Abstract

Lipid nanoparticles are a generic type of nanomaterial with broad applicability in medicine as drug delivery vehicles. Liposomes are a subtype of lipid nanoparticles and, as a therapeutic platform, can be loaded with a genetic material or pharmaceutical agents for use as drug treatments. An open question for these types of lipid nanoparticles is what factor(s) affect the long-term stability of the particles. The stability of the particle is of great interest to understand and predict the effective shelf-life and storage requirements. In this report, we detail a one-year study of liposome stability as a function of lipid composition, buffer composition/pH, and storage temperature. This was done in aqueous solution without freezing. The effect of lipid composition is shown to be a critical factor when evaluating stability of the measured particle size and number concentration. Other factors (i.e., storage temperature and buffer pH/composition) were shown to be less critical but still have some effect. The stability of these particles informs formulation and optimal storage requirements and assists with future developmental planning of a NIST liposome-based reference material. This work also highlights the complex nature of long-term soft particle storage in biopharmaceutical applications.
Introduction
Lipid-based nanoparticles of varied types are of great interest as clinical drug delivery vehicles.1−4 Two common types are liposomes and solid lipid nanoparticles. Liposomes consist of a lipid bilayer shell encapsulating an internal aqueous compartment, whereas solid lipid nanoparticles consist of a lipid bilayer encapsulating genetic material complexed with a cationic lipid. Liposomes are simple vesicles that can be implemented as a drug delivery vehicle.5−9 The ability to temporarily suppress or facilitate production of an arbitrary protein highlights both the utility and the broad use of this technological platform in treatment of disease. To better study lipid-based materials, there is a documented need for relevant reference materials that can be used for method development, validation, and instrument calibration.10 One of the challenges in developing a reference material to address this gap is ensuring that the material is sufficiently stable for long-term storage, study, and use. Only very recently has there been a report from Jukebek et al. examining the size stability of lipid nanoparticles and liposomes at −70 and 4 °C for up to 9 months.11 A recently reported body of work by Kirpotin demonstrates the drug stability of irinotecan contained in liposomes.12 This report describes and concludes that liposomes stored at low temperatures (≈4 °C) and at a basic pH demonstrated better stability due to decreased phospholipid hydrolysis. This is different from our work reported here, in that we used only empty liposomes as an initial case study for baseline stability measurements.
As an initial approach to long-term stability studies, we implemented liposomes as a relevant model particle here due to their simplicity and ease of preparation. The study of mechanisms of liposome degradation and stability may also be relevant for related types of lipid nanoparticle platforms in an unfrozen state. Liposomes have great utility, yet there are also many questions about the manufacture and implementation of these particles. There are open questions concerning inter-batch variability and the dependence of particle parameters on the formulation parameters.13,14 Some effort has been dedicated to understand the relationship between observed particle parameters such as the size and the formulation conditions implemented.15 A main approach to preparing these particles is through microfluidic mixing between aqueous and alcoholic streams in a controlled flow device.16,17 Particle size (and its associated homogeneity) are dependent upon the flow rate control parameters.18,19
Another key question is what determines particle stability, which is a critical requirement for possible reference material development.20−24 Prior work has examined the effects of parameters including solution pH, lyophilization, cryoprotectant choice (if frozen), and storage temperature on particle stability.25,26 However, most of these studies are of limited scope and/or duration (i.e., ≤6 months).25−28 To determine which factors control liposome stability, herein, we report a controlled systematic study of liposomes over the course of 1 year to assess the particle size and number concentration stability. Liposomes were prepared via controlled microfluidic mixing and were characterized orthogonally using asymmetric flow field flow fractionation with multi-angle light scattering (AF4-MALS) and particle tracking analysis (PTA), also referred to as nanoparticle tracking analysis (NTA). AF4-MALS has found expanding utility in measuring LNP samples in numerous studies.29−31 PTA has also been applied to characterization of LNPs.32−34 PTA measures both the particle size and number concentration.35
The particle size and particle number concentration of liposome samples were measured over the course of a 12-month period at three-month intervals while stored under selected conditions. In this work, the experimental parameters of lipid composition (including lipids more resistant to hydrolysis), buffer formulation/pH, and storage temperature (in liquid solution) were systematically varied. Frozen storage was not included here to avoid the complex influence of freeze–thaw cycles and cryoprotectants on the primary parameters of interest. At the conclusion of this study, selected liposome samples were characterized by cryogenic electron microscopy (cryoEM) and mass spectrometry to measure structure/size and identify lipid decomposition products, respectively. In general, lipid composition had the most significant effect on the particle size and number concentration over the 12-month course of the study. We also observed a less clear effect of buffer pH and storage temperature on particle stability in terms of size and number concentration. Overall, this study shows an approach to quantitatively assess storage conditions for these types of soft particles and demonstrates the critical impact of environmental conditions on liposome stability.
Materials and Methods
Certain commercial reagents and instrumentation are identified throughout to describe the experimental procedures with adequate detail. In no case does such an identification imply an endorsement by NIST nor does NIST suggest that the materials or equipment so identified are necessarily the best available for the purposes described herein.
Microfluidic Device Fabrication
A microfluidic mixer consisting of polydimethylsiloxane (PDMS) channels on a glass substrate was fabricated using standard microfabrication technology to facilitate preparation of liposomes. The full detailed procedures for the fabrication are available in the Supporting Information. Silicon masters were prepared for casting by heating to 100 °C in a silanization oven (YES-1224P, Yield Engineering Systems) and then fluorinated silane (tridecafluro-1, 1, 2, 2-tetrahydrooctyl)-1-trichlorosilane, United Chemical Technologies) was introduced in the vapor phase to form a release-promotion monolayer on the master. Functionalized masters were then placed into standard plastic Petri dishes (150 mm) for PDMS casting.
Standard PDMS (Sylgard 184, Dow Corning) was mixed in a 10:1 standard ratio of prepolymer to activator and poured gently over the master to cover it completely. The PDMS was degassed under partial vacuum for 1 h. After degassing, the masters covered with PDMS were transferred into a 60 °C oven for an overnight soft bake. Following the soft bake, PDMS castings of the device were cut out with a razor blade. A freshly cleaned glass slide was placed into a PDC-32 Plasma Cleaner (Harrick Plasma) along with the cast PDMS device and vacuum was applied. Following complete evacuation of the chamber, a small amount of air was bled into the chamber and the RF source set to the medium power setting. Upon visual confirmation of plasma in the chamber, the plasma treatment was continued for 60 s. Then, the RF source was switched off and the chamber was immediately returned to atmospheric pressure. The PDMS casting and the glass slide were promptly joined together and pressed firmly for 60 s to promote adhesion to prepare the fabricated devices, which were used to generate the liposomes as described below.
