Significance
The obligate intracellular bacterial pathogen, Coxiella burnetii, is the causative agent of the zoonotic infection termed Q fever. Agricultural outbreaks are an increasingly significant economic and public health burden, and understanding key features of C. burnetii pathogenesis will support the discovery of important targets for future interventions. Significantly, our study has found that a C. burnetii effector protein, MceF (Mitochondrial Coxiella effector protein F), regulates the location of the host protein, GPX4 (Glutathione Peroxidase 4), to protect the host cell from reactive oxygen species–induced cell death. Our findings provide insight into the host–pathogen interactions and demonstrate the utility of studying C. burnetii as a way to gain insight into the programmed cell death of human cells.
Keywords: Coxiella burnetii, GPX4, oxidative stress, Legionella pneumophila, Dot/Icm effector
Abstract
The causative agent of human Q fever, Coxiella burnetii, is highly adapted to infect alveolar macrophages by inhibiting a range of host responses to infection. Despite the clinical and biological importance of this pathogen, the challenges related to genetic manipulation of both C. burnetii and macrophages have limited our knowledge of the mechanisms by which C. burnetii subverts macrophages functions. Here, we used the related bacterium Legionella pneumophila to perform a comprehensive screen of C. burnetii effectors that interfere with innate immune responses and host death using the greater wax moth Galleria mellonella and mouse bone marrow–derived macrophages. We identified MceF (Mitochondrial Coxiella effector protein F), a C. burnetii effector protein that localizes to mitochondria and contributes to host cell survival. MceF was shown to enhance mitochondrial function, delay membrane damage, and decrease mitochondrial ROS production induced by rotenone. Mechanistically, MceF recruits the host antioxidant protein Glutathione Peroxidase 4 (GPX4) to the mitochondria. The protective functions of MceF were absent in primary macrophages lacking GPX4, while overexpression of MceF in human cells protected against oxidative stress–induced cell death. C. burnetii lacking MceF was replication competent in mammalian cells but induced higher mortality in G. mellonella, indicating that MceF modulates the host response to infection. This study reveals an important C. burnetii strategy to subvert macrophage cell death and host immunity and demonstrates that modulation of the host antioxidant system is a viable strategy to promote the success of intracellular bacteria.
Intracellular bacterial pathogens require a viable host cell for replication. As a result, programmed cell death is a common defense outcome of mammalian pathogen-sensing pathways. Metazoan-adapted intracellular pathogens, such as the zoonotic pathogen Coxiella burnetii, have developed strategies to block host cell death allowing the bacteria to replicate to large numbers while minimizing the inflammatory response to infection (1). In comparison, closely related accidental human pathogens, such as Legionella pneumophila, induce inflammatory host cell death leading to their rapid clearance (2).
C. burnetii is a gram-negative, obligate intracellular pathogen and the causative agent of human Q fever (3–6). During infection, C. burnetii has a natural tropism for alveolar macrophages where it establishes a unique intravacuolar replicative niche termed the Coxiella-containing vacuole (CCV). Phagocytosed C. burnetii are trafficked through the endocytic pathway to form the mature CCV, which has many hallmarks of a phagolysosome. An essential virulence factor of C. burnetii is the defective in organelle trafficking/intracellular multiplication (Dot/Icm) type 4B secretion system (T4BSS) which delivers proteins (termed effectors) across the bacterial and vacuole membranes into the host cell. Collectively, these effectors modulate host cell activity to facilitate C. burnetii replication and perturb host immune recognition (7). In the absence of a functional Dot/Icm T4BSS, C. burnetii is incapable of intracellular replication (8, 9). Approximately 130 C. burnetii T4BSS effectors have now been identified, and recent studies have demonstrated that many effectors make important contributions to the intracellular success of this pathogen (10).
L. pneumophila uses a functionally analogous Dot/Icm T4BSS to deliver over 300 novel effectors that allow the pathogen to avoid lysosomal fusion and establish the replicative Legionella-containing vacuole (11, 12). The ease with which L. pneumophila can be genetically manipulated has allowed this pathogen to serve as a surrogate system through which C. burnetii effector translocation can be verified (9, 11, 13–17). In addition, as L. pneumophila strongly activates the innate immune system, it provides a relevant biological tool within which to examine the impact of individual C. burnetii effectors on these processes (4).
Intracellular replication of C. burnetii is relatively slow, with an infectious cycle taking approximately 6 d. It is therefore imperative for C. burnetii to dampen the host response to infection, particularly programmed cell death, to maintain its replicative niche (18, 19). Several studies have demonstrated that C. burnetii infection results in efficient prevention of multiple types of host cell death. C. burnetii induces pro-survival pathways, such as extracellular signal-regulated kinase 1/2, Akt (a serine/threonine-specific protein kinase), and PKA (cAMP-dependent protein kinase) to upregulate antiapoptotic responses upon infection (20, 21). Multiple T4BSS effectors have been implicated in this antiapoptotic phenotype including AnkG, CaeA, and CaeB, which are all able to inhibit intrinsic host cell apoptosis when ectopically expressed in mammalian cells (19, 22–24). A recent investigation demonstrated that C. burnetii also employs the host membrane repair machinery (Endosomal Sorting Complex Required for Transport, ESCRT) to repair CCV membrane damage and subsequently avoid lysosome-dependent cell death (25). Finally, by monitoring caspase-1 activation by L. pneumophila expressing C. burnetii effectors, we previously found that the T4BSS effector IcaA inhibits the noncanonical activation of NLRP3 and subsequent pyroptosis (4).
In this study, the ability of 62 C. burnetii T4BSS effectors to influence the host response to L. pneumophila infection was examined both in vitro, with the measure of bone-marrow-derived macrophage (BMDM) cell death, and in vivo, using the greater wax moth Galleria mellonella and mouse infection models. We identified and validated 15 effectors that increase or decrease G. mellonella survival, demonstrating that multiple C. burnetii effectors can influence the interaction between L. pneumophila and eukaryotic host cells. The most striking phenotypes were obtained with L. pneumophila expressing CBU1543, which decreased G. mellonella death and inhibited macrophage cell death. We determined that CBU1543 encodes a mitochondrial effector, named Mitochondrial Coxiella effector protein F (MceF), that co-opts Glutathione Peroxidase 4 (GPX4) to mitochondria where it can aid in protection from reactive oxygen species–induced cell death. MceF contributes to the ability of C. burnetii to maintain host cell viability, and this study introduces GPX4 as a critical host component in the success of intracellular bacterial pathogens.
Results
C. burnetii T4BSS Effector CBU1543 Impacts L. pneumophila Infection Success In Vitro and In Vivo.
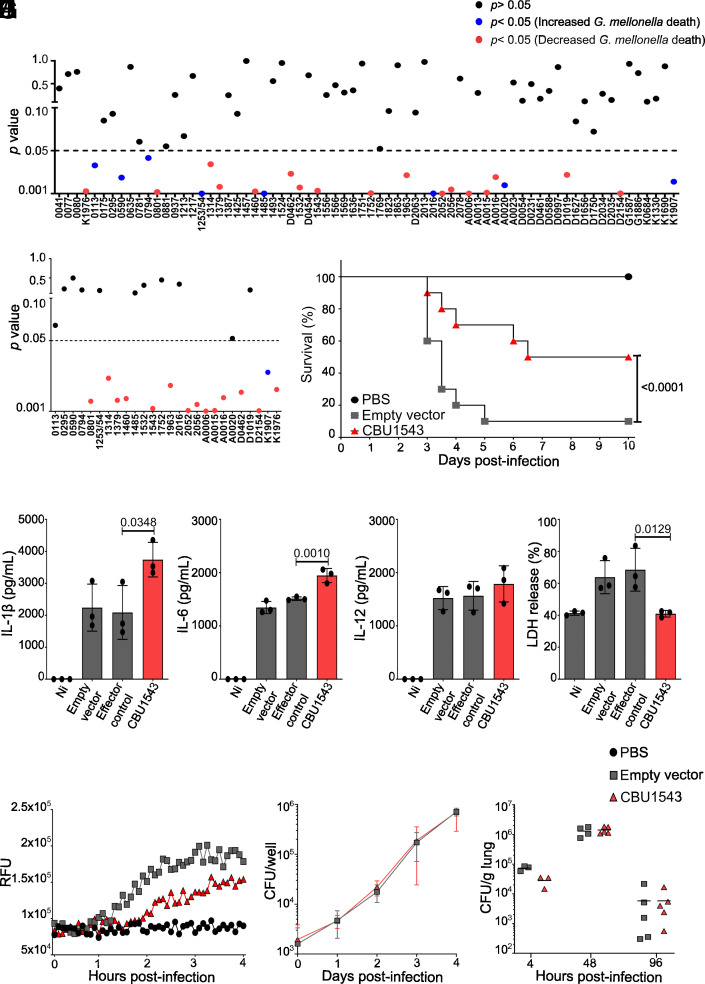
As an accidental pathogen, L. pneumophila is rapidly detected by infected mammalian cells leading to inflammatory host cell death. We employed this trait as a powerful and physiologically relevant model to identify C. burnetii effectors with the capacity to block this host cell response to infection. L. pneumophila JR32 ΔflaA was transformed with the expression plasmid pJB-CAT:3xFLAG to constitutively express individual C. burnetii T4BSS effectors. Seventy strains were engineered with 62 unique effectors and 4 effectors from both the RSA439 strain and another isolate that encodes a full-length version of the RSA439 pseudogene. Following confirmation that the 3xFLAG-tagged C. burnetii effectors were expressed, L. pneumophila strains were used to infect G. mellonella larvae. This inexpensive and rapid in vivo model allowed the evaluation of virulence with a simple survival readout (26–29). The P values comparing survival of G. mellonella infected with L. pneumophila pJB-CAT:3xFLAG [empty vector (EV)] compared to L. pneumophila expressing each C. burnetii effector are represented in Fig. 1A. Our initial screening identified 25 candidates (Fig. 1A) that were further validated and resulted in 15 effectors able to modulate G. mellonella survival (Fig. 1B). Only one effector (CBUK1907, full-length version of RSA439 AnkB) increased G. mellonella death, eliminating all the larvae in a shorter period (72 h) than the control (168 h) (Fig. 1 A and B, blue circle). In contrast, 14 effectors reproducibly induced increased survival, or delayed death, of the infected larvae (Fig. 1 A and B, red circles). Strikingly, this included CBUK1976 and CBUD0462 but not their truncated RSA439 homologues CBU0080 and CBU1524, respectively, providing insight into the effector-mediated virulence differences between C. burnetii isolates. Expression of 3xFLAGCBU1543 led to consistently high survival rates of G. mellonella with 50% of infected G. mellonella still alive 10 d postinfection, compared to only 10% with the EV (Fig. 1C). CBU1543 was confirmed as a C. burnetii Dot/Icm effector using the BlaM reporter assay (SI Appendix, Fig. S1 A and B). CBU1543 translocation was significantly less than another mitochondrial effector, MceA; however, a significant increase in signal was observed with addition of MnCl2 (30) (SI Appendix, Fig. S1A). This is consistent with mitochondrial pores enhancing CCF2-AM access to CBU1543 in the mitochondrial matrix, but providing no benefit to MceA, which is easily accessible on the mitochondrial outer membrane (OM) (31).
