Table 2, Supplemental Figure 9, Supplemental Table 11). Among non-Hispanic White participants without diabetes, the top independent variant was also rs36060036, intronic in UMOD, where every minor allele was associated with a 0.27%/yr slower decline in eGFR (P = 2.4×10−17; Table 2, Supplemental Table 12). No variants reached genome-wide significance among Black participants without diabetes (Supplemental Table 13). Results were similar for eGFR slope characterized on the absolute scale in ml/min per 1.73 m2 per year (Supplemental Table 14, Supplemental Table 15, Supplemental Table 16).
Figure 3.
Manhattan plot of the strength of association of genetic variants with eGFR decline (%/yr) in cross-ancestry analyses among individuals with CKD and with diabetes. The y axis represents −log10 P-values for a linear mixed model of genetic variant dosage on repeated log-transformed eGFR measurements among individuals with diabetes, adjusted for age, sex, and first ten principal components of ancestry, stratified by ethnicity, and then meta-analyzed for overall cross-ancestry results. The x axis indicates the chromosomal position of each SNP. A dotted red line marks the P = 1×10−8 threshold.
Candidate Genetic Variant Analysis
We next evaluated genetic variants with previously reported associations with decline in kidney function. All previously reported UMOD/PDILT lead variants were significantly associated with eGFR slope in our cohorts (Table 3). In addition, we replicated previously reported findings at PRKAG2, FGF5, and C15ORF54 in cross-ancestry analyses of individuals with and without diabetes (Supplemental Figures 11–13).
Table 3.
Association of candidate variants with eGFR decline (%/yr)
| Variant Characteristics | Previously Reported Effect Estimate (Gorski et al.) | Cross-Ancestry Estimate from Current Meta-Analysis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifier | Nearest Gene | Chr | Position | EA | OA | EAF | β a | P | EAF | β b | SEM | P | |
| rs34882080 | UMOD-PDILT | 16 | 20361441 | A | G | 0.82 | 0.092 | 3.3×10−62 | 0.80 | −0.27 | 0.03 | 7.3×10−24 | |
| rs77924615 | UMOD-PDILT | 16 | 20392332 | G | A | 0.80 | 0.099 | 3.8×10−69 | 0.78 | −0.30 | 0.03 | 4.9×10−27 | |
| rs10254101 | PRKAG2 | 7 | 151415536 | T | C | 0.28 | 0.037 | 1.8×10−14 | 0.29 | −0.10 | 0.02 | 8.2×10−6 | |
| rs1028455 | SPATA7 | 14 | 88829975 | T | A | 0.66 | 0.024 | 3.4×10−8 | 0.65 | −0.01 | 0.02 | 0.545 | |
| rs1458038 | FGF5 | 4 | 81164723 | C | T | 0.69 | 0.028 | 6.9×10−10 | 0.70 | −0.09 | 0.02 | 4.1×10−5 | |
| rs4930319 | OVOL1 | 11 | 65555458 | C | G | 0.33 | 0.028 | 5.3×10−10 | 0.35 | −0.03 | 0.02 | 0.098 | |
| rs434215 | TPPP | 5 | 699046 | A | G | 0.28 | 0.032 | 7.2×10−9 | 0.30 | −0.07 | 0.02 | 4.8×10−3 | |
| rs28857283 | C15ORF54 | 15 | 39224711 | G | A | 0.66 | 0.030 | 1.3×10−11 | 0.63 | −0.07 | 0.02 | 7.5×10−4 | |
| rs13095391 | ACVR2B | 3 | 38447232 | A | C | 0.50 | 0.025 | 4.0×10−8 | 0.53 | −0.03 | 0.02 | 0.181 | |
| rs9998485 | SHROOM3 | 4 | 77362445 | A | G | 0.47 | 0.027 | 9.8×10−9 | 0.50 | −0.05 | 0.02 | 0.014 | |
| rs1047891 | CPS1 | 2 | 211540507 | A | C | 0.29 | 0.029 | 1.2×10−9 | 0.33 | −0.03 | 0.02 | 0.209 | |
| rs2453533 | GATM | 15 | 45641225 | A | C | 0.42 | 0.029 | 1.7×10−11 | 0.40 | −0.02 | 0.02 | 0.412 | |
Because of the differing phenotype definitions between the two studies, a positive regression β from the analysis by Gorski et al. indicates a faster eGFR decline, whereas in our analyses a negative β indicates a faster eGFR decline. EA, effect allele; OA, other allele; EAF, effect allele frequency; β, regression estimate, difference in % decline/yr per additional copy of the effect allele; SEM, standard error of the mean.
