Abstract
Intravascular hemolysis results in the release of cell-free hemoglobin and heme in plasma. In sickle cell disease, the fragility of the sickle red blood cell leads to chronic hemolysis, which can contribute to oxidative damage and activation of inflammatory pathways. The scavenger proteins haptoglobin and hemopexin provide pathways to remove hemoglobin and heme, respectively, from the circulation. Heme also intercalates in membranes of blood cells and endothelial cells in the vasculature and associates with other plasma components such as albumin and lipoproteins. Hemopexin has a much higher affinity and can strip heme from the other pools and detoxify plasma from cell-free circulatory heme. However, due to chronic hemolysis, hemopexin is depleted in individuals with sickle cell disease. Thus, cell-free unbound heme is expected to accumulate in plasma. We developed a methodology for the accurate quantification of the fraction of heme, which is pathologically relevant in sickle cell disease, that does not appear to be sequestered to a plasma compartment. Our data show significant variation in the concentration of unbound heme, and rather unexpectedly, the size of the unbound fraction does not correlate to the degree of hemolysis, as measured by the concentration of bound heme. Very high heme concentrations (>150 µM) were obtained in some plasma with unbound concentrations that were several fold lower than in plasma with much lower hemolysis (<50 µM). These findings underscore the long-term effects of chronic hemolysis on the blood components and of the disruption of the essential equilibrium between release of hemoproteins/heme in the circulation and adaptative response of the scavenging/removal mechanisms. Understanding the clinical implications of this loss of response may provide insights into diagnostic and therapeutic targets in patients with sickle cell disease.
Keywords: Sickle cell disease, heme, hemopexin, hemolysis
Impact Statement
This study demonstrates the application of a novel method to distinguish between protein-bound and the pathologically relevant, unbound heme in plasma in individuals living with sickle cell disease (SCD). Existing methods for heme quantification do not distinguish between these separate heme fractions. Our findings may lead to additional applications of this acetone-based assay to further clarify the pathogenic role of heme and heme scavengers in SCD and other hemolytic disorders. Furthermore, accurate quantification of heme may allow investigation of novel therapeutic agents for SCD that target hemolysis and pathways that may be activated by circulating free heme.
Introduction
Heme is a complex of iron coordinated in a porphyrin ring and defines the hemoprotein family, including the essential hemoglobin (Hb). It plays a multitude of roles in biologic processes due to its ability to reversibly bind oxygen.1 –4 The insertion of heme into the heme-binding pockets of hemoproteins controls its reactivity, but once released, heme becomes highly cytotoxic due to the iron atom of the protoporphyrin IX ring. Due to its hydrophobic properties, cell-free heme not bound to hemoproteins readily intercalates into lipid bilayers and various cellular components, including red cell–derived microvesicles.5 –9 Iron acts as a Fenton’s reagent to catalyze the production of highly oxidative free radicals causing damage to proteins, cellular membrane lipids, organelles, and DNA. In these localized pools, targeted damage contributes to pathogenesis.10 –15 Heme also exerts toxic effects by activation of inflammation, upregulation of adhesion molecules, and recruitment of inflammatory leukocytes.9,10,16 –21 In blood, heme is bound to the α and β globin chains of Hb in the red blood cell (RBC).22,23 The release of Hb into plasma caused by intravascular hemolysis is common in hemolytic disorders and critical illness.15,24 –31 Oxidation of the heme iron from the ferrous (Fe2+) to the ferric (Fe3+) state generates met-Hb and the results in the release of unbound cell-free heme into the circulation. Released heme will partition into a variety of pools such as the membranes of erythrocytes, leukocytes, platelets, and endothelial cells. Heme will also bind to circulating plasma proteins such as albumin, lipoproteins, α-1-antitrypsin, α1-microglobulin, and scavenger proteins. Once these pools are saturated, the plasma fraction of unbound heme will increase (Figure 1). The degree to which heme releases from Hb depends on several factors including the pH of serum and availability of scavenging proteins. One such scavenging protein, hemopexin, a hepatically synthesized protein, binds heme with sufficient affinity to displace heme from the other plasma pools to neutralize its harmful effects.20,32 –35 In addition, haptoglobin, a scavenger protein also synthesized in the liver, binds cell-free Hb to neutralize and recycle iron.28,30,31 The pool of unbound heme may play an important role in pathogenesis of hemolytic disorders, such as SCD, a condition in which hemopexin is depleted by consumption. 28
Figure 1.

Heme pools in blood. Most of the approximately 8 mM of heme present in blood is bound to hemoglobin in red blood cells. Heme released from hemoglobin will disperse into a variety of hydrophobic pools including cell membranes and plasma proteins. Cell-free hemoglobin, released by hemolysis, is bound by haptoglobin to be recycled. These pools are in equilibrium with each other based on their abundance and binding capacity. Hemopexin with a high affinity for heme will remove heme from these pools for recycling. Heme is also metabolized by heme-oxygenase-1 into biliverdin, which is converted to bilirubin by biliverdin reductase.