Fig. 1.
CBU1543 perturbs L. pneumophila virulence in G. mellonella and modulates BMDM cell death. (A and B) Representative P values of survival curves for G. mellonella following infection with 105 L. pneumophila JR32 ΔflaA carrying pJB-CAT:3xFLAG (EV) compared to L. pneumophila JR32 ΔflaA individually expressing C. burnetii effectors. Results are shown as a representative of one (A) and two (B) independent biological replicates with 10 larvae per condition. P value > 0.05 represented by black circles, P value < 0.05 represented by blue circles = increased G. mellonella death, and red circles = decreased G. mellonella death, based on survival curve log-rank (Mantel-Cox) test. Some C. burnetii NMII RSA439 putative effector pseudogenes were evaluated using the “full length” homologs from other C. burnetii strains (“D” – Dugway; “G” - G Q212; “K” - K Q154). (C) The survival curve indicates viable G. mellonella larvae every 12 h postinfection with the indicated strains of L. pneumophila JR32 ΔflaA. The curve represents the averages of the results of three independent experiments (P value Mantel-Cox test of EV compared to CBU1543). (D–I) BMDMs from wild-type mice (C57BL/6) were not infected (Ni) or infected with L. pneumophila JR32 ΔflaA EV (control), effector control (expressing 3xFLAGCig57), or expressing 3xFLAGCBU1543 (red) at an MOI 10 for 9 h or as indicated. (D) IL-1β production, (E) IL-6 production, and (F) IL-12 production in infected BMDMs determined by ELISA. (G) Percentage of LDH release normalized to Triton X-100 treated cells. Bars represent the averages of the results of three independent experiments, and error bars are SD. Statistical analyses were performed by one-way ANOVA with the multiple comparison test of Bonferroni. (H) Pore membrane formation was fluorometrically assessed by propidium iodide (PI) uptake (RFUs, relative fluorescence units). (I) BMDMs were infected with the indicated L. pneumophila JR32 ΔflaA strains at an MOI 0.015, and bacterial replication was quantified by CFU of each well over 4 d postinfection. Curves represent the averages of four independent experiments, and error bars are SD. Statistical analyses were performed by two-way ANOVA with the multiple comparison test of Bonferroni. (J) C57BL/6 mice were infected intranasally with L. pneumophila JR32 ΔflaA strains at a dose of 105 bacteria/mouse. The mice were euthanized at 4, 48, or 96 h postinfection. Dilutions of the lung homogenates were added to BCYE agar plates for CFU determination. Each dot represents a single animal, and the horizontal lines represent the averages. When the P value was considered statistically significant (P < 0.05), its value was stated in the figure.
To examine the cellular response to infection with L. pneumophila expressing 3xFLAGCBU1543, BMDMs from C57BL/6 mice were infected with L. pneumophila JR32 ΔflaA pJB-CAT:3xFLAG-1543. Previous studies have shown that wild-type L. pneumophila activates the canonical inflammasomes (NLRP3, NAIP/NLRC4, and AIM2 inflammasomes) as well as the noncanonical inflammasome initiated by inflammatory caspases such as caspase 11, in mice (2). L. pneumophila lacking flagellin does not activate NAIP/NLRC4 pathways but still triggers the noncanonical NLRP3 inflammasome and triggers other forms of cell death (4, 32). Macrophage activity was monitored by quantifying IL-1β and noninflammasome-related cytokines, IL-6 and IL-12, in cell culture supernatants. Expression of 3xFLAGCBU1543 led to increased cellular release of IL-1β (Fig. 1D) and IL-6 but not IL-12 (Fig. 1 E and F). The increased production of IL-1β and IL-6 likely reflects the increased macrophage activity in the presence of MceF. Using both, LDH release 4 h postinfection (Fig. 1G) and monitoring membrane damage, via propidium iodide (PI) permeabilization, throughout infection (Fig. 1H), 3xFLAGCBU1543 was shown to provide host cell death protection with less BMDM death than L. pneumophila carrying the EV or expressing an unrelated C. burnetii effector (3xFLAGCig57).
To investigate whether this host cell death prevention would positively contribute to bacterial intracellular replication, BMDMs and C57BL/6 mice were infected with L. pneumophila JR32 ΔflaA EV or expressing 3xFLAGCBU1543, and colony-forming units (CFU) were quantified at the indicated time points. No differences in bacterial replication in macrophages or mice were observed (Fig. 1 I and J). This indicates that CBU1543 does not provide a significant advantage for intracellular replication of L. pneumophila in mice.
CBU1543 (MceF) Associates with the Mitochondrial Inner Membrane and Modulates mROS (Mitochondrial ROS) Production Induced by Rotenone.
To further elucidate the role of CBU1543, the localization of ectopically expressed 3xFLAGCBU1543 was examined in HeLa CCL2 cells. 3xFLAGCBU1543 demonstrated colocalization with host mitochondria (Fig. 2A). Given this distinct subcellular localization and following previous identification of mitochondrially localized C. burnetii effectors (31, 33), CBU1543 was subsequently renamed MceF.
Fig. 2.
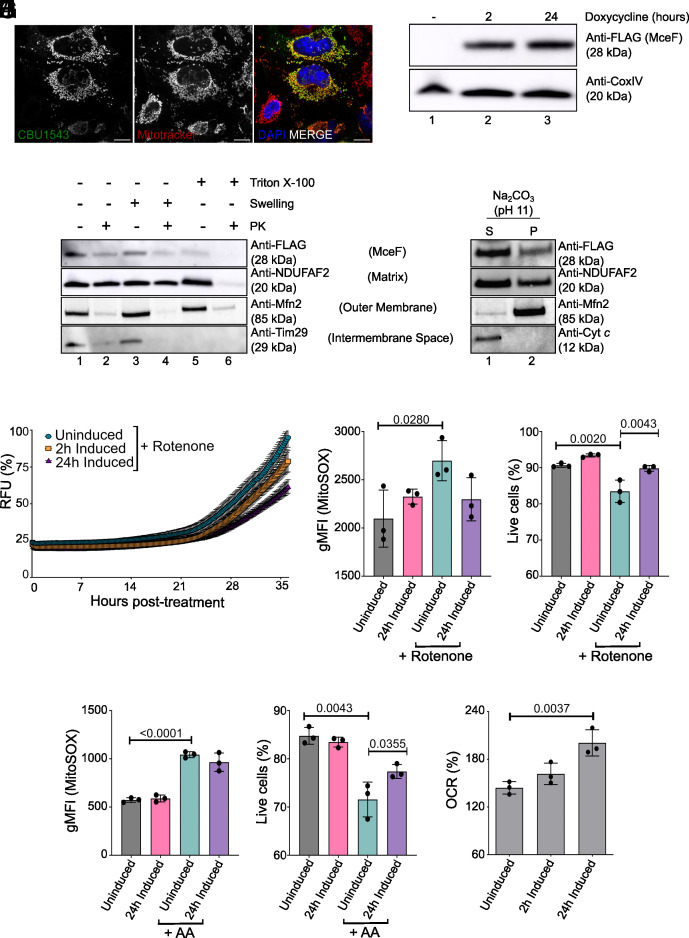
Mitochondrial localization of 3xFLAGCBU1543 (MceF) and its impact on cell death pathways and mitochondria metabolism. (A) HeLa CCL2 cells were transiently transfected with pcDNA4/TO:3xFLAG:mceF for 18 h. Cells were stained for FLAG, DNA (DAPI), and mitochondria (Mitotracker Red) and examined using confocal microscopy. (Scale bars: 15 µm.) (B) Isolated mitochondria (50 μg) from 3xFLAGMceF-expressing stable cell line doxycycline-induced for up to 24 h were analyzed by immunoblotting using the indicated antibodies. (C and D) Isolated mitochondria from 3xFLAGMceF stable cell line after 2 h of effector expression. (C) Mitochondria subfractionation: intact mitochondria (lanes 1 and 2), mitoplasts (generated by hypotonic swelling of the OM, lanes 3 and 4), and solubilized mitochondria (Triton X-100, lanes 5 and 6) were incubated with or without proteinase K (PK, 50 μg/mL) and analyzed by sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and western blotting using the indicated antibodies. (D) Alkaline extraction using 100 mM Na2CO3 (pH 11). The membrane (P) and soluble (S) fractions were separated by ultracentrifugation before SDS-PAGE and immunoblotting with the indicated antibodies. (E) The 3xFLAGMceF-expressing stable cell line was induced with doxycycline at the indicated times before cell death induction (1 µM Rotenone). Membrane integrity was assessed fluorometrically by the uptake of PI for the indicated times. RFUs, relative fluorescence units. Curves indicate the percentage of membrane pore formation normalized to Triton-X 100 treatment. (F–I) Quantification of the mitochondrial superoxide levels and cell viability in 3xFLAGMceF stable cell lines. Cells were induced with doxycycline for 24 h before mROS induction by 1 µM Rotenone for 20 h or by 0.1 µM AA for 4 h. Analysis was done by measuring MitoSOX Red dye (F and H) and Ghost Dye™ Red 780 (G and I) using a flow cytometer. Curves and bar graphics represent the averages of the results of three independent experiments, and error bars are SD. (J) The 3xFLAGMceF stable cell line was induced for effector expression for 2 and 24 h with doxycycline. Percentage of the spare respiratory capacity [(OCR3/OCR1) × 100)]. Statistical analyses were performed by one-way ANOVA with the multiple comparison test of Bonferroni. When the P value was considered statistically significant (P < 0.05), its value was stated in the figure.
