Abstract
Major depressive disorder (MDD) is known to cause significant disability. Genome-wide DNA methylation (DNAm) profiles can be used to estimate biological aging and as epigenetic clocks. However, information on epigenetic clocks reported in MDD patients is inconsistent. Since antidepressants are likely confounders, we evaluated biological aging using various DNAm-based predictors in patients with MDD who had never received depression medication. A publicly available dataset consisting of whole blood samples from untreated MDD patients (n = 40) and controls (n = 40) was used. We analyzed five epigenetic clocks (HorvathAge, HannumAge, SkinBloodAge, PhenoAge, and GrimAge), DNAm-based telomere length (DNAmTL), and DNAm-based age-related plasma proteins (GrimAge components), as well as DNAm-based white blood cell composition. The results indicate that patients with untreated MDD were significantly associated with epigenetic aging acceleration in HannumAge and GrimAge. Furthermore, a decrease in natural killer cells, based on DNAm, was observed in patients with untreated MDD.
Subject terms: Epigenetics, Psychiatric disorders
Introduction
Major depressive disorder (MDD) is a prevalent mental illness and is considered the leading cause of disability among psychiatric diseases worldwide, affecting an estimated 280 million individuals1. Depression has been linked to an increased risk of premature mortality from various comorbidities and all-cause mortality, including suicide2. Given that telomere shortening is a marker of cellular aging, depression has been found to be linked to accelerated biological aging in terms of telomere length3,4 and leads to neuropsychiatric disease-related aging such as dementia5,6. However, previous studies have shown that antidepressant medication prevents telomere shortening and the development of dementia6–8.
Recently, some researchers developed epigenetic clocks using genome-wide DNA methylation (DNAm) profiles to estimate biological aging9–13. Moreover, DNAm-based telomeres length (DNAmTL) and DNAm-based white blood cell compositions, which are associated with biological aging, have also been created10,14,15. The five epigenetic clocks, HorvathAge, HannumAge, SkinBloodAge, PhenoAge, and GrimAge as well as DNAmTL have been widely used9–14. Epigenetic clocks have been evaluated in various conditions, including psychiatric disorders, as follows: patients with coronavirus disease 201916, cancers17, cardiovascular disease18, diabetes19, MDD20–25, bipolar disorder26, schizophrenia27, post-traumatic stress disorder28, suicide29, and all-cause mortality30. The five epigenetic clocks have been utilized in the studies of various psychiatric disorders to provide an accurate estimation through various unique CpG sites31.
Epigenetic clocks have been previously evaluated in patients with MDD, but the results remain inconsistent20–23,25,32,33. Protsenko et al. reported that patients with untreated MDD exhibited greater epigenetic aging acceleration than healthy controls, using GrimAge23. Additionally, a previous study demonstrated that maternal prenatal antidepressant use can significantly decelerate offspring epigenetic age34. We hypothesized that depression is associated with accelerated biological aging based on DNAm, but that the use of depression medication may modulate changes in the methylation of CpG sites in patients with MDD. This could potentially confound the relationships between depression and biological aging markers. To verify our hypothesis, we analyzed five epigenetic clocks (HorvathAge, HannumAge, SkinBloodAge, PhenoAge, and GrimAge) and DNAmTL in patients with MDD who had never been treated with depression medication, as well as in control participants. We also evaluated the GrimAge components indicating age-predictive factors based on DNAm and DNAm-based white blood cell composition.
Results
Five epigenetic clocks, DNAmTL, and GrimAge components
Participants’ demographic data, including sex and age, are presented in Table 1. Supplementary Tables 1, 2, 3 summarize the correlations between chronological age and each epigenetic clock/GrimAge component/DNAm-based white blood cell composition (Fig. 1, and Supplementary Figs. 1, 2).
Table 1.
Demographics, DNAm-based age acceleration and telomere length acceleration, as well as GrimAge components, and DNAm-based white blood cell counts between patients with untreated MDD and controls.
| CTL (n = 40) | MDD (n = 40) | P-value | |
|---|---|---|---|
| Demographic characteristics | |||
| Sex (male/female) | 20/20 | 16/24 | 0.106a |
| Age (years old), median (IQR) | 24.0 (23.0, 26.25) | 43.5 (29.5, 52.0) | <0.001b |
| DNAmAge and DNAmTL accelerationc | |||
| AgeAccelHorvath, median (IQR) | −0.69 (−2.94, 1.49) | −0.07 (−2.42, 2.15) | 0.684b |
| AgeAccelHannum, median (IQR) | −1.86 (−5.75, 1.21) | 1.75 (−0.002, 3.72) | <0.001b |
| AgeAccelSkinBlood, median (IQR) | −0.10 (−3.67, 0.59) | 0.70 (−0.68, 2.19) | 0.0098b |
| AgeAccelPheno, median (IQR) | −1.30 (−5.19, 2.31) | 1.30 (−1.57, 3.36) | 0.0333b |
| AgeAccelGrim, median (IQR) | −0.97 (−2.51, 0.43) | 1.30 (−2.14, 3.61) | 0.013b |
| DNAmTLadjAge, median (IQR) | 0.04 (−0.14, 0.20) | −0.03 (−0.12, 0.07) | 0.0914b |
| GrimAge componentsd | |||
| DNAmADM [pg/ml] (IQR) | 278.378 (262.456, 291.612) | 317.582 (300.479, 337.206) | <0.001b |
| DNAmB2M [pg/ml] (IQR) | 1115022 (1071350, 1169213) | 1440094 (1225107, 1549209) | <0.001b |
| DNAmCystatinC [pg/ml] (IQR) | 504475 (486991, 515874) | 589892 (539036, 605227) | <0.001b |
| DNAmGDF-15 [pg/ml] (IQR) | 275.698 (226.861, 337.268) | 523.425 (378.453, 622.928) | <0.001b |
| DNAmLeptin [pg/ml] (IQR) | 4987.87 (−1390.06, 6282.70) | 5711.41 (554.882, 7582.69) | 0.012b |
| DNAmPAI-1 [pg/ml] (IQR) | 12689.9 (11927.0, 14153.7) | 15438.6 (13629.0, 17392.3) | <0.001b |
| DNAmTIMP-1 [pg/ml] (IQR) | 28674.7 (28312.8 29072.3) | 31910.2 (29996.9, 33285.2) | <0.001b |
| DNAmPACKYRS (IQR) | 5.08330 (2.68010, 6.73291) | 10.9202 (8.12618, 14.8601) | <0.001b |
| DNAm-based white blood cell compositione | |||
| CD8+ T cell (IQR) | 0.085 (0.058, 0.112) | 0.095 (0.082, 0.129) | 0.031b |
| Naive CD8+ T cell (IQR) | 292.1 (256.1, 321.7) | 230.7 (194.5, 271.4) | <0.001b |
| Exhausted CD8+ T cell (IQR) | 9.33 (7.184, 11.934) | 9.846 (8.739, 11.546) | 0.595b |
| CD4+ T cell (IQR) | 0.133 (0.097, 0.157) | 0.111(0.075, 0.142) | 0.058b |
| Naive CD4+ T cell (IQR) | 422.0 (331.4, 473.0) | 371.2 (323.2, 466.4) | 0.524b |
| Natural killer cell (IQR) | 0.130 (0.089, 0.163) | 0.030 (0.018, 0.049) | <0.001b |
| Monocyte (IQR) | 0.056 (0.048, 0.072) | 0.059 (0.050, 0.072) | 0.402b |
| Granulocyte (IQR) | 0.548 (0.490, 0.595) | 0.625 (0.554, 0.680) | <0.001b |
| B cell (IQR) | 0.045 (0.037, 0.058) | 0.049 (0.033, 0.063) | 0.642b |
| Plasmablast (IQR) | 1.817 (1.691, 1.962) | 1.831 (1.692, 1.936) | 0.792b |
aWe assessed p -value using the χ2-test.
