Abstract
A positive phenotypic characteristic of glucose-oxidizing acinetobacters was demonstrated with blood agar containing d-glucose. Glucose-oxidizing Acinetobacter baumannii, Acinetobacter genospecies 3, Acinetobacter lwoffii, and Acinetobacter genospecies 13 sensu Tjernberg and Ursing caused a unique brown discoloration of media supplemented with 5% blood (of horse, sheep, or human origin) and an aldose sugar (0.22 M d-glucose, d-galactose, d-mannose, d-xylose, or lactose). The browning effect was not observed when a ketose sugar (d-fructose or sucrose) was substituted for the aldose sugar or under high osmolarity in the presence of mannitol, glycerol, or sodium chloride. Other gram-negative nonfermenters (non-glucose-oxidizing acinetobacters, Pseudomonas aeruginosa, other Pseudomonas spp., Stenotrophomonas maltophilia, Flavobacterium spp., and Moraxella spp.) did not cause similar discoloration. This novel browning effect may serve as an alternative trait for identifying glucose-oxidizing acinetobacters.
Acinetobacters do not possess unique biochemical characteristics to allow for unambiguous phenotypic identification. The genus is characterized by properties it does not possess, i.e., being oxidase negative, nonmotile, and nonfermentative. In 1986 Bouvet and Grimont (5) identified 12 genospecies by DNA-DNA hybridization. Three years later Bouvet and Jeanjean (6) reported new proteolytic strains that they designated genospecies 13 to 17. In the same year Tjernberg and Ursing (17) published information on unrelated strains that were also assigned to genospecies 13 to 15. An extensive set of substrate assimilation tests has become necessary for the unequivocal identification of the genospecies of acinetobacters (19). However, even commercially available multitest biochemical systems may not be able to discriminate between certain genospecies, such as genospecies 1, 2, 3, and 13 sensu Tjernberg and Ursing (13TU) (3), and Gerner-Smidt et al. (9) have suggested that these genospecies should be referred to as the Acinetobacter calcoaceticus-Acinetobacter baumannii complex.
The endemic presence of acinetobacters in our locality has been reported previously (16). During specimen processing we found an acinetobacter isolate that grew as mucoid colonies on Sabouraud dextrose agar, a glucose-containing fungal medium. This observation, coupled with the report of glucose-oxidizing acinetobacters being capable of growing in 5% dextrose at room temperature (13), prompted us to initiate a study of the growth of acinetobacters on solid media with glucose as an ancillary nutrient. We report, for the first time, on the ability of glucose-oxidizing acinetobacters to cause a unique brown discoloration of blood agar into which glucose is incorporated. In an attempt to use this positive phenotype as a simple means of identification, we identified by PCR the genospecies of isolates that exhibited the browning effect, as well as those that did not. Since certain strains of acinetobacters produce gelatinase (2), lipase (2), and phospholipase C (12), tests to determine production of these extracellular enzymes in the presence of glucose were also performed.
Clinical isolates and experimental conditions.
Acinetobacters have been identified as oxidase negative, nonmotile, nonfermentative, gram-negative coccobacilli that grow well only under aerobic conditions (18). The ability to oxidize glucose was tested in Hugh and Leifson’s medium, and the organisms were reported as glucose-oxidizing or non-glucose-oxidizing Acinetobacter spp.
