Abstract
Burkholderia multivorans is the dominant Burkholderia pathogen recovered from lung infection in people with cystic fibrosis. However, as an understudied pathogen there are knowledge gaps in relation to its population biology, phenotypic traits and useful model strains. A phylogenomic study of B. multivorans was undertaken using a total of 283 genomes, of which 73 were sequenced and 49 phenotypically characterized as part of this study. Average nucleotide identity analysis (ANI) and phylogenetic alignment of core genes demonstrated that the B. multivorans population separated into two distinct evolutionary clades, defined as lineage 1 (n=58 genomes) and lineage 2 (n=221 genomes). To examine the population biology of B. multivorans , a representative subgroup of 77 B. multivorans genomes (28 from the reference databases and the 49 novel short-read genome sequences) were selected based on multilocus sequence typing (MLST), isolation source and phylogenetic placement criteria. Comparative genomics was used to identify B. multivorans lineage-specific genes – ghrB_1 in lineage 1 and glnM_2 in lineage 2 – and diagnostic PCRs targeting them were successfully developed. Phenotypic analysis of 49 representative B. multivorans strains showed considerable inter-strain variance, but the majority of the isolates tested were motile and capable of biofilm formation. A striking absence of B. multivorans protease activity in vitro was observed, but no lineage-specific phenotypic differences were demonstrated. Using phylogenomic and phenotypic criteria, three model B. multivorans CF strains were identified, BCC0084 (lineage 1), BCC1272 (lineage 2a) and BCC0033 lineage 2b, and their complete genome sequences determined. B. multivorans CF strains BCC0033 and BCC0084, and the environmental reference strain, ATCC 17616, were all capable of short-term survival within a murine lung infection model. By mapping the population biology, identifying lineage-specific PCRs and model strains, we provide much needed baseline resources for future studies of B. multivorans .
Keywords: Burkholderia multivorans, phylogenomics, phenotype, infection modelling, cystic fibrosis
Introduction
Cystic fibrosis (CF) is a hereditary genetic disorder affecting over 10 500 people in the UK [1]. Mutations in the CF transmembrane conductance regulator gene of people with CF results in several pathological features, with abnormal lung clearance, chronic respiratory infection and severe lung disease being major contributors to morbidity. Although Pseudomonas aeruginosa is the most prevalent CF pathogen, Burkholderia cepacia complex (Bcc) bacteria, a taxonomic group of closely related Burkholderia , emerged as virulent and transmissible CF lung infections in the 1990s [2]. For people with CF, infection with Bcc pathogens can contribute to severe lung function decline and the development of ‘cepacia syndrome’ [3], and those infected also have a lower survival rate after lung transplantation [4]. Whilst the Bcc have been reported at relatively low in prevalence in CF populations (<5 %) [4–6], they are of significant clinical consequence because they are hard to eradicate due to their intrinsic resistance to antibiotics, with certain strains being resistant to the 10 most administered antibiotics [7].
Burkholderia multivorans is a member of Bcc and is the most isolated Burkholderia species in the UK, with 56 % of all Burkholderia CF lung infection cases (n=361) attributed to the pathogen in 2017 [5]. Earlier surveys in the USA showed that B. multivorans accounted for 37 % of Burkholderia CF infections at the time [6] and the same dominance was observed in a Canadian study, with 45 % of 122 Burkholderia CF lung infection cases caused by this Bcc species [4]. The epidemiology of Burkholderia CF infections also shows that B. multivorans has become dominant due to reduced rates of Burkholderia cenocepacia infection, which is now the second most common Bcc species in multiple CF populations [4–6]. With strict infection control and the resulting absence of patient–patient transmission, the continuing emergence of B. multivorans in people with CF suggests that current infections arise sporadically from natural sources such as soil, the rhizosphere and water [2, 6, 8]. However, specific environmental reservoirs of B. multivorans remain elusive, with isolates rarely recovered from the natural environment [6, 8].
In contrast to this current epidemiological prevalence of B. multivorans , B. cenocepacia has been the most widely studied CF Burkholderia [9]. B. cenocepacia is generally considered to be the hypervirulent species within the Bcc [2] and can be separated into two genetic lineages (III-A and III-B) based on the recA gene [10, 11]. Recent genomic analysis of B. cenocepacia provided further evidence to show that the species should be split into at least two different species based on average nucleotide identity (ANI) differences [12]. The latter study argued for the name ‘Burkholderia servocepacia’ to be attributed to strains falling into the recA III-B grouping, but this proposition was invalid based on taxonomic and naming criteria. Burkholderia orbicola sp. nov. [13] has now been validly proposed as the species name for the genomic taxa represented by Burkholderia servocepacia. Overall, multiple studies have shown that epidemic and transmissible CF strains can be found in both B. cenocepacia and B. orbicola sp. nov. [9]. For example, B. cenocepacia III-A strains are associated with poor clinical outcome and major morbidity in several CF populations [2, 6], with the ET-12 strain being notable in virulence and prevalence, together with multiple other intercontinentally dispersed multilocus sequence types (MLSTs) [9]. Virulence factors such as the cable pilus, cenocepacia pathogenicity island and multiple quorum sensing-dependent pathogenicity traits, have also been characterized for B. cenocepacia [9].
In comparison, much less is known about the pathogenicity of B. multivorans in CF. The presence of non-mucoid isolates of Bcc bacteria have been shown to be correlated with greater decrease in lung function of infected individuals [14], and this mucoid variation in B. multivorans was associated with changes in metabolism, motility, biofilm formation and virulence [15]. Within-strain genomic evolution has been studied for multiple isolates recovered over 20 years from an individual with CF [16]. The average evolutionary substitution mutation rate for this single B. multivorans strain was low overall, at 2.4 mutations per year, with one intra-strain lineage evolving more rapidly than the others through non-synonymous mutations [16]. Alterations in the B. multivorans phenotype during chronic infection were linked to mutational changes in antimicrobial resistance, biofilm formation and LPS O-antigen presentation gene pathways [16]. Another study obtained genome sequences from 111 clonal isolates of B. multivorans from a single person with CF, as their lung disease progressed [17]. Statistically significant accumulations of mutations in loci contributing to increased antimicrobial resistance were seen in this single-strain evolutionary study [17]. Genomic comparison of B. multivorans isolates isogenic by MLST, but from CF infection and natural environmental sources, demonstrated that the same genomic lineages occur in these different niches and across different continents [18].
A comparison of multiple genetically distinct B. multivorans strains that includes both phenotypic and genomic characterization of the species has not yet been made. Our study aimed to unpick the phylogenomics and basic pathobiology of B. multivorans, as both a species and an understudied CF lung pathogen. Whole-genome sequencing (WGS) was used to characterize 73 genetically diverse B. multivorans strains drawn from multiple sources, MLST strain types and geographical regions. A further 210 B. multivorans genomes were obtained from publicly accessible databases and analysed phylogenetically. Twenty-eight of the database sequences were combined with 49 of the de novo genome-sequenced strains to produce a representative strain panel (n=77). The B. multivorans strain panel encompassed 61 unique MLST sequence types (STs; 5 novel), focusing on CF isolates (n=60) and including strains from the environment (n=8) and non-CF infection (n=8), with 1 isolate from an undetermined source. The phenotypic features of 49 representative strains selected from this panel were investigated by swimming and swarming motilities, biofilm formation, exopolysaccharide production and protease production, and 3 strains were also tested for survival in a mammalian respiratory inhalation lung infection model [19, 20]. From this analysis, an evolutionary split into two genetic lineages was shown for B. multivorans as a CF pathogen.
Methods
Bacterial strains and incubation conditions
The bacterial strains phenotypically studied and genome sequenced in this study were drawn from the Burkholderia strain collection at Cardiff University and additional recognized strain repositories [20, 21] (Table 1). A complete list of the 283 isolates and their genomes analysed within the study is provided in Table S1, available in the online version of this article. The isolates studied were recovered from a range of sources, including CF, chronic granulomatous disease (CGD), non-CF clinical infections (NON-CF), the natural environment (ENV) and the healthcare environment (ENVH). Stock cultures were stored at −80 °C in cryogenic vials by resuspension of fresh growth in tryptic soya broth (TSB; Oxoid) containing 8 % (v/v) dimethyl sulfoxide (Sigma-Aldrich). Culture purity was determined by plating frozen stocks onto tryptic soy agar (TSA) (Oxoid) and incubating plates for 24–48 h at 37 °C. Overnight cultures were made by taking a swab from a fresh TSA plate and transferring into 3 ml of TSB. Cultures were grown for 18–20 h at 37 °C using continuous shaking on a rotating platform set to 150 r.p.m.
Table 1.
