Significance
Cytokinins are phytohormones that play pivotal roles in regulating plant growth and numerous agricultural traits. To date, cytokinin biosynthesis was believed to occur exclusively within the cell. However, our studies using Oryza sativa have revealed that in addition to the intracellular pathway mediated by phosphoribohydrolase, the final biosynthetic step also occurs in the apoplastic space by the action of a distinct enzyme. This pathway, catalyzed by a cytokinin/purine riboside nucleosidase, plays a role in the deribosylation of apoplastic cytokinin precursors. Our study shows that loss-of-function of the gene affected panicle development. The identification of this pathway extends our spatial understanding of cytokinin metabolism to the extracellular space.
Keywords: apoplast, cell wall, cytokinin, nucleosidase, Oryza sativa
Abstract
In the final step of cytokinin biosynthesis, the main pathway is the elimination of a ribose-phosphate moiety from the cytokinin nucleotide precursor by phosphoribohydrolase, an enzyme encoded by a gene named LONELY GUY (LOG). This reaction accounts for most of the cytokinin supply needed for regulating plant growth and development. In contrast, the LOG-independent pathway, in which dephosphorylation and deribosylation sequentially occur, is also thought to play a role in cytokinin biosynthesis, but the gene entity and physiological contribution have been elusive. In this study, we profiled the phytohormone content of chromosome segment substitution lines of Oryza sativa and searched for genes affecting the endogenous levels of cytokinin ribosides by quantitative trait loci analysis. Our approach identified a gene encoding an enzyme that catalyzes the deribosylation of cytokinin nucleoside precursors and other purine nucleosides. The cytokinin/purine riboside nucleosidase 1 (CPN1) we identified is a cell wall–localized protein. Loss-of-function mutations (cpn1) were created by inserting a Tos17-retrotransposon that altered the cytokinin composition in seedling shoots and leaf apoplastic fluid. The cpn1 mutation also abolished cytokinin riboside nucleosidase activity in leaf extracts and attenuated the trans-zeatin riboside-responsive expression of cytokinin marker genes. Grain yield of the mutants declined due to altered panicle morphology under field-grown conditions. These results suggest that the cell wall–localized LOG-independent cytokinin activating pathway catalyzed by CPN1 plays a role in cytokinin control of rice growth. Our finding broadens our spatial perspective of the cytokinin metabolic system.
Cytokinins play essential roles in numerous aspects of plant growth and development. The primary activities include promoting cell proliferation and shoot development by interplay with auxin. Cytokinin activities are also involved in regulating agriculturally important traits, such as leaf senescence, abiotic and biotic stress responses, nitrogen-responsive growth promotion, and increased grain number (1–7). The basic structure and roles of cytokinin metabolic, transport, and signal transduction systems must be characterized to elucidate the molecular basis underlying these diverse activities.
The major pathways for cytokinin biosynthesis have been determined from studies in Arabidopsis thaliana and Oryza sativa. The initial step of de novo cytokinin biosynthesis is the production of the N6-(∆2-isopentenyl)adenine (iP) nucleotide precursor catalyzed by adenosine phosphate–isopentenyltransferase (8, 9). Conversion to the trans-zeatin (tZ) nucleotide precursor, which is hydroxylated at the prenyl side chain terminus, is subsequently catalyzed by CYP735A (10). The precursors are converted to nucleobase active forms by a cytokinin nucleotide phosphoribohydrolase encoded by a gene named LONELY GUY (LOG) (11). Phenotypic analyses of the loss-of-function mutants of these biosynthesis genes in Arabidopsis and O. sativa have shown that these three reactions play a central role in de novo cytokinin biosynthesis (12–16).
Before the discovery of LOG, another pathway in which dephosphorylation and deribosylation reactions sequentially occur was proposed for producing active cytokinin forms based on an analogy with purine metabolism (4, 17). These enzymes, nucleotidase and nucleosidase, were partially purified and characterized from wheat germ (18, 19). However, the corresponding genes were not identified, and little attention was paid to this LOG-independent cytokinin activation pathway.
Cytokinin riboside–type precursors, such as tZ riboside (tZR), are major cytokinin species in xylem sap (20–23), and tZR is responsible for root-to-shoot signaling to maintain normal shoot growth (14, 24). Tracer experiments showed that tZR entering cells is rapidly phosphorylated to tZR 5′-monophosphate by adenosine kinase (25) and then activated to tZ by LOG (16). In the Arabidopsis log1log2log3log4log5log7log8 septuple mutant, in which all functional LOG genes are mutated, growth is severely inhibited, yet a faint but measurable amount of active-form cytokinin is still detected (16). These lines of evidence imply that LOG-independent pathways occur and can play a role in cytokinin metabolism.
A variety of domesticated rice cultivars have been established by selecting those with useful traits caused by natural mutations (26). Many traits important for agricultural productivity are quantitative, and quantitative trait loci (QTL) analysis has been employed for identifying useful genes in diverse natural mutations among rice varieties (27, 28). Several of these traits are caused by changes in phytohormone action (29). For example, the gene responsible for the Green Revolution in rice encodes an enzyme in the gibberellin biosynthetic pathway; a reduction in the catalytic activity of this enzyme in rice confers a semi-dwarf trait (30–32). The gene responsible for increased grain number encodes cytokinin oxidase/dehydrogenase (CKX), a cytokinin-degrading enzyme. CKX activity increases cytokinin accumulation in inflorescence meristems, thereby promoting panicle branching and increasing grain number (33). These findings mean that it is also possible to identify natural mutations that alter endogenous levels of phytohormones by QTL analysis to verify the usefulness of the causal gene.
In this study, we analyzed the hormonal profiles of a rice chromosome segment substitution line (CSSL) population and searched for genes affecting endogenous hormone levels by QTL analysis. Our approach identified a gene encoding a nucleosidase that catalyzes the deribosylation of cytokinin and purine nucleosides involved in cytokinin metabolism in the apoplastic space. Furthermore, the loss-of-function mutation attenuated the expression of cytokinin-responsive genes upon cytokinin riboside precursor treatment and nitrogen resupply. Our experimental evidence sheds light on a LOG-independent pathway for cytokinin production during normal plant growth.
Results
Hormone Profiling of O. sativa Backcross Lines.
To find loci controlling the endogenous level of phytohormones, we analyzed the concentration of four major phytohormones, cytokinins, auxins, gibberellins, and abscisic acid, including their precursors and derivatives in the seedling shoots of a Sasanishiki x Habataki CSSL population (Dataset S1). QTL analysis detected a major QTL peak on chromosome 5 that affected the accumulation level of cis-zeatin riboside O-glucoside (cZROG), a glucoside form of a cytokinin riboside precursor (SI Appendix, Fig. S1). The Habataki allele had a positive effect on the accumulation level of cZROG. In the CSSLs, a putative causal locus was found in the overlapping region of the Habataki genome replaced in SL416 and SL417. We focused on this chromosome 5 locus to identify the causal gene.
Identification of the Causal Gene Affecting Cytokinin Ribosides.
