Abstract
Malaria is an important infectious disease that continues to claim hundreds of thousands of lives annually. The disease is caused by infection of host erythrocytes by apicomplexan parasites of the genus Plasmodium. The parasite contains three different apical organelles – micronemes, rhoptries and dense granules (DGs) – whose contents are secreted to mediate binding to and invasion of the host cell and the extensive remodelling of the host cell that occurs following invasion. Whereas the roles of micronemes and rhoptries in binding and invasion of the host erythrocyte have been studied in detail, the roles of DGs in Plasmodium parasites are poorly understood. They have been proposed to control host cell remodelling through regulated protein secretion after invasion, but many basic aspects of the biology of DGs remain unknown. Here we describe DG biogenesis timing for the first time, using RESA localization as a proxy for the timing of DG formation. We show that DG formation commences approximately 37 min prior to schizont egress, as measured by the recruitment of the DG marker RESA. Furthermore, using a bioinformatics approach, we aimed to predict additional cargo of the DGs and identified the J-dot protein HSP40 as a DG protein, further supporting the very early role of these organelles in the interaction of the parasite with the host cell.
Keywords: malaria, Plasmodium, apicomplexa, apical organelles, dense granules
Introduction
Malaria is caused by invasion, remodelling and lysis of host red blood cells by parasites of the genus Plasmodium. Invasion of the host cell is controlled by the regulated secretion of proteins from three specialized secretory organelles: micronemes, rhoptries and dense granules (DGs) (Fig. 1). The presence of three distinct apical organelles allows for compartmentalization of proteins with specific functions and temporal regulation of protein discharge for rapid and efficient invasion and modification of the host cell [1–5]. Micronemes are the first to secrete their contents [2] and primarily control host cell recognition, attachment and invasion through release of proteins such as erythrocyte binding antigen 175 (EBA-175) [6–8] and apical membrane antigen-1 (AMA-1) [9–12]. The rhoptries are second to discharge their contents, which include lipid whorls in addition to proteins [2, 13]; based on the timing of discharge it was thought that a subset of rhoptry proteins and the lipids secreted from the rhoptries initiate and support parasitophorous vacuole (PV) formation and invasion [5, 14, 15]. This has been supported by the finding that the rhoptry-associated protein (RAP) complex facilitates parasite growth and survival within the host cell after invasion, with conditional knockdown of RAP components resulting in structural deformity of the PV membrane (PVM), delayed intra-erythrocytic development and decreased parasitaemia in mouse models [16]. Although the content of apical organelles appears to be segregated along functional lines, studies in Plasmodium parasites and Toxoplasma parasites have revealed that cooperation between microneme and rhoptry proteins occurs to allow parasite invasion of the host erythrocyte [17, 18]. Binding of the microneme protein AMA1 and the rhoptry neck protein RON2 triggers formation of the moving junction complex (MJ) [5, 19]. The MJ functions as an interface between the membranes of the invading parasite and the host cell through which the parasite passes into the host cell during invasion. Blockage of the AMA1 binding site of RON2 inhibits formation of the MJ and parasitophorous vacuole [5, 20].
Fig. 1.
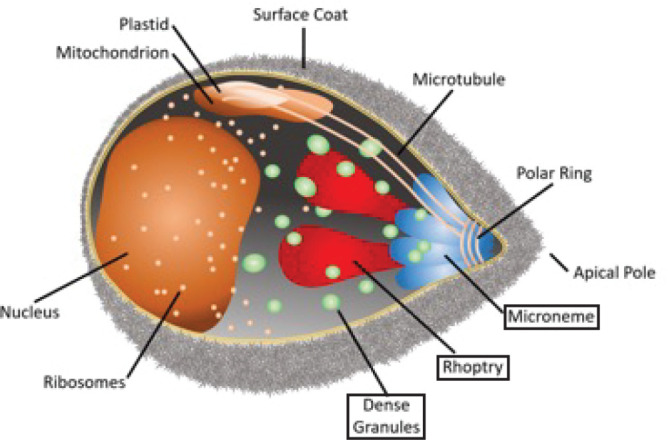
A Plasmodium falciparum merozoite. Indicated are the different organelles inside the parasite, with the names of the apical organelles in boxes.
DGs are the last organelle to secrete their contents [21–23] and are speculated to be required for the remodelling of the host cell after invasion, thereby allowing parasite survival and replication through the asexual stages [1, 24]. However, little is known about DGs in Plasmodium parasites; only a few DG proteins have been identified in Plasmodium falciparum, whilst over 40 have been described in the related apicomplexan parasite Toxoplasma gondii [25, 26], in which DGs have been studied more extensively than in Plasmodium parasites. The few known P. falciparum DG proteins enable transport of parasite effector proteins into the erythrocyte [27–29] or are exported and subsequently mediate alterations of the biochemical and biophysical properties of the host cell [30]. The only known contents of Plasmodium DGs are the five core proteins of the Plasmodium translocon of exported proteins (PTEX) – EXP2, HSP101, PTEX150, PTEX88 and TRX2 – an essential protein complex that transports parasite effector proteins across the PVM into the host cell [28, 31–34]; PV1, which interacts with exported proteins and the PTEX [35, 36]; EXP1, a transmembrane protein in the parasitophorous vacuole membrane [37–39] that has an important role in nutrient uptake across the PVM through its effect on the localization of EXP2 [27, 29, 37, 40]; P113, a glycosylphosphatidylinositol (GPI)-linked protein that associates with the PTEX and with PVM and exported proteins [41, 42]; LSA3, the function of which is unknown [43]; and RESA (ring-infected erythrocyte surface antigen) [44], which binds to and stabilizes spectrin tetramers below the erythrocyte surface, thereby reducing host cell deformability [22, 30, 45–47] (Table 1). The recently published Dense Granule Protein Database (DGPD) (http://dgpd.tlds.cc/DGPD/index/), which provides information on identified and predicted DG proteins of apicomplexan parasites, lists an additional four PHIST proteins and a ClpB1 orthologue as putative DG proteins [48], although the ClpB1 orthologue has also been identified previously as an apicoplast protein [49] (Table 1). Hence, based on the function of the known DG proteins, this organelle appears to facilitate the modification of the host cell into a hospitable environment for parasite growth. For example, the increased cell rigidity caused by RESA allows infected cells to avoid filtration by the spleen by sequestering cells in the microvasculature whilst also increasing heat shock resistance, allowing parasites to survive the increased temperatures of febrile episodes and inhibit further invasion by other merozoites [45–47]. The timing of DG protein secretion immediately following invasion, the extensive nature of erythrocyte remodelling at that time and the greater number of DG proteins found in other apicomplexan parasites all suggest that DGs of Plasmodium parasites probably contain many more exported proteins than those currently identified [50]. This theory is further supported by the late-stage transcriptional profiles of 90 genes encoding putative exported proteins containing a Plasmodium Export Element (PEXEL) that are predicted to be exported into the host erythrocyte [51]. This demonstrates that in asexual Plasmodium parasites, a large proportion of exported proteins (although not all) are synthesized very late in the intra-erythrocytic life cycle [52, 53].
Table 1.
