Abstract
Bacteria from the family Vibrionaceae have been implicated in mass mortalities of farmed Pacific oysters (Magallana gigas) in multiple countries, leading to substantial impairment of growth in the sector. In Ireland there has been concern that Vibrio have been involved in serious summer outbreaks. There is evidence that Vibrio aestuarianus is increasingly becoming the main pathogen of concern for the Pacific oyster industry in Ireland. While bacteria belonging to the Vibrio splendidus clade are also detected frequently in mortality episodes, their role in the outbreaks of summer mortality is not well understood. To identify and characterize strains involved in these outbreaks, 43 Vibrio isolates were recovered from Pacific oyster summer mass mortality episodes in Ireland from 2008 to 2015 and these were whole-genome sequenced. Among these, 25 were found to be V. aestuarianus (implicated in disease) and 18 were members of the V. splendidus species complex (role in disease undetermined). Two distinct clades of V. aestuarianus – clade A and clade B – were found that had previously been described as circulating within French oyster culture. The high degree of similarity between the Irish and French V. aestuarianus isolates points to translocation of the pathogen between Europe’s two major oyster-producing countries, probably via trade in spat and other age classes. V. splendidus isolates were more diverse, but the data reveal a single clone of this species that has spread across oyster farms in Ireland. This underscores that Vibrio could be transmitted readily across oyster farms. The presence of V. aestuarianus clades A and B in not only France but also Ireland adds weight to growing concern that this pathogen is spreading and impacting Pacific oyster production within Europe.
Keywords: Vibrio aestuarianus, Vibrio splendidus, summer mortality syndrome, Ireland, aquaculture, transmission
Outcome
Pacific oyster culture in Ireland has increasingly suffered from summer mass mortality events. Many of these mortalities in recent years have been associated with Vibrio aestuarianus; the role of another pathogen, Vibrio splendidus has, so far, remained inconclusive. Here we show that two clades of V. aestuarianus are circulating in Ireland, and that these are members of two clades that have previously caused extensive oyster die offs in France. Their discovery in Ireland is consistent with transport of infected oyster stock between the two countries. Although V. splendidus-like strains in Ireland were highly diverse, a small clonal group was detected that appears to have spread rapidly from a single source to disparate locations in Ireland. Combined, these findings highlight the appearance of a highly pathogenic Vibrio in Ireland, and the risk of transmission between interconnected oyster production industries in Europe.
Data Summary
Sequences generated in this study were deposited in the National Center for Biotechnology Information (NCBI) database. Accession number: PRJNA797364. Publicly accessed genomes are listed in Table S2. This article contains data hosted by Microreact.
Introduction
While the aquaculture industry has expanded rapidly in the past 50 years, oyster production has struggled to keep pace with other aquaculture products [1]. One of the significant factors constraining the development of oyster aquaculture has been infectious disease [1, 2]. Pacific oysters (Magallana gigas, formerly Crassostrea gigas) are an important farmed species [3, 4], with 620 000 tonnes produced on average each year worldwide between 2010 to 2019, worth an estimated USD $1.29 billion a year [5]. France is the major European producer (84 760 tonnes in 2019), although there are significant industries in other European countries, including Ireland (10 460 tonnes in 2019). In France and elsewhere, there have been increased reports of disease outbreaks responsible for the depletion of oyster stocks over the last decade [3]. These present major socioeconomic consequences for the future of the oyster farming industry [6].
Episodes of abnormal mortality of Pacific oysters affecting all age classes have been described globally since the 1950s. Mortality of larvae and spat has been linked to the presence of a number of pathogenic agents, including ostreid herpes virus 1 (OsHV-1), whilst the term summer mortality syndrome has been coined to describe those events of mixed aetiology in the summer months affecting older oysters where gonad maturation is present [7]. Studies have shown that the causes of summer mortality syndrome are complex, often involving a combination of physiological and environmental stress, alongside the presence of pathogens [8], particularly bacteria belonging to the genus Vibrio , including V. aestuarianus and V. splendidus [9].
In the summer of 2008, abnormally high mortality episodes affecting spat and juvenile Pacific oysters were reported in both France and Ireland. The losses were linked to the emergence of a new variant of OsHV-1, termed ostreid herpes virus 1 µVariant (OsHV-1 µVar) [10]. Both V. splendidus and V. aestuarianus were also detected during a number of these events, although their role in these events was never fully elucidated [7]. Between 2011 and 2013, a new mortality phenomenon began to emerge in France affecting principally adult Pacific oysters. During this period, the frequency of detection of V. aestuarianus in cases of adult mortality increased significantly from 30 % in 2011 to 77 % of cases in 2013, becoming the principle pathogen detected during summer mortality episodes in adult oysters in France [11].
The Pacific oyster industry in Ireland is heavily dependent on the importation of spat, which is predominantly sourced from France [12]. Hence, following the reports of increased detections of V. aestuarianus in cases of adult mortality in France, a monitoring programme and a retrospective study were instigated in Ireland to determine the extent of its distribution in Ireland. In this study, we characterize and compare 43 Vibrio isolates recovered from diseased Irish oysters from 2008 to 2015 using whole-genome sequencing.
We show, firstly, that a high proportion of these oyster die-offs are associated with the presence of V. aestuarianus isolates from two oyster-associated V. aestuarianus subsp. francensis clades, clade A and clade B previously shown to be a major cause of summer mortality syndrome in France [11]. Secondly, we showcase differences in gene content diversity in these clades. Thirdly we show that V. splendidus strains present in Irish oysters are diverse, but a small clonal group was detected in 2009 in multiple locations.
