Abstract
Background
We sought to develop a proteomics-based risk model for lung cancer and evaluate its risk-discriminatory performance in comparison with a smoking-based risk model (PLCOm2012) and a commercially available autoantibody biomarker test.
Methods
We designed a case-control study nested in 6 prospective cohorts, including 624 lung cancer participants who donated blood samples at most 3 years prior to lung cancer diagnosis and 624 smoking-matched cancer free participants who were assayed for 302 proteins. We used 470 case-control pairs from 4 cohorts to select proteins and train a protein-based risk model. We subsequently used 154 case-control pairs from 2 cohorts to compare the risk-discriminatory performance of the protein-based model with that of the Early Cancer Detection Test (EarlyCDT)-Lung and the PLCOm2012 model using receiver operating characteristics analysis and by estimating models’ sensitivity. All tests were 2-sided.
Results
The area under the curve for the protein-based risk model in the validation sample was 0.75 (95% confidence interval [CI] = 0.70 to 0.81) compared with 0.64 (95% CI = 0.57 to 0.70) for the PLCOm2012 model (Pdifference = .001). The EarlyCDT-Lung had a sensitivity of 14% (95% CI = 8.2% to 19%) and a specificity of 86% (95% CI = 81% to 92%) for incident lung cancer. At the same specificity of 86%, the sensitivity for the protein-based risk model was estimated at 49% (95% CI = 41% to 57%) and 30% (95% CI = 23% to 37%) for the PLCOm2012 model.
Conclusion
Circulating proteins showed promise in predicting incident lung cancer and outperformed a standard risk prediction model and the commercialized EarlyCDT-Lung.
Lung cancer is the leading cause of cancer death globally (1). In 2011, the National Lung Screening Trial in the United States demonstrated that screening high-risk individuals with low-dose computed tomography (LDCT) can reduce lung cancer mortality through early detection (2). This finding has since been replicated in several randomized trials (3-6). Currently, the US Preventive Services Task Force (USPSTF) guideline recommends annual LDCT screening for individuals aged 50-80 years who have smoked at least 20 pack-years and either currently smoke or have quit within the last 15 years (7). However, these eligibility criteria leave a large proportion of incident lung cancer cases ineligible for LDCT screening (8).
Risk biomarkers may be useful as a pre-LDCT screening eligibility test if they improve risk assessment. Proposed biomarkers span multiple domains including proteins, microRNAs, autoantibodies, and methylation of circulating tumor DNA (9-14). We recently reported that the commercialized autoantibody-based Early Cancer Detection Test (EarlyCDT)-Lung was not useful in predicting incident lung cancer based on prediagnostic samples from individuals with any history of regular smoking (15). However, our previous pilot study suggested that a panel of circulating proteins can improve lung cancer risk discrimination of a smoking-based risk model (16).
One of the key benefits of a risk-informative, biomarker-based prescreening test would be to identify individuals who are at high lung cancer risk despite not meeting current eligibility criteria. The current study aimed to evaluate if a preliminary proteomics-based, risk-prediction model can improve the lung cancer risk-discriminatory performance of the EarlyCDT-Lung autoantibody test and the PLCOm2012 model in a study sample reflecting the intended use population after age, sex, and smoking status is taken into account. We used prediagnostic samples and smoking-matched cancer free participants from 6 prospective cohorts participating in the Lung Cancer Cohort Consortium (LC3). This allowed us to study individuals from the entire spectrum of lung cancer risk experienced by the general population with a history of smoking.
Methods
Study design and study sample
This study focused on risk biomarkers used among individuals with a history of smoking in the general population, and we used prediagnostic blood samples from 6 population cohorts participating in the LC3 consortium. A detailed description and justification of the study design and the included cohorts were provided by Robbins et al. (17).
To train a preliminary protein-based prediction model, we used data from the discovery phase of the Integrative Analysis of Lung Cancer Etiology and Risk (INTEGRAL) project (18), including 478 case-control pairs from the Cancer Prevention Study II (CPS-II, USA, 115 case-control pairs), the Trøndelag Health Study (HUNT, Norway, 163 case-control pairs), the Melbourne Collaborative Cohort Study (MCCS, Australia, 108 case-control pairs), and the Singapore Chinese Health Study (SCHS, Singapore, 92 case-control pairs). To evaluate the risk-discriminative performance of the EarlyCDT-Lung and the preliminary protein-based risk model, we analyzed 154 case-control pairs from the European Investigation into Cancer and Nutrition (EPIC, Europe, 90 case-control pairs) and the Northern Sweden Health and Disease Study (NSHDS, Sweden, 64 case-control pairs).
We first identified incident lung cancer with a history of regular smoking (International Classification of Diseases code: C34) in each cohort who were diagnosed at most 3 years after donating their blood samples. For each lung cancer participant, 1 cancer free participant was randomly selected using incidence density sampling from risk sets consisting of all cohort participants alive and free of cancer (except nonmelanoma skin cancer) at the time of diagnosis of the index case. Matching criteria included cohort, study center, sex, date of blood collection, date of birth, smoking status, quit years in 2 categories for former smokers (<10 and ≥10 years since quitting), and intensity in 2 categories for current smokers (<15 and ≥15 cigarettes smoked per day). A detailed description was provided by Robbins et al. (17)
This study was approved by the Ethics Committee of the International Agency for Research on Cancer. The ethics approval title was “Biomarkers of lung cancer risk (LC3)” (No. 11-13). Informed consent from all participants was obtained in each cohort.
Proteomics assays
We used the Olink Proteomics discovery platform at the Olink core facility in Uppsala (Sweden) to measure circulating proteins. Relative concentrations of proteins were measured by quantitative polymerase chain reaction. Measurements expressed as normalized protein expression values on log-base-2 scale, which were derived from the cycle threshold (Ct) values obtained from the quantitative polymerase chain reaction. The INTEGRAL project measured 1161 proteins in the EPIC and NSHDS cohorts and between 392 and 484 proteins in the remaining 4 cohorts (17). For the present study, we only considered 302 proteins that were assayed on the 6 discovery cohorts, including the Cardiovascular III, Inflammation, Immuno-oncology, and Oncology II panels. We replaced protein values below the limit of detection with the limit of detection divided by the square root of 2 and rescaled each protein to a mean of 0 and a standard deviation of 1 within each cohort. We excluded proteins with missing values in greater than 10% of participants (Interleukin-2, IL2) and imputed missing values as mean values for the remaining proteins. Participants, including their paired participants, with missing data for greater than 10% of proteins were excluded (8 pairs: CPS, 1; HUNT, 1; MCCS, 4; SCHS, 2).
EarlyCDT-Lung assays
EarlyCDT-Lung was assayed at Umeå University (Sweden) using kits produced by Oncimmune (Nottingham, UK) according to the manufacturer’s protocol. The detailed protocol could be found in the previous article (15). In brief, 7 antigens (CAGE, GBU4-5, HuD, MAGE A4, NY-ESO-1, p53, and SOX2) and a control protein (VOL) were measured. The test results were classified as “no significant level,” “moderate level,” and “high level” according to the highest reading acquired among each of the 7 autoantibody markers.
