GATA2 deficiency is a complex multi-system disorder with high risk of developing myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML) with a nearly complete lifetime penetrance.1,2 GATA2 carriers show a highly variable expressivity, with some individuals developing early-onset MDS, while others remain asymptomatic throughout life. Although no prognostic biomarkers exist, it is likely that both cooperating genetic and epigenetic drivers shape the course of the disease.3 Despite advances in the identification of recurrent somatic mutations in a set of leukemia driver genes (i.e., STAG2, SETBP1, ASXL1 and ETV6), there are major gaps in understanding the molecular mechanisms associated with leukemic progression in GATA2 carriers.4 Moreover, DNA methylation alterations contribute to the initiation and expansion of leukemic clones and aberrant hypermethylation occurs in adult patients with MDS and AML.5,6 However, to date, a genome-wide DNA methylome analysis in GATA2 patients has not been performed.
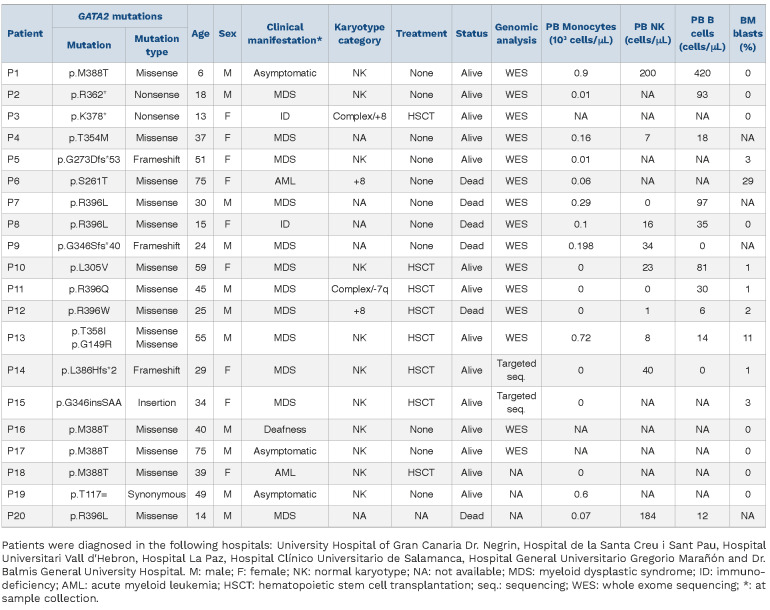
For this study, 20 clinically annotated GATA2 carriers from seven Spanish hospitals were enrolled (Table 1; Online Supplementary Figure S1). Median age at diagnosis was 36 (range, 6–75) years. The primary initial manifestation was MDS (n=12, 55%), followed by immunodeficiency (n=3, 15%), and AML (n=2, 10%). On cytogenetics, trisomy 8 was detected in three patients, complex karyotype in two patients, while 11 patients had normal karyotype (Online Supplementary Figure S1A-F). Based on DNA availability, somatic mutation profiling was performed in total peripheral blood (PB) or bone marrow (BM) of 17 GATA2 carriers. Somatic mutations in myeloid malignancy genes were identified in 71% (12/17) tested patients (Online Supplementary Figure S1F). This analysis confirmed the heterogeneity of acquired somatic mutations in GATA2 deficiency, with STAG2, ASXL1 and SETBP1 as recurrently affected genes. We obtained global DNA methylation profiles of patients who underwent complete genetic characterization by using the Infinium Human Methylation EPIC 850 K platform (Illumina). We profiled DNA of eight BM (P1, P3, P5, P6, P10, P11, P12, P13) and eight PB samples (P1, P4, P7, P8, P9, P13, P16 and P17) and compared them with a cohort of 12 (5 PB and 7 BM) age-matched healthy donor (HD) controls.
