Abstract
Sensors to measure the abundance and signaling of intracellular molecules are crucial for understanding their physiological functions. Although conventional fluorescent protein-based sensors have been designed, RNA-based sensors are promising imaging tools. Numerous RNA-based sensors have been developed. These sensors typically contain RNA G-quadruplex (RG4) motifs and thus may be suboptimal in living cells. Here we describe RNA-based sensors based on Pepper, a fluorogenic RNA without an RG4 motif. With Pepper, we engineered various sensors for metabolites, synthetic compounds, proteins and metal ions in vitro and in living cells. In addition, these sensors show high activation and selectivity, demonstrating their universality and robustness. In the case of sensors responding to S-adenosylmethionine (SAM), a metabolite produced by methionine adenosyltransferase (MATase), we showed that our sensors exhibited positively correlated fluorescence responding to different SAM levels. Importantly, we revealed the SAM biosynthesis pathway and monitored MATase activity and gene expression spatiotemporally in living individual human cells. Additionally, we constructed a ratiometric SAM sensor to determine the inhibition efficacy of a MATase inhibitor in living cells. Together, these sensors comprising Pepper provide a useful platform for imaging diverse cellular targets and their signaling pathway.
Graphical Abstract
Graphical Abstract.
INTRODUCTION
Genetically encoded sensors are imaging tools that allow cellular small molecules, proteins and ions to be imaged over time in living individual cells. These sensors can characterize the spatial and temporal distributions of the cellular targets and provide insights into how various signaling pathways regulate intracellular molecules (1). The conventional genetically encoded sensors are composed of a fluorescent protein flanking a target-binding domain. Target binding to the sensor triggers conformational changes and folds the fluorescent protein, thus inducing fluorescence (2). However, the lack of a target-binding protein domain prevents the application of fluorescent protein-based sensors (3,4). Additionally, the low signal-to-noise ratio of fluorescent protein-based sensors limits the sensitive detection of cellular targets (1,2,5).
In addition to fluorescent protein-based sensors, genetically encoded sensors can be composed of RNA (6). RNA-based sensors use a target-binding RNA to connect to a fluorogenic RNA aptamer, such as Spinach or Broccoli (7,8). In RNA-based sensors, the fluorophore-binding pocket of Spinach/Broccoli is connected to an RNA stem whose thermodynamic stability is regulated by target-binding RNA aptamers (6). Target binding induces a conformational change that regulates RNA folding, resulting in fluorescence (6). These genetically encoded RNA-based biosensors allow cellular targets to be imaged in real time.
However, these RNA-based biosensors widely use fluorogenic RNA aptamers containing RNA G-quadruplex (RG4) motifs (6,9–20). RG4-containing aptamers or biosensors may be suboptimal in living cells as RG4 may be unfolded or depleted by the cellular machinery (Supplementary Figure S1) (21–24). It is therefore optimal to develop RNA-based biosensors with non-RG4 motifs and robust optical performance for cell-based study.
Here, we describe an approach for generating RNA-based sensors with bright and stable fluorogenic RNA without RG4 motifs. These sensors are composed of Pepper, a recently reported fluorogenic RNA that activates the fluorescence of the otherwise non-fluorescent HBC fluorophore (25). Pepper exhibits several crucial merits. Pepper does not harbor the RG4 motif (26,27). Although there is no direct evidence or data to show that RG4-containing fluorogenic aptamers cannot function in cells, previous research suggests that RG4 motifs may be unfolded by cellular helicase (21,22) in living cells, which could inhibit the function of RG4-containing aptamers (22–24,28). Therefore, generating RNA-based sensors using Pepper or other RG4-free fluorogenic aptamers (29–33) would avoid this potential problem. Moreover, Pepper and HBC complexes showed an order of magnitude enhanced cellular fluorescence intensity and fluorescence turn-on ratio, one or two orders of magnitude enhanced affinity, ∼20°C increased Tm, expanded pH tolerance and a broad spectral range available for live cell studies relative to other fluorogenic RNA aptamers (25). In addition, Pepper has high stability since Pepper has a monomeric structure, and magnesium independence, without needing scaffold RNA and little interference with RNA compartmental localization (25). However, Pepper has not been designed as the RNA-based sensor for tracking devise targets, such as small molecules, proteins and ions, and their cellular signaling pathway.
We thus constructed Pepper-based sensors and imaged various targets in living cells, including signaling molecules, metabolites, small molecular drugs, proteins and metal ions, as well as the cellular signaling pathways. We first fused Pepper into an S-adenosylmethionine (SAM)-binding aptamer, developing an RNA-based sensor that is highly activated upon binding SAM. Using SAM sensors, we readily detected endogenous SAM and its dynamics with cell-to-cell heterogeneity. Additionally, we imaged the metabolic origin of SAM synthesis, and the activity and gene expression of methionine adenosyltransferase (MATase) in single living human cells. Moreover, we constructed a ratiometric SAM sensor to measure the half-maximal inhibitory concentration (IC50) of the MATase inhibitor AG-270. Furthermore, we showed that other target-binding aptamers, including tetracycline-binding, guanine-binding, protein-binding and metal ion-binding aptamers, can be converted into Pepper for intracellular sensing, demonstrating the universality of the non-RG4 RNA sensor tools. Importantly, Pepper-based sensors overcome the key defects of the previous Broccoli-based sensors which cannot accurately detect the target in vitro and in cells. Overall, Pepper provides a bright, stable, multicolor and non-RG4 platform for generating universal and robust RNA-based sensor tools with minimal cellular perturbations in living bacteria and human cells.