Methods
Bacterial isolation and initial characterization
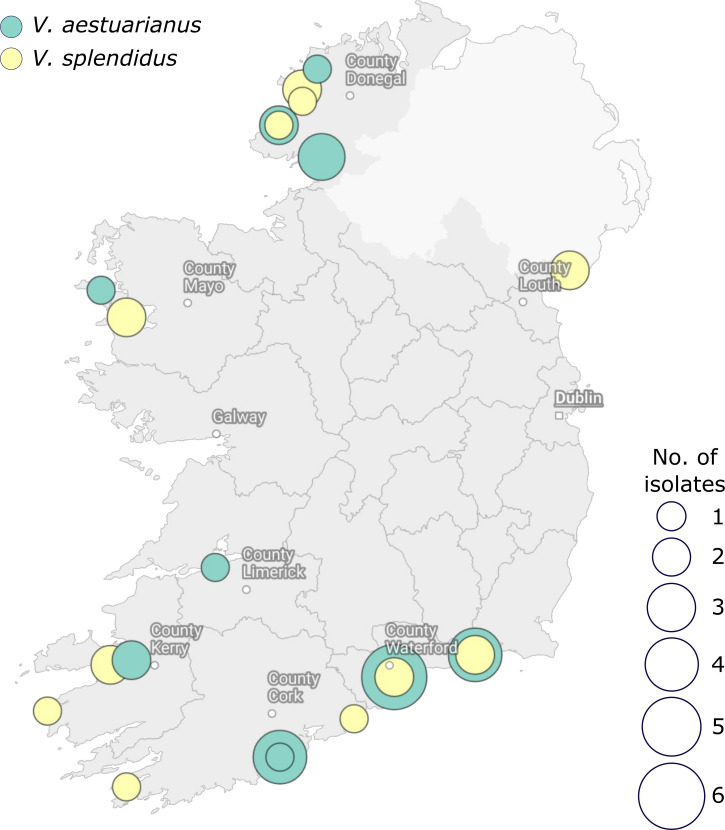
Forty-three Vibrio isolates obtained from oysters of varying age classes (Fig. 1 and Table 1) were collected from 22 sites around the Republic of Ireland between 2008 and 2015. Isolates were recovered from either haemolymph or crushed gill tissues and characterized. In most cases, isolates were recovered from sites where there were significant ongoing mortalities taking place (Table 1). They were then stored at −80 °C on cryovials using the protect storage system following the manufacturer’s instructions (Technical Service Consultants Ltd). To identify V. aestuarianus strains, primers developed for V. aestuarianus identification were used as described by McCleary and Henshilwood [13]. A 16S analysis was used to identify other Vibrio species as described by Lane et. al [14]. Primers used here were forward primer: 5′-AGAGTTTGATCCTGGCTCAG, reverse primer: 5′-GWATTACCGCGGCKGCTG.
Fig. 1.
Map of 43 strains sampled across Ireland; 24 V . splendidus and 18 V . aestuarianus isolates were collected from 23 locations. Pie charts indicate the proportion of each species sequenced from each location. These nodes are weighted by the number of isolates (scale, 1–6).
Table 1.
Vibrio strains selected for sequencing from Irish oysters
|
ID |
Year of extraction |
Site of extraction |
Tissue sample was extracted from |
Reported mortality rate % |
Age class |
Species |
|---|---|---|---|---|---|---|
|
16 025 |
2009 |
Loughros Beag |
Gill |
80–90 |
0+, 1+ |
|
|
16 028 |
2014 |
Woodstown |
Haemolymph |
60–70 |
3+ |
|
|
16 030 |
2014 |
Kinsale |
Haemolymph |
90 |
2+ |
|
|
16 033 |
2014 |
Dungarvan |
Gill |
10–20 |
0+, 1+ |
|
|
16 034 |
2014 |
Achill |
Gill |
10 |
0+ |
|
|
16 036 |
2015 |
Castlemaine |
Haemolymph |
40 |
2+ |
|
|
16 041 |
2015 |
Woodstown |
Gill |
1 |
1+ |
|
|
16 043 |
2015 |
Donegal Bay |
Haemolymph |
50–90 |
2+ |
|
|
16 044 |
2015 |
Dungarvan |
Haemolymph |
15–50 |
2+ |
|
|
16 048 |
2015 |
Dungarvan |
Haemolymph |
1 |
2+ |
|
|
16 049 |
2015 |
Dungarvan |
Haemolymph |
1.5 |
2+ |
|
|
16 050 |
2015 |
Woodstown |
Gill |
92 |
0+ |
|
|
16 053 |
2015 |
Kinsale |
Haemolymph |
70–80 |
2+ |
|
|
16 054 |
2010 |
Poularone Creek |
Gill |
30–40 |
0+ |
|
|
16 056 |
2015 |
Donegal Bay |
Haemolymph |
20 |
2+ |
|
|
16 057 |
2015 |
Dungarvan |
Haemolymph |
5.5 |
2+ |
|
|
16 058 |
2015 |
Dungarvan |
Haemolymph |
5 |
2+ |
|
|
16 059 |
2013 |
Kinsale |
Gill |
15–20 |
1+ |
|
|
16 060 |
2015 |
Donegal Bay |
Haemolymph |
2 |
2+ |
|
|
16 062 |
2015 |
Gweedore |
Haemolymph |
43 |
1+ |
|
|
16 063 |
2014 |
Kinsale |
Haemolymph |
90 |
1+ |
|
|
16 066 |
2013 |
Oysterhaven |
Gill |
50 |
0+, 1+, 2+ |
|
|
16 067 |
2015 |
Woodstown |
Haemolymph |
0 |
2+ |
|
|
16 070 |
2014 |
Woodstown |
Haemolymph |
60–70 |
1+ |
V. splendidus sensu stricto |
|
16 071 |
2014 |
Castlemaine |
Haemolymph |
30 |
2+ |
|
|
16 029 |
2009 |
Ballymacoda Bay |
Haemolymph |
20 |
0+ |
V. splendidus sensu stricto |
|
16 035 |
2009 |
Clew Bay |
Haemolymph |
10 |
0+ |
V. splendidus sensu stricto |
|
16 037 |
2013 |
Carlingford Lough |
Gill |
50 |
0+ |
V. splendidus sensu stricto |
|
16 051 |
2009 |
Clew Bay |
Haemolymph |
75 |
0+ |
V. splendidus sensu stricto |
|
16 052 |
2013 |
Dungloe Bay |
Haemolymph |
20 |
1+ |
V. splendidus sensu stricto |
|
16 065 |
2009 |
Dungloe Bay |
Haemolymph |
35 |
0+ |
V. splendidus sensu stricto |
|
16 069 |
2014 |
Woodstown Strand |
Haemolymph |
60 |
3+ |
V. splendidus sensu stricto |
|
16 072 |
2009 |
Clew Bay |
Haemolymph |
3–50 % |
0+ |
V. splendidus sensu stricto |
|
16 073 |
2009 |
Valentia River |
Haemolymph |
45 |
0+ |
V. splendidus sensu stricto |
|
16 074 |
2008 |
Dungarvan Harbour |
Haemolymph |
15 |
1+ |
V. splendidus sensu stricto |
|
16 061 |
2008 |
Dungarvan Harbour |
Haemolymph |
15 |
1+ |
V. splendidus sensu stricto |
|
16 077 |
2016 |
Woodstown Strand |
Haemolymph |
25 |
3+ |
V. splendidus sensu stricto |
|
16 078 |
2008 |
Castlemaine Harbour |
Haemolymph |
85 |
1+ |
V. splendidus sensu stricto |
|
16 079 |
2015 |
Trawenagh Bay |
Haemolymph |
70 |
1+ |
V. splendidus sensu stricto |
|
16 040 |
2009 |
Loughros Beag |
Haemolymph |
10 |
1+ |
V. splendidus sensu lato |
|
16 042 |
2013 |
Dunmanus Bay |
Haemolymph |
40 |
1+ |
V. splendidus sensu lato |
|
16 075 |
2010 |
Carlingford Lough |
Haemolymph |
30 |
0+ |
V. splendidus sensu lato |
DNA extraction and quantification
DNA was extracted from the isolates using the MasterPure Gram-positive DNA extraction kit (cat. no. MGP04100; Epicentre). The standard protocol was modified slightly to accommodate for the isolates being Gram-negative organisms. In summary, a 1 µl loopful of bacteria [previously sub-cultured onto seawater agar (SWA)] was placed into a 1.5 ml Eppendorf tube containing 1 ml 0.9 % saline. The solution was centrifuged at 1500 r.p.m., supernatant was removed and 150 µl TE buffer was added. Samples were vortexed to resuspend the pellet and 150 µl of a premade dilution of proteinase K in Gram-positive lysis solution was added to each sample, at a concentration of 1 µl proteinase K 150 µl−1 of Gram-positive lysis solution. The samples were vortexed and subsequently incubated at 65–70 °C for 15 min, which included vortexing every 5 min. Samples were cooled to 37 °C and then put on ice for 3–5 min, following which 175 µl of MPC protein precipitation reagent was added to each sample. Samples were vortexed and centrifuged at 1500 r.p.m. and 4 °C for 10 min. The supernatant was collected (pellets discarded) and 500 µl of isopropanol was added and samples were inverted 30–40 times and centrifuged again at 1500 r.p.m. and 4 °C for 10 min. The supernatant was removed, 70 % ethanol was added, and samples were centrifuged for a final time at 1500 r.p.m. and 4 °C for 5 min. Finally, the supernatant was removed, and samples were resuspended in 100 µl of molecular grade water and stored at −80 °C until future use. The extracted DNA was quantified using a Quantus fluorometer (Promega), and quality assessed using a NanoDrop ND-1000 Spectrophotometer (Thermo). Only those samples that passed the quality check were selected for high-throughput (Illumina) sequencing.
