Abstract
In Gram-positive bacteria such as Staphylococcus aureus and the coagulase-negative staphylococci (CoNS), the accessory gene regulator (agr) is a highly conserved but polymorphic quorum-sensing system involved in colonization, virulence and biofilm development. Signalling via agr depends on the interaction of an autoinducing peptide (AIP) with AgrC, a transmembrane sensor kinase that, once phosphorylated activates the response regulator AgrA. This in turn autoinduces AIP biosynthesis and drives target gene expression directly via AgrA or via the post-transcriptional regulator, RNAIII. In this review we describe the molecular mechanisms underlying the agr-mediated generation of, and response to, AIPs and the molecular basis of AIP-dependent activation and inhibition of AgrC. How the environment impacts on agr functionality is considered and the consequences of agr dysfunction for infection explored. We also discuss the concept of AIP-driven competitive interference between S. aureus and the CoNS and its anti-infective potential.
Keywords: agr, autoinducing peptides, inter-bacterial competition, quorum sensing, staphylococci, Staphylococcus aureus
Introduction
Coagulase-negative staphylococci (CoNS) such as Staphylococcus epidermidis and Staphylococcus hominis are primarily skin commensals while the coagulase-positive Staphylococcus aureus is not only a commensal colonizing human nares and skin but also a major opportunistic pathogen [1, 2]. S. aureus infections can be minor or invasive and life-threatening [3]. They may be acute or chronic ranging from skin, soft tissue and medical device-related to bacteraemia, endocarditis, osteomyelitis, food poisoning, septic arthritis, scalded skin and toxic shock syndromes [3]. S. aureus is also on the WHO ‘ESKAPE’ list of multi-antibiotic resistant priority pathogens given that treatment of S. aureus infections has been compounded by the emergence of multi-antibiotic-resistant strains [4]. These include vancomycin-resistant (VRSA and VISA) and methicillin (MRSA)-resistant isolates, which can be further sub-divided into hospital-acquired MRSA (HA-MRSA) and community-acquired (CA-MRSA) strains. S. aureus has often been reported as a co-infecting microbe in polymicrobial infections where it may be co-operative or competitive [5]. In the context of difficult-to-eradicate infections such as non-healing diabetic foot ulcers and in the lungs of individuals with cystic fibrosis (CF), co-infections of S. aureus with P. aeruginosa are indicative of much poorer clinical outcomes than in those infected with either species alone [6].
As both commensal and pathogen, S. aureus is capable of rapidly responding and adapting to fluctuating host and inanimate environments and switching between colonization and pathogenic modes. Such adaptation depends on local extracellular signals that include oxygen availability, temperature, pH, nutrient limitation, bacterial cell population density, other co-localizing microbes, antimicrobials and is associated with specific host tissue interactions, implanted medical devices, abiotic surfaces encountered in households or hospitals as well as particulate air pollution [7–9].
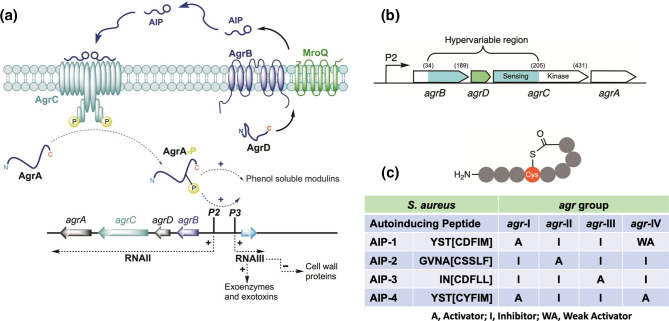
The versatility of S. aureus as a pathogen revolves around diverse cell-wall colonization factors, immune modulating agents and secreted exoproducts [3], many of which are controlled via quorum sensing (QS), a mechanism for synchronizing gene expression via self-generated diffusible signal molecules in a population-dependent manner [10]. In Gram-positive pathogens including the staphylococci, clostridia, enterococci and listeria, QS is mediated by the accessory gene regulator (agr) system [11, 12]. In Staphylococcus aureus , agr reciprocally regulates multiple cell-wall proteins (e.g. immunoglobulin and fibronectin-binding proteins) and exotoxins (e.g. haemolysins, enterotoxins, leucocidins, toxic shock syndrome toxin, exoenzymes (nucleases, proteases, lipases) and the phenol soluble modulins (PSMs), a family of short amphipathic peptides with cytolytic activity similar to δ-toxin [3] (Fig. 1). Staphylococcal surface proteins promote adherence to host tissues and aid immune evasion while exotoxins cause tissue damage and many function as super-antigens promoting the onset of shock-like syndromes [3, 13]. The S. aureus agr system is involved in endosomal escape, intracellular survival and replication [14–16]. With respect to biofilm development, agr contributes to initial attachment, structuring and dispersal [17].
Fig. 1.
The agr QS system in S. aureus negatively (-) regulates the production of capsular polysaccharides and multiple cell-wall proteins involved in host protein interactions including immunoglobulin, fibronectin and fibrinogen binding proteins. agr positively (+) regulates the expression of diverse virulence factor genes including those coding for tissue degrading exoenzymes, haemolysins, enterotoxins, exfoliative toxins leukocidins and phenol soluble modulins.
In acute animal models of skin, soft tissue, respiratory, arthritis and bone, S. aureus agr mutants are attenuated, highlighting a key role for QS-dependent regulation of virulence determinants at these infection sites [18]. Paradoxically, agr is required for skin colonization [19]. However, a functional agr system is dispensable for chronic, biofilm-related infections associated with, for example, implanted medical devices and cystic fibrosis [18]. Furthermore, allelic variation in agr genes contributes to intra- and inter-staphylococcal competition since the cognate agr signal molecule of one staphylococcal agr variant may inhibit agr in a strain possessing a different agr variant [20–22]. In this review we outline the intricate molecular mechanisms underlying the agr-mediated generation of, and response to, autoinducing peptide (AIP) signal molecules focusing primarily on S. aureus . We describe the specifics of AIP-activation and inhibition of the sensor kinase AgrC, how the environment impacts on agr functionality and explore the consequences of agr dysfunction for infection. We also discuss the concept of AIP-mediated competitive interference between S. aureus and the CoNS and its therapeutic potential for suppressing S. aureus skin disease and other infections.
The S. aureus agr system
In S. aureus the agr locus consists of two divergent transcriptional units, agrBDCA and the regulatory RNA effector, RNAIII controlled by the P2 and P3 promoters, respectively [20, 23]. (Fig. 2a). The P2 operon consists of four genes, agrBDCA, which are required for the activation of transcription from the P2 and P3 promoters while the P3 transcript, RNAIII, a 517-nucleotide transcript, is itself the primary effector for the agr response and also encodes δ-haemolysin [24]. AgrA and AgrC constitute a two-component system in which the transmembrane protein AgrC is the histidine sensor kinase and the cytoplasmic AgrA protein is the response regulator [23, 25]. The diffusible agr QS signal is an autoinducing peptide (AIP) (Fig. 2) derived via the AgrB-dependent proteolytic processing of the ribosomally synthesized AgrD pro-peptide. AIPs induce the trans-auto-phosphorylation of AgrC, which transfers the phosphate to a conserved Asp on AgrA. This binds to the agrP2 promoter upregulating agrBCDA, conferring a positive-feedback loop that autoinduces AIP production and drives target gene expression directly via AgrA or via the AgrA-dependent agrP3 promoter and the post-transcriptional regulator, RNAIII [24] (Fig. 2a). There are four allelic variants (I–IV) of the S. aureus agr locus with respect to a hypervariable region contained in the agrB, agrD and agrC genes (Fig. 2b, c). These reflect the amino acid sequence variations in the four S. aureus AIPs and explains their specificity for AgrC as activators of their cognate receptors but competitive inhibitors of the other AgrC variants [20, 26–31]. For example, AIP-1 is a competitive antagonist of the AIP-2/AgrC2 and AIP-3/AgrC3 interactions whereas AIP-2 and AIP-3 antagonize AIP-1/AgrC1 interactions (Fig. 2c). AIP-4, which differs from AIP-1 by a single amino acid residue (Asp is replaced by Tyr; Fig. 2c), is an agonist of both AgrC4 and AgrC1 but an inhibitor of AgrC2 and AgrC3 [28, 29]. Recently Raghuram et al. [32] developed a software tool (AgrVATE; github.com/VishnuRaghuram94/AgrVATE) to interrogate S. aureus agr variability and evolution. Analysis of ~40 000 S . aureus genomes revealed that the distribution of the four agr groups is ~60 % group I, ~22 % group II, ~14 % group III and ~2.5 % group IV [32]. This extensive in silico analysis did not uncover any novel agr groups, intermediate AIPs or AIPs acquired from CONS. Potential relationships between certain infectious diseases and agr group have been highlighted such as between toxic shock and scalded skin syndromes and agr groups III and IV, respectively [33]. However, the number of strains examined was relatively small and others [34] found no associations between agr-specific groups and infection type. A recent major study found that the distribution of agr groups in over 10 000 strains from blood, skin and the nasal cavity was similar to the general distribution of agr groups [32]. agr acts as an AIP-dependent autoinducible system such that mutation of any of the corresponding agrBDCA genes results in the loss of activity [20]. As a QS-dependent master virulence gene regulator, it is also subject to control via by a sophisticated interconnected network of regulators, which integrates diverse environmental and host cues via two component sensor regulator systems such as SaeRS, SrrAB and ArlRS, sigma factors (e.g. SigB) and the SarA family (e.g. SarA, SarR, SarS, MgrA and Rot). A detailed consideration of the molecular nature and function of these gene regulatory elements in controlling agr is beyond the scope of this review and the reader is referred to recent reviews [23, 24].
Fig. 2.
(a) Schematic of the staphylococcal agr quorum-sensing system. The agrBDCA locus is composed of two divergent transcripts, RNAII and RNAIII, driven by the agrP2 and agrP3 promoters, respectively. AgrD, the pro-peptide precursor of the autoinducing peptide (AIP) is processed at the cytoplasmic membrane by AgrB and MroQ such that AIPs are released extracellularly. AIPs bind to and activate the AgrC receptor, a membrane-bound histidine sensor kinase resulting in phosphorylation of the response regulator AgrA and activation of the agrP2 and agrP3 promoters. This drives the autoinduction circuitry to generate more AIP signal molecules and induces expression of virulence genes either indirectly via RNAIII or directly via the target gene promoters. (b) Schematic showing the hypervariable region (in green/cyan) of the agrBDCA locus incorporating agrD and giving rise to the different agr groups. Amino acid residues marking the beginning and ends of the variable regions are numbered (adapted from [142]). (c) Generalized AIP structure and summary table showing the amino acid sequences of the AIPs belonging to each of the four S. aureus agr groups and their cross-group activities. The brackets denote the amino acid residues within the macrocycle. In S. aureus , the AIP N-terminal tails have two, three or four amino acid residues.
AIP identification and quantification
In the staphylococcal agr system, AIP signal molecules are thiolactones, which have similar structures but different primary amino acid sequences. Each AIP has a common central Cys residue, the thiol of which is linked to the α-carboxyl group of the C-terminal residue forming a five residue, 16-membered macrocycle with an exocyclic N-terminus of variable length (Fig. 2c). These have been identified by liquid-chromatography (LC) mass spectrometry (MS) aided by the annotated AgrD amino acid sequence data [27, 28, 35, 36] or by chemo-selective trapping [37]. AIPs have been produced by solid-phase chemical synthesis [28, 38] and via protein engineering using mini-intein technology [39]. They can readily be detected at nanomolar concentrations in cell-free S. aureus culture supernatants using cell-based transcriptional reporter assays for AIP-dependent AgrC activation or inhibition. Several such reporter assays have been described in which the agrP2 or agrP3 promoter is fused to a reporter gene such as blaZ, gfp, lux or gluc to provide colorimetric, fluorescence or bioluminescence outputs [26, 31, 40–42]. The assays can be conducted using microtitre plates to quantify AIP levels, evaluate AIP structure activity relationships (SAR), pharmacological properties (agonist, inverse agonist, antagonist) or the functionality of mutations in AgrC receptor proteins. For example, Jensen et al. [31], deleted the chromosomal agr locus and replaced it with the luxCDABE operon under the control of the agrP3 promoter. The agrA and agrC genes were then introduced on a plasmid under the control of the agrP2 promoter. Since this reporter system is unable to produce AIPs and lacks the autoinduction pathway characteristic of the native agr QS system, it facilitates the in-depth pharmacological evaluation of AIP analogues without interference from an endogenously active agr system [31]. It also facilitates the introduction of the desired native or mutated agrC gene and requires no additional reagents. For example, using this reporter for AIP-1/AgrC1, an EC50 of 6±1 nM was derived from a dose–response curve and an EC50 of 9±1 nM for AIP-4/AgrC4. In contrast, AIP-1 is a very weak activator of AgrC4 (EC50 3542±997 nM) [31]. Comparable nanomolar EC50s have been reported by others for activation of AgrC by the cognate AIP [28, 29, 38] using alternative transcriptional reporters.
