Abstract
IS6110 restriction fragment length polymorphism typing is now established as the primary typing method for Mycobacterium tuberculosis. It has been assumed that the position of bands is random. Thus, the discrimination of the technique increases in proportion to the copy number. Two collections of M. tuberculosis were investigated to test this hypothesis. We identified 33 positions in isolates from a Tanzanian collection and 25 positions in isolates from a London, United Kingdom, collection where bands were significantly more likely to be present than would be expected by chance. These data suggest that band position is not random, and this possibility may have an impact on the interpretation of molecular epidemiological studies of M. tuberculosis.
The insertion sequence IS6110 is found in almost all isolates of Mycobacterium tuberculosis and is present in between 1 and over 20 copies per isolate (2). The positions of these copies have been used in an internationally standardized restriction fragment length polymorphism (RFLP) protocol to type M. tuberculosis (13), and the protocol is a powerful tool for unravelling questions of tuberculosis epidemiology. It has been particularly useful in investigating tuberculosis outbreaks in closed communities such as hospitals and prisons (4, 9, 11) and in detecting cross-infection by shared use of equipment or laboratory cross-contamination (9, 12). More recently population-based studies have been reported (3), and it has been possible to follow the spread of drug-resistant isolates in wider communities (1, 7, 14).
Mycobacterium paratuberculosis possesses an insertion sequence, IS900, which has a defined 5-bp target (AGGAG) at which it integrates into the bacterial chromosome (6). No similar target sequence has been reported for IS6110, although the direct repeat locus has been recognized as a hot spot for integration, being the common site for single copy strains (10). It has been speculated that this site represents the original point of entry for IS6110 and other copies arise from further transposition events (10). The sequence adjacent to this site has been determined and given the name IS6110 preferential locus (ipl) (8).
IS6110 typing is dependent on the premises that integration into the genome is random and that the discrimination of the technique increases in proportion to the copy number. The aim of this study was to test the hypothesis that integration of IS6110 into the genome of M. tuberculosis is random. If, alternatively, there were integration hot spots this would confound the interpretation of the many population-based IS6110 typing studies currently under way.
Two collections of M. tuberculosis isolates were investigated to ensure that results would not be confounded by a bias due to transmission of strains in a single population. One collection consisted of 207 strains isolated from patients with tuberculosis diagnosed in the Royal Free and St George’s Hospitals, London, United Kingdom, who had acquired their infection from a wide range of countries including the Indian subcontinent, Africa, Southern Europe, and the United Kingdom. The second was a group of 154 strains collected from the Northern Zone of Tanzania as part of a study of the interaction of human immunodeficiency virus and tuberculosis.
Analysis of IS6110 RFLP was performed by the International Standard Method (13). Fingerprints were compared by using GelCompar computer analysis software; this process normalized the gel image, dividing each track into 400 arbitrary divisions. The number of isolates with IS6110 bands located in each of the positions defined by the GelCompar program was plotted.
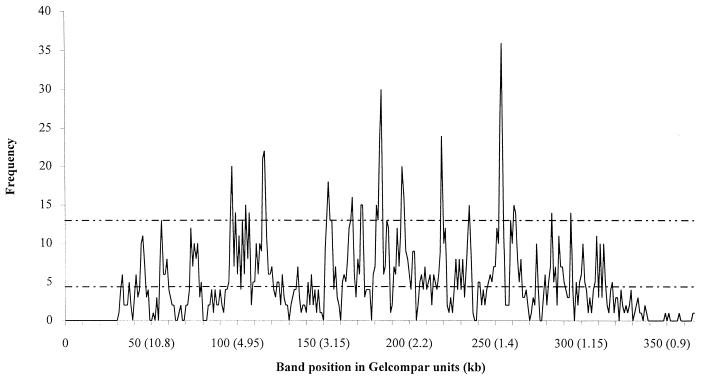
The number of IS6110 bands in each position is illustrated in Fig. 1 for the Tanzanian isolates, and a similar pattern was found for the London strains (Table 1). If IS6110 were randomly distributed in the M. tuberculosis genome, the graph line would be parallel to the x axis and have a value equal to the total number of IS6110 bands divided by the total number of positions and would vary around that mean. Distinct peaks were found in a number of positions.
