Abstract
In many arthropods, including insects responsible for transmission of human diseases, behaviors that include mating, aggregation, and aggression are triggered by detection of pheromones. Extracellular odorant binding proteins are critical for pheromone detection in many insects and are secreted into the fluid bathing the olfactory neuron dendrites. In Drosophila melanogaster, the odorant binding protein LUSH is essential for normal sensitivity to the volatile sex pheromone, 11-cis vaccenyl acetate (cVA). Using a genetic screen for cVA pheromone insensitivity, we identified ANCE-3, a homolog of human angiotensin converting enzyme that is required for detection of cVA pheromone. The mutants have normal dose–response curves for food odors, although olfactory neuron amplitudes are reduced in all olfactory neurons examined. ance-3 mutants have profound delays in mating, and the courtship defects are primarily but not exclusively due to loss of ance-3 function in males. We demonstrate that ANCE-3 is required in the sensillae support cells for normal reproductive behavior, and that localization of odorant binding proteins to the sensillum lymph is blocked in the mutants. Expression of an ance-3 cDNA in sensillae support cells completely rescues the cVA responses, LUSH localization, and courtship defects. We show the courtship latency defects are not due to effects on olfactory neurons in the antenna nor mediated through ORCO receptors, but instead stem from ANCE-3-dependent effects on chemosensory sensillae in other body parts. These findings reveal an unexpected factor critical for pheromone detection with profound influence on reproductive behaviors.
Keywords: olfaction, olfactory, mating, reproduction, ACE
Introduction
In mosquitoes, biting flies, and other insects including Drosophila melanogaster, odorants are detected by olfactory neurons located on the antenna and housed in chemosensory sensillae (Fig. 1a). Insect odorant receptors (Ors) are ligand-gated ion channels (Benton et al. 2006; Sato et al. 2008; Wicher et al. 2008). These receptors are thought to be heterotetramers of a common, broadly expressed co-receptor, Or co-receptor (ORCO), together with a member of a family of “tuning” receptor subunits responsible for odorant specificity. Expression of each tuning receptor subunit is restricted to subsets of olfactory neurons (Larsson et al. 2004; Hallem and Carlson 2006; Butterwick et al. 2018; Del Mármol et al. 2021). Additionally, several other receptor classes, including ionotropic receptors (Irs), gustatory receptors (Grs), and members of the pickpocket receptor family (Ppks) have been shown to mediate detection of food and pheromonal cues (Clyne et al. 2000; Kwon et al. 2007; Benton et al. 2009; Park and Kwon 2011; Thistle et al. 2012; Koh et al. 2014; Rimal and Lee 2018; Liu et al. 2020). Unlike vertebrate olfactory neurons, the dendrites of insect chemosensory neurons are compartmentalized in small groups with the dendrites of these clusters bathed in a common sensillum lymph within the shafts of the sensillae (Fig. 1a). This organization allows for differential expression of sensillum lymph factors secreted into different sensillae (reviewed in Ha and Smith 2009). Odorant binding proteins (OBPs) are not expressed by the neurons, but are secreted into the sensillum lymph by support cells. Different sensillae express different subsets of OBP family members (Galindo and Smith 2001; Larter et al. 2016). In Drosophila, OBPs have been shown to confer ligand sensitivity and influence response kinetics to a subset of odorants, affecting activation and deactivation kinetics (Xu et al. 2005; Scheuermann and Smith 2019). Exactly how invertebrate OBPs affect odorant responses is not clear, but involves direct interactions between binding proteins and odorants. Defects in pheromone and odorant detection have been observed in many insect species lacking specific OBP (Pophof 2004; Laughlin et al. 2008; Fawaz et al. 2014; Dong et al. 2017; Zhang et al. 2017; Dong et al. 2019; Scheuermann and Smith 2019; Mozuraitis et al. 2020; Chen et al. 2021; Diallo et al. 2021; Du and Chen 2021; Gao et al. 2021; Guo et al. 2021; Han W-K et al. 2022; Tian et al. 2023).
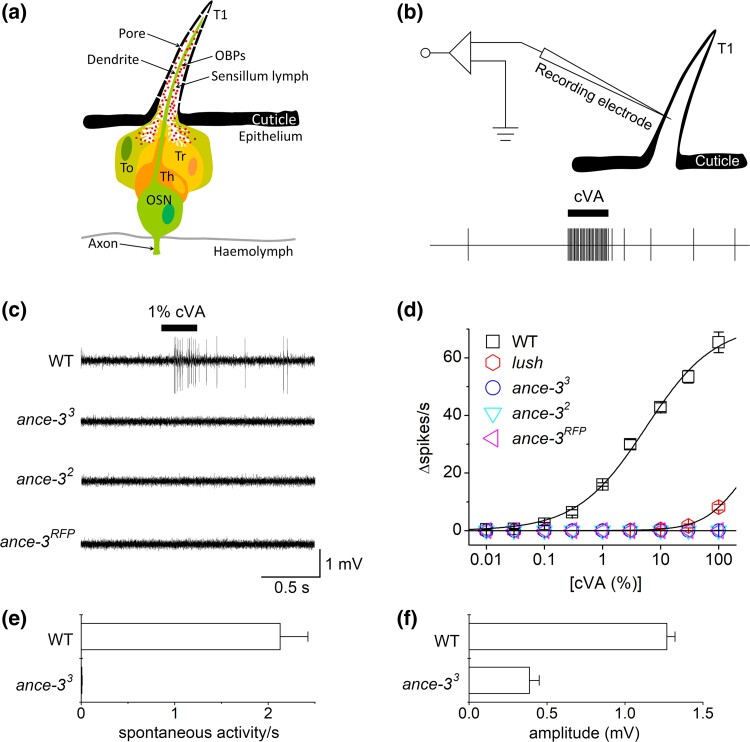
Fig. 1.
ance-3 mutants are defective for cVA responses. a) Cartoon of a single at 1 sensillum showing the Or67d neuron (OSN) projecting a dendrite into the sensillum lymph within the shaft of the sensillum. Tormogen and trichogen support cells (To, Tr) secrete OBPs including LUSH into the sensillum lymph. The thecogen support cell (Th) acts as a glial sheath cell wrapping the sensory neuron. b) Cartoon of the single sensillum recording setup. A sharp glass electrode is used to puncture the sensillum and measure spontaneous and cVA-induced action potentials from the neuron. c) Single sensillum recordings (SSR) from Or67d neurons in at1 sensillae. Responses from wild type (WT), ance-32, ance-33, and ance-3RFP mutant alleles. d) Dose–response curves for wild type and three ance-33 mutant alleles to various dilutions of cVA spotted on the stimulus filter paper. n = 28 for WT, n = 31 for ance-33, n = 13 for ance-32, n = 11 for ance-3RFP. No differences were observed between the alleles. e) ance-33 mutants are defective for spontaneous activity in cVA-sensitive Or67d neurons. The genotypes are significantly different (P = 4.75 × 10−7, Student's t-test, n = 28 for WT, n = 31 for ance-33, n = 13 for ance-32, n = 11 for ance-3RFP). f) SSR amplitudes are significantly reduced in ance-33 mutants compared to wild type (n = 10 for each genotype (P = 1.94 × 10−8, Student's t-test, n = 28 for WT, n = 31 for ance-33, n = 13 for ance-32, n = 11 for ance-3RFP).
Detection of the Drosophila male-specific sex pheromone, 11-cis vaccenyl acetate (cVA), is more complex than general food odorant detection, requiring additional components not required for detection of most food odorants (Ha and Smith 2009; Ronderos and Smith 2009). Genetic screens revealed that ORCO and the tuning receptor Or67d are essential for cVA detection, and Or67d is expressed in at1 sensillae neurons that are dedicated to cVA detection (Clyne et al. 1997; Ha and Smith 2006; Kurtovic et al. 2007; Jin et al. 2008). Additionally, SNMP1, a CD36 homolog (Benton et al. 2007; Jin et al. 2008; Li et al. 2014) and LUSH, a member of the odorant binding protein family (Xu et al. 2005) and are key @@components required for normal cVA pheromone detection. LUSH is specifically secreted into the sensillum lymph of trichoid sensillae (Kim et al. 1998; Xu et al. 2005), and lush mutants are defective for sensitivity to cVA (Xu et al. 2005; Ha and Smith 2006). Indeed, mis-expression of Or67d in trichoid sensillae neurons that normally express Or47b confers robust sensitivity to cVA to Or47b neurons, but only if LUSH is expressed in the lymph (Ha and Smith 2006). lush mutants respond to extremely high concentrations of cVA, indicating LUSH is a sensitizing factor. LUSH is thought to transport cVA molecules through the sensillum lymph to the olfactory neuron dendrites. However, there is a 400-fold reduction in spontaneous activity specific to Or67d neurons in lush mutants in the absence of cVA. This defect is reversed by infusing recombinant LUSH into the sensillum lymph, suggesting a possible role as a co-ligand or allosteric modulator (Xu et al. 2005; Trimmer et al. 2023). lush mutants have mating latencies that are 3-fold longer than wild-type controls (Xu et al. 2005; Billeter and Levine 2015). Here, using a genetic screen, we identify and characterize mutants in a new cVA-sensitivity factor, ANCE-3. ance-3 mutants fail to secrete odorant binding proteins into the sensillum lymph and have profound defects in courtship behavior beyond the defects in cVA detection.
Materials and methods
Drosophila stocks
w1118 controls, ASE5 GAL4, and nompA GAL4 stocks were obtained from the Bloomington Stock Center (Bloomington, IN, USA). lush GAL4 flies were generated by ligating the 3-kb lush promoter region (Kim et al. 1998) into pGATN (Brand and Perrimon 1993) and ligating the resulting GAL4 fusion into pCasper 4 (Pirrotta 1988) and generating transgenic flies (Spradling and Rubin 1982). ance-31, ance-32, and ance-33 were identified by screening second chromosome lines from the Zuker collection (Koundakjian et al. 2004) for insensitivity to air passed over 1% dilutions of cVA (Jin et al. 2008). nos-Cas9 flies used to generate CRISPR mutants were reported by Kondo and Ueda (2013). ance-33 and ance-3RFP mutants were backcrossed to w1118 for five generations to minimize differences in the genetic backgrounds and outcross any potential background genetic lesions, and these flies were used for electrophysiological and behavioral experiments. lush mutants were described in Kim et al. (1998) and Xu et al. (2005). Os-E/Os-F mutants were described in Scheuermann and Smith (2019), and Or65a, b, c triple mutants and the Or88a mutants were described in Pitts et al. (2016). Orco2 and Or47b mutants were the gift of Leslie Vosshall (Rockefeller). Or67dGAL4 flies (Kurtovic et al. 2007) were obtained from Barry Dickson (Queensland Brain Institute). UAS tdGFP reporter flies (Han C et al. 2011) were obtained from Robin Hiesinger.