MATERIALS AND METHODS
Reagents and equipment
All DNAs were ordered from Tsingke Biotechnology (Beijing, China). Fluorescence was measured on a FluoroMax + spectrofluorometer (Horiba Scientific) using FluorEssence software (v.3.9). Fluorescence imaging was acquired on an Olympus SpinSR10 microscope (Olympus). Data were plotted using Origin 2021 software. All RNA-binding fluorophores, including HBC, HBC620, BI, DFHO and tetramethyl rhodamine-dinitroaniline (TMR-DN) used in this study were synthesized as described previously (25,34,35).
Cell lines and transfection
HEK293T (ATCC-CRL-11268) and HeLa (ATCC-CCL-2) cells were cultured under standard tissue culture conditions (37°C and 5% CO2) in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen C11995500BT) containing 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 mg/ml streptomycin. Cells were screened for mycoplasma contamination by a mycoplasma detection kit (Vazyme D101). To transfect cells with plasmids, Lipofectamine 3000 (Invitrogen 92008) was used as the transfection reagent, and transfection was performed according to the manufacturer's instructions. Cells were seeded onto 24-well plates (Nest 801006) 1 day prior to transfection.
Preparation of RNA in vitro
For all Pepper-based sensor RNAs, double-stranded DNA (dsDNA) templates were designed to contain a 5′ T7 promoter to be used for in vitro transcription. dsDNA templates were prepared from single-stranded DNA oligos (Tsingke Biotechnology). DNA templates were amplified by polymerase chain reaction (PCR) using Taq DNA polymerase (Vazyme P112) and checked for quality using 1.5% agarose gel electrophoresis. PCRs were purified with the PCR purification kit (Axygen AP-PCR-250).
In vitro transcription reactions using the T7 High Yield RNA Transcription Kit (Vazyme DD4201) were carried out according to the manufacturer's instruction. Transcription reactions were terminated by treatment with RNase-free DNase I at 37°C for at least 15 min. An 80 μl aliquot of RNase-free water was added followed by an equal volume of water-saturated phenol–chloroform, and this was then centrifuged at 4°C, 12 000 rpm for 15 min. The supernatant liquid was aspirated with sodium acetate and 2 times the volume of anhydrous ethanol, and left to settle at –20°C for 30 min. Afterwards, the precipitate was washed with 75% anhydrous ethanol prepared with enzyme-free water and centrifuged for 5 min, and then dissolved in enzyme-free water.
In vitro characterization of the sensors
All concentrations quoted below are final concentrations in the measurement. In vitro purified RNA (1 μM) was mixed with HBC (10 μM) in the absence or presence of SAM (0.1 mM, Sigma-Aldrich A7007), tetracycline (0.1 mM, BBI A600504-0025), guanine (0.1 mM, Solarbio G8260) or silver acetate (CH3COOAg, 0.1 mM, Sigma-Aldrich 216674) in the buffer (40 mM HEPES, pH 7.4, 100 mM KCl and 1 mM MgCl2). After a 1 h incubation, the fluorescence of each sample was measured at 37°C using a spectrofluorometer with 485 nm excitation, 530 nm emission, 5 nm slit widths and 0.1 s integration time. The buffer containing HBC (10 μM) was measured as a background signal. The background signal was subtracted from the signal obtained from each RNA sample measurement. Pepper- or Broccoli-based RNA sensors were measured with a cognate fluorophore (10 μM HBC or 10 μM BI) in 40 mM HEPES pH 7.4, 100 mM KCl and 1 mM MgCl2 buffer. Corn-based RNA sensors were measured with DFHO (10 μM) in 40 mM HEPES pH 7.4, 100 mM KCl and 5 mM MgCl2 buffer.
For protein sensors, in vitro transcribed RNA (0.2 μM) was mixed with HBC (10 μM) in the absence or presence of streptavidin (100 μg/ml, ∼2 μM) in the buffer. After a 1 h incubation, the fluorescence signal of each sample was measured at room temperature (∼25°C) using a spectrofluorometer with 485 nm excitation, 530 nm emission, 5 nm slit widths and 0.1 s integration time.
The sequences of all Pepper-based sensors used in this study are shown in Supplementary Table S1.
Selectivity measurement of Pepper-based sensors
To test the selectivity of the Pepper-based SAM sensors, Pepper-based SAM sensor RNA (1 μM) was mixed with HBC (10 μM) in the presence of the target (0.1 mM) or its analogs in the buffer. The samples were incubated at 37°C for 1 h. The fluorescence signal of each sample was measured at 37°C using a spectrofluorometer with 505 nm excitation, 545 nm emission, 5 nm slit widths and 0.1 s integration time. Analogs of SAM include SAH (Aladdin S139501), adenosine (Coolaber CA1241) or methionine (Coolaber CM7211). Analogs of tetracycline include doxycycline (Solarbio ID0670), neomycin (Solarbio N8090), tobramycin (Solarbio T8810), gentamycin (Solarbio G8170), ampicillin (Solarbio A1170) and kanamycin (Solarbio K1030). Analogs of guanine including guanosine (MACKLIN G810366) and adenine (BBI A600013-0025) were used in guanine sensor selectivity measurement. Bovine serum albumin (BSA; Cusabio NP009501B), lysozyme (Cusabio NP004301C) and ovalbumin (Cusabio NP004201C) was used in streptavidin sensor selectivity measurement.