bWe assessed p -value using the Mann–Whitney U test.
cWe defined epigenetic age acceleration (AgeAccelHorvath, AgeAccelHannum, AgeAccelSkinBlood, AgeAccelPheno, and AgeAccelGrim) as the residual from regressing each DNAm age on the chronological age. We defined DNAmTLadjAge as the residual from regressing DNAmTL on chronological age.
dGrimAge components indicated DNAm-based age-predictive factors.
eThese white blood cell counts are predicted by DNAm.
If p-values are significant at <0.05, they are denoted in bold and in italics.
ADM adrenomedullin, B2M beta-2-microglobulin, CD4 cluster of differentiation 4, CD8 cluster of differentiation 8, CTL control, DNAm DNA methylation, DNAmPACKYRS DNA methylation-based smoking pack-years, DNAmTL DNA methylation-based telomere length, DNAmTLadjAge age-adjusted estimate of DNA methylation-based telomere length, GDF-15 growth differentiation factor 15, IQR interquartile range, MDD major depressive disorder, PAI-1 plasminogen activator inhibitor-1, TIMP-1 tissue inhibitor ofmetalloproteinases-1.
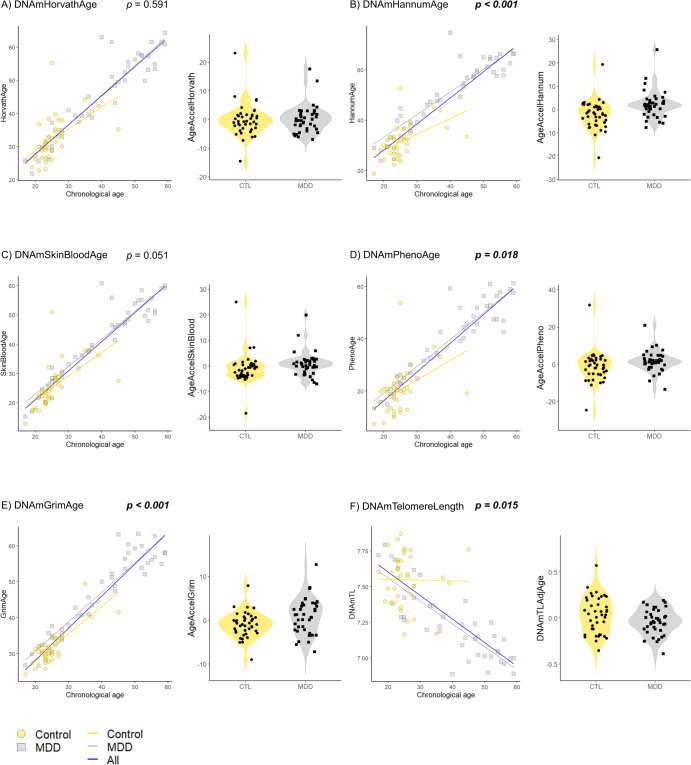
Fig. 1. Comparison of epigenetic age and DNAmTL between patients with untreated MDD and controls.
(A) HorvathAge, (B) HannumAge, (C) SkinBloodAge, (D) PhenoAge, (E) GrimAge, and (F) DNAmTL. The scatter plots indicate the epigenetic age and DNAmTL on the y-axis vs. the chronological age on the x-axis. We demonstrate the acceleration of epigenetic age and DNAmTLadjAge in patients with untreated MDD and in the control groups using the violin plots with data points. We refer p-values to multiple linear regression analysis. If p-values are significant at <0.05, they are denoted in bold and italics. CTL, control; DNAm, DNA methylation; DNAmTL, DNAm-based telomere length; DNAmTLadjAge, age-adjusted estimate of DNAm-based TL; MDD, major depressive disorder.
Patients with untreated MDD showed significant age acceleration compared with controls in AgeAccelHannum (p < 0.001), AgeAccelSkinBlood (p = 0.0098), AgeAccelPheno (p = 0.0333), and AgeAccelGrim (p = 0.013) (Fig. 1 and Table 1). After adjusting for confounding factors, such as age and sex, significant differences were observed in AgeAccelHannum (p < 0.001, R2 = 0.197), AgeAccelPheno (p = 0.018, R2 = 0.045), AgeAccelGrim (p < 0.001, R2 = 0.376), and DNAmTLadjAge (p = 0.015, R2 = 0.092) (Fig. 1 and Table 2).
Table 2.
Multiple regression analyses of DNAm-based Ages and TL acceleration, as well as GrimAge components, and DNAm-based white blood cell counts between patients with untreated MDD and controls.