Statistical analyses
Development of a preliminary protein-based risk prediction model
We used the development dataset to identify a set of risk informative proteins and train a preliminary protein-based risk model (CPS-II, HUNT, MCCS, and SCHS). We initially evaluated the association between each protein and lung cancer risk using logistic regression models with adjustment for matching factors (cohort, age, sex, year of blood collection, smoking status, smoking intensity, quit years for former smokers). The effective number of test method was used to account for multiple tests (19): we considered proteins associated with lung cancer risk if their P value was less than .05 divided by the number of principal components needed to explain 95% of the variance in proteins (ie, P < .05/nent). We subsequently applied the least absolute shrinkage and selection operator (LASSO) logistic regression model based on all proteins identified as being associated with lung cancer risk. First, we used tenfold cross-validation to confirm a suitable shrinkage parameter (λ). Second, we randomly generated 500 different datasets in which 75% of case-control pairs were included as the training set. For each training set, we applied the LASSO logistic regression models adjusted for matching factors, including age, sex, year of blood collection, smoking status, smoking intensity, and quit years for former smokers, as well as the PLCOm2012 model. We defined the final set of risk-informative proteins for inclusion in the preliminary protein-based risk model as those selected in at least 400 of the 500 training sets.
We subsequently developed a preliminary risk model in each training set using logistic regression, the protein-based risk model that included the selected proteins and matching factors. The remaining 500 (25%) case-control pairs were used to generate corresponding bootstrap-corrected risk discrimination estimates by averaging the area under curve (AUC) across the 500 random draws. The full development set was used to build the final model and generate the apparent AUCs as internal model validity metrics.
Discrimination analyses in an external validation sample
We applied the PLCOm2012 and the protein-based risk prediction models on the validation sample (EPIC and NSHDS combined) to estimate the AUC for each model, with adjustment for the matching factors. Differences between receiver operating characteristics curves were evaluated using paired comparison with the bootstrap method (R pROC package). To compare the validity of the EarlyCDT-Lung with the risk prediction models, we first calculated the sensitivity and specificity of EarlyCDT-Lung using the moderate threshold, which is recommended for clinical practice. We subsequently identified the model cutoff for each respective risk model that yielded the same specificity as the EarlyCDT-Lung. Differences between sensitivity estimates were evaluated using McNemar test. Stratified analyses were conducted by age, sex, smoking status, lead time, tumor-node-metastasis stage, eligibility by USPSTF screening criteria (7), and PLCOm2012 high-risk threshold (1.00%) (20).
Software used for statistical analyses
Statistical analyses were performed with the statistical software R version 4.0.4. The packages we used are listed in the Supplementary Methods (available online). All tests were 2-sided, and the cutoff to reject the null hypothesis was .05.
Results
Baseline characteristics
The final analysis included 624 lung cancer participants and 624 paired cancer-free participants with measurements of 301 proteins; 470 pairs of lung cancer participants and cancer-free participants were included from the development cohorts (HUNT, SCHS, CPS-II, and MCCS), and 154 pairs of lung cancer participants and cancer-free participants were included from the validation cohorts (EPIC and NSHDS). Compared with participants in the development cohorts, participants in the validation cohorts were younger and more frequently female and participants who currently smoke. According to the USPSTF lung cancer screening criteria (7), 60% of lung cancer participants and 50% of cancer-free participants in the development cohorts and 57% of lung cancer participants and 48% of cancer-free participants in the validation cohorts were eligible for screening. According to the PLCOm2012 model, the median 6-year risk of developing lung cancer was 2.4% for lung cancer participants and 1.5% for cancer-free participants in the development cohorts and 0.98% for lung cancer participants and 0.69% for cancer-free participants in the validation cohorts (Table 1).
Table 1.
Characteristics of the study participants
| Development set |
Validation set |
|||
|---|---|---|---|---|
| Characteristics | Lung cancer participants | Cancer-free participants | Lung cancer participants | Cancer-free participants |
| Total No. of participants | 470 | 470 | 154 | 154 |
| Female participants, No. (%) | 145 (30.9) | 145 (30.9) | 62 (40.3) | 62 (40.3) |
| Median age (Q1-Q3), y | 69 (64-74) | 69 (64-74) | 60 (53-60) | 59 (53-60) |
| Median BMI (Q1-Q3), kg/m2 | 25 (23-28) | 26 (23-29) | 25 (23-28) | 26 (23-29) |
| Prediagnosis lead time, y | ||||
| Mean (SD) | 1.5 (0.9) | — | 1.7 (0.9) | — |
| Median (Q1-Q3) | 1.5 (0.75-2.2) | — | 1.9 (0.98-2.5) | — |
| Smoking characteristics, No. (%) | ||||
| Former smokers | 246 (52.3) | 242 (51.5) | 54 (35.1) | 56 (36.4) |
| Current smokers | 224 (47.7) | 228 (48.5) | 100 (64.9) | 98 (63.6) |
| No. cigarettes smoked per day, median (Q1-Q3) | 20 (10-30) | 15 (9.5-20) | 15 (10-20) | 13 (9.4-19) |
| Years smoked, median (Q1-Q3) | 43 (34-50) | 40 (27-48) | 37 (31-43) | 35 (26-42) |
| Quit years, median (Q1-Q3)a | 15 (6.3-26) | 18 (6.8-32) | 11 (3.7-19) | 10 (5.4-24) |
| Participating cohorts, No. (%) | ||||
| CPS | 114 (24.3) | 114 (24.3) | — | — |
| HUNT | 162 (34.5) | 162 (34.5) | — | — |
| MCCS | 104 (22.1) | 104 (22.1) | — | — |
| SCHS | 90 (19.1) | 90 (19.1) | — | — |
| EPIC | — | — | 90 (58.4) | 90 (58.4) |
| NSHDS | — | — | 64 (41.6) | 64 (41.6) |
| TNM stage, No. (%) | ||||
| I-II | 49 (10.4) | — | 19 (12.3) | — |
| III | 73 (15.5) | — | 26 (16.9) | — |
| IV | 94 (20.0) | — | 30 (19.5) | — |
| Unknown or missing | 254 (54.1) | — | 79 (51.3) | — |
| Histology, No. (%) | ||||
| Adenocarcinoma | 161 (34.3) | — | 50 (32.5) | — |
| Small cell carcinoma | 74 (15.7) | — | 24 (15.6) | — |
| Squamous cell carcinoma | 103 (21.9) | — | 26 (16.9) | — |
| Other/NOS | 132 (28.1) | — | 54 (35.1) | — |
| Eligible for lung cancer screening, USPSTF, No. (%) | 279 (59.4) | 234 (49.8) | 87 (56.5) | 74 (48.1) |
| 6-year risk by PLCOm2012 model, median (Q1-Q3), % | 2.4 (1.1-4.6) | 1.5 (0.51-4.0) | 0.98 (0.46-1.9) | 0.69 (0.25-1.3) |
| Eligible for lung cancer screening, PLCOm2012, cutoff: 1.00%, No. (%) | 359 (76.4) | 287(61.1) | 76 (49.4) | 61 (39.6) |
Only former smokers. “—” signifies information is not available. BMI = body mass index; CPS = Cancer Prevention Study; EPIC = European Investigation into Cancer and Nutrition; HUNT = Trøndelag Health Study; MCCS = Melbourne Collaborative Cohort Study; NSHDS = Northern Sweden Health and Disease Study; SCHS = Singapore Chinese Health Study; TNM = tumor-node-metastasis; NOS = not otherwise specified; USPSTF = US Preventive Services Task Force; Q = quintile.