The ability to distinguish and quantify the presence of unbound cell-free heme in biological samples, like plasma, is compromised by its intrinsic property to non-specifically adhere to various biological components. We adapted an approach allowing for the quantification of the two cell-free heme fractions, protein-bound and unbound, in plasma. 36 We employed the properties of acetone, which results in lysis of membranes and precipitation of proteins, and acid which decouples heme from protein. Acidic acetone (AA) releases heme from all sources allowing quantification of total cell-free heme present in the sample. Neutral acetone (NA) treatment, however, leaves cell-free unbound heme in solution. The bound and unbound fractions can be calculated from these two pools. In this study, we aimed to validate this methodology and apply it to plasma from individuals with SCD and correlate the measurement with other known clinical and laboratory markers of disease severity. Since the toxic effects of free heme are manifest in various pathologies, including hemolytic diseases, malaria, atherosclerosis, sepsis, and stroke, our findings suggest that this approach will allow further understanding of the role of cell-free heme in pathogenesis, as well as the roles of haptoglobin and hemopexin which serve as targets for therapeutic intervention.
Materials and methods
Samples collection
Plasma was separated from whole blood, collected in EDTA (ethylenediaminetetraacetic acid) tubes, of healthy individuals and individuals with SCD at UCSF Benioff Children’s Hospital Oakland after obtaining informed consent. Samples were stored at −80°C until use. This study was approved by the UCSF institutional review board (IRB # 20-32015).
Acetone solutions
AA was prepared by mixing acetone with 3M HCl at a 9:1 (v/v) ratio. NA was prepared by buffering acetone with HBS (HEPES-buffered saline; 145 mM NaCl, 20 mM HEPES pH 7.0) at a 9:1 (v/v) ratio. Solutions were kept cold at −20°C.
Hemin, bilirubin, Hb preparation
A 10-mM hemin (ferric chloride heme) stock solution was prepared by dissolving 65 mg of bovine hemin (Sigma H9039) in 10 mL of 0.1 mM freshly made NaOH with constant stirring for 1 h at room temperature. The solution was then neutralized with 1N HCl and stored protected from light at 4°C. The solution was diluted to 0.1 mM immediately before use (see below). Human Hb (Sigma H7379) was dissolved in water at 0.2 mM and bilirubin (Sigma B4126) was dissolved in chloroform at 1 mM.
Detection and quantification
To accurately quantify heme extracted from plasma in either NA or AA solutions (see “Results” section), two heme standard curves were established in NA and in AA extracts obtained from a control plasma sample. Control plasma consisted of pooled plasma samples from individuals without SCD. Small volumes of plasma were aliquoted and kept frozen until use. Hemin was then added to the extracts in a 200 µL final volume at the concentrations indicated in Figure 2. These samples were then treated as for the experimental samples (see below). Experiments were performed without addition of heme, and the absorbance values at 400 nm were subtracted from the values obtained with the spiked heme samples. The corrected A400nm values were then plotted as a function of the concentration of heme in the samples. To establish an assay using sickle cell patient samples, the minimum volume of plasma sufficient to reliably allow quantification of heme over a wide range of samples was determined. Routinely, 40 and 160 µL of plasma were used for AA and NA extraction, respectively. Five volumes of AA or NA (chilled at −20°C) were added to plasma. After mixing by vortex for 5 s and incubation at 4°C for 60 min, the proteins were removed by centrifugation at 4°C and 16,000g for 25 min. The supernatants were collected and dried at room temperature with a SpeedVac centrifuge (Savant SpeedVac plus). The heme present in the pellet was resuspended in 100 µL dimethyl sulfoxide (DMSO) and solubilized for 20 min in a sonicator bath (Fisher Scientific Ultrasonic FS-9). Heme measurements were performed at 400 nm with a microplate reader (BioTek Synergy Neo2). To measure heme extraction from Hb, five volumes of AA or NA (chilled at −20°C) were added to increasing concentrations of Hb (0–50 µM) made in HBS buffer and treated with the same protocol as above to measure absorbance. Likewise, bilirubin dissolved in chloroform was spiked in plasma at concentrations of 0–250 M for extraction in AA and NA as above.
Figure 2.
Absorbance of heme under various extraction conditions. Panel A: Heme was added to neutral acetone (orange circles) and to a plasma extracted with neutral acetone (blue circles). Extracts were then dried, resuspended in DMSO, and measured at 400 nm, as described in the Methods section. The plot of the absorbance values (calculated with the molar extinction coefficient value, ε400nm of 170,000 M−1 cm−1 at 400 nm) of heme dissolved in DMSO40,41 is shown as a dashed line. The measured ε400nm values are presented in the table. Panels B and C: Recovery of heme added to acidic and neutral acetone extracts of plasma. Heme amounts were calculated using the ε400nm values obtained from standard curves performed under acidic or neutral conditions. The efficiencies of the extraction under acidic (Panel B) and neutral (Panel C) conditions are indicated in each of the panel as the linear relation of heme detected to heme added. Panel D: Values obtained in Panel C were plotted relative to the values obtained in Panel B. Quantification of heme under neutral and acidic condition shows a near perfect equivalence of 1. In all panels, the error bars represent the standard deviations of values obtained from three measurements.
Hemopexin in human plasma was determined by ELISA (enzyme-linked immunosorbent assay) according to the manufacturer instructions (KT-502; Kamiya Biomedical Company). Complete blood counts including reticulocyte counts were performed on an Advia 2120 (Siemens). Fetal hemoglobin (HbF) was quantified using high-performance liquid chromatography (HPLC) and HbF containing erythrocytes were measured using flow cytometry (Becton Dickinson, Fortessa). All heme and hemopexin measurements were made in triplicate. Calculations and statistical analysis (t test or two-way analysis of variance [ANOVA], as appropriate) were performed with GraphPad Prism 9.