| Phenotype | Age | Sex | Adjusted R2 | ||||
|---|---|---|---|---|---|---|---|
| Estimate | P-value | Estimate | P-value | Estimate | P-value | ||
| DNAmAge and DNAmTL accelerationa | |||||||
| AgeAccelHorvath | 0.80055684 | 0.591 | −0.01499872 | 0.803 | −1.69811179 | 0.150 | −0.00887 |
| AgeAccelHannum | 7.3704388 | <0.001 | −0.1807105 | 0.007 | −1.9114904 | 0.135 | 0.19710 |
| AgeAccelSkinBlood | 2.97353317 | 0.051 | −0.07232145 | 0.236 | −0.95934000 | 0.418 | 0.01747 |
| AgeAccelPheno | 4.717704 | 0.018 | −0.114084 | 0.153 | −1.734133 | 0.263 | 0.04549 |
| AgeAccelGrim | 3.56851612 | <0.001 | −0.07863095 | 0.020 | −3.77890619 | <0.001 | 0.37590 |
| DNAmTLAdjAge | −0.11769426 | 0.015 | 0.00270866 | 0.160 | 0.08750912 | 0.021 | 0.09222 |
| GrimAge componentsb | |||||||
| DNAmADM | 14.007312 | 0.00012 | 1.527318 | <0.001 | 20.452635 | <0.001 | 0.8329 |
| DNAmB2M | 53507.20 | 0.085 | 14468.58 | <0.001 | 18964.53 | 0.434 | 0.7744 |
| DNAmCystatinC | 14627.800 | 0.00536 | 3640.076 | <0.001 | −4098.721 | 0.310 | 0.8876 |
| DNAmGDF-15 | 58.962437 | 0.150 | 9.885624 | <0.001 | −1.349210 | 0.966 | 0.4982 |
| DNAmLeptin | 760.31512 | 0.235 | 36.74497 | 0.156 | 7059.14737 | <0.001 | 0.7330 |
| DNAmPAI-1 | 2319.41380 | 0.000587 | 32.76313 | 0.213 | −1446.18057 | 0.0057 | 0.3066 |
| DNAmTIMP-1 | 452.4535 | 0.000149 | 149.7430 | <0.001 | −229.8033 | 0.0185 | 0.9627 |
| DNAmPACKYRS | 3.3794458 | 0.123 | 0.3583494 | <0.001 | −8.5257449 | <0.001 | 0.4356 |
| DNAm-based white blood cell compositionc | |||||||
| CD8+ T cell | 0.04056 | 0.0013 | −0.00116 | 0.020 | 0.01363 | 0.157 | 0.1220 |
| Naive CD8+ T cell | −14.754 | 0.221 | −2.3271 | <0.001 | 4.4057 | 0.641 | 0.3856 |
| Exhausted CD8+ T cell | 0.03132 | 0.973 | 0.02144 | 0.572 | −0.27588 | 0.709 | −0.0304 |
| CD4+ T cell | −0.02286 | 0.146 | −0.00009 | 0.882 | 0.00582 | 0.635 | 0.0135 |
| Naive CD4+ T cell | 27.370 | 0.333 | −3.1214 | <0.001 | 33.357 | 0.136 | 0.0848 |
| Natural killer cell | −0.08937 | <0.001 | −0.00003 | 0.961 | −0.01051 | 0.383 | 0.4169 |
| Monocyte | 0.01141 | 0.023 | −0.00044 | 0.030 | −0.00633 | 0.106 | 0.0681 |
| Granulocyte | 0.04956 | 0.070 | 0.00155 | 0.159 | −0.00060 | 0.977 | 0.1246 |
| B cell | 0.00140 | 0.828 | 0.00006 | 0.819 | −0.00009 | 0.986 | −0.0358 |
| Plasmablast | 0.00564 | 0.914 | −0.00059 | 0.779 | −0.04562 | 0.268 | −0.0210 |
Dummy variables: phenotype, CTL = 0, and MDD = 1; sex, male = 0 and female= 1.
If p-values are significant at <0.05, they are denoted in bold and in italics.
R2 is the coefficient of determination.
aWe defined epigenetic age acceleration (AgeAccelHorvath, AgeAccelHannum, AgeAccelSkinBlood, AgeAccelPheno, and AgeAccelGrim) as the residual from regressing each DNAm age on the chronological age. We defined DNAmTLadjAge as the residual from regressing DNAmTL on chronological age.
bGrimAge components indicated DNAm-based age-predictive factors. For the GrimAge components, we corrected the significance level for multiple comparisons and the Bonferroni method defined the p-value as 0.05/8 = 0.00625 (eight GrimAge components).
If p-values are significant at <0.00625, they are denoted in bold and in italics.
cThese white blood cell counts were predicted using DNAm. For DNAm-based white blood cells, we adjusted the significance level for multiple comparisons and the Bonferroni method defined the p-value as 0.05/10 = 0.005 (ten white blood cell compositions).
If p-values are significant at <0.005, they are denoted in bold and in italics.
ADM adrenomedullin, B2M beta-2-microglobulin, CD4 cluster of differentiation 4, CD8 cluster of differentiation 8, CTL control, DNAm DNA methylation, DNAmPACKYRS DNA methylation-based smoking pack-years, DNAmTL DNA methylation-based telomere length, DNAmTLadjAge age-adjusted estimate of DNA methylation-based telomere length, GDF-15 growth differentiation factor 15, MDD major depressive disorder, PAI-1 plasminogen activator inhibitor-1, TIMP-1 tissue inhibitor ofmetalloproteinases-1.
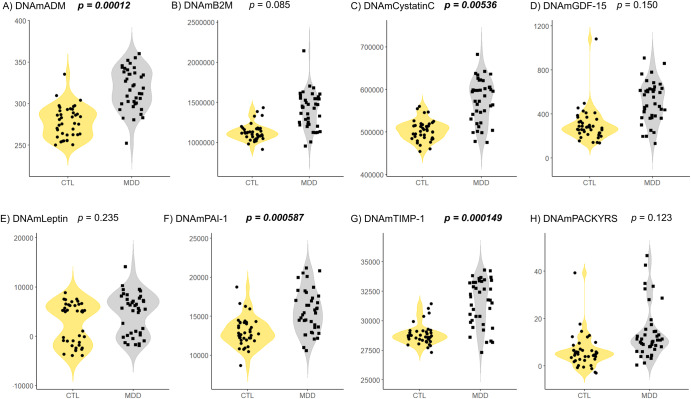
All GrimAge components were significantly different between patients with untreated MDD and controls (Fig. 2 and Table 1). After adjusting for confounding factors such as age and sex, DNAmADM (p = 0.00012, R2 = 0.833), DNAmCystatinC (p = 0.00536, R2 = 0.888), DNAmPAI-1 (p = 0.000587, R2 = 0.307), and DNAmTIMP-1 (p = 0.000149, R2 = 0.963) remained significantly increased in patients with untreated MDD compared to those in controls (Fig. 2 and Table 2). These results withstood the Bonferroni correction for multiple comparisons of eight GrimAge components (corrected significance was defined as p-value < 0.05/8 = 0.00625).
Fig. 2. Comparison of GrimAge components between patients with untreated MDD and controls.
(A) ADM, (B) B2M, (C) Cystatin C, (D) GDF-15, (E) Leptin, (F) PAI-1, (G) TIMP-1, and (H) PACKYRS. We show GrimAge components between patients with untreated MDD and control groups using the violin plots with data points. GrimAge components mean DNAm-based age-predictive factors. We refer p-values to multiple linear regression analysis. We corrected the significance level for multiple comparisons and the Bonferroni method defined the p-value as 0.05/8 = 0.00625. If p-values are significant at <0.00625, they are denoted in bold and italics. ADM, adrenomedullin; BMI, body mass index; B2M, beta-2-microglobulin; CTL, control; DNAm, DNA methylation; DNAmPACKYRS, DNA methylation-based smoking pack-years; GDF-15, growth differentiation factor 15; MDD, major depressive disorder; PACKYRS, smoking pack-years; PAI-1, plasminogen activator inhibitor-1; TIMP-1, tissue inhibitor ofmetalloproteinases-1.