Whole genome sequencing
Whole genome sequencing for ance-3 mutants was performed by the UTSW Genomics core facility (UTSW, Dallas, TX, USA). Briefly, genomic DNA was prepared from ance-3 mutants and the parental strain used to make the mutants by homogenization of adult flies, and DNA was isolated using phenol/chloroform extractions and ethanol precipitation. One hundred base-pair libraries were generated and sequenced using the paired-end method with an Illumina 2500 sequencer. These sequences have been deposited at NCBI through SRA (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA955298).
Generation of the ance-3RFP CRISPR allele
CRISPR sites flanking the first seven coding exons of ance-3 were identified using the CRISPR optimal target finder (Gratz et al. 2014). Oligonucletide primers encoding the chiRNAs were cloned into pU6-Bbs1-chiRNA plasmids following treatment with T4 polynucleotide kinase (NEB Ipswich, MA, USA) and annealing. One kilobase of homologous sequence upstream and downstream of these target sites were isolated using PCR, sequenced, and cloned sequentially into pHd-dsRED-attP. The two chi targeting plasmids (250 ng/ml) and the homologous recombination plasmid (500 ng/ml) were co-injected into 0- to 1-h-old nos-Cas9 Drosophila embryos (Kondo and Ueda 2013).
The sequences of the targeting and homology primers for the ance-3RFP allele are
-
Upstream target
5′-CTTCCAACGGCGACCTTGGCAGTG and
5′-AAACCACTGCCAAGGTCGCCGTTG
-
Downstream target
5′-CTTCGCAATTAAAGCGTCTAGCAG and
5′-AAACCTGCTAGACGCTTTAATTGC
-
Upstream homology
5′-GCGGCCGCTCGAGCTTAATCGATGCTC and
5′-CATATGTGCCAAGGTCGCCGTTGACAG
-
Downstream homology
5′-AGATCTCAGCGGCTGGTTATTAG and
5′-CTCGAGAGCCGAGTGCTGGGGCACAG.
ANCE, ANCE-3, and JHEdup cloning and ance-3 site-directed mutants
ance-3 cDNA was synthesized and codon optimized for Drosophila (Biomatik, Toronto). ance-3His, lacking the conserved histidine residues thought to coordinate zinc binding, was generated from the ance-3 cDNA by site-directed mutagenesis using the Q5 system (New England Biolabs) to change His554 and His558 to lysine codons (CAC to AAG in both cases) using the primers
5′-GGCCAAGATTCAGTATTTTCTGCAATACCG and
5′-ATTTCCTTATGCACGCTAATCAGGGA.
The ance-3QE mutant was similarly generated by mutating glutamine 582 to glutamate using the ance-3 cDNA as template with the primers
5′-CGCCTTTCACGAAGCCGTCGG and
5′-GGATTGGCTCCGTTGCGAAAG.
The ance cDNA was cloned from total fly mRNA isolated using Trizol (Ambion) and reverse-transcribed using Superscript 3 (Thermo-Fisher). First-strand cDNA was used as a template to amplify the ance cDNA using primers
5′-GAATTCAAAATGAGACTGTTTCTGCTAGCC-3′ and
5′-TCTAGATTATGATGAGACGCATTTATTTG.
cDNAs were sequenced and cloned into pUAS (Brand and Perrimon 1993), and transgenic flies were produced using standard methods (Spradling and Rubin 1982). The JHEdup cDNA was cloned from first-strand cDNA isolated from wild-type antennae (Trizol). DNA fragments were sequenced and cloned into pET28a (Novagen) for protein expression.
Antiserum, immunofluorescence, and Western blotting
Immunohistochemistry was performed on both sexes of Drosophila frozen head sections as previously described (Jin et al. 2008). ANCE-3 antiserum was generated by expressing a portion of the ance-3 cDNA encoding amino acids 23–210 in pET28a using the primers
5′-CATATGCCGCAACTGAACCTGCCGAC and
5′-GGATCCTTACAGCTCCTGTTGGTAGCGGCG.
For pET28a expression in bacteria, the region of the cDNA encoding JHEdup amino acids 47–220 was amplified, using the primers
5′-CATATGGGCATGGGCATTCCCTTTGCCCAG and
5′-CTCGAGGGGGCTAATCATGTGCAAGTGGGCAGA
and cloned and expressed in pET28a as described by the manufacturer (Novagen).
The denatured, bacterially expressed polypeptides were isolated using nickel agarose columns (Qiagen), eluted, dialyzed against water, and used to immunize rabbits.
Os-E and Os-F-specific antiserum were generated as previously described (Hekmat-Scafe et al. 1997) by expressing these cDNAs in bacteria using pET28a, affinity purifying each antiserum with the peptides used for immunization, followed by immunodepletion of cross-reacting antibodies by running anti-Os-E serum over Os-F columns, and vice versa.
Serum from ANCE-3 and JHEdup immunized rabbits was affinity-purified using the immunizing polypeptide bound to Affi-Gel15 columns (BioRad). Rabbit antibodies were detected with goat anti-rabbit secondary antibodies, congugated with Alexa 546, Alexa 568 or Alexa 488 (InVitrogen). Anti-chicken GFP was detected with goat anti-chicken antibodies (Molecular Probes). Confocal images were obtained using a Zeiss LSM 710 confocal microscope. Identical imaging settings were used for comparisons among genotypes. Western blots were performed on extracts from 20 antennae per genotype that were hand dissected and probe-sonicated in 30 ml of SDS page loading buffer, run on 14% polyacrylamide gels, and transferred to nitrocellulose membranes (Wattman Optitran BA-S 83) using semi-dry blotting (BioRad). Antiserum dilutions used were
Rabbit anti-ANCE-3, affinity purified: 1:100,
Rabbit anti-LUSH affinity purified (Western 1:500, Immunofluorescence 1:300),
Rabbit anti-Os-E affinity purified 1:1000,
Rabbit anti-Os-F affinity purified 1:500, and
Chicken anti-GFP (Aves labs GFP-1020) 1:1000.
ance-33 deletion primers
Primers to amplify the region spanning the ance-33 deletion are
5′-TAGCACTTGTTCAGCGTGGC and
5′-TTCGACGATCCCAAGCGATC
and produce a product of 610 base-pairs in wild type and 300 base-pairs in the deletion mutant.
Single sensillum recordings
Single sensillum recordings (SSR) were performed as previously described (Laughlin et al. 2008; Pitts et al. 2016). Filtered AC signals (200–3 kHz) were recorded and digitized for analysis (Autospike 32). Briefly, 3- to 5-day-old male or female flies were housed in fresh vials containing standard yeast molasses food individually or in small sex-specific groups prior to SSR recordings. Odorants used in single sensillum recordings were of the highest purity available (Sigma-Aldrich and Pherobank BV). For SSR, 30 ml of diluted or undiluted odorant was spotted on a small piece of Wattman filter paper (1.5 cm2), inserted into a 5.75-in Pasteur pipette and 300-ms pulses of air were passed over the filter into a constant stream of humidified air passing over the preparation (30 ml/s). Signals were amplified 1000× and fed into a computer via a 16-bit ADC and analyzed offline with AUTOSPIKE software (USB-IDAC system; Syntech, Hilversum, the Netherlands). Low cut-off filter setting was 200 Hz, and the high cut-off was 3 kHz. Action potentials were recorded by inserting a glass electrode near the base of a sensillum and a ground electrode in the head. Data analysis was performed according to de Bruyne et al. (2001). Signals were recorded for 20 or 30 s, starting 10 s before cVA stimulation. Spontaneous action potentials were counted 1 s before cVA stimulation and subtracted from spike numbers counted 1 s after cVA stimulation (Delta spikes). Recordings were performed from separate sensillae with a maximum of two sensillae recorded from any single fly. Differences between genotypes were tested for statistical differences using two-tailed Student's t-tests.
Behavioral assays and statistical analysis
Behavioral experiments were performed as described in Pitts et al. (2016). Male and female flies were collected at eclosion, housed separately in small groups, and stored at room temperature in standard vials. Single, 3- to 5-day-old, naïve virgin male and female flies were manually aspirated into a courtship chamber (2.3-cm diameter polystyrene wells containing 6 ml of 1% agarose in water overlayed with Wattman 3 filter paper discs) and covered with a glass coverslip. Videos were recorded using a Canon PowerShot 4000 HD camera or a GoPro HERO6. Copulation latency was calculated as the period from initial presentation of the target until copulation. Copulation latency data were tested for normality using Shapiro–Wilk's test. Data with a normal distribution were compared using one-way ANOVA with Tukey test to correct for multiple comparisons. Data that were not normally distributed were analyzed using the Kruskal–Wallis test with Dunn's correction. Courtship index (CI) in male–male assays were calculated as the percent of time target males spent performing courtship behaviors (chasing, wing extension, licking, attempted copulation) over a 30-minute period. Male CI data were analyzed using the Kruskal–Wallis test with Dunn's correction.
Locomotor activity of wild type and ance-33 males were measured as described in Sakai and Kitamoto (2006). Briefly, 16 individual, 3- to 5-day-old male flies for each genotype were placed into polycarbonate tubes (5-mm diameter, 65-mm length) containing a small amount of food on one end and placed into a DAM activity monitor (Trikinetics, Waltham, MA, USA). Locomotor activity was recorded for 24 h. The number of beam breaks in which the fly crossed the infrared beam was counted every 5 min. The average locomotor activity was determined as the sum of beam crosses for 16 flies over a 24-h period (288 5-min bins). Locomotor activity data were tested for significance using the two-tailed Student's t-test.
Results
Identification of ance-3 mutants in a screen for defective for cVA responses
We screened 2000 homozygous viable EMS mutagenized second chromosome lines (Koundakjian et al. 2004) for defective responses to 1% cVA, similar to our previous mutant screen for the third chromosome (Jin et al. 2008). Each mutagenized line carries a second chromosome with a unique array of mutations in genes not required for viability (Koundakjian et al. 2004). Each line was tested for electrophysiological responses from Or67d-expressing neurons to 1% cVA using single sensillum electrophysiological recordings (SSR, Fig. 1b). Among the mutants identified were three recessive, allelic lines (ance-31–3) that lacked responses to 1% cVA (Fig. 1c and d). cVA dose–response curves demonstrated that the mutants are insensitive to all pheromone concentrations tested (Fig. 1d). While cVA responses are generally absent in the mutants, even when applied at supra-physiological levels that can elicit small responses in lush mutants (Fig. 1d), we did observe very weak responses (Supplementary Figure 1c) to cVA in ∼10% of the mutants flies. In addition, we noted the spontaneous neuronal activity normally present in Or67d neurons is almost eliminated in ance-3 mutants (Fig. 1e), and the amplitudes of the action potentials in SSR recordings are approximately a third of wild type (Fig. 1f). These defects reveal the ance-3 mutants are defective for cVA detection, normal spontaneous action potential frequencies, and normal spike amplitudes in the cVA pheromone sensing neurons.
