Abstract
Gene functional descriptions offer a crucial line of evidence for candidate genes underlying trait variation. Conversely, plant responses to environmental cues represent important resources to decipher gene function and subsequently provide molecular targets for plant improvement through gene editing. However, biological roles of large proportions of genes across the plant phylogeny are poorly annotated. Here we describe the Joint Genome Institute (JGI) Plant Gene Atlas, an updateable data resource consisting of transcript abundance assays spanning 18 diverse species. To integrate across these diverse genotypes, we analyzed expression profiles, built gene clusters that exhibited tissue/condition specific expression, and tested for transcriptional response to environmental queues. We discovered extensive phylogenetically constrained and condition-specific expression profiles for genes without any previously documented functional annotation. Such conserved expression patterns and tightly co-expressed gene clusters let us assign expression derived additional biological information to 64 495 genes with otherwise unknown functions. The ever-expanding Gene Atlas resource is available at JGI Plant Gene Atlas (https://plantgeneatlas.jgi.doe.gov) and Phytozome (https://phytozome.jgi.doe.gov/), providing bulk access to data and user-specified queries of gene sets. Combined, these web interfaces let users access differentially expressed genes, track orthologs across the Gene Atlas plants, graphically represent co-expressed genes, and visualize gene ontology and pathway enrichments.
Graphical Abstract
Graphical Abstract.
INTRODUCTION
The flowering plant, Arabidopsis thaliana, has served as a model for functional genomics over the past two decades. While the goal of functionally characterizing each A. thaliana gene by the year 2010 (1) has yet to be fully realized, many large-scale studies, such as gene knock-out collections for reverse genetics, have tested the phenotypic effects of nearly half of A. thaliana protein-coding genes (2). These experimentally validated loci, and a massive set of predicted and curated gene functions form the foundation for gene characterization across 400M years of plant evolution.
Despite the potential for homology-based functional annotations across plants, putative gene functions in non-model plants are sparse, often containing a majority of genes with no functional descriptions. These knowledge gaps are undoubtedly due to the phylogenetic and functional scale of plant diversity. For the purpose of this paper, we define functional description as the role of a gene including its biological role, molecular function or any form of effect including its expression profile pertinent to an experimental condition or spatial location in an organism. At one extreme, DNA or protein sequences may have diverged so that no genes have obvious A. thaliana homologs. However, even with homology, assigning gene function to distantly related plants assumes function is evolutionarily conserved. This assumption is clearly violated in many situations: flowering plants have evolved diverse adaptive traits, specialized organs/tissues, and environmental responses, all of which are poorly captured by a single model organism. Further, gene neofunctionalization, subfunctionalization and gene cooption may invalidate direct superimposition of gene annotation from one species to another (3–5). The addition of other model species, including Brachypodium distachyon, Oryza sativa, and Physcomitrium patens, has helped fill gaps in homology-based functional annotations. However, 16.1–56.9% (M = 27.8; SD = 10.06) of protein coding genes across the plant phylogeny remain poorly characterized (Supplementary Figure S1) (6–8).
Incomplete gene functional annotations are not only due to an overreliance on few genetic model organisms, but also an inability to link experimental evidence across species. Centralized functional databases, which contain information generated from new experiments such as ongoing large-scale transcriptome projects and genome-wide association studies, could accelerate gene function discovery. However, to date, such centralized databases (e.g. Expression Atlas (9), Bgee (10) and PPRD (11)) serve primarily as a source of descriptive information within systems and require substantial downstream analysis to transfer information to new species. For example, it is currently possible to make queries about when, where, and in what conditions a particular gene is expressed; however, investigating gene functional roles or conducting cross-species expression analysis remains arduous, except when using a few available resources such as Bgee. Experimental consistency further complicates the utility of existing databases. Since these resources have been developed using curated datasets from public RNA-seq repositories, interpretation and integration across diverse studies is difficult because experimental and analytical protocols are rarely standardized. For example, different sample collection, RNA isolation, library construction protocols, and sequencing platforms can result in significant variation in sequence coverage and estimates of gene expression (12–15). This among-experiment variation reduces the accuracy and precision of comparisons across species and studies, which directly limits putative gene function inference from transcript abundance profiles.
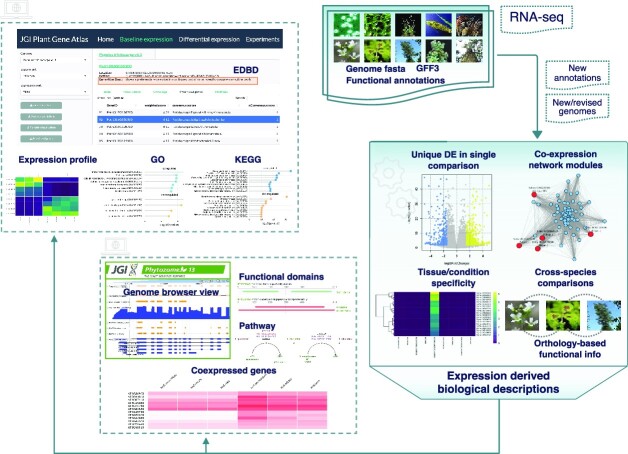
Here, we present an updateable large-scale dataset and a suite of experimental protocols to facilitate functional gene prediction across the diversity of plants. Crucially, we have standardized experimental conditions, tissue types, and analytical protocols that permit comprehensive analysis of gene expression across plants. We applied these conditions and collected 2090 tissue samples from 18 plant species spanning single-celled algae, bryophytes, and a diverse collection of flowering plants. This integrated dataset (1) forms a foundation to assign additional biological information to genes, (2) facilitates cross-species comparative transcriptomics within controlled environmental and laboratory conditions, and (3) permits high-powered tests of gene regulatory evolution across phylogenetically diverse plant genomes. To demonstrate this functionality, we cataloged the expression profiles of annotated genes, and built co-expressed clusters of genes that exhibited tissue/condition specific expression patterns including responses to changes in nitrogen (N) regimes, abiotic stressors, and developmental stages. We systematically assigned expression derived additional biological descriptors to an average of 40.6% (SD = 12.6) of annotated genes in the assessed genomes, 9.5% of which previously had no known function. The transcriptomic data and analytical resources are available to the research community at JGI Plant Gene Atlas (https://plantgeneatlas.jgi.doe.gov) and through Phytozome, the JGI Plant Portal, at https://phytozome.jgi.doe.gov/ (16).
MATERIALS AND METHODS
Plant growth and treatment conditions
Glycine max and Medicago truncatula
Plant seeds (G. max cv. Williams 82 and M. truncatula Jemalong A17) were surface-sterilized, transferred to pots containing 3:1 vermiculite perlite. 2 to 3 seedlings were planted in each pot and grown until plants were 4 weeks in a growth chamber under 16 h-light/8 h-dark conditions, 26–23°C temperature maintained at 250 μmol m−2s−1. Plants for nitrogen experiment were watered with nutrient solution containing either 10 mM KNO3 (NO3− plants) or 10 mM (NH4)3PO4 (NH4+ plants) or 10 mM urea (urea plants). We selected urea as a control condition for the counter ions, potassium and phosphate, as the best compromise. The nutrient solutions were renewed every 3 days. After 4 weeks, different tissues (leaf, stem, root, shoot, shoot tip, root tip, lateral roots, etc.) for N regimes and standard conditions were harvested. Plants under symbiotic conditions were watered with nutrient solution containing 0.5 mM NH4NO3 every other week. Subsequently, root nodules, roots, and trifoliate leaves under symbiotic conditions were collected and tissues from flower open and un-open were harvested from field grown plants.
Arabidopsis thaliana
Seeds (Columbia-0) were cold-stratified in water for 3 days and subsequently seeds were sown into 9 cm2 plastic pots (T.O. Plastics, Clearwater, FL, USA) filled with 2 parts Promix Biofungicide (Premier Tech, Riviére-du-Loup, QC, Canada) to 1 part Profile Field and Fairway (Profile, Buffalo Grove, IL, USA). Pots were placed in a growth chamber (22°C days/20°C nights, 14 h light at a photosynthetic photon flux density of 350 μmol m−2s−1), then thinned to 1 plant per pot containing Sunshine MVP potting mix (SunGro Horticulture) and transferred into a greenhouse at the University of Texas at Austin when rosettes had achieved 7–8 leaves. Plants supplemented with differing nitrogen source regimes (see Glycine max) were harvested after 30 days.
Brachypodium distachyon
Seeds (B. distachyon Bd21) were grown in Metro mix 360 soil in a growth chamber, under 12 h day and 12 h night conditions, maintained at 24°C/18°C, ∼50% relative humidity; 150 μmol m−2s−1. Plants were watered once a day or every two days depending on the size of plants and soil conditions and fertilized twice a week (Tuesday and Friday) using Jack's 15–16–17 at a concentration of 100 ppm. For the nitrogen source study, plants grown for 30 days under differing nitrogen source regimes (see Glycine max) were harvested.
For cold treatment experiment, Bd21 seeds were sown in soil without stratification. The germinated seeds were grown in a growth chamber under short day conditions (26°C 10 h light, 18°C 14 h dark) for 4 weeks and then moved to a cold room (4°C 10 h light, 4°C 14 h dark) for cold treatment. Whole shoots were harvested at different treatment time points and stored at –80°C for RNA extraction.
Chlamydomonas reinhardti
C. reinhardtii strain CC-1690 (also known as 21gr) was cultured at 24°C (agitated at 180 rpm at a photon flux density of 90 μmol m−2s−1 provided by cool white fluorescent bulbs at 4100 K and warm white fluorescent bulbs at 3000 K used in the ratio of 2:1) in tris acetate-phosphate (TAP) medium (17). For growth in differing nitrogen sources, TAP medium was supplemented with (NH4)3PO4 or KNO3, or urea (see Glycine max). Cultures of strain CC-1690 were inoculated to 1 × 105 cells ml−1 and collected for RNA at 1 × 106 cells ml−1, when the growth rates of all cultures were identical. For assessing the impact of cell density, cultures were inoculated at 1 × 104 cells ml−1 in replete medium and sampled at 5 × 105 cells ml−1 and at each doubling thereafter until the culture reached a final density of 8 × 106 cells ml−1.
Eucalyptus grandis
E. grandis samples were derived from tissues collected from clonal ramets of the genotype BRASUZ1 that was used to generate the E. grandis reference genome. Tissue samples were collected from three trees ca. 5 years old, and an adult tree ca. 8 years old at the time of sample collection, planted in experimental fields at Embrapa Genetic Resources and Biotechnology in Brasilia, Brazil (15.73 South, 47.90 West). RNA samples were prepared from adult leaves (completely developed), juvenile leaves (tender, thinner, not waxed), fruit buds, and developing cambium (from inside the tree bark). Plant material was collected from the field, immediately frozen in liquid nitrogen, and stored at -80°C until RNA extraction that followed an optimized CTAB-lithium chloride-based protocol (18).
Kalanchoë fedtschenkoi
Four-week-old K. fedtschenkoi plants (accession ORNL M2) were grown under a 250 mmol m−2 s−1 white light with a 12 h light (25°C)/12 h dark (18°C) cycle and were used as starting plant materials for eight different experiments (i.e. circadian, metabolite, temperature, drought, light intensity, light quality, nitrogen utilization, and standard tissue). The experiments were conducted under day/night temperature regime of 25°C/18°C except the temperature experiment and the circadian experiment. For the circadian experiment, two sets of plants were grown at a constant temperature of 25°C under two different lighting conditions: 1) a 12-h light (250 mmol m−2 s−1 white light)/12-h dark cycle for 16 days and 2) a 12-h light (250 mmol m−2 s−1 white light)/12-h dark cycle for 14 days followed by continuous lighting (100 μmol m−2 s−1) for four days. During the last two days of circadian experiment under each of the two lighting conditions, mature leaf samples (i.e. leaves 5–7 counting from the top of the plants) were collected every two hours over a 48 h period. For the metabolite experiment, plants were grown under an aerobic condition to prevent dark CO2 fixation and malate accumulation. This was accomplished by putting the plants in a sealed chamber with a closed air loop, through which air was continuously circulated. CO2 was subsequently continuously scrubbed from the air using a hydrated soda lime filter (LI-COR Biosciences, Lincoln NE) included in the loop. CO2 levels were monitored and maintained at an average of 3 ppm over the 12 h overnight aerobic treatment. Plants were removed from the aerobic condition just prior to the start of the daylight photoperiod. Mature leaves were harvested at 2 h intervals over the succeeding 24 h period (12 h light/12 h dark). For the temperature treatment, plants were grown under three different temperatures (8°C, 25°C and 37°C), respectively, for seven days. For drought treatments, plants were grown under three soil moisture conditions (40% ± 3% [control], 20% ± 3% [moderate drought] and 2% ± 3% [severe drought]), respectively, for 19 days. For the light intensity experiment, plants were grown under light intensity of 0 (darkness), 150 (low light) and 1000 (high light) mmol m−2 s−1 for 48 h. For the light quality experiment, plants were grown under blue light (270 mmol m−2 s−1), red light (280 mmol m−2 s−1), far-red light (280 mmol m−2 s−1) and constant darkness for 48 h. For the nitrogen utilization experiment, plants were treated with potassium nitrate (10 mM), ammonium phosphate (10 mM) and urea (10 mM), respectively, for four weeks. Immediately after the temperature, drought, light intensity, light quality and nitrogen utilization experiments, mature leaves were collected at two time points of dawn (2 h before the start of light period) and dusk (2 h before the start of dark period). For the nitrogen utilization experiment, root samples were also collected at dawn and dusk, respectively. For the standard tissue experiment, plants were grown in the greenhouse under a 12 h light/12 h dark cycle at Oak Ridge National Laboratory (Oak Ridge, TN) and five different tissue types (young leaf, young stem, mature stem, root, and flower) were collected at 10 am in the greenhouse.