White blood cell composition predicted based on DNAm
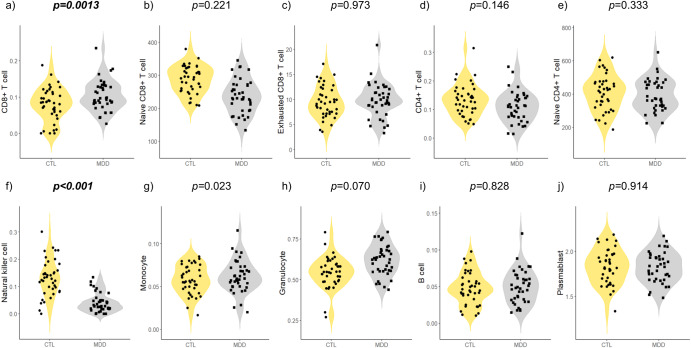
The analysis of DNAm-based white blood cell composition revealed significant differences in CD8+ T cells (p = 0.031), naive CD8+ T cells (p < 0.001), natural killer (NK) cells (p < 0.001), and granulocytes (p < 0.001) (Table 1). After adjusting for confounding factors such as age and sex, patients with untreated MDD were associated with an increase in CD8+ T cells (p = 0.0013, R2 = 0.122) and a decrease in NK cells (p < 0.001, R2 = 0.417) (Table 2 and Fig. 3). These results withstood the Bonferroni correction for multiple comparisons of 10 white blood cell compositions (corrected significance was defined as p-value < 0.05/10 = 0.005).
Fig. 3. Comparison of DNAm-based white blood cell composition among patients with untreated MDD and controls.
a cytotoxic CD8+ T cells, (b) naive CD8+ T cells, (c) exhausted CD8+ T cells, (d) helper CD4+ T cells, (e) naive CD4+ T cells, (f) natural killer cells, (g) monocytes, (h) granulocytes, (i) B cells, and (j) plasma blasts. Violin plots with dots indicate the DNAm-based white blood cell composition in patients with untreated MDD and control groups; p-values were evaluated using multiple linear regression analysis. We adjusted the significance level for multiple comparisons and the Bonferroni method defined the p-value as 0.05/10 = 0.005. If p-values are significant at <0.005, they are denoted in bold and italics. CD4, cluster of differentiation 4; CD8, cluster of differentiation 8; CTL, control; DNAm, DNA methylation; MDD, major depressive disorder.
Subgroup analysis of the participants younger than 40 years old
We conducted a subgroup analysis of the participants younger than 40 years old using the same methods and procedures as at the time of analysis for all ages (total analysis), whose age was not significantly different between the control and MDD groups (Age [years old], median [IQR]; CTL, 24.0 [23.0, 26.0]; MDD, 28.0 [24.0, 33.0]; p = 0.074) (Table 3 and Supplementary Table 4). Regarding the differences between groups in Table 1 and Table 3, there are no longer significant differences in the three GrimAge components (DNAmGDF-15, p = 0.0537; DNAmLeptin, p = 0.0776; DNAmPACKYRS, p = 0.0942) and one DNAm-based white blood cell composition (Naive CD8+ T cell, p = 0.215) (Table 3). The following results were significant in the total analysis and in the analysis of individuals under 40 after adjusting for confounding factors such as age and sex: AgeAccelHannum (p < 0.001, R2 = 0.191), AgeAccelGrim (p = 0.0016, R2 = 0.246), DNAmADM (p = 0.00287, R2 = 0.702), DNAmPAI-1 (p = 0.00304, R2 = 0.186), and DNAmTIMP-1 (p = 0.00089, R2 = 0.867), an increase in CD8+ T cells (p = 0.0012, R2 = 0.161) and a decrease in NK cells (p < 0.001, R2 = 0.361) (Supplementary Table 4).
Table 3.
Demographics, DNAm-based Age acceleration and telomere length acceleration, as well as GrimAge components, and DNAm-based white blood cell counts between patients with untreated MDD and controls younger than 40 years old.
| CTL (n = 39) | MDD (n = 17) | P-value | |
|---|---|---|---|
| Demographic characteristics | |||
| Sex (male/female) | 20/19 | 6/11 | 0.270a |
| Age (years old), median (IQR) | 24.0 (23.0, 26.0) | 28.0 (24.0, 33.0) | 0.074b |
| DNAmAge and DNAmTL accelerationc | |||
| AgeAccelHorvath, median (IQR) | −0.65 (−2.55, 1.51) | −1.79 (−3.14, 0.62) | 0.437b |
| AgeAccelHannum, median (IQR) | −1.77 (−5.22, 1.33) | 2.81 (0.62, 4.62) | <0.001b |
| AgeAccelSkinBlood, median (IQR) | −0.92 (−3.63, 0.62) | 1.24 (−0.21, 1.84) | 0.0053b |
| AgeAccelPheno, median (IQR) | −0.86 (−5.11, 2.41) | 0.96 (0.35, 4.33) | 0.0346b |
| AgeAccelGrim, median (IQR) | −0.97 (−2.42, 0.50) | 1.26 (0.10, 2.54) | 0.0056b |
| DNAmTLadjAge, median (IQR) | 0.04 (−0.15, 0.17) | −0.03 (−0.15, 0.12) | 0.202b |
| GrimAge componentsd | |||
| DNAmADM [pg/ml] (IQR) | 276.065 (262.423, 290.016) | 299.869 (287.251, 306.975) | <0.001b |
| DNAmB2M [pg/ml] (IQR) | 1114951 (1071275, 1167391) | 1218955 (1126147, 1254064) | 0.0037b |
| DNAmCystatinC [pg/ml] (IQR) | 503318 (486833, 514898) | 521895 (501579, 549532) | 0.0151b |
| DNAmGDF-15 [pg/ml] (IQR) | 272.530 (226.567, 329.455) | 365.640 (251.907, 419.807) | 0.0537b |
| DNAmLeptin [pg/ml] (IQR) | 4950.72 (−1525.32, 6312.50) | 5688.32 (−416.632, 7165.86) | 0.0776b |
| DNAmPAI-1 [pg/ml] (IQR) | 12743.0 (12051.9, 14162.8) | 14511.1 (13645.2, 16057.6) | 0.0012b |
| DNAmTIMP-1 [pg/ml] (IQR) | 28629.4 (28296.1, 29020.4) | 29691.4 (28608.7, 30386.9) | 0.0084b |
| DNAmPACKYRS (IQR) | 5.19338 (2.54860, 6.85158) | 8.09970 (4.03462, 10.90834) | 0.0942b |
| DNAm-based white blood cell compositione | |||
| CD8+ T cell (IQR) | 0.085 (0.057, 0.110) | 0.127 (0.090, 0.145) | 0.0026b |
| Naive CD8+ T cell (IQR) | 290.4 (256.1, 322.3) | 271.1 (242.7, 296.7) | 0.215b |
| Exhausted CD8+ T cell (IQR) | 9.47 (7.347, 12.009) | 9.159 (5.909, 10.368) | 0.447b |
| CD4+ T cell (IQR) | 0.132 (0.096, 0.160) | 0.106(0.079, 0.141) | 0.208b |
| Naive CD4+ T cell (IQR) | 418.9 (327.4, 468.0) | 456.6 (354.0, 476.2) | 0.264b |
| Natural killer cell (IQR) | 0.132 (0.094, 0.168) | 0.026 (0.017, 0.047) | <0.001b |
| Monocyte (IQR) | 0.057 (0.048, 0.073) | 0.060 (0.054, 0.080) | 0.228b |
| Granulocyte (IQR) | 0.548 (0.490, 0.590) | 0.623 (0.564, 0.645) | 0.0261b |
| B cell (IQR) | 0.046 (0.038, 0.060) | 0.045 (0.032, 0.061) | 1.000b |
| Plasmablast (IQR) | 1.814 (1.684, 1.964) | 1.857 (1.768, 1.946) | 0.888b |
aWe assessed p -value using the χ2-test.