Liposome Preparation and Formulation
Absolute ethanol (USP) was stored over 3 Å (0.3 nm pore size) molecular sieves for 1 week to ensure sufficient dryness. Small aliquots of ethanol were removed and filtered through a 0.45 μm filter to remove any particulates. A lipid mix was prepared gravimetrically consisting of a main structural choline-type lipid, cholesterol, and a polyethyleneglycol (PEG)-functionalized lipid for surface and steric stabilization at a typical molar ratio of 5:4:1, respectively. [Lipids included diphosphatidylcholine (DPPC) (Cat. No. 850355P, Avanti Polar Lipids), 18:0 Diether PC (Cat. No. 999991P, Avanti Polar Lipids), C16 PEG2000 Ceramide (Cat. No. 880180P, Avanti Polar Lipids), 18:0 PEG2000 PE (Cat. No. 880120P, Avanti Polar Lipids), C16 PEG2000 Ceramide (Cat. No. 880180P, Avanti Polar Lipids), and cholesterol (Sigma-Aldrich)]. The dried, filtered ethanol was added to the lipids and mixed thoroughly. The vial containing the lipid-ethanol mixture was placed in a warm water bath to promote dissolution. Once all the lipids were fully dissolved, they were filtered through a 0.02 μm Anotop filter and transferred into a glass-lined 15 mL conical centrifuge tube. Standard PBS (Sigma Aldrich) containing 3.08 mmol/L NaN3 was also filtered through a 0.02 μm anodized aluminum oxide syringe filter (Anotop) into a 15 mL conical centrifuge tube.
The solutions were loaded into a pneumatically actuated pump with an integrated flow controller (PneuWave Pump, CorSolutions). The fluid flows (75 μL/min aqueous, 25 μL/min alcohol) were connected to the prepared microfluidic devices, and an optical microscope was used to visualize the fluid interface. A standard volumetric flow rate ratio of 75:25 (aqueous:alcohol) was used for all samples and was held constant. After the alcohol/aqueous interface was stabilized, the initial 1 to 2 mL was discarded before collecting each sample. The aqueous and alcoholic solutions were switched out and purged as needed to prepare all samples using the desired combinations of lipids/buffers.
Asymmetric-Flow Field Flow Fractionation (AF4)
An Eclipse DualTec AF4 separation system (Wyatt Technology, Santa Barbara, CA) was coupled to a UV/vis diode array detector (Model 1260, Agilent Technologies, Santa Clara, CA), a HELEOS-II multiangle light scattering instrument (HELEOS-II, Wyatt Technology), and a differential refractive index detector (Optilab T-Rex, Wyatt Technology). The AF4 channel was a vendor-supplied “short” channel with 250 μm thick “wide” Mylar spacer and a 10 kDa nominal molecular-weight cutoff Ultracel regenerated cellulose (Millipore, Burlington, MA) ultrafiltration membrane served as the accumulation wall. Samples were introduced into the AF4 separation channel via an Agilent 1260 autosampler (Santa Clara, CA) with a focus position of 12% of the channel length. The focusing was accomplished by flowing 0.2 mL/min of buffer into the channel inlet and 1.3 mL/min of buffer through the channel outlet for 5 min. After the samples were introduced and focused against the ultrafiltration membrane, they were eluted from the column in a size selective manner with a channel flow of 1.0 mL/min while the cross flow was linearly ramped from 3.0 to 0 mL/min over 45 min. Post separation, the channel was rinsed for 5 min with 1.0 mL/min channel flow and 0 mL/min cross-flow and the injector “on” to rinse out the sample loop.
Multi-Angle Light Scattering Particle Quantification
The eluted fractions flowed into the HELEOS detector where the flow cell was illuminated with a plane-polarized laser (λ = 662 nm) and the scattering intensity was measured at 16 different angles simultaneously. The scattering data were fit with a coated sphere model (neglecting second virial coefficient effects), and the inner sphere refractive index was set to the experimentally measured value of 1.333, whereas the shell was set to 1.459 following a calculation based on the molar volumes. The shell thickness was empirically set to 8 nm by comparing the static light scattering data to the hydrodynamic data from the in situ dynamic light scattering measurement to define the optimized value. The model parameters were fit to the scattering intensity at each angle to derive the particle size. The scattering intensity at zero angle was extrapolated, and the particle number concentration was determined according to eq 1:
| 1 |
Here, R(0) is the extrapolated scatter intensity at 0° angle, λ0 is the laser wavelength, Vparticle is the volume of the particle, N is the particle number concentration in the slice, nshell, ncore/bulk are the refractive indices of the particle shell and the inner core/bulk fluid medium, respectively, and Vcore + shell and Vshell are the volume of the core plus the shell and the shell alone, respectively. The core and bulk refractive indices are effectively identical due to how the particles form. Extrapolation and calculations were done with ASTRA 7.3.2 software. The reported experimental error for the diameter was obtained by taking the reported error in the radius as calculated by the ASTRA software multiplying by a straight factor of exactly 2. The reported experimental error for the particle number concentration was obtained by taking the reported particle number concentration error as calculated by the ASTRA software and propagating the error using the uncertainty in the injection volume.
Particle Tracking Analysis
Particle tracking analysis (PTA) was performed using a commercial instrument (ZetaView PMX 110, Particle Metrix) with λ = 405 nm laser. Instrument performance (particle sizing) was verified using a 110 nm polystyrene bead standard (Microtrac). Particle sizes were calculated as sphere-equivalent hydrodynamic diameters. Samples were measured in triplicate at each time point, with the values reported as the mean of the three measurements ±1 standard deviation. In one case (S03, Day 0), only two replicates were acquired owing to a bubble in the measurement cell and insufficient material reserved for a repeat. For the particle size, the median diameter calculated for each replicate was used to generate the reported mean particle size.
Reported PTA particle number concentrations were corrected for dilution factors. Temperature of the sample cell during the measurements was controlled at either 20 or 25 °C. Each sample was diluted gravimetrically in two steps with the appropriate buffer (prefiltered through a 0.02 μm anodized aluminum oxide syringe filter (Anotop)), and three volumes of the diluted sample were measured and averaged to yield particle size distributions. Dilution factors ranged from 12,000 to 250,000 for the various liposome samples and measurement trials. For each measurement, videos of 90 frames were captured at 11 positions in the sample cell with a frame rate of 30 frames per second, a shutter speed of 50, and a camera sensitivity of 80. Video analysis was performed with the instrument ZetaView software (version 8.04.02 SP2). All video acquisition and analysis parameters were fixed based upon the initial measurements made in November 2019.
Cryogenic Electron Microscopy
A Vitrobot Mark IV system (Thermo Fisher Scientific) was used to blot and plunge-freeze grids into liquid ethane. Prior to sample application, the Vitrobot chamber was pre-equilibrated to 25 °C and 100% relative humidity. Liposome solution (3 μL) was applied to a grid and incubated for 1 min before blotting and plunge-freezing. Grids used for imaging were ultrathin carbon with a lacey carbon support (Ted Pella #01824G) or Quantifoil R3.5/1 (#N1-C19nCu20-01). Grids were imaged on a Glacios microscope (Thermo Fisher Scientific) at 200 kV with a K3 detector (Gatan) using parallel illumination. Images were collected as movies with a pixel size of .2317 nm, total dose of 4610 e–/nm2, and 92 frames. Movies were then motion-corrected using MotionCor2.36
Data Analysis
Datasets for liposome diameters and particle number concentrations were organized after the completion of the study based upon the liposome sample, measurement technique, and measurement parameter (diameter or particle number concentration). Analyses on the datasets were carried out in either Igor Pro 9 (Wavemetrics) or in Excel (Microsoft). In all cases, a p < 0.05 was used to indicate statistical significance.