The selected B. multivorans strain panel (n=77) including 49 phenotypically characterized strains sequenced in this study
|
Strain (and alternative strain name) |
Lineage |
Isolation source and geographical location |
ENA accession |
|---|---|---|---|
|
Sequenced and phenotypically studied | |||
|
BCC0006 |
1 |
CF |
ERR4672189 |
|
BCC0009 |
1 |
CGD |
ERR4672190 |
|
BCC0080 |
1 |
CF |
ERR4672267 |
|
BCC0084 |
1 |
CF |
ERR4672269 (complete genome: ERR10387434) |
|
BCC0101 |
1 |
CF |
ERR4672272 |
|
BCC0141 |
1 |
CF |
ERR4672273 |
|
BCC0303 |
1 |
CF |
ERR4672282 |
|
BCC0375 |
1 |
CF |
ERR4672284 |
|
BCC0381 |
1 |
NON |
ERR4672285 |
|
BCC0702 |
1 |
CF |
ERR4672759 |
|
BCC0737 |
1 |
CF |
ERR4676914 |
|
BCC0814 |
1 |
CF |
ERR4674025 |
|
BCC0865 |
1 |
CF |
ERR4674026 |
|
BCC0904 |
1 |
NON |
ERR4674027 |
|
BCC0921 |
1 |
CF |
ERR4674035 |
|
BCC1177 |
1 |
CF |
ERR4674031 |
|
BCC1190 |
1 |
CF |
ERR4674032 |
|
BCC1385 |
1 |
CF |
ERR4674033 |
|
BCC0047 |
2 a |
CF |
ERR4672260 |
|
BCC0066 |
2 a |
CF |
ERR4672262 |
|
BCC0074 |
2 a |
CF |
ERR4672264 |
|
BCC0188 |
2 a |
CF |
ERR4672274 |
|
BCC0225 |
2 a |
CF |
ERR4674034 |
|
BCC0264 |
2 a |
CF |
ERR4676953 |
|
BCC0266 |
2 a |
CF |
ERR4672280 |
|
BCC0317 |
2 a |
ENV |
ERR4672283 |
|
BCC0032 |
2b |
CF |
ERR4672191 |
|
BCC0033 |
2b |
CF |
ERR4672192 |
|
BCC0043 |
2b |
CF |
ERR4672194 |
|
BCC0065 |
2b |
NON |
ERR4672261 |
|
BCC0068 |
2b |
CF |
ERR4672263 |
|
BCC0075 |
2b |
CF |
ERR4672265 |
|
BCC0079 |
2b |
CF |
ERR4672266 |
|
BCC0082 |
2b |
CF |
ERR4672268 |
|
BCC0087 |
2b |
CF |
ERR4672270 |
|
BCC0096 |
2b |
CF |
ERR4672271 |
|
BCC0241 |
2b |
NON |
ERR4672275 |
|
BCC0246 |
2b |
CF |
ERR4672279 |
|
BCC0247 |
2b |
CF |
ERR4674976 |
|
BCC0269 |
2b |
CF |
ERR4672281 |
|
BCC0384 |
2b |
CF |
ERR4672589 |
|
BCC0493 |
2b |
CF |
ERR4672590 |
|
BCC0497 |
2b |
CF |
ERR4672598 |
|
BCC0710 |
2b |
CF |
ERR4672760 |
|
BCC1147 |
2b |
CF |
ERR4674028 |
|
BCC1148 |
2b |
CF |
ERR4674030 |
|
BCC1185 |
2b |
CF |
ERR4676921 |
|
BCC1272 |
2 a |
CF |
ERR4676913 (complete genome: ERR10387431) |
|
BCC1368 |
Other |
CF |
ERR4676903 |
|
Reference genomes from NCBI | |||
|
ATCC BAA-247 |
1 |
CF |
GCA_000959525.1 |
|
AU1185 |
1 |
NON |
GCA_001718755.1 |
|
AU10047 |
1 |
CF |
GCA_002981015.1 |
|
AU11358 |
1 |
CF |
GCA_002981015.1 |
|
AU28442 |
1 |
CF |
GCA_002981415.1 |
|
CGD2 |
1 |
CGD |
GCA_000182275.1 |
|
FDAARGOS_546 |
1 |
nd |
GCA_003938705.1 |
|
HI3534 |
1 |
ENVH |
GCA_001528605.1 |
|
R-20526 |
1 |
ENV |
GCA_001267755.1 |
|
ATCC 17616 |
2 a |
ENV |
GCA_000010545 |
|
AU10398 |
2 a |
CF |
GCA_002980695.1 |
|
AU15814 |
2 a |
CF |
GCA_002980895.1 |
|
AU17545 |
2 a |
CF |
GCA_002980995.1 |
|
AU18096 |
2 a |
CF |
GCA_002981145.1 |
|
AU28069 |
2 a |
CF |
GCA_002981845.1 |
|
CF2 |
2 a |
CF |
GCA_000286575.1 |
|
CGD1 |
2 a |
CGD |
GCA_000182255.1 |
|
DWS 42B-1 |
2 a |
ENV |
GCA_000756965.1 |
|
MSMB1272WGS |
2 a |
ENV |
GCA_001529925.1 |
|
MSMB1640WGS |
2 a |
ENV |
GCA_001718995.1 |
|
NKI379 |
2 a |
ENV |
GCA_001302465.1 |
|
AU4507 |
2b |
CF |
GCA_002981595.1 |
|
AU20929 |
2b |
CF |
GCA_002981635.1 |
|
AU21015 |
2b |
CF |
GCA_003048355.1 |
|
AU22892 |
2b |
CF |
GCA_002981295.1 |
|
AU24277 |
2b |
CF |
GCA_002981375.1 |
|
BMUL_CF170.0a |
2b |
CF |
GCA_003257435.1 |
|
D2214 |
2b |
CF |
GCA_000807815.1 |
Genome sequencing of B. multivorans
B. multivorans strains for genome sequencing were selected based on their source, geographical distribution and MLST-based genetic diversity [22, 23] (Tables 1 and S1). After revival and purity checking, 3 ml overnight cultures were subjected to DNA extraction using an automated Maxwell 16 Tissue DNA purification kit following the manufacturer’s instructions (Promega, UK). For long-read complete genome analysis, DNA was extracted using a DNA Wizard kit (Promega, UK). Upon extraction, each DNA sample was transferred into non-stick 1.5 ml microtubes and stored at −20 °C. DNA samples were checked for purity using the B. multivorans -specific recA primers, BCRBM1 and BCRBM2 [10], with PCR amplicons visualized on 1.5 % (w/v) agarose gels prior to Sanger sequence analysis to confirm that they were B. multivorans .
A total of 73 B. multivorans strains were subjected to short-read WGS using an Illumina MiSeq V2 platform within the Genome Hub at Cardiff School of Biosciences. Genomic reads were assembled and annotated using the shared Cloud Infrastructure for Microbial Genomics (CLIMB) computing facility [24]. Illumina reads were subjected to the Trim Galore v0.4.4 [25] wrapper script. This utilizes Cutadapt v1.9.1 [26] for automated quality and adapter trimming and FastQC v0.11.4 [27] for quality control. The MultiQC v1.7 [28] Python package was used to compile a single-file report and an interactive report for the samples, helping to streamline quality control screening. All genomes possessed sufficient quality to take forward for phylogenomic analyses (Table S2).
To assemble the bacterial genomes, we used the Unicycler v0.4.7 [29] assembly pipeline, which utilizes SPAdes [30] for optimizing and streamlining de novo assembly of the genome contigs. Complete genome sequence analysis was performed for the three selected model strains (BCC0033, BCC0084 and BCC1272) using long-read PacBio technology (carried out by Novogene, UK). The PacBio FASTQ reads were subjected to the Trycycler pipeline (v0.4.1) [29] and provide complete assemblies of four contigs (the three genomic replicons and a large plasmid in each strain). DNA sequence reads from the selected database genomes were also reassembled and all 283 B. multivorans genomes were subjected to Prokka v1.14.0 [31] to annotate the sequences and provide output files suitable for phylogenomic analysis. Accession numbers for the genome sequences obtained in this study are provided in Table 1.
Genomic taxonomy, phylogenomic and MLST analyis
To confirm the taxonomic identity of the B. multivorans genomes and filter out contaminating DNA, the Minikraken database from Kraken2 v2.08-beta [32] was used. QUAST v5.0 [33] was used to assess quality and respective statistics for the genomic assemblies. To confirm species taxonomy, the pairwise ANI was calculated for the B. multivorans genomes using the Python 3 module and the script PyANI v0.2.9 [34]. A 95 % threshold was used as an accepted standard to confirm that all strains were the same species in accordance with the Genomic Taxonomy database [35] and recent taxonomic analysis of Burkholderia genomes [36].
Phylogenomic and pan-genome analysis was performed as follows. The GFF annotated genome file outputs from Prokka [31] were evaluated in the Roary v3.12.0 pan genome pipeline [37] to assess the core and accessory genome of all 283 B. multivorans genomes. The command was performed using the default settings. Multiple Alignment using Fast Fourier Transform (MAFFT) [38] was used to create the Roary core gene alignment output file. Phylogenetic trees were built using maximum-likelihood (GTRGAMMA model) randomized accelerated maximum-likelihood (RAxML v8) [39], supported by 100 bootstraps. The B. dolosa AU0158 complete genome was initially used to root the phylogenetic trees as a closely related Bcc species; subsequent trees were rooted with the B. multivorans BCC1638 genome (Table 1). STs were determined for all B. multivorans strains using MLSTcheck and PubMLST blast schemes [40] (Table 2).
Table 2.
MLST alleles and sequence type (ST) for the 77-strain B. multivorans panel
|
Strain |
Isolation source |
MLST alleles |
ST* |
Clonal complex |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
atpD |
gltB |
gyrB |
recA |
lepA |
phaC |
trpB |
||||
|
ATCC 17616 |
ENV |
13 |
78 |
100 |
94 |
92 |
96 |
6 |
21 |
– |
|
BCC0006 |
CF |
11 |
60 |
251 |
81 |
37 |
96 |
5 |
650 |
– |
|
BCC0009 |
CGD |
9 |
223 |
445 |
81 |
137 |
35 |
215 |
1530 |
– |
|
BCC0032 |
CF |
13 |
151 |
168 |
139 |
142 |
100 |
132 |
191 |
– |
|
BCC0033 |
CF |
8 |
5 |
5 |
7 |
7 |
42 |
105 |
16 |
15 |
|
BCC0043 |
CF |
13 |
9 |
83 |
12 |
7 |
42 |
391 |
806 |
– |
|
BCC0047 |
CF |
13 |
62 |
695 |
110 |
45 |
14 |
452 |
1077 |
– |
|
BCC0065 |
NON |
8 |
5 |
5 |
7 |
7 |
42 |
105 |
16 |
15 |
|
BCC0066 |
CF |
336 |
61 |
97 |
11 |
64 |
96 |
104 |
880 |
– |
|
BCC0068 |
CF |
168 |
220 |
303 |
133 |
7 |
96 |
4 |
2213 |
– |
|
BCC0074 |
CF |
14 |
8 |
55 |
11 |
46 |
96 |
281 |
618 |
– |
|
BCC0075 |
CF |
13 |
7 |
6 |
10 |
224 |
42 |
415 |
899 |
– |
|
BCC0079 |
CF |
13 |
150 |
166 |
88 |
7 |
42 |
6 |
1792 |
– |
|
BCC0080 |
CF |
3 |
50 |
4 |
81 |
7 |
35 |
57 |
1964 |
– |
|
BCC0082 |
CF |
13 |
188 |
1424 |
165 |
200 |
96 |
884 |
2220 |
|
|
BCC0084 |
CF |
9 |
50 |
53 |
81 |
63 |
96 |
133 |
195 |
195 |
|
BCC0087 |
CF |
13 |
5 |
172 |
133 |
145 |
96 |
137 |
199 |
199 |
|
BCC0096 |
CF |
168 |
190 |
259 |
133 |
7 |
96 |
132 |
317 |
305 |
|
BCC0101 |
CF |
9 |
205 |
285 |
141 |
63 |
35 |
5 |
304 |
– |
|
BCC0141 |
CF |
9 |
50 |
84 |
141 |
37 |
96 |
7 |
1023 |
– |
|
BCC0188 |
CF |
125 |
154 |
171 |
140 |
144 |
14 |
136 |
196 |
– |
|
BCC0241 |
NON |
13 |
329 |
269 |
7 |
7 |
42 |
132 |
605 |
– |
|
BCC0225 |
CF |
14 |
8 |
55 |
11 |
46 |
96 |
281 |
618 |
– |
|
BCC0246 |
CF |
13 |
5 |
262 |
188 |
203 |
42 |
132 |
273 |
– |
|
BCC0247 |
CF |
8 |
5 |
5 |
7 |
7 |
42 |
105 |
16 |
15 |
|
BCC0264 |
CF |
13 |
61 |
264 |
184 |
144 |
42 |
6 |
274 |
274 |
|
BCC0266 |
CF |
13 |
152 |
1425 |
196 |
143 |
96 |
135 |
2228 |
– |
|
BCC0269 |
CF |
13 |
196 |
265 |
189 |
201 |
96 |
195 |
2219 |
– |
|
BCC0303 |
CF |
10 |
60 |
4 |
77 |
37 |
35 |
5 |
25 |
25 |
|
BCC0317 |
ENV |
13 |
63 |
53 |
80 |
61 |
96 |
56 |
22 |
– |
|
BCC0375 |
CF |
76 |
50 |
99 |
93 |
37 |
35 |
111 |
117 |
– |
|
BCC0381 |
NON |
9 |
75 |
54 |
93 |
63 |
35 |
66 |
18 |
– |
|
BCC0384 |
CF |
8 |
5 |
5 |
7 |
7 |
42 |
5 |
15 |
15 |
|
BCC0493 |
CF |
13 |
9 |
83 |
12 |
7 |
42 |
391 |
806 |
– |
|
BCC0497 |
CF |
13 |
9 |
83 |
12 |
7 |
42 |
7 |
26 |
26 |
|
BCC0702 |
CF |
9 |
50 |
169 |
81 |
409 |
96 |
81 |
2223 |
– |
|
BCC0710 |
CF |
8 |
5 |
5 |
7 |
7 |
42 |
5 |
15 |
15 |
|