Lupinus albus
RNA-seq data from cluster root samples were obtained from (19).
Panicum virgatum
Vegetatively propagated Alamo AP13 plants were grown in pre-autoclaved MetroMix 300 substrate (Sungro® Horticulture, http://www.sungro.com/) and grown in a walk-in growth chamber at 30/26°C day/night temperature with a 16 h photoperiod (250 μm−2 s−1) for four months. Tissues were harvested at six developmental stages, including leaf development (VLD: V2), stem elongation (STE: E2 and E4), and reproductive phases (REP: R2, S2, and S6) (20).
For P. virgatum photoperiod experiment, four switchgrass genotypes, AP13, WBC, AP13, and VS16 plants were vegetatively propagated and grown in one-gallon pots with a 6:1:1 mixture of Promix:Turface:Profile soil at a growth chamber at the University of Texas at Austin. After one-week maintenance with a 30/25°C day/night temperature and 14L/10D photoperiod, plants from each genotype were divided into two groups and received LD (16L/8D) or SD (8L/16D) treatment in separate growth chambers. Fully emerged young leaves were simultaneously harvested from three individuals as three biological replicates after three-week LD and SD treatments. We collected two leaf tissues (2 cm leaf tips and 2 cm leaf base) at two zeitgeber times (ZT1 and ZT17). All samples were immediately flash frozen in liquid nitrogen and stored at -80°C for DNA and RNA extraction.
Panicum hallii
The P. hallii FIL2 (var. filipes; Corpus Christi, TX; 27.65°N, 97.40°W) and P. hallii HAL2 (var. hallii; Austin, TX; 30.19°N, 97.87°W) were grown in 3.78 l pots at the University of TX Brackenridge Field Laboratory (Austin, Texas) in the greenhouse with mean daytime air temperature of 30°C and relative humidity of 65%. Plants supplemented with differing nitrogen source regimes (see Glycine max) were harvested after 30 days.
For P. hallii panicle samples, genotypes, HAL2 and FIL2, were grown in a growth chamber at University of Texas at Austin with 26°C day/22°C night temperature and 12 h photoperiod. Plants were grown in 3.5 inches square pots with a 6:1:1 mixture of Promix:Turface:Profile soil. Young panicle tissues were collected under a dissection microscope and the developmental stages were determined according to the lengths (0.1–0.2 cm for D1 stage, 0.5–1 cm for D2 stage, 4.5–5.5 cm for D3 stage, and 9–11 cm for D4 stage). Tissues for D1 and D2 stages were taken from at least fifty plants and pooled for each biological replicate. Tissues for D3 and D4 stages were taken from at least fifteen plants and pooled for each biological replicate. All samples were harvested at 17:00–18:00 of the day and immediately flash frozen in liquid nitrogen. Three biological replicates for each stage were stored at -80°C for DNA and RNA extraction (more details in (21)).
Physcomitrium patens
The protonemata cultures (ecotype Gransden was used for all samples except for two sporophyte sets) were systematically entrained by two successive weeks of culture prior to treatment to obtain a homogeneous culture as described in Perroud et al. (22). In brief, BCD (23) or Knop medium (24) were used to culture the moss. Solid medium (medium with 1% [w/v] agar) protonemal cultures were grown atop a cellophane film to allow tissue transfer for specific treatments (e.g. with hormones), and for ease of harvesting. Plates and flasks were cultivated at 22°C with a 16 h-light/8 h-dark regime under 60–80 μmol m−2s−1 white light (long-day conditions). All harvests were performed in the middle of the light photoperiod (+8 h of light in long day conditions) (22,25).
Populus trichocarpa
Populus trichocarpa (Nisqually-1) cuttings were potted in 4′ × 4″ × 5″ containers containing 1:1 mix of peat and perlite. Plants were grown under 16 h-light/8 h-dark conditions, maintained at 20–23°C and an average of 235 μmol m−2s−1 to generate tissue for (1) standard tissues and (2) nitrogen source study. Plants for standard tissue experiment were watered with McCown's woody plant nutrient solution and plants for nitrogen experiment were supplemented with either 10mM KNO3 (NO3− plants) or 10mM (NH4)3PO4 (NH4+ plants) or 10 mM urea (urea plants). Once plants reached leaf plastochron index 15 (LPI-15), leaf, stem, root, and bud tissues were harvested and immediately flash frozen in liquid nitrogen and stored at -80°C until further processing was done.
The plant material for the seasonal time course study was obtained from 2-year-old branches and apical buds (understood as the top bud of each branch) of 5-year-old hybrid poplar (Populus tremula × alba INRA 717 1B4) trees planted at the Centre for Plant Biotechnology and Genomics (CBGP) in Pozuelo de Alarcón, Madrid (3º49′W, 40º24′N), growing under natural conditions. Stem samples were collected weekly from 7 November 2014 to 9 April 2015. Buds were collected weekly from 13 January to 14 April 2015. For each time point, stem portions from 8 trees and 25 apical buds from 8 trees were pooled. RNA extraction was performed using the protocol described in (26). For the gene expression analysis, the weekly data were divided into groups named; fall, winter, and spring. Letter suffixes - ‘a, b, c, d, e’ were added to group names representing ‘early,’ ‘mid,’, ‘late’, ‘fortnight-1’ or ‘fortnight-2’ based on sampling dates within each season, following the Northern Meteorological Seasons dates.
Setaria italica and Setaria viridis
Seeds (S. italica B100 and S. viridis A10.1) were sown in flats (4 × 9 inserts/flat) containing Metro mix 360 soil and grown in a growth chamber, under 12 h day and 12 h night conditions, maintained at 31°C/22°C, 50–60% humidity; 450 μmol m−2s−1. Plants were watered once a day or every two days depending on the size of plants and soil conditions and fertilized twice a week (Tuesday and Friday) using Jack's 15–16–17 at a concentration of 100 ppm. For light treatment experiments, plants were grown under continuous monochromatic light, blue: 6 μmol m−2 s−1, red: 50 μmol m−2 s−1, far-red: 80 μmol m−2 s−1, respectively and watered with RO water every 3 days. Total aerial tissues were collected (at 9.30 AM) from 8-day old seedlings.
Sorghum bicolor
The reference line BTx623 was grown under 14 h day greenhouse conditions in topsoil to generate tissue for two separate experiments: (i) a nitrogen source study and (ii) a tissue by developmental stage timecourse. For the nitrogen source study, plants grown under differing nitrogen source regimes (see Glycine max) were harvested at 30 days after emergence (DAE). For the tissue by developmental stage timecourse, plants were harvested at the juvenile stage (8 DAE), the vegetative stage (24 DAE), at floral initiation (44 DAE), at anthesis (65 DAE), and at grain maturity (96 DAE) and leaf, root, stem and reproductive structures as described in McCormick et al. (27).
Sorghum bicolor var. Rio
Genetic material for S. bicolor var. Rio was obtained from a single seed source provided by W. Rooney at Texas A&M University. Plants were grown in greenhouse conditions and material for RNA extraction was collected at six biological stages: vegetative (5-leaf), floral initiation, flag leaf, anthesis, soft dough, and hard dough. Stages were identified based on biological characteristics defined in (28). At every stage, whole plants were harvested, and the topmost fully developed leaf and topmost internode were collected. During the first three stages, meristems were isolated from the topmost internode while floral and seed tissues were collected after plants had flowered. All tissues were immediately placed in RNA Later and stored at 4°C prior to RNA extraction. See also (29).
Sphagnum angustifolium (formally S. fallax)
S. angustifolium (collected from Marcell Experimental Forest, SPRUCE S1-Bog, 47.506639, −93.455897) were grown on BCD agar media pH 6.5, ambient temperature (20°C) and 350 μmol m−2 s−1 of photosynthetically active radiation (PAR) at a 12 h light/dark cycle for 2 months prior to initiation of experimental conditions. At 8 am on the morning of the treatments, Sphagnum plantlets were transferred to petri dishes with 15 ml of appropriate BCD liquid media and placed in a temperature-controlled growth cabinet. Excluding the dark treatment, all samples were kept under 350 PAR for the duration of the experiment. Morning treatment samples were harvested at noon. After each experiment the material was blotted dry, placed in a 15 mL Eppendorf tube, flash frozen in liquid nitrogen, and stored at -80°C until RNA extractions were completed.
For the control treatment, Sphagnum plants were placed in a 22.05 cm2 petri dish containing BCD media 6.5 pH and incubated in a growth cabinet at 20°C and ambient light 350 PAR. To test low pH gene expression, the sample was placed in a 22.05 cm2 petri dish containing 6.5 pH BCD media at 8 AM. Each hour, the pH was gradually decreased until the sample was transferred to 3.5 pH media at 11 a.m.. The samples were harvested at 12 p.m. This treatment was repeated for the high pH experiment except the sample was gradually brought from 6.5 to 9.0 pH. Temperature experiments were controlled in growth cabinets plantlets in 22.05 cm2 petri dishes containing 6.5 pH BCD media. The high temperature treatment began at 20°C and over three hours, temperature was gradually increased to 40°C. The low temperature treatment began at 20°C and over three hours, was gradually decreased to 6°C. To test water loss effects on gene expression, plantlets were placed on dry plates (no BCD media) for the duration of the experiment. Dark effect on gene expression was tested by placing plant material in a BCD filled petri dish in complete darkness from 8 AM to 12 PM. To evaluate gene expression that is present during immature growth stages, a sporophyte was collected from the mother of the S. angustifolium pedigree and germinated on solid Knop medium under axenic tissue culture conditions. After 10 days of growth, plantlets were predominantly within the thalloid protonemata with rhizoid stage and flash frozen in LN2 until RNA extraction using CTAB lysis buffer and Spectrum Total Plant RNA kit.
RNA extraction and sequencing
All tissues were immediately flash frozen in liquid nitrogen and stored at –80°C until further processing was done. Every harvest involved at least three independent biological replicates for each condition. High quality RNA was extracted mainly using standard Trizol-reagent based extraction (30), exceptions noted above under individual species. The integrity and concentration of the RNA preparations were checked initially using Nano-Drop ND-1000 (Nano-Drop Technologies) and then by BioAnalyzer (Agilent Technologies). Plate-based RNA sample prep was performed on the PerkinElmer Sciclone NGS robotic liquid handling system using Illumina's TruSeq Stranded mRNA HT sample prep kit utilizing poly-A selection of mRNA following the protocol outlined by Illumina in their user guide: http://support.illumina.com/sequencing/sequencing_kits/truseq_stranded_mrna_ht_sample_prep_kit.html, and with the following conditions: total RNA starting material was 1 μg per sample and 8 cycles of PCR was used for library amplification. The prepared libraries were then quantified by qPCR using the Kapa SYBR Fast Illumina Library Quantification Kit (Kapa Biosystems) and run on a Roche LightCycler 480 real-time PCR instrument. The quantified libraries were then prepared for sequencing on the Illumina HiSeq sequencing platform utilizing a TruSeq paired-end cluster kit, v4, and Illumina's cBot instrument to generate a clustered flow cell for sequencing. Sequencing of the flow cell was performed on the Illumina HiSeq2500 sequencer using HiSeq TruSeq SBS sequencing kits, v4, following a 2 × 150 indexed run recipe. The same standardized protocols were used to prevent introduction of any batch effects among samples throughout the project.
RNA-seq data normalization and differential gene expression analysis
Illumina RNA-seq 150 bp paired-end strand-specific reads were processed using custom Python scripts to trim adapter sequences and low-quality bases to obtain high quality (Q ≥ 25) sequence data. Reads shorter than 50 bp after trimming were discarded. The processed high-quality RNA-seq reads were aligned to current reference genomes (see Table 1) of Gene Atlas using GSNAP, a short read alignment program (31). HTSeq v1.99.2, a Python package was used to count reads mapped to annotated genes in the reference genome (32).
Table 1.
