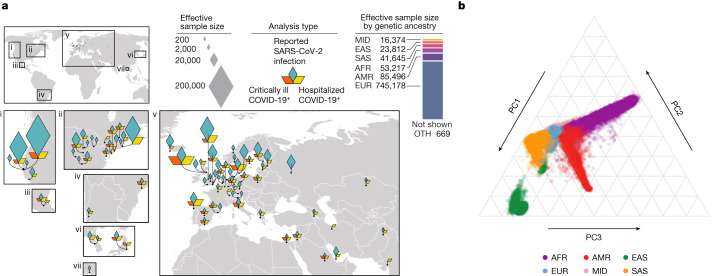
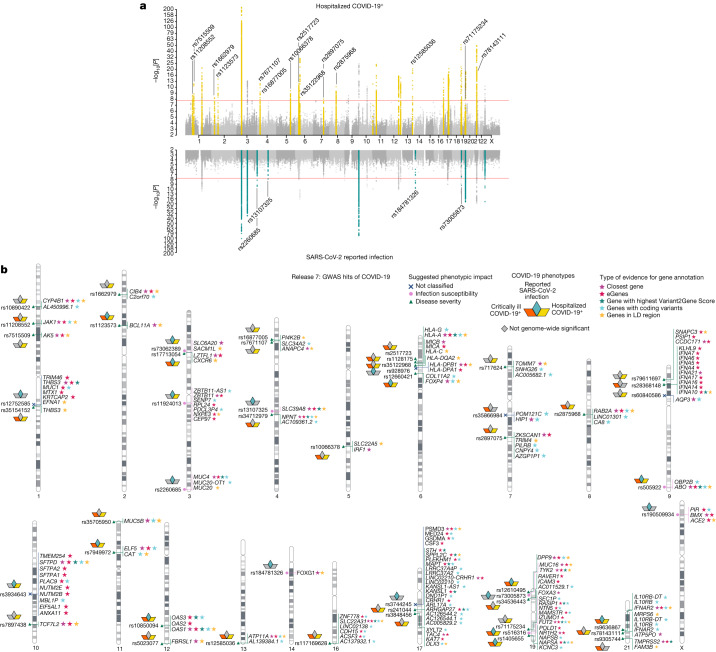
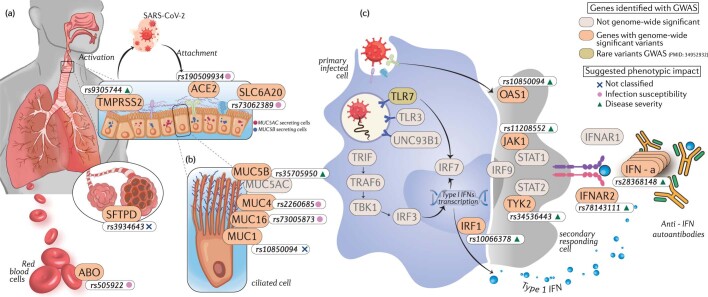
1Broad Institute of MIT and Harvard, Cambridge, MA USA
2University of California San Francisco, San Francisco, CA USA
3Institute for Molecular Medicine Finland (FIMM), University of Helsinki, Helsinki, Finland
4Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, MA USA
5Department of Psychiatry, School of Medicine, Yale University, New Haven, CT USA
6Veteran Affairs Connecticut Healthcare System, West Haven, CT USA
7Yale University, New Haven, CT USA
8Centre for Bioinformatics and Data Analysis, Medical University of Bialystok, Bialystok, Poland
9Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, MI USA
10Faculty of Industrial Sciences and Technology, Universiti Malaysia Pahang, Pahang, Malaysia
11Icahn School of Medicine at Mount Sinai, New York, NY USA
12Genolier Innovation Network and Hub, Swiss Medical Network, Genolier Healthcare Campus, Genolier, Switzerland
13Immediate, Independent Multidisciplinary Med & Biotech Research Association, Milan, Italy
14Clinical Research Unit of Nanoro, Institut de Recherche en Sciences de la Santé, Nanoro, Burkina Faso
15Sydney Brenner Institute for Molecular Biosciences, University of the Witwatersrand, Johannesburg, South Africa
16McGill University, Montreal, Quebec Canada
17University of Peurto Rico, San Juan, PR USA
18Institute for Biomedical Technologies, National Research Council, Milan, Italy
19University of Miami, Miami, FL USA
20Department of Biochemistry, All India Institute of Medical Sciences, Kalyani, India
21Department of Biosciences, School of Science and Technology, Nottingham Trent University, Nottingham, UK
22Department of Clinical Pathology, Faculty of Medicine, Mansoura University, Mansoura, Egypt
23Department of Clinical Pathology, Faculty of Medicine, Tanta University, Tanta, Egypt
24Neuron23, San Francisco, CA USA
25Vanderbilt University, Nasvhille, TN USA
26Stroke Pharmacogenomics and Genetics, Institut d’Investigació Biomèdica Sant Pau (IIBSANT PAU), Barcelona, Spain
27Department of Medicine, Surgery and Health Sciences, University of Trieste, Trieste, Italy
28Program in Medical and Population Genetics, Broad Institute of MIT and Harvard, Cambridge, MA USA
29Department of Genetics, University Medical Centre Groningen, University of Groningen, Groningen, The Netherlands
30Departamento de Tecnología Médica, Facultad de Ciencias de la Salud, Universidad de Tarapacá, Arica, Chile
31King’s College London, London, UK
32Naina Tech, New York, NY USA
33Vrije Universiteit Amsterdam, Amsterdam, The Netherlands
34Medical School, University of Pécs, Pécs, Hungary
35MNM Diagnostics, Poznan, Poland
36Genomics England, London, UK
37Heart Institute (InCor), University of Sao Paulo Medical School, Sao Paulo, Brazil
38Department of Physiology, All India Institute of Medical Sciences, Kalyani, India
39Montreal Heart Institute, Montreal, Quebec Canada
40Nantes Université, Ecole Centrale Nantes, INSERM, Center for Research in Transplantation and Translational Immunology, Nantes, France
41Department of Computer Science and Technology, University of Cambridge, Cambridge, UK
42William Harvey Research Institute, Barts and the London School of Medicine and Dentistry, Queen Mary University of London, London, UK
43University of Helsinki, Helsinki, Finland
44Department of Internal Medicine, Erasmus MC, University Medical Center Rotterdam, Rotterdam, The Netherlands
45Fonds de la Recherche Scientifique (FNRS) & Centre de Génétique Humaine, Hôpital Erasme, Université Libre de Bruxelles, Brussels, Belgium
46Department of Genetics, Hôpital Erasme, Université Libre de Bruxelles, Brussels, Belgium
47All India Institute of Medical Sciences Kalyani, Kalyani, India
48National Laboratory of Genomics for Biodiversity, CINVESTAV, Guanajuato, Mexico
49Queensland University of Technology, Brisbane, Queensland Australia
50Department of Human Genetics, McGill University, Montreal, Quebec Canada
51Lady Davis Institute, Jewish General Hospital, McGill University, Montreal, Quebec Canada
52Kyoto-McGill International Collaborative School in Genomic Medicine, Graduate School of Medicine, Kyoto University, Kyoto, Japan
53Japan Society for the Promotion of Science, Tokyo, Japan
54Division of Life Sciences, University of Northampton, Northampton, UK
55Root Deep, Boston, MA USA
56Instituto do Coração, Sao Paulo, Brazil
57Stanford University, Stanford, CA USA
58Department of Internal Medicine, Erasmus MC, University Medical Center Rotterdam, Rotterdam, The Netherlands
59Department of Epidemiology, Erasmus MC, University Medical Center Rotterdam, Rotterdam, The Netherlands
60Institute for Community Medicine, University Medicine Greifswald, Greifswald, Germany
61GI and Liver Section, Virgen del Rocío University Hospital and Department of Medicine, University of Seville, Institute of Biomedicine of Seville, Seville, Spain
62CIBEREHD, Instituto de Salud Carlos III (ISCIII), Seville, Spain
63Department of Health, Traunstein, Germany
64Rutgers Cancer Institute of New Jersey, New Brunswick, NJ USA
65Erasmus Medical Center, Rotterdam, The Netherlands
66Institute of Psychiatric Phenomics & Genomics, University of Munich, Munich, Germany
67Department of Psychiatry, University of Munich, Munich, Germany
68Institute of Virology, Technical University of Munich/Helmholtz Munich, Munich, Germany
69Institute of Human Genetics, University Hospital, Faculty of Medicine, University of Bonn, Bonn, Germany
70Department of Psychiatry, University Hospital, Faculty of Medicine, University of Bonn, Bonn, Germany
71Digital Health Center, Hasso Plattner Institute, University of Potsdam, Potsdam, Germany
72Institute of Biomedicine and Cancer Research Laboratories, Western Cancer Centre FICAN West, University of Turku, Turku, Finland
73IRCCS Istituto Giannina Gaslini, Genova, Italy
74Department of Neurosciences, Rehabilitation, Ophthalmology, Genetics, Maternal and Child Health, University of Genova, Genova, Italy
75Department of Biomedical Data Science, Stanford University, Stanford, CA USA
76Fundación Pública Andaluza para la Gestión de la Investigación en Salud de Sevilla, Seville, Spain
77Department of Pediatrics, Stanford University, Stanford, CA USA
78Department of Internal Medicine, Erasmus MC, University Medical Center Rotterdam, Rotterdam, The Netherlands
79University Medical Center Utrecht, Utrecht, The Netherlands
80Instituto de Investigación Interdisciplinaria & Escuela de Medicina, Universidad de Talca, Talca, Chile
81Programa de Genética Humana, ICBM and Chile Departamento de Oncología Básico Clínica, Universidad de Chile, Talca, Chile
82Department of Statistical Genetics, Osaka University Graduate School of Medicine, Osaka, Japan
83Department of Internal Medicine, International Islamic University Malaysia, Pahang, Malaysia
84Centro para el Desarrollo de la Investigación Científica (CEDIC), Asunción, Paraguay
8523andMe, Sunnyvale, CA USA
86Washington University School Of Medicine, St Louis, MO USA
87Department of Psychological & Brain Sciences, Washingon University in St Louis, St Louis, MO USA
88Institute of Biological Psychiatry, Mental Health Centre Sct Hans, Mental Health Services Capital Region of Denmark, Copenhagen, Denmark
89iPSYCH, The Lundbeck Foundation Initiative for Integrative Psychiatric Research, Aarhus, Denmark
90Laureate Institute for Brain Research, Tulsa, OK USA
91Department of Radiology, UCSD, San Diego, CA USA
92Nork Infection Clinical Hospital, Yerevan, Armenia
93Institute of Molecular Biology NAS RA, Yerevan, Armenia
94National Research Center for Cardiac Surgery, Astana, Kazakhstan
95City Infectious Disease Center, Astana, Kazakhstan
96National Center for Biotechnology, Astana, Kazakhstan
97Health Department of Astana, Astana, Kazakhstan
98MRC Integrative Epidemiology Unit, University of Bristol, Bristol, UK
99Division of Psychitary, University of Edinburgh, Edinburgh, UK
100Institute for Molecular Medicine Finland (FIMM), Helsinki, Finland
101University of Liege, GIGA-Institute, Liege, Belgium
102CHC Mont-Légia, Liege, Belgium
103BHUL (Liège Biobank), CHU of Liège, Liege, Belgium
104Institute for Molecular Medicine Finland, University of Helsinki, Helsinki, Finland
105Analytic & Translational Genetics Unit, Massachusetts General Hospital, Boston, MA USA
106Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA USA
107CHU of Liege, Liege, Belgium
108University of Liege, Liege, Belgium
109Centre de Génétique Humaine, Hôpital Erasme, Université Libre de Bruxelles, Brussels, Belgium
110Service de Médecine Interne, Hôpital Erasme, Université Libre de Bruxelles, Brussels, Belgium
111McGill Genome Centre and Department of Human Genetics, McGill University, Montréal, Quebec Canada
112Department of Human Genetics, Epidemiology, Biostatistics and Occupational Health, McGill University, Montreal, Quebec Canada
113Department of Twin Research, King’s College London, London, UK
114The Meakins-Christie Laboratories at the Research Institute of the McGill University Heath Centre Research Institute and Department of Medicine, Faculty of Medicine, McGill University, Montreal, Quebec Canada
115Research Centre of the Centre Hospitalier de l’Université de Montréal (CRCHUM), Montreal, Quebec Canada
116Centre Hospitalier de l’Université de Montréal (CHUM), Montreal, Quebec Canada
117Department of Epidemiology, Biostatistics and Occupational Health, McGill University, Montreal, Quebec Canada
118Department of Emergency Medicine, McGill University, Montreal, Quebec Canada
119Emergency Department, Jewish General Hospital, McGill University, Montreal, Quebec Canada
120McGill AIDS Centre, Department of Microbiology and Immunology, Lady Davis Institute for Medical Research, Jewish General Hospital, McGill University, Montreal, Quebec Canada
121Division of Infectious Diseases, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland
122Research Centre of the Centre Hospitalier de l’Université de Montréal and Université de Montréal, Montreal, Quebec Canada
123School of Rehabilitation, Faculty of Medicine and Health Sciences, Université de Sherbrooke, Sherbrooke, Canada
124Department of Medicine, Faculty of Medicine, Université de Montréal, Montreal, Quebec Canada
125Center for Immunity, Inflammation and Infectious Diseases, Montreal Clinical Research Institute (IRCM), Montreal, Quebec Canada
126Génome Québec, Montreal, Quebec Canada
127Département de Microbiologie et Infectiologie, Université de Sherbrooke, Sherbrooke, Quebec Canada
128Département de Médecine, Service d’Infectiologie, Centre de Recherche Clinique du Centre Hospitalier Universitaire de Sherbrooke, Sherbrooke, Quebec Canada
129Department of Epidemiology, Biostatistics and Occupational Health, McGill University, Montreal, Quebec Canada
130Center for Clinical Epidemiology, Lady Davis Institute, Jewish General Hospital, Montreal, Quebec Canada
131Department of Neurology and Neurosurgery, McGill University, Montreal, Quebec Canada
132Centre Intégré Universitaire de Santé et de Services Sociaux (CIUSSS) du Saguenay–Lac-Saint-Jean, Saguenay, Quebec Canada
133Department of Pharmacology-Physiology, Medicine and Health Sciences Faculty, Université de Sherbrooke, Sherbrooke, Quebec Canada
134Division of Respiratory Medicine, Department of Pediatrics, Sainte-Justine University Hospital Center, University of Montreal, Montreal, Quebec Canada
135Centre of Genomics and Policy, McGill University, Montreal, Quebec Canada
136Division of Genetic Medicine, Department of Medicine, Vanderbilt University Medical Center, Nashville, TN USA
137Vanderbilt Genetics Institute, Vanderbilt University Medical Center, Nashville, TN USA
138Institute of Human Genetics, University Hospital Bonn, Medical Faculty University of Bonn, Bonn, Germany
139Institute of Genomic Statistics and Bioinformatics, University Hospital Bonn, Medical Faculty University of Bonn, Bonn, Germany
140Department of Gastroenterology, Hepatology and Infectious Diseases, University Hospital Duesseldorf, Medical Faculty Heinrich Heine University, Duesseldorf, Germany
141Department of Gastroenterology, Hepatology and Infectiology, University Hospital Magdeburg, Magdeburg, Germany
142Clinic for Cardiology, Angiology and Internal Intensive Medicine, Medical Clinic I, RWTH Aachen University, Aachen, Germany