Known and predicted Plasmodium falciparum dense granule proteins
|
Category |
Gene ID |
Name |
Function |
Comment |
|---|---|---|---|---|
|
Known |
PF3D7_0102200 |
RESA |
Increases RBC rigidity |
Also identified in the Dense Granule Protein Database (DGPD) |
|
PF3D7_1105600 |
PTEX88 |
PTEX core component |
||
|
PF3D7_1436300 |
PTEX150 |
PTEX core component |
||
|
PF3D7_1116800 |
HSP101 |
PTEX core component |
Also in DGPD |
|
|
PF3D7_1345100 |
Trx2 |
PTEX core component |
||
|
PF3D7_1471100 |
EXP2 |
PTEX core component, nutrient pore |
Also in DGPD |
|
|
PF3D7_1129100 |
PV1 |
PTEX-associated PV protein |
||
|
PF3D7_1420700 |
P113 |
PTEX-associated PV protein |
||
|
PF3D7_1121600 |
EXP1 |
Integral PVM protein, regulates EXP2 pore activity |
Also in DGPD |
|
|
PF3D7_0220000 |
LSA3 |
Exported protein of unknown function |
||
|
DGPD predictions |
PF3D7_1149200 |
Ring-infected erythrocyte surface antigen (RESA 3) |
Unknown, essential |
Peak transcription mid-cycle |
|
PF3D7_1401600 |
PHISTb |
MEC domain-containing protein |
||
|
PF3D7_1252700 |
PHISTb |
Unknown function |
||
|
PF3D7_1201000 |
PHISTb |
Unknown function |
||
|
PF3D7_0532400 |
Lysine-rich membrane-associated PHISTb protein |
|||
|
PF3D7_0424600 |
PHISTb |
Peak transcription mid-cycle |
||
|
PF3D7_0201700 |
DnaJ protein, putative |
|||
|
PF3D7_0816600 |
Chaperone ClpB1 |
Apicoplast protein |
Despite the important role of DGs, many basic aspects of DG formation in Plasmodium parasites remain unknown, hampering the study of this important organelle. In this study we aimed to determine the timing of DG biogenesis and identify previously undescribed DG proteins. In order to investigate the timing of DG biogenesis, we generated parasites expressing RESA fused to mNeonGreen, allowing DG biogenesis to be observed by live video microscopy. RESA is the only currently known DG protein appropriate for use in DG formation imaging as the other known DG proteins are expressed earlier the intra-erythrocytic life cycle and localize to the PVM, where fluorescent signal would obscure signal from the forming DGs [54]. Using this knowledge of DG biogenesis, we applied a bioinformatics approach to identify additional DG proteins.
Methods
Parasite culture
P. falciparum erythrocytic stage parasites of strain 3D7 were cultured in human erythrocytes (UK National Blood Transfusion Service and Cambridge Bioscience) at 3 % haematocrit and 37 °C, 5 % CO2 in RPMI-1640 medium (Life Technologies) supplemented with 2.3 g l–1 sodium bicarbonate, 4 g l–1 dextrose, 5.957 g l–1 HEPES, 50 µM hypoxanthine, 0.5 % AlbuMax type II (Gibco) and 2 mM l-glutamine [complete RPMI (cRPMI)] according to established procedures [55].
Transfection
Parasites were transfected using the schizont method [56]. For each transfection, 20 µg of plasmid in 100 µl buffer was precipitated using 10 µl sodium acetate and 250 µl 100 % ethanol overnight at −80 °C. Precipitated DNA was washed with 70 % ethanol and air dried in a sterile environment before re-suspension in 10 µl sterile TE buffer. The DNA for the transfections was resuspended in 10 µl TE and 100 µl AMAXA P3 buffer solution was added.
To prepare the parasites for transfection, two T-75 flasks of 3D7 parasite cultures containing predominantly late-stage schizonts were synchronized tightly using two rounds of density gradient centrifugation using 70 % Percoll (GE Healthscience) and incubated at 37 °C with 25 nM of the parasite egress inhibitor ML10 for 1.5 h [57] to maximize numbers of late-stage segmented schizonts. The parasites were then washed with cRPMI to remove ML10 and resuspended in 5 ml cRPMI, which was distributed over five microcentrifuge tubes (one tube per transfection) and incubated at 37 °C for 18 min. The cells were pelleted, re-suspended in the DNA/AMAXA buffer solution and subsequently transferred to AMAXA transfection cuvettes and transfected using the LONZA Nucleofactor electroporation device using the P3 Primary Cell transfection reagent programme. The electroporated cell suspension was transferred to T-25 cell culture flasks containing 300 µl packed red blood cells (RBCs) in 2 ml cRPMI and the released merozoites were allowed to invade in a shaking incubator. After 30 min, 8 ml of medium was added to each flask and the cultures were transferred to a non-shaking incubator. After 24 h, WR99210 [a kind gift of Prof. David Baker, London School of Hygiene & Tropical Medicine (LSHTM)] was added to 2.5 nM to the parasites to select for transfectants. Transfected parasites were generally recovered after approximately 3 weeks. Integrants were selected with treatment with G418 (Generon) at 600 µg ml−1.
Plasmids
We aimed to produce a plasmid encoding a fusion of mNeonGreen (mNG) to the 3′ region of RESA (PF3D7_0102200), followed by sequence encoding the T2A skip peptide and a neomycin resistance marker, to allow for selection of parasites with integrated plasmid using G418 [58, 59]. First, the gene encoding GFP was removed from the plasmid pRESA-GFP [60] by digestion with PstI and MluI and replaced with a 705 bp insert encoding mNG that was amplified using primers CVO550 and CVO551 (Table S1, available in the online version of this article) and digested with PstI and MluI for insertion into the pRESA-GFP backbone, producing plasmid pTV001. The genes encoding the T2A peptide and G418 resistance marker were added by amplification of a fusion of these two genes from JT02-01-31 (a kind gift of James Thomas, LSHTM) using primers CVO576 and CVO577 (Table S1) and inserted at the 3′ end of the mNG fragment by digestion of pTV001 and the PCR fragment with MluI. The fragments were joined using In-Fusion (TaKaRa), producing plasmid pTV002, encoding a fusion of the 3′ 821 bp of the gene encoding RESA, the ORF encoding the T2A peptide and G418 the resistance marker.
Genomic DNA isolation
Erythrocytes were pelleted by centrifugation at 2000 g for 5 min and the resulting pellet was resuspended in an equal volume of 0.15 % saponin in PBS to release the parasites from the erythrocytes. Parasites were pelleted by centrifugation at 13 000 g for 5 min and the parasite DNA was isolated using the Monarch Genomic DNA Purification Kit (New England Biolabs).
Immunoblotting
Late-stage segmented schizonts were isolated from highly synchronized cultures by flotation on a 70 % Percoll gradient [61] and blocked with 25 nM ML10 until Giemsa smears showed only late-stage segmented schizonts. The schizonts were pelleted by centrifugation at 2000 g for 5 min, pellets were resuspended in SDS-PAGE gel-loading buffer and then heated to 98 °C for 20 min.
Proteins were separated by SDS-PAGE on 12.5 % gels at 200 V for 45 min and subsequently transferred to a nitrocellulose membrane at 0.1 A and 25 V for 50 min using a Bio Rad Transblot Turbo transfer system. The membranes were blocked with 5 % milk powder (Waitrose) dissolved in PBS-Tween for 1 h at room temperature before incubation with either anti-mNG primary antibody diluted 1 : 1000 in PBS-Tween or horseradish peroxidase (HRP)-linked anti-aldolase antibody (Abcam) diluted 1 : 5000 in PBS-Tween for 1 h. After extensive washing, the blot probed with the anti-mNG antibody (ChromoTek) was incubated with HRP-linked secondary antibody (Bio-Rad) for 1 h at room temperature and after extensive washing with PBS both blots were developed with Clarity ECL Western blotting substrate (Bio-Rad). Both were imaged on a Bio-Rad ChemiDoc and the images were further cropped and sized using Adobe Photoshop. Figures were produced using Adobe Illustrator.
Video microscopy
Highly synchronous late-stage schizonts were isolated on a 70 % Percoll gradient. Serial dilutions of schizonts in cRPMI containing 100 µM resveratrol were transferred to Ibidi poly-l-lysine µ-Slide VI0.4 channel slides. Slide wells were then sealed with petroleum jelly to prevent sample dehydration. Parasites were maintained at 37 °C during transport and transferred to a pre-heated Okolab Microscope Incubator Cage with Gas Micro-Environmental Chamber and Air Heater for live imaging. Imaging was carried out at 37 °C and in an atmosphere containing 5 % CO2 on a Nikon Eclipse TE fluorescence microscope equipped with a Hamamatsu ORCA-flash 4.0 digital camera C11440 controlled using Nikon NIS-Elements version 5.3 software. Schizonts were imaged every 5 min until egress had occurred in a majority of schizonts. Image data were analysed using NIS-Elements AR Analysis Software v. 4.51.01. Three replicate experiments recording DG formation to egress provided timing data for 47, 14 and 30 schizonts, respectively. Schizonts were numbered and timing from granular fluorescence patterning formation to egress was measured individually.
Statistics
Box-and-whisker plots, interquartile intervals and average of timing of DG formation to egress times were generated using GraphPad Prism 9.
