Abstract
In severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the non-structural protein NSP1 inhibits translation of host mRNAs by binding to the mRNA entry channel of the ribosome and, together with the 5′-untranslated region (UTR) of the viral mRNAs, allows the evasion of that inhibition. Here, we show that NSP1 mediates endonucleolytic cleavages of both host and viral mRNAs in the 5′UTR, but with different cleavage patterns. The first pattern is observed in host mRNAs with cleavages interspersed regularly and close to the 5′ cap (6–11 nt downstream of the cap). Those cleavage positions depend more on the position relative to the 5′ cap than on the sequence itself. The second cleavage pattern occurs at high NSP1 concentrations and only in SARS-CoV-2 RNAs, with the cleavages clustered at positions 45, 46 and 49. Both patterns of cleavage occur with the mRNA and NSP1 bound to the ribosome, with the SL1 hairpin at the 5′ end sufficient to protect from NSP1-mediated degradation at low NSP1 concentrations. We show further that the N-terminal domain of NSP1 is necessary and sufficient for efficient cleavage. We suggest that in the ribosome-bound NSP1 protein the catalytic residues of the N-terminal domain are unmasked by the remodelling of the α1- and α2-helices of the C-terminal domain.
Graphical Abstract
Graphical Abstract.
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a betacoronavirus from the Sarbecovirus subgenus that emerged in China in 2019 and is the causative agent of the COVID-19 pandemic (1,2). Its genomic single-stranded RNA is large (∼30 kb) and encodes non-structural, structural and additional auxiliary proteins necessary for the viral cycle. The SARS-CoV-2 genomic RNA has a positive-strand polarity, therefore the host ribosomes directly translate the non-structural proteins from the genomic RNA. These non-structural proteins are synthesized from open reading frame 1a (ORF1a) and ORF1ab to produce two large polyproteins that further undergo proteolytic cleavages by viral proteases to synthesize the non-structural proteins NSP1–NSP16. Among these, NSP1 is a major virulence factor (3). In coronavirus, NSP1 is a relatively conserved small multifunctional protein that has critical cytopathic effects that are essential for the infectious programme (4,5). During coronavirus infection, NSP1 provokes inhibition of translation of the host cellular mRNA (3), redirects the cellular translational machinery to viral transcripts (6–8), and induces a cell cycle arrest in the G0/G1 phase (9) and the specific degradation of host mRNAs (4). In the case of SARS-CoV-2, NSP1 is also a master down-regulator of the cellular antiviral responses induced by viral infection (10,11). The SARS-CoV-2 NSP1 also induces host translation shutdown. Thus, NSP1 binds to the host ribosome at the level of the mRNA entry channel by tight interactions between the ribosome and two helices α1 and α2 located in the C-terminal domain (CTD) of NSP1 (7,12–15). The binding of NSP1 induces a general host translation inhibition enabling the hijacking of the cellular translation machinery that is redirected to viral protein synthesis. For that purpose, translation of viral transcripts, which comprehend both genomic RNA and subgenomic RNAs, proceeds further in spite of the presence of NSP1 on the ribosome, through cooperation with the SL1 hairpin of all the viral transcripts (16), promoting what has been called NSP1 inhibition evasion (6–8). It was also suggested that SL1 protects the viral transcripts from RNA degradation (17). NSP1-mediated RNA degradation was demonstrated for SARS-CoV-1 (18,19). The cleavages are guided by the host ribosome and, since NSP1 has no RNase activity, the nuclease was suggested to be a host protein recruited by NSP1 on the ribosome that has not been identified so far (4,18–20). In the case of SARS-CoV-2, the mRNA degradation induced by SARS-CoV-2 NSP1 has not been clearly established so far. However, the presence of NSP1 in cells induces specific mRNA degradation among type I interferon (IFN) response-related mRNAs (21). Moreover, the cleavages occur only in the context of the host ribosome, and critical residues for the RNase activity were identified in the N-terminal domain (NTD) of NSP1 (8).
Here we show that SARS-CoV-2 NSP1 is in fact the endonuclease that has intrinsic cleavage activity without any host auxiliary factor. However, we demonstrate that this RNase activity is boosted and guided by the ribosome during translation. The cleavages occur exclusively in the 5′-untranslated region (UTR) of cellular mRNAs including β-globin mRNA and interferon-stimulated gene (ISG) mRNAs, suggesting that they occur during 5′–3′ scanning. The genomic viral RNA and the subgenomic nucleocapsid (N) mRNA are less sensitive to NSP1-induced cleavage although they are also cleaved at a high NSP1 concentration. However, the cleavage pattern is radically different on the viral transcript, which is due to the presence of SL1–SL2–SL3 hairpins on the viral RNAs.
MATERIALS AND METHODS
NSP1 overexpression and purification
Wild-type (wt) SARS-CoV-2 NSP1 and the mutants (Δ12, R99A, R124A K125A and K164A H165A) were produced as 6×His-GST-TEV-NSP1 proteins and purified to homogeneity as previously described (5,6). Briefly, the fusion proteins were expressed in Escherichia coli BL21 Rosetta (DE3) pLysS cells at 37°C to a cell density of OD600 = 0.6 and then induced with 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h at 37°C. The NSP1 proteins were first purified on an Ni-NTA Superflow resin (QIAGEN) followed by a second step on a Glutathione HiCap resin (QIAGEN). The purified fusion proteins were subjected to tobacco etch virus (TEV) protease cleavage overnight at 4°C (50/1 fusion/TEV molar ratio). Finally, the cleaved native NSP1 proteins were separated from the 6×His-GST domains using a last purification step on Ni-NTA resin that retained His-tagged glutathione S-transferase (GST) and TEV proteins. The pure NSP1 proteins were concentrated, and stored in buffer that contains 50% glycerol at −20°C.
In vitro transcription
All the mRNAs tested in this study were transcribed by run-off in vitro transcription with T7 RNA polymerase from DNA templates amplified by polymerase chain reaction (PCR) as previously described (22). A DNA fragment containing the first 900 nucleotides of SARS-COV-2 (accession no. MN908947.3) was used as a template for PCR amplifications and template synthesis for the SARS-CoV-2 genomic RNA and the subgenomic N RNA. For the other mRNAs, gene blocks were purchased from Integrated DNA Technology according to their sequences and used as PCR templates to generate the reporter mRNA by run-off transcription. The sequences used in this study were from Homo sapiens: β-globin mRNA (accession no. BC007075.1), ISG15 (accession no. NM_005101.4), ISG54 (accession no. NM_001547), interleukin-8 (IL-8) (accession no. BC013615), VIPERIN (accession no. AF442151), retinoic acid-inducible gene (RIG-I) (accession no. AF038963). The transcripts were purified by denaturing polyacrylamide gel electrophoresis (PAGE) and electro-eluted in Biotrap Schleicher&Schull apparatus. Sequences of the RNAs and primers are listed in Table 1.
Table 1.
Sequences of the RNAs and the primers used for PCR amplification
| Name | Sequence |
|---|---|
| RNAs | |
| β-Globin 5′UTR | GACAUUUGCUUCUGACACAACUGUGUUCACUAGCAACCUCAAACAGACACC |
| IL-8 | GAAGAAACCACCGGAAGGAACCAUCUCACUGUGUGUAAAC |
| ISG15 | GGCGGCUGAGAGGCAGCGAACUCAUCUUUGCCAGUACAGGAGCUUGUGCCGUGGCCCACAGCCCACAGCCCACAGCC |
| ISG54 | GGCAGAAGAGGAAGAUUUCUGAAGAGUGCAGCUGCCUGAACCGAGCCCUGCCGAACAGCUGAGAAUUGCACUGCAACC |
| CoV-sub-N | AUUAAAGGUUUAUACCUUCCCAGGUAACAAACCAACCAACUUUCGAUCUCUUGUAGAUCUGUUCUCUAAACGAACAAACUAAA |
| CoV-gen | AUUAAAGGUUUAUACCUUCCCAGGUAACAAACCAACCAACUUUCGAUCUCUUGUAGAUCUGUUCUCUAAACGAACUUUAAAAUCUGUGUGGCUGUCACUCGGCUGCAUGCUUAGUGCACUCACGCAGUAUAAUUAAUAACUAAUUACUGUCGUUGACAGGACACGAGUAACUCGUCUAUCUUCUGCAGGCUGCUUACGGUUUCGUCCGUGUUGCAGCCGAUCAUCAGCACAUCUAGGUUUCGUCCGGGUGUGACCGAAAGGUAAGAUGGAGAGCCUUGUCCCUGGUUUCAACGAGAAAAC |
| IGR | GCAAAAAUGUGAUCUUGCUUGUAAAUACAAUUUUGAGAGGUUAAUAAAUUACAAGUAGUGCUAUUUUUGUAUUUAGGUUAGCUAUUUAGCUUUACGUUCCAGGAUGCCUAGUGGCAGCCCCACAAUAUCCAGGAAGCCCUCUCUGCGGUUUUUCAGAUUAGUAGUCGAAAAACCUAAGAAAUUUACCU |
| Primers | |
| T7 β-globin | CAACAAATATTAATACGACTCACTATAGGACATTTGCTTCTGACACAACTGTGTTCA |
| T7 IL-8 | CAACAAATATTAATACGACTCACTATAGGAAGAAACCACCGGAAGGAACCATCTCACTGTGTGTAAACATGACTTCGAAAGTTTATGATCCAG |
| T7 ISG15 | CAACAAATATTAATACGACTCACTATAGGCGGCTGAGAGGCAGCGAACTCATCTTTGCCAGTACAGGAGCTTGTGCCGTGGCCCACAGCCCACAGCCCACAGCCATGACTTCGAAAGTTTATGATCCAG |
| T7 ISG54 | CAACAAATATTAATACGACTCACTATAGGCAGAAGAGGAAGATTTCTGAAGAGTGCAGCTGCCTGAACCGAGCCCTGCCGAACAGCTGAGAATTGCACTGCAACCATGACTTCGAAAGTTTATGATCCAG |
| T7 CoV | CAACAAATATTAATACGACTCACTATAGATTAAAGGTTTATACCTTCCCAGG |
| T7 IGR | CAACAAATATTAATACGACTCACTATAGGCAAAAATGTGATCTTGCTTG |
| Rev Renilla | ATGACTTCGAAAGTTTATGATCCAGAACAAAGGAAACAACAACAACAACAACAACAACAACAACAC |
RNA 5′ labelling
The purified RNA transcripts were labelled at their 5′ end by the addition of a radioactive cap. This was performed by the Vaccine Capping Enzyme with the ScriptCap m7G capping system from CELLSCRIPT in the presence 25 μCi of [32P]αGTP (>6000 Ci/mmol). The 5′-radiolabelled transcript was purified by denaturing PAGE and recovered by passive elution.