AgrD pro-peptide processing by the transmembrane protein, AgrB
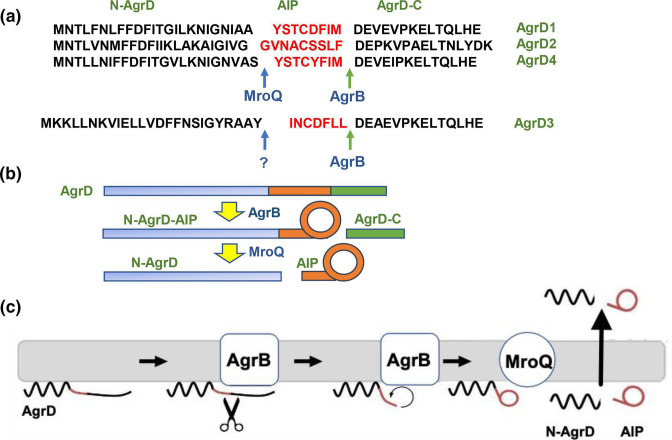
AIPs are derived from an internal fragment of an AgrD pro-peptide, which consists of 40–50 amino acid residues incorporating an N-terminal amphipathic leader (N-AgrD; 24–25 amino acids), a mid-region of 7–9 amino acid residues that constitutes the AIP and a charged C-terminal tail (AgrD-C; 14–15 amino acids) (Fig. 3). The generation of an extracellular AIP requires at least four membrane-associated steps (i) removal of AgrD-C, (ii) formation of the thiolactone macrocycle, N-AgrD-AIP (iii) cleavage of N-AgrD and (iv) export of AIP and N-AgrD (Fig. 3).
Fig. 3.
Schematic showing the processing of AgrD pro-peptides to generate the active cyclic AIP signal molecules. (a) Amino acid sequences of S. aureus AgrD1-D4 showing the AgrB cleavage site and the MroQ sites for AgrD1, D2 and D4. MroQ does not cleave AgrD3. (b) Processing of AgrD by AgrB and MroQ to release N-AgrD and the cyclic AIP. (c) Schematic showing the formation and release of the AIP and N-AgrD at the cytoplasmic membrane. Cleavage of AgrD by AgrB releases a 14 amino acid C-terminal peptide (AgrD-C), which is degraded in the cytoplasm. N-AgrD-AIP is cleaved by MroQ to release N-AgrD and the mature AIP. The mechanism by which the AIP and N-AgrD are exported is not known.
S. aureus agrB deletion mutants are unable to activate agr as they fail to produce AIPs. This is because AgrB, a unique transmembrane cysteine protease is required for processing the AgrD pro-peptide [27] via steps (i) and (ii) (Fig. 3). Although AgrB lacks homology with other proteins, the alignment of AgrB sequences from diverse Gram-positive bacteria has revealed multiple, highly conserved amino acid residues. However, the S. aureus AgrBs do exhibit some substrate specificity. AgrB1 is able to process AgrD1 and AgrD2 but not AgrD3 and vice versa. Chimeric AgrB1 and AgrB2 proteins have been constructed and regions putatively involved in AgrD group specificity identified [43].
Two of the invariant residues in AgrB homologues are Cys84 and His77. These appear to constitute a catalytic dyad that enables AgrB to cleave AgrD-C from AgrD. This is likely to result in the formation of an acyl-enzyme thioester intermediate, followed by peptidyl transfer to generate the thiolactone macrocycle via the internal Cys28 of AgrD so releasing N-AgrD-AIP (Fig. 3). Furthermore, an N-terminal amphipathic helix, the length of the C-terminal tail and certain C-terminal residues (e.g. E34 and L41; Fig. 3) of AgrD all appear to be essential for substrate recognition and processing by AgrB in bacterial cell-based assays [44–46].
When purified, the S. aureus AgrB protein forms stable dimers and is enzymatically active only when embedded in lipid bilayers [47]. It clearly drives a proteolytic cyclization reaction removing the C-terminal 14–15 amino acid residues of AgrD while reversibly catalysing the formation of a thiolactone ring, which undergoes slow irreversible hydrolysis to the corresponding linear form. The N-AgrD-AIP thiolactone is protected from ring opening by association with membrane phospholipids and the reaction is driven efficiently by the rapid degradation of the AgrD-C fragment [47], possibly via ClpP or ClpX in staphylococcal cells [48]
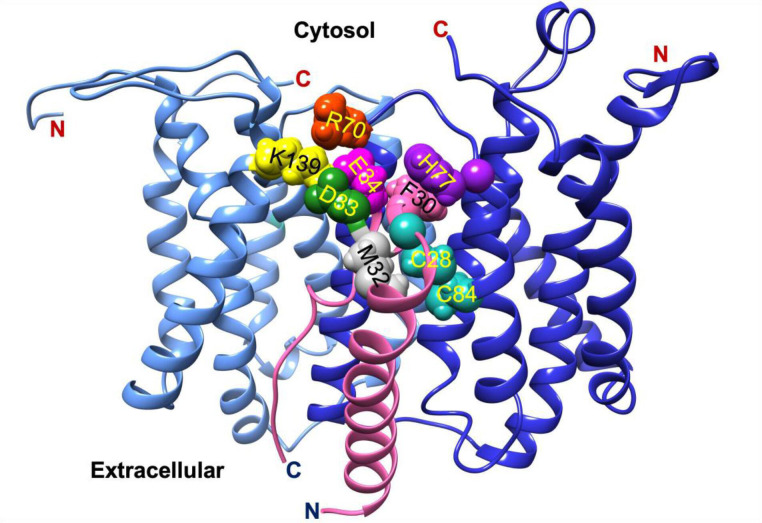
Topology predictions for AgrB initially proposed two different models; a six transmembrane domain model with both termini at cis [44, 49] and a four transmembrane domain with an additional half-transmembrane hairpin where both termini are at trans [50]. These contradictory models, which contain structurally implausible domains, have yet to be replaced by high-resolution crystallographic or NMR-derived structures. However, a recent molecular dynamics model supported by experimental circular dichroism data revealed a tightly packed six transmembrane domain helical topology for AgrB with both N- and C-termini on the same side and placing amino acid residues known to be important to function on the cis side near the polar/apolar interface [51]. These include the enzyme active site Cys84, which is located in the membrane interior providing access to the AgrD substrate. Additional molecular dynamics simulations of AgrB and AgrD in a membrane environment have provided a new model incorporating an AgrB dimer with crucially two non-equivalent AgrB sites in which one AgrB monomer facilitates insertion and positioning of AgrD in the correct orientation for catalytic processing by the second AgrB (Fig. 4) [51]. Whether the proposed (AgrB)2AgrD complex exists requires further experimental confirmation.
Fig. 4.
Conformation of membrane-embedded ternary complex AgrB2/AgrD after molecular dynamics simulations. The two AgrB proteins are inequivalent with AgrB-I (cornflower blue) guiding substrate AgrD (fuchsia) to the active site involving catalytic AgrB-II (blue). AgrD residues C28 and M32 are close to each other and to catalytic AgrB-II C84. Key interactions stabilizing the complex include AgrB-I K139-D33 and AgrB-I R70-E34 contacts with AgrD, and AgrB-II H77-F30 via π-interactions. Sidechains are colour-coded aqua (C28, C84 AgrB-II), grey (M32 AgrD), pink (F30 AgrD), purple (H77 AgrB-II), fuchsia (E34 AgrD), green (D33 AgrD), yellow (K139 AgrB-I) and orange (R70 AgrB-I). All four AgrB termini are on the cytosolic side (top) consistent with six transmembrane domain topology of AgrB. The (AgrB)2/AgrD complex orientation here is shown with cytosol at the top offering a better view of the cytosol-accessible active site (adapted from [51]).
N-terminal cleavage of AgrD requires a second transmembrane protease, MroQ
The identity of the transmembrane protease required for cleaving the N-terminal region from the N-AgrD-AIP thiolactone to release the AIP [step (iii) (Fig. 3)] has been something of an enigma. Expression of agrB and agrD from agr group I in Escherichia coli surprisingly resulted in the formation of extracellular AIP [45] indicating that E. coli must also possess an AgrD N-terminal cleaving protease. Kavanaugh et al. [52] developed a fluorescence assay based on a linear synthetic peptide designed to identify S. aureus peptidases that could potentially cleave the AgrD N-terminal amphipathic peptide. Evidence was provided in support of signal peptidase B (SpsB) as the missing transmembrane protease, which included the ability of SpsB peptide inhibitors to reduce agr reporter expression and AIP production in S. aureus . These data were also consistent with AIP production in E. coli , which has only one, essential signal peptidase that, in a temperature-sensitive mutant, can be genetically complemented by SpsB. However, more recent work indicates that in S. aureus a different transmembrane protein is the primary enzyme involved in removing AgrD N-terminal peptides [53, 54].
Upstream of the agr locus, a gene termed mroQ, the mutation of which results in the loss of virulence and significantly reduced extracellular AIP levels was identified [53, 54]. The translated product is predicted to have eight transmembrane-spanning helices (Fig. 5) and is a member of the CPBP (CAAX proteases and bacteriocin-processing enzymes) family of putative membrane metalloproteases encompassing over 5000 members that incorporate the misnamed ‘Abortive phage infection’ (Abi) domain [55]. CPBP superfamily members share three signature motifs and are found in both prokaryotes and eukaryotes. In the latter, prenylation is crucial for the function and membrane protein targeting [55]. In bacteria, only a few CPBP proteins have been studied experimentally but their functions in general have remained relatively elusive. SkkI for example from Lactobacillus plantarum functions as a bacteriocin receptor and immunity protein [56], whereas the group B streptococcal CPBP protein AbxI interacts with the histidine sensor kinase, CovS to control virulence gene expression [57]. For SkkI but not for AbxI proteolytic activity is necessary for function. Several CPBP proteins are present in S. aureus including MroQ, which contains the EEXXXR and FXXXH motifs considered necessary for enzyme activity. The critical residues for metalloprotease activity include the predicted MroQ catalytic site Glu141 and Glu142 and the His residues 180 and His213 required for zinc co-ordination [53] (Fig. 5). Replacement of either Glu or the His with Ala in these motifs variably reduced AIP production but S. aureus virulence was only attenuated in a mouse skin and soft-tissue infection model to the same level as an agr deletion mutant for Glu141Ala replacement [53]. In a cell-based assay, expression of agrD in an S. aureus mroQ mutant resulted in the accumulation of a membrane-associated AgrD intermediate but no AIPs were produced [58]. The amino acid sequence of MroQ does not vary with agr group suggesting that the same enzyme processes all four agr groups. However, in agr group III strains, deletion of mroQ did not prevent the production of an active AIP [58]. Whether AgrB and MroQ interact directly in S. aureus membranes to process AgrD in a manner analogous to that of the MroQ homologue SpdC (LyrA) with SagB is not known. SpdC and SagB (a membrane-bound N-acetylglucosaminidase) form a complex that functions as a peptidoglycan release factor. However, the conserved residues required for CAAX proteolytic activity are not required by SpdC [59, 60].
Fig. 5.
Topological model of transmembrane endopeptidase MroQ. The homology model of MroQ (unpublished) was obtained from I-TASSER [143] and annealed in atomistic MD simulations using NAMD [144] within a membrane patch built in CHARMM-GUI [145]. The model reveals an eight TMD topology – lateral view (left); and axial view (right). Residues Glu141 and Glu142 and His180 and His 213 required for MroQ functionality are shown within the membrane region where they have access to the hydrophobic AgrD substrate.
Biochemical confirmation that MroQ is able to remove the N-terminal peptide from N-AgrD-AIP was obtained by reconstituting the MBP-MroQ fusion proteins into proteoliposomes and incubating with N-AgrD-AIP thiolactones from agr groups I, II and III. The efficient generation of AIP-1 and AIP-2 was confirmed by LC MS/MS, although no cleavage of the agr group III substrate was observed. By including AgrB in the proteoliposomes with MroQ, complete reconstitution of AIP biosynthesis from AgrD was achieved [61]. These elegant biochemical studies together with the observation that SpsB is unable to cleave AIP biosynthetic intermediates in vitro confirmed that MroQ is the primary transmembrane protease involved in N-terminal cleavage of the AgrB processed AgrD thiolactone product. However, although the agr N-AgrD-AIP thiolactones from agr groups I and II were substrates for MroQ, it did not cleave the agr group III AgrD peptide thiolactone. Thus, there must be an alternative enzyme, which does not appear to be SpsB.
Both the N-AgrD peptide and AIP are released from the cells into the extracellular environment. N-AgrD has PSM toxin-like properties and is also amyloidogenic, capable of forming amyloid fibrils in S. aureus biofilms [62]. Gonzalez et al. [63] identified both formylated and non-formylated peptide variants derived from N-AgrD in S. aureus culture supernatants, which were cytotoxic for mammalian cell lines, modulated neutrophil chemotaxis and increased the size of murine skin lesions induced by an S. aureus agr mutant. Whether MroQ processes N-AgrD-AIP thiolactones on the cytoplasmic or external face of the cytoplasmic membrane has not yet been established. In either case it is not clear whether export occurs via MroQ, AgrB, an MroQ/AgrB complex or via dedicated transport mechanisms such as the ABC transporters PmtCD and AbcA [64]. These can independently export PSMs from either membrane or cytosolic environments.
The AgrCA two-component sensor regulator system
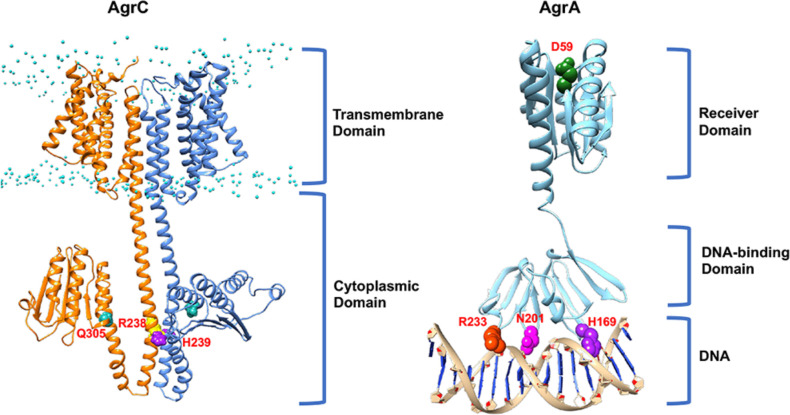
Once exported, AIPs are sensed via a two component system (TCS) in which AgrC is the histidine kinase sensor and AgrA, response regulator (Fig. 2a). AgrC belongs to the minor HPK10 histidine kinase protein sub-family [65] and has a modular architecture with an N-terminal sensory module incorporating an AIP-binding site linked via a helical linker to a C-terminal cytoplasmic module containing two subdomains [25] (Fig. 6). These are the dimerization and histidine phosphorylation (DHp) and the catalytic and ATP-binding (CA) subdomains [25]. The CA subdomain containing the G1 box lacks both the conserved DXGXG motif and the key Asp residue, which is replaced by Asn such that the binding affinity of AgrC for ATP and hence kinase activity is likely dependent on cellular energy levels [25, 66]. The interactions between AgrC and the activating AIP ligand are highly specific occurring at nanomolar affinities where the AIP non-cooperatively binds to AgrC in a 2 : 2 stoichiometry [25].