HeLa cells were engineered to express 3xFLAGMceF under the control of a doxycycline-inducible promoter. Isolated mitochondria from the 3xFLAGMceF-expressing cell line showed mitochondrial recruitment of 3xFLAGMceF even after just 2 h of doxycycline induction (Fig. 2B). This was also observed after 24 h of 3xFLAGMceF expression, and both induction times were used for further experiments. The submitochondrial localization of 3xFLAGMceF was examined using both hypo-osmotic swelling and carbonate extraction. 3xFLAGMceF was present in isolated mitochondria, and the 3xFLAG tag only became completely accessible to external protease (PK) when the inner membrane was disrupted (Fig. 2C, lane 6), indicating that the N terminus of the protein is partially protected inside the matrix (Fig. 2C, compare lanes 1-5). The OM protein Mfn2, the inner membrane protein Tim29 (large C terminus exposed to the intermembrane space, 34), and the matrix-located protein NDUFAF2 served as controls for mitochondrial fractionation. 3xFLAGMceF was present in both the soluble and insoluble fractions after carbonate treatment of isolated mitochondria. This localization is similar to the partially membrane-integrated matrix protein NDUFAF2, indicating that MceF is a partial membrane protein (Fig. 2D). Taken together, these data indicate that MceF is associated with the mitochondrial inner membrane.
Due to the observed cell death reduction in BMDMs infected with L. pneumophila JR32 ΔflaA 3xFLAGMceF (Fig. 1 G and H) and the mitochondrial localization of this effector (Fig. 2 A–D), we hypothesized that MceF may block cell death signaling at mitochondria. To examine this, the mitochondrial cell death inducer, rotenone, was employed. Through selective inhibition of the mitochondrial electron transport chain, rotenone leads to mROS-induced cell death (35, 36). 3xFLAGMceF expression was induced with doxycycline prior to rotenone treatment, and membrane integrity was assessed by PI uptake as an indicator of membrane damage caused by cell death (4, 37). Rotenone-induced cell death was delayed in the presence of 3xFLAGMceF, showing more than 40% reduction after 24 h of doxycycline induction in comparison to the nonexpressing 3xFLAGMceF cells (Fig. 2E). A specific mitochondrial superoxide indicator, MitoSOX, was used to measure the amount of induced mROS. After 24 h of doxycycline treatment, 3xFLAGMceF prevented the increase in mROS caused by rotenone, which was seen in uninduced cells (Fig. 2F), and subsequently, a decrease in cell death (Fig. 2G), reinforcing our previous results. Interestingly, when these cells were challenged with another selective inhibitor of the mitochondrial electron transport chain (Antimycin A – AA), cells expressing MceF showed decreased cell death (Fig. 2I), but not mROS accumulation (Fig. 2H). These results indicate that MceF can specifically minimize rotenone-induced mROS; however, MceF can still protect the host cell from reactive oxygen species–induced cell death.
The Cytoprotective Effects of MceF Enhance Mitochondrial Spare Respiratory Capacity (SRC) Which May Contribute to Modulating Cell Death.
Mitochondria play a pivotal role in the cellular energy supply, in regulating ROS, numerous cell death pathways, metabolism, cell proliferation, cell signaling, and the regulation of innate and adaptive immunity (38, 39). A decline in mitochondrial function, reflected by diminished electron transport chain activity, can condemn the cell to death due to metabolic catastrophe (39). One of the most informative tests of mitochondrial function is the quantification of cellular respiration in a real-time and live-cell assay (38). Quantification of oxygen consumption rates enables an analysis of the cell’s ability to synthesize adenosine triphosphate (ATP) through mitochondrial oxidative phosphorylation and glycolysis. To evaluate the impact of 3xFLAGMceF expression on mitochondrial function, the 3xFLAGMceF-expressing stable cell line was put through the Agilent Seahorse XF Mito Stress Test (SI Appendix, Fig. S2 A and B). 3xFLAGMceF-expressing cells were either uninduced or treated with doxycycline for 2 and 24 h before the start of the assay. No difference was observed for almost all parameters analyzed, including basal and maximal respiration, proton leak, ATP-linked respiration, nonmitochondrial respiration, and coupling efficiency (SI Appendix, Fig. S2 C–I). However, increased SRC was observed after 24 h of 3xFLAGMceF expression (Fig. 2J). Thus, MceF may contribute to an enhanced mitochondrial SRC and increase the ability of cells to respond to greater energy demands and oxidative stress, such as seen during infection.
MceF Co-Opts GPX4.
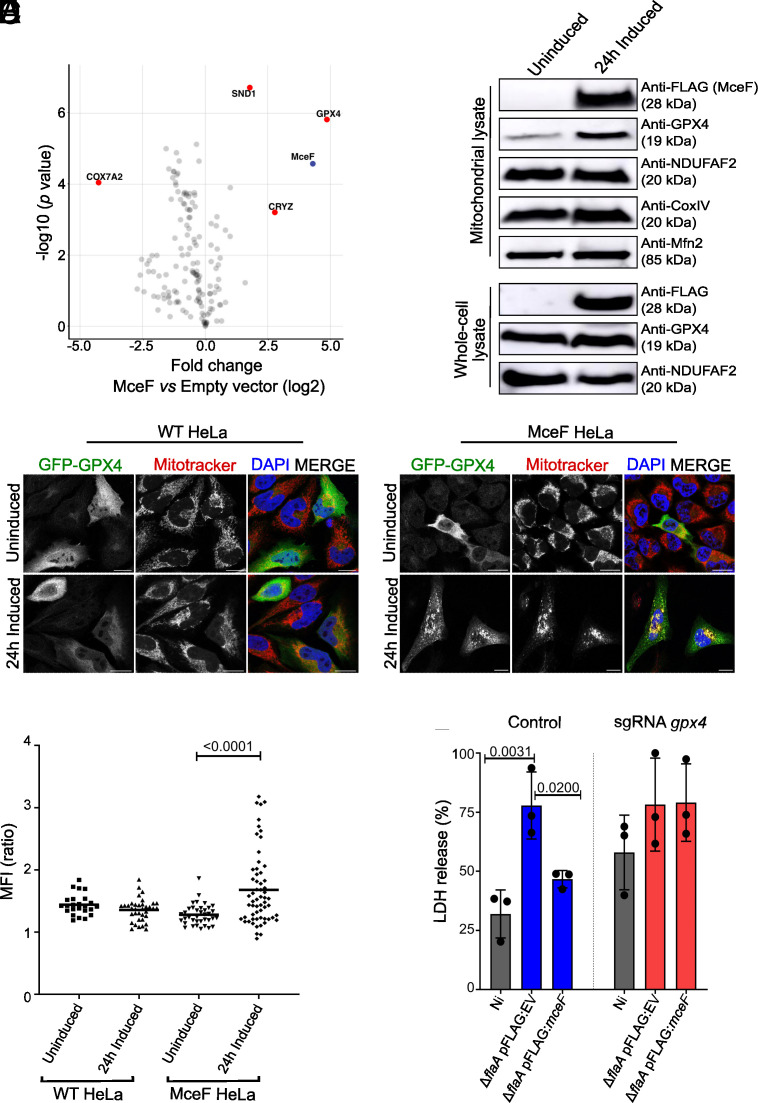
Next, we affinity-purified 3xFLAGMceF and any physically associated proteins using anti-FLAG resin and a stable cell line containing the EV as a negative control. Both cell lines were treated for 24 h with doxycycline prior to immunoprecipitation. Eluted samples were analyzed by label-free quantitative mass spectrometry to identify putative interacting partners of 3xFLAGMceF. We observed enrichment of GPX4 peptides in all four independent pull-downs of 3xFLAGMceF [log2 [fold change] of 3.5352 and -log10(P value) of 4.8762] (Fig. 3A), indicating that 3xFLAGMceF interacts with GPX4. This finding indicates that GPX4 is likely a host target through which MceF manipulates host cell viability.