Co-expression network analysis
A query of the www.malaria.tools database using RESA (PF3D7_0102200) as input gave an initial co-expression neighbourhood dataset of 59 proteins (Table S2). The dataset was further analysed using PlasmoDB.org; proteins lacking a signal sequence or transmembrane (TM) domain or annotated as having a function unlikely to be performed by secreted proteins were removed. The final list of proteins for further investigation comprised 23 proteins (Table 1).
Immunofluorescence assays
Smears of late-stage segmented 3D7 P. falciparum schizonts purified on a Percoll gradient were air dried and stored with desiccant beads at −20 °C. Parasites were fixed with acetone for 30 min, circled using an immuno-pen and subsequently blocked with 3 % BSA in PBS (blocking solution) for a further 30 min. Primary antibodies were diluted in blocking solution and applied to the smears which were incubated at room temperature for 1 h. Anti-HSP40 (PF3D7_0501100) antibodies (a kind gift of Prof. Catherine Braun-Breton, University of Montpellier) were used at a dilution of 1 : 250; anti-AMA1 and anti-Ron4 antibodies (kind gifts of Mike Blackman, Francis Crick Institute) were used at a dilution of 1 : 100 and 1 : 250, respectively, and the anti-RESA mAb 28/2 (obtained from the antibody facility at the Walter and Eliza Hall Institute of Medical Research) was used at a dilution of 1 : 500. The slides were then washed three times with PBS before application of the appropriate fluorophore-linked secondary antibodies and Hoechst 33 342 nuclear stain at 15 µg ml−1 in blocking solution and incubation for a further 45 min at room temperature. Following three washes with PBS, the slides were covered with Vectashield antifade mounting medium and sealed with nail polish. Parasites were imaged on a Nikon Eclipse TE fluorescence microscope equipped with a Hamamatsu ORCA-flash 4.0 digital camera C11440. The images were deconvolved using the Richardson–Lucy algorithm with 15 iterations using Nikon NIS-Elements version 5.3 software. The images where then separated, cropped, coloured and overlaid using FIJI software. Images were further cropped and sized using Photoshop. Figures were produced using Illustrator. Pearson’s coefficients were generated using the FIJI (ImageJ) Colocalize tool.
Results
Generation of a reporter strain for the study of DG formation
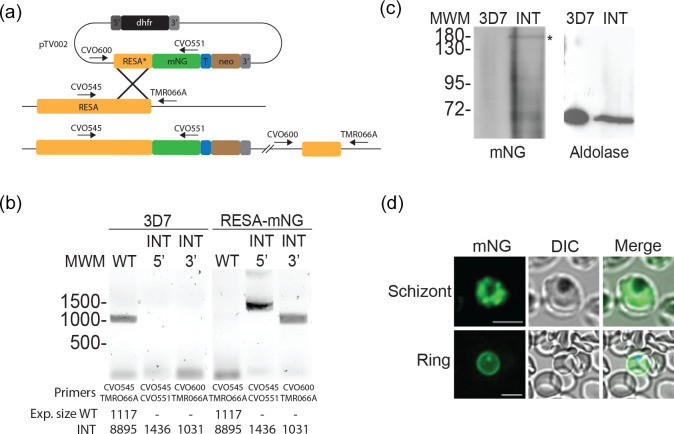
To study the biosynthesis of DGs and the movement of DG proteins through the secretory system, we fused the gene encoding the well-established DG marker RESA [22] with the gene encoding mNG. This fusion was introduced into 3D7 parasites and integration into the native RESA locus was selected for using selection-linked integration (SLI) [58] (Fig. 2a). Integration of the RESA-mNG gene fusion into the native RESA locus was verified using PCR and production of the expected mNG fusion protein product was verified by immunoblotting (Fig. 2b, c). The transgenic parasites were brightly fluorescent in the schizont stage and displayed the expected fluorescence around the periphery of the infected erythrocyte after the parasites had invaded, as seen previously with RESA-GFP fusions [62] (Fig. 2d).
Fig. 2.
Generation of an mNeonGreen-RESA-expressing parasite. (a) Integration strategy using selection-linked integration (SLI). mNG, mNeonGreen; T, T2A peptide; neo, amino 3′-glycosylphosphotransferase (neomycin resistance gene); 5′ and 3′, 5′ and 3′ regulatory regions, respectively. The plasmid pTV002, containing a fusion of the 3′ region of the RESA gene and the gene encoding mNG, was introduced into 3D7 parasites. Transfectants were selected with WR and integrants were subsequently selected with G418. (b) Integration PCR of genomic DNA from wild-type (3D7, left) and integrant (INT, right) parasites using the indicated primer pairs. See panel (a) for the binding sites of the primers. The expected sizes of the PCR products are indicated at the bottom. (c) Anti-mNeonGreen immunoblot of 3D7 (right) and RESA-mNG integrant parasite extracts (left). The band of the expected size is indicated (*). The same extracts were probed with anti-aldolase antibodies as loading control (right). (d) Live-cell fluorescence imaging of parasites expressing RESA-mNG. Top, schizont; bottom, ring. Bars, 5 µm.
Timing of DG biogenesis
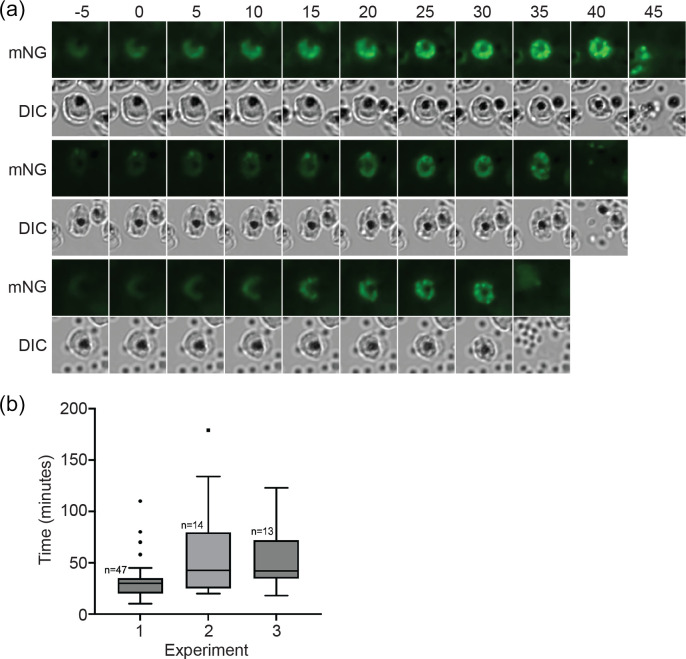
To determine the timing of DG biogenesis we observed the appearance of green fluorescence and the coalescence of the fluorescence into distinct organelles using live video microscopy imaging of the transgenic RESA-mNG-expressing parasites. As RESA expression initiates late in the erythrocytic cycle, as supported by our initial imaging experiments, we started imaging tightly synchronized late-stage schizonts from 46 to 47 h post-invasion at 5 min intervals. A 5 min interval was chosen to minimize the toxic effect of the laser exposure whilst providing an interval narrow enough to provide a reliable time frame. In three replicate experiments, schizonts were observed to undergo DG biogenesis, from the appearance of DG fluorescence to egress. The start of DG biosynthesis was set at the time of increasing fluorescence with the appearance of granular foci of fluorescence where one or more defined foci of fluorescence is visible (t=0) (Fig. 3a). The averaging of the times within the interquartile interval of each replicate revealed that DG formation occurs approximately 37 min prior to egress from the infected erythrocyte (Fig. 3b). Several outliers were observed in which the period from DG formation to egress was far greater than the majority of the other events (five outliers out of 74 total events). These outliers, which fell outside of the interquartile intervals, were not included in the calculation of average DG formation to egress timing. In the outliers in which the timing of DG formation to egress greatly exceeded the average, the cells were observed to reach a point at which egress would be expected to occur based on appearance by both fluorescence and digital interference contrast (DIC) microscopy, but egress appeared blocked or delayed. In these instances, DG fluorescence grew progressively brighter than in other cells as the RESA-mNG fusion continued to be produced and transported to the DGs; no other difference in appearance was observed. We had previously observed inhibited egress and eventual parasite death when imaging without the antioxidant resveratrol.
