Abstract
The diagnostic value of monitoring human cytomegalovirus (HCMV) late pp67 mRNA expression by nucleic acid sequence-based amplification (NASBA) after renal-allograft transplantation was evaluated. RNAs were isolated from 489 whole-blood specimens of 42 patients for the specific amplification of the late pp67 (UL65) mRNA. NASBA results were compared to results from the pp65 antigenemia assay, virus isolation by cell culture, and serology. The sensitivity value for NASBA proved to be higher than that for the antigenemia assay (50 versus 35%) for the detection of HCMV infection, while the sensitivity values of cell culture and NASBA were comparable (54 and 50%, respectively). NASBA detected the onset of HCMV infection simultaneously with cell culture and the antigenemia assay. Both the antigenemia assay and NASBA are very specific (100%) and highly predictive (100%) for the onset of HCMV infection. Antiviral therapy with ganciclovir resulted in negative results for cell culture, the antigenemia assay, and NASBA. In conclusion, monitoring HCMV pp67 mRNA expression by NASBA is a highly specific method for the detection of HCMV infection in renal-allograft recipients and is more sensitive than the antigenemia assay. Furthermore, NASBA can be used to monitor the progression of HCMV infections and the effect of antiviral therapy on viral activity.
Human cytomegalovirus (HCMV) belongs to the group of betaherpesviruses. Primary infection of immunocompetent individuals with HCMV usually does not lead to complications. In contrast, immunocompromised patients, like AIDS and transplantation patients, may suffer severe complications during primary infection and to a lesser degree during secondary infection. In fact, HCMV infections are the most common single cause of death in allograft recipients (9). Detection of HCMV at an early stage of infection is a prerequisite for effective preemptive antiviral therapy (22). Currently, several routine diagnostic tests are available for the direct or indirect detection of HCMV infection. Direct isolation of HCMV by cultivation of permissive human fibroblasts with samples from blood or urine is a sensitive method, but results are obtained rather late. The cytopathic effects (CPE) of replicating virus in the cells can be detected only after several days or weeks of cultivation. The presence of virus can be detected much earlier in cell cultures by detection of early-antigen fluorescent foci (DEAFF) with monoclonal antibodies directed against immediate-early 1 antigen of the virus (8). Although the cell culture assay was previously considered the “gold standard,” the HCMV antigenemia assay is more commonly used to monitor patients for HCMV disease. The antigenemia assay was developed by Van der Bij et al. (29) to detect the viral lower matrix protein pp65 (UL83) in blood leukocytes. By this immunocytochemical technique, test results can be obtained within 1 day, but it requires direct processing of fresh clinical specimens. The number of pp65-positive cells was shown to correlate with HCMV disease (26, 28). PCR is another method to detect the presence of HCMV. Although amplification of the viral genome by PCR proved to be a highly sensitive technique (up to 100%), it does not necessarily correlate with active infection, due to the possible amplification of latently present viral DNA and/or incomplete viral genomes (17, 24, 27). It was suggested that the detection of viral immediate-early and late mRNAs in blood leukocytes may improve HCMV diagnosis (1), as it directly reflects viral transcriptional activity. Indeed, reverse transcriptase PCR (RT-PCR) of viral late mRNA was found to have a high positive predictive value for HCMV disease, although sensitivity appeared to be low (6, 16, 21). The detection of mRNA by nucleic acid sequence-based amplification (NASBA) is a highly sensitive method. Unlike RT-PCR, NASBA allows detection of unspliced mRNA in a background of DNA (3). A qualitative and quantitative NASBA assay is currently available for the detection of human immunodeficiency virus (HIV) RNA (10, 30). The technique has now been used for the detection of HCMV RNA in blood leukocytes. In contrast to the methods just mentioned, serological diagnosis gives only indirect evidence of the presence of the virus and can be problematic because of the immunological disorders occurring in most patients at risk of developing HCMV infection (13, 23). However, detection of HCMV-specific antibodies can provide supportive evidence for the diagnosis of HCMV infection. Detection of immunoglobulin M (IgM) especially can be used as a diagnostic tool during both primary and secondary infections (7, 12).
Using a qualitative NASBA assay, we evaluated retrospectively the diagnostic value of monitoring late pp67 mRNA expression as a marker for HCMV infections after renal-allograft transplantation. The results of the NASBA assay were compared with the results of prospectively performed standard virological techniques, i.e., detection of the presence of infectious virus in blood by cell culture (CPE and DEAFF), testing for pp65 antigenemia, and serology.
MATERIALS AND METHODS
Patients.
From January 1995 to July 1996, a total of 60 kidney transplantations were carried out at the Department of Internal Medicine, University Hospital Maastricht. For this study, we selected 42 patients, from whom a minimum of four blood specimens were collected during the first three months after transplantation. Patients could be grouped according to the HCMV serostatus of the recipient before transplantation and the HCMV serostatus of the donor, which resulted in the following distribution of patients with different serogroups: 13 D+ and R+ patients, 8 D+ and R− patients, 13 D− and R+ patients, and 8 D− and R− patients, where D+ and D− indicate seropositive and -negative donors, respectively, and R+ and R− indicate seropositive and -negative recipients, respectively.