RNA degradation assay
RNA degradation assays were performed with cell-free translation extracts prepared from in-house-made rabbit reticulocyte lysates (RRLs) as previously described (6). Briefly, reactions were carried out in a 11 μl reaction with 5′-radiolabelled RNAs (50 000 cpm) incubated at 30°C for 5 min in the presence of RNasin (Promega) and 0.1–0.5 μM of pure recombinant NSP1: wt NSP1, NTD (1–127), R99A, R124A K125A and K164A H165A. The reactions were stopped by addition of 10 μl of formamide dye. For RNA degradation assay with pure proteins, the cleavages were performed with pure proteins in a specific buffer (20 mM Tris–HCl, pH 7.5, 0.25 mM spermidine, 2 mM dithiothreitol, 2.5 mM MgCl2) and incubated for 30 min at 37°C. The cleavage products were analysed by 12% denaturing PAGE. To map the cleavage sites, a denaturing RNase T1 ladder was migrated in parallel to the cleavage products.
In vitro translation with rabbit reticulocyte lysate
Translation reactions were performed in in-house-made RRL extracts as previously described (22). Reactions were incubated at 30°C for 60 min and included 100 and 200 nM of each RNA transcript and 5 μCi of [35S]Met (>1000 Ci/mmol). Aliquots of translation reactions were analysed by 10% sodium dodecylsulphate (SDS)–PAGE followed by phosphorimaging or by luciferase assays.
RESULTS AND DISCUSSION
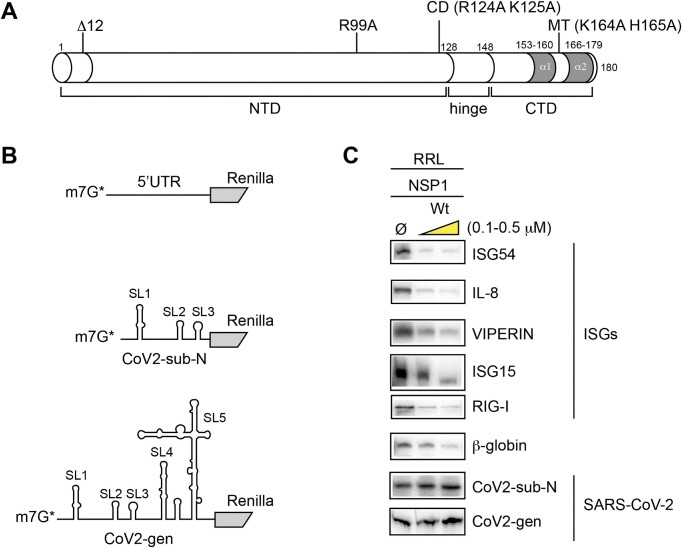
To investigate the mRNA degradation during SARS-CoV-2 infection, we purified recombinant wild-type NSP1 and mutated proteins (Figure 1A). We purified the mutant R99A which does not promote mRNA degradation but is still able to bind efficiently to the ribosome (8). We also purified two other mutants that were first described in SARS-CoV-1 (20), a double mutant that contains the substitutions K164A and H165A that are important for ribosome binding (mutant MT) and another double mutant containing the substitutions R124A and K125A that does not promote RNA degradation (mutant CD for cleavage deficient) (19,20,23). Since residues 124, 125, 164 and 165 are conserved between NSP1 from SARS-CoV-1 and SARS-CoV-2 (5), we hypothesized that the same mutations would induce the same phenotype in SARS-CoV-2 NSP1. Indeed, both mutants MT and CD have the expected effects on SARS-CoV-2 NSP1, namely the MT mutant does not bind to the host ribosome (12) and the CD mutant does not promote efficient RNA degradation (8). In addition, it has been shown that an N-terminally truncated variant of SARS-CoV-1 containing an NSP1 lacking the 12 N-terminal residues is associated with lower viral load, lower serum IFN-β production and, in general, less severe disease, suggesting that the N-terminus of NSP1 has critical functions for the viral cycle (24). Similarly, a variant of SARS-CoV-2 NSP1 lacking the 12 N-terminal amino acids is less efficient for promoting NSP1 evasion during viral translation, further confirming the functional importance of the N-terminus of NSP1 (5). This was also confirmed by expressing this truncated NSP1 variant in vivo (14). Therefore, we also purified a truncated version of NSP1 that lacks the first N-terminal 12 amino acids (Δ12). Purified recombinant NSP1 proteins were incubated in the presence of various model mRNAs. We constructed chimeric reporter mRNAs that contain variable 5′UTRs upstream of the Renilla coding sequence (Figure 1B). As standard model mRNAs, we used the H. sapiens β-globin 5′UTR, the SARS-CoV-2 5′-leader sequence of the genomic RNA (CoV2-gen) and the subgenomic viral RNA coding for nucleocapsid N (CoV2-sub-N), and the 5′UTRs of ISG, ISG15, ISG54, VIPERIN, IL-8 and RIG-I mRNAs.
Figure 1.
(A) Cartoon representing the peptide sequence of SARS-CoV-2 NSP1. The protein consists of three domains, a long NTD (1–116), a hinge domain (117–149) and a CTD (150–180). In this study, we purified recombinant NSP1 wt and four mutants. The amino acid substitutions and the positions of the mutations are indicated above the cartoon. In the CTD, two helices called α1 and α2 are shown in grey; they are required for ribosome binding. (B) The cartoon represents three types of RNA substrates. First, chimeric RNAs containing the 5′UTR of cellular mRNAs (ISG54, IL-8, VIPERIN, ISG15, RIG-I and β-globin) inserted upstream of the N-terminal part of the Renilla luciferase coding sequence, second chimeric RNA containing the 5′UTR of SARS-CoV-2 subgenomic RNA coding for the nucleocapsid (CoV2-sub-N) and third chimeric RNA containing the 5′UTR of the SARS-CoV-2 genomic RNA (Cov2-gen). (C) RNA degradation assay of various reporter mRNAs. The full-length 5′-labelled RNA transcripts are incubated for 5 min at 30°C in the presence of RRL in the absence (Ø) or presence of 0.1 and 0.5 μM wt NSP1 protein and analysed by denaturing 12% PAGE.
The mRNAs were radiolabelled at their 5′ end by the addition of a radioactive m7G cap. Then, the designed radioactive mRNAs were incubated in cell-free translation extracts made from rabbit reticulocytes in the presence of recombinant wt NSP1 protein (Figure 1C). All the tested mRNAs were degraded in the presence of wt NSP1 except CoV2-gen and CoV2-sub-N (Figure 1C). Interestingly, both SARS-CoV-2 RNAs are not degraded in the presence of NSP1 but rather are stabilized, indicating that the SARS-CoV-2 leader sequence contains protective elements that prevent degradation.