JGI Plant Gene Atlas species. Genome annotation versions of 18 diverse plants included in the current release
| Genome | Version | Project | Taxonomy ID | Source |
|---|---|---|---|---|
| Arabidopsis thaliana | TAIR10 | Gene Atlas | 3702 | phytozome.jgi.doe.gov/info/Athaliana_TAIR10 |
| Araport11 | phytozome.jgi.doe.gov/info/Athaliana_Araport11 | |||
| Brachypodium distachyon | v3.1 | Gene Atlas | 5143 | phytozome.jgi.doe.gov/info/Bdistachyon_v3_1 |
| Chlamydomonas reinhardtii | v5.6 | Gene Atlas | 3055 | phytozome.jgi.doe.gov/info/Creinhardtii_v5_6 |
| Eucalyptus grandis | v2.0 | Gene Atlas | 71 139 | phytozome.jgi.doe.gov/info/Egrandis_v2_0 |
| Glycine max | Wm82.a4.v1 | Gene Atlas | 3847 | phytozome.jgi.doe.gov/info/Gmax_Wm82_a4_v1 |
| Kalanchoë fedtschenkoi | v1.1 | Gene Atlas | 63 787 | phytozome.jgi.doe.gov/info/Kfedtschenkoi_v1_1 |
| Lupinus albus | v1.1 | Non-JGI | 3870 | phytozome.jgi.doe.gov/info/Lalbus_v1 |
| Medicago truncatula | Mt4.0v1 | Gene Atlas | 3880 | phytozome.jgi.doe.gov/info/Mtruncatula_Mt4_0v1 |
| Panicum hallii var. filipes | v3.1 | Gene Atlas | 907 226 | phytozome.jgi.doe.gov/info/Phallii_v3_1 |
| Panicum hallii var. hallii | v2.1 | Gene Atlas | 1 504 633 | phytozome.jgi.doe.gov/info/PhalliiHAL_v2_1 |
| Physcomitrium patens | v3.3 | Gene Atlas | 3218 | phytozome.jgi.doe.gov/info/Ppatens_v3_3 |
| Populus trichocarpa | v4.1 | Gene Atlas | 3694 | phytozome.jgi.doe.gov/info/Ptrichocarpa_v4_1 |
| Panicum virgatum | v5.1 | Gene Atlas | 38 727 | phytozome.jgi.doe.gov/info/Pvirgatum_v5_1 |
| Sorghum bicolor | v3.1.1 | Gene Atlas | 4558 | phytozome.jgi.doe.gov/info/Sbicolor_v3_1_1 |
| Sorghum bicolor var Rio | v2.1 | JGI-CSP | 4558 | phytozome.jgi.doe.gov/info/SbicolorRio_v2_1 |
| Sphagnum angustifolium | v1.1 | Gene Atlas | 76 341 | phytozome.jgi.doe.gov/info/Sfallax_v1_1 |
| Setaria italica | v2.2 | Gene Atlas | 4555 | phytozome.jgi.doe.gov/info/Sitalica_v2_2 |
| Setaria viridis | v2.1 | Gene Atlas | 4556 | phytozome.jgi.doe.gov/info/Sviridis_v2_1 |
Multiple steps for vetting libraries and identifying outliers were employed, including visualizing the multidimensional scaling plots to identify batch effects, if any, and outliers among the biological replicates were further identified based on Euclidean distance to the cluster center and the Pearson correlation coefficient, r ≥ 0.85. Libraries retained after QC and outlier-filtering steps were only considered for further analysis. Detected batch effects, if any, were accounted for using RUVSeq (v1.4.0) (33) with the residual RUVr approach. Fragments per kilobase of exon per million fragments mapped (FPKM) and transcripts per million (TPM) values were calculated for each gene by normalizing the read count data to both the length of the gene and the total number of mapped reads in the sample and considered as the metric for estimating gene expression levels (34,35). Genes with low expression were filtered out, by requiring ≥2 relative log expression normalized counts in at least two samples for each gene. Differential gene expression analysis was performed using the DESeq2 package (v1.30.1) (36) with adjusted P-value <0.05 using the Benjamini & Hochberg method and an log2 fold change >1 as the statistical cutoff for differentially expressed genes.
Co-expression network construction
Weighted gene co-expression networks were constructed using the WGCNA R package (v1.70.3) (37) with normalized expression data retained after filtering genes showing low expression levels (log2 values of expression < 2). Subsets of samples belonging to specific experiments such as N study, developmental stages, or stress treatment, were used to construct multiple networks for each species. Subsetting samples reduces the noise and increases the functional connectivity and specificity within modules. We followed standard recommended WGCNA network construction procedures for this analysis. Briefly, pairwise biweight mid-correlation correlations between each gene pair was weighted by raising them to power (β). To select proper soft-thresholding power, the network topology for a range of powers was evaluated using the WGCNA pickSoftThreshold function and its return value ‘powerEstimate’, the lowest power that reached the scale-free topology fit index above 0.9 which ensures an approximate scale-free topology of the resulting network was chosen. The pairwise weighted matrix was transformed into a topological overlap measure (TOM). And the TOM-based dissimilarity measure (1 – TOM) was used for hierarchical clustering and initial module assignments were determined by using a dynamic tree-cutting algorithm. Pearson correlations between each gene and each module eigengene, referred to as a gene's module membership, were calculated and module eigengene distance threshold of 0.25 was used to merge highly similar modules. These co-expression modules were assessed to determine their association with expression patterns distinct to a tissue or condition. Module eigengenes were associated with tissues or treatment conditions or developmental stages to gain insight into the role each module might play. These modules were visualized using igraph R package (v.1.2.6) (38) and in order to focus on relevant gene pair relationships, network depictions were limited to top 500 within-module gene-gene interactions as measured by topological overlap.
GO and KEGG pathway enrichment analysis
GO Ontology and annotations obtained from InterPro to GO mappings (v53.0) were used for GO enrichment analysis of differentially expressed genes (DEGs), co-expression modules and genes in tissue and condition specific clusters was performed using topGO (v.2.42.0) (39), an R Bioconductor package, to determine overrepresented GO categories across biological process (BP), cellular component (CC) and molecular function (MF) domains. Enrichment of GO terms was tested using Fisher's exact test with P< 0.05 considered as significant. KEGG (release v101.0) (40) pathway enrichment analysis was also performed on those gene sets based on hypergeometric distribution tests and pathways with P< 0.05 were considered as enriched.
Categorization of function descriptions
To identify genes with good (GGF) and poor (GPF) function descriptions, we assessed gene function descriptions obtained from the Phytozome which are generated from domain assignments and annotations provided by InterProScan. An augmented dictionary lookup approach that incorporates weighting for positive (amplifiers), negative (including attenuators), and adversative keywords adapted from sentiment analysis methodology was employed. We first generated a custom dictionary from gene function descriptions of all Gene Atlas plants. For this we tokenized descriptions into words and removed English stop words, if any, using tidytext (v0.3.2). Words including domain names such as PFAM, PANTHER, INTERPRO, ortholog, Arabidopsis and rice gene identifiers and duplicated words were discarded. Priority was set low for high frequency words in function descriptions like domain, binding, putative, uncharacterized, expressed, protein, and unknown to generate polarity data table with words and their assigned weights. We used a modified valence shifters data table containing words that alter, negate, intensify, or amplify the meaning of polarized words from lexicon package (v1.2.1). Combined with custom gene function description based polarity data table and valence shifters data, we obtained sentiment score using sentimentr (v.2.9.0) (https://cran.r-project.org/web/packages/sentimentr). We empirically determined the minimum cutoff for sentiment score to classify gene descriptions as good (score > 0.1999) and poor (score < 0.1999) function descriptors. To validate this approach, for each category of gene function descriptions we assessed the number of GO terms per gene below and above the mean number of GO terms in the genome, chi-squared tests determined that these numbers were significantly different with p-values close to zero for all genomes. We saw similar results with PFAM annotations which demonstrates that genes under poor function category have significantly fewer annotated domains while those under good function category have more; further suggesting that this approach could be used in lieu of functional annotations.
Identification of orthologous genes
OrthoFinder (v2.5.4) was used to identify orthologous genes across 18 Gene Atlas species using default parameters (41). OrthoFinder results were parsed to generate tables of orthologs for each species containing genes with one-to-one, one-to-many, and many-to-many orthologous relationships between species using rooted gene trees.
Gene ranking method
To rank and prioritize genes by their biological relevance, genes with distinct expression patterns identified based on i) tissue/condition specificity, ii) unique DE in a single contrast were given a score of 2 for each method i.e. a gene was assigned a score of 4 if it were identified by two methods. These scores were augmented with co-expression network analysis (described above). Genes in biologically relevant modules were ranked (score = 2) while hub genes in a co-expression module were ranked the highest (score = 4). Also, gene orthologs, considering one-to-one, one-to-many and many-to-many orthologous relationships, with consensus expression pattern in two or more plants were given additional scores based on the phylogenetic distance between species (42,43) i.e. larger the divergence time higher the score (million years ago/100) (Supplemental Data 1). Final ranking of the genes was calculated as the aggregate of individual scores.
Assigning expression derived biological descriptions
For each species we collated gene expression patterns into three lists: (i) DEGs with differential expression in a single contrast out of dozens of comparisons; (ii) genes with high expression proclivity towards a specific tissue or condition and (iii) genes within co-expression modules with coherent expression patterns. We assigned semi-automated descriptions of expression patterns using a combination of string replacements and a manually curated lookup table with a vocabulary similar to TAIR (www.arabidopsis.org) gene descriptions and GO terminology, where applicable. Additionally, orthologous function descriptions categorized as GGF derived from nearest phylogenetic neighbors of genes with previously poor function descriptions were grouped into a fourth list. These descriptions were appended with the gene identifier of the orthologous gene. Finally, expression based additional biological information was generated by concatenating multiple descriptions, if exists.
System design and implementation
All statistical analyses and visualizations were performed using the R 4.0.3 Statistical Software (R Development Core Team 2011) and its web interface was developed using shiny (v1.7.1). Currently, Gene Atlas is deployed on a CentOS Linux server by employing Docker (version 19.03.11), an open platform for developing and running applications.
RESULTS
Overview of the transcriptomic landscape of gene atlas plants
Scope of data generated
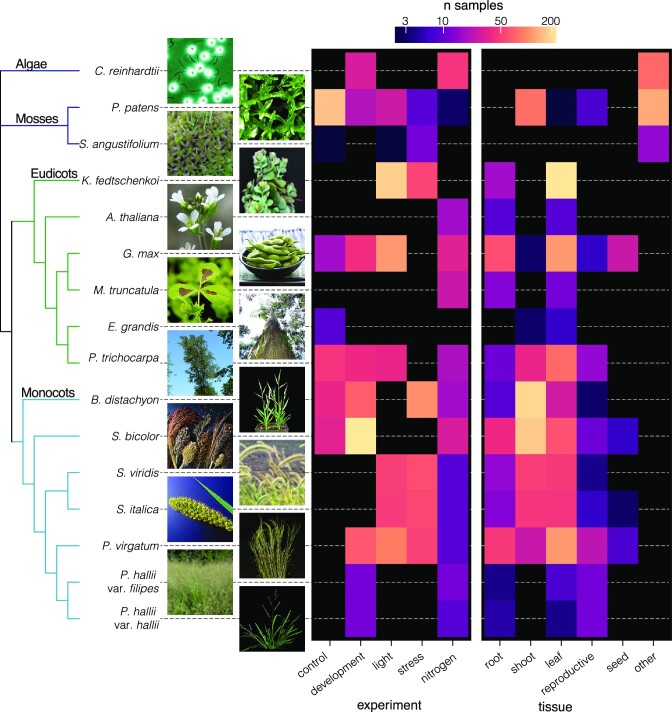
The JGI Gene Atlas is an updateable data store and analysis platform that currently contains 15.4 trillion sequenced RNA bases (Tb) and 2090 RNA-seq samples across 9 JGI plant ‘flagship’ genomes and 9 other reference plants (Table 1). JGI flagship species are important to DOE plant science mission to develop feedstocks for biofuels and understand plants response to environmental change. For each of the sequenced plants, we collected tissue samples representing appropriate developmental stages, growth conditions, tissues, and abiotic stresses (Figure 1). To reduce residual environmental variance, we followed standard growth conditions including light quality, quantity and duration, temperature, water, growth media, and nutrients. Experimental treatments were applied using standardized methods across all species (see Materials and Methods).
Figure 1.
The phylogenetic context and scope of Gene Atlas RNA-seq samples. The 16 genomes are ordered by their phylogenetic position, visualized on the left as a cladogram without branch lengths that was constructed from 10 single-copy orthologs. Tips are labeled with genome names and thumbnail photos. Photo credit given on Phytozome.
We also sought to limit among-experiment measurement and environmental variation by using identical molecular methods to extract (RNA integrity number, RIN ≥ 5 and at least 1 μg of total RNA) and sequence (Illumina stranded, paired-end 2 × 150 RNA-seq libraries) high-quality RNA. All samples were quality tested and sequenced at JGI. The resulting transcript abundance assays were highly correlated across biological replicates within conditions, tissues, and genotypes (Supplemental Data 2), which provides evidence that our gene expression measurements are highly accurate and robust.