Fig. 6.
Architecture of the AgrC and AgrA two-component sensor and response regulator proteins. Models of the homodimeric sensor kinase AgrC (left) (orange/cornflower blue) and response regulator AgrA (right) (cyan) associated with the agrP2 promoter site. Initial conformations of AgrC and AgrA were obtained from AlphaFold [146]. The AgrC dimer was built using ClusPro [147]. Lipid phosphates are shown to mark the membrane interface (P atoms are shown in cyan). His239, responsible for AgrC autophosphorylation, is shown in purple; Arg238 (yellow) and Gln305 (fuchsia) are important for the molecular ‘latch’ that stabilizes AgrC in the ‘off state’ [67]. The AgrA model was superimposed onto the structure of the AgrA C-terminal DNA-binding domain [PDB:3BS1] [71]. Asp59 (green) phosphorylation by AgrC is important for activation of AgrA. His169 (purple), Asn201 (fuchsia) and Arg233 (orange-red) are responsible for DNA recognition.
AgrC behaves like a rheostat, where activation of its membrane spanning domain following the binding of an AIP results in the twisting of a helical linker relative to the cytoplasmic domain and subsequent dimerization that results in AgrA phosphorylation [25]. The binding of a classical competitive antagonist such as AIP-3 to AgrC1 blocks rotation of the helical linker whereas AIP-2, an antagonist that behaves as an inverse agonist of AgrC-1 drives rotation of the helical linker in the opposite direction to that of the agonist AIP-1 [25].
A key non-covalent interaction between Arg238 and Gln305 that stabilizes AgrC in the ‘off’ state has also been identified [67] (Fig. 6). This ‘latch’ is proposed to lift following the binding of the cognate AIP to the extracellular AgrC sensor domain, thus mediating the structural changes that result in AgrC activation. Replacement of Arg238 with Ala renders AgrC constitutively active. The mechanism by which AIPs induce the structural changes in AgrC that lead to activation of kinase activity is not yet clear, although AIP-binding is likely to either induce or stabilize specific rotational conformations [67].
No full-length AgrC structure has yet been obtained and hence the nature of the AIP-binding site is currently not known. However, topology models predict that the N-terminal sensory module of AgrC contains six transmembrane-spanning helices and three extracellular loops (Fig. 6). Since the S. aureus agr system has undergone significant evolutionary divergence, the retention of functionality requires that changes in AgrD, which modify the AIP structures should be accompanied by compensatory changes in the AgrC receptor protein. Since AIPs are not internalized, the AIP-binding site is likely to be on the outer face of the transmembrane AgrC protein and therefore likely to involve the extracellular loops. AIP-1 and AIP-4 differ by a single amino acid; when the corresponding AgrC1 and AgrC4 are compared, only two of their three predicted AgrC extracellular loops exhibit amino acid differences [31, 68]. In loop 1, 7 out of 19 and in loop 2, 3 out of 9 amino acid residues differ. Replacement by site-specific mutagenesis of these in AgrC4 either singly or in combination with those from AgrC1 revealed that while differential recognition of AIP-1 and AIP-4 depends primarily on three amino acid residues in loop 2, loop 1 was essential for receptor activation by the cognate AIP [31, 68]. Furthermore, a single mutation in the AgrC1 loop 2 resulted in conversion of (Ala5)AIP-1, a non-native AgrC1 inhibitor, to an activator, essentially resulting in the forced evolution of a ‘new’ AIP group [31]. Taken together, these data suggest that extracellular loop two may constitute the AIP macrocycle-binding site while the exocyclic N-terminal amino acids interact with loop 1 to facilitate receptor activation. However, there is no direct evidence to demonstrate that the key amino acid residues in AgrC loop 2 are directly involved in AIP-binding. It is conceivable that they could act indirectly on the conformation/presentation of direct contact residues elsewhere on AgrC [68]. The specificity of AgrC1 could also be further broadened by replacement of Ile at position 171 with Lys in the third predicted AgrC extracellular loop. This site-specific AgrC1 mutant was activated at nM EC50 concentrations not only by the cognate AIP-1 but also by AIP-3, AIP-4 and (Ala5)AIP-1 [69]. Multiple AgrC mutants, which are constitutively active have also been isolated and map to both the last transmembrane helix of the sensor domain and to the histidine kinase domain [69].
Once AgrC has been trans-autophosphorylated, phospho-transfer to AgrA, a member of the LytTR class of transcriptional regulators, occurs via the AgrC DHp cytoplasmic phospho-transfer sub-domain. AgrA consists of an N-terminal receiver domain containing the conserved Asp residue that drives dimerization upon phosphorylation and a C-terminal DNA-binding domain [70] (Fig. 6). Dimerization of AgrA enhances DNA binding to the LytTR domain-binding sites of which there are two, 9 base pair, high affinity sites in the agrP2 promoter region separated by 12 bp and both a high- and a low-affinity LytTR binding site in the agrP3 promoter region [71]. This enables differential expression of RNAII and RNAIII to facilitate QS via autoinduction and to avoid premature expression or degradation of RNAIII [46, 72]. Related agr promoter region motifs have also been identified in genes directly regulated by AgrA such as those coding for PSMs [73].
AIP-mediated activation and inhibition of AgrC
Extensive AIP structure activity (SAR) studies using transcriptional reporters in S. aureus in combination with native AIPs and diverse AIP analogues have established the key molecular features for AgrC activation and inhibition [22, 28–30, 74–77] (Fig. 7). In general, minor differences in the AIP peptide sequence result in the complete loss of agonist activity. However competitive inhibition is highly tolerant of AIP sequence variation [22]. The macrocyclic ring is essential for AgrC activation as the corresponding linear peptides are inactive [26, 27, 78]. Similarly changes to the size of the macrocycle are not tolerated [79]. Removal of the three exocyclic N-terminal amino acids in AIP-1 also results in the loss of agonist activity as does replacement of the thiolactone S with N or with O to form the corresponding lactam (Fig. 7) and lactone, respectively [28, 29]. For AIP-1, replacement of each l-amino acid residue in turn with the corresponding d-isomer resulted in six out of the eight analogues exhibiting markedly lower activity. However, exchanging either the macrocycle Phe or the terminal Met residue with the corresponding d-isomer had little impact suggesting that the AIP-binding site in AgrC1 is able to accommodate these differences [28]. S-oxidation of the methionine thioether side-chain to form the methionyl sulphoxide derivative inactivated AIP-1 (Fig. 7), as did replacement of the Met with norLeu, Ser, Glu, Lys or Pro but not Ile, further emphasizing the critical role of the C-terminal thioether side-chain for AgrC1 activation [28, 29]. AIP-4, which also has a Met in the same position as AIP-1, is the only other S. aureus AIP capable of being inactivated via formation of the methionyl sulphoxide [28]. Substituting each AIP-1 amino acid residue in turn with Ala (Fig. 7), except for the central Cys, revealed that the only replacement showing increased activity (by ~2 fold) was for the exocyclic Ser [28]. The other AIP-1 Ala analogues were either inactive or exhibited reduced agonist activity, except that replacement of the endocyclic amino acid residue (Asp) located C-terminally to the central Cys with Ala converted AIP-1 from an activator to a potent low nanomolar IC50 cross-group inhibitor [28, 29] (Fig. 7) In AIP-4, the endocyclic Asp residue is replaced by Tyr (Fig. 2) and is therefore the critical determinant of AIP specificity for agr groups I and IV [28, 29]. Detailed SAR studies of activation and inhibition of the cognate AgrCs have also been undertaken for AIP-2 and AIP-3 with broadly similar findings to those reported for AIP-1 and AgrC1 [22, 29, 30, 38, 74–77]. These SAR data suggested that the macrocycle was required for receptor recognition and binding while the exocyclic region was necessary for receptor activation [22, 80]. Further refinement of these data via solution NMR structural analysis of the native AIP-3 peptide and a series of analogues revealed the importance of a hydrophobic ‘bulge’ formed by hydrophobic endocyclic residues and exocyclic tail contacts [22, 75]. These findings have highlighted the contribution of 3D conformation and the orientation of the AIP exocyclic tail relative to the macrocycle with respect to AgrC activation and inhibition.
Fig. 7.
SAR for S. aureus AIP-1 showing how minor modifications to the peptide sequence including Ala-scanning influence activity. Substitution of the Asp residue (d5 ) with Ala to give (Ala5)AIP-1 converted AIP-1 from an activator of AgrC1 to a potent cross group inhibitor. IC50 data from [29].
S. aureus agr heterogeneity and environmental inactivation
The agr system plays a key role in reciprocally regulating planktonic exotoxin producing, colonization and biofilm-associated lifestyles of S. aureus . It functions as a positive feedback loop displaying bimodal, heterogeneous behaviour that leads to the emergence of distinct subpopulations of S. aureus cells [81]. These subpopulations are characterized by the presence or absence of agr activity and by their relative numerical sizes consistent with bet-hedging strategies where some cells behave as individuals while others act co-operatively [82]. Such phenotypic heterogeneity has been also observed in the QS populations of other bacterial species where it may be transient and restricted to the early stages of activation with the population subsequently becoming homogeneous or heterogeneity may persist resulting in a bimodal, heterogeneous population [82].
In S. aureus the ratio of the agr ‘on’ to agr ‘off’ sub-population may be modified in response to environmental signals such as higher Mg2+ concentrations that increase cell wall rigidity by binding to teichoic acids and triggering the σB-mediated down-regulation of agr [81]. Other cell-wall changes have been observed to modify agr activation. For example, in some, but not all, HA-MRSA strains [83, 84] expression of mecA, which codes for penicillin-binding protein 2A, reduced agr expression and virulence in a mouse infection model. Expression of agr could be restored by partially digesting the cell wall, suggesting that MecA-induced changes in cell wall architecture potentially reduce accessibility of the AIP to AgrC. Regulatory interdependence of mecA and agrA has also been noted for certain CA-MRSA strains in which methicillin resistance is agr-regulated [85]. HA-MRSA and CA-MRSA strains appear to have similar kinetics of agr activation but the latter achieve much higher magnitudes [85].
For activation of agr, AIP levels must reach a critical threshold concentration. This is not fixed but will vary according to the relative rates of AIP production, accumulation, diffusion and inactivation, which in turn will also depend on the prevailing growth environment. Little difference in the EC50s for AgrCs activated by their cognate AIPs (all low nM) are apparent [28, 29, 38]. Exogenous provision of the cognate AIP at the time of inoculation overcomes the ‘quorum’, prematurely activating agr although there is a window within the first few hours of growth after which S. aureus does not respond [86, 87]. Genotype versus agr locus-dependent differences in agr dynamics have been investigated in the context of agr group divergence by constructing congenic S. aureus strains (8325–4 and Newman) each with a different agr group allele and carrying an agrP3-blaZ fusion [86]. These revealed differences in the timing and magnitude of agr activation with S. aureus agr group III cells showing the most delayed induction and lowest level of agr expression, which in turn was reflected in cell-wall protein and exotoxin production. Whether such differences impact on virulence in infection models has not yet been established.
The ability to switch agr on or off during different stages of infection of host cells and in different tissues is clearly advantageous when moving from an acute toxigenic to a chronic persistent lifestyle, where increasing production of surface adhesins, reducing exotoxin secretion and either taking up intracellular residence or forming a biofilm enables S. aureus to avoid host immune defences. In vivo evidence supporting this on/off agr switch was obtained by Wright et al. [88] using whole body luminescence imaging of S. aureus transformed with an agrP3-lux expression vector. This revealed early rapid activation of agr followed by several days without any agrP3 expression prior to renewed light output. While this switching of agr may have been due to transient agrP3 expression, it could alternatively have been a consequence of either exhaustion of the LuxAB and LuxCDE enzyme substrates FMNH2 and a long-chain fatty aldehyde or reduced oxygen availability in deeper tissues resulting in a lack of luciferase activity [89].
Although AIPs are not known to be degraded or inactivated by endogenous staphylococcal enzymes, the C-terminal Met of S. aureus AIP-1 is S-oxidized during aerobic growth in laboratory media to form the corresponding methionyl sulphoxide [28] (Fig. 7). This methionyl sulphoxide-containing compound is unable to activate or inhibit AgrC [28]. Whether its formation and accumulation impact on the timing of agr induction by reducing AIP-1 levels (or AIP-4 which is the only other S. aureus AIP with a terminal Met) below the activation threshold is not known. However, this S-oxidation reaction has in vivo relevance as it occurs in response to phagocyte derived reactive oxygen and nitrogen species such as hypochlorous acid (HOCl) and peroxynitrite (ONOO-) and results in down-regulation of agr and a concomitant reduction of virulence in a mouse skin infection model [15]. Oxidative stress can also down-regulate agr via AgrA modification as a consequence of disulphide bond formation between the redox-reactive Cys119 and Cys288 leading to dissociation of the modified AgrA from DNA [90]. In addition, Cys119 can undergo ‘CoAlation’, i.e. the covalent modification by co-enzyme A, which, under nutrient deprivation or oxidative stress conditions also reduces the affinity of AgrA for the agrP2 and agrP3 promoters [91]. During in vitro competition and evolution experiments, oxidative stress was observed to drive the emergence of agr mutants, which possess a fitness advantage only under aerobic growth conditions due to the reactive oxygen species generating capacity of PSMs and RNAIII-regulated factors [92]. Consequently, agr imposes an oxygen-dependent fitness cost such that hypoxia favours maintenance of QS and increased exotoxin production. This oxygen-driven tuning of the agr system may therefore exert a major influence on tissue-dependent disease progression during infection.