bWe assessed p -value using the Mann–Whitney U test.
cWe defined epigenetic age acceleration (AgeAccelHorvath, AgeAccelHannum, AgeAccelSkinBlood, AgeAccelPheno, and AgeAccelGrim) as the residual from regressing each DNAm age on the chronological age. We defined DNAmTLadjAge as the residual from regressing DNAmTL on chronological age.
dGrimAge components indicated DNAm-based age-predictive factors.
eThese white blood cell counts are predicted by DNAm.
If p-values are significant at <0.05, they are denoted in bold and in italics.
ADM adrenomedullin, B2M beta-2-microglobulin, CD4 cluster of differentiation 4, CD8 cluster of differentiation 8, CTL control, DNAm DNA methylation, DNAmPACKYRS DNA methylation-based smoking pack-years, DNAmTL DNA methylation-based telomere length, DNAmTLadjAge age-adjusted estimate of DNA methylation-based telomere length, GDF-15 growth differentiation factor 15, IQR interquartile range, MDD major depressive disorder, PAI-1 plasminogen activator inhibitor-1, TIMP-1 tissue inhibitor ofmetalloproteinases-1.
Discussion
In this study, we provide evidence of epigenetic aging using five epigenetic age clocks, DNAmTL, and GrimAge components, and DNAm-based white blood cell composition in patients with untreated MDD. We found that epigenetic aging acceleration was significantly related to patients with untreated MDD in HannumAge and GrimAge after adjusting for confounding factors such as age and sex. These results are consistent with those of a previous study using GrimAge23. Similarly, after adjusting for confounding factors, patients with untreated MDD showed an increase in three GrimAge components (DNAmADM, DNAmPAI-1, and DNAmTIMP-1) as well as an increase in CD8+ T cells and a decrease in NK cells.
Our findings revealed that patients with MDD who had never received depression medication presented with accelerated epigenetic aging using HannumAge and GrimAge epigenetic clocks. In one study, patients who had undergone treatment for MDD had accelerated epigenetic aging as calculated using methods similar to HorvathAge32 but showed no significant difference in the five epigenetic clocks21. A postmortem study found no significant differences in epigenetic aging acceleration in patients with MDD versus controls using HorvathAge25. Another study found accelerated epigenetic aging in MDD compared to controls using peripheral blood based on HorvathAge and GrimAge20,23. However, accelerated epigenetic aging differs depending on the patient background as well as on the epigenetic clocks and sample used, and further studies with detailed patient background data are required using the five epigenetic clocks.
In addition, we observed a tendency for shorter telomere length based on DNAmTL when we conducted the analysis through all-age. Telomeres are unique structures located on the terminals of chromosomes, required to protect chromatin from DNA damage35. Telomeres shorten through cell replication, and telomere shortening is considered to signify a “molecular clock” based on underlying cell aging, which can render the body more vulnerable to diseases related to aging35–37. Patients with chronic MDD presented with significantly shorter leukocyte telomere lengths (LTL) than healthy controls38. The number of depressive episodes has been reported to be associated with a shorter LTL39. Furthermore, patients with untreated MDD have significantly shorter telomeres compared to those of controls40. A previous study indicated that antidepressant medication may protect against telomere shortening8. Lu et al. demonstrated that DNAmTL exhibited a stronger correlation with chronological age than did LTL, and it outperformed LTL in predicting both morbidity and mortality14. We suggest that depression is associated with accelerated biological aging, leading to premature mortality from various diseases. Chronic stress leads to dysregulation of the hypothalamic-pituitary adrenal (HPA) axis, resulting in the development of depression and various organ disorders41. HPA axis dysregulation is associated with cardiovascular mortality and many cardiovascular disease risk factors such as obesity, hypercholesterolemia, hypertriglyceridemia, elevated blood pressure, and diabetes42. Although antidepressants may inhibit HPA activity, reduce levels of cortisol, and prevent the development of various diseases, further studies are needed to elucidate the mechanism by which antidepressants reduce epigenetic aging in patients with MDD43.
We found that DNAmADM, DNAmPAI-1, and DNAmTIMP-1 levels were increased and DNAmCystatinC is likely to increase in patients with untreated MDD, suggesting that these epigenetic markers may be useful in understanding the pathophysiology of MDD and as potential biomarkers. Previous studies have also reported an association between increased levels of DNAmCystatinC and MDD21,23, as well as higher serum cystatin C levels in individuals with depression44–46. Cystatin C inhibits endogenous cysteine protease activity, is an established marker of kidney function, and involved in immunomodulation and inflammation47,48. A large number of studies have also reported that inflammation is one of the pathogenic factors in MDD49–51. In addition, cystatin C is involved in the signal transduction pathway of interferon-gamma (IFN-γ), which is predominantly secreted by NK and T cells48.
We observed a marked decrease in DNAm-based NK cells in patients with untreated MDD. Chronic stress reduces NK cells in the bone marrow and blood of mice and has been reported to also decrease NK cell counts in humans52,53. Patients with MDD had lower levels of CD56+CD16− NK cells, which are the major subtype producing IFN-γ, as well as lower IFN-γ levels compared to healthy controls54–56. This suggests a possible association between NK cells, cystatin C, and the development of depression due to inflammation. In contrast, previous studies have shown that increased DNAm-based NK cells are related to suicide completion, which is one of the most severe phenotypes of depression29. Additionally, acute psychological stress has been found to increase NK cells57. These findings suggest that acute psychological stress leading to suicide increases NK cells, whereas chronic stress decreases NK cells. Further studies of NK cells may help us understand the severity of depression in patients with MDD.
A previous study showed that the severity of MDD is associated with a lower cluster of differentiation 4 (CD4)+/ cluster of differentiation 8 (CD8) ratio, which indicates that its severity is linked to a decrease in CD4+ cells and an increase in CD8+ cells, which is consistent with our results58. While the immune-inflammatory hypothesis of depression is supported by accumulating evidence, it remains inconclusive and requires further research49–51.
As for other GrimAge components that we found to be associated with MDD, ADM is thought to play a regulatory and protective role in the hypothalamic-pituitary-adrenal (HPA) axis activated by stressors59. Patients with MDD had significantly higher serum ADM concentrations than did healthy controls60. Moreover, patients with MDD had significantly higher serum levels of PAI-1 and TIMP-1 than did controls61–63. Although our findings are consistent with those of previous studies, further research is required to confirm reproducibility of our results with larger sample sizes using actual plasma and serum to elucidate the pathophysiology of MDD and develop biomarkers.