Dose–response curve measurements
Dose–response curves were determined by measuring fluorescence in the presence of fixed concentrations of RNA sensor (0.2 μM streptavidin sensor, 1 μM SAM sensor, tetracycline sensor, guanine sensor or Ag+ sensor) and fixed concentrations of HBC (10 μM) in relation to target concentration. Pepper-based sensor RNA was incubated for 1 h with a range of concentrations of target molecules in the buffer with HBC (10 μM) at 37°C. After a 1 h incubation, the fluorescence signal of each sample was measured using a spectrofluorometer at 37°C with an excitation wavelength of 485 nm, an emission wavelength of 530 nm, a slit width of 5 nm and an integration time of 0.1 s.
Microscopy and image processing
To image Escherichia coli cells, we coated confocal dishes with poly-l-lysine (Sangon Biotech A600751) for at least 4 h and rinsed them once in water. We incubated isopropyl-β-d-thiogalactopyranoside (IPTG)-induced E. coli with M9 minimal medium (Gibco A1374401) containing HBC (10 μM) on pre-treated confocal dishes. Live cell fluorescence images were acquired through a ×100 oil objective mounted on a microscope. Images were analyzed using Fiji software. During live cell imaging, conditions were maintained at 37°C.
To image mammalian cells, we coated 24-well confocal plates with poly-d-lysine (Sangon E607014) for at least 4 h and rinsed them once in water. One day after transfection, the cells were subcultured onto the pre-treated confocal plates. One hour before imaging, the cell medium was changed to phenol red-free DMEM (Biological Industries 06–1052-04–1ACS) supplemented with 10% FBS and HBC (10 μM). Live cell fluorescence images were acquired through a ×20 air objective (NA 0.75) mounted on the microscope. Images were analyzed using Fiji software. During live cell imaging, conditions were maintained at 37°C and 5% CO2.
Flow cytometry of Pepper-based sensor expressing cells
Two days after transfection of Pepper-based sensors, HEK293T cells were harvested and resuspended in phosphate-buffered saline (PBS) containing HBC (10 μM) and 4% FBS with or without cycloleucine (30 mM) or the cognate targets (tetracycline 0.1 mM, guanine 0.1 mM, CH3COOAg 20 μM) and kept on ice until analysis. Cells were analyzed using an LSRFortessa (BD Biosciences). Populations of cells were gated to avoid cell doublets detected by forward and side scattering, followed by gating for live cells by fluorescein isothiocyanate (FITC) fluorescence. Untransfected cells were used as a negative control for gating of fluorescence of Pepper-based sensors. Plots were generated using FlowJo software.
Determination of the IC50 of AG-270 with a ratiometric SAM sensor
To determine the activity of MATase, we constructed a plasmid expressing an F30 scaffold with a Pepper-based SAM sensor in one arm and RhoBAST in the other (Supplementary Table S2). Then 24-well confocal plates were coated with poly-d-lysine (Sangon E607014) for at least 4 h and rinsed once in water. HEK293T cells were transfected with plasmid expressing the ratiometric SAM sensor or control RNA. One day after transfection, the cells were subcultured onto the pre-treated confocal plates. In the meantime, different concentrations of AG-270 (5000, 1666.67, 555.56, 185.18, 61.73, 20.58, 6.86, 2.29 and 0.76 nM) were added to the medium. After incubation for 24 h, HBC (10 μM) and TMR-DN (100 nM) were added to the medium for imaging.
To process the image of the ratiometric SAM sensor, it is important to generate the green-to-red fluorescence ratio only for areas containing the cells. The red channel was used to create a binary mask to identify the cells. The region of interest (ROI) on this mask was regenerated in the images of both red and green channels. Then, areas outside the ROI were cleared. Finally, images containing a green-to-red fluorescence ratio were generated by dividing the masked green channel image by a masked red channel image. The ratio in the image is coded by 16-color look up tables.
To calculate the IC50 of AG-270, we counted the green-to-red fluorescence ratio signal of >20 cells at each pre-set concentration. The inhibition rate was calculated according to the following equation:
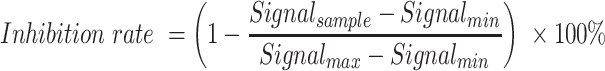 |
Subsequently, we plotted the logarithm of the concentration against the inhibition rate in Origin software and fitted it with a dose–response curve. The IC50 was acquired as the concentration corresponding to the 50% inhibition rate on the curve.
Detection of MATase expression by real-time PCR
Total RNA was isolated from HeLa cells using TRIzol reagent (Invitrogen 15596026) following the manufacturer's instructions. The quality and quantity of RNA were assessed using Nano drop (Thermo Fisher). Reverse transcription was performed using HiScript III 1st Strand cDNA Synthesis Kit (Vazyme R312) with 1 μg of RNA in a 20 μl system.