Illumina sequencing
Isolates were sequenced using an Illumina Miseq according to the standard protocols produced by the manufacturer. In brief, the DNA quantities were checked by fluorescence, diluted and prepared for sequencing with the Illumina Nextera XT library preparation kit, including optional 96-barcode adapters. Cleaned libraries were then sized-checked with an Agilent Technology 2100 Bioanalyzer using a high-sensitivity DNA chip and quantified by a Promega Quantus fluorometer using a OneDNA protocol. Finally, libraries were normalized, pooled and sequenced on the Miseq with Illumina V3 600 chemistry.
Quality check
Sequences were trimmed using Trimmomatic version 0.36, with the parameters :ILLUMINACLIP:*:2 : 30 : 10 MINLEN:36 SLIDINGWINDOW:4 : 20 TOPHRED64 [15]. FastQC version 0.11.7 was used to check the quality of trimmed reads, and to ensure that there were no significant contaminants [16].
Assembly and identification of open reading frames
Spades version 3.13.1 was used for assembly, with the parameters: -k 55,77,87,99,107,117,127 – careful –only assembler [17]. Prior to assembly, reads were downsampled to 100× coverage where needed. Reads were also merged using Flash v 1.2.11 with a minimum overlap of 10 bp, and maximum overlap equal to the maximum length of the reads per sample [18]. Contigs <500 bp were removed. In order to remove contigs with low coverage, reads were mapped to the assembly using bwa and SAMtools v1.8 was used to calculate coverage [19, 20]. Contigs with <10 % of the overall genome coverage, or at minimum 5× coverage, were removed. Assembled genomes were annotated using version 1.13 of Prokka, with the options: –addgenes –centre XXX –mincontiglen 200 –cdsrnaolap [21]. Quality assessment of assemblies was carried out using QUAST v4.6.3 [22]. QC scores for all reads and assemblies are provided in Table S1 (available in the online version of this article).
Accessing public genomes of V. splendidus and V. aestuarianus
We obtained publicly available WGS data for V. splendidus and V. aestuarianus in order to place the isolates from Irish oysters into broader phylogenetic contexts. Thirteen V. aestuarianus genomes were contributed by Goudenège et al. [23]. Assembled genomes of 102 isolates previously characterized as V. splendidus were downloaded from the National Center for Biotechnology Information (NCBI) database [24]. Information on each of these isolates can be found in Table S2. All subsequent genomic analysis was performed using datasets of 38 V . aestuarianus and 120 V . splendidus genomes.
Pangenome construction
A comprehensive pangenome of each species was constructed for using PIRATE [25], a toolbox for bacterial pangenomics analysis. Briefly, PIRATE produces putative gene families by first clustering protein-coding sequences using CD-HIT and carrying out an all-versus-all alignment of representative sequences for each cluster using Diamond’s blastp-like algorithm, filtering results that fall below a given threshold, and using the remaining bit scores to identify putative ‘gene families’ using the MCL algorithm. The process is then repeated, progressively filtering lower scoring matches, to identify the highest threshold at which genes are classified as belonging to a single family, following which there are some additional steps to identify orthologues, paralogues and fission loci. We used Phandango version 1.3.0 [26] to visualize the distribution of gene families within each population. Concatenated core genome alignments were built using PIRATE with default parameters. Individual gene family alignments were calculated using MAFFT for gene families present in at least 95 % of isolates with a maximum genome dosage (copy number) of 1.25, before being concatenated into a core genome alignment [27]. We used R version 3.2.3 [28] for statistical analysis and data visualization.
Core genome phylogeny
Based on a 2.56 Mb core genome alignment we constructed a bootstrapped phylogenetic tree for the 38 V . aestuarianus isolates using RAxML-NG version 0.9.0 [29] (tree building parameters: --model GTR+G seed 2 --tree pars{25},rand{25}, bootstrapping parameters: --model GTR+G --bs-trees 200). For the larger V. splendidus dataset, we constructed a neighbour-joining tree using RapidNJ (parameters: --bootstrap 100) [30] with a core genome of 2.97 Mb. Each tree was rooted at the midpoint using Figtree [31]. Phylogenies were visualized using Microreact [32]. The project URLs are https://microreact.org/project/gfAsh7KuduL4xuSTDaVU5r-vibrio-aestuarianus ( V. aestuarianus ) and https://microreact.org/project/eMABqKLAPcn2QG5NEnCVor-vibrio-splendidus ( V. splendidus ). Pairwise SNP distances between isolates in the core genome alignment were calculated using Disty McMatrixface 0.1.0, which calculates pairwise differences in a given alignment, ignoring Ns in a pairwise manner [33].