Difference in −1×eGFR decline, ml/min per 1.72 m2 per year, per copy of the effect allele, per Gorski et al. PMID 35716955.
Difference in the relative annual eGFR decline, %/yr, per copy of the effect allele.
APOL1 Risk Variants
The presence of two high-risk APOL1 variants was associated with a 1.3%/yr faster decline in eGFR among non-Hispanic Black participants without diabetes, relative to those with no high-risk variants (P = 2×10−11) (Supplemental Figure 14, Supplemental Table 17). Among non-Hispanic Black participants with diabetes, the presence of two high-risk APOL1 variants was associated with a 1.2%/yr faster decline in eGFR (P = 5.6×10−4) (Supplemental Figure 14, Supplemental Table 18).
Discussion
We undertook a large-scale GWAS meta-analysis of longitudinal change in eGFR among individuals with established CKD of African and European ancestry, stratified by diabetes status. Our data identified two loci associated with CKD progression and confirmed associations described with eGFR decline for UMOD/PDILT and APOL1 across the spectrum of eGFR, including those starting from a normal-range eGFR. In addition, we replicated previous findings in PRKAG2, FGF5, and C15ORF54.
The UMOD/PDILT region was the only one significantly associated at genome-wide significance with eGFR decline among diabetic participants with CKD in cross-ancestry analyses. The top variant in the region, rs77924615, residing in PDILT, has been previously identified as strongly associated with several kidney phenotypes. It was recently reported as the strongest variant associated with cross-sectional eGFR and ≥25% decline in creatinine-based eGFR among individuals from the general population without CKD.11 In analyses of kidney function from over a million individuals, the variant was associated with 19% lower odds of CKD (odds ratio; 95% confidence interval, 0.80 to 0.83).11 In addition, several lines of evidence support rs77924615 as a causal regulatory variant for UMOD, including associations with UMOD gene expression in tubular cells28,29 and uromodulin protein levels in urine.5 Furthermore, it has been recently shown that there is a differential effect of UMOD on eGFR that it is much larger in individuals with diabetes.30 Although this is not fully understood, it has been postulated that UMOD in the context of glucosuria has a large effect in the tubule-glomerular feedback and in hyperfiltration. Among individuals of African ancestry with diabetes and CKD, rs11624911, upstream of HEATR4, was associated with large differences in eGFR decline. This gene region has previously been reported to be associated with cross-sectional eGFR among individuals without diabetes10 and the variant rs11626972 in linkage disequilibrium with rs11624911 with urinary metabolite levels (X-13671) in CKD.31
We identified two gene regions associated with eGFR decline among individuals with nondiabetic CKD. The UMOD/PDILT region again was identified as the strongest signal in this population. In addition, the minor allele at the rs11592748 locus within the BICC1 gene region was associated with slower annual decline in eGFR. BICC1 is highly abundant in the proximal tubule in mice and in humans, is an RNA-binding protein, and is considered a key player in post-transcriptional gene regulation.29 Variants within the gene region have been associated with adult polycystic kidney disease and other cystic diseases of the kidney both in humans and animal models.32,33 In addition, Kraus et al. found two heterozygous loss-of-function mutations that lead to renal dysplasia in children.34 As expected, BICC1 has also been associated with other markers of kidney functions, such as blood urea nitrogen,5 or related kidney traits, such as serum urate levels.35–38
The largest GWAS of longitudinal change in eGFR, among 343,339 individuals from 62 studies with eGFR assessed twice longitudinally, identified 12 variants in 11 loci significantly associated with annual eGFR decline.26 Of these 12 variants, seven were associated with relative annualized eGFR decline in our cross-ancestry analyses, at a nominal P-value. The failure to replicate the remainder of previously reported associations between genetic variants and CKD progression is consistent with findings from attempted replications in other complex diseases.39 Some of the previous reports of association may have given false-positive results. However, more likely explanations for the discordance in findings on replication include differences among samples because of ancestral heterogeneity, disease heterogeneity, longer disease duration at initial examination, and phenotype definition and ascertainment.
