Abstract
Reverse transcription (RT)-PCR assays have been widely described for use in the diagnosis of human parainfluenza viruses (HPIVs) and other respiratory virus pathogens. However, these assays are mostly monospecific, requiring separate amplifications for each HPIV type. In the present work, we describe multiplex RT-PCR assays that detect and differentiate HPIV serotypes 1, 2, and 3 in a combined reaction. Specifically, a mixture of three pairs of primers to conserved regions of the hemagglutinin-neuraminidase gene of each HPIV serotype was used for primary amplification, yielding amplicons with similar sizes. For typing, a second amplification was performed with a mixture of nested primers, yielding amplicons with sizes easily differentiated by agarose gel electrophoresis. A modified single-amplification RT-PCR assay with fluorescence-labeled nested primers, followed by analysis of the labeled products on an automated sequencing gel, was also evaluated. Fifteen temporally and geographically diverse HPIV isolates from the Centers for Disease Control and Prevention archives and 26 of 30 (87%) previously positive nasopharyngeal specimens (8 of 10 positive for HPIV serotype 1 [HPIV1], 9 of 10 positive for HPIV2, and 9 of 10 positive for HPIV3) were positive and were correctly typed by both assays. Negative results were obtained with naso- or oropharyngeal specimens and/or culture isolates of 33 unrelated respiratory tract pathogens, including HPIV4, enterovirus, rhinovirus, respiratory syncytial virus, adenovirus, influenza virus, and Streptococcus pneumoniae. Our multiplex RT-PCR assays provide sensitive, specific, and simplified tools for the rapid diagnosis of HPIV infections.
Human parainfluenza viruses (HPIVs) are medically important respiratory pathogens and are second only to respiratory syncytial virus (RSV) as a major cause of lower respiratory tract (LRT) illness in infants and young children (4, 5, 15, 17, 18, 22). Although repeat infections in healthy older children and adults are typically less severe, serious LRT illness caused by HPIVs has been reported among immunocompromised individuals (1, 2, 12, 13, 26–28) and institutionalized elderly individuals (7).
Of the four recognized serotypes of HPIV (HPIV serotype 1 [HPIV1], HPIV2, HPIV3, and HPIV4) HPIV3 is most commonly associated with serious LRT illness, followed by HPIV1 and HPIV2; HPIV4 is rarely associated with serious illness (9). The use of classic diagnostic methods like viral isolation and serology can often result in a delay of several weeks before the results are available, and these methods are therefore less useful for making therapeutic decisions (4). Direct antigen detection methods are widely used for the rapid diagnosis of HPIV infections (11, 20, 21), but results can be variable (10), and some HPIV strains may be missed entirely by assays with specific monoclonal antibodies (24). To address these problems, reverse transcription (RT)-PCR assays that have been shown to provide rapid and sensitive detection of HPIV1 and HPIV3 have been developed (6, 14, 15). However, all of these methods are monospecific, requiring separate amplifications for each virus. As an alternative, multiplex PCR assays permit simultaneous amplification of several viruses in a single reaction (25). In this study, we describe novel multiplex RT-PCR assays for the detection and identification of HPIV1, -2, and -3.
MATERIALS AND METHODS
Virus isolates.
Cell culture isolates of reference strains of virus obtained from the Centers for Disease Control and Prevention archives were used to develop and standardize the RT-PCR assays. These included (i) HPIV prototype strains C-35 (HPIV1), Greer (HPIV2), and C-243 (HPIV3); (ii) 12 geographically and temporally diverse HPIV isolates from the United States, Canada, and Argentina (HPIV1 from Córdoba, Argentina [1967], Massachusetts [1985], Oregon [1974], and Nevada [1985]; HPIV2 from California [1980], Massachusetts [1971], Connecticut [1977], and Kansas [1982]; HPIV3 from Idaho [1977], Alabama [1979], Oregon [1981], and Manitoba, Canada [1981]); and (iii) individual isolates of HPIV4, RSV subtypes A and B, influenza A and B viruses, adenovirus, enterovirus, and rhinovirus.
Clinical samples.