Our study had several limitations. First, our sample size was small and only whole-blood samples were used. Second, the GrimAge components estimate plasma levels based on DNAm profiles and require verification using actual plasma. This is equally true for DNAm-based white blood cell composition. Third, we did not analyze other confounding factors related to the DNAm profile, such as the participant’s medical history, smoking history, and adverse childhood experiences, which impacted our results. Fourth, the control group was much younger than the MDD group. We did perform subgroup analysis of the participants younger than 40 years old, but this analysis halved MDD group and was targeted to people of a specific age. Although we adjusted our results for age as much as we could, a bias could still have remained and affected our conclusions. We also need to be cautious about interpreting the results of DNAmTL and DNAmCystatinC, where significant differences disappeared in the subgroup analysis limited to those under 40 years of age. Further study, with no age limit, matched for age and with an increased sample size, is required. Finally, we could not compare treated patients with MDD and untreated patients in the study to directly evaluate the effects of antidepressants based on the links between MDD and epigenetic age.
In conclusion, we investigated epigenetic aging using five epigenetic clocks, DNAmTL, GrimAge components, and DNAm-based white blood cell composition in patients with untreated MDD. We found a significant tendency for association between patients with untreated MDD and accelerated epigenetic aging, as determined by HannumAge and GrimAge. Additionally, our findings suggest that patients with untreated MDD are associated with alterations in several DNAm-based plasma proteins and two DNAm-based white blood cell composition, including decreased NK cells. It is possible that these findings provide insight into the pathophysiology and biomarkers of MDD, as well as unknown mechanisms of action of antidepressants.
Methods
Sample datasets (GSE201287)
In the present study, publicly available DNAm data was utilized from the Gene Expression Omnibus database. The GSE201287 dataset was contributed by Han and Liu et al. and comprised patients with untreated MDD (n = 40) and controls (n = 40), who gave written informed consent to attend the study approved by the Ethics Committee of the Chinese Academy of Medical Sciences and the Peking Union Medical College. All the subjects were of Chinese Han origin and recruited at the Department of Psychiatry, First Hospital of Shanxi Medical University, Taiyuan, China. All patients with MDD were in acute depressive episodes, with no history of psychiatric or psychological visits, and had not previously received treatment with medication. All patients were evaluated by at least two psychiatrists based on the Diagnostic and Statistical Manual of Mental Disorders Fourth Edition (DSM- IV). They used the Illumina Infinium 450k Human DNA methylation Beadchip data to obtain genome-wide DNA methylation profiles in peripheral blood samples (GEO Accession viewer (nih.gov))64.
Estimation via five epigenetic clocks, DNAmTL, and GrimAge components
Five epigenetic clocks (HorvathAge, HannumAge, SkinBloodAge, PhenoAge, and GrimAge) and DNAmTL were utilized via an online DNAm age calculator in the present study (https://horvath.genetics.ucla.edu/html/dnamage/)10. First-generation epigenetic clocks, HorvathAge and HannumAge, were developed by Horvath and Hannum et al.9,10. Horvath et al. developed SkinBloodAge as a revised clock using skin and blood cells to compensate for lower accuracy of first-generation epigenetic clocks in terms of fibroblasts11. Subsequently, PhenoAge, GrimAge, and DNAmTL have also been created, which have superior features in terms of predicting mortality and various health risks compared with first-generation epigenetic clocks12–14. GrimAge components comprise chronological age, sex, seven DNAm-based age-predictive plasma proteins (adrenomedullin [ADM], beta-2-microglobulin [B2M], cystatin C, growth differentiation factor-15 [GDF-15], leptin, plasminogen activation inhibitor-1 [PAI-1], and tissue inhibitor of metalloproteinases-1 [TIMP-1]), and DNAmethylation-based smoking pack-years (DNAmPACKYRS) in order to function as an epigenetic clock13. Epigenetic age acceleration (AgeAccelHorvath, AgeAccelHannum, AgeAccelSkinBlood, AgeAccelPheno, and AgeAccelGrim) means the residual from regressing each DNAm age on chronological age. Positive and negative values signify whether the epigenetic age is higher or lower comparison with the anticipated age (determined by chronological age). The age-adjusted estimate of DNAm-based TL (DNAmTLadjAge) is the residual calculated by regressing DNAmTL on the chronological age. Positive and negative values signify whether DNAmTLadjAge was longer or shorter than the anticipated DNAmTL (determined by chronological age), respectively. GrimAge components are described in this paper as follows: DNAmADM, DNAmB2M, DNAmCystatinC, DNAmGDF-15, DNAmleptin, DNAmPAI-1, DNAmTIMP-1, and DNAmPACKYRS13.
White blood cell composition predicted based on DNAm
Houseman et al. and Horvath developed the DNAm-based white blood cell composition10,15. We performed analysis of DNAm-based white blood cell composition using an online DNAm age calculator (https://horvath.genetics.ucla.edu/html/dnamage/)10. The DNAm-based white blood cell composition comprises a cytotoxic CD8+ T cells, naive CD8+ T cells, exhausted CD8+ T cells, helper CD4+ T cells, naive CD4+ T cells, NK cells, monocytes, granulocytes, B cells, and plasma blasts, which are estimated based on DNAm using Houseman’s and Horvath’s method10,15.
Statistical analysis
The data were analyzed using R version 4.2.2 (R Development Core Team, Vienna, Austria) and EZR software (version 1.61; Jichi Medical University, Saitama, Japan)65. We analyzed categorical variables and between-group differences in continuous variables using the χ2-test and Mann–Whitney U test, respectively. Regarding confounding factors, such as age and sex, we conducted multiple linear regression analyses. To examine the relationship between continuous variables, we performed Spearman’s rank correlation coefficient. Dummy variables were used where needed. Statistical significance was defined as a two-tailed p-value < 0.05.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
This work was partially supported by JSPS KAKENHI grant numbers JP18K15483, JP21K07520 (S.O.), JP17H04249, and JP21H02852 (A.H.).
Author contributions
S.O. and A.H. designed the study. S.O. and A.H. conducted the research. R.S., T.T., S.O., I.O., T.S., K.M., and T.H. collected data. R.S., T.T., I.O., T.S., K.M., and T.H. performed statistical analysis. T.T. investigated and visualized the data. R.S. and T.T. wrote and structured the first draft of the manuscript. S.O. and A.H. reviewed the manuscript. All authors contributed to and approved the final manuscript. R.S. and T.T. equally contributed to the work and are co-first authors.
Data availability
We utilized the GSE201287 dataset that is publicly available from the Gene Expression Omnibus database (GEO Accession viewer (nih.gov)), which has been used in previous studies64.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ryota Shindo, Takaki Tanifuji.
Supplementary information
The online version contains supplementary material available at 10.1038/s41514-023-00117-1.
