Abstract
Mechanistic studies are needed to understand how rotating shift work perturbs metabolic processing. We collected plasma samples (n = 196) from 49 males, rotating car factory shift workers at the beginning and end of a night-shift (22:00–06:00 h) and day-shift (06:00 h-14:00 h). Samples underwent targeted LC-MS/MS metabolomics and concentrations of 130 metabolites were log2-transformed and pareto-scaled. An elastic net selected the most influential metabolites for linear mixed models examining within-person variation in metabolite levels at night-shift end (06:00 h) compared to day-shift start (06:00 h). Quantitative enrichment analysis explored differentially enriched biological pathways between sample time points. We included 20 metabolites (amino acids, biogenic amines, acylcarnitines, glycerophospholipids) in mixed models. Night-shift was associated with changes in concentrations of arginine (geometric mean ratio [GMR] 2.30, 95%CI 1.25, 4.23), glutamine (GMR 2.22, 95%CI 1.53, 3.24), kynurenine (GMR 3.22, 95%CI 1.05, 9.87), lysoPC18:2 (GMR 1.86, 95%CI 1.11, 3.11), lysoPC20:3 (GMR 2.48, 95%CI 1.05, 5.83), PCaa34:2 (GMR 2.27, 95%CI 1.16, 4.44), and PCae38:5 (GMR 1.66, 95%CI 1.02, 2.68). Tryptophan metabolism, glutathione metabolism, alanine metabolism, glycine and serine metabolism, and urea cycle were pathways differing between shifts. Night shift work was associated with changes in metabolites and the perturbation of metabolic and biochemical pathways related to a variety of health outcomes.
Keywords: Circadian, rotating shift work, shift worker, metabolomics, metabolism, occupational health
Introduction
Rotating night shift work is an increasingly common work scheduling arrangement. Night shift work results in circadian misalignment where the circadian timing system is misaligned to the behavioral sleep/wake and feeding/fasting cycles. Circadian rhythms drive a multitude of biological processes in the human body. These rhythms are regulated by a central clock in the suprachiasmatic nuclei (SCN) in the brain and peripheral clocks in virtually all tissues (Mohawk et al. 2012; Potter and Wood 2020). Circadian rhythms are influenced primarily by the 24-h light dark cycle (Papantoniou et al. 2014), but can also be influenced by timing of sleep, meals, and physical activity (Gabriel and Zierath 2019; Gangwisch 2009; Potter and Wood 2020; Wehrens et al. 2017).
Circadian clocks control changes in metabolism across the 24 h day (Davies et al. 2014; Mohawk et al. 2012), and there is an established link between night shift work, disrupted circadian rhythms and cardiometabolic disorders (Hulsegge et al. 2019; McAlpine and Swirski 2016; Scheer et al. 2009). However, the mechanisms underlying associations between night shift work and cardiometabolic disorders remain unclear. Therefore, mechanistic studies are needed to provide insight into how rotating night shift work may perturb metabolic processing.
Metabolic profiling (metabolomics) has the ability to characterize metabolic phenotypes that may be associated with night shift work and disease pathology (Davies et al. 2014; Hancox et al. 2021). Leveraging metabolomic profiling allows for the investigation of genotype and environmental effects, integrating information derived from changes at the gene transcript level, the protein level and the posttranslational modification level together (Raamsdonk et al. 2001). The few studies that have incorporated metabolomics into analyses of night shift work have found that the normal rhythmicity of metabolite concentrations is absent or shifted following night shift work, indicating these changes may be due to the shifted behavioral cycles rather than the phase of the central SCN circadian clock (Kervezee et al. 2019, 2018; Skene et al. 2018). Moreover, these analyses only included data from simulated night shift studies, and data from few participants (n = 14 in Skene et al. 2018, n = 9 in Kervezee et al. 2019 and n = 8 in, 2018).
We undertook the present study to examine how shift work is related to changes in the plasma metabolomic profile within a population of male rotating shift workers who worked both day and night shifts in a slow backward rotation. We hypothesized that night shift work would up or down regulate metabolites relative to day shift work and be associated with biological pathway perturbations.
Methods
Study population
This study included 49 adult male backward rotating (counterclockwise) shift workers from the HORMONIT study who were working at a car factory in Barcelona, Spain (HORMONIT n.d.). Participants rotated through 3 weeks of night shifts (22:00–06:00 h), followed by 3 weeks of evening shifts (14:00–22:00 h) and 3 weeks of early morning shifts (06:00–14:00 h). All participants worked five-day work weeks from Monday-Friday followed by two days off on Saturday and Sunday. Participants were sampled on two days. One day of sample collection occurred during an early morning shift on the 2nd or 3rd week of the early morning shift rotation. The other day of sample collection occurred during a night shift, again on the 2nd or 3rd week of the night shift rotation. A venipuncture blood sample was taken at the beginning and end of each work shift on both sample collection days, resulting in a total of up to four blood samples collected for each study participant.
The study protocol was reviewed and approved by The Parc de Salut Mar Clinical Research Ethics committee (#2015/6351). All participants were given a leaflet with study information and signed an informed consent form. A total of 71 men volunteered for the study, of whom 7 were found to be ineligible and 8 withdrew prior to study start due. Ultimately, 56 participants were enrolled in the study, of whom 49 had complete metabolomic data and are included in the following analyses.
