Summary
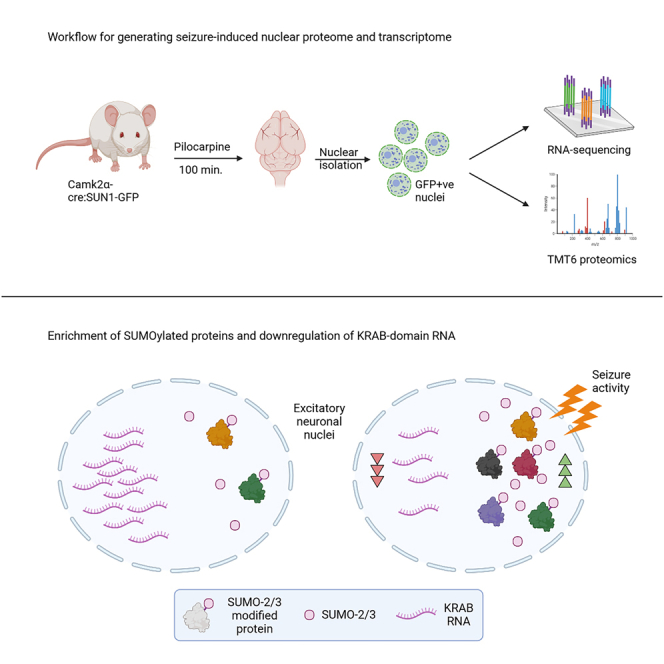
A single episode of pilocarpine-induced status epilepticus can trigger the development of spontaneous recurrent seizures in a rodent model for epilepsy. The initial seizure-induced events in neuronal nuclei that lead to long-term changes in gene expression and cellular responses likely contribute toward epileptogenesis. Using a transgenic mouse model to specifically isolate excitatory neuronal nuclei, we profiled the seizure-induced nuclear proteome via tandem mass tag mass spectrometry and observed robust enrichment of nuclear proteins associated with the SUMOylation pathway. In parallel with nuclear proteome, we characterized nuclear gene expression by RNA sequencing which provided insights into seizure-driven transcriptional regulation and dynamics. Strikingly, we saw widespread downregulation of zinc-finger transcription factors, specifically proteins that harbor Krüppel-associated box (KRAB) domains. Our results provide a detailed snapshot of nuclear events induced by seizure activity and demonstrate a robust method for cell-type-specific nuclear profiling that can be applied to other cell types and models.
Subject areas: Biological sciences, Neuroscience, Molecular neuroscience, Omics
Graphical abstract
Highlights
-
•
Generated seizure-induced nuclear proteome and transcriptome from excitatory neurons
-
•
Seizure nuclei are enriched with SUMO-associated proteins and regulated pathways
-
•
Downregulation of RNA-encoding KRAB domain zinc-finger proteins during seizures
Biological sciences; Neuroscience; Molecular neuroscience; Omics
Introduction
In epilepsy, intense bursts of neuronal activity that trigger seizures activate different intracellular signaling cascades that alter nuclear processes and shape transcriptional output. For a subset of cells in the epileptogenic zones, seizure-induced, activity-dependent modifications within the nucleus may result in persistent changes that fundamentally alter neuronal function and connectivity. Indeed, altered nuclear processes have been observed in studies of epilepsy. These include repositioning of chromosomes altering transcription dynamics,1,2,3 altered ribosome biogenesis,4 stimulation of transcription factor activity leading to neurodegeneration, inflammation or mossy fiber sprouting,5 epigenetic modifications which alter transcriptional outputs6,7 and others. Many of these studies of acquired epilepsy in rodent models provide a snapshot of one or two aspects of seizure-induced changes in the nucleus. To comprehensively capture nuclear changes in different brain cells during seizures, it is advantageous to use a single mouse model and system that is cross-compatible with multiple assays, platforms, and technologies.
High-throughput omics technologies can capture diverse changes in the brain during seizures across different time points and phases of the disorder. To date, several studies have profiled seizure-induced transcriptome,8,9,10,11,12,13,14,15 proteome,16,17,18,19,20,21,22 and epigenome.23,24,25,26 These studies are most commonly performed on resected epileptic brain tissues from patients or on specific brain regions—most commonly the hippocampus—in rodent models for acquired epilepsy. Collectively, these profiling studies have broadened our understanding of different brain states during progression of epilepsy, and a more comprehensive overview of activity-dependent changes in the brain can be found in the following reviews.27,28 Many early studies on epileptic tissues use bulk sequencing techniques which derive a shared signal from all cell types in the tissue. While these studies have provided valuable insights into activity-driven changes in broad regions of the brain, cell-type and sub-compartment-specific changes remain challenging to extract from these complex datasets. Nevertheless, progress has been made and recent advances in nucleic acid-sequencing technologies and combinatorial indexing have enabled researchers to study epilepsy-related changes at the single-cell level, even though issues with bias and lack of depth in coverage, as well as low capture efficiencies currently limit a deeper analysis of low-abundance transcripts and variations in RNA processing.10 While the depth, sensitivity, and specificity of transcriptome and epigenome studies have greatly improved, proteomics studies remain challenging as the technology still lags behind that of nucleic acid sequencing, and the development of single-cell proteomics is still a work in progress.29
We are interested in understanding how initial seizure activity drives different facets of nuclear signaling that lead to enduring changes which may contribute to the development of spontaneous recurrent seizures in rodents, as a model for the etiology of epilepsy. Given the challenges in proteomics, we required a method of cell-type-specific nuclear isolation that is particularly suited for mass spectrometry but also amenable for other types of -omics analyses. Using a transgenic mouse line that genetically tags the nuclei,30 we injected pilocarpine to induce seizures and isolated pure populations of neuronal nuclei for RNA-seq and LC-MS/MS analysis. This allowed us to capture population-specific changes in the nucleus while maintaining the depth and coverage afforded through bulk sequencing techniques. By focusing on the nucleus, we detected different pools of proteins and transcripts altered by seizure-induced activity, including the enrichment of all major subunits of the AP-1 complex, indicating this methodology is sufficiently sensitive to detect activity-dependent changes in the nucleus. In the proteome, we observed an enrichment of post-translational modification systems associated with the addition of small ubiquitin-like tags (UBLs). This enrichment is most striking for protein SUMOylation, with multiple proteins in the SUMOylation and associated pathways being detected in the dataset and a subset verified through immunoassays. In parallel to the proteome, RNA-seq data at the same time point shows a strong upregulation of genes involved in several different functions, including kinase/phosphatase activity relating to the MAPK pathways, and a downregulation of KRAB-domain genes likely through activation of THAP11/Ronin transcriptional repressor. Finally, we also compared intron- and exon-level differential gene expression between conditions, shedding light on seizure-driven transcriptional dynamics and post-transcriptional responses.
Results
Isolating excitatory neuronal nuclei from mock- and pilocarpine-injected SUN1-sfGFP mice
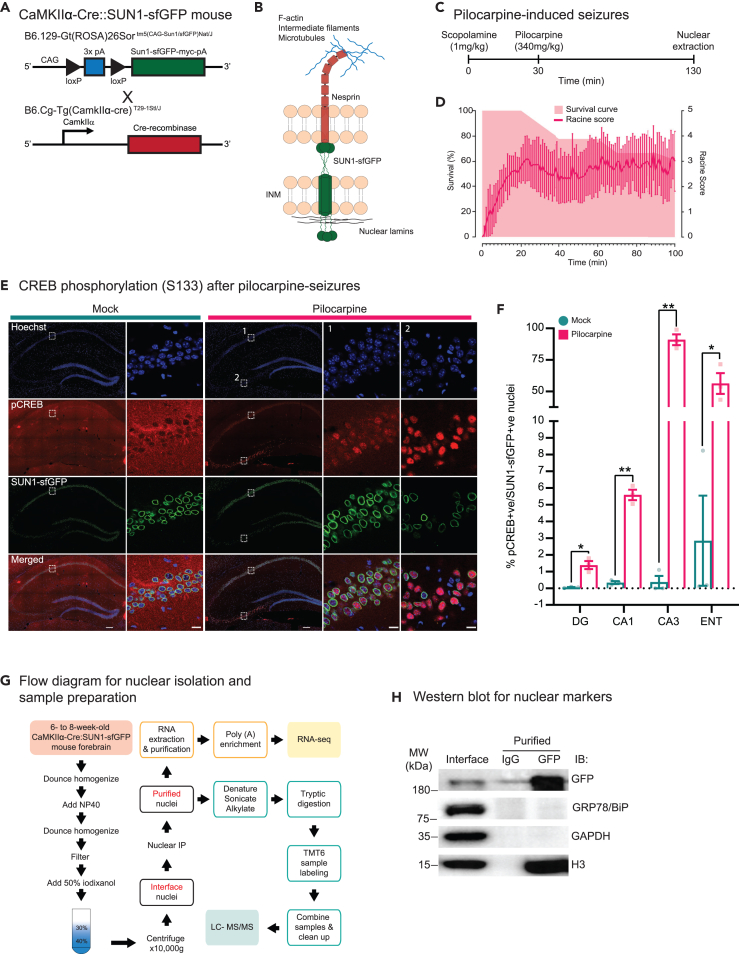
To examine how pilocarpine-induced seizures alter the nuclear proteome and transcriptome in excitatory neurons, we crossed B6.129-Gt(ROSA)26Sortm5(CAG−Sun1/sfGFP)Nat/J with B6.Cg-Tg(CamkIIα-cre)T29−1Stl/J to generate a transgenic mouse that expresses the fusion protein SUN1-sfGFP in excitatory neurons (Figure 1A). SUN1 is an inner nuclear membrane protein that is a part of the linker of nucleoskeleton and cytoskeleton (LINC) complex which functions to connect the nuclear lamina with the cytoskeleton in the cytoplasm (Figure 1B).31 As previously reported, breeding of mice with lox-stop-lox SUN1-sfGFP expression and CaMKIIα-Cre mice results in a robust SUN1-sfGFP labeling of nuclei in forebrain excitatory neurons and not in inhibitory neurons (Figure S1).30 We also observed widespread SUN1-sfGFP neuronal expression in multiple brain regions including the hippocampus and cortex (Figures S1 and S2).
Figure 1.
Pilocarpine-induced seizures of CaMKIIα-Cre:SUN1-sfGFP mice
(A) Transgenic mice used in the study and breeding plan for CaMKIIα-Cre:SUN1-sfGFP.
(B) Localization of SUN1-sfGFP and nesprin in nuclear membrane and their association with cytoskeletal elements in the nuclear periphery.
(C) Timeline of seizure induction by intraperitoneal administration of scopolamine and pilocarpine.
(D) Survival curve of pilocarpine-injected CaMKIIα-Cre:SUN1-sfGFP mice over the duration of the experiment. Seizure behaviors were video recorded and scored based on the Racine scale.
(E) Immunohistochemistry showing pCREB (S133) expression in CA1 (ROI#1) and CA3 (ROI#2) regions of the hippocampus in mock and pilocarpine-injected mice. SUN1-sfGFP (green); Hoechst (blue); pCREBS133 (red). Scale bars, 200 μm; 10 μm (magnified).
(F) Quantification of SUN1-sfGFP-positive and pCREB-positive neuronal nuclei (N = 3 pairs of animals; Total number of cells quantified = 14816 (mock)/12676 (pilocarpine) [DG], 2746 (mock)/2964 (pilocarpine) [CA1], 265 (mock)/276 (pilocarpine) [CA3], 3256 (mock)/3090 (pilocarpine) [ENT]. Paired t test with means ± SEM, p < 0.05 (∗), p < 0.01 (∗∗).
(G) Diagram of the nuclear isolation from the mouse forebrain.
(H) Western blot of nuclear extracts from interface stage and after purification with anti-GFP antibodies. Blots were labeled with antibodies against GFP, histone 3 (H3), GRP78/BiP, and GAPDH. IgG, Immunoglobulin G.
We subjected CaMKIIα-Cre:SUN1-sfGFP mice to a dose-response experiment with pilocarpine to determine the optimal concentration for maintaining seizures for a duration of 100 min (data not shown). Our goal was to capture activity-dependent changes in neuronal nuclei before widespread seizure-induced cell death.32,33 To that end, we used an intraperitoneal injection paradigm of scopolamine (1 mg/kg) for 30 min followed by pilocarpine (340 mg/kg) for 100 min before terminating the experiment (Figure 1C). Under this paradigm, mice exhibited symptoms of seizing with forelimb clonus and some rearing and falling. All mice were video-recorded, and severity of seizures was graded on a Racine scale with scores ranging between 2 and 4 (average of 3; Figure 1D). To validate the effectiveness of pilocarpine-induced seizures, forebrain sections were immunolabeled with antibodies to detect phosphorylation of cAMP response element-binding protein (CREB) (S133), a common marker to gauge activity-dependent transcription in neurons which is known to be elevated upon seizures.34,35 We observed widespread pCREB (S133) signal in excitatory neurons throughout the entire forebrain of animals injected with pilocarpine (Figures 1E and S2). Quantification of SUN1-sfGFP-positive nuclei in the hippocampus and cortex shows distinct regional differences, with CA3 region and entorhinal cortex (ENT) showing a more robust increase in CREB phosphorylation after pilocarpine injection (Figure 1F).
To extract nuclei, mice treated with either mock or pilocarpine injections were euthanized anesthesia free followed by extraction of the forebrain and midbrain structures (minus the olfactory bulb) and nuclear isolation using modified protocols for SUN1-sfGFP-expressing neurons (Figure 1G). We isolated on average, 4.4 x 106 SUN1-sfGFP-positive nuclei per mouse (79% yield) after immunopurification with magnetic beads conjugated with anti-GFP antibodies (Figures S3 and S4). Western blots indicated enrichment of nuclear markers and SUN1-sfGFP with negligible contamination from the endoplasmic reticulum or soluble cytoplasmic fractions indicated by the absence of GRP78/BiP and GAPDH, respectively (Figure 1H).
Profiling the nuclear proteome induced by seizures
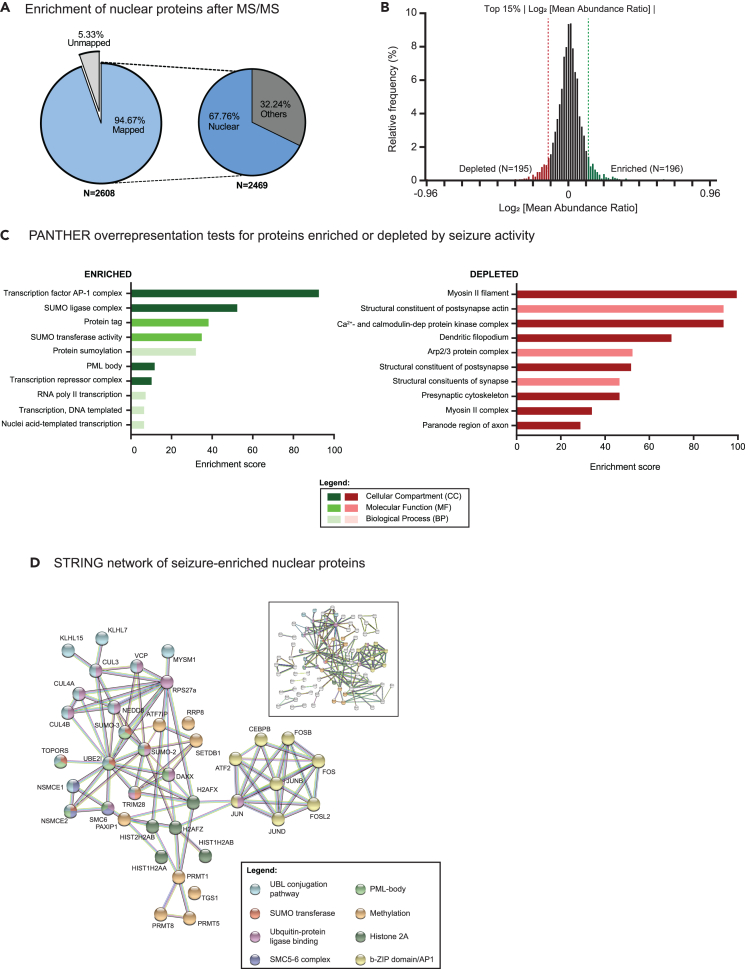
For the nuclear proteome, we identified N = 2608 unique proteins of which 94.67% (2469/2608) were mapped to the PANTHER database and 67.76% (1673/2469) of mapped proteins are annotated as nuclear proteins (GO:0005634; Figure 2A; Table S1). While the enrichment of nuclear proteins by sequencing indicates a successful isolation protocol, the actual number of nuclear proteins is likely to be underrepresented in the GO database as the annotations are incomplete. A histogram of the mean abundance ratio (MAR) for N = 2608 shows that the peak has a value close to 1 with a total of N = 1454 proteins having MAR >0, while N = 1154 have MAR <0 (Figure 2B).
Figure 2.
Nuclear proteome from seizure-induced forebrain excitatory neurons
(A) Chart showing percentage of mapped proteins annotated in the nucleus by PANTHER GO classification.
(B) Histogram showing relative frequency of log2 [mean abundance ratio] (MAR) between pilocarpine and mock-injected mice for N = 2608. The top 15% of total proteins with highest absolute log2MARs are highlighted in green for enriched (N = 196; log2MAR >0.1203) or red for depleted (N = 195; log2MAR < −0.1204) nuclear proteins.
(C) Top 10 GO terms ranked by enrichment scores across all three GO categories for proteins that are either enriched or depleted during seizures. GO classifications were performed separately for enriched or depleted pools of proteins.