The clinical samples used to evaluate the efficacies of the RT-PCR assays included 45 throat or nasopharyngeal swabs collected from pediatric patients at the Cardinal Glennon Children’s Hospital, St. Louis, Mo., and placed in viral transport medium; 10 specimens each were previously culture positive for HPIV1, -2, and -3, and the remaining 15 specimens were positive for other respiratory viruses, including 3 each for RSV, adenovirus, influenza A virus, enterovirus, and rhinovirus. Ten nasopharyngeal specimens from adults culture positive for Streptococcus pneumoniae and culture negative for respiratory viruses were also tested. All clinical samples had been stored at −70°C.
Primer design and preparation.
Previously published sequences of the HPIV hemagglutinin-neuraminidase gene (from 27 HPIV1 isolates, 2 HPIV2 isolates, and 10 HPIV3 isolates) obtained from GenBank were aligned by using the Wisconsin Analysis Package (version 8), Genetics Computer Group, Madison, Wis. External and nested primer sequences were chosen from conserved regions of the hemagglutinin-neuraminidase gene and were designed to achieve optimal performance in a multiplex reaction (Table 1). All primers were prepared on a model 394 ABI DNA synthesizer (Perkin-Elmer). Fluorescence-tagged primers were labeled at the 5′ end with 6-phosphoramidite (FAM; Glen Research). FAM-labeled primers were purified by reversed-phase high-performance liquid chromatography. All other primers were used without further purification.
TABLE 1.
Multiplex primers for HPIVs
| Amplification reaction and primer | Polaritya | Position (nucleotides)b | Sequence (5′→3′) |
|---|---|---|---|
| Primary | |||
| PIP1+ | HPIV1 + | 748–768 | CCTTAAATTCAGATATGTAT |
| PIP1− | HPIV1 − | 1206–1225 | GATAAATAATTATTGATACG |
| PIP2+ | HPIV2 + | 803–822 | AACAATCTGCTGCAGCATTT |
| PIP2− | HPIV2 − | 1291–1310 | ATGTCAGACAATGGGCAAAT |
| PIP3+ | HPIV3 + | 762–781 | CTGTAAACTCAGACTTGGTA |
| PIP3− | HPIV3 − | 1220–1239 | TTTAAGCCCTTGTCAACAAC |
| Nestedc | |||
| PIS1+ | HPIV1 + | 780–801 | CCGGTAATTTCTCATACCTATG |
| PIS1− | HPIV1 − | 1076–1096 | CTTTGGAGCGGAGTTGTTAAG |
| PIS2+ | HPIV2 + | 845–866 | CCATTTACCTAAGTGATGGAAT |
| PIS2− | HPIV2 − | 1027–1048 | GCCCTGTTGTATTTGGAAGAGA |
| PIS3+ | HPIV3 + | 884–905 | ACTCCCAAAGTTGATGAAAGAT |
| PIS3− | HPIV3 − | 966–986 | TAAATCTTGTTGTTGAGATTG |
RNA extraction.
RNA was extracted from the clinical samples and virus isolates as described previously (3). Briefly, 50 μl of each sample was treated with 200 μl of extraction buffer (4 M guanidinium thiocyanate, 0.5% N-lauryl sarcosine, 1 mM dithiothreitol, 25 mM sodium citrate, and 0.1 mg of glycogen per ml), followed by isopropanol and 70% ethanol precipitations.
RT.
The vacuum-dried RNA pellets were resuspended in 10 μl of hybridization buffer (300 mM NaCl, 5 mM Tris-HCl [pH 7.5], 1 mM EDTA) containing HPIV1F, HPIV2F, and HPIV3F each at a concentration of 2 μM and were denatured for 3 min at 94°C. The RNA template and primers were then allowed to hybridize for 45 min at 50°C. A 40-μl volume of RT buffer (10 mM Tris-HCl [pH 8.3]; 6 mM magnesium chloride; 1 mM dithiothreitol; dATP, dGTP, dCTP, and dTTP at a concentration of 1 mM each; and 40 U of RNase inhibitor [RNAsin; Boehringer Mannheim]) containing 50 U of avian myeloblastosis reverse transcriptase (Boehringer Mannheim) was then added, and the mixture was incubated for 1 h at 42°C. The enzyme was finally inactivated by incubation for 5 min at 92°C.
