Abstract
Long-term colonization of various body sites with a multidrug-resistant Pseudomonas aeruginosa clone (resistant to piperacillin, cefoperazone, ceftazidime, aztreonam, imipenem, cefepime, cefpirome, ofloxacin, ciprofloxacin, minocycline, and aminoglycosides) with subsequent severe infections in burn patients has not been reported previously. Thirty-nine isolates of multidrug-resistant P. aeruginosa (resistant to ceftazidime and at least three of the agents listed above) recovered from various clinical samples from three patients in an intensive care burn unit from April 1997 to May 1997 and seven preserved isolates recovered from six patients in other medical wards at National Taiwan University Hospital from April 1996 to May 1997 were studied for their epidemiological relatedness. The epidemic could be attributed to a multidrug-resistant P. aeruginosa clone belonging to serogroup O:F (serogroup O:4) by means of antimicrobial susceptibility testing, O serogrouping, and analysis of the randomly amplified polymorphic DNA patterns generated by arbitrarily primed PCR of the isolates. The epidemic strain persisted in the three patients for weeks to months; in the meantime, these patients had received multiple antimicrobial agents for the management of intervening episodes of invasive infections (bacteremia, ventilator-associated pneumonia, and/or catheter-related sepsis) caused by this strain, as well as concomitant infections due to other organisms. The strain had been isolated only once previously, from a burn patient who was on the unit in December 1996. The present report, describing a small outbreak due to P. aeruginosa, documents the fact that a single clone of multidrug-resistant P. aeruginosa can cause long-term persistence in different body sites of burn patients and that the colonization can subsequently result in various severe infections.
Despite advances in surgical care and the introduction of a wide variety of antimicrobial agents with antipseudomonal activities, life-threatening infections caused by Pseudomonas aeruginosa continue to be a common complication in burn patients and to contribute substantially to burn-related morbidity and mortality worldwide (6, 12, 13, 21, 22). Multidrug-resistant P. aeruginosa has frequently been reported as the cause of nosocomial outbreaks of infection in burn units or as colonizers of the wounds of burn patients (13, 21, 22). Also, the long-term colonization of more than one P. aeruginosa clone in the respiratory tracts of patients with cystic fibrosis or bronchiectasis has been well demonstrated by various genotypic and phenotypic methods (4, 9, 11, 17, 18). However, the situation in which a multidrug-resistant P. aeruginosa clone colonizes various body sites of burn patients for weeks and months and causes intervening episodes of severe infection has not been described previously.
In our intensive care burn unit, an outbreak of infection due to a P. aeruginosa strain was initially suspected on the basis of the identification of an unusual antibiotype (resistance to all antimicrobial agents routinely tested for activity against Pseudomonas species) among three isolates from two burn patients. In the course of the investigation we used epidemiological surveillance methods coupled with appropriate microbiological studies to determine the relatedness of the isolates with regard to antimicrobial susceptibility, serogroup, and molecular type. In the present report, we describe an outbreak of infections that was caused by one multidrug-resistant P. aeruginosa clone belonging to serogroup O:F as defined by The Serotyping Committee for the Japan Pseudomonas aeruginosa Society (Japanese Committee) (equivalent to U.S. serogroup O:4) and that was controlled by the maintenance of strict isolation of the patients.
MATERIALS AND METHODS
Background.
The intensive care burn unit of National Taiwan University Hospital, a 2,000-bed tertiary-care teaching hospital, is a referral center for northern Taiwan. The unit has eight single-bed intensive care rooms. On average, 120 thermally injured adults and children are treated in the burn unit each year. From 4 April to 10 April 1997, three unusual isolates of P. aeruginosa recovered from various clinical specimens from two patients (patients 2 and 4) (Table 1) were found to be resistant to 12 antimicrobial agents (cefoperazone, ceftazidime, aztreonam, piperacillin, ticarcillin-clavulanic acid, imipenem, minocycline, gentamicin, tobramycin, amikacin, ofloxacin, and ciprofloxacin) by the routine disk diffusion method (16). It was noted upon reviewing the microbiological records that only one P. aeruginosa strain with the same antibiotype had previously been isolated, and that was from a patient who had been hospitalized in the intensive care burn unit in December 1996.