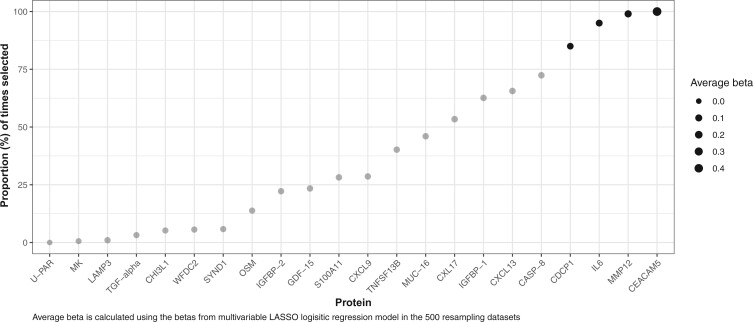
Selection of protein markers and training of protein-based risk models
We identified 22 proteins associated with lung cancer risk after correction for multiple testing in the development set. Four protein markers, including carcinoembryonic antigen-related cell adhesion molecule 5 (CEACAM5), macrophage metalloelastase (MMP12), interleukin 6 (IL6), and CUB domain-containing protein 1 (CDCP1), were selected by multivariable LASSO logistic regression in at least 400 of 500 training sets (Figure 1) and were, thus, included in the final protein-based risk models. Table 2 shows the β-coefficients for each model. Because the cancer-free participants were individually matched to the lung cancer participants by age, sex, and smoking status, all AUC estimates reflect the residual the risk-discriminatory performance of each risk model after accounting for the matching factors. The AUC for the PLCOm2012 model alone was 0.61 (95% confidence interval [CI] = 0.57 to 0.65). The training-sample AUC for the protein-based risk model was estimated at 0.72 (95% CI = 0.69 to 0.75; bootstrap-corrected AUC = 0.71) (Table 2).
Figure 1.
Proportion of proteins selected in 500 training datasets by LASSO logistic regression model. Proteins selected more than 400 times are marked as black. LASSO = least absolute shrinkage and selection operator; CEACAM5 = carcinoembryonic antigen-related cell adhesion molecule 5; MMP12 = macrophage metalloelastase; IL6 = interleukin 6; CDCP1 = CUB domain-containing protein 1; CASP-8 = caspase-8; CXCL13 = C-X-C motif chemokine 13; IGFBP-1 = insulin-like growth factor-binding protein 1; CXL17 = C-X-C motif chemokine 17; MUC-16 = mucin-16; TNFSF13B = tumor necrosis factor ligand superfamily member 13B; CXCL9 = C-X-C motif chemokine 9; S100A11 = Protein S100-A11; GDF-15 = growth/differentiation factor 15; IGFBP-2 = insulin-like growth factor-binding protein 2; OSM = oncostatin-M; SYND1 = syndecan-1; WFDC2 = WAP four-disulfide core domain protein 2; CHI3L1 = chitinase-3-like protein 1; TGF-alpha = transforming growth factor alpha; LAMP3 = lysosome-associated membrane glycoprotein 3; MK = midkine; U-PAR = urokinase plasminogen activator surface receptor.
Table 2.
β-coefficients and multivariable odds ratios (ORs) with 95% confidence intervals (CIs) for lung cancer risk factors in the development set
| PLCOm2012 modela |
Protein-based risk modela |
|||
|---|---|---|---|---|
| β | OR (95% CI) | β | OR (95% CI) | |
| Predictors in models | ||||
| Logit of PLCOm2012 model | 0.15 | 1.16 (1.08 to 1.26) | — | — |
| CEACAM5, per SD | — | — | 0.79 | 2.21 (1.81 to 2.74) |
| MMP12, per SD | — | — | 0.34 | 1.40 (1.18 to 1.67) |
| IL6, per SD | — | — | 0.24 | 1.27 (1.09 to 1.48) |
| CDCP1, per SD | — | — | 0.2 | 1.23 (1.05 to 1.44) |
| Model performanceb | ||||
| Apparent, AUC (95% CI) | 0.61 (0.57 to 0.65) | 0.72 (0.69 to 0.75) | ||
| Bootstrap-corrected, AUC (SD) | 0.60 (0.03) | 0.71 (0.03) | ||
Models were adjusted by matching factors: cohort, sex, year of blood collection, age, and smoking status (former smokers with <10 or ≥10 years since quitting and current smokers with <15 or ≥15 cigarettes smoked per day). “—” signifies no values were given because factors are not included in the corresponding models. AUC = area under the curve; CEACAM5 = carcinoembryonic antigen-related cell adhesion molecule 5; MMP12 = macrophage metalloelastase; IL6 = interleukin 6; CDCP1 = CUB domain-containing protein 1.
The AUC estimates reflect the residual risk-discriminatory performance of the risk models after accounting for age, sex, and smoking status (matching factors in the case-control study).
Risk-discriminative performance in the validation sample
When standardizing the risk score from the protein-based risk model (mean = 0 [1]), we found that the odds ratio (OR) for lung cancer was 3.43 (95% CI = 2.43 to 5.00) per 1 standard deviation increase (Table 3).
Table 3.
Odds ratios (ORs) and 95% confidence intervals (CIs) for lung cancer in relation to protein-based risk model in the validation set, scores of the model were scaled as mean = 0 (1)
| Groups | No. of lung cancer participants | No. of cancer-free participants | Protein-based risk modela |
|---|---|---|---|
| Per 1-SD increase | |||
| OR (95% CI) | |||
| Overall | 154 | 154 | 3.43 (2.43 to 5.00) |
| Age, y | |||
| Younger than 60 | 91 | 98 | 3.06 (2.02 to 4.88) |
| 60 or older | 63 | 56 | 3.47 (2.02 to 6.43) |
| Sex | |||
| Male | 92 | 92 | 2.81 (1.84 to 4.51) |
| Female | 62 | 62 | 5.07 (2.77 to 10.4) |
| Smoking status | |||
| Former | 54 | 56 | 6.12 (3.07 to 14.0) |
| Current | 100 | 98 | 2.78 (1.87 to 4.30) |
| Prediagnosed lead-time lung cancer participants and all cancer-free participants, y | |||
| <1.5 | 63 | 154 | 4.28 (2.70 to 7.30) |
| ≥1.5 | 91 | 154 | 3.28 (2.18 to 5.12) |
| TNM stage and all cancer-free participants | |||
| I-II | 19 | 154 | 6.83 (2.84 to 19.7) |
| III-IV | 56 | 154 | 4.91 (2.87 to 9.12) |
| Unknown or missing | 79 | 154 | 2.95 (1.96 to 4.65) |
| USPSTF 2020 | |||
| Yes | 87 | 74 | 3.19 (1.99 to 5.41) |
| No | 67 | 80 | 3.73 (2.21 to 6.90) |
| PLCOm2012, threshold: 1.00% | |||
| Yes, mean risk: 2.58% | 76 | 61 | 2.65 (1.65 to 4.48) |
| No, mean risk: 0.41% | 78 | 93 | 4.22 (2.53 to 7.61) |
Model was adjusted by matching factors: cohort, sex, year of blood collection, age, and smoking status (former smokers with <10 or ≥10 years since quitting, and current smokers with <15 or ≥15 cigarettes smoked per day). TNM = tumor-node-metastasis; USPSTF = US Preventive Services Task Force.