Exposure to air pollution and in particular particulates such as black carbon (BC) are associated with exacerbations of chronic respiratory disease [9]. Growth of S. aureus in BC prior to inoculation increased the adhesion to, and invasion of, human epithelial cells in vitro and murine respiratory tract colonization and pulmonary invasion in vivo [9]. Global transcriptional analysis revealed that numerous agr-regulated exoprotease, exotoxin and immune evasion genes were upregulated while certain adhesin and metabolic genes were repressed suggesting that that BC acts directly on the pathogen rather than exclusively on the host [9]. The mechanism by which BC controls this subset of the agr regulon has yet to be elucidated.
Host factors known to impact on the agr-driven switch from a colonizing to an invasive phenotype include serum proteins such the low-density (LDL) and very low-density (VLDL) particles associated apo-lipoprotein B (ApoB), which interfere with agr-dependent QS by specifically and reversibly sequestering S. aureus AIPs [93]. This ApoB-mediated downregulation of agr in serum can be circumvented using constitutively active AgrC variants [94]. All four S. aureus AIPs as well as the inactive methionyl oxide derivative of AIP-1 bind to ApoB via an interaction that is likely to be dependent on the hydrophobic thiolactone macrocycle as the (less hydrophobic) linear peptide corresponding to AIP-1 does not bind. The in vivo relevance of these findings is supported by the greater susceptibility of mice deficient in ApoB to invasive infection with a wild-type S. aureus strain compared with an isogenic agr deletion mutant [93].
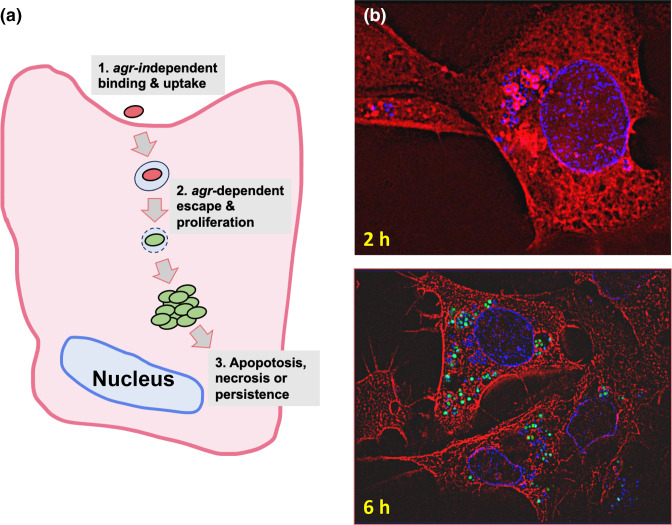
Although once considered to be an exclusively extracellular pathogen, S. aureus can internalize and survive in a variety of mammalian cells including endothelial, epithelial and professional phagocytic cells where they contribute to chronic and relapsing infections [95]. Quenching of agr-dependent QS in the bloodstream, for example, ensures that the S. aureus cell-wall proteins required for host cell attachment and internalization remain highly expressed [93]. Once inside an endosome or phagosome, S. aureus cells escape into the cytoplasm, kill the host cell, be killed or remain intracellular, protected from host defences and acting as a reservoir for persistent infections (Fig. 8). Within these intracellular vesicles, AIPs accumulate rapidly activating agr and leading to the expression of exotoxins such as α-haemolysin and the PSMs, which lyse the endosomal or phagosomal membrane [14, 16]. With respect to neutrophil phagosomes, AIP-mediated staphylococcal escape can be blocked by inactivation of AIP-1 and AIP-4 by S-oxidation via NAPDPH-derived oxygen radicals [15]. This intracellular escape process has been termed confinement or compartment sensing rather than QS since agr activation occurs in a single trapped bacterial cell rather than in a population [10, 96].
Fig. 8.
(a) Intracellular uptake of S. aureus into non-professional epithelial and endothelial cells is independent of agr expression. Once internalized, agr expression precedes endosomal escape by facilitating endosomal lysis via α-haemolysin or the PSMs. This enables S. aureus to replicate and persist within the cytoplasm protected from host immune defences and antibiotics or to lyse the host cells and establish further rounds of uptake and release leading to tissue destruction. (b) Fluorescence microscopy of S. aureus transformed with an agrP3-gfp reporter invading mammary epithelial cells and stained with DAPI (DNA; blue) and an anti-tubulin-Cy3 conjugate (red; microtubules). After 2 h incubation, intracellular staphylococcal cells are observed (white arrow) with red cell walls as the Cy3 antibody conjugate has bound to protein A showing that agr has not yet been activated. As the infection proceeds to 6 h agr expression has clearly been induced as observed by the high level of gfp expression (green) in the bacterial cells (adapted from [14]).
agr dysfunction and infection
Despite the relevance of agr to S. aureus virulence in animal infection models, agr defective mutants are commonly found in clinical samples from both asymptomatic nasal carriage and serious infections where the loss of agr functionality in both MRSA and methicillin-sensitive (MRSA) strains is associated with greater adaptability, persistent infections and poorer outcomes [87, 97–103]. In healthy individuals in the community, nasal carriage of agr mutants has been reported to be relatively low at ~9 % colonization. However, agr dysfunction has been strongly associated with hospitalization and antibiotic usage suggesting a trade-off between virulence and antibiotic resistance [100, 102]. S. aureus agr mutants exhibit enhanced survival in the presence of daptomycin because they shed phospholipids that neutralize the antibiotic extracellularly. In contrast, agr-functional strains released less phospholipid and secreted agr-regulated PSMs, which inhibited the phospholipid-daptomycin interaction [104].
Analysis of de novo mutations in >1000 S . aureus genomes from 105 infected patients with prior nasal colonization revealed that adaptive mutations in pathogenesis-associated genes including agr were enriched in infecting but not nasal-colonizing bacteria indicative of within host selection pressures [101]. Others have shown that events associated with agr inactivation result in agr-defective blood and nasal strain pairs that are enriched in mutations compared to pairs from wild-type controls [105]. These additional mutations outside the agr locus can contribute to diversification and adaptation during infection by agr mutants associated with poor patient outcomes [105].
In their analysis of >40 000 S . aureus genomes, Raghuram and co-workers [32] did not find any stable agr-defective strain lineages. This is consistent with previous suggestions that agr defective strains may be unable to establish and maintain circulating populations outside their original location [32, 99]. While the host factors responsible for driving the selection of agr dysfunctional mutants have not yet been identified, host resistance should not be overlooked as indicated by Thänert et al. [106] who compared susceptible (A/J) and resistant (C57BL/6) mouse strains with respect to S. aureus infection and virulence gene expression.
Strains with mutations in agr are capable of prolonged intracellular survival. Although they cannot escape phagosomes, they induce less cell death and so survive in greater numbers within host cells [107]. agr mutants can also persist via an alternative intracellular pathway in endothelial cells involving LC3+ vesicles [107]. Furthermore, sub-populations of slow-growing S. aureus small colony variants (SCVs) able to survive within host cells are frequently recovered from chronic and recurrent infections [108, 109]. These SCVs may be genetically stable with characteristic mutations in metabolic pathways or unstable and exhibiting increased expression of negative agr regulators (e.g. SigB, ArlRS and CodY). In both cases, a common characteristic of SCVs that arise from either altered electron transport or global regulatory pathway changes is reduced Agr activity.
These observations suggest that S. aureus populations progressing from colonization to infection at different body sites may be heterogenous with respect to agr expression, or consist of agr-functional cells or agr-defective mutants or mixed populations with the balance influencing the outcome. It is also apparent that agr mutant populations may contain a small fraction of phase variable cells capable of reverting to agr-functional cells [110]. This appears to arise at least in vitro via a mechanism involving a poly(A) tract alteration and a genetic duplication plus inversion event. This strategy has been suggested to act as cryptic insurance against host-mediated stress enabling the population to survive phagocytosis and sustain infection [110].
An explanation for why S. aureus agr mutants exhibit reduced virulence in animal models but are frequently isolated from clinical samples was suggested by Pollitt et al. [111] using a waxworm larvae infection model. They showed that agr-dependent QS is a beneficial social trait in which agr mutant ‘cheats’, which neither produce nor respond to AIPs exploit agr functional AIP-producing co-operators. However, while these data provide an explanation for mixed populations in toxigenic infections, they do not account for chronic, biofilm-associated clinical S. aureus infections where homogeneous agr-negative populations can rapidly emerge [112]. It should also be noted that S. aureus isolates exhibiting low levels of exotoxins are not necessarily agr mutants highlighting the likely existence of other, novel exotoxin regulators [113].
agr dysfunction – the molecular basis
DNA sequence analysis of agr-dysfunctional S. aureus clinical isolates has revealed frameshifts, insertions, deletions and substitutions in the agrBDCA operon [87, 97, 98, 113, 114]. In their analysis of over 40 000 S . aureus genomes Raghuram et al. [32] found that >5 % had agr operon frameshifts. These were almost exclusively found in agrA and agrC with the latter having accumulated the greatest number of different mutations. The insertion of an extra adenine at the 3′ end of agrA was the most common frameshift and is known to result in delayed agr activation and haemolysin production [115]. Interestingly, when comparing the agr histidine kinase (agrC) and response regulator (agrA) genes with other S. aureus TCSs (arl, kdp, nre, pho, srr and wal), the frequency of mutations in agr is highly enriched [32]. The frequent occurrence of common but independently acquired convergent mutations may be an adaptive response to specific host selective pressures.
Some of these naturally occurring changes have either been predicted or experimentally demonstrated to result in the complete loss of agr functionality. For example, Mairpady Shambat et al. [116] isolated an MRSA strain harbouring a single AgrC Y223C cytoplasmic domain substitution that switched the virulence phenotype from cytotoxic to colonizing that could be reversed by mutating back to C223Y. However, not all naturally occurring agr mutations are likely to be inactivating but may instead modify the timing and/or the strength of agr induction. Sloan et al. [87] identified a number of natural mutations associated with reduced cytotoxicity also linked to the cytoplasmic domain of AgrC. These delayed the onset and accumulation of AIPs and exhibited impaired AgrC-AIP responsiveness as revealed by the increased threshold for AgrC activation. Exotoxin production, in this case Panton–Valentin leucocidin production, could be restored by exogenous provision of the cognate AIP at the time of inoculation indicating that delayed activation of agr autoinduction and consequently failure to express RNAIII results in the lack of exotoxins. Molecular dynamics simulations from in silico engineered point mutations in the AgrC cytoplasmic domain revealed subtle changes that alter both domain conformation and relative domain orientation [87]. The efficiency of the rheostat-like behaviour of AgrC in which AIP binding induces the twisting of a helical linker relative to the cytoplasmic domain and subsequent dimerization [25] is likely to be impaired by these mutations. Consequently, a greater magnitude of ‘input’ into the rheostat-like mechanism will be required in order to produce the same response. In turn, this would impact on the efficiency of the agr autoinduction circuitry. Such conformational rearrangements of key functional subdomains in these AgrC cytoplasmic domain mutants highlight the cooperative responses of protein structures involving dimerization, ATP binding and phosphorylation, as well as sites involved in AgrA interactions. Whether increasing the threshold for agr activation offers a fitness advantage remains to be established.
Intra- and inter-species agr-mediated competitive interference among the Staphylococci
In S. aureus, strains belonging to one agr group produce an AIP that cross-inhibits each of the other three agr groups, giving rise to the concept of ‘competitive interference’ [27, 117]. In laboratory experiments in broth inoculated with mixed cultures, the agr group did not impact on competitiveness [117]. This is perhaps not surprising given that such interference is at the level of agr expression rather than growth. In vivo in a Manduca sexta (tobacco hornworm) infection model, despite the genetic diversity of the S. aureus strains tested, differences in the fitness of competing strains belonging to different agr groups were observed [117].
There have been very few documented reports of clinical samples containing mixed agr group isolates. In one case a patient was reported to have an S. aureus agr group 1 blood isolate and a group 2 wound isolate [97] while in another, a CF sputum sample contained genome sequences from both agr group 1 and 2 strains [32]. A comparison of consecutive and co-colonizing strains in healthy individuals or those with CF revealed that strain replacement was accompanied by a change in the agr group in 63 and 80 % of the two cohorts, respectively [118]. Co-colonizing strains from CF belonged to interfering agr groups in six of ten cases whereas for healthy individuals, nasal co-colonization with strains belonging to different agr groups was rarely observed [118]. However, in CF sputum, where agr is not expressed [119], no cross-group inhibitory AIPs will be synthesized making agr group competition unlikely.
Competitive interference however does occur during interactions between S. aureus and the CoNS, which produce a broad range of AIPs [37]. They constitute a diverse group of at least 38 different staphylococcal species that are primarily found on the healthy skin and mucous membranes of humans and other mammals, alongside other bacterial genera including corynebacteria, streptococci and micrococci [120, 121]. In a screen of culture supernatants prepared from 52 staphylococcal isolates representing 17 different species obtained from dogs, cows, horses, mink, cats, pigs and birds, Canovas et al. [21] identified 17 different species that inhibited S. aureus agr. Of 54 CoNS isolates obtained from 25 pig nasal swabs, the eight different species capable of inhibiting S. aureus agr included Staphylococcus hyicus, Staphylococcus simulans, Staphylococcus arlettae, Staphylococcus lentus and Staphylococcus chromogenes [2]. However, Staphylococcus sciuri and Staphylococcus xylosus were unable to inhibit S. aureus agr [2]. The presence of specific CoNS species in pig nares has been associated with reduced MRSA colonization and of relevance to intensive pig farming where the transmission of livestock associated MRSA from pigs to humans is a potential health risk [2].