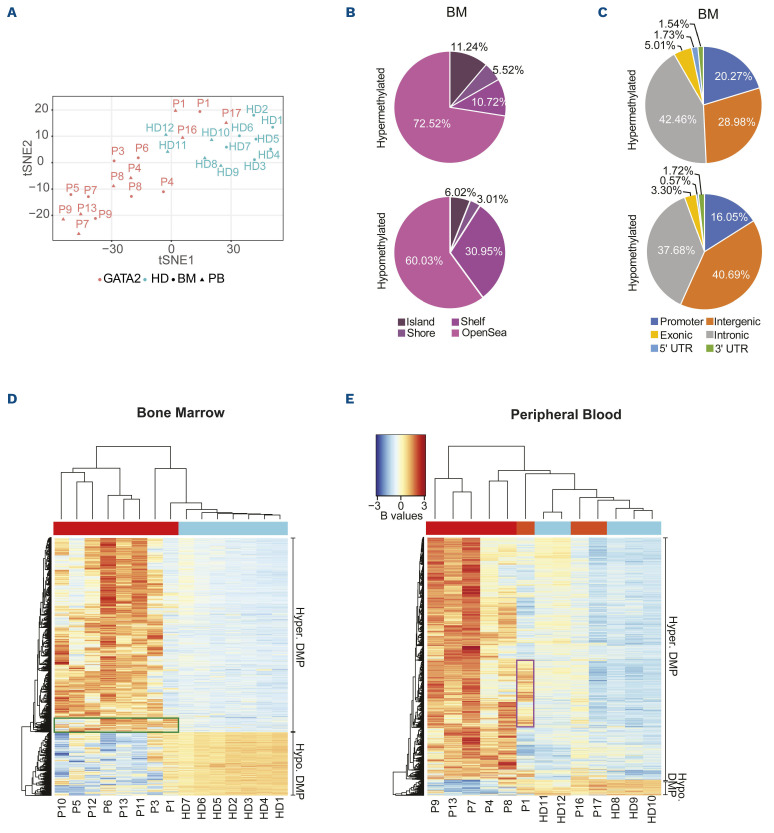
High-dimensional data visualization showed that the majority of GATA2 patients cluster tightly together and separately from HD. Asymptomatic carriers P1, P16 and P17 (at age 6, 40 and 75 years old, respectively) belonging to the same family, were encompassed to the HD group (Figure 1A). Additionally, we compared HD with asymptomatic GATA2 carriers alone, and we still observed intermixed samples (data not shown). Next, the DNA methylation changes were calculated between the GATA2 group and HD pairwise, revealing a DNA methylation pattern specific to GATA2 carriers. In detail, 2,834 differentially methylated positions (DMP) were identified in GATA2 BM samples and 1,406 DMP in PB samples (Online Supplementary Figure S2A). A descriptive analysis of the DMP distribution was performed using as a reference the probe distribution of the Infinium MethylEPIC array from distal to proximal CpG island regions (open sea, CpG shelf, CpG shore and CpG island) (Online Supplementary Figure 2B). Although previous studies showed that promoter-proximal methylation is negatively correlated with active gene expression;7 our analysis revealed that the majority of DMP are promoter-distant in both, BM (open sea: 60% hypomethylated DMP and 72.5% hypermethylated DMP) and PB samples (open sea: 52% hypomethylated and 82.3% hypermethylated) (Figure 1B; Online Supplementary Figure 2C).
Additionally, the DMP distribution using the neighboring gene as reference showed an enrichment in intergenic and intronic regions (Figure 1C; Online Supplementary Figure S2D-E). This observation was corroborated by a correlation analysis comparing the DMP distribution of GATA2 patients with the reference array (Online Supplementary Figure S2F). Overall, we observed that DNA methylation changes are enriched in gene-distant and intronic regions in GATA2 patients. Whether this discrepancy is a consequence of GATA2 deficiency, or arising solely from the MDS evolution, needs to be further investigated. Previous studies showed that endogenous GATA2 preferentially occupies sites distant to promoters in hematopoietic stem cells,8 hence the loss of DNA binding capacity of GATA2 mutant protein might result in aberrant DNA methylation of these loci.
