We compared metabolomic profiles of pheasantshell (Ortmanniana pectorosa) from a reoccurring mass mortality event in the Clinch River, Virginia and Tennessee, USA, and identified potential biomarkers and metabolic pathways indicative of health status.
Keywords: hemolymph, metabolites, mussel die-off, pheasantshell, Unionida
Abstract
Biologists monitoring freshwater mussel (order Unionida) populations rely on behavioral, often subjective, signs to identify moribund (“sick”) or stressed mussels, such as gaping valves and slow response to probing, and they lack clinical indicators to support a diagnosis. As part of a multi-year study to investigate causes of reoccurring mortality of pheasantshell (Ortmanniana pectorosa; synonym Actinonaias pectorosa) in the Clinch River, Virginia and Tennessee, USA, we analyzed the hemolymph metabolome of a subset of mussels from the 2018 sampling period. Mussels at the mortality sites were diagnosed in the field as affected (case) or unaffected (control) based on behavioral and physical signs. Hemolymph was collected in the field by non-lethal methods from the anterior adductor muscle for analysis. We used ultra-high-performance liquid chromatography with quadrupole time-of-flight mass spectroscopy to detect targeted and untargeted metabolites in hemolymph and compared metabolomic profiles by field assessment of clinical status. Targeted biomarker analysis found 13 metabolites associated with field assessments of clinical status. Of these, increased gamma-linolenic acid and N-methyl-l-alanine were most indicative of case mussels, while adenine and inosine were the best indicators of control mussels. Five pathways in the targeted analysis differed by clinical status; two of these, purine metabolism and glycerophospholipid metabolism, were also indicated in the untargeted analysis. In the untargeted nalysis, 22 metabolic pathways were associated with clinical status. Many of the impacted pathways in the case group were catabolic processes, such as degradation of amino acids and fatty acids. Hierarchical clustering analysis matched clinical status in 72% (18 of 25) of mussels, with control mussels more frequently (5 of 16) not matching clinical status. Our study demonstrated that metabolomic analysis of hemolymph is suitable for assessing mussel condition and complements field-based indicators of health.
Introduction
Freshwater mussels (Order Unionida) are vital sentinels of aquatic ecosystem health. As infaunal suspension feeders, unionids stabilize substrate, remove particulates and contaminants, interconvert nutrients and transfer energy to other trophic levels (Vaughn, 2010, 2018). North America has the most diverse unionid fauna in the world, with 298 recognized species (Williams et al., 2017), the greatest diversity of which occurs in the southeast USA (Haag, 2012). Freshwater mussels are one of the most critically imperiled faunal groups, both globally and in the USA. (Lydeard et al., 2004; Régnier et al., 2009). In North America, 70% of species are considered endangered, threatened or vulnerable, and 29 species have become extinct in the past 100 years (Haag and Williams, 2014); in 2021, eight additional species were proposed for removal from the US Federal Lists of Endangered Species due to extinction (86 FR 54298, 2021). Widespread habitat degradation (including pollution, siltation, river channelization and impoundment) is cited as the primary cause of extinction over the past century (Downing et al., 2010; Haag, 2019), coupled with more recent introductions of invasive molluscs, Corbicula and Dreissena spp. (Strayer et al., 1999; McMahon and Bogan, 2001; Ferreira-Rodríguez et al., 2018). Still, the causes of most mussel population declines are ill-defined (Downing et al., 2010).
Since the 1970s, episodic mass mussel mortality events have been documented in relatively pristine systems with no direct specific environmental changes or events, such as contaminants or drought (Haag, 2019). Investigations into causal agents of these mortality events have been limited in scope and without definitive answers (Neves, 1986; Starliper et al., 2011; Waller and Cope, 2019). Complicating these efforts is the lack of diagnostic tools for objective assessment of mussel health and discovery of causal factors of mortality. Field biologists monitoring freshwater mussel populations rely on behavioral, often subjective, symptoms to identify “sick” or stressed mussels, such as gaping valves or an extended foot with a slow response to stimulus. Field assessment of “sick” or moribund mussels is seldom accompanied by clinical findings to confirm or support the diagnosis, primarily because health assessment tools are lacking.
We investigated a reoccurring mussel mortality event in the Clinch River in southwest Virginia and northeast Tennessee, USA, that primarily affected pheasantshell (Ortmanniana pectorosa; synonym: Actinonaias pectorosa). The Clinch River is a biodiversity hotspot with 46 extant species of freshwater mussels (Jones et al., 2014), 20 of which are on the US Fish and Wildlife Service Lists of Endangered Species. Although mussel richness and abundance in the upper river in Virginia steadily fell from 1979 to 2014, mussel densities in the Tennessee portion of the upper Clinch River increased from 1979 to 2014 (Jones et al., 2018). Pheasantshell (O. pectorosa) was historically abundant in the Clinch River system (Jones et al., 2014), but beginning in 2016, field biologists began reporting a disproportionate number of moribund and dead individuals of this species in the shoals near the Virginia and Tennessee state line. Mortality reoccurred in the fall months of 2017 to 2019, resulting in a 50–90% decline of pheasantshell population at monitoring sites in the lower river (Richard et al., 2020). We conducted a multi-year study of mussels in the Clinch River to determine the role of infectious agents in pheasantshell mortality. We applied metagenomic analysis of hemolymph and compared unaffected (control) and affected (case) mussels in 2017 and 2018 to detect potential infectious agents associated with mussel mortality (Leis et al., 2019; Richard et al., 2020, 2021; Knowles et al., 2022). Infectious agents associated with the mortality period in the Clinch River included a novel virus, Clinch densovirus 1, (Richard et al., 2020), and the bacteria Yokenella regensburgei and Aeromonas salmonicida (Leis et al., 2019; Richard et al., 2021). Bacterial metagenomic analysis also found fewer sequence variants and greater bacterial load in case compared to control mussels (Richard et al., 2021). Currently, the primary cause of mortality remains unclear. As part of the investigation, we analyzed hemolymph metabolites of a subset of pheasantshell that were sampled in 2018 from four sites in the Clinch River. We compared metabolomic profiles of mussels that were field diagnosed as control and case and investigated how closely hemolymph metabolomes matched these field-based clinical assessments and thus might enhance non-lethal assessments of mussel health.
Metabolomics is the detection and analysis of biologically relevant molecules, and the metabolome is the total set of metabolites in a biological system (Wishart et al., 2007; Zamboni et al., 2015). Metabolomic profiles can be used to characterize the health status of individual mussels, and pathway analysis of metabolites can reveal mechanisms of disease (Liu et al., 2013; Nguyen et al., 2018, 2019). Metabolomics has been used in marine bivalve studies of disease (de Lorgeril et al., 2018; Nguyen et al., 2019), aquaculture conditions (Young et al., 2015; Hao et al., 2018; Tian et al., 2021), vitamin supplementation (Yang et al., 2019) and environmental stressors (Cappello et al., 2017; Dumas et al., 2020). Hemolymph is the circulatory fluid of bivalves and has multiple functions including immunity, gas exchange, osmoregulation, nutrient distribution and waste elimination (Machałowski and Jesionowski, 2020). The composition and concentration of hemolymph constituents reflect not only the metabolic products of various tissues but also those of infectious agents and other symbionts. Additional metabolites may be obtained from water and diet (phytoplankton, bacteria). Within this complex metabolome, those metabolites that have low variability and occur in most of the population can indicate stable condition of individual mussels, whereas shifts in levels of these metabolites from the status quo could signal stress. Metabolomic studies of freshwater mussels are limited in number and scope, particularly of hemolymph. Hemolymph chemistry values were determined using an automated clinical chemistry analyzer for Elliptio complanata from 19 populations to establish reference ranges for eight constituents (Gustafson et al., 2005). Whereas proton nuclear magnetic resonance (1H NMR) spectroscopy yielded poor detection of hemolymph metabolites (Hurley-Sanders et al., 2016), gas chromatography–mass spectrometry (GC–MS) and liquid chromatography–mass spectrometry (LC–MS) detected shifts in hemolymph metabolites in Amblema plicata in response to captivity, food limitation (Roznere et al., 2014) and relocation (Roznere et al., 2017).