Strengths of this study include the analysis of a large sample size with relatively long follow-up and excellent participant retention. The MVP cohort includes an ancestrally diverse population that is particularly susceptible to CKD because of a higher prevalence of diabetes and hypertension, making it an ideal group in which to identify genetic risk of CKD progression.40 Our study also has some important limitations. Although electronic health records provide a rich source of clinical data, the acquired creatinine data could be an inaccurate reflection of the entirety of the CKD spectrum. Outpatient testing of serum creatinine can be triggered by acute illnesses or monitoring of chronic diseases, which introduces potential bias. Missing eGFR values were introduced when participants were censored because of ESKD. The variable censoring and record length could have biased our study results, with healthier individuals contributing more data to the analysis. To help mitigate this bias, one could implement a joint analysis of longitudinal (eGFR) and survival (time to kidney failure or transplant) and develop weighting schemes on the basis of record lengths to avoid this type of selection bias. Such sophisticated statistical methods are computationally demanding and not currently available for GWASs. The number of participants in subgroups of African ancestry and diabetes was relatively small, limiting our power to detect subgroup-specific significant genome-wide associations. In addition, the findings of this study cannot be generalized to include people of other ethnic groups, such as those of Hispanic/Latino or Asian ancestry. A further important limitation, inherent to the MVP data set, is the large predominance of male participants (>95%). While women are included in this analysis, potential interactions between sex, phenotype, and genotype could not be addressed. Finally, because of statistical considerations, the genetic variants examined in our study were limited to those located on the autosomes. We may have missed important genetic associations with CKD progression that are located on the sex chromosomes.
Few new therapies have been developed to prevent or treat CKD progression over the past two decades, underscoring the need to identify and understand the mechanisms involved, and GWASs can potentially provide novel biological insights into disease pathophysiology. Our findings suggest that there is some, but not complete, overlap between the genetic architecture of CKD risk and CKD progression. It is possible that in the context of CKD, acquired changes in the metabolic milieu have larger relative roles in disease progression compared with genetic determinants or that genetic susceptibility to CKD progression has risk factors with smaller effect sizes. Nonetheless, GWASs using longitudinal data enabled the discovery of novel genetic variants that may play an important role in disease progression. Future work to understand the pathophysiology of CKD progression could be directed toward functional characterization of our findings, polygenic risk scores for risk stratification, and beyond GWASs toward an examination of gene-by-gene and gene-by-environment joint effects.
Supplementary Material
Footnotes
See related editorial, “Incorporating Linear Mixed Models into GWAS of Kidney Function Decline,” on pages 1473–1475.