Map-based cloning used backcross populations prepared by backcrossing SL416 to the recurrent Sasanishiki parent with cZROG accumulation level as the phenotype. Fine mapping using 2784 individuals from the recombinant backcross population (BC1F4) finally delimited the causal region to 22 kb (Fig. 1A). Gene prediction by the Rice Annotation Project (RAP) of the International Rice Genome Sequencing Project (IRGSP-1.0; https://rapdb.dna.affrc.go.jp), annotated three genes, Os05g0406000, Os05g0406100, and Os05g0406200, in this region (Fig. 1A). In addition, the MSU Rice Genome Annotation Project (RGAP; http://rice.uga.edu) predicted another gene, LOC_Os05g33644, at the margin of the candidate region. Reverse transcription (RT)-PCR analysis was performed using specific primers that could amplify almost the entire length of the gene using the parental line and the recombinant inbred lines with the same genotype as the parental lines. The expression of Os05g0406100 was very weak in individuals with the Habataki genotype in the candidate region, although expression of the other three genes was similar or below the detection level in both genotypes (Fig. 1B). This result suggested Os05g0406100 was the causal gene candidate.
Fig. 1.
Map base cloning of a QTL affecting cZROG content. (A) The mapping region in Habataki used to identify the causal gene. The marker names delimiting the causal region in each mapping step are indicated above the solid bars. The number of recombinants between the molecular markers is shown below the 85-kb high-resolution map. (B) Expression patterns of four genes in the candidate 22-kb region in Sasanishiki and Habataki genotypes. Total RNAs from seedling shoots of Sasanishiki, Habataki, and three recombinant inbred lines that have each parental genotype [a, b, and c for Habataki (Ha), and d, e, f for Sasanishiki (Sa)] were analyzed by RT-PCR. Actin1 (OsAct1) was used as the control. The details of markers and gene-specific primers for RT-PCR are shown in SI Appendix, Table S2.
To investigate further the relationship between the functionality of the Os05g0406100 gene and the endogenous cytokinin level, we screened the Nipponbare (NB) Tos17 retrotransposon insertion mutant library (34). We found two independent lines (NE8550 and NE8503) that had the transposon inserted into exon 7 of Os05g0406100 (SI Appendix, Fig. S2A). In the homozygous line, the full-length transcript of the Os05g0406100 coding region was not amplified by RT-PCR (SI Appendix, Fig. S2B). Comparative analysis of the cytokinin accumulation levels in seedlings showed that the levels of cytokinin ribosides, such as tZR, cis-zeatin riboside (cZR), cZROG, and tZR O-glucoside (tZROG), were significantly increased compared to NB in both mutant homozygotes (Fig. 2A and SI Appendix, Table S1). In contrast, the levels were lower than or similar to NB levels in the null segregates [8550(+/+) and 8503(+/+)], in which the Tos17-insertion in Os05g0406100 was missing. By contrast, one of the corresponding nucleobases, cZ O-glucoside (cZOG), was present at significantly lower levels in the Tos17-insertion mutants (Fig. 2B and SI Appendix, Table S1). The accumulation patterns of the riboside species are consistent with the changes in cZROG accumulation in the Habataki genotype in the Sasanishiki x Habataki backcross lines. These results strongly suggest that Os05g0406100 is the causal gene responsible for differences in the cytokinin riboside levels in the backcross lines.
Fig. 2.
Cytokinin concentrations in NB and the Tos17-insertion mutants of Os05g0406100. Cytokinin fractions were extracted and purified from 10-d-old seedling shoots of NB, the Tos17-insertion mutant of Os05g0406100 NE8550 and NE8503 [8550(−/−) and 8503(−/−), respectively], and the null segregant [8550(+/+) and 8503(+/+), respectively], and cytokinin concentrations were quantified. (A) Representative cytokinin riboside species, (B) The corresponding nucleobase species. Error bars represent the SD of values for four biological replicates. Different lowercase letters at the top of each column denote statistically significant differences by Tukey’s honestly significant difference test (HSD) (P < 0.05). FW, fresh weight. The fully quantified dataset is provided in SI Appendix, Table S1.
Os05g0406100 Functions as a Cytokinin/Purine Riboside Nucleosidase.
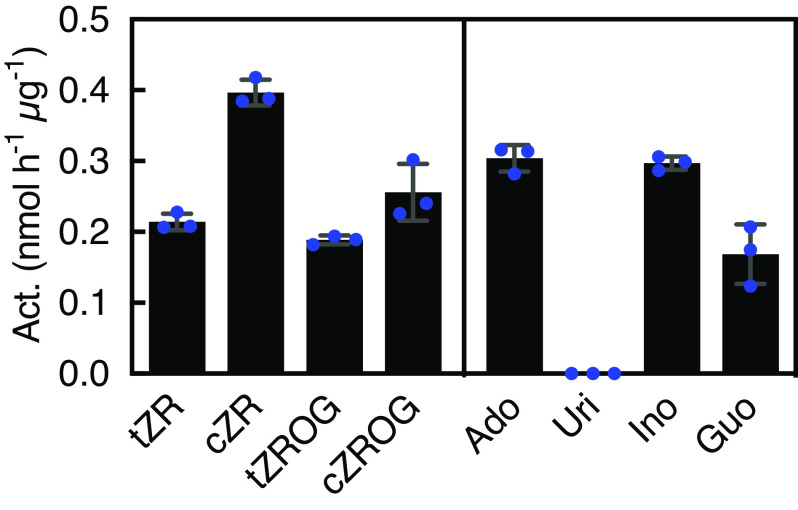
Os05g0406100 encodes a protein that is predicted to be an inosine/uridine-preferring nucleoside hydrolase domain–containing protein with two ribosidase domains within the polypeptide (SI Appendix, Fig. S3). In Arabidopsis, a ribosidase gene that functions in purine metabolism (35–37) and is an ortholog of Os05g0406100, At5g18860, is named NUCLEOSIDE HYDROLASE 3 (NSH3). NSH3’s putative function in cytokinin metabolism had not been characterized. To verify the enzymatic function of Os05g0406100, we overexpressed a His-tagged Os05g0406100 protein in Escherichia coli and used the purified preparation in an enzyme assay. The results revealed that the enzyme deribosylates cytokinin ribosides such as tZR, cZR, tZROG, and cZROG as substrates (Fig. 3). Purine nucleosides, such as adenosine, inosine, and guanosine were also deribosylated with efficiencies similar to cytokinin ribosides, whereas uridine, a pyrimidine nucleoside, was a very poor substrate. The Km values for tZR and adenosine were 10.1 ± 1.2 µM and 13.4 ± 3.2 µM, respectively, and the optimal pH value for tZR was 6.0 (SI Appendix, Fig. S4). These results indicate that Os05g0406100 is not a cytokinin riboside–specific enzyme but also accepts other purine nucleosides as substrates. Thus, Os05g0406100 was named cytokinin/purine riboside nucleosidase 1 (CPN1) before further characterizing its function. We also renamed NE8550(−/−) and NE8503(−/−) as cpn1-1 and cpn1-2, respectively.
Fig. 3.
Reactivity of Os05g0406100 (cytokinin/purine riboside nucleosidase) to possible substrates. The purified preparation of His-tagged Os05g0406100 recombinant protein was used in enzyme assays with 100 µM substrates of cytokinin ribosides and purine and pyrimidine nucleosides. Act., activity; tZR, trans-zeatin riboside; cZR, cis-zeatin riboside; tZROG, tZR-O-glucoside; cZROG, cZR-O-glucoside; Ado, adenosine; Uri, uridine; Ino inosine; Guo, guanosine. Identical amounts of protein from the purified preparation (0.3 µg) were used for the assays. Error bars represent the SD of values for three replicates.