Fig. 3.
MceF influences mitochondrial localization of GPX4 to reduce host cell death. (A) Label-free quantitative proteomic analysis of 4 independent replicates of an anti-FLAG immunoprecipitation from whole cell lysates of cells treated with doxycycline for 24 h to induce expression of 3xFLAGMceF or cells carrying the EV. Significantly enriched proteins were identified using Student’s t test with a false discovery rate (FDR) of 0.05 and S0 of 0.1. All detected mitochondrial proteins and MceF are shown as dots, and gene names of selected proteins were used as labels. The horizontal axis shows the log2(fold change) and the vertical axis shows -log10(P value). (B) One hundred μg of isolated mitochondria or of the whole cell lysate from 3xFLAGMceF stable cell line uninduced or induced for 24 h with doxycycline were analyzed by SDS-PAGE and western blotting using the indicated antibodies. (C) WT HeLa and MceF HeLa cells were treated with doxycycline for 24 h to induce expression of 3xFLAGMceF and then transiently transfected with GFP-GPX4 as indicated. Cells were stained for DNA (DAPI) and mitochondria (Mitotracker Red) and examined using confocal microscopy. (Scale bars: 15 µm.) (D) Ratio of the mean fluorescence intensity (MFI) of GFP-GPX4 within mitochondrial boundaries vs. cytosol. Image analyses were performed using Fiji ImageJ. (E) Cas9-BMDMs were transduced with the sgRNA lentivirus-containing supernatant (control and sgRNA gpx4) 4 d prior to the experiments. Cells were infected with an MOI 10 of L. pneumophila JR32 ΔflaA carrying pJB-CAT:3xFLAG (EV) or pJB-CAT:3xFLAG:mceF for 4 h before the supernatant was harvested for LDH release measurements. The percentage of LDH release was normalized to Triton X-100. Graphs represent the averages of three independent experiments and error bars are SD. Statistical analyses were performed by one-way ANOVA with the multiple comparison test of Bonferroni. When the P value was considered statistically significant (P < 0.05), its value was stated in the figure.
Mitochondria were isolated from untreated cells or cells treated with doxycycline for 24 h to induce 3xFLAGMceF expression, and equal amounts of mitochondrial protein were analyzed by western blot. GPX4 was notably increased in isolated mitochondria containing 3xFLAGMceF (Fig. 3B), but not in the whole-cell lysate, suggesting that the presence of 3xFLAGMceF facilitates mitochondrial localization rather than production of GPX4. Components of respiratory chain complexes I and IV (NDUFAF2 and CoxIV, respectively) and the mitochondrial OM protein Mfn2 were not affected by the expression of 3xFLAGMceF (Fig. 3B), indicating a specific impact on GPX4 at mitochondria. To confirm this altered localization, the localization of GFP-GPX4 was monitored in 3xFLAGMceF-expressing cells that were colabeled with Mitotracker Red (Fig. 3C). In the presence of 3xFLAGMceF, GFP-GPX4 was significantly recruited to mitochondria (Fig. 3D).
Finally, to investigate the role of GPX4 with MceF during infection in macrophages, BMDM GPX4 CRISPR-Cas9 knockout cells were generated (40, 41) (Fig. 3E). These cells, along with controls treated with the EV, were infected, at an MOI of 10, with L. pneumophila JR32 ΔflaA carrying pJB-CAT:3xFLAG (EV) or pJB-CAT:3xFLAG:mceF. After 4 h of infection, supernatants were collected for LDH release quantification. As previously demonstrated (Fig. 1G), 3xFLAGMceF expression by L. pneumophila inhibited cell death (measured by LDH release) when compared with bacteria encoding EV (P value = 0.02). This protective phenotype is lost in GPX4 CRISPR-Cas9 KO cells (P > 0.99, single guide RNA (sgRNA), GPX4) (Fig. 3E), providing further evidence that MceF functions through GPX4 to modulate host cell death. Interestingly, we observed that the lack of GPX4 in noninfected cells (Ni) was enough to double the release of LDH in comparison to the control (Ni GPX4 KO vs. Ni control cells) (Fig. 3E), confirming the critical role of GPX4 in promoting cell survival.
MceF Is Dispensable for Intracellular Replication of C. burnetii But It Contributes to G. mellonella Survival during Infection.
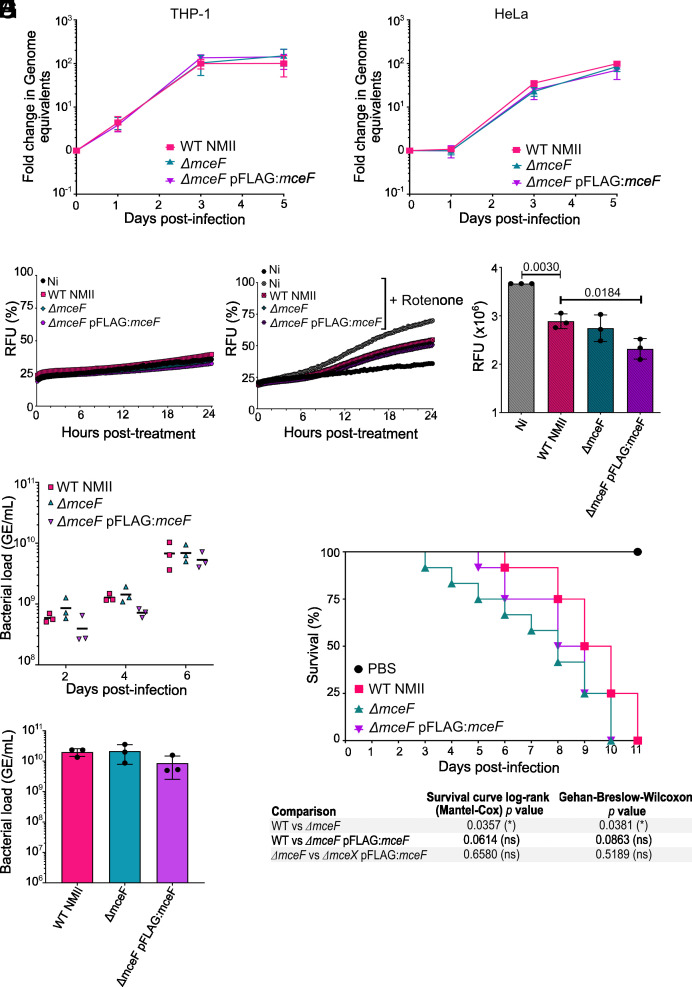
To evaluate the importance of MceF during C. burnetii infection, we generated a mceF mutant of C. burnetii RSA439 Nine Mile Phase II (WT NMII), replacing mceF with a kanamycin resistance cassette, as previously described (42). The mceF mutant was complemented with a plasmid constitutively expressing 3xFLAGMceF (referred to as ΔmceF pFLAG:mceF). THP-1 and HeLa cells were infected with WT NMII, ΔmceF, and ΔmceF pFLAG:mceF, and the number of genome equivalent (GE) of C. burnetii was quantified via qPCR at days 0, 1, 3, and 5 postinfection. All strains showed replication equivalent to that of the parental strain (Fig. 4 A and B), indicating that MceF is not required for C. burnetii intracellular replication in epithelial or macrophage-like cells.
Fig. 4.
Overexpression of MceF in vitro decreases rotenone-induced cell death and its absence reduces G. mellonella survival. (A and B) Intracellular replication of WT C. burnetii NMII, ΔmceF, and ΔmceF pFLAG:mceF in THP-1 macrophage-like cells (A) and HeLa cells (B) was evaluated at days 0, 1, 3, and 5 postinfection at an MOI 5. (C–E) THP-1 macrophage-like cells were infected with WT C. burnetii NMII, ΔmceF, and ΔmceF pFLAG:mceF at an MOI 5. After 3 d of infection, membrane pore formation was assessed for 24 h, without (C) or with (D) the addition of 1 µM rotenone. Curves indicate the percentage of membrane pore formation normalized to cells treated with Triton-X 100. (E) Area under the curve of D. (F–H) G. mellonella larvae were infected with C. burnetii WT NMII, ΔmceF, and ΔmceF pFLAG:mceF at 106 GE. (F) Bacterial load was quantified by qPCR collecting larvae while alive at days 2, 4, and 6. (G) From day 6 postinfection, dead larvae were collected for bacterial load quantification. (H) Survival curve indicates viable G. mellonella larvae every 24 h postinfection for 11 d with the indicated strains of C. burnetii NMII. A PBS control was also included. Graphs represent the averages of the results of three independent experiments, and error bars are SD. Statistical analyses were performed by one- or two-way ANOVA (bars or growth curve, respectively) with the multiple comparison test of Bonferroni. When the P value was considered statistically significant (P < 0.05), its value was stated in the figure. Ni–noninfected; RFU–relative fluorescence units.
To examine whether the absence of MceF would impact host cell death during C. burnetii infection, THP-1 cells were infected for 3 d and then either left untreated (Fig. 4C) or treated with rotenone (Fig. 4D), and PI uptake was measured every 10 min for 24 h. No difference was observed between WT NMII and ΔmceF-infected cells, which was expected given the redundancy of C. burnetii effectors that might interfere with host cell death. However, a small but significant reduction in rotenone-induced cell death was observed when MceF was overexpressed by the complemented strain compared to WT and ΔmceF-infected cells (Fig. 4E). These results indicate that during in vitro infection, overexpression of MceF reduces membrane damage generated by rotenone-induced cell death.