BCC0737 |
CF |
123 |
50 |
170 |
81 |
Novel |
35 |
5 |
Novel† |
– |
|
BCC0814 |
CF |
118 |
50 |
158 |
6 |
37 |
96 |
5 |
180 |
– |
|
BCC0865 |
CF |
9 |
142 |
161 |
81 |
137 |
96 |
66 |
181 |
– |
|
BCC0904 |
NON |
118 |
50 |
158 |
6 |
37 |
96 |
5 |
180 |
– |
|
BCC0921 |
CF |
9 |
142 |
161 |
81 |
137 |
96 |
66 |
181 |
– |
|
BCC1147 |
CF |
168 |
190 |
259 |
133 |
7 |
96 |
132 |
317 |
305 |
|
BCC1148 |
CF |
168 |
190 |
259 |
133 |
7 |
96 |
132 |
317 |
305 |
|
BCC1177 |
CF |
9 |
50 |
53 |
81 |
63 |
96 |
133 |
195 |
195 |
|
BCC1185 |
CF |
13 |
5 |
172 |
133 |
145 |
96 |
137 |
199 |
199 |
|
BCC1190 |
CF |
9 |
75 |
54 |
93 |
63 |
35 |
66 |
18 |
– |
|
BCC1272 |
CF |
13 |
78 |
100 |
94 |
92 |
96 |
6 |
21 |
– |
|
BCC1368 |
CF |
211 |
205 |
170 |
93 |
37 |
35 |
251 |
179 |
– |
|
BCC1385 |
CF |
7 |
270 |
4 |
81 |
137 |
35 |
5 |
847 |
– |
|
ATCC BAA-247 |
CF |
13 |
236 |
354 |
133 |
231 |
42 |
4 |
650 |
– |
|
AU1185 |
NON |
9 |
75 |
54 |
93 |
63 |
35 |
66 |
18 |
– |
|
AU4507 |
CF |
13 |
61 |
620 |
133 |
424 |
42 |
6 |
891 |
– |
|
AU10047 |
CF |
9 |
50 |
84 |
289 |
37 |
96 |
5 |
564 |
– |
|
AU10398 |
CF |
13 |
397 |
283 |
135 |
623 |
42 |
340 |
1512 |
– |
|
AU11358 |
CF |
9 |
223 |
445 |
81 |
37 |
35 |
215 |
646 |
– |
|
AU15814 |
CF |
193 |
234 |
325 |
185 |
239 |
42 |
256 |
418 |
– |
|
AU17545 |
CF |
13 |
334 |
483 |
309 |
355 |
42 |
340 |
623 |
– |
|
AU18096 |
CF |
13 |
334 |
483 |
309 |
355 |
42 |
340 |
603 |
– |
|
AU20929 |
CF |
13 |
328 |
475 |
7 |
239 |
96 |
334 |
715 |
– |
|
AU21015 |
CF |
13 |
329 |
259 |
133 |
46 |
96 |
132 |
622 |
– |
|
AU22892 |
CF |
13 |
333 |
482 |
133 |
10 |
96 |
4 |
190 |
190 |
|
AU24277 |
CF |
121 |
138 |
167 |
138 |
141 |
42 |
132 |
625 |
– |
|
AU28069 |
CF |
13 |
9 |
484 |
7 |
64 |
266 |
195 |
630 |
– |
|
AU28442 |
CF |
13 |
145 |
488 |
135 |
10 |
96 |
104 |
645 |
– |
|
BMUL CF170.0a |
CF |
13 |
236 |
354 |
133 |
231 |
42 |
4 |
783 |
– |
|
CF2 |
CF |
193 |
453 |
695 |
207 |
461 |
343 |
4 |
1079 |
– |
|
CGD1 |
CGD |
12 |
6 |
118 |
9 |
63 |
100 |
6 |
1762 |
|
|
CGD2 |
CGD |
11 |
75 |
251 |
141 |
37 |
35 |
7 |
442 |
– |
|
D2214 |
CF |
8 |
5 |
5 |
7 |
7 |
42 |
105 |
16 |
15 |
|
DWS 42B-1 |
ENV |
122 |
373 |
98 |
7 |
230 |
96 |
376 |
809 |
– |
|
FDAARGOS 546 |
nd |
10 |
153 |
315 |
93 |
37 |
96 |
66 |
355 |
355 |
|
HI3534 |
Other* |
7 |
332 |
170 |
81 |
63 |
35 |
5 |
620 |
– |
|
MSMB1272WGS |
ENV |
122 |
148 |
164 |
80 |
10 |
45 |
302 |
1088 |
– |
|
MSMB1640WGS |
ENV |
158 |
371 |
98 |
11 |
230 |
96 |
251 |
802 |
– |
|
NKI379 |
ENV |
13 |
786 |
166 |
11 |
239 |
42 |
715 |
1771 |
– |
|
R-20526 |
ENV |
9 |
50 |
169 |
81 |
409 |
96 |
133 |
836 |
– |
∗A novel MLST allele is indicated by ~ ahead of the allele number.
†Shared STs are colour coded with the same shading.
Assessment of swimming and swarming motilities
The motility of B. multivorans was measured using a modified method from Rashid and Kornberg [41]. Agar plates were prepared and dried on an even surface 24 h before use to ensure consistent moisture content, with each plate containing 20 ml. Agar concentrations were made using 0.3 % (w/v) Lysogeny Borth (LB) for swimming assays and 0.5 % (w/v) LB and 0.5 % (w/v) basal salts medium supplemented with 0.4 % (w/v) glucose (BSM-G) for swarming assays. Swimming motility was assessed by inoculating the agar, through to the base, with a sterile toothpick. Swarming motility was assessed by surface inoculation with a sterile toothpick. Plates were inverted and wrapped in sealed Petri dish bags to prevent drying. Plates were incubated at 37 °C and zones were measured at 24 h, averaging two perpendicular measurements. Each isolate was assigned a category: non-motile ≤5 mm, low motility 5–25 mm, intermediate motility 25–50 mm and high motility ≥50.0 mm.
Biofilm formation of B. multivorans
A crystal violet and 96-well PVC plate growth assay [42] was used to determine the biofilm mass formation of B. multivorans isolates. Overnight cultures were diluted to roughly 105 colony-forming units (c.f.u.) ml−1 in TSB in Falcon tubes. These were gently mixed using a vortex before transferring 100 µl into 96-well plates. The outer wells were left empty to prevent drying and B. multivorans biofilms were left to form over 24 h by static incubation of the plates at 37 °C. After removal of growth media and washing as described previously [42], biofilm biomass was stained with a solution of 0.1 % (w/v) crystal violet for 20 min. The plates were washed and allowed to dry and the absorbance at 570 nm was read for a 200 µl solubilization of the biomass stain in each well using 70 % ethanol.
Growth rate of B. multivorans
A Bioscreen C instrument (Labsystems, Finland) was used to determine the bacterial growth dynamics of B. multivorans isolates. Cultures (200 µl in TSB) were inoculated with approximately 106 c.f.u. ml−1 using an optical density-based standardization of fresh overnight liquid growth. Growth was monitored over 48 h with incubation at 37 °C. Well absorbance readings using a wideband filter (450–580 nm) were performed every 15 min after 10 s of medium shaking. A scatterplot analysis was performed in Microsoft Excel to visualize the growth curves. The data were further analysed using the GcFit function of the grofit package [43], which utilizes R statistical software [44] to output specific parameters of lag phase, maximum growth rate and maximum culture density.
Exopolysaccharide and protease production by B. multivorans
Exopolysaccharide (EPS) production of the B. multivorans strains was determined using yeast extract medium (YEM) agar as described by Zlosnik et al. [45]. The original protocol was used for the agar preparation, with no adaptations. B. multivorans was streaked for single colonies from freezer stocks onto the agar plates before incubation for 48 h at 37 °C. EPS was visually categorized into the following five groups based on the literature [45]: − (non-EPS producing), + (partially mucoid), ++ (low mucoidicity), +++ (medium-high mucoidicity) and ++++ (very high mucoidicity). B. multivorans protease production was assessed using a modified protocol from Morris et al. [46]. The lactose-free skimmed-milk agar was prepared as per the original protocol. Overnight cultures were diluted to ~107 c.f.u. ml−1. Aliquots of 10 µl culture were placed onto the protease media in triplicate. Plates were left to completely dry before being inverted and incubated at 37 °C for 24 h. Protease production was measured by taking the average of two perpendicular measurements of resulting colony and the zone of clearing around it (mm). A final protease production value was obtained by subtraction of colony size from the zone of clearing. P. aeruginosa LESB58 was used as a positive control for every protease assay.
Construction of B. multivorans fluorescent reporter strains
Electroporation was used to introduce the plasmid vector pIN301-eGFP [47] into the selected B. multivorans model strains as follows. Overnight cultures of B. multivorans (strains BCC0033, BCC0084, BCC1272 and ATCC 17616) were grown in TSB. These were diluted to an OD600 nm of 0.1 (~107 c.f.u. ml−1) in 3 ml TSB before incubation for approximately 4 h at 37 °C, with shaking at 150 r.p.m. This incubation step enabled the B. multivorans cultures to reach an OD600nm of ~1 and a 2 ml aliquot of culture was spun down in a centrifuge for 5 min at 4000 r.p.m. The pellet was washed twice with 2 ml sterile ddH2O before resuspending 30 µl of ddH2O. Ten nanogrammes of room temperature pIN301-eGFP DNA was added to the suspension, and the suspension was then transferred to a sterile 2 mm electroporation cuvette (Thermo Fisher). After electroporation using 2500 V, with a field capacity of 12.5 kV cm−1, 1 ml sterile TSB was used to recover the electroporated cells for 1 h at 37 °C with shaking at 150 r.p.m. The revived cultures were plated on TSA supplemented with 50 µg ml−1 chloramphenicol and incubated for 24 h at 37 °C before examination under UV light to confirm eGFP::pIN301 plasmid uptake. To confirm that the eGFP::pIN301 derivative was the same as the parental strain, genotyping using random amplification of polymorphic DNA (RAPD) PCR and primers 270 and 272 was performed as described elsewhere [48].
B. multivorans lineage-specific PCR primer design
A pan-genome wide association study (GWAS) approach [49] against the gene presence–absence output file determined via Roary analysis [37] was used to identify genes unique to each lineage. The GWAS traits were based solely on lineage grouping to identify the lineage-specific genes, and did not factor in other genomic or phenotypic variables. Four target genes were identified, and PCR primers designed for each as follows (Table S3). The genes were extracted from the B. multivorans strain panel genomes (Table 1) using Bedtools [50] and aligned using MAFFT [38]. Regions of within-lineage similarity were selected for primer design, and the resulting primer sequences checked for basic specificity using NCBI primer blast, and hairpin structures using the Oligoanalyzer tool (Integrated DNA Technologies). Forward and reverse primers for each gene, together with their genomic location are provided in Table S3; the information for the PCRs selected for testing is provided in Table 3. The PCR primers were synthesized (Eurofins Genomics) and optimized on the four B. multivorans model strains using a gradient PCR. Thermal cycling conditions of an initial denaturation (95 °C, 5 min), 30 cycles of denaturation (95 °C, 30 s), annealing (30 s; see Table 3 for temperature) and extension (72 °C, 30 s), followed by a final extension (72 °C, 10 min) were used. The PCRs were evaluated on the DNA from the 49 phenotypically characterized B. multivorans strain panel isolates (Table 1), with Burkholderia ambifaria and B. cenocepacia DNA used as negative controls. PCR products were separated by electrophoresis on a 1.2 % agarose gel and visualized using a UV transilluminator.