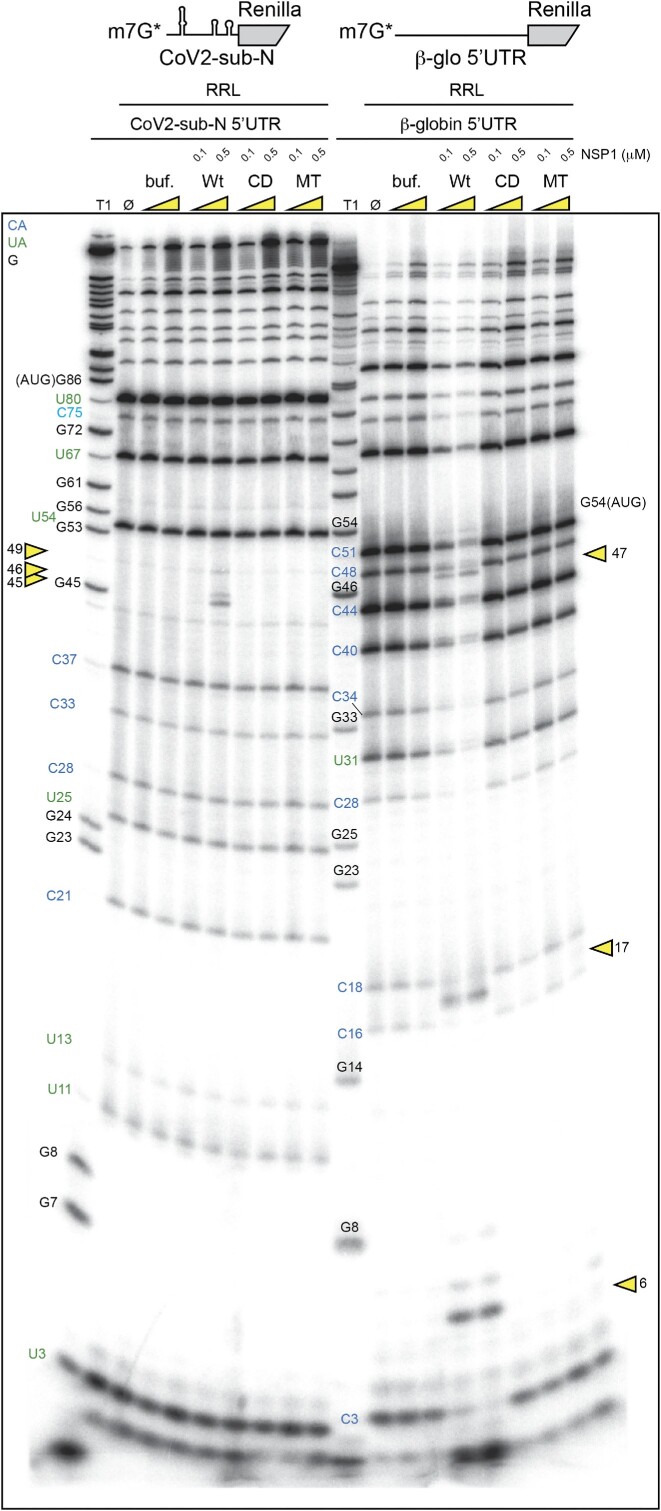
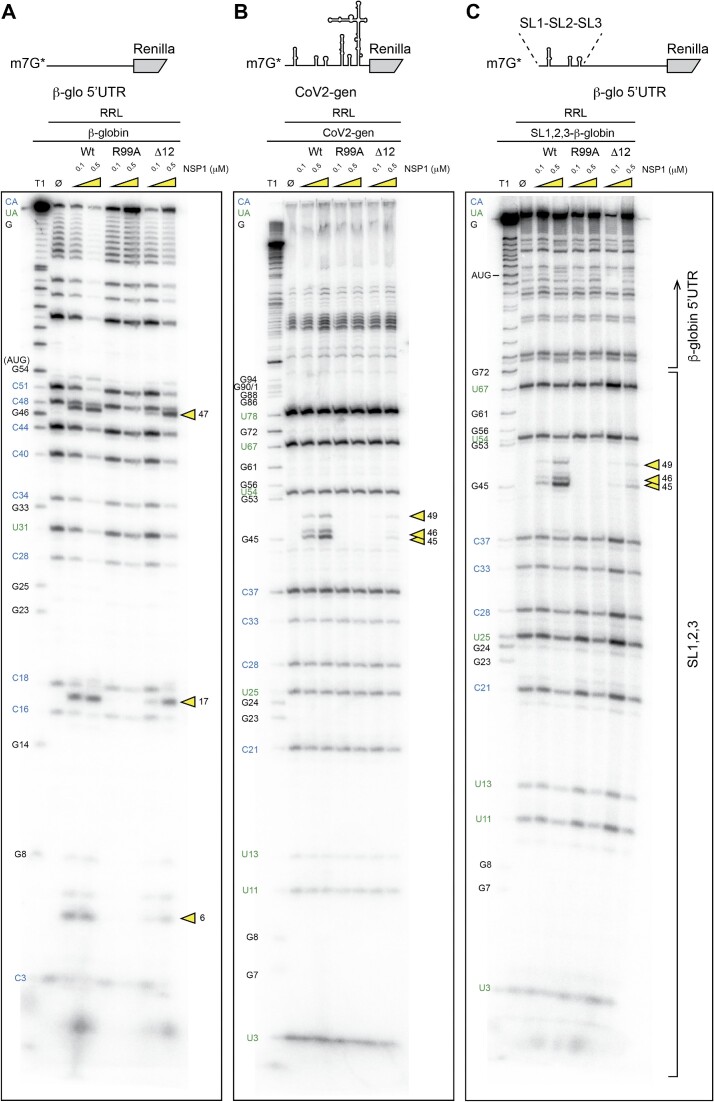
Next, we precisely determined the positions of the NSP1-induced cleavages by analysing the cleavage products by electrophoresis on denaturing polyacrylamide gels. We mapped the cleavage sites by comparison with a ladder obtained by RNase T1 digestion that cuts after the G residues (Figure 2). We found three major cleavage sites in the β-globin 5′UTR at positions 6, 16 and 47. All these cleavages are in the 5′UTR upstream of the AUG start codon. The cleavage pattern for CoV2-sub-N RNA is drastically different; three cleavages are clustered at positions 45, 46 and 49 (Figure 2). Furthermore, CoV2-sub-N RNA is only cleaved in the presence of much higher concentrations of NSP1 than β-globin mRNA, consistent with its greater resistance to degradation shown in Figure 1.
Figure 2.
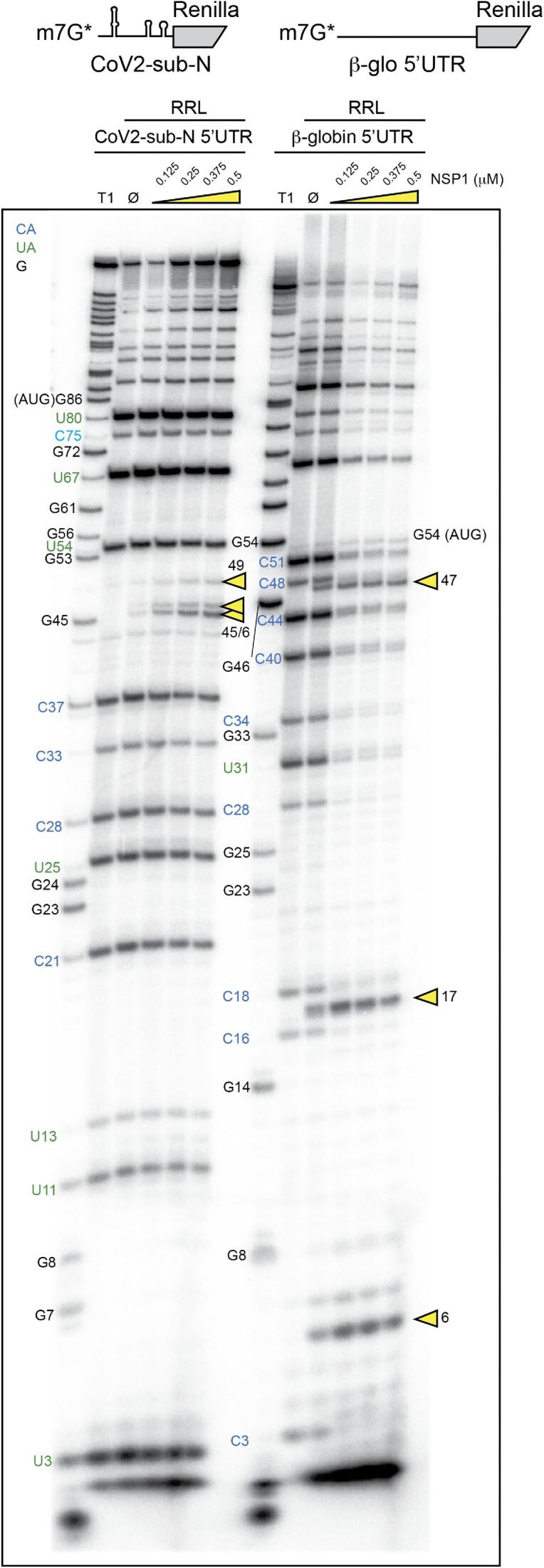
The cartoons represent the mRNAs used for the RNA degradation assay. SARS-CoV-2 subgenomic RNA (CoV2-sub-N) contains SL1–SL2–SL3 from the 5′UTR of the subgenomic RNA coding for the nucleocapsid N upstream of the N-terminal part of the Renilla luciferase coding sequence. The second RNA contains the 5′UTR of H. sapiens β-globin mRNA upstream of the N-terminal part of the Renilla luciferase coding sequence. Both RNAs have been radiolabelled at their 5′ end by the addition of a radioactive cap (*). The RNA degradation assay consists of incubating the radioactive RNAs in RRLs for 5 min at 30°C in the absence (Ø) or presence of 0.125, 0.25, 0.375 and 0.5 μM wt SARS-CoV-2 NSP1 and analysed by denaturing 12% PAGE. A T1 ladder made by RNase T1 digestion is loaded in parallel to map the cleavage site positions (T1). The cleavage sites are indicated by yellow arrowheads.