Results
Cell-free bound and unbound heme in plasma
The detection of a putative fraction of unbound heme in human plasma required both the removal of bound heme and the need to avoid depletion of the free form by techniques such as dialysis or filtration as well as its release from the bound pool under experimental conditions. Removal of bound heme by acetone precipitation of hemoproteins under neutral condition followed by the measurement of heme present in the acetone supernatant provided an efficient and sensitive quantification of “unbound” heme in plasma. Under an acidic condition37 –39 heme is released into the acetone environment and the denatured proteins, including the abundant globin chains, precipitate and are removed by centrifugation, resulting in the detection of the total heme content present in plasma. Both the NA and AA heme extracts were dried down and resuspended in a small volume of the solvent DMSO, allowing increased detection sensitivity. The fraction of bound heme can be calculated from the values of total and unbound heme. Heme extracted from various environments and methods is often quantified from the Soret band absorbance (λ ≈ 400–420 nm) using a single molar extinction coefficient value of ≈170,000 M−1 cm−1 which has been established with hemin dissolved in DMSO.40,41 However, the heme absorption spectrum is greatly influenced by various parameters, such as pH, water, and oligomerization.40,41 Although heme extracted under different conditions was dried and suspended in DMSO for measurements, we found significant differences in absorbance after recovery from acetone and acetone extract of plasma (Figure 2, panel A). The absorbance of heme in DMSO was also different when recovered from NA and AA. Thus, calibration standard curves were established under those two conditions to allow accurate quantification of extracted heme from the A400nm values (Figure 2, panels B and C). The efficiency of heme recovery from plasma under acidic condition was 90% (Figure 2(B)) and 99% under neutral condition (Figure 2(C)). Overall, there was only a 3% difference in heme quantification under these two conditions (Figure 2(D)). Using a small volume of 100 µL plasma, a reliable experimental detection limit of ⩾4 µM heme was achieved in both conditions and the procedure was reliable over a 10-fold concentration range, which eliminates the need of adjusting the volume of plasma for atypical samples with extremely low or high heme content. It should be noted that the plasma–acetone extracts are dried down and resuspended before analysis. Hence, the volume of plasma can be increased to allow detection of heme at even lower levels (<4 µM).
Detection of major heme source in plasma
As the result of intravascular hemolysis, cell-free Hb is a major source of heme in plasma. Bilirubin, a catabolic breakdown product of heme, is present in plasma samples from patients with chronic hemolysis. Heme and unconjugated bilirubin are tightly bound to globin chains and to serum albumin, respectively, and should not be detectable under the NA extraction condition. Increased concentrations of Hb and bilirubin were added to plasma and the results of the extraction under acidic and neutral conditions are presented in Figure 3. As expected, acidification resulted in the quantitative extraction of heme from Hb and of bilirubin from albumin. Heme quantification performed as described in Figure 2, resulted in the detection of four heme moieties per Hb. However, under neutral condition heme and bilirubin could only be detected when very high concentrations were added to plasma. These findings confirmed that heme and heme products bound to plasma hemoproteins remain bound under neutral condition and that detection under this condition represents the cell-free unbound fraction present in plasma. The efficient dissociation obtained under acidic condition confirmed that this condition provides the total amount of cell-free heme.
Figure 3.
Analysis of extraction procedure on bilirubin and hemoglobin. Panel A: Recovery of bilirubin added to plasma under acidic (total) and neutral (unbound) acetone extraction conditions, as described in Figure 2(B) and (C). Note that at concentrations up to 100 µM, no unbound bilirubin could be detected. At higher concentrations, bilirubin appeared to saturate the binding capacity of serum albumin and could be detected by extraction under neutral conditions. Panel B: Recovery of heme from hemoglobin under AA and NA extraction conditions. Extraction under neutral conditions could not release heme, whereas acidification resulted in the release of heme. Panel C: As presented in Figure 2(B), a standard curve was established under acidic condition to convert the absorbance values shown in panel B to the amount of heme extracted from hemoglobin. Heme values are presented plotted relative to the amount of hemoglobin extracted and established that four heme groups were recovered per molecule of Hb.
Detection of unbound heme in plasma
Analysis of a cohort of plasma samples from blood collected from individuals with SCD established that a wide range of total and unbound heme was detected after acetone extraction (Figure 4). The presence of a high amount of cell-free Hb was noticeable in some samples with very high total heme content (>100 µM), as indicated by the reddish color of the plasma. However, hemolysis did not bias the analysis since some plasma samples with no significant visible hemolysis had higher levels of unbound heme than the samples with higher hemolysis. The finding that the lysis of the RBCs and release of Hb in plasma were not predictive of the amount of unbound heme indicated that differences in plasma-binding capacity appeared to be a major critical factor influencing the partitioning of heme and its release into the circulation.
Figure 4.
Quantification of heme in plasma of sickle cell disease patients. The bar graphs represent the concentration of heme detected under acidic (total) and neutral (unbound) extraction conditions in the plasma of several SCD patients. Pictures of the plasma samples from each patient taken before extraction are shown below their respective total/unbound bars on the graphs. As indicated by the reddish color of the plasma, a high level of hemoglobin was noticeable in some samples (#4, 7, 8). Very high levels of total heme were detected in those samples, but it did not result in higher levels of unbound heme. Some samples (#3, 9) with lower total heme and no visible hemolysis had higher level of unbound heme than in the samples with high cell-free Hb (#4, 7, 8). The error bars represent the standard deviations of values obtained from three measurements.