Fig. 3.
Video microscopy of dense granule formation. (a) Highly synchronized RESA-mNG-expressing parasites were observed using live video microscopy at intervals of 5 min. Parasites were imaged on three separate occasions; each row is taken from a different experiment. The formation of punctate spots of fluorescence was designated as the start of dense granule formation (time point 0). mNG, mNeonGreen fluorescence; DIC, differential interference contrast. (b) Quantification of dense granule formation for the three different experiments. The boxes represent the interquartile range, where 50% of the data points are found. The horizontal line crossing the box represents the median. The y-axis represents the time (in minutes) from the first detection of clustering of RESA-mNG fluorescence in spots to egress of the parasites.
As P. falciparum rhoptries are formed predominantly between the second and fourth round of nuclear division and merozoite formation begins at the end of the fourth round of nuclear division [63], these experiments indicate that DGs are unusual among the apical organelles in that they are formed well after the final round of nuclear division – on average only 37 min prior to egress – a tiny fraction (1.3%) of the entire 48 h intra-erythrocytic life cycle.
Co-expression network analysis to identify candidate DG proteins
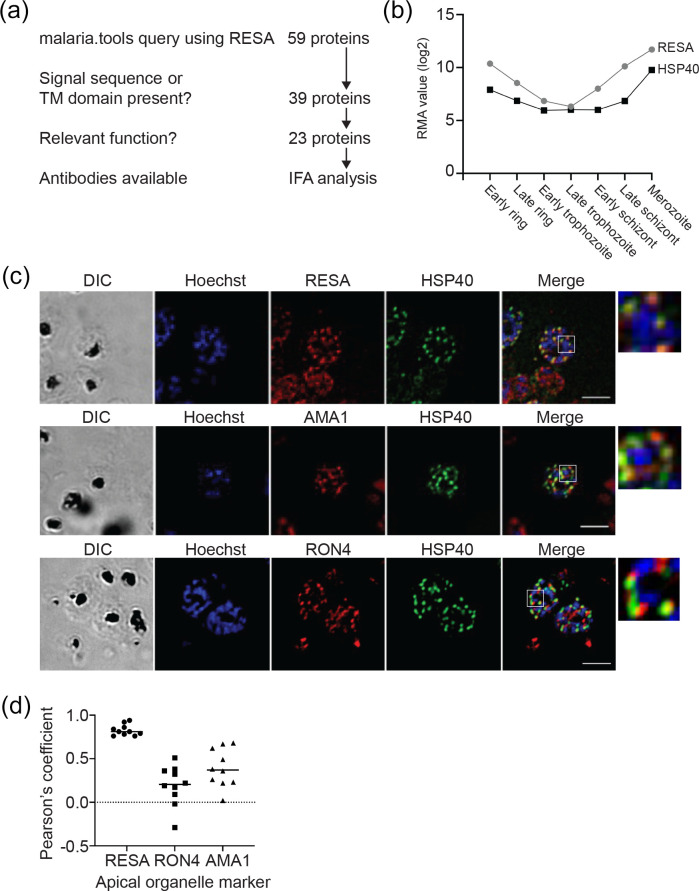
As our results above revealed that DGs are formed very late in the intra-erythrocytic cycle, we inferred that expression network analysis to find proteins with a late-stage expression peak around the time of DG formation could identify potential additional DG proteins for further investigation. We therefore searched for proteins with expression profiles similar to that of RESA using the malaria.tools database [64] (Fig. 4a). This provided a co-expression neighbourhood dataset comprising 59 proteins (Table S2). A query of Plasmodb.org of the identified proteins provided data on protein features, including TM domains, signal sequences and predicted export signals, mutant phenotype and rodent genetic modifications where available [65, 66]. Of the proteins in the original malaria.tools output, 39 (66 %) contained one TM domain or a signal sequence, indicating that they can enter the secretory pathway. The output also contained six RESA orthologues, which were included for further analysis. Proteins with an annotated function that indicates that they are not present in DGs were omitted.
Fig. 4.
Identification of additional dense granule proteins. (a) Outline of malaria.tools query using RESA (PF3D7_0102200). (b) Comparison of expression pattern of RESA and HSP40. Robust multi-array averaging (RMA) values of RESA (grey) and HSP40 (black) expression levels at different stages of the intra-erythrocytic life cycle are shown. Data were obtained from Le Roch et al. [54]. (c) Co-staining of P. falciparum parasites using anti-HSP40 and the dense granule marker RESA (top), the microneme marker AMA1 (middle) and the rhoptry neck marker RON4 (bottom). Samples were also stained with Hoechst 33 342 to visualize DNA. Panels on the far right show an expanded view of the region indicated in the white boxes. Bar, 5 µm. (d) Pearson’s correlation coefficient (PCC) of the colocalization of the anti-HSP40 staining and the staining using the indicated antibodies that recognize apical organelle markers. For each combination, the PCC was determined using ten clearly labelled merozoites in different schizonts.
Proteins were selected for further analysis on the criteria of having a previously described function with potential relevance to erythrocyte remodelling or protein transport and the presence of a signal sequence or TM domain capable of targeting the protein to the secretory pathway. Selection based on these criteria provided a list of 23 potential DG proteins (Table 2). Of the proteins that did not contain a TM domain or signal sequence, three were included in the final set based on their predicted function [v-SNARE, gametogenesis implicated protein (GIG) and DYN3/DrpC]. As v-SNAREs mediate fusion of vesicles and granules to target membranes [67–69], this v-SNARE was included to investigate a potential role in DG protein transport or fusion of DGs to the parasite membrane. GIG is implicated in gametocyte production and was included as it may point to gametocyte-specific alteration of the host cell [70]. The dynamin-like protein DrpC was included as tgDrpC is involved in vesicular transport and mitochondrial fission [71–73], and tgDrpB is required for formation of secretory organelles in T. gondii [74]. The role of DrpC in endocytosis has not been investigated. The final list of putative DG proteins comprised 23 proteins (Table 2). The previously identified DG proteins PTEX88 and LSA3 are present in the malaria.tools output, as well as several RESA N-terminal sequences, supporting the use of this technique as a tool for identifying DG proteins. However, as certain DG proteins, including EXP1 and EXP2, do not have this distinctive late-stage expression peak, they were not present in the output and it is likely that other DG proteins were not identified in this analysis for the same reason. Exported proteins are greatly overrepresented in the final list, and proteins for which mutant phenotypes have been described, for example PTP1 and PTP5, and members of the FIKK family, predominantly have roles in host cell remodelling and cytoprotection [75–77]. These characteristics are in keeping with the model that DGs are important mediators of host cell remodelling.
Table 2.