Using the same mRNAs, we searched for cleavages induced by the different NSP1s (Figure 3). Highly reproducible cleavage patterns were observed with wt NSP1 but not with the cleavage-deficient mutant CD, confirming that residues R124 and K125 are critical for RNA degradation as previously shown (8,20). This result clearly shows that the cell-free translation extract used here fully recapitulates NSP1-mediated RNA degradation. Similarly, no cleavages were detected with mutant MT, a mutant NSP1 that does not bind the ribosomes. Therefore, NSP1-mediated cleavages are only induced when NSP1 is bound to the ribosome, as already shown for SARS-CoV-1 (20). By comparing SARS-CoV-1 and SARS-CoV-2 NSP1, we found that two of the three cleavage sites are obtained with both proteins, indicating that both proteins have similar modes of action (Supplementary Figure S1). Interestingly, the cleavages found at high NSP1 concentration with SARS-CoV-2 NSP1 are not detected with SARS-CoV-1 NSP1, corroborating co-evolution between NSP1 and the 5′UTR as previously shown (5) (Supplementary Figure S2).
Figure 3.
The cartoons represent the mRNAs used for the RNA degradation assay. SARS-CoV-2 subgenomic RNA (CoV2-sub-N) contains SL1–SL2–SL3 from the 5′UTR of the subgenomic RNA coding for the nucleocapsid N upstream of the N-terminal part of the Renilla luciferase coding sequence. The second RNA contains the 5′UTR of H. sapiens β-globin mRNA upstream of the N-terminal part of the Renilla luciferase coding sequence. Both RNAs have been radiolabelled at their 5′ end by the addition of a radioactive cap (*). The RNA degradation assay consists of incubating the radioactive RNAs in RRLs for 5 min at 30°C in the absence (Ø) or presence of 0.1 and 0.5 μM wt or mutant CD (R124A K125A) and MT (K164A H165A) of SARS-CoV-2 NSP1 and analysed by denaturing 12% PAGE. A control lane with the storage buffer of NSP1 proteins is also loaded (buf). The black numbers indicate the position of G residues that are detected in the RNase T1 ladder (T1). The blue numbers indicate the position of the C residues from the spontaneous CA breaks that can be detected on the polyacrylamide gel and the green numbers indicate the position of the U residues from the spontaneous UA breaks that can be detected on the polyacrylamide gel. The cleavage sites are indicated by yellow arrowheads.
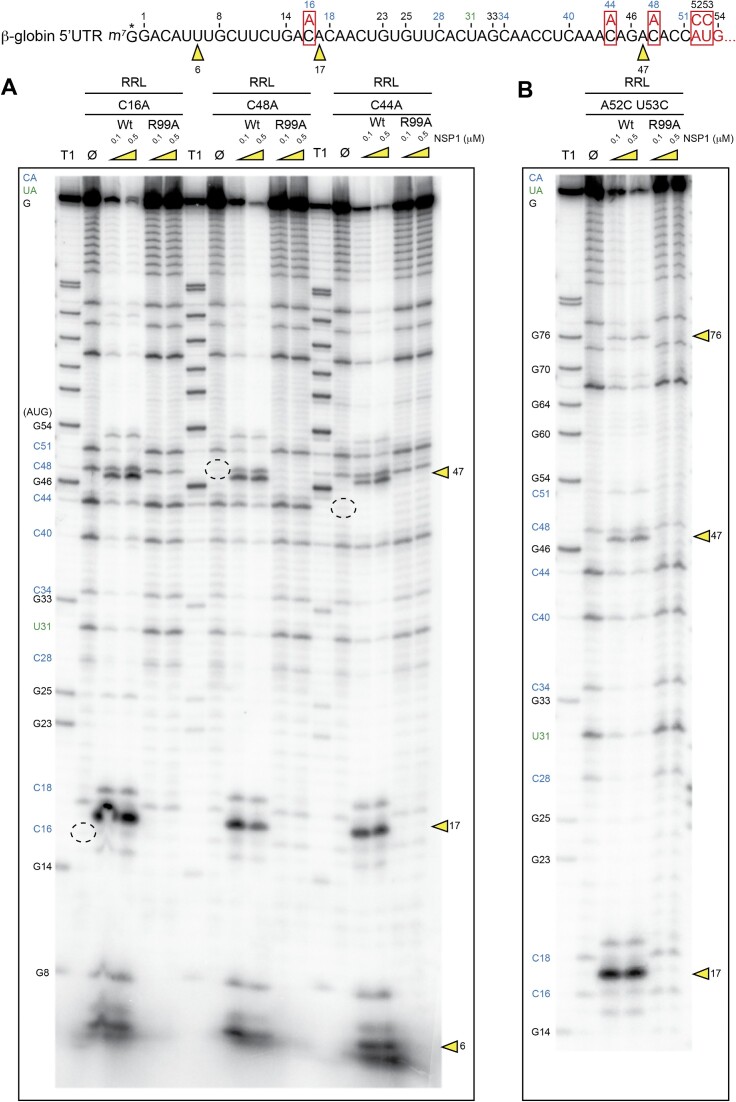
The cleavages that are found in the β-globin 5′UTR are located at or near CA residues. Since it is well established that CA dinucleotide steps are very susceptible to hydrolytic cleavages in RNA molecules (25–27), we examined the possibility that the NSP1-induced cleavages result from spontaneous hydrolytic cleavages in the RNA rather than genuine endonucleolytic cleavages. The electrophoretic analysis shown in Figure 4 effectively detects spontaneous cleavage sites as observed on RNAs incubated in cell-free translation extracts in the absence of NSP1 (Figure 4A, lanes Ø, blue numbers for CA cleavages and green numbers for UA cleavages). To exclude the possibility that the cleavages observed with NSP1 are spontaneous hydrolytic cleavages, possibly enhanced by the addition of recombinant NSP1, we introduced mutations in the mRNA in order to change the CA sequences into AA sequences which are more stable and resistant to hydrolysis. Indeed, these mutations stabilize the RNA since several spontaneous cleavages are no longer observed with C16A, C44A and C48A mutants (Figure 4A, dashed circles). In contrast, the NSP1 cleavages remain unchanged in the mutant RNAs, indicating that NSP1-mediated cleavages do not result from hydrolysis in the RNA but rather are specifically induced by NSP1 bound to the ribosome. In support of that conclusion, these cleavages are not observed with NSP1 mutant R99A, indicating that this critical residue at position 99 in NSP1 is required for efficient cleavage as previously reported (8). Moreover, mutations at the cleavage site (C16A) or in the flanking sequences of the cleavage site (C44A and C48A) do not alter the NSP1-induced cleavage pattern, indicating that the cleavage positions are not linked to a specific RNA sequence at the cleavage site, but rather to their position in the RNA. As already mentioned, all the cleavages sites were mapped in the 5′UTR of the β-globin mRNA. Therefore, we introduced a double mutation A52C U53G in the RNA that destroys the AUG start codon (AUG to CCG). In this RNA context, the 5′UTR is extended, with the appearance of an additional cleavage at position 76 in addition to the cleavage at position 47 (Figure 4B). This shows that the introduction of a longer 5′UTR induces an additional NSP1-mediated cleavage. These results indicate that the cleavages most probably occur during the 5′–3′ scanning step. To strengthen this hypothesis, we tested an RNA that does not promote scanning. We used uncapped cricket paralysis virus (CrPV) intergenic region (IGR) and did not detect any cleavage on this reporter mRNA, thereby corroborating that cleavages occur during the 5′–3′ scanning step (Supplementary Figure S3).
Figure 4.
The sequence of the 5′UTR of H. sapiens β-globin mRNA is shown. The four mutated RNAs have been radiolabelled at their 5′ end by the addition of a radioactive cap (*). The RNA degradation assay consists of incubating the radioactive RNAs in RRLs for 5 min at 30°C in the absence (Ø) or presence of 0.1 and 0.5 μM wt or R99A SARS-CoV-2 NSP1 and analysed by denaturing 12% PAGE. (A) Three mutated RNAs that change CA sequences to AA. (B) Mutant that changes the AUG codon to CCG and extends the length of the 5′UTR. The black numbers indicate the position of G residues that are detected in the RNase T1 ladder (T1). The blue numbers indicate the position of the C residues from the spontaneous CA breaks that can be detected on the polyacryamide gel and the green numbers indicate the position of the U residues from the spontaneous UA breaks that can be detected on the polyacryamide gel. The mutated residues are boxed. The cleavage sites are indicated by yellow arrowheads.
Next, we compared the NSP1 degradation pattern of the CoV2-gen RNA leader sequence with that of the CoV2-sub-N 5′UTR. In both RNAs tested, the same cleavages clustered at positions 45, 46 and 49 are found in the presence of wt NSP1 (Figure 5). Since both RNAs share only the 5′ part of the molecule containing the hairpins SL1, SL2 and SL3, we concluded that the 5′-proximal sequence of the RNA is critical to program the NSP1 cleavages and that the second segment of the RNA (SL4, SL5) has no impact on NSP1 degradation.
Figure 5.