Covariates
At each of the two sample collection points, participants responded to questionnaires collecting data on demographics, work-related aspects, lifestyle factors such as smoking, alcohol use, caffeine use and dietary habits, medical history and medication use and sleep-related information. We also collected information on daylight length (hours) on the days that blood samples were collected (using values available from the National Oceanic and Atmospheric Association calculator and inputting the latitude 41° 230 N and longitude 2° 100 E for Barcelona) (US Department of Commerce et al., 2005). Furthermore, participants wore an actigraphy device (Actigraph 2 GT3X+, USA) capable of measuring triaxial acceleration during the 24 h of the early morning shift collection period and during the 24 h of the night shift collection period to collect information on daily step counts and sleep (Madrid-Navarro et al. 2019). Finally, information on the urinary 6-sulfatoxymelatonin (aMT6s, the major melatonin metabolite) was collected from 24-h urine voids. Using the 24-h urine voids, we applied a cosinor analysis to plot the aMT6s level throughout the 24-h period of data collection and then extracted data on the aMT6s level corresponding to the clock hour of blood collection (Harding et al. 2022).
Outcomes
Participants provided blood samples at both the start (06:00 h) and end (14:00 h) of the early morning shift and the start (22:00 h) and end (06:00 h) of the night shift. Targeted metabolomic analysis was performed on a total of 196 plasma samples using the AbsoluteIDQ® p180 targeted metabolomics kit (Biocrates Life Sciences AG, Innsbruck, Austria), and a Waters Xevo TQ-S triple-quadrupole mass spectrometer coupled to an Acquity UPLC system (Waters Corporation, Milford, MA, USA). Methods for these analyses have been described in detail elsewhere (Honma et al. 2020; Skene et al. 2017). Briefly, plasma samples were prepared according to the manufacturer’s instructions adding several stable isotope–labelled standards to the samples prior to derivatization and extraction. Using reverse-phase UPLC-MS/MS (ultra-performance liquid chromatography/mass spectrometry) or FIA-MS/MS (flow injection analysis-MS/MS), 185 metabolites from 5 different compound classes (namely acylcarnitines, amino acids, biogenic amines, glycerophospholipids, and sphingolipids) could be quantified (Davies et al. 2014). Sample order was randomized and three levels of quality controls (QC) were run on each plate.
The metabolite levels in each QC were compared to the expected values and the percent coefficient of variation (CV%) was calculated. Metabolites where >25% of concentrations were below the limit of detection, out of range, or the QC2 coefficient of variance was >30%, were excluded (n = 50). Values < 1E −10 were replaced by 1E-10 for computational purposes. The remaining 130 quantified metabolites comprised 5 acylcarnitines, 21 amino acids, 8 biogenic amines, 13 lysophosphatidylcholines, 69 glycerophospholipids, and 14 sphingolipids.
Statistical analysis
Prior to undertaking statistical analyses, we applied log transformation (base 2) to all 130 metabolite values due to the log-normality of the data values. Metabolomics data are commonly normalized or scaled, which is done to reduce systematic variation and better ensure true biological variation can be revealed (Fan et al. 2019). We applied a pareto-scaling procedure utilizing Metaboanalyst 5.0 (Pang et al. 2021). Pareto-scaling was done to give each metabolite a similar distribution and reduce the influence of the larger metabolite values, as pareto-scaling will decrease larger fold changes more than smaller fold changes (Eriksson et al. 1999; van den Berg et al. 2006; Worley and Powers 2013). We examined basic differences for all 130 metabolites between the four time points using a one-way repeated measures ANOVA. We also examined differences in metabolite concentrations between the two time points that coincided in clock time (start of the day shift at 06:00 h and end of the night shift at 06:00 h) using paired t-tests. All p-values from the ANOVA and paired t-tests were corrected using a Benjamini Hochberg correction to control for the false discovery rate (FDR) due to multiple comparisons. To allow more potential metabolite differences to be found in these exploratory analyses, an FDR < 0.1 was considered significant. For all comparisons, we summarized the top metabolite differences between the time points using tables and box plots.
To pick the best subset of the 130 metabolites to include in subsequent mixed models investigating differences in metabolite levels between shifts, we fit an elastic net model including the two collection points that coincided in clock time (06:00 h at the beginning of the early morning shift, and at the end of the night shift). We used the R package glmnet to fit the elastic net, using the binomial family option (Friedman et al. 2010). For the elastic net, an elastic net mixing parameter, α, equal to 0.85, and a λ value equal to 0.05 were chosen using leave one out cross validation (LOOCV) and included in the model. Twenty metabolites were selected by the elastic net model, including amino acids (alanine, arginine, aspartate, citrulline, glutamine, phenylalanine), the biogenic amine kynurenine, acylcarnitines (carnitine and tetradecenoylcarnitine), and glycerophospholipids (lysoPCaC18:2, lysoPCaC20:3, lysoPCaC26:0, lysoPCaC28:0, PCaaC32:0, PCaaC34:2, PCaaC40:3, PCaeC30:0, PCaeC36:4, PCaeC38:1, and PCaeC38:5).
Using the 20 metabolites selected from the elastic net, we fit a linear mixed model to examine within-person differences in metabolite levels during the night shift compared to the day shift as the reference. The mixed model was adjusted for hours of daylight on the day of sample collection, aMT6s levels at the time of sample collection, 24-h step count, smoking, and duration of sleep in the 24 hours prior to sample collection. Beta coefficients were exponentiated and results are presented as Geometric mean ratio (GMR).