Maintenance of immunosuppression consisted of treatment with cyclosporin or tacrolimus together with low-dose prednisolone. In addition, some patients received azathioprine. Allograft rejection was assessed clinically and confirmed by needle core biopsy. Treatment for allograft rejection consisted of a 10-day course of rabbit anti-lymphocyte globulin (National Institute of Public Health and the Environment, Bilthoven, The Netherlands) or three doses of methylprednisolone.
HCMV infection was defined by positive results for at least two samples for one or more of the following items: cell culture, antigenemia, seroconversion of anti-HCMV IgM, and/or a significant rise (at least fourfold) in the anti-HCMV IgG level compared to the pretransplantation titer. HCMV infection could be further classified as a primary or secondary infection, depending on whether the patient was HCMV seronegative or seropositive before transplantation, respectively.
Ganciclovir (Cymevene; Hoffmann-La Roche Ltd., Basel, Switzerland) was given intravenously for 2 weeks, in case HCMV infection was detected and the patient showed clinical signs suggesting HCMV disease. The perioperative protocol did not include routinely prophylactic anti-HCMV therapy. If possible, the dosage of the immunosuppressive drug was diminished in case of HCMV infection. However, if a patient was treated with ATG or methylprednisolone and HCMV infection was indicated by positive laboratory tests, ganciclovir was given for 2 weeks intravenously, even when there were no clinical signs of HCMV disease.
Specimens.
Heparinized whole-blood and serum specimens were collected weekly during the clinical period after transplantation. After this period specimens were collected monthly or more frequently if the patient showed clinical symptoms of HCMV infection. Specimens were screened prospectively for the presence of HCMV by routine cell culture and the pp65 antigenemia assay. In addition, 1 ml of heparinized blood was added to 9 ml of NASBA lysis buffer (4.7 M guanidium thiocyanate, 46 mM Tris [pH 6.4], 20 mM EDTA, 1.2% [wt/vol] Triton X-100), and the mixture was stored at −70°C. From the 42 patients, a total of 489 blood specimens was collected.
Cell culture.
For the detection of infectious HCMV in blood, both conventional cell culture and DEAFF were performed as described previously (11). Briefly, leukocytes were separated from heparinized whole blood with dextran and screened for the presence of HCMV by inoculation of human embryo fibroblasts monolayers. Cell cultures were observed weekly during a period of 6 weeks for the appearance of a typical CPE of replicating HCMV. DEAFF was performed after 2 days of cultivation with the monoclonal antibody E13 (Biosoft, Paris, France) to the HCMV immediate-early 1 antigen. For analysis, DEAFF and CPE results were combined to give one outcome for the cell culture assay. Results are presented for either the day on which a positive specimen was obtained or the day on which a positive result could be reported to the clinic.
pp65 antigenemia assay.
The pp65 antigen was detected immunocytochemically essentially as described by Van der Bij et al. (29). Briefly, 45 × 104 leukocytes were isolated from heparinized whole blood with dextran and centrifuged onto a glass slide within 4 h of collection. The cells were subsequently fixed for 10 min in methanol and incubated with mouse monoclonal antibodies, namely, anti-pp65 IgG (produced in our laboratory). After incubation with horseradish peroxidase-coupled rabbit anti-mouse IgG (DAKO A/S, Glostrup, Denmark), the substrate 3-amino-9-ethylcarbazole was added to stain the pp65-positive cells. Finally, slides were examined microscopically. Results are expressed as numbers of pp65-positive cells per 15 × 104 leukocytes.
Serology.
Sera were tested retrospectively to obtain more information about viral activity, in addition to the antigenemia assay and cell culture results. For the qualitative detection of IgM against HCMV in serum, the IMx CMV assay (Abbott Laboratories, North Chicago, Ill.) was used, and for the semiquantitative detection of IgG against HCMV, the AXSym assay (Abbott Laboratories) was used. Both tests are based on microparticle enzyme immunoassay technology. Results from the IMx CMV assay were expressed as index values. Sera with index values of ≥0.500 were considered positive. Levels of anti-HCMV IgG were expressed as numbers of antibody units (AU) per milliliter. Sera with IgG levels of ≥15 AU/ml were considered positive. The detection limit of 250 AU/ml could be obtained for sera with high levels of IgG.
NASBA SC RNA.
A transcription vector was constructed to generate system control (SC) RNA, which served as a positive control during isolation, amplification, and detection of RNA in the NASBA procedure. The vector was constructed essentially as described by Morré et al. (18). Briefly, part of the pp67 region from strain AD169 (nucleotides 95920 to 96247) was cloned into the vector pG3O, downstream of a T7 RNA polymerase promoter region. pG3O is a pGEM3 derivative (Promega, Madison, Wis.) lacking the PstI and SphI sites in the multiple-cloning sequence. In addition, a new BamHI site was introduced after the BamHI site in the multiple-cloning sequence was removed. A PstI site downstream of position 96073 and an SphI site upstream of position 96094 were generated by PCR for the insertion of an additional DNA sequence. This 134-bp fragment, with 5′ PstI and 3′ SphI ends, comprised nucleotides 1015 to 1146 from the 5′ noncoding region of the HIV-1 pv22 sequence (19). This resulted in a plasmid from which the 20-bp wild-type HCMV sequence was deleted and replaced by the 134-bp HIV-1 pv22 fragment. This plasmid was linearized with BamHI, and in vitro RNA was generated by using T7 RNA polymerase (25). The RNA was treated with DNase I to remove plasmid DNA and purified on an anionic-exchange column (Qiagen, Leusden, The Netherlands). The recovered in vitro-generated SC RNA was quantified spectrophotometrically and stored at −70°C.