Putative Plasmodium falciparum dense granule proteins identified in malaria.tools using RESA as query
|
Name |
Alias |
Description |
piggyBac mutagenesis |
Rodent GM phenotype |
Annotation (and putative PEXEL sequence) |
|---|---|---|---|---|---|
|
PF3D7_0935600 |
GIG |
Gametogenesis implicated protein |
_ |
_ |
No transmembrane domain (TM), signal sequence or export signal |
|
PF3D7_0220000 |
LSA3 |
Known DG protein |
Dispensable |
_ |
Recessed signal sequence, TM at C terminus, RSLGE |
|
PF3D7_1201200 |
Plasmodium RESA N-terminal |
Plasmodium exported protein (PHISTa-like), unknown function |
Dispensable |
_ |
TM domain, recessed signal sequence, exported, RKLAD |
|
PF3D7_0702100 |
Plasmodium RESA N terminal |
Plasmodium exported protein (PHISTb), unknown function, pseudogene |
_ |
_ |
Exported, PEXEL-like sequences |
|
PF3D7_1016700 |
Plasmodium RESA N-terminal |
Unknown function |
_ |
_ |
TM domains, exported protein |
|
PF3D7_0424600 |
Plasmodium RESA N-terminal |
Unknown function |
Dispensable |
_ |
Exported protein |
|
PF3D7_1002100 |
PTP5 |
EMP1-trafficking protein |
Dispensable |
_ |
Recessed signal sequence, exported protein, RLLSE |
|
PF3D7_0501100 |
HSP40 |
Heat shock protein 40, type II |
Essential |
_ |
Recessed signal sequence, exported, RSLAE |
|
PF3D7_1218500 |
DYN3/DRPC |
Putative dynamin-like protein |
Essential |
_ |
No signal sequence or TM |
|
PF3D7_1016800 |
Plasmodium RESA N-terminal |
Plasmodium exported protein (PHISTc), unknown function |
Dispensable |
_ |
TM domain, exported |
|
PF3D7_1102800 |
ETRAMP11.2 |
Early transcribed membrane protein |
Essential |
_ |
Signal sequence and TM domain |
|
PF3D7_1102700 |
ETRAMP11.1 |
Early transcribed membrane protein |
Dispensable |
_ |
Signal sequence and TM domain |
|
PF3D7_1105600 |
PTEX88 |
Component of PTEX, known DG protein |
Essential |
Successful modification |
Signal sequence |
|
PF3D7_0424500 |
FIKK4.1 |
Serine threonine protein kinase |
Dispensable |
_ |
TM domain, exported |
|
PF3D7_0902500 |
FIKK9.6 |
Serine threonine protein kinase |
Dispensable |
_ |
No signal sequence or |
|
PF3D7_0202200 |
PTP1 |
EMP1 trafficking protein |
_ |
_ |
TM domains, exported |
|
PF3D7_0219700 |
GEXP20 |
Plasmodium exported protein (PHISTc), unknown function |
Essential |
_ |
Recessed signal sequence, exported |
|
PF3D7_0402100 |
Plasmodium RESA N-terminal |
Plasmodium exported protein (PHISTb), unknown function |
Dispensable |
_ |
Recessed signal sequence, exported |
|
PF3D7_0202500 |
ETRAMP2 |
Early transcribed membrane protein 2 |
Dispensable |
_ |
Signal sequence and TM domain |
|
PF3D7_0523000 |
MDR1 |
Multidrug resistance protein 1 |
Essential |
Non-successful modification |
TM domains |
|
PF3D7_0314100 |
v-SNARE |
Vesicle transport, putative |
Essential |
_ |
TM domain |
|
PF3D7_0425100 |
hyp6 |
Plasmodium exported protein, unknown function |
Dispensable |
_ |
Signal sequence, TM domain, predicted exported |
|
PF3D7_1001900 |
PFJ23-hyp16 |
Unknown function |
_ |
_ |
Recessed signal sequence, TM domains, exported |
Localization of putative DG proteins
To verify whether proteins identified in the bioinformatics screen are transported to DGs, we determined the localization of one of the potential DG proteins for which an antibody was available, HSP40, using immunofluorescence assays. We first compared the expression profile of HSP40 with that of RESA using previously published tramscriptomic data [54], which confirmed that the expression pattern of HSP40 is indeed very similar to that of RESA (Fig. 4b). Co-staining of late-stage schizonts with anti-HSP40 antibodies and microneme, rhoptry and DG markers (AMA1, RON4 and RESA, respectively) revealed strong overlap of the RESA and HSP40 signals in all late-stage segmented schizonts, indicating co-localization of the two proteins (Fig. 4c). Indeed, almost no HSP40 staining distinct from RESA staining was detected in late-stage segmented schizonts. Staining using antibodies against the rhoptry neck marker Ron4 and the microneme marker AMA1 exhibit greatly lower levels of colocalization with anti-HSP40 antibodies. In these samples HSP40 staining was detected distinctly adjacent to AMA1 and RON4 staining. Strong overlap with DG marker staining compared to the distinct staining patterns seen with microneme and rhoptry markers indicate that HSP40 localizes to DGs and not to the micronemes and rhoptries. This was further supported by Pearson’s correlation coefficient (PCC) for the antibody pairs HSP40–RESA (0.810), HSP40–AMA1 (0.370) and HSP40–RON4 (0.205) (Fig. 4d). This result identifies an additional DG protein and indicates that use of co-expression network analysis warrants further investigation as a tool for DG protein identification.
Discussion
Here we have shown that DGs in P. falciparum are formed extremely late in the erythrocytic life cycle – in our experiments DGs were detected on average 37 min prior to egress. However, as egress timing appears to be delayed by the oxidative stress induced by laser exposure and mNG fluorescence, this estimation of the start of DG biogenesis is likely to be an underestimation.
As DGs are formed very late in the intra-erythrocytic cycle and several known DG proteins, including RESA, consequently have a distinctive late-stage expression peak, we inferred that expression network analysis to find other proteins with a similarly distinctive late expression peak could identify potential DG proteins for further investigation. Through a search of the malaria.tools co-expression network platform using RESA as a query we identified genes with expression profiles similar to that of RESA, including heat shock protein HSP40 (Fig. 4b) [54]. We did indeed find that HSP40 co-localizes with RESA, indicating that HSP40 localizes to DGs in the late schizont stage [78]. This supports the use of co-expression network analysis as a method for DG protein prediction. As HSP40 is part of the J-dots, which are small parasite-derived structures found in the cytosol of infected erythrocytes that may be involved in the transport of parasite proteins through the host cell cytosol [79, 80], it indicates that at least some of the components of these structures are present in the DGs and exported to the host cell almost immediately after the parasite enters the host cell. Although the function of these structures remains unclear, it has been postulated that they may have a function in the transport of parasite proteins through the erythrocyte cytosol [81]. Hence, by releasing the component protein of J-dots in DGs, the parasite may enhance its ability to target these proteins to their proper intra-erythrocytic location almost immediately after invasion. This finding therefore may also further support the hypothesis that DGs probably contain many more exported proteins that have yet to be identified. This aligns with the timing of DG discharge immediately following invasion and the hypothesized role of DGs in erythrocyte remodelling. Interestingly, one protein identified in the malaria.tools investigation, PTP1, is necessary for correct formation of Maurer’s clefts and linking Maurer’s clefts to the cytoskeleton [77]. This may well indicate that Maurer’s clefts are formed very soon after invasion, using proteins that are exported immediately after DGs have been released.
Our bio-informatics approach identified the known DG proteins LSA3 and PTEX88 [34, 43]. However, several other DG proteins, including EXP1 and EXP2, were not included in the malaria.tools output, probably because they do not share the distinctive late-stage expression peak of RESA and instead are also expressed at other times throughout the life cycle. It is likely that other unidentified DG proteins will also be missing from the output owing to having expression profiles dissimilar to that of RESA. Combined, the listing of known and recently identified DG proteins, and the localization of the one protein that has been tested to date to the DGs, suggests that co-expression network analysis using malaria.tools works as a predictive method of identifying candidate proteins for further analysis. Further, exported proteins are over-represented within the dataset. As host cell effector proteins must be exported, the high number of exported proteins within the dataset accords with the understanding of the role of DGs as a secretory compartment containing proteins with roles in host cell modification that are released immediately after erythrocyte invasion. A recently developed database of predicted Plasmodium DG proteins also includes several PHIST proteins [48]. Whilst the function of many individual PHIST proteins remains unknown, the prediction of another family of exported proteins localizing to the DGs supports the theory of DG function in erythrocyte remodelling.
Three proteins with a transcriptional profile similar to that of RESA are of potential interest based on their putative function, despite lacking a signal sequence or TM domain: PF3D7_0314100, annotated as a v-SNARE, GIG and the dynamin-like protein DrpC. These proteins may serve important functions in the fusion of the DGs to the parasite plasma membrane, gametocyte-specific functions immediately after invasion and vesicular trafficking to the DGs, respectively [67–72, 82].
The endogenously tagged RESA line may be useful as a tool in future work aiming to identify factors that are involved in DG formation, sorting of proteins to the DGs and as a screen for drugs which act to inhibit DG formation or release.
This is the first description of the timing of DG biogenesis in Plasmodium parasites. This finding reveals that DGs are only present within the parasite for a very short portion (1.3 %) of the 48 h asexual cycle. We also find that the exported protein HSP40 is present in DGs. As HSP40 interacts with HSP70x to form a chaperone complex within the infected host cell, this finding further supports the hypothesis that DGs have an important role in host cell modification, in this instance through the chaperoning of parasite effector proteins [78]. These results provide further insight into how the parasite prepares for modification of the host cell.