TABLE 1.
Antibiotypes, O serogroups, and RAPD patterns of P. aeruginosa isolates from patients in a burn unit and other wards
| Patient no. | Isolate | Location | Source of isolate | Date of isolation (day/mo/yr) | O sero- groupa | Anti- biotype | RAPD pattern (M13/H5) |
|---|---|---|---|---|---|---|---|
| 1 | A | Burn unit | Wound (chest wall) | 27/12/1996 | F | I | I/I |
| 2 | B1 | Burn unit | Sputum | 8/1/1997 | B | II | II/II |
| B2 | Burn unit | Blood | 4/4/1997 | F | I | I/I | |
| B3 | Burn unit | Sputum | 21/4/1997 | F | I | I/I | |
| B4 | Burn unit | Sputum | 28/4/1997 | F | I | I/I | |
| 3 | C1 | Burn unit | Sputum | 14/4/1997 | F | I | I/I |
| C2 | Burn unit | Wound (right forearm) | 23/4/1997 | F | I | I/I | |
| C3 | Burn unit | Sputum | 28/4/1997 | F | I | I/I | |
| C4 | Burn unit | Sputum | 5/5/1997 | F | I | I/I | |
| C5 | Burn unit | CVCb tip | 5/5/1997 | F | I | I/I | |
| C6 | Burn unit | Blood | 5/5/1997 | F | I | I/I | |
| C7 | Burn unit | Urine | 12/5/1997 | F | I | I/I | |
| C8 | Burn unit | Sputum | 12/5/1997 | F | I | I/I | |
| C9 | Burn unit | Wound (right arm) | 12/5/1997 | F | I | I/I | |
| C10 | Burn unit | Wound (left arm) | 12/5/1997 | F | I | I/I | |
| C11 | Burn unit | Wound (left arm) | 15/5/1997 | F | I | I/I | |
| C12 | Burn unit | CVC tip | 15/5/1997 | F | I | I/I | |
| C13 | Burn unit | Nasal swab | 15/5/1997 | F | I | I/I | |
| C14 | Burn unit | Sputum | 19/5/1997 | F | I | I/I | |
| C15 | Burn unit | Stool | 23/5/1997 | F | I | I/I | |
| 4 | D1 | Burn unit | Wound (right elbow) | 10/4/1997 | F | I | I/I |
| D2 | Burn unit | Wound (left forearm) | 10/4/1997 | E | III | III/III | |
| D3 | Burn unit | Sputum | 14/4/1997 | F | I | I/I | |
| D4 | Burn unit | Wound (right forearm) | 14/4/1997 | F | I | I/I | |
| D5 | Burn unit | Wound (right arm) | 14/4/1997 | F | I | I/I | |
| D6 | Burn unit | Wound (left arm) | 14/4/1997 | F | I | I/I | |
| D7 | Burn unit | Wound (left arm) | 14/4/1997 | F | I | I/I | |
| D8 | Burn unit | Sputum | 21/4/1997 | F | I | I/I | |
| D9 | Burn unit | Wound (left leg) | 23/4/1997 | F | I | I/I | |
| D10 | Burn unit | Stool | 28/4/1997 | F | I | I/I | |
| D11 | Burn unit | Wound (left hand) | 29/4/1997 | F | I | I/I | |
| D12 | Burn unit | Sputum | 5/5/1997 | F | I | I/I | |
| D13 | Burn unit | Wound (neck) | 5/5/1997 | F | IV | IV/IV | |
| D14 | Burn unit | CVC tip | 6/5/1997 | F | I | I/I | |
| D15 | Burn unit | Sputum | 12/5/1997 | F | I | I/I | |
| D16 | Burn unit | Wound (neck) | 14/5/1997 | E | V | V/V | |
| D17 | Burn unit | Wound (right hand) | 14/5/1997 | F | I | I/I | |
| D18 | Burn unit | Nasal swab | 15/5/1997 | F | I | I/I | |
| D19 | Burn unit | Sputum | 19/5/1997 | N | VI | VI/VI | |
| D20 | Burn unit | Stool | 23/5/1997 | F | I | I/I | |
| 5 | E | Medical ward | Urine | 20/4/1996 | C | VII | VII/VII |
| 6 | F | Medical ward | Urine | 10/8/1996 | D | VIII | VIII/VIII |
| 7 | G1 | Medical ICUc | Sputum | 12/8/1996 | A | IX | IX/IX |
| G2 | Medical ICU | Urine | 12/8/1996 | A | IX | IX/IX | |
| 8 | H | Medical ward | Urine | 3/9/1996 | H | X | X/X |
| 9 | I | Medical ward | Drainage fluid | 16/9/1996 | I | XI | XI/XI |
| 10 | J | Medical ward | Sputum | 5/2/1997 | E | XII | XII/XII |
O serogroup as designated by Homma (7) (those designated by Lanyi and Bergan are given in parentheses): O:B (O:2), O:F (O:4), O:E (O:11), O:N (O:14), O:C (O:6), O:D (O:90), O:A (O:3), O:H (O:10), and O:I (O:1).
CVC, central venous catheter.
ICU, intensive care unit.
Epidemiological surveillance.