Finally, a role for MceF in C. burnetii virulence in vivo was examined using the G. mellonella model (26–29). G. mellonella larvae were infected with 106 GE of C. burnetii WT NMII, ΔmceF, or ΔmceF pFLAG:mceF strains. The survival and bacterial load of the infected larvae were monitored every 24 h for 11 d. A similar bacterial load was observed in larvae collected every 2 d until day 6 postinfection (Fig. 4F). After day 6, all strains started to kill the larvae (Fig. 4H), and dead larvae were also collected for bacterial load quantification. No difference in the bacterial load was observed (Fig. 4G). However, larvae infected with ΔmceF survived significantly less than larvae infected with the WT NMII (Fig. 4H). The complemented strain showed an intermediate phenotype, but not a significant difference to either WT NMII or ΔmceF (Fig. 4H). Consistently, these results showed that MceF can contribute to extending the survival of different host cells during in vitro and in vivo infection but does not interfere with bacterial replication in all models evaluated in this study.
Discussion
A distinct subset of human infections is caused by obligate intracellular bacterial pathogens that depend on their eukaryotic host to scavenge metabolites and to provide a niche for replication. Given this absolute dependence on their host, it is not surprising that recent studies have demonstrated that these organisms have evolved strategies to surpass and modulate the sophisticated programmed cell death pathways of eukaryotic host cells. Being able to maintain host viability facilitates intracellular bacterial replication and subsequent progression of infection. This evolutionary race in the host–pathogen context makes obligate intracellular bacterial pathogens a rich resource to facilitate our understanding of host–pathogen interactions (1, 7). By commandeering the T4BSS of L. pneumophila, we have developed a powerful model to individually examine the impact of C. burnetii effectors when delivered into cells in a physiologically relevant manner. Indeed, using the G. mellonella infection model, we demonstrated that 14 C. burnetii effectors can increase the G. mellonella tolerance of L. pneumophila demonstrating that these effectors have a measurable impact on infection outcome.
The effector CBU1543, renamed MceF, reduced L. pneumophila–induced cell death of BMDMs, and this cell death protection phenotype was recapitulated in epithelial cells engineered to express 3xFLAGMceF and treated with rotenone or AA. As strong inhibitors of complexes I and III of the mitochondrial respiratory chain, respectively, both lead to increased mROS production and subsequent cell death (43–46). Rotenone induces mROS from complex I by the nicotinamide adenine dinucleotide (NAD) + hydrogen (NADH) dehydrogenase located in the matrix side of the inner membrane and is usually dissipated in the mitochondria by matrix antioxidant defense, such as GPXs, catalases, and superoxide dismutases (43, 45). On the other hand, AA induces mROS from complex III in both cytoplasm and matrix (43). These differences in the mechanisms of action of each inhibitor may explain why MceF only significantly reduced the amount of mROS when treated with rotenone. Even so, MceF protected the cell from both forms of mROS-induced cell death.
Investigation of the human interactome of MceF identified GPX4 as a host target of MceF. In support of a functional interaction, ectopic expression of MceF promoted the recruitment of GFP-GPX4 to mitochondria. A functional relationship between GPX4 and MceF was further established in the context of infection by the loss of MceF-mediated protection against L. pneumophila–induced cell death in GPX4-deficient BMDMs. This provides a demonstration of effector-driven hijacking of the host antioxidant response to counter mROS-mediated damage.
Multiple forms of programmed cell death can be modulated by the recruitment of GPX4 to mitochondria. When recruited, GPX4 localizes in the inner membrane (47) and can protect mitochondria from oxidative damage (48, 49). Therefore, GPX4 is linked to the maintenance of oxidative phosphorylation complexes. Mitochondria are the major physiological source of ROS, which are generated during the TCA cycle and mitochondrial respiration. Lipid peroxidation occurs by an accumulation of oxidizing molecules, such as ROS, thereby altering lipid structure, activity, and physical properties (50). With its unique ability to reduce lipid peroxidation (48, 51, 52), GPX4 is known for its central role in regulating ferroptosis, an emerging form of regulated nonapoptotic cell death (50, 53). Here, we observed an enhanced SRC in the presence of MceF and reduced accumulation of rotenone-induced mROS, which may be linked to the increased abundance of mitochondrial GPX4. Enhancing oxidative phosphorylation contributes to consuming electrons by transferring them to oxygen and, consequently, fewer free radicals causing mitochondrial damage (48).
Oxidative stress–induced apoptosis is caused primarily by the peroxidation of cardiolipin (CL), a mitochondrion-specific phospholipid rich in polyunsaturated fatty acids (54, 55). CL is located exclusively in the mitochondrial inner membrane and preferentially binds to cytochrome c (54, 56). Peroxidation of CL in the mitochondria results in cytochrome c release and apoptosis, which can be avoided by the overexpression of GPX4 (54). Here, we demonstrated that the presence of MceF delays oxidative stress–induced apoptosis assessed by membrane integrity assays. To reinforce our hypothesis, AA-induced mROS is strongly linked to apoptosis due to the marked release of cytochrome c (46), but when MceF is present, less cell death was observed. Thus, it is reasonable to consider that MceF may also modulate AA-induced apoptosis.
Recently, it has been shown that lipid peroxidation drives Gasdermin-D cleavage and, consequently, pyroptosis, which is negatively regulated by GPX4 (57, 58). It is also known that mitochondrial dysfunction and mROS activate the NLRP3 inflammasome leading to pore formation and pyroptosis (59–61) and that GPX4 can protect mitochondria from oxidative damage (48, 49). We observed that BMDMs GPX4 CRISPR-Cas9 knockout cells released 50% more LDH in normal conditions than the controls, and overexpression of MceF failed to delay L. pneumophila-induced LDH release in the absence of GPX4. These results indicate that GPX4 is an essential antioxidant defense enzyme in myeloid cells and that MceF and host GPX4 together may also have a role in pyroptosis.
Due to its oxidative sensitivity (62) and its critical dependence on molecular iron (63), C. burnetii is most likely modulating host iron metabolism and oxidative stress during infection through multiple mechanisms. The bacterium itself relies on the short-chain dehydrogenase SdrA and de novo NAD biosynthesis for resistance to the oxidative phagolysosomal environment (64, 65). C. burnetii can also reduce oxidative stress by providing its own superoxide dismutase and catalase enzymes (66, 67). In addition, C. burnetii influences the host antioxidant response by blocking membrane localization of the cytosolic nicotinamide adenine dinucleotide phosphate hydrogen (NADPH) oxidase complex, through an unknown mechanism (68), and by inducing nuclear translocation of the antioxidant transcription factor Nrf2 which can upregulate GPX4 expression (69, 70). Here, we showed that the effector MceF increases mitochondrial recruitment of GPX4. Thus, we identified a mechanism by which an intracellular bacterial pathogen may be influencing multiple cell death signaling pathways. Commandeering a multifunctional antioxidant such as GPX4, C. burnetii can escape a range of host defense mechanisms.
The lack of MceF did not affect the intracellular replication of C. burnetii in macrophage or epithelial cell models of infection. However, we observed a measurable cytoprotective phenotype linked to MceF function during in vitro infection using an overexpression model. This likely reflects the impact of other C. burnetii effectors, including IcaA, AnkG, CaeA, CaeB, and other uncharacterized effectors, contributing to maintaining host cell viability (1). Indeed, in a recent study, we utilized mass spectrometry to identify several (MceA–MceE) other effectors that demonstrate mitochondrial localization during a native C. burnetii infection (31, 33). MceF was not identified in this study, indicating that it may be present in low abundance and/or there may be temporal control of its mitochondrial localization during infection. Examining the interplay between these effectors throughout the infection cycle may facilitate further understanding of how each effector contributes to the intracellular success of C. burnetii.
MceF did, however, influence the survival of G. mellonella. Overexpression of MceF increased the host tolerance of C. burnetii, while the lack of MceF reduced host tolerance when compared with the respective controls. Not surprisingly, insects also have antioxidant proteins, including copper–zinc superoxide dismutase, peroxiredoxin, and GPX (71–73). Thus, our results are suggesting that direct or indirect manipulation of lipid peroxidation is an essential and common mechanism to prolong the survival of a range of different hosts for C. burnetii, including ticks that play a role in the zoonotic cycle of C. burnetii.
This research demonstrated that GPX4, an antioxidant defense enzyme, is co-opted by C. burnetii to maintain a viable replicative niche. The effector protein, MceF, was identified as the driver of altered GPX4 subcellular localization and, consequently, i) protected mitochondria against oxidative damage and enhance oxidative phosphorylation capacity and ii) protected host cells against oxidative stress–induced apoptosis, pyroptosis, and ferroptosis. However, the biochemical mechanism utilized by MceF to influence GPX4 localization requires further investigation. Understanding this mechanism may contribute to the development of anti-inflammatory and cytoprotective strategies in lipid peroxidation–mediated diseases, such as Alzheimer’s and Parkinson’s which are characterized by high lipid peroxidation, and mitochondrial dysfunction. In this context, the biochemical function of MceF could be harnessed as an alternative way to redirect GPX4 to the mitochondria and reduce organelle damage.
Materials and Methods
Animals and BMDMs.
