Abstract
Klebsiella pneumoniae resistant to ceftazidime was isolated from six adult women and two neonates hospitalized between July and November 1993 in the Department of Obstetrics and Gynecology of Boucicaut Hospital (Paris, France). The epidemiological investigation revealed a notably short delay (less than 48 h) between admission and contamination of the six adults and peripartum transmission to the neonates. The only environmental source of ceftazidime-resistant K. pneumoniae was the ultrasonography coupling gel used in the emergency room. Phenotypic (biotyping and antibiotyping) and genotypic (plasmid profile and pulsed-field gel electrophoresis) analysis of all the clinical isolates indicated the spread of a single strain. It produced SHV-5 and TEM-1 β-lactamases, as demonstrated by isoelectric focusing and gene sequencing. The risk of cross-contamination in ultrasonography procedures is usually low and had not been associated so far with bacteria producing an extended-spectrum β-lactamase (ESBL). Furthermore, this is the first time an epidemic of an SHV-5 ESBL-producing member of the family Enterobacteriaceae has been reported from a French hospital.
Since they were identified in the early eighties, isolates of Klebsiella pneumoniae producing extended-spectrum β-lactamases (ESBLs) have been a major cause of concern worldwide, especially in France, where numerous outbreaks and a variety of enzymes have been reported (for references, see reference 15). Most ESBLs are plasmid-encoded enzymes derived from TEM- or SHV-type β-lactamases by one or more amino acid substitutions which confer resistance to broad-spectrum cephalosporins (12). Epidemic spread is likely to occur and therapeutic choices will be limited since many ESBL-producing strains are also resistant to aminoglycosides and to other antimicrobial agents. Risk factors for acquisition by patients of ESBL-producing Klebsiella include prolonged hospital stay, prior antimicrobial therapy, and treatment in an intensive care unit (22). The lower digestive tract of colonized patients is the main reservoir of these microorganisms, and cross-contamination is presumably hand carried by attending staff (7). In this work, we investigated a nosocomial outbreak of ESBL-producing K. pneumoniae in a department of obstetrics and gynecology. Resistance to β-lactams was genetically characterized. Genome fingerprinting allowed the identification of an unexpected reservoir (the ultrasonography coupling gel container of the emergency room) and two modes of transmission, i.e., ultrasound scanning (transvaginal ultrasonography) and peripartum colonization of neonates.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Eight ceftazidime-resistant K. pneumoniae isolates were obtained from clinical samples (isolates 93-1 to 93-8) and one was obtained from an ultrasonography coupling gel container (isolate 93-9) between July and November 1993 in the Department of Obstetrics and Gynecology of Boucicaut Hospital, Paris, France (Table 1). Four ESBL-producing isolates were added as controls for genome fingerprinting, two of them from neighboring hospitals (isolates L-51 and N-69) and the other two isolated from unrelated wards in our facility in July (B-93a) and October (B-93b) 1993. Nalidixic acid-resistant Escherichia coli K802N was used as a recipient in mating experiments. E. coli TG1 and plasmid pUC18 were used in cloning experiments as the host strain and vector, respectively. Bacteria were grown in Mueller-Hinton medium (Sanofi Diagnostics Pasteur) and on CHROMagar-Orientation agar (CHROMagar, Paris, France) (16) at 37°C. All identifications were confirmed by using the API 32E system (bioMérieux).
TABLE 1.
Characteristics of the eight patients contaminated with ceftazidime-resistant K. pneumoniae
| Patient data (no., sex, age [yr])a | Ward | Reason for admissionb | Datec of:
|
K. pneumoniae isolation
|
||||
|---|---|---|---|---|---|---|---|---|
| Admission | Ultrasound scan | Discharge | Date | Site | Strain no. | |||
| 1, F, 19 | Maternity | POL | 7/30 | 7/30 | 7/31 | 7/31 | Vagina | 93-1 |
| Maternity | Delivery | 8/18 | NDf | 8/22 | ||||
| 2, Md | Maternity | Birth | 8/19 (birth) | ND | 8/22 | 8/19 | Gastric fluid | 93-2 |
| 3, F, 29 | Gynecology | PID | 8/10 | 8/10 | 8/18 | 8/10 | Urine | 93-3 |
| 4, F, 23 | Maternity | Delivery | 9/18 | 9/18 | 9/20 | 9/19 | Vagina | 93-4 |
| 5, Fe | Maternity | Birth | 9/18 (birth) | ND | 9/20 | 9/18 | Meconium | 93-5 |
| 6, F, 35 | Gynecology | Cancer | 10/12 | 10/13 | 10/19 | 10/14 | Urine | 93-6 |
| 7, F, 37 | Gynecology | PID | 10/18 | 10/18 | 10/23 | 10/18 | Vagina | 93-7 |
| 8, F, 27 | Gynecology | PID | 11/17 | 11/17 | 11/21 | 11/18 | Rectum | 93-8 |
F, female; M, male.