Phage prediction
We used PHAge Search Tool (PHAST) [34] to identify potential phages in clade A isolate 12 142, French clade B isolate 01 308 and Irish clade B isolate 16 060 as representative genomes for each genome condition. Fasta assembly files were assessed using default PHAST parameters.
All bioinformatics was carried out using resources provided by MRC-CLIMB [35].
Results
V. aestuarianus: presence of two clades in Ireland
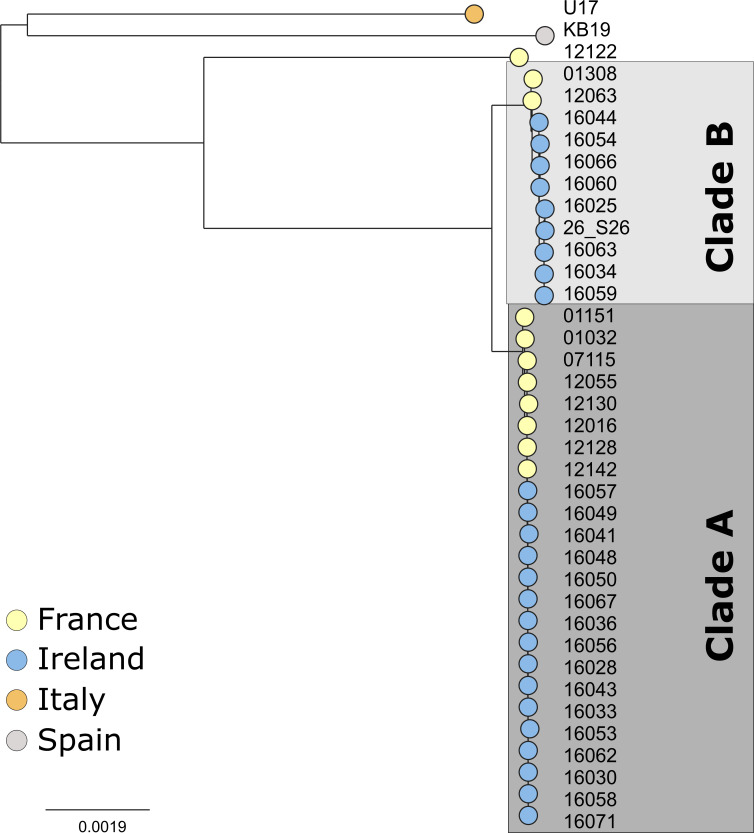
The core genome phylogeny of V. aestuarianus (Figs 2 and S2a) revealed that the French and Irish isolates were highly similar. The Irish isolates were resolved into the same two clades, A and B, previously reported to be circulating in French oyster culture [23]. Strains isolated from these two countries differ by 50 SNPs on average in clade A and 416 SNPs in clade B.
Fig. 2.
Core genome phylogeny of 38 V . aestuarianus isolates reveals two clades circulating in Ireland and France. A maximum-likelihood tree of 38 V . aestuarianus isolates constructed using a concatenated core genome alignment. The tree is rooted at the midpoint. The scale bar represents a mean of 0.0019 nucleotide substitutions per site. Tree tips are coloured by country of isolation. Isolates recovered in Ireland fall within two previously identified clades circulating in France, Clade A and clade B.
V. aestuarianus : gene content variation in each clade
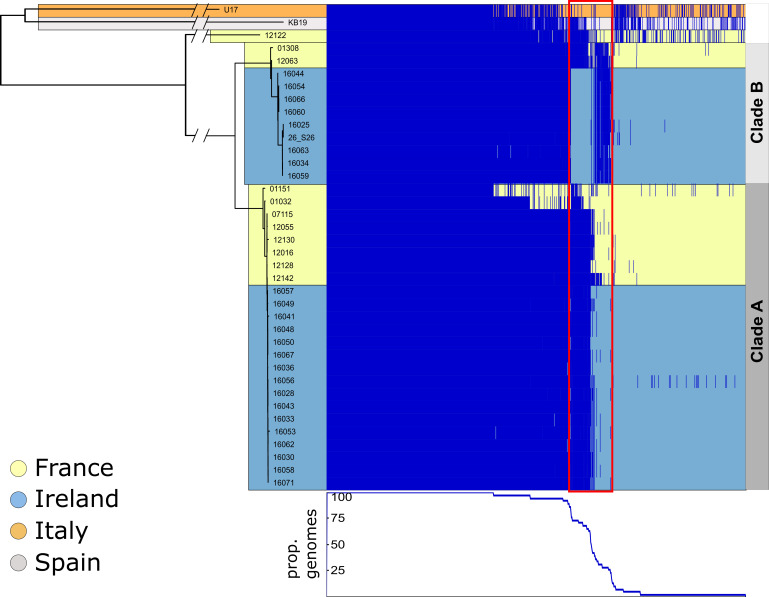
The pangenome of V. aestuarianus consists of 5650 gene families (Fig. 3). This includes 2746 core gene families present in at least 95 % of isolates, 1150 shared by 10–95% isolates and 1754 shared by a single isolate up to 10 % of isolates. Isolates 01151 and 01032 are missing many core genes due to poor-quality assemblies: these were excluded from further pangenome analyses.
Fig. 3.
Gene presence–absence of 38 V . aestuarianus . Presence–absence heatmap of the pangenome of 38 V . aestuarianus genomes generated by Phandango [26]. Dark blue blocks indicate the presence of a gene family. The proportion of genomes each gene family has been detected in is shown below the heatmap. Tree branches and heatmap rows are coloured by country of isolation. Tree lengths have been truncated, see Fig. 2 for true branch lengths. Indicated in a red box are multiple genes that differ between clade A and clade B. Isolates 01151 and 01032, French clade B isolates, notably contain most of these genes.
A set of 215 gene families present in all clade A isolates are absent in clade B isolates (Fig. 3). These genes are likely to have been horizontally acquired as mobile genetic elements (MGEs). To examine this, we checked the locations of these genes on the genome of the clade A isolate 12 142 and compared the GC content of these genes to the rest of the genome. The 215 genes resolved into 19 contiguous blocks of genes, each containing at least 2 genes (Table S3). The largest of these regions of contiguous genes contains 48 genes and has a GC content of 45.56 %, slightly higher than the genome average of 42.65 %. Another 13.5 kb region with 15 genes and a GC content of 43 % can be found 866 kb away from this region on the same contig. These two large gene regions have been identified as phages using PHAST (Table S4). The remaining contiguous blocks of genes are distributed across 11 contigs and contain mostly hypothetical proteins (108 of 130 genes). The presence of antitoxin- and phage-related proteins (YafN and IntA) suggests that many of these genes may lie on other uncharacterized mobile elements or plasmids.