FIG. 1.
Number of M. tuberculosis isolates with a band at a given position. The lower line represents the predicted mean number of bands per position (total number of bands divided by the number of band positions available equaled 4.4). The upper line represents the hot-spot cutoff line (see text for definition).
TABLE 1.
Positions of hot spots in Tanzanian and London isolates
| Approx. mol wt (kb) | Hot-spot position (Gelcompar units)
|
|
|---|---|---|
| Tanzanian isolates | London isolates | |
| 9.25 | 57 | |
| 5.25 | 97 | |
| 5.15 | 98 | 98 |
| 4.95 | 100 | 100 |
| 4.90 | 102 | |
| 4.80 | 104 | 104 |
| 4.75 | 105 | |
| 4.70 | 106 | 106 |
| 4.50 | 108 | 108 |
| 4.30 | 112 | |
| 4.10 | 116 | 116 |
| 4.05 | 117 | 117 |
| 3.20 | 153 | 153 |
| 3.15 | 154 | 154 |
| 3.10 | 155 | |
| 3.05 | 156 | 156 |
| 2.85 | 164 | |
| 2.8 | 166 | |
| 2.75 | 167 | |
| 2.70 | 168 | |
| 2.65 | 174 | |
| 2.60 | 175 | |
| 2.50 | 181 | |
| 2.45 | 182 | |
| 2.40 | 183 | |
| 2.40 | 184 | 184 |
| 2.35 | 185 | |
| 2.30 | 188 | |
| 2.20 | 197 | |
| 2.20 | 198 | |
| 2.20 | 200 | |
| 1.90 | 220 | 220 |
| 1.90 | 221 | |
| 1.70 | 236 | |
| 1.45 | 254 | 254 |
| 1.45 | 255 | |
| 1.45 | 259 | |
| 1.40 | 260 | 260 |
| 1.40 | 262 | |
| 1.40 | 263 | 263 |
| 1.25 | 284 | 284 |
| 1.20 | 295 | |
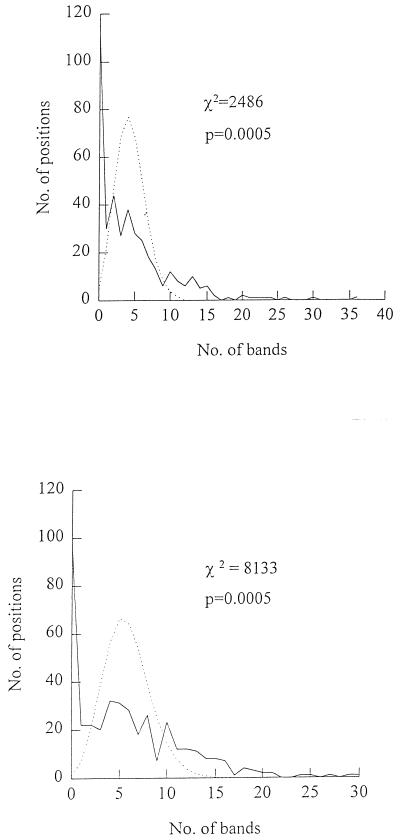
In Fig. 2 the number of positions with a given number of IS6110 bands is plotted for the Tanzanian and London isolates together with the expected number of positions with a given number of IS6110 bands and its Poisson distribution. A χ2 test was performed by calculating the expected number of positions with a given number of IS6110 bands and comparing this with the observed number. The results of this test for the Tanzanian isolates were χ2 = 2,486, df = 11, P < 0.0005, and for the London isolates the results were χ2 = 8,133, df = 12, P = 0.0005. This χ2 analysis demonstrates that the observed frequency distribution is significantly different from the predicted, random distribution for both populations, indicating that band positions are not randomly associated but that there are favored locations where more than the expected number of IS bands are found. By using these data it was possible to define hot spots for each collection of isolates as positions where the expected number of positions with a given number of IS6110 bands was less than 0.16 (i.e., >99% confidence interval), giving cutoff values of 13 and 16 IS bands per position for the Tanzanian and London isolates, respectively. These positions are listed in Table 1. For the Tanzanian isolates a total of 33 hot spots were identified, although 17 peaks were noted on the frequency distribution graphs as many hot spots are found in adjacent positions (Table 1). The majority of the hot spots are shared by the two populations we studied. It should be noted that these hot spots included the direct repeat locus which has already been defined and is located at hot-spot position 254 (10).
