Abstract
Faithful eucaryotic cell division requires spatio-temporal orchestration of multiple sequential events. To ensure the dynamic nature of these molecular and morphological transitions, a swift modulation of key regulatory pathways is necessary. The molecular process that most certainly fits this description is phosphorylation, the post-translational modification provided by kinases, that is crucial to allowing the progression of the cell cycle and that culminates with the separation of two identical daughter cells. In detail, from the early stages of the interphase to the cytokinesis, each critical step of this process is tightly regulated by multiple families of kinases including the Cyclin-dependent kinases (CDKs), kinases of the Aurora, Polo, Wee1 families, and many others. While cell-cycle-related CDKs control the timing of the different phases, preventing replication machinery errors, the latter modulate the centrosome cycle and the spindle function, avoiding karyotypic abnormalities typical of chromosome instability. Such chromosomal abnormalities may result from replication stress (RS) and chromosome mis-segregation and are considered a hallmark of poor prognosis, therapeutic resistance, and metastasis in cancer patients. Here, we discuss recent advances in the understanding of how different families of kinases concur to govern cell cycle, preventing RS and mitotic infidelity. Additionally, considering the growing number of clinical trials targeting these molecules, we review to what extent and in which tumor context cell-cycle-related kinases inhibitors are worth exploiting as an effective therapeutic strategy.
Subject terms: Oncogenes, Tumour-suppressor proteins, Cancer genetics, Kinases
Facts
G1 to S phase transition has been largely investigated to define the activation pattern of CDK4/6 and CDK2. However, a unifying model of how these kinases act to overcome the restriction point is still missing, mostly because of cell type-to-cell type variability as well as due to the employment of different reporters to monitor these dynamics.
Therapeutical targeting of CDKs has recently gained major attention after successful clinical employment of CDK4/6 inhibitors in breast cancer. Nevertheless, very little is known about the effect these inhibitors have in reshaping phosphatase activities, and how this could contribute to tumor resistance.
Despite having demonstrated promising results in vitro, DDR inhibitors still struggle to prove their therapeutic value in clinical trials, mostly as a consequence of burdensome side effects on patients. With DDR being so indispensable also for healthy tissues, it is tempting to speculate that these inhibitors will demonstrate their efficacy only in combinatorial treatments with compounds that enhance replication stress.
Open questions
The promiscuity of cyclins binding to CDKs suggests a plasticity in cell cycle rewiring that has been long overlooked, especially in the context of cancer therapy. How do cyclins behave when their respective CDKs are inhibited? Do they contribute to activating alternative CDKs to bypass the inhibition and promote tumor resistance?
Multiple phospho-proteomic screenings have expanded the known set of proteins targeted by DNA damage response (DDR) kinases, suggesting connections with many other cellular pathways, such as E3 ligases, splicing, and chromatin remodeling regulation. An attractive future line of research in system biology might focus on how this interplay occurs and whether it is necessary for functional checkpoints. Additionally, it might also be worth investigating whether or not the broad range of targets can explain the poor success of DDR inhibitors in clinics.
Similar to all kinases, cell cycle kinases use ATP as a substrate for phosphorylating their targets. Despite the evident interplay between cell cycle and energy production, very few studies have addressed this connection in cancer ontogenesis. Does metabolic rewiring influence the cell cycle through ATP biosynthesis, or is it instead influenced by the cell cycle transitions?
Introduction
In eukaryotic cells, two processes—the replication of genomic DNA and the ensuing segregation into daughter cells—occur across different cell cycle phases. These two processes are the core focus of cell cycle control. It is frequently believed that cancer cells move through the cell cycle uncontrollably and that the majority—if not all—cell cycle checkpoints must be altered for a cell to develop into cancer. A significant body of recent research, however, has produced compelling evidence that just particular parts of cell cycle control must be compromised for cancer cells to continue to proliferate. According to this research, the most relevant phase that needs to be altered for cancer cells to continue their uncontrolled division is their capacity to exit cell cycle [1–4]. Importantly, this also implies that most cell cycle regulatory mechanisms are necessary for the survival of cancer cells. These results highlight the essential distinction between the DNA damage checkpoint and RS checkpoint responses, which are designed, respectively, to inhibit the expansion and spread of DNA damage and replication of stress-induced DNA damage. Indeed, cancer cells frequently have a defective DNA damage checkpoint, allowing continued cell division despite the accumulation of genetic mistakes. Contrarily, genes implicated in the RS checkpoint are rarely altered in cancer cells, as many malignancies become increasingly reliant on checkpoint function to endure high levels of RS.
In this multifaceted context, cell cycle-related kinases represent the master regulators that provide swift and precise control, and their activity is typically dysregulated in cancer cells that proliferate too quickly. Multiple molecules that target these kinases have been successfully designed to control the proliferation and avoid karyotypic abnormalities of malignant cells; among them, a growing number have been clinically approved and many others are under clinical trial scrutiny.
In this Review, we provide a brief overview of cell cycle control pathways, and we explore in depth the current knowledge on how cell cycle regulatory kinases exert their roles in the numerous cell cycle checkpoints. Also, we highlight the elements of cell cycle control that cancer cells commonly lose control over, to continue dividing. Finally, we discuss our considerations and the general opinion for therapeutically targeting such kinases, in order to address these dependencies in cell cycle control and checkpoint mechanisms. The complexity of these processes makes it impossible to discuss all of them with high detail; rather, we will focus on just a few of them, while discussing about the others in general terms and guiding the reader to recently released reviews and articles.
The cell cycle
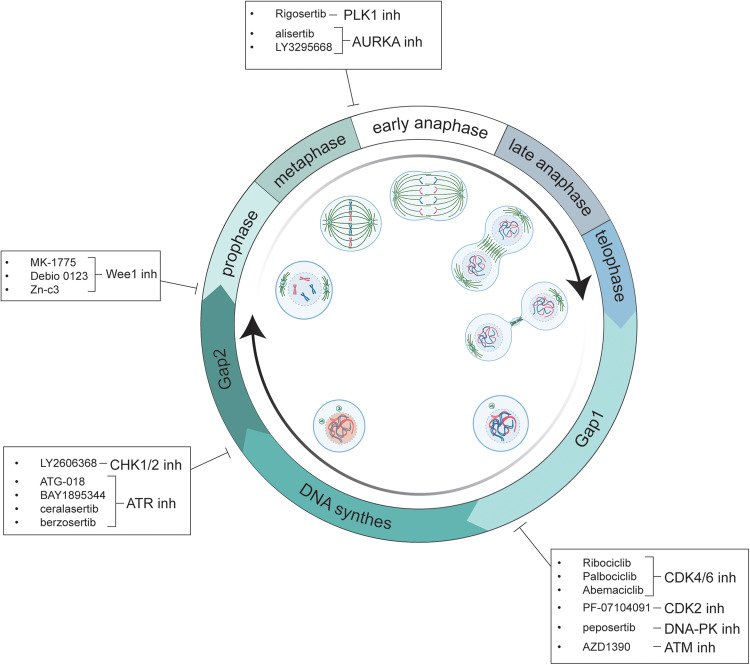
Cell division in unicellular and multicellular eukaryotes is governed by a sophisticated network of regulatory systems and balances to ensure that no errors occur before a cell is permitted to enter and advance through the cell cycle. The timely and precise duplication and segregation of the genomic DNA is the single objective of the intricate network of regulatory components that makes up the cell cycle (Fig. 1).
Fig. 1. Cell cycle-associated kinases inhibitors in clinical trials.
A significant number of inhibitors have been developed to specifically target kinases involved in cell cycle regulation and DNA replication. In each box, the active compounds that target cells undergoing the different cell cycle phases currently subjected to clinical trials (see Table 1). Figure created with BioRender.com.
Almost seventy years ago, Alma Howard and Stephen Pelc (1953) attributed, for the first time, defined time windows to cell division, and proposed two main separate phases of the mitotic cell cycle: the interphase and the M phase. The former is further subdivided into the S-phase (a period of DNA synthesis), the G1 or Gap1 - which occurs before the S-phase—and the G2 or Gap2, which takes place before the mitotic period. Gap phases were given names for the interphase intervals that separate the S phase from the M phase based on the obvious observation that these intervals exist between the two primary events, DNA duplication and segregation. In these two stages, the crucial choices to begin the cell cycle and to start the process that results in chromosomal segregation are made. Additionally, during G1, cells can leave the cell cycle and enter quiescence, or G0, a non-proliferative state.
Canonically, mitogen stimulation from the extracellular space perceived by cells in G0 induces the transcriptional increase of D-type Cyclins levels (D1, D2, D3), which in turn, stimulate their catalytic binding partners, Cyclin-Dependent Kinase 4 or 6 (CDK4 or CDK6). Contrarily, the progression is physiologically inhibited by growth factors withdrawal, replicative senescence, DNA damage and oncogene-induced stress that positively regulate the INK4 family of inhibitors (p16INK4A, p15INKB, p18INK4C, p19INK4D). The stability of Cyclin D-CDK4/6 is highly dependent on KIP/CIP proteins (p21, p27, p57), which serve as scaffold proteins for their interaction [5–9]. Simultaneously, KIP/CIP proteins are inhibitors of CDK2 and CDK1 activity [10]. Cyclin D-CDK4/6 kinases phosphorylate multiple cellular targets, the most important being the retinoblastoma protein (RB1) [11]. When hyperphosphorylated, RB1 is inhibited, releasing E2F transcriptional activity on genes that promote G1 to S phase transition. Among E2F transcribed genes, the two E-type Cyclins (E1 and E2) bind and activate CDK2, amplifying RB1 phosphorylation, thereby promoting the downstream E2F transcriptional activity [12]. Additionally, a systematic screen for substrates of cyclin D1-CDK4 and cyclin D3-CDK6 identified the Forkhead Box M1 (FOXM1) transcription factor as a shared target. CDK4 and 6 stabilize and trigger FOXM1 activity, shielding cancer cells from senescence [13].
This stage, also known as Restriction Point, is crucial since changes in the key regulators of the G1 to the S phase transition could allow cells to proliferate independently of mitogenic stimuli, unleashing tumorigenic growth.
During the transition, a part of the synthesized proteins contributes to the assembly of the replicon molecular machinery, composed of ORC1–6, CDC6, CDT1, and MCM2–7 DNA helicase. The consecutive series of phosphorylations primed by Cyclin E-CDK2 promotes the formation of the CMG complex (cdc45-MCM2–7-GIMS) that unwinds the DNA double-strand with its helicase activity. Subsequently, Cyclin A replaces Cyclin E as the catalytic binding partner of CDK2 and further stimulates DNA replication through the phosphorylation of targets such as cdc6. Interestingly, Cyclin A can trigger both CDK2 and CDK1, enabling the transition from S to G2-M phase and fostering Cyclin B1-CDK1 mitotic activity [14, 15]. When DNA damage occurs, the kinases ataxia telangiectasia mutated (ATM), ataxia telangiectasia and RAD3-related protein (ATR), and their downstream effectors checkpoint kinase 1 (CHK1) and checkpoint kinase 2 (CHK2) get triggered to promptly interrupt the cell cycle and enable the activity of the DNA damage repair machinery. Given their roles in preserving genomic integrity, mutations in the ATM-ATR cascade genes favor cancer development.
Beginning simultaneously with DNA replication, the centrosome cycle consists of duplication and maturation of the organelles that operate as the major microtubule-organizing center (MTOC). Once the mitosis started, the duplicated centrosomes support the formation of the mitotic spindle poles. Then, the maturation of each centrosome leads to the formation of its own aster of active microtubules originating from both poles of the mitotic spindle. In this dynamic context, serine-threonine protein kinases such as aurora A and polo-like kinase 1 (PLK1) [16] as well as CDK1 and CDK2, provide key regulatory activity.
When cells move from the G2 phase to the M, CDK1 is rapidly activated in combination with Cyclin A to speed up the start of mitosis via controlling chromosomal condensation and microtubule dynamics. Subsequently, Cyclin A degradation during nuclear envelope collapse allows the formation of Cyclin B-CDK1 complexes, indispensable for progressing through the M phase, supporting events such as cytoskeleton reorganization, centrosome maturation, spindle assembly, and chromosome separation [17]. Once chromosomal condensation in the late prophase took place, the ensuing equal segregation at the two spindle poles during anaphase allows both sister chromatids of each duplicated chromosome pair to be connected through their kinetochores to the MTOC. Should a chromosome be improperly linked to the mitotic spindle, the spindle assembly checkpoint (SAC) mediates the arrest at the anaphase. Specifically, the SAC, also known as the mitotic checkpoint, is a molecular hub that includes the mitotic arrest deficient protein 1 (MAD1), MAD2, monopolar spindle 1 (MPS1), budding uninhibited by benzimidazole 1 (BUB1), BUB3, and BUB1 homolog beta, mediates arrest (BUB1B), and aurora B. Altogether, they provide a signaling cascade that halts the activity of the APC complex, therefore inhibiting the mitotic progression. Restricted activity of the SAC leads to premature separation of the sister chromatids resulting in chromosomal instability. Last, cytokinesis requires the anaphase-promoting complex (APC/C) to degrade Cyclin B via the proteasome [18].
Cyclin-dependent-kinases overview
Cell cycle is a tightly coordinated process controlled by the oscillating activities of Cyclin-Dependent Kinases (CDKs). Their activity is positively modulated by Cyclins and inhibited by CDK inhibitors (CKIs) and fluctuation in their activation state can be regulated by transcriptional and post-translational modifications (typically phosphorylation), as well as the swift cyclic degradation of Cyclins and CKIs by the ubiquitin-proteasome system. While phosphorylation or association with CKIs ensures swift reversible variations during the process, ubiquitin-mediated degradation of key components of the cell cycle machinery provides directionality and irreversibility to cell cycle progression.
Historically, CDKs have been characterized by genetic and biochemical approaches in Saccharomyces pombe [19–21] in the late 80’s by the pioneering work of Nobel laureate Paul Nurse. His research proved the relevance of CDKs in facilitating cell cycle transitions and demonstrated that CDKs must associate with a regulatory subunit, named Cyclin, to exert their enzymatic function. In contrast with the promiscuous Cdc28, the central coordinator of the yeast cell cycle, mammalian CDKs display selectivity with one or few Cyclins acting as the regulatory subunit. The extraordinary degree of evolutionary divergence and specialization of the CDK family in mammals resulted in the division of CDKs into three cell-cycle-related subfamilies (CDK1, CDK4, and CDK5) and six transcriptional subfamilies (CDK7, CDK8, CDK9, CDK11, CDK12 and CDK20). Among the three cell-cycle-related subfamilies only the first two, CDK1 and CDK4, including respectively CDK1, −2, and −3 for the former and CDK4 and −6 for the latter, are known to directly participate in the cell division, while the other subfamily - CDK5 - participates in many different pathways, such as Wnt-dependent signaling or signal transduction in the primary cilium, therefore affecting cell cycle progression only secondarily. Being cell cycle kinases the distinct spotlight of this review, we will discuss only those CDKs that have been tightly linked to the core of cell cycle regulation. For other transcriptional subfamilies, as well as the CDK5 subfamily, we refer to more comprehensive reviews [22, 23].
Based on the sequence homology of the catalytic domain, CDKs belong to the CMGC superfamily, a group of kinases that also includes glycogen synthase kinase-3 (GSKs), members of the dual-specificity tyrosine-regulated kinase (DYRK) family, MAP kinases and CDK-like kinases. As other members of the CMGC group, CDKs are proline-directed serine/threonine-protein kinases that preferentially target the S/T-P-X-K/R sequence, due to the presence of a hydrophobic pocket close to the catalytic site that can accommodate the proline (position +1) [24].
The CDKs have a variable size ranging from 250 to more than 1500 amino acid residues. From a structural point of view, they have a two-lobed structure, with the amino-terminal lobe rich in beta-sheets, the carboxy-terminal one containing multiple α-helices, and the active site located in between. The N-terminal is characterized by a G-loop inhibitory element and a C-helix (containing the PSTAIRE sequence in CDK1), while the C-terminal includes a partially disordered activation domain, which extends from the DFG to the APE motifs and contains the phosphorylation-sensitive residue (e.g., T160 in CDK2) in the so-called T-loop. When bound to the respective Cyclin, different CDKs display partially non-redundant behaviors. For instance, CDK2 undergoes a conformational rearrangement that displaces the T-loop and exposes the catalytic cleft, making the threonine accessible for activating phosphorylation by CAK. Once phosphorylated, the heterodimer is stabilized, with this triggering its enzymatic activity [25]. Conversely, the Cyclin D binding to CDK4/6 does not force the kinase into an active conformation since the ATP-binding site of CDK4 is still inaccessible to its substrates. Interestingly, the contact site between CDK2 and Cyclin A comprises both the N-term and C-term lobes, while the contacts between CDK4 and Cyclin D are limited to the N-lobe [25, 26].