Table 3.
Lineage-specific B. multivorans target genes and PCR primer sequences
|
Lineage and target gene |
Primer name |
Primer sequence (5’ to 3’)* |
Primer length (bp) |
Position (replicon) |
Annealing temp. (oC) |
Product size (bp) |
|---|---|---|---|---|---|---|
|
Lineage 1 ghrB_1 |
GHRBBM1F |
CAAGCAACCGACCGAA AG |
18 |
4008677-4008694† |
53.0 |
744 |
|
GHRBBM1R |
GGAGACAG A ATCACGTTC |
18 |
4009403-4009420† (replicon 2) |
|||
|
Lineage 2 glnM_2 |
GLNMBM2F |
TG AA T G CCG GCCACGTA TG |
19 |
1792198-1792216‡ |
55.5 |
322 |
|
GLNMBM2R |
GACGCATACGACAG T TCC |
18 |
1791895-1791912‡ (replicon 1) |
*Mismatches for each primer sequence are highlighted in bold and underlined.
†Position found in complete genome of B. multivorans BCC0084.
‡Position relative to the complete genome ATCC 17616.
Murine lung infection modelling
A murine chronic lung infection model successfully applied to P. aeruginosa [19, 51] and B. ambifaria [20] was used to evaluate basic infection traits of three model B. multivorans strains. These included wild-type BCC0033 and ATCC 17616, and GFP-tagged derivatives BCC0084 eGFP::pIN301 and BCC0033 eGFP::pIN301. BALB/c female 6–8-week-old mice (Charles River, Margate, UK) were used for all experiments and randomly assigned to a cage of four mice by staff independent of the study. Mice were then housed in individually ventilated cages for 7 days before B. multivorans infection, to allow acclimatization. Overnight cultures of each B. multivorans strain were grown in TSB using a single-colony inoculation, and subcultured in fresh TSB supplemented with 20 % foetal bovine serum (FBS) for ~6 h to allow them to reach mid-exponential phase. Standardized suspensions of each B. multivorans strain were prepared, plated to determine viability and stored at −80 °C.
Murine infections were performed using a protocol from Green et al. [52], whereby the frozen B. multivorans stock suspensions were thawed at room temperature, harvested by centrifugation and resuspended in phosphate-buffered saline (PBS). For each B. multivorans strain, 24 mice were intranasally infected with ~107 c.f.u. ml−1 within a 50 µl suspension. This was performed under light anaesthesia using O2/isoflurane. The nasopharynx and lungs were removed, post-mortem, at 1, 3 and 5 days post-infection, before homogenization in 2 ml sterile PBS using a handheld tissue homogenizer (VWR). Tenfold serial dilutions of tissue homogenates were then prepared and plated onto B. cepacia selective agar (BCSA) (Oxoid, UK). B. multivorans viable cell counts were enumerated after incubation for up to 48 h at 37 °C. For each infection strain, the isolates at days 3 and 5 post-infection were pooled from the eight mouse replicate plates into one stock for the nasopharynx and one for the lungs. Genomic DNA was extracted from the post-infection isolate pools as described above and subjected to short-read Illumina sequencing (Novogene; Cambridge, UK). Genome sequences were then checked for quality and assembled as above. Snippy V3.2-dev was then used for SNP analysis [53].
Statistical analysis
The phenotypic analysis experiments were performed as three biological replicates unless stated otherwise. All statistical analysis was performed in R [44]. The data generated from the analyses within the study were considered to have non-normal distribution. This was checked using a q-q plot and Shapiro test in R. Therefore, the Kruskal–Wallis chi-squared test (two comparisons) or Dunn test with Benjamini–Hochberg correction (three or more comparisons) were used for statistical evaluation as stated.
Results
De novo genomic analysis of B. multivorans as a Bcc species
A total of 73 B. multivorans genomes were short-read sequenced as part of this study (49 shown in Table 1; additional strains in Table S2) and all possessed high-quality draft genome sequences (Table S2). The assembled contigs produced genomes that ranged in size from 6.02 to 7.1 Mb, with an average of 6.514 Mb and mean G+C content of 67.14 %. The number of predicted coding sequences (CDSs) ranged between 5975 and 7374, and between 43 and 67 RNA-encoding loci were identified per genome (Table S2). When the 73 strain genomes were combined with publicly available sequences to form the 283 master genome panel (Table S1), the genome metrics remained consistent, with a mean GC content of 67.04 %, sequence length of 6.5 Mb, N50 value of 338 304 and mean CDS number of 5814 found for B. multivorans.
Burkholderiales taxonomy has been extensively reclassified and continues to expand in terms of novel taxa. For example, recent phylogenomic analysis of seven genus Burkholderiales clades ( Burkholderia , Paraburkholderia , Trinickia , Caballeronia , Mycetohabitans , Robbsia and Pararobbsia ) predicted that 235 genomic species groups existed within a set of 4000+ genomes that encompassed 129 validly named species [36]. To gain insights into the B. multivorans species population biology and confirm the taxonomic classification of strains, ANI analysis was used as the current gold standard in bacterial genomic taxonomy [35]. Analysis was initially performed on the large dataset of 283 B. multivorans genomes (Table S1), with a sub-set of 77 strains representative of the genomic diversity selected for further analysis (Table 1; environmental, n=8; non-CF infection, n=8; CF, n=60; and 1 undetermined source).
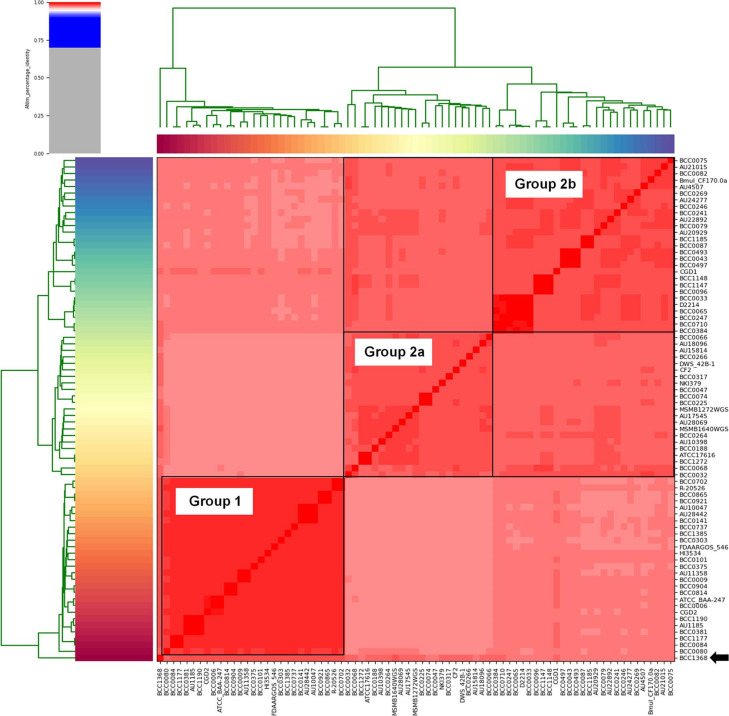
Using the species threshold of 95 % ANI [54] that has also proven to be appropriate for the majority of Burkholderia sensu lato genomic species [36], the B. multivorans isolate genomes (all 283 and the 77-strain panel) comprised a single genomic taxa (Fig. 1). The mean ANI for the 77 B. multivorans examined was 98.59 % and ranged from 97.24–100.00% identity. An ANI heatmap of the 77 strains demonstrated the presence of two prominent groups within the B. multivorans population that had further evolved towards more restricted identity (Fig. 1). These were designated ANI group 1 (n=28; mean ANI of 99 %) and ANI group 2 (n=49; mean ANI of 98 %). Further ANI sub-groupings were apparent within ANI group 2, designated 2a and 2b. The B. multivorans CF strain BCC1368 formed an outlying ANI group and was designated as ‘other’, but was still above the 95 % ANI threshold of the species (Fig. 1).
Fig. 1.
B. multivorans is a single genomic species comprisng two major ANI sub-groups. An ANI heatmap of the 77 sub-selected B. multivorans strains was generated using PyANI. The ANI percentage identity scale is shown (top left) with all red regions >95 % identity. The two major ANI groups, 1 and 2, are indicated with the further 2a and 2b sub-groups labelled. The outlier strain BCC1368 is indicated by the black arrow (bottom right) and was still >95 % ANI in terms of similarity with the other B. multivorans genomes (bottom right).
Pan-genome analysis [37] of the 283 B. multivorans genome dataset identified a total of 37 462 predicted genes. Within this were 30 738 accessory cloud genes and 6724 core genes (genes present in ≥95 % of the population and comprising 2476 shell-genes, 1250 soft-core genes and 2998 predicted core genes).
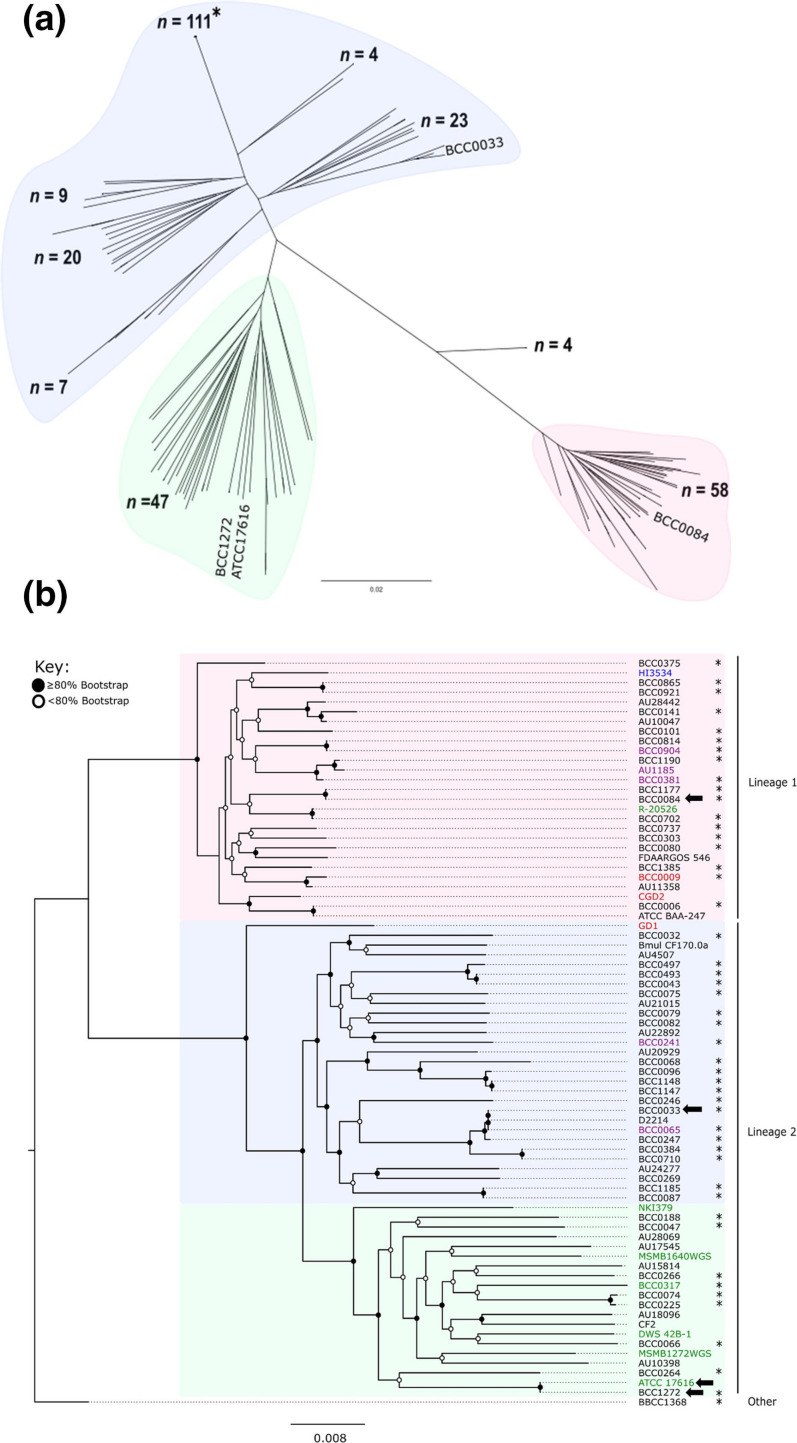
Core gene phylogenomics corroborates that B. multivorans has two major evolutionary lineages
To reconcile an evolutionary basis for the B. multivorans ANI population biology (Fig. 1), core gene phylogenies were analysed (Fig. 2). A master phylogeny was created from the 283 B. multivorans genomes using RAxML v8 [39] and alignment of 4319 core genes present in all samples (Fig. 2). The phylogenomic tree confirmed that the B. multivorans population structure comprised two major evolutionary lineages, with the greatest diversity and further sub-groupings apparent in lineage 2. The isolate source distribution for the 283 genomes was as follows (Table S1): CF, n=248; CGD, n=6; non-CF clinical infection (n=11); ENV, n=23; ENVH, n=1; and isolates of unknown source, n=2. CF strains were distributed throughout the phylogeny, with lineage 2 containing the majority of the CF strains (n=193) compared to lineage 1 (n=45); four CF strains, including the BCC1368 ANI outlier (Fig. 1), clustered within the ‘other’ B. multivorans lineage (Fig. 2a).