Disclosures
A. Bick reports Ownership Interest: TenSixteen Bio. C.P. Chung reports Advisory or Leadership Role: Clinical Pharmacology and Therapeutics, Arthritis Care and Research, and Clinical Rheumatology. A.M. Hung reports Consultancy: NHLBI consultant for Gene and life interaction grant; Research Funding: VHA CSR&D Merit “Genetics of Kidney Disease & Hypertension, Risk Prediction and Drug Response,” Vertex Grant to VUMC; and Advisory or Leadership Role: Co-Chair Million Veteran Program Publications & Presentation committee for Veterans Affairs, Co-chair Pharmacogenomics for COVID-19 Million Veteran Program, Journal of renal nutrition, Section Editor Clinical Nephrology, Standing member SRC HSR&D bioinformatics, Ad-hoc SRC NHLBI, Ad-hoc Scientific Review Committee CSR&D, Ad-Scientific Review Committee KNOD. T.A. Ikizler reports Consultancy: Abbott Renal Care, Fresenius–Kabi, La Renon, Nestle; Honoraria: Fresenius–Kabi, Abbott Renal Care, La Renon, Nestle; Patents or Royalties: Vanderbilt University Medical Center; and Advisory or Leadership Role: Kidney International. L. Lipworth reports Research Funding: EpidStrategies. M.E. Matheny reports Consultancy: NIH-VA-DoD Pain Management Grant Consortium (PMC3); and Advisory or Leadership Role: SMRB Study Section, VA HSR&D, Informatics & Methods Section, Steering Committee—Indianapolis VA HSR&D COIN Center, Steering Committee—VA HSR&D VIREC, Steering Committee—Salt Lake City VA HSR&D COIN Center, and VA ORD Million Veterans Program Executive Steering Committee. C. Robinson-Cohen reports Advisory or Leadership Role: Clinical Journal of the American Society of Nephrology Editorial Board Member, and Clinical Nephrology Genetics Section Editor. E.D. Siew reports Ownership Interest: Amazon stock, Apple stock; Patents or Royalties: Author for UptoDate (royalties); and Advisory or Leadership Role: Editorial board of CJASN. All remaining authors have nothing to disclose.
Funding
This work was supported by the CSR&D Merit award title Genetics of CKD and Hypertension–Risk Prediction and Drug Response in the MVP (CX001897 to AMH), the National Institutes of Diabetes and Digestive and Kidney Diseases grant R01DK122075 (to C. Robinson-Cohen) and by BIRCHWH funding K12HD043483 (to J.N. Hellwege). A subset of the data sets used for the analyses described was obtained from Vanderbilt University Medical Center's BioVU, which is supported by numerous sources: institutional funding, private agencies, and federal grants. These include the National Institutes of Health–funded Shared Instrumentation Grant S10RR025141 and Clinical and Translational Science Award grants UL1TR002243, UL1TR000445, and UL1RR024975. Genomic data are also supported by investigator-led projects that include U01HG004798, R01NS032830, RC2GM092618, P50GM115305, U01HG006378, U19HL065962, R01HD074711, and additional funding sources listed at https://victr.vanderbilt.edu/pub/biovu/. This research is also based on data from the Million Veteran Program, Veterans Health Administration's Office of Research and Development, Veterans Health Administration. This publication does not represent the views of the Department of Veteran Affairs or the US Government. Acknowledgment of the Million Veteran Program leadership and staff contributions can be found in the supplemental material titled “MVP core acknowledgments.”
Author Contributions
Conceptualization: Alexander Bick, J. Michael Gaziano, Jacklyn N. Hellwege, Adriana M. Hung, T. Alp Ikizler, Loren Lipworth, Cassianne Robinson-Cohen, Edward D. Siew, Ran Tao.
Data curation: Hua C. Chen, Cecilia P. Chung, J. Michael Gaziano, Jing He, Adriana M. Hung, Michael E. Matheny, Cassianne Robinson-Cohen, Bryce Rowan, Ran Tao, Wei-qi Wei, Otis D. Wilson, Neil S. Zheng.
Formal analysis: Hua C. Chen, Cecilia P. Chung, Jing He, Jacklyn N. Hellwege, Adriana M. Hung, Michael E. Matheny, Cassianne Robinson-Cohen, Bryce Rowan, Edward D. Siew, Ran Tao, Jefferson L. Triozzi, Otis D. Wilson, Neil S. Zheng.
Funding acquisition: J. Michael Gaziano, Adriana M. Hung, Cassianne Robinson-Cohen, Philip S. Tsao.
Investigation: Adriana M. Hung, Cassianne Robinson-Cohen, Edward D. Siew, Ran Tao, Jefferson L. Triozzi, Wei-qi Wei, Neil S. Zheng.
Methodology: Hua C. Chen, Adriana M. Hung, Michael E. Matheny, Cassianne Robinson-Cohen, Bryce Rowan, Ran Tao, Wei-qi Wei, Neil S. Zheng.