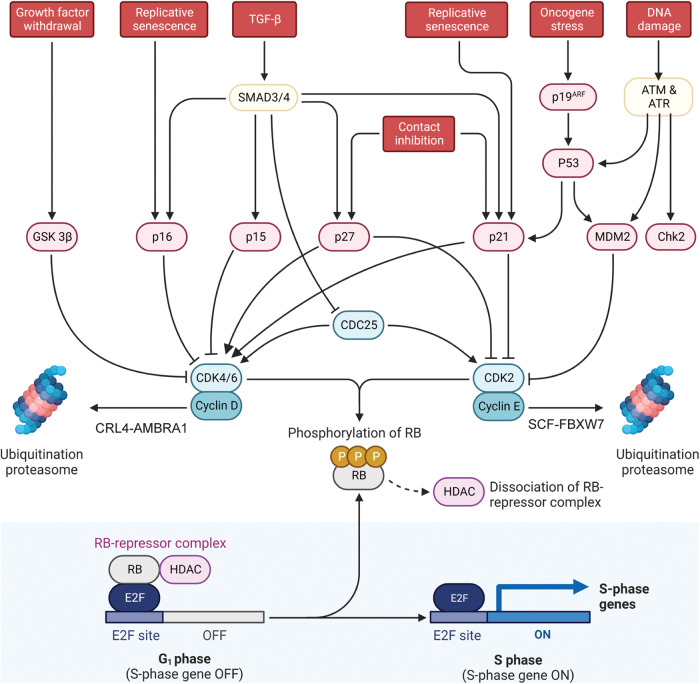
Physiologically, the prototypical mammalian cell cycle begins when growth-dependent CDKs activity (D-type Cyclin–CDK4/6) creates a decision timeframe, during which the cell can commit to initiate replication and begin a new cell cycle. However, the exact molecular mechanism, through which Rb is inhibited to promote G1 to S phase transition, is still a matter of debate. Different models could possibly explain how CDKs activity releases E2F transcriptional activity [27] (Fig. 2). Canonically, CDK4/6 first phosphorylates Rb on Ser807 and Ser811, priming it for subsequent phosphorylation of CDK2 to fully trigger E2F. This model is sustained by multiple evidence: the timing of cyclin expression through G1, with Cyclin D being expressed earlier than Cyclin E [28]; experiments of individual or combined functional inactivation of CDK4/6 and CDK2 activity[29]; and, finally, the E2F-dependent expression of Cyclin E, implying that it could be expressed if CDK4/6 phosphorylation partially inactivated Rb [30]. To add a degree of complexity to this pattern, Cyclins levels are periodically degraded through the proteasome to prevent CDKs/Cyclin holoenzymes hyperactivation. For instance, the E3-ligase substrate receptor, tumor suppressor and autophagy regulator AMBRA1 was recently found to be essential for Cyclin D degradation through CUL4-DDB1 E3 ligase complex [31–33]. Similarly, Cyclin E is recognized and ubiquitinated by SCF(FBXW7) [34], while Cyclin A and Cyclin B levels fluctuate according to APC/C activity [35].
Fig. 2. Molecular overview of G1/S transition.
The decision of a cell to enter the replicative phase is governed by a multifactorial network that controls faithfully the timing of the transition. The multiple processes involved converge all together on the de-repression of the E2F transcription factor, inducing the expression of S-phase related genes. Figure created with BioRender.com.
This modular and sequential activity was recently disputed by multiple pieces of evidence that sustain two alternative models. The former stems from the observation made by Nakashima et al. [36], that in synchronized cellsRb phosphorylation follows peculiar patterns, and that CDK4/6-Cyclin D activity provides only Rb mono-phosphorylation, whereas Rb hyperphosphorylation is coincident with the onset of CDK2 activity. Additionally, E2F transcriptional activity did not increase until the rise of CDK2 activity, suggesting that CDK4/6 are necessary for, but not sufficient to providing Rb inactivation, and that the sole CDK2 is responsible for E2F triggering. This model is further corroborated by the evidence that increasing the abundance of CDK4/6-Cyclin D complexes leads to CDK2 activation by sequestering its inhibitors p21 and p27. As a matter of fact, it was recently demonstrated that the CDK4/6 inhibitor palbociclib exerts its function not by directly inhibiting CDK4/6 activity toward Rb, but by increasing the abundance of p27 available to inhibit CDK2-Cyclin E complexes [37]. This may explain why the E2F-dependent Cyclin E expression, primarily regulated by Cyclin D/CDK4–6, is not necessary to induce Rb inhibition as in the first model. Finally, the latter disputes the conclusion that CDK2-Cyclin E plays any substantial role in inhibiting Rb during G1 [38, 39] and rather proposes that CDK4/6-Cyclin D is exclusively responsible for E2F activation. Indeed, by means of kinase-activity fluorescent reporters, the authors demonstrated that CDK4/6 activity precedes CDK2 activity and that once entered the S phase, cells require CDK2 function to maintain Rb inhibition [39]. Further expanding the complexity of this intricated network, a recent breakthrough discovery from the Cappell lab demonstrates that the choice to proliferate is totally reversible even beyond G1/S restriction point, at variance with previous knowledge. Also, the absence of mitogens along the entire interphase triggers cell cycle exit depending on CDK4/6 impairment, which is required for Cyclin A2/CDK2 activity at all stages of the cell cycle, not only in G1 [40].
Despite the crucial role CDK4/6 play in these contexts (Fig. 3), seminal genetic works established that cells could proliferate even without CDK4/6 activity [41]. Combined Cdk4 and Cdk6 ablation in mice, however, is embryonically lethal during the late stages of organogenesis despite displaying normal cell proliferation and apoptosis in most cell types. The main deficiencies are limited to the compromised maturation of different hematopoietic lineages. Supporting these observations, multiple non-consensus bindings between CDKs and cyclins occur to compensate for the absence of one of them. For instance, when Cdk4 and Cdk6 are absent, Cdk2 can bind D-type Cyclins. Another plausible explanation was provided only recently when the sequence of molecular events that support cell proliferation in their absence has been elucidated. Specifically, it was demonstrated that, when CDK4/6 are acutely inhibited, Cyclin E-CDK2 activation is delayed and heterogeneous within the same population, leading to a less effective G1/S transition than in the CDK4/6-initiated path. Additionally, CDK2 activity reaches its peak more slowly, and it is more susceptible to fluctuations, with this implying that CDK4/6 may also provide persistence to Cyclin E-CDK2 activity [42]. This was further explored recently by Arora et al., who demonstrated that acute inhibition of CDK2 forces CDK4/6 activity beyond G1, driving Cyclin A expression via Rb-E2F axis and enabling re-activation of CDK2 [43].
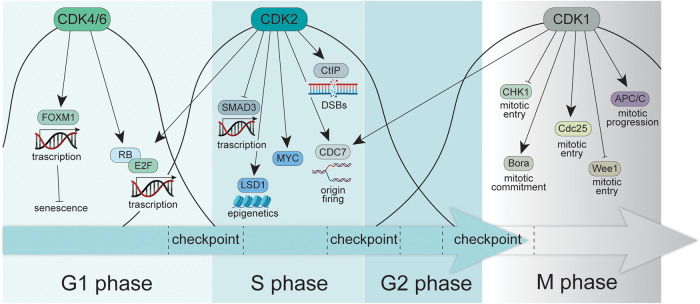
Fig. 3. Relevant CDKs phosphorylation targets throughout cell cycle.
Upon cell cycle entry, CDK4 and CDK6 kinases are activated allosterically by binding Cyclins D. Their catalitic activity mainly targets RB, in order to release E2F transcriptional activity and FOXM1 to suppress senescence. At the onset of the decision window between G1 and S phases, CDK2 activity arises and positively regulates multiple targets involved in DNA replication, repair and epigenetic control, while inhibiting SMAD3 transcriptional activity. Last, at the interface between G2 and M phase, CDK1 is activated and supports the mitotic process from the commitment to the completion, by phoshorylating - among others, targets such as CHK1, Bora, Cdc25, Wee1 and APC/C.
The intricacy of the CDKs network also affects the precise way cells perceive commitment to cell division. Historically, the restriction point in mammalian cells has been defined as the irreversible point of commitment to division, whose traversal makes growth factors unnecessary for the progression of the cell [44–46]. Indeed, while normal cells show conventional G1 restriction point, immortalized and transformed cell lines do not [47]. Interestingly, many of them are insensitive to the absence of mitogens even before completing mitosis in the preceding cell cycle [48]. A plausible explanation for this has been provided by Moser and colleagues, who demonstrated that only a subset of cells that possess high CDK2 activity is insensitive to growth factors and RS in the subsequent generation [49]. It is worth noting that another crucial contribution to this mother-to-daughter legacy is the DNA damage accumulated during the previous replication. This can lead to a p21-dependent entry of daughter cells into quiescence immediately after mitosis [50–52].
Beyond the restriction point, Cyclin E-CDK2 activity becomes predominant. Once the pre-replication complexes are formed upon DNA replication origins, CDK2 is thought to provide, in concert with the kinase CDC7, the signal that triggers origin firing. Very recently, this notion has been radically revisited by Suski and colleagues. They demonstrated that CDK1 is also active during G1/S transition and phosphorylates MCM proteins within pre-replication complex on CDC7-independent sites. Remarkably, they also found that, during S-phase entry, CDK1 associates with Cyclin B, and that these CDK1–Cyclin B complexes are catalytically active, demonstrating that this complex physiologically regulates two distinct transitions during the cell division cycle, and questioning the importance of CDK2 in this process [53]. This work reinforces previous observations made from genetic ablation studies, that revealed how mouse embryos lacking interphase Cdks (Cdk2, Cdk3, Cdk4 and Cdk6) undergo organogenesis and develop until midgestation. Nonetheless, proteomic screenings for CDK2 phosphorylation substrates revealed a diversified landscape [54], that not only affects cell cycle- related proteins but also histone modifiers (LSD1, DOT1L) [54], DDR regulators (CtIP, RAD54, XRCC1) [54, 55], RNA metabolism, Smad3 transcriptional activity [56] (Fig. 3).
Interestingly, CDK1 also shows promiscuity being able to bind all the G1/S phase transition Cyclins forming atypical active complexes [57]. Another milestone that proves how intricated and cryptic this field still is, also came from the lab of P. Nurse. By means of phosphoproteomic assays of in vivo CDK activity in fission yeast, they showed that S-CDK (S-phase) and M-CDK (mitotic phase) have redundant activities on similar substrates and that, in peculiar conditions, S-CDK can also drive mitosis [58]. In the context of M-CDK, ground-breaking quantitative mass spectrometry combined with small-molecule chemical inhibition of Cdk1 identified over 300 potential Cdk1 targets in yeast [59], underlying the plethora of processes in which this kinase takes part, ranging from DNA replication and repair [60] to mitotic spindle assembly. In contrast with its role in yeast, Cdk1 seems to be indispensable for the mammalian cell cycle, as its genetic substitution by Cdk2 leads to embryonic lethality and loss of meiotic function of Cdk2 [61].
In humans, at the interface between S and G2 phases CDK1 and CDK2 phosphorylate the tumor suppressor BRCA2 to control its interaction with RAD51, hence stimulating homologous recombination-dependent DNA repair. At the onset of mitosis, CDK1 inhibits both CHK1 and WEE1 kinases to prevent their activity and drive cells through prophase [62, 63]. Once entered mitosis, CDK1, in a complex with Cyclin A or B, triggers centrosome separation, nuclear envelope breakdown, and chromosome condensation [64]. During metaphase, the separation of the sister chromatids requires CDK1 activity to be switched off, to allow a large cysteine endopeptidase, called separase, to be activated [65]. The mechanism of CDK1-Cyclin B inhibition of separase was recently described and consists of the employment of pseudo-substrate motifs from intrinsically-disordered loops in the separase itself [66]. Additionally, prolonged inactivation of CDK1 is needed for chromosome decondensation, to regenerate the nuclear envelope, and for cytokinesis [67]. During anaphase, the E3 ubiquitin ligase APC/C, initially triggered by CDK1, promotes Cyclin B degradation to eventually turn off CDK1 activity and allow the mother cells to exit the cell cycle (Fig. 3). The ability of CDK1 to switch the order and timing of the phosphorylations on its multiple targets is dictated by Cks1, a phospho-adaptor subunit of the Cyclin- CDK1 complex. Interestingly, in contrast with the current consensus model of CDK1 as an exclusively proline-directed kinase, it has been recently shown that Cyclin A and Cks1 enhance non-proline-directed phosphorylation, preferably on sites with a +3 lysine residue [68].
As summarized above, despite initially being reputed essential for driving the cell cycle phases, interphase CDKs are only essential for the proliferation of specific cell types. On the other hand, tumors often display cell cycle defects that are likely to be mediated by unrestrained CDKs activity, which contributes to unscheduled proliferation as well as genomic instability. Among the interphase CDKs, a miscoding mutation (R24C) in CDK4 is present in a small set of melanoma patients, hindering the binding of Ink4 inhibitors. Furthermore, CDK4 amplifications are present in a number of malignant solid-tumor patients such as malignant gliomas, liposarcoma, non-small cell lung carcinoma, soft tissue sarcoma, lung adenocarcinoma and osteosarcoma [69]. Interestingly, genetically-engineered mouse models having R24C gain-of-function mutation induce endocrine neoplasia (insulinomas, and Leydig cell and pituitary tumors), epithelial hyperplasia (of the liver, gut and breast) and sarcomas [70, 71].
Although rarely mutated, sporadic cases with chromosomal translocation involving CDK6 were identified in some leukemias because of nearby translocations. Additionally, CDK6 is mutated in medulloblastoma, anaplastic ependymoma, high-grade gliomas, germ cell tumor and breast carcinoma, and esophageal carcinoma patients [69].
Even less frequent, CDK2 amplification is present in just 0.06% of AACR GENIE cases, with gastric adenocarcinoma, conventional glioblastoma multiforme, de-differentiated liposarcoma, lung adenocarcinoma, and with lung pleomorphic carcinoma having the greatest prevalence [69]. Recently, a comparative proteogenomic analysis has identified distinctive features of GBMs and LGGs, indicating CDK2 inhibitor might serve as a promising drug target for GBMs [72].
Despite being universally considered as potential oncogenes, the causal link between these mutations and tumor transformation is still missing, mostly because these mutations typically co-occur with other oncogenes/onco-suppressors dysregulation, and it is difficult to assess their actual contribution (see MDM2 or p53) to the oncogenesis process [73]. Nonetheless, the connection between CDKs activity and cancer is even more solid, taking into account mutations that directly contribute to CDK4/6 and CDK2 activation, such as overexpression of Cyclins (mainly D1 and E1), as well as loss of CKI (mainly INK4A, INK4B and KIP1). All together, these data highlight the relevance of CDKs in cancer ontogenesis and solidly argue for selective CDK inhibition as a therapeutic strategy against a plethora of malignant neoplasias (Fig. 4).
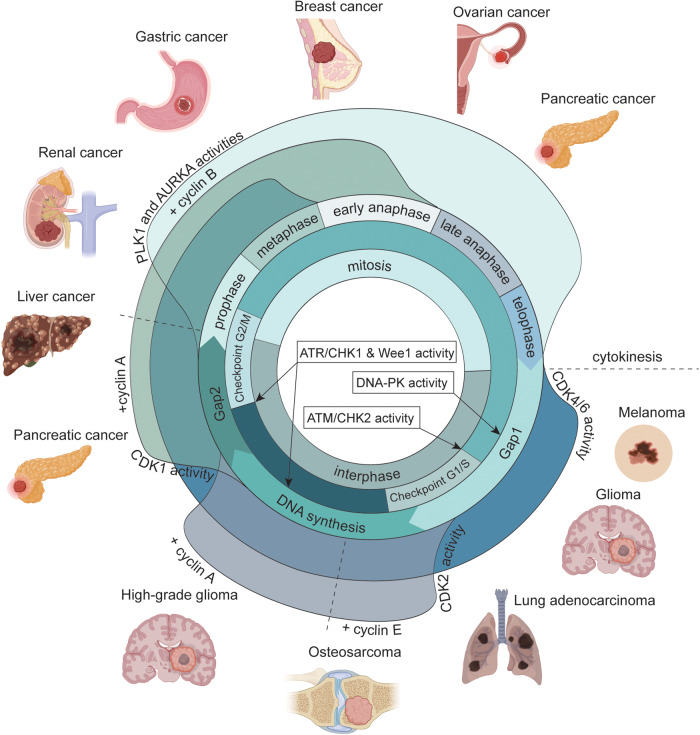
Fig. 4. Cell cycle-associated kinases activity waves and their related tumors.
Cell division in unicellular and multicellular eukaryotes is governed by a sophisticated network of regulatory systems and balances to ensure that no errors occur before a cell is permitted to enter and advance through the cell cycle. Cell cycle is divided into mitosis and interphase (inner circle), and within each of these cellular states multiple subphases are present. The timely and precise duplication and segregation of the genomic DNA is granted by multiple kinases activity (outer waves and inner circle boxes), that when deregulated contributes to the onset of several cancer types (in the figure the type of cancer is related by proximity to the respective kinase wave). Figure created with BioRender.com.
Cell cycle checkpoint kinases
G1/S, S and G2/M checkpoints
During cell cycle progression, DNA is constantly exposed to both endogenous and exogenous insults (i.e., oxidative stress, RS, inflammation, chemicals and radiation exposure), which can compromise DNA integrity, leading to chromosomes replication and segregation errors, DNA damage and eventually cancer transformation. In this context, eukaryotic cells have evolved a system of proteins, known as cell cycle checkpoint kinases, which participate in the so-called DNA damage response (DDR). The DDR pathway consists of a network of proteins including DNA damage sensors, transducers, mediators and effectors, which overall safeguards the integrity of the genome. Indeed, once the DNA damage signal is recognized by sensor proteins (such as γH2AX, PARP1 and the MRN complex), cell cycle checkpoint kinases transduce the signal, either arresting the cell cycle and promoting DNA repair, or in case of unrepairable damage leading to cell death. These kinases include the Ataxia Telangiectasia Mutated (ATM) kinase, the Ataxia Telangiectasia and Rad-3 related (ATR) kinase, the DNA-dependent protein kinase (DNA-PK), the Checkpoint Kinase 1 (CHK1), the Checkpoint Kinase 2 (CHK2) and the mitosis inhibitor protein kinase WEE1. ATM, ATR and DNA-PK belong to the family of the phosphatidylinositol 3-kinase related kinase (PIKK), and they all share a similar structure. In all cases, they have a catalytic domain at the C-terminal, flanked upstream by the FAT (FRAP-ATM-TRRAP) and the HEAT-repeats domains, and downstream by the PRD (PIKK regulatory domain) and the FATC (FAT C-terminal) domains. Moreover, the PIKKs preferentially phosphorylate a serine or threonine residue followed by a glutamine (S/T-Q) and they also contain S/T-Q motifs, reflecting their capacity of auto-phosphorylation. Conversely, CHK1, CHK2 and WEE1 are structurally different, although they all belong to the broader family of serine/threonine (Ser/Thr) protein kinases.