(D) STRING network for seizure-enriched nuclear proteins (N = 196; Interaction score >0.7 with disconnected nodes from hidden from network). Magnified network highlights nodes of interest from the entire network (boxed inset).
To find out if proteins changed in the nucleus during seizures are enriched for particular functions, we performed PANTHER over-representation tests separately on seizure-enriched and -depleted proteins using the top 15% altered proteins (Enriched: N = 196; log2MAR ≥0.1203; Depleted: N = 195; log2MAR ≤ −0.1204; Figure 2C). For proteins enriched after seizure activity, the top over-represented GO terms ranked according to enrichment scores across all GO categories include transcription factor AP-1 complex (GO:0035976), SUMO ligase complex (GO:0106068), protein tag (GO:0031386), SUMO transferase activity (GO:0019789), protein SUMOylation (GO:0016925), PML body (GO:0016605), transcription repressor complex (GO:0017053), and several terms associated with transcription (GO:0006366, GO:0006351, and GO:0097659; Figure 2C). Of note, we performed multiple over-representation tests at different thresholds (top 5%, 10%, 20% log2MAR) and found that many of the GO categories, particularly those associated with AP-1 complex and protein SUMOylation consistently showed high-enrichment scores (data not shown). Analysis with STRING indicates that most of the AP-1 complex subunits including FOS, FOSB, FOSL2, JUN, JUNB, and JUND (and to a lesser degree, ΔFOSB) are enriched during seizures (Figure 2D). This includes ATF2 and C/EBPβ, two bZIP transcription factors that interact with AP-1 complex through heterodimerization and undergo activity-dependent nuclear accumulation36,37 (Figure 2D). In addition to AP-1 transcription factor complex, PANTHER over-representation tests also show enrichment of multiple GO terms associated with protein SUMOylation and SUMO-related processes. SUMO is a small protein tag that is post-translationally attached to target proteins on a lysine residue, a process similar to ubiquitination. SUMOylated proteins participate in regulating a variety of cellular mechanisms including gene expression, DNA repair and subcellular localization of proteins.38,39 In our dataset, we found SUMO-2, SUMO-3, and several key SUMOylation enzymes elevated after seizures (Figure 2D). Furthermore, STRING analysis confirms that at least 7 proteins associated to promyelocytic leukemia (PML) nuclear bodies including DAXX, and at least 3 members of the structural maintenance of chromosomes 5 and 6 (SMC-5/6) complex are enriched in the nucleus after seizures with (SMC6, NSMCE1, and NSMCE2; Figure 2D). Both PML bodies and SMC-5/6 complex associate with multiple SUMO-related proteins and their functions are heavily regulated by protein SUMOylation.
It is noteworthy that, in addition to SUMOylation, several post-translational modification systems including methylation, ubiquitination, and ubiquitin-like proteins (classified under the enriched GO term “protein tag”) also responded to seizure activity. In particular, Cullin-RING ubiquitin ligases (CUL3, CUL4A, and CUL4B) and NEDD8, a protein tag much like SUMO, as well as several methyltransferases that target histones (SETDB1, PRMT1, PRMT5, PRMT8, and RRP8) are also enriched in the nucleus after seizures (Figure 2D). We also noted that a specific cluster of histone 2A-associated proteins are enriched during seizures (e.g., HIST2H2AB, HIST1H2AB, HIST1H2AA, H2AFZ, and H2AFX). The increase in nuclear proteins for SUMOylation, NEDDylation, ubiquitination, and methylation may indicate that exposure to pathological levels of activity during seizures triggers a global increase in post-translational modifications in neuronal nuclei.
While our study focused mainly on nuclear proteins enriched by activity, we also analyzed proteins depleted from the nucleus during seizures. Calcium- and calmodulin-dependent protein kinase complex (GO:0005954) had one of the highest over-representation scores, with all four members of the CaMKII family (CaMKIIα, CaMKIIβ, CaMKIIγ, and CaMKIIδ) ranked in the top 10 most-depleted proteins (Table S1). Other enriched GO terms include Arp2/3 protein complex (GO:0005885), myosin II filament (GO:0034314), structural constituent of post-synapse actin (GO:0099186), and dendritic filopodia (Figure 2C). STRING analysis of these activity-depleted proteins shows Arp2/3 actin nucleation complex (ACTR3, ARP1A, ARPC2, and ARPC3) and members of the myosin (MYH6, MYH9, MYH10, MYH11, MYH14, and MYO5A) and kinesin (KIF5A, KIF5B, KIF5C, and KLC2) motor protein families (Figure S6; Table S1). Many of these cytoskeletal proteins are found cell-wide in different subcellular compartments including axons, dendrites, synapses, and importantly, in the nucleus. The broader implications of cytoskeletal proteins having dual functions in the cytoplasm and nucleus will be elaborated in the discussion section.
Validation of nuclear proteins altered by seizures
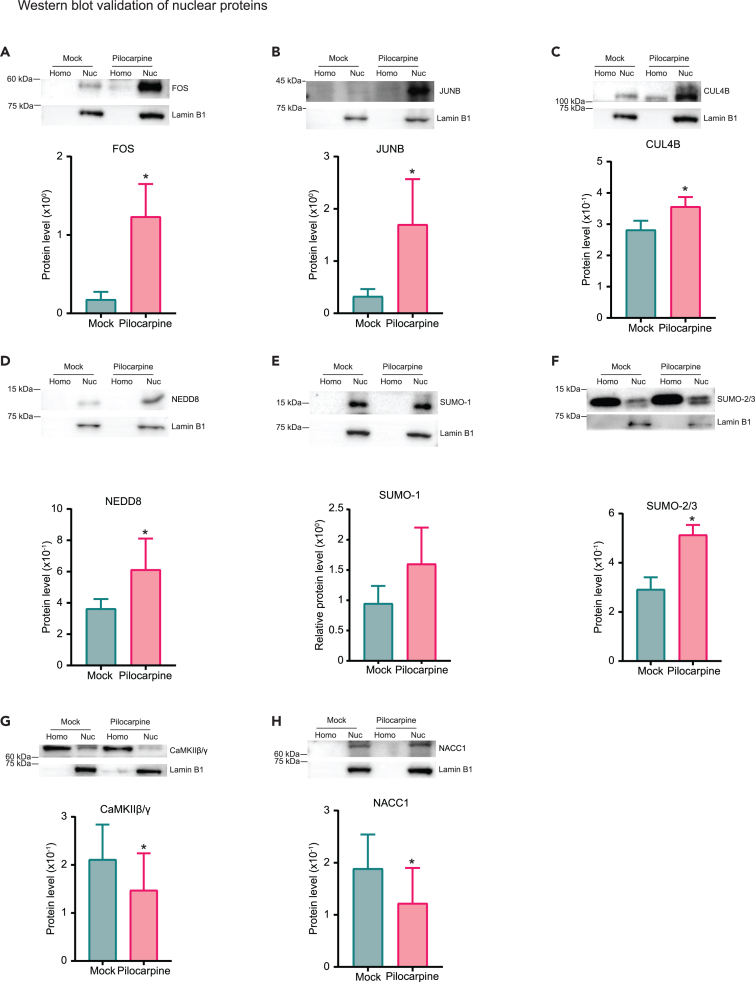
To validate the proteomic analysis, we performed western blots of different candidate proteins using an alternate nuclear extraction protocol from the mouse forebrain after mock or pilocarpine treatments. Unlike the INTACT-purified nuclei, the nuclear extracts for Western blots were prepared from wild-type mice using a sucrose cushion (Figure S7). This is to rule out the possibility that the overexpression of SUN1-sfGFP or the antibodies used for immuno-purification of the nuclei altered the sample composition. Overall, our Western blot data match most of the observed differences in the nuclear proteome. We found a strong increase in overall protein levels after seizures for two subunits of the AP-1 complex, FOS and JUNB (N = 6, p = 0.0313; Figures 3A and 3B). Similarly, NEDD8, CUL4B (N = 6, p = 0.0313), and SUMO-2/3 (N = 6, p = 0.0156) are all significantly increased in the nucleus upon seizures (Figures 3C, 3D, and 3F). However, SUMO-1 protein levels are below our threshold of enrichment (Figure 2B), and we included SUMO-1 in our validation studies to compare with SUMO2/3. In agreement with our LC-MS/MS results, SUMO-1 was not enriched in the nucleus after seizures (N = 6, p = 0.1234; Figure 3E). Finally, validation of seizure-depleted nuclear proteins NACC1 (N = 6, p = 0.0313) and CaMKIIβ/γ (N = 6, p = 0.0313) by Western blots also showed lower expression of both proteins from pilocarpine-induced animals (Figures 3G and 3H). Taken together, our Western blot results of selected nuclear proteins altered by seizures validate the proteomic data obtained through mass spectrometry sequencing.
Figure 3.
Western blot validation of selected nuclear-enriched and nuclear-depleted proteins in seizure-induced mice
(A–H) Western blots of nuclear proteins FOS, JUNB, CUL4B, NEDD8, SUMO-1, SUMO-2/3, CaMKIIβ/γ, and NACC1 in whole nuclear lysates from mock- and pilocarpine-injected animals. All proteins bands are reported as mean intensities after normalizing band intensity of interest against LaminB1. Homo, homogenate; Nuc, nuclear extract. N = 6 pairs of animals. All plots were analyzed with paired t-test or Wilcoxon test, and reported as means ± SEM, p < 0.05 (∗).
As the isolated forebrain nuclei for Western blots are heterogeneous, we examined protein changes at the single neuron level via IHC of brain sections and ICC of cultured hippocampal neurons. In pilocarpine-injected mice, FOS protein expression is elevated in forebrain sections including the hippocampal pyramidal layer as well as PIR. Both brain regions have been shown to be vulnerable to epileptogenic stimuli and damage from seizures (Figure S8).40,41 Interestingly, expression of nuclear FOS is region-specific at 100 min post-seizure induction, with CA1 hippocampal neurons recording the most robust changes (Figure S8). Like pilocarpine-induced seizures in animals, nuclear FOS expression is also increased in cultured hippocampal neurons in the presence of bicuculline (Bic), a GABAA receptor antagonist that induces action potential (AP) bursting in excitatory neurons (Figure S8). At 100 min post-injection, there is also an overall increase in JUNB expression in excitatory neurons in the hippocampus and PIR (Figure S8). The increase appears to be much more uniform across all brain regions quantified. Similar to FOS, JUNB expression in the nucleus is also significantly increased in hippocampal neurons stimulated with Bic (Figure S8).
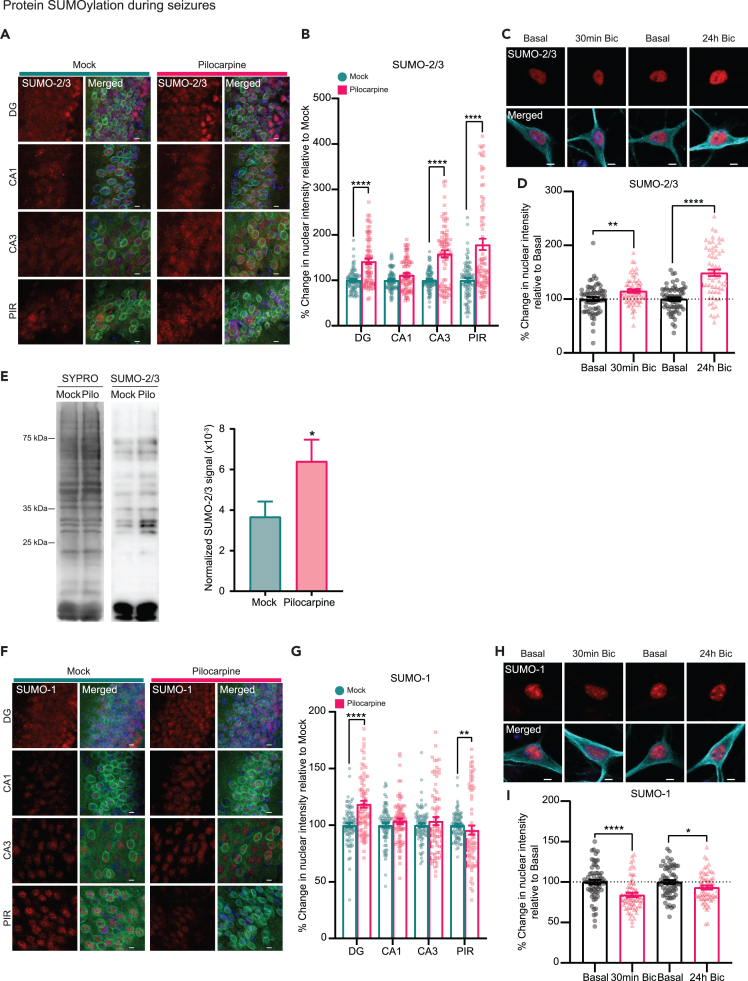
As SUMO2/3 levels are enhanced after seizures, we focused on verifying our observations in forebrain sections and cultured neurons. IHC of brain sections for mock- and pilocarpine-treated mice show a moderate elevation of nuclear SUMO-2/3 in different brain regions (DG, CA3, and PIR; Figures 4A, 4B, and S9). Similarly, in cultured hippocampal neurons, we observed an increase in nuclear SUMO-2/3 after exposure to Bic for either 30 min or 24 h. Notably, neurons treated with Bic for 24 h had on average a 1.5-fold increase in nuclear SUMO-2/3 as compared to basal levels (Figures 4C and 4D). The rise in SUMO-2/3 in the nucleus is also detected in the cytoplasm, though only after 24 h in the presence of Bic stimulation (Figure S10) and may contribute to hyper-SUMOylation of cytoplasmic and plasma membrane protein targets.42 Finally, to determine if the total SUMOylated protein level is elevated in animals that underwent seizures, we extracted nuclei from mock- or pilocarpine-treated mouse forebrains and performed western blots of the total nuclear extracts. Blots incubated with antibodies against SUMO-2/3 reveal an increase in SUMOylated proteins across different molecular weights (Figure 4E).
Figure 4.
Enhanced protein SUMOylation in neuronal nuclei during seizures and Bic-induced AP bursting in cultured hippocampal neurons
(A) Representative images of DG, CA1, and CA3 pyramidal layer neurons in the hippocampus, and piriform cortex (PIR) of mock- and pilocarpine-injected animals immunolabelled with anti-SUMO-2/3 antibodies. SUN1-sfGFP (green); Hoechst (blue); SUMO-2/3 (red). Scale bars, 10 μm.
(B) Quantification of SUMO-2/3 nuclear intensity in neurons shown in (A). Nuclear intensities for SUMO-2/3 were measured from SUN1-sfGFP-positive nuclei outlined by Hoechst and SUN1-GFP staining. All values for pilocarpine-treatments are normalized against mean nuclear intensity from mock-injected animals. N = 3 pairs of animals, n = 75 number of cells quantified across all sections.
(C) Representative images of hippocampal neurons (DIV14 to 15) stimulated with Bic (40 μM) to induce AP bursting for 30 min and 24 h. SUMO-2/3 (red); Hoechst (blue); MAP2 (cyan). Scale bars, 5 μm.
(D) Quantification of SUMO-2/3 staining in hippocampal neurons under basal condition or stimulated conditions with Bic. All experimental values were normalized against mean nuclear intensity of SUMO-2/3 at basal condition for each independent experiment. N = 3 independent experiments, n = 60 total number of cells quantified across experiments.
(E) Representative Western blot of total SUMOylated proteins by SUMO-2/3. Total lane intensities for SUMO-2/3 were normalized against total protein amount measured with SYPRO Ruby protein blot stain. N = 4 pairs of animals. Paired t-test with means ± SEM, p < 0.05 (∗).
(F) Representative images of DG, CA1, and CA3 pyramidal neurons of the hippocampus, and piriform cortex (PIR) of mock- and pilocarpine-injected animals immunolabeled with anti-SUMO-1 antibodies. SUN1-GFP (green); Hoechst (blue); SUMO-1 (red). Scale bars, 10 μm.
(G) Quantification of SUMO-1 nuclear intensities as described in (B). N = 3 pairs of animals, n = 75 number of cells quantified across all sections.
(H) Representative images of hippocampal neurons (DIV14 to 15) stimulated with Bic (40 μM) to induce AP bursting for 30 min and 24 h. SUMO-1 (red); Hoechst (blue); MAP2 (cyan). Scale bars, 5 μm.
(I) Quantification of SUMO-1 nuclear intensity in hippocampal neurons under basal or Bic stimulation as described in (D). N = 3 independent experiments, n = 60 total number of cells quantified across experiments. Unless otherwise stated, all statistics for IHC and ICC experiments were performed using Mann-Whitney test with bar graphs reporting means ± SEM, p < 0.05 (∗), p < 0.01 (∗∗), p < 0.0001 (∗∗∗∗).
To again compare with SUMO 2/3, we performed a similar IHC assay for SUMO-1. In contrast to SUMO-2/3, the majority of brain regions except for DG did not show an increase in SUMO-1 levels (Figures 4F and 4G). On the contrary, we noted a small decrease in the PIR of pilocarpine-treated animals (Figures 4F and 4G). Surprisingly, this decrease in SUMO-1 nuclear levels is also reflected in hippocampal neurons exposed to Bic for either 30 min or 24 h, with the former condition resulting in much lower SUMO-1 expression in the nucleus (Figures 4H and 4I). It is unclear why SUMO-1 levels decrease with activity. In the nuclear proteome, unlike SUMO-2/3, which is highly enriched in the nucleus during seizures, SUMO-1 levels are only mildly elevated. The implications of this observation will be further explored in the discussion section. In short, we confirm that SUMO-2/3 levels in neuronal nuclei are consistently elevated during seizures.