Next, we evaluated CPN activity in NB, cpn1-1, and cpn1-2 mutants using crude extracts from seedling shoots. When the substrate was tZR, tZ-forming activity was detected in NB but not in the two mutants (Fig. 4). The reactivity was recovered in the null segregant cpn1-1 (+/+). In contrast, adenine-forming activity was detected in all lines (Fig. 4). These results suggest that CPN1 catalyzes the in vivo deribosylation of nucleoside-type cytokinin precursors, such as tZR, and is almost fully responsible for the cytokinin riboside nucleosidase activity in this experimental condition. Since there was no significant difference between NB and the null segregant cpn1-1 (+/+), we used NB as the control line in the following experiments.
Fig. 4.
Nucleosidase activity in NB and the cpn1 mutants. Total proteins were extracted from 6-wk-old seedling shoots of NB, cpn1-1, cpn1-2, and the cpn1-1 null segregant (+/+) and were used to measure nucleosidase activities with tZR or adenosine (Ado) as substrates. Error bars represent the SD of values for three replicates. Different lowercase letters at the top of each column denote statistically significant differences by Tukey’s honestly significant difference test (HSD) (P < 0.05). FW, fresh weight; ND, not detected.
Expression Analysis at the Organ Level.
To identify the expression pattern of CPN1 at the organ level, we analyzed transcript abundance by RT-quantitative PCR (RT-qPCR) analysis in various organs of NB plants sampled at the heading stage. The results showed that CPN1 expression was relatively high in panicle branches, flag leaf blades, and sheaths, and lower leaf blades than in flowers before anthesis, nodes, and internodes (SI Appendix, Fig. S5A). In the younger seedling stage, CPN1 was strongly expressed in shoots compared to roots (SI Appendix, Fig. S5B).
CPN1 Localizes in the Apoplastic Space and Is Bound to the Cell Wall.
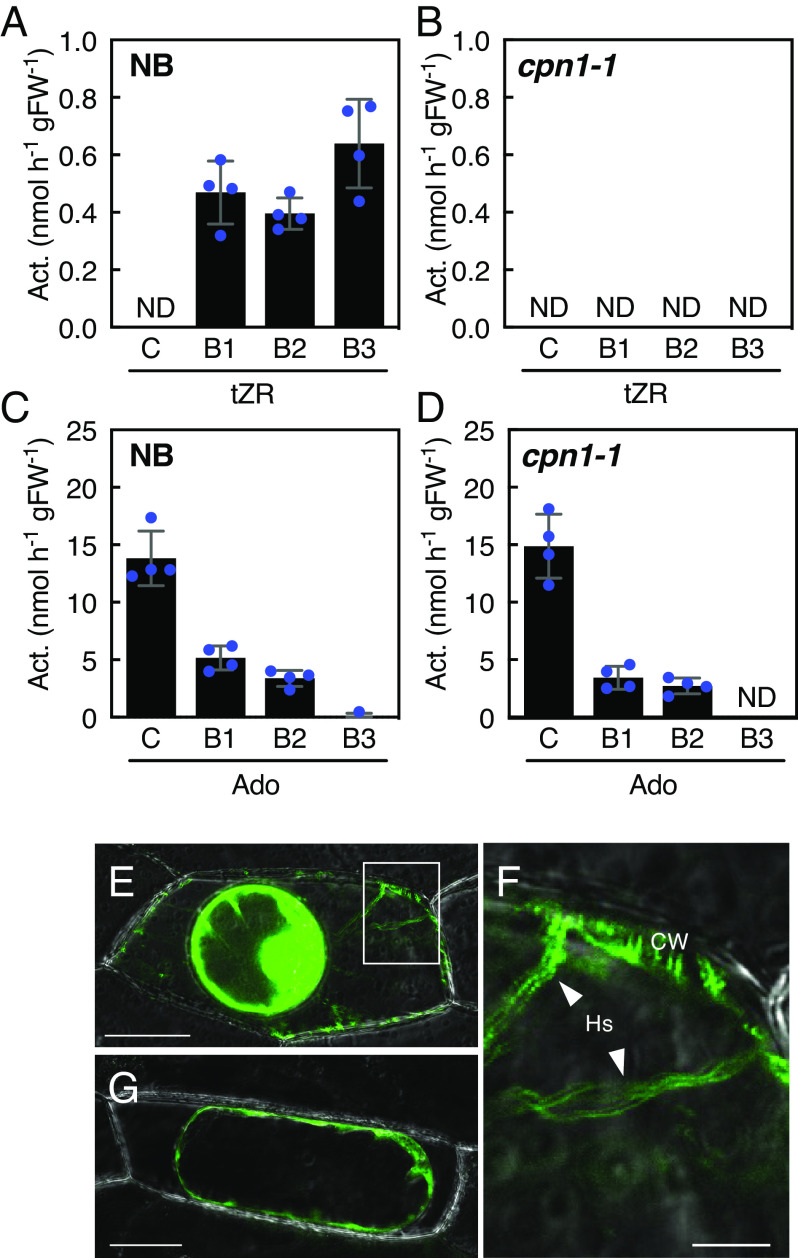
A crude rice leaf extract was fractionated, and the distribution of CPN activity in each fraction was analyzed to determine the subcellular localization of CPN. Since the Arabidopsis ortholog NSH3 is proposed to localize in the apoplast (37), we tested CPN’s intracellular and extracellular localization (Fig. 5 A–D). In NB, the nucleosidase activity using tZR as a substrate was below a detectable level in the cytosol fraction (C); however, nucleosidase activity was detected in fractions containing proteins that were weakly bound to the cell wall (B1), that bound hydrophobically to the cell wall (B2), and that strongly and electrostatically bound to the cell wall (B3). The activity was highest in the B3 fraction. In contrast, nucleosidase activity using adenosine as the substrate was strongest in fraction C, with lower activity in the B1, B2, and B3 fractions in this order. This adenosine-hydrolyzing activity pattern was consistent with that of glucose-6-phosphate dehydrogenase, a marker of cytosolic localization (SI Appendix, Fig. S6). On the other hand, cpn1-1 showed little change in adenosine-hydrolyzing activity, although tZR-hydrolyzing activity was below the detectable level in all fractions. These results indicated that CPN activity localizes to the apoplastic space, is bound to the cell wall, and is responsible for the cytokinin nucleosidase activity in rice shoots at this developmental stage.
Fig. 5.
Subcellular localization of CPN1. (A to D) Distribution of nucleosidase activity in fractions from a stepwise extraction. Three-week-old seedling leaves of NB (A and C) and cpn1-1 (B and D) were harvested and sequentially extracted using the method of Rast et al. (38). (C) a cytosolic protein fraction; B1, a protein fraction that weakly bound to cell walls; B2, a protein fraction that bound hydrophobically to cell walls; and B3, a protein fraction that strongly and electrostatically bound to cell walls. The fractions were analyzed by measuring nucleosidase activities with tZR (A and B) or adenosine (Ado) (C and D) as substrates. Error bars represent the SD values for four replicates of the enzyme assay. Act., activity; FW, fresh weight; ND, not detected. (E to G) Subcellular localization of CPN1:GFP proteins in onion epidermal cells. Chimeric constructs containing CaMV35Spro:GFP (G) or CaMV35Spro:CPN1-GFP proteins (E and F) were transiently expressed in onion epidermal cells by particle bombardment. GFP fluorescence was observed after plasmolysis. (F) is a magnified view of the white rectangular area in (E). CW, cell wall. The arrowheads indicate Hs. [Scale bars: 50 µm for (E and G) and 10 µm for (F).]