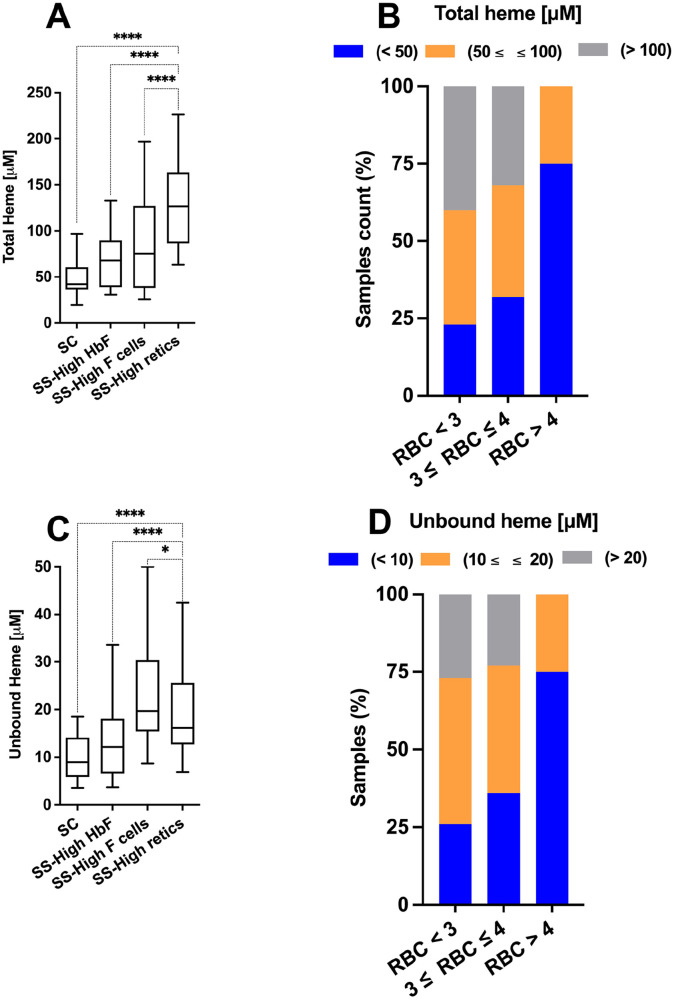
The heme measurement was stratified by beta globin genotype and other hematologic biomarkers of disease severity such as reticulocytosis and HbF production. Overall, plasma of the homozygous HbSS cohort, assumed to have a more severe phenotype than heterozygous HbSC on average, 42 had significantly increased plasma unbound heme compared with HbSC and HbAA (normal Hb) samples (Figure 5). In addition, samples from HbSS patients with high reticulocytosis, indicative of shorter RBC life span, also had increased plasma heme content compared with HbSS patients with high antisickling HbF, which is protective in SCD (Figure 6(A) and (C)). Total and unbound heme were also increased in patients with anemia as indicated by their lower RBC counts (Figure 6(B) and (D)). As confirmation of those findings, a significant trend of the accumulation of plasma heme as a function of increased erythropoiesis was observed (Figure 7).
Figure 5.

Release of heme in plasma of patients with sickle cell disease. Heme detected by extraction of plasma under neutral conditions of a cohort of SCD individuals (HbSC and HbSS) and control (HbAA) individuals is presented as a function of the hemoglobin type of the samples (HbSC, n = 20; HbSS, n = 76; HbAA, n = 24). Mean/median concentration (µM) values were HbSC 52/42, HbSS 104/87, HbAA 8.2/8.2. **** indicates P values of <0.0001.
Figure 6.
Analysis of plasma heme content in relation to hematology parameters I. Concentrations of total (panels A and B) and unbound heme (panels C and D) were determined in a cohort of plasma samples from SCD patients. The cohort was genotyped and analyzed for various hematology parameters (RBC count, % reticulocytes, % anti-HbF antibody positive RBC (F cells), % of HbF [HPLC]). The cohort was represented by HbSC (n = 20), HbSS with HbF level above 20% (n = 34), HbSS with %Fcells above 45% (n = 11), and HbSS with reticulocytes (retics) above 15% (n = 31). Note that few samples met several of the criteria but are only represented once in the analysis with priority given to the high retic category, followed by high HbF. Mean/median heme total concentration (µM) values were HbSC 52/42, HbSS-high HbF 72/68, HbSS-high Fcells 86/73, HbSS-high retic 126/145. Mean/median heme unbound concentration (µM) values were HbSC 9.8/8.9, HbSS-high HbF 14.4/12.1, HbSS-high Fcells 23.1/19.7, HbSS-high retic 20.4/16.1. In panels B and D, the counts of samples with the indicated concentrations of heme (total, panel B; unbound, panel D) were analyzed relative to the RBC counts of the blood samples: less than 3.106 cells/µL; from 3 to 4.106 cells/µL; more than 4.106 cells/µL. * and **** indicate P values of 0.02 and of <0.0001, respectively.
Figure 7.
Analysis of plasma heme content in relation to hematology parameters II. The concentrations of total heme (panels A and B) presented in Figure 6 were analyzed as a function of the reticulocyte count (panel A) and the percent of HbF (panel B). The inset in panel A shows the plot of unbound heme concentrations relative to reticulocyte counts. The mean values of total and unbound heme (µM) of the samples with less or more than 15% HbF, as indicated by the horizontal dashed line, are indicated in panel B. In panel C, the counts of samples with less than 3.106 RBC/µL and the indicated concentrations of total and unbound heme were analyzed in the samples with an HbF level of at least 15% and compared to the entire cohort.