We next tested whether ance-3 mutants are defective for responses to food odorants. We tested the basiconic olfactory neurons ab2A and ab3A for responses to ethyl acetate and ethyl butyrate, respectively (Hallem et al. 2004). We detected no difference in odorant response specificity or in overall sensitivity to odorants in these neurons between wild type and ance-3 mutants in dose–response curves (Supplementary Figure 2d and h). However, as we observed in the cVA sensing neurons, the amplitude of these food-odor responses is markedly reduced in the SSR recordings (Supplementary Figure 2b and f). These data reveal that the amplitudes of the food-sensing olfactory neuron responses are affected in the mutants, but there is no defect in odorant specificity or overall sensitivity in these food-sensing neurons.
ance-3 is lesioned in all three mutant alleles
We used whole genome sequencing to identify the lesions responsible for the defective cVA responses in the mutants. DNA from each homozygous mutant was sequenced, as was the parental (unmutagenized) control used for the mutagenesis screen (Koundakjian et al. 2004). The three mutant lines each have lesions in the ance-3 gene that were not present in the parental control. No other genes were lesioned in all three alleles. The ance-3 coding sequence is distributed over ∼55 kb of genomic DNA on the second chromosome at 34E4 (Fig. 2a). Four genes encoding putative proteins of unknown function are transcribed on the opposite strand within a large 36-kb intron between exon 7 and 8 of ance-3. We crossed the ance-3 alleles to Df(2L)BSC52, a deletion in the ance-3 region. The deletion failed to complement the cVA detection defects in ance-33 mutants, and the defects were as severe as the homozygous mutants, supporting the idea that ance-3 is the relevant locus and that the mutants are all strong hypomorphs or null alleles.
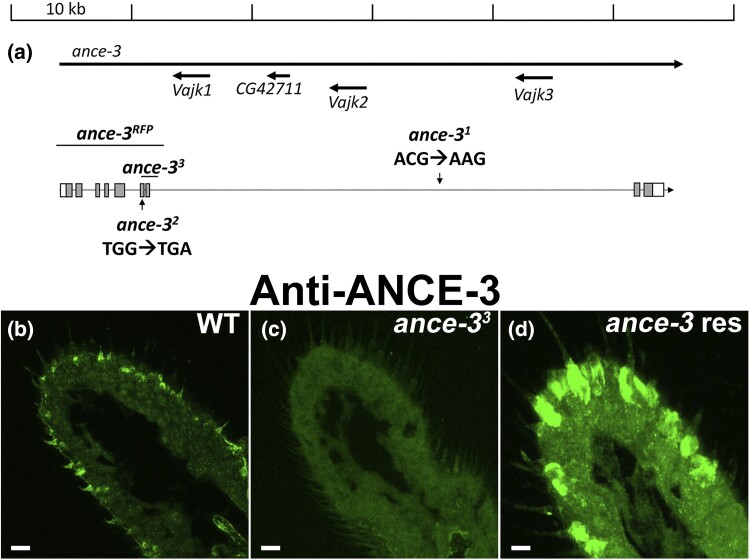
Fig. 2.
The ance-3 gene, which is expressed in antennal support cells, is lesioned in the mutants. a) Genomic map of the ance-3 locus. The ance-3 coding sequence is distributed over 55 kb of genomic DNA. Location of the lesions is depicted. ance-3RFP is a CRISPR Cas9 allele in which the first seven coding exons are replaced with 3xP3-dsRED. b) Anti-ANCE-3 antiserum reacted with wild-type antenna section detects protein in the antenna in a support cell pattern. Scale bar, 33 mm. c) Anti-ANCE-3 antiserum reacted with ance-33 mutants detects no protein in the mutant. Scale bar, 33 mm. d) Anti-ANCE-3 antiserum on ance-33 mutants expressing a wild-type ance-3 cDNA with lush GAL4 driver in the trichoid support cells. The strong lush promoter expresses high levels of ANCE-3. The brightness for panel c) is higher than b) and d), so the antenna is visible in the image. Scale bar, 8 mm.
Sequence analysis revealed that ance-31 has an intronic lesion in the large 36-kD intron that changes ACG to AAG, generating a potential splice acceptor. ance-32 has a TGG to TGA point mutation in exon 6 that introduces a stop codon in the ance-3 open reading frame at position 657 in the amino-acid sequence. This lesion is predicted to result in a truncation of the ANCE-3 protein product lacking the C-terminal 187 amino acids. ance-33 contains a deletion of 706 base-pairs that deletes most of exon 6 and all of exon 7 and is predicted to delete 57 amino acids of coding sequence and produce a frameshift in the remaining exons. Finally, we generated a CRISPR/Cas9 mutant allele by deleting the first seven coding exons of ance-3 (Fig. 2a). The CRISPR allele (ance-3RFP) has a mutant phenotype that is indistinguishable from the EMS mutants (Fig. 1c and d). Together, these data reveal that ANCE-3 is required for cVA detection and normal action potential amplitudes in chemosensory neurons.
ANCE-3 is related to angiotensin converting enzyme
ANCE-3 is predicted to encode an 844 amino-acid protein related to the vertebrate endothelial metaloproteinase, angiotensin converting enzyme (ACE; Caldwell et al. 1976). Human ACE is a central player in the renin–angiotensin system that regulates blood pressure by proteolytically processing angiotensin 1 to the bioactive form, angiotensin 2 (Caldwell et al. 1976). The zinc coordinating residues for metaloproteinases include an HEXXH motif and a glutamate residue located distal to the histidine motif (Hooper 1994). Six ACE homologs are encoded in the Drosophila genome, ANCE, ANCE-2, ANCE-3, ANCE-4, ANCE-5, and ACER (Coates et al. 2000). ANCE-3 is most similar to ANCE (29% amino-acid identity) and shares the zinc binding motif but lacks the conserved glutamate present in the proteolytically active ANCE and ACER enzymes (Houard et al. 1998; Coates et al. 2000).
ANCE-3 is required in the sensillae support cells for function
To elucidate the mechanism for ANCE-3 function on cVA pheromone responses, we began to dissect where ANCE-3 is required. ANCE-3 could fulfill any number of roles, ranging from regulating cell fate of the cells involved in cVA detection to acting as a signal transduction component. We first tested whether the cVA-sensing neurons are still present in ance-3 mutants and if they still express the Or67d tuning receptor. We used Or67dGAL4, a gene replacement knock-in (Kurtovic et al. 2007), to express GFP-tagged Or67d receptors (Benton et al. 2007). GAL4 expression is restricted to cVA-sensing, at1 sensillae neurons in these flies (Kurtovic et al. 2007). We examined expression of GFP-Or67d both in the wild type and ance-33 mutant backgrounds. Or67d is expressed equivalently in both genotypes, and a large fraction is localized to the olfactory neuron dendrites (Supplementary Figure 3). This reveals that ANCE-3 does not regulate Or67d cell fate, nor is it required for expression or localization of Or67d receptor subunits. Furthermore, when we expressed a wild-type ance-3 cDNA in the Or67d neurons in the ance-3 mutant background, this failed to restore cVA sensitivity (Fig. 3a and b). We conclude that cVA-sensing Or67d olfactory neurons or their precursors are not the site of ANCE-3 action.
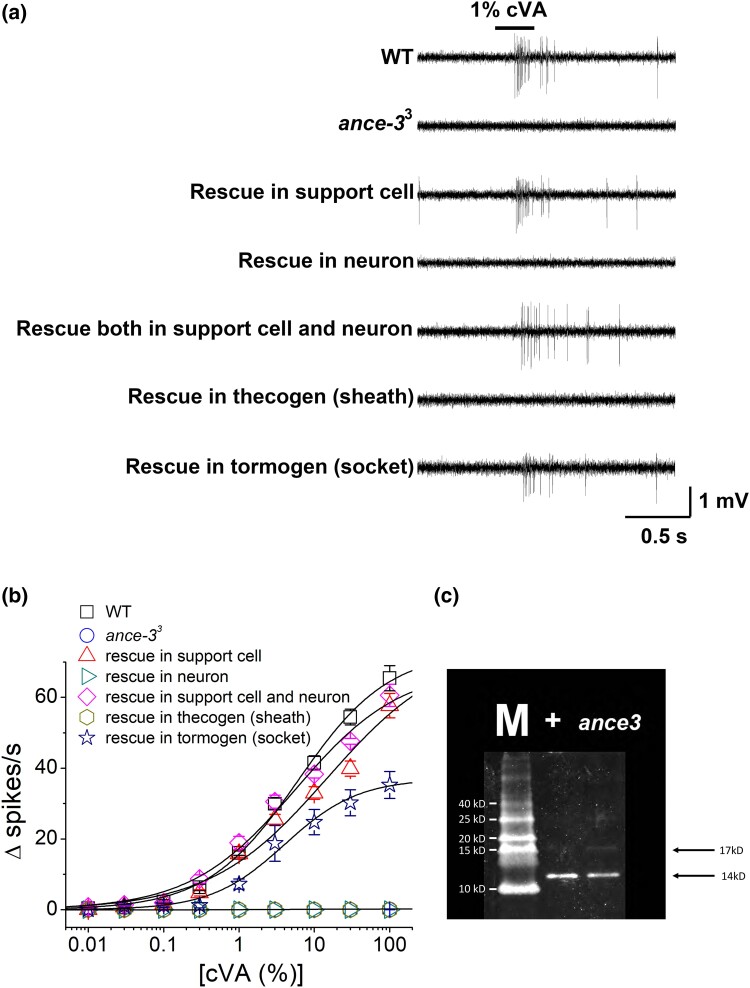
Fig. 3.
ANCE-3 is required in support cells. a) SSR traces from wild type, ance-33 mutants, and ance-33 mutants expressing a wild-type ance-3 transgene in the support cells or Or67d neurons. Support cell expression rescues cVA sensitivity, while expression in the Or67d neuron does not. cVA responses from ance-33 mutant flies expressing the transgene in both neurons and support cells are not different from expression in the support cells alone. ance-3 transgenic rescue with the tormogen socket cell-specific driver, ASE5 GAL4, rescues cVA responses from ance-33, but expression in the thecogen sheath cell driver, nompA GAL4, does not rescue. b) Dose–response curves to air passed over various dilutions of cVA from wild type (WT) and ance-33 mutants rescued with wild-type ance-3 cDNA expressed with different GAL4 drivers. (n = 28 for WT, n = 31 for ance-33, n = 17 for Rescue in support cell, n = 15 for Rescue in neuron, n = 13 for Rescue in both, n = 11 for Rescue in thecogen, and n = 12 for Rescue in tormogen. c) Western blot of antennal extracts from wild type (+) and ance-33 mutants (ance3). The mature LUSH protein with cleaved signal sequence is 14 kD. M, size markers.