To visualize the subcellular localization of CPN1, we constructed a chimeric gene fusing GFP to the C terminus of CPN1. Prior to creating the reporter construct, the translation initiation site was identified using the 5′-RACE method. This survey revealed that translation is initiated from another ATG codon upstream of the predicted translation start site identified in the RAP and RGAP databases. Thus, a full-length cDNA encoding 889 amino acids was cloned. We predicted the subcellular localization of the entire translated region using a signal peptide prediction program (SignalP 6.0; https://services.healthtech.dtu.dk/services/SignalP-6.0/) (39), which indicated a high likelihood of extracellular localization (Probability: 0.813348). The CPN1:GFP construct was transiently expressed in onion epidermal cells. After plasmolysis, the observed CPN1:GFP fluorescence signals gave a typical distribution pattern for cell wall proteins as previously reported (Fig. 5 E–G): fluorescence was detected in the cell wall domain and in Hechtian strands (Hs) connecting the cell wall to the plasma membrane (40, 41). These results support the hypothesis that CPN1 is a cell wall–localized protein.
CPN1 Is Involved in the Metabolism of Apoplastic Cytokinin Riboside.
To gain insight into the physiological role of CPN1, we collected apoplast fluid from leaf blades of NB, cpn1-1 and cpn1-2 and measured the concentration of cytokinins and cytokinin derivatives. The concentration of cytokinin ribosides such as tZR, cZR, and cZROG in cpn1-1 and cpn1-2 was significantly higher than in NB, whereas the concentration of the corresponding bases was unchanged (Fig. 6). This result suggests that CPN1 is involved in cytokinin riboside metabolism in the apoplastic space.
Fig. 6.
Cytokinin concentration of apoplastic fluids in NB and cpn1 mutant leaves. Apoplastic fluids were collected from fully developed, youngest leaves of 6-wk-old plants. Cytokinin fractions from the apoplastic fluids were purified and quantified. Error bars represent SDs of values for three or four replicates. Asterisks indicate significant differences compared to NB (**P < 0.05, Student’s t test). tZ, trans-zeatin; tZR, tZ riboside; cZ, cis-zeatin; cZR, cZ riboside; cZOG, cZ O-glucoside; cZROG, cZR-O-glucoside.
Notably, there was no remarkable difference in the concentration of cytokinin riboside species in the xylem sap of NB and the cpn1 mutants (SI Appendix, Fig. S7).
Involvement of CPN1 in Driving Cytokinin Signaling in Response to Nitrogen Nutrition.
To examine the possible involvement of CPN1 in cytokinin signaling, transcript abundance for cytokinin-inducible marker genes, OsRR1, OsRR6, and OsRR9/10, in 4-wk-old detached leaves of NB and cpn1-1 was analyzed after treatment with tZ or tZR (Fig. 7 A–C). Since the peak accumulation time differs for each OsRR gene, we used a treatment period with the highest expression level in the tZ treatment. In NB, OsRR6 and OsRR9/10 had similar induction levels in the tZ and tZR treatments (Fig. 7 B and C). In cpn1-1, however, the tZR-responsive induction of the genes was significantly reduced to about 65% of the tZ-responsive induction. The tZR-responsive expression level of OsRR1 was significantly lower than the tZ-responsive expression level in both NB and cpn1-1, but the magnitude of the difference was greater in cpn1-1 (Fig. 7A). These results suggest that the CPN1-mediated deribosylation in the apoplastic space could affect tZR-dependent cytokinin signaling in seedling leaves.
Fig. 7.
Inductive expression levels of cytokinin-inducible type-A RR genes in NB and cpn1 mutants. (A–C) Expression levels of type-A RR genes in NB and cpn1-1 leaves in response to tZ and tZR. Detached, fully developed youngest leaves of 4-wk-old plants were treated with solutions containing 50 nM tZ or tZR. Total RNAs were prepared after 60 min for OsRR1 analyses and after 30 min for the OsRR6 and OsRR9/10 analyses, followed by RT-qPCR analysis. The expression level in the tZR treatment is shown as a value relative to that of the tZ treatment, defined as 100. OsAct1 was used for normalization. Error bars represent the SD of three biological replicates. Asterisks indicate significant differences compared to tZ (**P < 0.05, Student’s t test). (D–F) Effect of inorganic nitrogen resupply on the expression of type-A RR genes in NB, cpn1-1, and cpn1-2. Total RNAs were prepared from leaves of nitrogen-starved plants that were supplemented with or without 1 mM NH4NO3 for the indicated period and followed by RT-qPCR analysis. The expression level is shown as the ratio of nitrogen resupply (+N) samples to nonsupply (−N) samples. OsAct1 was used for normalization. Error bars represent the SD of three or four biological replicates. Asterisks indicate significant differences compared to NB at each time point (**P < 0.05, Student’s t test).
The supply of a nitrogen source to roots up-regulates de novo cytokinin biosynthesis and translocation of tZR from roots to shoots via the xylem (5, 22, 42). We compared the induction level of cytokinin-responsive OsRR expression in leaves upon nitrogen resupply in NB and cpn1 mutants (Fig. 7 D–F). Since multiple processes intervene between nitrogen uptake by roots and cytokinin-responsive gene expression in the leaves, we compared the expression ratio upon nitrogen resupply to nontreatment after 3 h, 6 h, and 24 h. The results showed that the induction levels of the OsRRs in cpn1-1 and cpn1-2 were significantly reduced in 3 h and/or 6 h compared to that in NB. The difference in expression mainly recovered by 24 h, except for OsRR1 expression in cpn1-1. These results suggest that CPN1 is involved, at least partly, in driving leaf cytokinin signaling in response to nitrogen nutrition.
Effect of Loss-of-Function Mutations in CPN1 during Panicle Development.
We examined the effect of loss-of-function of CPN1 in morphological traits of field-grown plants. We focused on panicle architecture because CPN1 is expressed in panicle branches (SI Appendix, Fig. S5). In both cpn1-1 and cpn1-2, the panicle weight per plant was significantly smaller than for NB (Fig. 8A). When we inspected the gross morphology of the panicle, rachis length, grain number per panicle, and secondary panicle branch number were all significantly smaller than in NB, although the primary panicle branch number and the number of panicles did not change in the mutants (Fig. 8 B–E and SI Appendix, Fig. S8). These results suggest that CPN1 loss-of-function affects panicle development in field-grown conditions.
Fig. 8.
Comparison of panicle architecture in NB, cpn1-1, and cpn1-2. NB, cpn1-1, and cpn1-2 were grown in a paddy field and harvested after grain maturation. The panicle weight (A), rachis length (B), grain number (C), and the number of secondary panicle branches off the main panicle (D) for each plant were measured and are shown as box plots. Asterisks indicate significant differences compared to NB (**P < 0.05, Student’s t test, n = 13 or 14). (E) Typical panicle morphologies of NB, cpn1-1, and cpn1-2. (Scale bar: 5 cm.)