Two hundred isolates of acinetobacters collected between January 1990 and October 1995, including a positive control (A. baumannii ATCC 19606); 186 glucose-oxidizing isolates from blood (n = 119), wound (n = 30), and urinary (n = 21) and respiratory (n = 16) tract specimens; and 13 non-glucose-oxidizing isolates from blood (n = 9) and wound (n = 4) specimens, were subcultured for the following experiments: (i) acinetobacters were grown on Oxoid Columbia Agar Base (Unipath, Hants, United Kingdom) with or without 5% defibrinated horse, sheep, or human blood, in the presence or absence of 0.055 or 0.22 M (1 or 4% [wt/vol]) d-glucose, 0.22 M (3% [wt/vol]) d-galactose, 0.22 M (4% [wt/vol]) d-mannose, 0.22 M (3% [wt/vol]) d-xylose, 0.22 M (4% [wt/vol]) d-fructose, 0.22 M (7.5% [wt/vol]) lactose, 0.22 M (7.5% [wt/vol]) sucrose, 0.22 M (4% [wt/vol]) mannitol, 0.22 M (2% [wt/vol]) glycerol, or 0.34 M (2% [wt/vol]) sodium chloride (BDH Chemicals Ltd., Poole, United Kingdom), where the sugars and salt were dissolved in distilled water and filter sterilized before being added to the agar base; (ii) acinetobacters were cultured on plain horse blood agar at 44°C for 24 h; (iii) 47 clinical isolates of nonfermentative gram-negative bacilli (Pseudomonas aeruginosa, other Pseudomonas spp., Stenotrophomonas maltophilia, Flavobacterium spp., and Moraxella spp.) identified according to standardized protocols (10, 18) were grown on horse blood agar containing 0.22 M d-glucose; and (iv) acinetobacters were cultured on chocolate agar, gelatinase agar, DNase agar, and egg yolk agar with or without 0.22 M d-glucose. Serratia liquefaciens ATCC 27592, Serratia marcescens ATCC 8100, and Bacillus subtilis ATCC 6633 served as positive controls for gelatinase, DNase, and lecithinase production, respectively. All cultures, except those for gelatinase and DNase, were incubated at 37°C for 24 h unless otherwise specified. The plates for the gelatinase and DNase tests were kept at 25°C for 48 h before being flooded, respectively, with the Frazier reagent and 1 M HCl.
Identification of genospecies by PCR.
To assign acinetobacters to their respective genospecies, 42 of 199 clinical isolates were selected for PCR with primers G1 (5′-GAAGTCGTAACAAGG-3′) and L1 (5′-CAAGGCATCCACCGT-3′) (Gibco BRL, Life Technologies, Gaithersburg, Md.), which contain conserved sequences of the 16S to 23S rRNA spacer region, by modifications of protocols by Jensen et al. (11) and Nowak et al. (14). A fraction of a colony of each isolate was picked and suspended in a 50-μl reaction mixture containing GeneAmp PCR buffer (Perkin-Elmer) with 1.5 mM MgCl2, 100 μM (each of the four) deoxynucleoside triphosphates (Pharmacia), 1% Triton X-100, and 1.25 ng of each primer per μl. The resulting mixture was heated at 94°C for 5 min. After the addition of 2.5 U of Taq polymerase (Perkin-Elmer), each tube was transferred to a Bio-Rad gene cycler for PCR under the following conditions: 94°C for 1 min, 55°C for 7 min, and 72°C for 2 min for 25 cycles followed by a terminal extension at 72°C for 7 min. Negative controls containing the reagents but no DNA, as well as the positive control A. baumannii ATCC 19606, were included with each batch of test samples. PCR products were separated by polyacrylamide gel electrophoresis (5% polyacrylamide) at 80 V for 2 h. The genospecies of the acinetobacter strains were identified based on the uniquely sized primary PCR fragments of each strain’s DNA according to the method of Nowak et al. (14): A. baumannii (genospecies 2) possesses a band of 940 bp; Acinetobacter genospecies 3 possesses a band of 1,000 bp; and Acinetobacter lwoffii (genospecies 8 or 9) possesses a band of 1,300 bp. There were four acinetobacter isolates that showed a band of 1,100 bp, which did not fit into the classification scheme. Since these isolates also grew at 44°C, they were identified tentatively as Acinetobacter genospecies 13TU.
Results.