Fig. 2.
Core gene phylogenetic analysis of B. multivorans genomes corroborates the presence of two major lineages. (a) A core gene phylogeny of 283 B. multivorans strains was generated by aligning 4319 core genes using RAxML (100 bootstraps). The tree was rooted using BCC1368 (black arrow) and comprised an outgroup of four isolate genomes. Lineage 1 (red), 2a (green) and 2b (blue) groups are shaded. The position of the selected model B. multivorans strains is indicated by the strain names. The single strain group (n=111) represents sequential isolates of CF strain sequenced during chronic B. multivorans infection. (b) The core gene phylogeny of the 77-strain panel is also presented (also an alignment of the 4319 core genes using RAxML with 100 bootsraps). Nodes have been allocated a white circle to illustrate ≤80 % bootstrap or a black circle for ≥80 % bootstrap. The lineages are labelled (right) and isolates indicated with the asterisk were genome sequenced and studied phenotypically as part of this study (see Table 1). Isolate strain names are provided and the text colour denotes their source (black, CF; green, ENV; blue, ENVH, purple, NON CF; red, CGD) and the position of the model strains is indicated by the black arrows. The number of base substitutions per site is indicated by the scale bars on each respective phylogeny.
The selected sub-panel of 77 B. multivorans strains demonstrated the same phylogenomic population biology and two-lineage split (Fig. 2b). The greater diversity within lineage 2 strains was characterized by the longer branch length compared to lineage 1 strains, with the split into 2a and 2b sub-groups clearly observed (Fig. 2b). The total number of environmental isolates of B. multivorans was low in the larger 283 genome dataset (n=23; including n=2 ENVH strains) and a total of 20 environmental isolates clustered within lineage 2 (16 within the 2a subgroup and 4 in 2b; Table S1). The localization of six of these ENV lineage 2 genomes, and one lineage 1 ENV strain is shown in the core gene sub-panel phylogeny (Fig. 2b). Since lineage 2 isolates dominate the master genome collection (221 of 283; Table S1), finding 20 environmental isolate genomes within lineage 2 is not unexpected. Overall, these data corroborate previous findings that B. multivorans is a Bcc species that is rarely isolated from the natural environment [8] and further systematic study is required to identify sources for each lineage.
MLST has been a key epidemiological resource from which to understand Burkholderia infection on a global scale [55], with the Bcc MLST [23] database currently comprising over 4000 B. multivorans isolate profiles. Therefore, the phylogenomic divisions based on 4319 core genes were evaluated against the 7-gene phylogenies from the Bcc MLST strain typing scheme [23]. The MLSTcheck program [40] was implemented to derive an MLST allele profile and ST for the strain panel genomes (Table 2). Within the newly sequenced strains, this revealed 4 novel alleles [BCC0082 (2 alleles), BCC0266 and BCC0737] and 4 novel STs, with a total of 43 unique STs within the 77-strain panel (Table 2). Six different clonal complexes (CCs) were observed within the strain panel, with six strains part of CC1. This CC encompassed ST15 and ST16 B. multivorans strains, which had caused outbreaks of CF infection in several countries [22]. While phylogenetic analysis of the seven concatenated MLST alleles was able to resolve a two-lineage split within B. multivorans , a subset of strains clustered differently and flipped between the 2a and 2b subgroups (Fig. S1) that had been assigned by the core gene analysis (Fig. 2a). This demonstrated that the limited resolution of MLST would not be able to accurately cluster within lineage 2 strains but could assign them to the overall group. It also confirmed that recombination observed within the seven MLST loci [22] is a feature of B. multivorans .
Design and testing of B. multivorans lineage-specific PCRs
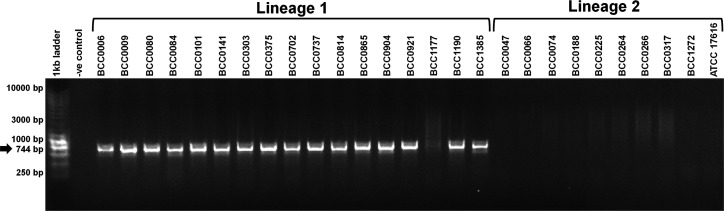
To enable rapid identification and future epidemiological surveillance of the B. multivorans lineages, PCR diagnostics were designed and evaluated as follows. Following a pan-GWAS analysis [49], three genes were identified as 100 % present and specific to lineage 1 strains: yiaJ_1, a predicted DNA-binding transcriptional repressor; ghrB_1, a putative glyoxylate/hydroxypyruvate reductase B; and naiP_3, a predicted niacin/nicotinamide transporter (Table S3). All three genes were encoded on the second chromosomal replicon when compared to the complete genome of the lineage 2 CF strain B. multivorans BCC0084. A single target gene, glnM_2, a putative glutamine ABC transporter permease, was specific to lineage 2 B. multivorans genomes and encoded on replicon 1, when correlated to the complete genome of strain ATCC 1716 (Table S3). After blast analysis of in silico primer specificity and consideration of mismatches in the primer designs (Table S3), the ghrB_1 and glnM_2 PCRs (Table 3) were tested against the panel of 49 phenotypically analysed strains (Table 1). Each PCR demonstrated specificity, with the correct amplicon size produced for strains of the target lineage, and they did not amplify the opposing B. multivorans lineage or control B. ambifaria and B. cenocepacia DNA (Fig. 3; a ghrB_1 PCR example).
Fig. 3.
Specificity of the ghrB_1 for identification of lineage 1 B. multivorans strains. The correct PCR amplicons (744 bp; see arrow on right) resulting from a ghrB_1 PCR on 18 lineage B. multivorans strains are shown (strain names are shown above each lane). No amplicon products were produced from the B. multivorans lineage 2 strains (10 shown on the gel) or the water negative control. Molecular size ladders (1 kb ladder) are shown with the relevant size DNA fragments labelled. Repeat PCR analysis of the degraded B. multivorans BCC1177 (lineage 1) DNA sample was successful. In addition, all testing of negative and positive strains for this PCR was reproducible.
B. multivorans phenotype is variable between strains
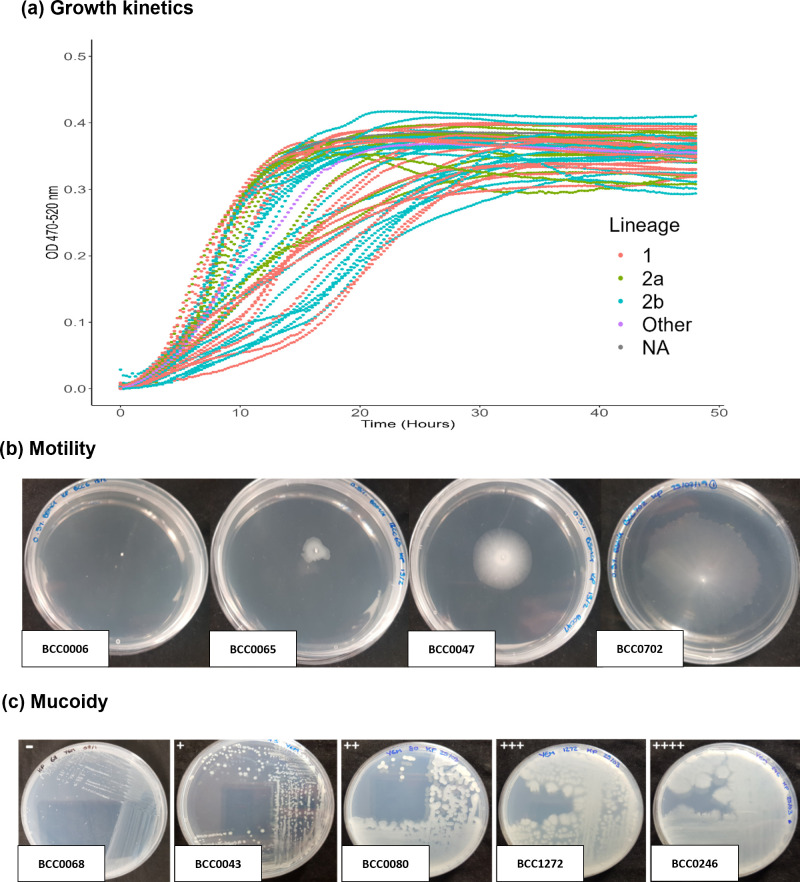
To examine the extent that the genomic lineages correlated to phenotypic differences in vitro, 49 representative strains (Table 1) were examined for growth kinetics, motility, biofilm formation, exopolysaccharide production and protease production. This collection comprised 18 lineage 1 strains, 30 lineage 2 strains (2 a, n=9; 2b, n=21) and the outlier B. multivorans BCC1368. Analysis of growth kinetics demonstrated that all B. multivorans strains produced typical sigmoidal growth curves in TSB but varied in their growth characteristics (Fig. 4a). In terms of maximum growth rate (collection mean=0.032 h−1), 11 strains (BCC0032, BCC0068, BCC0075, BCC0188, BCC0225, BCC0247, BCC0375, BCC00497, BCC0702, BCC0814 and BCC0865; 22 %) fell below the first quartile and were designated as slow growing (Table S4). Outliers for lag phase (collection mean=5.02 h) were BCC0303, BCC0269, BCC1185, BCC0493 and BCC0921 (mean=11.16 h), which possessed prolonged lag phases and small colony phenotypes on TSA (except for BCC0269) (Table S4). No statistically significant differences between B. multivorans lineages were identified for growth rate, maximal growth or lag phase (Fig. S2).