PCR amplification and product detection, two-step method.
For primary PCR amplification, 5 μl of cDNA prepared in the RT reaction was added to a PCR mixture containing 10 mM Tris-HCl (pH 8.3); 50 mM KCl; 3 mM MgCl; dATP, dCTP, dGTP, and dTTP each at a concentration of 200 μM; the primary amplification primers (Table 1) each at a concentration of 2 μM; and 1.25 U of Taq polymerase (AmpliTaq; Perkin-Elmer Cetus) in a final volume of 50 μl and the mixture was overlaid with mineral oil. Amplification was performed on a model 480 DNA Thermal Cycler (Perkin-Elmer Cetus) programmed for 35 cycles of 1 min of denaturation at 94°C, 1 min of annealing at 50°C, and 1 min of elongation at 72°C; an additional 2 min of denaturation preceded the first cycle, and elongation was extended to 5 min in the last cycle. For nested PCR, 1 μl of the primary amplification products was added to 49 μl of a new PCR mixture containing nested instead of primary reaction primers (Table 1), and the thermal cycle program annealing temperature was changed to 58°C. The PCR products were sized by gel electrophoresis on 2% agarose containing 0.5 g of ethidium bromide per ml in TBE (Tris-borate-EDTA) buffer and were visualized under UV light. Standard precautions were taken to avoid carryover contamination. Pipetting was performed with aerosol-resistant tips, and different biosafety cabinets were used for sample extraction and first amplification or nested amplification. Amplicon detection was performed in a different room.
PCR amplification and product detection, one-step method.
Primary amplification was performed with the nested primers directly with the RT products, but substituting FAM-labeled forward primers for the unlabeled forward primers. The labeled products were subjected to 6% acrylamide gel electrophoresis under denaturing conditions on an ABI PRISM 377 DNA sequencer (Perkin-Elmer Corp.) and were analyzed with GeneScan Analysis Software (version 2.0.2).
RESULTS
Viral isolates.
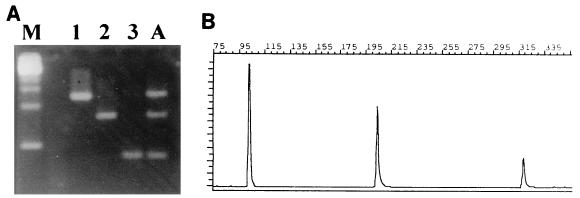
PCR products of the expected size were obtained with the three HPIV prototype strains when they were tested alone or in a mixed reaction (Fig. 1). Serial dilution experiments showed that the nested PCR was about 100-fold more sensitive than cell culture isolation for all three types. All reference HPIV isolates were detected and were correctly identified by both the one-step and the two-step procedures. All non-HPIV isolates were PCR negative.
FIG. 1.
Detection and typing of prototype HPIV1, HPIV2, and HPIV3 strains. (A) Lane M, molecular weight markers; lanes 1, 2, and 3, HPIV1, HPIV2, and HPIV3, respectively; lane A, mixture of all three HPIV types. (B) GeneScan Analysis Software profile of the mixture in lane A.
Clinical samples.