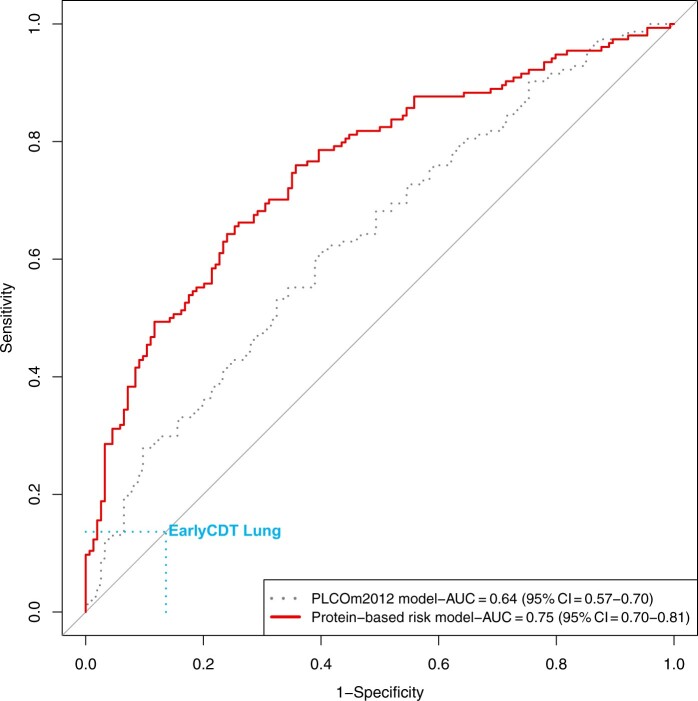
The overall AUC for the PLCOm2012 model in the validation sample was 0.64 (95% CI = 0.57 to 0.70), reflecting the residual risk-discriminatory performance after accounting for age, sex, and smoking status. When applying the protein-based risk models in the validation sample, the AUC was estimated at 0.75 (95% CI = 0.70 to 0.81, protein-based model vs PLCOm2012 model; Pdifference = .001) (Figure 2). The AUC estimates for the protein-based model appeared higher in strata with lower risk of lung cancer (Table 4). The risk-discriminatory performance for the protein biomarker-based risk model was higher in lung cancer participants who donated blood samples within 1.5 years of diagnosis (AUC = 0.79) compared with samples diagnosed after 1.5 years (AUC = 0.73).
Figure 2.
Comparison of ROC curves for the PLCOm2012 model and protein-based risk model in the validation set. The ROCs and associated AUC estimates reflect the residual risk-discriminatory performance of the risk models after accounting for age, sex, and smoking status (matching factors in the case-control study). AUC = area under the curve; EarlyCDT = Early Cancer Detection Test; ROC = receiver operating characteristics.
Table 4.
AUC of PLCOm2012 model and protein-based risk model in the validation seta
| Groups | No. of lung cancer participants | No. of cancer-free participants | PLCOm2012 model | Protein-based risk model |
|---|---|---|---|---|
| AUC (95% CI) | AUC (95% CI) | |||
| Overall | 154 | 154 | 0.64 (0.57 to 0.70) | 0.75 (0.70 to 0.81) |
| Age, y | ||||
| Younger than 60 | 91 | 98 | 0.62 (0.54 to 0.70) | 0.76 (0.69 to 0.83) |
| 60 and older | 63 | 56 | 0.65 (0.55 to 0.75) | 0.74 (0.65 to 0.83) |
| Sex | ||||
| Males | 92 | 92 | 0.65 (0.57 to 0.73) | 0.71 (0.63 to 0.78) |
| Females | 62 | 62 | 0.62 (0.52 to 0.72) | 0.81 (0.74 to 0.89) |
| Smoking status | ||||
| Former | 54 | 56 | 0.72 (0.62 to 0.81) | 0.81 (0.73 to 0.90) |
| Current | 100 | 98 | 0.58 (0.50 to 0.66) | 0.72 (0.65 to 0.79) |
| Prediagnosed lead-time lung cancer participants and all cancer-free participants, y | ||||
| <1.5 | 63 | 154 | 0.62 (0.53 to 0.70) | 0.79 (0.72 to 0.86) |
| ≥1.5 | 91 | 154 | 0.65 (0.58 to 0.72) | 0.73 (0.66 to 0.80) |
| TNM stage and all cancer-free participants | ||||
| I-II | 19 | 154 | 0.56 (0.42 to 0.71) | 0.76 (0.66 to 0.87) |
| III-IV | 56 | 154 | 0.61 (0.52 to 0.69) | 0.78 (0.71 to 0.86) |
| Unknown or missing | 79 | 154 | 0.67 (0.60 to 0.74) | 0.73 (0.66 to 0.80) |
| USPSTF 2020 | ||||
| Yes | 87 | 74 | 0.58 (0.49 to 0.67) | 0.72 (0.64 to 0.80) |
| No | 67 | 80 | 0.66 (0.58 to 0.75) | 0.78 (0.70 to 0.86) |
| PLCOm2012, threshold: 1.00% | ||||
| Yes, mean risk: 2.58% | 76 | 61 | 0.63 (0.54 to 0.73) | 0.70 (0.61 to 0.79) |
| No, mean risk: 0.41% | 78 | 93 | 0.63 (0.55 to 0.71) | 0.79 (0.71 to 0.86) |
The AUC estimates reflect the residual risk-discriminatory performance of the risk models after accounting for age, sex, and smoking status (matching factors). AUC = area under the curve; TNM = tumor-node-metastasis; USPSTF = US Preventive Services Task Force.
In the overall validation sample, the EarlyCDT-Lung gave positive results for 21 lung cancer participants and 21 cancer-free participants, yielding a sensitivity of 14% (95% CI = 8.2% to 19%) and a specificity of 86% (95% CI = 81% to 92%) (Table 5), which corresponds to a Youden index of 0. To allow direct comparisons between the models, we estimated the sensitivity according to the specificity defined by the EarlyCDT-Lung. The corresponding sensitivity (ie, at a specificity of 86%) for the protein-based model was 49% (95% CI = 41% to 57%; Pdifference in sensitivity to EarlyCDT = 4 × 10-10) and 30% (95% CI = 23% to 37%; Pdifference in sensitivity to protein-based model = 5 × 10-4) for the PLCOm2012 model. The sensitivity for the protein-based model was higher than that for EarlyCDT-Lung across all evaluated risk strata (Table 5).
Table 5.