On the other hand, when we compared shoot growth and apical meristem size at the vegetative phase, no differences were common among the mutant genotypes. (SI Appendix, Figs. S9 and S10).
Discussion
In this study, we identified a gene involved in apoplastic cytokinin metabolism by exploiting the diversity in natural variation among rice cultivars. QTL analysis has been generally used to search for causal genes using quantitative agronomic trait data, and useful genes have been successfully identified (27–29). Our results show that it is possible to identify genes using quantitative signaling molecule data, i.e., internal physiological information about plants. This approach has opened the possibility of discovering loci that are not easily manifested as visible growth phenotypes. cZROG, selected as the phenotype molecular species in this study, is one of the most abundant cytokinin derivatives in rice (43), is metabolically stable, and can be quantified with high accuracy. Thus, comprehensive hormone profiling of backcross lines, including glucosides and other conjugates, is effective for further gene identification and validation.
Os05g0406100, identified as the causal gene affecting the level of cZROG in Habataki x Sasanishiki CSSLs, encodes a cytokinin/purine riboside nucleosidase that reacts not only with cZROG but also with tZR and cZR (Fig. 3). In our hormone profiling of the CSSLs, no clear QTL peaks were detected for tZR and cZR. This finding is probably because the effect of other natural variations between Habataki and Sasanishiki compensated for the difference in the riboside levels. Also, despite Habataki being a higher grain-yielding cultivar than Sasanishiki (44), plants with the CPN1 Habataki allele accumulated higher levels of cZROG. This result seemingly contradicts the panicle architecture phenotype differences between NB and its cpn1 loss-of-function mutants. We do not have a clear explanation for this observation, but the CPN1 Habataki allele has a relatively weak effect. Other natural variations, such as Gn1a mentioned below, might negate the negative effect on panicle development.
The Gn1a locus encoding OsCKX2 is located on chromosome 1 in the Habataki genome and is involved in increasing grain number. Furthermore, the endogenous cytokinin level is high in the developing panicles of Habataki, in which OsCKX2 expression is lower, resulting in a higher grain number (33). However, no obvious cytokinin-related peaks corresponding to Gn1a were detected in our QTL analysis of young seedling shoots. This finding is probably because the expression profile of OsCKX2 might influence cytokinin levels during panicle development but have less influence during the seedling stage. In contrast, the CPN1 locus had a QTL peak in young seedlings, and the loss-of-function mutant cpn affected panicle morphology. The growth phenotypes of the cpn1 mutants must be interpreted cautiously, as a deficiency in both cytokinin and purine metabolism could affect the growth phenotypes. Nonetheless, these lines of evidence suggest that CPN1 is a gene that influences cytokinin and purine metabolism over long periods during rice growth and development. No obvious differences in shoot growth or apical meristem size were observed between NB and the mutants during vegetative growth (SI Appendix, Figs. S9 and S10). This result implies that the cpn1 mutation’s effects on rice growth are manifested after plants transition to the reproductive stage.
Our results strongly suggest that CPN1 is a cell wall–localized protein. The increased concentrations of cytokinin ribosides in apoplastic fluid in the cpn1 mutant (Fig. 6), the attenuated effect of tZR treatment on the induction of cytokinin-responsive gene expression in excised leaves, and the similar attenuation observed in leaves when nitrogen was resupplied to roots (Fig. 7) suggest that CPN1 is at least partly involved in the activation of cytokinin riboside precursors in the leaf cell wall space transported via the vascular system (SI Appendix, Fig. S11). This result means that in addition to the intracellular activation step by LOG, CPN1 plays a role in cytokinin activation by another extracellular pathway. Our finding extends the spatial scope of the cytokinin metabolic system. In contrast to the pronounced phenotypic consequences observed in the loss-of-function mutations of LOG (11, 16), the impact of the cpn1 mutation was relatively limited. We postulate that the cytosol serves as the primary site to produce active cytokinins, with a substantial portion of them being perceived by receptors localized on the endoplasmic reticulum membrane. Cytokinins perceived by plasma membrane receptors may play a limited role. Furthermore, LOG belongs to a gene family with diverse expression patterns across various organs and tissues (11, 15), whereas CPN1 appears to have a more specific function in organs such as leaves and panicles. This distinction may explain, in part, why the contribution of CPN1 to plant growth and development is relatively small compared to that of LOG. Given that translocation of tZR could be affected by nitrogen nutritional conditions (5, 22, 42), the CPN1-mediated LOG-independent pathway might have a significant role in cytokinin supply for environmentally responsive modulation of plant growth.
Crystal structure analysis of the CHASE domain of cytokinin receptors showed that cytokinin ribosides are very difficult to recognize as ligands. Cytokinin receptors are known to localize to the plasma membrane and the endoplasmic reticulum membrane (45–47), but the functional differentiation of the dual localization sites has not been elucidated (48–50). Our identification of CPN1 and the reduced response to tZR in its loss-of-function mutant provides evidence supporting the significance of nucleobase-type cytokinin reception on the plasma membrane.
The attenuation of inductive expression of cytokinin-responsive genes upon nitrogen resupply in the cpn1 mutants (Fig. 7 D–F) should be attributed to the deficiency of apoplastic cytokinin activation activity, not due to the decreased translocation of cytokinins from the root. Evidence that no essential differences in tZR in the xylem sap were observed between NB and the cpn1 mutants supports this idea (SI Appendix, Fig. S7). One plausible explanation for the lack of impact of the cpn1 mutation on the cytokinin profile of xylem sap is the predominant expression of CPN1 in aboveground organs (SI Appendix, Fig. S5). Although nitrogen supplements increase the translocation of tZR from the roots to shoots via xylem sap, the lack of conversion of tZR to tZ in leaf apoplasts of the cpn1 mutants should affect cytokinin signaling.
LOC_Os05g33644 is a gene adjacent to and structurally homologous to CPN1 (Fig. 1A and SI Appendix, Fig. S3). However, the expression level of LOC_Os05g33644 in the NB was less than 1% of that of CPN1 and about 3% in the cpn1 mutants (SI Appendix, Fig. S12). The function of this gene was not analyzed in this study. Since cytokinin ribosidase activity is below the detection limit in the cpn1 mutants, the contribution of this paralog gene, if any, is considered to be very limited.
Over 40 y ago, cytokinin riboside nucleosidase was partially purified from wheat germ, and a few properties were characterized (19). This enzyme has a molecular weight of approximately 60,000, an optimum pH of 4.7, and uses adenosine, tZR, and iPR as substrates but not guanosine or uridine. Although the substrate specificity differs from that of CPN1, the optimal pH implies that the nucleosidase studied in wheat germ is likely an extracellular enzyme.
One may think that the Km value of CPN1 for tZR (10.1 µM) appears high compared to the endogenous concentration of cytokinins. However, given that LOG, another cytokinin-activating enzyme, has Km values for its substrates within a similar range (11.7 µM for iPRMP and 22.0 µM for tZRMP) (11), this disparity in the values should not be regarded as a concern.