Here we apply LC–MS as a platform for targeted and untargeted metabolomics to characterize the metabolic profile of unaffected (control) and affected (case) mussels (based on field assessments of clinical status) and identify case-associated metabolites and pathways. Whether the infectious agents previously identified (e.g. densovirus, Y. regensburgei) in pheasantshell are the primary cause of mortality or indicative of environmental stressors that trigger mussel disease, defined as any deviation from normal function and structure, we expect metabolic differences between case and control mussels. Using both targeted and untargeted approaches to metabolite detection extended the scope of our analysis beyond that possible using a single approach. The targeted metabolomic results have higher confidence in the identification of specific metabolites, but the number of targeted metabolites is low, making affected pathway analysis difficult. The untargeted analysis relies on the MetaboAnalyst algorithm to identify potential metabolites based on molecular weight. The untargeted analysis detected many more metabolites than the targeted analysis and assigned those metabolites to specific pathways.
Materials and Methods
Field sampling
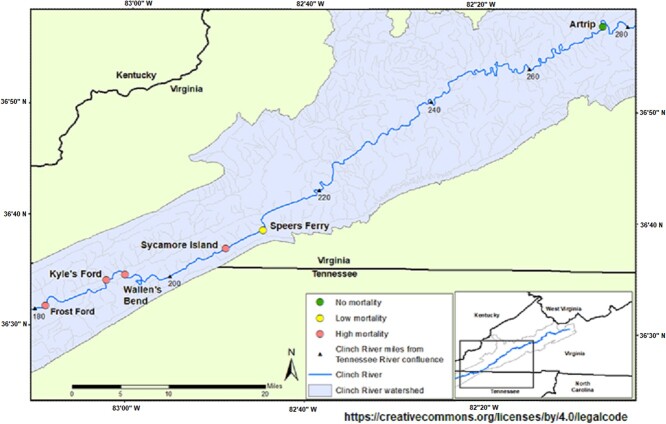
We sampled 30 pheasantshell from the Clinch River, including nine cases (from September and October of 2018) and 21 controls (from August, September and October 2018) at four sites (Fig. 1; Supplementary Table S1). Mussel samples were collected, and clinical status assessed as described by Richard et al. (2020). Briefly, during sampling, mussels were assigned to the control (apparently healthy, unaffected) group if they were firmly buried in the substrate, responded rapidly to tactile stimuli by valve closure or foot withdrawal and closed their valves strongly. Mussels were assigned to the case (moribund, affected) group if they were on the surface of the substrate and gaping, responded slowly to tactile stimuli and closed their valves weakly or held their valves slightly open. We sampled in August, before mortality was observed, and again in September and October when active mortality was occurring.
Figure 1.
Sampling locations on the Clinch River, Tennessee and Virginia, USA (adapted from Richard et al., 2020, original image published under a Creative Commons Attribution 4.0 International License).
To collect hemolymph, we gently opened the valves of each mussel with a sterile pediatric nasal speculum, disinfected the outer surface of the anterior adductor muscle with 70% isopropyl alcohol and extracted up to 1.0 ml of hemolymph (depending on the size of the mussel) from the anterior adductor muscle sinus using a 1-ml tuberculin syringe. We placed hemolymph in sterile tubes on dry ice in the field and then stored samples at −80°C until further analysis. We marked mussels with glue-on shellfish tags (Hallprint, Hindmarsh Valley, Australia) to prevent re-sampling during successive sampling events and then returned mussels to the shoals from which they were collected.
Tissue sample preparation
Hemolymph samples were processed in groups of 24 to limit processing time (Hernandes et al., 2017). After thawing, we removed 100 μl of hemolymph with 200 μl of cold methanol containing CUDA (CAS # 479413-68-8, 12-[[(cyclohexylamino)carbonyl]amino]-dodecanoic acid; Cayman Chemical, Ann Arbor, MI) as an internal standard. The samples were then vortexed for 30 s and centrifuged at 18 787g for 7 min at 4°C. We used a CentriVap micro-Infrared Vacuum Concentrator (Labconco, Kansas City, MO) at 1700 revolutions per minute (rpm) and 35°C to evaporate the methanol and increased temperature to 47.5°C to evaporate residual water/methanol. The samples were reconstituted in 125 μl of cold methanol, vortexed for 30 s and centrifuged at 18 787g for 7 min at 4°C. A sample volume of 100 μl was used for analysis.
Metabolomic analysis
Multiple methods have been developed for the extraction, detection, identification and quantification of the metabolome ( Liu and Locasale, 2017). We used a modified one-step extraction based on the modified Bligh–Dyer extraction method (Bligh and Dyer, 1959; Sitnikov et al., 2016; Satomi et al., 2017) and an ultra-high-performance liquid chromatography with quadrupole time-of-flight mass spectroscopy (HPLC-MS) to detect metabolomic signals in hemolymph (Putnam et al., 2017). We used both targeted and untargeted metabolomic analysis. The targeted analysis compared the detected signal mass, spectrum and retention time with a database of known standards, whereas the untargeted analysis examined all detectable metabolomic signals, both known and unidentified metabolites. The analytical method, quality control and data processing are described in the supplemental data.
Data processing
We used the Mass Spectrometry Metabolite Library from IROA Technologies (Mass Spectrometry Metabolite Library, IROA Technologies, Sea Grit, NJ) to identify targeted metabolites. The IROA Library also allowed us to ensure there was consistent chromatographic retention and peak shape over all samples. Targeted metabolites were identified by the injection of standards. The detected IROA Library standards were used to create a mass spectrum library containing information about each standard: retention, signal, m/z and chemical adduct (Supplementary Table S2). We annotated metabolites in the targeted analysis using the Human Metabolomic Database (HMDB—https://hmdb.ca/, release version 4.0)(Wishart et al., 2018) and Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.kegg.jp, release version 99.0, 99.1, and 100.0) (Hattori et al., 2003; Kanehisa et al., 2012). Data associated with the manuscript are available in the study by Putnam et al. (2022), “Metabolomic analysis of pheasantshell mussel (Ortmanniana pectorosa, order Unionida) from a mass mortality event in the Clinch River, Virginia and Tennessee, USA: U.S. Geological Survey data release”, https://doi.org/10.5066/P9ZT9F3S.
Statistical analyses
We used a multi-tiered statistical approach to identify relationships between field assessments of clinical status (case vs. control). First, we used a principal component analysis (PCA) to identify natural groupings of mussels by field assessments of clinical status based on the underlying structure of the data. We also used a robust PCA analysis to identify orthogonal outliers, good and bad leverage points. Good leverage points follow the trends found in all mussels, whereas bad leverage points do the exact opposite (Hubert et al., 2005; Alguraibawi et al., 2015). Orthogonal outliers are outside the normal plane of the PCA. The PCA analysis created a biplot of the samples based on the cosine similarity between two dimensional vectors in three dimensions. In addition, 95% confidence ellipses were added showing the Gaussian distribution of the normally distributed mussel metabolites. Second, we used volcano plots to detect important metabolites in a univariate analysis. The volcano plot analysis was completed by calculating the average abundance of each metabolite grouped into either a field assessment of case or control. The volcano plot analysis used the combined targeted and untargeted metabolomic data to show the influence of the field assessment. The P value was also calculated for each metabolite based on the field assessment. The fold change (FC) was calculated by taking the log2 of the average abundance for each metabolite for either the case or control mussels and then subtracting the control value from the case value. The volcano plot displays log2 FC for upregulated metabolites of the case mussels on the positive x-axis, and for the control mussels on the negative x-axis. Those metabolites with FC >1.0 and t test P value <0.1 were considered important for distinguishing case and control mussels. Metabolites with a P value between 0.05 and 0.10 show a weak relationship and when linked to FC >1.0 may offer further insight into differences between field assessment groups (Fan et al., 2020).
We used MetaboAnalyst (MetaboAnalyst, 2021) for the biomarker, targeted and untargeted pathway analysis and heat map analysis (Chong et al., 2019; Pang et al., 2021). All data used in the MetaboAnalyst platform had outliers filtered by calculating the relative standard deviation; data were normalized by the median, log transformed and scaled by Pareto scaling. All P values reported from MetaboAnalyst are adjusted P values. The biomarker analysis used area under the curve (AUC), FC of metabolite abundance and the false discovery rate t test between the field assessment (case vs. control) to rank metabolites as potential biomarkers. The IROA standard metabolites were assigned to targeted metabolic pathways using MetaboAnalyst. The untargeted pathway analysis in MetaboAnalyst uses an algorithm to identify metabolites based on known pathway and network data. The algorithm conducts multiple permutations to identify possible matches. These permutations are then used to create a gamma-distributed P value (Chen et al., 2014). The pathway analysis also uses an enrichment factor analysis to measure the number of pathway metabolites identified compared to expected values. We used the heat map tool to visualize the concentrations of the top 25 significant untargeted metabolites. Individual mussels were grouped in a hierarchical cluster by metabolite concentration. t Test values were used to compare mean values of the metabolites.
