Abstract
Periodontitis, a disease responsible for tooth loss worldwide, is characterized by chronic inflammation of the periodontium, eventually leading to destruction of periodontal ligaments and supporting alveolar bone. Spirochetes, identified by dark-field microscopy as being the most predominant bacteria in advanced lesions, are thought to play a causative role. Various spirochetal morphotypes were observed, but most of these morphotypes are as yet uncultivable. To assess the role of these organisms we designed oligonucleotide probes for the identification of both cultivable and so far uncultivable spirochetes in periodontitis patients. Subgingival plaque specimens taken from diseased sites (n = 200) and healthy control sites (n = 44) from 53 patients with rapidly progressive periodontitis (RPP) were submitted to direct in situ hybridization or dot blot hybridization after prior amplification with eubacterial primers. Spirochetes were found in all patients, but their distributions varied considerably. Parallel use of oligonucleotide probes specific for cultivable or so far uncultivable treponemes suggested the presence of novel yet unknown organisms at a high frequency. These uncultivable treponemes were visualized by fluorescence in situ hybridization, and their morphologies, sizes, and numbers could be estimated. All RPP patients included in this study harbored oral treponemes that represent either novel species, e.g., Treponema maltophilum, or uncultivable phylotypes. Therefore, it is necessary to include these organisms in etiologic considerations and to strengthen efforts to cultivate these as yet uncultivable treponemes.
Treponemes comprise a large group of spirochetes found in important infections such as syphilis or periodontal disease (12). While the role of Treponema pallidum in the pathogenesis of syphilis is well documented, the etiologic role of oral treponemes in periodontitis is postulated on the basis of the presence of elevated numbers of these organisms in periodontal lesions (9, 21). Spirochetes predominate in most patients who have chronic periodontal disease but who have not responded to therapy. Although various spirochetal morphotypes have been observed, most of these have not been cultured. Among the four cultivable oral treponema species presently recognized, Treponema denticola has been most frequently associated with chronic periodontal disease. However, it remains to be determined whether this organism is of etiologic relevance or merely the most easily cultured organism. Recent molecular genetic analyses revealed an unexpected diversity of treponema sequences in a subgingival plaque sample from a single periodontitis patient (5). More than 50 treponemal rRNA sequences were found, and these clustered into eight major taxonomic groups (groups I to VIII) exhibiting ≥92% sequence similarity. These groups could be divided into 23 phylotypes exhibiting ≥98% sequence homology, hence representing mostly novel yet uncultivable treponema species. In light of this observation it was necessary to reassess the etiologic role of oral treponemes in periodontal disease by applying methods that detect both cultivable and as yet uncultivable organisms. We synthesized a number of group- or phylotype-specific oligonucleotide probes to determine the frequency of known and novel organisms in subgingival plaque samples from 53 patients with rapidly progressive periodontitis (RPP) by dot blot or in situ hybridization.
MATERIALS AND METHODS
Clinical samples.
A total of 244 subgingival plaque specimens (200 specimens from deep periodontal pockets and 44 specimens from healthy control sites) from 53 RPP patients (average age, 34.7 years) were investigated. All patients were previously untreated. The patients received a diagnosis of RPP according to their advanced clinical and radiographical appearance in combination with their age and their history of periodontal disease, following the criteria of Page et al. (20). Patients who had chronic disease or patients who had received anti-inflammatory or antimicrobial therapy within the previous 6 months were excluded from the study. Subgingival plaque samples were taken from diseased sites with a probing pocket depth of ≥6 mm and bleeding on probing. Whenever possible a sample from an additional control site not clinically affected by the disease was selected. After supragingival plaque removal with a sterile curette and cotton pellet, three sterile paper points (ISO 35; Becht, Offenburg, Germany) were inserted into the pockets. After 10 s the paper points were removed and placed into 1 ml of reduced transport fluid (32) containing 25% glucose, transferred to the laboratory, and processed immediately.
Dark-field microscopy.
The total bacterial cell count in all samples was estimated by dark-field microscopy. The number of spirochetes was determined semiquantitatively as counts per microscopic field at ×1,000 magnification.
DNA extraction and amplification.