POL, premature onset of labor; PID, pelvic inflammatory disease.
Month/day.
Newborn of patient 1.
Newborn of patient 4.
ND, not done.
Epidemiological investigations.
On three different occasions between August and November 1993, rectal swabs or stool specimens from all patients hospitalized in the 56-bed Department of Obstetrics and Gynecology were analyzed. Environmental samples (n = 45) were collected from water sources, sinks, humidifiers, disinfectants, bedding, furniture, and finally from the ultrasonography equipment. Microbiological studies of the coupling gel and of the transvaginal ultrasound transducer probe were performed every 2 months over the following year. Two of the contaminated patients were seen again in 1996 and 1997, and they provided stool samples for analysis. All specimens were inoculated on selective agar (Drigalski agar; Sanofi Diagnostics Pasteur) supplemented with 4 μg of ceftazidime per ml except for control samples of the ultrasonography equipment which were inoculated on Mueller-Hinton agar without antibiotic.
Antibiotic susceptibility testing.
Disk diffusion tests were performed and interpreted according to the guidelines of the Comité de l’Antibiogramme de la Société Française de Microbiologie (1). The MICs of amoxicillin, cefotaxime, cefoxitin, and ceftazidime were determined on Mueller-Hinton agar by a dilution method with an inoculum of 105 CFU per spot, with or without clavulanic acid (2 μg/ml).
Genetic techniques.
Mating on filters was performed as described elsewhere (23). Transconjugants were selected on CHROMagar-Orientation agar plates supplemented with nalidixic acid (50 μg/ml) plus ceftazidime (8 μg/ml) or with nalidixic acid (50 μg/ml) plus gentamicin (50 μg/ml). E. coli transconjugants (pink colonies) were differentiated from spontaneous Klebsiella mutants (blue colonies). Frequency of transfer was expressed relative to the number of donor cells. Isolation of plasmid DNA from clinical isolates and E. coli transconjugants, restriction endonuclease digestion, ligation, molecular cloning, agarose gel electrophoresis, and other standard recombinant techniques were performed as described by Sambrook et al. (23). A supercoiled DNA ladder (2 to 16 kb; Gibco-BRL Life Technologies) and plasmids RP4 (54 kb), pCFF04 (85 kb), pIP112 (100.5 kb), pIP173 (125.8 kb), and pUD21 (170 kb) were used as plasmid DNA size markers.
Isoelectric focusing of β-lactamases.
Supernatants of sonicates of the clinical isolates and E. coli transconjugants were subjected to isoelectric focusing for 2 h by using a mini-isoelectric focusing cell 111 (Bio-Rad) and a gradient two-thirds of which consisted of polyampholytes with a pH range of 4 to 6 and one-third of which consisted of polyampholytes with a pH range of 3 to 10 (Bio-Rad). Extracts from TEM-1-, TEM-3-, SHV-1-, and SHV-5-producing strains were used as standards for pIs of 5.4, 6.3, 7.6, and 8.2, respectively. β-Lactamase activity was revealed by overlay with nitrocefin.
Molecular characterization of the β-lactamases.
Oligodeoxynucleotides OS5 (TTATCTCCCTGTTAGCCACC) and OS6 (GATTTGCTGATTTCGCTCGG) were used to amplify an internal fragment of about 90% of blaSHV genes (3). Oligodeoxynucleotides OT3 (ATGAGTATTCAACATTTCCG) and OT4 (CCAATGCTTAATCAGTGAGG) were used to amplify the entire sequence of blaTEM genes (2). PCRs were carried out as previously described (3) on plasmid DNA of the transconjugants. PCR products were purified, blunted, and then cloned into dephosphorylated SmaI-cut pUC18 by using the SureClone ligation kit (Pharmacia Biotech). Recombinant DNAs were introduced by transformation into E. coli TG1. In each case, the entire nucleotide sequence of three cloned amplicons obtained from independent PCRs were determined on both strands by using the Dye Primer Sequencing Kit on a Genetic ABI-PRISM 310 Sequencer Analyzer (Perkin-Elmer Applied Biosystem Division).