The protective effect of HbF on RBC heme release was also analyzed in the cohort of HbSS patients. Heme content was 40% lower in patients with HbF levels of >15% compared with patients with HbF <15% (Figure 7(B)). Even in patients with severe anemia, RBC count <3 × 106 cells/µL, the heme levels were lower when HbF levels were increased (Figure 7(C)).
The levels of unbound heme correlated well with the percent of unbound heme calculated relative to total heme concentration. This finding established that as heme accumulated in plasma, the unbound fraction also increased, which indicates a failure of plasma proteins in SCD blood to absorb heme as it accumulates (Figure 8(A)). Analysis of the cohort also showed a broad variability in the heme absorption and removal capacity. Differences in total heme concentration did not correlate with the levels of unbound heme. Furthermore, distinct populations of samples could be identified when unbound values were plotted relative to the total heme concentrations (Figure 8(B)). Whereas some patients had high levels of heme in plasma without free heme (high total/low unbound), others did not absorb heme even at much lower levels (low total/high unbound). Unexpectedly, an inverse relationship was observed between concentration of heme (total) and the size of the unbound fraction (% unbound) (Figure 8(C)). As also evident in the samples from patients with SCD presented in Figure 4, these findings further emphasized the major contributing role of cell-free Hb to the presence of heme and of the marked differences in the capacity of some patients’ plasma to remove heme from the circulation (high total/low %unbound) or inability to absorb or remove heme after release (low total/high %unbound).
Figure 8.
Analysis of unbound heme fraction in plasma of sickle patients. Panel A: Concentration of unbound heme of the sickle cohort was plotted relative to the unbound fraction, which was calculated as the percent of unbound heme relative to the total heme concentration. Panels B and C: Concentration of total heme was plotted relative to the concentration of unbound heme and to the unbound fraction. In panel B, samples are grouped (boxes) based on relative high or low total and unbound heme.
Plasma heme levels during vaso-occlusive episodes
Analysis of plasma heme in a cohort of SCD patients during an acute vaso-occlusive event (VOE) was also performed. Blood samples were obtained at baseline and on day 1 and day 3 of hospital admission for a VOE. In this cohort, patient ages ranged from 6 to 38 years and genotypes included 12 subjects with HbSS, 6 with HbSC, and 2 with HbSβ+ thalassemia. Although a broad variety of changes were observed (Supplemental Figure S1), small differences in plasma heme concentration were detected in the acute phase compared to the levels measured at baseline (Figure 9). Although the difference in mean values were not significant, an increased number of patients with higher unbound heme was detected during the acute phase (Figure 9(B)), and a 20% increase in unbound heme was measured on the first day of the VOE (means of 18.5–22.5 µM; P = 0.07)) (Figure 9(D)). To gain further insight into these changes, the levels of the heme scavenger protein hemopexin were measured. As expected, we observed a significant depletion of hemopexin in patients with SCD compared with expected serum concentration of 600–1200 µg/mL.31,34 Furthermore, individuals with the HbSS genotype had a 2.5-fold lower concentration of hemopexin compared to individuals with the HbSC genotype, though any changes observed between the steady state and acute phase were not statistically significant (Figure 10(A)). However, when hemopexin levels were analyzed in relation to the concentration of unbound heme, a twofold to threefold depletion of hemopexin was measured in samples with unbound heme values above the mean values of the cohort (Figure 10(B)). These findings are consistent with the interpretation that samples with above-average heme concentrations represent a combination of events leading to higher hemolysis, higher heme released, and deficiency in absorption and in removal (Figure 1).
Figure 9.
Heme plasma content of SCD patients in vaso-occlusive crisis. Concentrations of total and unbound heme in plasma samples of patients in VOE were measured at day 1 and day 3 and compared to baseline levels. Panels A and B: The percentage of samples with the indicated concentrations of heme were analyzed at baseline (n = 20), day 1 (n = 20), and day 3 (n = 10). Panels C and D: Note that the differences in unbound heme did not reach statistical significance but were more pronounced than total heme. Mean/median of total heme: baseline (68/59), day 1 (57/56), day 3 (55/65), and for unbound heme: baseline (18.5/14), day 1 (22.5/19.5), day 3 (17.5/18.5). ns, non-significant.
Figure 10.
Levels of the heme scavenger hemopexin and heme. Panel A: Concentrations of the plasma protein hemopexin were measured in the cohort of sickle patients presented in Figure 5. Panel B: The mean values of hemopexin concentration were calculated in samples with a heme concentration (total and unbound) below or above the mean values of the cohort (mean total, 93 µM; mean unbound 16.5 µM). Error bars represent standard error of the mean. ns, non-significant; * and **** indicate P values of <0.05 and <0.0001, respectively.
Discussion
When bound to appropriate hemoproteins, heme plays a significant role in human physiology including delivery of oxygen to tissues and electron transfer in mitochondrial membranes. Due to its lipophilic nature, when heme is released from proteins like Hb, it distributes between different cellular and extracellular pools, intercalates into cell membranes, and provides a focus point for the production of reactive oxygen species leading to activation of inflammation and cellular damage.5 –13,16 –18 Heme scavengers such as hemopexin mitigate this effect14,20,25,28 –32,35 by binding heme with high affinity and removing it from circulation. Hence, in the setting of chronic hemolysis, high levels of heme can lead to the relative depletion of hemopexin, as production does not keep up with removal. This in turn will lower the ability of hemopexin to effectively remove heme from the other pools in plasma and cellular damage will occur. In hemolytic disorders such as SCD, it has been hypothesized that when plasma heme-binding capacity is saturated, the unbound heme plays an important role in pathogenesis.8,11,14,17,29,34 There are several heme-binding proteins in plasma, most notably hemopexin and albumin. The levels of these proteins can be measured by commercial assays, but these assays cannot distinguish between protein that is available for heme binding and that which is saturated.