Aliquots of the plaque specimen of 100 μl were centrifuged at 13,000 × g for 10 min in a Labofuge 400 R centrifuge (Hereus, Hanau, Germany). The resulting bacterial pellets were placed in 100 μl of lysis buffer as described previously (5). No further purification of nucleic acids was performed. One microliter of bulk DNA was then added to the amplification mixture (final reaction volume, 100 μl) for in vitro amplification by PCR in a thermal cycler (Trioblock; Biometra, Göttingen, Germany) for 30 cycles of denaturation (1 min, 95°C), annealing (1 min, 56°C), and extension (1 min, 72°C). The broad-range eubacterial primers used for 16S rRNA gene amplification were TPU1 (5′-AGA GTT TGA TCM TGG CTC AG-3′; corresponding to positions 8 to 27 in the Escherichia coli 16S rRNA gene) (4) and RTU3 (5′-GWA TTA CCG CGG CKG CTG-3′; corresponding to complementary positions 519 to 536 in E. coli 16S rRNA) (4). Successful amplification was verified by agarose gel electrophoresis.
Oligonucleotide probes.
Oligonucleotide probes TRE I to TRE VII specific for all major phylogenetic clusters of oral treponemes were designed according to the phylogenetic tree retrieved from an earlier comparative 16S rRNA analysis (5). The sequences of these probes were as follows: TRE I, 5′-ACGCAAGCTCATCCTCAAG-3′; TRE II, 5′-GCTCTTTTCCTCATTTACCTTTAT-3′; TRE III, 5′-CCCCATCTTAAAGGTAGATCAC-3′; TRE IV, 5′-CGGTCACATTCGGTATTACCTACT-3′; TRE V, 5′-CCTTTATTCCGTGAGACCTTATC-3′; TRE VI, 5′-GTGGGCGCGTCGTCCACGCGTTAC-3′; and TRE VII, 5′-CCCATCCGAGAGGTACGTCATCCA-3′.
To assess specificity, the sequences of the probes were compared with those of all 16S rRNA entries at the EMBL and GeneBank databases currently (July 1997) accessible by using the program BLASTN of the Husar (version 4.0; Heidelberg Unix Sequence Analysis Resources) program package (DKFZ, Heidelberg, Germany). Probes TDEN, TVIN, TSOC, TPEC, and TMAL were designed to detect the known cultivable treponemes T. denticola, T. vincentii, T. socranskii, and T. pectinovorum or a novel, recently described species, T. maltophilum (34), respectively. The sequences of these probes were as follows: TDEN, 5′-CATGACTACCGTCATCAAAGAAGC-3′; TVIN, 5′-ATTGAGACTATTCGGTATTACCTGC-3′; TSOC, 5′-CATTGCTGCCTGCCGCTCGACTTG-3′; TPEC, 5′-CTCCAACTTATATGACCTTATCCG-3′; and TMAL, 5′-CTATTGTGCTTATTCATCAGGC-3′. All probes were checked for their practical use in hybridization experiments by using the program OLIGO (version 4.0). The probe EUB338, complementary to a region of the 16S rRNA gene conserved in the domain Bacteria, was used as a positive control (2).
Dot blot hybridization.
Dot blot hybridization of PCR-amplified plaque material was used to detect minute amounts of treponemes and to determine their presence in individual patients. After denaturation of PCR products, aliquots of 1 μl were spotted onto nylon membranes (Hybond N; Amersham, Buckinghamshire, United Kingdom) and were fixed by UV cross-linking (MWG Biotech, Ebersberg, Germany). A total of 34 products of amplified DNA from either recombinant clones retrieved from the original 16S rRNA gene library, known cultivable treponemes, or other putative periodontal pathogens were included as controls in all dot blot hybridizations. All probes were labeled nonisotopically with digoxigenin (DIG)-ddUTP (Boehringer Mannheim, Mannheim, Germany) and were detected by chemiluminescence according to the manufacturer’s recommendations. All hybridizations were performed at 54°C. Stringency washes were performed at temperatures of between 56 and 64°C with a washing buffer containing 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.2% sodium dodecyl sulfate (SDS) or 0.1× SSC–0.1% SDS, depending on the respective probe. DIG-labeled probes were detected with anti-DIG-alkaline phosphatase conjugates after adding the appropriate substrate according to the manufacturer’s recommendations. X-ray films were exposed to the membranes for 2 to 12 h. After stripping with 0.2 N NaOH–0.1% SDS (stripping buffer), identical membranes were used for multiple hybridization experiments with the probes mentioned above.