Results and Discussion
Microfluidic Mixer Preparation of Liposome Nanoparticles
Preparation of liposomes via microfluidic mixing is well-documented in the literature.37,38 In this approach, lipids dissolved in absolute ethanol are brought into contact with an aqueous solution and controlled perturbations facilitate rapid diffusion and interfacial mixing. The original design for the mixer consisted of a long 70 μm tall by 15 μm wide channel, which was a difficult structure to make from SU-8 as the curved sections are under stress and break easily during the PDMS removal. A structure that was wider than it was tall and having a large surface area in contact with the wafer would be much stronger and last longer. An ideal solution to this problem was to use a staggered herringbone mixer, which uses a wide channel (400 μm) with specially shaped herringbone grooves on the top.38,39 The schematic of the microfluidic device is shown in Figure 1, which also shows the design and features of the chevron-shaped perturbations fabricated into the device. In the lower part of Figure 1, the physical device can be seen with the inflow and outflow channels marked categorically. The fluid flow rate into the device is controlled by flow meters which adjust the pneumatic pressure with a closed-loop feedback strategy to maintain a stable mixing interface, ensuring more homogeneous particle formation.
Figure 1.

Schematic design of a microfluidic mixer device with the microscope image of the manufactured chevron pattern for perturbed flow along with schematic of device dimensions (upper left). Imaged fabricated device of PDMS on glass with inlets and outlet labeled on the device (lower left). Cryogenic EM image of liposome sample S10 showing mostly spherical particles with some pseudo-cylindrical particles (upper right). Cryogenic EM image of liposome sample S06 showing mostly spherical particles with a few multilamellar and elongated cylindrical particles (lower right).
Sample Array Preparation and Design of Experiment
To evaluate liposome stability over long time scales (≈12 months) in fluid solution without freezing and/or cryoprotectants, a variety of experimental factors were investigated including buffer composition, lipid composition and storage conditions. The complete array of factors and the factorial design are listed below in Table 1, showing sample buffer formulation and pH along with lipid composition. Samples stored in the cold were either transferred directly into the refrigerator (Rapid) or cooled slowly (Slow) at a rate of approximately 1 °C/min using a Mr. Frosty device (Thermo-Fisher Scientific, Catalog No. 5100-0001), a freezing container used to slowly cool and freeze cell culture preparations.
Table 1. Design of Experiment and Prepared Liposome Samples in Different Storage Conditions, along with Molecular Structures of the Lipids Used in This Studya.
“RT” refers to storage under ambient, room temperature conditions, generally around 22 °C.
The sample pH is expected to have an impact due to the pH-dependence of the hydrolysis reactions that lead to degradation of the lipids comprising the bilayer of the liposome. For this reason, we also evaluated formulations with ether-based rather than ester-based lipids, since the former would be theoretically less prone to hydrolysis in aqueous solution.40,41 In the same vein, the normal PEGylated lipid was substituted with a more theoretically stable ceramide-based lipid for the indicated samples below.
Based on the standard kinetic theory, a higher temperature should lead to more rapid degradation of the lipids; therefore, cold storage was explored as part of this work. The final dimension of exploration was the temperature change rate, as can be expected from fundamental chemical kinetics considerations. This was implemented by placing samples into a controlled-rate cooling container prior to transfer into the refrigerator, in comparison with an uncontrolled cooling by directly placing samples into the refrigerator. The samples were ramped the same way both in and out of cold storage: the controlled-rate cooling container was used to both cool the samples in the refrigerator and warm them back to room temperature before analyzing the samples at all time points post-initial.
We note that the PTA measurement and analysis parameters were optimized for initial measurements, and not changed through the course of the time series measurements. Run-to-run minor adjustment of the measurement parameters could introduce additional variability to the measurements, and so was not implemented. In this approach to monitor changes from the initial population of liposomes, the hypothesis was that any changes to the particle population would still yield a main population observable with the initial settings.
Particle Measurements and Comparisons: Particle Diameter Quantification
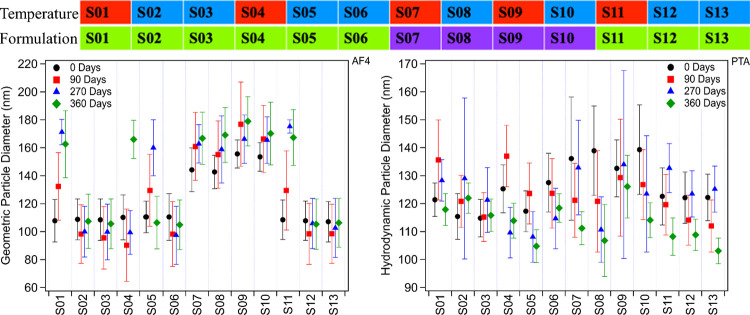
To evaluate stability of the liposomes over time, orthogonal particle measurements were made using AF4-MALS and PTA. These optical techniques effectively measure the same quantities but through different means. This orthogonal approach was carried out to assess the level of agreement/disagreement between the techniques as well as to give an idea of what intrinsic inter-method variation exists. AF4-MALS was carried out directly on the liposome preparations at each of the time points to measure both the geometric size as well as an overall particle number concentration. For the PTA instrument, samples were diluted by a factor of (2 to 9) × 104 to ensure the particle number concentrations would be within the optimal range. Measurements of the diluted samples were then corrected to give the original particle number concentration in the PTA measurements for direct comparison to the AF4-MALS data. Samples were assessed approximately every 90 days for both particle size and number concentration for 360 days, except for at 180 days due to limited access to the samples from the ongoing COVID-19 pandemic. The particle size measurements for each sample over time are plotted with their associated errors in Figure 2 for both the AF4 and PTA techniques. Note that the AF4-MALS calculates geometric diameter from the light scattering, whereas PTA calculates hydrodynamic diameter. The MALS measurement is based on angular dependence of the scattering, whereas the PTA measurement is based on Brownian motion of the particles over time.42 This fundamental aspect is reflected in the data labels in Figure 2.
Figure 2.
(L): Geometric particle diameter over time as measured by AF4-MALS. (R): Hydrodynamic particle diameter over time as measured by PTA. The data are color-coded by the approximate date on which they were collected, as referenced to the initial measurement (0 Days). Error bars represent the calculated experimental error (AF4) or 1 standard deviation (PTA). (Upper): Samples are color coded based on the experimental design shown in Table 1 to highlight systematic variations in storage temperature and lipid formulation.
While rich in data, the plots in Figure 2 (and Figure 3, see below) enable assessment of variability and time trends for individual samples, with quick reference to relevant conditions (Temperature and Formulation) and other samples. When examining the trends across the data what is immediately clear from the AF4 data is that samples using the standard PEGylated lipid stored at RT are inherently less stable than those stored at 4 °C. S01 and S11 both have large variations in average particle diameter across the time series, as does S05, which experienced rapid thermal cycling. The effect of pH seems to be minimal, at least in terms of this data set. The PTA data on the particle diameter is not perfectly correlated to the AF4 data, and samples of differing instability were observed. Overall, the PTA data show similar levels of variability in the measured particle diameters across all the samples over time, although the samples with largest variability vary between the two techniques. Individual measurements could show greater measured relative standard deviations (e.g., Day 0 measurements for S07, S08, and S10), but qualitative examination of the PTA data does not indicate clear sample-dependent variation as was the case for the AF4-measured samples. Samples in the pH 6.5 buffer showed low relative standard deviations across the time series, in general.
Figure 3.
(L): Calculated particle number concentrations (per mL basis) over time as measured by AF4-MALS. (R): Concurrently calculated particle number concentrations (per mL basis) over time as measured by PTA. The data are color-coded by the approximate date on which they were collected, as referenced to the initial measurement (0 Days). The ordinate axis is a logarithmic scale on both plots. Error bars represent the calculated experimental error (AF4) or 1 standard deviation (PTA). The calculated error bars on the AF4 measurements are too small to be seen.