After the infections with the multidrug-resistant P. aeruginosa strains were discovered, bacterial cultures for multidrug-resistant P. aeruginosa isolates (resistance to ceftazidime and at least three of the other agents listed above) were performed with various samples from all patients in the burn unit. These samples included burn wound, nasal swab, sputum, urine, and stool samples. Environmental samples for surveillance cultures were obtained from 10 April to 16 April 1997 to detect the presence of multidrug-resistant P. aeruginosa from various sources; for this purpose, 100-ml samples of water from sink faucets and swabs of sink surfaces in each patient’s room were obtained. Swab specimens from the hands and nares of the physicians and nurses in the unit were also collected for culture due to the concomitant occurrence of an outbreak due to oxacillin-resistant Staphylococcus aureus. All swab samples were inoculated directly onto blood agar plates (BBL Microbiology Systems, Cockeysville, Md.). Water specimens were processed as described previously (10). Samples from different body or environmental sites were collected once during the outbreak investigations. For a comparative study of the epidemiological relatedness of the isolates recovered in the burn unit, seven preserved multidrug-resistant isolates from six patients (patients 5 to 10) hospitalized on other medical wards geographically distant from the burn unit from April 1996 to May 1997 were also examined (Table 1).
Laboratory studies.
The isolation and identification of P. aeruginosa from all specimens except stool specimens were performed by following conventional procedures (3). The stool samples, which were screened for the presence of ceftazidime-resistant P. aeruginosa, were inoculated and streaked onto Trypticase-based 5% sheep blood agar (BBL Microbiology Systems), and a 30-μg ceftazidime disk (BBL Microbiology Systems) was placed in the first quadrant. After 18 to 20 h of incubation at 35°C, any colonies within the inhibition zone of the ceftazidime disk were subcultured and identified to the species level by conventional methods, as indicated previously (3). The susceptibilities of the Pseudomonas species to 12 antimicrobial agents were determined by the disk diffusion method at the Microbiology Laboratory (National Taiwan University Hospital), and this test was used as a first-pass screening test for defining multidrug-resistant isolates.
Determination of MICs.
The MICs of 12 antimicrobial agents for all the isolates were determined by the Etest (AB Biodisk, Solna, Sweden) according to the manufacturer’s guidelines (see Table 2). A bacterial suspension of growth from a Trypticase soy agar plate (BBL Microbiology Systems) was prepared in 5 ml of Mueller-Hinton broth (BBL Microbiology Systems), and the turbidity was adjusted so that it was equivalent to that of a 0.5 McFarland standard. The bacterial suspension was streaked onto a 150-mm-diameter plate containing Mueller-Hinton agar (BBL Microbiology Systems); the plate was later incubated at 35°C in ambient air for 16 to 18 h. The MIC was read on the basis of the interception of the elliptical zone of growth inhibition with the graded Etest strip. For defining the categories of susceptibility and resistance of the isolates (17), any Etest MIC that fell between twofold dilutions was rounded up to the next twofold dilution. A pan-drug-resistant strain was defined as one that was resistant to all 12 antimicrobial agents tested. Antibiotypes were considered identical if the MICs of all agents tested were identical or within a twofold dilution discrepancy.