143Department of Pneumology and Intensive Care Medicine, Faculty of Medicine, RWTH Aachen University, Aachen, Germany
144Department of Pneumology, Hannover Medical School, Hannover, Germany
145Department of Gastroenterology, Hepatology and Endocrinology, Hannover Medical School, Hannover, Germany
146Hannover Unified Biobank, Hannover Medical School, Hannover, Germany
147Department I of Internal Medicine, Faculty of Medicine and University Hospital of Cologne, University of Cologne, Cologne, Germany
148Center for Molecular Medicine Cologne (CMMC), University of Cologne, Cologne, Germany
149German Center for Infection Research (DZIF), partner site Bonn-Cologne, Cologne, Germany
150Institute for Human Genetics and Genomic Medicine, Medical Faculty, RWTH Aachen University, Aachen, Germany
151Department of Anesthesiology and Intensive Care Medicine, University Hospital Essen, University Duisburg-Essen, Essen, Germany
152Department of Child and Adolescent Psychiatry, University Hospital Essen, University of Duisburg-Essen, Essen, Germany
153Department of Infectious Diseases, University Hospital Essen, University Duisburg-Essen, Essen, Germany
154Department of Pneumology, Allergology and Respiratory Medicine, University Hospital Saarland, Homburg, Germany
155Center of Human and Molecular Biology, Department of Human Genetics, University Hospital Saarland, Homburg, Germany
156Department of Genetics & Epigenetics, Saarland University, Saarbrücken, Germany
157Institute of Innate Immunity, University Hospital Bonn, Bonn, Germany
158University of Toronto, Toronto, Ontario Canada
159Dalla Lana School of Public Health, University of Toronto, Toronto, Ontario Canada
160Lunenfeld-Tanenbaum Research Institute, Sinai Health, Toronto, Ontario Canada
161Simon Fraser University, Burnaby, British Columbia Canada
162Lady Davis Institute, Jewish General Hospital, McGill University, Montreal, Quebec Canada
163University of Waterloo, Waterloo, Ontario Canada
164The Hospital for Sick Children, Toronto, Ontario Canada
165Queen’s University, Kingston, Ontario Canada
166Mount Sinai Hospital, Sinai Health, Toronto, Ontario Canada
167St Michael’s Hospital, Unity Health Toronto, Toronto, Ontario Canada
168Princess Margaret Cancer Center, University Health Network, Toronto, Ontario Canada
169Ontario Institute for Cancer Research, Toronto, Ontario Canada
170Dynacare Laboratory, Toronto, Ontario Canada
171William Osler Health System, Brampton, Ontario Canada
172University Health Network, Toronto, Ontario Canada
173Mackenzie Health, Toronto, Ontario Canada
174Women’s College Hospital, Toronto, Ontario Canada
175University of British Columbia, Vancouver, British Columbia Canada
176BC Children’s Hospital, Vancouver, British Columbia Canada
177St Paul’s Hospital, Vancouver, British Columbia Canada
178University of Calgary, Calgary, Alberta Canada
179Alberta Children’s Hospital, Calgary, Alberta Canada
180British Columbia Cancer Research Centre, Vancouver, British Columbia Canada
181Toronto General Hospital, Toronto, Ontario Canada
182Division of Infectious Diseases, Department of Medicine, University of Ottawa, Ottawa, Ontario Canada
183Department of Biochemistry, Microbiology and Immunology, University of Ottawa Centre of Infection, Immunity and Inflammation (CI3), Ottawa, Ontario Canada
184The Ottawa Hospital Research Institute, Ottawa, Ontario Canada
185University of Montreal, Montreat, Quebec Canada
186Ste-Justine University Health Centre, Montreal, Quebec Canada
187University of Toronto Ontario, Toronto, Ontario Canada
188MitogenDx/Eve Technologies, Toronto, Ontario Canada
189Genome Sciences Centre, BC Cancer, Vancouver, British Columbia Canada
190Montreal Children’s Hospital, Montreal, Quebec Canada
191London Health Sciences Centre, London, Ontario Canada
192Kidney and Metabolism Program, St Michael’s Hospital, Unity Health Toronto, Toronto, Ontario Canada
193Sinai Health, Mount Sinai Hospital, Toronto, Ontario Canada
194Baycrest Health Sciences, Toronto, Ontario Canada
195Anschutz Medical Campus, University of Colorado, Aurora, CO USA
196Institute of Psychiatric Phenomics and Genomics (IPPG), University Hospital, LMU Munich, Munich, Germany
197Department of Genetic Epidemiology in Psychiatry, Central Institute of Mental Health, Medical Faculty Mannheim, University of Heidelberg, Mannheim, Germany
198Department of Psychiatry and Behavioral Sciences, Johns Hopkins University, Baltimore, MD USA
199Department of Psychiatry and Behavioral Sciences, SUNY Upstate Medical University, Syracuse, NY USA
200Department of Psychiatry and Psychotherapy, University Hospital, LMU Munich, Munich, Germany
201Institute of Virology, Technical University Munich/Helmholtz Zentrum München, Munich, Germany
202Department of Psychiatry and Psychotherapy, University Hospital Bonn, Medical Faculty University of Bonn, Bonn, Germany
203CIBER de Salud Mental, Instituto de Salud Carlos III, Barcelona, Spain
204Munich Clinic Schwabing, Academic Teaching Hospital, Ludwig-Maximilians University of Munich (LMU), Munich, Germany
205Department of Internal Medicine II, University Hospital rechts der Isar, Technical University of Munich, School of Medicine, Munich, Germany
206Institute of Clinical Chemistry and Pathobiochemistry, School of Medicine, Technical University of Munich, Munich, Germany
207TranslaTUM, Center for Translational Cancer Research, Technical University of Munich, Munich, Germany
208Department of Psychiatry and Psychotherapy, University Medical Center Goettingen, Goettingen, Germany
209German Center for Neurodegenerative Diseases (DZNE), Goettingen, Germany
210Neurosciences and Signaling Group, Institute of Biomedicine (iBiMED), Department of Medical Sciences, University of Aveiro, Agra do Crasto, Aveiro, Portugal
211Munich Site of the German Center for Infection Research (DZIF), Munich, Germany
212Institute of Computational Biology, Helmholtz Center Munich, Oberschleissheim, Germany
213Department of Hygiene and Epidemiology, University of Ioannina School of Medicine, Ioannina, Greece
2141st Department of Internal Medicine and Infectious Diseases Unit, University Hospital of Ioannina, Ioannina, Greece
215Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, London, UK
216Center for Evidence-Based Medicine, Department of Health Services, Policy and Practice, School of Public Health, Brown University, Providence, RI USA
217University of Ioannina School of Medicine, Ioannina, Greece
218Centre for Systems Biology, Biomedical Research Foundation of the Academy of Athens, Athens, Greece
219Institute of Clinical Molecular Biology, Christian-Albrechts-University, Kiel, Germany
220Novo Nordisk Foundation Center for Protein Research, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark
221Institut de Biotecnologia i de Biomedicina, Universitat Autònoma de Barcelona, Barcelona, Spain
222ICREA, Barcelona, Spain
223Institute of Biotechnology, Life Science Centre, Vilnius University, Vilnius, Lithuania
224Research Group for Evolutionary Immunogenomics, Max Planck Institute for Evolutionary Biology, Plön, Germany
225Research Unit for Evolutionary Immunogenomics, Department of Biology, University of Hamburg, Hamburg, Germany
226Department of Gastroenterology, Hospital Universitario Ramón y Cajal, University of Alcalá, Instituto Ramón y Cajal de Investigación Sanitaria (IRYCIS), Madrid, Spain
227Centro de Investigación Biomédica en Red en Enfermedades Hepáticas y Digestivas (CIBEREHD), Instituto de Salud Carlos III (ISCIII), Madrid, Spain
228Vall d’Hebron Institut de Recerca (VHIR), Vall d’Hebron Hospital Universitari, Barcelona, Spain
229Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Milan, Italy
230Department of Medicine, Møre & Romsdal Hospital Trust, Ålesund, Norway Geminicenter for Sepsis Research, Institute of Circulation and Medical Imaging (ISB), NTNU, Trondheim, Norway
231Microbiology Unit, Hospital Universitario Clinico San Cecilio, Granada, Spain
232Ibs.Granada Instituto de Investigación Biosanitaria, Granada, Spain
233Klinik für Innere Medizin I, Universitätsklinikum Schleswig-Holstein, Campus Kiel, Kiel, Germany
234Emergency Department, University Hospital Regensburg, Regensburg, Germany
235Department for Infectious Diseases and Infection Control, University Hospital Regensburg, Regensburg, Germany
236Department of Medicine I, Gastroenterology, Hepatology and Endocrinology, Medical University of Innsbruck, Innsbruck, Austria
237Christian Doppler Laboratory of Iron and Phosphate Biology at the Department of Medicine I, Medical University of Innsbruck, Innsbruck, Austria
238Institute of Clinical Medicine, University of Oslo, Oslo, Norway
239Department of Microbiology, Oslo University Hospital, Oslo, Norway
240Hospital Clinic, University of Barcelona and IDIBAPS, Barcelona, Spain
241European Foundation for the Study of Chronic Liver Failure (EF-CLIF), Barcelona, Spain
242GCAT—Genomes for Life, Germans Trias i Pujol Health Sciences Research Institute (IGTP), Badalona, Spain
243Genomes for Life—GCAT Laboratory, Germans Trias i Pujol Research Institute (IGTP), Spainermans Trias i Pujol Research Institute (IGTP), Badalona, Spain
244University of Milan, Milan, Italy
245Department of Liver and Gastrointestinal Diseases, Biodonostia Health Research Institute, Donostia University Hospital, University of the Basque Country (UPV/EHU), CIBEREHD, Ikerbasque, San Sebastian, Spain
246Digestive Diseases Unit, Virgen del Rocio University Hospital, Institute of Biomedicine of Seville, University of Seville, Seville, Spain
247University of Sevilla, Seville, Spain
248Instituto de Biomedicina de Sevilla (IBIS), Seville, Spain
249Hospital Universitario Virgen del Rocío de Seville, Seville, Spain
250Universitat Autònoma de Barcelona, Bellatera, Spain
251Liver Unit, Department of Internal Medicine, Hospital Universitari Vall d’Hebron, Vall d’Hebron Barcelona Hospital Campus, Barcelona, Spain
252Gastrointestinal Genetics Lab, CIC bioGUNE—BRTA, Derio, Spain
253Ikerbasque, Basque Foundation for Science, Bilbao, Spain
254Department of Liver and Gastrointestinal Diseases, Biodonostia Health Research Institute—Donostia University Hospital, University of the Basque Country (UPV/EHU), CIBEREHD, Ikerbasque, San Sebastian, Spain Ikerbasque, Basque Foundation for Science, Bilbao, Spain
255Department I of Internal Medicine, Faculty of Medicine and University Hospital Cologne, University of Cologne, Cologne, Germany
256University of Cologne, Faculty of Medicine and University Hospital Cologne, Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases (CECAD), Cologne, Germany
257European Reference Network on Hepatological Diseases (ERN RARE-LIVER), Fondazione IRCCS San Gerardo dei Tintori, Monza, Italy
258Division of Gastroenterology, Center for Autoimmune Liver Diseases, School of Medicine and Surgery, University of Milano-Bicocca, Milan, Italy
259Genomes for Life—GCAT lab, Germans Trias i Pujol Research Institute (IGTP), Badalona, Spain
260IRCCS Humanitas Research Hospital, Milan, Italy
261Department of Biomedical Sciences, Humanitas University, Pieve Emanuele, Milan, Italy
262Zentrum für Humangenetik Regensburg, Regensburg, Germany
263University Hospital Schleswig-Holstein (UKSH), Campus Kiel, Kiel, Germany
264Section for Gastroenterology, Department of Transplantation Medicine, Division for Cancer Medicine, Surgery and Transplantation, Oslo University Hospital Rikshospitalet, Oslo, Norway
265Research Institute for Internal Medicine, Division of Surgery, Inflammatory Diseases and Transplantation, Oslo University Hospital Rikshospitalet and University of Oslo, Oslo, Norway
266Norwegian PSC Research Center, Department of Transplantation Medicine, Division of Surgery, Inflammatory Diseases and Transplantation, Oslo University Hospital Rikshospitalet, Oslo, Norway
267Private University in the Principality of Liechtenstein, Triesen, Liechtenstein
268Division of Rheumatology, Inflammation and Immunity, Brigham and Women’s Hospital and Harvard Medical School, Boston, MA USA
269Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, MA USA
270Department of Biomedical Informatics, Harvard Medical School, Boston, MA USA
271Center for Data Sciences, Brigham and Women’s Hospital, Boston, MA USA
272Institut de Biotecnologia i de Biomedicina, Universitat Autònoma de Barcelona, Bellaterra, Barcelona, Spain