Fig. 4.
Phenotypic characteristic of B. multivorans . (a) The growth curves measured using a Bioscreen C instrument for each of the 49 B. multivorans panel strains (and 1 additional strain). The mean optical density of technical (n=3) and biological replicates (n=3) is plotted for every 15 min reading across 48 h. The key provides shows growth curves for the strains coloured by lineage; the growth rate data are provided in Table S4. (b) The motility of selected B. multivorans strains ranging from low to high motility on 0.5 % swarming BSM-G agar (BCC0006, non-motile; BCC0065, low motility; BCC0047, intermediate motility; BCC0702, high motility). This panel represents the motility categories (diameters) that were also observed on 0.3 % swimming agar. (c) A section of B. multivorans strain reflective of the EPS production scale seen after growth on YEM agar.
Motility on nutrient (TSA) versus minimal medium (BSM-G) was examined for swimming and swarming phenotypes. A consistent finding was that the majority of B. multivorans strains were motile on at least one type of agar (96 %; 47 of 49; Fig. 4b), but BCC0068 (a CF isolate) and BCC0904 (a non-CF infection isolate) were non-motile on all agar types (Table S5). Overall, a greater number of B. multivorans strains had the ability to swim (87 %) rather than swarm (80 %) on at least one medium type (Table S5). No statistically supported phenotypic differences were found between lineages in relation to motility (Fig. S3). The majority of B. multivorans strains (42 of 49; 86 %) were able to form biofilms in vitro within the 96-well PVC plate binding assay. A previous study [56] had shown strain ATCC 17616 to be a high biofilm former and BCC0010 (also known as strain C1962) to be a weak biofilm former. Three strains formed more biofilm than ATCC 17616 (BCC0047, BCC1147 and BCC1272), while seven B. multivorans strains had an average biofilm formation that was lower than that of BCC0010 (BCC0068, BCC0075, BCC0264, BCC0493, BCC0814, BCC0865 and BCC0921). The ability to form biofilms in vitro was not statistically linked to each lineage (Fig. S5).
Using the semi-quantitative YEM agar assay to determined exopolysaccharide production [45], the majority of B. multivorans tested (79 of 84; including all of the 49 panel strains in Table 1) had the ability to produce mucoid phenotypes on YEM agar (Fig. 4b). The non-mucoid phenotype was only observed within five strains (BCC0006, BCC0068, BCC0188, BCC0493 and BCC0497) and, interestingly, four of these strains also exhibited no or low motility on all agars (Table S5). All 49 B. multivorans (Table 1) strains were assessed for protease production using an updated assay [46], but none were found to secrete active proteases in vitro. In contrast, the positive control, P. aeruginosa strain LES B58, produced a clear halo of protease activity on all assays.
Selection of B. multivorans model CF strains
Using the resource of the extensive phylogenomic and phenotypic analyses obtained, three model B. multivorans CF strains were selected. The criteria used accounted for phylogenomic lineage and the possession of a phenotype reflective of the majority of B. multivorans strains. All the model strains possessed the following phenotypes, which were representative of B. multivorans as a species and also enabled further systematic research: (i) they grew well in vitro (Fig. S2); (ii) they were motile (see Fig. S3); (iii) they were capable of biofilm formation (Fig. S5); (iv) they had an absence of in vitro proteolytic activity; (v) they were amenable to transformation with a genetic reporter, pIN301-eGFP; and (v) they behaved reproducibly in all phenotypic testing.
The selected strains were: BCC0033 (also known as C5568) as a lineage 1 CF strain from Canada that was representative of the globally spread ST-16 and clonal complex 1 (Table 2); BCC0084 (also known as C6398), a lineage 2b CF strain from Canada (ST-195; Table 2); and BCC1272 (also known as AU0453), a lineage 2a CF strain from the USA (ST-21; Table 2). In addition to these three CF strains, the B. multivorans reference strain ATCC 17616 (BCC0011), a lineage 2 a soil isolate, was considered to be a fourth model strain because of its well-studied nature. Although ATCC 17616 was isolated from soil, CF isolate BCC1272 had the same MLST type, ST-21. Core gene phylogenomic analysis (Fig. 2b) and complete sequence analysis (Table 1) also showed that the soil and CF isolate were essentially identical at the genomic level. All four model strains were also shown to be genetically amenable to plasmid transformation by successful electroporation and reporter gene expression from plasmids pIN301-eGFP and pIN233-mCherry [47]. Finally, to ensure that the genomic resources for the model CF strains BCC033, BCC0084 and BCC1272 were substantive, they were subjected to complete genome sequencing (see Table 1 for accession numbers).
B. multivorans model strains were capable of murine respiratory infection
To understand whether the selected model B. multivorans were proficient in their ability to colonize the mammalian nasopharynx and lung, and therefore suitable for pathogenicity/therapeutic testing, they were examined in a murine model of respiratory infection [19, 20, 51]. A single experiment with statistical power to evaluate basic bacterial survival kinetics was carried out using the strains BCC0033 and its eGFP::pIN301 derivative (BCC033-GFP), BCC0084 eGFP::pIN301 (BCC084-GFP) and B. multivorans ATCC 17616. All of the initial B. multivorans stocks used for infection and the pooled isolates obtained from 3 and 5 days post-infection (the nasopharynx and lungs) were subjected to Illumina resequencing to confirm their genetic identity and evaluate whether short-term genomic evolution had occurred.
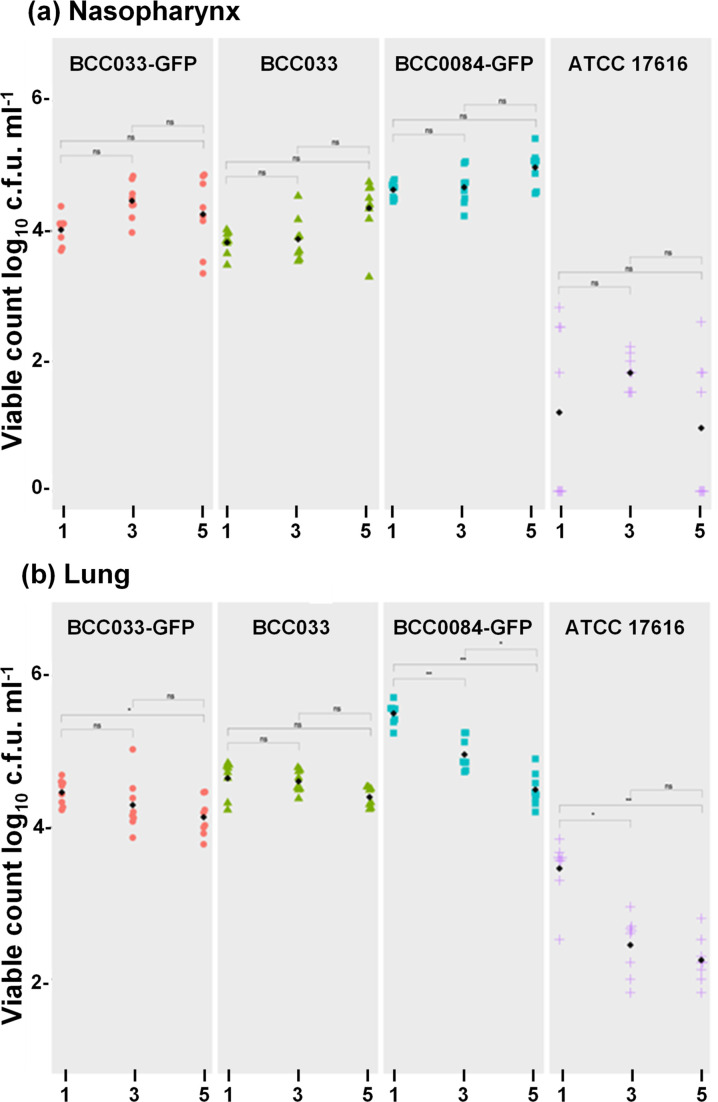
Intranasal infection with approximately 107 c.f.u. ml−1 of each B. multivorans strain resulted in colonization of the respiratory tract ranging from 102 to 105 log10 c.f.u. within both the nasopharynx and lungs, which persisted over the 5 day infection (Fig. 5). In rank order, BCC0084-GFP had the greatest rate of lung colonization (1.8×104 to 1.7×105 c.f.u. ml−1) over 5 days, followed by BCC0033 (1.3×104 to 2.5×104 c.f.u. ml−1), BCC0033-GFP (7.9×103 to 1.5×104 c.f.u. ml−1) and strain ATCC 17616, which possessed the lowest lung infection rate (1.1×102 to 1.8×103 c.f.u. ml−1) (Fig. 5).
Fig. 5.
B. multivorans model strains can persist within a mammalian respiratory infection model. Mouse lung and nasopharynx infection dynamics for the selected B. multivorans model strains are shown. Viable counts (c.f.u.) for the B. multivorans strains at days 1, 3 and 5 post-infection are shown with the within-strain statistical significance indicated for each time point. (a) Infection of the nasopharynx and (b) the lungs, with the individual (coloured) and median (black) c.f.u. for each tissue plotted.
Genome resequencing of the pooled isolates from the nasopharynx and lung demonstrated that infection isolates were essentially isogenic with each respective inoculated strain. Scaffolding of the short-read sequences to the complete genomes demonstrated that no major genomic rearrangements had occurred during the short-term infection. Overall, 242 SNP variants were observed to have accumulated amongst the 4 B. multivorans genomes as follows. In total, 72 (29.75 %) had annotated effects that were as follows: 4 conservative in-frame insertions, 4 disruptive in-frame deletions, 26 missense variants, 4 stop lost and splice region variants and 34 synonymous mutations. B. multivorans ATCC 17616 harboured the greatest number of SNPs (n=110). A total of 27 and 20 SNPs were found in the pooled lung isolates at days 3 and 5, respectively, and 23 and 40 SNPs in the nasopharynx, at the same respective time points. Strain BCC0033 harboured the fewest SNPs, with 27 identified (4 in lung day 3, 7 in lung day 5, 8 in the nasopharynx on days 3 and 5), followed by BCC0033-GFP, with 33 SNPs with a similar distribution (7 SNPs in lung day 3, 14 SNPs in lung day 5 and 6 SNPs in both nasopharynx days 3 and 5). BCC0084 eGFP::pIN301 had a total of 72 SNPs (19 in lung day 3, 17 in lung day 5, 16 in nasopharynx day 3, and 20 in nasopharynx day 5).
Discussion
A limited number of Burkholderia species have been subjected to in-depth population biology, phylogenomic and phenotypic analysis. B. multivorans has previously been investigated to MLST level, demonstrating the presence of globally distributed clonal complexes [22]. Using genomic analyses, we have taken epidemiological understanding a step further, identifying two evolutionary lineages within B. multivorans . Although no difference in the distribution of CF isolates across the two B. multivorans lineages was seen, it is interesting that the majority of globally distributed B. multivorans clonal complexes [22] resided in lineage 2b (Table 2). In comparison to B. cenocepacia [9], there are no currently defined model CF strains for B. multivorans . By combining the genomic findings with the common phenotypic features of B. multivorans , three model CF strains were identified as suitable for future studies alongside the well-characterized soil isolate ATCC 17616. The model strains (BCC0033, BCC0084 and ATCC17616) were all capable of in vivo infection in a murine model of respiratory tract infection, providing a future platform for virulence analysis and therapeutic screening. With straightforward PCR diagnostic probes also designed to rapidly identify each B. multivorans genomic lineage, clinical laboratories now have straightforward tools to evaluate their associated epidemiology.