Statistical analysis.
Statistical evaluation of the dot blot hybridization results was done by the chi-square test. The site-specific data were regarded as independent.
In situ hybridization.
In situ hybridization was performed to determine the frequency of occurrence of specific treponemes in a given subgingival plaque sample. For fixation of cells, 100 μl of a subgingival plaque sample suspension was pelleted at 5,200 × g for 10 min and washed twice with cold phosphate-buffered saline (PBS; pH 7.4). Finally, the cells were resuspended in 100 μl of PBS with 3.7% (vol/vol) formaldehyde. Fixed cells (2 μl) were spotted onto gelatin-coated [0.01% KCr(SO4)2 (wt/vol), 0.1% gelatin (wt/vol)] microscopic slides (Paul Marienfeld KG, Bad Mergentheim, Germany), air dried, and dehydrated in 50, 80, and 96% (vol/vol) ethanol.
Oligonucleotides identical to those used for the dot blot experiments were used for the in situ hybridizations. Fluorescence labeling was performed with 5-amino-propargyl-2′-deoxycytidine 5′-triphosphate coupled to Cy3 fluorescent dye (Cy3-dCTP; Amersham Life Sciences, Arlington Heights, Ill.) or fluorescein-12-dUTP (Boehringer Mannheim) and terminal transferase (Boehringer Mannheim).
For whole-cell hybridization, a 10-μl aliquot of the hybridization mix containing 20 mM Tris HCl, 0.9 M NaCl, 0.01% SDS, 0 to 20% formamide, and approximately 50 ng of the fluorescent probe were applied to each sample on microscopic slides. After 1 to 3 h of hybridization at 46°C in a moist chamber in the dark, each of the slides was washed for 30 min in preheated (46°C) washing buffer containing 20 mM Tris HCl, 0.01% SDS, and 0.9 to 0.225 M NaCl (depending on the formamide concentration used during hybridization) to ensure stringency. Finally, the slides were mounted with Citifluor AF1 (The Chemical Laboratory of the University of Kent, Kent, United Kingdom). The bacteria were observed with an Axioskop microscope (Zeiss, Jena, Germany) with the respective filter combinations (HQ filter sets F41-007 and F41-001; AHF Analysentechnik, Tübingen, Germany) at a magnification of ×1,000. For documentation, photomicrographs were taken with Kodak Ektachrome HC 400 film.
RESULTS
Dark-field microscopy.
Spirochetes were found in all 53 RPP patients. They were detected in 197 of 200 deep periodontal pockets and 19 of 44 control sites. Their numbers varied from none to about 10/microscopic field at healthy sites and about 1 to ≥102/microscopic field (×1,000 magnification; average of 10 microscopic fields) at diseased sites.
Dot blot hybridization.
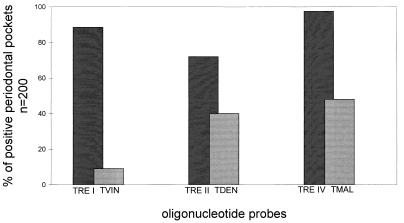
A strong hybridization signal was obtained with the eubacterium-specific probe EUB338 for all subgingival plaque specimens, indicating that PCR amplification was not hampered by the presence of inhibitors. Accordingly, all negative controls, i.e., samples that contained no DNA, showed no hybridization signal, suggesting a lack of contamination by carryover of amplified material. All 34 controls containing amplified DNA from either recombinant clones of the 16S rDNA gene library mentioned above, known cultivable treponemes, or relevant putative periodontal pathogens were detected only by the respective probe. No cross hybridization was observed (Fig. 1). With the exception of T. pectinovorum, which has not been found in any specimen, all other treponemal phylotypes were detected. All phylotypes except group VI organisms (P = 0.180) and T. vincentii (P = 0.040) were detected significantly more often in the deep periodontal pockets than in the respective control sites (P < 0.005). T. socranskii and group I and IV organisms were present in more than 85% of the deep subgingival pockets and in 96.2 and 100% of the patients, respectively (Table 1). In contrast, group III, V, VI, and VII oral treponemes were found in only 49.1, 20.8, 7.5, or 39.6% of the patients, respectively. Great discrepancy was observed for cultivable and as yet uncultivable treponemes of groups I and II. While T. vincentii, the only group I treponemal species cultivable so far, was found in 20.8% of the patients and in only 9% of all deep pockets and none of the control sites, probe TRE I detected treponemes in each patient and in 88.5% of diseased sites and 34.1% of control sites (Fig. 1 and 2). A similar discrepancy was observed for probes TRE II and TDEN. Although T. denticola was found in about 40% of diseased sites and 2.3% of healthy sites, as yet uncultivable treponemes detected by probe TRE II were found in 72% of affected sites and 15.9% of unaffected sites (Fig. 2).