273Institute for Medical Microbiology and Hospital Epidemiology, Hannover Medical School, Hannover, Germany
274Randaberg Municipality, Randaberg, Norway
275Department of Quality and Health Technology, Faculty of Health Sciences, University of Stavanger, Stavanger, Norway
276University Hospital Schleswig-Holstein, Campus Kiel, Kiel, Germany
277Research Department, Stavanger University Hospital, Stavanger, Norway
278Department of Genetics and Bioinformatics (HDGB), Division of Health Data and Digitalization, Norwegian Institute of Public Health, Oslo, Norway
279Centre for Genetics and Genomics Versus Arthritis, Centre for Musculoskeletal Research, Manchester Academic Health Science Centre, The University of Manchester, Manchester, UK
280Department of Intensive Care, Hospital Universitario Ramón y Cajal, Instituto Ramón y Cajal de Investigación Sanitaria (IRYCIS), University of Alcalá, Madrid, Spain
281Clinical Biochemistry Department, Donostialdea Integrated Health Organisation, Osakidetza Basque Health Service, San Sebastian, Spain
282Research Center and Memory Clinic, Ace Alzheimer Center Barcelona—Universitat Internacional de Catalunya, Barcelona, Spain
283CIBERNED, Network Center for Biomedical Research in Neurodegenerative Diseases, National Institute of Health Carlos III, Madrid, Spain
284Department of Biochemistry, University Hospital Vall d’Hebron, Barcelona, Spain
285Department of Acute Medicine, Oslo University Hospital, Oslo, Norway
286Division of Neurogenetics and Molecular Psychiatry, Department of Psychiatry and Psychotherapy, Faculty of Medicine and University Hospital Cologne, University of Cologne, Cologne, Germany
287Department of Neurodegenerative Diseases and Geriatric Psychiatry, Medical Faculty, University Hospital Bonn, Bonn, Germany
288German Center for Neurodegenerative Diseases (DZNE), Bonn, Germany
289Department of Psychiatry, Glenn Biggs Institute for Alzheimer’s and Neurodegenerative Diseases, San Antonio, TX USA
290Department of Anesthesiology and Intensive Care, University Hospital of North Norway, Tromsø, Norway
291Pediatrics and Tettamanti Center, Fondazione IRCCS San Gerardo dei Tintori and School of Medicine and Surgery, University of Milano-Bicocca, Monza, Italy
292Biochemistry Department, Echevarne Laboratory, Sant Cugat del Vallés, Barcelona, Spain
293Centre for Multidisciplinary Research in Health Science (MACH), University of Milan, Milan, Italy
294Gastroenterology Unit, Fondazione IRCCS Casa Sollievo della Sofferenza, San Giovanni Rotondo, Italy
295Microbiology Unit, Fondazione IRCCS San Gerardo, Monza, Italy
296Department of Infectious Diseases, Oslo University Hospital, Oslo, Norway
297Humanitas Gavazzeni-Castelli, Bergamo, Italy
298Microbiology Department, Hospital Universitari Vall d’Hebron, Barcelona, Spain
299Department of Respiratory Diseases, Hospital Universitario Ramón y Cajal, Instituto Ramón y Cajal de Investigación Sanitaria (IRYCIS), University of Alcalá, Centro de Investigación Biomédica en Red en Enfermedades Respiratorias (CIBERES), Madrid, Spain
300Department of Respiratory Medicine and Allergology, University Hospital, Goethe University, Frankfurt am Main, Germany
301Centro de Investigación Biomédica en Red de Enfermedades Respiratorias (CIBERES), Madrid, Spain
302Centro de Investigación Biomédica en Red de Epidemiología y Salud Pública (CIBERESP), Madrid, Spain
303Consejo Superior de Investigaciones Científicas, Seville, Spain
304Department of Infectious Diseases, Hospital Universitario Ramón y Cajal, Instituto Ramón y Cajal de Investigación Sanitaria (IRYCIS), University of Alcalá, Madrid, Spain
305Department of Transfusion Medicine and Haematology Laboratory, San Gerardo Hospital, Monza, Italy
306Respiratory Medicine & International Health, University of Lübeck, Lübeck, Germany
307Division of Clinical Infectious Diseases, Research Center Borstel, Borstel, Germany
308Clinical Tuberculosis Unit, German Center for Infection Research (DZIF), Borstel, Germany
309Internal Medicine Department, Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Milan, Italy
310Humanitas Clinical and Research Center, IRCCS, Milan, Italy
311Respiratory Service, Basurto University Hospital, Osakidetza Basque Health Service, Bilbao, Spain
312Department of Clinical and Molecular Medicine, Faculty of Medicine and Health Sciences, Norwegian University of Science and Technology, Trondheim, Norway
313Department of Medicine, Møre & Romsdal Hospital Trust, Ålesund, Norway
314Department of Internal Medicine II, Medical University of Innsbruck, Innsbruck, Austria
315Department of Anesthesiology and Critical Care, Hospital Universitario Ramón y Cajal, Instituto Ramón y Cajal de Investigación Sanitaria (IRYCIS), University of Alcalá, Madrid, Spain
316Immunohematology Department, Banc de Sang i Teixits, Autonomous University of Barcelona, Barcelona, Spain
317European Reference Network on Hepatological Diseases (ERN RARE-LIVER), San Gerardo Hospital, Monza, Italy
318Department of Medical Sciences, Università degli Studi di Torino, Turin, Italy
319Respiratory Service, Galdakao Hospital, Osakidetza Basque Health Service, Galdakao, Spain
320Biocruces Bizkaia Health Research Institute, Barakaldo, Spain
321Department of Infectious Diseases, E.O. Ospedali Galliera, Genova, Italy
322Accident & Emergency and Emergency Medicine Unit, San Gerardo Hospital, Monza, Italy
323Histocompatibilidad y Biologia Molecular, Centro de Transfusion de Madrid, Madrid, Spain
324Dino Ferrari Center, Department of Pathophysiology and Transplantation, University of Milan, Milan, Italy
325Neurology Unit, IRCCS Fondazione Ca’ Granda Ospedale Maggiore Policlinico, Milan, Italy
326Department of Pathophysiology and Transplantation, University of Milan, Milan, Italy
327Angelo Bianchi Bonomi Hemophilia and Thrombosis Center, Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Milan, Italy
328Respiratory Unit, Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Milan, Italy
329Department of Pathophysiology and Transplantation, Università degli Studi di Milano, Milan, Italy
330Internal Medicine Department, Virgen del Rocio University Hospital, Seville, Spain
331Biochemistry Department, University Hospital Vall d’Hebron, Barcelona, Spain
332Centre for Medical Sciences-CISMed, University of Trento, Trento, Italy
333Anesthesia and Intensive Care, Santa Chiara Hospital, APSS Trento, Trento, Italy
334Department of Anesthesiology, Intensive Care and Emergency, Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Milan, Italy
335Fondazione Grigioni per il Morbo di Parkinson and Parkinson Institute, ASST Gaetano Pini-CTO, Milan, Italy
336School of Medicine and Surgery, University of Milano-Bicocca, Milan, Italy
337Acute Geriatric Unit, IRCCS San Gerardo Foundation, Monza, Italy
338Neurointensive Care Unit, IRCCS San Gerardo dei Tintori, Monza, Italy
339Emergency Department, Anesthesia and Intensive Care, IRCCS San Gerardo dei Tintori, Monza, Italy
340Department of Anesthesiology, Intensive Care Medicine and Pain Therapy, University Hospital Frankfurt, Frankfurt am Main, Germany
341Acute Geriatric Unit, IRCCS San Gerardo Foundation Monza, Monza, Italy
342Department of Medical Microbiology, Clinic of Laboratory Medicine, St Olavs Hospital, Trondheim, Norway
343Department of Internal Medicine I, University Medical Center Schleswig-Holstein, Kiel, Germany
344Department of Infectious Diseases, St Olavs Hospital, Trondheim University Hospital, Trondheim, Norway
345Department of Clinical and Molecular Medicine, NTNU, Trondheim, Norway
346Institute of Parasitology and Biomedicine Lopez-Neyra, CSIC, Granada, Spain
347Institute for Cardiogenetics, University of Lübeck, Lübeck, Germany
348German Research Center for Cardiovascular Research, partner site Hamburg–Lübeck–Kiel, Lübeck, Germany
349University Heart Center Lübeck, Lübeck, Germany
350Respiratory ICU, Institut Clínic Respiratory, Hospital Clinic, University of Barcelona, and IDIBAPS, Barcelona, Spain
351Bioinformatics Area, Fundación Progreso y Salud and Institute of Biomedicine of Sevilla (IBIS), Seville, Spain
352Department of Research, Ostfold Hospital Trust, Gralum, Norway
353Department of Infectious Diseases, Hospital Universitario Clinico San Cecilio, Granada, Spain
354Infectious Diseases Service, Osakidetza, Biocruces Bizkaia Health Research Institute, Barakaldo, Spain
355Department of Research, St Olav Hospital, Trondheim University Hospital, Trondheim, Norway
356Novo Nordisk Foundation Center for Protein Research, Disease Systems Biology, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark
357Medical Department, Drammen Hospital, Vestre Viken Hospital Trust, Gjettum, Norway
358Research Center Borstel, BioMaterialBank Nord, Sülfeld, Germany
359German Center for Lung Research (DZL), Airway Research Center North (ARCN), Grosshansdorf, Germany
360Popgen 2.0 Network (P2N), Kiel, Germany
361Department of Clinical Science, University of Bergen, Bergen, Norway
362Pediatric Departement and Centro Tettamanti—European Reference Network (ERN) PaedCan, EuroBloodNet, MetabERN-University of Milano-Bicocca-Fondazione MBBM/Ospedale San Gerardo, San Gerardo, Italy
363Germans Trias i Pujol Research Institute (IGTP), Badalona, Spain
364Clinic of Medicine and Rehabilitation, Levanger Hospital, Nord-Trondelag Hospital Trust, Levanger, Norway
365Geminicenter for Sepsis Research, Institute of Circulation and Medical Imaging (ISB), NTNU, Trondheim, Norway
366HLA Laboratory, E.O. Ospedali Galliera, Genova, Italy
367ISGlobal, Barcelona, Spain
368Universitat Pompeu Fabra (UPF), Barcelona, Spain
369IMIM (Hospital del Mar Medical Research Institute), Barcelona, Spain
370Stefan-Morsch-Stiftung, Birkenfeld, Germany
371Osakidetza, OSI Donostialdea, Altza Primary Care, Biodonostia Health Research Institute, San Sebastián, Spain
372Center of Bioinformatics, Biostatistics and Bioimaging, School of Medicine and Surgery, University of Milano-Bicocca, Milan, Italy
373Biostatistics and Clinical Epidemiology, Fondazione IRCCS San Gerardo dei Tintori, Monza, Italy
374Department of Internal Medicine, Infectious Diseases, University Hospital Frankfurt & Goethe University Frankfurt, Frankfurt am Main, Germany
375Phase 1 Research Centre, ASST Monza, School of Medicine and Surgery, University of Milano-Bicocca, Milan, Italy
376Division of Intensive Care and Emergency Medicine, Department of Internal Medicine, Medical University Innsbruck, Innsbruck, Austria
377Department of Anesthesiology, Hospital Universitario Ramón y Cajal, Instituto Ramón y Cajal de Investigación Sanitaria (IRYCIS), Madrid, Spain
378University of Cologne, Faculty of Medicine and University Hospital Cologne, Clinical Trials Centre Cologne (ZKS Köln), Cologne, Germany
379Pulmonology Unit, Fondazione IRCCS San Gerardo dei Tintori, Monza, Italy
380Infectious Diseases Unit, Fondazione IRCCS San Gerardo dei Tintori, Monza, Italy
381Institute of Human Genetics, University of Bonn School of Medicine & University Hospital Bonn, Bonn, Germany
382Institute of Immunology, Christian-Albrechts-University of Kiel & UKSH Schleswig-Holstein, Kiel, Germany
383Intensive Care Department, Vall d’Hebron University Hospital, SODIR-VHIR research group, Barcelona, Spain
384Department of Medicine, Møre & Romsdal Hospital Trust, Molde, Norway
385Institute of Medical Virology, University Hospital Frankfurt, Goethe University, Frankfurt am Main, Germany
386German Centre for Infection Research (DZIF), external partner site Frankfurt, Frankfurt am Main, Germany
387Department of Neurology, Bezirksklinikum Regensburg, University of Regensburg, Regensburg, Germany
388Institute of Transfusionsmedicine, University Hospital Schleswig-Holstein (UKSH), Kiel, Germany
389School of Biological Sciences, Monash University, Clayton, Victoria Australia
390Department of Microbiology, University Hospital Vall d’Hebron, Barcelona, Spain
391Autonoma University of Barcelona, Barcelona, Spain
392Biochemistry Unit, Hospital Universitario Clinico San Cecilio, Granada, Spain
393Department of Infectious Diseases, University Hospital of North Norway, Tromsø, Norway
394Faculty of Health Sciences, UIT The Arctic University of Norway, Tromsø, Norway
395Catalan Institute of Oncology (ICO), Barcelona, Spain
396Bellvitge Biomedical Research Institute (IDIBELL), Barcelona, Spain