Profiling the nuclear transcriptome induced by seizures
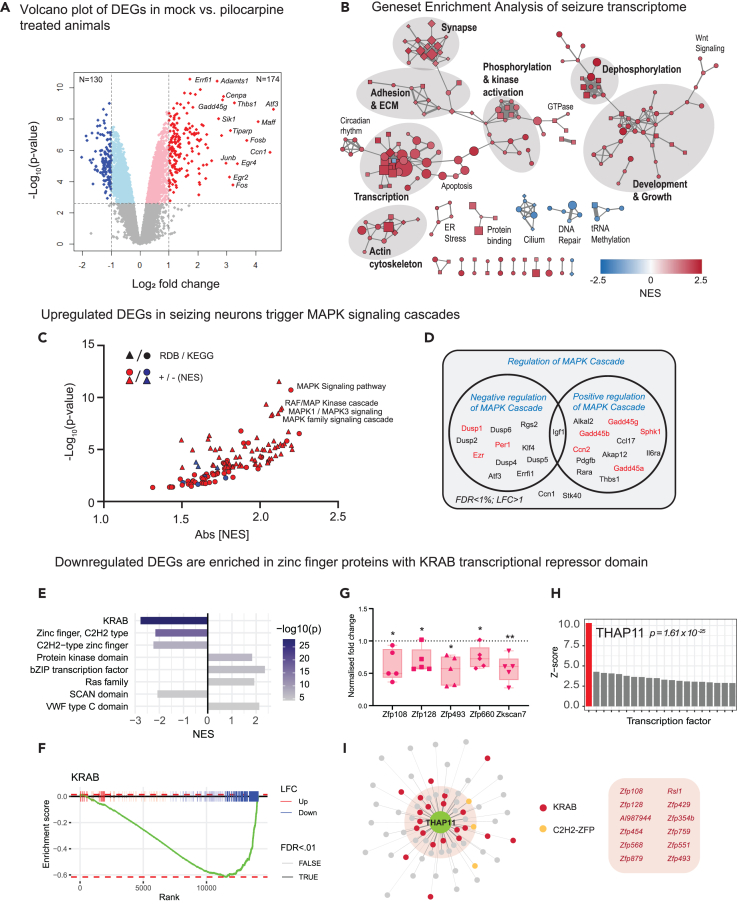
We next performed nuclear RNA-seq to analyze changes in gene expression and to compare with the nuclear proteome to look for common pathways altered by seizures. We also leveraged our experimental model to look at the pre-mRNA that is enriched in the nuclei—an RNA population that has not been rigorously examined in other seizure-induced transcriptomes. We extracted poly(A)-enriched RNA for sequencing from mock- and pilocarpine-injected animals using the same methodology outlined in Figure 1G. A total of 14,115 transcripts were detected, of which 89.08% were identified as protein coding (Figure S11). In total, N = 3130 protein-coding genes were differentially expressed during seizures (FDR <1%), with N = 174 strongly upregulated (LFC >1) and N = 130 strongly downregulated (LFC < −1; Figure 5A; Table S2). Induced genes include known activity-regulated primary response genes (with both rapid and delayed kinetics) and some secondary response genes (Figure S12).43 To examine functional enrichment of biological pathways, we performed gene set enrichment analyses (GSEA) of GO terms on expressed genes ranked by LFC (Table S3). The majority of enriched GO terms were upregulated and are associated with major cellular processes (i.e., transcription, growth/development, and kinase/phosphatase activities) or subcellular structures (i.e., actin cytoskeleton, synapse, and extracellular matrix). Smaller groups of GO terms also implicated alterations in circadian rhythms, Wnt signaling, GTPase activity, apoptosis, and ER stress during seizures (Figure 5B). Notably, some of the most strongly enriched GO terms in the network are associated with dephosphorylation of MAPK signaling cascades, including “negative regulation of ERK1 and ERK2 cascade” (N = 59, padj = 2.10e-6, NES = 2.42) and “inactivation of MAPK activity” (N = 16, padj = 5.77e-6, NES = 2.38; Figure 5B). GSEA analyses using both KEGG and reactome pathway databases also strongly implicated MAPK signaling pathways, with top terms including “KEGG-MAPK signaling pathways” (N = 216, NES = 2.19, padj = 3.54e-9), “RDB-RAF/MAP Kinase cascade” (N = 214, NES = 2.14, padj = 4.21e-7), “RDB-MAPK1/MAPK3 signaling” (N = 217, NES = 2.13, padj = 4.62e-7), and “RDB-MAPK family signaling cascade” (N = 238, NES = 2.11, padj = 8.54e-7; Figure 5C). A closer examination of strongly upregulated MAPK-regulating DEGs (FDR <0.01, LFC >1) uncovered multiple members of the dual-specificity phosphatases (Dusp1,2, 4–6) and growth arrest and DNA damage (Gadd45a, b, g) gene families robustly transcribed during seizures (Figure 5D).5,44,45,46 While transcripts belonging to the MAPK family are predominantly upregulated, we found equal numbers of genes classified as either positive or negative regulators of the MAPK cascade, indicating that seizures likely induce bidirectional regulation of MAPK pathways to accommodate cellular needs. Such is the case for MAPK stress response genes which include both positive (i.e., Gadd45g, Sphk1, and Ccn2) and negative (i.e., Dusp1, Ezr, and Per1) regulators of the pathway (labeled in red, Figure 5D). Our list of upregulated DEGs along with enrichment of MAPK-associated pathways during seizures is comparable to other studies, but our cell type-specific transcriptome profile suggests that many of these changes likely originate from transcription in excitatory neurons.5,44
Figure 5.
Altered gene expression in excitatory neuronal nuclei during seizures
(A) Volcano plot showing differential expression of protein-coding genes in mock-vs. pilocarpine-treated animals at 100 min. Thresholds for differential expression are indicated at false discovery rate (FDR) < 1% and log2 fold change (LFC) > ±1, at which N = 130 genes are downregulated and N = 174 are upregulated.
(B) Gene set enrichment analysis (GSEA) of the seizure-induced transcriptome (p < 0.05). Each node corresponds to an enriched GO term (circle: Biological Process (BP), square: Molecular Function (MF), diamond: Cellular Component (CC)) with node color representing nucleotide enrichment score (NES) and larger nodes having lower p values. Edges indicate gene set overlap, with thicker edges representing greater overlap. Clusters of related nodes were labeled manually. Disconnected nodes are not shown.
(C) GSEA of ReactomeDB (RDB) and KEGG pathways.
(D) DEGs involved in the regulation of MAPK cascade according to GO annotations. Genes in red regulate stress-activated MAPK signaling.
(E) GSEA of PFAM protein families.
(F) GSEA enrichment plot showing KRAB box genes ranked by LFC in the seizure vs. mock transcriptome (padj <0.01). Up and downregulated genes and DEGs are indicated. Enrichment score is plotted for each point in the ranked list, showing enriched downregulation of KRAB genes.
(G) qPCR validation of zinc-finger protein genes Zfp108, Zfp128, Zfp493, Zfp660, and Zkscan7 (N = 5 pairs of mock v. pilocarpine mice). Relative gene expression levels determined by normalizing against average ΔCt values for all mock-injected animals. All statistics were performed using one-sample t test with plots showing means ± SEM, p < 0.05 (∗), p < 0.01 (∗∗).
(H) Analysis of enriched transcription factor binding sites among downregulated DEGs using Pscan. Z-scores indicating target site enrichment for top 20 transcription factors.
(I) THAP11 target genes with maximum binding scores above background average. Top targets containing KRAB domains are listed.
While the majority of differentially expressed gene sets are upregulated during seizures, the GSEA GO analysis also revealed several downregulated processes, chief among them being cilium assembly, tRNA methylation, and DNA repair (Figure 5B). While DNA repair and tRNA methylation are nuclear-associated processes, recent studies also indicate that nuclear proteins such as nucleoporins are assembled at the cilium and that disruption of ciliary structure or assembly in neurons is linked to epilepsy.47,48,49,50 However, the most striking feature for downregulated DEGs came from GSEA analysis of protein domains with PFAM where we observed a dramatic downregulation of proteins harboring the Krüppel-associated box (KRAB) domain (Figures 5E and 5F). Out of a total of N = 254 unique KRAB domain-containing genes (PF01352) present in our dataset, approximately 83% (N = 210/254) of those genes show negative LFCs. Of these, we identified N = 105 as downregulated DEGs (FDR <0.01) as compared to N = 9 that were upregulated. Only two KRAB domain genes were strongly upregulated (LFC >1): zfp189 and zfp955a, with the former identified as a CREB target gene that plays a role in regulating transcriptional networks for resilience in the prefrontal cortex51. The KRAB domain is a zinc-finger motif that is found in approximately 1/3 of all C2H2 zinc-finger proteins and is known to function as a transcriptional repressor domain.52 We validated the downregulation of zinc-finger protein gene expression by performing quantitative PCR on a subset of zinc-finger protein DEGs identified in our profile from forebrain tissues of WT and seizure animals (Figure 5G) As controls, we also measured the upregulation of Fos and Junb expression (Figure S13). We then corroborated our observations by analyzing a previously published kainic acid-induced seizure RNA-seq dataset generated by Fernandez-Albert and colleagues (2019) and found that transcripts coding for KRAB domains are downregulated at 1 h post-induction but not at later time points (Figure S15). This downregulation can also be observed in ATAC-seq datasets, suggesting that these changes are linked to chromatin accessibility.53 Surprisingly, the suppression of KRAB domain genes is not confined to neurons undergoing chronic patterns of activity during seizures as this downregulation can also be detected in FOS-positive neurons following exposure to a novel environment.53,54
We next asked if transcriptional suppression during seizures is controlled by specific transcription factors by employing Pscan to hunt for over-represented transcription factor-binding site motifs among strongly downregulated DEGs (Table S6; FDR <0.01; LFC < −1).55 Binding sites for THAP11 (Ronin), a zinc-dependent transcriptional repressor, were by far the most enriched (Figure 5H). Strikingly, the THAP11 DNA-binding motif is found in KRAB domain genes, which represent 12 out of the top 16 genes with the strongest THAP11-binding scores in our dataset (Figure 5I). The consensus THAP11 DNA-binding site consists of a bipartite motif that is enriched within −250 to +50 from the transcription start site in our dataset (Figure S14). As expected, high-resolution confocal microscope localized THAP11 primarily in the nucleus (Figure S17). However, a brief exposure to Bic did not alter the nuclear-cytoplasmic ratio nor total protein concentration of THAP11 in cultured hippocampal neurons, suggesting that THAP11 activity may be regulated by post-translation modifications including protein SUMOylation (Figure S17).
Overall, we found that seizures robustly enhance and suppress the expression of different subsets of genes in excitatory neurons. While upregulated genes implicate MAPK signaling and transcriptional regulation, downregulated genes are enriched for zinc-finger proteins containing KRAB domains.
Exon-intron split analysis of seizure-induced nuclear transcriptome
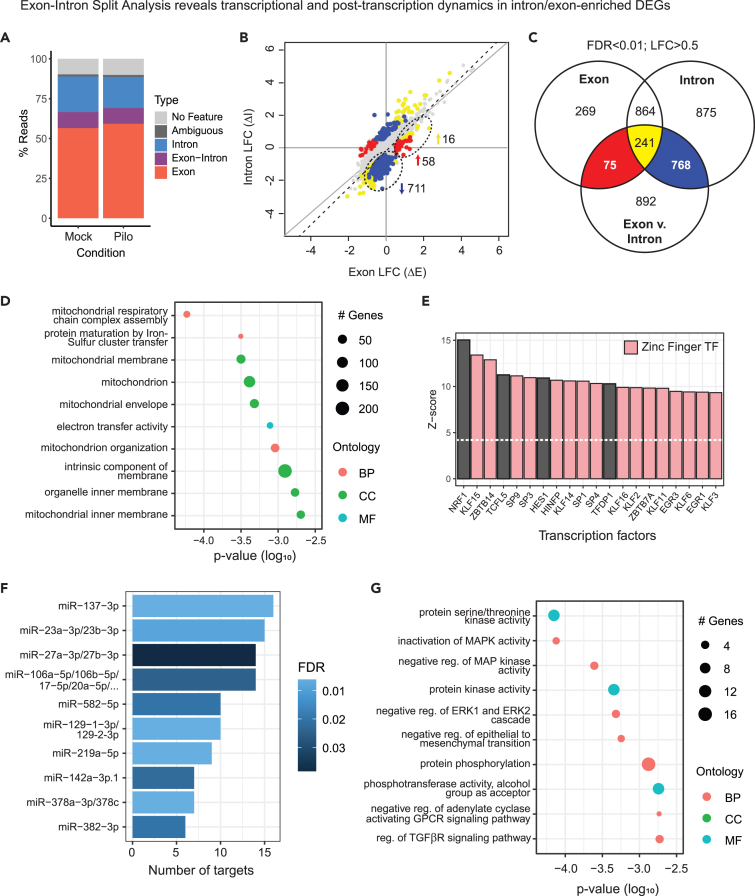
We noted that a large proportion of reads in our RNA-seq dataset were aligned to introns (∼20%; Figure 6A), a feature of nuclear RNA which contains a sizable fraction of pre-mRNAs. The vast majority (>95%) of expressed protein-coding genes had detectable intron signal (log2CPM >1) despite poly(A)-positive selection, enabling comparison of exon and intron expression profiles between conditions.56,57,58 Taking advantage of the cell type and compartment specificity of our dataset, we compared changes in gene expression during pilocarpine seizures at the exon and intron levels to shed light on transcriptional dynamics and post-transcriptional regulation.
Figure 6.
Differential gene expression at intron and exon levels reveals transcriptional dynamics and putative post-transcriptional regulation
(A) Percentage of reads uniquely aligned to exons, introns, intergenic regions, and ribosomal RNA (rRNA), aligned ambiguously or unaligned.
(B) Log fold changes (LFCs) in gene expression between mock and seizure in introns (ΔI) versus exons (ΔE) for protein-coding genes with >1 log2CPM at both levels (N = 11,389). Colored subsets of genes have significantly different ΔI vs. ΔE and are differentially expressed between mock and seizure in exons (red), introns (blue), or both (yellow). Subsets of interest are circled. Linear regression is indicated by the dashed line.
(C) Venn diagram of genes differentially expressed between mock and seizure in exons and introns, and genes with significantly different ΔI vs. ΔE, corresponding to subsets indicated on LFC plot.
(D) Top 10 terms in GO analysis of intron-specific downregulated DEGs showing number of significant genes in enriched terms and log10 p value (pink: Biological Process (BP), cyan: Molecular Function (MF), green: Cellular Component (CC)).
(E) Enriched transcription factor-binding sites for intron-specific downregulated DEGs using Pscan. Z-scores indicate target site enrichment for top 20 transcription factors (TFs), of which 16 are zinc-finger TFs.
(F) Enrichment of miRNA target sites in TargetScan database using MIENTURNET for DEGs with significantly greater ΔE than ΔI. Number of miRNA target genes is plotted against false discovery rate (FDR) for top 10 miRNAs by FDR.
(G) Top 10 terms in GO analysis of DEGs with significantly greater ΔE than ΔI (pink: BP, cyan: MF, green: CC).
Overall, fold changes in gene expression between mock and seizure conditions are similar at the exon and intron levels, with a correlation of r = 0.811 (N = 11,389) and slightly greater fold changes at the intron level on average (ΔI = 1116 ∗ ΔE - 0.034; Figure 6B). However, a parallel analysis of intronic and exonic reads in our dataset identified many additional DEGs at the intron level (N = 2748 compared to N = 1449 at the exon level, overlap N = 1105; FDR <0.01, LFC > ±0.5; Figures 6B and 6C; Table S4). As most introns are spliced during or shortly after transcription whereas mature mRNA may remain in the nucleus for some time,59 differential expression at the intron level can reflect different states of active transcription at the time of extraction.
We further probed differences in gene expression at the intronic and exonic levels using exon-intron split analysis58 and found N = 1,976 genes that had significantly different fold changes between conditions in introns vs. exons (FDR <0.01, LFC > ±0.5; Figures 6B and 6C). Of these, N = 768 were DE exclusively in introns and amazingly, over 90% of intron-specific DEGs were downregulated in seizure (N = 711). Transcriptional suppression may become evident at the intron level prior to significant changes in exon-level expression in nuclear poly(A)-positive RNA due to a more rapid depletion of the transient pool of partially spliced pre-mRNA compared to the exon-only mature mRNA population. Thus, we hypothesize that the intron-specific DEGs represent transcription occurring later or with slower kinetics relative to genes DE at both exon and intron levels. GO analysis indicates that many of these putatively delayed transcripts are involved in mitochondrial and respiratory functions (Figure 6D). A Pscan analysis of intron-specific downregulated DEGs showed enrichment of transcription factor-binding sites for NRF1, as well as KLF (Krüppel-Like Factors), SP, and ZBTB families of zinc-finger transcription factors, while THAP11 transcription factor-binding sites (TFBSs) were also weakly but significantly enriched (Figure 6E).
We also identified a smaller subpopulation of upregulated genes with substantially greater fold changes in exons than introns (N = 74), of which N = 58 were exclusively differentially expressed at the exon level. Previous research has shown that this expression pattern is indicative of post-transcriptional regulation.58 Indeed, we found that genes in this group were enriched for target sites of miRNAs previously implicated in epilepsy, including miR-137, miR-23a/b and miR-129 (Figure 6F). Intriguingly, exon-upregulated DEGs were also enriched for kinase activity and negative regulation of MAPK signaling (Figure 6G), suggesting that post-transcriptional processes such as miRNA interactions may contribute to the regulation of MAPK pathways during seizures.