PFGE analysis.
Genomic DNA was analyzed by pulsed-field gel electrophoresis (PFGE) after digestion with XbaI, using the contour-clamped homogenous electric field (CHEF) technique developed by Chu et al. (8). Briefly, overnight cultures were harvested and then resuspended in TE buffer (10 mM Tris, 1 mM EDTA [pH 7.5]) to an A600 of 2.0. The bacterial suspensions were mixed with an equal volume of low-melting-point 2% InCert agarose (FMC Bioproducts) and allowed to solidify in 100-μl molds. The agar inserts were then incubated for 48 h at 50°C with 0.5 M EDTA (pH 8), 1% (wt/vol) sodium dodecyl sulfate, and 50 mg of proteinase K (Appligene) per liter. After treatment with 1 mM phenylmethylsulfonyl fluoride (Sigma Chemicals) for 1 h, the inserts were washed twice for 30 min with TE buffer and then for 30 min with distilled water. DNA was cleaved overnight with restriction endonuclease XbaI according to the manufacturer’s recommendations (New England Biolabs). DNA fragments were separated by electrophoresis in 1% agarose gels (Seakem Gold Agarose; FMC Bioproducts) in 0.5× Tris-borate-EDTA buffer (23) with a CHEF apparatus (CHEF MAPPER; Bio-Rad) at 14°C and 6 V/cm and with alternating pulses at a 120° angle in a 7- to 20-s pulse time gradient for 28 h. DNAs from bacteriophage lambda concatemers were used as size markers. The gels were stained with ethidium bromide (0.25 mg/liter) and photographed under UV light. Restriction patterns were interpreted by the criteria proposed by Tenover et al. (26).
RESULTS
An outbreak of ceftazidime-resistant K. pneumoniae in a department of obstetrics and gynecology.
During a 4-month period, six adult and two neonate patients were colonized or infected with ceftazidime-resistant isolates of K. pneumoniae in the maternity ward (n = 4) and the gynecology ward (n = 4) of the Department of Obstetrics and Gynecology in the 380-bed Boucicaut Hospital (Table 1). The resistant isolates were responsible for urinary tract infection in patients 3 and 6 (both treated with norfloxacin) and for neonatal infection with fever and respiratory distress in patient 2 (treated with imipenem and amikacin). Other patients were considered as colonized only. All cases were diagnosed in less than 48 h after patient admission. The average duration of hospitalization was 5 days (range, 1 to 9 days), and there was no overlap of the periods of hospitalization of the six adult patients. None of them had a recent history of hospitalization or antibiotic treatment, except for patient 1 (alleged index case, transferred from another hospital). Patients 1 and 4 were pregnant women admitted before delivery. Their two neonates (patients 2 and 5) were shown to be infected (patient 2) or colonized (patient 5) with ceftazidime-resistant K. pneumoniae from samples taken immediately after they were born, suggesting vertical transmission before or during delivery.
An unusual source of contamination.
No digestive carriage of ceftazidime-resistant Klebsiella (defined as a positive culture of rectal swab or stools) was detected at any time during the epidemic in other patients in the Department of Obstetrics and Gynecology. Due to these negative results, cross-contamination by members of staff was ruled out, and investigations were directed towards possible environmental factors. Forty-four samples collected from water sources, sinks, humidifiers, disinfectants, bedding, and furniture did not reveal the presence of ceftazidime-resistant K. pneumoniae. Retrospective analysis of the history of contaminated patients during hospitalization demonstrated that the only feature the six adults had in common was the fact that they had undergone transvaginal ultrasonography (except for patient 4, who underwent standard ultrasound scanning) in the emergency room. The ultrasonography equipment was therefore investigated. On 20 November, a pure growth of ceftazidime-resistant K. pneumoniae (105 CFU/ml) was recovered from the ultrasonography coupling gel in a container located in the emergency room but not from any other part of the equipment. It appeared that gel bottles (250 ml, Sonecho gel; Echos Contacts, Eragny, France) were opened and emptied into a wide-mouthed container which was thought more convenient for coating the ultrasound transducer probe with gel before and during examination of patients. The transvaginal probe itself was not contaminated, probably because latex condoms were always used as sheaths. The cycle of contamination was broken once the container was removed, and single use of the same coupling gel was introduced. No other case has been reported since then. Patients 6 and 8 were readmitted in December 1996 and May 1997, respectively, and stool cultures performed at this time were free of ceftazidime-resistant members of the family Enterobacteriaceae. On control cultures carried out in 1994, the coupling gel and the transvaginal ultrasound transducer probe were always sterile.