397Universitat de Barcelona (UB), Barcelona, Spain
398Faculty of Medicine and University Hospital Cologne, Institute for Transfusion Medicine, University of Cologne, Cologne, Germany
399Departamento de Anatomía Patológica, Facultad de Medicina, Universidad de Chile, Santiago, Chile
400Servicio de Anatomía Patológica, Hospital Clínico de la Universidad de Chile, Santiago, Chile
401Laboratorio Clínico del Área Técnica de Biología Molecular, Hospital del Salvador, Providencia, Chile
402Departamento de Biotecnología, Universidad de Antofagasta, Antofagasta, Chile
403AUSTRAL-omics, Vicerrectoría de Investigación, Desarrollo y Creación Artística, Antofagasta, Chile
404Instituto de Ciencias Ambientales y Evolutivas, Facultad de Ciencias, Universidad Austral de Chile, Valdivia, Chile
405Escuela de Medicina, Universidad de Magallanes, Punta Arenas, Chile
406Genómica Evolutiva y Médica de Magallanes (GEMMa), Centro Asistencial, Docente y de Investigación (CADI-UMAG), Punta Arenas, Chile
407Interuniversity Center on Healthy Aging, Punta Arenas, Chile
408Instituto de Investigación Interdisciplinaria, Universidad de Talca, Talca, Chile
409Programa de Genética Humana, ICBM, Universidad de Chile, Santiago, Chile
410Departamento de Oncología Básico Clínica, Universidad de Chile, Santiago, Chile
411Departamento de Ciencias de la Computación, Facultadad de Ciencias Físicas y Matemáticas, Universidad de Chile, Santiago, Chile
412Laboratory of Chemical Carcinogenesis and Pharmacogenetics, Department of Basic and Clinical Oncology (DOBC), Faculty of Medicine, University of Chile, Santiago, Chile
413Latin American Network for Implementation and Validation of Clinical Pharmacogenomics Guidelines (RELIVAF-CYTED), Santiago, Chile
414Department of Pharmaceutical Sciences and Technology, Faculty of Chemical and Pharmaceutical Sciences, University of Chile, Santiago, Chile
415Department of Basic and Clinical Oncology (DOBC), Faculty of Medicine, University of Chile, Santiago, Chile
416University of Chile & Latin American Network for Implementation and Validation of Clinical Pharmacogenomics Guidelines (RELIVAF-CYTED), Santiago, Chile
417Departamento de Tecnología Médica, Facultad de Ciencias de la Salud, Universidad de Antofagasta, Antofagasta, Chile
418AUSTRAL-omics, Vicerrectoría de Investigación, Desarrollo y Creación Artística, Universidad Austral de Chile, Valdivia, Chile
419Clinical Biochemistry and Immunology Department, Faculty of Pharmacy, Universidad de Concepción, Concepción, Chile
420Department of Clinical Immunology, Zealand University Hospital, Køge, Denmark
421Department of Epidemiology Research, Statens Serum Institut, Copenhagen, Denmark
422Department of Clinical Immunology, Rigshospitalet, University of Copenhagen, Copenhagen, Denmark
423Department of Infectious Diseases, Copenhagen University Hospital (Rigshospitalet), Copenhagen, Denmark
424Department of Medical Endocrinology and Metabolism, Copenhagen University Hospital (Rigshospitalet), Copenhagen, Denmark
425Department of Hematology, Rigshospitalet, Copenhagen, Denmark
426Statens Serum Institut, Copenhagen, Denmark
427Department of Pediatrics, Faculty of Medicine, Mansoura University, Mansoura, Egypt
428Department of Surgery, Faculty of Medicine, Mansoura University, Mansoura, Egypt
429Stanley Center for Psychiatric Research & Program in Medical and Population Genetics, Broad Institute of MIT and Harvard, Cambridge, MA USA
430Chest Department, Faculty of Medicine, Mansoura University, Mansoura, Egypt
431Anesthesia, Surgical Intensive Care & Pain Management Department, Faculty of Medicine, Tanta University, Tanta, Egypt
432Department of Medical Biochemistry, Faculty of Medicine, Mansoura University, Mansoura, Egypt
433Department of Tropical Medicine, Faculty of Medicine, Mansoura University, Mansoura, Egypt
434Pediatric and Neonatology, Kafr Elzayat General Hospital, Kafr El-Zayat, Egypt
435Chest Department, Faculty of Medicine, Tanta University, Tanta, Egypt
436Pediatrics Department, Faculty of Medicine, Tanta University, Tanta, Egypt
437Anesthesia, Surgical Intensive Care & Pain Management Departmen, Faculty of Medicine, Mansoura University, Mansoura, Egypt
438Department of Population Health and Reproduction, School of Veterinary Medicine, University of California, Davis, Davis, CA USA
439Department of Anaethesia and Critical Care, Faculty of Medicine, Mansoura University, Mansoura, Egypt
440Department of Internal Medicine, Faculty of Medicine, Tanta University, Tanta, Egypt
441Faculty of Science, Tanta University, Tanta, Egypt
442Department of Internal Medicine, Erasmus MC, University Medical Center, Rotterdam, The Netherlands
443Department of Epidemiology, Erasmus MC, University Medical Center, Rotterdam, The Netherlands
444Department of Internal Medicine, Erasmus MC, Rotterdam, The Netherlands
445FinnGen, Helsinki, Finland
446Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland
447Public Health, Faculty of Medicine, University of Helsinki, Helsinki, Finland
448Finnish Institute for Health and Welfare (THL), Helsinki, Finland
449Clinical and Molecular Metabolism Research Program, Faculty of Medicine, University of Helsinki, Helsinki, Finland
450Department of Computational Biology, Institut Pasteur, Université de Paris, Paris, France
451Université Paris-Saclay, CEA, Centre National de Recherche en Génomique Humaine (CNRGH), Evry, France
452University of Bordeaux, INSERM, Bordeaux Population Health Center, UMR1219, Bordeaux, France
453Department of Neurology, Institute of Neurodegenerative Diseases, Bordeaux University Hospital, Bordeaux, France
454Laboratory of Human Genetics of Infectious Diseases, Necker Branch, University of Paris, INSERM U1163, Imagine Institute, Paris, France
455St Giles Laboratory of Human Genetics of Infectious Diseases, Rockefeller Branch, The Rockefeller University, New York, NY USA
456Department of Global Health, Institut Pasteur, Université de Paris, Paris, France
457Department of Nephrology, Transplantation, Dialysis and Apheresis, Bordeaux University Hospital, Bordeaux, France
458Immunoconcept CNRS UMR 5164, Bordeaux University, Bordeaux, France
459St Giles Laboratory of Human Genetics of Infectious Diseases, Rockefeller Branch, The Rockefeller University, New York, NY USA
460Université de Paris, INSERM, IAME UMR 1137, Paris, France
461UMR-CNRS 6249 Chrono-environnement, Université Bourgogne Franche-Comté, Besançon, France
462Human Evolutionary Genetics, Institut Pasteur, UMR 2000, CNRS, Paris, France
463Chair of Human Genomics and Evolution, Collège de France, Paris, France
464Human Evolutionary Genetics Unit, Institut Pasteur, Université Paris Cité, CNRS UMR2000, Paris, France
465Translational Immunology Laboratory, Institut Pasteur, Paris, France
466Catalan Institute of Oncology, Bellvitge Biomedical Research Institute, Consortium for Biomedical Research in Epidemiology and Public Health and University of Barcelona, Barcelona, Spain
467CIBER Epidemiología y Salud Pública (CIBERESP), Madrid, Spain
468Barcelona Supercomputing Center—Centro Nacional de Supercomputación (BSC-CNS), Life & Medical Sciences, Barcelona, Spain
469Programs in Metabolism and Medical and Population Genetics, Broad Institute of MIT and Harvard, Cambridge, MA USA
470Department of Medicine, Harvard Medical School, Boston, MA USA
471Diabetes Unit and Center for Genomic Medicine, Massachusetts General Hospital, Boston, MA USA
472CIBER de Enfermedades Infecciosas (CIBERINFEC), Barcelona, Spain
473Blizard Institute, Queen Mary University of London, London, UK
474School of Basic and Medical Biosciences, Faculty of Life Sciences and Medicine, King’s College London, London, UK
475Wolfson Institute of Population Health, Queen Mary University of London, London, UK
476Medical and Population Genomics, Wellcome Sanger Institute, Hinxton, UK
477Bradford Institute for Health Research, Bradford Teaching Hospitals National Health Service (NHS) Foundation Trust, Bradford, UK
478Genes & Health, Blizard Institute, Queen Mary University of London, London, UK
479Genentech, San Francisco, CA USA
480DNA Link, Seoul, Republic of Korea
481Seoul National University Hospital Gangnam Center, Seoul, Republic of Korea
482Division of Infectious Diseases, Department of Internal Medicine, Chungnam National University School of Medicine, Daejeon, Republic of Korea
483Department of Internal Medicine, School of Medicine, Kyungpook National University, Daegu, Republic of Korea
484Division of Infectious Diseases, Department of Internal Medicine, Incheon Medical Center, Incheon, Republic of Korea
485Department of Infectious Diseases, Keimyung University Dongsan Hospital, Keimyung University School of Medicine, Daegu, Republic of Korea
486Department of Internal Medicine, Pusan National University School of Medicine and Medical Research Institute, Pusan National University Hospital, Busan, Republic of Korea
487Division of Infectious Diseases, Department of Internal Medicine, Myongji Hospital, Goyang, Republic of Korea
488Institute for Health Promotion, Graduate School of Public Health, Yonsei University, Seoul, Republic of Korea
489Department of Precision Medicine, National Institute of Health, Cheongwon-gun, Republic of Korea
490Division of Genome Science, Department of Precision Medicine, National Institute of Health, Cheongwon-gun, Republic of Korea
491Baillie Gifford Pandemic Science Hub, Centre for Inflammation Research, The Queen’s Medical Research Institute, University of Edinburgh, Edinburgh, UK
492MRC Human Genetics Unit, Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital, Edinburgh, UK
493Roslin Institute, University of Edinburgh, Easter Bush, Edinburgh, UK
494William Harvey Research Institute, Queen Mary University of London, London, UK
495Intensive Care Unit, Royal Infirmary of Edinburgh, Edinburgh, UK
496Pain Service, NHS Tayside, Ninewells Hospital and Medical School, Dundee, UK
497Centre for Inflammation Research, The Queen’s Medical Research Institute, University of Edinburgh, Edinburgh, UK
498Wellcome Centre for Human Genetics, University of Oxford, Oxford, UK
499Institute for Molecular Bioscience, The University of Queensland, Brisbane, Queensland, Australia
500Biostatistics Group, Greater Bay Area Institute of Precision Medicine (Guangzhou), Fudan University, Guangzhou, China
501Centre for Global Health Research, Usher Institute of Population Health Sciences and Informatics, Edinburgh, UK
502Edinburgh Clinical Research Facility, Western General Hospital, University of Edinburgh, Edinburgh, UK
503School of Life Sciences, Westlake University, Hangzhou, China
504Westlake Laboratory of Life Sciences and Biomedicine, Hangzhou, China
505Great Ormond Street Hospital, London, UK
506Royal Hospital for Children, Glasgow, UK
507Centre for Tropical Medicine and Global Health, Nuffield Department of Medicine, University of Oxford, Oxford, UK
508Department of Anaesthesia and Intensive Care, The Chinese University of Hong Kong, Prince of Wales Hospital, Hong Kong, China
509Department of Critical Care Medicine, Queen’s University and Kingston Health Sciences Centre, Kingston, Ontario Canada
510Wellcome-Wolfson Institute for Experimental Medicine, Queen’s University Belfast, Belfast, UK
511Department of Intensive Care Medicine, Royal Victoria Hospital, Belfast, UK
512UCL Centre for Human Health and Performance, London, UK
513Clinical Research Centre at St Vincent’s University Hospital, University College Dublin, Dublin, Ireland
514National Heart and Lung Institute, Imperial College London, London, UK
515Imperial College Healthcare NHS Trust: London, London, UK
516Heart Institute, University of Sao Paulo, Sao Paulo, Brazil
517Intensive Care National Audit & Research Centre, London, UK
518NIHR Health Protection Research Unit for Emerging and Zoonotic Infections, Institute of Infection, Veterinary and Ecological Sciences, University of Liverpool, Liverpool, UK
519Respiratory Medicine, Alder Hey Children’s Hospital, Institute in The Park, University of Liverpool, Liverpool, UK
520Department of Medicine, University of Cambridge, Cambridge, UK
521Guys and St Thomas’ Hospital, London, UK
522James Cook University Hospital, Middlesbrough, UK
523Barts Health NHS Trust, London, UK
524Royal Stoke University Hospital, Stoke-on-Trent, UK
525North Middlesex University Hospital NHS trust, London, UK