Although rice has five homologous ribosidase genes, as does Arabidopsis, it is interesting that the ribosidase activity for cytokinin ribosides in the crude extract of the cpn1 mutant is below the detection limit (Fig. 4). This result suggests that among the multiple ribosidase isoenzymes present, only CPN1 has acquired reactivity toward cytokinin ribosides. Although the ribosidase family has been studied in Arabidopsis, no studies have reported its potential involvement in cytokinin metabolism (35, 37). NSH3, an Arabidopsis ortholog of CPN1, was shown to be involved in extracellular ATP metabolism and was implicated in the disease response (36). According to RiceXPro (51, 52)(https://ricexpro.dna.affrc.go.jp), a public database, Os05g0406100 also appears to have a similar physiological role, as its expression is up-regulated by jasmonic acid, like NSH3 in Arabidopsis. Thus, CPN1 might have a dual role in extracellular cytokinin and purine metabolism. Examining the function of orthologous enzyme genes in Arabidopsis from the perspective of their involvement in cytokinin metabolism would provide a universal understanding of the physiological significance of the apoplastic cytokinin activation pathway.
Materials and Methods
Plant Materials and Growth Conditions for Phytohormone QTL Analysis.
The populations consisted of 39 Sasanishiki × Habataki CSSLs (44) used for phytohormone profiling. Rice (O. sativa) seeds were germinated in distilled water at 30 °C overnight in the dark and then hydroponically grown in a growth chamber (Koitotron KG-201 SHL-D, Koito Industries, Tokyo) in tap water adjusted to pH 5.5 using HCl. Seedlings were provided a 12-h-light (30 °C)/ 12-h-dark (25 °C) photoperiod with a light intensity of 700 µmol m−2 s−1 for 9 d. The shoots were harvested and immediately frozen in liquid N.
Phytohormone Quantification.
Phytohormones were extracted and semipurified from about 100 mg fresh weight of root tissues as described previously (53). Cytokinins were quantified using an ultra-performance liquid chromatography (UPLC)-tandem quadrupole mass spectrometer (ACQUITY UPLC System/Xevo-TQS; Waters Corp.) with an octadecylsilyl (ODS) column (ACQUITY UPLC HSS T3, 1.8 µm, 2.1 mm × 100 mm, Waters Corp.). Other hormone species were analyzed as described previously (54).
QTL Analysis and Fine Mapping.
Segregation data for 166 markers and their linkage maps were used for QTL mapping. QTLs were detected using CSSL Finder version 0.9 (http://mapdisto.free.fr/CSSLFinder/). A selected CSSL, SL416, was backcrossed with Sasanishiki, and the progenies BC1F2, BC1F3, and BC1F4 were analyzed for fine mapping with the shoot cZROG concentration as the trait of interest. Marker information is shown in SI Appendix, Table S2.
Plant Materials Used to Isolate Retrotransposon Tos17-Inserted Mutant Lines.
Retrotransposon Tos17-inserted O. sativa NB lines (NE8503 and NE8550) were screened in silico by searching the flanking sequence database of the mutant panel (http://tos.nias.affrc.go.jp/~miyao/pub/tos17/) used for the Project for Rice Genome Research, in which mutant lines were generated by the random insertion of the endogenous retrotransposon Tos17 into the rice genome (34, 55). NB was used for the control line in the functional analysis of CPN1.
RT-qPCR.
Total RNA was extracted from frozen and ground rice tissues using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) with the RNase-Free-DNase Set (Qiagen) according to the supplier’s protocols. Total RNA (0.6 µg) was used to synthesize cDNA using the ReverTra Ace qPCR Master Mix with gDNA Remover (TOYOBO) according to the supplier’s protocol. One-fiftieth of the cDNA solution was used for each qPCR reaction with the THUNDERBIRD Next SYBER qPCR Mix (TOYOBO) and a real-time PCR system (Thermo Fisher, StepOnePlus). Expression levels were estimated using the relative quantification method (56), with Actin1 serving as the internal standard for normalization. Gene locus IDs and the specific primers used for amplification are listed in SI Appendix, Table S2.
Phylogenetic Analysis.
Alignments and phylogenetic reconstructions of Arabidopsis and O. sativa ribosidase family proteins were designed using the function “build” of ETE3 v3.1.1 (57) as implemented on GenomeNet (https://www.genome.jp/tools/ete/).
Overexpression of a Recombinant Enzyme Protein in E. coli.
The DNA fragment containing the entire reading frame of Os05g0406100 was amplified by RT-PCR using specific primers (SI Appendix, Table S2), followed by ligation into the XbaI/XhoI site of the pCOLD I vector (Takara Bio). The BL21 (DE3) strain harboring pG-Tf2 (Takara Bio) was used as the E. coli host. Protein expression was induced in modified M9 medium (9) for 20 h at 15 °C. Crude soluble proteins were extracted from the BL21 cells by sonication in extraction buffer (50 mM NaH2PO4-NaOH pH8.0, 300 mM NaCl, 10 mM imidazole, 1 mM MgCl2, 5 mM β-mercaptoethanol) containing a protease inhibitor cocktail (P8849, SIGMA). The recombinant proteins were purified by nickel affinity chromatography using Ni-NTA Superflow (QIAGEN) according to the manufacturer’s instructions. Purified recombinant proteins were stored at −80 °C in the presence of 15% (w/w) glycerol.
Extraction of Proteins and Enzyme Assay.
For the CPN enzyme assay using crude leaf extracts, about 0.3 g of leaves from 6-wk-old seedlings hydroponically grown in a greenhouse in a nutrient solution (58) were ground with a mortar and pestle in 0.6 mL of ice-cold extraction buffer (50 mM Na2HPO4/ KH2PO4 pH 7.0). The homogenate was centrifuged at 15,000 g at 4 °C for 5 min, and the activity of the resulting supernatant liquid was assayed. The reaction mixture (50 µL) to detect nucleosidase activity was prepared as follows: 50 mM Tris-acetate (pH 5.0), 20 µM substrate, and 20 µL of the crude extract. The mixture was incubated at 28 °C for 2.5 h, and the reaction was stopped by incubating the samples at 98 °C for 3 min. Five different substrates ranging in concentration from 5 to 100 µM were used to determine kinetic parameters. Citrate-phosphate buffer (Mcllvaine buffer) was used to prepare the reactions at pH 3.0 to 8.0. The reaction products were analyzed by an ACQUITY UPLC System/XEVO-TQS (Waters) with a pentafluorophenylpropyl column (Ascentis®Express F5, 15 cm × 2.1 mm, 2 µm, Merck) at a flow rate of 0.25 mL min−1 with linear gradients of solvent A (0.1% formic acid) and solvent B (acetonitrile) set according to the following profile: 0 min, 100% A + 0% B; 5 min, 99% A + 1% B; 13 min, 88% A + 12% B; 16 min, 40% A + 60% B; 21 min, 100% A + 0% B. Details of the mass spectrometry conditions are summarized in SI Appendix, Table S3.
Fractionation of Cell Walls from Rice Leaf Extracts.
About 0.3 g of leaves from 3-wk-old seedlings hydroponically grown in a greenhouse in a nutrient solution (58) were used for the fractionation with 3 volumes of 0.25 M Tris–HCl buffer (pH 8.0). Fractionation of cell walls from rice leaf extracts was conducted as described previously (38).
Analysis of GFP Fusion Proteins.