FIG. 2.
Frequency distribution of the number of positions with a given number of bands for isolates of M. tuberculosis collected in Tanzania (top) and London, United Kingdom (bottom). The dotted lines represent the predicted number of positions with a given number of bands calculated by using a Poisson distribution. The predicted mean value of the number of positions with a given number of bands is derived on the assumption that IS6110 is randomly associated. Therefore, the number is equal to the total number of bands divided by the total number of available positions (400).
The number of IS6110 bands which were located in hot spots was calculated and the data for the combined populations are tabulated in Table 2. These results demonstrate that the majority of strains with five or fewer copies have IS6110 bands which are located in hot spots and confirm the need for such strains to be investigated by an alternative technique such as polymorphic GC-rich RFLP or spoligotyping (2). High-copy-number strains had IS6110 bands in the low-copy-number hot-spot sites but the reverse was rarely the case (data not shown). Thus, it appears that there may be two subpopulations of M. tuberculosis with distinct collections of IS6110 integration sites. The first subpopulation is defined by strains that have low copy numbers, i.e., less than five bands, and the second is defined by strains that have high copy numbers. This division agrees with the observation of Yang et al., who noted the existence of an Asian subgroup of M. tuberculosis strains with low copy numbers (14). Alternatively, low-copy-number sites may be transcriptionally inactive, making it less likely that the transposase will be transcribed and trigger a transposition event. Thus, when rare transposition events to more active sites occur copy number will rise rapidly, leaving few strains with an intermediate number of IS6110 bands (5).
TABLE 2.
Percentage of IS6110 bands in hot spots by copy numbera
| Copy no. | IS6110 bands in hot spots (%) |
|---|---|
| 1 | 70.9 |
| 2 | 78.5 |
| 3 | 39.2 |
| 4 | 34.3 |
| 5 | 46.6 |
| 6 | 25.0 |
| 7 | 39.2 |
| 8 | 27.3 |
| 9 | 31.8 |
| 10 | 23.8 |
| 11 | 23.3 |
| 12 | 19.5 |
| 13 | 21.6 |
| 14 | 17.9 |
| 15 | 20.1 |
| 16 | 25.4 |
| 17 | 20.9 |
| 18 | 15.2 |
| 19 | 10.5 |
| 20 | 10.0 |
| 21 | 21.4 |
Data are for 154 M. tuberculosis isolates collected in Tanzania. Percentages are relative to total number of bands in isolates with different copy numbers.
In high-copy-number strains a significant proportion of IS6110 bands are located in hot spots, and this is likely to have an effect on the sensitivity of IS6110 RFLP typing for discriminating among isolates. In strains which have many hot-spot IS6110 bands, removal of these conserved IS6110 bands from the similarity calculation may enhance discrimination, weighting the calculation to band positions which are randomly associated.
Although it is unlikely that we will see a consensus sequence similar to the target for IS900 integration, detailed sequence analysis of IS6110 hot-spot regions will identify the characteristics of the genome which favor integration of this insertion sequence.
Acknowledgments
We gratefully acknowledge the help of Richard Morris with statistical advice, Nicki Hutchison for permission to use her RFLP data, and Anne Dickens for her technical assistance.
This work was supported by funds from The Special Trustees of the Royal Free Hospital (S.H.G.).