JGI Plant Gene Atlas data was initially released on Phytozome (version v1) with 887 RNA-seq libraries across 12 species. When new reference genome versions were available Gene Atlas data was updated for those species such as Populus trichocarpa (v3.1 to v4.1) and Setaria viridis (v1.1 to v2.1). The current release includes 1203 new RNA-seq samples and six new species (Kalanchoe fedtschenkoi, Panicum hallii var. hallii, Sphagnum fallax, Eucalyptus grandis, Sorghum bicolor var. Rio and Lupinus albus), demonstrating the flexibility and updateability of the JGI Gene Atlas with respect to new reference genomes. This updateability includes sequence data derived from other experiments and sequencing facilities. To illustrate this, we included S. bicolor ‘Rio’ (sweet sorghum, nsamples = 94) (29) from JGI’s Community Science Program project and Lupinus albus (white lupin) cluster root tissue (nsamples = 72) (19) data from a non-JGI project. A comprehensive list of all samples available so far is in Supplemental Data 3 and https://plantgeneatlas.jgi.doe.gov. Our custom pipeline to analyze expression levels of protein-coding genes is outlined in Supplementary Figure S2.
Developing a baseline of evolutionarily conserved gene expression
Across all 18 species, 47–87% (mean = 73%) of annotated genes were transcriptionally active (FPKM > 1) in JGI Plant Gene Atlas samples. To test for conserved and divergent expression levels across the 18 species, we applied the traditional method of comparing single-copy orthologs across species. While powerful, restricting tests to orthologs based on gene sequences can be problematic across evolutionarily diverged lineages. For example, given the phylogenetic distance and nested whole-genome duplications among our sampled species, we were only able to find 2066 one-to-one orthologous protein-coding genes (Supplemental Data 4) across just eight of the vascular plant genomes. Furthermore, such single-copy orthologs have evolutionarily conserved sequences and likely gene functions, permitting better homology-based functional descriptions (89.01% with good functional descriptions) than genome-wide averages (83.8%, Fisher's exact test odds = 1.607, P = 5.495e-12). Nonetheless, we observed 227 (10.98%) genes with 1:1 orthologs and consistent expression among species, but weak functional descriptions (Supplemental Data 5). Given the expected paucity of multi-genome single-copy orthologs, we also addressed the challenge of finding genes with similar expression across species by analyzing pairwise single-copy orthologs to a single reference genome, A. thaliana. Overall, we identified 6016 Arabidopsis orthologs that showed conserved expression patterns across multiple species. Surprisingly, these genes include 660 (11%) with little to no known functional description, making these genes rational targets for functional characterization studies (Supplemental Data 6). Identifying and improving the functional characterizations of such genes was one of the objectives of the Gene Atlas experiment. Genes with single-copy orthologs in A. thaliana and consistent expression were significantly enriched in transcription factors (n = 501, 8.3%; Fisher's exact test odds ratio = 1.507, P = 4.26e-13), suggesting that potential regulators of different biological processes are strongly conserved across the plant species (44). These observed evolutionarily conserved expression patterns inform functional details that complement direct sequence data comparisons.
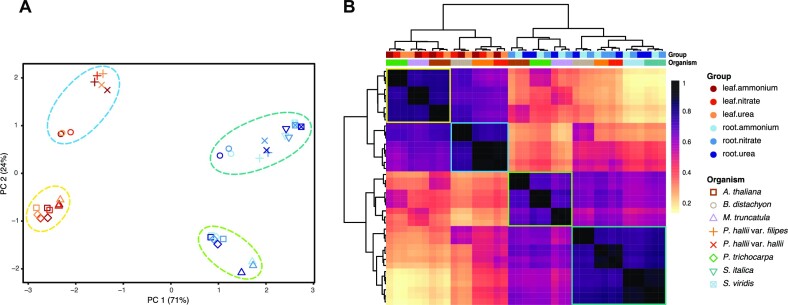
In contrast to these ortholog-constrained analyses, within-genome co-expression analyses are agnostic to orthology, which dramatically increases the number of genes that can be analyzed, providing a broader perspective on gene expression regulatory evolution. For example, multidimensional scaling and hierarchical clustering revealed that phylogenetically neighboring species have more similar expression profiles across tissues and nitrogen treatments than more distantly related species (Mantel R > 0.63, P < 0.04) (Figure 2). However, the phylogenetic signal of co-expression was dwarfed by variation among tissues, where far more of the total co-expression clustering across nitrogen source treatments was driven by patterns among tissues than genetic distance among species (tissues correlated with the first canonical correspondence analysis axis, which explains 41.46% of the variation), suggesting that genes in closely related species exhibit similar transcriptional profiles across tissues and conditions likely owing to the accumulation of evolutionarily conserved regulatory elements.
Figure 2.
Global patterns of gene expression across eight vascular plants. Multidimensional scaling based on the expression of 2066 single-copy orthologous genes in two tissues and three nitrogen treatment conditions show predominant clustering first by tissues and then by clade (mono-, dicots) (A). Hierarchical clustering based on Pearson correlation coefficients of log2 transformed normalized expression data (B).
In addition to genes showing conserved expression across species, differentially expressed genes (DEGs) within species across different developmental stages, tissues and varied environmental conditions were determined by defining biologically meaningful comparisons.
Patterns of tissue-specific gene expression across 18 species and >400M years of plant evolution
Tissue-specific expression complements global co-expression analyses by defining potential gene function associated with an organ or tissue. The major drawback of this approach results from morphological differences among species. For example, in Chlamydomonas, a single-celled organism, transcriptionally active genes in each condition represent expressed genes in the organism as a whole, whereas multicellular organisms exhibit gene expression variation across different cell subtypes. Furthermore, the mosses sampled here lack root systems, flowers, seeds or easily sampled reproductive organs. Even the far more closely related flowering plants have functionally divergent homologous structures, such as root nodules, panicles, florets, sepals, and rhizomes. As such, analysis of tissue-specific expression must be somewhat phylogenetically constrained and condensed into large-scale functional tissue types (Figure 1).
Our data suggest that large proportions of annotated genes (27–68%, M = 44.7; SD = 12.7) are commonly expressed (FPKM > 1) in multiple tissues (Supplemental Data 7), confirming that many genes serve multiple functions across tissues and environments and some such as housekeeping genes serve similar function across different tissues in different contexts. However, there was considerable among-tissue variation across species (ANOVA F = 70.01, df = 16, P < 2e-16) where gene expression is driven by variation among tissues or conditions (Supplemental Data 7). Such variably expressed genes may have evolved diverse functions depending on the regulatory environment across cell types.
Despite considerable across-tissue expression, we observed 220 218 (32.1%) of all genes with high expression specificity to a single tissue or condition. To identify genes exhibiting such strong tissue or condition specific expression, we used the Tau method (45) which accounts for the number of unique sample types and produces consistently robust results with highest correlation between datasets of varying sizes (46). Using this method, we identified genes specific to (1) reproductive and root tissue in S. italica, (2) leaf, inflorescence, and whole floret in switchgrass, (3) leaf, leaf blade, dry seed, and imbibed seed in S. bicolor, and (4) stem and flower related gene sets in Brachypodium. Of all the standard plant tissues, stem and leaf had the fewest uniquely expressed genes (two-tailed unpaired Welch's t-test, P = 8.338e-06) while roots followed by flower tissues were most unique (two-tailed unpaired Welch's t-test, P = 2.547e-10). Groups of genes with greater expression proclivity towards spores, protonema and leaflet were recognized in Physcomitrium; drought and high temperature in Sphagnum; and towards seed, root tip, lateral root, and nodules in soybean (Supplemental Data 8, 9). These gene sets were largely overrepresented in GO biological processes known for each tissue or condition (Supplemental Data 10). Genes and their promoter regions with such marked expression specificity represent valuable tissue-specific reporters and targets for plant genetic engineering applications.
Inferring additional biological information from patterns of gene expression
Variation in co-expression network topologies
Genes with similar expression patterns across diverse environmental conditions and tissues tend to serve similar biological functions across distantly related species and can be detected by co-expression clustering algorithms. For example, clusters of genes associated with a specific tissue or condition may be crucial for plant development or response to environmental cues. These strongly conserved tissue- and treatment-specific expression patterns facilitate biological gene function extrapolating from expression studies in one organism to close phylogenetic neighbors. To identify modules with such coherent expression patterns, we constructed 30 weighted gene co-expression networks (37) and 148 highly significant (min KME = 0.7, cut height = 0.25) co-expression modules within species and across different sets of tissues and conditions. Of these, 21 modules were significantly correlated with stress treatments (i.e. heat, cold, drought, salt, and wound stresses), 10 with N treatments, and 33 with other experimental conditions (Supplemental Data 11). Tissue-specific modules were also very common, e.g. root tissue-specific modules (n = 11), contained genes with GO terms enriched in responses to stimulus, oxidation-reduction process (47,48) and hydrogen peroxide metabolism (49) that are relevant to root functions (50–53). Leaf specific modules (n = 11) were enriched for phototropism, thylakoid membrane organization, pigment biosynthetic process, phototropism, and carbon fixation (Supplemental Data 12), suggesting that genes within the same module are associated with the same or interconnected biological functions.
Inferring transcription factor functions from co-expressed genes
‘Hub’ genes, which have the highest connectivity among neighboring genes within a module, are likely involved in preserving multi-gene regulatory variation and thus network integrity, potentially as trans-regulatory elements like transcription factors. We determined the top 10 most highly connected hub genes within each module. Across all the co-expression networks 87 hub genes belonged to transcription factor (TF) families (via PlantTFDB (54)) (Supplemental Data 13), a slight but not significant enrichment of TFs relative to the genomic background (% hub TFs = 6.21%, background TFs = 5.23%, Fisher's exact test odds ratio = 0.834, P = 0.104). TFs with many connections are presumed to be most influential in regulating the expression of modular genes in co-expression networks (55). Under this premise, we further explored the overrepresented TF families among the hub genes. Most represented TF families in N treatment modules were MYB, WRKY, and NAC. Similar observations were made by Canales et al. (56) from Arabidopsis root transcriptomic data generated under contrasting N conditions. As shown in previous studies (57–62), hub genes play key roles in orchestrating module behavior and provide a specific focus for investigations into trait or condition related modules.
Expression derived biological descriptions (EDBD)
To evaluate how well the predicted gene function descriptions of Gene Atlas plants illustrate validated gene functions, we categorized currently assigned functional descriptions available at Phytozome as genes with good (GGF) and poor (GPF) function descriptions using an augmented dictionary lookup approach that incorporates weighting for negative, positive, and adversative keywords. Overall, 16–56% of the functional descriptions are GPF across the plants, with a large percentage of such genes not having a known function (Supplementary Figure S1) (6,8). We then assigned EDBD to the two subsets, GGF and GPF, using results from tissue and condition specific expression groups, DEGs unique to a single contrast and co-expression network analysis along with ortholog function descriptions derived from nearest phylogenetic neighbors (see Materials and Methods).
Using this method, we added additional biological information to an average of 40.6% (SD = 12.6) of genes (excluding orthology based function descriptions) in these plant genomes (Table 2; Supplemental Data 14). Some of the same assigned descriptions were identified in as many as 13 different species. The frequency of these descriptors varied widely across species, within the top 100 the occurrence numbers ranged from 444 to 11 655 (tabulated in Supplemental Data 15). For example, in the case of S. bicolor, 5357 (15.65% of the total) genes lacked sequence homology-based function descriptions, 24 406 had good functional descriptions while overall 9723 had poor descriptions. Gene Atlas expression-based additional biological descriptions were assigned to a total of 20 259 genes, of which 14 891 (43.63% of total annotated genes) had good functional descriptions and 5368 (15.73%) had poor descriptions. To verify the reliability of the assigned functional associations, GO enrichment analysis of genes assigned with descriptions based on leaf and root samples was performed. We observed significant enrichment for photosynthesis, chloroplast organization, chlorophyll biosynthetic process and plastid translation (P< .05, Fisher's exact test) in leaf related EDBDs; and cell wall loosening (63), water transport and xyloglucan biosynthetic process (64) (P< 0.05, Fisher's exact test) in root related EDBDs. Similar analysis in Brachypodium genes with assigned descriptions based on abiotic stress experiments (i.e. cold, heat, drought, and salt stress) showed significant enrichment for regulation of cellular response to alkaline pH, response to cold, heat, and positive regulation of response to oxidative stress (P< 0.05, Fisher's exact test). Likewise, among genes annotated based on flower samples, specification of floral organ identity, fruit wall development and sporopollenin biosynthetic process were among the top enriched GO terms (P< 0.05, Fisher's exact test). These results indicate that the assigned descriptions show strong biological role predictability and the approach here aids in expanding our current understanding of plant gene functions.
Table 2.