A. baumannii ATCC 19606 and the majority of clinical isolates that oxidize glucose grew on horse blood agar containing 0.22 M glucose and produced a unique light-brown discoloration of the surrounding agar (Fig. 1). These results were reproducible with three different batches of media. The same browning effect could be demonstrated with a lower concentration of glucose (0.055 M) and was also evident on horse blood agar containing 0.22 M galactose, mannose, xylose, or lactose. The discoloration was present when sheep or human blood was used in place of horse blood but not on a similar agar base without the addition of blood. No discoloration was observed when acinetobacters grew on horse blood agar containing 0.22 M fructose, sucrose, mannitol, or glycerol or 0.34 M sodium chloride. Growth of acinetobacters on glucose-containing chocolate agar also did not cause any discoloration.
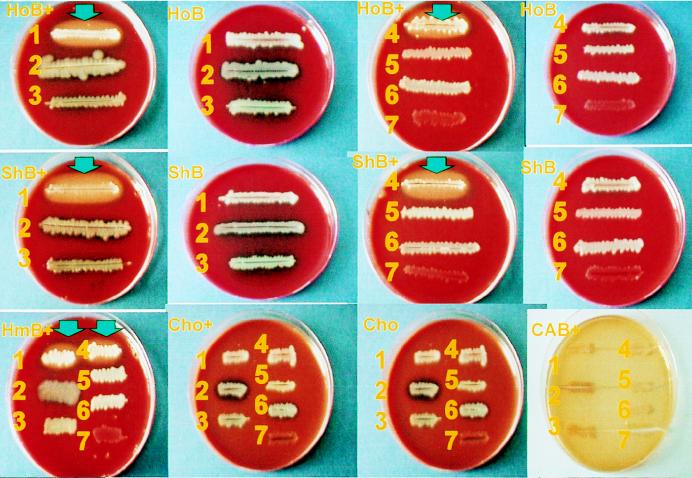
FIG. 1.
Browning effect of glucose-oxidizing acinetobacters on blood agar containing glucose. A. baumannii ATCC 19606 (1) and a clinical isolate of A. baumannii (4) caused a light-brown discoloration (indicated by arrows) on glucose-containing Columbia Agar Base with blood of horse (HoB+), sheep (ShB+), or human (HmB+) origin but not on plain agar with blood of horse (HoB), sheep (ShB), or human origin. The browning effect was not observed when other nonfermenters such as P. aeruginosa (2), Stenotrophomonas maltophilia (3), non-glucose-oxidizing A. lwoffii (5), Flavobacterium meningosepticum (6), and Moraxella osloensis (7) were cultured on glucose-containing blood agar. In addition, the discoloration was not seen on glucose-containing chocolate agar (Cho+) or Columbia Agar Base (CAB+) or on plain chocolate agar (Cho) and Columbia Agar Base.
Of the 186 glucose-oxidizing and 13 non-glucose-oxidizing clinical isolates that were tested for their ability to demonstrate the browning effect and growth at 44°C, the genospecies of 32 glucose-oxidizing and 10 non-glucose-oxidizing isolates were determined (Fig. 2). The majority of glucose-oxidizing isolates (93.5%) were positive for both characteristics (Table 1), and 25 of 29 isolates whose genospecies were identified were A. baumannii; the remaining 4 isolates were identified presumptively as Acinetobacter genospecies 13TU. Twelve glucose-oxidizing isolates (6.5%) could discolor the glucose-enriched horse blood agar but were not able to grow at 44°C, and three of these were selected randomly for identification of genospecies. Two isolates were Acinetobacter genospecies 3, and the other was A. lwoffii. There were three non-glucose-oxidizing isolates that showed no discoloration despite growth at 44°C, and all were identified as A. baumannii. The remaining 10 non-glucose-oxidizing isolates were negative for both characteristics, and all 7 isolates for which we determined genospecies were A. lwoffii.
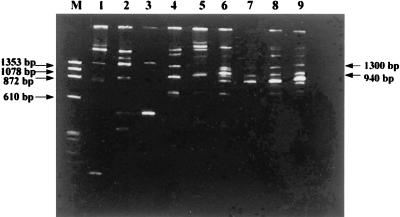
FIG. 2.
Representative PCR profiles of the two most common Acinetobacter genospecies, A. baumannii (lanes 4 to 9) and A. lwoffii (lanes 1 to 3), with unique bands of 940 and 1,300 bp, respectively. Lane M, HaeIII-digested φX174 DNA molecular size markers.