Real-time PCR analysis was conducted using Power SYBR Green PCR Master Mix (Applied Biosystems, 4309155) and carried out on a LightCycler 480 Real-Time PCR System (Roche Applied Science). Each target gene was amplified using the following primer sequences: glyceraldehyde phosphate dehydrogenase (GAPDH) forward primer 5′-TGGGTGTGAAACCATGAGAAGT-3′, reverse primer 5′-TGAGTCCTTCCACGATACCAA-3′; MAT1A forward primer 5′-GTGTGACCACTCTCTAAGTG-3′, reverse primer 5′-TGCCGGTCTTGCACACTGTC-3′.
The amplification protocol included an initial denaturation step at 95°C for 30 s, followed by 40 cycles of denaturation at 95°C for 10 s, and annealing and extension at 60°C for 15 s. A melt curve analysis was performed to verify the specificity of the amplification products.
Gene expression levels were calculated using the 2(–ΔΔCt) method (36), where the Ct values of the target gene were normalized to the Ct values of GAPDH and compared with a calibrator sample. Statistical analysis was performed using Origin 2021 software.
For quality control, three technical replicates were included, and no-template controls (NTCs) were used to monitor potential contamination or primer-dimer formation.
Live-cell imaging of Pepper-based sensors in E. coli
BL21 E. coli cells (Tsingke TSC-E01) were transformed with 40 ng of plasmid DNA expressing the Pepper-based sensor chimeras in pET28a under the control of a T7 promoter. Cells were plated, grown overnight and single colonies were picked for inoculation in Luria broth containing kanamycin (Solarbio K1030, 50 μg/ml). The broth was diluted to OD600 = 0.4, followed by the addition of 1 mM IPTG to the broth and shaking at 37°C for 2 h. A 100 μl aliquot of culture was then removed, centrifuged and resuspended in 2 ml of pH 6.0 M9 minimal medium containing HBC (2 μM), dispensed onto poly-l-lysine-coated confocal dishes (Nest 806001) and incubated at 37°C for 45 min.
To image SAM synthesis, cells were treated with 2 μl of 1000x methionine stock added to a final concentration of 50 mg/ml. Cells were imaged using the live-cell imaging conditions described above. Images were taken intermittently over a period of 3 h at 10 min intervals with an exposure time of 500 ms per image.
For tetracycline, guanine or Ag+ imaging, the target (0.1 mM) was added to the M9 medium and incubated with cells at 37°C for 45 min. Cells were imaged using the live-cell imaging conditions described above.
To image the streptavidin level, BL21 E. coli cells (Tsingke TSC-E01) were co-transformed with pET28a plasmid (carrying a kanamycin resistance gene) expressing a streptavidin sensor and pET21a plasmid (carrying an ampicillin resistance gene) expressing the streptavidin protein. Cells were plated and grown overnight on solid Luria broth plates containing both ampicillin (Solarbio A1170, 100 μg/ml) and kanamycin (Solarbio K1030, 50 μg/ml). Single colonies were picked for inoculation in Luria broth containing both ampicillin (100 μg/ml) and kanamycin (50 μg/ml). The broth was diluted to OD600 = 0.4, followed by the addition of 1 mM IPTG to the broth and shaking at 37°C for 2 h. A 100 μl aliquot of culture was then removed, centrifuged and resuspended in 2 ml of pH 6.0 M9 minimal medium containing HBC (2 μM), dispensed onto poly-l-lysine-coated confocal dishes and incubated at 37°C for 45 min. Cells were imaged using the live-cell imaging conditions described above.
Intracellular imaging in living mammalian cells
For intracellular SAM imaging, HEK293T or HeLa cells were transfected with Tornado plasmids encoding SAM sensors or Pepper aptamers. After 1 day of transfection, the cells were passaged onto coated glass-bottomed plates. After 3 days of transfection, cells were imaged using the live-cell imaging conditions described above.
For imaging SAM synthesis and consumption, cells expressing the SAM sensor were incubated with HBC-containing medium 1 h before imaging. DMEM lacking amino acids was supplemented with 1× minimal essential medium with non-essential amino acids (MEM NEAA) and other essential amino acids except methionine to generate the methionine-free medium. Immediately before imaging, we switched the medium to methionine-free DMEM. Cells were imaged for 4 h at 30 min intervals. Then, methionine (100 μM final) was reintroduced to the medium. Cells were imaged for another 4 h at 30 min intervals.
To monitor the activity of MATase, we added cycloleucine (Ark Pharm AK-29341, 30 mM) to the medium. Cells were then imaged for 2 h at 10 min intervals. We then withdrew the cycloleucine by changing the cell medium to fresh medium and continued to image the cells at 10 min intervals for 3 h.
To image the MATase transcription level in HeLa cells, we treated HeLa cells with SAHA (suberoylanilide hydroxamic acid; Yuanye Bio, S42929). Cells were imaged for 8 h at 30 min intervals.
For tetracycline or guanine imaging, HEK293T cells were transfected with Tornado plasmids encoding a tetracycline sensor or a guanine sensor. After 1 day of transfection, the cells were passaged onto coated glass-bottomed plates. After 3 days of transfection, we replaced the cell culture medium with phenol red-free DMEM (Biological Industries 06–1052-04–1ACS) containing HBC (10 μM) and tetracycline (BBI A600504) or guanine (Solarbio G8260) and incubated it at 37°C for 1 h. Cells were then imaged using the live-cell imaging conditions described above.