REFERENCES
- 1.Barnes P F, el-Hajj H, Preston-Martin S, Cave M D, Jones B E, Otaya M, Pogoda J, Eisenach K D. Transmission of tuberculosis among the urban homeless. JAMA. 1996;275:305–307. [PubMed] [Google Scholar]
- 2.Butcher P D, Hutchinson N A, Doran T J, Dale J W. The application of molecular techniques to the diagnosis and epidemiology of mycobacterial diseases. J Appl Bacteriol. 1996;81:53S–71S. [PubMed] [Google Scholar]
- 3.Chevrel-Dellagi D, Abderrahman A, Haltiti R, Koubaji H, Gicquel B, Dellagi K. Large-scale DNA fingerprinting of Mycobacterium tuberculosis strains as a tool for epidemiological studies of tuberculosis. J Clin Microbiol. 1993;31:2446–2450. doi: 10.1128/jcm.31.9.2446-2450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coronado V G, Beck-Sague C M, Hutton M D, Davis B J, Nicholas P, Villareal C, Woodley C L, Kilburn J O, Crawford J T, Freiden T R, Sinkowitz R L, Jarvis W R. Transmission of multidrug-resistant Mycobacterium tuberculosis among persons with human immunodeficiency virus infection in an urban hospital-epidemiologic and restriction fragment length polymorphism analysis. J Infect Dis. 1993;168:1052–1055. doi: 10.1093/infdis/168.4.1052. [DOI] [PubMed] [Google Scholar]
- 5.Dale J W. Mobile genetic elements in mycobacteria. Eur Resp J. 1995;20:633S–648S. [PubMed] [Google Scholar]
- 6.Doran T J, Tizard M, Millar D, Ford J, Sumar N, Loughlin M, Hermon-Taylor J. IS900 targets translation signals in Mycobacterium avium subsp. paratuberculosis to facilitate the expression of its hed gene. Microbiology. 1997;143:547–552. doi: 10.1099/00221287-143-2-547. [DOI] [PubMed] [Google Scholar]
- 7.Dwyer B, Jackson K, Raios K, Sievers A, Wilshire E, Ross B. DNA restriction fragment analysis to define an extended cluster of tuberculosis in homeless men and their associates. J Infect Dis. 1993;167:490–494. doi: 10.1093/infdis/167.2.490. [DOI] [PubMed] [Google Scholar]
- 8.Fang Z, Forbes K J. A Mycobacterium tuberculosis IS6110 preferential locus for insertion into the genome. J Clin Microbiol. 1997;35:479–481. doi: 10.1128/jcm.35.2.479-481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frieden T R, Woodley C L, Crawford J T, Lew D, Dooley S M. The molecular epidemiology of tuberculosis in New York City: the importance of nosocomial transmission and laboratory error. Tubercle Lung Dis. 1996;77:407–413. doi: 10.1016/s0962-8479(96)90112-4. [DOI] [PubMed] [Google Scholar]
- 10.Hermans P W M, van Soolingen D, Bik E M, de Haas P E W, Dale J W, van Embden J D A. Insertion element IS987 from Mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect Immun. 1991;59:2695–2705. doi: 10.1128/iai.59.8.2695-2705.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jereb J A, Burwen D R, Dooley S W, Haas W H, Crawford J T, Geiter L J, Edmond M B, Dowling J N, Shapiro R, Pasculle A W, Shanahan S L, Jarvis W R. Nosocomial outbreak of tuberculosis in a renal transplant unit: application of a new technique for restriction fragment length polymorphism analysis of Mycobacterium tuberculosis isolates. J Infect Dis. 1993;168:1219–1224. doi: 10.1093/infdis/168.5.1219. [DOI] [PubMed] [Google Scholar]
- 12.Small P M, McClenny N B, Singh S P, Schoolnik G K, Tompkins L S, Mickelsen P A. Molecular strain typing of Mycobacterium tuberculosis to confirm cross-contamination in the mycobacteriology laboratory and modification of procedures to minimize occurrence of false-positive cultures. J Clin Microbiol. 1993;31:1677–1682. doi: 10.1128/jcm.31.7.1677-1682.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Embden J D A, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P W M, Martin C, McAdam R, Shinick T, Small P M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standard methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Z H, Mtoni I, Chonde M, Mwasekaga M, Fuursted K, Askgard D S, Bennedsen J, de Haas P E W, van Soolingen D, van Embden J D A, Andersen A B. DNA fingerprinting and phenotyping of Mycobacterium tuberculosis isolates from human immunodeficiency virus (HIV)-seropositive and HIV-seronegative patients in Tanzania. J Clin Microbiol. 1995;33:1064–1069. doi: 10.1128/jcm.33.5.1064-1069.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]