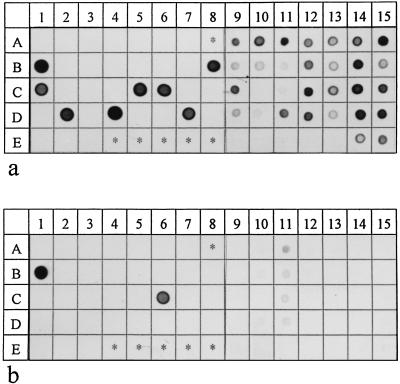
FIG. 1.
Dot blot hybridizations of identical membranes with group-specific probe TRE I (a) and species-specific probe TVIN (b). The strains were kindly provided by R. Mutters, Marburg, Germany (identified as RM); C. Wyss, Zürich, Switzerland (identified as CW); and B. Wilske, Munich, Germany (identified as BW). In columns 1 to 8 PCR products of the following strains were applied as controls: the putative oral pathogens Actinobacillus actinomycetemcomitans MCCM 02638 (A1) (RM), Capnocytophaga gingivalis MCCM 00858 (A2), (RM), Capnocytophaga ochracea MCCM 00238 (A3) (RM), Eubacterium lentum ATCC 25559T (A4) (RM), Fusobacterium nucleatum ATCC 25586T (A5) (RM), Porphyromonas gingivalis ATCC 33277 (A6) (RM), and Prevotella intermedia MCCM 00407 (A7) (RM); the cultivable treponema species T. vincentii ATCC 35580 (B1), T. denticola ATCC 35405T (B2), T. socranskii subsp. socranskii ATCC 35536 (B3), T. socranskii subsp. buccale ATCC 35534 (B4), T. maltophilum ATCC 51939T (B5) (CW), and T. phagedenis subsp. reiterii (B6) (BW); a clinical isolate (CW) (B8; highest degree of homology to clone NZM 3142), and T. pectinovorum ATCC 33768T (E1); group I recombinant clones NZM3D292 (C1), NZM3D464 (C5), NZM3112 (C6; sequence 100% homologue to probe TVIN), NZM3142 (D2), NZM3147 (D4), and NZM3166 (D7); group II recombinant clones NZM3106 (C7) and NZM3158 (D6); group III recombinant clones NZM3143 (D3), NZM3D298 (C3), and NZM3D527 (C4); group IV recombinant clones NZM3122 (C8), NZM3D505 (C2), and NZM3125 (D8); group V recombinant clones NZM3124 (D1) and NZM3155 (D5); the group VI recombinant clone NZM3104 (E2); and the group VII recombinant clone NZM3D384 (E3). In columns 9 to 15 PCR products from subgingival plaque samples were applied: lanes A to D, PCR products from deep periodontal pockets; lane E, PCR products from the respective controls.
TABLE 1.