Comparison of diagnostic performance of EarlyCDT-Lung, PLCOm2012 model, and protein-based risk model in the validation set
| Groups | No. of lung cancer participants | No. of cancer-free participants | EarlyCDT-Lung |
PLCOm2012 model |
Protein-based risk model |
||
|---|---|---|---|---|---|---|---|
| Sensitivity (95% CI) | P b | Sensitivitya (95% CI) | P c | Sensitivitya (95% CI) | |||
| Overall | 154 | 154 | 14% (8.2 to 19) | 4 × 10−10 | 30% (23 to 37) | 5 × 10−4 | 49% (41 to 57) |
| Age, y | |||||||
| Younger than 60 | 91 | 98 | 14% (7.1 to 21) | 6 × 10−6 | 26% (17 to 35) | .002 | 49% (39 to 60) |
| 60 and older | 63 | 56 | 13% (4.5 to 21) | 6 × 10−6 | 35% (23 to 47) | .06 | 51% (38 to 63) |
| Sex | |||||||
| Male | 92 | 92 | 12% (5.3 to 19) | 4 × 10−6 | 32% (22 to 41) | .05 | 46% (35 to 56) |
| Female | 62 | 62 | 16% (7.0 to 25) | 1 × 10−5 | 19% (9.5 to 29) | 4 × 10−5 | 56% (44 to 69) |
| Smoking status | |||||||
| Former | 54 | 56 | 19% (8.2 to 29) | 4 × 10−5 | 33% (21 to 46) | .02 | 56% (42 to 69) |
| Current | 100 | 98 | 11% (4.9 to 17) | 6 × 10−7 | 26% (17 to 35) | .004 | 46% (36 to 56) |
| Prediagnosed lead-time lung cancer participants and all cancer-free participants, y | |||||||
| <1.5 | 63 | 154 | 14% (5.6 to 23) | 3 × 10−6 | 29% (17 to 40) | .002 | 57% (45 to 69) |
| ≥1.5 | 91 | 154 | 13% (6.2 to 20) | 2 × 10−5 | 31% (21 to 40) | .06 | 44% (34 to 54) |
| TNM stage and all cancer-free participants | |||||||
| I-II | 19 | 154 | 21% (2.7 to 39) | .096 | 26% (6.5 to 46) | .21 | 47% (25 to 70) |
| III-IV | 56 | 154 | 11% (2.6 to 19) | 1 × 10−5 | 25% (14 to 36) | .02 | 52% (39 to 65) |
| Unknown or missing | 79 | 154 | 14% (6.3 to 22) | 2 × 10−5 | 34% (24 to 45) | .09 | 48% (37 to 59) |
| USPSTF 2020 | |||||||
| Yes | 87 | 74 | 14% (6.5 to 21) | 2 × 10−4 | 23% (14 to 32) | .02 | 40% (30 to 51) |
| No | 67 | 80 | 13% (5.3 to 22) | 3 × 10−7 | 37% (26 to 49) | .008 | 58% (46 to 70) |
| PLCOm2012, threshold: 1.00% | |||||||
| Yes, mean risk: 2.58% | 76 | 61 | 16% (7.6 to 24) | .01 | 38% (27 to 49) | .49 | 33% (22 to 43) |
| No, mean risk: 0.41% | 78 | 93 | 12% (4.4 to 19) | 3 × 10−7 | 26% (16 to 35) | 5 × 10−4 | 54% (43 to 65) |
Sensitivities for the PLCOm2012 model and protein-based risk model were estimated by adjusting the cutoff of each respective risk model that yielded the same specificity as the EarlyCDT-Lung, which was estimated at 86% in the overall smoking–matched control population and varied between 84% and 90% depending on the strata. EarlyCDT-Lung = Early Cancer Detection Test; TNM = tumor-node-metastasis; USPSTF = US Preventive Services Task Force
P value for the sensitivity difference between protein-based risk model and EarlyCDT-Lung at the same specificity level.
P value for the sensitivity difference between protein-based risk model and PLCOm2012 model at the same specificity level.
Discussion
We developed and externally evaluated a protein-based prediction tool derived from high-throughput proteomics data using prediagnostic samples. Our study sample captured the entire spectrum of lung cancer risk of individuals with a history of smoking. Based on lung cancer participants and cancer-free participants nested in 4 prospective cohorts, we developed a 4-marker protein-based model with subsequent validation in 2 independent prospective cohorts. The protein-based model performed well in the validation sample and outperformed the EarlyCDT-Lung and PLCOm2012 model in relevant strata.
The protein-based risk model demonstrated good discrimination between the lung cancer participants and smoking-matched cancer-free participants with an overall AUC of 0.75. Importantly, because the cancer-free participants were individually matched to the lung cancer participants by smoking, age, and sex, the risk-discriminative performance afforded by those important risk factors was accounted for by design, thus substantially attenuating our AUC estimates for PLCOm2012 model and biomarker-based models compared with that expected in a randomly selected study sample (21). Conversely, this study design provided more granularity for comparing AUC estimates between risk models. We found that the AUC for the protein-based risk model was higher in lung cancer participants who donated their blood closer to diagnosis (lead-time <1.5 years: 0.79; lead-time ≥1.5 years: 0.73) (Table 4). These results were expected considering the relatively short lead-time of up to 3 years and are in line with the hypothesis that the protein-to-lung cancer-risk associations reflect systemic response to a yet-to-be diagnosed lung cancer, rather than being generic risk markers (18).
The protein-based risk model included 4 specific markers: CEACAM5, MMP12, IL6, and CDCP1. CEACAM5 is an oncofetal protein, member of the immunoglobulin family that is usually overexpressed in several cancer types, including lung cancer. CEACAM5 has been applied in several biomarker panels for lung cancer prediction (22-24). MMP12 belongs to a family of zinc-dependent proteases that are involved in the degradation of extracellular matrix components and is secreted by inflammatory macrophages (25), which has been reported to be involved in the modulation of extracellular matrix during lung cancer metastasis (26). IL6 and CDCP1 are related to the immune system and inflammation (27). We have previously demonstrated that IL6, as well as CDCP1, is associated with lung cancer risk several years before cancer onset (27,28).
As recently reported by Wu et al. (15) and the German LDCT Lung Cancer Screening Intervention study (29), the poor performance of the EarlyCDT-Lung was notable. The sensitivity for the protein-based risk model was markedly higher (49%) than that of the EarlyCDT-Lung (14%) at the same specificity (86%). We observed some evidence that the protein-based risk model performed better in study participants at lower risk of lung cancer, whereas the EarlyCDT-Lung tended to perform better for study participants at higher risk (Tables 4 and 5), an observation that was also reported in the Danish study for the EarlyCDT-Lung (30). The EarlyCDT-Lung was developed in a series of 10 peer-reviewed studies (31-40), including prospective studies of patients on suspicion of lung cancer or heavy smokers (34,37). The poor performance of the EarlyCDT-Lung may, at least partly, reflect that it was developed and tested on high-risk individuals.
As highlighted previously, whereas our matched study design does not provide risk-discrimination metrics reflecting the expected model performance in a random study sample, the study design provides a valid and efficient means to compare the performance of different risk models. The final phase of the INTEGRAL project will use a case-cohort study design among people who ever smoked of more than 1500 lung cancer participants and 3000 cancer-free participants from the LC3 consortium to provide the absolute risk models that can be implemented in clinical practice to assess LDCT screening eligibility (17). We therefore emphasize that the protein-based models presented here are preliminary, and the final selection of proteins, model parameters, and model-based risk-discriminative performance may change in the forthcoming case-cohort analysis. The second potential concern is that the protein panels used in the study were selected from a discovery analysis in the EPIC and NSHDS studies, which may slightly bias the model performance in these cohorts. However, LASSO and round-robin sensitivity analysis indicated that this potential bias was minimal (Supplementary Figures 1 and 2; Supplementary Tables 2 and 3, available online). Another limitation concerns the sample size for stratified analysis. Whereas our overall study sample was relatively large considering the 3-year lead-time restriction, more than 50% of the incident lung cancer cases lacked data on clinical tumor-node-metastasis stage, thus limiting our ability to provide robust discrimination estimates by stage, as well as for histological subtypes (Supplementary Tables 4 and 5, available online). We also lacked information on chronic obstructive pulmonary disease, family history of cancer, and personal history of cancer; although we imputed for the current data (Supplementary Methods, Supplementary Table 1, available online), it may slightly decrease the risk discriminatory performance of the PLCOm2012 model. The key strength of our study is the direct comparison of 2 different marker panels in the same population with use of prediagnostic samples drawn up to 3 years prior to lung cancer diagnosis, along with the use of independent development and validation samples. Considering the 3-year lag-time restriction, our sample size was relatively large, and study samples originating from Europe, North America, Asia, and Australia ensured external validity of our findings.