Clade B isolates contain 92 gene families that are not shared with clade A, and the location of these genes was checked in clade B using isolate 16 060 as a representative genome. These are also largely hypothetical proteins (63 of 92) and are spread across 32 contigs in isolate 16 060, each carrying between 1 and 9 of these genes (Table S5). Genes related to two citrate fermentation operons that allow citrate to be used as an energy source in V. cholerae , citCDEFXG and citS-oadGAB-citAB [36], are only present in clade B isolates. citD-G and citX are all colocalized with citB and citA (also known as dpiA and dpiB). Genes oadA, oadB and oadG are found with citC and copies of citA and citX. No citS genes were detected in this species. Genes citA and citG, and one copy of citX are also found in one clade A genome: 12 142. vspR, a virulence gene repressor in Vibrio cholera [37], is also only found in Irish clade B genomes.
We also note that clade B strains isolated in France harbour both sets of genes, the 92 clade B genes and the 215 genes that are otherwise unique to clade A. This indicates that the clade B strains from Ireland included in this study have experienced extensive gene loss.
V. splendidus : widespread clonal group uncovered
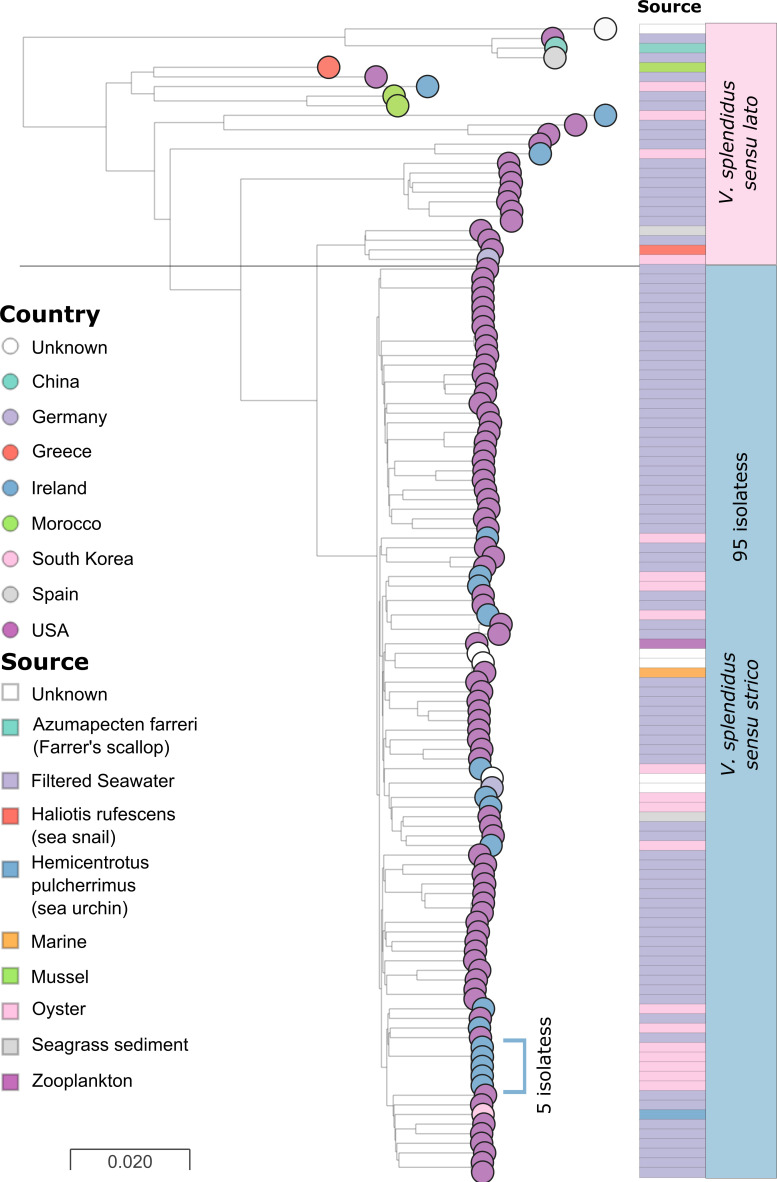
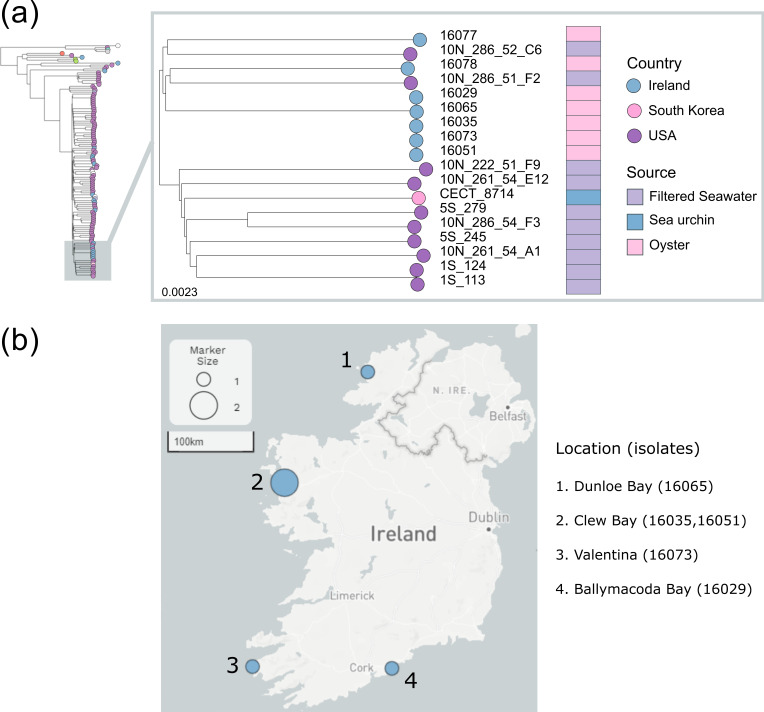
To place V. splendidus isolates appearing in Irish oysters within the population structure of this species, we compared these 18 strains to 102 publicly available V . splendidus genomes. The phylogeny of V. splendidus isolates revealed a large cluster of 95 isolates, accompanied by multiple more diverse lineages (Figs 4 and S2b). Here we have referred to the large clade of 95 genomes as V. splendidus sensu stricto, while more diverse lineages are referred to as V. splendidus sensu lato. Of the newly sequenced strains, 15 are found within V. splendidus sensu stricto, while three strains lie within the broader population. Although the publicly accessed genomes were all classified as V. splendidus species, a phylogenetic comparison with reference genomes within V. splendidus clade has shown that many of the more diverse V. splendidus sensu lato isolates in this dataset are likely to have been misclassified (Fig. 5). Instead, these isolates are expected to represent other species from the V. splendidus species complex. Thus, we have designated isolates 16 040, 16 042 and 16 075 as V. splendidus -like isolates [38]. A cluster of five isolates recovered from four separate locations in Ireland show high similarity within this population (Fig. 6). These isolates differ by 28 SNPs on average across the core genome alignment, whereas the remaining 10 Irish isolates within the V. splendidus sensu stricto cluster differ by 83, on average.