The most common CoNS skin species are S. epidermidis , S. hominis , Staphylococcus haemolyticus , Staphylococcus capitis , Staphylococcus lugdunensis and Staphylococcus warneri . Most CoNS are relatively harmless, beneficial commensals that actively contribute to shaping the skin microbiota and the cutaneous immune response, promoting tissue-repair and combatting the external threat posed by invading pathogens such as S. aureus and group A streptococci [1, 122–124]. However, certain CoNS species also have dual lifestyles as colonizers and opportunistic pathogens. S. epidermidis, regarded as a keystone skin commensal, is frequently responsible for blood stream infections associated with implanted medical devices including intravascular catheters, prosthetic vascular grafts and heart valves, cardiac devices and coronary stents [121]. Considering their genetic flexibility and integrity of the skin barrier, it has been proposed that the beneficial or harmful behaviour of S. epidermidis may vary depending on the specific strain and interactive context [123, 124]. Indeed, analysis of 1482 S . epidermidis genomes from the skin of five healthy individuals established that they had evolved from multiple founder lineages rather than a single colonizer [125]. S. saprophyticus , a ‘moderately’ pathogenic CoNS species found in the genito-urinary tract is the second most frequent cause of uncomplicated lower urinary tract infection in young women [121].
Among CoNS species, the agrBDCA genes are widespread but highly divergent and offer opportunities for competitive interference within and between species [126]. Given the diversity of CoNS colonizing the same environmental niche, there is considerable scope for competitive interference at both intra- and inter-species levels, promoting colonization and suppressing virulence factor production by both S. aureus and CoNS capable of adopting pathogenic lifestyles [1, 37]. Such control will ultimately depend not only on competitive agr interference but also on the production of antimicrobials by CoNS such as PSMs that selectively inhibit S. aureus and which may themselves be agr-regulated [122].
Based on their AgrD peptide sequences, most CoNS, in common with S. aureus can be divided into different agr subgroups; between two and six depending on the species (Table 1). In S. hominis there are six agr groups [127, 128] while of the four S. epidermidis agr groups, healthy human skin is commonly dominated by a large proportion of S. epidermidis agr group I strains together with smaller sub-populations of agr-II and -III strains [125, 129]. To date, CoNS species where chemical structures have been confirmed, produce AIPs that, in common with S. aureus , are usually between seven and nine amino acid residues and incorporate a thiolactone ring. The exceptions are S. intermedius that produces a lactone [130] and S. epidermidis which makes three AIPs with extended seven amino acid residue exocyclic tails [125, 129]. The S. intermedius AIP lactone is a functional autoinducer in which the switch from Cys to Ser may have arisen via spontaneous point mutation [130]. Neither AgrB1 nor AgrB2 from S. aureus could enzymatically process a Ser containing AgrD pro-peptide to generate the mature AIP nor could the S. aureus AIP-1 or AIP-2 lactones activate the S. aureus AgrC1 or AgrC2 receptors [28, 130]. Interestingly the readily S-oxidized C-terminal Met present in S. aureus AIP-1 and AIP-4 is very rarely found in other staphylococci except for S. argenteus and S. schweitzeri , which, respectively, make the same AIPs as S. aureus agr groups I and IV [37].
Table 1.
Competitive interference between AIPs produced by CoNS and the agr groups of S. aureus and S. epidermidis
|
Coagulase-negative staphylococci |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
|
Species |
Autoinducing peptide |
agrI |
agrII |
agrIII |
agrIV |
agrI |
agrII |
agrIII |
|
|
AIP-1 |
DSV-[CASYF] |
166 |
>1000 |
13 |
>1000 |
– |
9.64 |
34.3 |
|
|
AIP-2 |
NASKYNP-[CSNYL] |
– |
– |
– |
– |
13.9 |
– |
– |
|
|
AIP-3 |
NAAKYNP-[CASYL] |
– |
– |
– |
– |
2.13 |
|||
|
AIP |
YST-[CSYYF] |
0.6 |
0.26 |
0.2 |
9.0 |
– |
– |
– |
|
|
AIP-1 |
SYNV-[CGGYF] |
13 |
31 |
5 |
2910 |
NC |
34 |
16 |
|
|
AIP-2 |
SYSP-[CATYF] |
15 |
2109 |
3 |
1130 |
20 |
19 |
62 |
|
|
AIP-3 |
TYST-[CYGYF] |
11 |
4 |
6 |
NC |
4 |
3 |
3 |
|
|
AIP-4 |
TINT-[CGGYF] |
128 |
140 |
37 |
NC |
237 |
93 |
28 |
|
|
AIP-5 |
SQTV-CSGYF] |
43 |
59 |
4 |
3809 |
10 |
22 |
2 |
|
|
AIP-1 |
YSP-[CTNFF] |
10 |
4 |
13 |
146 |
3 |
19 |
– |
|
|
AIP-2 |
ANP-[CAMFY] |
2 |
30 |
2 |
2 |
– |
– |
– |
|
|
AIP-1 |
KYNP-[CLGFL] |
2.2 |
1.1 |
3.5 |
23 |
– |
– |
– |
|
|
AIP-2 |
KYYP-[CWGYF] |
1.6 |
1.5 |
11.5 |
40 |
– |
– |
– |
|
|
AIP-3 |
KYNP-[CWGYF] |
1.7 |
6 |
3.2 |
48 |
– |
– |
– |
|
|
AIP |
KYPF-[CIGYF] |
2.8 |
86 |
80 |
– |
– |
– |
– |
|
|
AIP-1 |
DI-[CNGYF] |
384 |
419 |
36.6 |
>1000 |
– |
– |
– |
|
|
AIP |
KINP-[CTVFF] |
3.3 |
350 |
4 |
180 |
– |
– |
– |
|
|
AIP |
SINP-[CTGFF] |
15 |
200 |
60 |
350 |
– |
– |
– |
|
|
AIP |
SFTP-[CTTYF] |
340 |
IA |
340 |
IA |
– |
– |
– |
|
|
AIP |
VIRG-[CTAFL] |
190 |
800 |
690 |
IA |
– |
– |
– |
|
The square brackets denote amino acid residues within the AIP macrocycle. IC50 values compiled from [37, 128, 133–136]; NC, not calculated, likely to be a weak inhibitor; IA, not active at highest dose tested; – not available.
In general, there is relatively little information on the target genes regulated via agr in different CoNS species. Given the difficulty in genetically manipulating CoNS, Severn et al. [128] used the AgrA inhibitor apicidin to transcriptionally profile genes regulated by an agr group-I S. hominis commensal and identified ~40 down- and ~7 up-regulated genes. Down-regulated genes coding for PSMs, a predicted lipase, acetoin production and multiple transcriptional regulators as well as agrB, agrD and RNAIII were identified [128]. The up-regulated S. hominis genes were of unknown function but likely to be involved in metabolism and sensing. In S. epidermidis , one of the most abundant CoNS skin colonizers in which a functional agr system enhances skin colonization [129], genes down-regulated in an RNAIII mutant included protease and lipase exoenzymes, PSMs, δ-haemolysin, and agr. The haemolytic CoNS species S. lugdunensis can cause severe human infections but its repertoire of agr-dependent virulence determinants have not been well characterized. In a recent study, Chin et al. [131] showed that the S. lugdunensis synergistic haemolysins (SLUSH), the metalloprotease lugdulysin, a urease and a number of ABC transporters were downregulated in an agr mutant as were unidentified factors required to protect the organism from host innate immune defences.
Studies of AIP-mediated activation of CoNS agr systems or of competitive interference between CoNS species, or between CoNS and S. aureus are usually undertaken initially using spent culture supernatants and screened using agrP3 reporter gene fusions based on activation or inhibition of S. aureus or CoNS agr groups [2, 21, 128, 130]. While these highlight the diversity of CoNS capable of inhibiting one or more S. aureus agr groups, such data requires validation via synthesis of the predicted AIP. This then facilitates quantitative determination (EC50 or IC50) of the agonist or antagonist activities of a specific AIP. In experiments employing S. epidermidis agrP3-gfp reporter fusions and spent culture supernatants [129], interference between S. epidermidis agr group I and groups II and III but not between agr groups II and III were observed. These data were subsequently confirmed using the corresponding synthetic AIPs [129, 132, 133] (Table 1). In contrast to AIP-1, both AIP-2 and AIP-3 have extended exocyclic tails that likely account for the differential activity. By quantifying δ-haemolysin as a read-out for agr inhibition in different S. aureus strains, S. epidermidis AIP-1 was reported to inhibit S. aureus agr groups I to III but not group IV. Conversely, S. aureus AIP-4 was the only S. aureus AIP found to inhibit S. epidermidis agr group I [132]. Using an agrP3 reporter, Yang et al., [133] observed that S. aureus AIP-2 but not AIP-1, AIP-3 or AIP-4 inhibited S. epidermidis agr group I [133]. S. hominis AIP-1 to −6 all inhibited S. aureus agr groups I, II and III with IC50s ranging from 3 to 140 nM except for AIP-2 which had little activity against S. aureus agr group II [126] (Table 1). The S. hominis AIPs were all active against the three S. epidermidis agr groups except for AIP-1, which lacked antagonistic activity towards S. epidermidis agr group I [128]. The strongest S. hominis intraspecies interactions were observed between agr groups I and II, which were the two most common S. hominis agr types identified on the skin of a group of 14 human volunteers and in database genome sequences [125, 128]. In common with S. epidermidis , none of the S. hominis AIPs were effective antagonists of S. aureus agr group IV [128] although the IC50s for the S. simulans AIPs were in the 20–40 nM range [134] (Table 1) S. simulans agr types also displayed varying degrees of resistance to cross-inhibition from S. aureus AIPs, with S. simulans agr III being weakly susceptible to antagonism by S. aureus AIP-1 and AIP-4 [134]. Intraspecies interactions between S. simulans agr variants may also occur since agr group I is inhibited by AIPs-2 and 3 whereas agr groups II and III are not subject to cross-inhibition. Two potent S. aureus cross-group inhibitors are S. warneri AIP-2 and the S. caprae AIP [135] both of which are potent antagonists of all four S. aureus agr groups [136] (Table 1). Although inter-species agr activation is rarely observed, the AIPs of Staphylococcus schleiferi and Staphylococcus hominis (AIP-3) both induce S. aureus agr group IV [37, 128] (Table 1).
Competitive interference and skin disease
S. aureus and especially CA-MRSA strains are the most common causes of skin and soft tissue infections in humans [1, 137, 138]. The severity of atopic dermatitis (AD), a chronic disease of unclear aetiology is characterized by dry, itchy and inflamed skin and by dysbiosis of the skin microbiota. S. aureus colonizes AD patients’ skin lesions and exacerbates disease by promoting inflammation and degradation of the skin barrier function. These have all been linked with S. aureus exotoxins, superantigens, exoproteases and PSMs [137] and also with an overabundance of S. epidermidis strains producing the cysteine protease EcpA [123, 139]. Metagenomic analysis of the AD skin microbiome revealed that an increase in the relative abundance of S. aureus in patients with active AD correlated with a lower CoNS AIPs to S. aureus ratio, thus reducing the ability of the CoNS to inhibit the S. aureus agr system [127]. In a further study involving 268 Japanese infants aged 1 to 6 months, 121 were colonized with S. aureus at 1 month irrespective of AD outcome [19]. However, colonization with S. aureus at 6 months increased the likelihood of developing AD. Selection for dysfunctional agr mutations primarily in agrC occurred in S. aureus strains from 6-month-old infants who did not develop AD. In an epicutaneous mouse inoculation model that induces agr and skin surface inflammation, the expression of a functional S. aureus agr system was found necessary for skin colonization and the development of AD-like inflammation [19]. There is therefore considerable interest in the therapeutic potential of commensal CoNS for suppressing S. aureus in AD either by production of potent S. aureus selective antimicrobials [122], via competitive interference [127] or both since antimicrobials such as the PSMs are regulated via agr.
The ability of a specific CONS species or the corresponding synthetic AIP to prevent or reduce skin colonization and damage by S. aureus CA-MRSA agr group I strain LAC has been extensively evaluated in murine epicutaneous (bacteria applied topically via gauze) and dermonecrosis (bacteria injected intradermally) skin models for S. hominis [127, 128], S. caprae [135], S. simulans [134] and S. warneri [136]. These in vivo investigations facilitated quantification of the dose-dependent efficacy of the CoNS AIPs or the corresponding CoNS strain with respect to reducing MRSA skin colonization, lesion morphologies and sizes, weight loss, bacterial burden and skin barrier integrity. S. hominis AIP-1 and AIP-2 both inhibited S. aureus agr group I activity on mouse skin and protected against epidermal damage by reducing skin lesion size, transepithelial water loss and inducing a productive host response without affecting S. aureus abundance [127, 128]. Co-challenge with S. hominis was not as effective as the S. hominis AIP-2 alone, which was suggested to be a consequence of lower levels of AIP-2 production on the skin by the human isolate employed or because mouse rather than human skin was used [128]. Similar results were obtained for MRSA co-infection with S. simulans [134], S. warneri [136] and S. caprae [135] and also in response to their respective AIPs. S. warneri AIP-2, which is more fivefold more potent in vitro than AIP-1, is one of the few naturally occurring AIPs that can inhibit S. aureus agr group IV [136]. The latter has been associated with scalded skin syndrome and the production of exfoliative toxins. In a dermonecrotic model, the S. warneri AIP-2 reduced skin lesion size and protected mice infected with an MRSA agr group IV strain from weight loss and skin damage [136]. Apart from S. caprae, none of the other CoNS AIPs reduced the MRSA agr group I bacterial burden during the course of infection to levels comparable to those observed for an MRSA agr deletion mutant [135]. This appears to be a consequence of the S. caprae AIP sensitizing MRSA to neutrophil-mediated clearance rather than a direct immunogenic effect of the AIP. Why this did not occur with the other CoNS AIPs is not clear, but may relate to the greater in vivo activity of the S. caprae AIP since significant protection was achieved at 10 µg per mouse [135] whereas comparable protection required 50 µg of the other CoNS AIPs [125, 128, 134, 136]. For each of the AIPs tested, significant protection was observed for a single AIP dose when administered at the time of infection although more subtle therapeutic effects were observed when AIPS were delivered later in the course of infection [127, 128, 134–136].