We generated a specific antiserum to the ANCE-3 protein and reacted this with wild type and ance-3 mutant individuals to gain additional insight into where ANCE-3 might function. Figure 2b shows that anti-ANCE-3 detects protein in the region of the antennal support cells in wild-type antennae, and this signal is absent in the ance-33 mutants (Fig. 2c). ANCE-3 is predicted to have a signal sequence (SignalP 5.0), but we did not detect ANCE-3 in the sensillum lymph. We expressed the ance-3 cDNA in trichoid support cells in the ance-33 mutant background using the lush promoter (Fig. 2d; Xu et al. 2005). In the antenna, the lush promoter drives expression specifically in support cells of the trichoid sensillae (Kim et al. 1998; Xu et al. 2005). Immuno-EM studies previously identified LUSH protein in the trichogen, thecogen, and the tormogen support cells of trichoid sensillae, but not in the neurons of these sensillae (Shanbhag et al. 2005). When expressed with the support cell-specific lush promoter, the ance-3 cDNA completely restored cVA sensitivity and normal action potential rates and amplitudes to the ance-33 mutant Or67d neurons (Fig. 3a and b).
Next, we tested ASE5 GAL4 and nompA GAL4 drivers to express the ance-3 cDNA in subsets of support cells in the ance-3 mutant background to assess rescue of cVA sensitivity. ASE5 GAL4, which is broadly expressed in the tormogen support cell of Drosophila sensillae (Barolo et al. 2000; Jeong et al. 2013; Larter et al. 2016), rescued cVA responses, although cVA sensitivity was less than wild type or lush GAL4 rescue (Fig. 3a and b). However, nompA GAL4, expressed in the thecogen (sheath) cells in adults (Chung et al. 2001), failed to rescue cVA sensitivity (Fig. 3a and b). This indicates ANCE-3 functions in the tormogen support cells that contribute to the sensillum lymph of at1 sensillae, but expression of ANCE-3 in the at1 thecogen sheath support cells is not sufficient for rescue of cVA sensitivity (Keil 1997).
Odorant binding proteins are not secreted in ance-3 mutants
While ANCE-3 lacks the catalytic glutamate residue, we cannot rule out the possibility it has protease activity. The only known cVA sensitivity factor that requires proteolytic processing is LUSH that is also expressed in the support cells that express ANCE-3. LUSH has a classic hydrophobic N-terminal signal peptide sequence that requires cleavage to generate the mature 14-kD protein (Xu et al. 2005). The uncleaved protein is predicted to be 17 kD (Kim et al. 1998). To test whether ANCE-3 functions in signal peptidase cleavage of LUSH, we ran Western blots of antennal extracts from wild type and ance-33 mutants and used anti-LUSH antiserum to look for abnormalities in LUSH molecular weight. Figure 3c shows the mature 14-kD LUSH product is the major form of LUSH in wild type and ance-33 mutant individuals (Kim et al. 1998). Based on this data, ANCE-3 is not required for LUSH signal peptide cleavage.
We next examined whether LUSH secretion from the support cells into the sensillum lymph is affected by loss of ANCE-3. Immunofluorescence of frozen antennal sections reacted with anti-LUSH antiserum revealed the presence of LUSH in the support cells and sensillum lymph in wild-type antenna, but remarkably, while present in the support cells, LUSH is not present in the sensillum lymph in ance-33 mutants (Fig. 4a and b).
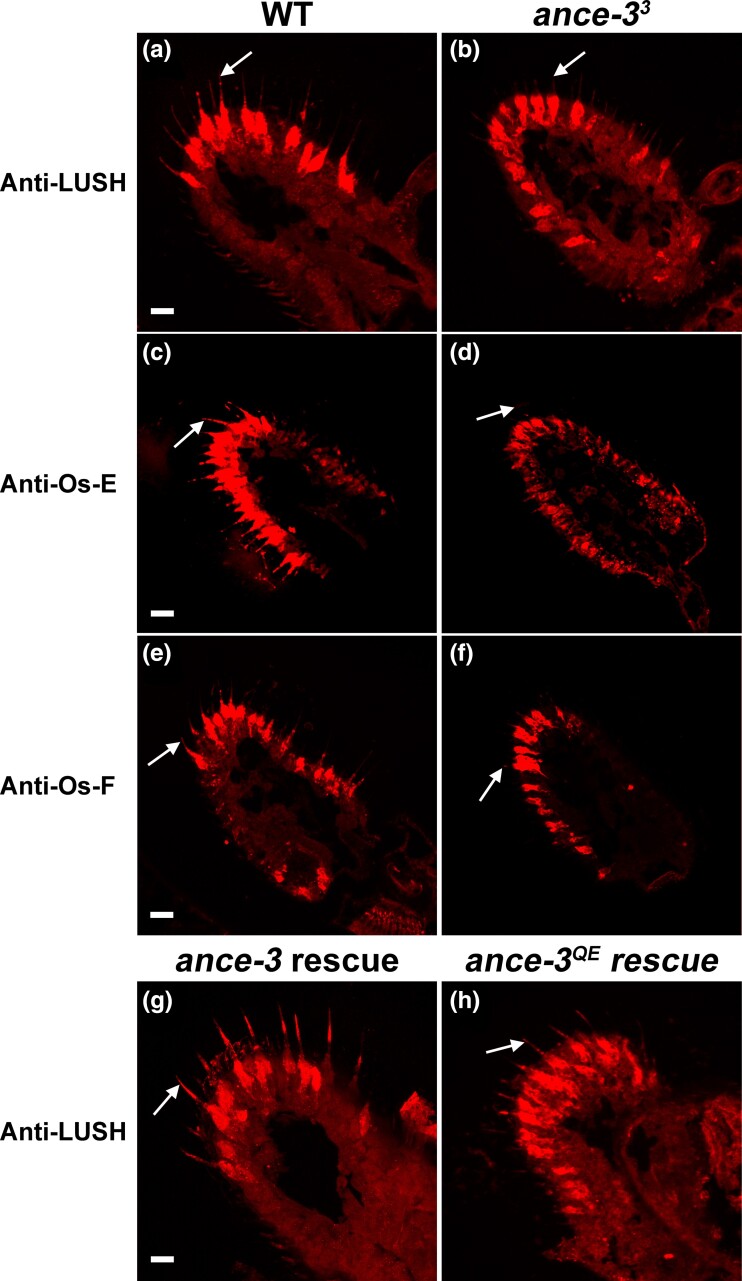
Fig. 4.
ance-3 mutants fail to secrete odorant binding proteins into the sensillum lymph. Frozen tissue sections from wild type (a, c, and e) or ance-33 mutant (b, d, f, g, and h) individuals reacted with anti-LUSH (a, b, g, and h), anti-Os-E (c and d), or anti-Os-F (e and f). Arrows indicate trichoid sensillae shafts. g) ance-33 mutants expressing a wild-type ance-3 cDNA with lush GAL4 reacted with anti-LUSH antiserum reveals restoration of LUSH secretion. h) ance-33 mutant antenna expressing ance-3QE with the lush GAL4 driver reacted with anti-LUSH antiserum partially restores LUSH secretion to ance-3 mutants. Scale bar, 10 mm.
To determine if ANCE-3 is required specifically for LUSH secretion, or if other OBPs are also mis-localized, we tested ance-3 mutants for secretion of Os-E and Os-F, two other OBPs expressed in trichoid sensillum lymph (Shanbhag et al. 2005). Os-E and Os-F are important for proper deactivation kinetics to a subset of odorants including farnesol by Or83c-expressing neurons (Scheuermann and Smith 2019). Anti-Os-E and anti-Os-F specific antiserum revealed that neither Os-E nor Os-F is present in the lymph in the ance-33 mutants (Fig. 4c–f). Transgenic expression of a wild-type ance-3 cDNA in ance-3 mutant support cells with the lush promoter restored LUSH secretion (Fig. 4g). Finally, to determine if ANCE-3 has a specific role in OBP localization, or if it has a more global effect on support cell secretion, we tested for secretion of JHEdup, an esterase normally secreted into the sensillum lymph of a subset of basiconic sensillae (Steiner et al. 2017). Antiserum to JHEdup localized the esterase in the support cells and the sensillum lymph of large basiconic sensillae, both in wild-type antennae and in the ance-3 mutants (Supplementary Figure 4). This suggests ANCE-3 has a role in OBP localization to the sensillum lymph, but is not required for secretion of all sensillum lymph proteins.
Ability of the ANCE paralog and ANCE-3 mutations in catalytic residues to rescue function
The closest D. melanogaster paralog to ANCE-3 is ANCE (Coates et al. 2000). To establish if this ANCE paralog can substitute for ANCE-3 if expressed in the trichoid support cells, we expressed an ance cDNA with the lush promoter in the ance-3 mutant background. Figure 5a and b shows that ANCE was unable to rescue the loss of ANCE-3 function on cVA sensitivity.
Fig. 5.
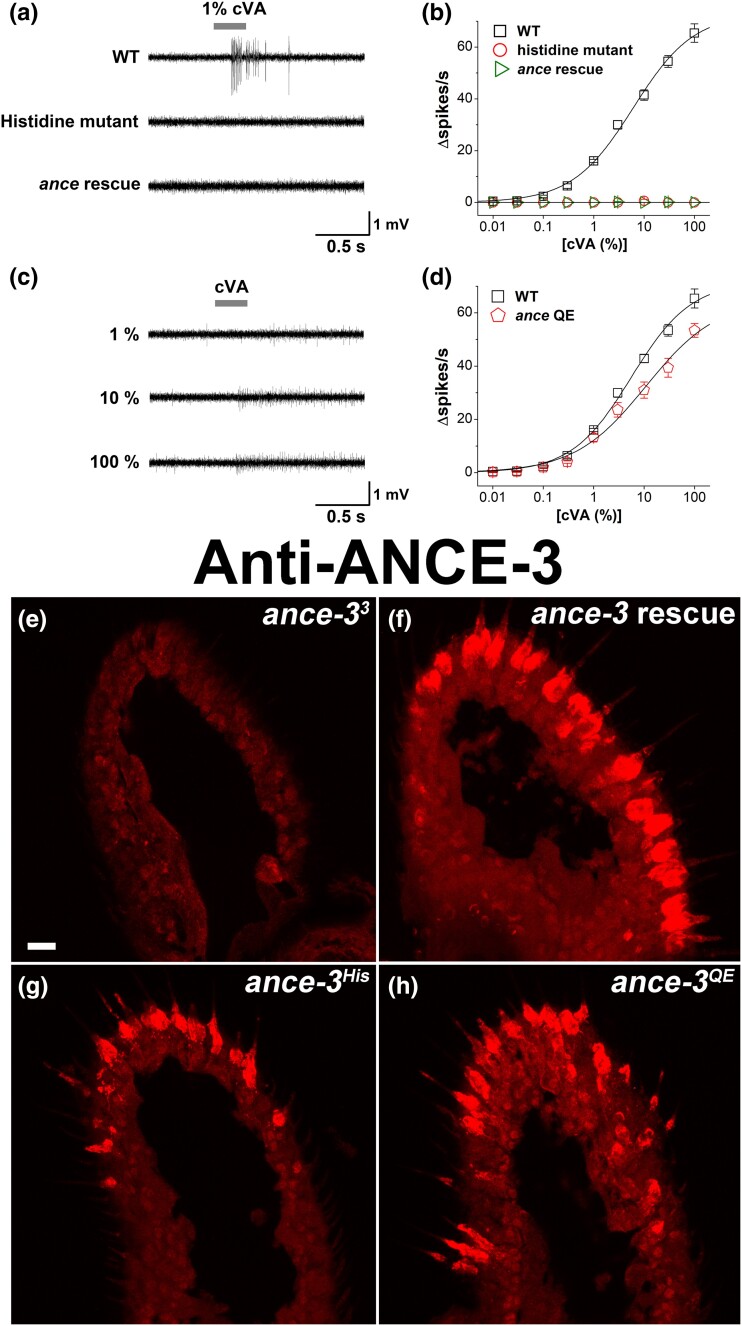
ANCE, ANCE-3His, and ANCE-3QE transgenic individuals for rescue of cVA sensitivity. a) SSR traces recorded from wild-type at1 sensillae, or sensillae from ance-33 mutants expressing ANCE-3His lacking the zinc coordinating histidines (Histidine mutant), or expressing ANCE, the closest paralog to ANCE-3 in the Drosophila genome (ance rescue). b) cVA dose–response curves for wild type (WT, black squares), and ance-33 mutants expressing the ance-3His mutant (open red circles) or the ance transgene rescue (ance rescue, green triangles). n = 28 for WT, n = 31 for ance-33, n = 9 for Histidine mutant, n = 12 for Ance rescue. c) Sample SSR traces induced by air passed over filters with 1%, 10%, or 100% cVA dilutions recorded from at1 sensillae from ance-33 mutants rescued with the ance-3QE construct. The ance-3QE rescue consistently restores cVA responses, even at low cVA concentrations, but has little effect on the reduced spike amplitude phenotype. d) cVA dose–response curves for wild type (WT, black squares) and ance-33 mutants expressing ance-3QE with the lush GAL4 driver (ance-3QE, red pentagons). ANCE-3QE significantly rescues ance-3 loss of function on cVA sensitivity, but the genotypes are significantly different at cVA applications above 10% (P < 0.05 for 10, 30, and 100%). n = 28 for WT, n = 8 for Ance QE. e–h) Anti-ANCE-3 antiserum reacted on ance-33 mutants expressing transgenic rescue constructs. e) ance-33 mutants. f) ance-33 mutants expressing wild type ance-3 cDNA with lush GAL4. g) ance-33 mutants expressing ance-3His with lush GAL4. h) ance-33 mutants expressing ance-3QE with lush GAL4. Scale bar, 7 mm.