Supplementary Data
Funding information
This work was supported by an MRC-LID studentship to T.V. and a Medical Research Council Career Development Award (MR/R008485/1) to C.v.O.
Acknowledgements
We are grateful to Dr James Thomas (LSHTM) for providing the JT02-01-31 plasmid containing the SLI fragment and Dr Gerhard Wunderlich (Institute of Biomedical Sciences, University of Sao Paulo) for providing plasmid pRESA-HA. We thank Prof. Catherine Braun-Breton (University of Montpellier) for providing the anti-HSP40 antibody, and Mike Blackman (Francis Crick Institute) for the anti-AMA1 and anti-RON4 antibodies. We thank Prof. David Baker for sharing WR99210. The authors acknowledge the facilities and the scientific and technical assistance of the LSHTM Wolfson Cell Biology Facility, with specific thanks to Liz McCarthy.
Conflicts of interest
The authors declare that there is no conflict of interest.
Footnotes
Abbreviations: DG, dense granule; DGPD, dense granule protein database; GIG, gametogenesis implicated protein; MJ, moving junction; mNG, mNeonGreen; PCC, Pearson's correlation coefficient; PEXEL, Plasmodium export element; PTEX, plasmodium translocon of exported proteins; PV, parasitophorous vacuole; PVM, parasitophorous vacuole membrane; SLI, selection-linked integration; TM, transmembrane.
Two supplementary tables are available with the online version of this article.
References
- 1.Carruthers VB, Sibley LD. Sequential protein secretion from three distinct organelles of Toxoplasma gondii accompanies invasion of human fibroblasts. Eur J Cell Biol. 1997;73:114–123. [PubMed] [Google Scholar]
- 2.Singh S, Alam MM, Pal-Bhowmick I, Brzostowski JA, Chitnis CE. Distinct external signals trigger sequential release of apical organelles during erythrocyte invasion by malaria parasites. PLoS Pathog. 2010;6:e1000746. doi: 10.1371/journal.ppat.1000746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kats LM, Cooke BM, Coppel RL, Black CG. Protein trafficking to apical organelles of malaria parasites - building an invasion machine. Traffic. 2008;9:176–186. doi: 10.1111/j.1600-0854.2007.00681.x. [DOI] [PubMed] [Google Scholar]
- 4.Gubbels MJ, Duraisingh MT. Evolution of apicomplexan secretory organelles. Int J Parasitol. 2012;42:1071–1081. doi: 10.1016/j.ijpara.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao J, Kaneko O, Thongkukiatkul A, Tachibana M, Otsuki H, et al. Rhoptry neck protein RON2 forms a complex with microneme protein AMA1 in Plasmodium falciparum merozoites. Parasitol Int. 2009;58:29–35. doi: 10.1016/j.parint.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Adams JH, Sim BKL, Dolan SA, Fang X, Kaslow DC, et al. A family of erythrocyte binding proteins of malaria parasites. Proc Natl Acad Sci. 1992;89:7085–7089. doi: 10.1073/pnas.89.15.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camus D, Hadley TJ. A Plasmodium falciparum antigen that binds to host erythrocytes and merozoites. Science. 1985;230:553–556. doi: 10.1126/science.3901257. [DOI] [PubMed] [Google Scholar]
- 8.Lee Sim BK, Toyoshima T, David Haynes J, Aikawa M. Localization of the 175-kilodalton erythrocyte binding antigen in micronemes of Plasmodium falciparum merozoites. Mol Biochem Parasitol. 1992;51:157–159. doi: 10.1016/0166-6851(92)90211-2. [DOI] [PubMed] [Google Scholar]
- 9.Peterson MG, Marshall VM, Smythe JA, Crewther PE, Lew A, et al. Integral membrane protein located in the apical complex of Plasmodium falciparum . Mol Cell Biol. 1989;9:3151–3154. doi: 10.1128/mcb.9.7.3151-3154.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Triglia T, Healer J, Caruana SR, Hodder AN, Anders RF, et al. Apical membrane antigen 1 plays a central role in erythrocyte invasion by Plasmodium species. Mol Microbiol. 2000;38:706–718. doi: 10.1046/j.1365-2958.2000.02175.x. [DOI] [PubMed] [Google Scholar]
- 11.Bannister LH, Hopkins JM, Dluzewski AR, Margos G, Williams IT, et al. Plasmodium falciparum apical membrane antigen 1 (PfAMA-1) is translocated within micronemes along subpellicular microtubules during merozoite development. J Cell Sci. 2003;116:3825–3834. doi: 10.1242/jcs.00665. [DOI] [PubMed] [Google Scholar]
- 12.Mitchell GH, Thomas AW, Margos G, Dluzewski AR, Bannister LH. Apical membrane antigen 1, a major malaria vaccine candidate, mediates the close attachment of invasive merozoites to host red blood cells. Infect Immun. 2004;72:154–158. doi: 10.1128/IAI.72.1.154-158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stewart MJ, Schulman S, Vanderberg JP. Rhoptry secretion of membranous whorls by Plasmodium falciparum merozoites. Am J Trop Med Hyg. 1986;35:37–44. doi: 10.4269/ajtmh.1986.35.37. [DOI] [PubMed] [Google Scholar]
- 14.Bannister LH, Mitchell GH. The fine structure of secretion by Plasmodium knowlesi merozoites during red cell invasion. J Protozool. 1989;36:362–367. doi: 10.1111/j.1550-7408.1989.tb05527.x. [DOI] [PubMed] [Google Scholar]
- 15.Ladda R, Aikawa M, Sprinz H. Penetration of erythrocytes by merozoites of mammalian and avian malarial parasites. 1969. J Parasitol. 2001;87:470–478. doi: 10.1645/0022-3395(2001)087[0470:POEBMO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 16.Ghosh S, Kennedy K, Sanders P, Matthews K, Ralph SA, et al. The Plasmodium rhoptry associated protein complex is important for parasitophorous vacuole membrane structure and intraerythrocytic parasite growth. Cell Microbiol. 2017;19:12733. doi: 10.1111/cmi.12733. [DOI] [PubMed] [Google Scholar]
- 17.Alexander DL, Mital J, Ward GE, Bradley P, Boothroyd JC. Identification of the moving junction complex of Toxoplasma gondii: a collaboration between distinct secretory organelles. PLoS Pathog. 2005;1:e17. doi: 10.1371/journal.ppat.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Besteiro S, Michelin A, Poncet J, Dubremetz JF, Lebrun M. Export of a Toxoplasma gondii rhoptry neck protein complex at the host cell membrane to form the moving junction during invasion. PLoS Pathog. 2009;5:e1000309. doi: 10.1371/journal.ppat.1000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamarque M, Besteiro S, Papoin J, Roques M, Vulliez-Le Normand B, et al. The RON2-AMA1 interaction is a critical step in moving junction-dependent invasion by apicomplexan parasites. PLoS Pathog. 2011;7:e1001276. doi: 10.1371/journal.ppat.1001276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Srinivasan P, Beatty WL, Diouf A, Herrera R, Ambroggio X, et al. Binding of Plasmodium merozoite proteins RON2 and AMA1 triggers commitment to invasion. Proc Natl Acad Sci. 2011;108:13275–13280. doi: 10.1073/pnas.1110303108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torii M, Adams JH, Miller LH, Aikawa M. Release of merozoite dense granules during erythrocyte invasion by Plasmodium knowlesi . Infect Immun. 1989;57:3230–3233. doi: 10.1128/iai.57.10.3230-3233.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Culvenor JG, Day KP, Anders RF. Plasmodium falciparum ring-infected erythrocyte surface antigen is released from merozoite dense granules after erythrocyte invasion. Infect Immun. 1991;59:1183–1187. doi: 10.1128/iai.59.3.1183-1187.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riglar DT, Richard D, Wilson DW, Boyle MJ, Dekiwadia C, et al. Super-resolution dissection of coordinated events during malaria parasite invasion of the human erythrocyte. Cell Host Microbe. 2011;9:9–20. doi: 10.1016/j.chom.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 24.de Koning-Ward TF, Dixon MWA, Tilley L, Gilson PR. Plasmodium species: master renovators of their host cells. Nat Rev Microbiol. 2016;14:494–507. doi: 10.1038/nrmicro.2016.79. [DOI] [PubMed] [Google Scholar]