Quantification and statistical analysis
All in vitro experiments were performed in three independent repeats. The data are represented as the mean ± standard deviation (SD) of three independent repeats in the plots (n = 3).
For the trajectory plot in Figure 3, Figure 4 and Supplementary Figure S7, the fluorescence intensity of the cell was calculated using Fiji by measuring the total fluorescence signal in an ROI divided by ROI area (μm2) and subtracting the background based on the fluorescence intensity of an untransfected cell. The ROI was defined as the cell cytoplasm area.
RESULTS
Strategy for designing RNA-based sensors with pepper
Prior strategies for the construction of RNA-based biosensors mainly took advantage of the formation of RG4-containing pockets by an allosteric effect triggered with the fused targeting-binding aptamer (Figure 1A; Supplementary Figure S2). For example, Spinach/Broccoli have been extensively constructed as RNA-based sensors for various targets (4,6,9–20,37). These Spinach/Broccoli-based sensors all contain RG4 motifs that can be regulated by target-binding RNA aptamers (Figure 1A; Supplementary Figure S2). This RG4-dependent design is also found in other fluorogenic RNA aptamer-based sensors. For example, our lab and the Jaffrey lab developed a series of Red Broccoli-, Spinach2- and Corn-based sensors (6,20,38,39). These sensors contain RG4 motifs in the fluorogenic aptamer domain.
Figure 1.
Identification of an insertion point on Pepper without an RG4 motif. (A) Diagram of the structure of Pepper and Broccoli aptamers. Broccoli contains an RG4 motif (red parallelograms and red circles) to induce the fluorescence of DFHBI or its derivatives. Pepper's conformation does not contain a G-quadruplex. (B) Pepper does not contain a potassium ion-dependent RG4 structure. In these experiments, we took advantage of the fact that RG4 is highly dependent on potassium to form its structure, while lithium fails to induce RG4 folding. Thus, we tested the potassium dependence for RNA aptamer-induced fluorescence. We diluted Pepper or Broccoli RNA (1 μM) in buffer containing K+ or Li+ (100 mM). Broccoli showed potassium dependence for its fluorescence. Notably, Pepper did not show potassium dependence, suggesting that Pepper does not contain an RG4 structure. The data are represented as the mean ± SD of three independent repeats. (C) The sequence and secondary structure of Pepper. The Pepper structure was predicted and generated based on Mfold software and the crystal structure (55). Pepper contains stem–loop 1 that can fuse the target-sensing RNA aptamer. Pepper was identified by SELEX. Because stem–loop 1 (boxed in blue) contains highly stable fixed sequences for every SELEX library (56), it may not directly interact with HBC (yellow). We speculated that stem–loop 1 plays a structural role and can be exploited in sensor construction. (D) Stem–loop 1 has a structural role in Pepper-induced HBC fluorescence. Stem–loop substitutions were performed to test the adaptability of Pepper for RNA sensors. The replacement stem–loop differs completely in sequence from the original stem–loop but retains a similar structure. We replaced the 6 bp stem with a duplex stem with a different sequence. The mismatched stem replaces the stem with five mismatched nucleotides to prevent duplex formation. Pepper is still able to induce fluorescence from HBC when stem–loop 1 is replaced with the paired stem. However, the mismatched stem reduces the fluorescence intensity to baseline. This suggests that stem–loop 1 is critical for fluorescence induction.
We wondered if fluorogenic aptamers without an RG4 motif could be reasonably constructed as an RNA-based sensor. Pepper is a recently reported RNA aptamer that does not harbor an RG4 motif (Figure 1A, B) (26,27). In addition, Pepper is bright, stable and colorful as an RNA tag that enables the imaging of diverse RNAs in living cells (25). However, Pepper has not been designed as an RNA-based sensor for various cellular targets, including intracellular small molecules, proteins and ions.
We next sought to design Pepper-based sensors. Similar to protein-based sensors and other RNA-based sensors (1,6,40), Pepper-based sensors should comprise three domains: Pepper as the fluorescent indicator, an RNA aptamer for specific binding to targets and a transducer domain that transmits the targeting-binding event to a fluorescence signal. The target-binding aptamer and transducer need to be inserted into a structurally critical stem of Pepper.
To identify the structurally critical stem, we studied the Systematic Evolution of Ligands by Exponential enrichment (SELEX) process and the crystal structure of Pepper (25,26). The SELEX library generated Pepper containing two 26 base random stretches separated by a 12 base fixed sequence (25). The 12 base fixed sequence with two bases forms a stable stem–loop structure (stem–loop 1) in Pepper (Figure 1C). Because the major part of stem–loop 1 was the fixed sequence in every library member, it may be less likely to make sequence-specific contacts with the HBC fluorophore. The crystal structure of Pepper supports that stem–loop 1 does not interact with HBC (26). We then assumed that stem–loop 1 in Pepper has a structural role in HBC binding that could be exploited in sensor design (26). To verify this idea, we mutated stem–loop 1 with different sequences. Mutagenesis revealed that stem–loop 1 has an essential structural role in Pepper fluorescence (Figure 1C, D). We thus used stem–loop 1 as an entry point for the insertion of targeting–binding aptamers, and designed various Pepper-based sensors for small molecules and proteins.