Quantitative enrichment analysis
To identify biological pathways that may be differentially enriched between time points, we utilized the MetaboAnalyst 5.0 Quantitative Enrichment Analysis module. With this module, we used enrichment analysis to look for groupings of metabolites in the same biological pathway that differed significantly between samples collected at the beginning of a day shift compared to the end of a night shift (both 06:00 h in clock time). To undertake the enrichment analysis, the log2-transformed concentrations for all 130 metabolites in our set were matched to their Human Metabolome Database identifier, uploaded to MetaboAnalyst 5.0, pareto scaling was applied, and their concentrations by shift schedule were compared to metabolite sets found in 99 Small Molecule Pathway Database (SMPDB) human metabolic pathways. Differences in pathways between shifts were quantified with Benjamini Hochberg FDRs, as an indication of how likely the pathway perturbation is related to true biologic variation, and not chance alone. The top five pathways found to be most different between day shift and night shift that included at least three metabolites from our set were further investigated to determine if the pathway may have biological relevance to shift work.
Results
Participants in this study were males with a mean age of 38 (SD ± 9) years. More than half of the population had a BMI ≥ 25 kg/m2 (53%). A large portion of participants reported smoking during their day (63%) or their night (59%) shift. Participants exercised more during their day shift than their night shift (Table 1).
Table 1.
Characteristics of HORMONIT participants (N = 49)
| Variable | Mean (SD) or % |
|---|---|
|
| |
| Age, years | 38 (9) |
| BMI kg/m2 | |
| < 25 | 47% |
| 25–30 | 35% |
| ≥ 30 | 18% |
| Education | |
| Primary | 29% |
| Professional | 71% |
| Smoking | |
| Smoking during day shift | 63% |
| Smoking during night shift | 59% |
| Minutes of daylight | |
| Minutes of daylight day | 719 (103) |
| Minutes of daylight night | 766 (114) |
| 6-Sulphatoxymelatonin level | |
| 6-Sulphatoxymelatonin mesor day shift | 7.7 (3.4) |
| 6-Sulphatoxymelatonin mesor night shift | 7.1 (4.0) |
| Daily step count | |
| Step count day shift | 20014 (3382) |
| Step count night shift | 18998 (3225) |
| Sleep duration | |
| Sleep duration day shift (h) | 6.0 (1.6) |
| Sleep duration night shift (h) | 6.1 (1.9) |
When examining differences in metabolite concentrations between the two time points that coincided in clock time (start of the day shift, 06:00 h and end of the night shift, 06:00 h), we found significant differences (FDR < 0.1) for several glycerophospholipids (lysoPCaC18:2, lysoPCaC20:4, PCaaC32:0, PCaaC34:2, and lysoPCaC20:3) and amino acids (alanine, citrulline, tryptophan, and phenylalanine) (Table 2). When visually examining the difference in concentration levels from the five metabolites with the most pronounced differences at the 06:00 h time points, the majority of metabolites appeared to be lower at the start of the day shift than they were at the end of the night shift (lysoPCaC18:2, lysoPCaC20:4, PCaaC32:0, and citrulline) (Figure 1).
Table 2.
Top 20 metabolites with the greatest differences in metabolite concentrations (μM) between the start of the day shift and end of the night shift (06:00 h).
| Metabolite | Class | P value | FDR | Concentration (μM) at start of day shift (06:00 h), median (IQR) | Concentration (μM) at end of night shift (06:00 h), median (IQR) |
|---|---|---|---|---|---|
|
| |||||
| alanine† | amino acid | 0.00 | 0.00 | 406.0 (346.0, 465.0) | 292.0 (255.0, 360.0) |
| lysoPCaC18:2† | glycerophospholipids | 0.00 | 0.00 | 60.2 (49.5, 76.4) | 82.4 (65.0, 102.0) |
| lysoPCaC20:4† | glycerophospholipids | 0.00 | 0.00 | 9.5 (7.5, 11.7) | 12.0 (10.2, 13.9) |
| PCaaC32:0 | glycerophospholipids | 0.00 | 0.06 | 9.7 (8.7, 10.8) | 10.9 (9.3, 13.0) |
| citrulline† | amino acid | 0.00 | 0.06 | 34.1 (28.7, 40.6) | 41.6 (33.8, 51.0) |
| PCaaC34:2 | glycerophospholipids | 0.00 | 0.06 | 149.0 (136.0, 160.0) | 159.0 (145.0, 168.0) |
| tryptophan† | amino acid | 0.00 | 0.06 | 72.0 (65.1, 86.2) | 85.1 (70.3,93.9) |
| lysoPCaC20:3 | glycerophospholipids | 0.00 | 0.06 | 3.7 (3.3, 4.5) | 4.5 (3.4, 5.9) |
| phenylalanine† | amino acid | 0.01 | 0.08 | 66.7 (57.0, 73.0) | 72.3 (63.4, 85.8) |
| PCaaC36:2 | glycerophospholipids | 0.01 | 0.12 | 127.0 (118.0, 135.0) | 134.0 (124.0, 143.0) |
| ornithine | amino acid | 0.01 | 0.12 | 60.5 (52.3, 69.7) | 66.3 (56.3, 83.5) |
| butyrylcarnitine | acylcarnitines | 0.01 | 0.12 | 0.20 (0.15, 0.23) | 0.22 (0.18, 0.27) |
| lysoPCaC28:1 | glycerophospholipids | 0.01 | 0.12 | 0.26 (0.21, 0.29) | 0.27 (0.23, 0.33) |
| lysoPCaC18:1 | glycerophospholipids | 0.02 | 0.16 | 31.3 (25.1, 38.0) | 34.3 (28.9, 42.5) |
| arginine† | amino acid | 0.02 | 0.16 | 90.7 (83.9, 107.0) | 104.0 (83.4, 124.0) |
| PCaaC34:1 | glycerophospholipids | 0.03 | 0.21 | 121.0 (113.0, 134.0) | 128.0 (119.0, 140.0) |
| SM(OH)C16:1 | sphingolipids | 0.03 | 0.21 | 3.7 (3.1, 4.5) | 4.0 (3.7, 4.6) |
| lysoPCaC26:1 | glycerophospholipids | 0.03 | 0.21 | 0.10 (0.08, 0.13) | 0.11 (0.10, 0.12) |
| SMC16:0 | sphingolipids | 0.03 | 0.21 | 135.0 (114.0, 151.0) | 149.0 (125.0, 165.0) |
| PCaeC42:5 | glycerophospholipids | 0.03 | 0.22 | 2.0 (1.7, 2.3) | 2.1 (1.7, 2.4) |
indicates that these metabolites were also included in the list of 20 top metabolite differences from the one-way repeated measures ANOVA analysis
Figure 1.