Here, we describe a method for the quantification of the unbound cell-free heme fraction in relation to the total cell-free heme in plasma. This assay may also be used for the assessment of the heme-binding/removal capacity of plasma as unbound heme will only be present when the various heme-binding sites have been saturated. We first show that total and unbound cell-free heme may be reliably measured after extraction with AA and NA, respectively. Then we use this method to determine heme measurements in plasma samples from individuals with SCD and correlated our findings with hematologic parameters known to be associated with disease severity. The measurement of total heme was similar to values measured using commercial kit-based assays in other cohorts.16,28 Our findings were consistent with the hypothesis that in individuals with more severe disease as expressed by hemolysis rate, reticulocytosis, lower RBC count, and lower HbF, heme is released and accumulates in the unbound pool. We also describe the impressive capacity of the blood system, even in chronic hemolytic disorders of such as SCD, to sequester free heme.
Whereas this assay provides reliable measurements requiring a small amount of plasma, limitations on biological relevance should be considered. In patients with chronic hemolysis, measurement of cell-free Hb could be included to assess its contribution to the total heme pool. Similarly, hemolysis from venipuncture and/or blood storage may impact the level of total heme measured in plasma. As shown above, however, the measurement of the unbound fraction of heme is not impaired by the presence of cell-free Hb. Regardless, care should be taken to collect and store blood and plasma to reduce ex vivo hemolysis. The method as described measures the presence of bound and unbound heme in plasma. When heme is released into plasma in vivo, heme will distribute between the plasma phase and the cellular membranes of the blood and vascular cells (Figure 1). The presence of heme in plasma likely reflects the presence of heme in these membranes as these pools are in equilibrium with each other. However, these membrane pools cannot be measured from plasma analysis. The endothelial membrane pools cannot be assessed in a blood draw. In addition, the RBCs represent a high membrane surface area to provide a binding pool for heme, but the overwhelming amount of heme derived from the RBC Hb extraction will make it difficult to correctly assess the heme in the RBC membrane. Finally, the method presented requires generation of a standard curve in heme-spiked acetone extracts of plasma for quantification. Though we were able to reproduce our standard curve using a large stock of pooled plasma and consistent techniques, this step in the method might represent some challenges for its implementation in a clinical setting.
In conclusion, we present a novel method to quantify total and unbound heme in biological samples. A direct application would be the assessment of heme in plasma to determine the heme-binding capacity of the plasma and to assess the therapeutic value of treatment with compounds like hemopexin and antisickling agents leading to an increase in RBC lifespan.11,20,26,29,33 This assay may be applied to understand the pathophysiologic impact of heme and heme-binding proteins in hemolytic disorders such as SCD and acute illness such as sepsis. In addition, these methods may be adapted to investigate the therapeutic potential of novel agents that target hemolysis and cell-free heme.
Supplemental Material
Supplemental material, sj-pdf-1-ebm-10.1177_15353702231157920 for Assessment of total and unbound cell-free heme in plasma of patients with sickle cell disease by Madhav Vissa, Sandra K Larkin, Elliott P Vichinsky, Frans A Kuypers and Eric Soupene in Experimental Biology and Medicine
Acknowledgments
We extend our gratitude to the clinical and laboratory staff at UCSF Benioff Children’s Hospital Oakland and to our patients.
Footnotes
Authors’ Contributions: All authors contributed to the design, conduct, and analysis of the study and generation of the manuscript. MV, ES, and FAK wrote the manuscript, MV, ES, and SKL conducted the experiments. EPV and FAK assisted with data analysis.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical Approval: Granted by the Institutional Review Board at the University of California, San Francisco.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: MV was supported for this project in part by funding from Global Blood Therapeutics, Inc. [ESR-P004].