Particle Measurements and Comparisons: Particle Number Concentration Quantification
Concomitant with the measured particle diameter, the overall particle number concentration (PNC) was also monitored and quantitated over time, as shown in Figure 3 for both AF4 and PTA measurements. The AF4 data show clear trends in the stability of the particle number concentrations over time.
(On the logarithmic scale, the same fractional change in PNC will display as the same vertical distance on the figure, regardless of the PNC absolute magnitude.) There is a clustering of samples 7, 8, 9, and 10, which corresponds to the samples of a different lipid composition/formulation. Concurrent examination of the PTA analysis of the number concentration shows a very similar trend, although S08 in the PTA analysis shows more variation than the same sample when examined via AF4. Interestingly, the samples prepared in a slightly acidic (pH ≈ 6.5) buffer also demonstrated relative stability over the time course: no significant monotonic trends in particle number concentration with relative standard deviations for each sample across the series <9% and S03 showing no significant differences in the particle number concentration for any time point. However, this was not observed by AF4 measurements and so the inference of stability at a slightly acidic pH cannot be made conclusively. The observed small error bars on the AF4 measurements originates as the measurement in the ASTRA software is only a general statistical measure of the error and does not account for any systematic error from the input quantities (refractive index, optical calibration constants, etc.).
Four samples, S07–S10, were observed to have more stable particle number concentrations by AF4. The PTA results show generally stable particle number concentrations for S07 and S09 with relative standard deviations across the time series <11%. These samples all have swapped out the more common DPPC lipid for the modified diether phosphatidylcholine. As this phosphatidylcholine comprises ≈50% (by mole fraction) of the overall lipid composition, it is not unexpected that this would have a significant effect on stability of the liposomes. The other main component is cholesterol, which (at ≈40%) is less susceptible to degradation than the esterified phospholipids like phosphatidylcholine.43 The remaining component of the lipid mix is a PEGylated phospholipid. As this comprises a small percentage (on a molar basis) of the overall lipid composition, its effects on the liposome stability were expected to be rather minimal.
It is also of interest to note that the same samples discussed above (S07–S10) show a larger particle size as measured across these four samples. While it is not clear why these particles are larger, it does match up quite well with the lower particle number concentrations observed for these same samples. As a finite amount of lipid is available for particle formation, an increase in the particle size will necessitate lower particle numbers to be formed during the microfluidic mixing. A fully detailed calculation and analysis is available in the SI to demonstrate the particle number concentrations expected based on two different particle diameters (i.e., observed diameters of 100 and 160 nm). In this calculation, the input of the lipid mass was adjusted to match the observed particle number concentrations. The calculated difference in particle number concentrations is approximately 2.56 times as many particles for the smaller diameter as for the larger. This matches up to the measured particle number concentrations and reinforces the underlying connection between the particle size and particle number concentration.
Intermeasurement Comparison Analysis
To directly compare the AF4 and PTA measurements across time, data from each measurement were plotted against each other, as seen in Figure 4. As mentioned previously, due to slight differences in the measured quantities, it was expected that the PTA particle diameter measurements would be slightly larger than the AF4 measurements on account of differences between hydrodynamic and geometric calculations of the relevant quantities. The expected discrepancy is generally observed in the plotted data of the two techniques (Figure 4). It was expected that the hydrodynamic size (as measured by PTA) would be larger than the geometric size (as measured by AF4). For some of the measurements seen in Figure 4 though, the AF4-measured diameters are larger than that of those measured by PTA. The refractive index of the lipid bilayer is highly critical in calculating the particle number concentration by AF4-MALS, but the size measurement is not as dependent on knowing the refractive index precisely. It is therefore not obvious why many of the samples show a larger geometric size than a hydrodynamic size.
Figure 4.
(L): Concordance between AF4 and PTA measured particle diameters, tracked over time. (R): Equivalent concordance data between the AF4 and PTA calculated particle number concentrations. The diagonal lines correspond to a 1:1 concordance.
A breakout analysis is also available in the SI (Figure S2/S3), which breaks the comparison down by each time point. What was not expected was the way in which samples tended to sort themselves into apparent clusters in terms of their location in the graphical data. The cluster space can be defined by time for a single sample. For example, S04 at 0 days, 90 days, and 270 days shows relatively limited movement for the diameter space before a larger movement in geometric diameter at 360 days. This aspect of the clustering phenomenon is apparent in Figure S2/S3 where the individual time point comparisons are presented across the time series. Clear clustering of groups of samples is demonstrable as well, even though these clusters change over time. In particular, the S07–S10 samples are clustered together in the particle size concordance by AF4 and PTA measurements (Figure 4/S2) and remain so across time even as clusters move across the plot over time. It can be inferred that these samples with a more chemically stable diether phosphatidylcholine lipid should both be similar in particle formation chemistry as well as similarly resistant to chemical degradation as this is the main structural lipid in these samples. Examining the same samples (S07–S10) in the particle number concentration concordance data (Figure 4/S3) is congruent with this explanation as the same clustering was observed, indicating that the underlying chemistry controls particle stability.
Time Series Analysis
Material stability can be assessed by looking at the measured properties as a function of time. As an initial overview of the data, samples were grouped by each storage parameter explored: storage temperature, lipid composition, and buffer. Figure 5 shows examples for this approach looking at measured liposome particle diameters. To assess the overall trends for the samples based upon storage conditions, the data from each group were fit with a linear model and 95% confidence intervals were calculated. The stability trends within each group were based upon whether the slope of the linear model was statistically significant. That is, a significant slope (either positive or negative) for p < 0.05 indicated changes in the samples over the time series, while an insignificant slope (p ≥ 0.05) suggested that the group corresponding to the storage parameter showed no general instability. The plots in Figure 5 (and Figure 6, see below) are separated by the measurement technique and storage parameter to provide a quick visual assessment of stability trends, with any significant slopes noted in the plot legend.
Figure 5.

Time series analysis of the particle size. Data are broken down by storage temperature (L), lipid composition (C), and buffer pH (R). Measurements from AF4 are plotted in the top row, while measurements from PTA are plotted in the bottom row. Error bars for each point represent the calculated experimental error (AF4) or 1 standard deviation (PTA). The solid lines show a linear model fit across all the data points in that group, with the dotted lines representative of the 95% confidence interval. The ∗ indicates a statistically significant slope for that factor.
Figure 6.

Time series analysis of the particle number concentration. Data are broken down by storage temperature (L), lipid composition (C), and buffer pH (R). Measurements from AF4 are plotted in the top row, while measurements from PTA are plotted in the bottom row. Error bars for each point represent the calculated experimental error (AF4) or 1 standard deviation (PTA). In some cases, error bars are smaller than the marker point size. The solid lines show a linear model fit across all the data points in that group, with the dotted lines representative of the 95% confidence interval. The ∗ indicates a statistically significant slope for that factor.
From these broad groupings, some trends are implied, although strong conclusions cannot be made. For comparison of storage at either room temperature (RT) or refrigerated, three of the four groups (treating AF4 and PTA measurements as separate groups) show significant slopes (significance indicated by p < 0.05 in all cases) over the time series. Only the AF4 measurements of the samples stored at 4 °C show stable diameters (i.e., a nonsignificant slope). Similarly, for the lipid composition, only the PTA measurements of the ester-based lipids showed stable diameters. Conversely, for buffer/pH, only one group (PTA, pH 7.4) showed a significant change over the time series.