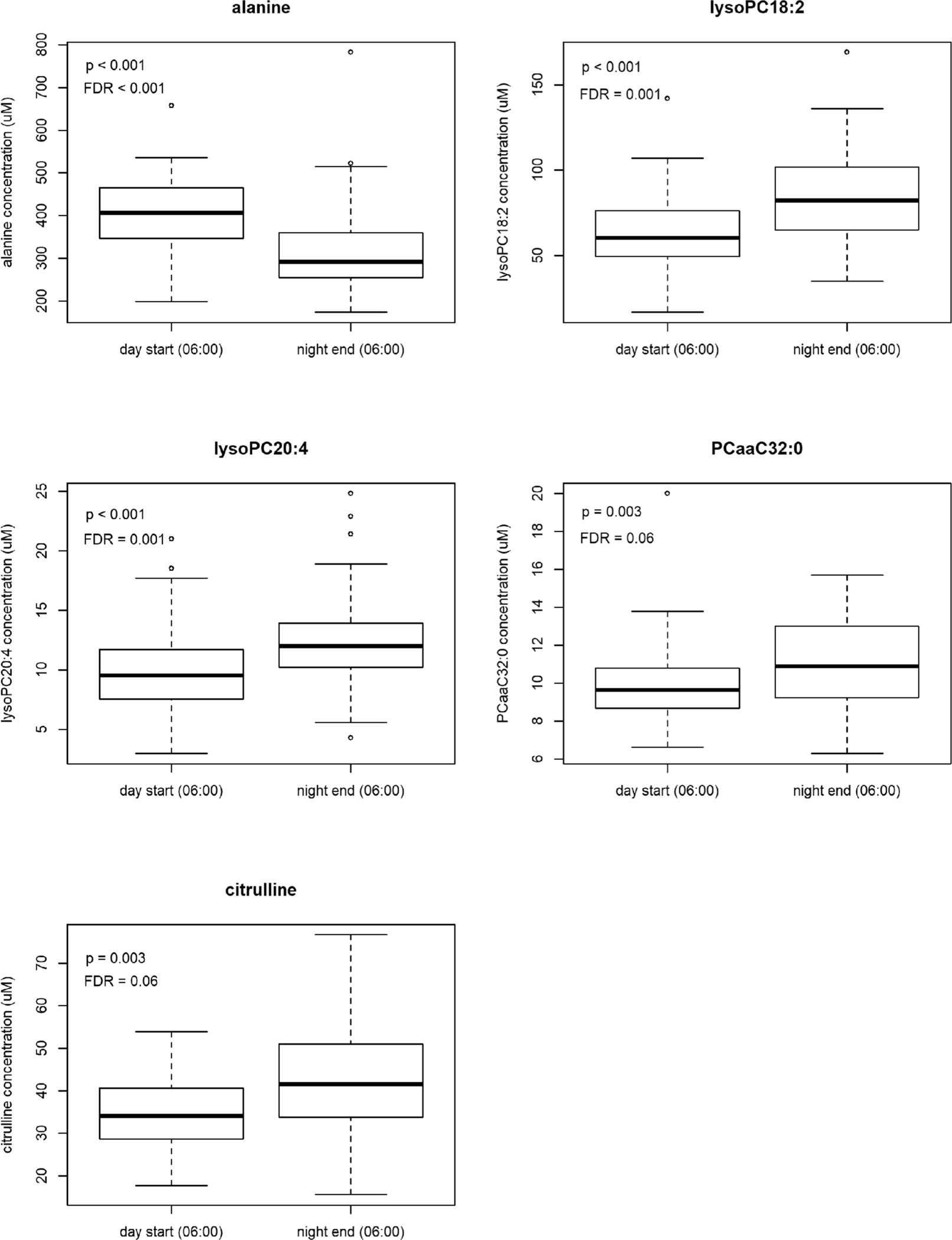
Boxplots showing raw metabolite concentrations (μM) at the start of the day shift and end of the night shift sampling time points that coincide in clock time (06:00 h) for the top 5 metabolite differences using t tests.
Results from the linear mixed models showed several metabolites with intra-individual differences in their concentrations at the end of the night shift (06:00 h) compared to the reference day shift sample (06:00 h) (Table 3). This includes the amino acids arginine (GMR 2.30, 95%CI 1.25, 4.23) and glutamine (GMR 2.22, 95% CI 1.53, 3.24), the biogenic amine kynurenine (GMR 3.22, 95%CI 1.05, 9.87), and the glycerophospholipids lysoPCaC18:2 (GMR 1.86, 95%CI 1.11, 3.11), lysoPCaC20:3 (GMR 2.48, 95%CI 1.05, 5.83), PCaaC34:2 (GMR 2.27, 95%CI 1.16, 4.44), PCaeC36:4 (GMR 2.04, 95%CI 1.00, 4.15), and PCaeC38:5 (GMR 1.66, 95%CI 1.02, 2.68). In all instances, these metabolites were higher at the end of the night shift period compared to the start of the day shift period at the same clock hour.