Given their pivotal role in genome surveillance and cell proliferation, the cell cycle checkpoint kinases are rarely deregulated in cancer, representing a survival advantage for cancer cells, and leading often to chemotherapy resistance. In contrast, when deregulated in precancerous lesions, these kinases may promote cancer transformation. Moreover, loss of ATM, ATR or CHK1 activity leads to early embryonic lethality in mice [74–76], revealing their essential role also in physiological conditions. Conversely, DNA-PK or CHK2 null mice show normal size at birth, with severe immunodeficiency in the case of DNA-PK loss [77] and associated radio-resistance in CHK2 defective mice [78], suggesting that these pathways might be somehow redundant or overlapping during embryonic development. In addition, it is well shown that mutations in any of the cell cycle checkpoint kinases dramatically increase the susceptibility to cancer transformation, and/or are very often associated with severe genetic disorders [79].
Depending on the type of damage and at which phase of the cell cycle it occurs, different pathways are activated, with the ATM/CHK2 axis regulating the G1/S checkpoint in response to DNA double-strand breaks (DSBs) or the ATR/CHK1/WEE1 axis controlling the S and the G2/M checkpoints upon RS and DNA single-strand breaks (SSBs). Instead, DNA-PK activation is less specific, being mainly triggered by DSBs, but also involved in theRS [80, 81]. While each kinase plays a specific role in cell cycle checkpoint activation, their interconnection is also crucial for the maintenance of genetic stability in response to DNA damage, as recently revealed by phosphoproteomics analysis [82]. Here we provide a summary of the main roles of the principal drivers of DDR, ATM, ATR and DNA-PK.
ATM has been first discovered in a patient affected by Ataxia Telangiectasia (AT) in 1995, and was found to encode a large protein of approximately 350 kDa [83]. AT is a severe genetic disorder characterized by cerebellar degeneration, immunodeficiency and cancer predisposition. Since then, many studies focused mainly on the role of ATM in AT pathogenesis and on DDR; however, new roles in redox homeostasis, proteostasis and mitochondrial metabolism have been also discovered.
In undamaged conditions, ATM exists as inactive dimers and it is canonically activated by DSBs through the interaction with the MRN (Mre11, Rad50 and Nbs1) complex, which induces ATM autophosphorylations and monomerization [84]. Ser 1981 is the first site autophosphorylated by ATM straight after DNA damage. However, autophosphorylation at other sites including Ser367, Ser1893 and Ser2996 has been reported in response to DSBs [85]. Upon activation, ATM elicits a cascade of reactions which includes phosphorylation and stabilization of p53 and CHK2, leading respectively to p21 activation and CDC25A inhibition, resulting in the arrest of the cell cycle at the G1/S transition. Indeed, on the one hand, p21 activation inhibits Cyclin A/CDK2 and Cyclin E/CDK2 complexes and, on the other hand, CDC25A inhibition prevents the removal of inhibitory phosphates on CDK2. In this scenario, DNA repair by homologous recombination (HR) can occur before DNA duplication in the S phase, in order to avoid genetic instability. In the past decades, a number of ATM substrates have been identified, including γH2AX, KAP1, BRCA1, ATF2, MDMX, RAD9A, CHK1 and many others, all together coordinating cell cycle arrest, DNA repair or apoptosis, as reviewed elsewhere [86–88] suggesting the central role of ATM in the DDR and as a tumor suppressor.
However, being ATM mutated in approximately 5% of all cancer, it might represent an advantage for those cancer cells carrying the wild-type form when exposed to DNA damage agents. In contrast, cancer patients where ATM mutation results in loss of function may benefit from PARP and ATR/CHK1 inhibitors co-treatments, such as in mantle cell lymphoma (MCL) or small cells lung cancer (SCLC), where ATM is mutated in 40 and 20% of the cases, respectively [89–95]. Indeed, in these contexts, cell cycle checkpoints might be overridden, resulting in DNA damage accumulation and cell death. Nevertheless, ATM mutation is not always predictive of higher sensitivity to the treatments mentioned above, as recently shown in neuroblastoma models [96].
In addition to its wide role in DDR, ATM is also crucial in the control of redox homeostasis, as shown in different in vivo and in vitro ATM-deficient models, as well as AT patients’ plasma, which has been shown to exhibit excessive accumulation of ROS [97–100]. In 2010, two seminal papers demonstrated ATM activation through the formation of disulfide bonds between its two monomers in response to oxidative stress. Indeed, in ATM-proficient cells, exposure to ROS directly activates ATM in the absence of DNA damage and the MRN complex, leading to the activation of TSC2 via the LKB1/AMPK pathway. This, in turn, causes the inhibition of the mTOR pathway and the induction of autophagy, contributing to the clearance of ROS-oxidized organelles and proteins [101, 102]. These and further studies clearly established ATM cytosolic functions, also showing its localization on peroxisomes and mitochondria where, in response to ROS, ATM induces pexophagy and mitophagy, respectively [102–105]. Importantly, ATM is also activated by hypoxia in an MRN complex-independent manner [106]. Under hypoxia conditions, ATM has been shown to phosphorylate mono-ubiquitinated H2AX (mUb-H2AX) by TRAF6, inducing HIF1α nuclear retention and stability and leading to the transcription of genes involved in glycolysis, cell survival, proliferation, and invasion, in different tumor contexts [107]. All together these studies outline the central role of ATM in sensing and coping with different types of stress, such as DNA damage and metabolic stress. As a tumor suppressor, ATM activity is beneficial during development and at early cancer stages, while it may promote cancer cells resistance to radiotherapy and chemotherapy at later stages. Thus, the evaluation of mono or combinatory treatments by using molecules targeting ATM should be carefully considered and adapted to the specific cancer context (Fig. 4).
ATR has been identified in 1996, as the human homolog of the fission yeast Mec1 and Rad3 proteins [108, 109]. Being active across the S and the G2/M checkpoints, ATR has the critical function of preserving DNA integrity during replication and before segregation. Therefore, any condition that perturbs the progression of the DNA replication fork causes activation of ATR, which in turn coordinates a cascade of reactions known as RS response. Regardless of the exogenous and endogenous stimuli that trigger RS, ATR is recruited to Replication Protein A (RPA)-coated single strand DNA (ssDNA), by its stable binding partner ATRIP (ATR interacting protein). However, to be fully activated, ATR requires further interaction with TopBP1 or ETAA1 proteins. Both contain an ATR activation domain that stimulates ATR enzymatic activity, but TopB1 interacts also with ATRIP [110], while ETAA1 directly binds RPA-coated ssDNA [111–114] suggesting that they can activate ATR in response to different sources of RS. Once activated, ATR phosphorylates CHK1 on Ser317 and Ser345, which in turn leads to the proteasomal degradation of CDC25A, a CDKs-specific phosphatase involved in the removal of inhibitory phosphates [115, 116]. Thus, CHK1 activation slows down or arrests cell cycle progression allowing DNA repair or, if the damage is too extensive, leads to senescence or programmed cell death. In response to RS, ATR activation safeguards DNA integrity in both S-phase and G2/M transition. In S-phase, ATR activation prevents fork collapse into DSBs by at least two distinct mechanisms: inhibiting new origin firing and regulating key fork remodeling enzymes. Indeed, ATR activation leads to the inhibition of CDK2, which is crucial for the loading of Cdc45 origin binding factor and the DNA polymerase recruitment on chromatin [117], therefore limiting new origin firing. This, in turn, prevents RPA exhaustion and contributes to the DNA replication fork stability [118]. Furthermore, ATR activation directly targets the SMARCAL1 helicase and promotes FANCD2 interaction with MCM helicase, suppressing fork collapse [119–121]. The importance of ATR in the intra-S checkpoint is emphasized in many studies, where it is shown that in ATR inhibition or ATR-deficiency contexts, cells fail to cope with RS, leading to fork collapse and DSBs [122–124]. Moreover, ATR plays a key role in the dNTP biosynthesis, regulating the expression of ribonucleoside-diphosphate reductase subunit M2 (RRM2), and in the sensing of nucleotide imbalance during unperturbed S-phase [125, 126]. On the other hand, in response to DNA damage, ATR mediates cyclin F degradation, thus preventing RRM2 degradation via SCF(cyclin F) [127].
To prevent DNA missegregation, ATR also controls the so-called G2/M checkpoint through its direct target CHK1, which phosphorylates CDC25C leading to inhibition of CDK1 and mitosis entry [76, 128]. Importantly, ATR activity is shown to be crucial not only upon DNA damage but also during embryonic development, in case of re-replication and stochastically in an unperturbed cell cycle, highlighting once again its central role in preserving genome integrity [74, 129–131]. Finally, ATR has been also described as the coordinator of an S/G2 checkpoint, controlling the phosphorylation status of the mitosis transcription factor FOXM1, although recent observations have pointed out the existence of an ATR-independent S/G2 checkpoint [132, 133].
As mentioned above, in addition to its best-characterized target CHK1, many different substrates of ATR have been discovered over the years. Most ATR substrates are proteins involved in DNA damage signaling, DNA replication and DNA repair, and many of them are shared with ATM [134], establishing a crosstalk between the two pathways. However, by large proteomic analysis, unique ATR substrates are identified, including 53BP1, NuMa1, Smc1, Pnk1 and others, as reported by Stokes and colleagues [135]. Moreover, a specific map of ATR signaling has been described in mouse testes, discovering that ATR controls CDK2 localization on meiotic chromosomes during spermatogenesis [136].
As described for ATM, also ATR is essential for proliferating cells, and its loss leads to E7.5-E8.5 embryonic lethality in mice [137]. Hypomorphic mutations of ATR, leading to low expression levels of the protein, are associated with the Seckel syndrome characterized by craniofacial abnormalities, microcephaly, and growth retardation [138]. However, in contrast to other DDR syndromes, Seckel syndrome is not associated with an increase in cancer predisposition. More in general, ATR is rarely mutated in cancer (2.7% of all cancer cases, as reported by the cBioportal), its activity being critical mainly for the survival of Myc- or Cyclin E-deregulated tumors, associated with oncogene-induced RS [139] (Fig. 4). For this reason, over the last two decades, many studies have explored the use of ATR inhibitors as a potential sensitizer of conventional cancer therapy, as nicely reviewed by Barnieh and colleagues [140].
The existence of the DNA-PK was first discovered by accident in 1985, as a kinase activated by dsDNA, and purified later by three different groups [141–143]. DNA-PK has a large catalytic subunit and is a crucial sensor of DSBs, recruited on the site of damage by the Ku heterodimer, a basket-shaped structure composed of the Ku70 and Ku80 subunits, in which dsDNA ends are accommodated. Once activated by DNA damage, DNA-PK promotes DNA repair by non-homologous end joining (NHEJ) throughout cell cycle. However, NHEJ is more active in G1 phase, where it is preferred to HR, since no homologous templates for recombination are available. In addition, it has been recently demonstrated an active role of DNA-PK in controlling replication fork dynamics, not only in the restart of stalled replication fork [144], but also in promoting fork reversal [145]. Furthermore, DNA-PK has also important functions in RNA processing, transcription regulation and inflammation [146–148] indicating a prominent role of this kinase in the surveillance of cell homeostasis.
Instead of other PIKKs, the main phosphorylation substrate of DNA-PK is itself. In fact, in its DNA-binding domain, it has two clusters of phosphorylation sites, the PQR and the ABCDE cluster, containing key residues that, when auto-phosphorylated, determine the activity of DNA-PK on the coordination of NHEJ. The most frequent sites of auto-phosphorylation are T2609 and S2056 residues, which are on the ABCDE and the PQR cluster, respectively. The phosphorylation of the first site has an important role in DNA end protection and in the recruitment of the nuclease Artemis, while the second leads to DNA-PK disassembly to allow end processing by other NEHJ factors [149, 150]. However, recent cryo-EM structural studies revealed the existence of a phosphorylation/dephosphorylation balance, especially in the ABCDE cluster, which is crucial for the shift between an inactive and an active conformation of DNA-PK, providing a new model of NHEJ regulation. While activated DNA-PK phosphorylates different NHEJ factors, such as Ku70, Ku80, Artemis, XLF, XRCC4 and PNKP, it is dephosphorylated by the phosphatase PP6, which provides new sites of phosphorylation keeping DNA-PK active [151]. Taken together, these studies demonstrate a tight autoregulation of DNA-PK in response to DNA damage, that contributes to the maintenance of genome stability and cell survival. However, it is worth noting that NHEJ is an error-prone DNA repair mechanism, which when it fails is bypassed by HR thanks to an elegant interconnection between DNA-PK, ATM and ATR.
As opposed to ATM and ATR, DNA-PK catalytic subunit loss does not result in embryonic lethality in mice [77], but as ATM and ATR, it is rarely mutated in cancer (4% of all cancer, as reported by the cBioportal), representing a possible advantage for tumor cells. Indeed, it has been shown that in ATM-deficient cells, DNA-PK plays a key role in preventing mitotic aberration and cell death, possibly compensating for ATM functions [152, 153]. Moreover, DNA-PK has been recently identified as a crucial regulator of the stemness properties of glioblastoma [154]. In contrast, DNA-PK dysregulation has been linked to a higher sensitivity to DNA-damaging agents, particularly to ionizing radiation (IR), making this kinase a strong therapeutic candidate for the treatment of tumor malignancies. Despite the sophisticated structure of the DNA-PK complex, many small-molecule inhibitors have been discovered over the years, some of which are currently undergoing clinical trials as single agents or in combination with chemotherapeutic drugs, as reviewed elsewhere [155].
Although DNA-PK has been mostly studied for its critical role in DDR, non-canonical functions are also emerging. DNA-PK is also found to be involved in aging and energy metabolism. One of the first hints of a DNA-PK role in aging, comes from the observation that it contributes to telomere maintenance and capping [156, 157]. Moreover, aging is characterized by metabolic and fitness decline, which can cause DNA breaks and activate DNA-PK. In this context, Park and colleagues have demonstrated that DNA-PK prevents the activation of the AMPK pathway, leading to dysfunctional mitochondrial biogenesis and energy metabolism, and to obesity and diabetes [158]. Other studies report additional DNA-PK roles, which are important for cellular homeostasis in both cancer and physiological condition, as nicely reviewed by Goodwin and Knudsen [159] (Fig. 4). Altogether, these findings indicate that our understanding of the multiple roles of DNA-PK is far from being complete and should foster our attention also on less explored angles, which might have important implication not only in cancer but also in aging-related diseases.
WEE1 kinase family is composed by three serine/threonine kinase members: PKMYT1 (membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase) and two WEE1 kinases (WEE1 and WEE1B). Despite the considerable conservation of their molecular structures, only PKMYT1 and WEE1 display a relevant role in eukaryotic somatic cell cycle regulation, while WEE1B activity is restricted to the maintenance of the meiotic arrest in oocytes during the germinal vesicle stage. Accordingly, the following section will discuss the first two, and therefore we refer the author to more detailed references for the third one [160, 161]. The structure of WEE1 kinases is comparable to other known kinases structures, with two lobes connected by a flexible hinge region. The N-terminal lobe is composed of five standard β-sheets and one α-helix, adjacent to the ATP-binding cleft. Also, at the N-terminal, a Glycin-rich loop, also known as P-loop, assumes different conformation depending on the bound ligand and the enzymatic activity of the kinase. The activation loop and the catalytic cleft are instead localized within the C-terminal domain. The catalytic cleft is separated into a front and back cleft, involving respectively the ATP-binding pocket, and the crucial residues for kinase regulation. Interestingly, it was proposed that WEE kinases do not need to be phosphorylated on the activation loop for their activation, since the residue that precedes the catalytic aspartate (WEE1: Asp426, PKMYT1: Asp233) is a nonpolar one (WEE1: Met425, PKMYT1: Leu232), instead of the typical arginine [162].
Both WEE1 and PKMYT1 target CDK1 to maintain it inhibited until the cell approaches mitosis.
WEE1 acts specifically to phosphorylate Tyr15, while PKMYT1 possesses a dual specificity for Tyr15 and Thr14 [163]. The reasons for such different specificity resides into a single differing residue in their P-loop controlling the Thr kinase activity of PKMYT1. Indeed, WEE1 possesses a Glu309 at the apex of this loop (substituted by the less bulky Ser120 in PKMYT1) that may impede a closer approach to CDK1 through steric hindrance [164].
WEE1 kinases also differ in their subcellular localization. While WEE1 localizes into the nucleus, PKMYT1 is manly associated to the endoplasmic reticulum and Golgi apparatus [165, 166].
Along the cell cycle, WEE1 family physiological roles span from the regulation of replication dynamics during S-phase (intra S-phase checkpoint) to the checkpoint function at the G2/M boundary. Indeed, similarly to CDK1, WEE1, but not PKMYT1 [165], exerts its activity also on Tyr15 of CDK2 [167, 168], therefore modulating S-phase dynamics. Additionally, across S-phase, WEE1 is also implicated in maintaining genome integrity by regulating Mus81-Eme1 endonuclease activity. Mus81 nuclease is constitutively active throughout the cell cycle, but can efficiently target its substrates only upon SLX4 association. To prevent unwanted processing of replicating chromosomes, WEE1 kinase restrains CDK1 and PLK1-mediated MUS81-SLX4 assembly during S phase [169, 170]. In late S-phase, WEE1 was also demonstrated to have an evolutionarily conserved epigenetic function by providing H2B Tyr37 phosphorylation. Such histone modification inhibits the transcription of multiple histone genes, therefore reducing the burden on the histone mRNA turnover machinery [171].
In the presence of DNA damage, ATM or ATR kinase pathways are differentially activated depending on the source of genotoxic stress. As previously discussed, while ATM is activated in response to DSBs, ATR is triggered by a broad range of genotoxic stresses that result in SSBs. ATR activation stimulates CHK1 activity that in turn phosphorylates WEE1 resulting in cell cycle arrest via CDK1 inactivation.
Then, at the onset of mitosis, through a negative feedback loop, CDK1 phosphorylates both WEE1 and PKMYT1, priming them for PLK1 and CK2 kinase-mediated phosphorylation. All together, they generate an unconventional phospho-degron that can be recognized by the ubiquitin ligase SCFβ-TrCP [172–174]. Once the mitotic process is completed, WEE1 is again activated by the FCP1 phosphatase [175] that is responsible for its dephosphorylation.