Project administration: Cecilia P. Chung, Adriana M. Hung, T. Alp Ikizler, Cassianne Robinson-Cohen, Ran Tao, Otis D. Wilson.
Resources: J. Michael Gaziano, Jacklyn N. Hellwege, Adriana M. Hung, T. Alp Ikizler, Cassianne Robinson-Cohen, Edward D. Siew, Philip S. Tsao, Otis D. Wilson.
Software: Adriana M. Hung, Ran Tao.
Supervision: Alexander Bick, Cecilia P. Chung, J. Michael Gaziano, Adriana M. Hung, T. Alp Ikizler, Loren Lipworth, Michael E. Matheny, Cassianne Robinson-Cohen, Edward D. Siew, Ran Tao, Philip S. Tsao, Wei-qi Wei.
Validation: Adriana M. Hung, Michael E. Matheny, Ran Tao.
Visualization: Hua C. Chen, Adriana M. Hung, Cassianne Robinson-Cohen, Ran Tao, Neil S. Zheng.
Writing – original draft: Hua C. Chen, Jing He, Adriana M. Hung, Cassianne Robinson-Cohen, Bryce Rowan, Ran Tao, Jefferson L. Triozzi.
Writing – review & editing: Alexander Bick, Cecilia P. Chung, J. Michael Gaziano, Jacklyn N. Hellwege, Adriana M. Hung, T. Alp Ikizler, Loren Lipworth, Michael E. Matheny, Cassianne Robinson-Cohen, Edward D. Siew, Ran Tao, Jefferson L. Triozzi, Philip S. Tsao, Wei-qi Wei, Otis D. Wilson, Neil S. Zheng.
Data Sharing Statement
The summary GWAS data can be accessed by request to the corresponding author and will be available at the GWAS Catalog (https://www.ebi.ac.uk/gwas/) upon publication.
Supplemental Material
This article contains the following supplemental material online at http://links.lww.com/JSN/E464, http://links.lww.com/JSN/E446, http://links.lww.com/JSN/E447, http://links.lww.com/JSN/E448, http://links.lww.com/JSN/E449, http://links.lww.com/JSN/E450, http://links.lww.com/JSN/E451, http://links.lww.com/JSN/E452, http://links.lww.com/JSN/E453, http://links.lww.com/JSN/E454, http://links.lww.com/JSN/E455, http://links.lww.com/JSN/E456, http://links.lww.com/JSN/E457, http://links.lww.com/JSN/E458, http://links.lww.com/JSN/E459, http://links.lww.com/JSN/E460, http://links.lww.com/JSN/E461, http://links.lww.com/JSN/E462, http://links.lww.com/JSN/E463.
Supplemental Figure 1. Quantile-Quantile plot of associations of single nucleotide polymorphisms with relative slope of eGFR, among (A) cross-ancestry analyses, (B) White participants, and (C) Black participants.
Supplemental Figure 2. Regional interrogation of the UMOD/PDILT locus, cross-ancestry analysis.
Supplemental Figure 3. Regional interrogation of the UMOD/PDILT locus among Black participants.
Supplemental Figure 4. Comparison between longitudinal and cross-sectional eGFR data.
Supplemental Figure 5. Regional interrogation of the BICC1 locus, cross-ancestry analysis. Plot shows −log10(P-value) on the left y axis and genomic location (in base pairs) on the x axis.
Supplemental Figure 6. Regional interrogation of the APOL1 locus among Black participants. Plot shows −log10(P-value) on the left y axis and genomic location (in base pairs) on the x axis.
Supplemental Figure 7. Regional interrogation of the UMOD/PDILT locus, cross-ancestry analysis among individuals with diabetes.
Supplemental Figure 8. Regional interrogation of the HEATR4 locus among Black individuals with diabetes.
Supplemental Figure 9. Regional interrogation of the UMOD/PDILT locus from transethnic analyses among individuals without diabetes.
Supplemental Figure 10. Regional interrogation of the BICC1 locus from cross-ancestry analyses among individuals without diabetes.