Results
The PCA results showed that two mussels were outside the normal metabolomic distribution (Supplementary Fig. S1), but both were retained because they showed variation within a population. One case mussel (Kyle’s Ford, October sample) influenced the shape of the ellipse for the normal distribution of the case mussels’ metabolites and was retained because it belonged to the affected classification.
The robust PCA analysis indicated additional mussels were either good leverage points (strong indicators of morbidity signs) or orthogonal outliers—unrelated signs (Supplementary Fig. S2). No bad leverage points were detected—mussels with metabolite distributions well outside the normal distribution. Three mussels were identified as good leverage points, meaning that they followed the trends of the normal observations. Two mussels were identified as case mussels and a third mussel was a control. These mussels may show the natural variability within the population. Four mussels were orthogonal outliers—one from a location well upstream from the mortality. Another orthogonal outlier was the mussel with the highest viral load and lowest field condition. A third orthogonal outlier was the only mussel collected at Wallen Bend. The fourth orthogonal outlier was a case mussel that may have been misdiagnosed, based on low viral loads, or have been within the natural variability of the population sampled.
We removed the five August mussels because of the small number of samples. Inclusion of August samples would increase the complexity of the dataset and lead to higher potential misclassification (Smith et al., 2014) without providing identification of differences between field diagnosis and temporal changes. The focus of this manuscript was the relationship between case and control and did not focus on the seasonality between a short period (August to October 2018).
Targeted analysis
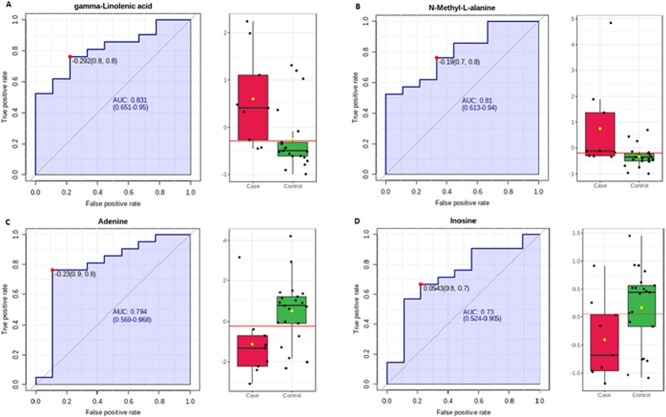
We detected 92 targeted metabolites (Supplementary Table S2). Volcano plot analysis (Fig. 3) indicated 17 targeted metabolites that were associated with field assessments of clinical status based on the adjusted P value (<0.1) and FC (≥1). We identified five metabolic pathways in our targeted analysis, which differed between case and control mussels based on the enrichment factor and P value. These included arginine/proline metabolism, purine metabolism, tryptophan metabolism, glycerophospholipid metabolism and arginine biosynthesis. Of these, purine metabolism and glycerophospholipid metabolism were also detected in the untargeted pathway analysis (Table 1). The targeted biomarker analysis found 13 metabolites that were associated with field assessment based on AUC, adjusted P value and FC (Table 2). Of these, increased gamma-linolenic acid (GLA) and N-methyl-l-alanine were most indicative of case mussels (Fig. 2A and B), while adenine and inosine were the best indicators of control mussels (Fig. 2C and D).
Figure 3.
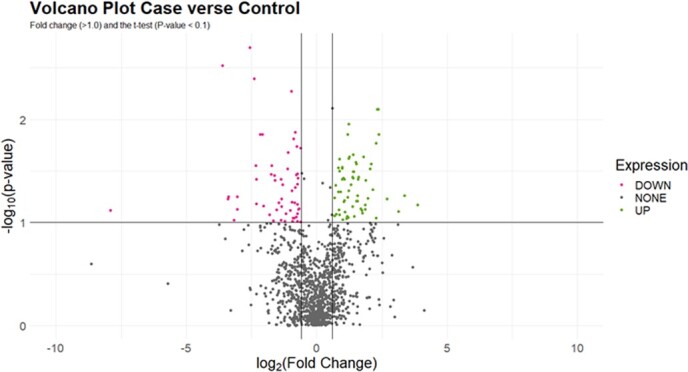
Volcano plots of both targeted and untargeted metabolites in hemolymph of pheasantshell (O. pectorosa) diagnosed as case or control. All metabolites were used in the volcano plot. Points in the upper left quadrant are metabolites with lower abundance in control mussels. Points in the upper right quadrant are metabolites with higher abundance in case mussels Metabolite differences between case and control mussels were deemed significant when FC (>1.0) and the t test (P < 0.1).
Table 1.
Impacted untargeted pathways associated with field assessment based on gamma P < 0.10
| Pathway | Total in pathway | Total detected | Significant hits | Gamma P |
|---|---|---|---|---|
| Glycerophospholipid metabolisma | 156 | 24 | 12 | >0.01 |
| Fatty acid activation | 74 | 23 | 9 | >0.01 |
| Phosphatidylinositol phosphate metabolism | 59 | 5 | 3 | 0.01 |
| Purine metabolisma | 80 | 13 | 5 | 0.01 |
| Ubiquinone biosynthesis | 10 | 2 | 2 | 0.01 |
| Androgen and estrogen biosynthesis and metabolism | 95 | 10 | 4 | 0.01 |
| Saturated fatty acids beta-oxidation | 36 | 7 | 3 | 0.02 |
| Methionine and cysteine metabolism | 94 | 7 | 3 | 0.02 |
| Prostaglandin formation from di-homo gamma-linoleic acid | 11 | 3 | 2 | 0.02 |
| Glycosphingolipid metabolism | 67 | 20 | 6 | 0.02 |
| Fatty acid oxidation | 35 | 8 | 3 | 0.02 |
| Butanoate metabolism | 34 | 8 | 3 | 0.02 |
| Glycine, serine, alanine and threonine metabolism | 88 | 13 | 4 | 0.03 |
| Linoleate metabolism | 46 | 18 | 5 | 0.03 |
| Caffeine metabolism | 1 | 5 | 2 | 0.04 |
| Urea cycle/amino group metabolism | 85 | 21 | 5 | 0.05 |
| Omega-3 fatty acid metabolism | 39 | 6 | 2 | 0.05 |
| Phytanic acid peroxisomal oxidation | 34 | 6 | 2 | 0.05 |
| Glycolysis and gluconeogenesis | 49 | 7 | 2 | 0.07 |
| Di-unsaturated fatty acid beta-oxidation | 26 | 7 | 2 | 0.07 |
| Carnitine shuttle | 72 | 33 | 7 | 0.07 |
| Hexose phosphorylation | 20 | 8 | 2 | 0.08 |
The gamma P cutoff of 0.1 was used to ensure weakly related pathways are retained for further investigation.
Also significant in target pathway analysis.
Table 2.
Targeted metabolites identified as potential biomarkers
| Metabolite name | HMDB ID | KEGG ID | Exact mass | Retention time (min) | AUC | log2 FC | P |
|---|---|---|---|---|---|---|---|
| GLA | HMDB0003073 | C06426 | 278.22 | 3.78 | 0.831 | 2.869 | 0.007 |
| Methyl jasmonate | HMDB0036583 | C11512 | 224.142 | 2.16 | 0.840 | −1.922 | 0.003 |
| Gamma-valerolactone | HMDB0029185 | C02240 | 100.0522 | 2.96 | 0.771 | −1.388 | 0.020 |
| Hexylamine | HMDB0032323 | C08306 | 101.121 | 5.49 | 0.750 | −4.970 | 0.040 |
| Betaine | HMDB0000043 | C00719 | 117.0785 | 0.52 | 0.750 | 1.927 | 0.037 |
| l-Homocystine | HMDB0000676 | C01817 | 268.0557 | 0.47 | 0.772 | 1.149 | 0.085 |
| Inosine | HMDB0000195 | C00294 | 268.081 | 0.57 | 0.729 | −1.731 | 0.054 |
| l-Isoleucine | HMDB0000172 | C00407 | 131.0944 | 0.52 | 0.729 | −1.379 | 0.073 |
| Adenosine | HMDB0000050 | C00212 | 267.0975 | 0.59 | 0.722 | −2.143 | 0.030 |
| l-Palmitoylcarnitine | HMDB0000222 | C02990 | 399.2375 | 1.41 | 0.715 | 1.557 | 0.054 |
| Stachyose | HMDB0003553 | C01613 | 666.4989 | 2.31 | 0.715 | 1.501 | 0.063 |
| Caprylic acid | HMDB0000482 | C06423 | 144.1147 | 3.52 | 0.708 | −1.158 | 0.082 |
| l-Valine | HMDB0000883 | C00183 | 117.0791 | 0.48 | 0.701 | 1.928 | 0.087 |
The criteria for inclusion as a potential biomarker included AUC >0.7, P < 0.1, and log2 FC >1.00. The P value cut off was selected to ensure weakly related biomarkers are retained for further investigation. Positive FC values indicate higher metabolite level in case mussels, while a negative FC value indicates higher metabolite level in the control mussels. HMDB ID in Human Metabolomic Database (https://hmdb.ca/) and KEGG ID in Kyoto Encyclopedia of Genes and Genomes (http://www.kegg.jp).