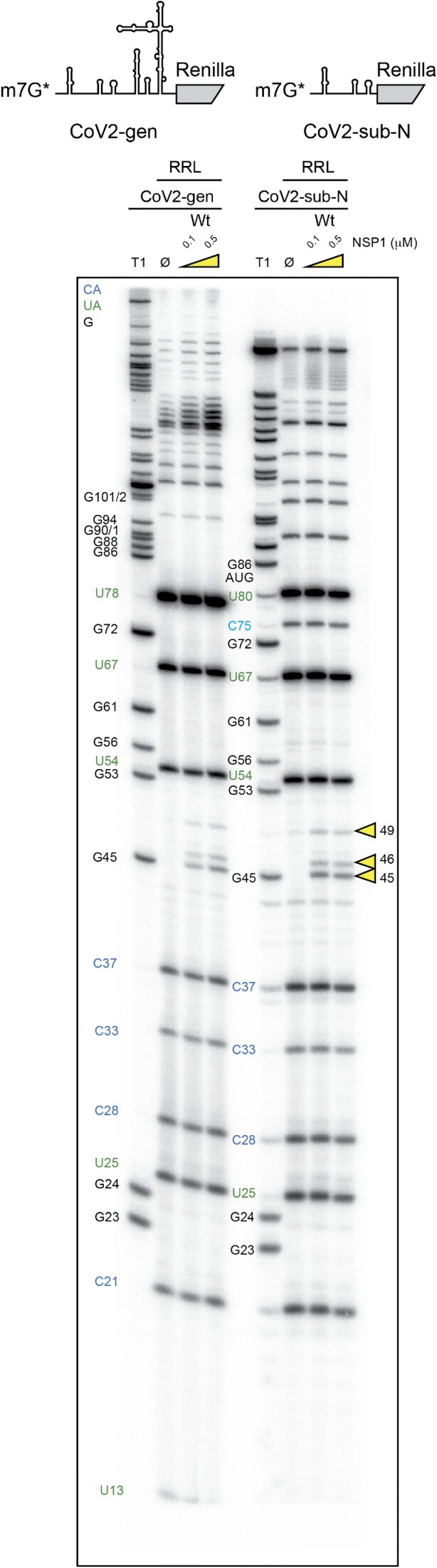
The cartoons represent the mRNAs used for the RNA degradation assay. SARS-CoV-2 genomic RNA (CoV2-gen) contains SL1–SL2–SL3–SL4–SL5 from the 5′UTR of the genomic RNA upstream of the N-terminal part of the Renilla luciferase coding sequence. The second RNA contains SL1–SL2–SL3 from the 5′UTR of the subgenomic RNA coding for the nucleocapsid N upstream of the N-terminal part of the Renilla luciferase coding sequence. Both RNAs have been radiolabelled at their 5′ end by the addition of a radioactive cap (*). The RNA degradation assay consists of incubating the radioactive RNAs in RRLs for 5 min at 30°C in the absence (Ø) or presence of 0.1 and 0.5 μM wt or R99A SARS-CoV-2 NSP1 and analysed by denaturing 12% PAGE. The black numbers indicate the position of G residues that are detected in the RNase T1 ladder (T1). The blue numbers indicate the position of the C residues from the spontaneous CA breaks that can be detected on the polyacryamide gel and the green numbers indicate the position of the U residues from the spontaneous UA breaks that can be detected on the polyacryamide gel. The cleavage sites are indicated by yellow arrowheads.
We then compared the degradation pattern of the β-globin mRNA with that obtained with CoV2-gen leader RNA. As previously mentioned, the NSP1-mediated cleavage patterns are drastically different (Figures 3 and 6A, B). The three cleavages detected in the β-globin 5′UTR are observed with wt and Δ12 NSP1 but are not observed with R99A. In the case of SARS-CoV-2 leader RNA, the clustered cleavages are found only with the wt NSP1 but not with Δ12, suggesting another distinct feature. Since the Δ12 NSP1 mutant is not able to promote efficient evasion of translation inhibition, possibly due to altered interaction with SL1 (5), this result suggests that SL1 might also be required for specific degradation of viral transcripts by NSP1. As previously mentioned, and shown in Figure 5, the role of 5′-proximal sequences is critical to program NSP1-mediated cleavages. To further corroborate this statement, we compared the cleavage patterns of β-globin RNA (Figure 6A) and the CoV2 genomic RNA (Figure 6B) with a chimeric β-globin RNA containing, at the 5′ end, the SL1, SL2 and SL3 hairpins upstream of the β-globin 5′UTR (Figure 6C). The observed degradation pattern was identical to that detected with the SARS-CoV-2 genomic leader RNA (Figure 6B, C). This confirms that the 5′-proximal sequence of the mRNA is involved in the NSP1 cleavage activity programming. Consistent with this statement, the chimeric RNA was not cleaved by the Δ12 NSP1 mutant, a specific feature that has only been observed with SARS-CoV-2 RNAs (Figure 6B, C) in contrast to the β-globin RNA that is efficiently cleaved by the Δ12 variant (Figure 6A). To further examine the influence of SL1, we generated reporter mRNAs containing a complete deletion of SL1 in CoV2-sub-N and a mutant that contains four mutations in the apical part of SL1 in CoV2-gen. Both mutated reporters are degraded in the presence of NSP1 (Supplementary Figure S4). More precisely, the degradations are mapped close to the 5′ cap (Supplementary Figures S5 and S6). Altogether, these experiments demonstrate that the SL1 hairpin at the 5′ end is sufficient to protect from NSP1-mediated degradation.
Figure 6.
The cartoons represent the mRNAs used for the RNA degradation assay. (A) The first RNA contains the 5′UTR of H. sapiens β-globin mRNA upstream of the N-terminal part of the Renilla luciferase coding sequence. (B) SARS-CoV-2 genomic RNA (CoV2-gen) contains SL1–SL2–SL3–SL4–SL5 from the 5′UTR of the genomic RNA upstream of the N-terminal part of the Renilla luciferase coding sequence. (C) The third RNA is a chimeric RNA that contains SL1–SL2–SL3 from the 5′UTR of the subgenomic RNA coding for the nucleocapsid N upstream of the 5′UTR of H. sapiens β-globin mRNA followed by the N-terminal part of the Renilla luciferase coding sequence. The three RNAs have been radiolabelled at their 5′ end by the addition of a radioactive cap (*). The RNA degradation assay consists of incubating the radioactive RNAs in RRLs for 5 min at 30°C in the absence (Ø) or presence of 0.1 and 0.5 μM wt or R99A SARS-CoV-2 NSP1 and analysed by denaturing 12% PAGE. The black numbers indicate the position of G residues that are detected in the RNase T1 ladder (T1). The blue numbers indicate the position of the C residues from the spontaneous CA breaks that can be detected on the polyacryamide gel and the green numbers indicate the position of the U residues from the spontaneous UA breaks that can be detected on the polyacryamide gel. The cleavage sites are indicated by yellow arrowheads.
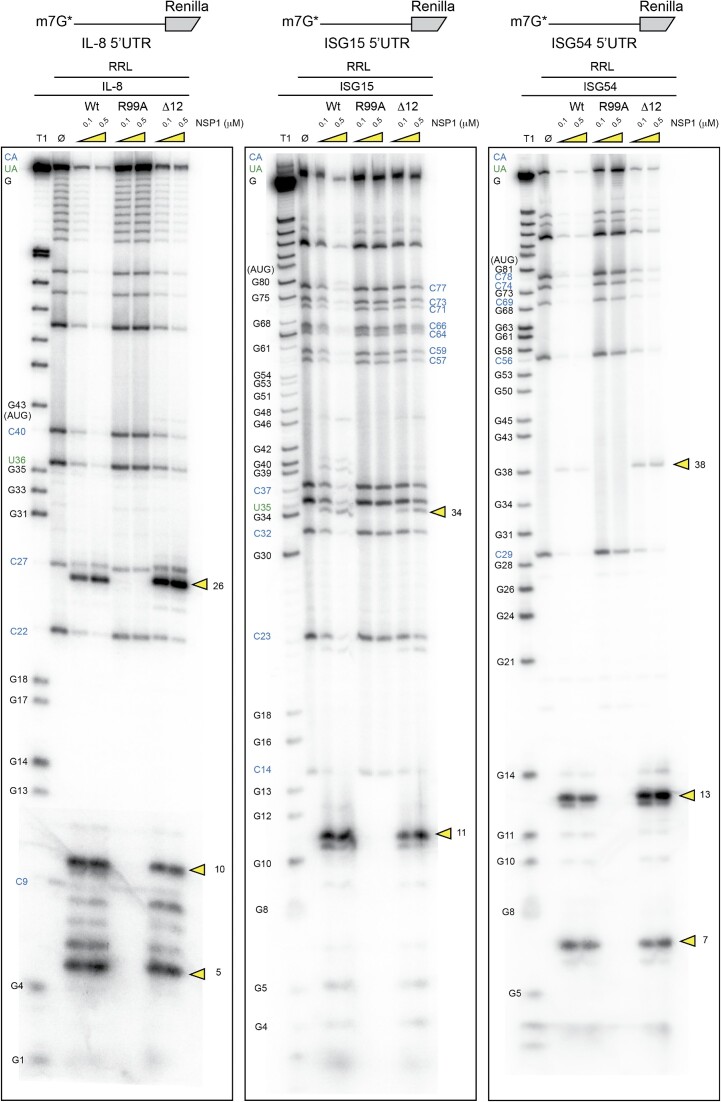
Next, we examined other mRNAs that are physiologically relevant to the viral cycle. It has been shown that NSP1 has a critical cytopathic effect on the antiviral response by promoting the specific degradation of ISG mRNAs (21). Therefore, we tested the IL-8, ISG15 and ISG54 mRNAs for specific NSP1-mediated cleavages (Figure 7). Three cleavages were found in IL-8 at positions 5, 11 and 26. For ISG15, only two cleavages at positions 11 and 34 were detected and for ISG54, three cleavages at positions 7, 13 and 38. All these cleavages are NSP1 specific; they are induced by both wt and Δ12 NSP1 but not by R99A as expected. These mRNAs show similar cleavage patterns to β-globin mRNAs and do not require the N-terminus of NSP1 since the Δ12 NSP1 mutant promotes efficient cleavages.