Being gatekeepers of G2 arrest, WEE1 and PKMYT1 are rarely mutated in cancer patients, and little is known about the functional consequences of such mutations. However, to sustain cancer cell proliferation despite the high genomic instability, WEE1 is expressed at high levels in various cancer types including breast cancers [176], leukemia [177, 178], melanoma [179], and adult and pediatric brain tumors [180–183], where it has been correlated with a worse prognosis in patients (Fig. 4). At variance with that, a number of studies have reported an opposite relationship between WEE1 expression and prognosis in non-small cell lung cancer (NSCLC) and colon cancer [184, 185].
Mitotic checkpoint kinases
PLK1 (Polo-like kinase 1) is a Ser/Thr kinase belonging to the Polo-Like Kinase family, which has been first identified by genetic screens in yeast and Drosophila, as a family of five crucial mitotic regulators [186, 187]. PLKs share a similar structure containing an N-terminal kinase domain (KD) and two C-terminal Polo-box domains (PBD), which are important for the kinase subcellular localization. Importantly, the KD and the PBD can interact and inhibit each other. PLK1 is the founder of the family, and it controls a variety of processes throughout the cell cycle, including mitotic entry, centrosome maturation, chromosome segregation and cytokinesis [188–194]. Therefore, unlike other cell cycle checkpoint kinases, PLK1 is inhibited rather than activated during DDR, in order to prevent cell division in the presence of unresolved DNA damage. Indeed, both ATM and ATR have been shown to inhibit PLK1 in response to DNA damage by direct phosphorylation, or indirectly promoting PLK1 dephosphorylation by PP2A [195–197]. These data and other observations highlight once again the existence of a fine interplay among various cell cycle kinases, in which each plays a delicate role in safeguarding the genome. However, due to the redundancy of some functions, both normal and cancer cells have evolved the ability to survive also when deficient in one of these regulators, showing an extraordinary adaptive and flexible behavior.
In this regard, PLK1 is activated by other kinases only when the DNA has been properly replicated and/or the DNA damage has been repaired. The crucial residues for such activation are Thr210 and Ser137, both localized on the kinase domain of PLK1 [198]. While the dynamics of the former phosphorylation are largely elucidated, the function of the latter is still not completely clear. Indeed, two contrasting studies report that PLK1 is phosphorylated on Ser137 in late mitosis or prior mitosis [198, 199] opening the question of whether this phosphorylation has other mitosis-independent functions not yet discovered. However, these studies demonstrated that when both sites are phosphorylated, the catalytic activity is enhanced. By contrast, different studies consistently suggest a detailed model, by which Thr210 phosphorylation is mediated by the kinase AuroraA and its cofactor Bora, with this representing a crucial step for mitosis onset. According to this model, Bora is activated by CDK1/Cyclin A in G2 phase, and binds to PLK1 in the cytosol, where PLK1 is kept in an inactive conformation by the interaction of its PBD and KD domains. Bora binding leads to a conformational change, which exposes a nuclear localization sequence (NLS) of PLK1 and induces its translocation to the nucleus. Importantly, NLS disruption leads to G2 arrest, indicating that PLK1 nucleus translocation is crucial for mitosis onset. At the G2/M transition, AuroraA phosphorylates PLK1 at Thr210, inducing mitosis entry, while Bora is degraded through phosphorylation by PLK1 [17, 200–203]. Recently, this model has been further detailed by the observation that Bora activation triggers PLK1 dimerization in G2, inducing a conformational change that makes accessible the Thr210 for AuroraA-dependent phosphorylation, in late G2. Thr210 phosphorylation leads in turn to the dissociation of the dimer and the exposure of the NLS, which allows PLK1 nuclear translocation, supporting mitosis entry [204]. Once activated, PLK1 sustains CDK1/Cyclin B activation by degrading the mitotic inhibitor WEE1 and inhibiting the kinasePKMYT1, which both catalyze inhibitory phosphorylation of CDK1. Moreover, PLK1 directly phosphorylates Cyclin B and CDC25C promoting their translocation to the nucleus and thus further supporting CDK1 activity [17, 173, 174, 200, 205]. Although we have here summarized the role of PLK1 in the entry into mitosis, it is important to note that its function does not end with the initiation of mitosis, but its activity is rather required in every mitotic stage up to cytokinesis, as well as during S phase. Indeed, a variety of phosphoproteomic studies demonstrated that PLK1 can phosphorylate thousands of sites on hundreds of proteins, hence regulating a plethora of biological processes and pathways [206–210].
As mentioned above, PLK1 regulates centrosome maturation by directly targeting CEP192 and Cenexin, both involved in the recruitment of pericentriolar material, including the γ-Tubulin Ring Complex (γ-TuRC), essential for microtubules nucleation [211, 212]. Moreover, PLK1 is recruited on the kinetochore in metaphase, where it phosphorylates KNL-1 and MPS-1, executing the so-called spindle assembly checkpoint (SAC) to control whether chromosomes are correctly aligned and to prevent premature mitosis exit [213]. In anaphase, most mitotic proteins are degraded by the ubiquitin ligase APC complex. PLK1 promotes APC complex activation by phosphorylation-dependent degradation of EME1, a major APC complex inhibitor [214]. Finally, PLK1 localizes in the midzone during cytokinesis, interacting with several proteins and phosphorylating abscission-factors such as CEP55, to properly coordinate abscission [215]. Although PLK1 is mostly studied for its role in mitosis regulation, its involvement is also reported in DNA replication, DDR, and genome stability as well as in processes beyond cell cycle, such as autophagy, epithelial-mesenchymal transition and inflammation. For a more comprehensive description of these functions, we refer the readers to a recent review by Iliaki and colleagues [216].
Considering the pleiotropic role of PLK1 in regulating many fundamental processes for cell survival and proliferation, it is reasonable to believe it may function as a pro-oncogenic factor. This is further supported by the fact that PLK1 is frequently overexpressed in human cancer, including breast cancer and gastric adenocarcinoma, and its expression correlates with poor prognosis. In this regard, PLK1 can negatively regulate tumor suppressors such as TP53 and PTEN, as well as stabilize tumor-promoting factors, including the oncogene MYCN [217–219]. Paradoxically, PLK1 can also act as a tumor suppressor. Interestingly, PLK1 loss is embryonic lethal in mice, indicating its crucial function in cell division. However, PLK1 heterozygotes mice are healthy at birth, but they develop tumors with a three-fold higher incidence than the wild-type counterpart, suggesting that PLK1 might have tumor-suppressing properties in a haploinsufficient context [220]. Finally, as other kinases mentioned in this text, PLK1 is rarely mutated in cancer, as reported in TCGA, PanCancer Atlas dataset, with this pointing out once again the essentiality of this kinase in both physiological and cancer conditions. For these reasons, targeting PLK1 as a therapeutic strategy for cancer treatment has been considered a double-edge sword for a long time. On the one hand, targeting PLK1 in cancer would be detrimental for cancer progression, but on the other hand, this may result in toxicity also in normal healthy cells, thus limiting PLK1 inhibitors usage in cancer patients. Nevertheless, both academia and industry have put efforts into the development of specific PLK1 inhibitors, and nowadays there are two classes of small molecules available, one targeting the ATP binding domain and the other the PBD. Some of these inhibitors have also entered in clinical trials (Table 1) for patients with different tumors, but their therapeutic index is not yet satisfactory [221, 222]. Moreover, the efficacy of PLK1 inhibitors has been shown to be also dependent on the tumor genetics (Fig. 4). For instance, patients with colon cancer harboring APC mutation have a better prognosis when PLK1 expression is higher [223]. Instead, PLK1 inhibition has a strong anti-tumor activity in patient-derived xenograft of metastatic breast cancer with CCND1 amplification and Palbociclib resistance [224]. Therefore, the therapeutic index of PLK1 inhibitors must be carefully evaluated, especially regarding the systemic toxicity and the tumor context, making the race toward a more personalized medicine extremely urgent.
Table 1.
Clinically relevant cell cycle kinase inhibitors.
| Targeted kinase | Tumor | Inhibitor | Clinical trials (NCT nUMBER) | Phase | Status |
|---|---|---|---|---|---|
| CDK4/6 | Liposarcoma (CDK4/6 amp) | Abemaciclib | NCT04967521 [251] | Phase 3 | Recruiting |
| Sarcoma (CDK4 overexpression/amp) |
Palbociclib Palbociclib |
Phase 2 Phase 2 |
Recruiting Completed |
||
| Solid tumors (CDK4/6 amp) |
Abemaciclib Palbociclib |
NCT01037790 [252, 255] |
Phase 2 Phase 2 |
Recruiting Completed |
|
| Metastatic breast cancer (HR + ; HER2-) | Abemaciclib + Aromatase Inhibitor / Abemaciclib + Fulvestrant | NCT05362760 | Phase 4 | Recruiting | |
| Glioblastoma (CDK4/6 amp) |
Abemaciclib Palbociclib |
Phase 2 Phase 2 |
Active Terminated |
||
| Cancer with brain metastasis | Abemaciclib | NCT02308020 [258] | Phase 2 | Completed | |
| Non-Small cell lung cancer |
Abemaciclib Abemaciclib + Osimertib |
NCT02152631 [259, 260] |
Phase 3 Phase 2 |
Active Unknown |
|
|
Squamous cell lung cancer (CDK4 amp) |
Palbociclib + Docetaxel | NCT02785939 | Phase 2/3 | Completed | |
|
Esophagogastric cancer (CDK6 amp) |
Abemaciclib | NCT03292250 | Phase 2 | Completed | |
| CDK2 | Breast cancer and solid tumors | PF-07104091 | NCT05262400 | Phase 2 | Recruiting |
| ATR | Solid tumors | Berzosertib | NCT03718091 [262] | Phase 2 | Completed |
| Ovarian cancer | Berzosertib + gemcitabine | NCT02595892 [263] | Phase 2 | Active | |
| Solid tumors (relapsed or refractory) | BAY1895344 | NCT05071209 | Phase 1/2 | Active | |
| Non-small cell lung cancer | Ceralasertib | NCT05450692 | Phase 3 | Recruiting | |
| Advanced solid tumors and Hematological Malignancies | ATG-018 | NCT05338346 | Phase 1 | Recruiting | |
| DNA-PK | Rectal cancer | Peposertib | NCT03770689 | Phase 1/2 | Completed |
| CHK1/2 | Solid tumors | LY3295368 | NCT02873975 | Phase 2 | Completed |
| Small cell lung cancer | LY3295368 | NCT02735980 | Phase 2 | Completed | |
| Ovarian Carcinoma, Endometrial Adenocarcinoma, and Urothelial Carcinoma | LY2606368 | NCT05548296 | Phase1/2 | Recruiting | |
| WEE1 | Triple-negative Metastatic Breast Cancer | MK-1775 | NCT03012477 | Phase 2 | Completed |
| Adenocarcinoma of the Pancreas | MK-1775 | NCT02037230 [264] | Phase 1/2 | Completed | |
| Advanced solid tumors | Debio 0123 | NCT05109975 | Phase 1 | Recruiting | |
| High-Grade Serous Ovarian, Fallopian Tube or Primary Peritoneal Cancer (CycE Driven) | ZN-C3 | NCT05128825 | Phase 2 | Recruiting | |
| Uterine Serous Carcinoma | ZN-C3 | NCT04814108 | Phase 2 | Recruiting | |
| PLK1 | Pancreatic cancer | Rigosertib + Gemcitabine | NCT01360853 [265] | Phase 3 | Completed |
| Myelodysplastic Syndromes | Rigosertib | NCT01241500 [266] | Phase 3 | Completed | |
| AURORA A | Prostate cancer | Alisertib | NCT01799278 [267] | Phase 2 | Completed |
| Acute Myeloid Leukemia | Alisertib | NCT02560025 [268] | Phase 2 | Completed | |
| Cancer Patients (KRAS G12C mutated) | LY2606368 | NCT04956640 | Phase 1 | Recruiting |
In a wide range of tumor malignancies, several kinase inhibitors that target cell cycle regulators are currently undergoing clinical trials for cancer therapy, both as single and combined treatment.
AURORA kinases (Ipl1-like kinases) are a family of proteins that in mammals consists of multiple Ser/Thr kinase crucial for mitotic entry and spindle regulation, namely Aurora A (AURKA), Aurora B (AURKB) and Aurora C (AURKC). On the other hand, Ipl1 is the unique S. cerevisiae representative of a family that diverges into two Ipl1-like kinases (Aurora-A and Aurora-B) in Drosophila, C. elegans and X. laevis.
Initially discovered in 1993 during a search for S. cerevisiae mitotic mutants that failed to undergo typical chromosomal segregation, Ipl1 kinases were then independently identified in cell cycle studies in Xenopus laevis and Drosophila melanogaster [225–229]. In mammals, the three paralogs share very high similarity in sequence, with 71% of identity between Aurora A and B in the carboxyterminal catalytic cleft. Residing also in the C-terminal, the degradation box (A-box/D-box/KEN-box) of Aurora kinases is crucial for its recognition and degradation by APC/C. Conversely, the three Auroras differ in the length and sequence of the amino-terminal domain [230, 231]. In humans, AURKA, AURKB and AURKC map on chromosomes 20q13.2, 17p13.1, and 19q13.43
As often observed in kinases, Aurora kinases are regulated by phosphorylation of a conserved residue in the catalytic T-loop residues, namely Thr288 (AURKA), Thr232 (AURKB), and Thr195 (AURKC), respectively. These residues reside in a RRXS/TY motif, that is recognized and phosphorylated by protein kinase A (PKA). Additionally, the consensus motif has been also identified as an auto-phosphorylation site [232], which is able to boost the catalytic activity by up to 157 times [233]. The reshaping in the folding of the active site caused by such events is necessary but not sufficient for full catalytic activation. For instance, to be fully activated, AURKA depends on the binding to an allosteric activator at the N-terminal called Tpx2 [233, 234]. This microtubule-binding protein once released by RAN-GTP from importins, actively positions AURKA on microtubules to support mitotic spindle assembly. Recently, it was reported that the kinase Bora (see also above) exerts a similar function, thanks to a 100 amino acid region encompassing two short Tpx2-like motifs and a phospho-Serine-Proline motif at Serine 112, that activate cytoplasmic AURKA during mitotic commitment.
Despite showing striking structural and sequence similarities, Aurora kinases exhibit entirely diverse subcellular localization and mitotic functions. For instance, AURKA is initially associated with pericentriolar material at the centrosome from the conclusion of the S-phase, when centrosome duplication occurs, it is then distributed to the pole proximal ends of spindle microtubules, and finally is degraded upon mitotic exit [235]. Conversely, AURKB is widely dispersed into the nucleus until prometaphase, when it travels to the centromeres to form a complex with two other proteins, inner centromere protein (INCENP) and survivin, and it behaves as chromosomal “passenger”. Being a “passenger” protein, AURKB associates with centromeric heterochromatin at the onset of mitosis and gradually moves to the midbody, where it remains until the end of cytokinesis. Very little is known about AURKC apart from that is tipically active in meiotic cells, where it also acts as a chromosomal passenger [236, 237].
Once completed their functions, Aurora kinases are inactivated by dephosphorylation, which is typically carried out by protein phosphatase 1 (PP1). Then, anaphase-promoting complex/cyclosome (APC/C) detects Aurora kinases in late mitosis, and they are subsequently destroyed via the proteasome.
The extensive and thorough description of the three Aurora kinases` function is far beyond the scope of this review, and for that reason we will focus on the one that over the years has been mostly associated with cancer ontogenesis, while referring the reader to other recently issued review and articles on the remaining two [238–240].
As we previously discussed, AURKA in ensuring centrosome functionality and bipolar spindle assembly. AURKA functions in cancer contexts have been widely explored and both oncogenic and tumor suppressor functions have been elucidated. As for the former, AURKA is often amplified in breast, colorectal, ovarian, pancreatic, gastric, bladder, cervical, and head and neck cancer. Similarly, the overexpression mediated by constitutive phosphorylation on S51 that inhibits its APC/C proteasomal degradation contributes to head and neck cancer formation. Further confirmation of its oncogenic abilities was obtained in immortalized rodent fibroblasts, where AURKA overexpression was able per se to induce oncogenic transformation and tetraploidy by boosting chromosome instability [230, 241, 242]. Elegant studies have contributed to the characterization of the functional link between AURKA aberrant expression and oncogenesis.
Additional oncogenic events were reported to contribute to AURKA overexpression. For instance, HER-2 oncogenic signaling, a commonly altered feature of breast cancer, induces AURKA phosphorylation, thereby increasing its stability [243]. TP53 deficiency affects AURKA levels both transcriptionally and post-translationally. In the former regulation, by decreasing p21 expression, hence positively affecting CDK2-E2F3 axis, a well-known AURKA transcriptional activator. In the latter, by downregulating FBXW7, a member of the SCF E3 ligase complex known to ubiquitinate AURKA [244]. Alternatively, AURKA kinase activity directly affects p53 phosphorylation status on S215 and S315, abrogating its transactivation activity [245, 246] and increasing its Mdm2-mediated degradation [247]. Similarly to p53, AURKA exerts its kinase activity also on another well-established tumor suppressor, BRCA1. Aberrant AURKA-mediated phosphorylation on its S305 overrides G2/M checkpoint, leading to centrosome amplification and CIN.
Conversely, acting upstream of AURKA, loss-of-function mutation in the mitotic checkpoint protein Chfr induces aberrant AURKA kinase activity, by impairing its ubiquitination and proteasomal degradation [248].
The first evidence that AURKA could also possess a tumor suppressor function stems from the identification of two coding single nucleotide polymorphisms (SNPs) in AURKA (91 T > A (encoding F31I) and 169 G > A (encoding V57I)), that are associated with reduced kinase activity and increased risk of developing esophageal cancer [249]. In line with that, haploinsufficiency in Aurka+/− mice develops a higher incidence of spontaneous tumor formation [250].
In sum, despite the causal relationship between AURKA amplification and oncogenic formation remains still unresolved (since its genomic aberration typically affects a large chromosomal locus), multiple cues strongly demonstrate that deregulated AURKA activity is a driving force towards genomic instability and tumor progression (Fig. 4).