526King’s College Hospital, London, UK
527Charing Cross Hospital, St Mary’s Hospital and Hammersmith Hospital, London, UK
528The Royal Liverpool University Hospital, Liverpool, UK
529John Radcliffe Hospital, Oxford, UK
530Addenbrooke’s Hospital, Cambridge, UK
531Nottingham University Hospital, Nottingham, UK
532St George’s Hospital, London, UK
533BHRUT (Barking Havering)—Queens Hospital and King George Hospital, London, UK
534Royal Infirmary of Edinburgh, Edinburgh, UK
535Kingston Hospital, London, UK
536Queen Alexandra Hospital, Portsmouth, UK
537Royal Gwent Hospital, Newport, UK
538Royal Blackburn Teaching Hospital, Blackburn, UK
539Stepping Hill Hospital, Stockport, UK
540Northumbria Healthcare NHS Foundation Trust, North Shields, UK
541Countess of Chester Hospital, Chester, UK
542Pinderfields General Hospital, Wakefield, UK
543Ninewells Hospital, Dundee, UK
544Croydon University Hospital, Croydon, UK
545Morriston Hospital, Swansea, UK
546Queen Elizabeth University Hospital, Glasgow, UK
547Broomfield Hospital, Chelmsford, UK
548Heartlands Hospital, Birmingham, UK
549Royal Sussex County Hospital, Brighton, UK
550York Hospital, York, UK
551Queen Elizabeth Hospital, Birmingham, UK
552Royal Glamorgan Hospital, Pontyclun, UK
553Barnet Hospital, London, UK
554Wythenshawe Hospital, Manchester, UK
555Medway Maritime Hospital, Gillingham, UK
556Royal Berkshire NHS Foundation Trust, Reading, UK
557Whiston Hospital, Prescot, UK
558The Royal Oldham Hospital, Manchester, UK
559Chesterfield Royal Hospital Foundation Trust, Chesterfield, UK
560Aberdeen Royal Infirmary, Aberdeen, UK
561Royal Devon and Exeter Hospital, Exeter, UK
562Glasgow Royal Infirmary, Glasgow, UK
563Blackpool Victoria Hospital, Blackpool, UK
564Southampton General Hospital, Southampton, UK
565Ashford and St Peter’s Hospital, Chertsey, UK
566Derriford Hospital, Plymouth, UK
567East Surrey Hospital, Redhill, UK
568Poole Hospital, Poole, UK
569Royal Alexandra Hospital, Paisley, UK
570St James’s University Hospital and Leeds General Infirmary, Leeds, UK
571Bedford Hospital, Bedford, UK
572Southport and Formby District General Hospital, Ormskirk, UK
573The Tunbridge Wells Hospital and Maidstone Hospital, Kent, UK
574Queen Elizabeth Hospital, Woolwich, London, UK
575North Manchester General Hospital, Manchester, UK
576Royal Victoria Infirmary, Newcastle Upon Tyne, UK
577Hull Royal Infirmary, Hull, UK
578Manchester Royal Infirmary, Manchester, UK
579Royal Derby Hospital, Derby, UK
580Aintree University Hospital, Liverpool, UK
581Fairfield General Hospital, Bury, UK
582Norfolk and Norwich University hospital (NNUH), Norwich, UK
583Milton Keynes University Hospital, Milton Keynes, UK
584Good Hope Hospital, Birmingham, UK
585Queen Elizabeth Hospital Gateshead, Gateshead, UK
586Royal Bolton Hospital, Bolton, UK
587Tameside General Hospital, Ashton Under Lyne, UK
588Salford Royal Hospital, Manchester, UK
589Great Ormond St Hospital and UCL Great Ormond St Institute of Child Health NIHR Biomedical Research Centre, London, UK
590Southmead Hospital, Bristol, UK
591William Harvey Hospital, Ashford, UK
592Arrowe Park Hospital, Wirral, UK
593Royal Hampshire County Hospital, Winchester, UK
594Bradford Royal Infirmary, Bradford, UK
595Glan Clwyd Hospital, Bodelwyddan, UK
596Royal Bournemouth Hospital, Bournemouth, UK
597Bristol Royal Infirmary, Bristol, UK
598University Hospital North Durham, Darlington, UK
599Darlington Memorial Hospital, Darlington, UK
600Basildon Hospital, Basildon, UK
601University College Hospital, London, UK
602Whittington Hospital, London, UK
603Western General Hospital, Edinburgh, UK
604Ipswich Hospital, Ipswich, UK
605Hereford County Hospital, Hereford, UK
606Sunderland Royal Hospital, Sunderland, UK
607Queens Hospital Burton, Burton-On-Trent, UK
608Musgrove Park Hospital, Taunton, UK
609The Royal Papworth Hospital, Cambridge, UK
610University Hospital Lewisham, London, UK
611The Princess Alexandra Hospital, Harlow, UK
612University Hospital of Wales, Cardiff, UK
613West Middlesex Hospital, Isleworth, UK
614Royal Albert Edward Infirmary, Wigan, UK
615Stoke Mandeville Hospital, Aylesbury, UK
616Royal Lancaster Infirmary, Lancaster, UK
617Basingstoke and North Hampshire Hospital, Basingstoke, UK
618Worthing Hospital, Worthing, UK
619St Richard’s Hospital, Chichester, UK
620The Alexandra Hospital, Redditch and Worcester Royal Hospital, Worcester, UK
621Royal Cornwall Hospital, Truro, UK
622Watford General Hospital, Watford, UK
623Macclesfield District General Hospital, Macclesfield, UK
624Royal Surrey County Hospital, Guildford, UK
625Rotherham General Hospital, Rotherham, UK
626Craigavon Area Hospital, Craigavon, UK
627King’s Mill Hospital, Nottingham, UK
628Dumfries and Galloway Royal Infirmary, Dumfries, UK
629Prince Charles Hospital, Merthyr Tydfil, UK
630Ysbyty Gwynedd, Bangor, UK
631Royal Preston Hospital, Preston, UK
632The Great Western Hospital, Swindon, UK
633Lincoln County Hospital, Lincoln, UK
634University Hospital of North Tees, Stockton on Tees, UK
635Glangwili General Hospital, Camarthen, UK
636Southend University Hospital, Westcliff-on-Sea, UK
637Lister Hospital, Stevenage, UK
638Diana Princess of Wales Hospital, Grimsby, UK
639West Suffolk Hospital, Bury St Edmunds, UK
640Victoria Hospital, Kirkcaldy, UK
641Calderdale Royal Hospital, Halifax, UK
642Huddersfield Royal Infirmary, Huddersfield, UK
643Dorset County Hospital, Dorchester, UK
644Russell’s Hall Hospital, Dudley, UK
645Royal United Hospital, Bath, UK
646St Mary’s Hospital, Newport, UK
647George Eliot Hospital NHS Trust, Nuneaton, UK
648Yeovil Hospital, Yeovil, UK
649Forth Valley Royal Hospital, Falkirk, UK
650Frimley Park Hospital, Camberley, UK
651Chelsea & Westminster NHS Foundation Trust, London, UK
652Queen Elizabeth the Queen Mother Hospital, Margate, UK
653Royal Brompton Hospital, London, UK
654Darent Valley Hospital, Dartford, UK
655University Hospital Crosshouse, Kilmarnock, UK
656University Hospital Wishaw, Wishaw, UK
657University College Dublin, St Vincent’s University Hospital, Dublin, Ireland
658The Queen Elizabeth Hospital, King’s Lynn, UK
659Walsall Manor Hospital, Walsall, UK
660Princess Royal Hospital, Brighton, UK
661Barnsley Hospital, Barnsley, UK
662Warrington General Hospital, Warrington, UK
663Royal Victoria Hospital, Belfast, UK
664Royal Hallamshire Hospital and Northern General Hospital, Sheffield, UK
665Harefield Hospital, London, UK
666Cumberland Infirmary, Carlisle, UK
667Eastbourne District General Hospital, Eastbourne, UK
668Conquest Hospital, Saint Leonards-on-sea, UK
669Salisbury District Hospital, Salisbury, UK
670Airedale General Hospital, Keighley, UK
671Leicester Royal Infirmary, Leicester, UK
672Peterborough City Hospital, Peterborough, UK
673Hinchingbrooke Hospital, Huntingdon, UK
674Colchester General Hospital, Colchester, UK
675Princess Royal Hospital, Telford and Royal Shrewsbury Hospital, Shrewsbury, UK
676University Hospital Monklands, Airdrie, UK
677Wrexham Maelor Hospital, Wrexham, UK
678New Cross Hospital, Wolverhampton, UK
679University Hospital Hairmyres, East Kilbride, UK
680Warwick Hospital, Warwick, UK
681Sandwell General Hospital and City Hospital, Birmingham, UK
682Royal Manchester Children’s Hospital, Manchester, UK
683Gloucestershire Royal Hospital, Gloucester, UK
684University Hospitals Coventry & Warwickshire NHS Trust, Coventry, UK
685Torbay Hospital, Torquay, UK
686Pilgrim Hospital, Lincoln, UK
687Prince Philip Hospital, Lianelli, UK
688Princess of Wales Hospital, Llantrisant, UK
689Northampton General Hospital NHS Trust, Northampton, UK
690The Christie NHS Foundation Trust, Manchester, UK
691James Paget University Hospital NHS Trust, Great Yarmouth, UK
692Birmingham Children’s Hospital, Birmingham, UK
693Withybush General Hospital, Haverfordwest, UK
694Northwick Park Hospital, London, UK
695North Devon District Hospital, Barnstaple, UK
696Scunthorpe General Hospital, Scunthorpe, UK
697Royal Free Hospital, London, UK
698Raigmore Hospital, Inverness, UK
699West Cumberland Hospital, Whitehaven, UK
700Furness General Hospital, Barrow-in-Furness, UK
701Liverpool Heart and Chest Hospital, Liverpool, UK
702Scarborough General Hospital, Scarborough, UK
703Bronglais General Hospital, Aberystwyth, UK
704Alder Hey Children’s Hospital, Liverpool, UK
705Borders General Hospital, Melrose, UK
706Leighton Hospital, Cheshire, UK
707Kent & Canterbury Hospital, Canterbury, UK
708Harrogate and District NHS Foundation Trust, Harrogate, UK
709The Royal Marsden Hospital, London, UK
710Ealing Hospital, Southall, UK
711St John’s Hospital Livingston, Livingston, UK
712Wexham Park Hospital, Slough, UK
713Sheffield Children’s Hospital, Sheffield, UK
714Homerton University Hospital Foundation NHS Trust, London, UK
715National Hospital for Neurology and Neurosurgery, London, UK
716The Royal Alexandra Children’s Hospital, Brighton, UK
717Golden Jubilee National Hospital, Clydebank, UK
718Illumina Cambridge, Cambridge, UK
719Test and Trace, The Health Security Agency, Department of Health and Social Care, London, UK
720Imperial College London, London, UK
721NIHR Clinical Research Network (CRN), North West London Core Team, London, UK
722Cambridge University Hospitals NHS Foundation Trust, Cambridge, UK
723Biostatistics Group, State Key Laboratory of Biocontrol, School of Life Sciences, Sun Yat-sen University, Guangzhou, China
724Department of Infectious Diseases, Leiden University Medical Center, Leiden, The Netherlands
725Genotek, Moscow, Russia
726Helix, San Mateo, CA USA
727Center for Genomic Medicine, Desert Research Institute, Reno, NV USA
728Renown Health, Reno, NV USA
729Gujarat Biotechnology Research Centre (GBRC), Department of Science and Technology (Government of Gujarat), Gandhinagar, India
730Commissionerate of Health Medical Services and Medical Education Gandhinagar, Gandhinagar, India
731Department of Microbiology, B.J. Medical College and Civil hospital, Institute of Medical Post-Graduate Studies and Research, Ahmedabad, India
732Department of Medicine, B.J. Medical College and Civil hospital, Institute of Medical Post-Graduate Studies and Research, Ahmedabad, India
733Department of Community Medicine, Government Medical College, Surat, India
734Department of Microbiology, Government Medical College, Bhavnagar, India
735Department of General Medicine, GMERS Medical College & Hospital, Gotri, Vadodara, India
736Department of Community & Family Medicine, All India Institute of Medical Sciences, Rajkot, India
737National Laboratory of Genomics for Biodiversity (LANGEBIO), Advanced Genomics Unit (UGA), CINVESTAV, Irapuato, Guanajuato, Mexico
738Instituto Nacional de Enfermedades Respiratorias Ismael Cosío Villegas, Tlalpan, Mexico City, Mexico
739Departamento de Bioquímica, Facultad de Medicina, Universidad Nacional Autónoma de México, Coyoacán, Mexico City, Mexico
740Instituto Nacional de Salud Pública, Cuernavaca, Mexico
741International Laboratory for Human Genome Research (LIIGH), UNAM Juriquilla, Queretaro, Mexico
742Unidad de Biología Molecular y Medicina Genómica, Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán/ Instituto de Investigaciones Biomédicas, UNAM, Mexico City, Mexico
743Instituto Nacional de Medicina Genómica, Mexico City, Mexico
744Instituto Nacional de Pediatría, Mexico City, Mexico
745Escuela Superior de Medicina, Instituto Politécnico Nacional, Mexico City, Mexico
746Tecnologico de Monterrey, Escuela de Medicina y Ciencias de la Salud, Monterrey, Mexico
747Division of Pulmonary Medicine, Department of Medicine, Keio University School of Medicine, Tokyo, Japan
748Department of Statiatical Genetics, Osaka University Graduate School of Medicine, Suita, Japan
749Laboratory of Statistical Immunology, Immunology Frontier Research Center (WPI-IFReC), Osaka University, Suita, Japan
750Integrated Frontier Research for Medical Science Division, Institute for Open and Transdisciplinary Research Initiatives, Osaka University, Suita, Japan
751Division of Health Medical Intelligence, Human Genome Center, the Institute of Medical Science, The University of Tokyo, Tokyo, Japan
752Laboratory of Viral Infection I, Department of Infection Control and Immunology, Ōmura Satoshi Memorial Institute & Graduate School of Infection Control Sciences, Kitasato University, Tokyo, Japan