- 25.Ming P, Mingjun L, Longjiao L, Yongle S, Lun H, et al. Identification of novel dense-granule proteins in Toxoplasma gondii by two proximity-based Biotinylation approaches. J Proteome Res. 2018;18:319–330. doi: 10.1021/acs.jproteome.8b00626. [DOI] [PubMed] [Google Scholar]
- 26.Griffith MB, Pearce CS, Heaslip AT. Dense granule biogenesis, secretion, and function in Toxoplasma gondii . J Eukaryot Microbiol. 2022;69:e12904. doi: 10.1111/jeu.12904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho C-M, Beck JR, Lai M, Cui Y, Goldberg DE, et al. Malaria parasite translocon structure and mechanism of effector export. Nature. 2018;561:70–75. doi: 10.1038/s41586-018-0469-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elsworth B, Matthews K, Nie CQ, Kalanon M, Charnaud SC, et al. PTEX is an essential nexus for protein export in malaria parasites. Nature. 2014;511:587–591. doi: 10.1038/nature13555. [DOI] [PubMed] [Google Scholar]
- 29.Garten M, Nasamu AS, Niles JC, Zimmerberg J, Goldberg DE, et al. EXP2 is a nutrient-permeable channel in the vacuolar membrane of Plasmodium and is essential for protein export via PTEX. Nat Microbiol. 2018;3:1090–1098. doi: 10.1038/s41564-018-0222-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pei X, Guo X, Coppel R, Bhattacharjee S, Haldar K, et al. The ring-infected erythrocyte surface antigen (RESA) of Plasmodium falciparum stabilizes spectrin tetramers and suppresses further invasion. Blood. 2007;110:1036–1042. doi: 10.1182/blood-2007-02-076919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beck JR, Muralidharan V, Oksman A, Goldberg DE. PTEX component HSP101 mediates export of diverse malaria effectors into host erythrocytes. Nature. 2014;511:592–595. doi: 10.1038/nature13574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charnaud SC, Kumarasingha R, Bullen HE, Crabb BS, Gilson PR. Knockdown of the translocon protein EXP2 in Plasmodium falciparum reduces growth and protein export. PLoS One. 2018;13:e0204785. doi: 10.1371/journal.pone.0204785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Koning-Ward TF, Gilson PR, Boddey JA, Rug M, Smith BJ, et al. A newly discovered protein export machine in malaria parasites. Nature. 2009;459:945–949. doi: 10.1038/nature08104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bullen HE, Charnaud SC, Kalanon M, Riglar DT, Dekiwadia C, et al. Biosynthesis, localization, and macromolecular arrangement of the Plasmodium falciparum translocon of exported proteins (PTEX) J Biol Chem. 2012;287:7871–7884. doi: 10.1074/jbc.M111.328591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morita M, Nagaoka H, Ntege EH, Kanoi BN, Ito D, et al. PV1, a novel Plasmodium falciparum merozoite dense granule protein, interacts with exported protein in infected erythrocytes. Sci Rep. 2018;8:3696. doi: 10.1038/s41598-018-22026-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chu T, Lingelbach K, Przyborski JM. Genetic evidence strongly support an essential role for PfPV1 in intra-erythrocytic growth of P. falciparum . PLoS One. 2011;6:e18396. doi: 10.1371/journal.pone.0018396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Günther K, Tümmler M, Arnold HH, Ridley R, Goman M, et al. An exported protein of Plasmodium falciparum is synthesized as an integral membrane protein. Mol Biochem Parasitol. 1991;46:149–157. doi: 10.1016/0166-6851(91)90208-n. [DOI] [PubMed] [Google Scholar]
- 38.Iriko H, Ishino T, Otsuki H, Ito D, Tachibana M, et al. Plasmodium falciparum exported protein 1 is localized to dense granules in merozoites. Parasitol Int. 2018;67:637–639. doi: 10.1016/j.parint.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 39.Simmons D, Woollett G, Bergin-Cartwright M, Kay D, Scaife J. A malaria protein exported into a new compartment within the host erythrocyte. EMBO J. 1987;6:485–491. doi: 10.1002/j.1460-2075.1987.tb04779.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mesén-Ramírez P, Bergmann B, Tran TT, Garten M, Stäcker J, et al. EXP1 is critical for nutrient uptake across the parasitophorous vacuole membrane of malaria parasites. PLoS Biol. 2019;17:e3000473. doi: 10.1371/journal.pbio.3000473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyazaki S, Chitama B-YA, Kagaya W, Lucky AB, Zhu X, et al. Plasmodium falciparum SURFIN4.1 forms an intermediate complex with PTEX components and Pf113 during export to the red blood cell. Parasitol Int. 2021;83:102358. doi: 10.1016/j.parint.2021.102358. [DOI] [PubMed] [Google Scholar]
- 42.Bullen HE, Sanders PR, Dans MG, Jonsdottir TK, Riglar DT, et al. The Plasmodium falciparum parasitophorous vacuole protein P113 interacts with the parasite protein export machinery and maintains normal vacuole architecture. Mol Microbiol. 2022;117:1245–1262. doi: 10.1111/mmi.14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morita M, Takashima E, Ito D, Miura K, Thongkukiatkul A, et al. Immunoscreening of Plasmodium falciparum proteins expressed in a wheat germ cell-free system reveals a novel malaria vaccine candidate. Sci Rep. 2017;7:46086. doi: 10.1038/srep46086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aikawa M, Torii M, Sjölander A, Berzins K, Perlmann P, et al. Pf155/RESA antigen is localized in dense granules of Plasmodium falciparum merozoites. Exp Parasitol. 1990;71:326–329. doi: 10.1016/0014-4894(90)90037-d. [DOI] [PubMed] [Google Scholar]
- 45.Da Silva E, Foley M, Dluzewski AR, Murray LJ, Anders RF, et al. The Plasmodium falciparum protein RESA interacts with the erythrocyte cytoskeleton and modifies erythrocyte thermal stability. Mol Biochem Parasitol. 1994;66:59–69. doi: 10.1016/0166-6851(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 46.Silva MD, Cooke BM, Guillotte M, Buckingham DW, Sauzet J-P, et al. A role for the Plasmodium falciparum RESA protein in resistance against heat shock demonstrated using gene disruption. Mol Microbiol. 2005;56:990–1003. doi: 10.1111/j.1365-2958.2005.04603.x. [DOI] [PubMed] [Google Scholar]
- 47.Diez-Silva M, Park Y, Huang S, Bow H, Mercereau-Puijalon O, et al. Pf155/RESA protein influences the dynamic microcirculatory behavior of ring-stage Plasmodium falciparum infected red blood cells. Sci Rep. 2012;2:614. doi: 10.1038/srep00614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu H, Lu Z, Feng H, Chen G, Wang Y, et al. DGPD: a knowledge database of dense granule proteins of the Apicomplexa. Database. 2022;2022:baac085. doi: 10.1093/database/baac085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.El Bakkouri M, Pow A, Mulichak A, Cheung KLY, Artz JD, et al. The Clp chaperones and proteases of the human malaria parasite Plasmodium falciparum . J Mol Biol. 2010;404:456–477. doi: 10.1016/j.jmb.2010.09.051. [DOI] [PubMed] [Google Scholar]
- 50.Rezaei F, Sharif M, Sarvi S, Hejazi SH, Aghayan S, et al. A systematic review on the role of GRA proteins of Toxoplasma gondii in host immunization. J Microbiol Methods. 2019;165:105696. doi: 10.1016/j.mimet.2019.105696. [DOI] [PubMed] [Google Scholar]
- 51.Marti M, Good RT, Rug M, Knuepfer E, Cowman AF. Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science. 2004;306:1930–1933. doi: 10.1126/science.1102452. [DOI] [PubMed] [Google Scholar]
- 52.Tomavo S. Evolutionary repurposing of endosomal systems for apical organelle biogenesis in Toxoplasma gondii . Int J Parasitol. 2014;44:133–138. doi: 10.1016/j.ijpara.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 53.Hallée S, Counihan NA, Matthews K, de Koning-Ward TF, Richard D. The malaria parasite Plasmodium falciparum Sortilin is essential for merozoite formation and apical complex biogenesis. Cell Microbiol. 2018;20:e12844. doi: 10.1111/cmi.12844. [DOI] [PubMed] [Google Scholar]