Measurements of PNC were treated similarly as shown for the three parameters in Figure 6. For PNC, most groups showed general stability (nonsignificant slopes), with the exceptions being the ether composition group measured by AF4 and the pH 8.5 buffer group measured by PTA. Overall, the broad groups analysis suggests that pH 6.5 may offer a stable liposome environment as has been reported in the literature.44 For the pH 6.5 samples, no significant slopes were observed in either diameter or PNC as measured by either AF4 or PTA. Other conditions yielded a mix of trends, indicating that a more in-depth examination of the individual sample trends was needed.
To explore the relative changes in individual samples as a function of elapsed time, two approaches were used. These analyses are available in the SI. In the first approach (Figure S4, liposome diameter), the change is calculated on a point-to-point basis. That is, the change from 0 to 90 days is plotted as the 90-day time point, and then, the further change from 90 to 270 days is also plotted as the 270-day time point. A way of setting the initial measurement at 0 days as a pseudo-reference point and then measuring the change from 0 days is also possible and was plotted in the SI (Figure S5). Several samples were highly variable, with changes in the (20 to 40%) range. The most stable samples of the group had modest percentage changes of approximately (5 to 10%) over the course of one year. The samples showing stability in diameter for both AF4 and PTA measurements included all samples stored at 4 °C in pH 6.5 buffer, as well as several of the samples using ether-based linkages in the lipids that were stored in pH 7.4 buffer. Sample S09, specifically, was stored at room temperature in pH 7.4 buffer and had <15% change in diameter from Day 0 to Day 360 as measured by either AF4 or PTA.
As was done with the particle diameter measurements, the relative change at each measured point in time for the particle number concentrations is also available in the SI (Figure S6/S7). It is curious to note that for AF4, the initial change (from 0 to 90 days) appears overall to be quite large and then tends to drop off at the following time points. This suggests that the liposomes undergo an Ostwald-like ripening process where the liposomes are initially unstable and therefore undergo changes and shifts initially until they settle into a stable configuration. These changes appear to be complete after the initial 90-day period. This can be reasonably expected as these samples were kept in the liquid state, and it is known that liposomes exchange lipids with each other throughout the fluid solution medium. It is reasonable that repeated collisions and exchanges of the lipids, as documented in the literature, over some period of weeks to months can change the overall particle size and number concentration until the samples reach stable equilibrium states.45,46 Overall, for both AF4 and PTA measurements, the samples using ether-based linkages showed relatively minor changes over the time series, with sample S09 showing particularly small changes in PNC (<20% Day 0 to Day 360). Samples S07 and S10 also seemed to demonstrate relatively good stability (<41% change in PNC, Day 0 to Day 360).
Post Time-Series Analysis: Mass Spectrometry and Cryogenic Electron Microscopy
It was clear from the particle metrology that the particle samples changed over time. As a means of assessing liposome quality and degradation, cryogenic electron microscopy (cryoEM) and lipid mass spectrometry were used to characterize the particles at the end point of the study. Liposomes like those used here are particularly amenable to this type of electron microscopy.47 The measured diameters as seen in the images were approximately 100 to 140 nm (Figure 1, right panels), as expected based on generic size control via microfluidic mixing. In the EM images shown in Figure 1, most of the particles are unilamellar approximate spheres. However, some elongated pseudo-cylinder particles are observable as well as a few multilamellar particles. This imaging was done approximately 18 months after the initial preparation of the particles. Therefore, the morphology demonstrated in Figure 1 is a post-study measurement, carried out approximately 6 months after the conclusion of the study. It is also key to understand that the primary measurements carried out in this study are ensemble average measurements, which provide only an average size and/or particle number concentration based on a very large ensemble of particles that are probed in these types of measurements. All the liposome samples used in this study were generated via the microfluidic device of PDMS on glass.
One hypothesis of this study was that breakdown/degradation of the lipids would lead to the observed changes. To probe this, mass spectrometry of selected samples was used to characterize possible breakdown products. The samples were analyzed at the ≈18 month mark to examine the lipid composition at that time by using mass spectrometry. The data are available in the SI where the selected samples were analyzed, and the mass spectra are presented (Figures S8–S12). One of the observed molecules was lysophosphatidylcholine, which is the known hydrolysis product of DPPC.48,49 This arises due to hydrolysis and cleavage of the fatty acid side chains. This product was expected as the liposomes are susceptible to hydrolysis in aqueous solution. In general, it is expected that most of the fatty acid groups are inaccessible to the solvent as they should be contained within the internal bilayer. However, published work in the literature has shown that the lipid bilayer is not static but is instead dynamic.50,51 As the lipid molecules move around, water molecules would then be able to react with the ester groups, thus hydrolyzing off the fatty acid groups and giving the observed lysophosphatidylcholine. Another observed product was oleamide. This is the breakdown product of the hydrolyzed fatty acid side chains. Oleamide is a C16 molecule that interestingly shows up in every measured sample. Upon initial examination, this seems unlikely as several samples contain DPPC as the main structural lipid, which has C18 side chains. However, these samples contain a phosphoethanol PEG2000 lipid used for steric bulk. This lipid does contain C16 alkyl fatty acid groups and therefore is the likely source of the observed oleamide breakdown product.
Liposome Stability Factor Analysis
As a means of developing a rudimentary statistical measure of variation in the data, analysis of variance (ANOVA) was carried out on the PTA particle diameter and particle number concentration data. In this analysis, for each sample, the means of the particle diameter or particle number concentration at each time point were compared to assess statistically significant variations. ANOVA was done for the PTA data but not AF4 data since the PTA data were collected in triplicate, but the AF4 data were not. The results of the ANOVA are reported in the SI and show statistically significant differences among the populations means for almost all particle diameter measurements (Table S1) except for sample S09. A similar analysis for the particle number concentration (Table S2) shows a similar result with all samples exhibiting statistically significant differences, except for sample S03. The analysis indicates that most of the samples show statistically significant variations in the liposome size and number concentration over the time course of this study. For the two samples showing relative stability (S09 for diameter, S03 for particle number concentration), it is unclear why and difficult to attribute to any particulate study parameter. It is possible that some complex interaction between lipid composition, buffer pH, and storage temperature is responsible for the two noted samples’ apparent stability, but this study cannot account for it. We note that while the mean diameter or PNC may show statistically significant differences from time point to time point, however, all but one of the samples (S10) showed overall nonsignificant slopes for either diameter or PNC over the course of the time series. Several of the samples showed general stability (i.e., insignificant slopes) for both parameters: S01, S03, and S09. These correspond to samples stored in the pH 6.5 buffer and one of the ether-based lipid samples, which have been noted above as appearing stable.