Figure 2.
Four targeted metabolites identified as potential biomarkers based on AUC. GLA (A) and N-methyl-l-alanine (B) abundances were relatively greater in case vs. control mussels. Adenine (C) and inosine (D) abundances were slightly greater in control versus case mussels.
Untargeted analysis
The volcano plot analysis was used to visualize metabolites indicative of field assessment (Fig. 3). We detected significant differences in 94 metabolites between case and control mussels. Twenty-two metabolic pathways were associated with field assessment (Table 1). Many of the impacted pathways were catabolic processes, such as degradation of amino acids and fatty acids. Eleven pathways were related to lipid and fatty acid metabolism. The untargeted biomarker analysis identified 22 metabolites as potential biomarkers (AUC >0.7, FC ≥│1.5│, P ≤ 0.05; Supplementary Table S3). We used a more stringent FC and adjusted P value requirement for the untargeted biomarkers to select those metabolites with the greater potential to indicate disease.
Overall, the heat map showed a distinct separation of case and control mussels based on concentrations of 25 significant metabolites (Fig. 4). Seven of 25 (28%) mussels differed in overall metabolite concentrations between their classification of case or control. Two case mussels were categorized as controls and separated as a pair from other case mussels. Five control mussels were grouped with case mussels. Three of these clustered together and were collected from the same location in October. We created a heat map to verify the metabolites related to case vs. control were also not related to a temporal change between September and October (Supplementary Fig. S3). The Supplementary Fig. S3 heat map shows how metabolites are related to the temporal change during the sampling period.
Figure 4.
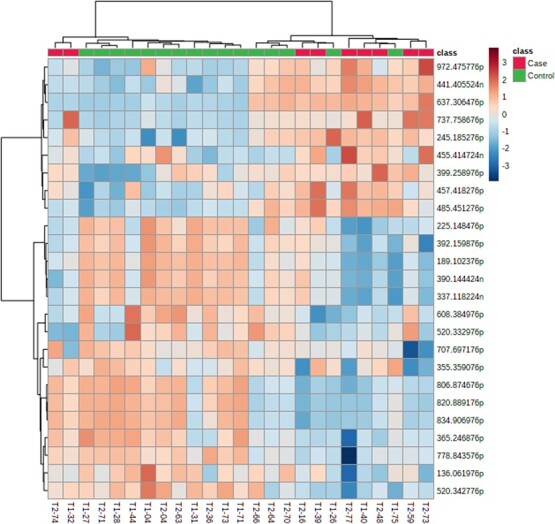
Heat map of 25 significant metabolites and hierarchical cluster of case and control pheasantshell (O. pectorosa) mussels by metabolite concentration. The horizontal axis is the unique mussel identification T1 = September, T2 = October sample. The vertical axis is the mass and mode of detection (p = positive, n = negative).
Discussion
Targeted analysis identified 13 biomarkers, based on AUC, adjusted P value and FC criteria, and five pathways that differed between case and control groups (Table 2). Of the 13 biomarkers, inosine and adenosine, both biomarkers in the purine metabolism pathway, were the most robust indicators of the control group. Purine metabolism and the associated metabolites inosine, adenosine and adenine were less abundant in case mussels. Purine metabolism was indicated as significant in both the targeted and untargeted pathway analyses. Both adenine and inosine were categorized as “stable” metabolites in mussels in unperturbed Indiana streams (Waller et al., 2023). Inosine and other nucleotide metabolites decreased during captivity of A. plicata (Roznere et al., 2014). The shifts in purine pathways that were observed in our study and by Roznere et al. (2014) support the potential use of nucleotide metabolites as biomarkers of mussel health.
GLA was the biomarker most associated with case mussels. The untargeted analysis also found that the pathway for prostaglandin formation from di-homo gamma-linoleic acid was impacted. GLA is an omega-6 fatty acid and plays a role in maintaining membrane fluidity and in immune responses (Brett and Müller-Navarra, 1997; Filimonova et al., 2016). GLA modulates immune function through conversion to di-homo-GLA and synthesis of prostaglandins, a class of eicosanoids. Eicosanoids are signaling molecules that can play a role in immune response to infectious agents (bacteria, fungus, virus) and inflammation processes. The higher presence of GLAs in case mussels may be indicative of inflammation or a mounting immune response, both of which underlie the potential significance of the GLA biomarker. GLA is also an abundant fatty acid in some cyanobacteria (Napolitano, 1999; Gugger et al., 2002). Studies on zooplankton Branchinella kugenumaensis (Yang et al., 2016) and golden mussel, Limnoperna fortunei (Mytilidae) (Hernando et al., 2021), found GLA was a potential biomarker for Microcystis aeruginosa as a food source in primary consumers. The source of GLA in our mussels did not appear to be related to cyanobacteria ingestion. Metagenomic analysis of hemolymph from the same mussels used in the present study found cyanobacteria were associated with controls and were absent from moribund mussels (Richard et al., 2021).
Other biomarkers that were elevated in case mussels included betaine, l-homocystine, palmitoylcarnitine and l-valine. Each of these is also part of one or more pathways that were significant in the untargeted pathway analysis. Betaine is an osmolyte and found in pathways for glycine, serine and threonine metabolism and glycerophospholipid metabolism. l-Homocystine is a metabolite of cysteine and methionine metabolism. Palmitoylcarnitine is a long-chain fatty acid ester of carnitine and functions to transfer fatty acids into the mitochondria for oxidation (i.e. carnitine shuttle pathway). These biomarkers and associated pathways indicate increased activity of catabolic pathways in case mussels, a potential response to a stressor.
The biomarker analysis also identified several metabolites that may be exogenous, (e.g. methyl jasmonate, stachyose and hexylamine). These metabolites may point to a xenobiotic exposure (Liu et al., 2021), microbial activity (Aldridge and Rhee, 2014), food sources (Erban et al., 2019) or potential misidentification of a similar compound (Alseekh et al., 2021). For example, methyl jasmonate, which is derived from linolenic acid, could have been mistaken for a breakdown product of GLA. Alternatively, methyl jasmonate is a metabolite of microorganisms including bacteria discovered in sponges (Bibi et al., 2020), a bacterium isolated from halophyte Prosopis strombulifera (Piccoli et al., 2011), and fungus (Piccoli et al., 2011; Eng et al., 2021). Further work would be required to confirm the identification of these metabolites and to determine their biological significance in unionids. Even if unidentified, these biomarkers may have value for future health classification analyses.
Targeted pathway analysis found arginine biosynthesis and arginine/proline metabolism were different between case and control mussels. The majority of arginine in mollusk tissue is in the phosphagen form as phosphoarginine and is released to generate ATP, similar to creatine phosphate in mammals, in the initial stages of anaerobic metabolism (Bishop et al., 1983). Arginine metabolism produces proline and polyamines, such as acetylputrescine (Supplementary Table S2), the latter of which was lower in abundance in case mussels (Supplementary Table S2). Roznere et al. (2014) reported decreased levels of polyamine metabolites when mussels were brought into captivity. Polyamines are important for cell division and growth, and decreased levels may indicate that resources are diverted away from these processes. The major role of arginine in anaerobic metabolism and the association with field assessment make it noteworthy for further investigation as a biomarker of health.