Figure 7.
The cartoons represent the mRNAs used for the RNA degradation assay. The first RNA contains the 5′UTR of H. sapiens IL-8 mRNA upstream of the N-terminal part of the Renilla luciferase coding sequence. The second RNA contains the 5′UTR of H. sapiens ISG15 mRNA upstream of the N-terminal part of the Renilla luciferase coding sequence. The third RNA contains the 5′UTR of H. sapiens ISG54 mRNA upstream of the N-terminal part of the Renilla luciferase coding sequence. The three RNAs have been radiolabelled at their 5′ end by the addition of a radioactive cap (*). The RNA degradation assay consists of incubating the radioactive RNAs for 5 min at 30°C in RRLs in the absence (Ø) or presence of 0.1 and 0.5 μM wt or R99A SARS-CoV-2 NSP1 and analysed by denaturing 12% PAGE. The black numbers indicate the position of G residues that are detected in the RNase T1 ladder (T1). The blue numbers indicate the position of the C residues from the spontaneous CA breaks that can be detected on the polyacryamide gel and the green numbers indicate the position of the U residues from the spontaneous UA breaks that can be detected on the polyacryamide gel. The cleavage sites are indicated by yellow arrowheads.
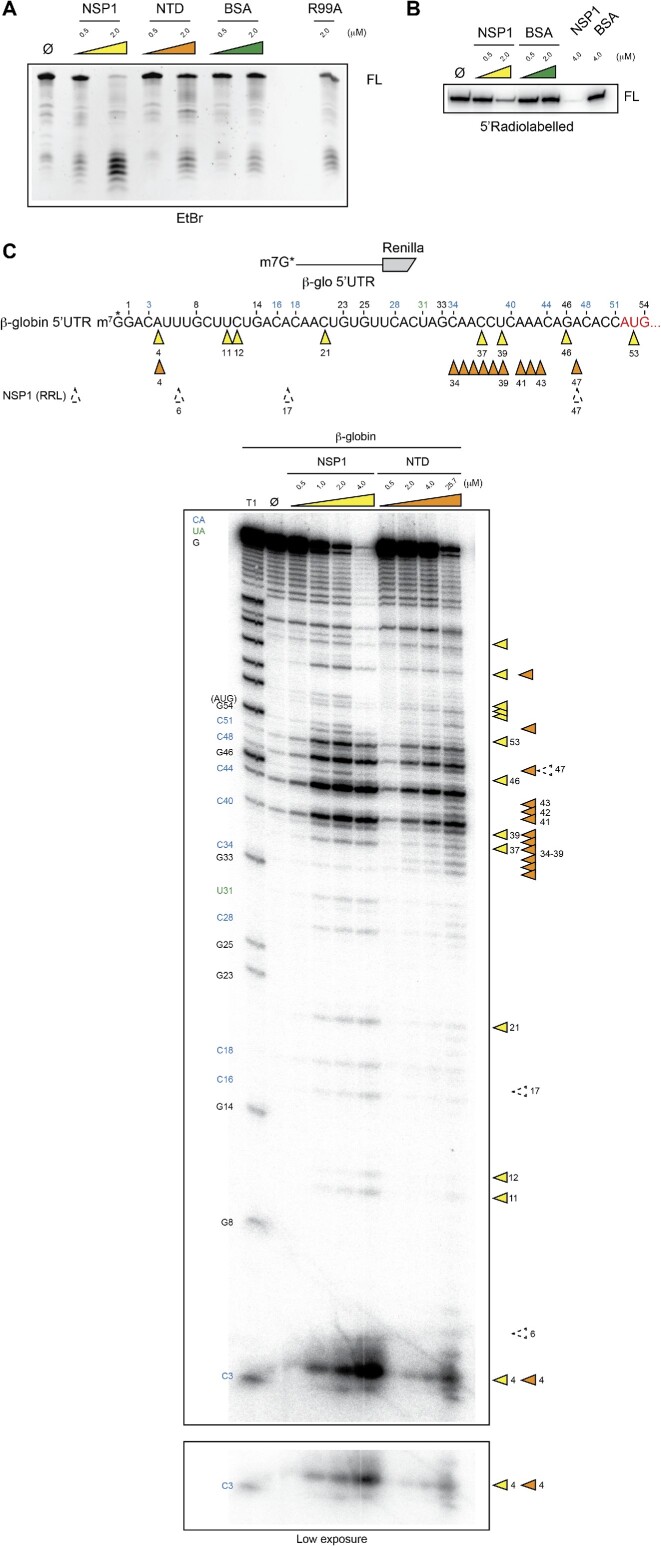
We further investigated the cleavage activity of NSP1 in its free state with pure protein. Interestingly, we could detect degradation of the β-globin 5′UTR reporter with 0.5 and 2 μM free NSP1 (Figure 8A). We also tested the isolated NTD of NSP1 and found a nuclease activity although with a lower efficiency. No RNA degradation was detected with bovine serum albumin (BSA) at the same concentrations. Importantly, the absence of cleavage with mutant R99A strongly suggests that the cleavage is performed by NSP1 and not by a putative contaminant that could have been co-purified. Next, we mapped the cleavages using 5′ radiolabelled RNA substrates and confirmed NSP1-specific degradation (Figure 8B). The mapping of the cleavages indicates endonucleolytic activity with partial overlap of the cleavage sites between NSP1 and the NTD (Figure 8C). Here again, a higher concentration of NTD is required for efficient cleavage. Furthermore, the cleavage sites detected with free NSP1 are located at distinct positions from those of the cleavages detected when NSP1 is bound to the ribosome (Figure 8C, dashed arrows). These experiments demonstrate that NSP1 has intrinsic endonuclease activity located in its NTD. When NSP1 is carried by the pre-initiation complex during the scanning step, the cleavages are regularly interspersed and restricted to the 5′UTR. In its free state, NSP1 retains endonucleolytic activity but the cleavages are distributed along the whole mRNA even in the coding sequence beyond the AUG start codon. This suggests that AUG recognition by the pre-initiation complex possibly induces NSP1 remodelling that prevents further cleavages in the coding sequence.
Figure 8.
(A) RNA degradation assay in the presence of pure wt NSP1 (0.5–2 μM), the isolated NTD (0.5–2 μM), BSA (0.5–2 μM) or R99A NSP1 (2 μM). A 500 ng aliquot of unlabelled RNA that contains the 5′UTR of H. sapiens β-globin mRNA upstream of the N-terminal part of the Renilla luciferase coding sequence was incubated in the presence of the proteins for 30 min at 37°C and analysed by denaturing 6% PAGE. The full-length RNA (FL) and the degradation products are visualized by ethidium bromide staining (B) RNA degradation assay in the presence of pure wt NSP1 (0.5–2 μM), the isolated NTD (0.5–2 μM) or BSA (0.5–2 μM). 5′-Capped radiolabelled RNA that contains the 5′UTR of H. sapiens β-globin mRNA upstream of the N-terminal part of the Renilla luciferase coding sequence was incubated in the presence of the proteins for 30 min at 37°C and analysed by denaturing 12% PAGE. (C) The cartoons represent the mRNAs used for the RNA degradation assay. The RNA contains the 5′UTR of H. sapiens β-globin mRNA upstream of the N-terminal part of the Renilla luciferase coding sequence. The RNA has been radiolabelled at the 5′ end by the addition of a radioactive cap (*). The RNA degradation assay consists of incubating the radioactive RNAs for 30 min at 37°C in the absence (Ø) or presence of pure wt NSP1 (0.5–4 μM) or the isolated NTD (0.5–25.7 μM) and analysed by denaturing 12% PAGE. The black numbers indicate the positions of the G residues that are detected in the RNase T1 ladder (T1). The blue numbers indicate the position of the C residues from the spontaneous CA breaks that can be detected on the polyacryamide gel and the green numbers indicate the position of the U residues from the spontaneous UA breaks that can be detected on the polyacryamide gel. The NSP1 cleavage sites are indicated by yellow arrowheads and NTD cleavage sites are indicated by orange arrowheads. The cleavage sites that are detected when NSP1 is bound on the ribosome are indicated by dashed arrows.