Fig. 4.
V. splendidus core genome phylogeny. Neighbour-joining core genome phylogeny of 18 V. splendidus isolates sequenced here and 102 V . splendidus isolates accessed on the NCBI coloured by country of isolation. The scale bar represents an estimated dissimilarity of 2 % of the alignment. The tree is annotated with the source of isolation. Publicly available samples largely come from the USA and were sampled in seawater. The population structure of the dataset includes a large cluster of 95 genomes (purple). Isolates from Ireland are distributed throughout this population. However, one cluster of five highly similar isolates can be identified (blue).
Fig. 5.
Phylogeny of V. splendidus species complex. A reference genome (in bold) for 13 species belonging to the V. splendidus species complex were combined with the 120 V . splendidus genomes used previously. Above is a neighbour-joining tree constructed using a core genome alignment. V. splendidus sensu stricto, containing a V. splendidus reference strain, is collapsed, and represents 96 isolates. Eleven of the publicly accessible genomes identified as V. splendidus species on NCBI that do not fall within V. splendidus sensu stricto are more similar to V. splendidus -like reference genomes. Similarly, three genomes isolated in Ireland – 16075, 16 040 and 16 042 seen in blue – are not found within the V. splendidus sensu stricto clade and are likely V. splendidus -like species.
Fig. 6.
Subtree of V. splendidus reveals a cluster of five highly similar isolates in Ireland. (a) A subtree of 19 V . splendidus isolates from the tree shown in Fig. 4, including 5 Irish isolates with high similarity. Tree tips are coloured by country and annotated with the source of isolation. (b) Map of the five related V. splendidus strains shows these isolates were not recovered in the same locations. Nodes are weighted by the number of isolates per location.
The pangenome of this dataset contains 18 891 gene families, with a core genome of 3513 genes (95–100 % of isolates) and 13 270 rare accessory genes (0–10 % of isolates) (Fig. S1). Forty-two gene families are unique to the five Irish clonal group isolates. These include 18 genes dispersed within a 35.6 kb region, including a trio of resistance-related genes: cobalt–zinc–cadmium resistance protein, czcA; multidrug resistance protein, mdtA; and outer membrane protein oprM. Multiple genes related to stress response and signalling are also found in this region, including nreB oxygen sensor histidine kinase; cmpR a transcriptional activator involved in CO2 stress [39]; htpG a chaperone protein involved in general stress responses [40]; a putative signalling protein; and pdeB, a gene implicated in biofilm formation [41].
Discussion
In Ireland, V. aestuarianus has been detected in oyster mortality events reported to the Marine Institute in 2001, 2003, 2006 and 2007, and more frequently in mortality events in spat from 2008 onwards, which had previously been attributed to OsHV-1 [42] (D. Cheslett, personal communication). Whilst mortality in adult oysters was only infrequently reported in Ireland prior to 2012, the frequency of reports and the detection of V. aestuarianus increased in line with those seen in France, particularly from 2015 onwards, following massive mortality events countrywide in 2015 [42, 43]. The trend of increased detections mirrored that in France; although the timeline of increased detections was later than that reported in France [11–13, 43]. The predominant pathogen detected in cases of adult and half-grown mortality in Ireland was V. aestuarianus , whilst that in spat was OsHV-1µVar. However, other bacteria, particularly other Vibrio sp., have also been isolated, mainly in conjunction with OsHV-1 and V. aestuarianus . Here by applying whole-genome sequencing we have characterized Vibrio strains that might play a major role in Irish oyster mortality events.
Two V. aestuarianus clades linked with oyster mortalities in both Ireland and France
Our results show that all V. aestuarianu s strains detected in oysters in Ireland are members of two V. aestuarianus subsp. francensis clades, A and B, which have previously been detected in France [23]. SNP analysis revealed a high level of identity between the Irish and French V. aestuarianus isolates, suggesting that the clades causing disease outbreaks in France are also responsible for disease outbreaks across Ireland. There is a significant trade in live oysters between France and Ireland [12, 44], which has likely facilitated the movement of pathogens between rearing areas. However, broader genomic surveillance of V. aestuarianus associated with oyster mortalities is needed to uncover the exact distribution of each clade outside of these key M. gigas-producing countries.
A recent study involving the sequencing of V. aestuarianus strains across Europe showed that these two oyster-associated clades have now been found in multiple countries within Europe [45]. Their findings suggest that the emergence of these clades is the result of adaptation to a new environmental niche within which they have become oyster-specialist pathogens. The authors found low genomic diversity within each clade. Thus, the high genetic identity between Irish and French strains does not necessarily indicate a direct transmission chain between these two countries. While the data assessed here cannot be used to evaluate fine-scale transmission events between Ireland and France in V. aestuarianu s, we advocate for further whole-genome sequencing efforts within and across interconnected oyster-producing countries in Europe and elsewhere to help capture the spread and evolution of these emerging infectious clades [46].
Evidence of gene loss in Irish clade B strains
Our data revealed a large number of gene families that are found in French but not Irish clade B isolates. This difference in genome content may suggest that a clade B strain was introduced once to Ireland, and that the founder population lost or previously lacked those genes. Although some of these genes were revealed to be on phages, the mechanisms of gene loss of the remaining 152 non-consecutive gene families in these otherwise highly related strains has not been determined. It is possible that this rapid genome reduction may have conferred a selective advantage to the Irish strains [47]. Given that these Irish strains are only compared to two strains from France, more extensive sequencing of clade B isolates across a wider range of affected regions is needed to evaluate the full diversity of the clade and determine whether this gene loss is exclusive to these Irish strains.