We used a traditional adjusted P value (<0.05), AUC and FC values in the untargeted biomarker analysis and found 22 additional unidentified metabolites that could be indicators of health classification (Supplementary Table S3). Although we did not identify these specific metabolites, the untargeted biomarkers were useful in construction and comparison of metabolomic profiles and pathway analysis of case and control mussels. Those specific metabolites that show the most potential as biomarkers can be identified using the exact molecular weight and retention time.
The pathway analysis distinguished trends and relationships between all detected metabolite signals. It also provided a broader, holistic view of the mussel’s health status and metabolic changes. The untargeted pathway analysis found multiple pathways that differed by field-based clinical assessment. Generally, pathways associated with cases were catabolic and oxidative pathways (Table 1). Furthermore, 11 pathways related to lipid and fatty acid metabolism were greater in case mussels. As lipids are key sources of energy, the number of these pathways that were detected as affected in the untargeted analysis indicates case mussels were consuming energy resources at a higher rate than control mussels. A key outcome of the untargeted pathway analysis was identifying pathways to investigate in future targeted analyses. For example, glycerophospholipid and ubiquinone pathways differed between case and control mussels, and metabolites within these pathways may be selected for future targeted analysis. Both are important for formation and maintenance of cell membranes. Glycerophospholipids are a major component of cell membranes and are critical for membrane stability as well as cell proliferation (Hu et al., 2022). Glycerophospholipid metabolism may be altered in an immune or inflammatory response. Ubiquinone (coenzyme Q) has multiple roles in cellular metabolism and is an antioxidant important for maintaining membrane integrity (Turunen et al., 2004). Determining shifts in specific metabolites within these pathways may provide insight on the mechanism of stress or disease that is contributing to mussel mortality.
Hierarchical clustering showed separation of case and control mussels but also indicated discrepancies with current methods to categorize mussel clinical status in the field. Heat map classification supported field assessment of 72% (18 of 25) of mussels in our study; however, 5 of 16 control mussels clustered with case mussels, while 2 of the 9 case mussels clustered with controls. The health status of a mussel in the field is most often based on behavioral response indicators, such as gaping valves, extended foot and slow response to probing, which vary widely and are highly subjective. For example, females that are gravid often move to the substrate surface and may be agape (Haag, 2012) and slow to respond, all behavioral signs of stress. In contrast, mussels may bury and die in place without outward signs of stress. Ten mussels in our study were gravid females, seven in the control group and three in the case group. Of these, three grouped with the other group in the cluster analysis (two control and one case, 30% misclassified), similar to the percentage for all mussels. Pheasantshell spawn in late summer and females become gravid starting in August and September (Ortmann, 1921). Gravid females may allocate energy substrates to larval development (Gustafson et al., 2005; Hornbach et al., 2019), resulting in a different metabolic profile relative to non-gravid females and adults. However, metabolomic analysis of two lampsiline species from three stable streams found considerable overlap in metabolite levels between gravid and non-gravid females, and sex-related differences were less important than site and species (Waller et al., 2023).
Metabolite composition may not shift strongly in early, asymptomatic stages of a disease (Wagner et al., 2015). Mussels that were field assessed as controls during active mortality may have been in this early stage and not yet showing physical or behavioral signs of disease. Increased sampling intensity and higher resolution field-based measures of clinical status may identify/detect a gradient of metabolite alterations that corresponds to the progression of disease.
Although few studies have followed seasonal changes in metabolites, we expect temporal variability in metabolite composition and pathways (Melvin et al., 2018; Whipple et al., 2021). We initially found differences between August and October control mussels in pathways for fatty acid biosynthesis and amino acid metabolism that may reflect shifts in diet. However, our sample size in August was limited and precluded further evaluation of temporal changes. Furthermore, viruses are also known to promote fatty acid synthesis and glycolytic metabolism, suggesting that these altered pathways could possibly be the result of viral infection (Sanchez and Lagunoff, 2015; Thaker et al., 2019). Notably, several viruses were identified from moribund mussels during the autumnal mortality event (Richard et al., 2020).
Metabolites in hemolymph are more transient and occur in lower concentration than those in other tissues. For example, metabolites in hemolymph were less detectable than in digestive gland, foot, adductor muscle, gill and mantle tissue using 1 (not superscripted)H NMR (Hurley-Sanders et al., 2016). However, other researchers have successfully detected a range of metabolites in hemolymph using HPLC–MS and GC–MS analyses (e.g. Roznere et al., 2014 and 2017; Nguyen et al., 2019; Nguyen and Alfaro, 2020). Despite the limitations of hemolymph metabolite analysis, hemolymph collection is generally non-lethal and less invasive than foot or mantle biopsy, a key consideration when working with imperiled fauna. In addition, hemolymph is relatively sterile compared to other tissues and potentially less contaminated by exogenous metabolites. Overall, we found that hemolymph analysis uncovered metabolomic differences that supported field-based clinical assessments of most mussels in our study.
Conclusions
Determining the health status of freshwater mussels in situ is especially challenging as no defined and verified clinical signs of disease have been established, resulting in health assessments that are based on subjective behavioral observations. As used herein, metabolomics was a real-time quantitative assessment tool to supplement field diagnosis of mussel mortality. A definitive cause of mussel mortality in the Clinch River has not been verified, but both targeted and untargeted analysis detected metabolic differences among case and control mussels. The targeted biomarker analysis found 13 metabolites associated with field assessments of clinical status. Of these, increased GLA and N-methyl-l-alanine were most indicative of case mussels, while adenine and inosine were the best indicators of control mussels. Five pathways in the targeted analysis differed by clinical status; two of these, purine metabolism and glycerophospholipid metabolism, were also indicated in the untargeted analysis. In the untargeted analysis, 22 metabolic pathways were associated with clinical status.
Metabolomic analysis of hemolymph presents a relatively non-lethal, minimally invasive method to discern organismal status, which could be incorporated into monitoring programs for imperiled mussel populations. However, routine use of metabolomics is hindered by the limited database on freshwater mussel metabolism. Only a few studies have investigated metabolic shifts in response to specific stressors in freshwater mussels [e.g. captivity/starvation (Roznere et al., 2014); relocation (Roznere et al., 2017)]. To identify potential causes of disease, controlled studies that expose mussels to different stressors and then characterize biomarkers and metabolomic pathway shifts associated with the stressor will be important. Temporal sampling of hemolymph metabolite levels within and across mussel populations will also be important for characterizing seasonal variation in metabolism. We investigated metabolite levels in the mussel species most affected by mortality in the Clinch River in 2018 (i.e. pheasantshell). Additional sampling of unaffected species may help discern differences in species resistance to stressors. Further understanding of interspecies variation, or similarity, in metabolism may lead to use of common species as surrogates for rare and imperiled mussel species. Conservation efforts for freshwater mussels have increased around the globe and would benefit from a variety of tools to assess the real-time health status of individual mussels to supplement long-term assessment tools (growth, recruitment), especially the ones that are non-lethal and can portend a stressor before mortality ensues.
Supplementary Material
Acknowledgments
We thank three anonymous reviewers for their constructive review of the manuscript. We thank the US Fish and Wildlife Service (USFWS) Virginia Field Office for logistic support during field sampling, and the USFWS La Crosse Fish Health Center, USFWS Midwest Fisheries Center and US Geological Survey National Wildlife Health Center for assistance with sample processing and analyses.
Contributor Information
Joel G Putnam, Conagen, Inc., 15 Deangelo Drive, Bedford, MA 01730, USA.
John N Steiner, US Geological Survey, Upper Midwest Environmental Science Center, 2630 Fanta Reed Road, La Crosse WI 54603, USA.
Jordan C Richard, US Fish and Wildlife Service, Southwestern Virginia Field Office, 330 Cummings Street, Abingdon, VA 24210, USA; Department of Pathobiological Sciences, University of Wisconsin-Madison, 1656 Linden Drive, Madison WI 53706, USA.
Eric Leis, US Fish and Wildlife Service, Midwest Fisheries Center, La Crosse Fish Health Center, 555 Lester Ave., Onalaska, WI 54650, USA.
Tony L Goldberg, Department of Pathobiological Sciences, University of Wisconsin-Madison, 1656 Linden Drive, Madison WI 53706, USA; Global Health Institute, University of Wisconsin-Madison, 1300 University Avenue, Madison, WI 53706, USA.
Christopher D Dunn, Department of Pathobiological Sciences, University of Wisconsin-Madison, 1656 Linden Drive, Madison WI 53706, USA.