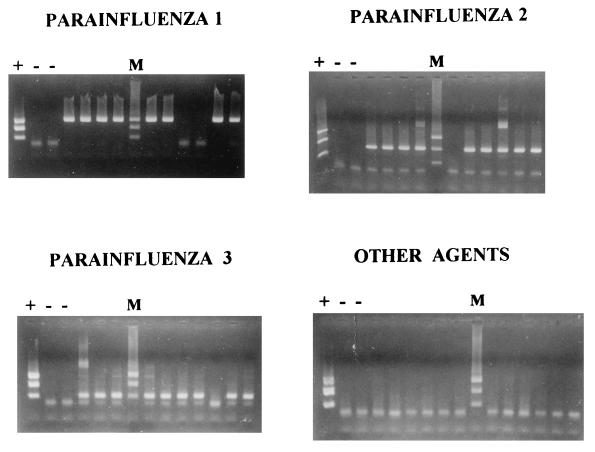
Of the clinical samples from which HPIVs had previously been isolated, 8 of 10 (80%) samples containing HPIV1, 9 of 10 (90%) samples containing HPIV2, and 9 of 10 (90%) samples containing HPIV3 were RT-PCR positive and were correctly typed by both methods (Fig. 2). Only three of them were detected after the first amplification in the two-step method. HPIV was reisolated from one of the four clinical samples that were negative by RT-PCR. The isolate was positive for HPIV-1 by RT-PCR, consistent with the original isolate identification. None of the 25 control specimens containing other respiratory viruses or bacterial pathogens were positive. No discrepancies were found between the one-step and two-step procedures.
FIG. 2.
Results of nested RT-PCR with clinical samples. No discrepancies with the results obtained by the method with GeneScan Analysis Software were found (data not shown).
DISCUSSION
RT-PCR assays for the detection of HPIVs have been described, but they have been limited to the detection of HPIV3 (14, 15) or HPIV1 and HPIV3 (6), without distinguishing between the two serotypes. In contrast, our multiplex assays both detect and differentiate all three medically important HPIV serotypes and provide a sensitive and specific means of identifying HPIVs directly from clinical specimens; 87% (26 of 30) of the clinical samples culture positive for HPIVs were correctly identified, whereas none of the specimens containing a diverse group of other respiratory virus and bacterial pathogens were positive.
Although our primers were designed to react with temporally and geographically diverse HPIV isolates, published HPIV sequence data were limited, and therefore unanticipated strain sequence variation in the primer regions could result in occasional false-negative results. This may account for our failure to amplify virus from three of the initially culture-positive clinical samples. However, we believe that the most likely explanation for our false-negative results with these specimens was that the samples were subjected to at least two freeze-thaw cycles between the initial isolation and RT-PCR procedures, possibly causing a decrease in virus titer and RNA degradation. Further testing of diverse fresh clinical samples by RT-PCR will be required to provide a better estimate of the sensitivity and specificity of this assay in a diagnostic setting.
By combining the reactions into fewer assays, the multiplex design is able to achieve substantial savings in time and reagent costs compared with those required for monospecific methods. The two-step multiplex procedure can be readily adapted to laboratories familiar with RT-PCR procedures by conventional DNA detection methods. The one-step procedure offers several important advantages for laboratories with access to the required equipment. PCR products labeled by incorporation of fluorescence-labeled primers can be detected and sized (or typed) with sensitivity comparable to that of nested PCR, but without the added risk of DNA contamination associated with nested PCR procedures; moreover, the GeneScan Analysis Software makes it possible to size bands with single-base-pair accuracy (19), minimizing the chance of detecting an incorrect but similarly sized product.
In summary, the multiplex RT-PCR assays described here proved to be very effective for the detection and rapid identification of HPIVs. Since primers for other respiratory viruses can be added to the reaction mixture, the efficiencies of these assays can potentially be further improved. However, additional studies will be required to determine the extent to which these assays can complement or replace conventional diagnostic methods.
ACKNOWLEDGMENT
This study was performed at the Centers for Disease Control and Prevention with funding support from the Spanish Ministry of Science and Education (grant PR95-32; Estancias de Investigadores Españoles en Centros de Investigación Extranjeros).