References
- 1.Theo Vos, Stephen S Lim & Cristiana Abbafati. et al. (GBD 2019 Diseases and Injuries Collaborators). Global burden of 369 diseases and injuries in 204 countries and territories, 1990-2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet396, 1204–1222 (2020). [DOI] [PMC free article] [PubMed]
- 2.Lepine JP, Briley M. The increasing burden of depression. Neuropsychiatr. Dis. Treat. 2011;7:3–7. doi: 10.2147/NDT.S19617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Darrow SM, et al. The association between psychiatric disorders and telomere length: a meta-analysis involving 14,827 persons. Psychosom. Med. 2016;78:776–787. doi: 10.1097/PSY.0000000000000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ridout KK, Ridout SJ, Price LH, Sen S, Tyrka AR. Depression and telomere length: a meta-analysis. J. Affect. Disord. 2016;191:237–247. doi: 10.1016/j.jad.2015.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richmond-Rakerd LS, D’Souza S, Milne BJ, Caspi A, Moffitt TE. Longitudinal associations of mental disorders with dementia: 30-year analysis of 1.7 million New Zealand citizens. JAMA Psychiatry. 2022;79:333–340. doi: 10.1001/jamapsychiatry.2021.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang, L. et al. Depression, depression treatments, and risk of incident dementia: a prospective cohort study of 354,313 participants. Biol. Psychiatry10.1016/j.biopsych.2022.08.026 (2022). [DOI] [PubMed]
- 7.Bartels C, et al. Impact of SSRI therapy on risk of conversion from mild cognitive impairment to alzheimer’s dementia in individuals with previous depression. Am. J. Psychiatry. 2018;175:232–241. doi: 10.1176/appi.ajp.2017.17040404. [DOI] [PubMed] [Google Scholar]
- 8.Wolkowitz OM, et al. Resting leukocyte telomerase activity is elevated in major depression and predicts treatment response. Mol. Psychiatry. 2012;17:164–172. doi: 10.1038/mp.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hannum G, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell. 2013;49:359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;10:R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horvath S, et al. Epigenetic clock for skin and blood cells applied to Hutchinson Gilford Progeria Syndrome and ex vivo studies. Aging. 2018;10:1758–1775. doi: 10.18632/aging.101508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levine ME, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging. 2018;10:573–591. doi: 10.18632/aging.101414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu AT, et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging. 2019;11:303–327. doi: 10.18632/aging.101684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu AT, et al. DNA methylation-based estimator of telomere length. Aging. 2019;11:5895–5923. doi: 10.18632/aging.102173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Houseman, E. A. et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics13, 10.1186/1471-2105-13-86 (2012). [DOI] [PMC free article] [PubMed]
- 16.Cao X, et al. Accelerated biological aging in COVID-19 patients. Nat. Commun. 2022;13:2135. doi: 10.1038/s41467-022-29801-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kresovich JK, et al. Epigenetic mortality predictors and incidence of breast cancer. Aging. 2019;11:11975–11987. doi: 10.18632/aging.102523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lind, L., Ingelsson, E., Sundstrom, J., Siegbahn, A. & Lampa, E. Methylation-based estimated biological age and cardiovascular disease. Eur. J. Clin. Invest.48, 10.1111/eci.12872 (2018). [DOI] [PubMed]
- 19.Fraszczyk E, et al. DNA methylation trajectories and accelerated epigenetic aging in incident type 2 diabetes. Geroscience. 2022;44:2671–2684. doi: 10.1007/s11357-022-00626-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whalley, H. C. et al. Accelerated epigenetic ageing in major depressive disorder. https://www.biorxiv.org/content/10.1101/210666v1 (2017).
- 21.Tanifuji, T. et al. Epigenetic clock analysis reveals increased plasma cystatin C levels based on DNA methylation in major depressive disorder. Psychiatry Res.10.1016/j.psychres.2023.115103 (2023). [DOI] [PubMed]
- 22.Rampersaud R, et al. Dimensions of childhood adversity differentially affect biological aging in major depression. Transl. Psychiatry. 2022;12:431. doi: 10.1038/s41398-022-02198-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Protsenko E, et al. "GrimAge," an epigenetic predictor of mortality, is accelerated in major depressive disorder. Transl. Psychiatry. 2021;11:193. doi: 10.1038/s41398-021-01302-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo A, et al. Epigenetic aging is accelerated in alcohol use disorder and regulated by genetic variation in APOL2. Neuropsychopharmacology. 2020;45:327–336. doi: 10.1038/s41386-019-0500-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Z, He Y, Ma X, Chen X. Epigenetic age analysis of brain in major depressive disorder. Psychiatry Res. 2018;269:621–624. doi: 10.1016/j.psychres.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 26.Okazaki S, et al. Decelerated epigenetic aging associated with mood stabilizers in the blood of patients with bipolar disorder. Transl. Psychiatry. 2020;10:129. doi: 10.1038/s41398-020-0813-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okazaki S, et al. Epigenetic clock analysis of blood samples from Japanese schizophrenia patients. NPJ. Schizophr. 2019;5:4. doi: 10.1038/s41537-019-0072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katrinli S, et al. Evaluating the impact of trauma and PTSD on epigenetic prediction of lifespan and neural integrity. Neuropsychopharmacology. 2020;45:1609–1616. doi: 10.1038/s41386-020-0700-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okazaki S, et al. Accelerated extrinsic epigenetic aging and increased natural killer cells in blood of suicide completers. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2020;98:109805. doi: 10.1016/j.pnpbp.2019.109805. [DOI] [PubMed] [Google Scholar]
- 30.Marioni RE, et al. DNA methylation age of blood predicts all-cause mortality in later life. Genome. Biol. 2015;16:1–12. doi: 10.1186/s13059-015-0584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryan CP. "Epigenetic clocks": Theory and applications in human biology. Am. J. Hum. Biol. 2021;33:e23488. doi: 10.1002/ajhb.23488. [DOI] [PubMed] [Google Scholar]
- 32.Han LKM, et al. Epigenetic aging in major depressive disorder. Am. J. Psychiatry. 2018;175:774–782. doi: 10.1176/appi.ajp.2018.17060595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu C, et al. Association between depression and epigenetic age acceleration: a co-twin control study. Depress Anxiety. 2022;39:741–750. doi: 10.1002/da.23279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKenna BG, et al. Maternal prenatal depression and epigenetic age deceleration: testing potentially confounding effects of prenatal stress and SSRI use. Epigenetics. 2021;16:327–337. doi: 10.1080/15592294.2020.1795604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collins K, Mitchell JR. Telomerase in the human organism. Oncogene. 2002;21:564–579. doi: 10.1038/sj.onc.1205083. [DOI] [PubMed] [Google Scholar]