The factors examined in this study were the lipid composition, buffer pH, sample storage temperature, and the temperature ramp (if kept under cold conditions). An overall look at the particle size and number concentration measurements shows the clearest effect of the lipids used in the mixture. This is particularly notable when examining longitudinal plots of the data (available in the SI in Figures S13 and S14) when looking at trends in the particle number concentrations specifically. To compare the magnitude of changes between different lipid composition (ether-linked lipids: group S07 to S10) and buffers (groups S01 to S03, S04 to S06, S11 to S13), the average of the absolute measurement-to-measurement changes for all samples within each group were calculated (see also, Figures S4 and S6 for individual samples). The samples using the ether-based main structural lipid (group S07 to S10) were markedly more stable when measured over the time course of a year in fluid solution. These samples as a group showed <11% change in PNC on average from measurement-to-measurement as measured by AF4-MALS and PTA (excluding S08 for PTA, which seemed to be an outlier at ≈100% average change). The other groupings based upon ester-linked lipids in varied buffers (groups S01 to S03, S04 to S07, and S11 to S13) tended to show higher average measurement-to-measurement variations, ranging from 43 to 93% for AF4. As noted above, groups S01 to S03 appeared relatively stable by PTA (<7% average change), but the other sample groups showed larger average changes of 26 and 38%. The diameter measurements were treated similarly. These measurements generally showed smaller overall average changes measurement-to-measurement. For both AF4 and PTA, the average for any group was <10%, except for group S04 to S06 measured by AF4, which showed an average change of ≈17%. As explained above, the apparent stability of the group S07 to S10 was expected as the ether linkages are much less prone to hydrolysis in the aqueous buffers used here.
Other than the observation above that the samples stored at pH 6.5 show some general stability, the buffer pH has a less clear effect on the observed stability. Looking across the AF4 data for individual samples in terms of particle size and particle number concentration, there is no obvious trend in the data. In isolated parts of the data (ex. PTA-measured particle size), there does appear to be an overall effect, i.e., the samples in higher pH (≈8.5) buffer do show greater instability over the time series. There seems to be an interaction between storage temperature and buffer pH that is too complex to be captured fully by the study reported here, although it is certainly of interest in future work.
The effect of storage temperature becomes more apparent when examining the longitudinal data (Figure S13/S14). The samples stored at room temperature (S01, S04, S11) can be observed to show more stochastic variation when examining both the AF4 and PTA data in terms of particle diameter, but not particle number concentration. There is thus a general trend that samples stored at room temperature (≈22 °C) tended to be less stable relative to the cold-storage samples. This is expected due to increased kinetic rates of hydrolysis at elevated temperatures. However, the samples with the diether PC in place of DPPC do not show a clear trend based on the storage temperature. Therefore, we conclude that while the lipid composition is of paramount importance and the storage temperature does seem to matter, the buffer pH is less relevant (at least for the buffer pH levels tested here). Consideration of these factors is critical in consideration of solution liposome stability over a time course of at least a year.
Conclusions
This work provides evidence of the impact of various parameters of interest on the long-term stability of liposome particles in fluid solution. The findings will guide efforts toward possible development of a liposome dispersion that has sufficient stability to serve as a reference material relevant to nanomedicine. We demonstrate the strong effect that lipid composition/formulation has on particle stability as measured over a one-year timeframe. The effects of solution pH/buffer composition and storage temperature are less conclusive, but still of note. It is of utility to store such particles at cooler temperatures, if possible. As a caveat, the samples stored at the lower temperature (≈4 °C) were repeatedly cycled in and out of the temperature for measurements. This not only allows for understanding thermal cycling effects but also may complicate the analysis as an additional variable. Future work should address and isolate this as a parameter. This work also demonstrates that orthogonal characterization of these types of soft biological material is essential for rigorous analysis and understanding of how material properties vary over time. A caveat for the conclusions we draw here is that these were empty liposomes without a payload, and as recent work highlights, the payload strongly determines the overall stability.12 The conclusions presented here only apply to this simplistic case as due to liposome-drug interactions a loaded molecule or therapeutic will necessarily affect the stability of the particles. The U.S. Food and Drug Administration (FDA) has released recommended guidance for characterization of liposome-based drug products.52 It would be of interest to incorporate the guidelines laid out by the FDA into further explorations of liposome stability, particularly as we endeavor to generate a reference material. This work focused on the physicochemical attributes of the liposomes—but as the FDA document delineates—another primary focus should be structural measurement of the lipid stability over time to evaluate degradation. This measurement of the lipids as components of the liposome with a more direct measurement should be carried out to enhance understanding of the processes affecting overall particle stability. Future avenues of work for this type of endeavor would necessarily include implementing cryoprotectants and freezing the samples between each measurement to examine effects of freeze–thaw cycles as well as long-term effects of very cold (−80 °C) storage as is currently being implemented for therapeutic vectors such as the COVID-19 mRNA vaccines. Another key question is how the particle morphology evolves over time if there is some Ostwald ripening-like process which occurs in solution. Future measurements should examine the particle morphology as a function of time to assess the appropriateness of the hollow sphere type model, which is commonly implemented in the typical quantitative analysis for these types of soft, biological particles.
Acknowledgments
This work was carried out in part in the NIST Center for Nanoscale Science and Technology. CryoEM measurements were performed at the Maryland Center for Advanced Molecular Analysis (M-CAMA) located in the Institute for Bioscience and Biotechnology Research.
Data Availability Statement
Data contained in this manuscript are freely available in a publicly accessible repository at the National Institute of Standards and Technology. It is available under the following DOI: https://doi.org/10.18434/mds2-2702.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.langmuir.3c01270.