753Department of Surgery, Keio University School of Medicine, Tokyo, Japan
754Department of Organoid Medicine, Keio University School of Medicine, Tokyo, Japan
755Department of Infectious Diseases, Keio University School of Medicine, Tokyo, Japan
756Department of Respiratory Medicine and Clinical Immunology, Osaka University Graduate School of Medicine, Suita, Japan
757Department of Immunopathology, Immunology Frontier Research Center (WPI-IFReC), Osaka University, Suita, Japan
758Institute of Research, Tokyo Medical and Dental University, Tokyo, Japan
759Department of Insured Medical Care Management, Tokyo Medical and Dental University Hospital of Medicine, Tokyo, Japan
760Genome Medical Science Project (Toyama), National Center for Global Health and Medicine, Tokyo, Japan
761Division of Gastroenterology and Hepatology, Department of Medicine, Keio University School of Medicine, Tokyo, Japan
762M&D Data Science Center, Tokyo Medical and Dental University, Tokyo, Japan
763Kyoto University, Department of Pathology and Tumor Biology Institute for the Advanced Study of Human Biology (WPI-ASHBi), Kyoto University, Kyoto, Japan
764Department of Medicine, Center for Hematology and Regenerative Medicine, Karolinska Institute, Stockholm, Sweden
765Department of Emergency and Critical Care Medicine, Keio University School of Medicine, Tokyo, Japan
766Department of Anesthesiology, Keio University School of Medicine, Tokyo, Japan
767Department of Laboratory Medicine, Keio University School of Medicine, Tokyo, Japan
768Division of Infection Control and Prevention, Osaka University Hospital, Suita, Japan
769Department of Biomedical Ethics and Public Policy, Osaka University Graduate School of Medicine, Suita, Japan
770Center for Genomic Medicine, Kyoto University Graduate School of Medicine, Kyoto, Japan
771Department of Pulmonary Medicine, Faculty of Medicine, University of Tsukuba, Tsukuba, Japan
772Department of Neurosurgery, Faculty of Medicine, The University of Tokyo, Tokyo, Japan
773Laboratory of Immune Regulation, Department of Microbiology and Immunology, Osaka University Graduate School of Medicine, Suita, Japan
774Medical Innovation Promotion Center, Tokyo Medical and Dental University, Tokyo, Japan
775Clinical Research Center, Tokyo Medical and Dental University Hospital of Medicine, Tokyo, Japan
776Department of Medical Informatics, Tokyo Medical and Dental University Hospital of Medicine, Tokyo, Japan
777Respiratory Medicine, Tokyo Medical and Dental University, Tokyo, Japan
778Clinical Laboratory, Tokyo Medical and Dental University Hospital of Medicine, Tokyo, Japan
779Department of Pathology and Tumor Biology, Kyoto University, Kyoto, Japan
780Department of Respiratory Medicine, Juntendo University Faculty of Medicine and Graduate School of Medicine, Tokyo, Japan
781Department of Emergency and Disaster Medicine, Juntendo University Faculty of Medicine and Graduate School of Medicine, Tokyo, Japan
782Department of Cardiovascular Biology and Medicine, Juntendo University Faculty of Medicine and Graduate School of Medicine, Tokyo, Japan
783Department of Respiratory Medicine, Tokyo Women’s Medical University, Tokyo, Japan
784Department of General Medicine, Tokyo Women’s Medical University, Tokyo, Japan
785Department of Respiratory Medicine, Saitama Cardiovascular and Respiratory Center, Saitama, Japan
786Kawasaki Municipal Ida Hospital, Kawasaki, Japan
787Internal Medicine, Saitama Medical Center, JCHO (Japan Community Health Care Organization), Saitama, Japan
788Saitama City Hospital, Saitama, Japan
789Division of Infection Control, Eiju General Hospital, Tokyo, Japan
790Department of Pulmonary Medicine, Eiju General Hospital, Tokyo, Japan
791Department of Respiratory Medicine, Osaka Saiseikai Nakatsu Hospital, Osaka, Japan
792Department of Infection Control, Osaka Saiseikai Nakatsu Hospital, Osaka, Japan
793Department of Infectious Diseases, Tosei General Hospital, Seto, Japan
794Fukujuji Hospital, Kiyose, Japan
795Department of Emergency and Critical Care Medicine, Tokyo Women’s Medical University Medical Center East, Tokyo, Japan
796Department of Medicine, Tokyo Women’s Medical University Medical Center East, Tokyo, Japan
797Department of Pediatrics, Tokyo Women’s Medical University Medical Center East, Tokyo, Japan
798Japan Community Health Care Organization Kanazawa Hospital, Kanazawa, Japan
799Division of Pulmonary Medicine, Department of Internal Medicine, Federation of National Public Service Personnel Mutual Aid Associations, Tachikawa Hospital, Tachikawa, Japan
800Department of Respiratory Medicine, Japan Organization of Occupational Health and Safety, Kanto Rosai Hospital, Kawasaki, Japan
801Department of General Internal Medicine, Japan Organization of Occupational Health and Safety, Kanto Rosai Hospital, Kawasaki, Japan
802Department of Emergency and Critical Care Medicine, Kansai Medical Univrsity General Medical Center, Kirakata, Japan
803Department of Respiratory Medicine, Kitasato University Kitasato Institute Hospital, Tokyo, Japan
804Ishikawa Prefectural Central Hospital, Kanazawa, Japan
805Internal Medicine, Sano Kosei General Hospital, Sano, Japan
806Saiseikai Yokohamashi Nanbu Hospital, Yokohama, Japan
807Kanagawa Cardiovascular and Respiratory Center, Yokohama, Japan
808Saiseikai Utsunomiya Hospital, Utsunomiya, Japan
809Department of Respiratory Medicine, KKR Sapporo Medical Center, Sapporo, Japan
810Internal Medicine, Internal Medicine Center, Showa University Koto Toyosu Hospital, Tokyo, Japan
811Department of Respiratory Medicine, Toyohashi Municipal Hospital, Toyohashi, Japan
812Keiyu Hospital, Yokohama, Japan
813Department of Rheumatology, National Hospital Organization Hokkaido Medical Center, Sapporo, Japan
814Department of Respiratory Medicine, National Hospital Organization Tokyo Medical Center, Tokyo, Japan
815Department of Allergy, National Hospital Organization Tokyo Medical Center, Tokyo, Japan
816Department of General Internal Medicine and Infectious Diseases, National Hospital Organization Tokyo Medical Center, Tokyo, Japan
817Japanese Red Cross Musashino Hospital, Musashino, Japan
818Department of Respiratory Medicine, Tohoku University Graduate School of Medicine, Sendai, Japan
819Department of Internal Medicine, Division of Respiratory Medicine, Nihon University School of Medicine, Tokyo, Japan
820Department of Emergency and Critical Care Medicine, St Marianna University School of Medicine, Kawasaki, Japan
821Division of General Internal Medicine, Department of Internal Medicine, St Marianna University School of Medicine, Kawasaki, Japan
822National Hospital Organization Kanazawa Medical Center, Kanazawa, Japan
823Division of Infectious Diseases and Respiratory Medicine, Department of Internal Medicine, National Defense Medical College, Saitama, Japan
824Department of Emergency and Critical Care Medicine, Faculty of Medicine, Fukuoka University, Fukuoka, Japan
825Department of Infection Control, Fukuoka University Hospital, Fukuoka, Japan
826Tokyo Saiseikai Central Hospital, Tokyo, Japan
827Department of Internal Medicine, Fukuoka Tokushukai Hospital, Kasuga, Japan
828Department of Infectious Disease and Clinical Research Institute, National Hospital Organization Kyushu Medical Center, Fukuoka, Japan
829Department of Respirology, National Hospital Organization Kyushu Medical Center, Division of Respirology, Rheumatology, and Neurology, Department of Internal Medicine, Kurume University School of Medicine, Fukuoka, Japan
830Department of Infectious Disease, National Hospital Organization Kyushu Medical Center, Fukuoka, Japan
831Matsumoto City Hospital, Matsumoto, Japan
832Uji-Tokushukai Medical Center, Uji, Japan
833Department of Respiratory Medicine, Nagoya University Graduate School of Medicine, Nagoya, Japan
834Department of Respiratory Medicine, Fujisawa City Hospital, Fujisawa, Japan
835Sapporo City General Hospital, Sapporo, Japan
836Department of Emergency and Critical Care Medicine, Chiba University Graduate School of Medicine, Chiba, Japan
837Division of Respiratory Medicine, Social Welfare Organization Saiseikai Imperial Gift Foundation, Saiseikai Kumamoto Hospital, Kumamoto, Japan
838Department of Anesthesiology and Intensive Care Medicine, Kyoto Prefectural University of Medicine, Kyoto, Japan
839Ome Municipal General Hospital, Ome, Japan
840Hanwa Daini Hospital, Osaka, Japan
841Department of Respiratory Internal Medicine, St Marianna University School of Medicine, Yokohama-City Seibu Hospital, Yokohama, Japan
842Division of Hematology, Department of Internal Medicine, St Marianna University Yokohama-City Seibu Hospital, Yokohama, Japan
843Division of Pulmonary Medicine, Department of Medicine, Tokai University School of Medicine, Tokai University School of Medicine, Tokyo, Japan
844Division of Pulmonary Medicine, Department of Medicine, Tokai University School of Medicine, Tokyo, Japan
845National Hospital Organization Kumamoto Medical Center, Kumamoto, Japan
846Department of Respiratory Medicine, Tokyo Medical University Hospital, Tokyo, Japan
847Department of Respiratory Medicine, Japanese Red Cross Medical Center, Tokyo, Japan
848JA Toride Medical Hospital, Toride, Japan
849Japan Organization of Occupational Health and Safety Okayama Rosai Hospital, Okayama, Japan
850Emergency and Disaster Medicine, Graduate School of Medicine, Gifu University School of Medicine, Gifu, Japan
851Niigata University, Niigata, Japan
852National Hospital Organization Kyoto Medical Center, Kyoto, Japan
853Research Institute for Diseases of the Chest, Graduate School of Medical Sciences, Kyushu University, Fukuoka, Japan
854Department of Medicine and Biosystemic Science, Kyushu University Graduate School of Medical Sciences, Fukuoka, Japan
855Department of Emergency and Critical Care Medicine, Tsukuba University, Tsukuba, Japan
856Department of Nephrology, Faculty of Medicine, University of Tsukuba, Tsukuba, Japan
857Department of Hematology, Faculty of Medicine, University of Tsukuba, Tsukuba, Japan
858National Hospital Organization Tokyo Hospital, Tokyo, Japan
859Fujioka General Hospital, Fujioka, Japan
860Division of Respiratory Medicine and Allergology, Department of Medicine, School of Medicine, Showa University, Tokyo, Japan
861Department of Pulmonary Medicine, Fukushima Medical University, Fukushima, Japan
862Kansai Electric Power Hospital, Osaka, Japan
863Kumamoto City Hospital, Kumamoto, Japan
864Department of Emergency and Critical Care Medicine, Tokyo Metropolitan Police Hospital, Tokyo, Japan
865Department of Respiratory Medicine, International University of Health and Welfare Shioya Hospital, Narita, Japan
866Department of Clinical Laboratory, International University of Health and Welfare Shioya Hospital, Narita, Japan
867National Hospital Organization Saitama Hospital, Saitama, Japan
868Department of Respiratory Medicine, Gunma University Graduate School of Medicine, Maebashi, Japan
869Department of Orthopedic Surgery, Tokyo Medical University, Ibaraki Medical Center, Tokyo, Japan
870Department of Internal Medicine, Kiryu Kosei General Hospital, Kiryu, Japan
871Daini Osaka Police Hospital, Osaka, Japan
872Genome Medical Science Project (Konodai), National Center for Global Health and Medicine, Chiba, Japan
873Ministry Of Health, Amman, Jordan
874School of Dentistry, The University of Jordan, Amman, Jordan
875School of Medicine, The University of Jordan, Amman, Jordan
876Cell Therapy Center, The University of Jordan, Amman, Jordan
877King Hussein Cancer Center, Amman, Jordan
878King Abdullah University Hospital, Amman, Jordan
879Gardens Hospital, Amman, Jordan
880Centre for Heart Lung Innovation, University of British Columbia, Vancouver, British Columbia Canada
881Division of Respiratory Medicine, Faculty of Medicine, University of British Columbia, Vancouver, British Columbia Canada
882Institut Universitaire de Cardiologie et de Pneumologie de Québec—Université Laval, Quebec City, Quebec Canada
883Department of Genetics and Genomic Sciences, Icahn School of Medicine at Mount Sinai, New York, NY USA
884Global Health, University of Washington, Seattle, WA USA
885Monoceros Bio, San Diego, CA USA
886Department of Pathology and Medical Biology Groningen, University Medical Centre Groningen, University of Groningen, Groningen, The Netherlands