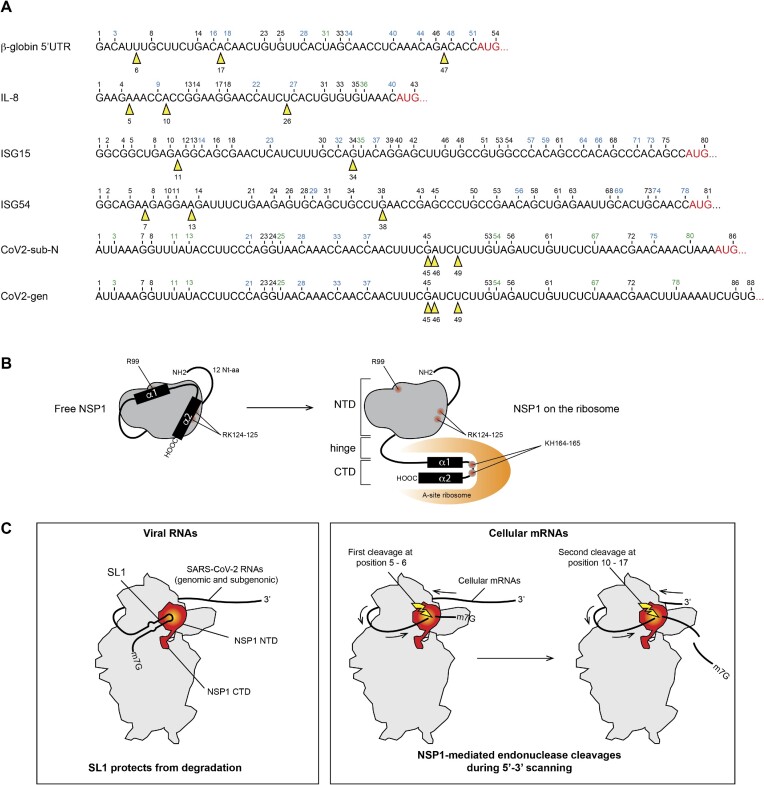
Altogether, we show that NSP1 can promote RNA degradation in the 5′UTR of mRNAs. We detected two distinct cleavage patterns (Figure 9A). The first cleavage pattern is observed with β-globin model mRNA and the cellular mRNAs tested (ISG mRNAs). The first type of cleavage usually occurs very close to the 5′ cap (6–11 nt downstream of the cap) and other cuts are regularly interspersed in the 5′UTR. The position of the cleavages does not seem to depend on the sequence itself but rather on the position relative to the 5′ cap. In agreement with this, we found a cleavage in H. sapiens β-globin mRNA at position 16. A cleavage at the same position has also been found in the rabbit globin mRNA in the presence of SARS-CoV-1 NSP1 (18). Since human and rabbit β-globin mRNAs do not have the same sequence, this indicates that the cleavage position is more dependent on the distance to the 5′ cap than on the sequence itself. The conservation of the cleavage positions most probably relies on the conservation of resides R99, R124 and K125 in both SARS-CoV-1 and SARS-CoV-2 NSP1 (5).
Figure 9.
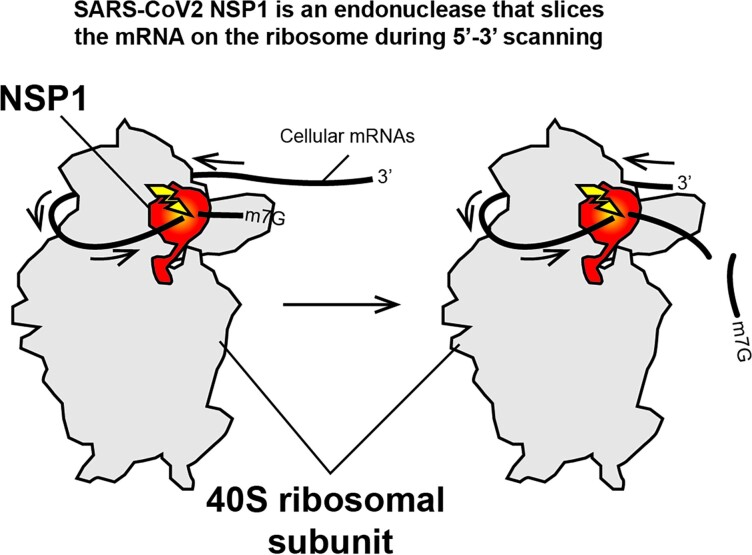
(A) The sequences of the 5′UTR tested in this study are shown. The black numbers correspond to the position of G residues that are detected by RNase T1 digestion in the T1 ladder. The positions of NSP1-mediated cleavages are shown by yellow arrowheads. (B) Cartoon showing the NSP1 topology. In a free state, helices α1 and α2 are positioned on the N-terminal globular domain (grey) of NSP1, thereby masking residues R99, R124 and K125. NSP1 binding to the ribosome induces an important structural rearrangement; helices α1 and α2 interact specifically with the ribosome mRNA channel, which liberates the residues R99, R124 and K125 that are critical for endonuclease activity. (C) Model of endonuclease cleavage of NSP1 on the ribosome. NSP1 (in red) binds to the 40S ribosomal subunit (in grey). The NTD binds to the beak of the 40S subunit and the CTD binds in the mRNA channel and inhibits translation. The SL1 hairpin that is present on viral RNAs interacts directly or indirectly with NSP1 and thereby blocks the endonuclease cleavage site that is located in the NTD (in yellow). Cellular mRNAs do not contain SL1; they undergo endonucleolytic cleavages during 5′–3′ scanning. The first cleavage (shown by a yellow flash) occurs at position 5–6, then during subsequent sliding of the RNA, the second cleavage occurs at position 10–17.
The second cleavage pattern is drastically different. It is found only in SARS-CoV-2 RNAs with cleavages clustered at positions 45, 46 and 49, and only at higher concentrations of NSP1 than for cellular mRNAs (Figure 9A). In addition, SARS-CoV-2 RNAs are not cleaved by the Δ12 NSP1 variant, in contrast to the other mRNAs that are cleaved with the same efficiency as with the wt NSP1. Based on these observations, we propose the model for NSP1-mediated cleavages shown in Figure 9B. The present results confirm that residues R99A, R124 and K125 are critical for efficient endonuclease activity as previously shown (8,18). The cleavages are observed only when NSP1 is bound to the ribosome, and a mutant of NSP1 unable to bind to the ribosome cannot promote mRNA cleavage. Structural studies showed that the structure of NSP1 can be drastically remodelled (28,29). Indeed, while the C-terminal part of the molecule is clearly stable and visible in the NSP1-40S structure solved by cryo-electron microscopy, the N-terminus cannot be observed because of too high flexibility (12–14). In contrast, in the free NSP1 structures, the NTD is well ordered while the first 12 residues and the whole CTD are usually deleted to obtain crystals (30,31). Fortunately, a recent nuclear magentic resonance (NMR) structure of the full-length NSP1 has been solved (28). The structure shows that the NTD of NSP1 is well structured and partially masked by the ∼50 amino acid long C-terminal disordered tail. The tail interacts with the RNA-binding surface, and residues R99, R124 and K125 are not accessible in the free NSP1 state (28). Since these residues are critical for efficient cleavage on the ribosome, we propose the following model (Figure 9B). In its free state, NSP1 residues R99, R124 and K125 are hidden by the C-terminal disordered domain. Binding to the ribosome induces dramatic allosteric remodelling of the NSP1 structure, thereby liberating the NTD and inducing the folding of the two helices of the CTD for efficient ribosome interaction (28). In this configuration, the residues R99, R124 and K125 are in the required configuration to promote efficient cleavage. In this model, the presence of the ribosome is required both to remodel the structure of the NSP1 and to confer its properties, i.e. its ability to bind to the ribosome via its NTD and CTD and to bind to mRNA via its NTD residues. The release of residues R99, R124 and K125, which participate directly or indirectly in the endonuclease activity, completes the interaction pattern of the three partners, ribosome, NSP1 and mRNA. The participation of a cellular RNase as an additional partner responsible for cleavage has been suggested but never confirmed, while an RNase activity of NSP1 could not be characterized so far. Here, we demonstrate that NSP1 is indeed the cleavage factor and the NTD is necessary and sufficient for efficient cleavage. Although we cannot totally exclude a co-purified RNase contamination, the fact that the R99A mutant does not cleave at 2 μM strongly suggests that the cleavage activity is brought by NSP1 on its own (Figure 8A). In addition, since the N-terminal extension is necessary for efficient cleavage of SARS-CoV-2 RNA, we hypothesize that this extension interacts with SL1 of the mRNA leader sequence (Figure 9C). This is in good agreement with previous experiments showing that the NTD is essential for NSP1 function and that it co-evolved with SL1 in coronaviruses, suggesting a direct contact between SL1 and the 12 N-terminal amino acids of NSP1 (5). This interaction maintains the RNA in an appropriate orientation to induce clustered cleavages at high concentration of NSP1. Variants of SARS-CoV-2 with truncated NSP1 lacking these residues are associated with a lower viral load, lower serum IFN-β and less severe disease (32,33), which further suggests that these residues are critical for the optimal viral cycle. In the case of cellular mRNAs, the NTD is not required and no interactions with the mRNAs are made. Since the cleavages are interspaced regularly and only present in the 5′UTR, we propose that the cleavages occur during 5′–3′ scanning (Figure 9C). After cleavages, the resulting mRNAs lack a 5′ cap, can no longer be translated and will be rapidly further degraded by the host degradation machinery.
Concerning the cleavages that are detected in the SARS-CoV-2 5′UTR at a high concentration of NSP1 (0.5 μM), we hypothesize that they are potentially used for regulation purposes at the end of the viral cycle, when the NSP1 concentration is high. We determined the impact of these cleavages on translation efficiency by constructing truncated mRNAs mimicking the cleaved RNAs. We found that the RNA with the largest deletion (Δ49) was still efficiently translated, even in the absence of a cap (Supplementary Figure S7). Moreover, the cleavage products are no longer sensitive to NSP1 inhibition, most probably because they do not carry SL1. The exact role of these cleavages in the viral RNA will require more investigations to determine the impact on the viral cycle.