These findings provide the tantalizing prospect of employing CoNS or AIPs as a means of limiting skin infections caused by S. aureus especially in the context of multi-antibiotic resistant MRSA. Further work will need to consider asymptomatic mouse skin models, the differences between human and mouse microflora as well as innate immune and healing responses. Given that skin is colonized by diverse commensals producing many different AIPs and antimicrobials, their overall impact is likely to be complex [125]. As yet, there is little information on competitive interference when multiple CoNS strains and AIPs are involved or even other commensal bacteria such as the Corynebacterium species, which are capable of inhibiting S. aureus agr groups I to III via an unknown mechanism [140]. In a recent, double-blinded, randomized phase I clinical trial, the safety of an S. hominis strain delivered topically to forearm skin was evaluated over 7 days. Promisingly, the trial met its primary endpoint of safety with those treated with S. hominis experiencing fewer AD-associated adverse events and a reduction in S. aureus [141].
Concluding remarks
The function and dysfunction via inhibition or mutation of the agr system play key roles in the adaptive behaviour and fitness of S. aureus in different host environments, in response to antibiotics and during encounters with other staphylococcal species especially on the skin. Considerable progress in understanding the molecular mechanisms underlying agr-dependent QS has been gained although there is still relatively little high-resolution information on the 3D structures of AgrC, AgrB and MroQ or on the nature of the respective AIP and AgrD substrate-binding sites on each of these transmembrane proteins. Whether MroQ processes its thiolactone substrate on the inner or outer face of the cytoplasmic membrane and how AIPs are exported remain to be elucidated. Given the diversity of AIPs leading to extensive competitive interference, it is perhaps surprising that there is little cross-activation within or between staphylococcal species. Whether AIP-driven interference occurs between staphylococci and other Gram-positive bacteria that harbour agr systems especially in complex microbial communities is not yet known. We also have little understanding of the selective pressures that led to the emergence of different agr groups or indeed why four agr groups evolved in S. aureus with no evidence of intermediates. Given the rise of multi-antibiotic-resistant MRSA, the agr system does offer multiple druggable molecular targets for inhibitors including AgrA, AgrB and AgrC [18]. However, despite the in vitro identification of highly active AIP analogues and non-peptidic agr inhibitors much more in vivo work will be required before any ‘hits’ with the appropriate pharmacological and pharmacokinetic properties can enter human clinical trials. These are likely to be complicated by the association between agr dysfunction, biofilm formation and chronic infections. Nevertheless, employing CoNS strains or antagonistic AIPs as a means of limiting S. aureus skin infections appears to offer a realistic therapeutic opportunity [141].
Funding information
Work on staphylococcal agr system in the authors’ laboratories was funded by the Medical Research Council UK (Grant No. MR/N010477/1).
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: agr, accessory gene regulator; AIP, auto-inducing peptide; BC, black carbon; CA, ATP-binding; CA-MRSA, community-acquired MRSA; CF, cystic fibrosis; CoNS, coagulase-negative staphylococci; DHp, histidine phosphotransfer; HA-MRSA, hospital-acquired MRSA; LC, liquid chromatography; LDL, low density lipoprotein; MRSA, methicillin resistant Staphylococcus aureus; MS, mass spectrometry; NMR, nuclear magnetic resonance; PSM, phenol soluble modulin; QS, quorum sensing; SAR, structure activity relationship; SCV, small colony variant; SLUSH, Staphylococcus lugdunensis synergistic haemolysins; TCS, two component system; TMD, transmembrane domain; VISA, vancomycin intermediate Staphylococcus aureus; VLDL, very low density lipoprotein; VRSA, vancomycin resistant Staphylococcus aureus.
References
- 1.Parlet CP, Brown MM, Horswill AR. Commensal Staphylococci influence Staphylococcus aureus skin colonization and disease. Trends Microbiol. 2019;27:497–507. doi: 10.1016/j.tim.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peng P, Baldry M, Gless BH, Bojer MS, Espinosa-Gongora C, et al. Effect of co-inhabiting coagulase negative Staphylococci on S. aureus agr quorum sensing, host factor binding, and biofilm formation. Front Microbiol. 2019;10:2212. doi: 10.3389/fmicb.2019.02212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheung GYC, Bae JS, Otto M. Pathogenicity and virulence of Staphylococcus aureus . Virulence. 2021;12:547–569. doi: 10.1080/21505594.2021.1878688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chambers HF, Deleo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol. 2009;7:629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nair N, Biswas R, Götz F, Biswas L. Impact of Staphylococcus aureus on pathogenesis in polymicrobial infections. Infect Immun. 2014;82:2162–2169. doi: 10.1128/IAI.00059-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orazi G, O’Toole GA. “It takes a village”: mechanisms underlying antimicrobial recalcitrance of polymicrobial biofilms. J Bacteriol. 2019;202:e00530-19. doi: 10.1128/JB.00530-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kavanaugh JS, Horswill AR. Impact of environmental cues on Staphylococcal quorum sensing and biofilm development. J Biol Chem. 2016;291:12556–12564. doi: 10.1074/jbc.R116.722710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balasubramanian D, Harper L, Shopsin B, Torres VJ. Staphylococcus aureus pathogenesis in diverse host environments. Pathog Dis. 2017;75:ftx005. doi: 10.1093/femspd/ftx005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Purves J, Hussey SJK, Corscadden L, Purser L, Hall A, et al. Air pollution induces Staphylococcus aureus USA300 respiratory tract colonization mediated by specific bacterial genetic responses involving the global virulence gene regulators Agr and Sae. Environ Microbiol. 2022;24:4449–4465. doi: 10.1111/1462-2920.16076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams P, Winzer K, Chan WC, Cámara M. Look who’s talking: communication and quorum sensing in the bacterial world. Philos Trans R Soc Lond B Biol Sci. 2007;362:1119–1134. doi: 10.1098/rstb.2007.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gray B, Hall P, Gresham H. Targeting agr- and agr-like quorum sensing systems for development of common therapeutics to treat multiple gram-positive bacterial infections. Sensors. 2013;13:5130–5166. doi: 10.3390/s130405130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darkoh C, DuPont HL, Norris SJ, Kaplan HB. Toxin synthesis by Clostridium difficile is regulated through quorum signaling. mBio. 2015;6:e02569. doi: 10.1128/mBio.02569-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nizet V. Understanding how leading bacterial pathogens subvert innate immunity to reveal novel therapeutic targets. J Allergy Clin Immunol. 2007;120:13–22. doi: 10.1016/j.jaci.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 14.Qazi SN, Counil E, Morrissey J, Rees CE, Cockayne A, et al. agr expression precedes escape of internalized Staphylococcus aureus from the host endosome. Infect Immun. 2001;69:7074–7082. doi: 10.1128/IAI.69.11.7074-7082.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rothfork JM, Timmins GS, Harris MN, Chen X, Lusis AJ, et al. Inactivation of a bacterial virulence pheromone by phagocyte-derived oxidants: new role for the NADPH oxidase in host defense. Proc Natl Acad Sci U S A. 2004;101:13867–13872. doi: 10.1073/pnas.0402996101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pang YY, Schwartz J, Thoendel M, Ackermann LW, Horswill AR, et al. agr-dependent interactions of Staphylococcus aureus USA300 with human polymorphonuclear neutrophils. J Innate Immun. 2010;2:546–559. doi: 10.1159/000319855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schilcher K, Horswill AR. Staphylococcal biofilm development: structure, regulation, and treatment strategies. Microbiol Mol Biol Rev. 2020;84:e00026-19. doi: 10.1128/MMBR.00026-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otto M. Critical assessment of the prospects of quorum-quenching therapy for Staphylococcus aureus infection. Int J Mol Sci. 2023;24:4025. doi: 10.3390/ijms24044025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakamura Y, Takahashi H, Takaya A, Inoue Y, Katayama Y, et al. Staphylococcus agr virulence is critical for epidermal colonization and associates with atopic dermatitis development. Sci Transl Med. 2020;12:eaay4068. doi: 10.1126/scitranslmed.aay4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Novick RP, Geisinger E. Quorum sensing in Staphylococci . Annu Rev Genet. 2008;42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 21.Canovas J, Baldry M, Bojer MJ, Andersen PS, Grzeskowiak PK, et al. Cross-talk between Staphylococcus aureus and other Staphylococcal species via the agr quorum sensing. Syst Front Microbiol. 2016;7:1733. doi: 10.1021/acs.jmedchem.9b00798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horswill AR, Gordon CP. Structure-activity relationship studies of small molecule modulators of the staphylococcal accessory gene regulator. J Med Chem. 2020;63:2705–2730. doi: 10.1021/acs.jmedchem.9b00798. [DOI] [PubMed] [Google Scholar]
- 23.Jenul C, Horswill AR. Regulation of Staphylococcus aureus virulence. Microbiol Spectr. 2019;7 doi: 10.1146/annurev-micro-102215-095708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bronesky D, Wu Z, Marzi S, Walter P, Geissmann T, et al. Staphylococcus aureus RNAIII and its regulon link quorum sensing, stress responses, metabolic adaptation, and regulation of virulence gene expression. Annu Rev Microbiol. 2016;70:299–316. doi: 10.1146/annurev-micro-102215-095708. [DOI] [PubMed] [Google Scholar]
- 25.Wang B, Zhao A, Novick RP, Muir TW. Activation and inhibition of the receptor histidine kinase AgrC occurs through opposite helical transduction motions. Molecular Cell. 2014;53:929–940. doi: 10.1016/j.molcel.2014.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ji G, Beavis RC, Novick RP. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc Natl Acad Sci U S A. 1995;92:12055–12059. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ji G, Beavis R, Novick RP. Bacterial interference caused by autoinducing peptide variants. Science. 1997;276:2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- 28.McDowell P, Affas Z, Reynolds C, Holden MTG, Wood SJ, et al. Structure, activity and evolution of the group I thiolactone peptide quorum-sensing system of Staphylococcus aureus . Mol Microbiol. 2001;41:503–512. doi: 10.1046/j.1365-2958.2001.02539.x. [DOI] [PubMed] [Google Scholar]
- 29.Lyon GJ, Wright JS, Muir TW, Novick RP. Key determinants of receptor activation in the agr autoinducing peptides of Staphylococcus aureus . Biochemistry. 2002;41:10095–10104. doi: 10.1074/jbc.M109989200. [DOI] [PubMed] [Google Scholar]
- 30.Lyon GJ, Wright JS, Christopoulos A, Novick RP, Muir TW. Reversible and specific extracellular antagonism of receptor-histidine kinase signaling. J Biol Chem. 2002;277:6247–6253. doi: 10.1074/jbc.M109989200. [DOI] [PubMed] [Google Scholar]
- 31.Jensen RO, Winzer K, Clarke SR, Chan WC, Williams P. Differential recognition of Staphylococcus aureus quorum-sensing signals depends on both extracellular loops 1 and 2 of the transmembrane sensor AgrC. J Mol Biol. 2008;381:300–309. doi: 10.1016/j.jmb.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 32.Raghuram V, Alexander AM, Loo HQ, Petit RA, Goldberg JB, et al. Species-wide phylogenomics of the Staphylococcus aureus Agr operon revealed convergent evolution of frameshift mutations. Microbiol Spectr. 2022;10:e0133421. doi: 10.1128/spectrum.01334-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jarraud S, Mougel C, Thioulouse J, Lina G, Meugnier H, et al. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect Immun. 2002;70:631–641. doi: 10.1128/IAI.70.2.631-641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lange J, Heidenreich K, Higelin K, Dyck K, Marx V, et al. Staphylococcus aureus pathogenicity in cystic fibrosis patients-results from an observational prospective multicenter study concerning virulence genes, phylogeny, and gene plasticity. Toxins. 2020;12:279. doi: 10.3390/toxins12050279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalkum M, Lyon GJ, Chait BT. Detection of secreted peptides by using hypothesis-driven multistage mass spectrometry. Proc Natl Acad Sci. 2003;100:2795–2800. doi: 10.1073/pnas.0436605100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Junio HA, Todd DA, Ettefagh KA, Ehrmann BM, Kavanaugh JS, et al. Quantitative analysis of autoinducing peptide I (AIP-I) from Staphylococcus aureus cultures using ultrahigh performance liquid chromatography-high resolving power mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2013;930:7–12. doi: 10.1016/j.jchromb.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gless BH, Bojer MS, Peng P, Baldry M, Ingmer H, et al. Identification of autoinducing thiodepsipeptides from staphylococci enabled by native chemical ligation. Nat Chem. 2019;11:463–469. doi: 10.1038/s41557-019-0256-3. [DOI] [PubMed] [Google Scholar]