Wild-type ANCE-3 lacks the conserved glutamate found in other metaloproteinases (Coates et al. 2000), but retains the HEXXH motif important for coordinating zinc ions that are essential for metaloproteinase activity (Menach et al. 2013). To determine if the conserved histidines are important for ANCE-3 function, we mutated histidines 554 and 558 to lysines and called this protein ANCE-3His. We used the lush promoter to express ANCE-3His protein in the ance-33 mutant background. The ANCE-3His protein is expressed (Fig. 5g). However, this construct failed to rescue the cVA sensitivity or amplitude defects (Fig. 5a and b).
We noted that both mosquitoes and Tsetse flies have ANCE-3 homologs. The Tsetse fly Glossina fuscipes, the vector for Trypanosoma brucei (African sleeping sickness), has a highly conserved ANCE-3 homolog that shares the glutamine substitution for the glutamate observed in Drosophila ANCE-3. However, for the homologs in the mosquito disease vectors Aedes egyptii and Anopholes gambiae, the catalytic glutamate is present. To determine if ANCE-3 still functions in Drosophila with the glutamine replaced with glutamate, we replaced the codon for glutamine 582 with one for glutamate yielding the ANCE-3QE protein. The ance-3QE mutant is expressed (Fig. 5h), and significantly restores cVA sensitivity to ance-3 loss of function mutants when expressed with the lush promoter (Fig. 5c and d). ANCE-3QE also restores LUSH secretion (Fig. 4h). However, the action potential amplitude defect is not rescued by ANCE-3QE (Fig. 5c).
Mating behavior is severely compromised in ance-3 mutants
ance-3 mutant flies appear morphologically normal, do not have any obvious defects in general behavior, and have normal locomotor behavior (Fig. 6e). Given that cVA detection is strongly affected, we examined mating behavior in wild type and ance-3 mutants to assess the in vivo biological importance of ANCE-3. Courtship behavior results from complex interactions between male and female flies, and is characterized by a number of steps with characteristic behaviors, including orienting, tapping, wing vibration, licking, and copulation (Greenspan and Ferveur 2000). Wild-type flies rapidly progress through the courtship steps and typically copulate in 7–10 min (Fig. 6a). cVA detection is known to affect courtship behaviors (Jallon et al. 1981; Kurtovic et al. 2007; Datta et al. 2008; Griffith and Ejima 2009; Billeter and Levine 2015), and females detect cVA emitted from single males at a distance of 1 cm and less (Laughlin et al. 2008).
Fig. 6.
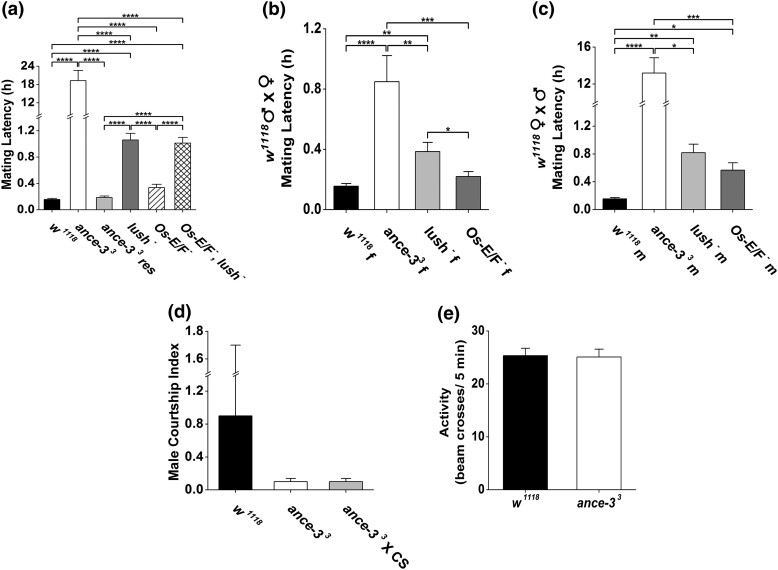
ance-3 mutants have defective courtship behaviors. a) Time to copulation (Mating Latency) for single pairs of wild type, ance-33 mutants, ance-33 mutants expressing a wild-type ance-3 transgene with lush GAL4 (ance-33 res), lush mutants, or double mutants defective for expression of Os-E, Os-F odorant binding proteins (Scheuermann and Smith 2019), and triple mutants lacking LUSH, Os-E, and Os-F odorant binding proteins (Os-E/Os-F−, lush−). n = 10 for each genotype. **** genotypes different at P < 0.0001, one-way ANOVA with Tukey test. If no bar with asterixis, not significantly different between genotypes. b) Time to copulation (Mating Latency) for virgin females of different genotypes with wild-type males. n = 10 for each genotype. **P < 0.01, ***P < 0.001, ****P < 0.0001, by Kruskal–Wallis test with Dunn's correction. If no bar with asterixis, not significantly different between genotypes. c) Time to copulation (Mating Latency) for wild-type virgin females crossed to males of different genotypes. n = 10 for each genotype. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, by Kruskal–Wallis test with Dunn's correction. If no bar with asterixis, not significantly different between genotypes. d) Male–male courtship index. n = 10 for each genotype. Genotypes are not significantly different by Kruskal–Wallis test with Dunn's correction. Hypersexual behavior in w1118 has been observed previously (Krstic et al. 2013). e) Locomotor behavior is not significantly different for wild type and ance-33 mutants by two-tailed Student's t-test. n = 16 for each genotype.
Using video recordings of single pairs, we measured the time to copulation for wild type, lush mutants, and ance-3 mutants (Fig. 6a). Wild-type flies mated within 7–10 min, while lush mutants have greater mating latency (Billeter and Levine 2015), averaging 60 min to copulation (Fig. 6a). If the loss of LUSH protein in the sensillum lymph is the sole factor responsible for the defective copulation latency in ance-3 mutants, we predicted the mating latency for the ance-3 mutants would be similar to lush mutants. However, ance-3 mutants are profoundly defective for courtship and copulation. The average time to copulation for ance-3 mutant pairs is ∼19 h (Fig. 6a). By crossing wild-type animals with ance-3 mutants of the opposite sex, we found the copulation latency defect is largely due to the requirement for ANCE-3 in males, as ance-3 mutant males paired with wild-type females have a similar delay in copulation (Fig. 6c). However, ANCE-3 also is important in females, because wild-type males paired with ance-3 mutant females also have significantly longer copulation latencies than wild-type controls (Fig. 6b). We observed no abnormal courtship behaviors in male–male assays with ance-3 mutants (Fig. 6e). To rule out a possible additive effect of missing LUSH, Os-E, and Os-F, we tested triple mutants lacking all three OBPs. The courtship latencies in the triple mutants are not different from lush mutants alone (Fig. 6d).
Driving the ance-3 cDNA in support cells with the lush GAL4 driver or the ASE5 GAL4 driver reversed the cVA detection defects in ance-3 mutants, but nompA GAL4 did not (Fig. 3a and b). Surprisingly, all three GAL4 drivers rescued the copulation latency defects, revealing the mating latency defects are not due to cVA detection deficits alone (Fig. 7a). We also examined the ability of ANCE-3QE to rescue copulation latency. ANCE-3QE largely restores LUSH secretion and cVA sensitivity to ance-3 mutants (Fig. 5d), and also partially rescues mating defects, reducing the average copulation latency to 4 h (Fig. 7b).
Fig. 7.
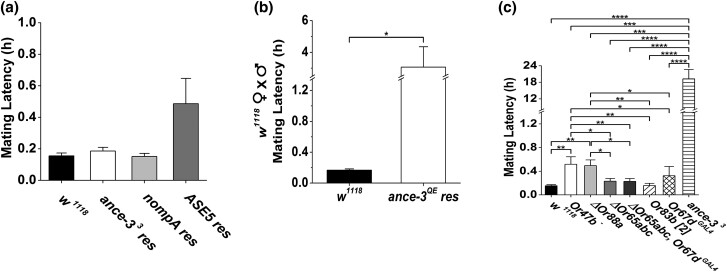
Trichoid neuron receptor mutant mating latencies are less severe than ance-3 mutants. a) Time to copulation (Mating Latency) for wild type (w1118), ance-3 cDNA rescue of ance-33 mutants with lush GAL4 (ance-33 res), nompA GAL4 (nompA res), and ASE5 GAL4 (ASE5 res). All rescue copulation latency. n = 10 for each genotype. No differences between genotypes using one-way ANOVA with Tukey test. b) Time to copulation (Mating Latency) for lush GAL4 expression of ANCE-3QE partially rescues copulation latency in ance-33 males from 19 to 4 h. n = 15 for each genotype. Genotypes are different by two-tailed Student's t-test. c) Time to copulation (Mating Latency) for single pairs of wild type, Or47b receptor mutants (Or47b−), Or88a receptor mutants (DOr88a), Or65abc deletion mutants (DOr65abc), Or65abc; Or67dGAL4 double mutants (DOr65abc; Or67dGAL4), Orco mutants (Or83b2), Or67d receptor mutants (Or67dGAL4), and ance-33 mutants. n = 10 for each genotype. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, significantly different by Kruskal–Wallis test with Dunn's correction. If no bar with asterixis, not significantly different between genotypes.