Supplementary Material
ACKNOWLEDGEMENTS
Our laboratory is funded by the Agence Nationale pour la Recherche, the Fondation pour la Recherche Médicale (project CoronaIRES), the University of Strasbourg and the Centre National de la Recherche Scientifique. We would also like to thank Cecilia Arraiano for fruitful discussions on our project.
Contributor Information
Yann Tardivat, Université de Strasbourg, Institut de Biologie Moléculaire et Cellulaire, Architecture et Réactivité de l’ARN, CNRS UPR9002, 2, allée Konrad Roentgen, F-67084 Strasbourg, France.
Piotr Sosnowski, Université de Strasbourg, Institut de Biologie Moléculaire et Cellulaire, Architecture et Réactivité de l’ARN, CNRS UPR9002, 2, allée Konrad Roentgen, F-67084 Strasbourg, France.
Antonin Tidu, Université de Strasbourg, Institut de Biologie Moléculaire et Cellulaire, Architecture et Réactivité de l’ARN, CNRS UPR9002, 2, allée Konrad Roentgen, F-67084 Strasbourg, France.
Eric Westhof, Université de Strasbourg, Institut de Biologie Moléculaire et Cellulaire, Architecture et Réactivité de l’ARN, CNRS UPR9002, 2, allée Konrad Roentgen, F-67084 Strasbourg, France.
Gilbert Eriani, Université de Strasbourg, Institut de Biologie Moléculaire et Cellulaire, Architecture et Réactivité de l’ARN, CNRS UPR9002, 2, allée Konrad Roentgen, F-67084 Strasbourg, France.
Franck Martin, Université de Strasbourg, Institut de Biologie Moléculaire et Cellulaire, Architecture et Réactivité de l’ARN, CNRS UPR9002, 2, allée Konrad Roentgen, F-67084 Strasbourg, France.
DATA AVAILABILITY
The authors confirm that the data supporting the findings of this study are available within the article and/or its supplementary data.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Centre National de la Recherche Scientifique; Fondation pour la Recherche Médicale [project CoronaIRES]; Agence Nationale pour la Recherche, [ANR-17-CE12-0025-01 and ANR-20-COVI-0078]; and the University of Strasbourg.
Conflict of interest statement. E.W. is an Executive Editor of NAR.
REFERENCES
- 1. Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L.et al.. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020; 579:270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Worobey M. Dissecting the early COVID-19 cases in Wuhan. Science. 2021; 374:1202–1204. [DOI] [PubMed] [Google Scholar]
- 3. Eriani G., Martin F.. Viral and cellular translation during SARS-CoV-2 infection. FEBS Open Bio. 2022; 12:1584–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nakagawa K., Makino S.. Mechanisms of coronavirus NSP1-mediated control of host and viral gene expression. Cells. 2021; 10:300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sosnowski P., Tidu A., Eriani G., Westhof E., Martin F.. Correlated sequence signatures are present within the genomic 5’UTR RNA and NSP1 protein in coronaviruses. RNA. 2022; 28:729–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tidu A., Janvier A., Schaeffer L., Sosnowski P., Kuhn L., Hammann P., Westhof E., Eriani G., Martin F.. The viral protein NSP1 acts as a ribosome gatekeeper for shutting down host translation and fostering SARS-CoV-2 translation. RNA. 2021; 27:253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Banerjee A.K., Blanco M.R., Bruce E.A., Honson D.D., Chen L.M., Chow A., Bhat P., Ollikainen N., Quinodoz S.A., Loney C.et al.. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell. 2020; 183:1325–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mendez A.S., Ly M., González-Sánchez A.M., Hartenian E., Ingolia N.T., Cate J.H., Glaunsinger B.A.. The N-terminal domain of SARS-CoV-2 NSP1 plays key roles in suppression of cellular gene expression and preservation of viral gene expression. Cell Rep. 2021; 37:109841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shen Z., Zhang G., Yang Y., Li M., Yang S., Peng G.. Lysine 164 is critical for SARS-CoV-2 Nsp1 inhibition of host gene expression. J. Gen. Virol. 2021; 102:001513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Minkoff J.M., TenOever B.. Innate immune evasion strategies of SARS-CoV-2. Nat. Rev. Microbiol. 2023; 21:178–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kumar A., Ishida R., Strilets T., Cole J., Lopez-Orozco J., Fayad N., Felix-Lopez A., Elaish M., Evseev D., Magor K.E.et al.. SARS-CoV-2 nonstructural protein 1 inhibits the interferon response by causing depletion of key host signaling factors. J. Virol. 2021; 95:e0026621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thoms M., Buschauer R., Ameismeier M., Koepke L., Denk T., Hirschenberger M., Kratzat H., Hayn M., MacKens-Kiani T., Cheng J.et al.. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020; 369:1249–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schubert K., Karousis E.D., Jomaa A., Scaiola A., Echeverria B., Gurzeler L.A., Leibundgut M., Thiel V., Mühlemann O., Ban N.. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat. Struct. Mol. Biol. 2020; 27:959–966. [DOI] [PubMed] [Google Scholar]
- 14. Yuan S., Peng L., Park J.J., Hu Y., Devarkar S.C., Dong M.B., Shen Q., Wu S., Chen S., Lomakin I.B.et al.. Nonstructural protein 1 of SARS-CoV-2 is a potent pathogenicity factor redirecting host protein synthesis machinery toward viral RNA. Mol. Cell. 2020; 80:1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lapointe C.P., Grosely R., Johnson A.G., Wang J., Fernández I.S., Puglisi J.D.. Dynamic competition between SARS-CoV-2 NSP1 and mRNA on the human ribosome inhibits translation initiation. Proc. Natl Acad. Sci. USA. 2021; 118:e2017715118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miao Z., Tidu A., Eriani G., Martin F.. Secondary structure of the SARS-CoV-2 5′-UTR. RNA Biol. 2021; 18:447–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Finkel Y., Gluck A., Nachshon A., Winkler R., Fisher T., Rozman B., Mizrahi O., Lubelsky Y., Zuckerman B., Slobodin B.et al.. SARS-CoV-2 uses a multipronged strategy to impede host protein synthesis. Nature. 2021; 594:240–245. [DOI] [PubMed] [Google Scholar]
- 18. Huang C., Lokugamage K.G., Rozovics J.M., Narayanan K., Semler B.L., Makino S.. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. 2011; 7:e1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kamitani W., Huang C., Narayanan K., Lokugamage K.G., Makino S.. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat. Struct. Mol. Biol. 2009; 16:1134–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lokugamage K.G., Narayanan K., Huang C., Makino S.. Severe acute respiratory syndrome coronavirus protein nsp1 is a novel eukaryotic translation inhibitor that represses multiple steps of translation initiation. J. Virol. 2012; 86:13598–13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lei X., Dong X., Ma R., Wang W., Xiao X., Tian Z., Wang C., Wang Y., Li L., Ren L.et al.. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020; 11:3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Martin F., Barends S., Jaeger S., Schaeffer L., Prongidi-Fix L., Eriani G.. Cap-assisted internal initiation of translation of histone H4. Mol. Cell. 2011; 41:197–209. [DOI] [PubMed] [Google Scholar]
- 23. Tanaka T., Kamitani W., DeDiego M.L., Enjuanes L., Matsuura Y.. Severe acute respiratory syndrome coronavirus nsp1 facilitates efficient propagation in cells through a specific translational shutoff of host mRNA. J. Virol. 2012; 86:11128–11137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wathelet M.G., Orr M., Frieman M.B., Baric R.S.. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J. Virol. 2007; 81:11620–11633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fuchs E., Falschlunger C., Micura R., Breuker K.. The effect of adenine protonation on RNA phosphodiester backbone bond cleavage elucidated by deaza-nucleobase modifications and mass spectrometry. Nucleic Acids Res. 2019; 47:7223–7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Soukup G.A., Breaker R.R.. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999; 5:1308–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kaukinen U., Lyytikäinen S., Mikkola S., Lönnberg H.. The reactivity of phosphodiester bonds within linear single-stranded oligoribonucleotides in strongly dependent on the base sequence. Nucleic Acids Res. 2002; 30:468–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang Y., Kirkpatrick J., Lage S.z., Carlomagno T.. Structural insights into the activity regulation of full-length non-structural protein 1 from SARS-CoV-2. Structure. 2023; 31:128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mishra N., Kant R., Leung D.W., Gross M.L., Amarasinghe G.K.. Biochemical and HDX mass spectral characterization of the SARS-CoV-2 Nsp1 protein. Biochemistry. 2023; 62:1744–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Clark L.K., Green T.J., Petit C.M.. Structure of nonstructural protein 1 from SARS-CoV-2. J. Virol. 2021; 95:e02019-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Semper C., Watanabe N., Savchenko A.. Structural characterization of nonstructural protein 1 from SARS-CoV-2. Iscience. 2021; 24:101903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Benedetti F., Snyder G.A., Giovanetti M., Angeletti S., Gallo R.C., Ciccozzi M., Zella D. Emerging of a SARS-CoV-2 viral strain with a deletion in nsp1. J. Transl. Med. 2020; 18:329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lin J.w., Tang C., Wei H.c., Du B., Chen C., Wang M., Zhou Y., Yu M.x., Cheng L., Kuivanen S.et al.. Genomic monitoring of SARS-CoV-2 uncovers an Nsp1 deletion variant that modulates type I interferon response. Cell Host Microbe. 2021; 29:489–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and/or its supplementary data.