More generally, we would argue that any biomarker intended for use in informing screening eligibility should be developed with the aim to identify the large number of lung cancer cases who are currently not eligible for LDCT screening. Including prediagnostic samples from low-risk people is crucially important throughout the development and validation of such a biomarker. To this end, the preliminary protein-based prediction model assessed in our study had promising performance characteristics that warrant further evaluation in a larger study sample. Ultimately, this should include an evaluation of whether the benefits in risk-discriminative performance outweighs the cost and inconvenience inherent to using a blood-based risk-assessment tool compared with a standard risk model such the PLCOm2012.
We developed a preliminary protein-based model and externally validated its performance in discriminating future lung cancer participants and smoking matched cancer-free participants. Based on prediagnostic blood samples drawn up to 3 years prior to lung cancer diagnosis from population cohorts, we found that the protein-based risk model showed promising risk-discriminative performance, both in comparison with the EarlyCDT-Lung and the PLCOm2012 risk model.
Supplementary Material
Acknowledgements
The Trøndelag Health Study (HUNT) is a collaboration between HUNT Research Centre (Faculty of Medicine and Health Sciences, Norwegian University of Science and Technology NTNU), Trøndelag County Council, Central Norway Regional Health Authority, and the Norwegian Institute of Public Health.
The authors express sincere appreciation to all Cancer Prevention Study-II participants and to each member of the study and biospecimen management group. The authors would like to acknowledge the contribution to this study from central cancer registries supported through the Centers for Disease Control and Prevention’s National Program of Cancer Registries and cancer registries supported by the National Cancer Institute’s Surveillance Epidemiology and End Results Program.
The Singapore Chinese Health Study was supported by the US National Institutes of Health Grant No. R01CA080205, R01CA144034 and UM182876.
Melbourne Collaborative Cohort Study (MCCS) cohort recruitment was funded by VicHealth and Cancer Council Victoria. The MCCS was further augmented by Australian National Health and Medical Research Council grants 209057, 396414, and 1074383 and by infrastructure provided by Cancer Council Victoria.
We thank the Biobank Research Unit at Umeå University, Västerbotten Intervention Programme, the Northern Sweden MONICA study, the Mammography Study and Region Västerbotten for providing data and samples and acknowledge the contribution from Biobank Sweden, supported by the Swedish Research Council (VR 2017-00650).
The coordination of EPIC was financially supported by Direction Générale de la Santé (French Ministry of Health) (Grant GR-IARC-2003-09-12-01), the European Commission (Directorate General for Health and Consumer Affairs), International Agency for Research on Cancer (IARC) and by the Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London with additional infrastructure support provided by the NIHR Imperial Biomedical Research Centre (BRC). The national cohorts are supported by: Danish Cancer Society (Denmark); Ligue Contre le Cancer, Institut Gustave Roussy, Mutuelle Générale de l’Education Nationale, Institut National de la Santé et de la Recherche Médicale (INSERM) (France); German Cancer Aid, German Cancer Research Center (DKFZ), German Institute of Human Nutrition Potsdam-Rehbruecke (DIfE), Federal Ministry of Education and Research (BMBF) (Germany); Associazione Italiana per la Ricerca sul Cancro-AIRC-Italy, Compagnia di SanPaolo and National Research Council (Italy); Dutch Ministry of Public Health, Welfare and Sports (VWS), Netherlands Cancer Registry (NKR), LK Research Funds, Dutch Prevention Funds, Dutch ZON (Zorg Onderzoek Nederland), World Cancer Research Fund (WCRF), Statistics Netherlands (The Netherlands); Health Research Fund (FIS) - Instituto de Salud Carlos III (ISCIII), Regional Governments of Andalucía, Asturias, Basque Country, Murcia and Navarra, and the Catalan Institute of Oncology—ICO (Spain); Swedish Cancer Society, Swedish Research Council and County Councils of Skåne and Västerbotten (Sweden); Cancer Research UK (14136 to EPIC-Norfolk; C8221/A29017 to EPIC-Oxford), Medical Research Council (1000143 to EPIC-Norfolk; MR/M012190/1 to EPIC-Oxford) (United Kingdom).
The funders had no role in study design, data analysis, data interpretation, or writing of this report.
The preliminary results of this study have been presented as a poster in IASLC 2022 World Conference on Lung Cancer (WCLC 2022) (https://doi.org/10.1016/j.jtho.2022.07.162).
Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organization, the authors alone are responsible for the views expressed in this article, and they do not necessarily represent the decisions, policy, or views of the International Agency for Research on Cancer/World Health Organization.
Contributor Information
Xiaoshuang Feng, Genomic Epidemiology Branch, International Agency for Research on Cancer, Lyon, France.
Wendy Yi-Ying Wu, Department of Radiation Sciences, Oncology, Umea University, Umea, Sweden.
Justina Ucheojor Onwuka, Genomic Epidemiology Branch, International Agency for Research on Cancer, Lyon, France.
Zahra Haider, Department of Radiation Sciences, Oncology, Umea University, Umea, Sweden.
Karine Alcala, Genomic Epidemiology Branch, International Agency for Research on Cancer, Lyon, France.
Karl Smith-Byrne, Cancer Epidemiology Unit, University of Oxford, Oxford, UK.
Hana Zahed, Genomic Epidemiology Branch, International Agency for Research on Cancer, Lyon, France.
Florence Guida, Environment and Lifestyle Epidemiology Branch, International Agency for Research on Cancer, Lyon, France.
Renwei Wang, Division of Cancer Control and Population Sciences, UPMC Hillman Cancer Center, University of Pittsburgh, Pittsburgh, PA, USA.
Julie K Bassett, Cancer Epidemiology Division, Cancer Council Victoria, Melbourne, VIC, Australia.
Victoria Stevens, Rollins School of Public Health, Emory University, Atlanta, GA, USA.
Ying Wang, American Cancer Society, Atlanta, GA, USA.
Stephanie Weinstein, Metabolic Epidemiology Branch, Division of Cancer Epidemiology and Genetics, National Cancer Institute, Rockville, MD, USA.
Neal D Freedman, Metabolic Epidemiology Branch, Division of Cancer Epidemiology and Genetics, National Cancer Institute, Rockville, MD, USA.
Chu Chen, Program in Epidemiology, Division of Public Health Sciences, Fred Hutchinson Cancer Center, Seattle, WA, USA.
Lesley Tinker, Women’s Health Initiative Clinical Coordinating Center, Fred Hutchinson Cancer Research Center, Seattle, WA, USA.
Therese Haugdahl Nøst, Department of Community Medicine, University of Tromsø, The Arctic University of Norway, Tromsø, Norway.
Woon-Puay Koh, Healthy Longevity Translational Research Programme, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, Singapore; Singapore Institute for Clinical Sciences, Agency for Science Technology and Research (A*STAR), Singapore, Singapore.
David Muller, Division of Genetic Medicine, Imperial College London School of Public Health, London, UK.
Sandra M Colorado-Yohar, Department of Epidemiology, Murcia Regional Health Council, IMIB-Arrixaca, Murcia, Spain; Centro de Investigación Biomédica en Red de Epidemiología y Salud Pública (CIBERESP), Madrid, Spain; Research Group on Demography and Health, National Faculty of Public Health, University of Antioquia, Medellín, Colombia.
Rosario Tumino, Hyblean Association for Epidemiological Research, AIRE ONLUS Ragusa, Ragusa, Italy.
Rayjean J Hung, Prosserman Centre for Population Health Research, Lunenfeld-Tanenbaum Research Institute, Sinai Health, Toronto, Canada; Dalla Lana School of Public Health, University of Toronto, Toronto, Canada.