Interestingly, unsupervised analysis of hypermethylated DMP highlighted the presence of a hypermethylated DMP subcluster across all the GATA2 BM samples (hereinafter subcluster A), including P1, the asymptomatic GATA2 carrier (Figure 1D). The matching PB of P1 revealed a sub-cluster of hypermethylated DMP (hereinafter subcluster B) as affected GATA2 patients (Figure 1E). Importantly, the 2-year longitudinal follow-up of P1 showed the evolution to MDS with multilineage dysplasia (MDS-MLD) and monosomy 7 without secondary mutations. In contrast, the two asymptomatic GATA2 carriers P16 and P17 (father and grandfather of P1) had a DNA methylation profile comparable with the HD group (Figure 1E). This observation suggests the presence of a likely early aberrant DNA methylation at specific loci in GATA2 carriers that might have a potential use in early detection of patients at risk for impending myeloid transformation.
Table 1.
Patient clinical characteristics and genetic landscape.
After the gene annotation of the 131 DMP of the subcluster A (Online Supplementary Figure S2G), 118 genes were associated to hypermethylated DMP (Online Supplementary Figure S2H), including MECOM, which epigenetic regulation has been reported in AML.9 The top candidate was PROMININ1 (PROM1/CD133) with four hypermethylated DMP upstream of its promoter region (Online Supplementary Figure S2I). Interestingly, the aberrant methylation status of the PROM1 promoter has already been described in various cancers including AML.10
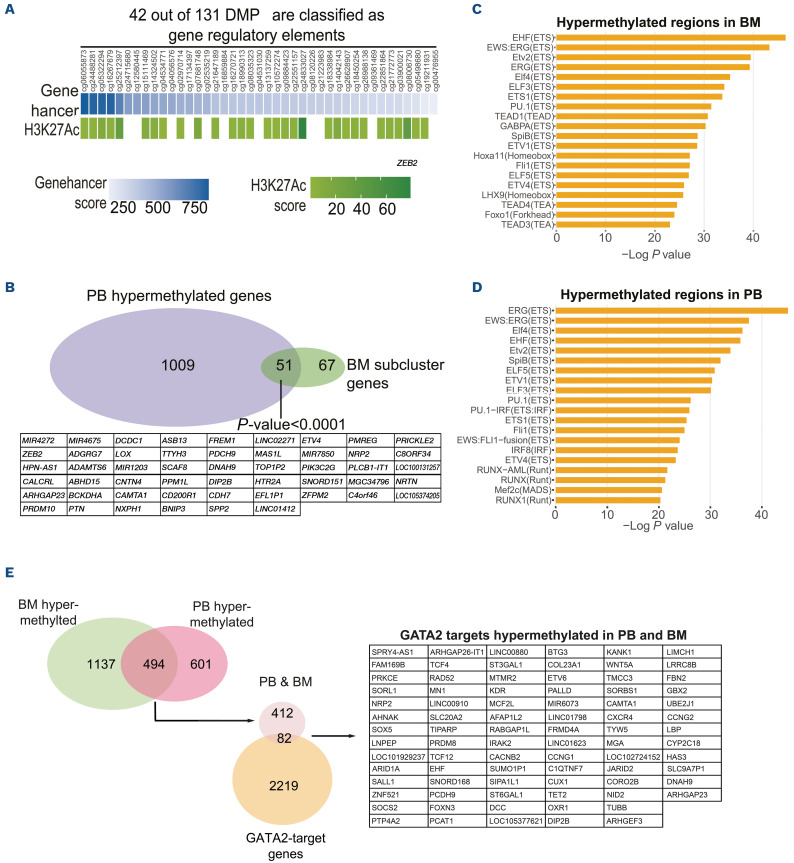
Moreover, 42 of 131 hypermethylated DMP subclusters are classified as gene regulatory elements; and most of them are enriched for H3K27ac, a chromatin mark associated with enhancer activity (Figure 2A).