Summary of assigned expression derived biological descriptions (EDBD) to Gene Atlas. Number of annotated genes and the percentage of genes with good function descriptions (GGF), poor function descriptions (GPF) categorized using an augmented dictionary lookup approach that incorporates weighting for negative, positive, and adversative keywords and percentage of genes assigned with expression derived biological descriptions
| Genome | No. genes | % GGF | % Assigned (GGF) | % GPF | % Assigned (GPF) | No. assigned | % Assigned (total) |
|---|---|---|---|---|---|---|---|
| A. thaliana Araport11 | 27 655 | 91.33 | 51.32 | 8.67 | 3.22 | 15 082 | 54.54 |
| B. distachyon | 34 310 | 72.56 | 34.17 | 27.44 | 13.19 | 16 247 | 47.35 |
| C. reinhardtii | 17 741 | 43.08 | 12.6 | 56.92 | 15.98 | 5070 | 28.58 |
| E. grandis | 36 349 | 79.74 | 28.63 | 20.26 | 3.997 | 11 858 | 32.62 |
| G. max | 52 872 | 80.37 | 47.25 | 19.63 | 11.33 | 30 971 | 58.58 |
| K. fedtschenkoi | 30 964 | 82.01 | 39.56 | 17.99 | 7.644 | 14 615 | 47.20 |
| M. truncatula | 50 894 | 67.94 | 23.66 | 32.06 | 5.285 | 14 731 | 28.94 |
| P. halliivar.filipes | 33 805 | 72.65 | 34.97 | 27.35 | 8.656 | 14 746 | 43.62 |
| P. hallii var. hallii | 33 263 | 73.36 | 31.16 | 26.64 | 7.946 | 13 007 | 39.10 |
| P. patens | 32 926 | 55.44 | 19.35 | 44.56 | 14.06 | 11 003 | 33.42 |
| P. trichocarpa | 34 699 | 82.31 | 39.57 | 17.69 | 7.997 | 16 507 | 47.57 |
| P. virgatum | 80 278 | 69.2 | 39.73 | 30.8 | 14.15 | 43 251 | 53.88 |
| S. bicolor | 34 129 | 71.51 | 43.63 | 28.49 | 15.73 | 20 259 | 59.36 |
| S. bicolorRio | 35 490 | 69.16 | 15.04 | 30.84 | 4.765 | 7029 | 19.81 |
| S. fallax | 25 100 | 78.31 | 32.36 | 21.69 | 9.183 | 10 427 | 41.54 |
| S. italica | 34 584 | 77 | 39.37 | 23 | 10.78 | 17 344 | 50.15 |
| S. viridis | 38 334 | 70.43 | 35.99 | 29.57 | 12.74 | 18 680 | 48.73 |
| L. albus | 38 258 | 78.17 | 11.01 | 21.83 | 2.415 | 5138 | 13.43 |
To help investigators target important genes for additional functional studies, we ranked the biological relevance of genes using a scoring methodology based on expression patterns of genes identified using tissue/condition specificity, differential expression, co-expression, hub status in a co-expression module and consensus expression across species. Gene orthologs with similar expression profiles in two or more species were given additional scores derived from the phylogenetic distance, i.e. larger the divergence time higher the score (see Methods). We identified a total of 1797 top ranked genes (top 5% with highest aggregate score) across Gene Atlas plants (1316 have orthologs in ≥5 plants; 47 of which have orthologs in ≥10 of evaluated plants) that have poor functional information but with the potential to improve our understanding of plant biology and form a list of prioritized targets for future experimental investigations (Table 3; Supplemental Data 16).
Table 3.
Prioritized top ranked genes with poor functional descriptions for future experimental investigations. Genes were given scores based on expression patterns identified from i) unique differential expression in a single contrast, ii) tissue/condition specific expression and iii) biologically relevant co-expression modules (each given a score of 2) while hub genes in a co-expression module were given a score of 4. Gene orthologs with similar expression profiles were given additional scores derived from the phylogenetic distance. Total score was calculated as the aggregate of individual scores. Top ranked genes (two per species) are represented here
| Score | ||||||||
|---|---|---|---|---|---|---|---|---|
| Organism | Gene ID | Differential expression | Condition specific expression | Co-expression | Hub gene | Consensus expression | Total | Arabidopsis orthologs |
| A. thaliana Araport11 | AT5G02090 | 2 | 2 | 2 | 0 | 18.73 | 24.73 | AT2G20080 |
| AT2G44010 | 0 | 2 | 2 | 0 | 19.2 | 23.2 | AT4G28840 | |
| B. distachyon | Bradi1g38210 | 0 | 2 | 2 | 0 | 10.7 | 14.7 | AT2G42760 |
| Bradi2g23445 | 0 | 0 | 2 | 0 | 11.69 | 13.69 | AT5G02090; AT2G37750 | |
| C. reinhardtii | Cre02.g078550 | 0 | 2 | 2 | 4 | 0 | 8 | |
| Cre02.g092700 | 0 | 2 | 2 | 4 | 0 | 8 | ||
| E. grandis | Eucgr.F01122 | 0 | 2 | 2 | 0 | 17.12 | 21.12 | |
| Eucgr.B00604 | 0 | 2 | 2 | 0 | 16.57 | 20.57 | AT5G08050 | |
| G. max | Glyma.16G013600 | 0 | 0 | 2 | 0 | 20.84 | 45.68 | AT3G14280 |
| Glyma.13G227500 | 0 | 0 | 2 | 0 | 20.28 | 44.56 | AT1G33055 | |
| K. fedtschenkoi | Kaladp0965s0006 | 2 | 0 | 2 | 0 | 26.23 | 30.23 | AT2G30230; AT1G06980 |
| Kaladp0065s0016 | 0 | 0 | 2 | 0 | 19.79 | 21.79 | AT4G28840; AT2G20080 | |
| M. truncatula | Medtr3g031140 | 0 | 2 | 2 | 0 | 27.28 | 31.28 | AT2G30230; AT1G06980 |
| Medtr2g079300 | 2 | 2 | 2 | 0 | 18.11 | 24.11 | ||
| P. hallii var. filipes | Pahal.3G090000 | 2 | 2 | 2 | 0 | 13.2 | 19.2 | AT5G02160 |
| Pahal.7G305700 | 2 | 2 | 2 | 0 | 12.05 | 18.05 | AT4G21445 | |
| P. hallii var. hallii | PhHAL.5G229300 | 2 | 2 | 2 | 0 | 11.532 | 17.532 | AT5G62770; AT3G27880; AT1G23710; AT1G70420 |
| PhHAL.3G160400 | 0 | 0 | 2 | 0 | 14.55 | 16.55 | ||
| P. patens | Pp3c11_15500 | 2 | 2 | 2 | 4 | 0 | 10 | |
| Pp3c13_2427 | 0 | 2 | 2 | 4 | 0 | 8 | ||
| P. trichocarpa | Potri.018G084100 | 2 | 2 | 2 | 0 | 19.79 | 25.79 | AT4G28840; AT2G20080 |
| Potri.003G193400 | 2 | 0 | 2 | 0 | 20.89 | 24.89 | ||
| P. virgatum | Pavir.5NG404000 | 0 | 0 | 2 | 0 | 13.64 | 15.64 | |
| Pavir.2NG640501 | 0 | 2 | 2 | 0 | 9.72 | 13.72 | AT5G13720 | |
| S. bicolor | Sobic.009G229000 | 0 | 0 | 2 | 4 | 13.35 | 19.35 | AT4G28840; AT2G20080 |
| Sobic.001G118400 | 2 | 2 | 2 | 0 | 10.44 | 16.44 | AT1G73885 | |
| S. bicolorRio | SbRio.08G154700 | 0 | 2 | 2 | 0 | 12.31 | 16.31 | AT5G08050 |
| SbRio.10G134000 | 0 | 2 | 2 | 0 | 12.05 | 16.05 | AT4G01150 | |
| S. angustifolium | Sphfalx02G142200 | 2 | 2 | 2 | 4 | 0 | 10 | |
| Sphfalx11G077900 | 2 | 2 | 0 | 0 | 4.5 | 8.5 | AT3G03341 | |
| S. italica | Seita.9G407600 | 2 | 0 | 2 | 0 | 12.7053 | 16.7053 | AT1G63410; AT3G14260 |
| Seita.9G436900 | 0 | 2 | 2 | 0 | 12.5741 | 16.5741 | AT2G30230; AT1G06980 | |
| S. viridis | Sevir.1G151100 | 2 | 0 | 2 | 0 | 13.2732 | 17.2732 | AT1G12320; AT1G62840; AT3G60780; AT2G45360 |
| Sevir.5G247600 | 0 | 0 | 2 | 0 | 13.2732 | 15.2732 | AT5G62770; AT3G27880; AT1G23710; AT1G70420 | |
Case studies exploring plant gene atlas
Here we present results from two experiments that assay (1) transcriptional changes in sorghum across five developmental stages, and (2) plant responses to different N sources and availabilities. These studies exemplify some of the experiments available in this resource.
Transcription modulation across developmental stages
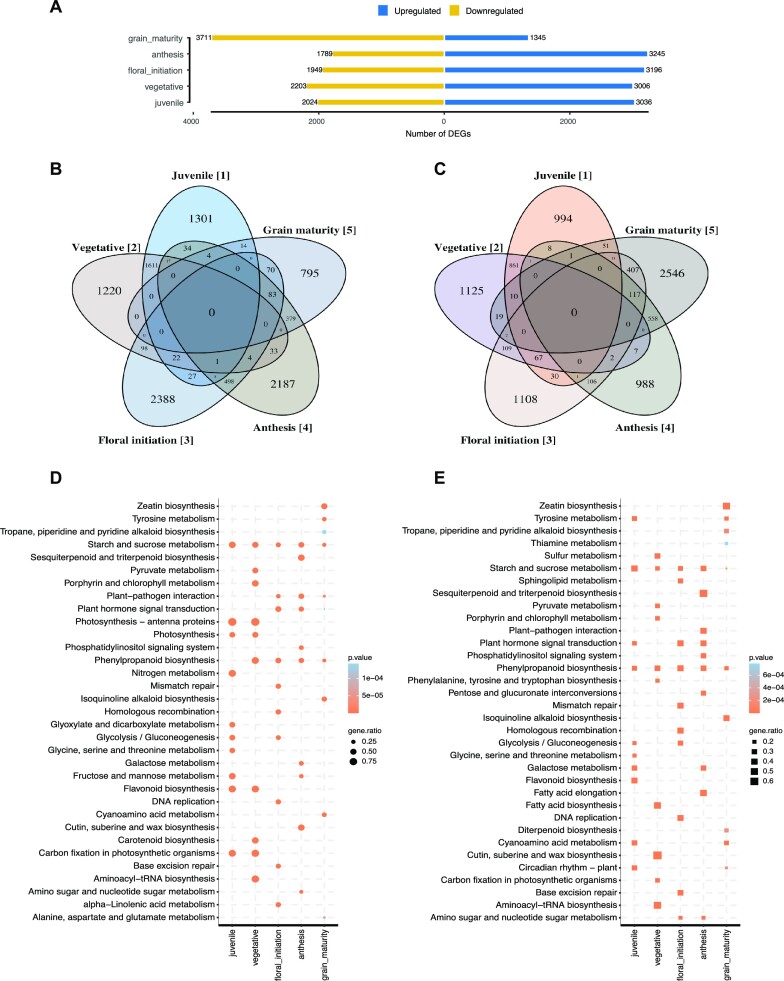
Developmental time-courses represent a particularly powerful experiment to understand gene function and the dynamics of transcript abundance. As an example of such a time course, we evaluated the regulation of gene expression in leaf tissue in five developmental stages of Sorghum bicolor (juvenile, vegetative, floral initiation, anthesis and grain maturity). Overall, we identified 13 992 unique DEGs (n total annotated genes = 34 211) across the five developmental stages (Figure 3A, B, C). KEGG pathway enrichments of up-regulated differentially expressed genes were largely consistent with physiological expectations: photosynthesis, carbohydrate and N metabolism terms were overrepresented in juvenile/vegetative stages (P <0.05, hypergeometric test), floral initiation/anthesis stages were enriched in reproductive organ development and hormone signal transduction, and grain maturity stage were enriched for amino acid metabolism and transport, and zeatin and tyrosine metabolism (Figure 3D, E). We observed the enrichment pattern to be reversed among downregulated genes in different stages, e.g. plant-pathogen interaction and plant hormone signal transduction were suppressed in juvenile and vegetative stages whereas photosynthesis, carbohydrate and N metabolism related pathways were among those suppressed in late developmental stages (Supplementary Figure S3). These overrepresented pathways among DEGs at each stage illustrate the key biological events over the growing season, e.g. as juveniles the S. bicolor are collecting energy to increase the biomass, and as they flower and mature, they express defense mechanisms, and finally, with grain maturity, they reduce photosynthesis and slow down nutrient acquisition. The S. bicolor dataset provides an example of high-resolution characterization of gene expression changes and insight into the molecular responses of the plant across developmental stages represented by the Gene Atlas dataset.