Microfabrication materials and methods description; geometric calculation of particle number concentrations based on the particle size; geometric particle diameter over time data plotted with identical ordinate scaling; total plot of particle size measurements with included error bars, along with a breakout of each time point; total plot of all particle number measurements with included error bars, along with a breakout of each time point; percentage change for particle diameter data as calculated on a point-to-point basis; percentage change for particle diameter data as calculated by using Day 0 as a pseudo-reference point; percentage change for particle number concentration data as calculated on a point-to-point basis; percentage change for particle number concentration data as calculated by using Day 0 as a pseudo-reference point; mass spectrometry of the lipids of Sample S01 at 180 days post-study; mass spectrometry of the lipids of Sample S06 at 180 days post-study; mass spectrometry of the lipids of Sample S07 at 180 days post-study; mass spectrometry of the lipids of Sample S10 at 180 days post-study; mass spectrometry of the lipids of Sample S11 at 180 days post-study; ANOVA results for the PTA particle diameter (α = 0.05); ANOVA results for PTA particle number concentration (α = 0.05); longitudinal time series analysis of particle diameter measured over time; and longitudinal time series analysis of particle number concentration measured over time (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Duan Y.; Dhar A.; Patel C.; Khimani M.; Neogi S.; Sharma P.; Kumar N. S.; Vekariya R. L. A Brief Review on Solid Lipid Nanoparticles: Part and Parcel of Contemporary Drug Delivery Systems. RSC Adv. 2020, 10, 26777–26791. 10.1039/D0RA03491F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X.; Zaks T.; Langer R.; Dong Y. Lipid Nanoparticles for mRNA Delivery. Nat. Rev. Mater. 2021, 6, 1078–1094. 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra V.; Bansal K. K.; Verma A.; Yadav N.; Thakur S.; Sudhakar K.; Rosenholm J. M. Solid Lipid Nanoparticles: Emerging Colloidal Nano Drug Delivery Systems. Pharmaceutics 2018, 10, 191. 10.3390/pharmaceutics10040191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenchov R.; Bird R.; Curtze A. E.; Zhou Q. Lipid Nanoparticles-From Liposomes to mRNA Vaccine Delivery, a Landscape of Research Diversity and Advancement. ACS Nano 2021, 15, 16982–17015. 10.1021/acsnano.1c04996. [DOI] [PubMed] [Google Scholar]
- Saraf S.; Jain A.; Tiwari A.; Verma A.; Panda P. K.; Jain S. K. Advances in Liposomal Drug Delivery to Cancer: An Overview. J. Drug Delivery Sci. Technol. 2020, 56, 101549 10.1016/j.jddst.2020.101549. [DOI] [Google Scholar]
- Man F.; Gawne P. J.; de Rosales R. T. M. Nuclear Imaging of Liposomal Drug Delivery Systems: A Critical Review of Radiolabelling Methods and Applications in Nanomedicine. Adv. Drug Delivery Rev. 2019, 143, 134–160. 10.1016/j.addr.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hammadi M. M.; Arias J. L. An Update on Liposomes in Drug Delivery: A Patent Review. Expert Opin. Ther. Pat. 2019, 29, 891–907. 10.1080/13543776.2019.1679767. [DOI] [PubMed] [Google Scholar]
- Olusanya T. O. B.; Ahmad R. R. H.; Ibegbu D. M.; Smith J. R.; Elkordy A. A. Liposomal Drug Delivery Systems and Anticancer Drugs. Molecules 2018, 23, 907. 10.3390/molecules23040907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quach; Mock J. N.; Scholpa N. E.; Eggert M. W.; Payré C.; Lambeau G.; Arnold R. D.; Cummings B. S. Role of the Phospholipase A2 Receptor in Liposome Drug Delivery in Prostate Cancer Cells. Mol. Pharmaceutics 2014, 11, 3443–3451. 10.1021/mp500174p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halamoda-Kenzaoui B.; Holzwarth U.; Roebbe G.; Bogni A.; Bremer-Hoffman S. Mapping of the Available Standards Against the Regulatory Needs for Nanomedicines. Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol. 2018, 11, e1531 10.1002/wnan.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubek Z. J.; Chen S.; Zaifman J.; Tam Y. Y. C.; Zou S. Lipid Nanoparticle and Liposome Reference Materials: Assessment of Size Homogeneity and Long-Term −70 °C and 4 °C Storage Stability. Langmuir 2023, 39, 2509–2519. 10.1021/acs.langmuir.2c02657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirpotin D. B.; Hayes M. E.; Noble C. O.; Huang Z. R.; Wani K.; Moore D.; Kesper K.; O’Brien D.; Drummond D. C. Drug Stability and Minimized Acid-/Drug-Catalyzed Phospholipid Degradation in Liposome Irinotecan. J. Pharm. Sci. 2023, 112, 416–434. 10.1016/j.xphs.2022.11.025. [DOI] [PubMed] [Google Scholar]
- Puri A.; Loomis K.; Smith B.; Lee J.-H.; Yavlovich A.; Heldman E.; Blumenthal R. Lipid-Based Nanoparticles as Pharmaceutical Drug Carriers: From Concepts to Clinic. Crit. Rev. Ther. Drug Carrier Syst. 2009, 26, 523–580. 10.1615/CritRevTherDrugCarrierSyst.v26.i6.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roces C. B.; Lou G.; Jain N.; Abraham S.; Thomas A.; Halbert G. W.; Perrie Y. Manufacturing Considerations for the Development of Lipid Nanoparticles Using Microfluidics. Pharmaceutics 2020, 12, 1095. 10.3390/pharmaceutics12111095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakney A. K.; McKay P. F.; Yus B. I.; Aldon Y.; Shattock R. J. Inside Out: Optimization of Lipid Nanoparticle Formulations for Exterior Complexation and In Vivo Delivery of saRNA. Gene Ther. 2019, 26, 363–372. 10.1038/s41434-019-0095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn A.; Reiner J. E.; Vreeland W. N.; DeVoe D. L.; Locascio L. E.; Gaitan M. Preparation of Nanoparticles by Continuous-Flow Microfluidics. J. Nanopart. Res. 2008, 10, 925. 10.1007/s11051-007-9340-5. [DOI] [Google Scholar]
- Leung A. K. K.; Tam Y. Y. C.; Chen S.; Hafez I. M.; Cullis P. R. Microfluidic Mixing: A General Method for Encapsulating Macromolecules in Lipid Nanoparticle Systems. J. Phys. Chem. B 2015, 119, 8698–8706. 10.1021/acs.jpcb.5b02891. [DOI] [PubMed] [Google Scholar]
- Hood R. R.; DeVoe D. L.; Atencia J.; Vreeland W. N.; Omiatek D. M. A Facile Route to the Synthesis of Monodisperse Nanoscale Liposomes Using 3D Microfluidic Hydrodynamic Focusing in a Concentric Capillary Array. Lab Chip 2014, 14, 2403–2409. 10.1039/C4LC00334A. [DOI] [PubMed] [Google Scholar]
- Kimura N.; Maeki M.; Sato Y.; Note Y.; Ishida A.; Tani H.; Harashima H.; Tokeshi M. Development of the iLiNP Device: Fine Tuning the Lipid Nanoparticle Size Within 10 nm for Drug Delivery. ACS Omega 2018, 3, 5044–5051. 10.1021/acsomega.8b00341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armengol X.; Estelrich J. Physical Stability of Different Liposome Compositions Obtained by Extrusion Method. J. Microencapsulation 1995, 12, 525–535. 10.3109/02652049509006783. [DOI] [PubMed] [Google Scholar]
- Briuglia M.-L.; Rotella C.; McFarlane A.; Lamprou D. A. Influence of Cholesterol on Liposome Stability and on In Vitro Drug Release. Drug Delivery Transl. Res. 2015, 5, 231–242. 10.1007/s13346-015-0220-8. [DOI] [PubMed] [Google Scholar]
- Heurtault B.; Saulnier P.; Pech B.; Proust J.-E.; Benoit J.-P. Physico-Chemical Stability of Colloidal Lipid Particles. Biomaterials 2003, 24, 4283–4300. 10.1016/S0142-9612(03)00331-4. [DOI] [PubMed] [Google Scholar]