In vitro characterization of the Pepper-based sensor
We first asked if a small molecule-binding aptamer inserted into stem–loop 1 can modulate Pepper fluorescence intensity. To test this, we chose SAM-III riboswitch aptamer binding to SAM, a metabolite that acts as a methyl donor for nearly all cellular methylation reactions, and regulates the activities of DNA, RNA and proteins (41,42). We previously found that this SAM-III riboswitch can regulate RG4 folding in Spinach/Broccoli (Supplementary Figure S2), and induces fluorescence when fused via a single transducer domain (6,13,39). In this study, we reasoned that the SAM-III riboswitch could regulate Pepper folding and fluorescence.
To generate a Pepper-based SAM sensor, we fused the SAM-binding aptamer to stem–loop 1 in Pepper via a transducer domain (Figure 2A). The transducer domain is an RNA duplex that is unhybridized in the absence of SAM, thus disrupting the Pepper structure. However, SAM addition causes the SAM aptamer to become stabilized, which then brings the strands of the transducer together, resulting in the duplex formation and subsequent folding of Pepper. To optimize the SAM sensor, we tested different transducer domains, each with different thermodynamic stability (Figure 2B). Among these sensors, the SAM sensor containing transducer 2 exhibits the largest fold increase in fluorescence (19.3-fold), and high selectivity (Figure 2C, D). We therefore used transducer 2 for all subsequent experiments on the SAM sensor.
Figure 2.
Development of a Pepper-based SAM sensor with high activation, selectivity and accuracy. (A) Modular design of Pepper-based SAM sensors. Pepper-based SAM sensors are composed of three domains: Pepper (green), transducer (red) and SAM-binding aptamer (blue). The transducer module is composed of two strands that form a weakly base-paired stem. SAM binding induces the allosteric conformation of the SAM-binding aptamer, then stabilizes the helix structure of the transducer domain, thus enabling Pepper to bind HBC (black) and induce HBC fluorescence. (B) The overall sequences of Pepper-based SAM sensor variants for sensor optimization. (C) Optimization of thge SAM sensor's transducer. In vitro transcribed SAM sensor RNA with different transducers (transducers 1–6) was incubated with HBC (10 μM) in the presence of SAM (0.1 mM) or vehicle at 37°C for 1 h, and fluorescence was measured. The optimal transducer domain (transducer 2, indicated in the black box) was chosen with the highest signal-to-background (∼19.3-fold). (D) The Pepper-based SAM sensor is only activated by SAM but not by related molecules. We incubated SAM sensor (1 μM)–HBC (10 μM) and 0.1 mM SAM or its analogs for 1 h at 37°C and subsequently measured the fluorescence. These data indicate that the SAM sensor exhibits high specificity toward SAM. (E) Fluorescence of Pepper-based or Broccoli-based SAM sensors toward SAM with gradually increasing concentration. We incubated Pepper- or Broccoli-based SAM sensors (1 μM) using SAM with different concentrations and subsequently measured the fluorescence. Pepper: Ex = 485 nm, Em = 530 nm; Broccoli: Ex = 470 nm, Em = 505 nm. The data are represented as the mean ± SD of three independent repeats.
We next asked whether SAM sensor fluorescence could reflect SAM concentration. SAM physiological concentrations within different cell types are heterogeneous, typically at the tens to hundreds of micromolar level (9–11). In some cases, the SAM physiological concentration has been reported to reach the millimolar level in cells (12). We then tested the SAM concentration–response fluorescence with the Pepper-based SAM sensor and the reported Broccoli-based SAM sensors (13).
Pepper-based sensors give gradual increases in fluorescence with increasing SAM concentrations ranging from 1 to 500 μM (Figure 2E). When the SAM concentration exceeds 500 μM, the fluorescence signal of Pepper-based sensors saturates. Similarly, Broccoli-based SAM sensors showed fluorescence increases with increasing SAM concentration, and a good linearity between 0.1 and 100 μM. However, when the SAM concentration is >500 μM, Broccoli-based SAM sensors showed an obvious artificial decrease in fluorescence (Figure 2E). This may be because SAM diminishes Broccoli fluorescence (Supplementary Figure S3). These data indicate that the Pepper-based sensor showed the corresponding response to SAM at a physiological concentration. In addition, unlike Broccoli-based sensors, the Pepper-based sensor is less prone to inhibition by high concentrations of SAM.
Imaging SAM and its biosynthesis pathway with cell-to-cell heterogeneity
We next imaged SAM in living human cells with the Pepper-based SAM sensor. To do so, we expressed the SAM sensor as a circular RNA via a Tornado (Twister-optimized RNA for durable overexpression) expressing system in living HEK293T cells (13). The Tornado system recently reported by the Jaffrey lab allows RNA-based sensors to be accumulated to micromolar levels in living mammalian cells, similar to protein-based sensors (13). After treating with HBC (10 μM), we observed strong fluorescence in SAM sensor-expressing HEK293T cells with confocal microscopy and flow cytometry (Figure 3B; Supplementary Figures S4A, E and S5). These data indicate that a Pepper-based SAM sensor can image endogenous SAM in living human cells (43).