Mice used in this study (C57BL/6–Jax 000664) were bred and maintained in an institutional animal facility at the Medical School of Ribeirao Preto/University of Sao Paulo, Brazil (FMRP/USP–approved protocol number 196/2016). BMDMs were obtained as previously described (74). For all in vitro experiments, the plates were centrifuged at 300 × g for 5 min, RT, after cell plating and infection. For the in vivo experiments, all mice were matched by sex and age (all were at least 8 wk old at the time of infection). Mice were infected intranasally with 105 bacteria contained in 40 μL of phosphate buffered saline (PBS), as previously described (75, 76). At the indicated time points, the lungs were harvested and macerated for 30 s in 5 mL of autoclaved distilled water using a tissue homogenizer (Power Gen 125; Thermo Scientific). Dilutions were plated on buffered charcoal yeast extract (BCYE) +10 μg/mL of streptomycin, and plates were incubated for 4 d at 37 °C for CFU counting.
Tissue Culture Cell Lines.
HeLa human cervical carcinoma cells (CCL2, American Type Culture Collection - ATCC) were cultured in Dulbecco’s Modified Eagle’s Media GlutaMAX™ (DMEM, Gibco) supplemented with 10% heat-inactivated fetal calf serum (FCS, Gibco). During infection, cultures were maintained in DMEM + 5% FCS. THP-1 human monocytic cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 with GlutaMAX™ (Gibco) supplemented with 10% FCS. All cell lines and primary cells were maintained at 37 °C, 5% CO2.
Bacterial Culture and Preparation.
Detailed methods for bacterial culture and construction of the library of L. pneumophila expressing C. burnetii effectors are found in SI Appendix.
Genetic Manipulation of C. burnetii.
C. burnetii plasmids (pJB-CAT:3xFLAG and pJC-CAT) were transformed into C. burnetii as previously described (77). The cbu1543 deletion strain was created through homologous recombination and sucrose counterselection in accordance with a previously described method (42, 78). The replacement of cbu1543 with a kanamycin resistance gene was confirmed by PCR.
Replication of L. pneumophila in BMDMs.
Experiments to quantify bacterial CFU in macrophages were made in 24-well plates. A total of 2 × 105 BMDMs/well were plated in RPMI + 10% FBS and incubated overnight. The medium was replaced with the bacterial suspension in RPMI + 10% FBS with a multiplicity of infection (MOI) of 0.015 for 1 h before being replaced again by fresh RPMI + 10% FBS. At the indicated time points, the supernatants were collected, cells were lysed with autoclaved distilled water, and the lysate was added to the supernatants. Dilutions were plated on BCYE and incubated for 4 d for CFU counting.
ELISA and Cell Death Measurement by LDH Release.
In vitro cytokine production and lactate dehydrogenase (LDH) release were analyzed in the cell-free supernatants harvested from 1 × 105 BMDMs/well from 96-well-plates infected with the relevant strain at MOI 10. For LDH analysis, supernatants were collected up to 9 h postinfection, while for cytokines, supernatants were collected 9 h postinfection. Plates were centrifuged at 300 × g for 5 min at room temperature (RT), and supernatants were harvested and processed according to the manufacturer’s instructions (IL-1β, IL-6, and IL-12 BD Biosciences) and CytoTox 961 Non-Radioactive Cytotoxicity Assay (Promega, Wisconsin, USA). The optical density (OD) was measured at 450 nm (enzyme-linked immunoassay - ELISA) and 490 nm (LDH) with a SpectraMax plate reader (Molecular Devices, California, USA).
Pore Formation Assay.
For estimation of cell membrane pore formation, 1 × 105 macrophages/well or 5 × 104 HeLa cells/well were plated on black, clear-bottom 96-well plates (Corning) in RPMI or DMEM containing 10% fetal bovine serum (FBS) or FCS (as mentioned above) and incubated overnight or differentiated as required. The medium was replaced with the bacterial suspension (estimated to reach the indicated MOI) or with rotenone (1 µM; Sigma) (48, 79) in RPMI or DMEM without Phenol Red with 6 μL/mL of PI (Invitrogen). PI was excited at 538 nm, and emission was measured at 617 nm with a SpectraMax plate reader (Molecular Devices, California, USA) or with a CLARIOstar®Plus plate reader (BMG LABTECH) every 10 min for the indicated times. Cells treated with 1.3% Triton-X100 were used as a total cell death control for normalization.
C. burnetii Infections.
Detailed methods for C. burnetii of tissue culture cells and G. mellonella are provided in SI Appendix.
Immunofluorescence.
Prior to fixation, cells were incubated with MitoTrackerTM Red CMXRos (ThermoFisher Scientific) according to the manufacturer’s protocol. HeLa cells were fixed for 5 min at −20 °C with 1/1 (v/v) Methanol (Sigma-Aldrich)/Acetone (Sigma-Aldrich). Cells were then blocked using PBS + 2% BSA, before being incubated with mouse anti-FLAG (Sigma-Aldrich, 1:250). After three washes in PBS, cells were incubated with relevant secondary antibodies (Goat AlexaFluor-488 Mouse IgG, ThermoFisher Scientific, 1:2,000). PBS containing DAPI (Life Technologies) at 1:10,000 was added to cells before additional washes in PBS. Coverslips were mounted onto glass slides with ProLong Gold reagent (Life Technologies). All samples were imaged using a Zeiss LSM700 instrument with Zen software, and images were analyzed using Fiji ImageJ (80).
Mitochondrial Isolation and Mitochondrial Procedures.
Isolation of mitochondria from tissue culture cells was performed by differential centrifugation as described previously (31, 33, 34, 81). Harvested cells were washed once with ice-cold PBS and centrifuged at 800 × g for 5 min at 4 °C. The pellets were resuspended in ice-cold solution A (70 mM sucrose, 220 mM mannitol, 20 mM HEPES-KOH (pH 7.6), 1 mM ethylenediamine tetraacetic acid (EDTA), 0.1 mM PMSF, and 2 mg/mL BSA) and homogenized in a handheld glass Douce homogenizer. The homogenate was centrifuged at 600 × g for 5 min at 4 °C to remove cellular and nuclear debris. The supernatant was centrifuged at 12,000 × g for 10 min at 4 °C, and the pellet (crude mitochondrial fraction) was resuspended in ice-cold solution B (70 mM sucrose, 220 mM mannitol, 20 mM HEPES-KOH (pH 7.6), and 1 mM EDTA). Samples were quantified using a BCA kit (Pierce) following the manufacturer’s instructions.
For mitochondrial subfractionation experiments, mitochondrial pellets (30 to 50 µg of mitochondrial protein) were resuspended in ice-cold solution B, ice-cold swelling buffer (10 mM HEPES-KOH pH 7.4), or ice-cold solubilization buffer (0.5% [v/v] Triton-X 100). Samples were split and either left untreated or treated with proteinase K (PK—50 µg/mL) for 10 min on ice, followed by the addition of 1 mM PMSF for 5 min. For sodium carbonate extraction, mitochondrial pellets (30 to 50 µg of mitochondrial protein) were resuspended in freshly prepared Na2CO3 (100 mM, pH 11 to 12). Samples were incubated on ice for 30 min with occasional mixing and subsequently ultracentrifuged at 100,000 × g for 30 min. The soluble (supernatant) and insoluble (pellet) fractions were separated. Mitochondrial subfractionation and carbonate extraction samples were trichloroacetic acid (TCA) precipitated and analyzed by immunoblotting (detailed experimental methods for immunoblotting are described in SI Appendix).
Immunoprecipitation, LC-MS Analysis of 3xFLAGMceF Interactome, and Proteomic analysis.
Detailed methods for immunoprecipitation of 3xFLAGMceF, sample preparation, liquid chromatography-mass spectrometry (LC-MS) conditions, and data analysis are contained in SI Appendix. The resulting MS data and search results have been deposited into the PRIDE ProteomeXchange Consortium repository (82–84).
Measurement of Mitochondrial Respiration and m ROS.
Detailed experimental methods for mitochondrial respiration and measurement of mROS are described in SI Appendix.
CRISPR/Cas9 Manipulation of BMDMs.
Two double-stranded guide RNA (gRNA) targeting gpx4 were designed online using the webserver (http://crispr.mit.edu), and oligonucleotide pairs for each gRNA were synthesized (SI Appendix, Table S1). The LentiGuide-puro (Addgene) plasmid was digested with BsmBI (New England Biolabs, USA) and annealed to each gRNA. For lentiviral production, HEK293 cells were transfected with LentiGuide-puro gRNA and packing plasmids psPAX2 (Addgene) and pMD2.G (Addgene). Sixty hours posttransfection, lentivirus-containing supernatant from HEK293 cells was used to transduce BMDMs. For the generation of BMDM deficient in gpx4, tibias and femurs were removed from Cas9 transgenic mice and bone marrow was flushed as above. On the third day of differentiation, lentivirus-containing supernatant was added to the culture, and after 48 h, puromycin (10 µg/mL) was added for the selection. Cells were used 4 d after puromycin selection.
Statistical Analysis.