Since expression of the ance-3 transgene with the lush promoter restored normal mating behaviors to ance-3 mutants, the source of the mating defects must be in cells expressing this driver. We sought to identify the sensillae responsible for the mating latency defects. In the antenna, only trichoid sensillae express this driver, and only four classes of trichoid olfactory neurons are exposed to LUSH. These include Or67d neurons in at1 sensillae, and the three at4 sensillae neuron classes expressing Or47b receptors, Or88a receptors, or neurons co-expressing Or65a, Or65b, and Or65c (Couto et al. 2005). To establish if any of these neurons mediate mating latency, we tested the receptor mutants for each of these neuron classes for mating latency defects. None showed the striking copulation delays we observe in ance-3 mutants (Fig. 7c). Furthermore, mutants defective for Orco expression (Larsson et al. 2004) have mating latencies that are not different from wild-type controls (Fig. 7c). This suggests receptors other than Or members are important for the mating latency defects in ance-3 mutants. To evaluate lush GAL expression in more detail, we crossed the lush GAL4 driver to UAS tdGFP, encoding a membrane-localized GFP (Han C et al. 2011) to look for additional sites of expression. We found the lush GAL4 driver is expressed in multiple chemosensory sensillae in all leg segments of front, middle, and hind legs in both sexes, as well as in chemoreceptor sensillae along the anterior wing margins (Supplementary Figure 5). We suggest that chemosensory neurons in one or both locations function in mate recognition, and require ANCE-3 function in the corresponding support cells for this behavior.
Discussion
ance-3 mutants have prolonged mating latency
The courtship behavior defect in ance-3 mutants is surprising in its severity. The defect is early in the courting progression, as ance-3 mutants rarely engage in any courtship steps, consistent with a defect in chemical detection of mating partners (Greenspan and Ferveur 2000). Our data demonstrate that ANCE-3 is required for rapid progression through the courtship behavior program by influencing chemosensory neurons indirectly through a role in support cells.
There are several possible mechanisms that could explain why ance-3 mutants have such a strong effect on mating latency. There may be specific odorant binding protein members expressed in the legs or wings that are critical for sensitizing chemosensory neurons to contact or volatile mating pheromone ligands, similar to the role for LUSH and cVA detection (Xu et al. 2005). Supporting this notion, the OBPs we examined are not secreted in the absence of ANCE-3, three independent support cell drivers expressing an ance-3 cDNA restore function, and ANCE-3QE rescue experiments partially restore both LUSH secretion and mating latency. The failure to secrete OBPs may be more important for mating latency than the reduced action potential amplitudes in ance-3 mutants in the chemosensory neurons, because ANCE-3QE does not rescue spike sizes, but does partially restore mating latency. Identifying OBPs co-expressed with ANCE-3 in leg cells using single-cell transcriptomes, combined with lush GAL4-expressed RNAi constructs, is one way to explore this possibility. Indeed, a number of OBPs are known to be expressed in the wing and tarsi chemosensory sensillae, including OBP19b, OBP56d, and OBP57b (Galindo and Smith 2001). Alternatively, there may be factors other than OBPs secreted from support cells that are important for chemosensory neuron function that require ANCE-3. Support for this idea comes from the small amplitude action potentials in all chemosensory neurons in ance-3 mutants, and the demonstration that ance-3 mutants have more severe cVA sensitivity defects than lush mutants (Fig. 1d). Future studies will be required to determine exactly how ANCE-3 affects support cell function to subsequently affect chemosensory neuron function and courtship behavior.
We have narrowed the location of the critical chemosensory neurons important for mating latency. The lush GAL4 driver completely restores normal mating latency to ance-3 mutants when used to drive a wild-type ance-3 cDNA. We have eliminated the four classes of trichoid olfactory neurons in the antenna as the cause for the prolonged mating latency. Mutants in Or67d, Or47b, Or65a-c, and Or88a, the receptor genes defining these classes, do not show prolonged mating latency. Indeed, Orco mutants also have normal mating latency, indicating Or receptors as a whole are not responsible for the mating latency defects observed in ance-3 mutants. This suggests chemosensory sensillae expressing ANCE-3 in the support cells on the anterior wing margin or legs might play a role in mate identification. Previous reports revealed that surgical removal of the wings does not significantly affect male courtship behaviors (Averhoff and Richardson 1974). This points to chemosensory neurons on the legs as critical for detecting pheromonal cues important for rapid progression through courtship. While LUSH itself might be important for detecting pheromones in the legs, loss of LUSH alone does not explain the protracted courtship, because lush mutants have relatively modest delays in courtship. Indeed, neither LUSH, Os-E, nor Os-F are likely involved, as the triple mutant has relatively normal courtship behavior. Therefore, if loss of OBP expression is a factor in ANCE-3-dependent mating latency, other OBP members must be important.
Interestingly, expression of a wild-type ance-3 cDNA in ance-3 mutants using the nompA GAL4 driver, expressed in the sheath supporting cells, failed to rescue cVA pheromone sensitivity in ance-3 mutants, but did rescue mating latency defects. Some OBPs have been shown to be expressed by the sheath cells (Larter et al. 2016). Perhaps ANCE-3 is required for expression of OBPs in tarsi support cells, possibly including thecogen sheath cells that are important for sensitizing of one or more non-ORCO receptors to pheromone ligands.
ance-3 mutants are 120 times slower to copulate than wild-type flies and 30 times slower than lush mutants. Indeed, ance-3 mutants are among the most severe courtship defects reported (Hall 1994). Other Drosophila mutants known to have delayed time to copulation include dissatisfaction mutants that are defective for expression of a steroid hormone receptor, and take over 20 min to copulate, primarily due to female unresponsiveness, but also have egg laying defects (Finley et al. 1998). courtless mutants have reduced expression of a ubiquitin-conjugating enzyme and most do not court, but these flies are also defective for sperm production (Orgad et al. 2000). fruitless mutants, defective for male-specific variants of this transcription factor, are defective for courtship due to fate determination effects on sex-specific neurons (Ryner et al. 1996; von Philipsborn et al. 2014).
Chemosensory-specific mutants that affect courtship include the tarsi pickpocket receptors ppk23 and ppk29, and loss of these receptors delays courtship initiation and reduces discrimination between the sexes (Thistle et al. 2012). However, while assayed in a different manner, the delays appear less severe than those we observed in ance-3 mutants, and ance-3 mutants have no defects in male–male courtship behavior. To date, OBPs have only been shown to affect responses to ligands detected by Or/ORCO receptors (Ha and Smith 2006; Kurtovic et al. 2007; Larter et al. 2016; Scheuermann and Smith 2019). However, they may sensitize neurons expressing different classes of OBPs to pheromone ligands.
ANCE-3 is required for cVA detection and OBP secretion
Our genetic screen identified ANCE-3 as a new cVA pheromone sensitivity factor. Food-sensing neurons retain normal selectivity and sensitivity to odorant ligands. However, cVA pheromone detection is strongly affected, and most ance-3 mutants fail to respond to any cVA concentration. ANCE-3 functions in support cells, and members of the OBP family, including LUSH, are not secreted normally into the sensillum lymph in the mutants. At least one other secreted sensillum lymph factor, JHEdup, is secreted in ance-3 mutants, suggesting only a subset of secreted proteins require ANCE-3 function. This selective effect on secretion likely explains why ANCE-3 mutants are not lethal. One attractive possibility is that ANCE-3 is a chaperone for folding or transport of factors that include OBPs. Determining the subcellular location of ANCE-3 in support cells and identifying the site where OBP secretion is blocked in ance-3 mutants would provide insight into its role in OBP secretion.
ANCE-3 affects olfactory neuron amplitudes
ance-3 mutant olfactory neurons have small amplitude action potentials. This defect is reverted in Or67d neurons by expressing a wild-type ance-3 cDNA with the lush GAL4 driver, indicating a non-cell autonomous effect on olfactory neuron function. No OBP mutant reported to date is associated with small action potentials in the cognate sensillae neurons. This implies the small spike amplitude phenotype is not associated with loss of OBPs. One possibility is that ANCE-3 affects one or more ion transporters in the support cells that are important for the proper ionic composition of the sensillum lymph. The basis for small amplitude action potentials in ance-3 mutants is unclear, and will require future study.
Structure/function analysis
Drosophila ANCE, ACER, and ANCE-5 paralogs retain the conserved catalytic residues of the zinc metaloproteinase family, including the HEXXH motif and a conserved glutamic acid residue for zinc coodination (Hooper 1994). ANCE and ACER have been shown to be functional proteases (Houard et al. 1998). ANCE-2, ANCE-3, and ANCE-4 lack subsets of these putative zinc binding motifs, although ANCE-2 and ANCE-3 retain the HEXXH motif (Coates et al. 2000). Mutation of these conserved histidines in ANCE-3 eliminated rescue of cVA sensitivity in ance-3 mutants. We postulate that these residues may be important for proper folding of ANCE-3.
The closest Drosophila paralog to ANCE-3 is ANCE. Expression of an ance cDNA in the support cells also failed to rescue loss of ance-3. This demonstrates functional specialization among the ANCE paralogs. It would be interesting to determine if these paralogs have roles in secretion for other Drosophila proteins. Conserved ANCE-3 homologs are present in insect disease vectors including Anopholes and Aedes mosquitoes and Tsetse flies (Glossina morsitans). Volatile sex pheromones have recently been reported in this insect vector (Ebrahim et al. 2023). The Tsetse ANCE-3 homolog is 81% similar to the Drosophila protein, and has both the conserved zinc-binding histidines in ANCE-3, as well as the glutamine substitution present in Drosophila ANCE-3 (International Glossina Genome 2014). This supports the idea that ANCE-3 homologs in both fly species have a similar function.
In Anopholes mosquitoes, the ance-3 homolog AnoACE7 has a similar genomic organization and zinc binding motif, but retains the catalytic glutamate. In the neutral protease from Bacillus stearotherophilus, another metaloproteinase family member, substitution of the equivalent glutamate for glutamine abolishes proteolytic activity (Kubo et al. 1992). This raises the possibility that the Drosophila and Glossina ANCE-3 proteins are not proteases. If ANCE-3 has a similar role in mosquitoes as we have shown for Drosophila, does reverting the glutamine back to glutamate still function? We expressed Drosophila ANCE-3QE in support cells, and showed this partially rescued cVA sensitivity, LUSH secretion, and mating latency, but failed to restore spike size amplitudes. It is not clear why the small spike phenotype is not rescued by ANCE-3QE. Perhaps the glutamine substitution has evolved to function more efficiently. While the function of these homologs in insect vector species is unknown, it is tempting to speculate that they perform a role similar to what we have demonstrated for ANCE-3 in Drosophila. Generating lesions in these genes in the disease vectors in the future will provide insight into this question.