Rose Agbalog, US Fish and Wildlife Service, Southwestern Virginia Field Office, 330 Cummings Street, Abingdon, VA 24210, USA.
Susan Knowles, US Geological Survey, National Wildlife Health Center, 6006 Schroeder Rd., Madison, WI 53711, USA.
Diane L Waller, US Geological Survey, Upper Midwest Environmental Science Center, 2630 Fanta Reed Road, La Crosse WI 54603, USA.
Author Contributions
J.C.R., E.L., D.L.W., S.K., J.G.P. and T.L.G. designed the study; J.C.R., R.A. and T.L.G. performed field work; J.G.P. and J.N.S. performed laboratory analysis; J.G.P., J.N.S. and D.L.W. analyzed and interpreted the data; all authors contributed to writing, reviewing and improving the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest. Any use of trade, firm or product names is for descriptive purposes only and does not imply endorsement by the US Government or the authors. The findings and conclusions in this article are those of the author(s) and do not necessarily represent the views of the US Fish and Wildlife Service.
Ethics and Animal Welfare Statement
We obtained biological samples in accordance with all federal, state and local laws and policies.
Funding
This work was funded by the US Geological Survey, Ecological Missions Area, Midcontinent Region, regional flex funds and the University of Wisconsin-Madison John D. MacArthur Professorship Chair.
Data Availability
Data associated with the manuscript are available in the study by Putnam et al. (2022), “Metabolomic analysis of pheasantshell mussel (O. pectorosa; Order Unionida) from a mass mortality event in the Clinch River, Virginia and Tennessee, USA: U.S. Geological Survey data release”, https://doi.org/10.5066/P9ZT9F3S.
Supplementary Material
Supplementary material is available at Conservation Physiology online.
References
- 86 FR 54298 (2021) Endangered and Threatened Wildlife and Plants; Removal of 23 Extinct Species from the Lists of Endangered and Threatened Wildlife and Plants. 86 Fed. Reg. 54298 (Sept. 30, 2021) (to be codified at 50 C.F.R. 17).
- Aldridge BB, Rhee KY (2014) Microbial metabolomics: innovation, application, insight. Curr Opin Microbiol 19: 90–96. 10.1016/j.mib.2014.06.009. [DOI] [PubMed] [Google Scholar]
- Alguraibawi M, Midi H, Imon AHM (2015) A new robust diagnostic plot for classifying good and bad high leverage points in a multiple linear regression model. Math Probl Eng 2015Article ID 279472, 12 pages, 2015,: 1–12. 10.1155/2015/279472. [DOI] [Google Scholar]
- Alseekh S, Aharoni A, Brotman Y, Contrepois K, D’Auria J, Ewald J, Ewald JC, Fraser PD, Giavalisco P, Hall RDet al. (2021) Mass spectrometry-based metabolomics: a guide for annotation, quantification and best reporting practices. Nat Methods 18: 747–756. 10.1038/s41592-021-01197-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibi F, Yasir M, Al-Sofyani A, Naseer MI, Azhar EI (2020) Antimicrobial activity of bacteria from marine sponge Suberea mollis and bioactive metabolites of Vibrio sp. EA348. Saudi J Biol Sci 27: 1139–1147. 10.1016/j.sjbs.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop SH, Ellis LL, Burcham JM (1983) Amino acid metabolism in molluscs. In Hochachka PW, ed, Metabolic Biochemistry and Molecular Biomechanics. Academic Press, San Diego, CA, pp. 243–327 [Google Scholar]
- Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37: 911–917. 10.1139/y59-099. [DOI] [PubMed] [Google Scholar]
- Brett M, Müller-Navarra D (1997) The role of highly unsaturated fatty acids in aquatic foodweb processes. Freshw Biol 38: 483–499. 10.1046/j.1365-2427.1997.00220.x. [DOI] [Google Scholar]
- Cappello T, Fernandes D, Maisano M, Casano A, Bonastre M, Bebianno MJ, Mauceri A, Fasulo S, Porte C (2017) Sex steroids and metabolic responses in mussels Mytilus galloprovincialis exposed to drospirenone. Ecotoxicol Environ Saf 143: 166–172. 10.1016/j.ecoenv.2017.05.031. [DOI] [PubMed] [Google Scholar]
- Chen Z, Yang W, Liu Q, Yang JY, Li J, Yang MQ (2014) A new statistical approach to combining p-values using gamma distribution and its application to genome-wide association study. BMC Bioinform 15: 1–7. 10.1186/1471-2105-15-S17-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J, Wishart DS, Xia J (2019) Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr Protoc Bioinformatics 68: e86. 10.1002/cpbi.86. [DOI] [PubMed] [Google Scholar]
- Downing JA, Van Meter P, Woolnough DA (2010) Suspects and evidence: a review of the causes of extirpation and decline in freshwater mussels. Anim Biodivers Conserv 33: 151–185. 10.32800/abc.2010.33.0151. [DOI] [Google Scholar]
- Dumas T, Bonnefille B, Gomez E, Boccard J, Castro NA, Fenet H, Courant F (2020) Metabolomics approach reveals disruption of metabolic pathways in the marine bivalve Mytilus galloprovincialis exposed to a WWTP effluent extract. Sci Total Environ 712: 136551. 10.1016/j.scitotenv.2020.136551. [DOI] [PubMed] [Google Scholar]
- Eng F, Marin JE, Zienkiewicz K, Gutiérrez-Rojas M, Favela-Torres E, Feussner I (2021) Jasmonic acid biosynthesis by fungi: derivatives, first evidence on biochemical pathways and culture conditions for production. PeerJ 9: e10873. 10.7717/peerj.10873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erban A, Fehrle I, Martinez-Seidel F, Brigante F, Más AL, Baroni V, Wunderlin D, Kopka J (2019) Discovery of food identity markers by metabolomics and machine learning technology. Sci Rep 9: 9697. 10.1038/s41598-019-46113-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S, Shahid M, Jin P, Asher A, Kim J (2020) Identification of metabolic alterations in breast cancer using mass spectrometry-based metabolomic analysis. Metabolites 10: 170. 10.3390/metabo10040170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira-Rodríguez N, Sousa R, Pardo I (2018) Negative effects of Corbicula fluminea over native freshwater mussels. Hydrobiologia 810: 85–95. 10.1007/s10750-016-3059-1. [DOI] [Google Scholar]
- Filimonova V, Gonçalves F, Marques JC, De Troch M, Gonçalves AMM (2016) Fatty acid profiling as bioindicator of chemical stress in marine organisms: a review. Ecol Indic 67: 657–672. 10.1016/j.ecolind.2016.03.044. [DOI] [Google Scholar]
- Gugger M, Lyra C, Henriksen P, Couté A, Humbert J-F, Sivonen K (2002) Phylogenetic comparison of the cyanobacterial genera Anabaena and Aphanizomenon. Int J Syst Evol Microbiol 52: 1867–1880. 10.1099/00207713-52-5-1867. [DOI] [PubMed] [Google Scholar]