- 38.Mayville P, Ji G, Beavis R, Yang H, Goger M, et al. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc Natl Acad Sci. 1999;96:1218–1223. doi: 10.1073/pnas.96.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malone CL, Boles BR, Horswill AR. Biosynthesis of Staphylococcus aureus autoinducing peptides by using the synechocystis DnaB mini-intein. Appl Environ Microbiol. 2007;73:6036–6044. doi: 10.1128/AEM.00912-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malone CL, Boles BR, Lauderdale KJ, Thoendel M, Kavanaugh JS, et al. Fluorescent reporters for Staphylococcus aureus . J Microbiol Methods. 2009;77:251–260. doi: 10.1016/j.mimet.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murray EJ, Williams P. Detection of Agr-type autoinducing peptides produced by Staphylococcus aureus . Methods Mol Biol. 2018;1673:89–96. doi: 10.1007/978-1-4939-7309-5_7. [DOI] [PubMed] [Google Scholar]
- 42.Blower I, Tong C, Sun X, Murray E, Luckett J, et al. Gaussia Luciferase as a Reporter for Quorum Sensing in Staphylococcus aureus . Sensors. 2020;20:4305. doi: 10.3390/s20154305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang L, Ji G. Identification of a Staphylococcal AgrB segment(s) responsible for group-specific processing of AgrD by gene swapping. J Bacteriol. 2004;186:6706–6713. doi: 10.1128/JB.186.20.6706-6713.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qiu R, Pei W, Zhang L, Lin J, Ji G. Identification of the putative Staphylococcal AgrB catalytic residues involving the proteolytic cleavage of AgrD to generate autoinducing peptide. J Biol Chem. 2005;280:16695–16704. doi: 10.1074/jbc.M411372200. [DOI] [PubMed] [Google Scholar]
- 45.Thoendel M, Horswill AR. Identification of Staphylococcus aureus AgrD residues required for autoinducing peptide biosynthesis. J Biol Chem. 2009;284:21828–21838. doi: 10.1074/jbc.M109.031757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang B, Muir TW. Regulation of virulence in Staphylococcus aureus: molecular mechanisms and remaining puzzles. Cell Chem Biol. 2016;23:214–224. doi: 10.1016/j.chembiol.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang B, Zhao A, Novick RP, Muir TW. Key driving forces in the biosynthesis of autoinducing peptides required for Staphylococcal virulence. Proc Natl Acad Sci. 2015;112:10679–10684. doi: 10.1073/pnas.1506030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frees D, Qazi SNA, Hill PJ, Ingmer H. Alternative roles of ClpX and ClpP in Staphylococcus aureus stress tolerance and virulence. Mol Microbiol. 2003;48:1565–1578. doi: 10.1046/j.1365-2958.2003.03524.x. [DOI] [PubMed] [Google Scholar]
- 49.Zhang L, Gray L, Novick RP, Ji G. Transmembrane topology of AgrB, the protein involved in the post-translational modification of AgrD in Staphylococcus aureus . J Biol Chem. 2002;277:34736–34742. doi: 10.1074/jbc.M205367200. [DOI] [PubMed] [Google Scholar]
- 50.Thoendel M, Horswill AR. Random mutagenesis and topology analysis of the autoinducing peptide biosynthesis proteins in Staphylococcus aureus . Mol Microbiol. 2013;87:318–337. doi: 10.1111/mmi.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bardelang P, Murray EJ, Blower I, Zandomeneghi S, Goode A, et al. Conformational analysis and interaction of the Staphylococcus aureus transmembrane peptidase AgrB with its AgrD propeptide substrate. Front Chem. 2023;11:1113885. doi: 10.3389/fchem.2023.1113885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kavanaugh JS, Thoendel M, Horswill AR. A role for type I signal peptidase in Staphylococcus aureus quorum sensing. Mol Microbiol. 2007;65:780–798. doi: 10.1111/j.1365-2958.2007.05830.x. [DOI] [PubMed] [Google Scholar]
- 53.Cosgriff CJ, White CR, Teoh WP, Grayczyk JP, Alonzo F. Control of Staphylococcus aureus quorum sensing by a membrane-embedded peptidase. Infect Immun. 2019;87:e00019-19. doi: 10.1128/IAI.00019-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marroquin S, Gimza B, Tomlinson B, Stein M, Frey A, et al. MroQ is a novel abi-domain protein that influences virulence gene expression in Staphylococcus aureus via modulation of agr activity. Infect Immun. 2019;87:e00002-19. doi: 10.1128/IAI.00002-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pei J, Mitchell DA, Dixon JE, Grishin NV. Expansion of type II CAAX proteases reveals evolutionary origin of γ-secretase subunit APH-1. J Mol Biol. 2011;410:18–26. doi: 10.1016/j.jmb.2011.04.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kjos M, Snipen L, Salehian Z, Nes IF, Diep DB. The Abi proteins and their involvement in bacteriocin self-immunity. J Bacteriol. 2010;192:2068–2076. doi: 10.1128/JB.01553-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Firon A, Tazi A, Da Cunha V, Brinster S, Sauvage E, et al. The Abi-domain protein Abx1 interacts with the CovS histidine kinase to control virulence gene expression in group B Streptococcus . PLoS Pathog. 2013;9:e1003179. doi: 10.1371/journal.ppat.1003179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stock MR, Fang L, Johnson KR, Cosgriff C, Teoh WP, et al. Characterization of MroQ-dependent maturation and export of the Staphylococcus aureus accessory gene regulatory system autoinducing peptide. Infect Immun. 2022;90:e0026322. doi: 10.1128/iai.00263-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Willing S, Schneewind O, Missiakas D. Regulated cleavage of glycan strands by the murein hydrolase SagB in S. aureus involves a direct interaction with LyrA (SpdC) J Bacteriol. 2021;203:e00014-21. doi: 10.1128/JB.00014-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schaefer K, Owens TW, Page JE, Santiago M, Kahne D, et al. Structure and reconstitution of a hydrolase complex that may release peptidoglycan from the membrane after polymerization. Nat Microbiol. 2021;6:34–43. doi: 10.1038/s41564-020-00808-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao A, Bodine SP, Xie Q, Wang B, Ram G, et al. Reconstitution of the S. aureus agr quorum sensing pathway reveals a direct role for the integral membrane protease MroQ in pheromone biosynthesis. Proc Natl Acad Sci U S A. 2022;119:e2202661119. doi: 10.1073/pnas.2202661119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schwartz K, Sekedat MD, Syed AK, O’Hara B, Payne DE, et al. The AgrD N-terminal leader peptide of Staphylococcus aureus has cytolytic and amyloidogenic properties. Infect Immun. 2014;82:3837–3844. doi: 10.1128/IAI.02111-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gonzalez DJ, Corriden R, Akong-Moore K, Olson J, Dorrestein PC, et al. N-terminal ArgD peptides from the classical Staphylococcus aureus Agr system have cytotoxic and proinflammatory activities. Chem Biol. 2014;21:1457–1462. doi: 10.1016/j.chembiol.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dickey SW, Burgin DJ, Huang S, Maguire D, Otto M. Two transporters cooperate to secrete amphipathic peptides from the cytoplasmic and membranous milieus. Proc Natl Acad Sci. 2023;120:e2211689120. doi: 10.1073/pnas.2211689120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grebe TW, Stock JB. The histidine protein kinase superfamily. Adv Microb Physiol. 1999;41:139–227. doi: 10.1016/s0065-2911(08)60167-8. [DOI] [PubMed] [Google Scholar]
- 66.Srivastava SK, Rajasree K, Fasim A, Arakere G, Gopal B. Influence of the AgrC-AgrA complex on the response time of Staphylococcus aureus quorum sensing. J Bacteriol. 2014;196:2876–2888. doi: 10.1128/JB.01530-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xie Q, Zhao A, Jeffrey PD, Kim MK, Bassler BL, et al. Identification of a molecular latch that regulates Staphylococcal virulence. Cell Chem Biol. 2019;26:548–558. doi: 10.1016/j.chembiol.2019.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Geisinger E, George EA, Chen J, Muir TW, Novick RP. Identification of ligand specificity determinants in AgrC, the Staphylococcus aureus quorum-sensing receptor. J Biol Chem. 2008;283:8930–8938. doi: 10.1074/jbc.M710227200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Geisinger E, Muir TW, Novick RP. agr receptor mutants reveal distinct modes of inhibition by staphylococcal autoinducing peptides. Proc Natl Acad Sci. 2009;106:1216–1221. doi: 10.1073/pnas.0807760106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nikolskaya AN, Galperin MY. A novel type of conserved DNA-binding domain in the transcriptional regulators of the AlgR/AgrA/LytR family. Nucleic Acids Res. 2002;30:2453–2459. doi: 10.1093/nar/30.11.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sidote DJ, Barbieri CM, Wu T, Stock AM. Structure of the Staphylococcus aureus AgrA LytTR domain bound to DNA reveals a beta fold with an unusual mode of binding. Structure. 2008;16:727–735. doi: 10.1016/j.str.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reynolds J, Wigneshweraraj S. Molecular insights into the control of transcription initiation at the Staphylococcus aureus agr operon. J Mol Biol. 2011;412:862–881. doi: 10.1016/j.jmb.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 73.Queck SY, Jameson-Lee M, Villaruz AE, Bach T-HL, Khan BA, et al. RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus . Mol Cell. 2008;32:150–158. doi: 10.1016/j.molcel.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tal-Gan Y, Stacy DM, Foegen MK, Koenig DW, Blackwell HE. Highly potent inhibitors of quorum sensing in Staphylococcus aureus revealed through a systematic synthetic study of the group-III autoinducing peptide. J Am Chem Soc. 2013;135:7869–7882. doi: 10.1021/ja3112115. [DOI] [PubMed] [Google Scholar]
- 75.Tal-Gan Y, Ivancic M, Cornilescu G, Cornilescu CC, Blackwell HE. Structural characterization of native autoinducing peptides and abiotic analogues reveals key features essential for activation and inhibition of an AgrC quorum sensing receptor in Staphylococcus aureus . J Am Chem Soc. 2013;135:18436–18444. doi: 10.1021/ja407533e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tal-Gan Y, Ivancic M, Cornilescu G, Blackwell HE. Characterization of structural elements in native autoinducing peptides and non-native analogues that permit the differential modulation of AgrC-type quorum sensing receptors in Staphylococcus aureus . Org Biomol Chem. 2016;14:113–121. doi: 10.1039/c5ob01735a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tal-Gan Y, Ivancic M, Cornilescu G, Yang T, Blackwell HE. Highly stable, amide-bridged autoinducing peptide analogues that strongly inhibit the AgrC quorum sensing receptor in Staphylococcus aureus . Angew Chem Int Ed Engl. 2016;55:8913–8917. doi: 10.1002/anie.201602974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Otto M, Süssmuth R, Vuong C, Jung G, Götz F. Inhibition of virulence factor expression in Staphylococcus aureus by the Staphylococcus epidermidis agr pheromone and derivatives. FEBS Lett. 1999;450:257–262. doi: 10.1016/s0014-5793(99)00514-1. [DOI] [PubMed] [Google Scholar]
- 79.Johnson JG, Wang B, Debelouchina GT, Novick RP, Muir TW. Increasing AIP macrocycle size reveals key features of agr activation in Staphylococcus aureus . Chembiochem. 2015;16:1093–1100. doi: 10.1002/cbic.201500006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chan WC, Coyle BJ, Williams P. Virulence regulation and quorum sensing in Staphylococcal infections: competitive AgrC antagonists as quorum sensing inhibitors. J Med Chem. 2004;47:4633–4641. doi: 10.1021/jm0400754. [DOI] [PubMed] [Google Scholar]
- 81.García-Betancur J-C, Goñi-Moreno A, Horger T, Schott M, Sharan M, et al. Cell differentiation defines acute and chronic infection cell types in Staphylococcus aureus . Elife. 2017;6:e28023. doi: 10.7554/eLife.28023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mukherjee S, Bassler BL. Bacterial quorum sensing in complex and dynamically changing environments. Nat Rev Microbiol. 2019;17:371–382. doi: 10.1038/s41579-019-0186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rudkin JK, Edwards AM, Bowden MG, Brown EL, Pozzi C, et al. Methicillin resistance reduces the virulence of healthcare-associated methicillin-resistant Staphylococcus aureus by interfering with the agr quorum sensing system. J Infect Dis. 2012;205:798–806. doi: 10.1093/infdis/jir845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Grundstad ML, Parlet CP, Kwiecinski JM, Kavanaugh JS, Crosby HA, et al. Quorum sensing, virulence, and antibiotic resistance of USA100 methicillin-resistant Staphylococcus aureus isolates. mSphere. 2019;4:e00553-19. doi: 10.1128/mSphere.00553-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cheung GYC, Wang R, Khan BA, Sturdevant DE, Otto M. Role of the accessory gene regulator agr in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. Infect Immun. 2011;79:1927–1935. doi: 10.1128/IAI.00046-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Geisinger E, Chen J, Novick RP. Allele-dependent differences in quorum-sensing dynamics result in variant expression of virulence genes in Staphylococcus aureus . J Bacteriol. 2012;194:2854–2864. doi: 10.1128/JB.06685-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sloan TJ, Murray E, Yokoyama M, Massey RC, Chan WC, et al. Timing is everything: impact of naturally occurring Staphylococcus aureus AgrC cytoplasmic domain adaptive mutations on autoinduction. J Bacteriol. 2019;201:e00409-19. doi: 10.1128/JB.00409-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wright JS, Jin R, Novick RP. Transient interference with Staphylococcal quorum sensing blocks abscess formation. Proc Natl Acad Sci U S A. 2005;102:1691–1696. doi: 10.1073/pnas.0407661102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Winson MK, Swift S, Hill PJ, Sims CM, Griesmayr G, et al. Engineering the luxCDABE genes from Photorhabdus luminescens to provide a bioluminescent reporter for constitutive and promoter probe plasmids and mini-Tn 5 constructs. FEMS Microbiol Lett. 1998;163:193–202. doi: 10.1111/j.1574-6968.1998.tb13045.x. [DOI] [PubMed] [Google Scholar]