Figure 3.
Differentially expressed gene comparison across five developmental stages in Sorghum bicolor. Numbers of differentially expressed genes across developmental stages (A). Venn diagrams of up-regulated (B) and down-regulated genes that are unique and shared between developmental stages (C). Top 10 KEGG metabolic pathway enrichments (P <0.05, hypergeometric test) of up-regulated differentially expressed genes in each of the five developmental stages (D) and upregulated genes unique to each stage (E). ‘gene.ratio’ represents the ratio of number of DEGs over the number of genes annotated specific to the pathway.
Transcriptional responses to different N sources
Tissue-specific gene expression regulatory responses to environmental cues are often evolutionarily conserved. These conserved responses offer a framework to test hypotheses about gene function as it relates to environmental sensitivity. A particularly powerful experiment adjusts the amount and type of necessary resource available. Drought, light and nutrient availability manipulations have provided strong evidence for gene function across the diversity of plants (65–69). In addition to providing evidence for the function of specific candidate genes’ responses to environmental stimuli, highly controlled manipulations, like our nitrogen source experiments, offer a framework to compare the relative roles of gene families and molecular pathways.
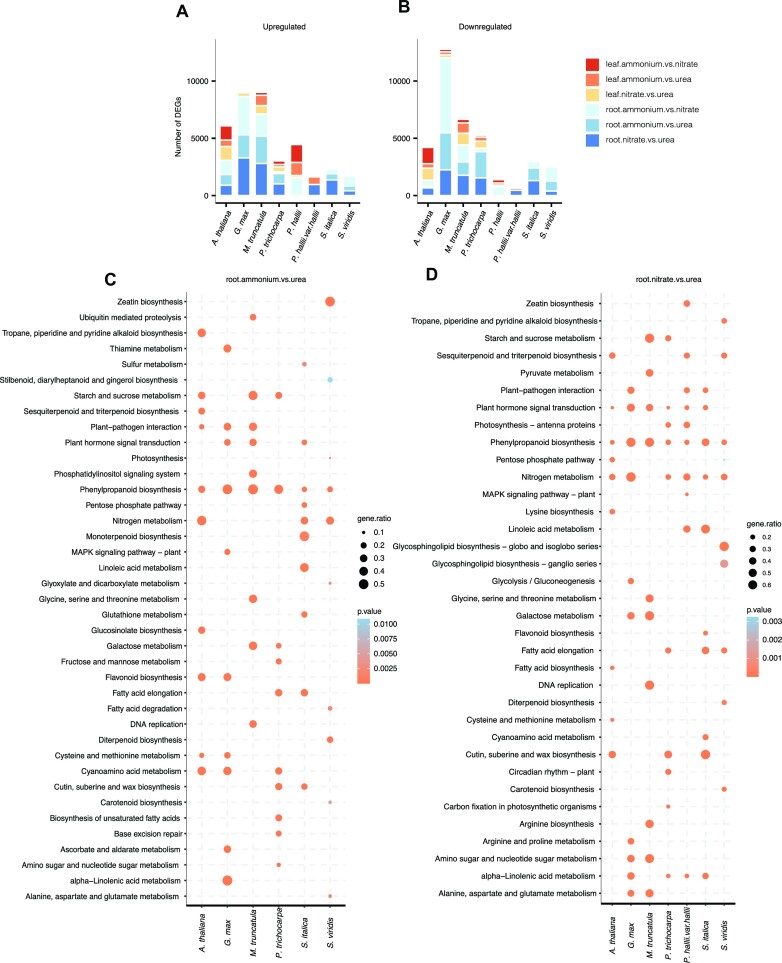
To understand gene expression underpinnings of N metabolism, we contrasted transcript abundance in aboveground and root tissues of each Gene Atlas species (where available, see Figure 1) grown on N from three sources: urea, ammonium (NH4+), and nitrate (NO3−) (Supplemental Data 17). Since our experiments had similar statistical power and biological replicates among species and conditions, the total number of DEGs is a strong indicator of the transcriptional effects of different N sources. The most striking patterns were those related to tissue-specific gene expression variation within genotypes (Figure 4A, B). For example, the root transcriptome was more responsive than aboveground tissues in all eudicot genotypes (Mann–Whitney U-test, P = 5e-04) except Arabidopsis (two-tailed unpaired Welch's t-test, P = 0.4526). We observed consistent enrichments of N metabolism pathway genes among differentially expressed genes between treatments across many species, which demonstrates that this experiment elicits molecular responses of genes with homologs in genetic model species.
Figure 4.
Transcriptional response of Gene Atlas plants towards NH4+ and NO3− compared to urea as the sole nitrogen source in root and leaf tissues. Numbers of genes differentially upregulated (A) and numbers of genes differentially downregulated in response to changing nitrogen regime (B). Top 10 KEGG metabolic pathway enrichments (P <0.05, hypergeometric test) in up-regulated differentially expressed genes in roots from Gene Atlas plants in ammonia versus urea (C) and nitrate versus urea treatment comparisons (D). ‘gene.ratio’ represents the ratio of number of DEGs over the number of genes annotated specific to the pathway.
Despite the power of discovering enriched groups of genes with similar and expected functional annotations, a major goal of the Gene Atlas is to provide a framework to discover novel biological roles of genes and their interactions. As such, we were excited to find starch and sucrose metabolism, and phenylpropanoid biosynthesis pathways overrepresented in upregulated DEGs. Indeed, many of the DEGs we identified in pairwise comparisons between N-sources are not directly involved in N metabolism. For example, genes associated with plant-pathogen interaction, plant hormone signal transduction, and carbohydrate metabolism were abundant (Figure 4C, D, Supplementary Figure S4). Similar observations were reported previously in Sorghum genotypes with varying N-stress tolerance subjected to N-limiting conditions (70). Notably, nitrogen and amino acid metabolism-related pathways were overrepresented mainly in DEGs in nitrate vs. urea comparison. Such comparisons highlight differences in plant's response to NO3− as a sole N source compared to NH4+ at the metabolic level.
Using the JGI plant gene atlas resource
JGI Plant Gene Atlas data are currently hosted at two portals:
JGI Plant Gene Atlas (https://plantgeneatlas.jgi.doe.gov), a dedicated portal provides bulk access to the data. For each species, baseline expression page provides access to expression values (FPKM or TPM) across available samples along with expression derived biological descriptions. It allows users to explore the expression of highly variable genes across samples and query expression profiles of a single gene to multiple gene sets. Users can also explore expression profiles of orthologous genes across other Gene Atlas plants. Differential expression page displays DEGs for each comparison while allowing users to specify criteria, log2 fold-change and adjusted p-value, for differential expression. Additionally, plots representing the GO and KEGG pathway enrichments in the set of differentially expressed genes can be visualized. Detailed documentation about using this resource is included under the ‘Help’ tab on the portal.
JGI’s plant portal, Phytozome (phytozome.jgi.doe.gov) currently provides detailed functional annotations, genome, transcript, coding and peptide sequences, protein homologs, plant family information and additionally genome browser view of gene models on the gene report page. It also provides users with an efficient interactive graphical representation of co-expressed genes.
DISCUSSION
Here, we analyzed the transcriptional landscape of 18 plants from 2090 RNA-seq datasets. To the best of our knowledge, it is the largest compendium of plant transcriptome data generated following standardized protocols across diverse plant species. These datasets enable JGI’s efforts to improve genome annotations especially related to conserved biological processes across the diversity of plants. Comparing orthologs among common gene sets between species allowed us to pinpoint and rank biologically relevant and evolutionarily conserved genes, demonstrating the potential of cross-species analysis from the transcriptome resource generated in this study.
Furthermore, our results documented plant responses to varying N resources at the organ level and highlights differences among plants at the clade level. Transcriptional patterns in different developmental stages of Sorghum revealed important biological events at each stage in its life cycle and provides in-depth look at genes involved in those processes. These and other analyses highlight shared and varied gene expression regulatory evolution across plants.
The Gene Atlas datasets, along with the additional expression derived biological descriptions, are valuable resources to the plant research community and provide targets, unknown or poorly described TFs, hub genes, and conserved genes, for functional studies that directly improve gene functional descriptions. We acknowledge that these functional associations are not definitive evidence of their functions, but we anticipate that they will be useful in directing future functional characterization experiments. We will continue to expand the Gene Atlas through new datasets from JGI’s Community Science Program and standardized procedures to increase the specificity of these biological descriptions. We strongly believe that results from this study and additional custom analyses on this resource will aid researchers in better understanding of roles of genes in their own experiments and get a better handle on biological processes at the system level.
Supplementary Material
ACKNOWLEDGEMENTS
Thanks to Daniel S. Rokhsar for involvement in the early formulation of Gene Atlas and helpful discussions.
Author contributions: A.S. and C.P. conducted transcriptome data analyses. J.T.L. conducted metadata analysis. MSH carried out soybean and Medicago experiments. J.G., K.B., M.A., M.K., M.W., A.L., Jenifer J, L.S., K.A., M.Z., C.D. performed RNA-seq library preparation and sequencing. J.K. and S.D.G. conducted Chlamydomonas experiments. J.C., J.P., and D.G. maintains the data repository at Phytozome. J.W.J. and C.P. assembled genomes. S.S. conducted genome annotation at JGI. I.T-J., and M.U. conducted Panicum virgatum experiments. S.S.J. conducted Populus experiments and Sphagnum RNA extractions. J-G.C. and Y.Y. conducted Populus hormone treatment experiments. D.C. and M.P. conducted Populus seasonal time course experiments. H.J., C.S., P.H., J.S., C.L., A.M. and S.C. conducted Setaria experiments and sample preparations. L.L. conducted Brachypodium cold treatment experiments. A.A.C. conducted Sphagnum experiments. B.W. conducted Sorghum N-treatment experiments. R.H. conducted Kalanchoe experiments. M.R.P. conducted Eucalyptus experiments. K.S. conducted validation experiments for N-treatment. E.V.S. and X.W. conducted Arabidopsis, P. hallii and P. virgatum photoperiod experiments and sample preparations. A.M. conducted P. virgatum drought stress experiments and sample preparation. P.F.P. and F.B.H. analyzed Physcomitrium data. P.F.P., M.H. and S.A.R. provided Physcomitrium samples. D.G., D.T., D.J.W., E.A.C., E.A.K., G.S., G.A.T., I.A., I.B., J.S., J.M., J.P.V., S.A.R., S.S.M., T.P.B., T.E.J., T.C.M., X.Y. and Y.T. are principal investigators (alphabetical order). All authors read and approved the final manuscript. A.S., J.T.L. and J.S. prepared the manuscript with input from all authors.
Notes
Present address: Md Shakhawat Hossain, Texas A&M AgriLife Research and Department of Soil and Crop Sciences, Texas A&M University, College Station, TX, USA.
Present address: Fabian B. Haas, Department of Algal Development and Evolution, Max Planck Institute for Biology Tübingen, Tübingen, Germany.
Present address: Manuel Hiss, GSK Vaccines GmbH, Emil-von-Behring-Str, Marburg, Germany.
Present address: Pierre-François Perroud, Université Paris-Saclay, INRAE, AgroParisTech, Institut Jean-Pierre Bourgin (IJPB) Versailles, France.
Present address: Sean D. Gallaher and Sabeeha S. Merchant, Department of Plant and Microbial Biology and Department of Molecular and Cell Biology, QB3, University of California, Berkeley, Berkeley, CA, USA.
Present address: Eugene V. Shakirov, Department of Biological Sciences, Marshall University, Huntington, WV, USA.
Present address: Andrew Muchlinski, Firmenich, San Diego, CA, USA.
Present address: Christine Shyu, Bayer Crop Science, St. Louis, MO, USA.
Present address: Pu Huang, BASF Corporation, Durham, NC, USA.
Present address: Jose Sebastian, Department of Biological Sciences, Indian Institute of Science Education and Research, Berhampur (IISER BPR), Odisha, India.
Present address: Carol Laiben, Propper Asset Management, 17 Research Park Drive, St. Charles, MO, USA.
Present address: Alyssa Medlin, Saginaw High School, 800 N Blue Mound Rd, Saginaw, TX, USA.
Present address: Sankalpi Carey, Guardant Health Inc, 3100 Hanover Street, Palo Alto, CA, USA.
Present address: Elizabeth A. Cooper, Department of Bioinformatics and Genomics, University of North Carolina at Charlotte, Charlotte, NC, USA.
Present address: Michael Udvardi, Center for Crop Science, Queensland Alliance for Agriculture and Food Innovation, The University of Queensland, St. Lucia, Brisbane, Australia.
Present address: Stefan A. Rensing, Faculty of Chemistry and Pharmacy, University of Freiburg, Freiburg, Germany.