A single clone of V. splendidus highlights transmission potential
V. splendidus clade strains were frequently detected in Irish oyster mortalities, although the role they played in disease is uncertain. Here we showed that these isolates were mostly distinct strains within a highly diverse species complex. V. splendidus is a highly diverse species and opportunistic pathogen [48, 49]. Given this, we would expect isolates associated with disease in Ireland to be largely unrelated, unless they happened to be isolated in the same location at one time or had recently been introduced through a common source. In 2009, a clonal group of highly similar isolates was found in multiple locations across Ireland (Fig. 6). In all cases, samples were taken where mortality was occurring in recently introduced French oyster seed. Both OsHV-1 µVar and V. splendidus were detected, suggesting that these isolates may be linked through the source of oyster seed. While this clonal group may have proliferated across Irish waters in 2009, given that such events have not been described in this species to date, it is much more likely that it was spread to multiple farms through a common source. Indeed, at least four of these isolates were found in sites which at that time contained stock from the same hatchery in France. The occurrence of this highly related clonal group of V. splendidus across multiple sites in the same year signifies the presence of transmission routes available to important oyster pathogens between production facilities.
Perspectives
Pacific oyster summer mortality events in Ireland are shown here to be associated with two V. aestuarianus clades and a variety of strains within the V. splendidus complex. Notably, the two V. aestuarianus clades in Ireland have been described elsewhere in Europe, as clade A and B [23, 45]. Novel lineages were not detected, which underscores the importance of these two clades in Pacific oyster summer mortalities. The occurrence of a probable transmission event of V. splendidus across Ireland emphasizes the capacity for the spread of potentially pathogenic Vibrio within the oyster industry. Further genomic surveillance studies, which can build on this one, are needed within countries experiencing summer mortality syndrome and countries with which they frequently trade. This could lead to a fuller picture of the proliferation and evolution of this emerging pathogen worldwide and to better measures to prevent or deal with its future spread.
Supplementary Data
Funding information
This project was funded by the Department for Environment and Rural Affairs (DEFRA). N.M.C. was funded by the University of Bath and Raoul and Catherine Hughes. J.C.L.T. was funded by The University of Bath.
Acknowledgements
The authors would like to thank Dr Sion Bayliss and Dr Harry Thorpe for their contributions to the bioinformatics scripts used in this study. We would also like to acknowledge the contribution of the staff at the Fish Health Unit, Marine Institute, Ireland. Many thanks to Dr Georg Engelhard for his generous guidance on the manuscript. Finally, the authors would like to extend gratitude to Dr Marie-Agnès Travers and colleagues at Ifremer for contributing data to this study.
Author contributions
N.C.: formal analysis, investigation, methodology, software, visualization, writing – original draft, writing – review and editing. C.O.T.: data curation, investigation, writing – original draft, writing – review and editing. J.T.: investigation, methodology, writing – original draft, writing – review and editing. D.R.: data curation, formal analysis, investigation, methodology, visualization, writing – original draft, writing – review and editing. M.G.: investigation. T.B.: conceptualization, methodology. A.W.J.: investigation. A.W.: investigation. E.F.: conceptualization, methodology, supervision, writing – original draft, writing – review and editing. D.C.: conceptualization, supervision, writing – original draft, writing – review and editing. D.V.J.: conceptualization, funding acquisition, resources, supervision, writing – original draft, writing – review and editing.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: OsHV-1 µVar, Osteid herpes virus-1 µVariant; SNP, single nucleotide polymorphism; V. aestuarianus, Vibrio aestruarianus; V. splendidus, Vibrio splendidus.
All supporting data, code and protocols have been provided within the article or through supplementary data files.
Two supplementary figures and five supplementary tables are available with the online version of this article.
References
- 1.Botta R, Asche F, Borsum JS, Camp EV. A review of global oyster aquaculture production and consumption. Marine Policy. 2020;117:103952. doi: 10.1016/j.marpol.2020.103952. [DOI] [Google Scholar]
- 2.Jennings S, Stentiford GD, Leocadio AM, Jeffery KR, Metcalfe JD, et al. Aquatic food security: insights into challenges and solutions from an analysis of interactions between fisheries, aquaculture, food safety, human health, fish and human welfare, economy and environment. Fish Fish. 2016;17:893–938. doi: 10.1111/faf.12152. [DOI] [Google Scholar]
- 3.Barbosa Solomieu V, Renault T, Travers M-A. Mass mortality in bivalves and the intricate case of the Pacific oyster, Crassostrea gigas . J Invertebr Pathol. 2015;131:2–10. doi: 10.1016/j.jip.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 4.Salvi D, Mariottini P. Revision shock in Pacific oysters taxonomy: the genus Magallana (formerly Crassostrea in part) is well-founded and necessary. Zool J Linn Soc. 2021;192:43–58. doi: 10.1093/zoolinnean/zlaa112. [DOI] [Google Scholar]
- 5.Food and Agriculture Organization of the United Nations FAO Yearbook. fishery and aquaculture statistics 2019/FAO Annuaire. Statistiques des Pêches et de L’Aquaculture 2019/FAO Anuario. Estadísticas de Pesca Y Acuicultura 2019. Food Agriculture Org. 2021 [Google Scholar]
- 6.Girard S, Pérez Agúndez JA. The effects of the oyster mortality crisis on the economics of the shellfish farming sector: preliminary review and prospects from a case study in Marennes-Oleron Bay (France) Mar Policy. 2014;48:142–151. doi: 10.1016/j.marpol.2014.03.024. [DOI] [Google Scholar]
- 7.EFSA Panel on Animal Health and welfare (AHAW) Scientific opinion on the increased mortality events in Pacific oysters, Crassostrea gigas . EFSA J. 2010;8:1894. doi: 10.2903/j.efsa.2010.1894. [DOI] [Google Scholar]
- 8.Wendling CC, Wegner KM. Relative contribution of reproductive investment, thermal stress and Vibrio infection to summer mortality phenomena in Pacific oysters. Aquaculture. 2013;412–413:88–96. doi: 10.1016/j.aquaculture.2013.07.009. [DOI] [Google Scholar]
- 9.Lemire A, Goudenège D, Versigny T, Petton B, Calteau A, et al. Populations, not clones, are the unit of vibrio pathogenesis in naturally infected oysters. ISME J. 2015;9:1523–1531. doi: 10.1038/ismej.2014.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Segarra A, Pépin JF, Arzul I, Morga B, Faury N, et al. Detection and description of a particular Ostreid herpesvirus 1 genotype associated with massive mortality outbreaks of Pacific oysters, Crassostrea gigas, in France in 2008. Virus Res. 2010;153:92–99. doi: 10.1016/j.virusres.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 11.European Union Reference Laboratory 2014 Annual meeting of the National reference Laboratories for Mollusc diseases. 2014;22 [Google Scholar]
- 12.Collins E, McCleary S, Morrissey T, Geary M, Connor CO, et al. Sporulating Haplosporidium nelsoni in Crassostrea gigas in a production Bay in Ireland. Bulletin of the EAFP. 2018;38:4–11. [Google Scholar]
- 13.McCleary S, Henshilwood K. Novel quantitative TaqMan® MGB real-time PCR for sensitive detection of Vibrio aestuarianus in Crassostrea gigas . Dis Aquat Organ. 2015;114:239–248. doi: 10.3354/dao02869. [DOI] [PubMed] [Google Scholar]
- 14.Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, et al. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andrews S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Cambridge, United Kingdom: Babraham Bioinformatics, Babraham Institute; 2010. [Google Scholar]
- 17.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 22.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goudenège D, Travers MA, Lemire A, Petton B, Haffner P, et al. A single regulatory gene is sufficient to alter Vibrio aestuarianus pathogenicity in oysters. Environ Microbiol. 2015;17:4189–4199. doi: 10.1111/1462-2920.12699. [DOI] [PubMed] [Google Scholar]
- 24.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–5. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bayliss SC, Thorpe HA, Coyle NM, Sheppard SK, Feil EJ. PIRATE: a fast and scalable pangenomics toolbox for clustering diverged orthologues in bacteria. Gigascience. 2019;8:giz119. doi: 10.1093/gigascience/giz119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hadfield J, Croucher NJ, Goater RJ, Abudahab K, Aanensen DM, et al. Phandango: an interactive viewer for bacterial population genomics. Bioinformatics. 2017 doi: 10.1101/119545. [DOI] [PMC free article] [PubMed]
- 27.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.R Core Team R: A language and environment for statistical computing. 2013.