We next monitored the consumption and biosynthesis of SAM in living cells. Methionine is the substrate for SAM biosynthesis in cells, as methionine can be converted to SAM by MATase (Figure 3A; Supplementary Figures S6 and S7A) (44). We detected Pepper fluorescence easliy when SAM sensor-expressing cells were cultured in complete medium containing methionine. On switching cells to a methionine-free medium, we observed a drop in fluorescence (Figure 3B, C).
Figure 3.
Monitoring the SAM biosynthesis pathway in individual human cells. (A) Methionine is the metabolic origin of SAM (57). (B) Single-cell imaging of SAM dynamics and its metabolic origin using the Pepper-based SAM sensor. To determine endogenous SAM and its metabolic origin, individual HEK293T cell expressing the SAM sensor (top row) or Pepper (bottom row) were imaged. Methionine depletion led to a reduction in SAM levels in 4 h and methionine (100 μM) addition led to fluorescence recovery in 4 h. Images were acquired with a green fluorescent protein (GFP) filter at 10 min time intervals. Exposure time, 500 ms. Scale bar, 20 μm. (C) Methionine addition produces significant metabolic subtypes and cellular heterogeneity based on SAM production. After methionine addition to methionine-starved cells, cells can be broadly classified into three subtypes based on the rate of SAM production (n = 60 cells in total). For type I cells (top left, red lines, n = 23 cells), when methionine was added, SAM levels rapidly increased to the original level, and continued to rise gradually thereafter; the SAM level at 8 h is higher than that of the initial state (t = 0 h). Type II cells (top right, green lines, n = 7 cells) showed a moderate SAM consumption and synthesis ability. Type III cells (bottom left, blue lines, n = 30 cells) exhibited a gradual increase in SAM level and reached a plateau upon methionine addition. Notably, the final SAM levels at t = 8 h are lower than at t = 0 h. Cells expressing circular Pepper (bottom right) showed no obvious change in response to methionine depletion or addition.
To further verify that the decrease in fluorescence was caused by SAM consumption when there was a lack of methionine, we then added methionine (100 μM) to the medium to see if the fluorescence would recover. By reintroducing methionine, cellular fluorescence was restored in 4 h (Figure 3B, C). With the SAM sensor, we found that HEK293T cells exhibit three patterns of SAM synthesis rates. For type I cells, SAM levels rapidly increase to the original level, and continue to rise gradually thereafter. Type I cells could have a weaker SAM consumption ability but a higher SAM synthesis ability, and the SAM level in Type I cells at 8 h is higher than that of the initial state (t = 0 h). Type II cells showed a moderate SAM consumption and synthesis ability. Type III cells exhibited a gradual increase in SAM level and reached a plateau, which is lower than the SAM level at the initial state in fluorescence (t = 0 h) (Figure 3B, C). In addition, we also observed SAM biosynthesis in bacterial cells expressing the SAM sensor (Supplementary Figure S7). These data indicate that the SAM sensor enables real-time detection of SAM consumption and biosynthesis in living cells.
Monitoring MATase activity and gene expression
We next monitored MATase activity in living human cells. Cycloleucine is able to inhibit MATase activity and decrease the endogenous SAM level (Figure 4A) (45). We added cycloleucine (30 mM) to the medium of HEK293T cells expressing the SAM sensor. The cellular fluorescence decreased to a minimum within 2 h upon cycloleucine treatment (Figure 4B, C). We then removed the cycloleucine by replacing the medium with a fresh cycloleucine-free medium. Cellular fluorescence was recovered within 3 h after cycloleucine removal (Figure 4B, C). As a control, we used the original Pepper aptamer, to which SAM was unable to bind and thus modulate fluorescence. The fluorescence of cells expressing Pepper was unaffected by cycloleucine treatment and removal (Figure 4B, C). Overall, these data show that the SAM sensor enables real-time monitoring of MATase activity in living individual human cells.
Figure 4.
Monitoring MATase activity and gene expression in real-time. (A) MATase is the synthetase of SAM. The catalytic activity of MATase (indicated with a yellow ellipse) can be inhibited by cycloleucine (right) (45). Furthermore, MATase gene expression is activated by SAHA (left) in cancer cells (46). A change in MATase activity and expression influences SAM synthesis, reflected by the fluorescence of the SAM sensor. (B) Imaging of MATase activity with the SAM sensor. HEK293T cells expressing the SAM sensor (top row) or Pepper (bottom row) were imaged with HBC (10 μM). Cells were treated with cycloleucine (30 mM) at 0 min, which inhibits MATase activity and blocks SAM biosynthesis. HEK293T cells expressing the SAM sensor exhibited a drop in fluorescence, and recovery after washing out cycloleucine. Images were acquired with a microscope at 10 min time intervals. Exposure time, 500 ms. Scale bar, 20 μm. (C) SAM trajectory plots of individual cells on cycloleucine addition and removal. We measured the fluorescence of eight cells to generate SAM trajectory plots of individual HEK293T cells. Cells expressing circular Pepper showed no change in fluorescence following treatment with cycloleucine. The SAM dynamics exhibited in the trajectory plots imply that the addition of cycloleucine inhibits the process of SAM synthesis by MATase. (D) Monitoring the MATase expression level in living cells. MATase is expressed at a low level in HeLa cells, while elevated MATase expression can be induced by SAHA. After expressing the SAM sensor in HeLa cells, we treated the cells with SAHA (100 μM) and observed enhanced fluorescence in the cells. Images were acquired with a microscope at 30 min time intervals. Image acquisition time, 500 ms. Scale bar, 20 μm. (E) Trajectory plots of the cellular SAM level on SAHA addition. After 8 h of SAHA addition, cells expressing the SAM sensor generally showed enhanced fluorescence (n = 8 cells from three separate experiments). (F) SAHA induces the expression of MATase mRNA. We extracted RNA from HeLa cells after 8 h of SAHA treatment and performed quantitative PCR to detect the expression of MATase mRNA. Relative gene expression levels were calculated using the 2(–ΔΔCt) method (36), where the Ct values of the target gene were normalized to the Ct values of the reference gene GAPDH and compared with a control sample. The results showed that SAHA highly induced MATase mRNA expression.