Nucleic acid isolation for NASBA.
For analysis, 1 ml of the lysed-whole-blood suspension, which equals 100 μl of whole-blood input sample, was used. Twenty microliters of SC RNA containing 3,000 copies of pp67 SC RNA in 1.0 mM Tris buffer, pH 8.5, was added. Nucleic acid isolation was performed essentially as described by Boom et al. (2). Briefly, lysates were incubated with 50 μl of activated silica (0.4 g of suspension in 0.1 N HCl per ml). Subsequently, the silica particles carrying adsorbed nucleic acids were washed twice with 1 ml of wash buffer (5.25 M guanidium thiocyanate, 50 mM Tris [pH 6.4]), twice with 1 ml of 70% ethanol, and once with 1 ml of acetone. Finally, after the silica was dried at 56°C for 10 min, the nucleic acids were eluted in 50 μl of 1.0 mM Tris buffer, pH 8.5, and stored at −20°C.
Qualitative NASBA.
The primers CMVpp67-1.2 and CMVpp67-2.4 were designed to amplify part of the mRNA encoding pp67 (Table 1). NASBA reactions were carried out as described by Kievits et al. (10), with minor modifications. Reactions were performed with a 20-μl reaction mixture containing 40 mM Tris (pH 8.5; Sigma, St. Louis, Mo.); 12 mM MgCl2 (Sigma); 70 mM KCl (Baker, Concord, Ontario, Canada); 15% (vol/vol) dimethyl sulfoxide (Sigma); 5 mM dithiothreitol (Sigma); 1 mM each deoxynucleoside triphosphate (Pharmacia, Uppsala, Sweden); 2 mM ATP, CTP, and UTP; 1.5 mM GTP (Pharmacia); 0.5 mM ITP (Boehringer, Mannheim, Germany); 2 μg of bovine serum albumin (Boehringer); 0.08 U of RNase H (Pharmacia); 32 U of T7 RNA polymerase (Pharmacia); 6.4 U of avian myeloblastosis RT (Seikagaku, Ijamsville, Md.); 0.2 μM each primer; and 5 μl of the isolated nucleic acids. The NASBA reaction mixtures were incubated for 5 min at 65°C, before the enzymes were added, to allow destabilization of secondary RNA structures and subsequently cooled down for 5 min to 41°C to allow primer annealing. After the enzymes were added, reactions mixtures were incubated at 41°C for 90 min and subsequently stored at −20°C.
TABLE 1.
Sequences of NASBA primers and probes used, respectively, for amplification and detection of pp67 WT and SC RNAs
| Oligonucleotide | Function | Sequence |
|---|---|---|
| CMVpp67-1.2 | T7 primer | 5′-AATTCTAATACGACTCACTATAGGGAGAGGGTCGATTCAGACTGA-3′ |
| CMVpp67-2.4 | Primer 2 | 5′-CTGGAGATATATGTTGACCA-3′ |
| CMVpp67CAP1 | Capture probe for WT and SC RNAs | 5′-GGATTCGGACTTTCCGTTCGA-3′ |
| CMVpp67ECL1 | ECL probe for WT RNA | 5′-CCAAAAAGCTAGCCGTCACG-3′ |
| SC-ECL1 | ECL probe for SC RNA | 5′-GAAAAAAGCACAGCAAGCAGCAGG-3′ |
ECL-based detection.
A twofold dilution of the amplification products was made in detection diluent (1.0 mM Tris [pH 8.5], 0.2 g of 2-methylisothiazolone HCl per liter). Subsequently, 5 μl of the diluted amplification products (wild type [WT] and SC) was incubated for 30 min at 41°C with 0.084 μM pp67 mRNA-specific biotine probe (CMVpp67CAP1) (Table 1) bound to 5 μg of streptavidin-coated magnetic beads (mean diameter ± standard deviation, 2.8 ± 0.2 μm; Dynal, Great Neck, N.Y.) and 3 × 1011 molecules of an electrochemiluminescence (ECL) complex (tris[2,2-bipyridine]ruthenium [II])-labeled oligonucleotide probe, specific for either pp67 WT RNA (CMVpp67ECL1) or SC RNA (SC-ECL1), in a total volume of 25 μl of 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). As negative controls, detection diluent was also incubated with the different bead-oligonucleotide and ECL complex-probe mixtures for WT and SC RNA detection. During incubation, tubes were agitated every 10 min to keep the beads in suspension. Subsequently, 300 μl of assay buffer solution (100 mM tripropylamine, pH 7.5) was added and the tubes were placed in an ECL detection instrument (NASBA QR system, model 2000; Organon Teknika B.V., Boxtel, The Netherlands). In order to check the instrument and to be able to compare ECL signals from different runs, a tube with reference solution containing 66.67 μg of streptavidin-coated magnetic beads per ml in assay buffer solution was included. ECL signals for WT and SC RNA were adjusted with the values obtained for the reference solution [(ECL signal for reference solution/40 × 103) × ECL signal for WT or SC RNA] and for background signals (i.e., negative controls). ECL signals higher than 600 (1.5% of 40 × 103) were considered positive.