Conclusions, open questions, and perspectives
Although rarely mutated, cell cycle kinases appear to be frequently deregulated in cancer, and this might be mainly due to altered transcriptional regulation by oncogenes. Indeed, there is a plethora of transcription factors that, when mutated or epigenetically altered, function as oncogenic factors with a crucial role in the initiation/progression of cancer as well as in invasion and chemo-resistance. Most of these factors (e.g., Myc, Ras, E2F, Wnt, etc) are difficult to target. Therefore, many studies have focused on the limitation of their effects, targeting their substrates. However, cell cycle kinase inhibition has in many cases yet to be carefully evaluated with regard to tumor genetics. To this end, the capacity of sequencing in public health systems should be increased, to better tailor the treatment according to the patient.
Both Academia and Industry have made a lot of efforts in the development of a variety of small-molecule inhibitors targeting cell cycle kinases with some of them entered in advanced clinical trials (Table 1). However, many inhibitors showed limitations in their application, due to low specificity and/or to the severity of adverse effects. One of the reasons is that most kinase inhibitors are designed against the ATP-binding site, making them less specific. However, nowadays there are new classes of inhibitors binding other residues on the kinases and inhibiting the protein activation with different mechanisms.
Although this increased specificity, it still does not prevent the chemotherapy-acquired resistance often seen in cancer therapy, due to the rise of kinase mutations. Hence, future studies should pay attention to the development of more specific and well-tolerated inhibitors, focusing on new classes of non-ATP competitive molecules and new models of chemotherapy resistance. Advanced high-throughput cell-based screening using natural compound libraries might lead to the discovery not only of new kinase inhibitors but also of new physiological functions of kinases.
As we have reported in this review, most cell cycle checkpoint kinases have roles beyond their canonical one in DDR. However, ATR is mainly associated with the response to RS and ssDNA breaks, opening the question of whether this kinase may also have some functions, not yet uncovered. Future studies on this direction should be encouraged. Moreover, since most of the here mentioned kinases work together with binding partners, such as Cyclins for CDKs, ATRIP for ATR, MNR complex for ATM and Ku70/Ku80 for DNA-PK, additional functions independent from such binding proteins should also be investigated.
Supplementary information
Author contributions
GM, VC and FC conceived and conceptualized the work, GM and VC wrote the manuscript and FC edited it.
Funding
GM is/has been supported by AIRC Fellowship (Ref. 26727) and by the Danish Cancer Society (R302-A17590). FC labs are supported by AIRC IG23543, Novo Nordisk 0070834, KBVU R231-A14034 and R325-A19075, PRIN 2017FS5SHL and 2020PKLEPN from the Italian Ministry of Research, RF-2019-12369700 from the Italian Ministry of Health, and Danmarks Grundforskningsfond (DNRF125).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Giacomo Milletti, Valeria Colicchia.
Contributor Information
Giacomo Milletti, Email: gimi@cancer.dk.
Francesco Cecconi, Email: francesco.cecconi@unicatt.it.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-023-01196-z.
References
- 1.Adikes RC, Kohrman AQ, Martinez MAQ, Palmisano NJ, Smith JJ, Medwig-Kinney TN, et al. Visualizing the metazoan proliferation-quiescence decision in vivo. Elife. 2020;9:e63265. doi: 10.7554/eLife.63265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matthews HK, Bertoli C, de Bruin RAM. Cell cycle control in cancer. Nat Rev Mol Cell Biol. 2022;23:74–88. doi: 10.1038/s41580-021-00404-3. [DOI] [PubMed] [Google Scholar]
- 3.Pienta KJ, Hammarlund EU, Brown JS, Amend SR, Axelrod RM. Cancer recurrence and lethality are enabled by enhanced survival and reversible cell cycle arrest of polyaneuploid cells. Proc Natl Acad Sci USA. 2021;118:e2020838118. doi: 10.1073/pnas.2020838118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Segeren HA, van Rijnberk LM, Moreno E, Riemers FM, van Liere EA, Yuan R, et al. Excessive E2F transcription in single cancer cells precludes transient cell-cycle exit after DNA damage. Cell Rep. 2020;33:108449. doi: 10.1016/j.celrep.2020.108449. [DOI] [PubMed] [Google Scholar]
- 5.Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–7. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 6.Lim S, Kaldis PCdks. cyclins and CKIs: roles beyond cell cycle regulation. Development. 2013;140:3079–93. doi: 10.1242/dev.091744. [DOI] [PubMed] [Google Scholar]
- 7.LaBaer J, Garrett MD, Stevenson LF, Slingerland JM, Sandhu C, Chou HS, et al. New functional activities for the p21 family of CDK inhibitors. Genes Dev. 1997;11:847–62. doi: 10.1101/gad.11.7.847. [DOI] [PubMed] [Google Scholar]
- 8.Cheng M, Olivier P, Diehl JA, Fero M, Roussel MF, Roberts JM, et al. The p21(Cip1) and p27(Kip1) CDK 'inhibitors' are essential activators of cyclin D-dependent kinases in murine fibroblasts. EMBO J. 1999;18:1571–83. doi: 10.1093/emboj/18.6.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fassl A, Geng Y, Sicinski P. CDK4 and CDK6 kinases: from basic science to cancer therapy. Science. 2022;375:eabc1495. doi: 10.1126/science.abc1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 2004;18:2699–711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- 11.Malumbres M, Barbacid M. To cycle or not to cycle: a critical decision in cancer. Nat Rev Cancer. 2001;1:222–31. doi: 10.1038/35106065. [DOI] [PubMed] [Google Scholar]
- 12.Hwang HC, Clurman BE. Cyclin E in normal and neoplastic cell cycles. Oncogene. 2005;24:2776–86. doi: 10.1038/sj.onc.1208613. [DOI] [PubMed] [Google Scholar]
- 13.Anders L, Ke N, Hydbring P, Choi YJ, Widlund HR, Chick JM, et al. A systematic screen for CDK4/6 substrates links FOXM1 phosphorylation to senescence suppression in cancer cells. Cancer Cell. 2011;20:620–34. doi: 10.1016/j.ccr.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malumbres M, Hunt SL, Sotillo R, Martin J, Odajima J, Martin A, et al. Driving the cell cycle to cancer. Adv Exp Med Biol. 2003;532:1–11. doi: 10.1007/978-1-4615-0081-0_1. [DOI] [PubMed] [Google Scholar]
- 15.Geng Y, Yu Q, Sicinska E, Das M, Schneider JE, Bhattacharya S, et al. Cyclin E ablation in the mouse. Cell. 2003;114:431–43. doi: 10.1016/S0092-8674(03)00645-7. [DOI] [PubMed] [Google Scholar]
- 16.Asteriti IA, De Mattia F, Guarguaglini G. Cross-talk between AURKA and Plk1 in mitotic entry and spindle assembly. Front Oncol. 2015;5:283. doi: 10.3389/fonc.2015.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vigneron S, Sundermann L, Labbe JC, Pintard L, Radulescu O, Castro A, et al. Cyclin A-cdk1-dependent phosphorylation of bora is the triggering factor promoting mitotic entry. Dev Cell. 2018;45:637–50.e637. doi: 10.1016/j.devcel.2018.05.005. [DOI] [PubMed] [Google Scholar]
- 18.Martinez-Alonso D, Malumbres M. Mammalian cell cycle cyclins. Semin Cell Dev Biol. 2020;107:28–35. doi: 10.1016/j.semcdb.2020.03.009. [DOI] [PubMed] [Google Scholar]
- 19.Lee MG, Nurse P. Complementation used to clone a human homologue of the fission yeast cell cycle control gene cdc2. Nature. 1987;327:31–5. doi: 10.1038/327031a0. [DOI] [PubMed] [Google Scholar]
- 20.Moreno S, Nurse P. Regulation of progression through the G1 phase of the cell cycle by the rum1+ gene. Nature. 1994;367:236–42. doi: 10.1038/367236a0. [DOI] [PubMed] [Google Scholar]
- 21.Johnston LH. Cell cycle control of gene expression in yeast. Trends Cell Biol. 1992;2:353–7. doi: 10.1016/0962-8924(92)90041-K. [DOI] [PubMed] [Google Scholar]
- 22.Dhavan R, Tsai LH. A decade of CDK5. Nat Rev Mol Cell Biol. 2001;2:749–59. doi: 10.1038/35096019. [DOI] [PubMed] [Google Scholar]
- 23.Malumbres M. Cyclin-dependent kinases. Genome Biol. 2014;15:122. doi: 10.1186/gb4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Echalier A, Endicott JA, Noble ME. Recent developments in cyclin-dependent kinase biochemical and structural studies. Biochim Biophys Acta. 2010;1804:511–9. doi: 10.1016/j.bbapap.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Morris MC, Gondeau C, Tainer JA, Divita G. Kinetic mechanism of activation of the Cdk2/cyclin A complex. Key role of the C-lobe of the Cdk. J Biol Chem. 2002;277:23847–53. doi: 10.1074/jbc.M107890200. [DOI] [PubMed] [Google Scholar]
- 26.Takaki T, Echalier A, Brown NR, Hunt T, Endicott JA, Noble ME. The structure of CDK4/cyclin D3 has implications for models of CDK activation. Proc Natl Acad Sci USA. 2009;106:4171–6. doi: 10.1073/pnas.0809674106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rubin SM, Sage J, Skotheim JM. Integrating old and new paradigms of G1/S control. Mol Cell. 2020;80:183–92. doi: 10.1016/j.molcel.2020.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harbour JW, Luo RX, Dei Santi A, Postigo AA, Dean DC. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell. 1999;98:859–69. doi: 10.1016/S0092-8674(00)81519-6. [DOI] [PubMed] [Google Scholar]
- 29.Lundberg AS, Weinberg RA. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol Cell Biol. 1998;18:753–61. doi: 10.1128/MCB.18.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeGregori J, Kowalik T, Nevins JR. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol. 1995;15:4215–24. doi: 10.1128/MCB.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maiani E, Milletti G, Nazio F, Holdgaard SG, Bartkova J, Rizza S, et al. AMBRA1 regulates cyclin D to guard S-phase entry and genomic integrity. Nature. 2021;592:799–803. doi: 10.1038/s41586-021-03422-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaikovsky AC, Li C, Jeng EE, Loebell S, Lee MC, Murray CW, et al. The AMBRA1 E3 ligase adaptor regulates the stability of cyclin D. Nature. 2021;592:794–8. doi: 10.1038/s41586-021-03474-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simoneschi D, Rona G, Zhou N, Jeong YT, Jiang S, Milletti G, et al. CRL4(AMBRA1) is a master regulator of D-type cyclins. Nature. 2021;592:789–93. doi: 10.1038/s41586-021-03445-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, et al. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294:173–7. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- 35.Zachariae W, Nasmyth K. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 1999;13:2039–58. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]
- 36.Narasimha AM, Kaulich M, Shapiro GS, Choi YJ, Sicinski P, Dowdy SF. Cyclin D activates the Rb tumor suppressor by mono-phosphorylation. Elife. 2014;3:e02872. doi: 10.7554/eLife.02872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guiley KZ, Stevenson JW, Lou K, Barkovich KJ, Kumarasamy V, Wijeratne TU, et al. p27 allosterically activates cyclin-dependent kinase 4 and antagonizes palbociclib inhibition. Science. 2019;366:6471. doi: 10.1126/science.aaw2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chung M, Liu C, Yang HW, Koberlin MS, Cappell SD, Meyer T. Transient hysteresis in CDK4/6 activity underlies passage of the restriction point in G1. Mol Cell. 2019;76:562–73.e564. doi: 10.1016/j.molcel.2019.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang HW, Cappell SD, Jaimovich A, Liu C, Chung M, Daigh LH, et al. Stress-mediated exit to quiescence restricted by increasing persistence in CDK4/6 activation. Elife. 2020;9:e44571. doi: 10.7554/eLife.44571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cornwell JA, Crncec A, Afifi MM, Tang K, Amin R, Cappell SD. Loss of CDK4/6 activity in S/G2 phase leads to cell cycle reversal. Nature. 2023;619:363–70. doi: 10.1038/s41586-023-06274-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malumbres M, Sotillo R, Santamaria D, Galan J, Cerezo A, Ortega S, et al. Mammalian cells cycle without the D-type cyclin-dependent kinases Cdk4 and Cdk6. Cell. 2004;118:493–504. doi: 10.1016/j.cell.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 42.Liu C, Konagaya Y, Chung M, Daigh LH, Fan Y, Yang HW, et al. Altered G1 signaling order and commitment point in cells proliferating without CDK4/6 activity. Nat Commun. 2020;11:5305. doi: 10.1038/s41467-020-18966-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arora M, Moser J, Hoffman TE, Watts LP, Min M, Musteanu M, et al. Rapid adaptation to CDK2 inhibition exposes intrinsic cell-cycle plasticity. Cell. 2023;186:2628–43.e2621. doi: 10.1016/j.cell.2023.05.013. [DOI] [PubMed] [Google Scholar]
- 44.Pardee AB, Dubrow R, Hamlin JL, Kletzien RF. Animal cell cycle. Annu Rev Biochem. 1978;47:715–50. doi: 10.1146/annurev.bi.47.070178.003435. [DOI] [PubMed] [Google Scholar]
- 45.Johnson A, Skotheim JM. Start and the restriction point. Curr Opin Cell Biol. 2013;25:717–23. doi: 10.1016/j.ceb.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pennycook BR, Barr AR. Restriction point regulation at the crossroads between quiescence and cell proliferation. FEBS Lett. 2020. 10.1002/1873-3468.13867 [DOI] [PubMed]
- 47.Schwarz C, Johnson A, Koivomagi M, Zatulovskiy E, Kravitz CJ, Doncic A, et al. A precise cdk activity threshold determines passage through the restriction point. Mol Cell. 2018;69:253–64.e255. doi: 10.1016/j.molcel.2017.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spencer SL, Cappell SD, Tsai FC, Overton KW, Wang CL, Meyer T. The proliferation-quiescence decision is controlled by a bifurcation in CDK2 activity at mitotic exit. Cell. 2013;155:369–83. doi: 10.1016/j.cell.2013.08.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moser J, Miller I, Carter D, Spencer SL. Control of the Restriction Point by Rb and p21. Proc Natl Acad Sci USA. 2018;115:E8219–E8227. doi: 10.1073/pnas.1722446115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arora M, Moser J, Phadke H, Basha AA, Spencer SL. Endogenous replication stress in mother cells leads to quiescence of daughter cells. Cell Rep. 2017;19:1351–64. doi: 10.1016/j.celrep.2017.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barr AR, Cooper S, Heldt FS, Butera F, Stoy H, Mansfeld J, et al. DNA damage during S-phase mediates the proliferation-quiescence decision in the subsequent G1 via p21 expression. Nat Commun. 2017;8:14728. doi: 10.1038/ncomms14728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang HW, Chung M, Kudo T, Meyer T. Competing memories of mitogen and p53 signalling control cell-cycle entry. Nature. 2017;549:404–8. doi: 10.1038/nature23880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suski JM, Ratnayeke N, Braun M, Zhang T, Strmiska V, Michowski W, et al. CDC7-independent G1/S transition revealed by targeted protein degradation. Nature. 2022;605:357–65. doi: 10.1038/s41586-022-04698-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chi Y, Carter JH, Swanger J, Mazin AV, Moritz RL, Clurman BE. A novel landscape of nuclear human CDK2 substrates revealed by in situ phosphorylation. Sci Adv. 2020;6:eaaz9899. doi: 10.1126/sciadv.aaz9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buis J, Stoneham T, Spehalski E, Ferguson DO. Mre11 regulates CtIP-dependent double-strand break repair by interaction with CDK2. Nat Struct Mol Biol. 2012;19:246–52. doi: 10.1038/nsmb.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matsuura I, Denissova NG, Wang G, He D, Long J, Liu F. Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature. 2004;430:226–31. doi: 10.1038/nature02650. [DOI] [PubMed] [Google Scholar]
- 57.Santamaria D, Barriere C, Cerqueira A, Hunt S, Tardy C, Newton K, et al. Cdk1 is sufficient to drive the mammalian cell cycle. Nature. 2007;448:811–5. doi: 10.1038/nature06046. [DOI] [PubMed] [Google Scholar]
- 58.Basu S, Greenwood J, Jones AW, Nurse P. Core control principles of the eukaryotic cell cycle. Nature. 2022;607:381–6. doi: 10.1038/s41586-022-04798-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Holt LJ, Tuch BB, Villen J, Johnson AD, Gygi SP, Morgan DO. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–6. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–92. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Satyanarayana A, Berthet C, Lopez-Molina J, Coppola V, Tessarollo L, Kaldis P. Genetic substitution of Cdk1 by Cdk2 leads to embryonic lethality and loss of meiotic function of Cdk2. Development. 2008;135:3389–3400. doi: 10.1242/dev.024919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harvey SL, Charlet A, Haas W, Gygi SP, Kellogg DR. Cdk1-dependent regulation of the mitotic inhibitor Wee1. Cell. 2005;122:407–20. doi: 10.1016/j.cell.2005.05.029. [DOI] [PubMed] [Google Scholar]
- 63.Enomoto M, Goto H, Tomono Y, Kasahara K, Tsujimura K, Kiyono T, et al. Novel positive feedback loop between Cdk1 and Chk1 in the nucleus during G2/M transition. J Biol Chem. 2009;284:34223–30. doi: 10.1074/jbc.C109.051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Malumbres M, Barbacid M. Mammalian cyclin-dependent kinases. Trends Biochem Sci. 2005;30:630–41. doi: 10.1016/j.tibs.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 65.Stemmann O, Zou H, Gerber SA, Gygi SP, Kirschner MW. Dual inhibition of sister chromatid separation at metaphase. Cell. 2001;107:715–26. doi: 10.1016/S0092-8674(01)00603-1. [DOI] [PubMed] [Google Scholar]