Table 3.
Mixed model results from the start of the day shift and end of the night shift sampling time points that coincide in clock time (06:00 h).
| Metabolite | Class | Minimally adjusteda |
Fully adjustedb |
||||||
|---|---|---|---|---|---|---|---|---|---|
| GMR | 95%CI | P value | GMR | 95%CI | P value | ||||
|
| |||||||||
| alanine | amino acid | 0.98 | 0.54 | 1.79 | 0.95 | 0.70 | 0.25 | 1.98 | 0.51 |
| arginine | amino acid | 1.71 | 1.15 | 2.54 | 0.01 | 2.30 | 1.25 | 4.23 | 0.01 |
| aspartate | amino acid | 0.99 | 0.93 | 1.06 | 0.79 | 0.97 | 0.88 | 1.07 | 0.56 |
| citrulline | amino acid | 2.08 | 1.30 | 3.33 | 0.00 | 1.94 | 0.92 | 4.08 | 0.08 |
| glutamine | amino acid | 1.64 | 0.85 | 3.18 | 0.14 | 2.22 | 1.53 | 3.24 | 0.00 |
| phenylalanine | amino acid | 1.26 | 0.64 | 2.47 | 0.50 | 1.66 | 0.63 | 4.37 | 0.30 |
| kynurenine | biogenic amine | 1.69 | 0.75 | 3.83 | 0.21 | 3.22 | 1.05 | 9.87 | 0.04 |
| carnitine | acylcarnitines | 1.16 | 0.57 | 2.39 | 0.68 | 1.06 | 0.38 | 2.95 | 0.91 |
| tetradecenoylcarnitine | acylcarnitines | 0.60 | 0.34 | 1.08 | 0.09 | 0.84 | 0.33 | 2.12 | 0.71 |
| lysoPCaC18:2 | glycerophospholipids | 1.70 | 1.11 | 2.59 | 0.02 | 1.86 | 1.11 | 3.11 | 0.02 |
| lysoPCaC20:3 | glycerophospholipids | 1.77 | 1.06 | 2.94 | 0.03 | 2.48 | 1.05 | 5.83 | 0.04 |
| lysoPCaC26:0 | glycerophospholipids | 1.03 | 0.59 | 1.80 | 0.92 | 0.65 | 0.28 | 1.53 | 0.33 |
| lysoPCaC28:0 | glycerophospholipids | 1.90 | 1.15 | 3.14 | 0.01 | 0.74 | 0.30 | 1.80 | 0.51 |
| PCaaC32:0 | glycerophospholipids | 1.45 | 0.88 | 2.40 | 0.15 | 0.96 | 0.43 | 2.13 | 0.91 |
| PCaaC34:2 | glycerophospholipids | 1.54 | 0.54 | 4.44 | 0.42 | 2.27 | 1.16 | 4.44 | 0.02 |
| PCaaC 40:3 | glycerophospholipids | 0.80 | 0.59 | 1.08 | 0.15 | 0.93 | 0.48 | 1.81 | 0.82 |
| PCaeC30:0 | glycerophospholipids | 2.18 | 1.33 | 3.55 | 0.00 | 1.06 | 0.34 | 3.26 | 0.92 |
| PCaeC36:4 | glycerophospholipids | 1.00 | 0.64 | 1.57 | 0.99 | 2.04 | 1.00 | 4.15 | 0.05 |
| PCaeC38:1 | glycerophospholipids | 1.19 | 0.77 | 1.83 | 0.43 | 1.38 | 0.67 | 2.86 | 0.38 |
| PCaeC38:5 | glycerophospholipids | 1.33 | 0.92 | 1.92 | 0.12 | 1.66 | 1.02 | 2.68 | 0.04 |
Adjusted for daylight,
adjusted for daylight, aMT6s level, step count in last 24 hours, smoking in last 24 hours and sleep duration during last 24 hours
We included the top five pathways that contained at least three of the metabolites in our metabolite set. These included: tryptophan metabolism, glutathione metabolism, alanine metabolism, glycine and serine metabolism, and the urea cycle. Table 4 outlines the pathways that were found to differ between day shift and night shift, including information on the biological relevance of the pathway.
Table 4.
Findings from enrichment analysis. Using normalized and scaled data from the start of the day shift and end of the night shift sampling time points that coincide in clock time (06:00 h).
| Pathway name | Total Compound | Hits | Raw P | FDR | Biologic relevance |
|---|---|---|---|---|---|
|
| |||||
| Tryptophan Metabolism | 60 | 5 | 0.00 | 0.03 | TRP regulates immunity, neuronal function, metabolic stress, intestinal homeostasis |
| Glutathione Metabolism | 21 | 3 | 0.00 | 0.03 | GSH provides antioxidant defense, nutrient metabolism, regulates cellular events |
| Alanine Metabolism | 17 | 3 | 0.00 | 0.03 | ALA is used for protein biosynthesis, provides energy under fasting conditions |
| Glycine and Serine Metabolism | 59 | 7 | 0.01 | 0.08 | GLY/SER provide essential precursors for proteins, nucleic acids, lipids |
| Urea Cycle | 29 | 7 | 0.02 | 0.15 | Converts excess ammonia (from amino acid catabolism) into urea in the liver cells |
Discussion
We used a repeated design to compare metabolomic profiles among individuals working a night shift rotation compared to a day shift rotation in a real-world setting, and found that night shift work perturbed metabolism. In particular, we found that the levels of several amino acids, several glycerophospholipids and one biogenic amine were higher during the night shift than the day shift after at least one week of adaptation.