Statistical analysis.
Statistical analysis was carried out by the Wilcoxon matched-pair signed-rank test. P values of ≤0.05 were considered significant.
RESULTS
Incidence of HCMV infection and disease.
A group of 42 patients was monitored for the occurrence of HCMV infection and HCMV disease after renal transplantation. Seropositive patients receiving a kidney from a seropositive donor showed the highest incidence of HCMV infection (85% [11 of 13 patients]). For patients of the D+ and R− serogroup and those of the D− and R+ serogroup, the incidence of HCMV infection was 75% (6 of 8 patients) and 69% (9 of 13 patients), respectively. HCMV was not detected by any of the assays in blood samples from patients from the D− and R− serogroup. For four patients, HCMV disease was diagnosed according to the criteria of Metselaar (15), namely, fever for at least three consecutive days, leukocytopenia, thrombocytopenia, liver abnormalities, and organ involvement, and was confirmed by concomitant positive cell culture and/or antigenemia assay results. Three of these patients belonged to the D+ and R− serogroup (38% [3 of 8 patients]), while one belonged to the D+ and R+ serogroup (8% [1 of 13 patients]). All patients with HCMV disease underwent antiviral therapy with ganciclovir. Two additional patients received ganciclovir when HCMV infection was detected during treatment for rejection.
Detection of HCMV infection.
HCMV infection could be classified as primary or secondary, depending on whether the patient was seronegative or seropositive before transplantation, respectively. In each patient with a primary infection (n = 6), HCMV was detected by cell culture, the antigenemia assay, and NASBA. In 20 patients, a secondary infection was detected. Forty percent of these infections were detected by cell culture, whereas 35% were detected by NASBA. The antigenemia assay appeared to be the least-sensitive assay (15%).
A total of 489 blood specimens was collected from the 42 patients. In one sample, NASBA repeatedly failed to amplify both WT RNA and the added SC RNA. This result was considered invalid. Antigenemia assay results could be compared to NASBA results for 173 samples. In this study, we found 56% of the positive antigenemia results to be positive also by NASBA (Table 2). Of 141 antigenemia assay-negative samples, 134 were NASBA negative (95%). Cell culture and NASBA results could be compared for 458 of the 489 samples. Of 36 positive cell culture results, 44% were found to be positive by NASBA. There was 96% agreement of negative cell culture results with negative NASBA results.
TABLE 2.
pp67 NASBA results compared to antigenemia assay and cell culture results
| Test | Result | No. of test samplesb | No. (%) of test samples with indicated result by pp67 NASBA
|
|
|---|---|---|---|---|
| Positive | Negative | |||
| Antigenemia assay | Positive | 32 | 18 (56) | 14 (44) |
| Negative | 141 | 7 (5) | 134 (95) | |
| No result | 315 | 11 (4) | 304 (96) | |
| Cell culturea | Positive | 36 | 16 (44) | 20 (56) |
| Negative | 422 | 19 (5) | 403 (95) | |
| No result | 30 | 1 (3) | 29 (97) | |
DEAFF and CPE results were combined.
The total number of test samples was 488 (1 sample was invalid for NASBA).
Table 3 shows sensitivities, specificities, and positive and negative predictive values (PPV and NPV) of NASBA, the antigenemia assay, and cell culture for HCMV infection (see Materials and Methods for the criteria used to diagnose HCMV infection). The sensitivity values of NASBA (50%) and cell culture (54%) are comparable, while the sensitivity value of NASBA proved to be higher than that of the antigenemia assay: 50% versus 35%. Comparable results were obtained for the NPV. Specificity and PPV of cell culture for HCMV infection were 94 and 93%, respectively, whereas for NASBA and the antigenemia assay they were as high as 100%.
TABLE 3.
Diagnostic values for the detection of HCMV infection by pp67 NASBA, cell culture, and the antigenemia assaya
| Test | % Sensitivity | % Specificity | % PPV | % NPV |
|---|---|---|---|---|
| pp67 NASBA | 50 | 100 | 100 | 55 |
| Cell culture | 54 | 94 | 93 | 56 |
| Antigenemia assay | 35 | 100 | 100 | 49 |
See Materials and Methods for the criteria used to diagnose HCMV infection.
Time of detection of HCMV infection.