Before constructing plasmids for particle bombardment, 5′-RACE was performed to identify the translation initiation codon using the SMARTer RACE 5′/3′ kit (Clontech) with gene-specific primers (SI Appendix, Table S2). The full-length coding region was amplified by PCR and fused to the amino terminus of sGFP(S65T) within the pTH2 vector (59), whose expression is controlled by the cauliflower mosaic virus 35S promoter, with NEBuilderHiFi DNA Assembly (NEB). The resulting DNA constructs were introduced into onion epidermal cells by particle bombardment (Bio-Rad; PDU-1000/He). The 35Spro:GFP empty vector was used as the control marker for cytosol and nuclei. Transient expression was observed by confocal laser scanning fluorescence microscopy (Leica; SP5) after overnight incubation.
Collection of Apoplastic Fluid from Rice Leaves.
NB and cpn1 plants were hydroponically grown in a greenhouse in a nutrient solution (58) for 6 wk. Apoplastic fluid was collected from the fully developed youngest and second youngest leaves using a previously described method (60). Three leaves were used for each sample, and the fluid was collected by centrifugation as described previously (61). Cytoplasmic contamination was evaluated by testing for glucose-6-phosphate dehydrogenase (G6PDH) activity in the collected fluid (60).
Collection of Xylem Sap.
NB and cpn1 plants were hydroponically grown in a greenhouse in a nutrient solution (58) for 7 wk. Shoots were cut about 15 mm above the root/shoot transition with a razor blade, and the root-pressure exudate was collected using a micropipette for 2 h. Exudate samples were used to analyze cytokinin levels.
Cytokinin Treatment Experiment.
The youngest, fully developed leaves of 4-wk-old NB and cpn1-1 grown in a greenhouse were cut underwater at the laminar joint with a razor blade. The basal ends were placed vertically in 50-mL conical tubes that contained a 20-mL solution of 50 nM tZ or tZR. The mock treatment was 0.01% (v/v) DMSO, equivalent to the DMSO concentration in the cytokinin treatment. Treated leaves and the controls were incubated for 30 min and 60 min.
Nitrogen Response of OsRR Genes.
NB, cpn1-1, and cpn1-2 seedlings were hydroponically grown in a greenhouse in a nutrient solution (58) for 3 wk. Seedlings were then transferred to tap water (pH 5.5 adjusted with HCl) and further grown for 1 wk under this nitrogen starvation condition. The nitrogen source was resupplied by adding 1 mM NH4NO3, and leaves were harvested at the indicated times. The controls were not resupplied with nitrogen.
Evaluation of Morphological Traits.
NB, cpn1-1, and cpn1-2 were grown under natural conditions in a paddy field at the Field Science Center at Nagoya University, Togo, Aichi, Japan. The seeds were germinated in a seedbed in early May and transplanted to the field in late June 2021. Plants were harvested in October, and the agricultural traits were measured.
Observation of Shoot Apical Meristems.
Shoot apices were excised from 9-wk-old NB and cpn1-1 and prefixed with 4% (w/v) paraformaldehyde and 2% (v/v) glutaraldehyde in 50 mM sodium cacodylate buffer (pH7.4) overnight at 4 °C and postfixed with 1% (v/v) osmium tetroxide at room temperature for 3 h. The samples were dehydrated using a graded ethanol series and embedded in Epok 812 resin (Oken, Tokyo, Japan). Microtome sections (1 µm thick) were stained with 0.05% (w/v) toluidine blue and observed using an upright light microscope (BX53, Olympus).
Sequence data from this article were deposited with the DDBJ/EMBL/GenBank data libraries under accession numbers LC746295 for CPN1 (Os06g0406100).
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Acknowledgments
We are grateful to Drs Motoyuki Ashikari, Takatoshi Kiba, Fanny Bellegarde, Mimi Hashimoto (Nagoya University), Misato Ohtani (The University of Tokyo), and Kazuhiko Nishitani (Kanagawa University) for their helpful support and discussions.
Author contributions
M.K., N.M., T.Y., and H.S. designed research; M.K., N.M., K.M., M. Yoshino., A.I., M.O., A. Surjana, T.K., N.T.-K., K.T., T.A., A. Shomura, and T.H. performed research; A.M. and H.H. contributed new reagents/analytic tools; M.K., N.M., M.O., M. Yano., T.Y., T.H., and H.S. analyzed data; and M.K. and H.S. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
Sequence data from this article were deposited with the DDBJ/EMBL/GenBank data libraries under accession numbers LC746295 (62) for CPN1 (Os06g0406100). All other data are included in the article and/or supporting information.
Supporting Information
References
- 1.Chen L., Zhao J., Song J., Jameson P. E., Cytokinin dehydrogenase: A genetic target for yield improvement in wheat. Plant Biotechnol. J. 18, 614–630 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cortleven A., et al. , Cytokinin action in response to abiotic and biotic stresses in plants. Plant Cell Environ. 42, 998–1018 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Hwang I., Sheen J., Müller B., Cytokinin signaling networks. Annu. Rev. Plant Biol. 63, 353–380 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Sakakibara H., Cytokinins: Activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 57, 431–449 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Sakakibara H., Cytokinin biosynthesis and transport for systemic nitrogen signaling. Plant J. 105, 421–430 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Schaller G. E., Bishopp A., Kieber J. J., The yin-yang of hormones: Cytokinin and auxin interactions in plant development. Plant Cell 27, 44–63 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wybouw B., De Rybel B., Cytokinin–A developing story. Trends Plant Sci. 24, 177–185 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Kakimoto T., Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate:ATP/ADP isopentenyltransferases. Plant Cell Physiol. 42, 677–685 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Takei K., Sakakibara H., Sugiyama T., Identification of genes encoding adenylate isopentenyltransferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana. J. Biol. Chem. 276, 26405–26410 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Takei K., Yamaya T., Sakakibara H., Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyse the biosynthesis of trans-Zeatin. J. Biol. Chem. 279, 41866–41872 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Kurakawa T., et al. , Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445, 652–655 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Miyawaki K., et al. , Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 103, 16598–16603 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsumoto-Kitano M., et al. , Cytokinins are central regulators of cambial activity. Proc. Natl. Acad. Sci. U.S.A. 105, 20027–20031 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiba T., Takei K., Kojima M., Sakakibara H., Side-chain modification of cytokinins controls shoot growth in Arabidopsis. Dev. Cell 27, 452–461 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Kuroha T., et al. , Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell 21, 3152–3169 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tokunaga H., et al. , Arabidopsis lonely guy (LOG) multiple mutants reveal a central role of the LOG-dependent pathway in cytokinin activation. Plant J. 69, 355–365 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Mok D. W. S., Mok M. C., Cytokinin metabolism and action. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 89–118 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Chen C.-M., Kristopeit S. M., Metabolism of cytokinin: Dephosphorylation of cytokinin ribonucleotide by 5’-nucleotidases from wheat germ cytosol. Plant Physiol. 67, 494–498 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen C.-M., Kristopeit S. M., Metabolism of cytokinin: Deribosylation of cytokinin ribonucleoside by adenosine nucleosidase from wheat germ cells. Plant Physiol. 68, 1020–1023 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirose N., et al. , Regulation of cytokinin biosynthesis, compartmentalization and translocation. J. Exp. Bot. 59, 75–83 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Osugi A., et al. , Systemic transport of trans-zeatin and its precursor have differing roles in Arabidopsis shoots. Nat. Plants 3, 17112 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Kamada-Nobusada T., Makita N., Kojima M., Sakakibara H., Nitrogen-dependent regulation of de novo cytokinin biosynthesis in rice: The role of glutamine metabolism as an additional signal. Plant Cell Physiol. 