Supplemental Figure 11. Regional interrogation of the PRKAG2 locus from cross-ancestry analyses. Plot shows −log10(P-value) on the left y axis and genomic location (in base pairs) on the x axis.
Supplemental Figure 12. Regional interrogation of the FGF5 locus from cross-ancestry analyses.
Supplemental Figure 13. Regional interrogation of the C15ORF54 locus from cross-ancestry analyses.
Supplemental Figure 14. Violin plots of distribution of relative eGFR by APOL1 inheritance model among Black participants without (A) and with (B) diabetes.
Supplemental Figure 15. Manhattan plot of the strength of association of genetic variants with eGFR decline (%/yr) among White individuals with CKD.
Supplemental Figure 16. Manhattan plot of the strength of association of genetic variants with eGFR decline (%/yr) among Black individuals with CKD.
Supplemental Table 1. Associations of single nucleotide polymorphisms with relative slope of eGFR, cross-ancestry analyses (P < 5×10−8).
Supplemental Table 2. Associations of single nucleotide polymorphisms with relative slope of eGFR, among White individuals (P < 5×10−6).
Supplemental Table 3. Associations of single nucleotide polymorphisms with relative slope of eGFR, among Black individuals (P < 5×10−6).
Supplemental Table 4. Associations of independent genetic variants with relative slope of eGFR in summary statistics by Gorski et al. (PMID 35716955).
Supplemental Table 5. Associations of single nucleotide polymorphisms with relative slope of eGFR, among individuals with diabetes, cross-ancestry analyses (P < 5×10−6).
Supplemental Table 6. Associations of single nucleotide polymorphisms with relative slope of eGFR, among White individuals with diabetes (P < 5×10−6).
Supplemental Table 7. Associations of single nucleotide polymorphisms with relative slope of eGFR, among Black individuals with diabetes (P < 5×10−6).
Supplemental Table 8. Associations of single nucleotide polymorphisms with absolute slope of eGFR, among individuals with diabetes, cross-ancestry analyses (P < 5×10−6).
Supplemental Table 9. Associations of single nucleotide polymorphisms with absolute slope of eGFR, among White individuals with diabetes (P < 5×10−6).
Supplemental Table 10. Associations of single nucleotide polymorphisms with absolute slope of eGFR, among Black individuals with diabetes (P <5×10−6).
Supplemental Table 11. Associations of single nucleotide polymorphisms with relative slope of eGFR, among individuals without diabetes, cross-ancestry analyses (P < 5×10−6).
Supplemental Table 12. Associations of single nucleotide polymorphisms with relative slope of eGFR, among White individuals without diabetes (P < 5×10−6).
Supplemental Table 13. Associations of single nucleotide polymorphisms with relative slope of eGFR, among Black individuals without diabetes (P <5×10−6).
Supplemental Table 14. Associations of single nucleotide polymorphisms with absolute slope of eGFR, among individuals without diabetes, cross-ancestry analyses (P < 5×10−6).
Supplemental Table 15. Associations of single nucleotide polymorphisms with absolute slope of eGFR, among White individuals without diabetes (P < 5×10−6).
Supplemental Table 16. Associations of single nucleotide polymorphisms with absolute slope of eGFR, among Black individuals without diabetes (P < 5×10−6).
Supplemental Table 17. Association of APOL1 high-risk variants with eGFR slope among Black individuals without diabetes at baseline.
Supplemental Table 18. Association of APOL1 high-risk variants with eGFR slope among Black individuals with diabetes at baseline.
MVP core acknowledgments: Acknowledgment of the Million Veteran Program leadership and staff contributions can be found in the supplementary material entitled.