In this study, 26 of the 42 patients (62%) developed signs of HCMV infection. The onset of HCMV infection was detected by NASBA at a median of 36 days after transplantation (range, 21 to 75 days). For the antigenemia assay and cell culture, detection occurred at 35 days (range, 28 to 69 days) and 65 days (range, 27 to 103 days), respectively. Results of NASBA and the antigenemia assay were found to be positive for HCMV infection simultaneously in four cases, while results of the antigenemia assay indicated positivity earlier than those of NASBA in three cases. However, this difference appeared not to be statistically significant (P = 0.11). Results of NASBA were found to be positive before those of cell culture were found to be positive in seven cases, while results of cell culture were earlier in three cases. This difference also appeared not to be significant (P = 0.11). In five cases, results of the antigenemia assay were found to be positive before positive cell culture results could be reported to the clinic. Although this appears to be significant (P = 0.04), the numbers are low.
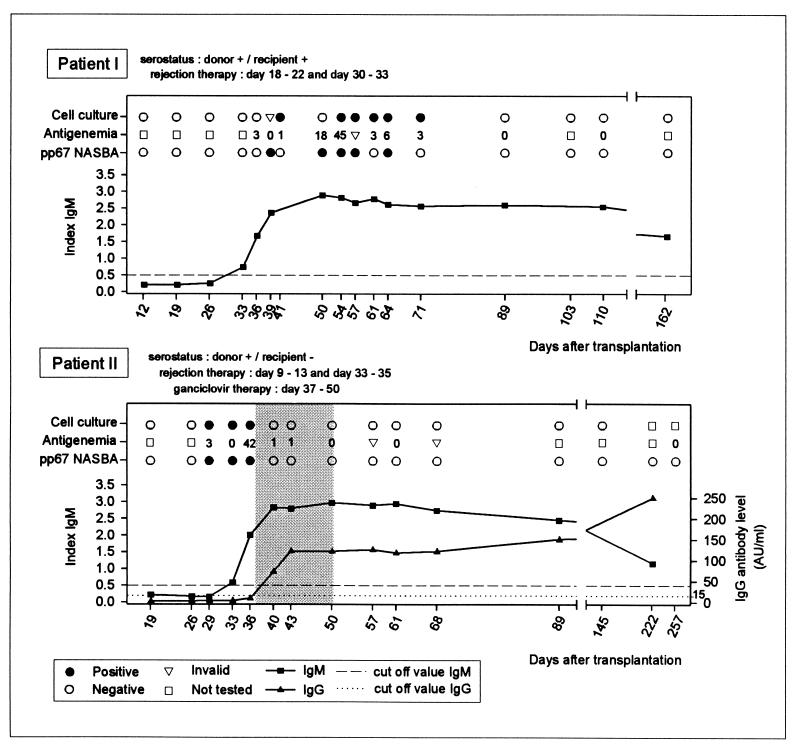
Figure 1 shows representative examples of NASBA results, compared to antigenemia assay, cell culture, and serology results. Patient I (D+ and R+) was treated twice for rejection with methylprednisolone from days 18 to 22 and days 30 to 33. The onset of HCMV infection was first detected at day 36 by the antigenemia assay, followed by NASBA at day 39. The first cell culture-positive result (CPE) could be reported to the clinic at day 76 (specimen was obtained at day 41). Serology confirmed the onset of a secondary infection, with a rise in IgM levels from day 33. No significant rise in IgG levels was detected because assay results were as high as the detection limit of 250 AU/ml during the tested period (results not shown). HCMV disease was not diagnosed, and the patient did not receive antiviral therapy with ganciclovir. By NASBA, pp67 mRNA was detected up to day 64, while positive antigenemia assay and cell culture specimens were obtained up to day 71. Patient II (D+ and R−) was also treated twice for rejection with methylprednisolon from days 9 to 13 and days 33 to 35. The onset of HCMV infection was detected by the antigenemia assay and NASBA at day 29. A cell culture-positive specimen was also obtained at day 29, although this result was not known before day 56. Serology results indicated the onset of a primary infection, with seroconversion of IgM and IgG at days 33 and 40, respectively. HCMV was detected during the period that the patient was treated for rejection. In addition, the patient had fever at day 37, indicative of HCMV infection. Therefore, antiviral therapy with ganciclovir was started at day 37 and maintained until day 50, although HCMV disease was not diagnosed for this patient. During that period, the samples became antigenemia assay, cell culture, and NASBA negative.
FIG. 1.
Detection of HCMV in patients after renal-allograft transplantation by pp67 NASBA, antigenemia, cell culture, and serological assays. HCMV infection was diagnosed for both patient I and patient II after a period of therapy for allograft rejection. Antiviral therapy with ganciclovir was initiated for patient II (indicated by the shaded area), because the patient had fever and HCMV was detected during a period of rejection therapy with methylprednisolone. However, the patient did not meet the criteria for diagnosis of HCMV disease. Antigenemia results are expressed as numbers of positive cells/15 × 104 leukocytes. DEAFF and CPE results were combined to give one outcome for the cell culture assay. Cell culture results are given for the day on which the sample was taken from the patient.