Several B. cepacia complex species have recently been observed to contain unexpected genomic diversity, resulting in the identification of novel genomic taxa within them. For example, the historical recA gene-based lineage originally identified in B. cenocepacia as III-B [11] was identified as a separate genomic taxa [12] and subsequently proposed as the new species B. orbicola sp. nov. [13]. B. gladioli , the third most common Burkholderia CF pathogen seen in the USA [6], was thought to comprise several pathovars, but genomic analyses demonstrated that five distinct evolutionary clades existed within this single genomic species [21]. Further, bongkrekic acid toxin-producing strains (clades 1a, 1b and 1c) occurring as CF lung infections were identified for the first time within B. gladioli. Finally, across Burkholderia species as a whole, multiple novel genomic taxa have been identified, with only approximately half of these having formal species names [36]. Our phylogenomic analysis of B. mutlivorans shows that this important CF pathogen does not harbour further genomic taxa (Fig. 1), but does comprise two major evolutionary lineages (Fig. 2). As with the two genomic groups observed in the major CF pathogen P. aeruginosa [57, 58], the pathogenic significance of these B. multivorans lineages remains to be determined.
We identified that B. multivorans strains possess highly variable phenotypes, with no direct linkage to their genomic lineage. However, what was consistent was that most strains from CF infection were motile and able to form biofilms in vitro, but lacked the ability to produce proteases on growth media. An absence of B. multivorans protease activity in vitro is in stark contrast to other CF airway pathogens such as B. cenocepacia [59]. A lack of proteolytic activity and an absence of homologues for the virulence-linked B. cenocepacia zmpA metalloprotease was observed in a limited study of eight B. multivorans strains [59]. Our data corroborate and extend this finding to B. multivorans as a species, with no zmpA homologues identified in our taxonomically confirmed (Fig. 1) genomic datasets. The B. multivorans genomes did encode multiple other putative protease genes, including metalloproteases, but further study is required to understand their expression and function. When investigating the B. multivorans growth rate in vitro, two strain groups were apparent, splitting the isolates into approximately two groups, those that reached stationary phase by 24 h versus those reaching this growth stage at 30 h (Fig. 4a). Reduced B. multivorans growth rates have previously been observed in CF infection [16] and is also the case for P. aeruginosa chronic lung infection isolates [60]. All 11 B. multivorans strains identified as slow growers had been recovered from CF infection, suggesting that this is also a pathogenic adaptation that the species makes during chronic infection.
Overall, screening a collection of B. multivorans demonstrated that the majority of strains retained motility as a core phenotype. This contrasts with P. aeruginosa, where isolates from chronic CF lung infection are known to become non-motile [61], but correlates with longitudinal analysis of B. cepacia complex isolates, where only swimming motility was examined [62]. Non-swimming B. multivorans were rare among the collection of isolates screened (14 %) and loss of swimming motility was previously suggested to not be a common adaptive feature of chronically infecting CF strains [62]. Silva et al. [16] examined 22 longitudinal isolates recovered from an individual with CF spanning 20 years and showed decreased swimming motility of this single strain that was likely due to mutations accumulating in the cyclic di-GMP (c-di-GMP) metabolism pathway. Loss of motility has been observed in invasive B. cenocepacia strains that were isolated from the bloodstream of CF individuals suffering from acute ‘cepacia syndrome’ [63]. Of the genetically diverse isolates screened in our study, only B. multivorans strain BCC0068 (a CF isolate) was non-motile on all motility agar types, while BCC0006 showed no swarming motility (Fig. 4b), but retained limited swimming ability (Table S5). For B. cepacia complex species, it has been shown that infection with nonmucoid strains correlates to an increased lung function decline, as compared to infection with mucoidal variants [14]. Only five nonmucoid B. multivorans variants were identified in our study, but all of the nonmucoid strains exhibited no or limited motility, as had been observed in other studies [15, 62, 64].
A useful finding from the B. multivorans strains examined using the murine respiratory infection model was that they demonstrated similar initial levels of lung and nasopharynx colonization to P. aeruginosa strain LESB65 (between log 2 and 4 c.f.u. in each tissue) [19, 51]. This is substantially greater than the low level of colonization (<1000 c.f.u/tissue) observed for the B. cepacia complex species, B. ambifaria , in the same murine infection model [20]. The limited ability of B. ambifaria to colonize the mammalian respiratory tract correlates to the species epidemiology in CF, where it has historically been rarely seen [6] or, more recently, not observed [5] compared to B. multivorans , which was the dominant CF Burkholderia in both epidemiological studies. The B. multivorans CF strain BCC0084-GFP (lineage 1) was the most adept colonizer of both the mouse lung and nasopharynx, with the environmentally derived ATCC 17616 showing the lowest colonization rate (Fig. 5); however, this was still greater, in terms of infectivity, than B. ambifaria [20]. Investigating the genomic differences between B. multivorans and B. ambifaria would be an interesting future study to help understand why B. multivorans is capable of murine (Fig. 5) and CF lung infection rates [5]. Additionally, further systematic studies of B. cepacia complex species in this murine model of infection [19, 20, 51] will need to be carried out to establish their comparative pathogenicity, but promisingly, it is clear the model is a good system for studying B. multivorans .
In summary, although B. multivorans possesses a highly variable phenotype, it is genomically one species harbouring two major lineages. At this stage in our analyses, no differences between B. multivorans lineages have been observed. However, with the identification of representative model strains reflecting each lineage and the conserved species phenotypes, as well as PCR primers to rapidly identify each lineage, in-depth studies of B. multivorans as a CF pathogen can now be undertaken.
Supplementary Data
Funding information
K.M.P. was funded by PhD studentship award from the Medical Research Council GW4 Biomed Doctoral Training Program (project no. BV19101122). E.M. acknowledges additional CF lung infection research support from the US Cystic Fibrosis Foundation (grant MAHENT20G0). A.E.G. and D.R.N. were supported by a Sir Henry Dale fellowship, awarded to D.R.N. and funded by Wellcome and the Royal Society (204457/Z/16/Z). E.M. and D.R.N. note support of their research from a UK Strategic Research Centre (SRC). ‘An evidence-based preclinical framework for the development of antimicrobial therapeutics in cystic fibrosis’ (PIPE-CF; Project No. SRC 022), funded by UK Cystic Fibrosis Trust and US Cystic Fibrosis Foundation. T.R.C. acknowledges support from the MRC for the CLIMB-BIG-DATA resource (grant reference: MR/T030062/1).
Author contributions
The CReDIT contributor roles taxonomy was used to recognize author contributions as follows. Conceptualization: E.M., T.R.C., K.M.P. Methodology: K.M.P., E.M., A.E.G., D.R.N. Software: K.M.P., T.R.C. Validation: K.M.P., E.M., A.E.G., D.R.N. Formal analysis: K.M.P., A.E.G., D.R.N . Investigation: K.M.P., E.M., A.E.G., D.R.N. Resources: E.M., T.R.C., D.R.N. Data curation: K.M.P. Writing – original draft preparation: K.M.P., E.M. Writing – review and editing: all authors. Visualization: K.M.P., E.M. Supervision: E.M., D.R.N. Project administration: E.M., D.R.N. Funding acquisition: E.M., T.R.C., D.R.N., K.M.P.
Conflicts of interest
The authors do not have any conflicts of interest to disclose in relation to this study.
Ethical statement
Murine infection modelling was performed with full ethical permission at the University of Liverpool under project licence PP2072053 (approved by the UK Home Office and the University of Liverpool Animal Welfare and Ethical Review Board).
Footnotes
Abbreviations: ANI, average nucleotide identity; BALB/c, Bagg Albino; BCC, Burkholderia cepacia complex; BCSA, Burkholderia cepacia Selective Agar; BLAST, Basal Local Alignment Search Tool; BSM-G, Basal salts media with glycerol; CC, clonal complex; CF, cystic fibrosis; CGD, chronic granulomas disease; CLIMB, cloud infrastructure for microbial bioinformatics; ENV, environmental; ENVH, environmental (hospital); EPS, exopolysaccharide; LB, Lysogeny Broth; MAFFT, multiple alignment using fast fourier transform; MLST, multilocus sequence typing; NCBI, National Centre of Biotechnology and Information; non-CF, non-cystic fibrosis infection; PacBio, pacific biosciences; pubMLST, public databases for MLST; RAPD, random amplification of polymorphic DNA; RAxML, randomized accelerated maximum likelihood; ST, sequence type; TSA, tryptone soya agar; TSB, tryptone soya broth; WGS, whole genome sequencing; YEM, yeast extract mannitol.
Six supplementary figures and six supplementary tables are available with the online version of this article.