- Gustafson LL, Stoskopf MK, Showers W, Cope G, Eads C, Linnehan R, Kwak TJ, Andersen B, Levine JF (2005) Reference ranges for hemolymph chemistries from Elliptio complanata of North Carolina. Dis Aquat Organ 65: 167–176. 10.3354/dao065167. [DOI] [PubMed] [Google Scholar]
- Haag WR (2012) North American Freshwater Mussels: Natural History, Ecology, and Conservation. Cambridge University Press, Cambridge, United Kingdom, p. 505.ISBN 978-0-521-19938-4(hardback) [Google Scholar]
- Haag WR (2019) Reassessing enigmatic mussel declines in the United States. Freshw Mollusk Biol Conserv 22: 43. 10.31931/fmbc.v22i2.2019.43-60. [DOI] [Google Scholar]
- Haag WR, Williams JD (2014) Biodiversity on the brink: an assessment of conservation strategies for North American freshwater mussels. Hydrobiologia 735: 45–60. 10.1007/s10750-013-1524-7. [DOI] [Google Scholar]
- Hao R, Wang Z, Yang C, Deng Y, Zheng Z, Wang Q, Du X (2018) Metabolomic responses of juvenile pearl oyster Pinctada maxima to different growth performances. Aquaculture 491: 258–265. 10.1016/j.aquaculture.2018.03.050. [DOI] [Google Scholar]
- Hattori M, Okuno Y, Goto S, Kanehisa M (2003) Heuristics for chemical compound matching. Genome Inform 14: 144–153. 10.11234/gi1990.14.144. [DOI] [PubMed] [Google Scholar]
- Hernandes VV, Barbas C, Dudzik D (2017) A review of blood sample handling and pre-processing for metabolomics studies. Electrophoresis 38: 2232–2241. 10.1002/elps.201700086. [DOI] [PubMed] [Google Scholar]
- Hernando M, De Troch M, Rosa F, Giannuzzi L (2021) Fatty acid response of the invasive bivalve Limnoperna fortunei fed with Microcystis aeruginosa exposed to high temperature. Comp Biochem Physiol C Toxicol Pharmacol 240: 108925. 10.1016/j.cbpc.2020.108925. [DOI] [PubMed] [Google Scholar]
- Hornbach DJ, Hove MC, MacGregor KR, Kozarek JL, Sietman BE, Davis M (2019) A comparison of freshwater mussel assemblages along a land-use gradient in Minnesota. Aquat Conserv Mar Freshw Ecosyst 29: 1826–1838. 10.1002/aqc.3167. [DOI] [Google Scholar]
- Hu Z, Feng J, Song H, Zhou C, Yang M-J, Shi P, Yu Z-L, Guo Y-J, Li Y-R, Zhang T (2022) Metabolic response of Mercenaria mercenaria under heat and hypoxia stress by widely targeted metabolomic approach. Science of The Total Environment 809: 151172. 10.1016/j.scitotenv.2021.151172. [DOI] [PubMed] [Google Scholar]
- Hubert M, Rousseeuw PJ, Vanden Branden K (2005) ROBPCA: a new approach to robust principal component analysis. Dent Tech 47: 64–79. 10.1198/004017004000000563. [DOI] [Google Scholar]
- Hurley-Sanders JL, Stoskopf MK, Nelson SAC, Showers W, Law JM, Gracz HS, Levine JF (2016) Tissue extraction methods for metabolic profiling of a freshwater bivalve, Elliptio complanata (Lightfoot, 1786). Am Malacol Bull 33: 185–194. 10.4003/006.033.0209. [DOI] [Google Scholar]
- Jones J, Ahlstedt S, Ostby B, Beaty B, Pinder M, Eckert N, Butler R, Hubbs D, Walker C, Hanlon Set al. (2014) Clinch River freshwater mussels upstream of Norris Reservoir, Tennessee and Virginia: a quantitative assessment from 2004 to 2009. J Am Water Resour Assoc 50: 820–836. 10.1111/jawr.12222. [DOI] [Google Scholar]
- Jones J, Lane T, Ostby B, Braven B, Ahlstedt S, Butler R, Hubbs D, Walker C (2018) Collapse of the Pendleton Island mussel fauna in the Clinch River, Virginia: setting baseline conditions to guide recovery and restoration. Freshw Mollusk Biol Conserv 21: 36–56. . [DOI] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M (2012) KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 40: D109–D114. 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles S, Leis EM, Richard JC, Cole R, Agbalog RE, Putnam JG, Goldberg TL, Waller DL (2022) A novel gonadotropic microsporidian parasite (Microsporidium clinchi n. sp.) infecting a declining population of pheasantshell mussels (Actinonaias pectorosa) (Unionidae) from the Clinch River, USA. Parasitologia 2: 1–12. 10.3390/parasitologia2010001. [DOI] [Google Scholar]
- Leis E, Erickson S, Waller D, Richard J, Goldberg T (2019) A comparison of bacteria cultured from unionid mussel hemolymph between stable populations in the Upper Mississippi River basin and populations affected by a mortality event in the Clinch River. Freshw Mollusk Biol Conserv 22: 70–80. . [DOI] [Google Scholar]
- Liu KH, Lee CM, Singer G, Bais P, Castellanos F, Woodworth MH, Ziegler TR, Kraft CS, Miller GW, Li Set al. (2021) Large scale enzyme based xenobiotic identification for exposomics. Nat Commun 12: 5418. 10.1038/s41467-021-25698-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Locasale JW (2017) Metabolomics: a primer. Trends Biochem Sci 42: 274–284. 10.1016/j.tibs.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhao J, Wu H, Wang Q (2013) Metabolomic analysis revealed the differential responses in two pedigrees of clam Ruditapes philippinarum towards Vibrio harveyi challenge. Fish Shellfish Immunol 35: 1969–1975. 10.1016/j.fsi.2013.09.037. [DOI] [PubMed] [Google Scholar]
- Lorgeril J, Lucasson A, Petton B, Toulza E, Montagnani C, Clerissi C, Vidal-Dupiol J, Chaparro C, Galinier R, Escoubas J-Met al. (2018) Immune-suppression by OsHV-1 viral infection causes fatal bacteraemia in Pacific oysters. Nat Commun 9: 4215. 10.1038/s41467-018-06659-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydeard C, Cowie RH, Ponder WF, Bogan AE, Bouchet P, Clark SA, Cummings KS, Frest TJ, Gargominy O, Herbert DGet al. (2004) The global decline of nonmarine mollusks. Bioscience 54: 321. 10.1641/0006-3568(2004)054[0321:TGDONM]2.0.CO;2. [DOI] [Google Scholar]
- Machałowski T, Jesionowski T (2020) Hemolymph of molluscan origin: from biochemistry to modern biomaterials science. Applied Physics A 127. 10.1007/s00339-020-04166-1. [DOI] [Google Scholar]
- McMahon RF, Bogan AE (2001) 11 – Mollusca: Bivalvia. In Thorp JH, Covich AP, eds, Ecology and Classification of North American Freshwater Invertebrates, Ed2nd. Academic Press, San Diego, CA, pp. 331–429 [Google Scholar]
- Melvin SD, Lanctôt CM, Doriean NJC, Carroll AR, Bennett WW (2018) Untargeted NMR-based metabolomics for field-scale monitoring: temporal reproducibility and biomarker discovery in mosquitofish (Gambusia holbrooki) from a metal(loid)-contaminated wetland. Environ Pollut 243: 1096–1105. 10.1016/j.envpol.2018.09.071. [DOI] [PubMed] [Google Scholar]
- Metabo Analyst (2021). MetaboAnalyst 5.0—user-friendly, streamlined metabolomics data analysis. Version 5. https://www.metaboanalyst.ca/home.xhtml (date accessed December 12, 2021).