887GRIAC Research Institute, University Medical Centre Groningen, University of Groningen, Groningen, The Netherlands
888Department of Pulmonary Diseases, University Medical Center Groningen, University of Groningen, Groningen, The Netherlands
889Center for Genomic Medicine, Massachusetts General Hospital, Boston, MA USA
890Institute of Epidemiology and Preventive Medicine, National Taiwan University, Taipei, Taiwan
891Master of Public Health Program, National Taiwan University, Taipei, Taiwan
892Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital, Boston, MA USA
893Brigham and Women’s Hospital, Boston, MA USA
894Psychiatric and Neurodevelopmental Genetics Unit, Center for Genomic Medicine, Massachusetts General Hospital, Boston, MA USA
895Department of Neurology, Massachusetts General Hospital, Boston, MA USA
896Division of General Internal Medicine, Massachusetts General Hospital and Department of Medicine, Harvard Medical School and Program in Medical and Population GeneticsBroad Institute, Boston, MA USA
897Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Broad Institute of MIT and Harvard, Harvard Medical School, Boston, MA USA
898Department of Human Genetics, David Geffen School of Medicine at UCLA, Los Angeles, CA USA
899Departamento de Neurología y Psiquiatría, Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Tlalpan, Mexico City, Mexico
900Dirección de Medicina, Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Tlalpan, Mexico City, Mexico
901Infectología, Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Tlalpan, Mexico City, Mexico
902Clínica de Obesidad, Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Tlalpan, Mexico City, Mexico
903Unidad de Investigación en Enfermedades Metabólicas (UIEM), Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Tlalpan, Mexico City, Mexico
904Tecnologico de Monterrey, The Institute for Obesity Research, Nuevo León, Monterrey, Mexico
905Tecnologico de Monterrey, Escuela de Medicina y Ciencias de la Salud, Monterrey, Mexico
906Departamento de Endocrinología y Metabolismo de Lípidos, Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Tlalpan, Mexico City, Mexico
907Departamento de Psiquiatría y Salud Mental, Facultad de Medicina, Universidad Nacional Autónoma de México, Mexico City, Mexico
908Unidad de Biología Molecular y Medicina Genómica, Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán/Instituto de Investigaciones Biomédicas UNAM, Tlalpan, México
909Universidad Nacional Autónoma de México, Mexico City, Mexico
910Institute for Precision Health, David Geffen School of Medicine at UCLA, Los Angeles, CA USA
911Unidad de Biología Molecular y Medicina Genómica, Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, CDMX, Tlalpan, México
912Universidad Autónoma Metropolitana, Mexico City, Mexico
913Departamento de Medicina Genómica y Toxicología Ambiental, Instituto de Investigaciones Biomédicas, UNAM, Mexico City, Mexico
914Department of Computer Science, School of Engineering, UCLA, Los Angeles, CA USA
915Unidad de Biología Molecular y Medicina Genómica, Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Tlalpan, Mexico City, Mexico
916Laboratorio de Microbiología, Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Tlalpan, Mexico City, Mexico
917Laboratorio de Neuropsicología y Cognición, Facultad de Psicología, Universidad Nacional Autónoma de México, Mexico City, Mexico
918Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Tlalpan, Mexico City, Mexico
919Facultad de Psicología, Universidad Nacional Autónoma de México, Mexico City, Mexico
920Facultad de Medicina, Universidad Nacional Autónoma de México, Mexico City, Mexico
921Laboratorio de Neuropsicología, Facultad de Medicina, Universidad Nacional Autónoma de México, UNAM, Coyoacán, Mexico City, Mexico
922Research Division, School of Medicine, Universidad Nacional Autónoma de México, Mexico City, Mexico
923Department of Computational Medicine, David Geffen School of Medicine at UCLA, Los Angeles, CA USA
924Department of Pathology and Laboratory Medicine, David Geffen School of Medicine at UCLA, Los Angeles, CA USA
925Division of Immunology, Allergy and Rheumatology, Department of Pediatrics, UCLA, Los Angeles, CA USA
926Department of Microbiology, Immunology, and Molecular Genetics, UCLA, Los Angeles, CA USA
927Department of Neurology, David Geffen School of Medicine at UCLA, Los Angeles, CA USA
928Department of Psychiatry and Biobehavioral Sciences, Semel Institute, David Geffen School of Medicine at UCLA, Los Angeles, CA USA
929Center for Autism Research and Treatment, Semel Institute, David Geffen School of Medicine at UCLA, Los Angeles, CA USA
930Institute for Precision Health, Los Angeles, CA USA
931Dirección de Nutrición, Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Tlalpan, Mexico City, Mexico
932Deutsches Herzzentrum der Charité, Universitätsmedizin, Berlin, Germany
933Charite Universitätsmedizin Berlin, Berlin, Germany
934Department of Tropical Medicine, Bernhard Nocht Institute for Tropical Medicine, Hamburg, Germany
935Department of Medicine I, University Medical Centre Hamburg-Eppendorf, Hamburg, Germany
936Qatar Genome Program, Qatar Foundation Research, Development and Innovation, Qatar Foundation, Doha, Qatar
937Directorate of Research, Shifa Tameer-e-Millat University, Islamabad, Pakistan
938Shifa Tameer-e-Millat University, Islamabad, Pakistan
939Department of Vascular Surgery, Shifa International Hospital, Islamabad, Pakistan
940Department of Infectious Diseases, Shifa International Hospital, Islamabad, Pakistan
941Shifa International Hospital, Islamabad, Pakistan
942Department of Pathology, Shifa International Hospital, Islamabad, Pakistan
943MNM Bioscience, Cambridge, MA USA
944Faculty of Physics, Adam Mickiewicz University, Poznań, Poland
945Department of Medical Chemistry and Laboratory Medicine, Poznan University of Medical Sciences, Poznan, Poland
946Central Clinical Hospital of Ministry of the Interior and Administration in Warsaw, Warsaw, Poland
947Institute of Genetics and Biotechnology, Faculty of Biology, University of Warsaw, Warsaw, Poland
948Genomics Division, Instituto Tecnológico y de Energías Renovables, Santa Cruz de Tenerife, Spain
949Research Unit, Hospital Universitario N.S. de Candelaria, Santa Cruz de Tenerife, Spain
950Centre for Biomedical Network Research on Respiratory Diseases (CIBERES), Instituto de Salud Carlos III, Madrid, Spain
951Facultad de Ciencias de la Salud, Universidad Fernando Pessoa Canarias, Las Palmas de Gran Canaria, Spain
952Centro Nacional de Genotipado (CEGEN), Universidade de Santiago de Compostela, Santiago de Compostela, Spain
953Centre for Biomedical Network Research on Rare Diseases (CIBERER), Instituto de Salud Carlos III, Madrid, Spain
954Fundación Pública Galega de Medicina Xenómica, Sistema Galego de Saúde (SERGAS) Santiago de Compostela, Santiago de Compostela, Spain
955Instituto de Investigación Sanitaria de Santiago (IDIS), Santiago de Compostela, Spain
956Centro Singular de Investigación en Medicina Molecular y Enfermedades Crónicas (CIMUS), Universidade de Santiago de Compostela, Santiago de Compostela, Spain
957Hospital General Santa Bárbara de Soria, Soria, Spain
958Biocruces Bizkai HRI, Bizkaia, Spain
959Cruces University Hospital, Osakidetza, Bizkaia, Spain
960Fundació Docència I Recerca Mutua Terrassa, Barcelona, Spain
961Hospital General de Occidente, Zapopan, Mexico
962Centro Universitario de Tonalá, Universidad de Guadalajara, Tonalá, Mexico
963Centro de Investigación Multidisciplinario en Salud, Universidad de Guadalajara, Tonalá, Mexico
964Laboratorio de Vigilancia Molecular Aplicada, Escola Tecnica de Saúde, Pará, Brazil
965Genetics Postgraduate Program, Federal University of Pernambuco, Recife, Brazil
966Cardiovascular Genetics Center, Institut d’Investigació Biomèdica Girona (IDIBGI), Girona, Spain
967Medical Science Department, School of Medicine, University of Girona, Girona, Spain
968Centre for Biomedical Network Research on Cardiovascular Diseases (CIBERCV), Instituto de Salud Carlos III, Madrid, Spain
969Cardiology Service, Hospital Josep Trueta, Girona, Spain
970Institute of Biomedicine of Seville (IBiS), Consejo Superior de Investigaciones Científicas (CSIC), University of Seville, Virgen del Rocio University Hospital, Seville, Spain
971Departamento de Medicina, Hospital Universitario Virgen del Rocío, Universidad de Sevilla, Seville, Spain
972Centre for Biomedical Network Research on Epidemiology and Public Health (CIBERESP), Instituto de Salud Carlos III, Madrid, Spain
973Instituto de Biomedicina de Sevilla, Seville, Spain
974Osakidetza, Cruces University Hospital, Bizkaia, Spain
975Centre for Biomedical Network Research on Diabetes and Metabolic Associated Diseases (CIBERDEM), Instituto de Salud Carlos III, Madrid, Spain
976University of Pais Vasco, UPV/EHU, Bizkaia, Spain
977Hospital Universitario Río Hortega, Valladolid, Spain
978Servicio de Medicina intensiva, Complejo Hospitalario Universitario de A Coruña (CHUAC), Sistema Galego de Saúde (SERGAS), A Coruña, Spain
979Instituto Aragonés de Ciencias de la Salud (IACS), Zaragoza, Spain
980Instituto Investigación Sanitaria Aragón (IIS-Aragon), Zaragoza, Spain
981Unidad Diagnóstico Molecular. Fundación Rioja Salud, La Rioja, Spain
982IDIVAL, Santander, Spain
983Universidad de Cantabria, Santander, Spain
984Hospital U M Valdecilla, Santander, Spain
985Hospital Universitario de Getafe, Servicio de Genética, Madrid, Spain
986IIS La Fe, Plataforma de Farmacogenética, Valencia, Spain
987Departamento de Farmacología, Universidad de Valencia, Valencia, Spain
988Facultad de Ciencias, Universidad de los Andes, Bogotá, Colombia
989SIGEN Alianza Universidad de los Andes—Fundación Santa Fe de Bogotá, Bogotá, Colombia
990Hospital General de Segovia, Medicina Intensiva, Segovia, Spain
991Instituto de Biomedicina (IBIOMED), Universidad de León, León, Spain
992Departamento Patologia y Laboratorios, Fundación Santa Fe de Bogota, Bogotá, Colombia
993Unidad de Genética y Genómica Islas Baleares, Islas Baleares, Spain
994Unidad de Diagnóstico Molecular y Genética Clínica, Hospital Universitario Son Espases, Islas Baleares, Spain
995Faculdade de Medicina, Universidade de Brasília, Brasília, Brazil
996Programa de Pós-Graduação em Ciências Médicas, Universidade de Brasília, Brasília, Brazil
997Programa de Pós-Graduação em Ciências da Saúde, Universidade de Brasília, Brasília, Brazil
998Unidad Cuidados Intensivos, Hospital El Bierzo, León, Spain
999Medicina Interna, Hospital Universitario Mostoles, Madrid, Spain
1000Universidad Francisco de Vitoria, Madrid, Spain
1001Departamento de Analises Clinicas e Toxicologicas, Universidade Federal do Rio Grande do Norte, Natal, Brazil
1002Programa de Pós-Graduação em Biologia Animal, Universidade de Brasília, Brasília, Brazil
1003Programa de Pós-Graduação Profissional em Ensino de Biologia, Universidade de Brasília, Brasília, Brazil
1004Department of Child and Adolescent Psychiatry, Institute of Psychiatry and Mental Health, Hospital General Universitario Gregorio Marañón (IiSGM), Madrid, Spain
1005Centre for Biomedical Network Research on Mental Health (CIBERSAM), Instituto de Salud Carlos III, Madrid, Spain
1006School of Medicine, Universidad Complutense, Madrid, Spain
1007Núcleo de Pesquisas em Oncologia, Universidade Federal do Pará, Belém, Brazil
1008Infectious Diseases, Microbiota and Metabolism Unit, Center for Biomedical Research of La Rioja (CIBIR), Logroño, Spain
1009Inditex, A Coruña, Spain
1010GENYCA, Madrid, Spain
1011Clinica Comfamiliar Risaralda, Pereira, Colombia
1012Neurometabolic Diseases Laboratory, Bellvitge Biomedical Research Institute (IDIBELL), L’Hospitalet de Llobregat, Spain
1013Catalan Institution of Research and Advanced Studies (ICREA), Barcelona, Spain
1014Unidad de Infección Viral e Inmunidad, Centro Nacional de Microbiología (CNM), Instituto de Salud Carlos III (ISCIII), Madrid, Spain
1015Centro de Investigación Biomédica en Red de Enfermedades Infecciosas (CIBERINFEC), Instituto de Salud Carlos III, Madrid, Spain
1016Unidad de Cuidados Intensivos, Hospital Clínico Universitario de Santiago (CHUS), Sistema Galego de Saúde (SERGAS), Santiago de Compostela, Spain
1017Department of Preventive Medicine and Public Health, School of Medicine, Universidad Autónoma de Madrid, Madrid, Spain