- 90.Sun F, Liang H, Kong X, Xie S, Cho H, et al. Quorum-sensing agr mediates bacterial oxidation response via an intramolecular disulfide redox switch in the response regulator AgrA . Proc Natl Acad Sci USA. 2012;109:9095–9100. doi: 10.1073/pnas.1200603109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Baković J, Yu BYK, Silva D, Baczynska M, Peak-Chew SY, et al. Redox Regulation of the Quorum-sensing Transcription Factor AgrA by Coenzyme A. Antioxidants. 2021;10:841. doi: 10.3390/antiox10060841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.George SE, Hrubesch J, Breuing I, Vetter N, Korn N, et al. Oxidative stress drives the selection of quorum sensing mutants in the Staphylococcus aureus population. Proc Natl Acad Sci U S A. 2019;116:19145–19154. doi: 10.1073/pnas.1902752116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Peterson MM, Mack JL, Hall PR, Alsup AA, Alexander SM, et al. Apolipoprotein B is an innate barrier against invasive Staphylococcus aureus infection. Cell Host Microbe. 2008;4:555–566. doi: 10.1016/j.chom.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.James EH, Edwards AM, Wigneshweraraj S. Transcriptional downregulation of agr expression in Staphylococcus aureus during growth in human serum can be overcome by constitutively active mutant forms of the sensor kinase AgrC . FEMS Microbiol Lett. 2013;349:153–162. doi: 10.1111/1574-6968.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Strobel M, Pförtner H, Tuchscherr L, Völker U, Schmidt F, et al. Post-invasion events after infection with Staphylococcus aureus are strongly dependent on both the host cell type and the infecting S. aureus strain. Clin Microbiol Infect. 2016;22:799–809. doi: 10.1016/j.cmi.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 96.Carnes EC, Lopez DM, Donegan NP, Cheung A, Gresham H, et al. Confinement-induced quorum sensing of individual Staphylococcus aureus bacteria. Nat Chem Biol. 2010;6:41–45. doi: 10.1038/nchembio.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Traber KE, Lee E, Benson S, Corrigan R, Cantera M, et al. agr function in clinical Staphylococcus aureus isolates. Microbiology. 2008;154:2265–2274. doi: 10.1099/mic.0.2007/011874-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shopsin B, Drlica-Wagner A, Mathema B, Adhikari RP, Kreiswirth BN, et al. Prevalence of agr dysfunction among colonizing Staphylococcus aureus strains. J Infect Dis. 2008;198:1171–1174. doi: 10.1086/592051. [DOI] [PubMed] [Google Scholar]
- 99.Shopsin B, Eaton C, Wasserman GA, Mathema B, Adhikari RP, et al. Mutations in agr do not persist in natural populations of methicillin-resistant Staphylococcus aureus . J Infect Dis. 2010;202:1593–1599. doi: 10.1016/j.tim.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 100.Painter KL, Krishna A, Wigneshweraraj S, Edwards AM. What role does the quorum-sensing accessory gene regulator system play during Staphylococcus aureus bacteremia? Trends Microbiol. 2014;22:676–685. doi: 10.1016/j.tim.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 101.Young BC, Wu C-H, Gordon NC, Cole K, Price JR, et al. Severe infections emerge from commensal bacteria by adaptive evolution. Elife. 2017;6:e30637. doi: 10.7554/eLife.30637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee SO, Lee S, Lee JE, Song K-H, Kang CK, et al. Dysfunctional accessory gene regulator (agr) as a prognostic factor in invasive Staphylococcus aureus infection: a systematic review and meta-analysis. Sci Rep. 2020;10:10:20697. doi: 10.1038/s41598-020-77729-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lee JE, Lee S, Park S, Lee SO, Lee SH. Impact of agr Functionality on the outcome of patients with methicillin-susceptible Staphylococcus aureus bacteremia. Microbiol Spectr. 2021;9:e0011621. doi: 10.1128/Spectrum.00116-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pader V, Hakim S, Painter KL, Wigneshweraraj S, Clarke TB, et al. Staphylococcus aureus inactivates daptomycin by releasing membrane phospholipids. Nat Microbiol. 2016;2:16194. doi: 10.1038/nmicrobiol.2016.194. [DOI] [PubMed] [Google Scholar]
- 105.Altman DR, Sullivan MJ, Chacko KI, Balasubramanian D, Pak TR, et al. Genome Plasticity of agr-Defective Staphylococcus aureus during clinical infection. Infect Immun. 2018;86:e00331-18. doi: 10.1128/IAI.00331-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Thänert R, Goldmann O, Beineke A, Medina E. Host-inherent variability influences the transcriptional response of Staphylococcus aureus during in vivo infection. Nat Commun. 2017;8:14268. doi: 10.1038/ncomms14268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Siegmund A, Afzal MA, Tetzlaff F, Keinhörster D, Gratani F, et al. Intracellular persistence of Staphylococcus aureus in endothelial cells is promoted by the absence of phenol soluble modulins. Virulence. 2021;12:1186–1198. doi: 10.1128/microbiolspec.GPP3-0069-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Proctor R, Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, et al. Respiration and small colony variants of Staphylococcus aureus . Microbiol Spectr. 2019;7 doi: 10.1128/microbiolspec.GPP3-0069-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tuchscherr L, Löffler B, Proctor RA. Persistence of Staphylococcus aureus: multiple metabolic pathways impact the expression of virulence factors in small-colony variants (SCVs) Front Microbiol. 2020;11:1028. doi: 10.3389/fmicb.2020.01028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gor V, Takemura AJ, Nishitani M, Higashide M, Medrano Romero V, et al. Finding of Agr phase variants in Staphylococcus aureus . mBio. 2019;10:e00796-19. doi: 10.1128/IAI.01216-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pollitt EJ, West SA, Crusz SA, Burton-Chellew MN, Hamushan SP. Cooperation, quorum sensing, and evolution of virulence in Staphylococcus aureus . Infect Immun. 2014;82:1045–1051. doi: 10.1002/ctm2.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.He L, Zhang F, Jian Y, Lv H, Hamushan M, et al. Key role of quorum-sensing mutations in the development of Staphylococcus aureus clinical device-associated infection. Clin Transl Med. 2022;12:e801. doi: 10.1099/mic.0.001119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Laabei M, Peacock SJ, Blane B, Baines SL, Howden BP, et al. Significant variability exists in the cytotoxicity of global methicillin-resistant Staphylococcus aureus lineages. Microbiology. 2021;167:001119. doi: 10.1099/mic.0.001119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Smyth DS, Kafer JM, Wasserman GA, Velickovic L, Mathema B, et al. Nasal carriage as a source of agr-defective Staphylococcus aureus bacteremia. J Infect Dis. 2012;206:1168–1177. doi: 10.1093/infdis/jis483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Traber K, Novick R. A slipped-mispairing mutation in AgrA of laboratory strains and clinical isolates results in delayed activation of agr and failure to translate delta- and alpha haemolysins. Mol Microbiol. 2006;59:1519–1530. doi: 10.1038/srep31360. [DOI] [PubMed] [Google Scholar]
- 116.Mairpady Shambat S, Siemens N, Monk IR, Mohan DB, Mukundan S, et al. A point mutation in AgrC determines cytotoxic or colonizing properties associated with phenotypic variants of ST22 MRSA strains. Sci Rep. 2016;6:31360. doi: 10.1128/JB.00700-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fleming V, Feil E, Sewell A, Day N, Buckling A, et al. Agr interference between clinical Staphylococcus aureus strains in an insect model of virulence. J Bacteriol. 2006;188:7686–7688. doi: 10.1086/376450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Goerke C, Kümmel M, Dietz K, Wolz C. Evaluation of intraspecies interference due to agr polymorphism in Staphylococcus aureus during infection and colonization. J Infect Dis. 2003;188:250–256. doi: 10.1128/IAI.68.3.1304-1311.2000. [DOI] [PubMed] [Google Scholar]
- 119.Goerke C, Campana S, Bayer MG, Döring G, Botzenhart K, et al. Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile In vitro . Infect Immun. 2000;68:1304–1311. doi: 10.1128/IAI.68.3.1304-1311.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Byrd AL, Belkaid Y, Segre JA. The human skin microbiome. Nat Rev Microbiol. 2018;16:143–155. doi: 10.1128/CMR.00109-13. [DOI] [PubMed] [Google Scholar]
- 121.Becker K, Heilmann C, Peters G. Coagulase-negative Staphylococci . Clin Microbiol Rev. 2014;27:870–926. doi: 10.1126/scitranslmed.aah4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nakatsuji T, Chen TH, Narala S, Chun KA, Two AM, et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci Transl Med. 2017;9:eaah4680. doi: 10.1371/journal.ppat.1009026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Brown MM, Horswill AR. Staphylococcus epidermidis - Skin friend or foe? PLoS Pathog. 2020;16:e1009026. doi: 10.1038/s41579-022-00780-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Severn MM, Horswill AR. Staphylococcus epidermidis and its dual lifestyle in skin health and infection. Nat Rev Microbiol. 2023;21:97–111. doi: 10.1016/j.cell.2020.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhou W, Spoto M, Hardy R, Guan C, Fleming E, et al. Host-specific evolutionary and transmission dynamics shape the functional diversification of Staphylococcus epidermidis in human skin. Cell. 2020;180:454–470. doi: 10.1126/scitranslmed.aat8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dufour P, Jarraud S, Vandenesch F, Greenland T, Novick RP, et al. High genetic variability of the agr locus in Staphylococcus species. J Bacteriol. 2022;184:1180–1186. doi: 10.1128/mbio.00930-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Williams MR, Costa SK, Zaramela LS, Khalil S, Todd DA, et al. Quorum sensing between bacterial species on the skin protects against epidermal injury in atopic dermatitis. Sci Transl Med. 2019;11:eaat8329. doi: 10.1128/JB.01882-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Severn MM, Williams MR, Shahbandi A, Bunch ZL, Lyon LM, et al. The ubiquitous human skin commensal Staphylococcus hominis protects against opportunistic pathogens. mBio. 2022;13:e0093022. doi: 10.1128/JB.187.9.3139-3150.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Olson ME, Todd DA, Schaeffer CR, Paharik AE, Van Dyke MJ, et al. Staphylococcus epidermidis agr quorum-sensing system: signal identification, cross talk, and importance in colonization. J Bacteriol. 2014;196:3482–3493. doi: 10.1128/JB.01882-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ji G, Pei W, Zhang L, Qiu R, Lin J, et al. Staphylococcus intermedius produces a functional agr autoinducing peptide containing a cyclic lactone. J Bacteriol. 2005;187:3139–3150. doi: 10.1128/AAC.00172-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chin D, Flannagan RS, Tuffs SW, Chan JK, McCormick JK, et al. Staphylococcus lugdunensis uses the agr regulatory system to resist killing by host innate immune effectors. Infect Immun. 2022;90:e0009922. doi: 10.1128/IAI.69.3.1957-1960.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Otto M, Echner H, Voelter W, Götz F. Pheromone cross-inhibition between Staphylococcus aureus and Staphylococcus epidermidis . Infect Immun. 2001;69:1957–1960. doi: 10.1016/j.chom.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yang T, Tal-Gan Y, Paharik AE, Horswill AR, Blackwell HE. Structure-function analyses of a Staphylococcus epidermidis autoinducing peptide reveals motifs critical for AgrC type receptor modulation. ACS Chem Biol. 2016;11:1982–1991. doi: 10.1016/j.jid.2022.05.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Brown MM, Kwiecinski JM, Cruz LM, Shahbandi A, Todd DA, et al. Novel peptide from commensal Staphylococcus simulans blocks methicillin-resistant Staphylococcus aureus quorum sensing and protects host skin from damage. Antimicrob Agents Chemother. 2020;64:e00172-20. doi: 10.1021/acschembio.6b00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Paharik AE, Parlet CP, Chung N, Todd DA, Rodriguez EI, et al. Coagulase-negative Staphylococcal strain prevents Staphylococcus aureus colonization and skin infection by blocking quorum sensing. Cell Host Microbe. 2017;22:746–756. doi: 10.1016/j.chom.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Severn MM, Cho Y-SK, Manzer HS, Bunch ZL, Shahbandi A, et al. The commensal Staphylococcus warneri makes peptide inhibitors of MRSA quorum sensing that protect skin from atopic or necrotic damage. J Invest Dermatol. 2022;142:3349–3352. doi: 10.1016/j.jid.2022.05.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Liu H, Archer NK, Dillen CA, Wang Y, Ashbaugh AG, et al. Staphylococcus aureus epicutaneous exposure drives skin inflammation via IL-36-mediated T cell responses. Cell Host Microbe. 2021;147:955–966. doi: 10.1016/j.jaci.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Geoghegan JA, Irvine AD, Foster TJ. Staphylococcus aureus and atopic dermatitis: a complex and evolving relationship. Trends Microbiol. 2018;26:484–497. doi: 10.3389/fmicb.2016.01230. [DOI] [PubMed] [Google Scholar]
- 139.Cau L, Williams MR, Butcher AM, Nakatsuji T, Kavanaugh JS, et al. Staphylococcus epidermidis protease EcpA can be a deleterious component of the skin microbiome in atopic dermatitis. J Allergy Clin Immunol. 2021;147:955–966. doi: 10.1038/s41591-021-01256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ramsey MM, Freire MO, Gabrilska RA, Rumbaugh KP, Lemon KP. Staphylococcus aureus shifts toward commensalism in response to Corynebacterium Species. Front Microbiol. 2016;7:1230. doi: 10.1021/cr100370n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nakatsuji T, Hata TR, Tong Y, Cheng YJ, Shafiq F, et al. Development of a human skin commensal microbe for bacteriotherapy of atopic dermatitis and use in a phase 1 randomized clinical trial. Nat Med. 2021;27:700–709. doi: 10.1038/s41594-021-00714-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Thoendel M, Kavanaugh JS, Flack CE, Horswill AR. Peptide signaling in the Staphylococci . Chem Rev. 2011;111:117–151. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1002/jcc.20945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 146.Buel GR, Walters KJ. Can AlphaFold2 predict the impact of missense mutations on structure? Nat Struct Mol Biol. 2022;29:1–2. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, et al. The ClusPro web server for protein-protein docking. Nat Protoc. 2017;12:255–278. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]