Present address: Sabeeha S. Merchant, Environmental Genomics and Systems Biology, Lawrence Berkeley National Laboratory, Berkeley, CA, USA.
Contributor Information
Avinash Sreedasyam, HudsonAlpha Institute for Biotechnology, Huntsville, AL, USA.
Christopher Plott, HudsonAlpha Institute for Biotechnology, Huntsville, AL, USA.
Md Shakhawat Hossain, Division of Plant Science and Technology, C.S. Bond Life Science Center, University of Missouri, Columbia, MO, USA.
John T Lovell, HudsonAlpha Institute for Biotechnology, Huntsville, AL, USA; Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA, USA.
Jane Grimwood, HudsonAlpha Institute for Biotechnology, Huntsville, AL, USA.
Jerry W Jenkins, HudsonAlpha Institute for Biotechnology, Huntsville, AL, USA.
Christopher Daum, Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA, USA.
Kerrie Barry, Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA, USA.
Joseph Carlson, Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA, USA.
Shengqiang Shu, Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA, USA.
Jeremy Phillips, Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA, USA.
Mojgan Amirebrahimi, Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA, USA.
Matthew Zane, Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA, USA.
Mei Wang, Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA, USA.
David Goodstein, Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA, USA.
Fabian B Haas, Plant Cell Biology, Faculty of Biology, University of Marburg, Karl-von-Frisch-Str, Marburg, Germany.
Manuel Hiss, Plant Cell Biology, Faculty of Biology, University of Marburg, Karl-von-Frisch-Str, Marburg, Germany.
Pierre-François Perroud, Plant Cell Biology, Faculty of Biology, University of Marburg, Karl-von-Frisch-Str, Marburg, Germany.
Sara S Jawdy, Center for Bioenergy Innovation, Oak Ridge National Laboratory, Oak Ridge, TN, USA.
Yongil Yang, Center for Bioenergy Innovation, Oak Ridge National Laboratory, Oak Ridge, TN, USA.
Rongbin Hu, Center for Bioenergy Innovation, Oak Ridge National Laboratory, Oak Ridge, TN, USA.
Jenifer Johnson, Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA, USA.
Janette Kropat, Department of Chemistry and Biochemistry and Institute for Genomics and Proteomics, University of California, Los Angeles, CA, USA.
Sean D Gallaher, Department of Chemistry and Biochemistry and Institute for Genomics and Proteomics, University of California, Los Angeles, CA, USA.
Anna Lipzen, Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA, USA.
Eugene V Shakirov, Department of Integrative Biology, University of Texas at Austin, Austin, TX, USA.
Xiaoyu Weng, Department of Integrative Biology, University of Texas at Austin, Austin, TX, USA.
Ivone Torres-Jerez, Noble Research Institute, Ardmore, OK, USA.
Brock Weers, Department of Biochemistry and Biophysics, Texas A&M University, College Station, TX, USA.
Daniel Conde, Centro de Biotecnología y Genómica de Plantas, Universidad Politécnica de Madrid, Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA-CSIC), Madrid, Spain.
Marilia R Pappas, Laboratório de Genética Vegetal, EMBRAPA Recursos Genéticos e Biotecnologia, EPQB Final W5 Norte, Brasília, Brazil.
Lifeng Liu, Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA, USA.
Andrew Muchlinski, Department of Biological Sciences, Virginia Tech, Blacksburg, VA, USA.
Hui Jiang, Donald Danforth Plant Science Center, St. Louis, MO, USA.
Christine Shyu, Donald Danforth Plant Science Center, St. Louis, MO, USA.
Pu Huang, Donald Danforth Plant Science Center, St. Louis, MO, USA.
Jose Sebastian, Donald Danforth Plant Science Center, St. Louis, MO, USA.
Carol Laiben, Donald Danforth Plant Science Center, St. Louis, MO, USA.
Alyssa Medlin, Donald Danforth Plant Science Center, St. Louis, MO, USA.
Sankalpi Carey, Donald Danforth Plant Science Center, St. Louis, MO, USA.
Alyssa A Carrell, Biosciences Division, Oak Ridge National Laboratory, Oak Ridge, TN, USA.
Jin-Gui Chen, Biosciences Division, Oak Ridge National Laboratory, Oak Ridge, TN, USA.
Mariano Perales, Centro de Biotecnología y Genómica de Plantas, Universidad Politécnica de Madrid, Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA-CSIC), Madrid, Spain; Departamento de Biotecnología-Biología Vegetal, Escuela Técnica Superior de Ingeniería Agronómica, Alimentaria y de Biosistemas, Universidad Politécnica de Madrid, Madrid, Spain.
Kankshita Swaminathan, HudsonAlpha Institute for Biotechnology, Huntsville, AL, USA.
Isabel Allona, Centro de Biotecnología y Genómica de Plantas, Universidad Politécnica de Madrid, Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA-CSIC), Madrid, Spain; Departamento de Biotecnología-Biología Vegetal, Escuela Técnica Superior de Ingeniería Agronómica, Alimentaria y de Biosistemas, Universidad Politécnica de Madrid, Madrid, Spain.
Dario Grattapaglia, Laboratório de Genética Vegetal, EMBRAPA Recursos Genéticos e Biotecnologia, EPQB Final W5 Norte, Brasília, Brazil.
Elizabeth A Cooper, Advanced Plant Technology Program, Clemson University, Clemson, SC, USA.
Dorothea Tholl, Department of Biological Sciences, Virginia Tech, Blacksburg, VA, USA.
John P Vogel, Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA, USA.
David J Weston, Biosciences Division, Oak Ridge National Laboratory, Oak Ridge, TN, USA.
Xiaohan Yang, Center for Bioenergy Innovation, Oak Ridge National Laboratory, Oak Ridge, TN, USA.
Thomas P Brutnell, McClintock LLC, St. Louis, MO, USA.
Elizabeth A Kellogg, Donald Danforth Plant Science Center, St. Louis, MO, USA.
Ivan Baxter, Donald Danforth Plant Science Center, St. Louis, MO, USA.
Michael Udvardi, Noble Research Institute, Ardmore, OK, USA.
Yuhong Tang, Noble Research Institute, Ardmore, OK, USA.
Todd C Mockler, Donald Danforth Plant Science Center, St. Louis, MO, USA.
Thomas E Juenger, Department of Integrative Biology, University of Texas at Austin, Austin, TX, USA.
John Mullet, Department of Biochemistry and Biophysics, Texas A&M University, College Station, TX, USA.
Stefan A Rensing, Plant Cell Biology, Faculty of Biology, University of Marburg, Karl-von-Frisch-Str, Marburg, Germany.
Gerald A Tuskan, Center for Bioenergy Innovation, Oak Ridge National Laboratory, Oak Ridge, TN, USA.
Sabeeha S Merchant, Department of Chemistry and Biochemistry and Institute for Genomics and Proteomics, University of California, Los Angeles, CA, USA.
Gary Stacey, Division of Plant Science and Technology, C.S. Bond Life Science Center, University of Missouri, Columbia, MO, USA.
Jeremy Schmutz, HudsonAlpha Institute for Biotechnology, Huntsville, AL, USA; Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA, USA.
DATA AVAILABILITY
The RNA-seq data that support the findings of this study are available from the NCBI Sequence Read Archive (SRA) under accessions provided in Supplemental Data 2. To enable exploration of the transcriptome datasets for JGI Plant Gene Atlas v2.0, the data are hosted on Gene Atlas portal (https://plantgeneatlas.jgi.doe.gov) and JGI’s Phytozome plant portal.
Documentation for data processing and downloadable data are available in the ‘Methods’ section (https://plantgeneatlas.jgi.doe.gov). Custom code used in our analyses are publicly available at https://github.com/asreedasyam/geneatlas (permanent doi: https://doi.org/10.6084/m9.figshare.23635635.v2).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The work (proposal: 10.46936/10.25585/60000843) conducted by the U.S. Department of Energy Joint Genome Institute (https://ror.org/04xm1d337); a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy [DE-AC02-05CH11231]; the Populus work was partially supported by the BioEnergy Science Center (BESC) and Center for Bioenergy Innovation (CBI); BESC and CBI are Bioenergy Research Centers supported by the Office of Biological and Environmental Research in the US Department of Energy Office of Science; specific funding for the soybean transcriptome atlas was provided by a grant from the United Soybean Board (to G.S.); the switchgrass work was carried out under the support of the BioEnergy Science Center (BESC, a U.S. Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science, U.S. Department of Energy) and funded by the Samuel Roberts Noble Foundation; B.H. work was funded in part by U.S. DOE the Office of Biological and Environmental Research [DE-SC0012629[; the Chlamydomonas work is supported by the US Department of Energy Grant [DE-FC02-02ER63421]; National Institutes of Health (NIH) [R24 GM092473 to S.S.M.]; the Eucalyptus work was supported by the Brazilian Federal District Research Foundation (FAP-DF) NEXTREE grant; the Panicum hallii work was supported by the DOE Office of Science, Office of Biological and Environmental Research (BER) [DE-SC0008451, DE-SC0021126 to T.E.J.]; the sorghum work by JM laboratory was funded in part by the DOE Great Lakes Bioenergy Research Center [DOE BER grant DE-SC0018409]; the Setaria work was funded by the DOE Office of Science [DE-SC0018277, DE-SC0008769]; the Kalanchoë work was partially supported by the DOE Office of Science, Genomic Science Program [DE-SC0008834]; research related to Sphagnum was funded by DOE BER Early Career Research Program; Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the US DOE [DE-AC05-00OR22725]. Funding for open access charge: Office of Science of the U.S. Department of Energy [DE-AC02-05CH11231].
Conflict of interest statement. None declared.