REFERENCES
- 1.Apalsch A M, Green M, Ledesmamedina J, Nour B, Wald E R. Parainfluenza and influenza virus infections in pediatric organ transplant recipients. Clin Infect Dis. 1995;21:394–399. doi: 10.1093/clinids/20.2.394. [DOI] [PubMed] [Google Scholar]
- 2.Arola M, Ruuskanen O, Ziegler T, Salmi T T. Respiratory virus infections during anticancer treatment in children. Pediatr Infect Dis J. 1995;14:690–694. doi: 10.1097/00006454-199508000-00008. [DOI] [PubMed] [Google Scholar]
- 3.Casas I, Powell L, Klaper P E, Cleator G M. New method for the extraction of viral RNA and DNA from cerebrospinal fluid for use in the polymerase chain reaction assay. J Virol Methods. 1995;53:25–36. doi: 10.1016/0166-0934(94)00173-e. [DOI] [PubMed] [Google Scholar]
- 4.Collins P L, Chanock R M, McIntosh K. Parainfluenza viruses. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publications; 1996. pp. 1205–1241. [Google Scholar]
- 5.Gardner P S, Court S D M, Brocklebank S J M, Dowham M A P S, Weightman D. Virus cross infection in paediatric wards. Br Med J. 1973;2:571–575. doi: 10.1136/bmj.2.5866.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilbert L L, Dakhama A, Bone B M, Thomas E E, Hegele R G. Diagnosis of viral respiratory tract infections in children by using a reverse transcription-PCR panel. J Clin Microbiol. 1996;34:140–143. doi: 10.1128/jcm.34.1.140-143.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glasgow K W, Tamblyn S E, Blair G. A respiratory outbreak due to parainfluenza virus type 3 in a home for the aged—Ontario. Can Communicable Dis Rep. 1995;21:57–61. [PubMed] [Google Scholar]
- 8.Gorman W L, Gill D S, Scroggs R A, Portner A. The hemagglutinin-neuraminidase glycoproteins of human parainfluenza virus type 1 and Sendai virus have high structure-function similarity with limited antigenic cross-reactivity. Virology. 1990;175:211–221. doi: 10.1016/0042-6822(90)90201-2. [DOI] [PubMed] [Google Scholar]
- 9.Hendley J O. Parainfluenza viruses. In: Mandell G L, Douglas R G, Bennett J E, editors. Principles and practice of infectious diseases. 3rd ed. New York, N.Y: Churchill Livingstone; 1990. pp. 1255–1260. [Google Scholar]
- 10.Henrickson K J. Human parainfluenza viruses. In: Lennette E H, Lennette D A, Lennette E T, editors. Diagnostic procedures for viral, rickettsial, and chlamydial infections. 7th ed. Washington, D.C: American Public Health Association; 1995. pp. 481–494. [Google Scholar]
- 11.Hierholzer J C, Johansson K H, Anderson L J, Tsou C J, Halonen P E. Comparison of monoclonal time-resolved fluoroimmunoassay with monoclonal capture-biotinylated detector enzyme immunoassay for adenovirus antigen detection. J Clin Microbiol. 1989;25:1662–1667. doi: 10.1128/jcm.25.9.1662-1667.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jarvis W R, Middleton P J, Gelfand E W. Parainfluenza pneumonia in severe combined immunodeficiency disease. J Pediatr. 1979;94:423–425. doi: 10.1016/s0022-3476(79)80590-9. [DOI] [PubMed] [Google Scholar]
- 13.Josephs S, Kim H W, Brandt C O, Parrot R H. Parainfluenza 3 virus and other common respiratory pathogens in children with human immunodeficiency virus infection. Pediatr Infect Dis J. 1988;7:207–209. [PubMed] [Google Scholar]
- 14.Karron R A, Froehlich J L, Bobo L, Belshe R B, Yolken R. Rapid detection of parainfluenza virus type 3 RNA in respiratory specimens: use of reverse transcription-PCR-enzyme immunoassay. J Clin Microbiol. 1994;32:484–488. doi: 10.1128/jcm.32.2.484-488.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karron R A, O’Brien K L, Frohelich J L, Brown V A. Molecular epidemiology of a parainfluenza type 3 virus outbreak on a pediatric ward. J Infect Dis. 1993;167:1441–1445. doi: 10.1093/infdis/167.6.1441. [DOI] [PubMed] [Google Scholar]