DISCUSSION
In this study, we evaluated the diagnostic value of monitoring late pp67 mRNA expression by NASBA as a marker for HCMV infection. The expression of HCMV late mRNA, such as pp67 (UL65), in circulating blood leukocytes is considered to directly reflect HCMV replication and dissemination in the infected host and should cease upon effective blockage of viral polymerase by antiviral agents, such as ganciclovir (14). The mRNA encoding the viral structural protein pp67 is one of the most abundant late transcripts (5). It was suggested that pp67 may have important functions in the replication cycle of HCMV, such as protein kinase activity, DNA binding, and possible transcriptional activity of immediate-early genes (4). In contrast to what occurs in RT-PCR, the unspliced pp67 mRNA can be specifically amplified by NASBA in a background of DNA (3).
We evaluated the sensitivity of pp67 NASBA by testing blood specimens collected from 42 patients after renal-allograft transplantation. HCMV infection is defined in Materials and Methods. According to this definition, the combined results for cell culture (CPE and DEAFF), the antigenemia assay, and serology are considered the gold standard for the calculation of the sensitivity, specificity, and PPV and NPV of the NASBA assay. For 26 patients (62%) HCMV infection was diagnosed according to these criteria, and 4 of these patients (15%) developed HCMV disease.
Table 2 shows that 44% of antigenemia assay-positive samples were NASBA negative. It should be noted, however, that for all these samples, only weakly positive antigenemia assay results had been obtained (1 to 3 positive cells/15 × 104 leukocytes). Furthermore, for all patients in whom HCMV infection was detected by the antigenemia assay, the results of NASBA were also found to be positive. Importantly, for four patients, HCMV infection was detected by NASBA but not by the antigenemia assay, which resulted in a higher sensitivity value for the NASBA assay than for the antigenemia assay (50 versus 35%). The sensitivity values of cell culture and the NASBA assay for HCMV infection were comparable (54 and 50%, respectively), although 56% of cell culture-positive samples were NASBA negative (Table 2). However, cell cultures were maintained for a minimum of 21 days before being reported as CPE negative, or for 2 days in case of DEAFF, while the NASBA assay can be performed in 1 day. In addition, there appears to be no significant difference between the moment at which positive cell culture results can be reported to the clinic and the moment at which HCMV infection is detected by NASBA. Furthermore, results of NASBA and the antigenemia assay were also found positive simultaneously. It should be noted, however, that the results of the statistical analysis in this study are based on low numbers.
pp67 NASBA was also evaluated as a prognostic marker of HCMV disease in thoracic organ transplant recipients (20). A PPV of 100% was found for HCMV disease. Also, NASBA-positive results occurred in 84% of cases before a clinically significant number of antigenemia assay-positive results occurred (>10 positive cells/2 × 105 leukocytes). In our study, only four patients developed HCMV disease. HCMV infection in these patients was detected by all assays.
Six patients received ganciclovir to treat HCMV infection. Figure 1 shows the data for one of these patients (patient II). Ganciclovir therapy was started at day 37, while the patient was being treated for rejection and had fever suggesting HCMV infection, confirmed by concomitant positive antigenemia assay results (the first cell culture-positive result was not known before day 56). Results of NASBA were found to be positive simultaneously with those of the antigenemia assay, which demonstrates that pp67 NASBA results can be conclusive enough for the clinician to initiate antiviral therapy. Antiviral therapy was sustained for 2 weeks. During that period, leukocytes became pp65 antigen negative. Comparatively, pp67 mRNA expression was no longer detected. Thus, reduction of viral activity can also be monitored by pp67 NASBA, which was expected, because ganciclovir inhibits replication by acting on the viral DNA polymerase (14) while late mRNAs, including pp67 mRNA, are transcribed only after replication of the viral genome. Patient I was not treated with ganciclovir. However, negative cell culture and antigenemia assay results after day 71 indicated reduction of viral activity. Again, NASBA results were comparable to cell culture and antigenemia assay results.
Meyer-König et al. (16) and Bitsch et al. (1) used RT-PCR to detect immediate-early and late pp150 mRNAs in the pheripheral blood leukocytes of renal-allograft recipients. Bitsch et al. found immediate-early mRNA to be present in almost every patient with HCMV infection. Late pp150 mRNA was detected only in leukocytes from a patient with HCMV disease who showed very high numbers of pp65-antigen-positive cells. Meyer-König et al. also found the presence of HCMV mRNA to correlate positively with the number of pp65-antigen-positive cells. However, their RT-PCR assay was not superior to the pp65 antigenemia test for diagnosis and monitoring of HCMV disease.
The NASBA assay offers some practical advantages compared to other tests. For example, 100 μl of whole blood is sufficient to perform one NASBA assay. The blood sample does not need to be processed before uptake in the NASBA lysis buffer. In addition, it is possible to store whole-blood samples in NASBA lysis buffer at −70°C until they are needed for further processing for nucleic acid isolation. For the antigenemia assay, leukocytes have to be isolated from the whole-blood sample within a few hours after it is taken from the patient. The pp67 NASBA assay will be commercially available as a ready-to-use kit, named the NucliSens CMV pp67 assay kit (Organon Teknika B.V.) and will contain all primer and enzyme mixes needed. One person can perform the NASBA assay for 20 blood samples in 1 day, using the kit and the ECL detection instrument. In the near future it will also be possible to use an automated extraction instrument for isolation of nucleic acids from blood samples, with which more blood samples can be handled in 1 day.