REFERENCES
- 1. Koornneef M., Meinke D.. The development of Arabidopsis as a model plant. Plant J. 2010; 61:909–921. [DOI] [PubMed] [Google Scholar]
- 2. Berardini T.Z., Reiser L., Li D., Mezheritsky Y., Muller R., Strait E., Huala E.. The Arabidopsis information resource: making and mining the “gold standard” annotated reference plant genome. Genesis. 2015; 53:474–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li C., Li Q.G., Dunwell J.M., Zhang Y.M.. Divergent evolutionary pattern of starch biosynthetic pathway genes in grasses and dicots. Mol. Biol. Evol. 2012; 29:3227–3236. [DOI] [PubMed] [Google Scholar]
- 4. Nicotra A.B., Atkin O.K., Bonser S.P., Davidson A.M., Finnegan E.J., Mathesius U., Poot P., Purugganan M.D., Richards C.L., Valladares F.et al.. Plant phenotypic plasticity in a changing climate. Trends Plant Sci. 2010; 15:684–692. [DOI] [PubMed] [Google Scholar]
- 5. Raissig M.T., Matos J.L., Anleu Gil M.X., Kornfeld A., Bettadapur A., Abrash E., Allison H.R., Badgley G., Vogel J.P., Berry J.A.et al.. Mobile MUTE specifies subsidiary cells to build physiologically improved grass stomata. Science. 2017; 355:1215–1218. [DOI] [PubMed] [Google Scholar]
- 6. Gollery M., Harper J., Cushman J., Mittler T., Girke T., Zhu J.-K., Bailey-Serres J., Mittler R., Fay J.C., Wu C.I.et al.. What makes species unique? The contribution of proteins with obscure features. Genome Biol. 2006; 7:R57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gollery M., Harper J., Cushman J., Mittler T., Mittler R.. POFs: what we don’t know can hurt us. Trends Plant Sci. 2007; 12:492–496. [DOI] [PubMed] [Google Scholar]
- 8. Rhee S.Y., Mutwil M.. Towards revealing the functions of all genes in plants. Trends Plant Sci. 2014; 19:212–221. [DOI] [PubMed] [Google Scholar]
- 9. Moreno P., Fexova S., George N., Manning J.R., Miao Z., Mohammed S., Muñoz-Pomer A., Fullgrabe A., Bi Y., Bush N.et al.. Expression Atlas update: gene and protein expression in multiple species. Nucleic Acids Res. 2022; 50:D129–D140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bastian F.B., Roux J., Niknejad A., Comte A., Fonseca Costa S.S., de Farias T.M., Moretti S., Parmentier G., de Laval V.R., Rosikiewicz M.et al.. The Bgee suite: integrated curated expression atlas and comparative transcriptomics in animals. Nucleic Acids Res. 2021; 49:D831–D847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yu Y., Zhang H., Long Y., Shu Y., Zhai J.. Plant Public RNA-seq Database: a comprehensive online database for expression analysis of ∼45 000 plant public RNA-Seq libraries. Plant Biotechnol. J. 2022; 20:806–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Levin J.Z., Yassour M., Adiconis X., Nusbaun C., Thompson D.A., Friedman N., Gnirke A., Regev A.. Comprehensive comparative analysis of strand specific RNA sequencing methods. Nat. Methods. 2010; 7:709–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ross M.G., Russ C., Costello M., Hollinger A., Lennon N.J., Hegarty R., Nusbaum C., Jaffe D.B., Tsujimoto T., Jeromin A.et al.. Characterizing and measuring bias in sequence data. Genome Biol. 2013; 14:R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sudmant P.H., Alexis M.S., Burge C.B.. Meta-analysis of RNA-seq expression data across species, tissues and studies. Genome Biol. 2015; 16:287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yu Y., Fuscoe J.C., Zhao C., Guo C., Jia M., Qing T., Bannon D.I., Lancashire L., Bao W., Du T.et al.. A rat RNA-Seq transcriptomic BodyMap across 11 organs and 4 developmental stages. Nat. Commun. 2014; 5:3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N.et al.. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012; 40:D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boyle N.R., Page M.D., Liu B., Blaby I.K., Casero D., Kropat J., Cokus S.J., Hong-Hermesdorf A., Shaw J., Karpowicz S.J.et al.. Three acyltransferases and nitrogen-responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas. J. Biol. Chem. 2012; 287:15811–15825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Inglis P.W., Pappas M.d.C.R., Resende L.V., Grattapaglia D.. Fast and inexpensive protocols for consistent extraction of high quality DNA and RNA from challenging plant and fungal samples for high-throughput SNP genotyping and sequencing applications. PLoS One. 2018; 13:e0206085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hufnagel B., Marques A., Soriano A., Marquès L., Divol F., Doumas P., Sallet E., Mancinotti D., Carrere S., Marande W.et al.. High-quality genome sequence of white lupin provides insight into soil exploration and seed quality. Nat. Commun. 2020; 11:492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moore K.J., Moser L.E., Vogel K.P., Waller S.S., Johnson B.E., Pedersen J.F.. Describing and quantifying growth stages of perennial forage grasses. Agron. J. 1991; 83:1073–1077. [Google Scholar]
- 21. Weng X., Song H., Sreedasyam A., Haque T., Zhang L., Chen C., Yoshinaga Y., Williams M., O’Malley R.C., Grimwood J.et al.. Transcriptome and DNA methylome divergence of inflorescence development between 2 ecotypes in Panicum hallii. Plant Physiol. 2023; 192:2374–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Perroud P.-F., Haas F.B., Hiss M., Ullrich K.K., Alboresi A., Amirebrahimi M., Barry K., Bassi R., Bonhomme S., Chen H.et al.. The Physcomitrella patens gene atlas project: large-scale RNA-seq based expression data. Plant J. 2018; 95:168–182. [DOI] [PubMed] [Google Scholar]
- 23. Cove D.J., Perroud P.-F., Charron A.J., McDaniel S.F., Khandelwal A., Quatrano R.S.. The moss Physcomitrella patens: a novel model system for plant development and genomic studies. Cold Spring Harb. Protoc. 2009; 2009:pdb.emo115. [DOI] [PubMed] [Google Scholar]
- 24. Reski R., Abel W.O.. Induction of budding on chloronemata and caulonemata of the moss, Physcomitrella patens, using isopentenyladenine. Planta. 1985; 165:354–358. [DOI] [PubMed] [Google Scholar]
- 25. Fernandez-Pozo N., Haas F.B., Meyberg R., Ullrich K.K., Hiss M., Perroud P.-F., Hanke S., Kratz V., Powell A.F., Vesty E.F.et al.. PEATmoss (Physcomitrella Expression Atlas Tool): a unified gene expression atlas for the model plant Physcomitrella patens. Plant J. 2020; 102:165–177. [DOI] [PubMed] [Google Scholar]
- 26. Ibañez C., Ramos A., Acebo P., Contreras A., Casado R., Allona I., Aragoncillo C.. Overall alteration of circadian clock gene expression in the chestnut cold response. PLoS One. 2008; 3:e3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mccormick R.F., Truong S.K., Sreedasyam A., Jenkins J., Shu S., Sims D., Kennedy M., Amirebrahimi M., Weers B.D., Mckinley B.et al.. The Sorghum bicolor reference genome: improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization. Plant J. 2017; 93:338–354. [DOI] [PubMed] [Google Scholar]
- 28. Vanderlip R.L., Reeves H.E.. Growth stages of sorghum [Sorghum bicolor, (L.) Moench.]. Agron. J. 1972; 64:13–16. [Google Scholar]
- 29. Cooper E.A., Brenton Z.W., Flinn B.S., Jenkins J., Shu S., Flowers D., Luo F., Wang Y., Xia P., Barry K.et al.. A new reference genome for Sorghum bicolor reveals high levels of sequence similarity between sweet and grain genotypes: implications for the genetics of sugar metabolism. BMC Genomics. 2019; 20:420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li Z., Trick H.N.. Rapid method for high-quality RNA isolation from seed endosperm containing high levels of starch. BioTechniques. 2005; 38:872. [DOI] [PubMed] [Google Scholar]
- 31. Wu T.D., Nacu S.. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010; 26:873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Anders S., Pyl P.T., Huber W.. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015; 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Risso D., Ngai J., Speed T.P., Dudoit S.. Normalization of RNA-seq data using factor analysis of control genes or samples (RUVSeq). Nat. Biotechnol. 2014; 32:896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li B., Dewey C.N.. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011; 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Trapnell C., Williams B.a., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L.. Transcript assembly and abundance estimation from RNA-Seq reveals thousands of new transcripts and switching among isoforms. Nat. Biotechnol. 2011; 28:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Langfelder P., Horvath S.. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008; 9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Csardi G., Nepusz T.. The igraph software package for complex network research. InterJournal,Complex Syst. 2006; 1695:1–9. [Google Scholar]
- 39. Alexa A., Rahnenfuhrer J.. topGO: enrichment analysis for gene ontology. 2022; R package version 2.42.0.
- 40. Kanehisa M., Goto S.. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000; 28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Emms D.M., Kelly S.. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019; 20:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zeng L., Zhang Q., Sun R., Kong H., Zhang N., Ma H.. Resolution of deep angiosperm phylogeny using conserved nuclear genes and estimates of early divergence times. Nat. Commun. 2014; 5:4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kumar S., Stecher G., Suleski M., Hedges S.B.. TimeTree: a Resource for Timelines, Timetrees, and Divergence Times. Mol. Biol. Evol. 2017; 34:1812–1819. [DOI] [PubMed] [Google Scholar]
- 44. Keightley P.D., Hill W.G.. Variation maintained in quantitative traits with mutation–selection balance: pleiotropic side-effects on fitness traits. Proc. R. Soc. Lond. B Biol. Sci. 1990; 242:95–100. [Google Scholar]
- 45. Yanai I., Benjamin H., Shmoish M., Chalifa-Caspi V., Shklar M., Ophir R., Bar-Even A., Horn-Saban S., Safran M., Domany E.et al.. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics. 2005; 21:650–659. [DOI] [PubMed] [Google Scholar]
- 46. Kryuchkova-Mostacci N., Robinson-Rechavi M.. A benchmark of gene expression tissue-specificity metrics. Brief. Bioinform. 2017; 18:205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Manzano C., Pallero-Baena M., Casimiro I., De Rybel B., Orman-Ligeza B., Van Isterdael G., Beeckman T., Draye X., Casero P., Del Pozo J.C. The emerging role of reactive oxygen species signaling during lateral root development. Plant Physiol. 2014; 165:1105–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Passaia G., Queval G., Bai J., Margis-Pinheiro M., Foyer C.H.. The effects of redox controls mediated by glutathione peroxidases on root architecture in Arabidopsis thaliana. J. Exp. Bot. 2014; 65:1403–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ma F., Wang L., Li J., Samma M.K., Xie Y., Wang R., Wang J., Zhang J., Shen W.. Interaction between HY1 and H2O2 in auxin-induced lateral root formation in Arabidopsis. Plant Mol. Biol. 2014; 85:49–61. [DOI] [PubMed] [Google Scholar]
- 50. Bruex A., Kainkaryam R.M., Wieckowski Y., Kang Y.H., Bernhardt C., Xia Y., Zheng X., Wang J.Y., Lee M.M., Benfey P.et al.. A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genet. 2012; 8:e1002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kogawara S., Yamanoshita T., Norisada M., Kojima K.. Steady sucrose degradation is a prerequisite for tolerance to root hypoxia. Tree Physiol. 2014; 34:229–240. [DOI] [PubMed] [Google Scholar]
- 52. Li W., Lan P.. Re-analysis of RNA-seq transcriptome data reveals new aspects of gene activity in Arabidopsis root hairs. Front. Plant Sci. 2015; 6:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Loreti E., Poggi A., Novi G., Alpi A., Perata P.. A genome-wide analysis of the effects of sucrose on gene expression in Arabidopsis seedlings under anoxia. Plant Physiol. 2005; 137:1130–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jin J., Tian F., Yang D.-C., Meng Y.-Q., Kong L., Luo J., Gao G.. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017; 45:D1040–D1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mukhtar M.S., Carvunis A.-R., Dreze M., Epple P., Steinbrenner J., Moore J., Tasan M., Galli M., Hao T., Nishimura M.T.et al.. Independently Evolved Virulence Effectors Converge onto Hubs in a Plant Immune System Network. Science. 2011; 333:596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Canales J., Moyano T.C., Villarroel E., Gutiérrez R.A.. Systems analysis of transcriptome data provides new hypotheses about Arabidopsis root response to nitrate treatments. Front. Plant Sci. 2014; 5:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ghazalpour A., Doss S., Zhang B., Wang S., Plaisier C., Castellanos R., Brozell A., Schadt E.E., Drake T.A., Lusis A.J.et al.. Integrating genetic and network analysis to characterize genes related to mouse weight. PLoS Genet. 2006; 2:e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Horvath S., Zhang B., Carlson M., Lu K.V., Zhu S., Felciano R.M., Laurance M.F., Zhao W., Qi S., Chen Z.et al.. Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:17402–17407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liu W., He G., Deng X.W.. Biological pathway expression complementation contributes to biomass heterosis in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2021; 118:e2023278118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Miller J.A., Oldham M.C., Geschwind D.H.. A systems level analysis of transcriptional changes in Alzheimer's disease and normal aging. J. Neurosci. 2008; 28:1410–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Torkamani A., Dean B., Schork N.J., Thomas E.A.. Coexpression network analysis of neural tissue reveals perturbations in developmental processes in schizophrenia. Genome Res. 2010; 20:403–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Voineagu I., Wang X., Johnston P., Lowe J.K., Tian Y., Horvath S., Mill J., Cantor R.M., Blencowe B.J., Geschwind D.H.. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011; 474:380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Somssich M., Khan G.A., Persson S.. Cell wall heterogeneity in root development of Arabidopsis. Front. Plant Sci. 2016; 7:1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Peña M.J., Kong Y., York W.S., O’Neill M.A.. A galacturonic acid-containing xyloglucan is involved in Arabidopsis root hair tip growth. Plant Cell. 2012; 24:4511–4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Faye J.M., Akata E.A., Sine B., Diatta C., Cisse N., Fonceka D., Morris G.P.. Quantitative and population genomics suggest a broad role of stay-green loci in the drought adaptation of sorghum. Plant Genome. 2022; 15:e20176. [DOI] [PubMed] [Google Scholar]
- 66. Zhang F., Wu J., Sade N., Wu S., Egbaria A., Fernie A.R., Yan J., Qin F., Chen W., Brotman Y.et al.. Genomic basis underlying the metabolome-mediated drought adaptation of maize. Genome Biol. 2021; 22:260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Huang J., Zhao X., Chory J.. The Arabidopsis Transcriptome Responds Specifically and Dynamically to High Light Stress. Cell Rep. 2019; 29:4186–4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Swift J., Alvarez J.M., Araus V., Gutiérrez R.A., Coruzzi G.M.. Nutrient dose-responsive transcriptome changes driven by Michaelis–Menten kinetics underlie plant growth rates. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:12531–12540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Li Y., Wang M., Teng K., Dong D., Liu Z., Zhang T., Han L.. Transcriptome profiling revealed candidate genes, pathways and transcription factors related to nitrogen utilization and excessive nitrogen stress in perennial ryegrass. Sci. Rep. 2022; 12:3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gelli M., Duo Y., Konda A., Zhang C., Holding D., Dweikat I., Maunder A.B., Edwards G.E., Franceschi V.R., Voznesenskaya E.V.et al.. Identification of differentially expressed genes between sorghum genotypes with contrasting nitrogen stress tolerance by genome-wide transcriptional profiling. BMC Genomics. 2014; 15:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data that support the findings of this study are available from the NCBI Sequence Read Archive (SRA) under accessions provided in Supplemental Data 2. To enable exploration of the transcriptome datasets for JGI Plant Gene Atlas v2.0, the data are hosted on Gene Atlas portal (https://plantgeneatlas.jgi.doe.gov) and JGI’s Phytozome plant portal.
Documentation for data processing and downloadable data are available in the ‘Methods’ section (https://plantgeneatlas.jgi.doe.gov). Custom code used in our analyses are publicly available at https://github.com/asreedasyam/geneatlas (permanent doi: https://doi.org/10.6084/m9.figshare.23635635.v2).