Overall, our bulk sequencing of nuclear mRNA enabled us to examine different mature and immature RNA populations and identify pools of transcripts that are differentially altered at the exon and intron levels, indicating differences in transcriptional or post-transcriptional regulation in excitatory neurons.
Nuclear proteome and transcriptome co-expression analysis
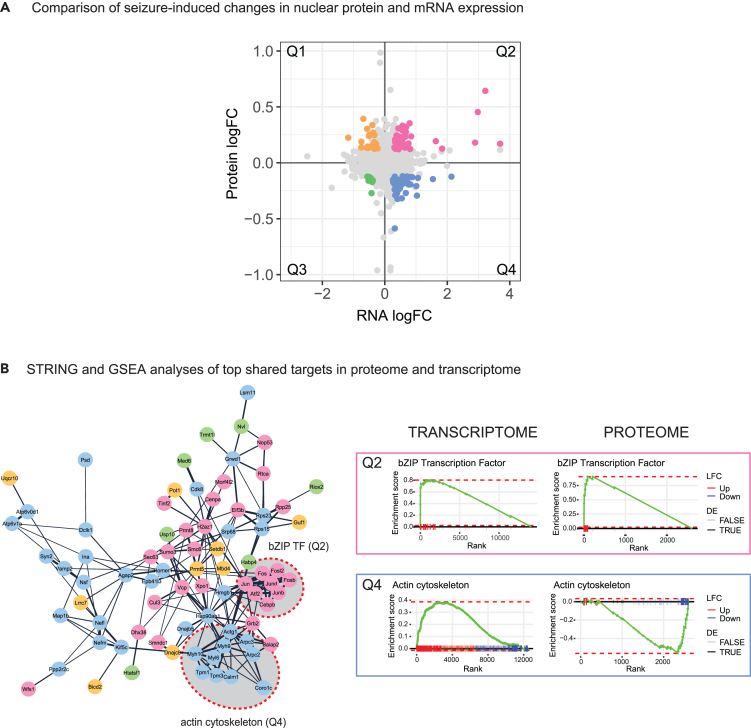
Finally, we compared the nuclear proteome and transcriptome to understand the relationship between seizure-driven changes in the nuclear proteome and altered gene expression. Subunits of the AP-1 complex and its bZIP interacting partners showed the strongest association between datasets, being significantly upregulated in both the nuclear proteome and transcriptome. These genes are known to undergo activity-dependent transcription shortly after onset of seizures and are then rapidly translated in the cytoplasm before being imported into the nucleus (Figures 7A and 7B; Table S5). In addition, we found that several proteins associated with UBL conjugation and the DNA damage response (DDR) are upregulated in both the proteome and transcriptome (Figure 7B, Q2). We also identified subsets of seizure-affected proteins with discordant regulation in the transcriptome, including a group of proteins that is depleted from the nucleus while showing an upregulation in mRNA expression. This group of proteins is associated with actin (Arpc2, Arpc3, Tpm1, Actg1, and Coro1c) and myosin (Myl6, Myh9, and Myh10). There is strong experimental evidence that cytoskeletal proteins function in the nucleus.60,61 For example, Arpc2 and Arpc3 are major components of the Arp2/3 actin complex that can modulate transcription and modify chromatin (Figures 7A and 7B, Q4),62 while several myosin isoforms including Myh9 and Myl6 are detected in the nucleus and regulate gene expression.63,64 A possible explanation for this discordant protein-RNA relationship is that loss of actin-cytoskeletal proteins triggers an increase in gene expression to replenish the depleted pool of proteins. Finally, we identified a small subset of nuclear-enriched proteins that are downregulated at the mRNA level and linked to DNA repair and methylation (Pot1, Prmt5, Mdb4, and Setdb1; Figures 5B and 7A, Q1), an observation that applies more broadly across the datasets as GSEA plots confirm that, overall, proteins associated with DNA repair and methylation are enriched in neuronal nuclei while corresponding gene expression is downregulated (Figure S18). While the biological relevance of this protein (Up)-mRNA (Down) coordination is unclear, changes in protein stability may account for this observation.
Figure 7.
Comparison between nuclear protein and mRNA levels during seizures
(A) Plot comparing log2 fold changes (LFCs) in the nuclear transcriptome and proteome (N = 2509). Genes with FDR <1% in the transcriptome and LFC in the top 15% of the proteome are highlighted and colored according to quadrant. Quadrants are labeled Q1-4.
(B) STRING protein interaction network of DE genes/proteins with annotated interactions (confidence ≥0.4). Node color corresponds to quadrant. Subsets of highly connected nodes in Q2 (bZIP transcription factors) and Q4 (actin cytoskeleton) are circled and adjacent GSEA plots for the corresponding gene sets in the transcriptome and proteome are presented.
Discussion
SUN1-sfGFP mouse can be used to detect activity-induced changes in the nucleus
We have demonstrated that the CaMKIIα-Cre:SUN1-sfGFP mouse can be used to identify cell-type-specific changes in the nuclear proteome following seizures and potentially other activity-induced changes in the brain. A previous report using this mouse model profiled a kainic acid-induced seizure transcriptome but did not isolate the nuclear proteome.53 Functional classification of the enriched proteins revealed several pathways altered by seizures. Two groups stood out: AP-1 transcription factor complex and post-translational protein modification systems. AP-1 is a dimeric transcription factor complex that consists of different FOS and JUN subunits. These subunits belong to the family of bZIP transcription factors, many of which are IEGs that accumulate in the nucleus upon neuronal activity.65,66,67 The activity-dependent upregulation of FOS mRNA and protein, and to a lesser extent JUNB, during different behavioral responses as well as during seizures have been reported.54,68 However, the AP-1 complex subunits are rarely detected in most proteomic studies.16,18,20,21,69,70,71 Our approach for sequencing the nuclear proteome and mRNA detected enrichment of all core members of the AP-1 complex during seizures, including ΔFOSB which is a splice isoform of FOSB, and closely related transcription factors CEBPB and ATF2. The enrichment in both datasets also identifies AP-1 complex subunits as the only family of transcription factors that is strongly upregulated in the nucleus at both the protein and mRNA levels during the initial period of seizure activity. It is also noteworthy that IHC showed clear differences in patterns of FOS and JUNB expression and even pCREB activation across brain regions during seizures, which may indicate that individual subunits respond to neuronal activity differently, thereby generating distinct transcriptional outputs.
Enhanced nuclear SUMOylation may indicate activation of DNA damage response pathways
The enrichment of proteins that regulate post-translational modification systems, along with upregulation of a subset of stress-responsive genes in the MAPK signaling cascade, suggests that seizures likely cause substantial DNA damage that triggers the host repair mechanisms. In particular, the role of SUMOylation and ubiquitination in DNA damage response (DDR) is well-documented. In DDR, SUMOylated proteins are reported to accumulate at double-strand break sites where they are ubiquitinated by a specific class of SUMO-targeted ubiquitin ligases for proteasomal degradation, a step that is crucial for the repair process and one that links SUMOylation with proteasomal degradation.72,73 Other examples include activation of the recombinational DNA repair pathway mediated by SMC-5/6 complex through SUMOylation. The E3 SUMO ligase NSMCE2 (also known as MMS21 or NSE2) is part of the SMC-5/6 complex and its SUMOylation activity is enhanced by DNA binding.74,75,76 In addition, SMC-5/6-NSMCE2 is also known to protect telomeres through SUMOylation of the Shelterin complex, of which 2 members of the complex, POT1 and TINF2 are also found highly enriched in our nuclear proteome.77 The role of protein SUMOylation in DNA repair also extends to other nuclear assemblies such as PML nuclear bodies which are assembled in response to external stressors, as is the case during seizures.78,79,80,81 As with SMC-5/6-NSMCE2, several core components of the PML nuclear bodies are highly enriched in the nucleus upon seizure. Interestingly, apart from Sumo3, nuclear-enriched proteins in the SUMOylation pathway are not upregulated at the RNA level, suggesting that accumulation is not due to increased transcription but may instead be regulated by protein turnover, nucleocytoplasmic shuttling, or protein synthesis from a pre-existing pool of transcripts. While other studies have shown that brain traumas including hypothermia, brain ischemia, cardiac ischemia, and reperfusion injury can result in massive increases in SUMO and SUMO-conjugated proteins,82,83,84,85,86 we believe our study is the first to show global changes in nuclear protein SUMOylation following seizures in rodent models. We also note that while SUMO-2/3 nuclear levels are consistently enhanced during seizures across all our assays, SUMO-1 nuclear levels remain largely unaltered with Western blots and immunohistochemistry of different brain regions showing no increase in nuclear levels. Intriguingly, in the PIR as well as in neuronal cultures, increase in neural activity resulted in a drop of SUMO-1 levels. As the PIR is a critical node for epileptiform activity,87,88 we speculate whether a partial redundancy in protein SUMOylation between the two isoforms may explain the brain region-specific rise of SUMO-2/3 and concurrent decline of SUMO-1 in the nucleus.89 Further work must be done to uncover the complex relationship between SUMO-1 and SUMO-2/3 during seizures and epileptogenesis.
Beyond SUMOylation, other UBLs enriched in our dataset also participate in DDR. NEDD8 is a substrate for NEDDylation, a post-translational covalent modification similar to SUMOylation that enhances Cullin-RING E3 ubiquitin ligase function90—another family of proteins that is found enriched in our dataset. The localized activity of the E3 ubiquitin ligases CUL3, CUL4A, and CUL4B at the site of DNA damage is known to promote double-strand break repair either through ubiquitination of local substrate proteins or histones.91,92 In alignment with the enrichment of post-translational mechanisms in the nucleus that are associated with DDR, transcriptome profiles also indicate an upregulation of multiple transcripts linked to cellular stress responses including Dusp1 and Gadd45a, -b, and -g. The expression of these genes, which form part of a larger MAPK signaling network, is partially responsible for switching on cellular survival responses, including DDR, when exposed to harmful stimuli.93,94,95
Activation of SUMOylation can suppress transcriptional outputs
While post-translational systems are integral for DNA repair, their presence can also alter transcription through modifications of transcription factors, histones, and DNA. In the case of seizures, evidence for transcriptional suppression can be found in both our sequencing datasets. Our RNA-seq analysis shows many more genes significantly downregulated in seizures when pre-mRNA is included in the assessment. Among the downregulated genes, we find evidence for a broad activation of zinc-finger transcriptional repressors along with a dramatic reduction in KRAB-ZFP gene expression. Furthermore, GSEA analyses of both genes and proteins indicate an enrichment of transcriptional repressors (Figure S19). Coincidentally, it is also widely reported in the literature that the vast majority of SUMOylated transcription factors reduce or repress transcription.96 SUMOylation represses transcription by a variety of methods, including enhancing transcription factor recruitment of HDACs where the deacetylation of histones can potentially reduce transcriptional activation at selected genomic loci during seizure.23,25,97,98 Like HDACs, SUMOylated transcription factor is also known to preferentially recruit repressive cofactors99 or to prevent other PTMs such as phosphorylation or acetylation from activating transcription.100,101 Indeed, we note that the transcriptional repressor THAP11 contains a highly conserved protein SUMOylation site in the coding region (Figure S17). Collectively, a rise in nuclear SUMOylation in response to prolonged neural activity might signify a cellular defense mechanism to halt runaway activity-dependent transcription.
Downregulation of zinc-finger protein gene expression linked to Zn2+ influx during seizures
Among the downregulated DEGs, THAP11-binding site is greatly over-represented in the upstream promoter and is likely a master transcriptional suppressor for KRAB-ZFPs during seizures. That THAP11 itself is also a zinc-finger transcriptional repressor is not surprising since there is evidence that zinc-finger transcription factors regulate the expression of many other zinc-finger genes.102 Furthermore, the involvement of Zn2+ in coordinating the activities of transcription factors that drive the downregulation of intron-specific DEGs is striking. This is true even for NRF1 activity which is modulated by Zn2+ and forms a complex with zinc-finger proteins to regulate gene expression required for mitochondrial biogenesis.103,104,105 We postulate that a massive influx of intracellular Zn2+—an excitotoxic event that is known to occur during epileptic seizures or traumatic brain injury—is the reason for the surge in zinc-finger-mediated repression.106,107,108,109 That said, the downregulation of KRAB-domain genes in neurons is also detected in rodents exposed to a novel environment and though the magnitude of the response may differ from seizures, it indicates that this activity-dependent phenomenon might not be limited to only pathological levels of neuronal activity.
Nuclear transcriptome enables intron-exon analysis of transcription dynamics during seizures
By isolating nuclear RNA from excitatory neurons, we can interpret differential changes in intron and exon expression levels and provide additional details of gene expression regulation and transcriptional dynamics during seizures. A major finding using this approach is the discovery of enhanced gene silencing at the intron level which we attribute to subsequent waves of transcriptional repression, supported by strong enrichment of specific transcription factor-binding sites. We have also ruled out the possibility that loss of intron signal is due to activity-dependent excision of stably retained introns.57 By examining coverage statistics and individual coverage profiles110 in combination with alternative isoform annotations,111 we found that intron signal was altered throughout the gene body for most DEGs with very little evidence for differential IR in our dataset (data not shown). Nevertheless, we cannot rule out that transcriptional suppression might be obscured at the exon level, such as by changes in mRNA stability or nuclear retention.
Using EISA, we also identified enhanced upregulation of genes involved in kinase activity at the exon level and enrichment of binding sites for miRNAs previously reported to have altered expression in epilepsy.112,113,114,115 For example, miR-137 is reduced in human temporal lobe epilepsy and mouse chronic epilepsy models, and suppression of miR-137 in mouse hippocampus was found to increase seizure frequency and severity as well as neuronal excitability.115 A reduction of microRNA levels would result in elevation of their target genes specifically at the exon level and in our study, these genes include negative regulators of MAPK signaling Dusp1, Dusp4, and Spred2, suggesting that post-transcriptional mechanisms might contribute to regulation of MAPK signaling during seizures.
Coordination between protein and mRNA expression during seizures
We profiled neuronal nuclei at 100 min following seizure induction, a time point at which both protein and mRNA levels of canonical activity-induced proteins such as the AP-1 complex subunits are elevated. In addition to the AP-1 complex, we observed several functional clusters of proteins and mRNAs with coordinated activity-dependent responses. The clusters of interest include actin-associated proteins (proteins DOWN; mRNA UP), proteins linked to UBL conjugation and DDR (protein UP; mRNA UP), and DNA repair and methylation (protein UP; mRNA DOWN). Overall, the correlation between protein and mRNA levels in our data is not significant. The weak relationship between protein and RNA expression has been described across different biological systems and is usually attributed to post-transcriptional processes that impact steady-state protein levels (e.g., protein turnover and RNA degradation),116,117,118,119 as well as the dynamics of transcription and translation. In the future, priority should be given to investigations of PTMs affecting various nuclear processes during epilepsy, particularly protein SUMOylation.
Downregulated cytoskeletal proteins have dual nucleus-cytoplasm localization and function
In our analysis of proteins depleted in the nucleus after seizures, we isolated multiple cytoskeletal elements, molecular motors, and scaffolding proteins. While we verify that SUN1-sfGFP expression is limited to the nucleus, we cannot rule out the possibility that other subcellular compartments such as the synapse, cytoskeletal, and scaffolding components located in the perinuclear space that associates with SUN1-KASH complex will co-purify with the nucleus during the INTACT protocol. Certainly, we have identified multiple nuclear proteins that are strongly depleted in the nucleus including NACC1 and members of the CaMKII family of proteins, many of which have splice isoforms encoding nuclear localization sequences.120 Furthermore, an increasing number of cytoskeletal and scaffolding proteins such as actin (e.g., ACTB and ACTG), actin-associated proteins (e.g., ARPC2, ARPC3, and ACTR3), myosin (e.g., MYL6 and MYL9), nuclear/synapse scaffolds (ADD1, CORO1, etc.) are shown to possess dual functions in the cytoplasm and in the nucleus.61,62,63,64,121,122,123,124 These intriguing findings on activity-dependent changes in nuclear cytoskeletal elements certainly warrant additional investigation.
Overall, our study has uncovered novel changes in both proteome and transcriptome of forebrain excitatory neuronal nuclei during seizures. We have also demonstrated the robustness of this mouse model and nuclear isolation techniques that can be used for multiple -omics platforms to look at changes in specific cell types including astrocytes and other inhibitory neurons using various Cre-inducible mouse lines.