- 16.Kawano M, Bando H, Yuasa T, Kondo K, Tsurudome M, Komada H, Nishio M, Ito Y. Sequence determination of the hemagglutinine-neuraminidase (HN) gene of human parainfluenza type 2 virus and the construction of a phylogenetic tree for HN proteins of all the paramyxoviruses that are infectious to humans. Virology. 1990;174:308–313. doi: 10.1016/0042-6822(90)90081-2. [DOI] [PubMed] [Google Scholar]
- 17.Meissner H C, Murray S A, Kieman M A, Snydman D A, McIntosh K. Simultaneous outbreak of respiratory syncytial virus and parainfluenza virus type 3 in a newborn nursery. J Pediatr. 1984;104:680–684. doi: 10.1016/s0022-3476(84)80943-9. [DOI] [PubMed] [Google Scholar]
- 18.Mufson M A, Mocega H E, Krause H E. Acquisition of parainfluenza 3 virus infection by hospitalized children. I. Frequencies, rates, and temporal data. J Infect Dis. 1973;128:141–147. doi: 10.1093/infdis/128.2.141. [DOI] [PubMed] [Google Scholar]
- 19.Pecharatana S, Pickett M A, Watt P J, Ward M E. Genotyping ocular strains of Chlamydia trachomatis by single-tube nested PCR. PCR Methods Appl. 1993;3:200–204. doi: 10.1101/gr.3.3.200. [DOI] [PubMed] [Google Scholar]
- 20.Sarkkinen H K, Halonen P E, Salmi A A. Type specific detection of parainfluenza viruses by enzyme-immunoassay and radioimmunoassay in nasopharyngeal specimens of patients with acute respiratory disease. J Gen Virol. 1981;56:49–57. doi: 10.1099/0022-1317-56-1-49. [DOI] [PubMed] [Google Scholar]
- 21.Shen K, Zhaori G, Zweygberg-Wirgart B, Ying M, Grandien M, Wahren B, Linde A. Detection of respiratory viruses in nasopharyngeal secretions with immunofluorescence technique for multiplex screening—an evaluation of the Chemicon assay. Clin Diagn Virol. 1996;6:147–154. doi: 10.1016/0928-0197(96)00229-2. [DOI] [PubMed] [Google Scholar]
- 22.Singh-Naz N, Willy M, Riggs N. Outbreak of parainfluenza virus type 3 in a neonatal nursery. Pediatr Infect Dis J. 1990;9:31–33. doi: 10.1097/00006454-199001000-00007. [DOI] [PubMed] [Google Scholar]
- 23.Storey D G, Côté M J, Dimock K, Yong Kang C. Nucleotide sequence of the coding and flanking regions of the human parainfluenza virus 3 hemagglutinin-neuraminidase gene: comparison with other paramyxoviruses. Intervirology. 1987;27:69–80. doi: 10.1159/000149722. [DOI] [PubMed] [Google Scholar]
- 24.Swierkosz E M, Erdman D D, Bonnot T, Schneiderheinze C, Waner J L. Isolation and characterization of a naturally occurring parainfluenza 3 virus variant. J Clin Microbiol. 1995;33:1839–1841. doi: 10.1128/jcm.33.7.1839-1841.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tenorio A, Echevarría J E, Casas I, Echevarría J M, Tabarés E. Detection and typing of human herpesviruses by multiplex polymerase chain reaction. J Virol Methods. 1993;44:261–269. doi: 10.1016/0166-0934(93)90061-u. [DOI] [PubMed] [Google Scholar]
- 26.Wendt C H, Weisdorf D J, Jordan M C, Balfour H H, Hertz M I. Parainfluenza virus respiratory infection after bone marrow transplantation. N Engl J Med. 1992;326:921–926. doi: 10.1056/NEJM199204023261404. [DOI] [PubMed] [Google Scholar]
- 27.Wendt C H, Hertz M I. Respiratory syncytial virus and parainfluenza virus infections in the immunocompromised host. Semin Respir Infect. 1995;10:224–231. [PubMed] [Google Scholar]
- 28.Whimbey E, Vartivarian S E, Champlin R E, Elting L S, Luna M, Bodey G P. Parainfluenza virus infection in adult bone marrow transplant recipients. Eur J Clin Microbiol Infect Dis. 1993;12:699–701. doi: 10.1007/BF02009383. [DOI] [PubMed] [Google Scholar]