An outbreak due to the spreading of a single strain.
All the outbreak isolates exhibited the same biotype and antibiotic resistance profile. They were resistant to cefotaxime and ceftazidime (MICs, 8 and 128 μg/ml, respectively) and susceptible to cefoxitin (MIC, 4 μg/ml). Addition of clavulanic acid (2 μg/ml) restored the activity of cefotaxime and ceftazidime, suggesting the production of an ESBL. All isolates were resistant to gentamicin, tobramycin, were intermediately resistant to kanamycin, and were susceptible to amikacin, a phenotype most probably related to the production of an aminoglycoside 3-N-acetyltransferase type II (29). The isolates were also resistant to sulfonamides, trimethoprim, and tetracycline but susceptible to imipenem and quinolones.
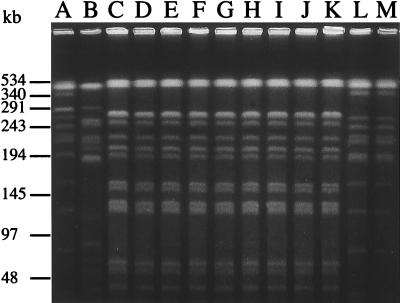
All isolates demonstrated the same plasmid profile, with four plasmids of ca. 130, 110, 5, and 3 kb (data not shown). Digestion of genomic DNA with XbaI produced 10 to 15 fragments in the size range of 40 to 500 kb (Fig. 1). According to the interpretative criteria of Tenover et al. (26), DNA restriction patterns of all the outbreak isolates (Fig. 1, lanes C to K) were genotypically indistinguishable, i.e., they had the same number of bands and the corresponding bands were of the same apparent size. Isolates from Laënnec and Necker hospitals (Fig. 1, lanes A and B) were genotypically unrelated both to each other and to the outbreak isolates, i.e., they showed changes consistent with more than three independent genetic events. Endemic isolates from Boucicaut Hospital (Fig. 1, lanes L and M), although unrelated to the epidemic strain, were genotypically indistinguishable.
FIG. 1.
PFGE patterns of XbaI-restricted DNA from ESBL-producing K. pneumoniae. Lanes: A, clinical strain L-51 from Laënnec Hospital; B, clinical strain N-69 from Necker Hospital; C to J, clinical strains 93-1, 93-2, 93-3, 93-4, 93-5, 93-6, 93-7, and 93-8, isolated from the outbreak in the Department of Obstetrics and Gynecology; K, strain 93-9, isolated from the ultrasonography coupling gel container; L and M, clinical strains B-93a and B-93b, isolated from other wards of Boucicaut Hospital in July and October 1993.
Identification of the β-lactamases.
Isoelectric focusing demonstrated that the outbreak isolates displayed two main bands of β-lactamase activity with pIs of 5.4 and 8.2 and a fainter one with a pI of 7.6 probably corresponding to the specific K. pneumoniae chromosomal β-lactamase (data not shown). Conjugation experiments were carried out, and E. coli transconjugant BC-1 was selected on nalidixic acid-ceftazidime plates with a transfer frequency of 10−6. BC-1 was resistant to broad-spectrum cephalosporins (ceftazidime MIC, 128 μg/ml) but not to aminoglycosides, tetracycline, sulfonamides, or trimethoprim. It harbored only the 130-kb plasmid and showed only the pI 8.2 band (therefore corresponding to the ESBL activity) on isoelectric focusing. To demonstrate that other resistance determinants were also transferable, conjugation was repeated, this time with nalidixic acid-gentamicin selection. Transconjugant BC-2 was obtained with a frequency of 10−4. It harbored only the 110-kb plasmid and displayed a single band of pI 5.4 on isoelectric focusing. BC-2 was resistant to aminoglycosides, sulfonamides, trimethoprim, tetracycline, and ampicillin (MICs, ≥512 μg/ml) but susceptible to broad-spectrum cephalosporins, indicating the presence of a broad-spectrum β-lactamase (probably TEM-1 in consideration of its pI of 5.4).