TABLE 2.
In vitro susceptibilities of 12 selected P. aeruginosa isolates to 12 antimicrobial agents
| Isolate | MIC (μg/ml)a
|
Anti- biotype | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PP | CP | TZ | AT | IP | AK | NC | MC | OF | CI | PM | CR | ||
| A | >256 | >256 | >256 | 48 | >32 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | I |
| B1 | >256 | >256 | >256 | 64 | 6 | 8 | 24 | >256 | >256 | >256 | >256 | >256 | II |
| D2 | 24 | 64 | 6 | 4 | 4 | 24 | 32 | >256 | 0.19 | 2 | 4 | 4 | III |
| D13 | >256 | 96 | 4 | 4 | 4 | 12 | >256 | >256 | 0.75 | 4 | 2 | 4 | IV |
| D16 | >256 | 128 | 3 | 4 | 3 | 12 | >256 | >256 | 4 | 16 | 2 | 4 | V |
| D19 | >256 | >256 | 128 | 24 | 6 | 64 | 12 | 2 | 0.094 | 0.75 | 64 | 64 | VI |
| E | >256 | >256 | >256 | >256 | >32 | 16 | >256 | 24 | >256 | >256 | >256 | >256 | VII |
| F | >256 | >256 | >256 | 48 | >32 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | VIII |
| G1 | >256 | >256 | >256 | 96 | 3 | 16 | 16 | >256 | 0.19 | 1 | 128 | 64 | IX |
| H | >256 | >256 | 24 | 24 | >32 | 192 | >256 | >256 | >256 | >256 | 16 | 32 | X |
| I | >256 | >256 | 128 | 24 | >32 | 8 | 16 | >256 | >256 | >256 | 64 | 64 | XI |
| J | 48 | 96 | 48 | 12 | 2 | 24 | 96 | >256 | 2 | 24 | 24 | 32 | XII |
PP, piperacillin; CP, cefoperazone; TZ, ceftazidime; AT, aztreonam; IP, imipenem; AK, amikacin; NC, netilmicin; MC, minocycline; OF, ofloxacin; CI, ciprofloxacin; PM, cefepime; CR, cefpirome.
Determination of O serogroup.
Group antisera against 14 O-serogroup antigens, designated A to N, were purchased from Denka Seiken Co., Ltd. (Tokyo, Japan). The O serogroup of each isolate was determined by using the slide agglutination method described previously (7). These 14 O serogroups alphabetically designated by the Japanese committee corresponded to those previously designated numerically by Lanyi and Bergan (7).
RAPD patterns.
The randomly amplified polymorphic DNA (RAPD) patterns generated by arbitrarily primed PCR (AP-PCR) were determined essentially as described before (8). Amplification was performed in a PTC-100 thermocycler (MJ Research Inc., Watertown, Mass.) and consisted of the following steps: predenaturation at 94°C for 4 min and 40 cycles of 1 min at 94°C, 1 min at 35°C, and 2 min at 72°C, with a final extension for 5 min at 72°C. Two arbitrary oligonucleotide primers were used: M13 (5′-TTATGTAAAACGACGGCCAGT-3′; Gibco BRL Products, Gaithersburg, Md.) and H5 (5′-AGTCGTCCCC-3′; OPERON Technologies, Inc., Alameda, Calif.). The amplification products were separated by electrophoresis in 1.5% agarose gels. A 1-kb ladder (Gibco BRL Products) was used in each gel as a DNA fragment size marker. The AP-PCR analyses of these isolates were done in duplicate. Isolates with RAPD patterns that differed by one or more discrete bands were considered different; otherwise, the isolates were considered identical.