The data were analyzed using GraphPad Prism 9.3 software. The statistical significance was calculated using Student’s t test or ANOVA with SD (SD) as indicated in the figure legends. Differences were considered statistically significant when P was <0.05, as indicated in the figures.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We would like to kindly thank Prof. John Silke (Walter and Eliza Hall Institute of Medical Research, Australia) for providing lentiviral plasmids, and Prof. Robert Heinzen (Rocky Mountain Laboratories, NIH) for supplying C. burnetii genomic DNA and expression plasmids. We thank the team at the Biological Optical Microscopy Platform (University of Melbourne) and the Melbourne Cytometry Platform (Doherty Institute node, University of Melbourne) who provided invaluable technical expertise and maintenance for the confocal microscope and cytometry used in this study. The Australian Genome Research Facility provided sequencing services in Melbourne. We thank Prof Mike Ryan for gifting to the Stojanovski Lab the Mfn2 and NDUFAF2 antibodies. We thank the Laboratory Managers Maira Nakamura and Livia Marangoni Alfaya for technical support. R.K.L. was a graduate student at the Basic and Applied Immunology Program from the Medical School of Ribeirão Preto, University of São Paulo, Brazil, and recipient of the fellowships (2016/24275-7 and 2018/236898) from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP). This work was supported by grants from the Center for Research on Inflammatory Diseases (CRID/FAPESP, grant 2013/08216-2), FAPESP (grants 2014/04684-4, 2019/11342-6, and 2016/07870-9) and NHMRC Ideas Grant 2010841. G.H.G. and D.S.Z. are 1A and 1B Research Fellows from CNPq, respectively.
Author contributions
R.K.L., D.R.T., Y.W.L., G.H.G., D.S.Z., and H.J.N. designed research; R.K.L., D.R.T., W.A., Y.W.L., L.L.S., D.P.A.M., T.M.S., J.C., L.F.F., L.L., L.E.B., N.E.S., and H.J.N. performed research; R.K.L., G.H.G., D.S., D.S.Z., and H.J.N. contributed new reagents/analytic tools; R.K.L., D.R.T., L.E.B., D.S., N.E.S., D.S.Z., and H.J.N. analyzed data; and R.K.L., D.S.Z., and H.J.N. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Dario S. Zamboni, Email: dszamboni@fmrp.usp.br.
Hayley J. Newton, Email: hayley.newton@monash.edu.
Data, Materials, and Software Availability
Proteomics data have been deposited in PRIDE ProteomeXchange Consortium repository (PXD030191; reviewer_pxd030191@ebi.ac.uk Password: Hr6uHdoK) (84).
Supporting Information
References
- 1.Loterio R. K., Zamboni D. S., Newton H. J., Keeping the host alive–Lessons from obligate intracellular bacterial pathogens. Pathog. Dis. 79, 1–15 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Mascarenhas D. P. A., Zamboni D. S., Inflammasome biology taught by Legionella pneumophila. J. Leukoc. Biol. 101, 841–849 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Fernandes T. D., et al. , Murine alveolar macrophages are highly susceptible to replication of Coxiella burnetii phase II in vitro. Infect. Immun. 84, 2439–2448 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cunha L. D., et al. , Inhibition of inflammasome activation by Coxiella burnetii type IV secretion system effector IcaA. Nat. Commun. 6, 1–13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moffatt J. H., Newton P., Newton H. J., Coxiella burnetii: Turning hostility into a home. Cell Microbiol. 17, 621–631 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Armstrong M. R., Mccarthy K. L., Horvath R. L., A contemporary 16-year review of Coxiella burnetii infective endocarditis in a tertiary cardiac center in Queensland, Australia. Infect. Dis. 50, 1–8 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Pechstein J., Schulze-Luehrmann J., Lührmann A., Coxiella burnetii as a useful tool to investigate bacteria-friendly host cell compartments. Int. J. Med. Microbiol. 308, 77–83 (2017), 10.1016/j.ijmm.2017.09.010. [DOI] [PubMed] [Google Scholar]
- 8.Beare P. A., et al. , Dot/Icm type IVB secretion system requirements for Coxiella burnetii growth in human macrophages. mBio 2, 1–10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carey K. L., Newton H. J., Lu A., Roy C. R., The Coxiella burnetii dot/Icm system delivers a unique repertoire of type IV effectors into host cells and is required for intracellular replication. PLoS Pathog. 7, 1–18 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lührmann A., Newton H. J., Bonazzi M., Beginning to understand the role of the Type IV secretion system effector proteins in Coxiella burnetii pathogenesis. Curr. Top Microbiol. Immunol. 413, 243–268 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Zamboni D. S., McGrath S., Rabinovitch M., Roy C. R., Coxiella burnetii express type IV secretion system proteins that function similarly to components of the Legionella pneumophila Dot/Icm system. Mol. Microbiol. 49, 965–976 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Coers J., Monahan C., Roy C. R., Modulation of phagosome biogenesis by Legionella pneumophila creates an organelle permissive for intracellular growth. Nat. Cell Biol. 1, 451–453 (1999). [DOI] [PubMed] [Google Scholar]
- 13.Cerqueira D. M., Pereira M. S. F., Silva A. L. N., Cunha L. D., Zamboni D. S., Caspase-1 but not caspase-11 is required for NLRC4-mediated pyroptosis and restriction of infection by flagellated legionella species in mouse macrophages and in vivo. J. Immunol. 195, 2303–2311 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Weber M. M., et al. , Identification of Coxiella burnetii type IV secretion substrates required for intracellular replication and Coxiella-containing vacuole formation. J. Bacteriol. 195, 3914–3924 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lifshitz Z., et al. , Identification of novel Coxiella burnetii Icm/Dot effectors and genetic analysis of their involvement in modulating a mitogen-activated protein kinase pathway. Infect. Immun. 82, 3740–3752 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lifshitz Z., et al. , Computational modeling and experimental validation of the Legionella and Coxiella virulence-related type-IVB secretion signal. Proc. Natl. Acad. Sci. U.S.A. 110, E707–E715 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen C., et al. , Large-scale identification and translocation of type IV secretion substrates by Coxiella burnetii. Proc. Natl. Acad. Sci. U.S.A. 107, 21755–21760 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Broederdorf L. J., Voth D. E., Cheating death: A Coxiella effector prevents apoptosis. Front. Microbiol. 2, 1–2 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cordsmeier A., Wagner N., Lührmann A., Berens C., Defying death–How Coxiella burnetii copes with intentional host cell suicide. Yale J. Biol. Med. 92, 619–628 (2019). [PMC free article] [PubMed] [Google Scholar]
- 20.Voth D. E., Heinzen R. A., Sustained activation of Akt and Erk1/2 is required for Coxiella burnetii antiapoptotic activity. Infect. Immun. 77, 205–213 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lührmann A., Roy C. R., Coxiella burnetii inhibits activation of host cell apoptosis through a mechanism that involves preventing cytochrome c release from mitochondria. Infect. Immun. 75, 5282–5289 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lührmann A., Nogueira C. V., Carey K. L., Roy C. R., Inhibition of pathogen-induced apoptosis by a Coxiella burnetii type IV effector protein. Proc. Natl. Acad. Sci. U.S.A. 107, 18997–19001 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klingenbeck L., Eckart R. A., Berens C., Lührmann A., The Coxiella burnetii type IV secretion system substrate CaeB inhibits intrinsic apoptosis at the mitochondrial level. Cell Microbiol. 15, 675–687 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Cordsmeier A., et al. , The Coxiella burnetii T4SS effector protein AnkG hijacks the 7SK small nuclear ribonucleoprotein complex for reprogramming host cell transcription. PLoS Pathog. 18, e1010266 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Radulovic M., Bongiovanni A., Schink K. O., Nähse V., ESCRT-mediated lysosome repair precedes lysophagy and promotes cell survival. EMBO J. 37, 1–15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Norville I. H., et al. , Galleria mellonella as an alternative model of Coxiella burnetii infection. Microbiology (N. Y.) 160, 1175–1181 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Martinez E., et al. , Coxiella burnetii effector CvpB modulates phosphoinositide metabolism for optimal vacuole development. Proc. Natl. Acad. Sci. U.S.A. 113, E3260–E3269 (2016), 10.1073/pnas.1522811113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohler L. J., et al. , Effector protein cig2 decreases host tolerance of infection by directing constitutive fusion of autophagosomes with the Coxiella-containing vacuole. mBio 7, 1–14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuba M., et al. , Eira is a novel protein essential for intracellular replication of Coxiella burnetii. Infect. Immun. 88, 1–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rama Rao K. V., Norenberg M. D., Manganese induces the mitochondrial permeability transition in cultured astrocytes. J. Biol. Chem. 279, 32333–32338 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Fielden L. F., et al. , A farnesylated Coxiella burnetii effector forms a multimeric complex at the mitochondrial outer membrane during infection. Infect. Immun. 85, 1–15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pereira M. S. F., et al. , Activation of NLRC4 by flagellated bacteria triggers caspase-1-dependent and -independent responses to restrict Legionella pneumophila replication in macrophages and in vivo. J. Immunol. 187, 6447–6455 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Fielden L. F., et al. , Proteomic identification of Coxiella burnetii effector proteins targeted to the host cell mitochondria during infection. Mol. Cell. Proteomics 20, 1–19 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang Y., et al. , Tim29 is a novel subunit of the human TIM22 translocase and is involved in complex assembly and stability. Elife 5, 1–23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heinz S., et al. , Mechanistic investigations of the mitochondrial complex i inhibitor rotenone in the context of pharmacological and safety evaluation. Sci. Rep. 7, 1–13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lima-Junior D. S., Mineo T. W. P., Calich V. L. G., Zamboni D. S., Dectin-1 activation during leishmania amazonensis phagocytosis prompts Syk-dependent reactive oxygen species production to trigger inflammasome assembly and restriction of parasite replication. J. Immunol. 