A 796-bp DNA fragment was amplified with oligonucleotides OS5 and OS6 from transconjugant BC-1 (but not from transconjugant BC-2) and then cloned and sequenced. The sequence was identical to that of the SHV-5 β-lactamase gene published by Billot-Klein et al. (5). An 858-bp DNA fragment was amplified with OT3 and OT4 from transconjugant BC-2 (but not from transconjugant BC-1) and then cloned and sequenced. The sequence was identical to that of blaTEM-1A of plasmid pBR322 (25), confirming that the broad-spectrum β-lactamase of pI 5.4 encoded by the 110-kb plasmid was indeed TEM-1.
DISCUSSION
In the present article, we report an outbreak of ESBL-producing K. pneumoniae in a department of obstetrics and gynecology with contamination of patients in less than 48 h following admission; ESBL-producing bacteria are usually acquired only after several days or weeks of hospitalization (22). Nosocomial outbreaks due to ESBL-producing members of the family Enterobacteriaceae have been mostly described for intensive care units, chronic care facilities, and nursing homes (22), not for obstetrics or gynecology wards, where short duration of hospitalization and low antibiotic selective pressure probably contribute to the low rates of nosocomial infections.
In addition, we demonstrate that two full-term neonates were contaminated by the multiresistant strain before or during delivery. Contamination of neonates with ESBL-producing bacteria was reported in intensive care units and was associated with prematurity, low weight, and prolonged hospitalization (6, 9, 28). This is the first time that mother-to-infant vertical transmission has been described for such bacteria.
PFGE has been shown to be an excellent tool for typing ESBL-producing K. pneumoniae strains (11). In this work, the technique permitted confirmation that the outbreak was due to a single strain, unrelated to the endemic strain isolated during the same period from other wards in the facility (Fig. 1). PFGE also enabled the identification of the source of contamination as the coupling gel used for ultrasonography in the emergency room. Given that (i) the only common feature in the hospital history of the adult patients was ultrasonography performed on arrival in the emergency room, (ii) there was no overlap of the periods of hospitalization, and (iii) elimination of the contaminated gel stopped the outbreak, we considered that the gel was the only reasonable source of contamination. Investigations showed that the multiresistant K. pneumoniae strain was able to survive for at least 4 weeks in the gel (data not shown). This finding is in keeping with previous reports suggesting that coupling gels can support bacterial growth (17, 20, 27). However, infection hazard in ultrasonography has rarely been documented. Staphylococcus aureus and Burkholderia cepacia cross-infections related to ultrasound scanning of infected surgical wounds (19) and transrectal ultrasound-guided biopsy of the prostate (13), respectively, have been reported. Theoretical risk of contamination in obstetric ultrasonography was recently emphasized by Storment et al. (24), who questioned the efficacy of latex condoms as sheaths for transvaginal probes, but here we present the first clinical report of cross-contamination due to transvaginal ultrasonography. Because of the unusual mode of transmission, infection control measures such as isolation of colonized patients, use of gloves, and hand washing did not halt the outbreak, although they were probably effective in preventing a wider dissemination of the multiresistant strain.
Sequencing of the blaSHV gene showed that the outbreak strain produced SHV-5 and not one of the recently described β-lactamases that have the same pI of 8.2, i.e., SHV-9, SHV-10 (21), SHV-11, and SHV-12 (18). SHV-5 is one of the most frequently encountered ESBLs produced by epidemic K. pneumoniae strains worldwide (15), but it has not been associated so far with any of the numerous outbreaks described in France, despite sporadic cases. TEM-1 β-lactamase was also produced by the epidemic clone, and we showed that the β-lactamase genes were located on different transferable plasmids. Furthermore, all other associated resistance determinants were present uniquely on the plasmid that carried the blaTEM gene, in contrast with other studies in which aminoglycoside and sulfonamide resistance determinants were carried on the same plasmid as the blaSHV-5 gene (4, 10, 14).
To summarize, this outbreak showed unique features, i.e., very early cross-contamination, lack of recognized risk factors of patients, transmission of the multiresistant strain by a soiled ultrasonography coupling gel, and intrapartum contamination of two neonates. It highlights the necessity of evaluation of nosocomial risks associated with procedures with good safety records such as ultrasonography.