Christopher I Amos, Institute for Clinical and Translational Research, Baylor College of Medicine, Houston, TX, USA.
Xihong Lin, Department of Biostatistics, Harvard T.H. Chan School of Public Health, Boston, MA, USA; Program in Medical and Population Genetics, Broad Institute of Harvard and MIT, Cambridge, MA, USA; Department of Statistics, Harvard University, Cambridge, MA, USA.
Xuehong Zhang, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA, USA.
Alan A Arslan, Department of Population Health, New York University School of Medicine, New York, NY, USA.
Maria-Jose Sánchez, Centro de Investigación Biomédica en Red de Epidemiología y Salud Pública (CIBERESP), Madrid, Spain; Escuela Andaluza de Salud Pública (EASP), Granada, Spain; Instituto de Investigación Biosanitaria ib, Granada, Spain; Department of Preventive Medicine and Public Health, University of Granada, Granada, Spain.
Elin Pettersen Sørgjerd, HUNT Research Centre, Department of Public Health and Nursing, Norwegian University of Science and Technology, Levanger, Norway.
Gianluca Severi, Inserm, Université Paris-Saclay, Villejuif, France.
Kristian Hveem, HUNT Research Centre, Department of Public Health and Nursing, Norwegian University of Science and Technology, Levanger, Norway.
Paul Brennan, Genomic Epidemiology Branch, International Agency for Research on Cancer, Lyon, France.
Arnulf Langhammer, HUNT Research Centre, Department of Public Health and Nursing, Norwegian University of Science and Technology, Levanger, Norway; Levanger Hospital, Nord-Trøndelag Hospital Trust, Levanger, Norway.
Roger L Milne, Cancer Epidemiology Division, Cancer Council Victoria, Melbourne, VIC, Australia; Centre for Epidemiology and Biostatistics, The University of Melbourne, Melbourne, VIC, Australia; Precision Medicine, School of Clinical Sciences at Monash Health, Monash University, Clayton, VIC, Australia.
Jian-Min Yuan, Division of Cancer Control and Population Sciences, UPMC Hillman Cancer Center, University of Pittsburgh, Pittsburgh, PA, USA; Department of Epidemiology, Graduate School of Public Health, University of Pittsburgh, Pittsburgh, PA, USA.
Beatrice Melin, Department of Radiation Sciences, Oncology, Umea University, Umea, Sweden.
Mikael Johansson, Department of Radiation Sciences, Oncology, Umea University, Umea, Sweden.
Hilary A Robbins, Genomic Epidemiology Branch, International Agency for Research on Cancer, Lyon, France.
Mattias Johansson, Genomic Epidemiology Branch, International Agency for Research on Cancer, Lyon, France.
Data availability
Access to data from the Lung Cancer Cohort Consortium (LC3) is governed by the LC3 Access Policy, which is available at the following link: https://www.iarc.who.int/wp-content/uploads/2021/12/LC3_Access_Policy.pdf. Interested investigators are encouraged to contact Dr Johansson or Dr Robbins.
Author contributions
Xiaoshuang Feng, PhD (Data curation; Formal analysis; Methodology; Writing – original draft; Writing – review & editing), Rayjean Hung, PhD (Writing – review & editing), Christopher Amos, PhD (Writing – review & editing), Xihong Lin, PhD (Writing – review & editing), Xuehong Zhang, PhD (Writing – review & editing), Alan Arslan, MD (Writing – review & editing), Maria-Jose Sánchez, MD, PhD (Writing – review & editing), Elin Sørgjerd, PhD (Writing – review & editing), Gianluca Severi, PhD (Writing – review & editing), Kristian Hveem, MD, PhD (Writing – review & editing), Paul Brennan, PhD (Writing – review & editing), Arnulf Langhammer, MD, PhD (Writing – review & editing), Roger Milne, PhD (Writing – review & editing), Jian-Min Yuan, MD, PhD (Writing – review & editing), Beatrice Melin, MD, PhD (Conceptualization; Data curation; Writing – review & editing), Mikael Johansson, MD, PhD (Conceptualization; Methodology; Writing – review & editing), Rosario Tumino, PhD (Writing – review & editing), Sandra Colorado-Yohar, MPH, PhD (Writing – review & editing), David Muller, PhD (Writing – review & editing), Woon-Puay Koh, MBBS, PhD (Writing – review & editing), Wendy Yi-Ying Wu, PhD (Data curation; Formal analysis; Writing – review & editing), Justina Onwuka, PhD (Data curation; Writing – review & editing), Zahra Haider, PhD (Data curation; Writing – review & editing), Karine Alcala, MS (Data curation; Writing – review & editing), Karl Smith-Byrne, DPhil (Data curation; Methodology; Writing – review & editing), Hana Zahed, MS (Data curation; Methodology; Writing – review & editing), Florence Guida, PhD (Data curation; Methodology; Writing – review & editing), Hilary A. Robbins, PhD (Conceptualization; Supervision; Writing – review & editing), Renwei Wang, MD (Writing – review & editing), Victoria Stevens, PhD (Writing – review & editing), Ying Wang, PhD (Writing – review & editing), Stephanie Weinstein, PhD (Writing – review & editing), Neal Freedman, PhD, MPH (Writing – review & editing), Chu Chen, PhD (Writing – review & editing), Lesley Tinker, PhD, RD (Writing – review & editing), Therese Nøst, PhD (Writing – review & editing), Julie Bassett, MSc, PhD (Writing – review & editing), and Mattias Johansson, PhD (Conceptualization; Investigation; Methodology; Supervision; Writing – review & editing).
Funding
This study was supported by the US NCI (INTEGRAL program U19 CA203654 and R03 CA245979), l’Institut National Du Cancer (2019-1-TABAC-01, INCa, France), the Cancer Research Foundation of Northern Sweden (AMP19-962), an early detection of cancer development grant from Swedish Department of Health ministry, and Cancer Research UK [C18281/A29019]. RJH is supported by the Canada Research Chair of the Canadian Institute of Health Research.
Conflicts of interest
The authors declare no competing interests.