From our study it can be concluded that NASBA of late pp67 mRNA is more sensitive than the antigenemia assay for the detection of HCMV infection in renal-allograft recipients. Both the antigenemia assay and pp67 NASBA are very specific and highly predictive of the onset of HCMV infection. Furthermore, pp67 NASBA can be used to monitor progression of HCMV infections and the effect of antiviral therapy, with results comparable to those of the antigenemia assay. Alternatively, detection and monitoring of immediate-early or early antigen mRNA expression by NASBA may provide an additional protocol for early identification of infection. Such studies are in progress.
ACKNOWLEDGMENT
We thank Kees Vink for reading the manuscript critically and for helpful discussions.
REFERENCES
- 1.Bitsch A, Kirchner H, Dupke R, Bein G. Cytomegalovirus transcripts in peripheral blood leukocytes of actively infected transplant patients detected by reverse transcription-polymerase chain reaction. J Infect Dis. 1993;167:740–743. doi: 10.1093/infdis/167.3.740. [DOI] [PubMed] [Google Scholar]
- 2.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Compton J. Nucleic acid sequence-based amplification. Nature. 1991;350:91–92. doi: 10.1038/350091a0. [DOI] [PubMed] [Google Scholar]
- 4.Davis M G, Huang E-S. Nucleotide sequence of a human cytomegalovirus DNA fragment encoding a 67-kilodalton phosphorylated viral protein. J Virol. 1985;56:7–11. doi: 10.1128/jvi.56.1.7-11.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis M G, Mar E-C, Wu Y-M, Huang E-S. Mapping and expression of a human cytomegalovirus major viral protein. J Virol. 1984;52:129–135. doi: 10.1128/jvi.52.1.129-135.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gozlan J, Laporte J P, Lesage S, Labopin M, Najman A, Gorin N C, Petit J C. Monitoring of cytomegalovirus infection and disease in bone marrow recipients by reverse transcription-PCR and comparison with PCR and blood and urine cultures. J Clin Microbiol. 1996;34:2085–2088. doi: 10.1128/jcm.34.9.2085-2088.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffiths P D, Stagno S, Pass R F, Smith R J, Alford C A., Jr Infection with cytomegalovirus during pregnancy: specific IgM antibodies as a marker of recent primary infection. J Infect Dis. 1982;145:647–653. doi: 10.1093/infdis/145.2.647. [DOI] [PubMed] [Google Scholar]
- 8.Griffiths P D, Stirk P R, Ganczakowski M, Panjwani D D, Ball M G, Blacklock H A, Prentice H G. Rapid diagnosis of cytomegalovirus infection in immunocompromised patients by detection of early antigen fluorescent foci. Lancet. 1984;ii:1242–1244. doi: 10.1016/s0140-6736(84)92797-1. [DOI] [PubMed] [Google Scholar]
- 9.Ho M. Advances in understanding cytomegalovirus infection after transplantation. Transplant Proc. 1994;26:7–11. [PubMed] [Google Scholar]
- 10.Kievits T, van Gemen B, van Strijp D, Schukkink R, Dircks M, Adriaanse H, Malek L, Sooknanan R, Lens P. NASBA isothermal enzymatic in vitro nucleic acid amplification optimized for the diagnosis of HIV-1 infection. J Virol Methods. 1991;35:273–286. doi: 10.1016/0166-0934(91)90069-c. [DOI] [PubMed] [Google Scholar]
- 11.Kraat Y J, Christiaans M H L, Nieman F H M, van de Berg P M, van Hooff J P, Bruggeman C A. Risk factors for cytomegalovirus infection and disease in renal transplant recipients: HLA-DR7 and triple therapy. Transplant International. 1994;7:362–367. doi: 10.1007/BF00336713. [DOI] [PubMed] [Google Scholar]
- 12.Kraat Y J, Stals F S, Christiaans M H, Lazzarotto T, Landini M P, Bruggeman C A. IgM antibody detection of ppUL80A and ppUL32 by immunoblotting: an early parameter for recurrent cytomegalovirus infection in renal transplant recipients. J Med Virol. 1996;48:289–294. doi: 10.1002/(SICI)1096-9071(199603)48:3<289::AID-JMV13>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 13.Landini M P, Mirolo G, Coppolecchia P, Re M C, La Placa M. Serum antibodies to individual cytomegalovirus structural polypeptides in renal transplant recipients during viral infection. Microbiol Immunol. 1986;30:683–695. doi: 10.1111/j.1348-0421.1986.tb02994.x. [DOI] [PubMed] [Google Scholar]
- 14.Matthews, T., and R. Boehme. 1988. Antiviral activity and mechanism of action of ganciclovir. Rev. Infect. Dis. 10(Suppl. 3):S490–S494. [DOI] [PubMed]
- 15.Metselaar H J. Diagnosis and prevention of cytomegalovirus infection after organ transplantation. Ph.D. thesis. Rotterdam, The Netherlands: University of Rotterdam; 1990. [Google Scholar]