TABLE 1.
Prevalence of Acinetobacter genospecies exhibiting particular phenotypic characteristicsa
| Hugh and Leifson’s medium | Browning | Growth at 44°C | Total no. of isolates | No. of isolates for PCR genospecies identification | No. of isolates of genospeciesb:
|
|||
|---|---|---|---|---|---|---|---|---|
| 2 | 3 | 8 or 9 | 13TU | |||||
| Glucose-oxidizing | + | + | 174 | 29 | 25 | 0 | 0 | 4 |
| + | − | 12 | 3 | 0 | 2 | 1 | 0 | |
| Non-glucose-oxidizing | − | + | 3 | 3 | 3 | 0 | 0 | 0 |
| − | − | 10 | 7 | 0 | 0 | 7 | 0 | |
Acinetobacter isolates were tested for their abilities to oxidize glucose in Hugh and Leifson’s medium, cause browning of blood agar containing 0.22 M glucose, and grow on plain blood agar when they are incubated at 44°C.
Genospecies 2 (A. baumannii), genospecies 3 (unnamed), and genospecies 8 or 9 (A. lwoffii) were identified according to the method Nowak et al. (14). The presumptive identification of genospecies 13TU was as described in the text.
Browning of glucose-enriched horse blood agar was not observed with other nonfermentative gram-negative bacilli such as P. aeruginosa (n = 18), Stenotrophomonas maltophilia (n = 11), Pseudomonas spp. (n = 8), Flavobacterium spp. (n = 7), and Moraxella spp. (n = 3) (Fig. 1).
To further elucidate the nature of the discoloration, growth of the acinetobacters on other media used for the routine detection of gelatinase, DNase, and lecithinase was tested in the presence and absence of glucose. The gelatinase and DNase agars were not cleared; therefore, the discoloration on glucose-enriched blood agar was not caused by a gelatinase or a DNase. There was turbidity on egg yolk agar containing glucose. However P. aeruginosa (n = 5) also caused turbidity. Thus, the observed turbidity probably resulted from a nonspecific reaction(s).
Discussion.
A browning of the surrounding agar was observed when glucose-oxidizing acinetobacters were cultured on blood agar containing an aldose sugar (Fig. 1). This effect was not exhibited under high osmolarity or in the presence of ketose sugars. The browning effect is unlikely to be useful as a phenotypic trait for identifying the genospecies of acinetobacters, as it was not genospecies specific. Some strains of A. baumannii and A. lwoffii tested positive, while others of the same genospecies tested negative (Table 1). If the ability of an acinetobacter to cause browning is related to its ability to oxidize glucose, our work corroborates the report of Bouvet and Bouvet (4), who showed that each of these genospecies comprises glucose-oxidizing and non-glucose-oxidizing strains.
The assignment of genospecies to acinetobacters was based on the unique fragment size(s) of the spacer regions between amplified 16S and 23S rRNAs (11, 14). Fragments of other sizes were also present among strains of the same genospecies (Fig. 2), which may reflect heterogeneity of spacer regions within that genospecies. This finding was consistent with the previously reported (14) presence of weak and varied secondary amplification products in individual acinetobacter genospecies.
Although the browning effect could not be used for identification of genospecies, the test for browning may still be useful as one of the few positive laboratory tests for the easy detection of glucose-oxidizing acinetobacters (n = 186). Furthermore, this test allows us to differentiate nonfermenters that do cause browning from nonfermenters that do not cause browning, such as non-glucose-oxidizing acinetobacters (n = 13), as well as members of other genera like Pseudomonas, Stenotrophomonas, Flavobacterium, and Moraxella (n = 47) (Fig. 1). Hence, blood agar containing glucose may serve as an alternative to Hugh and Leifson’s medium. Further testing with a larger number of clinical isolates is required to assess the feasibility of using the browning trait for identifying glucose-oxidizing acinetobacters.