Limitations of the study
While our nuclear proteome detected enrichment of nuclear proteins, we are unable to quantify how much of the nuclear protein content was lost during purification or if there is any movement of proteins through the nuclear membrane via nuclear pore complexes. While we detect an overall enhancement of SUMO-2/3-modified protein signal from Western blots, our studies have yet to identify specific targets of SUMO-2/3-modified proteins in the nucleus which will require additional experiments using more sensitive techniques.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| pCREB | Cell Signaling Technology | #9198 |
| GRP78/BiP | Gifted from Dr. Y. Lian (NTU) | Not applicable. |
| GAPDH | Proteintech | Cat#60004-1-Ig |
| H3 | Cell Signaling Technology | Cat#9715 |
| LaminB1 | Abcam | Cat#16048 |
| FOS | Cell Signaling Technology | Cat#2250 |
| GFP | Thermo Fisher Scientific | Cat#11122 |
| GFP | NeuroMab | Cat#75-131 |
| JUNB | Santa Cruz Biotechnology | Cat#8051 |
| CUL4B | Sigma-Aldrich | Cat#011880 |
| NEDD8 | St John’s Laboratory | Cat#115482 |
| SUMO1 | Abcam | Cat#32058 |
| SUMO2/3 | Proteintech | Cat#11251-1-AP |
| SUMO-2/3 | Cell Signaling Technology | Cat#4971 |
| CaMKIIβ/γ | St John’s Laboratory | Cat#91990 |
| NACC1 | Santa Cruz Biotechnology | Cat#376216 |
| GAD67 | Merk Millipore | Cat##5406 |
| THAP11 | Proteintech | Cat#23030-1-AP |
| MAP2 | PhosphoSolutions | Cat#1100 |
| Alexa Fluor® 488 goat anti-mouse IgG (H + L) | Invitrogen | Cat#11001 |
| Alexa Fluor® 555 goat anti-mouse IgG (H + L) | Invitrogen | Cat#21422 |
| Alexa Fluor® 488 goat anti-rabbit IgG (H + L) | Invitrogen | Cat#11034 |
| Alexa Fluor® 555 goat anti-rabbit IgG (H + L) | Invitrogen | Cat#21428 |
| Alexa Fluor ® 647 goat anti-chicken IgY (H + L) | Invitrogen | Cat#21449 |
| Goat anti-mouse light chain specific horseradish peroxidase (HRP)-conjugated |
EMD Millipore | Cat#AP200P |
| Goat anti-rabbit IgG (H + L) HRP-conjugated | Bethyl Laboratories | Cat#A120-201P |
| VeriBlot for IP detection reagent (HRP) | Abcam | Cat#131366 |
| Chemicals, peptides, and recombinant proteins | ||
| HBSS | Gibco | Cat#14170112 |
| Trypsin | Life Technologies | Cat#15090046 |
| DNAse | Sigma Aldrich | Cat#D5025 |
| Trypsin Inhibitor | Sigma Aldrich | Cat#T9128 |
| Neurobasal®-A medium | Gibco | Cat#10888022 |
| Gentamycin | Gibco | Cat#15750045 |
| FBS | Thermo Fisher Scientific | Cat#26140079 |
| B27 | Gibco | Cat#17504044 |
| GlutaMAXTM | Gibco | Cat#35050061 |
| Poly-D-Lysine | Sigma Aldrich | Cat#P7886 |
| 1(S),9(R)-(−)-Bicuculline methiodide | Sigma Aldrich | Cat#14343 |
| TTX | Tocris | Cat#1078 |
| Scopolamine methyl nitrate | Sigma Aldrich | Cat#S2250 |
| Pilocarpine hydrochloride | Sigma Aldrich | Cat#P6503 |
| IGEPAL®-CA 630 | Sigma Aldrich | Cat#I8896 |
| Dynabeads™ Protein G | Invitrogen | Cat#10-004-D |
| SYPRO® Ruby | Molecular Probes | Cat#S11791 |
| TRIzol™ Reagent | Invitrogen | Cat#15-596-018 |
| PowerUpTM SYBRTM Green | Applied Biosystems | Cat#A25741 |
| Critical commercial assays | ||
| Pierce BCA Protein Assay Kit | Thermo Fisher Scientific | Cat#23227 |
| PicoPure RNA isolation kit | Thermo Fisher Scientific | Cat#KIT0204 |
| RevertAid First strand cDNA synthesis kit | Thermo Fisher Scientific | Cat#K1622 |
| Deposited data | ||
| Mass Spectometry data | ProteomeXchange via PRIDE | Accession ID: PXD030637 |
| RNA-sequencesing data | Gene Expression Omnibus (GEO) | Accession ID: GSE205454 |
| Experimental models: Organisms/strains | ||
| Mice: B6; 129-Gt(ROSA)26Sortm5(CAG−Sun1/sfGFP)Nat/J | The Jackson Laboratory | Strain #021039 |
| Mice: B6.Cg-Tg(CamkIIα-cre)T29−1Stl/J | The Jackson Laboratory | Strain #005359 |
| Mice: C57BL6/J | The Jackson Laboratory via In Vivos Singapore | Strain #000664 |
| Oligonucleotides | ||
| Fos Forward: CACACAGGACTTTTGCGC Reverse: GACACGGTCTTCACCATTCC |
IDT | Generated internally, not applicable |
| Hprt Forward: TGTTGTTGGATATGCCCTTG Reverse: GGCCACAGGACTAGAACACC |
IDT | Generated internally, not applicable |
| Junb Forward: TCACGACGACTCTTACGCAG Reverse: CCTTGAGACCCCGATAGGGA |
IDT | Generated internally, not applicable |
| Zfp108 Forward: GCCCATCAGAGAGTTCATACC Reverse: CCCACACTCCACACACATATAC |
IDT | Generated internally, not applicable |
| Zfp128 Forward: CCCTGGATACTGCTCAACATAC Reverse: TCAGGGCTTCTCCATGTTAATAC |
IDT | Generated internally, not applicable |
| Zfp493 Forward: CCAGAGAATTCACACAGGAGAG Reverse: GCTGTTTAAGGGTGGAGGAATA |
IDT | Generated internally, not applicable |
| Zfp660 Forward: CAGGCAGACATCTCAGGTTATT Reverse: GTAACGATAAGCTCTCCCACAC |
IDT | Generated internally, not applicable |
| Zkscan7 Forward: GTGGCTATGGTGGAGGATTT Reverse: CTCAGTGCTCCCATCTTCTTT |
IDT | Generated internally, not applicable |
| Software and algorithms | ||
| Image Lab | Bio-Rad | https://www.bio-rad.com/en-sg/product/image-lab-software?ID=KRE6P5E8Z |
| Zen Blue | Zeiss | https://www.micro-shop.zeiss.com/en/us/softwarefinder/software-categories/zen-blue/ |
| ImageJ v1.53c | Fiji-ImageJ | https://imagej.nih.gov/ij/ |
| IMARIS v9.7.2 | Oxford Instruments | https://imaris.oxinst.com/ |
| GraphPad Prism v9.1.2 | GraphPad | https://www.graphpad.com/features |
| PANTHER v16 | PANTHER maintained by P. Thomas laboratory | http://www.pantherdb.org/ |
| STRING v11 | STRING Consortium | https://string-db.org/ |
| R v4.1.1 | R Foundation | https://www.r-project.org/ |
| Cytoscape v3.8.2 | Cytoscape Team | https://cytoscape.org/ |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Toh Hean Ch'ng (thchng@ntu.edu.sg).
Materials availability
-
•
No new unique reagents were generated in this study.
Experimental model and study participant details
Animals
All animal studies were carried out in accordance with protocols approved by Nanyang Technological University Institutional Animal Care and Use Committee (IACUC: E0019 and A18091). CaMKIIα-Cre:SUN1-sfGFP mice, were generated by crossing homozygous B6; 129-Gt(ROSA)26Sortm5(CAG−Sun1/sfGFP)Nat/J and B6.Cg-Tg(CamkIIα-cre)T29−1Stl/J mice (The Jackson Laboratory) as described by30 (Figure 1A). 6- to 8-week-old male CaMKIIα-Cre:SUN1-sfGFP or wild-type C57BL6/J mice were used for in vivo experiments. Mouse hippocampal neurons were isolated from P0 to P2 wild-type pups and maintained for 14 to 16 days in vitro (DIV) prior to any pharmacological treatment.
Primary hippocampal cultures
Unless otherwise stated, all culture-based experiments were carried out using mature hippocampal neurons (DIV14 to 16). Both male and female pups were used to prepare the cultures. Briefly, hippocampi were isolated from P0-2 wild-type C57BL6/J pups in ice-cold 1x HBSS (1 mM sodium pyruvate, 1% HEPES; Gibco). Once dissected, the hippocampi were trypsinized (HBSS, 0.25% trypsin, 1.26 mM CaCl2 and 0.48 mg/mL DNase) for 10 min and the enzymatic digestion stopped with trypsin inhibitor (1 mg/mL; Sigma-Aldrich). Hippocampal tissue was triturated in 1 mL of Neurobasal-A medium (B27, GlutaMAX, 0.01% FBS, 200 ng/mL gentamycin; Gibco) and number of cells in the suspension was counted. ∼65,000 cells were seeded on Poly-D-Lysine-coated (0.1 mg/mL; Sigma-Aldrich) 12 mm glass coverslips (Marienfeld Superior). Cells were maintained in an incubator (humidified, 37°C, 5% CO2) for 14–16 days until maturity and used for pharmacological treatments and immunocytochemistry. For treatment of hippocampal neurons, the following pharmacological agents were used: 1(S),9(R)-(−)-Bicuculline methiodide (Bic, 40 μM; Sigma-Aldrich), tetrodotoxin (TTX, 1 μM; Tocris). Mature hippocampal neurons were incubated with these pharmacological agents in conditioned media and maintained in an incubator (humidified, 37°C, 5% CO2) for an appropriate amount of time before paraformaldehyde (PFA) fixation and immunocytochemistry.
Method details
Pilocarpine seizure model
To induce seizures, the following pharmacological agents were used for intraperitoneal injections (IP): (−) scopolamine methyl nitrate (1 mg/kg; Sigma-Aldrich) and pilocarpine hydrochloride (340 mg/kg; Sigma-Adrich). Appropriate volumes of diluent (0.9% NaCl; B. Braun) were used for mock injections. Briefly, mice were injected with scopolamine 30 min prior to pilocarpine or mock injections. Pilocarpine-injected mice were allowed to develop acute seizures for 100 min. Mice with seizure behaviors which fell within the range of 2–4 on the Racine scale were used for downstream experiments (Racine, 1972). For immunohistochemistry, animals were anesthetized by intraperitoneal delivery of pentobarbital (0.3 mL/40 g) and perfused with ice-cold PBS followed by 4% PFA diluted in 0.1 M phosphate buffer (PB). Following that, brains were removed and post-fixed with 4% PFA for 2–3 h and dehydrated in 30% sucrose in 0.1 M PB for 48 h at 4°C before cryosectioning. Coronal forebrain slices (40 μm thickness) were sectioned and stored in a cryoprotective solution (30% ethylene glycol, 30% glycerol, 20 mM PB) at −20°C until use. For RNA extraction, forebrains were isolated and flash-frozen in liquid nitrogen before storing at −80°C until use.
Nuclear extraction and purification
INTACT protocol was modified from (Mo et al., 2015). Unless otherwise stated, all sample processing steps were performed on ice. Briefly, after seizures, mice were euthanized by cervical dislocation and the brains were harvested and placed in ice-cold homogenization buffer (HB; 0.25 M sucrose, 25 mM KCl, 5 mM MgCl2, 20 mM tricine, pH 7.8) supplemented with cOmplete, EDTA-free protease inhibitor cocktail (Roche). Forebrains were isolated and homogenized in a 5 mL Potter-Elvehjem tissue grinder with 10 strokes of the pestle (Corning). 0.3% IGEPAL-CA 630 (Sigma) in PBS was then added to the homogenate and further homogenized with another 5 strokes of the pestle. Homogenates were then filtered through a 40 μm cell strainer (Falcon) and mixed 1:1 with 50% OptiPrep Density Gradient Medium in dilution buffer (DB, 150 mM KCl, 30 mM MgCl2, 120 mM tricine, pH 7.8) before being layered over an iodixanol gradient composed of 30% and 40% OptiPrep in HB. Samples were centrifuged at 10,000 g for 18 min with SW28 swinging bucket rotor at a deceleration setting of 5 (Beckman Coulter). After centrifugation, nuclei were extracted from the interface between 30% and 40% of the OptiPrep gradient. To immunoprecipitate SUN1-sfGFP-positive nuclei by INTACT, immunoprecipitation buffer (IPB, 0.25 M sucrose, 25 mM KCl, 5 mM MgCl2, 20 mM tricine, 0.4% IGEPAL-CA 630, pH 7.8) was added to nuclear extracts (approximately 4–5 million nuclei) with 3 μg of anti-IgG or anti-GFP antibodies used for each reaction and incubated for 30 min at 4°C. Pre-washed Dynabeads Protein G (Invitrogen) were added for another 40 min before nuclei were isolated with extensive and repeated binding steps to enhance binding efficiency. After the final resuspension, the Dynabeads-bound nuclei were pelleted and washed with IPB and prepared for mass spectrometry or immunoblotting.
For nuclear extraction with a sucrose cushion, filtrates were mixed 2.3:1 with sucrose cushion buffer (SCB, 1.8 M sucrose, 10 mM tris-HCl, 1.5 mM MgCl2, pH 6.9) and layered over SCB. Samples were centrifuged at 30,000 g for 45 min with a deceleration setting of 5 using an SW41 Ti swinging bucket rotor (Beckman Coulter). Recovered nuclear pellets were resuspended in SCB (∼100 μL) and processed for immunoblotting.
Immunoblotting (IB)
Nuclei obtained from INTACT and sucrose cushion extractions were lysed by adding RIPA buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton X-100). Protein concentrations of the nuclear lysates were measured using Pierce BCA protein assay kit (Thermo Fisher Scientific). SDS-PAGE, transfer and immunoblotting were performed using standard protocols. For signal detection, membranes were treated with either SuperSignal West Pico PLUS Chemiluminescent Substrate or SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific) for chemiluminescence imaging with ChemiDoc MP imaging system using Image Lab software (Rio-Rad). For immunoblots probed for total SUMOylated proteins using anti-SUMO-2/3 antibody, membranes were pre-stained with SYPRO Ruby protein blot stain (Molecular Probes) according to manufacturer’s instructions prior to addition of antibodies.
Immunocytochemistry (ICC)
For ICC, hippocampal neurons cultured on glass coverslips were fixed with 3.2% paraformaldehyde (PFA) in Tyrode’s buffer (140 mM NaCl, 10 mM HEPES, 5 mM KCl, 3 mM CaCl2, 1 mM MgCl2, 10 mM glucose) for 10 min. After fixing, the coverslips were washed with PBS and permeabilized with PBST solution (PBS, 0.1% Triton X-100) for 5 min, washed with PBS and blocked in 10% goat serum in PBS (Invitrogen) for 1 h. Coverslips were incubated with primary antibodies (at least 3 h or 4°C overnight) followed by secondary antibodies in the presence of Hoechst nuclear dye (1 h) diluted in blocking solution. All coverslips were mounted onto glass slides with Aqua-Poly/Mount (PolySciences). Unless otherwise specified, all images of single neurons were taken using Zeiss LSM800 confocal system (63x 1.4NA oil objective). Primary hippocampal neurons were randomly sampled across the coverslip.
Immunohistochemistry (IHC)
For IHC, unless otherwise specified, all washing steps and treatments were performed in 24-well cell culture plates (Corning). Free-floating brain sections were washed with PBS for 5 min, permeabilized with pre-warmed PBST for 5 min and incubated with pre-warmed antigen retrieval solution (12.1 μM Tris, 0.37 μM EDTA, 10% Triton X-100) for 10–15 min at 37°C. After antigen retrieval, the sections were washed with PBST at 37°C and then again at room temperature before blocking with 5% goat serum in PBST for 1 h. Sections were incubated with primary antibodies overnight at 4°C followed by extensive washes with PBST before incubation with secondary antibodies and Hoechst dye for 1 h. Stained sections were mounted onto POLYSINE adhesion slides (Thermo Fisher Scientific) with Aqua-Poly/Mount. For the validation of candidate proteins (FOS, JUNB, SUMO-1 and SUMO-2/3), whole brain sections were imaged on Zeiss LSM800 confocal system (40x 1.3NA or 63x 1.4NA oil objectives).
LC-MS/MS sample preparation, TMT6-labeling, and analysis
For nuclear proteome, nuclei immunopurified by the INTACT method from 2 pairs of mock- and pilocarpine-induced mice per independent experiment were combined after the washing steps, resuspended in appropriate volumes of SDS-based buffer and boiled. A total of 3 independent experiments were carried out. All samples were sonicated twice (10 min) with a Bioruptor (Diagenode Diagnostics) before acetone precipitation of the proteins for 4 h at −20°C. Protein pellets were solubilized in 8 M urea 50 mM triethylammonium bicarbonate (TEAB) buffer and final protein concentration was determined with Pierce BCA protein assay kit. For each sample, 100 μg of protein was reduced with 10 mM DTT for 30 min at 60°C and alkylated with 55 mM iodoacetamide (IAA) for 30 min at room temperature. The samples were diluted 7-fold with 50 mM TEAB followed by digestion with sequencing-grade modified trypsin (Promega) at 37°C overnight. The tryptic peptides were then used for tandem mass tag (TMT) labeling with TMTsixplex Isobaric Label Reagent Set (Thermo Fisher Scientific) according to manufacturer’s protocol. The 3 independent biological replicates were labeled as mock replicate 1 (M1) with TMT-126; pilocarpine replicate 1 (P1) with TMT-127; M2 with TMT-128; P2 with TMT-129; M3 with TMT-130 and P3 with TMT-131, respectively. All 6 samples of labeled peptides were then pooled and desalted using a C18 Sep-Pak 200 mg cartridge (Waters) before drying in a vacuum concentrator. The TMT-labeled sample was then subjected to high-performance liquid chromatography (HPLC) fractionation. Briefly, dried peptides were reconstituted in 200 μL of mobile phase A (10 mmol/L ammonium hydroxide in water) and fractionated using a XBridge BEH C18 column (130 Å, 3.5 μm, 4.6 × 250 mm; Waters) on a Prominence ultra-fast liquid chromatograph (UFLC) system (Shimadzu) with UV monitoring of peptide intensities at 280 nm. Peptide separation was performed at 1 mL/min using a 60 min gradient as follows: 0–5% B for 3 min, 5–35% B for 40 min, 35–70% B for 12 min, and 70-0% B for 5 min. Fractions were collected at intervals of 1 min and combined by concatenation. Concatenated fractions were then completely dried in the vacuum concentrator. Following that, dried sample fractions were reconstituted in 3% acetonitrile (ACN)/0.1% formic acid (FA) before injection to LC-MS/MS using a Dionex UltiMate 3000 RSLCnano system coupled with a Q Exactive tandem mass spectrometer (Thermo Fisher Scientific). Spray was generated using an EASY-Spray ion source (Thermo Fisher Scientific) working at 1.5 kV. Peptide separation was performed using a PepMap C18 column (100 Å, 3 μm; Thermo Fisher Scientific) maintained at 35°C. Separation of peptides was performed over a 60 min gradient with mobile phase A (0.1% FA in water) and mobile phase B (0.1% FA in 100% ACN) as follows: 3−30% B for 40 min, 30−50% B for 5 min, 50−80% B for 3 min, 80% B for 3 min, and finally maintained isocratic at 3% B for 10 min. Q Exactive data acquisition was performed in positive ion mode using Xcalibur version 3.0.63 (Thermo Fisher Scientific) with alternating full Fourier transform MS (FT-MS; 350–1600 m/z range, resolution of 70,000 at m/z 200, 1 micro-scan/spectrum) and FT-MS/MS (resolution of 35,000) for the 10 most intense ions with charge > +2 and isolated within a 2 Da window. Fragmentation of ions was performed by high energy collisional dissociation fragmentation mode using 28% normalized collision energy. A threshold of 500 counts was enabled. For full FT-MS and FT-MS/MS, automatic gain control was set to 5×106 and 2×105, respectively.