- 29.Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35:4453–4455. doi: 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crandall KA, Lagergren J. Algorithms in Bioinformatics. Berlin, Heidelberg: Springer; 2008. Rapid neighbour-joining; pp. 113–122. [DOI] [Google Scholar]
- 31.Rambaut A. FigTree. 2016.
- 32.Argimón S, Abudahab K, Goater RJE, Fedosejev A, Bhai J, et al. Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb Genom. 2016;2:e000093. doi: 10.1099/mgen.0.000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ranjith KM, Brinda K, Arjun U, Hegde NG, Nath R. Double phase transition in the triangular antiferromagnet Ba3CoTa2O9 . J Phys Condens Matter. 2017;29:115804. doi: 10.1088/1361-648X/aa57be. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. PHAST: a fast phage search tool. Nucleic Acids Res. 2011;39:W347–52. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Connor TR, Loman NJ, Thompson S, Smith A, Southgate J, et al. CLIMB (the Cloud Infrastructure for Microbial Bioinformatics): an online resource for the medical microbiology community. Microb Genom. 2016;2:e000086. doi: 10.1099/mgen.0.000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu M, Hao G, Li Z, Zhou Y, Garcia-Sillas R, et al. CitAB two-component system-regulated citrate utilization contributes to Vibrio cholerae competitiveness with the gut microbiota. Infect Immun. 2019;87:e00746-18. doi: 10.1128/IAI.00746-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davies BW, Bogard RW, Young TS, Mekalanos JJ. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell. 2012;149:358–370. doi: 10.1016/j.cell.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pérez-Cataluña A, Lucena T, Tarazona E, Arahal DR, Macián MC, et al. An MLSA approach for the taxonomic update of the Splendidus clade, a lineage containing several fish and shellfish pathogenic Vibrio spp. Syst Appl Microbiol. 2016;39:361–369. doi: 10.1016/j.syapm.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 39.Omata T, Gohta S, Takahashi Y, Harano Y, Maeda S. Involvement of a CbbR homolog in low CO2-induced activation of the bicarbonate transporter operon in cyanobacteria. J Bacteriol. 2001;183:1891–1898. doi: 10.1128/JB.183.6.1891-1898.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grudniak AM, Markowska K, Wolska KI. Interactions of Escherichia coli molecular chaperone HtpG with DnaA replication initiator DNA. Cell Stress Chaperones. 2015;20:951–957. doi: 10.1007/s12192-015-0623-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chao L, Rakshe S, Leff M, Spormann AM. PdeB, a cyclic Di-GMP-specific phosphodiesterase that regulates Shewanella oneidensis MR-1 motility and biofilm formation. J Bacteriol. 2013;195:3827–3833. doi: 10.1128/JB.00498-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.European Union Reference Laboratory Annual Meeting and Seventh Combined Technical Workshop of the National Reference Laboratories for Mollusc Diseases. Ifremer. 2016
- 43.European Union Reference Laboratory Report of the 2020 Annual Meeting of National Reference Laboratories for Mollusc Diseases (IFREMER). Ifremer. 2020.
- 44.Roux FL, Wegner KM, Baker-Austin C, Vezzulli L, Osorio CR, et al. The emergence of Vibrio pathogens in Europe: ecology, evolution, and pathogenesis (Paris, 11–12th March 2015) Front Microbiol. 2015;6 doi: 10.3389/fmicb.2015.00830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mesnil A, Jacquot M, Garcia C, Tourbiez D, Canier L, et al. Emergence and clonal expansion of Vibrio aestuarianus lineages pathogenic for oysters in Europe. Mol Ecol. 2023;32:2869–2883. doi: 10.1111/mec.16910. [DOI] [PubMed] [Google Scholar]
- 46.Bayliss SC, Verner-Jeffreys DW, Bartie KL, Aanensen DM, Sheppard SK, et al. The promise of whole genome pathogen sequencing for the molecular epidemiology of emerging aquaculture pathogens. Front Microbiol. 2017;8:121. doi: 10.3389/fmicb.2017.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koskiniemi S, Sun S, Berg OG, Andersson DI. Selection-driven gene loss in bacteria. PLoS Genet. 2012;8:e1002787. doi: 10.1371/journal.pgen.1002787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Le Roux F, Zouine M, Chakroun N, Binesse J, Saulnier D, et al. Genome sequence of Vibrio splendidus: an abundant planctonic marine species with a large genotypic diversity. Environ Microbiol. 2009;11:1959–1970. doi: 10.1111/j.1462-2920.2009.01918.x. [DOI] [PubMed] [Google Scholar]
- 49.Gay M, Renault T, Pons A-M, Le Roux F. Two vibrio splendidus related strains collaborate to kill Crassostrea gigas: taxonomy and host alterations. Dis Aquat Organ. 2004;62:65–74. doi: 10.3354/dao062065. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.