199, 2055–2068 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Rieger A. M., Nelson K. L., Konowalchuk J. D., Barreda D. R., Modified annexin V/propidium iodide apoptosis assay for accurate assessment of cell death. J. Visualized Exp. 50, 3–6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yépez V. A., et al. , OCR-Stats: Robust estimation and statistical testing of mitochondrial respiration activities using seahorse xf analyzer. PLoS One 13, 1–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang D., Kang R., Berghe T. V., Vandenabeele P., Kroemer G., The molecular machinery of regulated cell death. Cell Res. 29, 347–364 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clausen T. M., et al. , SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell 183, 1043–1057 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alix E., et al. , The tumour suppressor TMEM127 is a Nedd4-family E3 ligase adaptor required by salmonella SteD to ubiquitinate and degrade MHC class II molecules. Cell Host. Microbe. 28, 54–68 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.P. Beare, R. Heinzen, Gene inactivation in Coxiella burnetii. Host-bacteria interactions: Methods and protocols. Methods Mol. Biol. 1197, 329–345 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Javadov S., The calcium-ROS-pH triangle and mitochondrial permeability transition: Challenges to mimic cardiac ischemia-reperfusion. Front. Physiol. 6, 83 (2015). 10.3389/fphys.2015.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang L., et al. , Mitochondrial respiratory chain inhibitors involved in ROS production induced by acute high concentrations of iodide and the effects of sod as a protective factor. Oxid Med. Cell Longev. 2015, 217670 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Q., Vazquez E. J., Moghaddas S., Hoppel C. L., Lesnefsky E. J., Production of reactive oxygen species by mitochondria: Central role of complex III. J. Biol. Chem. 278, 36027–36031 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Woo H. P., Yong W. H., Suhn H. K., Sung Z. K., An ROS generator, antimycin A, inhibits the growth of HeLa cells via apoptosis. J. Cell Biochem. 102, 98–109 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Liang H., et al. , Glutathione peroxidase 4 differentially regulates the release of apoptogenic proteins from mitochondria. Free Radic. Biol. Med. 47, 312–320 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cole-Ezea P., Swan D., Shanley D., Hesketh J., Glutathione peroxidase 4 has a major role in protecting mitochondria from oxidative damage and maintaining oxidative phosphorylation complexes in gut epithelial cells. Free Radic. Biol. Med. 53, 488–497 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Liang H., et al. , Gpx4 protects mitochondrial ATP generation against oxidative damage. Biochem. Biophys. Res. Commun. 356, 893–898 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Gaschler M. M., Stockwell B. R., Lipid peroxidation in cell death. Biochem. Biophys. Res. Commun. 482, 419–425 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nomura K., Imai H., Koumura T., Kobayashi T., Nakagawa Y., Mitochondrial phospholipid hydroperoxide glutathione peroxidase inhibits the release of cytochrome c from mitochondria by suppressing the peroxidation of cardiolipin in hypoglycaemia-induced apoptosis. Biochem. J. 351, 183–193 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maiorino M., Conrad M., Ursini F., GPx4, lipid peroxidation, and cell death: Discoveries, rediscoveries, and open issues. Antioxid. Redox Signal 29, 61–74 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Dixon S. J., et al. , Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nomura K., Imai H., Koumura T., Arai M., Nakagawa Y., Mitochondrial phospholipid hydroperoxide glutathione peroxidase suppresses apoptosis mediated by a mitochondrial death pathway. J. Biol. Chem. 274, 29294–29302 (1999). [DOI] [PubMed] [Google Scholar]
- 55.Tang D., Ferroptosis in Health and Disease (Springer Nature, Switzerland, 2019), 10.1007/978-3-030-26780-3. [DOI] [Google Scholar]
- 56.Ran Q., et al. , Transgenic mice overexpressing glutathione peroxidase 4 are protected against oxidative stress-induced apoptosis. J. Biol. Chem. 279, 55137–55146 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Kang R., et al. , Lipid peroxidation drives gasdermin D-mediated pyroptosis in lethal polymicrobial sepsis article lipid peroxidation drives gasdermin D-mediated pyroptosis in lethal polymicrobial sepsis. Cell Host. Microbe 24, 97–108.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu H., Santo A., Jia Z., Li Y., GPx4 in bacterial infection and polymicrobial sepsis: Involvement of ferroptosis and pyroptosis. React. Oxyg. Species 7, 154–160 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou R., Yazdi A. S., Menu P., Tschopp J., A role for mitochondria in NLRP3 inflammasome activation. Nature 469, 221–226 (2011). [DOI] [PubMed] [Google Scholar]
- 60.Chung C., et al. , Mitochondrial oxidative phospholiration complex regulates NLRP3 inflammasome activation and predicts patient survival in nasopharyngeal carcinoma. Mol. Cell Proteomics 19, 142–154 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Evavold C. L., et al. , Control of gasdermin D oligomerization and pyroptosis by the Ragulator-Rag-mTORC1 pathway. Cell 184, 4495–4511.e19 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zamboni D. S., Rabinovitch M., Nitric oxide partially controls. Am. Soc. Mcrobiol. 71, 1225–1233 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sanchez S. E., Omsland A., Critical role for molecular iron in Coxiella burnetii replication and viability. Am. J. Public Health 5, 1–17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bitew M. A., et al. , De novo NAD synthesis is required for intracellular replication of Coxiella burnetii, the causative agent of the neglected zoonotic disease Q fever. J. Biol. Chem. 293, 18636–18645 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bitew M. A., et al. , SdrA, an NADP(H)-regenerating enzyme, is crucial for Coxiella burnetii to resist oxidative stress and replicate intracellularly. Cell Microbiol. 22, 1–16 (2019). [DOI] [PubMed] [Google Scholar]
- 66.Akporiaye E. T., Baca O. G., Superoxide anion production and superoxide dismutase and catalase activities in Coxiella burnetii. J. Bacteriol. 154, 520–523 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brennan R. E., Kiss K., Baalman R., Samuel J. E., Cloning, expression, and characterization of a Coxiella burnetii Cu/Zn superoxide dismutase. BMC Microbiol. 15, 1–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Siemsen D. W., Kirpotina L. N., Jutila M. A., Quinn M. T., Inhibition of the human neutrophil NADPH oxidase by Coxiella burnetii. Microbes. Infect. 11, 671–679 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Winchell C. G., et al. , Coxiella burnetii subverts p62/sequestosome 1 and activates Nrf2 signaling in human macrophages. Infect. Immun. 86, 1–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dodson M., Castro-Portuguez R., Zhang D. D., NRF2 plays a critical role in mitigating lipid peroxidation and ferroptosis. Redox Biol. 23, 101107 (2019). 10.1016/j.redox.2019.101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Büyükgüzel E., Evidence of oxidative and antioxidative responses by Galleria mellonella larvae to malathion. J. Econ. Entomol. 102, 152–159 (2009). [DOI] [PubMed] [Google Scholar]
- 72.Büyükgüzel E., Kalender Y., Exposure to streptomycin alters oxidative and antioxidative response in larval midgut tissues of Galleria mellonella. Pestic. Biochem. Physiol. 94, 112–118 (2009). [Google Scholar]
- 73.Brennan L. J., Keddie B. A., Braig H. R., Harris H. L., The endosymbiont Wolbachia pipientis induces the expression of host antioxidant proteins in an Aedes albopictus cell line. PLoS One 3, 1–8 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marim F. M., Silveira T. N., Lima D. S., Zamboni D. S., A method for generation of bone marrow-derived macrophages from cryopreserved mouse bone marrow cells. PLoS One 5, 1–8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gonçalves A. V., et al. , Gasdermin-D and caspase-7 are the key caspase-1/8 substrates downstream of the NAIP5/NLRC4 inflammasome required for restriction of Legionella pneumophila. PLoS Pathog. 15, e1007886 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mascarenhas D. P. A., et al. , Inhibition of caspase-1 or gasdermin-D enable caspase-8 activation in the Naip5/NLRC4/ASC inflammasome. PLoS Pathog. 13, 1–28 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Newton H. J., et al. , A screen of Coxiella burnetii Mutants reveals important roles for dot/Icm effectors and host autophagy in vacuole biogenesis. PLoS Pathog. 10, 1–16 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Beare P. A., Larson C. L., Gilk S. D., Heinzen R. A., Two systems for targeted gene deletion in Coxiella burnetii. Appl. Environ. Microbiol. 78, 4580–4589 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lima-Junior D. S., et al. , Inflammasome-derived IL-1β production induces nitric oxide-mediated resistance to Leishmania. Nat. Med. 19, 909–915 (2013). [DOI] [PubMed] [Google Scholar]
- 80.Schindelin J., et al. , Fiji–An open platform for biological image analysis. Nat. Methods 9, 1–15 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Johnston A. J., et al. , Insertion and assembly of human Tom7 into the preprotein translocase complex of the outer mitochondrial membrane. J. Biol. Chem. 277, 42197–42204 (2002). [DOI] [PubMed] [Google Scholar]
- 82.Vizcaíno J. A., et al. , 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Perez-Riverol Y., et al. , The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Loterio R., Scott N., Newton H., CBU1543 immunoprecipation (CBU1543 vs empty vector). PRICE Proteomics Identifications Database. http://www.ebi.ac.uk/pride/archive/projects/PXD030191. Deposited 5 December 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Proteomics data have been deposited in PRIDE ProteomeXchange Consortium repository (PXD030191; reviewer_pxd030191@ebi.ac.uk Password: Hr6uHdoK) (84).