In our mixed model, which included the confounders of daylight, aMT6s levels, daily step count, smoking, and sleep duration, two non-essential amino acids, arginine and glutamine, were found to be significantly different between the day and night shift. The former is important in immune function, has a putative role in preventing or treating circulatory diseases, is produced and used by the body when fatigued, and is important in the formation of urea (PubChem n.d.-a); the latter is important for the regulation of ammonia levels in the body, and is necessary for a range of vital functions, including muscle growth, immune system support, and synthesizing neurotransmitters, nucleotides, and nucleic acid (PubChem n.d.-b). Five glycerophospholipids, a class of glycerol-based phospholipids that are the key component of biological membranes, and important for signal processing and induction (PubChem n.d.-c), also differed significantly between day and night shift. In a previous simulation study by Skene et al. (2018) (3-day simulated day shift sleeping period 22:00 h-06:00 h, 3-night simulated night shift sleeping period 10:00 h-18:00 h), similar differences in the levels of several amino acids and glycerophospholipids after simulated night shift work were observed, providing evidence that the circadian rhythm of many metabolites shifted as a result of night shift work. In the same study, Skene et al. (2018) also found an alteration in kynurenine levels associated with night-shift work, a biogenic amine found to be significantly different in our mixed model.
Several pathways that were identified as being perturbed in our dataset from the quantitative enrichment analysis have also been found to be perturbed in other shift work and sleep studies, adding to the evidence of their potential disruption due to working at night. Bhat et al. (2020) and Davies et al. (2014) reported on tryptophan metabolism being altered in both rats and humans experiencing sleep deprivation, with sleep deprivation also increasing production and accumulation of kynurenic acid, which is the byproduct of tryptophan metabolism and is related to adverse neurological outcomes (Bhat et al. 2020; Davies et al. 2014). In a simulated shift work study by Skene et al., serine, glycine, alanine, and glutathione pathways were found to be enriched in day workers compared to night workers, similar to our finding in rotating shift workers (2018).
A strength of this study was the sampling strategy employed. By design, we sampled the same participants both when they were in their 2nd or 3rd week of day shift and again when they were in their 2nd or 3rd week of night shift work (repeated design). While some individuals are capable of faster and more complete adaptation to night shift work than others, we expected that during the 2nd or 3rd week of the night shift, the majority of participants would exhibit metabolite rhythms that were nearly opposite to the “normal” daytime conditions due to the shifting of their central SCN clock and their peripheral clocks (Pickel and Sung 2020; Skene et al. 2018). Although we are unable to comment on the rhythmicity of metabolites in our study (having only collected samples at two time points on any given day) we did observe that the concentrations of several metabolites were significantly altered in the night shift period compared to the same clock hour in a day shift period, even with presumed biological adaptation to the night shift.
When we examined metabolite levels at each of the four sample time points, we noted that for the majority of metabolites, the concentration decreased over the working day during the day shift and similarly during the night shift. Interestingly, for the majority of metabolites, the metabolite concentrations at the start of the night shift were higher than at the start of the day shift (Supplemental Figure 1). To hypothesize the reason for this finding, both internal and external factors must be considered. Internal biological cues, related to circadian rhythms or diurnal variation could drive differences in the metabolites that are tested at differing clock times (06:00 h versus 22:00 h). However, external factors must also be considered. Prior to starting a 06:00 h early morning shift, participants likely wake up and go straight to work. In contrast, participants may have eaten, engaged in physical activity, or a variety of other social activities in the hours leading up to the night shift start (22:00 h). Prior research has shown that feeding is an external cue that exerts one of the strongest influences on the synchronization of peripheral clocks both in rodents and humans (Damiola et al., 2000; Pickel and Sung 2020; Wehrens et al. 2017) and that feeding and meal timing results in acute changes of some metabolite levels. In addition, time of sleep onset, sleep duration and other lifestyle habits influence metabolite levels (Dollet and Zierath 2019). With our study design, we are unable to separate out how these varying external cues may impact metabolite levels. While measured perturbations to the metabolomic profile may be due to lifestyle habits, timing/quality of meals, or physical activity, these are downstream effects of night shift work. Therefore, our findings provide further evidence that night shift work, either through changing dietary, lifestyle, sleeping, and other habits, and/or through disruption to internal biological rhythms, perturbs the metabolomic profile.
This study has several strengths including its inclusion of a larger cohort of real-world rotating shift workers. Prior metabolomic studies included a smaller sample size and shift work was simulated in controlled laboratory conditions. We also have repeated measures that controls for individual differences, with data on metabolite levels available at four different time points per participant, combined with data on many covariates and potential external cues, allowing us to further account for factors that may influence acute changes in plasma metabolite levels. This study also has limitations including the fact that we had only four time points where we were able to determine metabolite concentrations (two samples per shift). Therefore, we are unable to say if our findings truly reflect a shift in the timing of the metabolite rhythms, or if our results can be explained fully or in part by acute effects of the feeding/fasting state, sleep deprivation or other lifestyle behaviors on the day of sampling (Davies et al. 2014). Despite our careful efforts in taking two samples during each shift, discrete sampling cannot track shifted peripheral clock rhythms. While we asked participants to report eating times during the day and night shifts when urine samples were collected, unfortunately these data were not able to be included due to high degree of missingness and lack of clarity on whether the times reported were AM or PM. Recognizing that feeding/fasting state and timing of meals can be related to metabolite rhythms, collecting more complete meal data will be an important area to improve for future studies.