Distribution of cultivable and uncultured oral treponemes in RPP patients
| Oligonucleotide probe group and probe | % Positive patientsa
|
|
|---|---|---|
| Deep periodontal pockets (n = 53) | Control sites (n = 44) | |
| Group-specific probes | ||
| TRE I | 100.0 | 34.1 |
| TRE II | 86.8 | 15.9 |
| TRE III | 49.1 | 6.8 |
| TRE IV | 100.0 | 47.7 |
| TRE V | 20.8 | 0.0 |
| TRE VI | 7.5 | 0.0 |
| TRE VII | 39.6 | 2.3 |
| Species-specific probes | ||
| TVIN | 20.8 | 0.0 |
| TDEN | 62.3 | 4.5 |
| TMAL | 66.0 | 9.1 |
| TSOC | 96.2 | 20.5 |
| TPEC | 0.0 | 0.0 |
At least one positive site, as analyzed by dot blot hybridization.
FIG. 2.
Presence of cultivable versus uncultivable oral treponemes in RPP patients revealed by dot blot hybridization with group-specific probes (probes TRE I, TRE II, and TRE IV) and species-specific probes (probes TVIN, TDEN, and TMAL).
In situ hybridization.
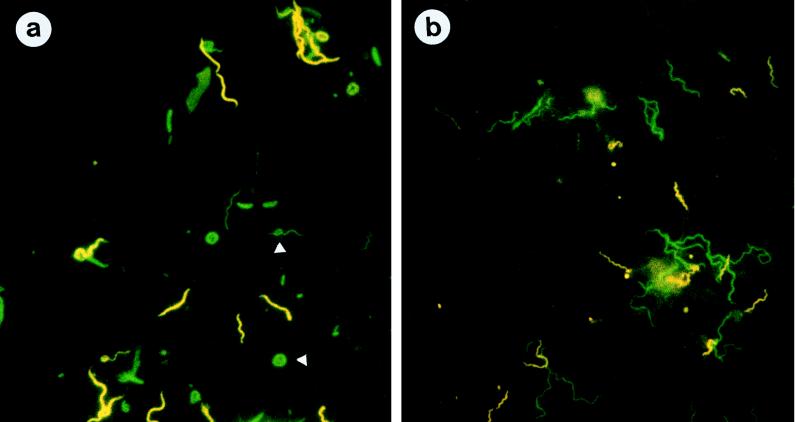
The results of in situ hybridization experiments with oligonucleotide probes and patient specimens identical to those used for dot blot hybridization complemented the dot blot hybridization results and indicated that these organisms are present in high proportions in subgingival plaque samples and thus represent the predominant flora. Figure 3a shows a microphotograph from representative subgingival plaque material after simultaneous hybridization with the fluorescein isothiocyanate (FITC)-labeled probe EUB338 and the Cy3-labeled probe TRE I. With the FITC filter set the diversity of the microbial community in the periodontal plaque sample could be observed, since bacteria of all different morphologies and sizes were stained (green). With the Cy3 filter combination, group I treponemes appeared as large, thick spirochetes (yellow). Because in this sample T. vincentii, the only cultivable species of group I, was not detected by dot blot hybridization, we assume that these spirochetes are as yet uncultivable. In situ hybridization of the same patient material with TRE IFITC and TRE IICy3 revealed that group II treponemes are rather small and thin with many waves and occurred less frequently than group I treponemes in this specimen (Fig. 3b).
FIG. 3.
Fluorescence in situ hybridization of subgingival plaque material from an RPP patient. (a) Microphotograph showing simultaneous hybridization with EUB338FITC (green) and TRE ICy3 (yellow). The eubacterial probe reveals the different morphotypes of subgingival plaque bacteria and the spherical bodies of the treponemes (arrows), as described by Wecke et al. (33). (b) Microphotograph showing hybridization with TRE IFITC (green) and TRE IICy3 (yellow). Note the different morphologies of the treponemes detected with the group-specific probes.