- Sopyan I.; Sunan K. I.; Gozali D. A Review: A Novel of Efforts to Enhance Liposome Stability as Drug Delivery Approach. Syst. Rev. Pharm. 2020, 11, 555–562. [Google Scholar]
- Yu J. Y.; Chuesiang P.; Shin G. H.; Park H. J. Post-Processing Techniques for the Improvement of Liposome Stability. Pharmaceutics 2021, 13, 1023. 10.3390/pharmaceutics13071023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball R. L.; Bajaj P.; Whitehead K. A. Achieving Long-Term Stability of Lipid Nanoparticles: Examining the Effect of pH, Temperature, and Lyophilization. Int. J. Nanomed. 2017, 12, 305–315. 10.2147/IJN.S123062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P.; Hou X.; Yan J.; Du S.; Xue Y.; Li W.; Xiang G.; Dong Y. Long-Term Storage of Lipid-Like Nanoparticles for mRNA Delivery. Bioact. Mater. 2020, 5, 358–363. 10.1016/j.bioactmat.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng T.-X.; Liang D.-S.; Guo F.; Peng H.; Xu Y.-C.; Luo N.-P.; Zhang X.-Y.; Zhong H.-J. Enhanced Storage Stability of Solid Lipid Nanoparticles by Surface Modification of Comb-Shaped Amphiphilic Inulin Derivatives. Colloids Surf., B 2019, 181, 369–378. 10.1016/j.colsurfb.2019.05.061. [DOI] [PubMed] [Google Scholar]
- Thompson A. K.; Haisman D.; Singh H. Physical Stability of Liposomes Prepared from Milk Fat Globule Membrane and Soya Phospholipids. J. Agric. Food Chem. 2006, 54, 6390–6397. 10.1021/jf0605695. [DOI] [PubMed] [Google Scholar]
- Écija-Arenas A.; Román-Pizarro V.; Fernández-Romero J. M. Separation and Characterization of Liposomes using Asymmetric Flow Field-Flow Fractionation with Online Multi-Angle Light Scattering Detection. J. Chromatogr. A 2021, 1636, 461798 10.1016/j.chroma.2020.461798. [DOI] [PubMed] [Google Scholar]
- Monteiro L. O. F.; Malachias Â.; Pound-Lana G.; Magalhães-Paniago R.; Mosqueira V. C. F.; Oliveira M. C.; de Barros A. L. B.; Leite E. A. Paclitaxel-Loaded pH-Sensitive Liposome: New Insights on Structural and Physicochemical Characterization. Langmuir 2018, 34, 5728–5737. 10.1021/acs.langmuir.8b00411. [DOI] [PubMed] [Google Scholar]
- Caputo F.; Arnould A.; Bacia M.; Ling W. L.; Rustique E.; Texier I.; Mello A. P.; Couffin A.-C. Measuring Particle Size Distribution by Asymmetric Flow Field Flow Fractionation: A Powerful Method for the Preclinical Characterization of Lipid-Based Nanoparticles. Mol. Pharmaceutics 2019, 16, 756–767. 10.1021/acs.molpharmaceut.8b01033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozo A. J.; Cox M. H.; Devitt A.; Rothnie A. J.; Goddard A. D. Biophysical Analysis of Lipidic Nanoparticles. Methods 2020, 180, 45–55. 10.1016/j.ymeth.2020.05.001. [DOI] [PubMed] [Google Scholar]
- Guimarães D.; Cavaco-Paulo A.; Noguiera E. Design of Liposomes as Drug Delivery System for Therapeutic Applications. Int. J. Pharm. 2021, 601, 120571 10.1016/j.ijpharm.2021.120571. [DOI] [PubMed] [Google Scholar]
- Singh P.; Bodycomb J.; Travers B.; Tatarkiewicz K.; Travers S.; Matyas G. R.; Beck Z. Particle Size Analysis of Polydisperse Liposome Formulations with a Novel Multispectral Advanced Nanoparticle Tracking Technology. Int. J. Pharm. 2019, 566, 680–686. 10.1016/j.ijpharm.2019.06.013. [DOI] [PubMed] [Google Scholar]
- Hole P.Particle Tracking Analysis (PTA). In Characterization of Nanoparticles, 1st ed.; Hodoroaba I. V.-D., Unger W., Shard A, Eds.; Elsevier, 2020; pp 79–96. [Google Scholar]
- Zheng S. Q.; Palovcak E.; Armache J.-P.; Verba K. A.; Cheng Y.; Agard D. A. MotionCor2: Anisotropic Correction of Beam-Induced Motion for Improved Cryo-Electon Microscopy. Nat. Methods 2017, 14, 331–332. 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn A.; Stavis S. M.; Hong J. S.; Vreeland W. N.; DeVoe D. L.; Gaitan M. Microfluidic Mixing and Formation of Nanoscale Lipid Vesicles. ACS Nano 2010, 4, 2077–2087. 10.1021/nn901676x. [DOI] [PubMed] [Google Scholar]
- Stroock A. D.; Dertinger S. K. W.; Ajdari A.; Mezić I.; Stone H. A.; Whitesides G. M. Chaotic Mixer for Microchannels. Science 2002, 295, 647–651. 10.1126/science.1066238. [DOI] [PubMed] [Google Scholar]
- Hadjigeorgiou A. G.; Boudouvis A. G.; Kokkoris G. Thorough Computational Analysis of the Staggered Herrinbone Micromixer Reveals Transport Mechanisms and Enables Efficiency-Based Improved Design. Chem. Eng. J. 2021, 414, 128775 10.1016/j.cej.2021.128775. [DOI] [Google Scholar]
- Comisar C. M.; Hunter S. E.; Walton A.; Savage P. E. Effect of pH on Ether, Ester, and Carbonate Hydrolysis in High-Temperature Water. Ind. Eng. Chem. Res. 2008, 47, 577–584. 10.1021/ie0702882. [DOI] [Google Scholar]
- Bender M. L.; Ginger R. D.; Unik J. P. Activation Energies of the Hydrolysis of Esters and Amides Involving Carbonyl Oxygen Exchange. J. Am. Chem. Soc. 1958, 80, 1044–1048. 10.1021/ja01538a006. [DOI] [Google Scholar]
- Wyatt P. J. Measurement of Special Nanoparticle Structures by Light Scattering. Anal. Chem. 2014, 86, 7171–7183. 10.1021/ac500185w. [DOI] [PubMed] [Google Scholar]
- Stefanidis D.; Jencks W. P. General Base Catalysis of Ester Hydrolysis. J. Am. Chem. Soc. 1993, 115, 6045–6050. 10.1021/ja00067a020. [DOI] [Google Scholar]
- Grit M.; Underberg W. J. M.; Crommelin D. J. A. Hydrolysis of Saturated Soybean Phosphatidylcholine in Aqueous Liposome Dispersions. J. Pharm. Sci. 1993, 82, 362–366. 10.1002/jps.2600820405. [DOI] [PubMed] [Google Scholar]
- Liu J.; Jiang X.; Ashley C.; Brinker C. J. Electrostatically Mediated Liposome Fusion and Lipid Exchange with a Nanoparticle-Supported Bilayer for Control of Surface Charge, Drug Containment, and Delivery. J. Am. Chem. Soc. 2009, 131, 7567–7569. 10.1021/ja902039y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler H. Exchange and Interactions Betwen Lipid Layers at the Surface of a Liposome Solution. Biochim. Biophys. Acta, Biomembr. 1979, 555, 316–336. 10.1016/0005-2736(79)90171-8. [DOI] [PubMed] [Google Scholar]
- Tonggu L.; Wang L. Cryo-EM Sample Preparation Method for Extremely Low Concentration Liposomes. Ultramicroscopy 2020, 208, 112849 10.1016/j.ultramic.2019.112849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grit M.; de Smidt J. H.; Struijke A.; Crommelin D. J. A. Hydrolysis of Phosphatidylcholine in Aqueous Liposome Dispersions. Int. J. Pharm. 1989, 50, 1–6. 10.1016/0378-5173(89)90173-7. [DOI] [Google Scholar]
- Steinbrecher U. P.; Pritchard P. H. Hydrolysis of Phosphatidylcholine During LDL Oxidation is Mediated by Platelet-Activating Factor Acetylhydrolase. J. Lipid Res. 1989, 30, 305–315. 10.1016/S0022-2275(20)38359-0. [DOI] [PubMed] [Google Scholar]
- Ebersberger L.; Schindler T.; Kirsch S. A.; Pluhackova K.; Schambony A.; Seydel T.; Böckmann R. A.; Unruh T. Lipid Dynamics in Membranes Slowed Down by Transmembrane Proteins. Front. Cell Dev. Biol. 2020, 8, 579388 10.3389/fcell.2020.579388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch R. L.; Brown F. L. H.; Haran G. Correlated Diffusion in Lipid Bilayers. Proc. Natl. Acad. Sci. U. S. A. 2021, 118, e2113202118 10.1073/pnas.2113202118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liposome Drug Products Guidance for Industry . Services; U. S. Department of Health and Human Services, Ed., 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data contained in this manuscript are freely available in a publicly accessible repository at the National Institute of Standards and Technology. It is available under the following DOI: https://doi.org/10.18434/mds2-2702.