Since the browning effect was associated with the presence of aldose sugars, one possible explanation is that discoloration occurred as a result of oxidation of sugars by an aldose dehydrogenase, glucose dehydrogenase (GDH). Present information on acinetobacter GDH is derived mainly from studies of A. calcoaceticus sensu stricto, a glucose-oxidizing genospecies commonly found in soil. A. calcoaceticus contains soluble and membrane-bound GDH (8, 15), whereas other oxidative bacteria contain the membrane-bound enzyme exclusively. As the browning effect was observed only with glucose-oxidizing acinetobacters, discoloration may develop as a consequence of the action of soluble GDH, as other membrane-bound GDH-producing organisms such as Pseudomonas aeruginosa did not discolor the glucose-enriched blood agar. Furthermore, the browning effect was observed when acinetobacters were grown in the presence of lactose, and it is known that the soluble GDH, but not the membrane-bound GDH, converts dissaccharides such as lactose to their corresponding acids (7).
The browning was not caused by enzymes such as lecithinase, lipase, and gelatinase. Thus, the nature of browning on glucose-incorporated blood agar remains to be clarified. On the other hand Affeldt and Rockwood (1) have demonstrated that glucose-oxidizing acinetobacters cause browning in liquid media. A brown coloration developed in the culture filtrate when glucose-oxidizing A. baumannii, but not non-glucose-oxidizing A. lwoffii, grew in a sodium acetate and ammonium dihydrogen phosphate basal salts medium (containing 30 mM NaC2H3O2 · 3H2O, 18 mM NH4H2PO4, 40 mM K2HPO4, 22 mM KH2PO4, and 0.83 mM MgSO4 · 7H2O [pH 7.0]). Affeldt and Rockwood have suggested that the Maillard reaction, the nonenzymatic glycation between reducing sugars and amino groups of proteins, may be responsible for the observed brown color, as chromatographic analysis of the brown freeze-dried supernatant fluid yielded hexose sugars (glucose and galactose), amino hexose (galactosamine), and amino acids (ornithine, serine, threonine, alanine, valine, and leucine or isoleucine).
In conclusion, the unique browning of acinetobacters as demonstrated on blood agar containing glucose may be a useful phenotypic trait for the identification of glucose-oxidizing acinetobacters and for their differentiation from other non-glucose-oxidizing nonfermenters. The nature of the browning remains to be elucidated.
Acknowledgments
This work was supported by the Committee of Research and Conference Grants, The University of Hong Kong.
We thank Janice Y. C. Lo for comments on an earlier version of the manuscript; W. C. Yam, S. K. Lau, and H. Y. Ngan for photographic assistance; and K. M. Ng, W. B. Lai, Y. L. Chiu, Y. L. Chow, and M. L. Cheung for their interest in this study.
REFERENCES
- 1.Affeldt M M, Rockwood S W. Browning of acetate medium by Herellea vaginicola (Achromobacter anitratus) Can J Microbiol. 1970;16:325–330. doi: 10.1139/m70-058. [DOI] [PubMed] [Google Scholar]
- 2.Baumann P, Doudoroff M, Stanier R Y. A study of the Moraxella group. II. Oxidative-negative species (genus Acinetobacter) J Bacteriol. 1968;95:1520–1541. doi: 10.1128/jb.95.5.1520-1541.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernards A T, van der Toorn J, van Boven C P A, Dijkshoorn L. Evaluation of the ability of a commercial system to identify Acinetobacter genomic species. Eur J Clin Microbiol Infect Dis. 1996;15:303–308. doi: 10.1007/BF01695662. [DOI] [PubMed] [Google Scholar]
- 4.Bouvet P J M, Bouvet O M M. Glucose dehydrogenase activity in Acinetobacter species. Res Microbiol. 1989;140:531–540. doi: 10.1016/0923-2508(89)90085-5. [DOI] [PubMed] [Google Scholar]