- 66.Yu J, Raia P, Ghent CM, Raisch T, Sadian Y, Cavadini S, et al. Structural basis of human separase regulation by securin and CDK1-cyclin B1. Nature. 2021;596:138–42. doi: 10.1038/s41586-021-03764-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Potapova TA, Daum JR, Pittman BD, Hudson JR, Jones TN, Satinover DL, et al. The reversibility of mitotic exit in vertebrate cells. Nature. 2006;440:954–8. doi: 10.1038/nature04652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Al-Rawi A, Kaye E, Korolchuk S, Endicott JA, Ly T. Cyclin A and Cks1 promote kinase consensus switching to non-proline-directed CDK1 phosphorylation. Cell Rep. 2023;42:112139. doi: 10.1016/j.celrep.2023.112139. [DOI] [PubMed] [Google Scholar]
- 69.Consortium APG. AACR Project GENIE: powering precision medicine through an international consortium. Cancer Discov. 2017;7:818–31. doi: 10.1158/2159-8290.CD-17-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sotillo R, Garcia JF, Ortega S, Martin J, Dubus P, Barbacid M, et al. Invasive melanoma in Cdk4-targeted mice. Proc Natl Acad Sci USA. 2001;98:13312–7. doi: 10.1073/pnas.241338598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rane SG, Cosenza SC, Mettus RV, Reddy EP. Germ line transmission of the Cdk4(R24C) mutation facilitates tumorigenesis and escape from cellular senescence. Mol Cell Biol. 2002;22:644–56. doi: 10.1128/MCB.22.2.644-656.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Y, Luo R, Zhang X, Xiang H, Yang B, Feng J, et al. Proteogenomics of diffuse gliomas reveal molecular subtypes associated with specific therapeutic targets and immune-evasion mechanisms. Nat Commun. 2023;14:505. doi: 10.1038/s41467-023-36005-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Knudsen ES, Kumarasamy V, Nambiar R, Pearson JD, Vail P, Rosenheck H, et al. CDK/cyclin dependencies define extreme cancer cell-cycle heterogeneity and collateral vulnerabilities. Cell Rep. 2022;38:110448. doi: 10.1016/j.celrep.2022.110448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000;14:397–402. doi: 10.1101/gad.14.4.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Daniel JA, Pellegrini M, Lee BS, Guo Z, Filsuf D, Belkina NV, et al. Loss of ATM kinase activity leads to embryonic lethality in mice. J Cell Biol. 2012;198:295–304. doi: 10.1083/jcb.201204035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000;14:1448–59. doi: 10.1101/gad.14.12.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kurimasa A, Ouyang H, Dong LJ, Wang S, Li X, Cordon-Cardo C, et al. Catalytic subunit of DNA-dependent protein kinase: impact on lymphocyte development and tumorigenesis. Proc Natl Acad Sci USA. 1999;96:1403–8. doi: 10.1073/pnas.96.4.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Takai H, Naka K, Okada Y, Watanabe M, Harada N, Saito S, et al. Chk2-deficient mice exhibit radioresistance and defective p53-mediated transcription. EMBO J. 2002;21:5195–205. doi: 10.1093/emboj/cdf506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lapenna S, Giordano A. Cell cycle kinases as therapeutic targets for cancer. Nat Rev Drug Discov. 2009;8:547–66. doi: 10.1038/nrd2907. [DOI] [PubMed] [Google Scholar]
- 80.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–11. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 81.Lin YF, Shih HY, Shang ZF, Kuo CT, Guo J, Du C, et al. PIDD mediates the association of DNA-PKcs and ATR at stalled replication forks to facilitate the ATR signaling pathway. Nucleic Acids Res. 2018;46:1847–59. doi: 10.1093/nar/gkx1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schlam-Babayov S, Bensimon A, Harel M, Geiger T, Aebersold R, Ziv Y, et al. Phosphoproteomics reveals novel modes of function and inter-relationships among PIKKs in response to genotoxic stress. EMBO J. 2021;40:e104400. doi: 10.15252/embj.2020104400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–53. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 84.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–4. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 85.Kozlov SV, Graham ME, Jakob B, Tobias F, Kijas AW, Tanuji M, et al. Autophosphorylation and ATM activation: additional sites add to the complexity. J Biol Chem. 2011;286:9107–19. doi: 10.1074/jbc.M110.204065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee JH, Paull TT. Cellular functions of the protein kinase ATM and their relevance to human disease. Nat Rev Mol Cell Biol. 2021;22:796–814. doi: 10.1038/s41580-021-00394-2. [DOI] [PubMed] [Google Scholar]
- 87.Marechal A, Zou L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol. 2013;5:a012716. doi: 10.1101/cshperspect.a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Phan LM, Rezaeian AH. ATM: main features, signaling pathways, and its diverse roles in dna damage response, tumor suppression, and cancer development. Genes (Basel) 2021;12:845. doi: 10.3390/genes12060845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jette NR, Radhamani S, Arthur G, Ye R, Goutam S, Bolyos A, et al. Combined poly-ADP ribose polymerase and ataxia-telangiectasia mutated/Rad3-related inhibition targets ataxia-telangiectasia mutated-deficient lung cancer cells. Br J Cancer. 2019;121:600–10. doi: 10.1038/s41416-019-0565-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lloyd RL, Wijnhoven PWG, Ramos-Montoya A, Wilson Z, Illuzzi G, Falenta K, et al. Combined PARP and ATR inhibition potentiates genome instability and cell death in ATM-deficient cancer cells. Oncogene. 2020;39:4869–83. doi: 10.1038/s41388-020-1328-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schmitt A, Knittel G, Welcker D, Yang TP, George J, Nowak M, et al. ATM deficiency is associated with sensitivity to PARP1- and ATR inhibitors in lung adenocarcinoma. Cancer Res. 2017;77:3040–56. doi: 10.1158/0008-5472.CAN-16-3398. [DOI] [PubMed] [Google Scholar]
- 94.Williamson CT, Kubota E, Hamill JD, Klimowicz A, Ye R, Muzik H, et al. Enhanced cytotoxicity of PARP inhibition in mantle cell lymphoma harbouring mutations in both ATM and p53. EMBO Mol Med. 2012;4:515–27. doi: 10.1002/emmm.201200229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wilson Z, Odedra R, Wallez Y, Wijnhoven PWG, Hughes AM, Gerrard J, et al. ATR Inhibitor AZD6738 (ceralasertib) exerts antitumor activity as a monotherapy and in combination with chemotherapy and the PARP inhibitor olaparib. Cancer Res. 2022;82:1140–52. doi: 10.1158/0008-5472.CAN-21-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Di Giulio S, Colicchia V, Pastorino F, Pedretti F, Fabretti F, Nicolis di Robilant V, et al. A combination of PARP and CHK1 inhibitors efficiently antagonizes MYCN-driven tumors. Oncogene. 2021;40:6143–52. doi: 10.1038/s41388-021-02003-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu N, Stoica G, Yan M, Scofield VL, Qiang W, Lynn WS, et al. ATM deficiency induces oxidative stress and endoplasmic reticulum stress in astrocytes. Lab Invest. 2005;85:1471–80. doi: 10.1038/labinvest.3700354. [DOI] [PubMed] [Google Scholar]
- 98.Pietrucha B, Heropolitanska-Pliszka E, Maciejczyk M, Car H, Sawicka-Powierza J, Motkowski R, et al. Comparison of selected parameters of redox homeostasis in patients with ataxia-telangiectasia and nijmegen breakage syndrome. Oxid Med Cell Longev. 2017;2017:6745840. doi: 10.1155/2017/6745840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Quick KL, Dugan LL. Superoxide stress identifies neurons at risk in a model of ataxia-telangiectasia. Ann Neurol. 2001;49:627–35. doi: 10.1002/ana.1005. [DOI] [PubMed] [Google Scholar]
- 100.Reichenbach J, Schubert R, Schindler D, Muller K, Bohles H, Zielen S. Elevated oxidative stress in patients with ataxia telangiectasia. Antioxid Redox Signal. 2002;4:465–9. doi: 10.1089/15230860260196254. [DOI] [PubMed] [Google Scholar]
- 101.Alexander A, Cai SL, Kim J, Nanez A, Sahin M, MacLean KH, et al. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc Natl Acad Sci USA. 2010;107:4153–8. doi: 10.1073/pnas.0913860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Guo Z, Kozlov S, Lavin MF, Person MD, Paull TT. ATM activation by oxidative stress. Science. 2010;330:517–21. doi: 10.1126/science.1192912. [DOI] [PubMed] [Google Scholar]
- 103.Fang EF, Kassahun H, Croteau DL, Scheibye-Knudsen M, Marosi K, Lu H, et al. NAD(+) replenishment improves lifespan and healthspan in ataxia telangiectasia models via mitophagy and DNA repair. Cell Metab. 2016;24:566–81. doi: 10.1016/j.cmet.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lee JH, Mand MR, Kao CH, Zhou Y, Ryu SW, Richards AL, et al. ATM directs DNA damage responses and proteostasis via genetically separable pathways. Sci Signal. 2018;11:eaan5598. doi: 10.1126/scisignal.aan5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang J, Tripathi DN, Jing J, Alexander A, Kim J, Powell RT, et al. ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat Cell Biol. 2015;17:1259–69. doi: 10.1038/ncb3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bencokova Z, Kaufmann MR, Pires IM, Lecane PS, Giaccia AJ, Hammond EM. ATM activation and signaling under hypoxic conditions. Mol Cell Biol. 2009;29:526–37. doi: 10.1128/MCB.01301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rezaeian AH, Li CF, Wu CY, Zhang X, Delacerda J, You MJ, et al. A hypoxia-responsive TRAF6-ATM-H2AX signalling axis promotes HIF1alpha activation, tumorigenesis and metastasis. Nat Cell Biol. 2017;19:38–51. doi: 10.1038/ncb3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bentley NJ, Holtzman DA, Flaggs G, Keegan KS, DeMaggio A, Ford JC, et al. The Schizosaccharomyces pombe rad3 checkpoint gene. EMBO J. 1996;15:6641–51. doi: 10.1002/j.1460-2075.1996.tb01054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cimprich KA, Shin TB, Keith CT, Schreiber SL. cDNA cloning and gene mapping of a candidate human cell cycle checkpoint protein. Proc Natl Acad Sci USA. 1996;93:2850–5. doi: 10.1073/pnas.93.7.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mordes DA, Glick GG, Zhao R, Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 2008;22:1478–89. doi: 10.1101/gad.1666208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bass TE, Luzwick JW, Kavanaugh G, Carroll C, Dungrawala H, Glick GG, et al. ETAA1 acts at stalled replication forks to maintain genome integrity. Nat Cell Biol. 2016;18:1185–95. doi: 10.1038/ncb3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Feng S, Zhao Y, Xu Y, Ning S, Huo W, Hou M, et al. Ewing tumor-associated antigen 1 interacts with replication protein A to promote restart of stalled replication forks. J Biol Chem. 2016;291:21956–62. doi: 10.1074/jbc.C116.747758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Haahr P, Hoffmann S, Tollenaere MA, Ho T, Toledo LI, Mann M, et al. Activation of the ATR kinase by the RPA-binding protein ETAA1. Nat Cell Biol. 2016;18:1196–207. doi: 10.1038/ncb3422. [DOI] [PubMed] [Google Scholar]
- 114.Lee YC, Zhou Q, Chen J, Yuan J. RPA-binding protein ETAA1 Is an ATR activator involved in DNA replication stress response. Curr Biol. 2016;26:3257–68. doi: 10.1016/j.cub.2016.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Busino L, Donzelli M, Chiesa M, Guardavaccaro D, Ganoth D, Dorrello NV, et al. Degradation of Cdc25A by beta-TrCP during S phase and in response to DNA damage. Nature. 2003;426:87–91. doi: 10.1038/nature02082. [DOI] [PubMed] [Google Scholar]
- 116.Sorensen CS, Syljuasen RG, Falck J, Schroeder T, Ronnstrand L, Khanna KK, et al. Chk1 regulates the S phase checkpoint by coupling the physiological turnover and ionizing radiation-induced accelerated proteolysis of Cdc25A. Cancer Cell. 2003;3:247–58. doi: 10.1016/S1535-6108(03)00048-5. [DOI] [PubMed] [Google Scholar]
- 117.Costanzo V, Shechter D, Lupardus PJ, Cimprich KA, Gottesman M, Gautier J. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol Cell. 2003;11:203–13. doi: 10.1016/S1097-2765(02)00799-2. [DOI] [PubMed] [Google Scholar]
- 118.Toledo LI, Altmeyer M, Rask MB, Lukas C, Larsen DH, Povlsen LK, et al. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 2013;155:1088–103. doi: 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
- 119.Couch FB, Bansbach CE, Driscoll R, Luzwick JW, Glick GG, Betous R, et al. ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev. 2013;27:1610–23. doi: 10.1101/gad.214080.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lossaint G, Larroque M, Ribeyre C, Bec N, Larroque C, Decaillet C, et al. FANCD2 binds MCM proteins and controls replisome function upon activation of s phase checkpoint signaling. Mol Cell. 2013;51:678–90. doi: 10.1016/j.molcel.2013.07.023. [DOI] [PubMed] [Google Scholar]
- 121.Sijacki T, Alcon P, Chen ZA, McLaughlin SH, Shakeel S, Rappsilber J, et al. The DNA-damage kinase ATR activates the FANCD2-FANCI clamp by priming it for ubiquitination. Nat Struct Mol Biol. 2022;29:881–90. doi: 10.1038/s41594-022-00820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ammazzalorso F, Pirzio LM, Bignami M, Franchitto A, Pichierri P. ATR and ATM differently regulate WRN to prevent DSBs at stalled replication forks and promote replication fork recovery. EMBO J. 2010;29:3156–69. doi: 10.1038/emboj.2010.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Moiseeva T, Hood B, Schamus S, O'Connor MJ, Conrads TP, Bakkenist CJ. ATR kinase inhibition induces unscheduled origin firing through a Cdc7-dependent association between GINS and And-1. Nat Commun. 2017;8:1392. doi: 10.1038/s41467-017-01401-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ragland RL, Patel S, Rivard RS, Smith K, Peters AA, Bielinsky AK, et al. RNF4 and PLK1 are required for replication fork collapse in ATR-deficient cells. Genes Dev. 2013;27:2259–73. doi: 10.1101/gad.223180.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Buisson R, Boisvert JL, Benes CH, Zou L. Distinct but concerted roles of ATR, DNA-PK, and Chk1 in countering replication stress during S phase. Mol Cell. 2015;59:1011–24. doi: 10.1016/j.molcel.2015.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Diehl FF, Miettinen TP, Elbashir R, Nabel CS, Darnell AM, Do BT, et al. Nucleotide imbalance decouples cell growth from cell proliferation. Nat Cell Biol. 2022;24:1252–64. doi: 10.1038/s41556-022-00965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.D'Angiolella V, Donato V, Forrester FM, Jeong YT, Pellacani C, Kudo Y, et al. Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell. 2012;149:1023–34. doi: 10.1016/j.cell.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Peng CY, Graves PR, Thoma RS, Wu Z, Shaw AS, Piwnica-Worms H. Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science. 1997;277:1501–5. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- 129.Daigh LH, Liu C, Chung M, Cimprich KA, Meyer T. Stochastic endogenous replication stress causes ATR-triggered fluctuations in CDK2 activity that dynamically adjust global DNA synthesis rates. Cell Syst. 2018;7:17–27.e13. doi: 10.1016/j.cels.2018.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lin JJ, Dutta A. ATR pathway is the primary pathway for activating G2/M checkpoint induction after re-replication. J Biol Chem. 2007;282:30357–62. doi: 10.1074/jbc.M705178200. [DOI] [PubMed] [Google Scholar]
- 131.Moiseeva TN, Yin Y, Calderon MJ, Qian C, Schamus-Haynes S, Sugitani N, et al. An ATR and CHK1 kinase signaling mechanism that limits origin firing during unperturbed DNA replication. Proc Natl Acad Sci USA. 2019;116:13374–83. doi: 10.1073/pnas.1903418116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Saldivar JC, Hamperl S, Bocek MJ, Chung M, Bass TE, Cisneros-Soberanis F, et al. An intrinsic S/G(2) checkpoint enforced by ATR. Science. 2018;361:806–10. doi: 10.1126/science.aap9346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zonderland G, Vanzo R, Gadi SA, Martin-Doncel E, Coscia F, Mund A, et al. The TRESLIN-MTBP complex couples completion of DNA replication with S/G2 transition. Mol Cell. 2022;82:3350–65.e3357. doi: 10.1016/j.molcel.2022.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–6. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 135.Stokes MP, Rush J, Macneill J, Ren JM, Sprott K, Nardone J, et al. Profiling of UV-induced ATM/ATR signaling pathways. Proc Natl Acad Sci USA. 2007;104:19855–60. doi: 10.1073/pnas.0707579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sims JR, Faca VM, Pereira C, Ascencao C, Comstock W, Badar J, et al. Phosphoproteomics of ATR signaling in mouse testes. Elife. 2022;11:e68648. doi: 10.7554/eLife.68648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.de Klein A, Muijtjens M, van Os R, Verhoeven Y, Smit B, Carr AM, et al. Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr Biol. 2000;10:479–82. doi: 10.1016/S0960-9822(00)00447-4. [DOI] [PubMed] [Google Scholar]