1018IdiPaz (Instituto de Investigación Sanitaria Hospital Universitario La Paz), Madrid, Spain
1019IMDEA-Food Institute, CEI UAM+CSIC, Madrid, Spain
1020Complejo Asistencial Universitario de León, León, Spain
1021Servicio de Análisis Clínicos e Inmunología, Hospital Universitario Virgen de las Nieves, Granada, Spain
1022Departamento Bioquímica, Biología Molecular e Inmunología III, Universidad de Granada, Granada, Spain
1023Genomics of Complex Diseases Unit, Research Institute of Hospital de la Santa Creu i Sant Pau, IIB Sant Pau, Barcelona, Spain
1024Servicio de Anestesiologia y Reanimación, Hospital Clinico Universitario de Valladolid, Valladolid, Spain
1025Departamento de Cirugía, Universidad de Valladolid, Valladolid, Spain
1026Hospital de Niños Ricardo Gutierrez, Buenos Aires, Argentina
1027Fundación Universitaria de Ciencias de la Salud, Bogotá, Colombia
1028Familial Cancer Clinical Unit, Spanish National Cancer Research Centre, Madrid, Spain
1029Life Science Research Center, Universidad Simón Bolívar, Barranquilla, Colombia
1030Instituto Murciano de Investigación Biosanitaria (IMIB-Arrixaca), Murcia, Spain
1031Sección Genética Médica—Servicio de Pediatría, Hospital Clínico Universitario Virgen de la Arrixaca, Servicio Murciano de Salud, Murcia, Spain
1032Departamento Cirugía, Pediatría, Obstetricia y Ginecología, Facultad de Medicina, Universidad de Murcia (UMU), Murcia, Spain
1033Grupo Clínico Vinculado, Centre for Biomedical Network Research on Rare Diseases (CIBERER), Instituto de Salud Carlos III, Madrid, Spain
1034Department of Genetics & Genomics, Instituto de Investigación Sanitaria-Fundación Jiménez Díaz University Hospital—Universidad Autónoma de Madrid (IIS-FJD, UAM), Madrid, Spain
1035Human Genotyping-CEGEN Unit, Spanish National Cancer Research Centre, Madrid, Spain
1036Instituto de Genética Médica y Molecular (INGEMM), Hospital Universitario La Paz-IDIPAZ, Madrid, Spain
1037ERN-ITHACA—European Reference Network, Madrid, Spain
1038Unit of Infectious Diseases, Hospital Universitario 12 de Octubre, Instituto de Investigación Sanitaria Hospital 12 de Octubre (imas12), Madrid, Spain
1039Spanish Network for Research in Infectious Diseases (REIPI RD16/0016/0002), Instituto de Salud Carlos III, Madrid, Spain
1040Pediatric Neurology Unit, Department of Pediatrics, Navarra Health Service Hospital, Pamplona, Spain
1041Navarra Health Service, NavarraBioMed Research Group, Pamplona, Spain
1042Neumología, Hospital Universitario Virgen Macarena, Seville, Spain
1043Spanish National Cancer Research Center, CNIO Biobank, Madrid, Spain
1044Universidad Católica San Antonio de Murcia (UCAM), Murcia, Spain
1045Servicio de Medicina Interna-Unidad de Enfermedades Infecciosas, Hospital Universitario de Salamanca-IBSAL, Salamanca, Spain
1046Universidad de Salamanca, Salamanca, Spain
1047Hospital Universitario Mutua Terrassa, Barcelona, Spain
1048Xenética Cardiovascular, Instituto de Investigación Sanitaria de Santiago (IDIS), Santiago de Compostela, Spain
1049Servicio de Medicina Interna, Hospital Universitario de Salamanca-IBSAL, Salamanca, Spain
1050Oncology and Genetics Unit, Instituto de Investigacion Sanitaria Galicia Sur, Xerencia de Xestion Integrada de Vigo-Servizo Galego de Saúde, Vigo, Spain
1051Departament of Microgravity and Translational Regenerative Medicine, Otto von Guericke University, Magdeburg, Germany
1052Unidad de Genética, Hospital Universitario Mostoles, Madrid, Spain
1053Preventive Medicine Department, Instituto de Investigacion Sanitaria Galicia Sur, Xerencia de Xestion Integrada de Vigo-Servizo Galego de Saúde, Vigo, Spain
1054Servicio de Cardiología, Hospital Universitario de Salamanca-IBSAL, Salamanca, Spain
1055Instituto Regional de Investigación en Salud-Universidad Nacional de Caaguazú, Caaguazú, Paraguay
1056Urgencias Hospitalarias, Complejo Hospitalario Universitario de A Coruña (CHUAC), Sistema Galego de Saúde (SERGAS), A Coruña, Spain
1057Grupo de Investigación en Interacciones Gen-Ambiente y Salud (GIIGAS), Instituto de Biomedicina (IBIOMED), Universidad de León, León, Spain
1058Ministerio de Salud Ciudad de Buenos Aires, Buenos Aires, Argentina
1059Data Analysis Department, Instituto de Investigación Sanitaria-Fundación Jiménez Díaz University Hospital, Universidad Autónoma de Madrid (IIS-FJD, UAM), Madrid, Spain
1060Department of Immunology, Hospital Universitario 12 de Octubre, Madrid, Spain
1061Transplant Immunology and Immunodeficiencies Group, Instituto de Investigación Sanitaria Hospital 12 de Octubre (imas12), Madrid, Spain
1062Department of Immunology, Ophthalmology and ENT, Universidad Complutense de Madrid, Madrid, Spain
1063Unidad de Investigación Médica en Enfermedades Infecciosas y Parasitarias, Centro Médico Nacional Siglo XXI, Instituto Mexicano del Seguro Social (IMSS), Mexico City, Mexico
1064Centro de Investigación Biomédica en Red de Enfermedades Infecciosas (CIBERINFEC), Instituto de Salud Carlos III (ISCIII), Madrid, Spain
1065Departamento de Ensino e Pesquisa, Hospital Ophir Loyola, Belém, Brazil
1066Allergy Unit, Hospital Infanta Elena, Madrid, Spain
1067Instituto de Investigación Sanitaria-Fundación Jiménez Díaz University Hospital—Universidad Autónoma de Madrid (IIS-FJD, UAM), Madrid, Spain
1068Faculty of Medicine, Universidad Francisco de Vitoria, Madrid, Spain
1069Hospital Universitario Infanta Leonor, Madrid, Spain
1070Complutense University of Madrid, Madrid, Spain
1071Gregorio Marañón Health Research Institute (IiSGM), Madrid, Spain
1072Haemostasis and Thrombosis Unit, Hospital de la Santa Creu i Sant Pau, IIB Sant Pau, Barcelona, Spain
1073Servicio de Hematologia y Hemoterapia, Hospital Clinico Universitario de Valladolid, Valladolid, Spain
1074University Hospital of Burgos, Burgos, Spain
1075Department of Clinical Immunology, Aarhus University Hospital, Aarhus, Denmark
1076Department of Clinical Immunology, Copenhagen University Hospital—Rigshospitalet, Copenhagen, Denmark
1077deCODE genetics, Reykjavik, Iceland
1078Department of Finance Copenhagen Business School, Copenhagen, Denmark
1079Danish Headache Center, Department of Neurology, Copenhagen University Hospital, Rigshospitalet—Glostrup, Copenhagen, Denmark
1080Centre for Cancer Research, Danish Cancer Society, Copenhagen, Denmark
1081Department of Dermatology, Zealand University hospital, Roskilde, Denmark
1082Department of Biomedicine, Aarhus University, Aarhus, Denmark
1083Institute of Biological Psychiatry, Mental Health Centre Sct. Hans, Copenhagen University Hospital, Roskilde, Denmark
1084deCODE Genetics, Reykjavik, Iceland
1085Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark
1086Department of Clinical Immunology, Odense University Hospital, Odense, Denmark
1087Department of Neuroscience, Karolinska Institutet, Stockholm, Sweden
1088Max Planck Institute for Evolutionary Anthropology, Leipzig, Germany
1089Stanley Center for Psychiatric Research & Program in Medical and Population Genetics, Cambridge, UK
1090Anaesthesiology and Intensive Care Medicine, Department of Surgical Sciences, Uppsala University, Uppsala, Sweden
1091Integrative Physiology, Department of Medical Cell Biology, Uppsala University, Uppsala, Sweden
1092Hedenstierna Laboratory, CIRRUS, Anaesthesiology and Intensive Care Medicine, Department of Surgical Sciences, Uppsala University, Uppsala, Sweden
1093Division Anesthesiology and Intensive Care, CLINTEC, Karolinska Institutet, Stockholm, Sweden
1094Department of Genetic Epidemiology, University of Regensburg, Regensburg, Germany
1095Institute of Medical Microbiology and Hygiene, Molecular Microbiology (Virology), University of Regensburg, Regensburg, Germany
1096Virologisches Institut—Klinische und Molekulare Virologie, Universitätsklinikum Erlangen, Friedrich-Alexander Universität Erlangen-Nürnberg (FAU), Erlangen, Germany
1097Institute of Clinical Microbiology and Hygiene, University Hospital, Regensburg, Germany
1098Department of Computer Science, School of Engineering, University of California Los Angeles, Los Angeles, CA USA
1099University of California Los Angeles, Los Angeles, CA USA
1100Department of Psychiatry and Biobehavioral Sciences, David Geffen School of Medicine at University of California Los Angeles, Los Angeles, CA USA
1101Division of Immunology, Allergy and Rheumatology, Department of Pediatrics, Department of Microbiology, Immunology and Molecular Genetics, University of California Los Angeles, Los Angeles, CA USA
1102Departments of Neurology, Psychiatry and Human Genetics, Center for Autism Research and Treatment, Institute for Precision Health, Los Angeles, CA USA
1103Department of Pathology and Laboratory Medicine, Human Genetics, David Geffen School of Medicine at UCLA, Los Angeles, CA USA
1104Bioinformatics IDP, University of California Los Angeles, Los Angeles, CA USA
1105Department of Neurology, David Geffen School of Medicine at University of California Los Angeles, Los Angeles, CA USA
1106Departments of Human Genetics and Urology, David Geffen School of Medicine at University of California Los Angeles, Los Angeles, CA USA
1107Big Data Institute, Nuffield Department of Population Health, University of Oxford, Li Ka Shing Centre for Health Information and Discovery, Oxford, UK
1108Genomics PLC, Oxford, UK
1109Nuffield Department of Medicine, Experimental Medicine Division, University of Oxford, John Radcliffe Hospital, Oxford, UK
1110Public Health England, Field Service, Addenbrooke’s Hospital, Cambridge, UK
1111Public Health England, Data and Analytical Services, National Infection Service, London, UK
1112Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX USA
1113Program in Bioinformatics and Integrative Genomics, Harvard Medical School, Boston, MA USA
1114Program in Biological and Biomedical Sciences, Harvard Medical School, Boston, MA USA
1115Center for Economic and Social Research, University of Southern California, Los Angeles, CA USA
1116Department of Economics, University of Southern California, Los Angeles, CA USA
1117Institute for Molecular Medicine Finland (FIMM), Univerisity of Helsinki, Helsinki, Finland
1118Wellcome Centre for Human Genetics, University of Oxford, Oxford, UK
1119Department of Clinical Research and Leadership, George Washington University, Washington, DC USA
1120Institute for Biomedical Research of Barcelona (IIBB), National Spanish Research Council (CSIC), Madrid, Spain
1121Institut d’Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS), Barcelona, Spain
1122Institute of Biomedicine of Valencia (IBV), CSIC, Valencia, Spain
1123Network Center for Biomedical Research on Neurodegenerative Diseases CIBERNED, Instituto de Salud Carlos III (ISCIII), Valencia, Spain
1124Institute for Biomedical Research of Barcelona (IIBB), National Spanish Research Council (CSIC), Madrid, Spain
1125Department of Neurology, Hospital Universitari MútuaTerrassa, Fundació Docència i Recerca MútuaTerrassa, Terrassa, Spain
1126Department of Molecular and Cell Biology, Centro Nacional de Biotecnología (CNB-CSIC), Campus Universidad Autónoma de Madrid, Madrid, Spain
1127Instituto de Física de Cantabria (IFCA-CSIC), Santander, Spain
1128Hospital Clínic, IDIBAPS, Barcelona, Spain
1129Hospital Clínic, Barcelona, Spain
1130Hospital Clínic, IDIBAPS, School of Medicine, University of Barcelona Barcelona, Barcelona, Spain
1131IDIBAPS, Barcelona, Spain
1132IIBB-CSIC, Barcelona, Spain
1133Servicio de Salud del Principado de Asturias, Oviedo, Spain
1134Hospital Mutua de Terrassa, Terrassa, Spain
1135Vall d’Hebron Research Institute, Vall d’Hebron Barcelona Hospital Valle Hebrón Campus, Barcelona, Spain
1136Unidad de Excelencia Instituto de Biomedicina y Genética Molecular (IBGM, Universidad de Valladolid-CSIC), Valladolid, Spain
1137Hospital Clínico Universitario de Valladolid (SACYL), Valladolid, Spain
1138Department of Neurology, University Hospital of Albacete, Albacete, Spain
1139Research Unit, University Hospital of Albacete, Albacete, Spain
1140Department of Neurology, Biomedical Research Institute Sant Pau (IIB Sant Pau), Hospital de la Santa Creu i Sant Pau, Barcelona, Spain
1141Hospital Universitario Ramón Y Cajal, IRYCIS, Madrid, Spain
1142Institute de Biomedicine of Seville, IBiS/Hospital Universitario Virgen del Rocío/CSIC/University of Seville & Department of Neurology, Hospital Universitario Virgen Macarena, Seville, Spain