References
- 1.Go AS, Chertow GM, Fan D, McCulloch CE, Hsu C-Y. Chronic kidney disease and the risks of death, cardiovascular events, and hospitalization. N Engl J Med. 2004;351(13):1296–1305. doi: 10.1056/NEJMoa041031 [DOI] [PubMed] [Google Scholar]
- 2.Denker M Boyle S Anderson AH, et al. Chronic renal insufficiency cohort study (CRIC): overview and summary of selected findings. Clin J Am Soc Nephrol. 2015;10(11):2073–2083. doi: 10.2215/CJN.04260415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pattaro C Teumer A Gorski M, et al. Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nat Commun. 2016;7(1):10023. doi: 10.1038/ncomms10023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee J, Lee Y, Park B, Won S, Han JS, Heo NJ. Genome-wide association analysis identifies multiple loci associated with kidney disease-related traits in Korean populations. PLoS One. 2018;13(3):e0194044. doi: 10.1371/journal.pone.0194044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wuttke M Li Y Li M, et al. A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat Genet. 2019;51(6):957–972. doi: 10.1038/s41588-019-0407-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graham SE Nielsen JB Zawistowski M, et al. Sex-specific and pleiotropic effects underlying kidney function identified from GWAS meta-analysis. Nat Commun. 2019;10(1):1847. doi: 10.1038/s41467-019-09861-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorski M van der Most PJ Teumer A, et al. 1000 Genomes-based meta-analysis identifies 10 novel loci for kidney function. Sci Rep. 2017;7(1):45040. doi: 10.1038/srep45040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahajan A Rodan AR Le TH, et al. Trans-ethnic fine mapping highlights kidney-function genes linked to Salt sensitivity. Am J Hum Genet. 2016;99(3):636–646. doi: 10.1016/j.ajhg.2016.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wojcik GL Graff M Nishimura KK, et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature. 2019;570(7762):514–518. doi: 10.1038/s41586-019-1310-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hellwege JN Velez Edwards DR Giri A, et al. Mapping eGFR loci to the renal transcriptome and phenome in the VA Million Veteran Program. Nat Commun. 2019;10(1):3842. doi: 10.1038/s41467-019-11704-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorski M Jung B Li Y, et al. Meta-analysis uncovers genome-wide significant variants for rapid kidney function decline. Kidney Int. 2021;99(4):926–939. doi: 10.1016/j.kint.2020.09.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson AH Xie D Wang X, et al. Novel risk factors for progression of diabetic and nondiabetic CKD: findings from the chronic renal insufficiency cohort (CRIC) study. Am J Kidney Dis. 2021;77(1):56–73.e1. doi: 10.1053/j.ajkd.2020.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parsa A Kanetsky PA Xiao R, et al. Genome-wide association of CKD progression: the chronic renal insufficiency cohort study. J Am Soc Nephrol. 2017;28(3):923–934. doi: 10.1681/ASN.2015101152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaziano JM Concato J Brophy M, et al. Million Veteran Program: a mega-biobank to study genetic influences on health and disease. J Clin Epidemiol. 2016;70:214–223. doi: 10.1016/j.jclinepi.2015.09.016 [DOI] [PubMed] [Google Scholar]
- 15.Ritchie MD Denny JC Crawford DC, et al. Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet. 2010;86(4):560–572. doi: 10.1016/j.ajhg.2010.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pendergrass S, Dudek SM, Roden DM, Crawford DC, Ritchie MD. Visual integration of results from a large DNA biobank (BioVU) using synthesis-view. Pac Symp Biocomput. 2011:265–275. doi: 10.1142/9789814335058_0028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loh PR Danecek P Palamara PF, et al. Reference-based phasing using the haplotype reference Consortium panel. Nat Genet. 2016;48(11):1443–1448. doi: 10.1038/ng.3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Auton A Abecasis GR Altshuler DM, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Browning BL, Browning SR. Genotype imputation with millions of reference samples. Am J Hum Genet. 2016;98(1):116–126. doi: 10.1016/j.ajhg.2015.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang H Hui Q Lynch J, et al. Harmonizing genetic ancestry and self-identified race/ethnicity in genome-wide association studies. Am J Hum Genet. 