DISCUSSION
As with most other mixed infections, conceptual and technical difficulties were encountered in searches for the etiologic agents of periodontal infections (7, 11, 18, 31). In the past, dark-field microscopy and culture-based methods have been used to link the presence of elevated numbers of one or more organisms with the existence of disease. When subgingival plaques were analyzed by dark-field microscopy, spirochetes usually represented between 10 and 60% of the total bacterial count (3, 13–16, 28). Not only was there great variation in the distribution of spirochetes among individual patients but there was also significant intraindividual variation, with proportions ranging from 10 to 50% in different lesions of the same patient (8). Despite its ease and elegance, dark-field microscopy is not very useful for etiologic analyses since spirochetes cannot be specified by this method. In contrast, use of the predominant cultivable organisms approach allowed the biochemical identification and taxonomic characterization of bacteria growing in pure culture. Unfortunately, there was great discrepancy between the numbers of spirochetal morphotypes seen by dark-field microscopy and the rather small numbers of cultivable oral treponema species. In addition, depending on the culture conditions, the highest recoveries averaged about 1% of the total cultivable microbiota (17, 19, 27, 34). Although it may be biased to recognize not the etiologically relevant but merely the bacteria that are most easily cultured, the predominant cultivable organisms approach is still widely accepted. Furthermore, this approach does not distinguish between overgrowth of the opportunistic organisms that colonize niches created by the underlying disease and increases in the proportions of the true pathogens that cause periodontitis. Riviere and coworkers (23–26) applied immunofluorescence microscopy with monoclonal antibodies raised against T. pallidum to the detection of yet uncultured treponemes. They found tissue-invasive, so-called pathogen-related oral spirochetes (PROS), which occurred at a high frequency (23–26). However, we showed recently that PROS do not represent a single treponema species but rather represent a heterogeneous group of spirochetes clustering in the group I oral treponemes, of which T. vincentii is currently the only cultivable species (6). In the study presented here, yet uncultured group I isolates were found at a high frequency, but T. vincentii was found in only 9% of the samples, suggesting that most of the group I phylotypes and possibly PROS-positive treponemes have not been cultured so far. A discrepancy not as pronounced as that for T. vincentii and group I treponemes has been observed for T. denticola and group II spirochetes: T. denticola (29), which has often been found at frequencies of as high as 90% in other studies (27, 29), was detected in only 40% of the patient’s specimens, but as yet uncultured group II treponemes were present in 72% of the patient’s periodontal pockets. Group IV treponemes, including the novel species T. maltophilum, were found in each patient and 97.5% of all samples of subgingival plaque material. Positive hybridization signals with treponeme-specific probes in samples in which no spirochetes had been observed by dark-field microscopy are explained by the greater sensitivity of PCR amplification (<102 organisms/ml) compared to that of microscopy.
All treponemes described here predominated at diseased sites but were detected only infrequently at periodontally healthy sites, indicating that they indeed may be of etiologic relevance. However, whether the spirochetes found at control sites belong to the resident flora or originated from spillover periodontal lesion sulcus fluid carrying a high load of the respective treponemal phylotypes cannot be answered by the results of this study. It is intriguing to speculate that some of the treponemes found at healthy sites may later induce periodontal disease.
Highly sensitive molecular biology-based genetic methods may detect pathogenic microorganisms even in the absence of disease, thus questioning the specificity of the parasite-disease association demanded by Koch’s postulates (10). Therefore, molecular geneticists, periodontists, and oral microbiologists have formulated guidelines for establishing causal relationships between microbes and disease (10, 30, 31). Large prospective molecular biology-based epidemiological studies that should also include so-called periodontitis-resistant populations (1, 22) will be required to unravel the etiology of periodontal disease.
In conclusion, sequence-based identification in combination with dot blot and in situ hybridization analyses provide strong evidence for the causal role of treponemes in periodontal disease and may be of value in the development of new diagnostic and therapeutic strategies in order to potentially manage these ubiquitous and important infections.
ACKNOWLEDGMENTS
This study has been supported by a grant (01KI9318) from the Bundesministerium für Bildung und Forschung (U.B.G.) and the Körber European Research Award (U.B.G.).