- 5.Bouvet P J M, Grimont P A D. Taxonomy of the genus Acinetobacter with the recognition of Acinetobacter baumannii sp. nov., Acinetobacter haemolyticus sp. nov., Acinetobacter johnsonii sp. nov., and Acinetobacter junii sp. nov. and emended descriptions of Acinetobacter calcoaceticus and Acinetobacter lwoffii. Int J Syst Bacteriol. 1986;36:228–240. [Google Scholar]
- 6.Bouvet P J M, Jeanjean S. Delineation of new proteolytic genomic species in the genus Acinetobacter. Res Microbiol. 1989;140:291–299. doi: 10.1016/0923-2508(89)90021-1. [DOI] [PubMed] [Google Scholar]
- 7.Cleton-Jansen A M, Goosen N, Wenzel T J, van de Putte P. Cloning of the gene encoding quinoprotein glucose dehydrogenase from Acinetobacter calcoaceticus: evidence for the presence of a second enzyme. J Bacteriol. 1988;170:2121–2125. doi: 10.1128/jb.170.5.2121-2125.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duine J A, Frank J, Jzn R, van der Meer R A. Different forms of quinoprotein aldose-(glucose)-dehydrogenase in Acinetobacter calcoaceticus. Arch Microbiol. 1982;131:27–31. doi: 10.1007/BF00451494. [DOI] [PubMed] [Google Scholar]
- 9.Gerner-Smidt P, Tjernberg I, Ursing J. Reliability of phenotypic tests for identification of Acinetobacter species. J Clin Microbiol. 1991;29:277–282. doi: 10.1128/jcm.29.2.277-282.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilligan P H. Pseudomonas and Burkholderia. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C: American Society for Microbiology; 1995. pp. 509–519. [Google Scholar]
- 11.Jensen M A, Webster J A, Straus N. Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphisms. Appl Environ Microbiol. 1993;59:945–952. doi: 10.1128/aem.59.4.945-952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lehmann V. Hemolytic activity of various strains of Acinetobacter. Acta Pathol Microbiol Scand Sect B. 1973;81:427–432. doi: 10.1111/j.1699-0463.1973.tb02226.x. [DOI] [PubMed] [Google Scholar]
- 13.Maki D G, Martin W T. National epidemic of septicemia caused by contaminated infusion products. IV. Growth of microbial pathogens in fluids for intravenous infusion. J Infect Dis. 1975;131:267–272. doi: 10.1093/infdis/131.3.267. [DOI] [PubMed] [Google Scholar]
- 14.Nowak A, Burkiewicz A, Kur J. PCR differentiation of seventeen genospecies of Acinetobacter. FEMS Microbiol Lett. 1995;126:181–188. doi: 10.1111/j.1574-6968.1995.tb07414.x. [DOI] [PubMed] [Google Scholar]
- 15.Olsthoorn A J J, Duine J. Production, characterization, and reconstitution of recombinant quinoprotein glucose dehydrogenase (soluble type; EC 1.1.99.17) apoenzyme of Acinetobacter calcoaceticus. Arch Biochem Biophys. 1996;336:42–48. doi: 10.1006/abbi.1996.0530. [DOI] [PubMed] [Google Scholar]
- 16.Siau H, Yuen K Y, Wong S S Y, Ho P L, Luk W K. The epidemiology of acinetobacter infections in Hong Kong. J Med Microbiol. 1996;44:340–347. doi: 10.1099/00222615-44-5-340. [DOI] [PubMed] [Google Scholar]
- 17.Tjernberg I, Ursing J. Clinical strains of Acinetobacter classified by DNA-DNA hybridization. APMIS. 1989;97:595–605. doi: 10.1111/j.1699-0463.1989.tb00449.x. [DOI] [PubMed] [Google Scholar]
- 18.von Graevenitz A. Acinetobacter, Alcaligenes, Moraxella, and other nonfermentative gram-negative bacteria. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C: American Society for Microbiology; 1995. pp. 520–532. [Google Scholar]
- 19.Weaver R E, Actis L A. Identification of Acinetobacter species. J Clin Microbiol. 1994;32:1833. doi: 10.1128/jcm.32.7.1833-.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]