Supplementary Material
Acknowledgements
We thank Helmut Kramer for comments on the manuscript, Allan-Hermann Pool for assistance with bioinformatics, and Joan Conway for bridge funding.
Contributor Information
Tal Soo Ha, Department of Biomedical Science, Daegu University, 201 Daegudae-ro, Gyeongsan-si, Gyeongbuk, 38453 Republic of Korea.
Samarpita Sengupta, Department of Pharmacology, University of Texas Southwestern Medical Center, 5323 Harry Hines Blvd., Dallas, TX 75390-9111, USA; Department of Physician Assistant Studies, School of Health Professions, University of Texas Southwestern Medical Center, 5323 Harry Hines Blvd., Dallas, TX 75390-9111, USA.
Jordan Powell, Department of Pharmacology, University of Texas Southwestern Medical Center, 5323 Harry Hines Blvd., Dallas, TX 75390-9111, USA.
Dean P Smith, Department of Pharmacology, University of Texas Southwestern Medical Center, 5323 Harry Hines Blvd., Dallas, TX 75390-9111, USA; Department of Neuroscience, University of Texas Southwestern Medical Center, 5323 Harry Hines Blvd., Dallas, TX 75390-9111, USA; O’Donnell Brain Institute, University of Texas Southwestern Medical Center, 5323 Harry Hines Blvd., Dallas, TX 75390-9111, USA.
Data availability
Fly stocks and plasmids are available upon request. ance-33 and ance-3RFP have been submitted to the Bloomington stock center (stock number 97376 and 97375, respectively). Genomic DNA sequences for the ance-3 mutants are available through NCBI (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA955298). The authors affirm that all data necessary for confirming the conclusions are present within the article and figures.
Supplemental material available at GENETICS online.
Funding
This work was supported in part by NIH R01 DC015230 and bridge funding support from UT Southwestern Medical Center.
Author contributions
SS identified the ance-3 mutants in the genetic screen. JP performed the immunocytochemistry and behavioral experiments and statistical analysis and generated transgenic and CRISPR mutant flies. TSH performed the electrophysiological experiments and data analysis. DPS directed the research and wrote the manuscript.
Literature cited
- Averhoff WW, Richardson RH. Pheromonal control of mating patterns in Drosophila melanogaster. Behav Genet. 1974;4(3):207–225. doi: 10.1007/BF01074155. [DOI] [PubMed] [Google Scholar]
- Barolo S, Walker RG, Polyanovsky AD, Freschi G, Keil T, Posakony JW. A notch-independent activity of suppressor of hairless is required for normal mechanoreceptor physiology. Cell. 2000;103(6):957–969. doi: 10.1016/S0092-8674(00)00198-7. [DOI] [PubMed] [Google Scholar]
- Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4(2):e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136(1):149–162. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Vosshall LB. An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature. 2007;450(7167):289–293. doi: 10.1038/nature06328. [DOI] [PubMed] [Google Scholar]
- Billeter J-C, Levine JD. The role of cVA and the odorant binding protein LUSH in social and sexual behavior in Drosophila melanogaster. Front Ecol Evol. 2015;3:1–14. doi: 10.3389/fevo.2015.00075. [DOI] [Google Scholar]
- Brand A, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118(2):401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Butterwick JA, Del Marmol J, Kim KH, Kahlson MA, Rogow JA, et al. Cryo-EM structure of the insect olfactory receptor Orco. Nature. 2018;560(7719):447–452. doi: 10.1038/s41586-018-0420-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell PRB, Seegal BC, Hsu KC, Das M, Soffer RL. Angiotensin-converting enzyme: vascular endothelial localization. Science. 1976;191(4231):1050–1051. doi: 10.1126/science.175444. [DOI] [PubMed] [Google Scholar]
- Chen X, Lei Y, Li H, Xu L, Yang H, et al. CRISPR/Cas9 mutagenesis abolishes odorant-binding protein BdorOBP56f-2 and impairs the perception of methyl eugenol in Bactrocera dorsalis (Hendel). Insect Biochem Mol Biol. 2021;139:103656. doi: 10.1016/j.ibmb.2021.103656. [DOI] [PubMed] [Google Scholar]
- Chung YD, Zhu J, Han Y, Kernan MJ. Nompa encodes a PNS-specific, ZP domain protein required to connect mechanosensory dendrites to sensory structures. Neuron. 2001;29(2):415–428. doi: 10.1016/S0896-6273(01)00215-X. [DOI] [PubMed] [Google Scholar]
- Clyne PJ, Grant A, O'Connell R, Carlson JR. Odorant response of individual sensilla on the Drosophila antenna. Invert Neurosci. 1997;3(2–3):127–135. doi: 10.1007/BF02480367. [DOI] [PubMed] [Google Scholar]
- Clyne PJ, Warr CG, Carlson JR. Candidate taste receptors in Drosophila. Science. 2000;287(5459):1830–1834. doi: 10.1126/science.287.5459.1830. [DOI] [PubMed] [Google Scholar]
- Coates D, Isaac RE, Cotton J, Siviter R, Williams TA, et al. Functional conservation of the active sites of human and Drosophila angiotensin I-converting enzyme. Biochemistry. 2000;39(30):8963–8969. doi: 10.1021/bi000593q. [DOI] [PubMed] [Google Scholar]
- Couto A, Alenius M, Dickson BJ. Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr Biol. 2005;15(17):1535–1547. doi: 10.1016/j.cub.2005.07.034. [DOI] [PubMed] [Google Scholar]
- Datta SR, Vasconcelos ML, Ruta V, Luo S, Wong A, et al. The Drosophila pheromone cVA activates a sexually dimorphic neural circuit. Nature. 2008;452(7186):473–477. doi: 10.1038/nature06808. [DOI] [PubMed] [Google Scholar]
- De Bruyne M, Foster K, Carlson J R. Odor coding in the Drosophila antenna. Neuron. 2001;30(2):537–552. 10.1016/S0896-6273(01)00289-6. PMID:1139501 [DOI] [PubMed] [Google Scholar]
- Del Mármol J, Yedlin MA, Ruta V. The structural basis of odorant recognition in insect olfactory receptors. Nature. 2021;597(7874):126–131. doi: 10.1038/s41586-021-03794-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diallo S, Shahbaaz M, Makwatta JO, Muema JM, Masiga D, et al. Antennal enriched odorant binding proteins are required for odor communication in Glossina f. fuscipes. Biomolecules. 2021;11(4):541. doi: 10.3390/biom11040541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong XT, Liao H, Zhu GH, Khuhro SA, Ye ZF, et al. CRISPR/Cas9-mediated PBP1 and PBP3 mutagenesis induced significant reduction in electrophysiological response to sex pheromones in male Chilo suppressalis. Insect Sci. 2019;26(3):388–399. doi: 10.1111/1744-7917.12544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Chen J. The odorant binding protein, SiOBP5, mediates alarm pheromone olfactory recognition in the red imported fire ant, Solenopsis invicta. Biomolecules. 2021;11(11):1595. doi: 10.3390/biom11111595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahim SAM, Dweck HKM, Weiss BL, Carlson JR. A volatile sex attractant of tsetse flies. Science. 2023;379(6633):eade1877. doi: 10.1126/science.ade1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawaz EY, Allan SA, Bernier UR, Obenauer PJ, Diclaro JW. Swarming mechanisms in the yellow fever mosquito: aggregation pheromones are involved in the mating behavior of Aedes aegypti. J Vector Ecol. 2014;39(2):347–354. doi: 10.1111/jvec.12110. [DOI] [PubMed] [Google Scholar]
- Finley KD, Edeen PT, Foss M, Gross E, Ghbeish N, et al. Dissatisfaction encodes a tailless-like nuclear receptor expressed in a subset of CNS neurons controlling Drosophila sexual behavior. Neuron. 1998;21(6):1363–1374. doi: 10.1016/S0896-6273(00)80655-8. [DOI] [PubMed] [Google Scholar]
- Galindo K, Smith DP. A large family of divergent odorant-binding proteins expressed in gustatory and olfactory sensilla. Genetics. 2001;159(3):1059–1072. doi: 10.1093/genetics/159.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao SS, Li RM, Xue S, Zhang YC, Zhang YL, et al. Odorant binding protein C17 contributes to the response to Artemisia vulgaris oil in Tribolium castaneum. Front Toxicol. 2021;3:627470. doi: 10.3389/ftox.2021.627470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, et al. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics. 2014;196(4):961–971. doi: 10.1534/genetics.113.160713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan RJ, Ferveur JF. Courtship in Drosophila. Annu Rev Genet. 2000;34(1):205–232. doi: 10.1146/annurev.genet.34.1.205. [DOI] [PubMed] [Google Scholar]
- Griffith LC, Ejima A. Multimodal sensory integration of courtship stimulating cues in Drosophila melanogaster. Ann N Y Acad Sci. 2009;1170(1):394–398. doi: 10.1111/j.1749-6632.2009.04367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Guo PP, Sun YL, Huang LQ, Wang CZ. Contribution of odorant binding proteins to olfactory detection of (Z)-11-hexadecenal in Helicoverpa armigera. Insect Biochem Mol Biol. 2021;131:103554. doi: 10.1016/j.ibmb.2021.103554. [DOI] [PubMed] [Google Scholar]
- Ha TS, Smith DP. A pheromone receptor mediates 11-cis-vaccenyl acetate-induced responses in Drosophila. J Neurosci. 2006;26(34):8727–8733. doi: 10.1523/JNEUROSCI.0876-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha TS, Smith DP. Odorant and pheromone receptors in insects. Front Cell Neurosci. 2009;3:10. doi: 10.3389/neuro.03.010.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JC. The mating of a fly. Science. 1994;264(5166):1702–1714. doi: 10.1126/science.8209251. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Carlson JR. Coding of odors by a receptor repertoire. Cell. 2006;125(1):143–160. doi: 10.1016/j.cell.2006.01.050. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Ho MG, Carlson JR. The molecular basis of odor coding in the Drosophila antenna. Cell. 2004;117(7):965–979. doi: 10.1016/j.cell.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Han C, Jan LY, Jan YN. Enhancer-driven membrane markers for analysis of nonautonomous mechanisms reveal neuron–glia interactions in Drosophila. Proc Natl Acad Sci U S A. 2011;108(23):9673–9678. doi: 10.1073/pnas.1106386108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W-K, Yang Y-X, Si Y-X, Wei Z-Q, Liu S-R, Liu X-L, Yan Q, Dong S-L. Involvement of GOBP2 in the perception of a sex pheromone component in both larval and adult Spodoptera litura revealed using CRISPR/Cas9 mutagenesis. Insect Biochem Mol Biol. 2022;141:103719. doi: 10.1016/j.ibmb.2022.103719. [DOI] [PubMed] [Google Scholar]
- Hekmat-Scafe D, Steinbrecht R, Carlson JR. Coexpression of two odorant binding homologs in Drosophila: implications for olfactory coding. J Neurosci. 1997;17(5):1616–1624. doi: 10.1523/JNEUROSCI.17-05-01616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper NM. Families of zinc metalloproteases. FEBS Lett. 1994;354(1):1–6. doi: 10.1016/0014-5793(94)01079-X. [DOI] [PubMed] [Google Scholar]