REFERENCES
- 1.Africa C W, Parker J R, Reddy J. Bacteriological studies of subgingival plaque in a periodontitis resistant population. I. Darkfield microscopic studies. J Periodontol Res. 1985;20:1–7. doi: 10.1111/j.1600-0765.1985.tb00403.x. [DOI] [PubMed] [Google Scholar]
- 2.Amann R I, Binder B J, Olson R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armitage G C, Dickinson W R, Jenderseck R S, Levine S M, Chambers D W. Relationship between the percentage of subgingival spirochetes and the severity of periodontal disease. J Periodontol. 1982;53:550–556. doi: 10.1902/jop.1982.53.9.550. [DOI] [PubMed] [Google Scholar]
- 4.Brosius J, Palmer L, Kennedy J P, Noller H F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci USA. 1987;75:4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi B K, Paster B J, Dewhirst F E, Göbel U B. Diversity of cultivable and uncultivable oral spirochetes from a patient with severe destructive periodontitis. Infect Immun. 1994;62:1889–1895. doi: 10.1128/iai.62.5.1889-1895.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi B K, Wyss C, Göbel U B. Phylogenetic analysis of pathogen-related oral spirochetes (PROS) J Clin Microbiol. 1996;34:1922–1925. doi: 10.1128/jcm.34.8.1922-1925.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christersson L A, Zambon J J, Genco R J. Dental bacterial plaques. Nature and role in periodontal disease. J Clin Periodontol. 1991;18:441–446. doi: 10.1111/j.1600-051x.1991.tb02314.x. [DOI] [PubMed] [Google Scholar]
- 8.Evian C J, Rosenberg E S, Listgarten M A. Bacterial variability within diseased periodontal sites. J Periodontol. 1982;53:595–598. doi: 10.1902/jop.1982.53.10.595. [DOI] [PubMed] [Google Scholar]
- 9.Fiehn, N.-K. 1989. Small-sized oral spirochetes and periodontal disease. Acta Pathol. Microbiol. Scand. 97(Suppl. 7):1–31. [PubMed]
- 10.Fredricks D N, Relman D A. Sequence-based identification of microbial pathogens: a reconsideration of Koch’s postulates. Clin Microbiol Rev. 1996;9:18–33. doi: 10.1128/cmr.9.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kornman K S. Nature of periodontal diseases: assessment and diagnosis. J Periodontol Res. 1987;22:192–204. doi: 10.1111/j.1600-0765.1987.tb01566.x. [DOI] [PubMed] [Google Scholar]
- 12.Lancet Spirochetes in periodontal disease. Lancet. 1991;338:1177–1178. . (Editorial.) [PubMed] [Google Scholar]
- 13.Liljenberg B, Lindhe J. Juvenile periodontitis. Some microbiological, histopathological and clinical characteristics. J Clin Periodontol. 1980;7:48–61. doi: 10.1111/j.1600-051x.1980.tb01948.x. [DOI] [PubMed] [Google Scholar]
- 14.Lindhe J, Liljenberg B, Listgarten M A. Some microbiological and histopathological features of periodontal diseases in man. J Periodontol. 1980;51:264–269. doi: 10.1902/jop.1980.51.5.264. [DOI] [PubMed] [Google Scholar]
- 15.Listgarten M A, Hellden L. Relative distribution of bacteria at clinically healthy and periodontally diseased sites in humans. J Clin Periodontol. 1978;5:115–132. doi: 10.1111/j.1600-051x.1978.tb01913.x. [DOI] [PubMed] [Google Scholar]
- 16.Listgarten M A, Levin S. Positive correlation between the proportions of subgingival spirochetes and motile bacteria and susceptibility of human subjects to periodontal deterioration. J Clin Periodontol. 1981;8:122–138. doi: 10.1111/j.1600-051x.1981.tb02352.x. [DOI] [PubMed] [Google Scholar]
- 17.Loesche W J. The role of spirochetes in periodontal disease. Adv Dent Res. 1988;2:275–283. doi: 10.1177/08959374880020021201. [DOI] [PubMed] [Google Scholar]
- 18.Moore W E C. Microbiology of periodontal disease. J Periodontol Res. 1987;22:335–341. doi: 10.1111/j.1600-0765.1987.tb01595.x. [DOI] [PubMed] [Google Scholar]