RESULTS
Characteristics of the patients.
During the 2 months (April and May 1997) of investigation, 16 patients were treated in the intensive care burn unit. Table 1 presents the sources and dates of recovery of the 47 isolates of P. aeruginosa included in this study: 40 from various clinical samples from four patients in the burn unit and 7 from six patients in other medical wards. The four burn patients all had more than a 60% total-body-surface-area burn wound and had been exposed to various invasive procedures (central venous catheter insertion and ventilator use) during their stays in the burn unit. None of these patients received hydrotherapy. All of these patients had been treated with various β-lactams, including those with antipseudomonal activity (piperacillin, ceftazidime, and imipenem), and aminoglycosides before the acquisition of P. aeruginosa infection or colonization. Other isolates (oxacillin-resistant S. aureus, Klebsiella pneumoniae, Acinetobacter baumannii, or yeast isolates) were recovered from burn wounds of all of these patients during their hospital stays. In addition to the colonization or infection of the burn wounds of the four patients by the epidemic multidrug-resistant P. aeruginosa strain, this organism caused other complicated infections: patient 2 had bacteremia and ventilator-associated pneumonia, and patients 3 and 4 both had central venous catheter-related sepsis and ventilator-associated pneumonia. After the notification of the presence of the multidrug-resistant P. aeruginosa strain, these four burn patients received a wide array of antimicrobial agents, including broad-spectrum cephalosporins, aminoglycosides, and ciprofloxacin, which were prescribed for the treatment of concomitant infections caused by pathogens other than P. aeruginosa. Patient 2 died of respiratory failure due to a fulminant pneumonia caused by oxacillin-resistant S. aureus. Patients 3 and 4 both survived, although strains of P. aeruginosa were repeatedly isolated from different clinical specimens during the course of care in the intensive care burn unit (Table 1).
Susceptibility testing.
Table 2 presents the Etest MICs of 12 antimicrobial agents for 12 of the 47 P. aeruginosa isolates tested. For all isolates from patients 2 (all isolates except isolate B1), 3 (isolates C1 to C15), and 4 (all isolates except isolates D2, D13, D16, and D19), the MICs of all antimicrobial agents tested except aztreonam were identical to those for isolate A; for aztreonam we found no more than one twofold dilution discrepancy in the MICs (MICs, 32 to 64 μg/ml) for these isolates. A total of 12 antibiotypes were found (Table 2): six antibiotypes were found among the 40 isolates recovered in the burn unit and the other six were discovered among the seven isolates from patients on medical wards (Table 1).
O serogrouping.
Nine O serogroups among the 40 isolates recovered from patients in the burn unit and six O serogroups among the seven isolates from patients on medical wards were defined. The epidemic multidrug-resistant strains all belonged to serogroup O:F (serogroup O:4). Isolates belonging to the same antibiotype belonged to identical serogroups. However, isolates D2, D16, and J, which belonged to the same serogroup (serogroup O:E), had different antibiotypes. Isolates G1 and G2 from patient 7 had the same antibiotype and both belonged to O serogroup A.
RAPD patterns.
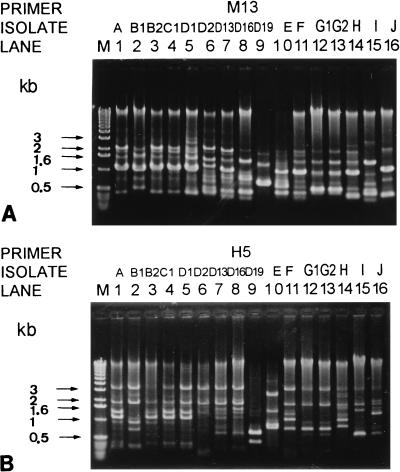
AP-PCR with two primers (primers M13 and H5) indicated that the 47 isolates recovered from the 10 patients had 12 RAPD patterns (Fig. 1). All of the epidemic multidrug-resistant serogroup O:F P. aeruginosa isolates possessed identical RAPD patterns. Isolates with the same antibiotype and belonging to the same O serogroup also had the same RAPD patterns.