ACKNOWLEDGMENTS
We are grateful to Claire Poyart and Nicolas Fortineau for providing strains and helpful comments and to Édouard Bingen for critical review of the manuscript.
REFERENCES
- 1.Acar J, Bergogne-Bérézin E, Chardon H, Courvalin P, Dabernat H, Drugeon H, Dubreuil L, Duval J, Flandrois J P, Goldstein F, Meyran M, Morel C, Philippon A, Sirot J, Thabaut A. Comité de l’Antibiogramme de la Société Française de Microbiologie. Communiqué 1994. Pathol Biol. 1994;42:1–8. [Google Scholar]
- 2.Arlet G, Brami G, Decré D, Flippo A, Gaillot O, Lagrange P H, Philippon A. Molecular characterisation by PCR-restriction fragment length polymorphism of TEM β-lactamases. FEMS Microbiol Lett. 1995;134:203–208. doi: 10.1111/j.1574-6968.1995.tb07938.x. [DOI] [PubMed] [Google Scholar]
- 3.Arlet G, Rouveau M, Philippon A. Substitution of alanine for aspartate at position 179 in the SHV-6 extended-spectrum β-lactamase. FEMS Microbiol Lett. 1997;152:163–167. doi: 10.1016/s0378-1097(97)00196-1. [DOI] [PubMed] [Google Scholar]
- 4.Bauerfeind A, Rosenthal E, Eberlein E, Holley M, Schweighart S. Spread of Klebsiella pneumoniae producing SHV-5 beta-lactamase among hospitalized patients. Infection. 1993;21:18–22. doi: 10.1007/BF01739303. [DOI] [PubMed] [Google Scholar]
- 5.Billot-Klein D, Gutmann L, Collatz E. Nucleotide sequence of the SHV-5 β-lactamase gene of a Klebsiella pneumoniae plasmid. Antimicrob Agents Chemother. 1990;34:2439–2441. doi: 10.1128/aac.34.12.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bingen E H, Desjardins P, Arlet G, Bourgeois F, Mariani-Kurkdjian P, Lambert-Zechovsky N Y, Denamur E, Philippon A, Elion J. Molecular epidemiology of plasmid spread among extended broad-spectrum β-lactamase-producing Klebsiella pneumoniae isolates in a pediatric hospital. J Clin Microbiol. 1993;31:179–184. doi: 10.1128/jcm.31.2.179-184.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brun-Buisson C, Legrand P, Philippon A, Montravers F, Ansquer M, Duval J. Transferable enzymatic resistance to third-generation cephalosporins during nosocomial outbreak of multiresistant Klebsiella pneumoniae. Lancet. 1987;ii:302–306. doi: 10.1016/s0140-6736(87)90891-9. [DOI] [PubMed] [Google Scholar]
- 8.Chu G, Vollrath D, Davis R W. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science. 1986;234:1582–1585. doi: 10.1126/science.3538420. [DOI] [PubMed] [Google Scholar]
- 9.Coovadia Y M, Johnson A P, Bhana R H, Hutchinson G R, George R C, Hafferjee I E. Multiresistant Klebsiella pneumoniae in a neonatal nursery: the importance of maintenance of infection control policies and procedures in the prevention of outbreaks. J Hosp Infect. 1992;22:197–205. doi: 10.1016/0195-6701(92)90044-m. [DOI] [PubMed] [Google Scholar]
- 10.French G L, Shannon K P, Simmons N. Hospital outbreak of Klebsiella pneumoniae resistant to broad-spectrum cephalosporins and β-lactam–β-lactamase inhibitor combinations by hyperproduction of SHV-5 β-lactamase. J Clin Microbiol. 1996;34:358–363. doi: 10.1128/jcm.34.2.358-363.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gouby A, Neuwirth C, Bourg G, Bouziges N, Carles-Nurit M J, Despaux E, Ramuz M. Epidemiological study by pulsed-field gel electrophoresis of an outbreak of extended-spectrum β-lactamase-producing Klebsiella pneumoniae in a geriatric hospital. J Clin Microbiol. 1994;32:301–305. doi: 10.1128/jcm.32.2.301-305.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacoby G A, Medeiros A A. More extended-spectrum β-lactamases. Antimicrob Agents Chemother. 1991;35:1697–1704. doi: 10.1128/aac.35.9.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keizur J J, Lavin B, Leidich R B. Iatrogenic urinary tract infection with Pseudomonas cepacia after transrectal ultrasound guided needle biopsy of the prostate. J Urol. 1993;149:523–526. doi: 10.1016/s0022-5347(17)36135-9. [DOI] [PubMed] [Google Scholar]