Additionally, this study only included men. It should also be noted that the targeted metabolomics kit and the reverse-phase UPLC-MS/MS platform used is restricted to analyzing five metabolite classes, being enriched for glycerophospholipids. Considering the established link between night shift work, obesity and cardiometabolic dysfunction, this metabolomics platform was appropriate. We found several lipids were altered, but caution is advised in not over-interpreting the influence of night shift on lipid levels because of the features of the metabolomics kit chosen. Although reverse-phase UPLC-MS/MS is considered the gold standard in metabolomics due its reproducibility and ease of use, polar metabolites, for example, sugars, organic acids, and nucleotides/nucleosides cannot be measured. To measure polar metabolites a hydrophilic interaction liquid chromatography (HILIC) system comprised of UPLC coupled to a triple-quadrupole mass spectrometer operating in both positive and negative electrospray ionization modes would be needed.
In summary, we found that night shift work was associated with changes in the levels of several amino acids, glycerophospholipids and one biogenic amine, and the perturbation of primary metabolic and biochemical pathways. These changes are related to a variety of health outcomes ranging from cardiometabolic and cancer-related, to neurologic and kidney or liver function. Although additional research is needed, particularly among larger diverse populations and with biological samples collected during several time points throughout a shift, these findings provide insight into additional mechanistic pathways that may explain the associations between night shift work and many common non-communicable diseases.
Supplementary Material
Acknowledgements
The authors thank Emilia Molinero, Mayte Martín Bustamante and Elena Juanola Pages from the Generalitat de Catalunya for all of their support in planning this study. The authors also thank Ms. Holly-May Lewis and the Metabolomics Core Facility at the University of Surrey. In addition, the authors thank the study participants for their important contributions.
Funding
The study was partially supported by the Instituto de Salud Carlos III-FEDER [PI14/00444]. ISGlobal acknowledges support from the Spanish Ministry of Science and Innovation through the “Centro de Excelencia Severo Ochoa 2019–2023” Program [CEX2018-000806-S], and support from the Generalitat de Catalunya through the CERCA Program.
Footnotes
Disclosure statement
JMN, PS and AT work at the Occupational Health service of the car factory, which was the setting of the present study. Within the HORMONIT study working group they express their own views and do not represent the company. All other authors have no disclosures.
Data availability statement
The data underlying this analysis is available at: https://github.com/bakermarissa/HORMONIT
References
- Bhat A, Pires AS, Tan V, Babu Chidambaram S, Guillemin GJ. 2020. Effects of sleep deprivation on the tryptophan metabolism. Int J Tryptophan Res. 13:1178646920970902. doi: 10.1177/1178646920970902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. 2000. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 14:2950–61. doi: 10.1101/gad.183500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies SK, Ang JE, Revell VL, Holmes B, Mann A, Robertson FP, Cui N, Middleton B, Ackermann K, Kayser M, et al. 2014. Effect of sleep deprivation on the human metabolome. Proc Natl Acad Sci U S A. 111:10761–66. doi: 10.1073/pnas.1402663111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollet L, Zierath JR. 2019. Interplay between diet, exercise and the molecular circadian clock in orchestrating metabolic adaptations of adipose tissue. J Physiol. 597:1439–50. doi: 10.1113/JP276488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson L, Johansson E, Kettapeh-Wold S, Wold S. 1999. Scaling: introduction to multi-and megavariate data analysis using projection methods (PCA & PLS) Umetrics. Umea, Sweden. [Google Scholar]
- Fan S, Kind T, Cajka T, Hazen SL, Tang WHW, Kaddurah-Daouk R, Irvin MR, Arnett DK, Barupal DK, Fiehn O. 2019. Systematic error removal using random forest for normalizing large-scale untargeted lipidomics data. Anal Chem. 91:3590–96. doi: 10.1021/acs.analchem.8b05592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. 2010. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 33:1–22. doi: 10.18637/jss.v033.i01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel BM, Zierath JR. 2019. Circadian rhythms and exercise—re-setting the clock in metabolic disease. Nature Reviews. Endocrinology. 15:197–206. doi: 10.1038/s41574-018-0150-x [DOI] [PubMed] [Google Scholar]
- Gangwisch JE. 2009. Epidemiological evidence for the links between sleep, circadian rhythms and metabolism. Obesity Rev. 10:37–45. doi: 10.1111/j.1467-789X.2009.00663.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancox TPM, Skene DJ, Dallmann R, Dunn WB. 2021. Tick-tock consider the clock: the influence of circadian and external cycles on time of day variation in the human metabolome—a review. Metabolites. 11:328. doi: 10.3390/metabo11050328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding BN, Castaño-Vinyals G, Palomar-Cros A, Papantoniou K, Espinosa A, Skene DJ, Middleton B, Gomez-Gomez A, Navarrete JM, Such P, et al. 2022. Changes in melatonin and sex steroid hormone production among men as a result of rotating night shift work - the HORMONIT study. Scand J Work Environ Health. 48:41–51. doi: 10.5271/sjweh.3991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma A, Revell VL, Gunn PJ, Davies SK, Middleton B, Raynaud FI, Skene DJ. 2020. Effect of acute total sleep deprivation on plasma melatonin, cortisol and metabolite rhythms in females. Eur J Neurosci. 51:366–78. doi: 10.1111/ejn.14411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- HORMONIT. n.d. ISGlobal. [Cited June 23, 2022]. https://www.isglobal.org/-/hormonit-molecular-epidemiological-study-on-hormonal-changes-circadian-disruption-night-shift-workers