- 19.Moore W E C, Holdeman E V, Smibert S M, Hash D E, Burmeister J A, Ranney R R. Bacteriology of severe periodontitis in young adult humans. Infect Immun. 1982;38:1137–1148. doi: 10.1128/iai.38.3.1137-1148.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Page R C, Altmann L C, Ebersole J L, Vandesteen G E, Dahlberg W H, Williams B L, Osterberg S K. Rapidly progressive periodontitis. A distinct clinical condition. J Periodontol. 1983;54:197–209. doi: 10.1902/jop.1983.54.4.197. [DOI] [PubMed] [Google Scholar]
- 21.Penn C-W. Pathogenicity and molecular biology of treponemes. Rev Med Microbiol. 1991;2:68–75. [Google Scholar]
- 22.Reddy J, Africa C W, Parker J R. Darkfield microscopy of subgingival plaque of an urban black population with poor oral hygiene. J Clin Periodontol. 1986;13:578–582. doi: 10.1111/j.1600-051x.1986.tb00850.x. [DOI] [PubMed] [Google Scholar]
- 23.Riviere G R, Wagoner M A, Baker-Zander S A, Weisz K S, Adams D F, Simonson L, Lukehart S A. Identification of spirochetes related to Treponema pallidum in necrotizing ulcerative gingivitis and chronic periodontitis. N Engl J Med. 1991;325:539–543. doi: 10.1056/NEJM199108223250803. [DOI] [PubMed] [Google Scholar]
- 24.Riviere G R, Weisz K S, Adams D F. Relative proportions of pathogen-related oral spirochetes (PROS) and Treponema denticola in supragingival and subgingival plaque from patients with periodontitis. J Periodontol. 1991;63:131–136. doi: 10.1902/jop.1992.63.2.131. [DOI] [PubMed] [Google Scholar]
- 25.Riviere G R, Weisz K S, Adams D F, Thomas D D. Pathogen-related oral spirochetes from dental plaque are invasive. Infect Immun. 1991;59:3377–3380. doi: 10.1128/iai.59.10.3377-3380.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riviere G R, Weisz K S, Simonson L G, Lukehart S A. Pathogen-related spirochetes identified within gingival tissue from patients with acute necrotizing ulcerative gingivitis. Infect Immun. 1991;59:2653–2657. doi: 10.1128/iai.59.8.2653-2657.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salvador S L, Syed S A, Loesche W J. Comparison of three dispersion procedures for quantitative recovery of cultivable species of subgingival spirochetes. J Clin Microbiol. 1987;25:2230–2232. doi: 10.1128/jcm.25.11.2230-2232.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Savitt E D, Socransky S S. Distribution of certain subgingival microbial species in selected periodontal conditions. J Periodontol Res. 1984;19:111–123. doi: 10.1111/j.1600-0765.1984.tb00800.x. [DOI] [PubMed] [Google Scholar]
- 29.Simonson L G, Goodman C H, Bial J J, Morton H E. Quantitative relationship of Treponema denticola to severity of periodontal disease. Infect Immun. 1988;56:726–728. doi: 10.1128/iai.56.4.726-728.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Socransky S S. Microbiology of periodontal disease—present status and future considerations. J Periodontol. 1977;48:497–504. doi: 10.1902/jop.1977.48.9.497. [DOI] [PubMed] [Google Scholar]
- 31.Socransky S S, Haffajee A D, Smith G L F, Dzink J L. Difficulties encountered in the search for the etiologic agents of destructive periodontal diseases. J Clin Periodontol. 1987;14:588–593. doi: 10.1111/j.1600-051x.1987.tb01520.x. [DOI] [PubMed] [Google Scholar]
- 32.Syed S A, Loesche W J. Survival of human dental plaque flora in various transport media. Appl Microbiol. 1972;24:638–644. doi: 10.1128/am.24.4.638-644.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wecke J, Wolf V, Fath S, Bernimoulin J P. The occurrence of treponemes and their spherical bodies on polytetrafluoroethylene membranes. Oral Microbiol Immunol. 1995;10:278–283. doi: 10.1111/j.1399-302x.1995.tb00154.x. [DOI] [PubMed] [Google Scholar]
- 34.Wyss C, Choi B K, Schüpbach P, Guggenheim B, Göbel U B. Treponema maltophilum sp. nov., a small oral spirochete isolated from human periodontal lesions. Int J Syst Bacteriol. 1996;46:745–752. doi: 10.1099/00207713-46-3-745. [DOI] [PubMed] [Google Scholar]