- 14.Legakis N J, Tzouvelekis L S, Hatzoudis G, Tzelepi E, Gourkou A, Pitt T L, Vatopoulos A C. Klebsiella pneumoniae infections in Greek hospitals. Dissemination of plasmids encoding an SHV-5 type β-lactamase. J Hosp Infect. 1995;31:177–187. doi: 10.1016/0195-6701(95)90064-0. [DOI] [PubMed] [Google Scholar]
- 15.Medeiros, A. A. 1997. Evolution and dissemination of beta-lactamases accelerated by generations of β-lactam antibiotics. Clin. Infect. Dis. 24(Suppl. 1):19–45. [DOI] [PubMed]
- 16.Merlino J, Siarakas S, Robertson G J, Funnel G R, Gottlieb T, Bradbury R. Evaluation of CHROMagar Orientation for differentiation and presumptive identification of gram-negative bacilli and Enterococcus species. J Clin Microbiol. 1996;34:1788–1793. doi: 10.1128/jcm.34.7.1788-1793.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muradali D, Gold W L, Phillips A, Wilson S. Can ultrasound probes and coupling gel be a source of nosocomial infection in patients undergoing sonography? An in vivo and in vitro study. Am J Roentgenol. 1995;164:1521–1524. doi: 10.2214/ajr.164.6.7754907. [DOI] [PubMed] [Google Scholar]
- 18.Nüesch-Inderbinen M T, Kayser F H, Hächler H. Survey and molecular genetics of SHV β-lactamases in Enterobacteriaceae in Switzerland: two novel enzymes, SHV-11 and SHV-12. Antimicrob Agents Chemother. 1997;41:943–949. doi: 10.1128/aac.41.5.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Doherty A J, Murphy P G, Curran R A. Risk of Staphylococcus aureus transmission during ultrasound investigation. J Ultrasound Med. 1989;8:619–620. doi: 10.7863/jum.1989.8.11.619. [DOI] [PubMed] [Google Scholar]
- 20.Patterson S L, Monga M, Silva J B, Bishop K D, Blanco J D. Microbiologic assessment of the transabdominal ultrasound transducer head. South Med J. 1996;89:503–504. doi: 10.1097/00007611-199605000-00011. [DOI] [PubMed] [Google Scholar]
- 21.Prinarakis E E, Miriagou V, Tzelepi E, Gazouli M, Tzouvelekis L S. Emergence of an inhibitor-resistant β-lactamase (SHV-10) derived from an SHV-5 variant. Antimicrob Agents Chemother. 1997;41:838–840. doi: 10.1128/aac.41.4.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quinn, J. P. 1994. Clinical significance of extended-spectrum β-lactamases. Eur. J. Clin. Microbiol. Infect. Dis. 13(Suppl. 1):39–42. [DOI] [PubMed]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Storment J M, Monga M, Blanco J D. Ineffectiveness of latex condoms in preventing contamination of the transvaginal ultrasound transducer head. South Med J. 1997;90:206–208. doi: 10.1097/00007611-199702000-00007. [DOI] [PubMed] [Google Scholar]
- 25.Sutcliffe J G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci USA. 1978;75:3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tesch C, Fröschle G. Sonography machines as a source of infection. Am J Roentgenol. 1997;168:567–568. doi: 10.2214/ajr.168.2.9016251. [DOI] [PubMed] [Google Scholar]
- 28.Venezia R A, Scarano F J, Preston K E, Steele L M, Root T P, Limberger R, Archinal W, Kacica M A. Molecular epidemiology of an SHV-5 extended-spectrum beta-lactamase in Enterobacteriaceae isolated from infants in neonatal intensive care unit. Clin Infect Dis. 1995;21:915–923. doi: 10.1093/clinids/21.4.915. [DOI] [PubMed] [Google Scholar]
- 29.Vliegenthart J S, Ketelaar-van Gaalen P A G, van de Klundert J A M. Nucleotide sequence of the aacC2 gene, a gentamicin resistance determinant involved in a hospital epidemic of multiply resistant members of the family Enterobacteriaceae. Antimicrob Agents Chemother. 1989;33:1153–1159. doi: 10.1128/aac.33.8.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]