- Hulsegge G, Picavet HSJ, van der Beek AJ, Verschuren WMM, Twisk JW, Proper KI. 2019. Shift work, chronotype and the risk of cardiometabolic risk factors. Eur J Public Health. 29:128–34. doi: 10.1093/eurpub/cky092 [DOI] [PubMed] [Google Scholar]
- Kervezee L, Cermakian N, Boivin DB. 2019. Individual metabolomic signatures of circadian misalignment during simulated night shifts in humans. PLoS Biol. 17:e3000303. doi: 10.1371/journal.pbio.3000303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kervezee L, Cuesta M, Cermakian N, Boivin DB. 2018. Simulated night shift work induces circadian misalignment of the human peripheral blood mononuclear cell transcriptome. Proc Natl Acad Sci U S A. 115:5540–45. doi: 10.1073/pnas.1720719115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid-Navarro CJ, Puertas Cuesta FJ, Escamilla-Sevilla F, Campos M, Ruiz Abellán F, Rol MA, Madrid JA. 2019. Validation of a device for the ambulatory monitoring of sleep patterns: a pilot study on Parkinson’s disease. Front Neurol. 10:356. doi: 10.3389/fneur.2019.00356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlpine CS, Swirski FK. 2016. Circadian influence on metabolism and inflammation in atherosclerosis. Circ Res. 119:131–41. doi: 10.1161/CIRCRESAHA.116.308034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohawk JA, Green CB, Takahashi JS. 2012. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci. 35:445–62. doi: 10.1146/annurev-neuro-060909-153128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Z, Chong J, Zhou G, de Lima Morais DA, Chang L, Barrette M, Gauthier C, Jacques P-É, Li S, Xia J. 2021. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 49:W388–W396. doi: 10.1093/nar/gkab382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papantoniou K, Pozo OJ, Espinosa A, Marcos J. Castano-Vinayls G, Basagana X, Calduch Ribas F, Mirabent J, Martin J, Carenys G, Reyes Martin C, Middleton B, Skene DJ, Kogevinas M. 2014. Circadian variation of melatonin, light exposure, and diurnal preference in day and night shift workers of both sexesmelatonin, light exposure, and night shift. Cancer Epidemiology, Biomarkers & Prevention. 23:1176–1186. doi: 10.1158/1055-9965.EPI-13-1271 [DOI] [PubMed] [Google Scholar]
- Pickel L, Sung H-K. 2020. Feeding rhythms and the circadian regulation of metabolism. Front Nutr. 7:39. doi: 10.3389/fnut.2020.00039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter GDM, Wood TR. 2020. The future of shift work: circadian biology meets personalised medicine and behavioural science. Front Nutr. 7:116. doi: 10.3389/fnut.2020.00116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- PubChem. n.d.-a. Arginine. [Cited June 23, 2022]. https://pubchem.ncbi.nlm.nih.gov/compound/Arginine
- PubChem. n.d.-b. Glutamine. [Cited June 23, 2022]. https://pubchem.ncbi.nlm.nih.gov/compound/glutamine
- PubChem. n.d.-c. Glycerophospholipid biosynthesis. [Cited June 23, 2022]. https://pubchem.ncbi.nlm.nih.gov/pathway/Reactome:R-HSA-1483206
- Raamsdonk LM, Teusink B, Broadhurst D, Zhang N, Hayes A, Walsh MC, Berden JA, Brindle KM, Kell DB, Rowland JJ, et al. 2001. A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nat Biotechnol. 19:45–50. doi: 10.1038/83496 [DOI] [PubMed] [Google Scholar]
- Scheer FAJL, Hilton MF, Mantzoros CS Shea SA. 2009. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci U S A. 106:4453–58. doi: 10.1073/pnas.0808180106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene DJ, Middleton B, Fraser CK, Pennings JLA, Kuchel TR, Rudiger SR, Bawden CS, Morton AJ. 2017. Metabolic profiling of presymptomatic Huntington’s disease sheep reveals novel biomarkers. Sci Rep. 7:43030. doi: 10.1038/srep43030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene DJ, Skornyakov E, Chowdhury NR, Gajula RP, Middleton B, Satterfield BC, Porter KI, Van Dongen HPA, Gaddameedhi S. 2018. Separation of circadian- and behavior-driven metabolite rhythms in humans provides a window on peripheral oscillators and metabolism. Proc Natl Acad Sci U S A. 115:7825–30. doi: 10.1073/pnas.1801183115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- US Department of Commerce, NOAA, & Global Monitoring Laboratory. 2005. ESRL Global Monitoring Laboratory - Global Radiation and Aerosols. [cited May 14, 2022] https://gml.noaa.gov/grad/solcalc/
- van den Berg RA, Hoefsloot HCJ, Westerhuis JA, Smilde AK, van der Werf MJ. 2006. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genomics. 7:142. doi: 10.1186/1471-2164-7-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrens SMT, Christou S, Isherwood C, Middleton B, Gibbs MA, Archer SN, Skene DJ, Johnston JD. 2017. Meal timing regulates the human circadian system. Curr Biol. 27:1768–1775.e3. doi: 10.1016/j.cub.2017.04.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley B, Powers R. 2013. Multivariate analysis in metabolomics. Curr Metabolomics. 1:92–107. doi: 10.2174/2213235X11301010092 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this analysis is available at: https://github.com/bakermarissa/HORMONIT