ORCID iDs: Madhav Vissa  https://orcid.org/0000-0002-1088-5856
https://orcid.org/0000-0002-1088-5856
Frans A Kuypers  https://orcid.org/0000-0003-0731-1007
https://orcid.org/0000-0003-0731-1007
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Bettati S, Viappiani C, Mozzarelli A. Hemoglobin, an “evergreen” red protein. Biochim Biophys Acta 2009;1794:1317–24 [DOI] [PubMed] [Google Scholar]
- 2. Eaton WA, Henry ER, Hofrichter J, Bettati S, Viappiani C, Mozzarelli A. Evolution of allosteric models for hemoglobin. IUBMB Life 2007;59: 586–99 [DOI] [PubMed] [Google Scholar]
- 3. Henry ER, Bettati S, Hofrichter J, Eaton WA. A tertiary two-state allosteric model for hemoglobin. Biophys Chem 2002;98:149–64 [DOI] [PubMed] [Google Scholar]
- 4. Kurtz DM., Jr. Oxygen-carrying proteins: three solutions to a common problem. Essays Biochem 1999;34:85–100 [DOI] [PubMed] [Google Scholar]
- 5. Beckman JD, Abdullah F, Chen C, Kirchner R, Rivera-Rodriguez D, Kiser ZM, Nguyen A, Zhang P, Nguyen J, Hebbel RP, Belcher JD, Vercellotti GM. Endothelial TLR4 expression mediates vaso-occlusive crisis in sickle cell disease. Front Immunol 2020;11:613278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Belcher JD, Chen C, Nguyen J, Milbauer L, Abdulla F, Alayash AI, Smith A, Nath KA, Hebbel RP, Vercellotti GM. Heme triggers TLR4 signaling leading to endothelial cell activation and vaso-occlusion in murine sickle cell disease. Blood 2014;123:377–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Camus SM, De Moraes JA, Bonnin P, Abbyad P, Le Jeune S, Lionnet F, Loufrani L, Grimaud L, Lambry JC, Charue D, Kiger L, Renard JM, Larroque C, Le Clesiau H, Tedgui A, Bruneval P, Barja-Fidalgo C, Alexandrou A, Tharaux PL, Boulanger CM, Blanc-Brude OP. Circulating cell membrane microparticles transfer heme to endothelial cells and trigger vasoocclusions in sickle cell disease. Blood 2015;125:3805–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gouveia Z, Carlos AR, Yuan X, Aires-da-Silva F, Stocker R, Maghzal GJ, Leal SS, Gomes CM, Todorovic S, Iranzo O, Ramos S, Santos AC, Hamza I, Gonçalves J, Soares MP. Characterization of plasma labile heme in hemolytic conditions. FEBS J 2017;284:3278–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Loomis Z, Eigenberger P, Redinius K, Lisk C, Karoor V, Nozik Grayck-E, Ferguson SK, Hassell K, Nuss R, Stenmark K, Buehler P, Irwin DC. Hemoglobin induced cell trauma indirectly influences endothelial TLR9 activity resulting in pulmonary vascular smooth muscle cell activation. PLoS ONE 2017;12:e0171219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nader E, Romana M, Connes P. The red blood cell-inflammation vicious circle in sickle cell disease. Front Immunol 2020;11:454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vallelian F, Buehler PW, Schaer DJ. Hemolysis, free hemoglobin toxicity and scavenger protein therapeutics. Blood 2022;140:1837–44 [DOI] [PubMed] [Google Scholar]
- 12. Wagener FA, Abraham NG, van Kooyk Y, de Witte T, Figdor CG. Heme-induced cell adhesion in the pathogenesis of sickle-cell disease and inflammation. Trends Pharmacol Sci 2001;22:52–4 [DOI] [PubMed] [Google Scholar]
- 13. Wagener FA, Eggert A, Boerman OC, Oyen WJ, Verhofstad A, Abraham NG, Adema G, van Kooyk Y, de Witte T, Figdor CG. Heme is a potent inducer of inflammation in mice and is counteracted by heme oxygenase. Blood 2001;98:1802–11 [DOI] [PubMed] [Google Scholar]
- 14. Yalamanoglu A, Deuel JW, Hunt RC, Baek JH, Hassell K, Redinius K, Irwin DC, Schaer DJ, Buehler PW. Depletion of haptoglobin and hemopexin promote hemoglobin-mediated lipoprotein oxidation in sickle cell disease. Am J Physiol Lung Cell Mol Physiol 2018;315:L765–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Detterich JA, Liu H, Suriany S, Kato RM, Chalacheva P, Tedla B, Shah PM, Khoo MC, Wood JC, Coates TD, Milne GL, Oh JY, Patel RP, Forman HJ. Erythrocyte and plasma oxidative stress appears to be compensated in patients with sickle cell disease during a period of relative health, despite the presence of known oxidative agents. Free Radic Biol Med 2019;141:408–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Adisa OA, Hu Y, Ghosh S, Aryee D, Osunkwo I, Ofori-Acquah SF. Association between plasma free haem and incidence of vaso-occlusive episodes and acute chest syndrome in children with sickle cell disease. Br J Haematol 2013;162:702–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Frimat M, Boudhabhay I, Roumenina LT. Hemolysis derived products toxicity and endothelium: model of the second hit. Toxins (Basel) 2019;11:660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guarda CCD, Santiago RP, Fiuza LM, Aleluia MM, Ferreira JRD, Figueiredo CVB, Yahouedehou SCMA, Oliveira RM, Lyra IM, Gonçalves MS. Heme-mediated cell activation: the inflammatory puzzle of sickle cell anemia. Expert Rev Hematol 2017;10:533–41 [DOI] [PubMed] [Google Scholar]