- 138.Mokrani-Benhelli H, Gaillard L, Biasutto P, Le Guen T, Touzot F, Vasquez N, et al. Primary microcephaly, impaired DNA replication, and genomic instability caused by compound heterozygous ATR mutations. Hum Mutat. 2013;34:374–84. doi: 10.1002/humu.22245. [DOI] [PubMed] [Google Scholar]
- 139.Macheret M, Halazonetis TD. Intragenic origins due to short G1 phases underlie oncogene-induced DNA replication stress. Nature. 2018;555:112–6. doi: 10.1038/nature25507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Barnieh FM, Loadman PM, Falconer RA. Progress towards a clinically-successful ATR inhibitor for cancer therapy. Curr Res Pharm Drug Discov. 2021;2:100017. doi: 10.1016/j.crphar.2021.100017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Carter T, Vancurova I, Sun I, Lou W, DeLeon S. A DNA-activated protein kinase from HeLa cell nuclei. Mol Cell Biol. 1990;10:6460–71. doi: 10.1128/mcb.10.12.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lees-Miller SP, Chen YR, Anderson CW. Human cells contain a DNA-activated protein kinase that phosphorylates simian virus 40 T antigen, mouse p53, and the human Ku autoantigen. Mol Cell Biol. 1990;10:6472–81. doi: 10.1128/mcb.10.12.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Jackson SP, MacDonald JJ, Lees-Miller S, Tjian R. GC box binding induces phosphorylation of Sp1 by a DNA-dependent protein kinase. Cell. 1990;63:155–65. doi: 10.1016/0092-8674(90)90296-Q. [DOI] [PubMed] [Google Scholar]
- 144.Ying S, Chen Z, Medhurst AL, Neal JA, Bao Z, Mortusewicz O, et al. DNA-PKcs and PARP1 bind to unresected stalled DNA replication forks where they recruit XRCC1 to mediate repair. Cancer Res. 2016;76:1078–88. doi: 10.1158/0008-5472.CAN-15-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Dibitetto D, Marshall S, Sanchi A, Liptay M, Badar J, Lopes M, et al. DNA-PKcs promotes fork reversal and chemoresistance. Mol Cell. 2022;82:3932–42.e3936. doi: 10.1016/j.molcel.2022.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Goodwin JF, Kothari V, Drake JM, Zhao S, Dylgjeri E, Dean JL, et al. DNA-PKcs-mediated transcriptional regulation drives prostate cancer progression and metastasis. Cancer Cell. 2015;28:97–113. doi: 10.1016/j.ccell.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Harding SM, Benci JL, Irianto J, Discher DE, Minn AJ, Greenberg RA. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature. 2017;548:466–70. doi: 10.1038/nature23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Shao Z, Flynn RA, Crowe JL, Zhu Y, Liang J, Jiang W, et al. DNA-PKcs has KU-dependent function in rRNA processing and haematopoiesis. Nature. 2020;579:291–6. doi: 10.1038/s41586-020-2041-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Jiang W, Crowe JL, Liu X, Nakajima S, Wang Y, Li C, et al. Differential phosphorylation of DNA-PKcs regulates the interplay between end-processing and end-ligation during nonhomologous end-joining. Mol Cell. 2015;58:172–85. doi: 10.1016/j.molcel.2015.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Liu L, Chen X, Li J, Wang H, Buehl CJ, Goff NJ, et al. Autophosphorylation transforms DNA-PK from protecting to processing DNA ends. Mol Cell. 2022;82:177–89.e174. doi: 10.1016/j.molcel.2021.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chen X, Xu X, Chen Y, Cheung JC, Wang H, Jiang J, et al. Structure of an activated DNA-PK and its implications for NHEJ. Mol Cell. 2021;81:801–810 e803. doi: 10.1016/j.molcel.2020.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Shang ZF, Huang B, Xu QZ, Zhang SM, Fan R, Liu XD, et al. Inactivation of DNA-dependent protein kinase leads to spindle disruption and mitotic catastrophe with attenuated checkpoint protein 2 Phosphorylation in response to DNA damage. Cancer Res. 2010;70:3657–66. doi: 10.1158/0008-5472.CAN-09-3362. [DOI] [PubMed] [Google Scholar]
- 153.Tomimatsu N, Mukherjee B, Burma S. Distinct roles of ATR and DNA-PKcs in triggering DNA damage responses in ATM-deficient cells. EMBO Rep. 2009;10:629–35. doi: 10.1038/embor.2009.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Fang X, Huang Z, Zhai K, Huang Q, Tao W, Kim L, et al. Inhibiting DNA-PK induces glioma stem cell differentiation and sensitizes glioblastoma to radiation in mice. Sci Transl Med. 2021;13:eabc7275. doi: 10.1126/scitranslmed.abc7275. [DOI] [PubMed] [Google Scholar]
- 155.Damia G. Targeting DNA-PK in cancer. Mutat Res. 2020;821:111692. doi: 10.1016/j.mrfmmm.2020.111692. [DOI] [PubMed] [Google Scholar]
- 156.Gilley D, Tanaka H, Hande MP, Kurimasa A, Li GC, Oshimura M, et al. DNA-PKcs is critical for telomere capping. Proc Natl Acad Sci USA. 2001;98:15084–8. doi: 10.1073/pnas.261574698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ruis BL, Fattah KR, Hendrickson EA. The catalytic subunit of DNA-dependent protein kinase regulates proliferation, telomere length, and genomic stability in human somatic cells. Mol Cell Biol. 2008;28:6182–95. doi: 10.1128/MCB.00355-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Park SJ, Gavrilova O, Brown AL, Soto JE, Bremner S, Kim J, et al. DNA-PK promotes the mitochondrial, metabolic, and physical decline that occurs during aging. Cell Metab. 2017;25:1135–46.e1137. doi: 10.1016/j.cmet.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Goodwin JF, Knudsen KE. Beyond DNA repair: DNA-PK function in cancer. Cancer Discov. 2014;4:1126–39. doi: 10.1158/2159-8290.CD-14-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Oh JS, Susor A, Conti M. Protein tyrosine kinase Wee1B is essential for metaphase II exit in mouse oocytes. Science. 2011;332:462–5. doi: 10.1126/science.1199211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Nakanishi M, Ando H, Watanabe N, Kitamura K, Ito K, Okayama H, et al. Identification and characterization of human Wee1B, a new member of the Wee1 family of Cdk-inhibitory kinases. Genes Cells. 2000;5:839–47. doi: 10.1046/j.1365-2443.2000.00367.x. [DOI] [PubMed] [Google Scholar]
- 162.Schmidt M, Rohe A, Platzer C, Najjar A, Erdmann F, Sippl W. Regulation of G2/M transition by inhibition of WEE1 and PKMYT1 kinases. Molecules. 2017;22:2045. doi: 10.3390/molecules22122045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mueller PR, Coleman TR, Kumagai A, Dunphy WG. Myt1: a membrane-associated inhibitory kinase that phosphorylates Cdc2 on both threonine-14 and tyrosine-15. Science. 1995;270:86–90. doi: 10.1126/science.270.5233.86. [DOI] [PubMed] [Google Scholar]
- 164.Endicott JA, Noble ME, Tucker JA. Cyclin-dependent kinases: inhibition and substrate recognition. Curr Opin Struct Biol. 1999;9:738–44. doi: 10.1016/S0959-440X(99)00038-X. [DOI] [PubMed] [Google Scholar]
- 165.Booher RN, Holman PS, Fattaey A. Human Myt1 is a cell cycle-regulated kinase that inhibits Cdc2 but not Cdk2 activity. J Biol Chem. 1997;272:22300–6. doi: 10.1074/jbc.272.35.22300. [DOI] [PubMed] [Google Scholar]
- 166.Liu F, Stanton JJ, Wu Z, Piwnica-Worms H. The human Myt1 kinase preferentially phosphorylates Cdc2 on threonine 14 and localizes to the endoplasmic reticulum and Golgi complex. Mol Cell Biol. 1997;17:571–83. doi: 10.1128/MCB.17.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Gu Y, Rosenblatt J, Morgan DO. Cell cycle regulation of CDK2 activity by phosphorylation of Thr160 and Tyr15. EMBO J. 1992;11:3995–4005. doi: 10.1002/j.1460-2075.1992.tb05493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wu CL, Kirley SD, Xiao H, Chuang Y, Chung DC, Zukerberg LR. Cables enhances cdk2 tyrosine 15 phosphorylation by Wee1, inhibits cell growth, and is lost in many human colon and squamous cancers. Cancer Res. 2001;61:7325–32. [PubMed] [Google Scholar]
- 169.Duda H, Arter M, Gloggnitzer J, Teloni F, Wild P, Blanco MG, et al. A mechanism for controlled breakage of under-replicated chromosomes during mitosis. Dev Cell. 2016;39:740–55. doi: 10.1016/j.devcel.2016.11.017. [DOI] [PubMed] [Google Scholar]
- 170.Dominguez-Kelly R, Martin Y, Koundrioukoff S, Tanenbaum ME, Smits VA, Medema RH, et al. Wee1 controls genomic stability during replication by regulating the Mus81-Eme1 endonuclease. J Cell Biol. 2011;194:567–79. doi: 10.1083/jcb.201101047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Mahajan K, Fang B, Koomen JM, Mahajan NP. H2B Tyr37 phosphorylation suppresses expression of replication-dependent core histone genes. Nat Struct Mol Biol. 2012;19:930–7. doi: 10.1038/nsmb.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Watanabe N, Arai H, Nishihara Y, Taniguchi M, Watanabe N, Hunter T, et al. M-phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCFbeta-TrCP. Proc Natl Acad Sci USA. 2004;101:4419–24. doi: 10.1073/pnas.0307700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Toyoshima-Morimoto F, Taniguchi E, Shinya N, Iwamatsu A, Nishida E. Polo-like kinase 1 phosphorylates cyclin B1 and targets it to the nucleus during prophase. Nature. 2001;410:215–20. doi: 10.1038/35065617. [DOI] [PubMed] [Google Scholar]
- 174.Nakajima H, Toyoshima-Morimoto F, Taniguchi E, Nishida E. Identification of a consensus motif for Plk (Polo-like kinase) phosphorylation reveals Myt1 as a Plk1 substrate. J Biol Chem. 2003;278:25277–80. doi: 10.1074/jbc.C300126200. [DOI] [PubMed] [Google Scholar]
- 175.Visconti R, Palazzo L, Della Monica R, Grieco D. Fcp1-dependent dephosphorylation is required for M-phase-promoting factor inactivation at mitosis exit. Nat Commun. 2012;3:894. doi: 10.1038/ncomms1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Iorns E, Lord CJ, Grigoriadis A, McDonald S, Fenwick K, Mackay A, et al. Integrated functional, gene expression and genomic analysis for the identification of cancer targets. PLoS One. 2009;4:e5120. doi: 10.1371/journal.pone.0005120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Tibes R, Bogenberger JM, Chaudhuri L, Hagelstrom RT, Chow D, Buechel ME, et al. RNAi screening of the kinome with cytarabine in leukemias. Blood. 2012;119:2863–72. doi: 10.1182/blood-2011-07-367557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Porter CC, Kim J, Fosmire S, Gearheart CM, van Linden A, Baturin D, et al. Integrated genomic analyses identify WEE1 as a critical mediator of cell fate and a novel therapeutic target in acute myeloid leukemia. Leukemia. 2012;26:1266–76. doi: 10.1038/leu.2011.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Magnussen GI, Holm R, Emilsen E, Rosnes AK, Slipicevic A, Florenes VA. High expression of Wee1 is associated with poor disease-free survival in malignant melanoma: potential for targeted therapy. PLoS One. 2012;7:e38254. doi: 10.1371/journal.pone.0038254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Mir SE, De Witt Hamer PC, Krawczyk PM, Balaj L, Claes A, Niers JM, et al. In silico analysis of kinase expression identifies WEE1 as a gatekeeper against mitotic catastrophe in glioblastoma. Cancer Cell. 2010;18:244–57. doi: 10.1016/j.ccr.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Mueller S, Hashizume R, Yang X, Kolkowitz I, Olow AK, Phillips J, et al. Targeting Wee1 for the treatment of pediatric high-grade gliomas. Neuro Oncol. 2014;16:352–60. doi: 10.1093/neuonc/not220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Music D, Dahlrot RH, Hermansen SK, Hjelmborg J, de Stricker K, Hansen S, et al. Expression and prognostic value of the WEE1 kinase in gliomas. J Neurooncol. 2016;127:381–9. doi: 10.1007/s11060-015-2050-4. [DOI] [PubMed] [Google Scholar]
- 183.Harris PS, Venkataraman S, Alimova I, Birks DK, Balakrishnan I, Cristiano B, et al. Integrated genomic analysis identifies the mitotic checkpoint kinase WEE1 as a novel therapeutic target in medulloblastoma. Mol Cancer. 2014;13:72. doi: 10.1186/1476-4598-13-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Yoshida T, Tanaka S, Mogi A, Shitara Y, Kuwano H. The clinical significance of Cyclin B1 and Wee1 expression in non-small-cell lung cancer. Ann Oncol. 2004;15:252–6. doi: 10.1093/annonc/mdh073. [DOI] [PubMed] [Google Scholar]
- 185.Backert S, Gelos M, Kobalz U, Hanski ML, Bohm C, Mann B, et al. Differential gene expression in colon carcinoma cells and tissues detected with a cDNA array. Int J Cancer. 1999;82:868–74. doi: 10.1002/(SICI)1097-0215(19990909)82:6<868::AID-IJC16>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 186.Hartwell LH, Smith D. Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S. cerevisiae. Genetics. 1985;110:381–95. doi: 10.1093/genetics/110.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Sunkel CE, Glover DM. polo, a mitotic mutant of Drosophila displaying abnormal spindle poles. J Cell Sci. 1988;89:25–38. doi: 10.1242/jcs.89.1.25. [DOI] [PubMed] [Google Scholar]
- 188.Arnaud L, Pines J, Nigg EA. GFP tagging reveals human Polo-like kinase 1 at the kinetochore/centromere region of mitotic chromosomes. Chromosoma. 1998;107:424–9. doi: 10.1007/s004120050326. [DOI] [PubMed] [Google Scholar]
- 189.Lane HA, Nigg EA. Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J Cell Biol. 1996;135:1701–13. doi: 10.1083/jcb.135.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Neef R, Preisinger C, Sutcliffe J, Kopajtich R, Nigg EA, Mayer TU, et al. Phosphorylation of mitotic kinesin-like protein 2 by polo-like kinase 1 is required for cytokinesis. J Cell Biol. 2003;162:863–75. doi: 10.1083/jcb.200306009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Petronczki M, Glotzer M, Kraut N, Peters JM. Polo-like kinase 1 triggers the initiation of cytokinesis in human cells by promoting recruitment of the RhoGEF Ect2 to the central spindle. Dev Cell. 2007;12:713–25. doi: 10.1016/j.devcel.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 192.Seong YS, Kamijo K, Lee JS, Fernandez E, Kuriyama R, Miki T, et al. A spindle checkpoint arrest and a cytokinesis failure by the dominant-negative polo-box domain of Plk1 in U-2 OS cells. J Biol Chem. 2002;277:32282–93. doi: 10.1074/jbc.M202602200. [DOI] [PubMed] [Google Scholar]
- 193.Sumara I, Gimenez-Abian JF, Gerlich D, Hirota T, Kraft C, de la Torre C, et al. Roles of polo-like kinase 1 in the assembly of functional mitotic spindles. Curr Biol. 2004;14:1712–22. doi: 10.1016/j.cub.2004.09.049. [DOI] [PubMed] [Google Scholar]
- 194.van Vugt MA, Bras A, Medema RH. Polo-like kinase-1 controls recovery from a G2 DNA damage-induced arrest in mammalian cells. Mol Cell. 2004;15:799–811. doi: 10.1016/j.molcel.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 195.Lee HJ, Hwang HI, Jang YJ. Mitotic DNA damage response: Polo-like kinase-1 is dephosphorylated through ATM-Chk1 pathway. Cell Cycle. 2010;9:2389–98. doi: 10.4161/cc.9.12.11904. [DOI] [PubMed] [Google Scholar]
- 196.Smits VA, Klompmaker R, Arnaud L, Rijksen G, Nigg EA, Medema RH. Polo-like kinase-1 is a target of the DNA damage checkpoint. Nat Cell Biol. 2000;2:672–6. doi: 10.1038/35023629. [DOI] [PubMed] [Google Scholar]
- 197.van Vugt MA, Smits VA, Klompmaker R, Medema RH. Inhibition of Polo-like kinase-1 by DNA damage occurs in an ATM- or ATR-dependent fashion. J Biol Chem. 2001;276:41656–60. doi: 10.1074/jbc.M101831200. [DOI] [PubMed] [Google Scholar]
- 198.Jang YJ, Ma S, Terada Y, Erikson RL. Phosphorylation of threonine 210 and the role of serine 137 in the regulation of mammalian polo-like kinase. J Biol Chem. 2002;277:44115–20. doi: 10.1074/jbc.M202172200. [DOI] [PubMed] [Google Scholar]
- 199.van de Weerdt BC, van Vugt MA, Lindon C, Kauw JJ, Rozendaal MJ, Klompmaker R, et al. Uncoupling anaphase-promoting complex/cyclosome activity from spindle assembly checkpoint control by deregulating polo-like kinase 1. Mol Cell Biol. 2005;25:2031–44. doi: 10.1128/MCB.25.5.2031-2044.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Gheghiani L, Loew D, Lombard B, Mansfeld J, Gavet O. PLK1 activation in late G2 sets up commitment to mitosis. Cell Rep. 2017;19:2060–73. doi: 10.1016/j.celrep.2017.05.031. [DOI] [PubMed] [Google Scholar]
- 201.Kachaner D, Garrido D, Mehsen H, Normandin K, Lavoie H, Archambault V. Coupling of Polo kinase activation to nuclear localization by a bifunctional NLS is required during mitotic entry. Nat Commun. 2017;8:1701. doi: 10.1038/s41467-017-01876-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Seki A, Coppinger JA, Jang CY, Yates JR, Fang G. Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science. 2008;320:1655–8. doi: 10.1126/science.1157425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Taniguchi E, Toyoshima-Morimoto F, Nishida E. Nuclear translocation of plk1 mediated by its bipartite nuclear localization signal. J Biol Chem. 2002;277:48884–8. doi: 10.1074/jbc.M206307200. [DOI] [PubMed] [Google Scholar]
- 204.Raab M, Matthess Y, Raab CA, Gutfreund N, Dotsch V, Becker S, et al. A dimerization-dependent mechanism regulates enzymatic activation and nuclear entry of PLK1. Oncogene. 2022;41:372–86. doi: 10.1038/s41388-021-02094-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Yuan J, Eckerdt F, Bereiter-Hahn J, Kurunci-Csacsko E, Kaufmann M, Strebhardt K. Cooperative phosphorylation including the activity of polo-like kinase 1 regulates the subcellular localization of cyclin B1. Oncogene. 2002;21:8282–92. doi: 10.1038/sj.onc.1206011. [DOI] [PubMed] [Google Scholar]