Because tissue-specific DNA methylation patterns might alter the methylation results, we overlapped the 118 BM-hypermethylated genes with the 1,060 PB-hypermethyl-ated genes. We found 51 commonly hypermethylated common genes, implying that PB samples, at least partially reflect the dysregulated pattern observed in the BM (Figure 2B). Additionally, the 205 PB-hypermethylated subcluster B genes were crossed with 118 BM-hypermethylated subcluster A genes, observing an overlapp of 30 genes, indicating that the aberrant DNA methylation profile is detectable in both BM and PB samples (Online Supplementary Figure S2H). Aberrant epigenetic changes have been associated with alterations of transcription factor (TF) genomic binding capacities.11,12 Therefore, we assessed whether the hypermethylated and hypomethylated DMP identified in PB and BM samples were enriched in specific TF DNA binding motives using Hypergeometric Optimization of Motif EnRichment (HOMER). This analysis revealed in the hypermethylated DMP a significant enrich ment in TF motives of the ETS family, such as ETV2, ETV6, ELF5 and PU.1 among others (Figure 2C, D), which are known to play a role in MDS.13 The hypomethylated DMP showed an enrichment in TF of the bZIP family preferentially (Online Supplementary Figure S3A, B). In order to evaluate whether the GATA2 binding sites are linked to DNA methylation in GATA2 deficiency, we integrated the hypermethylated genes in both PB and BM samples with a GATA2 chromatin immunoprecipitation sequencing (ChiPseq) dataset from our laboratory (GSE107639).14 This analysis revealed the presence of 82 of 494 hypermethylated genes that are also GATA2 targets (Figure 2E). On the contrary, the intersection of GATA2-regulated genes with the hypomethylated genes in BM and PB did not show any relevant gene enrichment (Online Supplementary Figure S3C). Additionally, the common hypermethylated genes of PB and BM were crossed with K562 myeloid leukemia GATA2 ChIPseq dataset (GSE18868),8 observing that 204 of 494 hypermethylated genes are GATA2 targets and 51 of 82 genes are in common between Castaño et al.14 and Fujiwara et al.8 datasets (Online Supplementary Figure S3D, E). Gene ontology analysis of the 82 hypermethylated GATA2 target genes showed an enrichment in transcriptional regulation and cell differentiation. In silico analysis of the upstream regulators inferred that ETV6, TCF12, MGA, and SOX5 are cooperative TF of the GATA2 gene regulatory network (Online Supplementary Figure 3F). Together, our data suggest that GATA2 deficiency is associated with aberrant DNA methylation in GATA2 target genes.
Figure 1.
Unsupervised hierarchical clustering and the heat map visualization of differentially methylated CpG sites of GATA2 patients. (A) T-distributed stochastic neighbor embedding (t-SNE) showing the distribution of GATA2 patient (P) data (in red) and healthy donors (HD) (in blue), based on DNA methylation profile. Bone marrow (BM) samples are represented with circles and peripheral blood (PB) samples are represented with triangles. (B) Differentially methylated probes (DMP) distribution of BM samples, hypermethylated (above) and hypomethylated (below). CpG island distance, island (eggplant), shore (lollipop), shelf (mauve) and open sea (fandango). (C) DMP distribution of BM samples, hypermethylated (above) and hypomethylated (below). Promoter (dark blue), intergenic (orange), intronic (grey), exonic (yellow), 5’ UTR (light blue) and 3’ UTR (green). (D) Heatmap of DMP BM vs. HD samples. The DMP in common among all GATA2 patients (subcluster A) are squared in green. Scale β values from -3 (blue/hypomethylated) to +3 (red/hypermethylated). (E) Heatmap of DMP of PB vs. HD samples. The hypermethylated DMP in P1 sample (subcluster B), which are in common with affected GATA2 patients, are squared in fuchsia. Scale β values from -3 (blue/hypomethylated) to +3 (red/hypermethylated).