54, 1881–1893 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morris S. E., Turnbull C. G. N., Murfet I. C., Beveridge C. A., Mutational analysis of branching in pea. Evidence that Rms1 and Rms5 regulate the same novel signal. Plant Physiol. 126, 1205–1213 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiba T., et al. , The trans-zeatin-type side-chain modification of cytokinins controls rice growth. Plant Physiol. 192, 2457–2474 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schoor S., et al. , Adenosine kinase contributes to cytokinin interconversion in Arabidopsis. Plant Physiol. 157, 659–672 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mussurova S., Al-Bader N., Zuccolo A., Wing R. A., Potential of platinum standard reference genomes to exploit natural variation in the wild relatives of rice. Front. Plant Sci. 11, 1–8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miura K., Ashikari M., Matsuoka M., The role of QTLs in the breeding of high-yielding rice. Trends Plant Sci. 16, 319–326 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Ashikari M., Matsuoka M., Identification, isolation and pyramiding of quantitative trait loci for rice breeding. Trends Plant Sci. 11, 344–350 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Deveshwar P., Prusty A., Sharma S., Tyagi A. K., Phytohormone-mediated molecular mechanisms involving multiple genes and QTL govern grain number in rice. Front. Genet. 11, 586462 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spielmeyer W., Ellis M. H., Chandler P. M., Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc. Natl. Acad. Sci. U.S.A. 99, 9043–9048 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monna L., et al. , Positional cloning of rice semidwarfing gene, sd-1: Rice “green revolution gene” encodes a mutant enzyme involved in gibberellin synthesis. DNA Res. 9, 11–17 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Sasaki A., et al. , A mutant gibberellin-synthesis gene in rice. Nature 416, 701–702 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Ashikari M., et al. , Cytokinin oxidase regulates rice grain production. Science 309, 741–745 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Miyao A., et al. , Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell 15, 1771–1780 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jung B., et al. , Uridine-ribohydrolase is a key regulator in the uridine degradation pathway of Arabidopsis. Plant Cell 21, 876–891 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daumann M., Fischer M., Niopek-Witz S., Girke C., Möhlmann T., Apoplastic nucleoside accumulation in arabidopsis leads to reduced photosynthetic performance and increased susceptibility against Botrytis cinerea. Front. Plant Sci. 6, 1158 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jung B., Hoffmann C., Möhlmann T., Arabidopsis nucleoside hydrolases involved in intracellular and extracellular degradation of purines. Plant J. 65, 703–711 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Rast D. M., Baumgartner D., Mayer C., Hollenstein G. O., Cell wall-associated enzymes in fungi. Phytochemistry 64, 339–366 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Teufel F., et al. , SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 401023-1025 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scott A., Wyatt S., Tsou P.-L., Robertson D., Strömgren Allen N., Model system for plant cell biology: GFP imaging in living onion epidermal cells. Biotechniques 26, 1125–1132 (1999). [DOI] [PubMed] [Google Scholar]
- 41.Yoneda A., Ohtani M., Katagiri D., Hosokawa Y., Demura T., Hechtian strands transmit cell wall integrity signals in plant cells. Plants 9, 604 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takei K., Sakakibara H., Taniguchi M., Sugiyama T., Nitrogen-dependent accumulation of cytokinins in root and the translocation to leaf: Implication of cytokinin species that induces gene expression of maize response regulator. Plant Cell Physiol. 42, 85–93 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Osugi A., Sakakibara H., Q&A: How do plants respond to cytokinins and what is their importance? BMC Biol. 13, 102 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ando T., et al. , Genetic dissection and pyramiding of quantitative traits for panicle architecture by using chromosomal segment substitution lines in rice. Theor. Appl. Genet. 116, 881–890 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Lomin S. N., Yonekura-Sakakibara K., Romanov G. A., Sakakibara H., Ligand-binding properties and subcellular localization of maize cytokinin receptors. J. Exp. Bot. 62, 5149–5159 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wulfetange K., et al. , The cytokinin receptors of arabidopsis are located mainly to the endoplasmic reticulum. Plant Physiol. 156, 1808–1818 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caesar K., et al. , Evidence for the localization of the Arabidopsis cytokinin receptors AHK3 and AHK4 in the endoplasmic reticulum. J. Exp. Bot. 62, 5571–5580 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Romanov G. A., Lomin S. N., Schmülling T., Cytokinin signaling: From the ER or from the PM? That is the question! New Phytol. 218, 41–53 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Antoniadi I., et al. , Cell-surface receptors enable perception of extracellular cytokinins. Nat. Commun. 11, 4284 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zürcher E., Liu J., Di Donato M., Geisler M., Müller B., Plant development regulated by cytokinin sinks. Science 353, 1027–1030 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Sato Y., et al. , RiceXPro version 3.0: Expanding the informatics rerefrefsource for rice transcriptome. Nucleic Acids Res. 41, 1206–1213 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sato Y., et al. , RiceXPro: A platform for monitoring gene expression in japonica rice grown under natural field conditions. Nucleic Acids Res. 39, 1141–1148 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kojima M., et al. , Highly sensitive and high-throughput analysis of plant hormones using MS-probe modification and liquid chromatographytandem mass spectrometry: An application for hormone profiling in Oryza sativa. Plant Cell Physiol. 50, 1201–1214 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kojima M., Sakakibara H., Highly sensitive high-throughput profiling of six phytohormones using MS-probe modification and liquid chromatography-tandem mass spectrometry. Methods Mol. Biol. 918, 151–164 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Hirochika H., Sugimoto K., Otsuki Y., Tsugawa H., Kanda M., Retrotransposons of rice involved in mutations induced by tissue culture. Proc. Natl. Acad. Sci. U.S.A. 93, 7783–7788 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Livak K. J., Schmittgen T. D., Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 57.Huerta-Cepas J., Serra F., Bork P., ETE 3: Reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 33, 1635–1638 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kamachi K., Yamaya T., Mae T., Ojima K., A role for glutamine synthetase in the remobilization of leaf nitrogen during natural senescence in rice leaves. Plant Physiol. 96, 411–417 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Niwa Y., A Synthetic green fluorescent protein gene for plant biotechnology. Plant Biotechnol. 20, 1–11 (2003). [Google Scholar]
- 60.Nouchi I., et al. , Overcoming the difficulties in collecting apoplastic fluid from rice leaves by the infiltration-centrifugation method. Plant Cell Physiol. 53, 1659–1668 (2012). [DOI] [PubMed] [Google Scholar]
- 61.Gentzel I., Giese L., Zhao W., Alonso A. P., Mackey D., A simple method for measuring apoplast hydration and collecting apoplast contents. Plant Physiol. 179, 1265–1272 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sakakibara H., et al. , Oryza sativa Japonica Group Nipponbare CPN1 mRNA for cytokinin/purine riboside nucleosidase1, complete cds. DDBJ/EMBL/GenBank. https://www.ncbi.nlm.nih.gov/nuccore/LC746295. Deposited 12 January 2023. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Data Availability Statement
Sequence data from this article were deposited with the DDBJ/EMBL/GenBank data libraries under accession numbers LC746295 (62) for CPN1 (Os06g0406100). All other data are included in the article and/or supporting information.