Sample preparation for RNA sequencing and data analysis
Nuclei immunopurified by the INTACT method from N = 64 pairs of mock- and pilocarpine-induced mice were lysed in 1 mL of TRIzol Reagent (Invitrogen) and stored at −80°C until RNA extraction. Total RNA was extracted, and column-purified with PicoPure RNA isolation kit (Applied Biosystems) according to manufacturer’s instructions. Purified RNA was then sent for directional poly(A)-positive mRNA library preparation and sequencing of >45M paired-end 150 bp reads per sample on an Illumina NovaSeq 6000 (NovogeneAIT Genomics, Singapore). All raw RNA-seq data can be downloaded from the Gene Expression Omnibus (GEO) web portal (Accession ID: GSE205454).
RNA extraction and cDNA synthesis
Flash-frozen forebrains were thawed at room temperature and transferred to a 17 mL Potter-Elvehjem tissue grinder (Corning). 3 mL of TRIzol Reagent (Invitrogen) was added to the tissue grinder and the forebrains were homogenized with 10 strokes of the pestle. Homogenized samples were incubated at room temperature for 5 min. After incubation, homogenates were aliquoted equally into separate 1.5 mL tubes and centrifuged at 12, 000 g at 4°C for 10 min. Insoluble fraction in the homogenate was removed by transferring the supernatant to fresh 1.5 mL tubes. To each tube containing 1 mL of supernatant, 200 μL of chloroform (Fisher Chemical) was added. Samples were then mixed by shaking vigorously for 1 min and incubated at room temperature for 5 min. Next, samples were centrifuged at 11, 600 g at 4°C for 15 min before the colorless aqueous phase for each sample was transferred to a fresh 1.5 mL tube. An equal volume of pure ethanol (Sigma-Aldrich) was added dropwise to the aqueous phase with intermittent mixing by inversion of the tube. Total RNA was then extracted, and column-purified with PicoPure RNA isolation kit (Applied Biosystems) according to manufacturer’s instructions. RNA concentrations were measured using the NanoDropTM OneC instrument (Thermo Fisher Scientific). 2 μg of RNA from each sample was reverse transcribed with the RevertAid First Strand cDNA synthesis kit (Thermo Fisher Scientific) in a 20 μL mixture using the VeritiTM 96-well thermal cycler (Applied Biosystems). The conditions for cDNA synthesis were 42°C for 60 min followed by 70°C for 5 min to terminate the reaction. cDNA samples were then stored in −20°C until quantitative (qPCR).
Quantitative PCR (qPCR)
All qPCR reactions and analyses were performed with the QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems). qPCR reactions were performed in triplicates with each reaction consisting of 30 ng of cDNA, 100 nM of forward and reverse primers (Table S7), and appropriate amount of PowerUp SYBR Green Master Mix (Applied Biosystems). No reverse transcriptase samples were included as negative controls. Relative gene expression levels were determined using the comparative Ct method (Livak and Schmittgen, 2001). ΔCt values were obtained by normalizing the mean Ct values for each target gene to the mean Ct value for housekeeping gene hypoxanthine-guanine phosphoribosyltransferase 1 (Hprt1). ΔCt values were further normalized to the mean ΔCt value for all samples from mock-injected animals to obtain the ΔΔCt values.
Quantification and statistical analysis
Immunoassay quantification
All immunoblot quantifications were done with Image Lab software. Bands of each candidate protein were manually outlined with boxes such that the area quantified are the same across lanes. Band intensities were then normalized against lamin B1 as a loading control. For SUMOylated proteins, the total intensity for each lane was normalized against total loaded protein determined with SYPRO Ruby stain. For immunocytochemistry quantification of nuclear and cytoplasmic intensities, mean pixel intensities in the respective subcellular compartments were obtained based on somatic (MAP2) or nuclear (Hoechst) marker staining of neurons. Quantification of all nuclear and cytoplasmic intensities from cultures were done with ImageJ 1.53c. All data across conditions were normalized against the average basal values for each independent experiment. For immunohistochemistry quantification SUN1-sfGFP-positive excitatory neuronal nuclei were randomly selected from the DG, CA1, CA3 of the hippocampus, as well as the piriform cortex (PIR) for quantification with ImageJ 1.53c. A SUN1-sfGFP-positive nucleus is defined by the observation of a Hoescht-stained nucleus outlined with a distinct ring in Channel 488. The ring-like pattern of SUN1-sfGFP also ensures that the nucleus quantified is in focus in the images. Mean nuclear intensities for the candidate proteins were recorded from these SUN1-sfGFP-positive nuclei. Separately, for the quantification of pCREB-positive/SUN-sfGFP-positive nuclei, brain sections (1 slice per animal with N = 3 animals) were imaged with Zeiss Axioscan (20× objective). Randomly selected regions of interest (ROIs; CA1, DG, Cortex: 1895 × 1984 pixels; CA3: 856 × 298 pixels) were analyzed with IMARIS 9.7.2 using filters and size thresholds to outline nuclei. The number of pCREB-positive and SUN-sfGFP-positive nuclei were quantified and the percentages of SUN1-sfGFP-positive nuclei expressing pCREB in mock- and pilocarpine-induced mice were calculated.
Statistics
All graphs for quantification were plotted using GraphPad Prism 9.1.2 with the error bars being presented as mean ± SEM. For ICC and IHC experiments, statistical significance for two-group comparisons was determined by t-test (parametric) or Mann-Whitney test (non-parametric). For IB experiments, statistical significance for comparisons between two correlated groups was determined by paired t-test (parametric) or Wilcoxon test (non-parametric). Shapiro-Wilk test and D'Agostino & Pearson test were conducted for small (n < 30) and large sample sizes, respectively, prior to any statistical test. Significance level (alpha) was set at 0.05 and the resulting p values for significantly different data were indicated as follows: p < 0.05 (∗), p < 0.01 (∗∗), p < 0.001 (∗∗∗), p < 0.0001 (∗∗∗∗).
Proteomics analysis
The acquired data were processed using Proteome Discoverer version 2.1 (PD2.1; Thermo Fisher Scientific). The MS/MS spectra in raw data files were directly converted to Mascot generic file (mgf) format for protein identification by searching against UniProt mouse protein database using Mascot algorithm (Matrix Sciences). The raw data files were also imported into PD2.1 and searched for the MS/MS spectra using SEQUEST and Mascot algorithms with PD2.1 standard workflow. Subsequent data analysis was performed using R in RStudio version 1.4.1717. A total of 14603 peptides were identified by PD2.1 and variance stabilizing normalization (VSN) was performed on the PD2.1-determined abundance ratio to reduce pooled variance across all samples using the package NormalyzerDE version 1.10.0 (Figure S5) (Chawade et al., 2014; Willforss et al., 2019). In total, PD2.1 identified N = 2948 “master proteins”, of which N = 2608 proteins with TMT ratios in all three independent replicates and q-value <0.05 were used for further analysis. Proteins enriched or depleted in the nucleus were identified based on the abundance ratio between conditions for paired samples for each identified protein. Gene ontology (GO) analysis was performed using PANTHER version 16.0 and protein-protein interaction analysis was performed using STRING version 11.5(Mi et al., 2021; Szklarczyk et al., 2015). All MS data are uploaded to ProteomeXchange via PRIDE (Accession ID: PXD030637).
RNA sequencing analysis
Pre-processing
Sequenced reads were pre-processed using Trimmomatic (v0.39)125 to remove adapter sequences and low-quality or short-length reads and then aligned to the mouse genome (GENCODE M24) using STAR (v2.7.1a).126 Reads aligned to exons were counted using HTSeq (v0.11.2)127 using the GENCODE primary assembly annotation. These steps were performed on the Gekko high-performance computing cluster.
Differential expression
Gene count matrices were analyzed using R 4.1.1 (R Core Team, 2021). First, genes with <1 log2 counts per million (CPM) in more than 4 samples were removed, retaining N = 14,115 expressed genes, and then trimmed mean of M values (TMM) normalization was applied. Differential expression between Mock and Seizure conditions was analyzed using edgeR quasi-likelihood F-test with robust dispersion estimation.128,129,130 Thresholds of false discovery rate (FDR) < 1% and log2 fold change (LFC) > ±1 were used to identify differentially expressed genes (DEGs).
Gene set enrichment analyses
Gene set enrichment analyses (GSEA) were conducted on expressed genes or proteins ranked by LFC between conditions using the fgsea R package.131 The gene sets used for analysis were Gene Ontology, PFAM, ReactomeDB and KEGG pathways.132,133,134,135 Terms with <5 or >800 annotated genes were excluded. Individual enrichment plots were generated for gene sets of interest. Enriched GO terms (padj <0.05; N = 327) were mapped in Cytoscape (v3.8.2),136 following a similar approach to Merico et al.137 Each node represents a GO term and edges indicate Jaccard similarity as a measure of gene set overlap (minimum 0.1). Edge thickness was mapped to Jaccard coefficient, node color to nucleotide enrichment score (NES) and node size to -log10 adjusted p value. Prefuse force-directed layout was applied with minor manual adjustments for clarity, and clusters were identified and labeled manually.
Transcription factor binding motif enrichment
Pscan55 was used to analyze over-representation of regulatory transcription factor (RTF) binding site motifs within gene promoters (−950 to +50 from transcription start site [TSS]) using the Jaspar 2020 non-redundant database.138 For the top candidate RTF (THAP11), genes with binding scores greater than the average background score were plotted in Cytoscape, indicating genes with KRAB or C2H2 domains annotated in PFAM. Edge width and edge length (using the edge-weighted spring-embedded layout) were mapped to binding score.
Analyses of intronic reads
Aligned reads were re-quantified using the INSPEcT R package139 with UCSC mm10 knownGene annotation and ‘prioritizeExons’ set to false. Data were filtered to retain protein-coding genes with reads in both exon and intron regions and differential expression analyses were conducted on exons and introns separately as described above. Exon-intron split analysis (EISA) was then performed using default parameters to determine differential expression between exon and intron levels.58 Data were again filtered to retain genes with average log2 CPM >1 and DEGs were determined using thresholds of LFC > ±0.5 and FDR <0.01 for each test. DEG subsets of interest were analyzed using Pscan (as described above), MIENTURNET140 with TargetScan database (v7.2)141 and topGO.142 For topGO, Fisher’s exact test was used to compare GO term enrichment in the subset of interest to all expressed genes. Gene coverage profiles were generated using scripts described in Lee et al., 2020.110
Acknowledgments
The authors would like to acknowledge the contributions of S. Langley (NTU LKCMedicine, Singapore), A. Chen (Scintillon Institute, USA) for discussion and/or comments on the manuscript. Meng Wei from the School of Biological Sciences Mass Spectrometry Core Facility in Nanyang Technological University, Singapore for sample preparation and protein sequencing. This research is supported by Singapore Ministry of Education Academic Research Fund Tier 1 (MOE2018-T1-002-033), Academic Research Fund Tier 3 (MOE2017-T3-1-002) and Nanyang Technological University NAP Start-up funds.
Author contributions
H.R.S., J.G., and T.H.C. conceived and designed the experiments for this study. Experiments and analysis for nuclear proteome were performed by H.R.S., J.G., S.K.S., V.A.B., C.L., and T.H.C. and transcriptome by J.G., T.H.C., and H.R.S. The paper was written by H.R.S., J.G., and T.H.C.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: August 23, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107707.
Supplemental information
Data and code availability
-
•
No new code was generated in this study.