References
- 1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209-249. [DOI] [PubMed] [Google Scholar]
- 2. Aberle DR, Adams AM, Berg CD, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365(5):395-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Paci E, Puliti D, Lopes Pegna A, et al. ; for the ITALUNG Working Group. Mortality, survival and incidence rates in the ITALUNG randomised lung cancer screening trial. Thorax. 2017;72(9):825-831. [DOI] [PubMed] [Google Scholar]
- 4. Rota M, Pizzato M, La Vecchia C, et al. Efficacy of lung cancer screening appears to increase with prolonged intervention: results from the MILD trial and a meta-analysis. Ann Oncol. 2019;30(7):1040-1043. [DOI] [PubMed] [Google Scholar]
- 5. Becker N, Motsch E, Trotter A, et al. Lung cancer mortality reduction by LDCT screening-results from the randomized German LUSI trial. Int J Cancer. 2020;146(6):1503-1513. [DOI] [PubMed] [Google Scholar]
- 6. de Koning HJ, van der Aalst CM, de Jong PA, et al. Reduced lung-cancer mortality with volume CT screening in a randomized trial. N Engl J Med. 2020;382(6):503-513. [DOI] [PubMed] [Google Scholar]
- 7. Krist AH, Davidson KW, Mangione CM, et al. ; for the US Preventive Services Task Force. Screening for lung cancer: US Preventive Services Task Force recommendation statement. JAMA. 2021;325(10):962-970. [DOI] [PubMed] [Google Scholar]
- 8. Landy R, Young CD, Skarzynski M,. et al. Using prediction models to reduce persistent racial and ethnic disparities in the draft 2020 USPSTF lung cancer screening Guidelines. J Natl Cancer Inst. 2021;113(11):1590-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pine SR, Mechanic LE, Enewold L, et al. Increased levels of circulating interleukin 6, interleukin 8, C-reactive protein, and risk of lung cancer. J Natl Cancer Inst. 2011;103(14):1112-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shiels MS, Katki HA, Hildesheim A, et al. Circulating inflammation markers, risk of lung cancer, and utility for risk stratification. J Natl Cancer Inst. 2015;107(10):djv199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sozzi G, Boeri M, Rossi M, et al. Clinical utility of a plasma-based miRNA signature classifier within computed tomography lung cancer screening: a correlative MILD trial study. J Clin Oncol. 2014;32(8):768-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Montani F, Marzi MJ, Dezi F, et al. miR-test: a blood test for lung cancer early detection. J Natl Cancer Inst. 2015;107(6):djv063. [DOI] [PubMed] [Google Scholar]
- 13. Cohen JD, Li L, Wang Y, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359(6378):926-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hulbert A, Jusue-Torres I, Stark A, et al. Early detection of lung cancer using DNA promoter hypermethylation in plasma and sputum. Clin Cancer Res. 2017;23(8):1998-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wu WY, Haider Z, Feng X, et al. Assessment of the EarlyCDT-Lung test as an early biomarker of lung cancer in ever-smokers: a retrospective nested case-control study in two prospective cohorts. Intl J Cancer 2022;152(9):2002-2010. doi: 10.1002/ijc.34340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guida F, Sun N, Bantis LE, et al. ; for the Integrative Analysis of Lung Cancer Etiology and Risk (INTEGRAL) Consortium for Early Detection of Lung Cancer. Assessment of lung cancer risk on the basis of a biomarker panel of circulating proteins. JAMA Oncol. 2018;4(10):e182078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Robbins HA, Alcala K, Moez EK,. et al. Design and methodological considerations for biomarker discovery and validation in the Integrative Analysis of Lung Cancer Etiology and Risk (INTEGRAL) Program. Ann Epidemiol. 2023;77:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. The Lung Cancer Cohort Consortium (LC3)., et al. The blood proteome of imminent lung cancer diagnosis. Nat Commun. 2023. DOI: 10.1038/s41467-023-37979-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Galwey NW. A new measure of the effective number of tests, a practical tool for comparing families of non-independent significance tests. Genet Epidemiol. 2009;33(7):559-568. [DOI] [PubMed] [Google Scholar]
- 20. Meza R, Jeon J, Toumazis I, et al. Evaluation of the benefits and harms of lung cancer screening with low-dose computed tomography: modeling study for the US Preventive Services Task Force. JAMA. 2021;325(10):988-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tammemägi MC, Katki HA, Hocking WG, et al. Selection criteria for lung-cancer screening. N Engl J Med. 2013;368(8):728-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tu Y, Wu Y, Lu Y, et al. Development of risk prediction models for lung cancer based on tumor markers and radiological signs. J Clin Lab Anal. 2021;35(3):e23682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Doseeva V, Colpitts T, Gao G, et al. Performance of a multiplexed dual analyte immunoassay for the early detection of non-small cell lung cancer. J Transl Med. 2015;13:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu L, Teng J, Zhang L, et al. The combination of the tumor markers suggests the histological diagnosis of lung cancer. Biomed Res Int. 2017;2017:2013989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nagase H, Woessner JF Jr. Matrix metalloproteinases. J Biol Chem. 1999;274(31):21491-21494. [DOI] [PubMed] [Google Scholar]
- 26. Hofmann HS, Hansen G, Richter G, et al. Matrix metalloproteinase-12 expression correlates with local recurrence and metastatic disease in non-small cell lung cancer patients. Clin Cancer Res. 2005;11(3):1086-1092. [PubMed] [Google Scholar]
- 27. Dagnino S, Bodinier B, Guida F, et al. Prospective identification of elevated circulating CDCP1 in patients years before onset of lung cancer. Cancer Res. 2021;81(13):3738-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brenner DR, Fanidi A, Grankvist K, et al. Inflammatory cytokines and lung cancer risk in 3 prospective studies. Am J Epidemiol. 2017;185(2):86-95. [DOI] [PubMed] [Google Scholar]
- 29. González Maldonado S, Johnson T, Motsch E, et al. Can autoantibody tests enhance lung cancer screening?-an evaluation of EarlyCDT(®)-Lung in context of the German Lung Cancer Screening Intervention Trial (LUSI). Transl Lung Cancer Res. 2021;10(1):233-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Borg M, Wen SWC, Nederby L, et al. Performance of the EarlyCDT® Lung test in detection of lung cancer and pulmonary metastases in a high-risk cohort. Lung Cancer. 2021;158:85-90. [DOI] [PubMed] [Google Scholar]
- 31. Murray A, Chapman CJ, Healey G, et al. Technical validation of an autoantibody test for lung cancer. Ann Oncol. 2010;21(8):1687-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Boyle P, Chapman CJ, Holdenrieder S, et al. Clinical validation of an autoantibody test for lung cancer. Ann Oncol. 2011;22(2):383-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lam S, Boyle P, Healey GF, et al. EarlyCDT-Lung: an immunobiomarker test as an aid to early detection of lung cancer. Cancer Prev Res (Phila). 2011;4(7):1126-1134. [DOI] [PubMed] [Google Scholar]
- 34. Chapman CJ, Healey GF, Murray A, et al. EarlyCDT®-lung test: improved clinical utility through additional autoantibody assays. Tumour Biol. 2012;33(5):1319-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Macdonald IK, Murray A, Healey GF, et al. Application of a high throughput method of biomarker discovery to improvement of the EarlyCDT(®)-Lung Test. PLoS One. 2012;7(12):e51002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Healey GF, Lam S, Boyle P, et al. Signal stratification of autoantibody levels in serum samples and its application to the early detection of lung cancer. J Thorac Dis. 2013;5(5):618-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jett JR, Peek LJ, Fredericks L, et al. Audit of the autoantibody test, EarlyCDT®-lung, in 1600 patients: an evaluation of its performance in routine clinical practice. Lung Cancer. 2014;83(1):51-55. [DOI] [PubMed] [Google Scholar]
- 38. Jett J, Healey G, Macdonald I, et al. P2.13-013 determination of the detection lead time for autoantibody biomarkers in early stage lung cancer using the UKCTOCS cohort. J Thorac Oncol. 2017;12(11):S2170. [Google Scholar]
- 39. Massion PP, Healey GF, Peek LJ, et al. Autoantibody signature enhances the positive predictive power of computed tomography and nodule-based risk models for detection of lung cancer. J Thorac Oncol. 2017;12(3):578-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sullivan FM, Mair FS, Anderson W, et al. ; for The Early Diagnosis of Lung Cancer Scotland (ECLS) Team. Earlier diagnosis of lung cancer in a randomised trial of an autoantibody blood test followed by imaging. Eur Respir J 2020;57(1):2000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Access to data from the Lung Cancer Cohort Consortium (LC3) is governed by the LC3 Access Policy, which is available at the following link: https://www.iarc.who.int/wp-content/uploads/2021/12/LC3_Access_Policy.pdf. Interested investigators are encouraged to contact Dr Johansson or Dr Robbins.