- 19. Kucukal E, Man Y, Quinn E, Tewari N, An R, Ilich A, Key NS, Little JA, Gurkan UA. Red blood cell adhesion to ICAM-1 is mediated by fibrinogen and is associated with right-to-left shunts in sickle cell disease. Blood Adv 2020;4:3688–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vinchi F, Costa da Silva M, Ingoglia G, Petrillo S, Brinkman N, Zuercher A, Cerwenka A, Tolosano E, Muckenthaler MU. Hemopexin therapy reverts heme-induced proinflammatory phenotypic switching of macrophages in a mouse model of sickle cell disease. Blood 2016;127: 473–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. White J, Lancelot M, Gao X, McGraw BJ, Tabb C, 2nd, Hines P. Cross-sectional analysis of adhesion in individuals with sickle cell disease using a standardized whole blood adhesion bioassay to VCAM-1. Blood Cells Mol Dis 2020;81:102397. [DOI] [PubMed] [Google Scholar]
- 22. Antonini E, Brunori M. Hemoglobin. Annu Rev Biochem 1970;39: 977–1042 [DOI] [PubMed] [Google Scholar]
- 23. Hill R, Holden HF. The preparation and some properties of the globin of oxyhaemoglobin. Biochem J 1926;20:1326–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gbotosho OT, Kapetanaki MG, Kato GJ. The worst things in life are free: the role of free heme in sickle cell disease. Front Immunol 2020;11:561917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ghosh S, Adisa OA, Chappa P, Tan F, Jackson KA, Archer DR, Ofori-Acquah SF. Extracellular hemin crisis triggers acute chest syndrome in sickle mice. J Clin Invest 2013;123:4809–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Immenschuh S, Vijayan V, Janciauskiene S, Gueler F. Heme as a target for therapeutic interventions. Front Pharmacol 2017;8:146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Oh JY, Williams A, Patel RP. The role of redox-dependent mechanisms in heme release from hemoglobin and erythrocyte hemolysates. Arch Biochem Biophys 2019;662:111–20 [DOI] [PubMed] [Google Scholar]
- 28. Santiago RP, Guarda CC, Figueiredo CVB, Fiuza LM, Aleluia MM, Adanho CSA, Carvalho MOS, Pitanga TN, Zanette DL, Lyra IM, Nascimento VML, Vercellotti GM, Belcher JD, Goncalves MS. Serum haptoglobin and hemopexin levels are depleted in pediatric sickle cell disease patients. Blood Cells Mol Dis 2018;72:34–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schaer DJ, Buehler PW, Alayash AI, Belcher JD, Vercellotti GM. Hemolysis and free hemoglobin revisited: exploring hemoglobin and hemin scavengers as a novel class of therapeutic proteins. Blood 2013;121:1276–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schaer DJ, Vinchi F, Ingoglia G, Tolosano E, Buehler PW. Haptoglobin, hemopexin, and related defense pathways-basic science, clinical perspectives, and drug development. Front Physiol 2014;5:415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Muller-Eberhard U, Javid J, Liem HH, Hanstein A, Hanna M. Plasma concentrations of hemopexin, haptoglobin and heme in patients with various hemolytic diseases. Blood 1968;32:811–5 [PubMed] [Google Scholar]
- 32. Griffiths S, Clark J, Adamides AA, Ziogas J. The role of haptoglobin and hemopexin in the prevention of delayed cerebral ischaemia after aneurysmal subarachnoid haemorrhage: a review of current literature. Neurosurg Rev 2020;43:1273–88 [DOI] [PubMed] [Google Scholar]
- 33. Gentinetta T, Belcher JD, Brugger-Verdon V, Adam J, Ruthsatz T, Bain J, Schu D, Ventrici L, Edler M, Lioe H, Patel K, Chen C, Nguyen J, Abdulla F, Zhang P, Wassmer A, Jain M, Mischnik M, Pelzing M, Martin K, Davis R, Didichenko S, Schaub A, Brinkman N, Herzog E, Zurcher A, Vercellotti GM, Kato GJ, Hobarth G. Plasma-derived hemopexin as a candidate therapeutic agent for acute vaso-occlusion in sickle cell disease: preclinical evidence. J Clin Med 2022;11:630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Muller-Eberhard U, English EC. Purification and partial characterization of human hemopexin. J Lab Clin Med 1967;70:619–26 [PubMed] [Google Scholar]
- 35. Tolosano E, Fagoonee S, Morello N, Vinchi F, Fiorito V. Heme scavenging and the other facets of hemopexin. Antioxid Redox Signal 2010;12:305–20 [DOI] [PubMed] [Google Scholar]
- 36. Espinas NA, Kobayashi K, Takahashi S, Mochizuki N, Masuda T. Evaluation of unbound free heme in plant cells by differential acetone extraction. Plant Cell Physiol 2012;53:1344–54 [DOI] [PubMed] [Google Scholar]
- 37. Fanelli AR, Antonini E, Caputo A. Studies on the structure of hemoglobin. I. Physicochemical properties of human globin. Biochim Biophys Acta 1958;30:608–15 [DOI] [PubMed] [Google Scholar]
- 38. Mc QE, Beniams HN. Preparation of globin from human hemoglobin. Proc Soc Exp Biol Med 1954;86:627–8 [DOI] [PubMed] [Google Scholar]
- 39. Rossi-Fanelli A, Antonini E, Caputo A. Pure native globin from human hemoglobin: preparation and some physico-chemical properties. Biochim Biophys Acta 1958;28:221. [DOI] [PubMed] [Google Scholar]
- 40. Collier GS, Pratt JM, De Wet CR, Tshabalala CF. Studies on haemin in dimethyl sulphoxide/water mixtures. Biochem J 1979;179:281–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Itoh T, Yamada T, Kodera Y, Matsushima A, Hiroto M, Sakurai K, Nishimura H, Inada Y. Hemin (Fe(3+))– and heme (Fe(2+))–smectite conjugates as a model of hemoprotein based on spectrophotometry. Bioconjug Chem 2001;12:36. [DOI] [PubMed] [Google Scholar]
- 42. Piel FB, Steinberg MH, Rees DC. Sickle cell disease. N Engl J Med 2017;377:305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-ebm-10.1177_15353702231157920 for Assessment of total and unbound cell-free heme in plasma of patients with sickle cell disease by Madhav Vissa, Sandra K Larkin, Elliott P Vichinsky, Frans A Kuypers and Eric Soupene in Experimental Biology and Medicine