FIG. 1.
RAPD patterns of 16 isolates of P. aeruginosa generated by AP-PCR with two primers, M13 (A) and H5 (B). Lanes: M, molecular size marker; 1 to 16, the isolates designated in Table 1.
Infection control measures.
All staff members in the intensive care burn unit were notified of the need for strict compliance with the recommended isolation procedures during the outbreak investigation period (12). Samples from health care workers (three physicians and two nurses) were culture positive for oxacillin-resistant S. aureus, but all of the samples were negative for P. aeruginosa. None of the samples from environmental sites were positive for P. aeruginosa. Except for one patient (patient 3) who was found to be harboring the epidemic multidrug-resistant strain shortly after (2 days) the unit became aware of the outbreak, no additional cases of infection with the clone occurred. Environmental decontamination measures were not undertaken. The physicians in the unit were not told to restrict their use of broad-spectrum cephalosporins, aztreonam, or imipenem or to change the dosages of antimicrobial agents administered. The follow-up cultures of samples from various body sites (wound, sputum, and stool samples) from the two surviving patients 2 months after the completion of the investigation did not reveal the presence of the epidemic strain.
DISCUSSION
The present study, in which 40 isolates of multidrug-resistant P. aeruginosa recovered from four patients in an intensive care burn unit were characterized phenotypically and genotypically, disclosed two important points. First, more than one clone of multidrug-resistant P. aeruginosa can colonize not only burn wounds but also other sites of the bodies of burn patients. Second, a single clone of multidrug-resistant P. aeruginosa can persist in different body sites of burn patients for weeks and months and can subsequently cause various severe infections.
Previous studies suggest that the selective pressure from the use of antimicrobial agents is a major determinant for the emergence of resistant strains (3, 15, 21). The subinhibitory antibiotic concentration in burn wounds, due to the administration of an inappropriate dosage of a β-lactam antibiotic or the regular administration of an aminoglycoside in combination with a β-lactam, provides optimal conditions for the selection and persistence of multidrug-resistant P. aeruginosa strains and their subsequent local invasion and hematogenous dissemination in burn patients (1, 21). Successful control of an outbreak caused by a multiresistant clone of P. aeruginosa by increasing the daily dosage of antimicrobial agents that are active in vitro was reported previously (21), although it has been recommended that this approach not be overused. The concomitant use of multiple antimicrobial agents, including those with good activity against P. aeruginosa, might have contributed to the emergence of the epidemic strain in patient 1. This also contributed to the long-term colonization and the subsequent spread of the epidemic multidrug-resistant strain in our burn patients.
In addition to the use of auxanograms and antibiograms, various conventional typing systems, including serotyping and pyocin and phage typing, have been extensively used to study the epidemiology of P. aeruginosa infections (20). However, the usefulness of these methods is limited by the high frequency of both nonserotypeable and polyagglutinable strains and the low reproducibility of pyocin typing, especially for strains from cystic fibrosis patients (20). Among the many DNA-based typing methods, restriction endonuclease analysis achieved by pulsed-field gel electrophoresis may be the most discriminatory (2, 5, 6, 9, 14, 19). However, fingerprinting by AP-PCR is documented as having discriminatory power comparable to that of pulsed-field gel electrophoresis for the epidemiological study of P. aeruginosa strains from cystic fibrosis patients (9). In the present study, all isolates with identical antibiotypes which all belonged to serogroup O:F (serogroup O:4) had identical RAPD patterns, suggesting the clonal origin of the epidemic strain. The good concordance of the discriminatory power of the antibiotyping and RAPD patterns for the outbreak-related and non-outbreak-related strains of multidrug-resistant P. aeruginosa might be due to the limited number of isolates studied.