- 206.Grosstessner-Hain K, Hegemann B, Novatchkova M, Rameseder J, Joughin BA, Hudecz O, et al. Quantitative phospho-proteomics to investigate the polo-like kinase 1-dependent phospho-proteome. Mol Cell Proteom. 2011;10:M111 008540. doi: 10.1074/mcp.M111.008540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Johnson JM, Hebert AS, Drane QH, Lera RF, Wan J, Weaver BA, et al. A Genetic Toggle for Chemical Control of Individual Plk1 Substrates. Cell Chem Biol. 2020;27:350–362 e358. doi: 10.1016/j.chembiol.2020.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Kettenbach AN, Schweppe DK, Faherty BK, Pechenick D, Pletnev AA, Gerber SA. Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci Signal. 2011;4:rs5. doi: 10.1126/scisignal.2001497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Lera RF, Potts GK, Suzuki A, Johnson JM, Salmon ED, Coon JJ, et al. Decoding Polo-like kinase 1 signaling along the kinetochore-centromere axis. Nat Chem Biol. 2016;12:411–8. doi: 10.1038/nchembio.2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Santamaria A, Wang B, Elowe S, Malik R, Zhang F, Bauer M, et al. The Plk1-dependent phosphoproteome of the early mitotic spindle. Mol Cell Proteom. 2011;10:M110 004457. doi: 10.1074/mcp.M110.004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Aljiboury A, Mujcic A, Curtis E, Cammerino T, Magny D, Lan Y, et al. Pericentriolar matrix (PCM) integrity relies on cenexin and polo-like kinase (PLK)1. Mol Biol Cell. 2022;33:br14. doi: 10.1091/mbc.E22-01-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Meng L, Park JE, Kim TS, Lee EH, Park SY, Zhou M, et al. Bimodal interaction of mammalian polo-like kinase 1 and a centrosomal scaffold, Cep192, in the regulation of bipolar spindle formation. Mol Cell Biol. 2015;35:2626–40. doi: 10.1128/MCB.00068-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.von Schubert C, Cubizolles F, Bracher JM, Sliedrecht T, Kops G, Nigg EA. Plk1 and Mps1 cooperatively regulate the spindle assembly checkpoint in human cells. Cell Rep. 2015;12:66–78. doi: 10.1016/j.celrep.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 214.Hansen DV, Loktev AV, Ban KH, Jackson PK. Plk1 regulates activation of the anaphase promoting complex by phosphorylating and triggering SCFbetaTrCP-dependent destruction of the APC Inhibitor Emi1. Mol Biol Cell. 2004;15:5623–34. doi: 10.1091/mbc.e04-07-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Bastos RN, Barr FA. Plk1 negatively regulates Cep55 recruitment to the midbody to ensure orderly abscission. J Cell Biol. 2010;191:751–60. doi: 10.1083/jcb.201008108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Iliaki S, Beyaert R, Afonina IS. Polo-like kinase 1 (PLK1) signaling in cancer and beyond. Biochem Pharm. 2021;193:114747. doi: 10.1016/j.bcp.2021.114747. [DOI] [PubMed] [Google Scholar]
- 217.Ando K, Ozaki T, Yamamoto H, Furuya K, Hosoda M, Hayashi S, et al. Polo-like kinase 1 (Plk1) inhibits p53 function by physical interaction and phosphorylation. J Biol Chem. 2004;279:25549–61. doi: 10.1074/jbc.M314182200. [DOI] [PubMed] [Google Scholar]
- 218.Liu XS, Song B, Elzey BD, Ratliff TL, Konieczny SF, Cheng L, et al. Polo-like kinase 1 facilitates loss of Pten tumor suppressor-induced prostate cancer formation. J Biol Chem. 2011;286:35795–35800. doi: 10.1074/jbc.C111.269050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Xiao D, Yue M, Su H, Ren P, Jiang J, Li F, et al. Polo-like kinase-1 regulates myc stabilization and activates a feedforward circuit promoting tumor cell survival. Mol Cell. 2016;64:493–506. doi: 10.1016/j.molcel.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 220.Lu LY, Wood JL, Minter-Dykhouse K, Ye L, Saunders TL, Yu X, et al. Polo-like kinase 1 is essential for early embryonic development and tumor suppression. Mol Cell Biol. 2008;28:6870–6. doi: 10.1128/MCB.00392-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Chiappa M, Petrella S, Damia G, Broggini M, Guffanti F, Ricci F. Present and future perspective on PLK1 inhibition in cancer treatment. Front Oncol. 2022;12:903016. doi: 10.3389/fonc.2022.903016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Liu Z, Sun Q, Wang X. PLK1, a potential target for cancer therapy. Transl Oncol. 2017;10:22–32. doi: 10.1016/j.tranon.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Raab M, Sanhaji M, Matthess Y, Horlin A, Lorenz I, Dotsch C, et al. PLK1 has tumor-suppressive potential in APC-truncated colon cancer cells. Nat Commun. 2018;9:1106. doi: 10.1038/s41467-018-03494-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Montaudon E, Nikitorowicz-Buniak J, Sourd L, Morisset L, El Botty R, Huguet L, et al. PLK1 inhibition exhibits strong anti-tumoral activity in CCND1-driven breast cancer metastases with acquired palbociclib resistance. Nat Commun. 2020;11:4053. doi: 10.1038/s41467-020-17697-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Chan CS, Botstein D. Isolation and characterization of chromosome-gain and increase-in-ploidy mutants in yeast. Genetics. 1993;135:677–91. doi: 10.1093/genetics/135.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Paris J, Philippe M. Poly(A) metabolism and polysomal recruitment of maternal mRNAs during early Xenopus development. Dev Biol. 1990;140:221–4. doi: 10.1016/0012-1606(90)90070-Y. [DOI] [PubMed] [Google Scholar]
- 227.Andresson T, Ruderman JV. The kinase Eg2 is a component of the Xenopus oocyte progesterone-activated signaling pathway. EMBO J. 1998;17:5627–37. doi: 10.1093/emboj/17.19.5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Roghi C, Giet R, Uzbekov R, Morin N, Chartrain I, Le Guellec R, et al. The Xenopus protein kinase pEg2 associates with the centrosome in a cell cycle-dependent manner, binds to the spindle microtubules and is involved in bipolar mitotic spindle assembly. J Cell Sci. 1998;111:557–72. doi: 10.1242/jcs.111.5.557. [DOI] [PubMed] [Google Scholar]
- 229.Glover DM, Leibowitz MH, McLean DA, Parry H. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell. 1995;81:95–105. doi: 10.1016/0092-8674(95)90374-7. [DOI] [PubMed] [Google Scholar]
- 230.Bischoff JR, Anderson L, Zhu Y, Mossie K, Ng L, Souza B, et al. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 1998;17:3052–65. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Carmena M, Earnshaw WC. The cellular geography of aurora kinases. Nat Rev Mol Cell Biol. 2003;4:842–54. doi: 10.1038/nrm1245. [DOI] [PubMed] [Google Scholar]
- 232.Cheeseman IM, Anderson S, Jwa M, Green EM, Kang J, Yates JR, 3rd, et al. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–72. doi: 10.1016/S0092-8674(02)00973-X. [DOI] [PubMed] [Google Scholar]
- 233.Dodson CA, Bayliss R. Activation of Aurora-A kinase by protein partner binding and phosphorylation are independent and synergistic. J Biol Chem. 2012;287:1150–7. doi: 10.1074/jbc.M111.312090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Ruff EF, Muretta JM, Thompson AR, Lake EW, Cyphers S, Albanese SK, et al. A dynamic mechanism for allosteric activation of Aurora kinase A by activation loop phosphorylation. Elife. 2018;7:e32766. doi: 10.7554/eLife.32766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Marumoto T, Honda S, Hara T, Nitta M, Hirota T, Kohmura E, et al. Aurora-A kinase maintains the fidelity of early and late mitotic events in HeLa cells. J Biol Chem. 2003;278:51786–95. doi: 10.1074/jbc.M306275200. [DOI] [PubMed] [Google Scholar]
- 236.Gopalan G, Chan CS, Donovan PJ. A novel mammalian, mitotic spindle-associated kinase is related to yeast and fly chromosome segregation regulators. J Cell Biol. 1997;138:643–56. doi: 10.1083/jcb.138.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Bernard M, Sanseau P, Henry C, Couturier A, Prigent C. Cloning of STK13, a third human protein kinase related to Drosophila aurora and budding yeast Ipl1 that maps on chromosome 19q13.3-ter. Genomics. 1998;53:406–9. doi: 10.1006/geno.1998.5522. [DOI] [PubMed] [Google Scholar]
- 238.Willems E, Dedobbeleer M, Digregorio M, Lombard A, Lumapat PN, Rogister B. The functional diversity of Aurora kinases: a comprehensive review. Cell Div. 2018;13:7. doi: 10.1186/s13008-018-0040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Lens SM, Voest EE, Medema RH. Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat Rev Cancer. 2010;10:825–41. doi: 10.1038/nrc2964. [DOI] [PubMed] [Google Scholar]
- 240.Quartuccio SM, Schindler K. Functions of Aurora kinase C in meiosis and cancer. Front Cell Dev Biol. 2015;3:50. doi: 10.3389/fcell.2015.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A, et al. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20:189–93. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 242.Anand S, Penrhyn-Lowe S, Venkitaraman AR. AURORA-A amplification overrides the mitotic spindle assembly checkpoint, inducing resistance to Taxol. Cancer Cell. 2003;3:51–62. doi: 10.1016/S1535-6108(02)00235-0. [DOI] [PubMed] [Google Scholar]
- 243.D'Assoro AB, Liu T, Quatraro C, Amato A, Opyrchal M, Leontovich A, et al. The mitotic kinase Aurora–a promotes distant metastases by inducing epithelial-to-mesenchymal transition in ERalpha(+) breast cancer cells. Oncogene. 2014;33:599–610. doi: 10.1038/onc.2012.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Fujii Y, Yada M, Nishiyama M, Kamura T, Takahashi H, Tsunematsu R, et al. Fbxw7 contributes to tumor suppression by targeting multiple proteins for ubiquitin-dependent degradation. Cancer Sci. 2006;97:729–36. doi: 10.1111/j.1349-7006.2006.00239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Wu CC, Yang TY, Yu CT, Phan L, Ivan C, Sood AK, et al. p53 negatively regulates Aurora A via both transcriptional and posttranslational regulation. Cell Cycle. 2012;11:3433–42. doi: 10.4161/cc.21732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Liu Q, Kaneko S, Yang L, Feldman RI, Nicosia SV, Chen J, et al. Aurora-A abrogation of p53 DNA binding and transactivation activity by phosphorylation of serine 215. J Biol Chem. 2004;279:52175–82. doi: 10.1074/jbc.M406802200. [DOI] [PubMed] [Google Scholar]
- 247.Katayama H, Sasai K, Kawai H, Yuan ZM, Bondaruk J, Suzuki F, et al. Phosphorylation by aurora kinase A induces Mdm2-mediated destabilization and inhibition of p53. Nat Genet. 2004;36:55–62. doi: 10.1038/ng1279. [DOI] [PubMed] [Google Scholar]
- 248.Yu X, Minter-Dykhouse K, Malureanu L, Zhao WM, Zhang D, Merkle CJ, et al. Chfr is required for tumor suppression and Aurora A regulation. Nat Genet. 2005;37:401–6. doi: 10.1038/ng1538. [DOI] [PubMed] [Google Scholar]
- 249.Kimura MT, Mori T, Conroy J, Nowak NJ, Satomi S, Tamai K, et al. Two functional coding single nucleotide polymorphisms in STK15 (Aurora-A) coordinately increase esophageal cancer risk. Cancer Res. 2005;65:3548–54. doi: 10.1158/0008-5472.CAN-04-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Lu LY, Wood JL, Ye L, Minter-Dykhouse K, Saunders TL, Yu X, et al. Aurora A is essential for early embryonic development and tumor suppression. J Biol Chem. 2008;283:31785–90. doi: 10.1074/jbc.M805880200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Thiel JT, Daigeler A, Kolbenschlag J, Rachunek K, Hoffmann S. The role of CDK pathway dysregulation and its therapeutic potential in soft tissue sarcoma. Cancers (Basel) 2022;14:3380. doi: 10.3390/cancers14143380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.McAndrew NP, Dickson MA, Clark AS, Troxel AB, O'Hara MH, Colameco C, et al. Early treatment-related neutropenia predicts response to palbociclib. Br J Cancer. 2020;123:912–8. doi: 10.1038/s41416-020-0967-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Dickson MA, Schwartz GK, Keohan ML, D'Angelo SP, Gounder MM, Chi P, et al. Progression-free survival among patients with well-differentiated or dedifferentiated liposarcoma treated with CDK4 inhibitor palbociclib: a phase 2 clinical trial. JAMA Oncol. 2016;2:937–40. doi: 10.1001/jamaoncol.2016.0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Dickson MA, Tap WD, Keohan ML, D'Angelo SP, Gounder MM, Antonescu CR, et al. Phase II trial of the CDK4 inhibitor PD0332991 in patients with advanced CDK4-amplified well-differentiated or dedifferentiated liposarcoma. J Clin Oncol. 2013;31:2024–8. doi: 10.1200/JCO.2012.46.5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Karasic TB, O'Hara MH, Teitelbaum UR, Damjanov N, Giantonio BJ, d'Entremont TS, et al. Phase II trial of palbociclib in patients with advanced esophageal or gastric cancer. Oncologist. 2020;25:e1864–e1868. doi: 10.1634/theoncologist.2020-0681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Liu H, Qiu W, Sun T, Wang L, Du C, Hu Y, et al. Therapeutic strategies of glioblastoma (GBM): the current advances in the molecular targets and bioactive small molecule compounds. Acta Pharm Sin B. 2022;12:1781–804. doi: 10.1016/j.apsb.2021.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Taylor JW, Parikh M, Phillips JJ, James CD, Molinaro AM, Butowski NA, et al. Phase-2 trial of palbociclib in adult patients with recurrent RB1-positive glioblastoma. J Neurooncol. 2018;140:477–83. doi: 10.1007/s11060-018-2977-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Tolaney SM, Sahebjam S, Le Rhun E, Bachelot T, Kabos P, Awada A, et al. A phase II study of abemaciclib in patients with brain metastases secondary to hormone receptor-positive breast cancer. Clin Cancer Res. 2020;26:5310–9. doi: 10.1158/1078-0432.CCR-20-1764. [DOI] [PubMed] [Google Scholar]
- 259.Goldman JW, Mazieres J, Barlesi F, Dragnev KH, Koczywas M, Goskel T, et al. A randomized phase III study of abemaciclib versus erlotinib in patients with stage IV non-small cell lung cancer with a detectable KRAS mutation who failed prior platinum-based therapy: JUNIPER. Front Oncol. 2020;10:578756. doi: 10.3389/fonc.2020.578756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Goldman JW, Shi P, Reck M, Paz-Ares L, Koustenis A, Hurt KC. Treatment Rationale and Study Design for the JUNIPER Study: A Randomized Phase III Study of Abemaciclib With Best Supportive Care Versus Erlotinib With Best Supportive Care in Patients With Stage IV Non-Small-Cell Lung Cancer With a Detectable KRAS Mutation Whose Disease Has Progressed After Platinum-Based Chemotherapy. Clin Lung Cancer. 2016;17:80–4. doi: 10.1016/j.cllc.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 261.La Monica S, Fumarola C, Cretella D, Bonelli M, Minari R, Cavazzoni A, et al. Efficacy of the CDK4/6 dual inhibitor abemaciclib in EGFR-mutated NSCLC cell lines with different resistance mechanisms to osimertinib. Cancers (Basel) 2020;13:6. doi: 10.3390/cancers13010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Hattinger CM, Patrizio MP, Magagnoli F, Luppi S, Serra M. An update on emerging drugs in osteosarcoma: towards tailored therapies? Expert Opin Emerg Drugs. 2019;24:153–71. doi: 10.1080/14728214.2019.1654455. [DOI] [PubMed] [Google Scholar]
- 263.Konstantinopoulos PA, Cheng SC, Wahner Hendrickson AE, Penson RT, Schumer ST, Doyle LA, et al. Berzosertib plus gemcitabine versus gemcitabine alone in platinum-resistant high-grade serous ovarian cancer: a multicentre, open-label, randomised, phase 2 trial. Lancet Oncol. 2020;21:957–68. doi: 10.1016/S1470-2045(20)30180-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Cuneo KC, Morgan MA, Sahai V, Schipper MJ, Parsels LA, Parsels JD, et al. Dose escalation trial of the wee1 inhibitor adavosertib (AZD1775) in combination with gemcitabine and radiation for patients with locally advanced pancreatic cancer. J Clin Oncol. 2019;37:2643–50. doi: 10.1200/JCO.19.00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.O'Neil BH, Scott AJ, Ma WW, Cohen SJ, Aisner DL, Menter AR, et al. A phase II/III randomized study to compare the efficacy and safety of rigosertib plus gemcitabine versus gemcitabine alone in patients with previously untreated metastatic pancreatic cancer. Ann Oncol. 2015;26:1923–9. doi: 10.1093/annonc/mdv264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Garcia-Manero G, Fenaux P, Al-Kali A, Baer MR, Sekeres MA, Roboz GJ, et al. Rigosertib versus best supportive care for patients with high-risk myelodysplastic syndromes after failure of hypomethylating drugs (ONTIME): a randomised, controlled, phase 3 trial. Lancet Oncol. 2016;17:496–508. doi: 10.1016/S1470-2045(16)00009-7. [DOI] [PubMed] [Google Scholar]
- 267.Beltran H, Oromendia C, Danila DC, Montgomery B, Hoimes C, Szmulewitz RZ, et al. A phase II trial of the aurora kinase a inhibitor alisertib for patients with castration-resistant and neuroendocrine prostate cancer: efficacy and biomarkers. Clin Cancer Res. 2019;25:43–51. doi: 10.1158/1078-0432.CCR-18-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Brunner AM, Blonquist TM, DeAngelo DJ, McMasters M, Fell G, Hermance NM, et al. Alisertib plus induction chemotherapy in previously untreated patients with high-risk, acute myeloid leukaemia: a single-arm, phase 2 trial. Lancet Haematol. 2020;7:e122–e133. doi: 10.1016/S2352-3026(19)30203-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.