-
•
Mass spectrometry data are found at ProteomeXchange (PXD030637) and RNA sequencing data have been deposited at GEO (GSE205454).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Borden J., Manuelidis L. Movement of the X chromosome in epilepsy. Science. 1988;242:1687–1691. doi: 10.1126/science.3201257. [DOI] [PubMed] [Google Scholar]
- 2.Skupien-Jaroszek A., Walczak A., Czaban I., Pels K.K., Szczepankiewicz A.A., Krawczyk K., Ruszczycki B., Wilczynski G.M., Dzwonek J., Magalska A. The interplay of seizures-induced axonal sprouting and transcription-dependent Bdnf repositioning in the model of temporal lobe epilepsy. PLoS One. 2021;16 doi: 10.1371/journal.pone.0239111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walczak A., Szczepankiewicz A.A., Ruszczycki B., Magalska A., Zamlynska K., Dzwonek J., Wilczek E., Zybura-Broda K., Rylski M., Malinowska M., et al. Novel higher-order epigenetic regulation of the Bdnf gene upon seizures. J. Neurosci. 2013;33:2507–2511. doi: 10.1523/JNEUROSCI.1085-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vashishta A., Slomnicki L.P., Pietrzak M., Smith S.C., Kolikonda M., Naik S.P., Parlato R., Hetman M. RNA Polymerase 1 Is Transiently Regulated by Seizures and Plays a Role in a Pharmacological Kindling Model of Epilepsy. Mol. Neurobiol. 2018;55:8374–8387. doi: 10.1007/s12035-018-0989-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lösing P., Niturad C.E., Harrer M., Reckendorf C.M.Z., Schatz T., Sinske D., Lerche H., Maljevic S., Knöll B. SRF modulates seizure occurrence, activity induced gene transcription and hippocampal circuit reorganization in the mouse pilocarpine epilepsy model. Mol. Brain. 2017;10:30. doi: 10.1186/s13041-017-0310-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hauser R.M., Henshall D.C., Lubin F.D. The Epigenetics of Epilepsy and Its Progression. Neuroscientist. 2018;24:186–200. doi: 10.1177/1073858417705840. [DOI] [PubMed] [Google Scholar]
- 7.Kobow K., Blümcke I. Epigenetics in epilepsy. Neurosci. Lett. 2018;667:40–46. doi: 10.1016/j.neulet.2017.01.012. [DOI] [PubMed] [Google Scholar]
- 8.Hansen K.F., Sakamoto K., Pelz C., Impey S., Obrietan K. Profiling status epilepticus-induced changes in hippocampal RNA expression using high-throughput RNA sequencing. Sci. Rep. 2014;4:6930. doi: 10.1038/srep06930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conte G., Parras A., Alves M., Ollà I., De Diego-Garcia L., Beamer E., Alalqam R., Ocampo A., Mendez R., Henshall D.C., et al. High concordance between hippocampal transcriptome of the mouse intra-amygdala kainic acid model and human temporal lobe epilepsy. Epilepsia. 2020;61:2795–2810. doi: 10.1111/epi.16714. [DOI] [PubMed] [Google Scholar]
- 10.Pfisterer U., Petukhov V., Demharter S., Meichsner J., Thompson J.J., Batiuk M.Y., Asenjo-Martinez A., Vasistha N.A., Thakur A., Mikkelsen J., et al. Identification of epilepsy-associated neuronal subtypes and gene expression underlying epileptogenesis. Nat. Commun. 2020;11:5038. doi: 10.1038/s41467-020-18752-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dixit A.B., Banerjee J., Srivastava A., Tripathi M., Sarkar C., Kakkar A., Jain M., Chandra P.S. RNA-seq analysis of hippocampal tissues reveals novel candidate genes for drug refractory epilepsy in patients with MTLE-HS. Genomics. 2016;107:178–188. doi: 10.1016/j.ygeno.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 12.Xi Z.Q., Xiao F., Yuan J., Wang X.F., Wang L., Quan F.Y., Liu G.W. Gene expression analysis on anterior temporal neocortex of patients with intractable epilepsy. Synapse. 2009;63:1017–1028. doi: 10.1002/syn.20681. [DOI] [PubMed] [Google Scholar]
- 13.Arion D., Sabatini M., Unger T., Pastor J., Alonso-Nanclares L., Ballesteros-Yáñez I., García Sola R., Muñoz A., Mirnics K., DeFelipe J. Correlation of transcriptome profile with electrical activity in temporal lobe epilepsy. Neurobiol. Dis. 2006;22:374–387. doi: 10.1016/j.nbd.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 14.Laurén H.B., Lopez-Picon F.R., Brandt A.M., Rios-Rojas C.J., Holopainen I.E. Transcriptome analysis of the hippocampal CA1 pyramidal cell region after kainic acid-induced status epilepticus in juvenile rats. PLoS One. 2010;5 doi: 10.1371/journal.pone.0010733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okamoto O.K., Janjoppi L., Bonone F.M., Pansani A.P., da Silva A.V., Scorza F.A., Cavalheiro E.A. Whole transcriptome analysis of the hippocampus: toward a molecular portrait of epileptogenesis. BMC Genom. 2010;11:230. doi: 10.1186/1471-2164-11-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Canto A.M., Matos A.H.B., Godoi A.B., Vieira A.S., Aoyama B.B., Rocha C.S., Henning B., Carvalho B.S., Pascoal V.D.B., Veiga D.F.T., et al. Multi-omics analysis suggests enhanced epileptogenesis in the Cornu Ammonis 3 of the pilocarpine model of mesial temporal lobe epilepsy. Hippocampus. 2021;31:122–139. doi: 10.1002/hipo.23268. [DOI] [PubMed] [Google Scholar]
- 17.do Canto A.M., Vieira A.S., H B Matos A., Carvalho B.S., Henning B., Norwood B.A., Bauer S., Rosenow F., Gilioli R., Cendes F., Lopes-Cendes I. Laser microdissection-based microproteomics of the hippocampus of a rat epilepsy model reveals regional differences in protein abundances. Sci. Rep. 2020;10:4412. doi: 10.1038/s41598-020-61401-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pires G., Leitner D., Drummond E., Kanshin E., Nayak S., Askenazi M., Faustin A., Friedman D., Debure L., Ueberheide B., et al. Proteomic differences in the hippocampus and cortex of epilepsy brain tissue. Brain Commun. 2021;3:fcab021. doi: 10.1093/braincomms/fcab021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keck M., van Dijk R.M., Deeg C.A., Kistler K., Walker A., von Rüden E.L., Russmann V., Hauck S.M., Potschka H. Proteomic profiling of epileptogenesis in a rat model: Focus on cell stress, extracellular matrix and angiogenesis. Neurobiol. Dis. 2018;112:119–135. doi: 10.1016/j.nbd.2018.01.013. [DOI] [PubMed] [Google Scholar]
- 20.Bitsika V., Duveau V., Simon-Areces J., Mullen W., Roucard C., Makridakis M., Mermelekas G., Savvopoulos P., Depaulis A., Vlahou A. High-Throughput LC-MS/MS Proteomic Analysis of a Mouse Model of Mesiotemporal Lobe Epilepsy Predicts Microglial Activation Underlying Disease Development. J. Proteome Res. 2016;15:1546–1562. doi: 10.1021/acs.jproteome.6b00003. [DOI] [PubMed] [Google Scholar]
- 21.Keren-Aviram G., Dachet F., Bagla S., Balan K., Loeb J.A., Dratz E.A. Proteomic analysis of human epileptic neocortex predicts vascular and glial changes in epileptic regions. PLoS One. 2018;13 doi: 10.1371/journal.pone.0195639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X.Y., Yang J.L., Chen L.J., Zhang Y., Yang M.L., Wu Y.Y., Li F.Q., Tang M.H., Liang S.F., Wei Y.Q. Comparative proteomics and correlated signaling network of rat hippocampus in the pilocarpine model of temporal lobe epilepsy. Proteomics. 2008;8:582–603. doi: 10.1002/pmic.200700514. [DOI] [PubMed] [Google Scholar]
- 23.Miller-Delaney S.F.C., Bryan K., Das S., McKiernan R.C., Bray I.M., Reynolds J.P., Gwinn R., Stallings R.L., Henshall D.C. Differential DNA methylation profiles of coding and non-coding genes define hippocampal sclerosis in human temporal lobe epilepsy. Brain. 2015;138:616–631. doi: 10.1093/brain/awu373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiese K., Jablonski J., Hackenbracht J., Wrosch J.K., Groemer T.W., Kornhuber J., Blümcke I., Kobow K. Epigenetic control of epilepsy target genes contributes to a cellular memory of epileptogenesis in cultured rat hippocampal neurons. Acta Neuropathol. Commun. 2017;5:79. doi: 10.1186/s40478-017-0485-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobow K., Kaspi A., Harikrishnan K.N., Kiese K., Ziemann M., Khurana I., Fritzsche I., Hauke J., Hahnen E., Coras R., et al. Deep sequencing reveals increased DNA methylation in chronic rat epilepsy. Acta Neuropathol. 2013;126:741–756. doi: 10.1007/s00401-013-1168-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dębski K.J., Pitkanen A., Puhakka N., Bot A.M., Khurana I., Harikrishnan K.N., Ziemann M., Kaspi A., El-Osta A., Lukasiuk K., Kobow K. Etiology matters - Genomic DNA Methylation Patterns in Three Rat Models of Acquired Epilepsy. Sci. Rep. 2016;6 doi: 10.1038/srep25668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conboy K., Henshall D.C., Brennan G.P. Epigenetic principles underlying epileptogenesis and epilepsy syndromes. Neurobiol. Dis. 2021;148 doi: 10.1016/j.nbd.2020.105179. [DOI] [PubMed] [Google Scholar]
- 28.do Canto A.M., Donatti A., Geraldis J.C., Godoi A.B., da Rosa D.C., Lopes-Cendes I. Neuroproteomics in Epilepsy: What Do We Know so Far? Front. Mol. Neurosci. 2020;13 doi: 10.3389/fnmol.2020.604158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vistain L.F., Tay S. Single-Cell Proteomics. Trends Biochem. Sci. 2021;46:661–672. doi: 10.1016/j.tibs.2021.01.013. [DOI] [PubMed] [Google Scholar]
- 30.Mo A., Mukamel E.A., Davis F.P., Luo C., Henry G.L., Picard S., Urich M.A., Nery J.R., Sejnowski T.J., Lister R., et al. Epigenomic Signatures of Neuronal Diversity in the Mammalian Brain. Neuron. 2015;86:1369–1384. doi: 10.1016/j.neuron.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Razafsky D., Hodzic D. Bringing KASH under the SUN: the many faces of nucleo-cytoskeletal connections. J. Cell Biol. 2009;186:461–472. doi: 10.1083/jcb.200906068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Covolan L., Smith R.L., Mello L.E. Ultrastructural identification of dentate granule cell death from pilocarpine-induced seizures. Epilepsy Res. 2000;41:9–21. doi: 10.1016/s0920-1211(00)00122-4. [DOI] [PubMed] [Google Scholar]
- 33.do Nascimento A.L., dos Santos N.F., Campos Pelágio F., Aparecida Teixeira S., de Moraes Ferrari E.A., Langone F. Neuronal degeneration and gliosis time-course in the mouse hippocampal formation after pilocarpine-induced status epilepticus. Brain Res. 2012;1470:98–110. doi: 10.1016/j.brainres.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 34.Sakamoto K., Karelina K., Obrietan K. CREB: a multifaceted regulator of neuronal plasticity and protection. J. Neurochem. 2011;116:1–9. doi: 10.1111/j.1471-4159.2010.07080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beaumont T.L., Yao B., Shah A., Kapatos G., Loeb J.A. Layer-Specific CREB Target Gene Induction in Human Neocortical Epilepsy. J. Neurosci. 2012;32:14389–14401a. doi: 10.1523/jneurosci.3408-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopez-Bergami P., Lau E., Ronai Z. Emerging roles of ATF2 and the dynamic AP1 network in cancer. Nat. Rev. Cancer. 2010;10:65–76. doi: 10.1038/nrc2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hong S., Friedman A.D. C/EBP and AP-1 Proteins Form Leucine Zipper Heterodimers and Reduce C/EBP Homodimer Formation During Monopoiesis. Blood. 2010;116:2606. doi: 10.1182/blood.V116.21.2606.2606. [DOI] [Google Scholar]
- 38.Cubeñas-Potts C., Matunis M.J. SUMO: a multifaceted modifier of chromatin structure and function. Dev. Cell. 2013;24:1–12. doi: 10.1016/j.devcel.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 2004;18:2046–2059. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- 40.Holmes G.L., Ben-Ari Y. The Neurobiology and Consequences of Epilepsy in the Developing Brain. Pediatr. Res. 2001;49:320–325. doi: 10.1203/00006450-200103000-00004. [DOI] [PubMed] [Google Scholar]
- 41.Vaughan D.N., Jackson G.D. The piriform cortex and human focal epilepsy. Front. Neurol. 2014;5:259. doi: 10.3389/fneur.2014.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qi Y., Wang J., Bomben V.C., Li D.P., Chen S.R., Sun H., Xi Y., Reed J.G., Cheng J., Pan H.L., et al. Hyper-SUMOylation of the Kv7 potassium channel diminishes the M-current leading to seizures and sudden death. Neuron. 2014;83:1159–1171. doi: 10.1016/j.neuron.2014.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tyssowski K.M., Gray J.M. The neuronal stimulation-transcription coupling map. Curr. Opin. Neurobiol. 2019;59:87–94. doi: 10.1016/j.conb.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kirchner A., Dachet F., Loeb J.A. Identifying targets for preventing epilepsy using systems biology of the human brain. Neuropharmacology. 2020;168 doi: 10.1016/j.neuropharm.2019.107757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Henshall D.C., Sinclair J., Simon R.P. Relationship between seizure-induced transcription of the DNA damage-inducible gene GADD45, DNA fragmentation, and neuronal death in focally evoked limbic epilepsy. J. Neurochem. 1999;73:1573–1583. doi: 10.1046/j.1471-4159.1999.0731573.x. [DOI] [PubMed] [Google Scholar]
- 46.Zhu R.L., Graham S.H., Jin J., Stetler R.A., Simon R.P., Chen J. Kainate induces the expression of the DNA damage-inducible gene, GADD45, in the rat brain. Neuroscience. 1997;81:707–720. doi: 10.1016/s0306-4522(97)00205-4. [DOI] [PubMed] [Google Scholar]
- 47.Del Viso F., Huang F., Myers J., Chalfant M., Zhang Y., Reza N., Bewersdorf J., Lusk C.P., Khokha M.K. Congenital Heart Disease Genetics Uncovers Context-Dependent Organization and Function of Nucleoporins at Cilia. Dev. Cell. 2016;38:478–492. doi: 10.1016/j.devcel.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marquez J., Bhattacharya D., Lusk C.P., Khokha M.K. Nucleoporin NUP205 plays a critical role in cilia and congenital disease. Dev. Biol. 2021;469:46–53. doi: 10.1016/j.ydbio.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kirschen G.W., Liu H., Lang T., Liang X., Ge S., Xiong Q. The radial organization of neuronal primary cilia is acutely disrupted by seizure and ischemic brain injury. Front. Biol. 2017;12:124–138. doi: 10.1007/s11515-017-1447-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loucks C.M., Park K., Walker D.S., McEwan A.H., Timbers T.A., Ardiel E.L., Grundy L.J., Li C., Johnson J.L., Kennedy J., et al. EFHC1, implicated in juvenile myoclonic epilepsy, functions at the cilium and synapse to modulate dopamine signaling. Elife. 2019;8 doi: 10.7554/eLife.37271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lorsch Z.S., Hamilton P.J., Ramakrishnan A., Parise E.M., Salery M., Wright W.J., Lepack A.E., Mews P., Issler O., McKenzie A., et al. Stress resilience is promoted by a Zfp189-driven transcriptional network in prefrontal cortex. Nat. Neurosci. 2019;22:1413–1423. doi: 10.1038/s41593-019-0462-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lupo A., Cesaro E., Montano G., Zurlo D., Izzo P., Costanzo P. KRAB-Zinc Finger Proteins: A Repressor Family Displaying Multiple Biological Functions. Curr. Genom. 2013;14:268–278. doi: 10.2174/13892029113149990002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fernandez-Albert J., Lipinski M., Lopez-Cascales M.T., Rowley M.J., Martin-Gonzalez A.M., Del Blanco B., Corces V.G., Barco A. Immediate and deferred epigenomic signatures of in vivo neuronal activation in mouse hippocampus. Nat. Neurosci. 2019;22:1718–1730. doi: 10.1038/s41593-019-0476-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lacar B., Linker S.B., Jaeger B.N., Krishnaswami S.R., Barron J.J., Kelder M.J.E., Parylak S.L., Paquola A.C.M., Venepally P., Novotny M., et al. Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat. Commun. 2016;7 doi: 10.1038/ncomms11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zambelli F., Pesole G., Pavesi G. Pscan: finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Res. 2009;37:W247–W252. doi: 10.1093/nar/gkp464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ameur A., Zaghlool A., Halvardson J., Wetterbom A., Gyllensten U., Cavelier L., Feuk L. Total RNA sequencing reveals nascent transcription and widespread co-transcriptional splicing in the human brain. Nat. Struct. Mol. Biol. 2011;18:1435–1440. doi: 10.1038/nsmb.2143. [DOI] [PubMed] [Google Scholar]
- 57.Mauger O., Lemoine F., Scheiffele P. Targeted Intron Retention and Excision for Rapid Gene Regulation in Response to Neuronal Activity. Neuron. 2016;92:1266–1278. doi: 10.1016/j.neuron.2016.11.032. [DOI] [PubMed] [Google Scholar]
- 58.Gaidatzis D., Burger L., Florescu M., Stadler M.B. Analysis of intronic and exonic reads in RNA-seq data characterizes transcriptional and post-transcriptional regulation. Nat. Biotechnol. 2015;33:722–729. doi: 10.1038/nbt.3269. [DOI] [PubMed] [Google Scholar]
- 59.Weil D., Boutain S., Audibert A., Dautry F. Mature mRNAs accumulated in the nucleus are neither the molecules in transit to the cytoplasm nor constitute a stockpile for gene expression. RNA. 2000;6:962–975. doi: 10.1017/s1355838200000479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davidson P.M., Cadot B. Actin on and around the Nucleus. Trends Cell Biol. 2021;31:211–223. doi: 10.1016/j.tcb.2020.11.009. [DOI] [PubMed] [Google Scholar]
- 61.Viita T., Vartiainen M.K. From Cytoskeleton to Gene Expression: Actin in the Nucleus. Handb. Exp. Pharmacol. 2017;235:311–329. doi: 10.1007/164_2016_27. [DOI] [PubMed] [Google Scholar]
- 62.Yoo Y., Wu X., Guan J.L. A novel role of the actin-nucleating Arp2/3 complex in the regulation of RNA polymerase II-dependent transcription. J. Biol. Chem. 2007;282:7616–7623. doi: 10.1074/jbc.M607596200. [DOI] [PubMed] [Google Scholar]
- 63.Ye G., Yang Q., Lei X., Zhu X., Li F., He J., Chen H., Ling R., Zhang H., Lin T., et al. Nuclear MYH9-induced CTNNB1 transcription, targeted by staurosporin, promotes gastric cancer cell anoikis resistance and metastasis. Theranostics. 2020;10:7545–7560. doi: 10.7150/thno.46001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xie X., Wang X., Liao W., Fei R., Wu N., Cong X., Chen Q., Wei L., Wang Y., Chen H. MYL6B, a myosin light chain, promotes MDM2-mediated p53 degradation and drives HCC development. J. Exp. Clin. Cancer Res. 2018;37:28. doi: 10.1186/s13046-018-0693-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morgan J.I., Curran T. Proto-oncogene transcription factors and epilepsy. Trends Pharmacol. Sci. 1991;12:343–349. doi: 10.1016/0165-6147(91)90594-i. [DOI] [PubMed] [Google Scholar]
- 66.Sonnenberg J.L., Macgregor-Leon P.F., Curran T., Morgan J.I. Dynamic alterations occur in the levels and composition of transcription factor AP-1 complexes after seizure. Neuron. 1989;3:359–365. doi: 10.1016/0896-6273(89)90260-2. [DOI] [PubMed] [Google Scholar]