Routine culture of samples from burn patients for surveillance and microbiological typing of P. aeruginosa is not recommended because of the high frequency of burn wound colonization and the organism’s polyclonal origin (13). In addition, surveillance cultures of environmental samples are notorious for being nonproductive and for yielding results which usually are not clinically relevant. In the present study, the outbreak-related strain could be isolated from various body sites other than burn wounds and subsequently caused invasive infections. It is suggested that if a multidrug-resistant P. aeruginosa strain is isolated and an infection caused by this strain is documented, extensive cultures of samples from all body sites of all patients hospitalized in the same unit and an environmental survey for the strain, as well as the use of isolation precautions, may be crucial for the early control of an ongoing outbreak.
The route of transfer of the O:F strain was not identified. Previous studies suggested that surveys for the identification of reservoirs of P. aeruginosa in burn units performed by obtaining samples for culture have yielded samples from sinks and hydrotherapy equipment that are positive by culture (21, 22). However, only hydrotherapy equipment is strongly linked to the epidemic of P. aeruginosa burn wound infections (22). Consequently, dispersal of P. aeruginosa from colonized or infected patients in the burn unit will result in further contamination of the environment of the unit as well as the hands of medical personnel. In the present investigation, all cultures of the environmental samples and hands and nasal swabs from medical personnel were negative for the epidemic strain. P. aeruginosa colonization of the rectums of burn patients is considered to be a potential source of subsequent burn wound infection. Only two of the cultures of stool samples obtained from the patients examined in the present study yielded the epidemic strain during surveillance of the outbreak. Whether the epidemic P. aeruginosa strains from the two patients were acquired from the alimentary tract or were merely contaminants in the stool specimens from nearby infected skin is unclear, because stool or rectal swab specimens were not obtained before the first positive epidemic strain was recovered from other body sites.
The present study, in which phenotypic and molecular biology-based studies were performed, highlights the fact that a single clone of multidrug-resistant P. aeruginosa can persist at different body sites of burn patients for weeks and months and can subsequently cause various severe infections. The eradication of the pan-drug-resistant strains from these patients is difficult. The extensive use of broad-spectrum cephalosporins in the unit was probably responsible for the emergence and selection of this multidrug-resistant strain.
REFERENCES
- 1.Aronoff S C, Shlaes D M. Factors that influence the evolution of β-lactam resistance in β-lactamase-inducible strains of Enterobacter cloacae and Pseudomonas aeruginosa. J Infect Dis. 1987;155:936–941. doi: 10.1093/infdis/155.5.936. [DOI] [PubMed] [Google Scholar]
- 2.Bennekov T, Colding H, Ojeniyi B, Bentzon M W, Høiby N. Comparison of ribotyping and genome fingerprinting of Pseudomonas aeruginosa isolates from cystic fibrosis patients. J Clin Microbiol. 1996;34:202–204. doi: 10.1128/jcm.34.1.202-204.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilligan P H. Pseudomonas and Burkholderia. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C: American Society for Microbiology; 1995. pp. 509–519. [Google Scholar]
- 4.Godard C, Plesiat P, Michel-Briand Y. Persistence of Pseudomonas aeruginosa strains in seven cystic fibrosis patients followed for 20 months. Eur J Med. 1993;2:117–120. [PubMed] [Google Scholar]
- 5.Grundmann H, Schneider C, Hartung D, Daschner F D, Pitt T L. Discriminatory power of the three DNA-based typing techniques for Pseudomonas aeruginosa. J Clin Microbiol. 1995;33:528–534. doi: 10.1128/jcm.33.3.528-534.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holder I A. P. aeruginosa burn infections: pathogenesis and treatment. In: Campa M, editor. Pseudomonas aeruginosa is an opportunistic pathogen. New York, N.Y: Plenum Press; 1993. pp. 275–295. [Google Scholar]