Figure 2.
Regulatory element analysis of hypermethylated genomic regions identify in GATA-mutant patients. (A) Ranked representation of the regulatory function of the hypermethylated position based on GeneHancer (blue) score and H3K27ac enrichment of those genomic positions (green). (B) Venn diagram of the total hypermethylated genes in peripheral blood (PB) (n=1,060) vs. subcluster of hypermethylated genes in bone marrow (BM) (n=118), the genes of the intersection are 51 genes. P value=1.572541e-14 (hypergeometric distribution test). (C) Hypergeometric Optimization of Motif EnRichment (HOMER) analysis using hypermethylated differentially methylated probes in BM samples. Enriched motifs found are predominantly from the ETS family including ETV and PU.1. (D) HOMER analysis using hypermethylated DMP in peripheral blood (PB) samples. Enriched motifs found are predominantly from the ETS family and IRF family. (E) Top row: Venn diagram of the neighboring gene of hypermethylated DMP in the BM samples (n=1,631) compared to the neighboring gene of hypermethylated DMP in the PB samples (n=1,095), intersection 494 genes. Bottom row: GATA2-regulated genes (n=2301) obtained from the intersection of 2 GATA2 chromatin immunoprecipitation sequencing (ChIPseq) data GSE107639.15 The crossed of top intersection (n=494) vs. GATA2-regulated genes (n=2301), the intersection gives 82 GATA2 hypermethylated targets both in PB and BM.
Finally, we compared our BM hypermethylation data with publicly available methylation profiles of 184 pediatric AML patients, TARGET 2018.15 Interestingly, 50% (4/8) of our GATA2 patients clustered together with AML samples (which had a known GATA2 mutation-negative status), showing a similar methylation pattern with AML samples (Online Supplementary Figure S3G). This points to the possibility that some aberrant methylation signatures observed in our GATA2 patients might be directly linked to the AML transformation and thus arise independently of the underlying GATA2 germline mutation. Future genome-wide association studies are warranted to address this question in depth.
In conclusion, we identified an aberrant DNA hypermethylated signature in GATA2 deficiency. Specifically, we described the presence of a subset of aberrant hypermethylated genes present in GATA2 carriers at early (and not yet symptomatic) disease stage, which could be potentially used as predictors of disease progression. In this context, the implementation of customized methyl-ation-specific assays might be instrumental to validate our findings in larger cohorts of patients and to test its clinical prognostic utility. Finally, a collaborative effort will be essential to increase the number patients with this rare yet high-risk MDS/AML predisposition syndrome, allowing for comprehensive genetic and epigenetic analyses to understand the impact of the secondary hits and/or aberrant DNA methylation on the disease progression.
Supplementary Material
Acknowledgments
We thank Francesca di Giorgio, Loris Mularoni, Chiara Mazzanti, Joan Pera, Dolly Viviana Fiallo-Suárez, Adela Escudero-Lopez, María Díez-Campelo, Teresa González, Montserrat Arnan, Francesc Solé, Amaia Vilas Zornoza, Montse Rovira (Hospital Clinic, Barcelona), Paola Romecin and Laura Palomo, for technical support.