2019;105(4):763–772. doi: 10.1016/j.ajhg.2019.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abraham G, Inouye M. Fast principal component analysis of large-scale genome-wide data. PLoS One. 2014;9(4):e93766. doi: 10.1371/journal.pone.0093766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inker LA Schmid CH Tighiouart H, et al. Estimating glomerular filtration rate from serum creatinine and cystatin C. N Engl J Med. 2012;367(1):20–29. doi: 10.1056/NEJMoa1114248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pruim RJ Welch RP Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8(1):1826. doi: 10.1038/s41467-017-01261-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorski M Rasheed H Teumer A, et al. Genetic loci and prioritization of genes for kidney function decline derived from a meta-analysis of 62 longitudinal genome-wide association studies. Kidney Int. 2022;102(3):624–639. doi: 10.1016/j.kint.2022.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stanzick KJ Li Y Schlosser P, et al. Discovery and prioritization of variants and genes for kidney function in >1.2 million individuals. Nat Commun. 2021;12(1):4350. doi: 10.1038/s41467-021-24491-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gadegbeku CA Gipson DS Holzman LB, et al. Design of the Nephrotic Syndrome Study Network (NEPTUNE) to evaluate primary glomerular nephropathy by a multidisciplinary approach. Kidney Int. 2013;83(4):749–756. doi: 10.1038/ki.2012.428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu C Huang S Park J, et al. Renal compartment-specific genetic variation analyses identify new pathways in chronic kidney disease. Nat Med. 2018;24(11):1721–1731. doi: 10.1038/s41591-018-0194-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winkler TW Rasheed H Teumer A, et al. Differential and shared genetic effects on kidney function between diabetic and non-diabetic individuals. Commun Biol. 2022;5(1):580. doi: 10.1038/s42003-022-03448-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlosser P Li Y Sekula P, et al. Genetic studies of urinary metabolites illuminate mechanisms of detoxification and excretion in humans. Nat Genet. 2020;52(2):167–176. doi: 10.1038/s41588-019-0567-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lian P Li A Li Y, et al. Loss of polycystin-1 inhibits Bicc1 expression during mouse development. PLoS One. 2014;9(3):e88816. doi: 10.1371/journal.pone.0088816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seufert L, Benzing T, Ignarski M, Muller RU. RNA-binding proteins and their role in kidney disease. Nat Rev Nephrol. 2021;18(3):153–170. doi: 10.1038/s41581-021-00497-1 [DOI] [PubMed] [Google Scholar]
- 34.Kraus MR Clauin S Pfister Y, et al. Two mutations in human BICC1 resulting in Wnt pathway hyperactivity associated with cystic renal dysplasia. Hum Mutat. 2012;33(1):86–90. doi: 10.1002/humu.21610 [DOI] [PubMed] [Google Scholar]
- 35.Boocock J Leask M Okada Y, et al. Genomic dissection of 43 serum urate-associated loci provides multiple insights into molecular mechanisms of urate control. Hum Mol Genet. 2020;29(6):923–943. doi: 10.1093/hmg/ddaa013 [DOI] [PubMed] [Google Scholar]
- 36.Sakaue S Kanai M Tanigawa Y, et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat Genet. 2021;53(10):1415–1424. doi: 10.1038/s41588-021-00931-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tin A Marten J Halperin Kuhns VL, et al. Target genes, variants, tissues and transcriptional pathways influencing human serum urate levels. Nat Genet. 2019;51(10):1459–1474. doi: 10.1038/s41588-019-0504-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sinnott-Armstrong N Tanigawa Y Amar D, et al. Genetics of 35 blood and urine biomarkers in the UK Biobank. Nat Genet. 2021;53(2):185–194. doi: 10.1038/s41588-020-00757-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet. 2001;29(3):306–309. doi: 10.1038/ng749 [DOI] [PubMed] [Google Scholar]
- 40.Selim AJ Berlowitz DR Fincke G, et al. The health status of elderly veteran enrollees in the Veterans Health Administration. J Am Geriatr Soc. 2004;52(8):1271–1276. doi: 10.1111/j.1532-5415.2004.52355.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The summary GWAS data can be accessed by request to the corresponding author and will be available at the GWAS Catalog (https://www.ebi.ac.uk/gwas/) upon publication.