References
- 1.UK-Cystic-Fibrosis-Registry UK Cystic Fibrosis Registry 2020 Annual Data Report. 2021. https://www.cysticfibrosis.org.uk/sites/default/files/2022-05/2020%20Annual%20data%20report%20-%20Version%204.pdf
- 2.Mahenthiralingam E, Urban TA, Goldberg JB. The multifarious, multireplicon Burkholderia cepacia complex. Nat Rev Microbiol. 2005;3:144–156. doi: 10.1038/nrmicro1085. [DOI] [PubMed] [Google Scholar]
- 3.Ledson MJ, Gallagher MJ, Corkill JE, Hart CA, Walshaw MJ. Cross infection between cystic fibrosis patients colonised with Burkholderia cepacia . Thorax. 1998;53:432–436. doi: 10.1136/thx.53.5.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zlosnik JEA, Zhou G, Brant R, Henry DA, Hird TJ, et al. Burkholderia species infections in patients with cystic fibrosis in British Columbia, Canada. 30 years’ experience. Ann Am Thorac Soc. 2015;12:70–78. doi: 10.1513/AnnalsATS.201408-395OC. [DOI] [PubMed] [Google Scholar]
- 5.Kenna DTD, Lilley D, Coward A, Martin K, Perry C, et al. Prevalence of Burkholderia species, including members of Burkholderia cepacia complex, among UK cystic and non-cystic fibrosis patients. J Med Microbiol. 2017;66:490–501. doi: 10.1099/jmm.0.000458. [DOI] [PubMed] [Google Scholar]
- 6.Lipuma JJ. The changing microbial epidemiology in cystic fibrosis. Clin Microbiol Rev. 2010;23:299–323. doi: 10.1128/CMR.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aaron SD, Ferris W, Henry DA, Speert DP, Macdonald NE. Multiple combination bactericidal antibiotic testing for patients with cystic fibrosis infected with Burkholderia cepacia . Am J Respir Crit Care Med. 2000;161:1206–1212. doi: 10.1164/ajrccm.161.4.9907147. [DOI] [PubMed] [Google Scholar]
- 8.Peeters C, Depoorter E, Praet J, Vandamme P. Extensive cultivation of soil and water samples yields various pathogens in patients with cystic fibrosis but not Burkholderia multivorans . J Cyst Fibros. 2016;15:769–775. doi: 10.1016/j.jcf.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 9.Drevinek P, Mahenthiralingam E. Burkholderia cenocepacia in cystic fibrosis: epidemiology and molecular mechanisms of virulence. Clin Microbiol Infect. 2010;16:821–830. doi: 10.1111/j.1469-0691.2010.03237.x. [DOI] [PubMed] [Google Scholar]
- 10.Mahenthiralingam E, Bischof J, Byrne SK, Radomski C, Davies JE, et al. DNA-Based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J Clin Microbiol. 2000;38:3165–3173. doi: 10.1128/JCM.38.9.3165-3173.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vandamme P, Holmes B, Coenye T, Goris J, Mahenthiralingam E, et al. Burkholderia cenocepacia sp. nov.--a new twist to an old story. Res Microbiol. 2003;154:91–96. doi: 10.1016/S0923-2508(03)00026-3. [DOI] [PubMed] [Google Scholar]
- 12.Wallner A, King E, Ngonkeu ELM, Moulin L, Béna G. Genomic analyses of Burkholderia cenocepacia reveal multiple species with differential host-adaptation to plants and humans. BMC Genomics. 2019;20:803. doi: 10.1186/s12864-019-6186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morales-Ruíz L-M, Rodríguez-Cisneros M, Kerber-Díaz J-C, Rojas-Rojas F-U, Ibarra JA, et al. Burkholderia orbicola sp. nov., a novel species within the Burkholderia cepacia complex. Arch Microbiol. 2022;204:178. doi: 10.1007/s00203-022-02778-0. [DOI] [PubMed] [Google Scholar]
- 14.Zlosnik JEA, Costa PS, Brant R, Mori PYB, Hird TJ, et al. Mucoid and nonmucoid Burkholderia cepacia complex bacteria in cystic fibrosis infections. Am J Respir Crit Care Med. 2011;183:67–72. doi: 10.1164/rccm.201002-0203OC. [DOI] [PubMed] [Google Scholar]
- 15.Silva IN, Ferreira AS, Becker JD, Zlosnik JEA, Speert DP, et al. Mucoid morphotype variation of Burkholderia multivorans during chronic cystic fibrosis lung infection is correlated with changes in metabolism, motility, biofilm formation and virulence. Microbiology. 2011;157:3124–3137. doi: 10.1099/mic.0.050989-0. [DOI] [PubMed] [Google Scholar]
- 16.Silva IN, Santos PM, Santos MR, Zlosnik JEA, Speert DP, et al. Long-term evolution of Burkholderia multivorans during a chronic cystic fibrosis Infection reveals shifting forces of selection. mSystems. 2016;1:e00029-16. doi: 10.1128/mSystems.00029-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diaz Caballero J, Clark ST, Wang PW, Donaldson SL, Coburn B, et al. A genome-wide association analysis reveals a potential role for recombination in the evolution of antimicrobial resistance in Burkholderia multivorans . PLoS Pathog. 2018;14:e1007453. doi: 10.1371/journal.ppat.1007453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peeters C, Cooper VS, Hatcher PJ, Verheyde B, Carlier A, et al. Comparative genomics of Burkholderia multivorans, a ubiquitous pathogen with a highly conserved genomic structure. PLoS One. 2017;12:e0176191. doi: 10.1371/journal.pone.0176191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bricio-Moreno L, Sheridan VH, Goodhead I, Armstrong S, Wong JKL, et al. Evolutionary trade-offs associated with loss of PmrB function in host-adapted Pseudomonas aeruginosa . Nat Commun. 2018;9:2635. doi: 10.1038/s41467-018-04996-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mullins AJ, Murray JAH, Bull MJ, Jenner M, Jones C, et al. Genome mining identifies cepacin as a plant-protective metabolite of the biopesticidal bacterium Burkholderia ambifaria . Nat Microbiol. 2019;4:996–1005. doi: 10.1038/s41564-019-0383-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones C, Webster G, Mullins AJ, Jenner M, Bull MJ, et al. Kill and cure: genomic phylogeny and bioactivity of Burkholderia gladioli bacteria capable of pathogenic and beneficial lifestyles. Microb Genom. 2021;7:mgen000515. doi: 10.1099/mgen.0.000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baldwin A, Mahenthiralingam E, Drevinek P, Pope C, Waine DJ, et al. Elucidating global epidemiology of Burkholderia multivorans in cases of cystic fibrosis by multilocus sequence typing. J Clin Microbiol. 2008;46:290–295. doi: 10.1128/JCM.01818-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baldwin A, Mahenthiralingam E, Thickett KM, Honeybourne D, Maiden MCJ, et al. Multilocus sequence typing scheme that provides both species and strain differentiation for the Burkholderia cepacia complex. J Clin Microbiol. 2005;43:4665–4673. doi: 10.1128/JCM.43.9.4665-4673.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Connor TR, Loman NJ, Thompson S, Smith A, Southgate J, et al. CLIMB (the Cloud Infrastructure for Microbial Bioinformatics): an online resource for the medical microbiology community. Microb Genom. 2016;2:e000086. doi: 10.1099/mgen.0.000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galore! KFT. Trim Galore! 2017.
- 26.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2009;17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 27.Andrews S. FastQC: a quality control tool for high throughput sequence data. 2009. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 28.Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 32.Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pritchard L. Pyani: python module for average nucleotide identity analyses. 2014.
- 35.Parks DH, Chuvochina M, Rinke C, Mussig AJ, Chaumeil P-A, et al. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. 2022;50:D785–D794. doi: 10.1093/nar/gkab776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mullins AJ, Mahenthiralingam E. The hidden genomic diversity, specialized metabolite capacity, and revised taxonomy of Burkholderia sensu lato. Front Microbiol. 2021;12:726847. doi: 10.3389/fmicb.2021.726847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Page AJ, Taylor B, Keane JA. Multilocus sequence typing by blast from de novo assemblies against PubMLST. J Open Source Softw. 2016;1:118. doi: 10.21105/joss.00118. [DOI] [Google Scholar]
- 41.Rashid MH, Kornberg A. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa . Proc Natl Acad Sci. 2000;97:4885–4890. doi: 10.1073/pnas.060030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O’Toole GA. Microtiter dish biofilm formation assay. J Vis Exp. 2011;2011:47. doi: 10.3791/2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kahm M, Hasenbrink G, Lichtenberg-Frate H, Ludwig J, Kschischo M. Grofit: Fitting biological growth curves. Nat Prec. 2010 doi: 10.1038/npre.2010.4508.1. [DOI] [Google Scholar]
- 44.R-Core-Team R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. [Google Scholar]
- 45.Zlosnik JEA, Hird TJ, Fraenkel MC, Moreira LM, Henry DA, et al. Differential mucoid exopolysaccharide production by members of the Burkholderia cepacia complex. J Clin Microbiol. 2008;46:1470–1473. doi: 10.1128/JCM.02273-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morris LS, Evans J, Marchesi JR. A robust plate assay for detection of extracellular microbial protease activity in metagenomic screens and pure cultures. J Microbiol Methods. 2012;91:144–146. doi: 10.1016/j.mimet.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 47.Mesureur J, Feliciano JR, Wagner N, Gomes MC, Zhang L, et al. Macrophages, but not neutrophils, are critical for proliferation of Burkholderia cenocepacia and ensuing host-damaging inflammation. PLoS Pathog. 2017;13:e1006795. doi: 10.1371/journal.ppat.1006795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mahenthiralingam E, Campbell ME, Henry DA, Speert DP. Epidemiology of Burkholderia cepacia infection in patients with cystic fibrosis: analysis by randomly amplified polymorphic DNA fingerprinting. J Clin Microbiol. 1996;34:2914–2920. doi: 10.1128/jcm.34.12.2914-2920.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brynildsrud O, Bohlin J, Scheffer L, Eldholm V. Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol. 2016;17:262. doi: 10.1186/s13059-016-1132-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quinlan AR. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr Protoc Bioinformatics. 2014;47:11. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fothergill JL, Neill DR, Loman N, Winstanley C, Kadioglu A. Pseudomonas aeruginosa adaptation in the nasopharyngeal reservoir leads to migration and persistence in the lungs. Nat Commun. 2014;5:4780. doi: 10.1038/ncomms5780. [DOI] [PubMed] [Google Scholar]
- 52.Green AE, Howarth D, Chaguza C, Echlin H, Langendonk RF, et al. Pneumococcal colonization and virulence factors identified via experimental evolution in infection models. Mol Biol Evol. 2021;38:2209–2226. doi: 10.1093/molbev/msab018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seemann T. Snippy: fast bacterial variant calling from NGS reads. 2018. https://github.com/tseemann/snippy
- 54.Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caraher E, Duff C, Mullen T, Mc Keon S, Murphy P, et al. Invasion and biofilm formation of Burkholderia dolosa is comparable with Burkholderia cenocepacia and Burkholderia multivorans . J Cyst Fibros. 2007;6:49–56. doi: 10.1016/j.jcf.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 57.Freschi L, Vincent AT, Jeukens J, Emond-Rheault J-G, Kukavica-Ibrulj I, et al. The Pseudomonas aeruginosa pan-genome provides new insights on its population structure, horizontal gene transfer, and pathogenicity. Genome Biol Evol. 2019;11:109–120. doi: 10.1093/gbe/evy259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weiser R, Green AE, Bull MJ, Cunningham-Oakes E, Jolley KA, et al. Not all Pseudomonas aeruginosa are equal: strains from industrial sources possess uniquely large multireplicon genomes. Microb Genom. 2019;5:e000276. doi: 10.1099/mgen.0.000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gingues S, Kooi C, Visser MB, Subsin B, Sokol PA. Distribution and expression of the ZmpA metalloprotease in the Burkholderia cepacia complex. J Bacteriol. 2005;187:8247–8255. doi: 10.1128/JB.187.24.8247-8255.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rau MH, Hansen SK, Johansen HK, Thomsen LE, Workman CT, et al. Early adaptive developments of Pseudomonas aeruginosa after the transition from life in the environment to persistent colonization in the airways of human cystic fibrosis hosts. Environ Microbiol. 2010;12:1643–1658. doi: 10.1111/j.1462-2920.2010.02211.x. [DOI] [PubMed] [Google Scholar]
- 61.Mahenthiralingam E, Campbell ME, Speert DP. Nonmotility and phagocytic resistance of Pseudomonas aeruginosa isolates from chronically colonized patients with cystic fibrosis. Infect Immun. 1994;62:596–605. doi: 10.1128/iai.62.2.596-605.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zlosnik JEA, Mori PY, To D, Leung J, Hird TJ, et al. Swimming motility in a longitudinal collection of clinical isolates of Burkholderia cepacia complex bacteria from people with cystic fibrosis. PLoS One. 2014;9:e106428. doi: 10.1371/journal.pone.0106428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kalferstova L, Kolar M, Fila L, Vavrova J, Drevinek P. Gene expression profiling of Burkholderia cenocepacia at the time of cepacia syndrome: loss of motility as a marker of poor prognosis? J Clin Microbiol. 2015;53:1515–1522. doi: 10.1128/JCM.03605-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Coutinho CP, Dos Santos SC, Madeira A, Mira NP, Moreira AS. Long-term colonization of the cystic fibrosis lung by Burkholderia cepacia complex bacteria: epidemiology, clonal variation, and genome-wide expression alterations. Front Cell Inf Microbio. 2011;1:12. doi: 10.3389/fcimb.2011.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.