- 67.Morris T.A., Jafari N., DeLorenzo R.J. Chronic DeltaFosB expression and increased AP-1 transcription factor binding are associated with the long term plasticity changes in epilepsy. Brain Res. Mol. Brain Res. 2000;79:138–149. doi: 10.1016/s0169-328x(00)00112-1. [DOI] [PubMed] [Google Scholar]
- 68.Jaeger B.N., Linker S.B., Parylak S.L., Barron J.J., Gallina I.S., Saavedra C.D., Fitzpatrick C., Lim C.K., Schafer S.T., Lacar B., et al. A novel environment-evoked transcriptional signature predicts reactivity in single dentate granule neurons. Nat. Commun. 2018;9:3084. doi: 10.1038/s41467-018-05418-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Leitner D.F., Mills J.D., Pires G., Faustin A., Drummond E., Kanshin E., Nayak S., Askenazi M., Verducci C., Chen B.J., et al. Proteomics and Transcriptomics of the Hippocampus and Cortex in SUDEP and High-Risk SUDEP Patients. Neurology. 2021;96:e2639–e2652. doi: 10.1212/WNL.0000000000011999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sadeghi N., Arrigoni F., D'Angelo M.G., Thomas C., Irfanoglu M.O., Hutchinson E.B., Nayak A., Modi P., Bassi M.T., Pierpaoli C. Tensor-based morphometry using scalar and directional information of diffusion tensor MRI data (DTBM): Application to hereditary spastic paraplegia. Hum. Brain Mapp. 2018;39:4643–4651. doi: 10.1002/hbm.24278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sadeghi L., Rizvanov A.A., Dabirmanesh B., Salafutdinov I.I., Sayyah M., Shojaei A., Zahiri J., Mirnajafi-Zadeh J., Khorsand B., Khajeh K., Fathollahi Y. Proteomic profiling of the rat hippocampus from the kindling and pilocarpine models of epilepsy: potential targets in calcium regulatory network. Sci. Rep. 2021;11:8252. doi: 10.1038/s41598-021-87555-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sarangi P., Zhao X. SUMO-mediated regulation of DNA damage repair and responses. Trends Biochem. Sci. 2015;40:233–242. doi: 10.1016/j.tibs.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Su S., Zhang Y., Liu P. Roles of Ubiquitination and SUMOylation in DNA Damage Response. Curr. Issues Mol. Biol. 2020;35:59–84. doi: 10.21775/cimb.035.059. [DOI] [PubMed] [Google Scholar]
- 74.Varejão N., Ibars E., Lascorz J., Colomina N., Torres-Rosell J., Reverter D. DNA activates the Nse2/Mms21 SUMO E3 ligase in the Smc5/6 complex. EMBO J. 2018;37 doi: 10.15252/embj.201798306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao X., Blobel G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. USA. 2005;102:4777–4782. doi: 10.1073/pnas.0500537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Andrews E.A., Palecek J., Sergeant J., Taylor E., Lehmann A.R., Watts F.Z. Nse2, a Component of the Smc5-6 Complex, Is a SUMO Ligase Required for the Response to DNA Damage. Mol. Cell Biol. 2005;25:185–196. doi: 10.1128/MCB.25.1.185-196.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Potts P.R., Yu H. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat. Struct. Mol. Biol. 2007;14:581–590. doi: 10.1038/nsmb1259. [DOI] [PubMed] [Google Scholar]
- 78.Lallemand-Breitenbach V., de Thé H. PML nuclear bodies. Cold Spring Harbor Perspect. Biol. 2010;2:a000661. doi: 10.1101/cshperspect.a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yalçin Z., Selenz C., Jacobs J.J.L. Ubiquitination and SUMOylation in Telomere Maintenance and Dysfunction. Front. Genet. 2017;8:67. doi: 10.3389/fgene.2017.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sahin U., Ferhi O., Jeanne M., Benhenda S., Berthier C., Jollivet F., Niwa-Kawakita M., Faklaris O., Setterblad N., de Thé H., Lallemand-Breitenbach V. Oxidative stress-induced assembly of PML nuclear bodies controls sumoylation of partner proteins. J. Cell Biol. 2014;204:931–945. doi: 10.1083/jcb.201305148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hall M.H., Magalska A., Malinowska M., Ruszczycki B., Czaban I., Patel S., Ambrożek-Latecka M., Zołocińska E., Broszkiewicz H., Parobczak K., et al. Localization and regulation of PML bodies in the adult mouse brain. Brain Struct. Funct. 2016;221:2511–2525. doi: 10.1007/s00429-015-1053-4. [DOI] [PubMed] [Google Scholar]
- 82.Yang W., Ma Q., Mackensen G.B., Paschen W. Deep hypothermia markedly activates the small ubiquitin-like modifier conjugation pathway; implications for the fate of cells exposed to transient deep hypothermic cardiopulmonary bypass. J. Cerebr. Blood Flow Metabol. 2009;29:886–890. doi: 10.1038/jcbfm.2009.16. [DOI] [PubMed] [Google Scholar]
- 83.Wang L., Ma Q., Yang W., Mackensen G.B., Paschen W. Moderate hypothermia induces marked increase in levels and nuclear accumulation of SUMO2/3-conjugated proteins in neurons. J. Neurochem. 2012;123:349–359. doi: 10.1111/j.1471-4159.2012.07916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang W., Sheng H., Warner D.S., Paschen W. Transient global cerebral ischemia induces a massive increase in protein sumoylation. J. Cerebr. Blood Flow Metabol. 2008;28:269–279. doi: 10.1038/sj.jcbfm.9600523. [DOI] [PubMed] [Google Scholar]
- 85.Yang W., Paschen W. SUMO proteomics to decipher the SUMO-modified proteome regulated by various diseases. Proteomics. 2015;15:1181–1191. doi: 10.1002/pmic.201400298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hotz P.W., Wiesnet M., Tascher G., Braun T., Müller S., Mendler L. Profiling the Murine SUMO Proteome in Response to Cardiac Ischemia and Reperfusion Injury. Molecules. 2020;25:5571. doi: 10.3390/molecules25235571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ryu B., Nagappan S., Santos-Valencia F., Lee P., Rodriguez E., Lackie M., Takatoh J., Franks K.M. Chronic loss of inhibition in piriform cortex following brief, daily optogenetic stimulation. Cell Rep. 2021;35 doi: 10.1016/j.celrep.2021.109001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Barone P., Morelli M., Cicarelli G., Cozzolino A., DeJoanna G., Campanella G., DiChiara G. Expression of c-fos protein in the experimental epilepsy induced by pilocarpine. Synapse. 1993;14:1–9. doi: 10.1002/syn.890140102. [DOI] [PubMed] [Google Scholar]
- 89.Vertegaal A.C.O., Andersen J.S., Ogg S.C., Hay R.T., Mann M., Lamond A.I. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol. Cell. Proteomics. 2006;5:2298–2310. doi: 10.1074/mcp.M600212-MCP200. [DOI] [PubMed] [Google Scholar]
- 90.Enchev R.I., Schulman B.A., Peter M. Protein neddylation: beyond cullin-RING ligases. Nat. Rev. Mol. Cell Biol. 2015;16:30–44. doi: 10.1038/nrm3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dong Z., Chen W., Chen C., Wang H., Cui W., Tan Z., Robinson H., Gao N., Luo B., Zhang L., et al. CUL3 Deficiency Causes Social Deficits and Anxiety-like Behaviors by Impairing Excitation-Inhibition Balance through the Promotion of Cap-Dependent Translation. Neuron. 2020;105:475–490.e6. doi: 10.1016/j.neuron.2019.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hannah J., Zhou P. Distinct and overlapping functions of the cullin E3 ligase scaffolding proteins CUL4A and CUL4B. Gene. 2015;573:33–45. doi: 10.1016/j.gene.2015.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liebermann D.A., Hoffman B. Gadd45 in stress signaling. J. Mol. Signal. 2008;3:15. doi: 10.1186/1750-2187-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Canovas B., Nebreda A.R. Diversity and versatility of p38 kinase signalling in health and disease. Nat. Rev. Mol. Cell Biol. 2021;22:346–366. doi: 10.1038/s41580-020-00322-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu Y.X., Wang J., Guo J., Wu J., Lieberman H.B., Yin Y. DUSP1 is controlled by p53 during the cellular response to oxidative stress. Mol. Cancer Res. 2008;6:624–633. doi: 10.1158/1541-7786.MCR-07-2019. [DOI] [PubMed] [Google Scholar]
- 96.Rosonina E., Akhter A., Dou Y., Babu J., Sri Theivakadadcham V.S. Regulation of transcription factors by sumoylation. Transcription. 2017;8:220–231. doi: 10.1080/21541264.2017.1311829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kobow K., Ziemann M., Kaipananickal H., Khurana I., Mühlebner A., Feucht M., Hainfellner J.A., Czech T., Aronica E., Pieper T., et al. Genomic DNA methylation distinguishes subtypes of human focal cortical dysplasia. Epilepsia. 2019;60:1091–1103. doi: 10.1111/epi.14934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ouyang J., Shi Y., Valin A., Xuan Y., Gill G. Direct binding of CoREST1 to SUMO-2/3 contributes to gene-specific repression by the LSD1/CoREST1/HDAC complex. Mol. Cell. 2009;34:145–154. doi: 10.1016/j.molcel.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hua G., Paulen L., Chambon P. GR SUMOylation and formation of an SUMO-SMRT/NCoR1-HDAC3 repressing complex is mandatory for GC-induced IR nGRE-mediated transrepression. Proc. Natl. Acad. Sci. USA. 2016;113:E626–E634. doi: 10.1073/pnas.1522821113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Van Rechem C., Boulay G., Pinte S., Stankovic-Valentin N., Guérardel C., Leprince D. Differential regulation of HIC1 target genes by CtBP and NuRD, via an acetylation/SUMOylation switch, in quiescent versus proliferating cells. Mol. Cell Biol. 2010;30:4045–4059. doi: 10.1128/MCB.00582-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Escobar-Ramirez A., Vercoutter-Edouart A.S., Mortuaire M., Huvent I., Hardivillé S., Hoedt E., Lefebvre T., Pierce A. Modification by SUMOylation Controls Both the Transcriptional Activity and the Stability of Delta-Lactoferrin. PLoS One. 2015;10 doi: 10.1371/journal.pone.0129965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.O'Geen H., Squazzo S.L., Iyengar S., Blahnik K., Rinn J.L., Chang H.Y., Green R., Farnham P.J. Genome-wide analysis of KAP1 binding suggests autoregulation of KRAB-ZNFs. PLoS Genet. 2007;3:e89. doi: 10.1371/journal.pgen.0030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dehaene H., Praz V., Lhôte P., Lopes M., Herr W. THAP11F80L cobalamin disorder-associated mutation reveals normal and pathogenic THAP11 functions in gene expression and cell proliferation. PLoS One. 2020;15 doi: 10.1371/journal.pone.0224646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wei D.W., Gui L.S., Raza S.H.A., Zhang S., Khan R., Wang L., Guo H.F., Zan L.S. NRF1 and ZSCAN10 bind to the promoter region of the SIX1 gene and their effects body measurements in Qinchuan cattle. Sci. Rep. 2017;7:7867. doi: 10.1038/s41598-017-08384-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vahidi Ferdowsi P.N.,R., Adulcikas J., Sohal S.S., Myers S. Zinc Modulates Several Transcription-Factor Regulated Pathways in Mouse Skeletal Muscle Cells. Molecules. 2020;25:5908. doi: 10.3390/molecules25215098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Côté A., Chiasson M., Peralta M.R., 3rd, Lafortune K., Pellegrini L., Tóth K. Cell type-specific action of seizure-induced intracellular zinc accumulation in the rat hippocampus. J. Physiol. 2005;566:821–837. doi: 10.1113/jphysiol.2005.089458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Qian J., Xu K., Yoo J., Chen T.T., Andrews G., Noebels J.L. Knockout of Zn transporters Zip-1 and Zip-3 attenuates seizure-induced CA1 neurodegeneration. J. Neurosci. 2011;31:97–104. doi: 10.1523/JNEUROSCI.5162-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kindermann B., Döring F., Pfaffl M., Daniel H. Identification of genes responsive to intracellular zinc depletion in the human colon adenocarcinoma cell line HT-29. J. Nutr. 2004;134:57–62. doi: 10.1093/jn/134.1.57. [DOI] [PubMed] [Google Scholar]
- 109.Suh S.W., Thompson R.B., Frederickson C.J. Loss of vesicular zinc and appearance of perikaryal zinc after seizures induced by pilocarpine. Neuroreport. 2001;12:1523–1525. doi: 10.1097/00001756-200105250-00044. [DOI] [PubMed] [Google Scholar]
- 110.Lee S., Zhang A.Y., Su S., Ng A.P., Holik A.Z., Asselin-Labat M.L., Ritchie M.E., Law C.W. Covering all your bases: incorporating intron signal from RNA-seq data. NAR Genom. Bioinform. 2020;2:lqaa073. doi: 10.1093/nargab/lqaa073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Howe K.L., Achuthan P., Allen J., Allen J., Alvarez-Jarreta J., Amode M.R., Armean I.M., Azov A.G., Bennett R., Bhai J., et al. Ensembl 2021. Nucleic Acids Res. 2021;49:D884–D891. doi: 10.1093/nar/gkaa942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Song Y.J., Tian X.B., Zhang S., Zhang Y.X., Li X., Li D., Cheng Y., Zhang J.N., Kang C.S., Zhao W. Temporal lobe epilepsy induces differential expression of hippocampal miRNAs including let-7e and miR-23a/b. Brain Res. 2011;1387:134–140. doi: 10.1016/j.brainres.2011.02.073. [DOI] [PubMed] [Google Scholar]
- 113.Sun Y., Wang X., Wang Z., Zhang Y., Che N., Luo X., Tan Z., Sun X., Li X., Yang K., et al. Expression of microRNA-129-2-3p and microRNA-935 in plasma and brain tissue of human refractory epilepsy. Epilepsy Res. 2016;127:276–283. doi: 10.1016/j.eplepsyres.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 114.Wang G.Y., Luan Z.L., Che N.W., Yan D.B., Sun X.W., Zhang C., Yin J. Inhibition of microRNA-129-2-3p protects against refractory temporal lobe epilepsy by regulating GABRA1. Brain Behav. 2021;11 doi: 10.1002/brb3.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang W., Guo Y., He L., Chen C., Luo J., Ma Y., Li J., Yang Y., Yang Q., Du C., et al. Overexpression of miRNA-137 in the brain suppresses seizure activity and neuronal excitability: A new potential therapeutic strategy for epilepsy. Neuropharmacology. 2018;138:170–181. doi: 10.1016/j.neuropharm.2018.06.010. [DOI] [PubMed] [Google Scholar]
- 116.Perl K., Ushakov K., Pozniak Y., Yizhar-Barnea O., Bhonker Y., Shivatzki S., Geiger T., Avraham K.B., Shamir R. Reduced changes in protein compared to mRNA levels across non-proliferating tissues. BMC Genom. 2017;18:305. doi: 10.1186/s12864-017-3683-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Vogel C., Marcotte E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chen G., Gharib T.G., Huang C.C., Taylor J.M.G., Misek D.E., Kardia S.L.R., Giordano T.J., Iannettoni M.D., Orringer M.B., Hanash S.M., Beer D.G. Discordant protein and mRNA expression in lung adenocarcinomas. Mol. Cell. Proteomics. 2002;1:304–313. doi: 10.1074/mcp.m200008-mcp200. [DOI] [PubMed] [Google Scholar]
- 119.Lan P., Li W., Schmidt W. Complementary proteome and transcriptome profiling in phosphate-deficient Arabidopsis roots reveals multiple levels of gene regulation. Mol. Cell. Proteomics. 2012;11:1156–1166. doi: 10.1074/mcp.M112.020461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zalcman G., Federman N., Romano A. CaMKII Isoforms in Learning and Memory: Localization and Function. Front. Mol. Neurosci. 2018;11:445. doi: 10.3389/fnmol.2018.00445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wei M., Fan X., Ding M., Li R., Shao S., Hou Y., Meng S., Tang F., Li C., Sun Y. Nuclear actin regulates inducible transcription by enhancing RNA polymerase II clustering. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aay6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kapoor P., Shen X. Mechanisms of nuclear actin in chromatin-remodeling complexes. Trends Cell Biol. 2014;24:238–246. doi: 10.1016/j.tcb.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schrank B.R., Aparicio T., Li Y., Chang W., Chait B.T., Gundersen G.G., Gottesman M.E., Gautier J. Nuclear ARP2/3 drives DNA break clustering for homology-directed repair. Nature. 2018;559:61–66. doi: 10.1038/s41586-018-0237-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Maly I.V., Hofmann W.A. In: Myosins: A Superfamily of Molecular Motors. Coluccio L.M., editor. Springer International Publishing; 2020. Myosins in the Nucleus; pp. 199–231. [DOI] [Google Scholar]
- 125.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Anders S., Pyl P.T., Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhou X., Lindsay H., Robinson M.D. Robustly detecting differential expression in RNA sequencing data using observation weights. Nucleic Acids Res. 2014;42:e91. doi: 10.1093/nar/gku310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lun A.T.L., Chen Y., Smyth G.K. It's DE-licious: A Recipe for Differential Expression Analyses of RNA-seq Experiments Using Quasi-Likelihood Methods in edgeR. Methods Mol. Biol. 2016;1418:391–416. doi: 10.1007/978-1-4939-3578-9_19. [DOI] [PubMed] [Google Scholar]
- 131.Korotkevich G., Sukhov V., Budin N., Shpak B., Artyomov M.N., Sergushichev A. Fast Gene Set Enrichment Analysis. bioRxiv. 2021 doi: 10.1101/060012. Preprint at. [DOI] [Google Scholar]
- 132.Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., Tosatto S.C.E., Paladin L., Raj S., Richardson L.J., et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021;49:D412–D419. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jassal B., Matthews L., Viteri G., Gong C., Lorente P., Fabregat A., Sidiropoulos K., Cook J., Gillespie M., Haw R., et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020;48:D498–D503. doi: 10.1093/nar/gkz1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Merico D., Isserlin R., Stueker O., Emili A., Bader G.D. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Fornes O., Castro-Mondragon J.A., Khan A., van der Lee R., Zhang X., Richmond P.A., Modi B.P., Correard S., Gheorghe M., Baranašić D., et al. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2020;48:D87–D92. doi: 10.1093/nar/gkz1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.de Pretis S., Kress T., Morelli M.J., Melloni G.E.M., Riva L., Amati B., Pelizzola M. INSPEcT: a computational tool to infer mRNA synthesis, processing and degradation dynamics from RNA- and 4sU-seq time course experiments. Bioinformatics. 2015;31:2829–2835. doi: 10.1093/bioinformatics/btv288. [DOI] [PubMed] [Google Scholar]
- 140.Licursi V., Conte F., Fiscon G., Paci P. MIENTURNET: an interactive web tool for microRNA-target enrichment and network-based analysis. BMC Bioinf. 2019;20:545. doi: 10.1186/s12859-019-3105-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Alexa A., Rahnenfuhrer J. topGO: Enrichment Analysis for Gene Ontology. R package version 2.46.0; 2021. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
No new code was generated in this study.
-
•
Mass spectrometry data are found at ProteomeXchange (PXD030637) and RNA sequencing data have been deposited at GEO (GSE205454).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.