- 7.Homma J Y. Designation of the thirteen O-group antigens of Pseudomonas aeruginosa: an amendment for the tentative proposal in 1976. Jpn J Exp Med. 1982;52:317–320. [PubMed] [Google Scholar]
- 8.Hsueh P R, Teng L J, Ho S W, Hsieh W C, Luh K T. Clinical and microbiological characteristics of Flavobacterium indologenes infections associated with indwelling devices. J Clin Microbiol. 1996;34:1908–1913. doi: 10.1128/jcm.34.8.1908-1913.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kersulyte D, Struelens M J, Deplano A, Berg D E. Comparison of arbitrarily primed PCR and macrorestriction (pulsed-field gel electrophoresis) typing of Pseudomonas aeruginosa strains from cystic fibrosis patients. J Clin Microbiol. 1995;33:2216–2219. doi: 10.1128/jcm.33.8.2216-2219.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maki D G C, Alvarado J, Hassemer C A, Zilz M A. Relationship of the intimate hospital environment to endemic nosocomial infection. N Engl J Med. 1982;307:1562–1566. doi: 10.1056/NEJM198212163072507. [DOI] [PubMed] [Google Scholar]
- 11.Martin C, Ichou M A, Massicot P, Goudeau A, Quentin R. Genetic diversity of Pseudomonas aeruginosa strains isolated from patients with cystic fibrosis revealed by restriction fragment length polymorphism of the rRNA gene region. J Clin Microbiol. 1995;33:1461–1466. doi: 10.1128/jcm.33.6.1461-1466.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayhall C G. Nosocomial burn wound infections. In: Mayhall G C, editor. Hospital epidemiology and infection control. Baltimore, Md: The Williams & Wilkins Co.; 1996. pp. 225–236. [Google Scholar]
- 13.McManus A T, Mason A D, McManus W F, Pruitt B A. Twenty-five year review of Pseudomonas aeruginosa bacteremia in a burn center. Eur J Clin Microbiol. 1985;4:219–223. doi: 10.1007/BF02013601. [DOI] [PubMed] [Google Scholar]
- 14.Mifsud A J, Watine J, Picard B, Charet J C, Solignac-Bourrel C, Pitt T L. Epidemiologically related and unrelated strains of Pseudomonas aeruginosa serotype O12 cannot be distinguished by phenotypic and genotypic typing. J Hosp Infect. 1997;36:105–116. doi: 10.1016/s0195-6701(97)90116-x. [DOI] [PubMed] [Google Scholar]
- 15.Murray S A, Snydman D R. Investigation of an epidemic of multi-drug-resistant Pseudomonas aeruginosa. Infect Control. 1982;82:456–460. doi: 10.1017/s0195941700056575. [DOI] [PubMed] [Google Scholar]
- 16.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility tests. Approved standard M2-A6. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 17.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A4. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 18.Peterson S S, Koch C, Høiby N, Rosendal K. An epidemic spread of multiresistant Pseudomonas aeruginosa in a cystic fibrosis centre. J Antimicrob Chemother. 1986;17:505–516. doi: 10.1093/jac/17.4.505. [DOI] [PubMed] [Google Scholar]
- 19.Pitt T L, Livermore D M, Pitcher D, Vatopoulos A C, Legakis N J. Multiresistant serotype O:12 Pseudomonas aeruginosa: evidence for a common strain in Europe. Epidemiol Infect. 1989;103:565–576. doi: 10.1017/s095026880003096x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poh C I, Yeo C C. Recent advances in typing of Pseudomonas aeruginosa. J Hosp Infect. 1993;24:175–181. doi: 10.1016/0195-6701(93)90047-4. [DOI] [PubMed] [Google Scholar]
- 21.Richard P, Floch R L, Chamoux C, Pannier M, Espaze E, Richet H. Pseudomonas aeruginosa outbreak in a burn unit: role of antimicrobials in the emergence of multiply resistant strains. J Infect Dis. 1994;170:377–383. doi: 10.1093/infdis/170.2.377. [DOI] [PubMed] [Google Scholar]
- 22.Tredget E E, Shankowsky H A, Joffe A M, Inkson T I, Volpel K, Paranchych W, Kibsey P C, Alton J D M, Buke J F. Epidemiology of infections with Pseudomonas aeruginosa in burn patients: the role of hydrotherapy. Clin Infect Dis. 1992;15:941–949. doi: 10.1093/clind/15.6.941. [DOI] [PubMed] [Google Scholar]