- 16.Meyer-Köning U, Serr A, von Laer D, Kirste G, Wolff C, Haller O, Neumann Haefelin D, Hufert F T. Human cytomegalovirus immediate early and late transcripts in peripheral blood leukocytes: diagnostic value in renal transplant recipients. J Infect Dis. 1995;171:705–709. doi: 10.1093/infdis/171.3.705. [DOI] [PubMed] [Google Scholar]
- 17.Miller M J, Bovey S, Pado K, Bruckner D A, Wagar E A. Application of PCR to multiple specimen types for diagnosis of cytomegalovirus infection: comparison with cell culture and shell vial assay. J Clin Microbiol. 1994;32:5–10. doi: 10.1128/jcm.32.1.5-10.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morré S A, Sillekens P, Jacobs M V, van Aarle P, de Blok S, van Gemen B, Walboomers J M, Meijer C J, van den Brule A J. RNA amplification by nucleic acid sequence-based amplification with an internal standard enables reliable detection of Chlamydia trachomatis in cervical scrapings and urine samples. J Clin Microbiol. 1996;34:3108–3114. doi: 10.1128/jcm.34.12.3108-3114.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muesing M A, Smith D H, Cabradilla C D, Benton C V, Lasky L A, Capon D J. Nucleic acid structure and expression of the human AIDS/lymphadenopathy retrovirus. Nature (London) 1995;313:450–458. doi: 10.1038/313450a0. [DOI] [PubMed] [Google Scholar]
- 20.Oldenburg N, Lam K M C, Top B, Khan M A, Tacken N, Mikhail G W, Banner N R, Yacoub M. Program and abstracts of the 6th International Cytomegalovirus Workshop. 1997. A novel strategy to monitor CMV immediate early and late gene expression as prognostic markers of CMV disease in thoracic organ transplant recipients using NASBA, abstr. 58; p. A-29. [Google Scholar]
- 21.Patel R, Smith T F, Espy M, Portela D, Wiesner R H, Krom R A, Paya C V. A prospective comparison of molecular diagnostic techniques for the early detection of cytomegalovirus in liver transplant recipients. J Infect Dis. 1995;171:1010–1014. doi: 10.1093/infdis/171.4.1010. [DOI] [PubMed] [Google Scholar]
- 22.Patel R, Snydman D R, Rubin R H, Ho M, Pescovitz M, Martin M, Paya C V. Cytomegalovirus prophylaxis in solid organ transplant recipients. Transplantation. 1996;61:1279–1289. doi: 10.1097/00007890-199605150-00001. [DOI] [PubMed] [Google Scholar]
- 23.Rasmussen L, Kelsall D, Nelson R, Carney W, Hirsch M, Winston D, Preisaitis J, Merican T C. Virus-specific IgG and IgM antibodies in normal and immunocompromised subjects infected with cytomegalovirus. J Infect Dis. 1982;145:191–199. doi: 10.1093/infdis/145.2.191. [DOI] [PubMed] [Google Scholar]
- 24.Roseff S D, Rockis M, Keiser J F, Caparas M M, Comerford J, Sandin R L, Garrett C T. Optimization for detection of cytomegalovirus by the polymerase chain reaction (PCR) in clinical samples. J Virol Methods. 1993;42:137–146. doi: 10.1016/0166-0934(93)90027-o. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.The T H, Van der Bij W, Van den Berg A P, Van der Giessen M, Weits J, Sprenger H G, Van Son W J. Cytomegalovirus antigenemia. Rev Infect Dis. 1990;12:S737–S744. [PubMed] [Google Scholar]
- 27.The T H, van der Ploeg M, van den Berg A P, Vlieger A M, van der Giessen M, van Son W J. Direct detection of cytomegalovirus in peripheral blood leukocytes—a review of the antigenemia assay and polymerase chain reaction. Transplantation. 1992;54:193–198. doi: 10.1097/00007890-199208000-00001. [DOI] [PubMed] [Google Scholar]
- 28.Van den Berg A P, Ven der Bij W, Van Son W J, Anema J, Van der Giessen M, Schirm J, Tegzess A M, The T H. Cytomegalovirus antigenemia as a useful marker of symptomatic cytomegalovirus infection after renal transplantation—a report of 130 consecutive patients. Transplantation. 1989;48:991–995. doi: 10.1097/00007890-198912000-00019. [DOI] [PubMed] [Google Scholar]
- 29.Van der Bij W, Schirm J, Torensma R, van Son W J, Tegzess A M, The T H. Comparison between viremia and antigenemia for detection of cytomegalovirus in blood. J Clin Microbiol. 1988;26:2531–2535. doi: 10.1128/jcm.26.12.2531-2535.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Gemen B, van Beuningen R, Nabbe A, van Strijp D, Jurriaans S, Lens P, Kievits T. A one-tube quantitative HIV-1 RNA NASBA nucleic acid amplification assay using electrochemiluminescent (ECL) labelled probes. J Virol Methods. 1994;49:157–167. doi: 10.1016/0166-0934(94)90040-x. [DOI] [PubMed] [Google Scholar]