- Napolitano GE (1999) Fatty acids as trophic and chemical markers in freshwater ecosystems. In Arts MT, Wainman BC, eds, Lipids in Freshwater Ecosystems. Springer, New York, pp. 21–44 [Google Scholar]
- Neves D (1986) Information sought on mussel die-off. Fisheries 11: 1–72. 10.1577/1548-8446-11-1. [DOI] [Google Scholar]
- Nguyen TV, Alfaro AC (2020) Applications of omics to investigate responses of bivalve haemocytes to pathogen infections and environmental stress. Aquaculture 518: 734488. 10.1016/j.aquaculture.2019.734488. [DOI] [Google Scholar]
- Nguyen TV, Alfaro AC, Young T, Merien F (2019) Tissue-specific immune responses to Vibrio sp. infection in mussels (Perna canaliculus): a metabolomics approach. Aquaculture 500: 118–125. 10.1016/j.aquaculture.2018.09.061. [DOI] [Google Scholar]
- Nguyen TV, Alfaro AC, Young T, Ravi S, Merien F (2018) Metabolomics study of immune responses of New Zealand Greenshell™ mussels (Perna canaliculus) infected with pathogenic Vibrio sp. Marine Biotechnol 20: 396–409. 10.1007/s10126-018-9804-x. [DOI] [PubMed] [Google Scholar]
- Ortmann AE (1921) The anatomy of certain mussels from the upper Tennessee. Nautilus 34: 81–91. [Google Scholar]
- Pang Z, Chong J, Zhou G, Lima Morais DA, Chang L, Barrette M, Gauthier C, Jacques P-É, Li S, Xia J (2021) MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res 49: W388–W396. 10.1093/nar/gkab382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccoli P, Travaglia C, Cohen A, Sosa L, Cornejo P, Masuelli R, Bottini R (2011) An endophytic bacterium isolated from roots of the halophyte Prosopis strombulifera produces ABA, IAA, gibberellins A1 and A3 and jasmonic acid in chemically-defined culture medium. Plant Growth Regul 64: 207–210. 10.1007/s10725-010-9536-z. [DOI] [Google Scholar]
- Putnam JG, Nelson JE, Leis EM, Erickson RA, Hubert TD, Amberg JJ (2017) Using silver and bighead carp cell lines for the identification of a unique metabolite fingerprint from thiram-specific chemical exposure. Chemosphere 168: 1477–1485. 10.1016/j.chemosphere.2016.11.046. [DOI] [PubMed] [Google Scholar]
- Putnam JP, Steiner JN, Richard J, Leis E, Goldberg T, Dunn CD, Agbalog R, Knowles S, Waller DL (2022) Metabolomic analysis of pheasantshell mussel (Ortmanniana pectorosa; order Unionida) from a mass mortality event in the Clinch River, Virginia and Tennessee, USA: U.S. Geological Survey data release. 10.5066/P9ZT9F3S. [DOI]
- Régnier C, Fontaine B, Bouchet P (2009) Not knowing, not recording, not listing: numerous unnoticed mollusk extinctions. Conserv Biol 23: 1214–1221. 10.1111/j.1523-1739.2009.01245.x. [DOI] [PubMed] [Google Scholar]
- Richard JC, Campbell LJ, Leis EM, Agbalog RE, Dunn CD, Waller DL, Knowles S, Putnam JG, Goldberg TL (2021) Mussel mass mortality and the microbiome: evidence for shifts in the bacterial microbiome of a declining freshwater bivalve. Microorganisms 9: 1976. 10.3390/microorganisms9091976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard JC, Leis E, Dunn CD, Agbalog R, Waller D, Knowles S, Putnam J, Goldberg TL (2020) Mass mortality in freshwater mussels (Actinonaias pectorosa) in the Clinch River, USA, linked to a novel densovirus. Sci Rep 10: 14498. 10.1038/s41598-020-71459-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roznere I, Watters GT, Wolfe BA, Daly M (2014) Nontargeted metabolomics reveals biochemical pathways altered in response to captivity and food limitation in the freshwater mussel Amblema plicata. Comp Biochem Physiol D Genom Proteom 12: 53–60. 10.1016/j.cbd.2014.09.004. [DOI] [PubMed] [Google Scholar]
- Roznere I, Watters GT, Wolfe BA, Daly M (2017) Effects of relocation on metabolic profiles of freshwater mussels: metabolomics as a tool for improving conservation techniques. Aquat Conserv Mar Freshw Ecosyst 27: 919–926. 10.1002/aqc.2776. [DOI] [Google Scholar]
- Sanchez EL, Lagunoff M (2015) Viral activation of cellular metabolism. Virology 479-480: 609–618. 10.1016/j.virol.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satomi Y, Hirayama M, Kobayashi H (2017) One-step lipid extraction for plasma lipidomics analysis by liquid chromatography mass spectrometry. J Chromatogr B 1063: 93–100. 10.1016/j.jchromb.2017.08.020. [DOI] [PubMed] [Google Scholar]
- Sitnikov DG, Monnin CS, Vuckovic D (2016) Systematic assessment of seven solvent and solid-phase extraction methods for metabolomics analysis of human plasma by LC-MS. Sci Rep 6: 1–11. 10.1038/srep38885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MR, Martinez T, Giraud-Carrier C (2014) An instance level analysis of data complexity. Mach Learn 95: 225–256. 10.1007/s10994-013-5422-z. [DOI] [Google Scholar]
- Starliper CE, Powell J, Garner JT, Schill WB (2011) Predominant bacteria isolated from moribund Fusconaia ebena ebonyshells experiencing die-offs in Pickwick Reservoir, Tennessee River, Alabama. J Shellfish Res 30: 359–366. 10.2983/035.030.0223. [DOI] [Google Scholar]
- Strayer DL, Caraco NF, Cole JJ, Findlay S, Pace ML (1999) Transformation of freshwater ecosystems by bivalves: a case study of zebra mussels in the Hudson River. Bioscience 49: 19–27. 10.2307/1313490. [DOI] [Google Scholar]
- Thaker SK, Ch’ng J, Christofk HR (2019) Viral hijacking of cellular metabolism. BMC Biol 17: 59. 10.1186/s12915-019-0678-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Xu T, Li Y, Liu Y, Liu J (2021) An untargeted LC-MS metabolomics approach to the metabolic profiles of bottom cultured scallops (Mizuhopecten yessoensis) subjected to mechanical shock in early post-harvest handling. Aquaculture 533: 736061. 10.1016/j.aquaculture.2020.736061. [DOI] [Google Scholar]
- Turunen M, Olsson J, Dallner G (2004) Metabolism and function of coenzyme Q. Biochimica et Biophysica Acta (BBA) - Biomembranes 1660: 171–199. 10.1016/j.bbamem.2003.11.012. [DOI] [PubMed] [Google Scholar]
- Vaughn CC (2010) Biodiversity losses and ecosystem function in freshwaters: emerging conclusions and research directions. Bioscience 60: 25–35. 10.1525/bio.2010.60.1.7. [DOI] [Google Scholar]
- Vaughn CC (2018) Ecosystem services provided by freshwater mussels. Hydrobiologia 810: 15–27. 10.1007/s10750-017-3139-x. [DOI] [Google Scholar]
- Wagner ND, Lankadurai BP, Simpson MJ, Simpson AJ, Frost PC (2015) Metabolomic differentiation of nutritional stress in an aquatic invertebrate. Physiol Biochem Zool 88: 43–52. 10.1086/679637. [DOI] [PubMed] [Google Scholar]
- Waller DL, Cope WG (2019) The status of mussel health assessment and a path forward. Freshw Mollusk Biol Conserv 22: 26. 10.31931/fmbc.v22i2.2019.26-42. [DOI] [Google Scholar]
- Waller DL, Putnam M, Steiner JN, Fisher B, Burcham GN, Oliver J, Smith SB, Erickon R, Remek A, Bodeker N (2023) Targeted metabolomics characterizes metabolite occurrence and variability in stable freshwater mussel populations. Conserv Physiol 11: 1–19. 10.1093/conphys/coad040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipple B, Agar J, Zhao J, Pearce DA, Kovács AD (2021) The acidified drinking water-induced changes in the behavior and gut microbiota of wild-type mice depend on the acidification mode. Sci Rep 11: 2877. 10.1038/s41598-021-82570-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JD, Bogan AE, Butler RS, Cummings KS, Garner JT, Harris JL, Johnson NA, Watters GT (2017) A revised list of the freshwater mussels (Mollusca: Bivalvia: Unionida) of the United States and Canada. Freshw Mollusk Biol Conserv 20: 33. 10.31931/fmbc.v20i2.2017.33-58. [DOI] [Google Scholar]
- Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vázquez-Fresno R, Sajed T, Johnson D, Li C, Karu Net al. (2018) HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res 46: D608–D617. 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney Set al. (2007) HMDB: the human metabolome database. Nucleic Acids Res 35: D521–D526. 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Du X, Hao R, Wang Q, Deng Y, Sun R (2019) Effect of vitamin D3 on immunity and antioxidant capacity of pearl oyster Pinctada fucata martensii after transplantation: insights from LC–MS-based metabolomics analysis. Fish Shellfish Immunol 94: 271–279. 10.1016/j.fsi.2019.09.017. [DOI] [PubMed] [Google Scholar]
- Yang D, Nam S, Hwang S-J, An K-G, Park Y-S, Shin K-H, Park S (2016) Fatty acid biomarkers to verify cyanobacteria feeding abilities of herbivorous consumers. J Freshwater Ecol 31: 77–91. 10.1080/02705060.2015.1025304. [DOI] [Google Scholar]
- Young T, Alfaro A, Villas-Bôas S (2015) Identification of candidate biomarkers for quality assessment of hatchery-reared mussel larvae via GC/MS-based metabolomics. N Z J Mar Freshw Res 49: 87–95. 10.1080/00288330.2014.958504. [DOI] [Google Scholar]
- Zamboni N, Saghatelian A, Patti GJ (2015) Defining the metabolome: size, flux, and regulation. Mol Cell 58: 699–706. 10.1016/j.molcel.2015.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data associated with the manuscript are available in the study by Putnam et al. (2022), “Metabolomic analysis of pheasantshell mussel (O. pectorosa; Order Unionida) from a mass mortality event in the Clinch River, Virginia and Tennessee, USA: U.S. Geological Survey data release”, https://doi.org/10.5066/P9ZT9F3S.