Funding Statement
Funding: This work was supported by ERA PerMed GATA2-HuMo Funding Mechanism in Spain: Acció instrumental de SLT011/18/00006 of the Department of Health of the Government of Catalonia to AG and AB; in Hungary: ED-18-1-2019-001 grant from the National Research, Development and Innovation Office to CB; and in Germany: German Federal Ministry of Education and Research (BMBF) 2018-123/01KU1904 to MWW.ÚNKP-21-2-I-SE-21 and Hungarian National Academy of Scientist Education grant to KL, TKP2021-NVA-15. TKP2021-EGA-24 and EU's Horizon 2020 research and innovation program under grant agreement no. 739593 and Elixir Hungary to CB. The Spanish Ministry of Economy, Industry, CERCA/Generalitat de Catalunya and Fundació Josep Carreras-Obra Social la Caixa and the Deutsche Josep Carreras Leukämie-Stiftung (DJCLS15R/2021) to PM: Asociación Española contra el cancer (AECC, PRYGN211192BUEN) and Health Institute Carlos III (PI20/00822) to CB. Competitiveness (MINECO PID2020-15591RB-100), La Marató de TV3 (202001-32), FPS Grant 2018 by Fondazione Pisana per la Scienza ONLUS and CERCA Programme/ Generalitat de Catalunya for institutional support to AG, and BMBF MyPred 01GM1911A to MWW. OM-B Is supported by 101029927-scGATA2track (H2020-MSCA-IF-2020) and KOG-202109-01162. AL is the recipient of the APOSTD2021/212 fellowship from the Generalitat Valenciana. Funding for this project was provided in part by an EHA Research Grant award granted by the European Hematology Association (KOG-202109-01162). This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No 101029927. We thank CERCA Programme / Generalitat de Catalunya for institutional support.
References
- 1.Kotmayer L, Romero-Moya D, Marin-Bejar O, et al. GATA2 deficiency and MDS/AML: experimental strategies for disease modelling and future therapeutic prospects. 2022;199(4):482-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Y, Wu J, Qin T, et al. Comparison of the revised 4th (2016) and 5th (2022) editions of the World Health Organization classification of myelodysplastic neoplasms. Leukemia. 2022;36(12):2875-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al Seraihi AF, Rio-Machin A, Tawana K, et al. GATA2 monoallelic expression underlies reduced penetrance in inherited GATA2-mutated MDS/AML. Leukemia. 2018;32(11):2502-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.West RR, Calvo KR, Embree LJ, et al. ASXL1 and STAG2 are common mutations in GATA2 deficiency patients with bone marrow disease and myelodysplastic syndrome. Blood Adv. 2022;6(3):793-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Figueroa ME, Abdel-Wahab O, Lu C, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Figueroa ME, Skrabanek L, Li Y, et al. MDS and secondary AML display unique patterns and abundance of aberrant DNA methylation. Blood. 2009;114(16):3448-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Futscher BW, Oshiro MM, Wozniak RJ, et al. Role for DNA methylation in the control of cell type specific maspin expression. Nat Genet. 2002;31(2):175-179. [DOI] [PubMed] [Google Scholar]
- 8.Fujiwara T, O'Geen H, Keles S, et al. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol Cell. 2009;36(4):667-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Groschel S, Sanders MA, Hoogenboezem R, et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell. 2014;157(2):369-381. [DOI] [PubMed] [Google Scholar]
- 10.Gopisetty G, Xu J, Sampath D, Colman H, Puduvalli VK. Epigenetic regulation of CD133/PROM1 expression in glioma stem cells by Sp1/myc and promoter methylation. Oncogene. 2013;32(26):3119-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Gomez A, Li T, de la Calle-Fabregat C, et al. Targeting aberrant DNA methylation in mesenchymal stromal cells as a treatment for myeloma bone disease. Nat Commun. 2021;12(1):421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blattler A, Farnham PJ. Cross-talk between site-specific transcription factors and DNA methylation states. J Biol Chem. 2013;288(48):34287-34294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao J, Li D, Seo J, Allen AS, Gordan R. Quantifying the impact of non-coding variants on transcription factor-DNA binding. Res Comput Mol Biol. 2017;10229:336-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castano J, Aranda S, Bueno C, et al. GATA2 promotes hematopoietic development and represses cardiac differentiation of human mesoderm. Stem Cell Reports. 2019;13(3):515-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolouri H Farrar JE Triche T, Jr.et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat Med. 2018;24(1):103-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.