- Houard X, Williams TA, Michaud A, Dani P, Isaac RE, et al. The Drosophila melanogaster-related angiotensin-I-converting enzymes Acer and Ance–distinct enzymic characteristics and alternative expression during pupal development. Eur J Biochem. 1998;257(3):599–606. doi: 10.1046/j.1432-1327.1998.2570599.x. [DOI] [PubMed] [Google Scholar]
- International Glossina Genome Genome sequence of the tsetse fly (Glossina morsitans): vector of African trypanosomiasis. Science. 2014;344(6182):380–386. doi: 10.1126/science.1249656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jallon J-M, Antony C, Benamar O. Un anti-aphrodisiaque produit par les males Drosophila melanogaster et transere aux femalles lors de la copulation. C. R. Academie Science Paris. 1981;292:1147–1149. [Google Scholar]
- Jeong YT, Shim J, Oh SR, Yoon HI, Kim CH, et al. An odorant-binding protein required for suppression of sweet taste by bitter chemicals. Neuron. 2013;79(4):725–737. doi: 10.1016/j.neuron.2013.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Ha TS, Smith DP. SNMP is a signaling component required for pheromone sensitivity in Drosophila. Proc Natl Acad Sci U S A. 2008;105(31):10996–11001. doi: 10.1073/pnas.0803309105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil TA. Comparative morphogenesis of sensilla: a review. Int J Insect Morphol Embryol. 1997;26(3–4):151–160. doi: 10.1016/S0020-7322(97)00017-2. [DOI] [Google Scholar]
- Kim MS, Repp A, Smith DP. LUSH odorant-binding protein mediates chemosensory responses to alcohols in Drosophila melanogaster. Genetics. 1998;150(2):711–721. doi: 10.1093/genetics/150.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh TW, He Z, Gorur-Shandilya S, Menuz K, Larter NK, et al. The Drosophila IR20a clade of ionotropic receptors are candidate taste and pheromone receptors. Neuron. 2014;83(4):850–865. doi: 10.1016/j.neuron.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Ueda R. Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics. 2013;195(3):715–721. doi: 10.1534/genetics.113.156737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koundakjian EJ, Cowan DM, Hardy RW, Becker AH. The Zuker collection: a resource for the analysis of autosomal gene function in Drosophila melanogaster. Genetics. 2004;167(1):203–206. doi: 10.1534/genetics.167.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krstic D, Boll W, Noll M. Influence of the White locus on the courtship behavior of Drosophila males. PLoS One. 2013;8(10):e77904. doi: 10.1371/journal.pone.0077904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo M, Mitsuda Y, Takagi M, Imanaka T. Alteration of specific activity and stability of thermostable neutral protease by site-directed mutagenesis. Appl Environ Microbiol. 1992;58(11):3779–3783. doi: 10.1128/aem.58.11.3779-3783.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtovic A, Widmer A, Dickson BJ. A single class of olfactory neurons mediates behavioural responses to a Drosophila sex pheromone. Nature. 2007;446(7135):542–546. doi: 10.1038/nature05672. [DOI] [PubMed] [Google Scholar]
- Kwon JY, Dahanukar A, Weiss LA, Carlson JR. The molecular basis of CO2 reception in Drosophila. Proc Natl Acad Sci U S A. 2007;104(9):3574–3578. doi: 10.1073/pnas.0700079104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, Vosshall LB. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43(5):703–714. doi: 10.1016/j.neuron.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Larter NK, Sun JS, Carlson JR. Organization and function of Drosophila odorant binding proteins. Elife. 2016;5. doi: 10.7554/eLife.e20242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughlin JD, Ha TS, Jones DN, Smith DP. Activation of pheromone-sensitive neurons is mediated by conformational activation of pheromone-binding protein. Cell. 2008;133(7):1255–1265. doi: 10.1016/j.cell.2008.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Ni JD, Huang J, Montell C. Requirement for Drosophila SNMP1 for rapid activation and termination of pheromone-induced activity. PLoS Genet. 2014;10(9):e1004600. doi: 10.1371/journal.pgen.1004600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Wang Y, Tian Y, Zhang J, Zhao J, Guo A. The receptor channel formed by ppk25, ppk29 and ppk23 can sense the Drosophila female pheromone 7,11-heptacosadiene. Genes Brain Behav. 2020;19(2):e12529. doi: 10.1111/gbb.12529. [DOI] [PubMed] [Google Scholar]
- Menach E, Hashida Y, Yasukawa K, Inouye K. Effects of conversion of the zinc-binding motif sequence of thermolysin, HEXXH, to that of dipeptidyl peptidase III, HEXXXH, on the activity and stability of thermolysin. Biosci Biotechnol Biochem. 2013;77(9):1901–1906. doi: 10.1271/bbb.130360. [DOI] [PubMed] [Google Scholar]
- Mozuraitis R, Hajkazemian M, Zawada JW, Szymczak J, Palsson K, et al. Male swarming aggregation pheromones increase female attraction and mating success among multiple African malaria vector mosquito species. Nat Ecol Evol. 2020;4(10):1395–1401. doi: 10.1038/s41559-020-1264-9. [DOI] [PubMed] [Google Scholar]
- Orgad S, Rosenfeld G, Greenspan RJ, Segal D. Courtless, the Drosophila UBC7 homolog, is involved in male courtship behavior and spermatogenesis. Genetics. 2000;155(3):1267–1280. doi: 10.1093/genetics/155.3.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Kwon JY. A systematic analysis of Drosophila gustatory receptor gene expression in abdominal neurons which project to the central nervous system. Mol Cells. 2011;32(4):375–381. doi: 10.1007/s10059-011-0128-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V. Vectors for P-mediated transformation in Drosophila. In: Rodriguez RL, Denhart DT, editors. Vectors: A Survey of Molecular Cloning Vectors and Their Uses. Boston: Butterworths; 1988. p. 437–456. [DOI] [PubMed] [Google Scholar]
- Pitts S, Pelser E, Meeks J, Smith D. Odorant responses and courtship behaviors influenced by at4 neurons in Drosophila. PLoS One. 2016;11(9):e0162761. doi: 10.1371/journal.pone.0162761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pophof B. Pheromone-binding proteins contribute to the activation of olfactory receptor neurons in the silkmoths Antheraea polyphemus and Bombyx mori. Chem Senses. 2004;29(2):117–125. doi: 10.1093/chemse/bjh012. [DOI] [PubMed] [Google Scholar]
- Rimal S, Lee Y. The multidimensional ionotropic receptors of Drosophila melanogaster. Insect Mol Biol. 2018;27(1):1–7. doi: 10.1111/imb.12347. [DOI] [PubMed] [Google Scholar]
- Ronderos DS, Smith DP. Diverse signaling mechanisms mediate volatile odorant detection in Drosophila. Fly (Austin). 2009;3(4):290–297. doi: 10.4161/fly.9801. [DOI] [PubMed] [Google Scholar]
- Ryner LC, Goodwin SF, Castrillon DH, Anand A, Villella A, et al. Control of male sexual behavior and sexual orientation in Drosophila by the fruitless gene. Cell. 1996;87(6):1079–1089. doi: 10.1016/S0092-8674(00)81802-4. [DOI] [PubMed] [Google Scholar]
- Sakai T, Kitamoto T. Differential roles of two major brain structures, mushroom bodies and central complex, for Drosophila male courtship behavior. J Neurobiol. 2006;66(8):821–834. doi: 10.1002/neu.20262. [DOI] [PubMed] [Google Scholar]
- Sato K, Pellegrino M, Nakagawa T, Nakagawa T, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452(7190):1002–1006. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- Scheuermann EA, Smith DP. Odor-specific deactivation defects in a Drosophila odorant-binding protein mutant. Genetics. 2019;213(3):897–909. doi: 10.1534/genetics.119.302629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag SR, Smith DP, Steinbrecht RA. Three odorant-binding proteins are co-expressed in the sensilla trichodea of Drosophila melanogaster. Arthropod Struct Dev. 2005;34(2):153–165. doi: 10.1016/j.asd.2005.01.003. [DOI] [Google Scholar]
- Spradling AC, Rubin GM. Transposition of cloned P elements into Drosophila germline chromosomes. Science. 1982;218(4570):341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- Steiner C, Bozzolan F, Montagne N, Maibeche M, Chertemps T. Neofunctionalization of “juvenile hormone esterase duplication” in Drosophila as an odorant-degrading enzyme towards food odorants. Sci Rep. 2017;7(1):12629. doi: 10.1038/s41598-017-13015-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thistle R, Cameron P, Ghorayshi A, Dennison L, Scott K. Contact chemoreceptors mediate male–male repulsion and male–female attraction during Drosophila courtship. Cell. 2012;149(5):1140–1151. doi: 10.1016/j.cell.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Z, Li R, Cheng S, Zhou T, Liu J. The Mythimna separata general odorant binding protein 2 (MsepGOBP2) is involved in the larval detection of the sex pheromone (Z)-11-hexadecenal. Pest Manag Sci. 2023;79(6):2005–2016. doi: 10.1002/ps.7373. [DOI] [PubMed] [Google Scholar]
- Trimmer C, Arroyave R, Vuilleumier C, Wu L, Dumer A, et al. Allosteric modulation of a human odorant receptor. Curr Biol. 2023;33(8):1523–1534.e4. doi: 10.1016/j.cub.2023.03.016. [DOI] [PubMed] [Google Scholar]
- von Philipsborn AC, Jorchel S, Tirian L, Demir E, Morita T, et al. Cellular and behavioral functions of fruitless isoforms in Drosophila courtship. Curr Biol. 2014;24(3):242–251. doi: 10.1016/j.cub.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicher D, Schafer R, Bauernfeind R, Stensmyr MC, Heller R, et al. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature. 2008;452(7190):1007–1011. doi: 10.1038/nature06861. [DOI] [PubMed] [Google Scholar]
- Xu P, Atkinson R, Jones DN, Smith DP. Drosophila OBP LUSH is required for activity of pheromone-sensitive neurons. Neuron. 2005;45(2):193–200. doi: 10.1016/j.neuron.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Zhang R, Wang B, Grossi G, Falabella P, Liu Y, et al. Molecular basis of alarm pheromone detection in aphids. Curr Biol. 2017;27(1):55–61. doi: 10.1016/j.cub.2016.10.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Fly stocks and plasmids are available upon request. ance-33 and ance-3RFP have been submitted to the Bloomington stock center (stock number 97376 and 97375, respectively). Genomic DNA sequences for the ance-3 mutants are available through NCBI (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA955298). The authors affirm that all data necessary for confirming the conclusions are present within the article and figures.
Supplemental material available at GENETICS online.