- 36.Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 37.Blasco MA. Telomeres and human disease: ageing, cancer and beyond. Nat. Rev. Genet. 2005;6:611–622. doi: 10.1038/nrg1656. [DOI] [PubMed] [Google Scholar]
- 38.Wolkowitz OM, et al. Leukocyte telomere length in major depression: correlations with chronicity, inflammation and oxidative stress–preliminary findings. PLoS ONE. 2011;6:e17837. doi: 10.1371/journal.pone.0017837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simon NM, et al. Telomere shortening and mood disorders: preliminary support for a chronic stress model of accelerated aging. Biol. Psychiatry. 2006;60:432–435. doi: 10.1016/j.biopsych.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 40.Garcia-Rizo C, et al. Abnormal glucose tolerance, white blood cell count, and telomere length in newly diagnosed, antidepressant-naive patients with depression. Brain Behav. Immun. 2013;28:49–53. doi: 10.1016/j.bbi.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tian, Y. E. et al. Evaluation of brain-body health in individuals with common neuropsychiatric disorders. JAMA Psychiatry10.1001/jamapsychiatry.2023.0791 (2023). [DOI] [PMC free article] [PubMed]
- 42.Jokinen J, Nordstrom P. HPA axis hyperactivity and cardiovascular mortality in mood disorder inpatients. J. Affect. Disord. 2009;116:88–92. doi: 10.1016/j.jad.2008.10.025. [DOI] [PubMed] [Google Scholar]
- 43.McKay MS, Zakzanis KK. The impact of treatment on HPA axis activity in unipolar major depression. J. Psychiatr. Res. 2010;44:183–192. doi: 10.1016/j.jpsychires.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 44.Sun T, Chen Q, Li Y. Associations of serum cystatin C with depressive symptoms and suicidal ideation in major depressive disorder. BMC Psychiatry. 2021;21:576. doi: 10.1186/s12888-021-03509-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, et al. Cystatin C and risk of new-onset depressive symptoms among individuals with a normal creatinine-based estimated glomerular filtration rate: a prospective cohort study. Psychiatry Res. 2019;273:75–81. doi: 10.1016/j.psychres.2019.01.009. [DOI] [PubMed] [Google Scholar]
- 46.Huang Y, Huang W, Wei J, Yin Z, Liu H. Increased serum cystatin C levels were associated with depressive symptoms in patients with type 2 diabetes. Diabetes Metab Syndr Obes. 2021;14:857–863. doi: 10.2147/DMSO.S295088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evangelopoulos AA, et al. Association between serum cystatin C, monocytes and other inflammatory markers. Intern. Med. J. 2012;42:517–522. doi: 10.1111/j.1445-5994.2011.02500.x. [DOI] [PubMed] [Google Scholar]
- 48.Zi M, Xu Y. Involvement of cystatin C in immunity and apoptosis. Immunol. Lett. 2018;196:80–90. doi: 10.1016/j.imlet.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Colasanto M, Madigan S, Korczak DJ. Depression and inflammation among children and adolescents: a meta-analysis. J. Affect. Disord. 2020;277:940–948. doi: 10.1016/j.jad.2020.09.025. [DOI] [PubMed] [Google Scholar]
- 50.Jokela M, Virtanen M, Batty GD, Kivimaki M. Inflammation and specific symptoms of depression. JAMA Psychiatry. 2016;73:87–88. doi: 10.1001/jamapsychiatry.2015.1977. [DOI] [PubMed] [Google Scholar]
- 51.Kiecolt-Glaser JK, Derry HM, Fagundes CP. Inflammation: depression fans the flames and feasts on the heat. Am. J. Psychiatry. 2015;172:1075–1091. doi: 10.1176/appi.ajp.2015.15020152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ishikawa Y, et al. Repeated social defeat stress induces neutrophil mobilization in mice: maintenance after cessation of stress and strain-dependent difference in response. Br. J. Pharmacol. 2021;178:827–844. doi: 10.1111/bph.15203. [DOI] [PubMed] [Google Scholar]
- 53.Maydych V, et al. Impact of chronic and acute academic stress on lymphocyte subsets and monocyte function. PLoS ONE. 2017;12:e0188108. doi: 10.1371/journal.pone.0188108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suzuki H, et al. Altered populations of natural killer cells, cytotoxic T lymphocytes, and regulatory T cells in major depressive disorder: Association with sleep disturbance. Brain Behav. Immun. 2017;66:193–200. doi: 10.1016/j.bbi.2017.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim YK, et al. Differences in cytokines between non-suicidal patients and suicidal patients in major depression. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2008;32:356–361. doi: 10.1016/j.pnpbp.2007.08.041. [DOI] [PubMed] [Google Scholar]
- 56.Cooper MA, Fehniger TA, Caligiuri MA. The biology of human natural killer-cell subsets. Trends Immunol. 2001;22:633–640. doi: 10.1016/s1471-4906(01)02060-9. [DOI] [PubMed] [Google Scholar]
- 57.Breen MS, et al. Acute psychological stress induces short-term variable immune response. Brain Behav. Immun. 2016;53:172–182. doi: 10.1016/j.bbi.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 58.Zhou D, et al. A novel joint index based on peripheral blood CD4+/CD8+ T cell ratio, albumin level, and monocyte count to determine the severity of major depressive disorder. BMC Psychiatry. 2022;22:248. doi: 10.1186/s12888-022-03911-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khan S, Michaud D, Moody TW, Anisman H, Merali Z. Effects of acute restraint stress on endogenous adrenomedullin levels. NeuroReport. 1999;10:2829–2833. doi: 10.1097/00001756-199909090-00024. [DOI] [PubMed] [Google Scholar]
- 60.Akpinar A, Yaman GB, Demirdas A, Onal S. Possible role of adrenomedullin and nitric oxide in major depression. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2013;46:120–125. doi: 10.1016/j.pnpbp.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 61.Jiang H, et al. Plasminogen activator inhibitor-1 in depression: results from animal and clinical studies. Sci. Rep. 2016;6:30464. doi: 10.1038/srep30464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lahlou-Laforet K, et al. Relation of depressive mood to plasminogen activator inhibitor, tissue plasminogen activator, and fibrinogen levels in patients with versus without coronary heart disease. Am. J. Cardiol. 2006;97:1287–1291. doi: 10.1016/j.amjcard.2005.11.062. [DOI] [PubMed] [Google Scholar]
- 63.Chan MK, et al. Blood-based immune-endocrine biomarkers of treatment response in depression. J. Psychiatr. Res. 2016;83:249–259. doi: 10.1016/j.jpsychires.2016.08.020. [DOI] [PubMed] [Google Scholar]
- 64.Xiu J, et al. Elevated BICD2 DNA methylation in blood of major depressive disorder patients and reduction of depressive-like behaviors in hippocampal Bicd2-knockdown mice. Proc. Natl Acad. Sci. USA. 2022;119:e2201967119. doi: 10.1073/pnas.2201967119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kanda Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant. 2013;48:452–458. doi: 10.1038/bmt.2012.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We utilized the GSE201287 dataset that is publicly available from the Gene Expression Omnibus database (GEO Accession viewer (nih.gov)), which has been used in previous studies64.