- 54.Le Roch KG, Zhou Y, Blair PL, Grainger M, Moch JK, et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- 55.Crabb BS, Rug M, Gilberger TW, Thompson JK, Triglia T, et al. Transfection of the human malaria parasite Plasmodium falciparum . Methods Mol Biol. 1999;270:267–276. doi: 10.1385/1-59259-793-9:263. [DOI] [PubMed] [Google Scholar]
- 56.Collins CR, Hackett F, Strath M, Penzo M, Withers-Martinez C, et al. Malaria parasite cGMP-dependent protein kinase regulates blood stage merozoite secretory organelle discharge and egress. PLoS Pathog. 2013;9:e1003344. doi: 10.1371/journal.ppat.1003344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ressurreição M, Thomas JA, Nofal SD, Flueck C, Moon RW, et al. Use of a highly specific kinase inhibitor for rapid, simple and precise synchronization of Plasmodium falciparum and Plasmodium knowlesi asexual blood-stage parasites. PLoS One. 2020;15:e0235798. doi: 10.1371/journal.pone.0235798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Birnbaum J, Flemming S, Reichard N, Blancke Soares A, Mesén-Ramírez P, et al. Selection linked integration (SLI) for endogenous gene tagging and knock sideways in Plasmodium falciparum parasites. Protoc Exch. 2017 [Google Scholar]
- 59.Birnbaum J, Flemming S, Reichard N, Soares AB, Mesén-Ramírez P, et al. A genetic system to study Plasmodium falciparum protein function. Nat Methods. 2017;14:450–456. doi: 10.1038/nmeth.4223. [DOI] [PubMed] [Google Scholar]
- 60.de Azevedo MF, Gilson PR, Gabriel HB, Simões RF, Angrisano F, et al. Systematic analysis of FKBP inducible degradation domain tagging strategies for the human malaria parasite Plasmodium falciparum . PLoS One. 2012;7:e40981. doi: 10.1371/journal.pone.0040981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rivadeneira EM, Wasserman M, Espinal CT. Separation and concentration of schizonts of Plasmodium falciparum by Percoll gradients. J Protozool. 1983;30:367–370. doi: 10.1111/j.1550-7408.1983.tb02932.x. [DOI] [PubMed] [Google Scholar]
- 62.Rug M, Wickham ME, Foley M, Cowman AF, Tilley L. Correct promoter control is needed for trafficking of the ring-infected erythrocyte surface antigen to the host cytosol in transfected malaria parasites. Infect Immun. 2004;72:6095–6105. doi: 10.1128/IAI.72.10.6095-6105.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Margos G, Bannister LH, Dluzewski AR, Hopkins J, Williams IT, et al. Correlation of structural development and differential expression of invasion-related molecules in schizonts of Plasmodium falciparum . Parasitology. 2004;129:273–287. doi: 10.1017/S0031182004005657. [DOI] [PubMed] [Google Scholar]
- 64.Tan QW, Mutwil M. Malaria.tools-comparative genomic and transcriptomic database for Plasmodium species. Nucleic Acids Res. 2020;48:D768–D775. doi: 10.1093/nar/gkz662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Amos B, Aurrecoechea C, Barba M, Barreto A, Basenko EY, et al. VEuPathDB: the eukaryotic pathogen, vector and host bioinformatics resource center. Nucleic Acids Res. 2022;50:D898–D911. doi: 10.1093/nar/gkab929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.David S. PlasmoDB: an integrative database of the Plasmodium falciparum genome. Tools for accessing and analyzing finished and unfinished sequence data. Nucleic Acids Res. 2001;29:66–69. doi: 10.1093/nar/29.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dhara M, Yarzagaray A, Makke M, Schindeldecker B, Schwarz Y, et al. V-SNARE transmembrane domains function as catalysts for vesicle fusion. Elife. 2016;5:e17571. doi: 10.7554/eLife.17571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen YA, Scheller RH. SNARE-mediated membrane fusion. Nat Rev Mol Cell Biol. 2001;2:98–106. doi: 10.1038/35052017. [DOI] [PubMed] [Google Scholar]
- 69.Wang T, Li L, Hong W. SNARE proteins in membrane trafficking. Traffic. 2017;18:767–775. doi: 10.1111/tra.12524. [DOI] [PubMed] [Google Scholar]
- 70.Gardiner DL, Dixon MWA, Spielmann T, Skinner-Adams TS, Hawthorne PL, et al. Implication of a Plasmodium falciparum gene in the switch between asexual reproduction and gametocytogenesis. Mol Biochem Parasitol. 2005;140:153–160. doi: 10.1016/j.molbiopara.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 71.Heredero-Bermejo I, Varberg JM, Charvat R, Jacobs K, Garbuz T, et al. TgDrpC, an atypical dynamin-related protein in Toxoplasma gondii, is associated with vesicular transport factors and parasite division. Mol Microbiol. 2019;111:46–64. doi: 10.1111/mmi.14138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Melatti C, Pieperhoff M, Lemgruber L, Pohl E, Sheiner L, et al. A unique dynamin-related protein is essential for mitochondrial fission in Toxoplasma gondii . PLoS Pathog. 2019;15:e1007512. doi: 10.1371/journal.ppat.1007512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Breinich MS, Ferguson DJP, Foth BJ, van Dooren GG, Lebrun M, et al. A dynamin is required for the biogenesis of secretory organelles in Toxoplasma gondii . Curr Biol. 2009;19:277–286. doi: 10.1016/j.cub.2009.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Spielmann T, Gras S, Sabitzki R, Meissner M. Endocytosis in Plasmodium and Toxoplasma parasites. Trends Parasitol. 2020;36:520–532. doi: 10.1016/j.pt.2020.03.010. [DOI] [PubMed] [Google Scholar]
- 75.Davies H, Belda H, Broncel M, Ye X, Bisson C, et al. An exported kinase family mediates species-specific erythrocyte remodelling and virulence in human malaria. Nat Microbiol. 2020;5:848–863. doi: 10.1038/s41564-020-0702-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maier AG, Rug M, O’Neill MT, Brown M, Chakravorty S, et al. Exported proteins required for virulence and rigidity of Plasmodium falciparum-infected human erythrocytes. Cell. 2008;134:48–61. doi: 10.1016/j.cell.2008.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rug M, Cyrklaff M, Mikkonen A, Lemgruber L, Kuelzer S, et al. Export of virulence proteins by malaria-infected erythrocytes involves remodeling of host actin cytoskeleton. Blood. 2014;124:3459–3468. doi: 10.1182/blood-2014-06-583054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu Q, Liang C, Zhou L. Structural and functional analysis of the Hsp70/Hsp40 chaperone system. Protein Sci. 2020;29:378–390. doi: 10.1002/pro.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Petersen W, Külzer S, Engels S, Zhang Q, Ingmundson A, et al. J-dot targeting of an exported HSP40 in Plasmodium falciparum-infected erythrocytes. Int J Parasitol. 2016;46:519–525. doi: 10.1016/j.ijpara.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 80.Külzer S, Rug M, Brinkmann K, Cannon P, Cowman A, et al. Parasite-encoded Hsp40 proteins define novel mobile structures in the cytosol of the P. falciparum-infected erythrocyte. Cell Microbiol. 2010;12:1398–1420. doi: 10.1111/j.1462-5822.2010.01477.x. [DOI] [PubMed] [Google Scholar]
- 81.Behl A, Kumar V, Bisht A, Panda JJ, Hora R, et al. Cholesterol bound Plasmodium falciparum co-chaperone “PFA0660w” complexes with major virulence factor “PfEMP1” via chaperone “PfHsp70-x.”. Sci Rep. 2019;9:2664. doi: 10.1038/s41598-019-39217-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bai M-J, Wang J-L, Elsheikha HM, Liang Q-L, Chen K, et al. Functional characterization of dense granule proteins in Toxoplasma gondii RH strain using CRISPR-Cas9 system. Front Cell Infect Microbiol. 2018;8:300. doi: 10.3389/fcimb.2018.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.