We next asked if the SAM sensor can monitor the MATase gene expression level in HeLa cancer cells. MATase mRNA is expressed at a low level in cancer cells, due to the histone acetylation proximal to the promoter region of the MATase gene (46). Thus, inhibition of histone acetylation may induce the expression of MATase mRNA, leading to SAM accumulation. We expressed SAM sensor in HeLa cells and treated the cells with SAHA, a histone acetylation inhibitor, to up-regulate the expression of MATase mRNA (Figure 4A). A significant increase in cellular fluorescence was observed 8 h after the addition of SAHA (Figure 4D, E). Quantitative reverse transcription–PCR (RT–qPCR) verified that MATase mRNA expression was elevated after SAHA treatment (Figure 4F). These data suggest that the SAM sensor can indicate intracellular epigenetic regulation of MATase expression in cancer cells.
Last, we wondered about Pepper-based sensors exhibiting multicolor modulation. The Pepper aptamer can bind different HBC analogs and produce fluorescence of varying colors (25). For example, Pepper binds HBC620 to produce red fluorescence at a wavelength of 620 nm (Supplementary Figure S8A) (25). We suspected that this multicolor property could be extended to the Pepper-based sensor. To test this, we imaged cells expressing the SAM sensor with HBC620. SAM-dependent red fluorescence emissions were observed under the red fluorescence filter (Supplementary Figure S8B, Ct). These data indicate that the Pepper-based sensor exhibits color modulation, which is especially crucial when used for ratiometric or multitarget sensing with other orthogonal aptamers or aptamer-based sensors.
Assessment of MATase inhibitor efficacy using a ratiometric SAM sensor
To accurately investigate MATase activity, we next sought to design a ratiometric SAM sensor. The signal of the ratiometric sensor is independent of the RNA expression level, thus enabling quantitative assessment of MATase activity. We chose another non-RG4 fluorogenic RNA aptamer, RhoBAST (47), as an internal reference in the ratiometric SAM sensor. RhoBAST binds specifically TMR-DN to emit red fluorescence, making RhoBAST an orthogonal fluorogenic aptamer to the green fluorescent Pepper.
We next developed a ratiometric SAM sensor with the Pepper-based SAM sensor and RhoBAST. To facilitate the proper folding of each aptamer, we inserted the Pepper-based SAM sensor and RhoBAST into two arms of an F30 scaffold (Figure 5A) (48). After incubation with SAM, the ratiometric SAM sensor showed a significant increase in green fluorescence, while the red fluorescence was not affected (Figure 5B). These results demonstrate that RhoBAST can be used as a normalizer in the ratiometric SAM sensor.
Figure 5.
Pepper-based ratiometric SAM sensor. (A) Constructing the ratiometric SAM sensor. The Pepper-based SAM sensor (green and blue lines, left arm of F30) and RhoBAST (indicated by red lines, right arm of F30) were inserted into two arms of F30 scaffold (black lines). SAM (gray) binding activates the green fluorescence of the Pepper-based SAM sensor (green circle). The constitutive red fluorescence of RhoBAST:TMR-DN (red circle) is used for normalization, thus eliminating the confounding effect of cell-to-cell variation in the sensor expression level. (B) SAM activates Pepper fluorescence in the ratiometric SAM sensor without affecting RhoBAST. The ratiometric SAM sensor was incubated with HBC (10 μM) and TMR-DN (100 nM) in the buffer for 1 h in the presence or absence of SAM (0.1 mM). Pepper fluorescence was subsequently measured at Ex = 485 nm, Em = 530 nm, and RhoBAST fluorescence at Ex = 564 nm, Em = 590 nm. Pepper-based SAM sensor fluorescence showed a similar response signal to that in Figure 2, while RhoBAST fluorescence showed little change in the presence of SAM. The data are represented as the mean ± SD of three independent repeats. (C) Live-cell imaging of the ratiometric SAM sensor. We expressed the ratiometric SAM sensor in HEK293T cells, and the fluorescent signal can be easily identified and distinguished in different channels. Image acquisition time, 300 ms. Scale bar, 20 μm. (D) Detection of the inhibition efficacy of AG-270 on MATase. We incubated cells expressing the ratiometric SAM sensor with a series of concentrations of AG-270 for 24 h before adding HBC (10 μM) and TMR-DN (100 nM) to the medium for imaging. Inhibition ratios were then calculated from the green-to-red fluorescent ratio of at least 20 cells at each concentration. The IC50 of AG-270 was calculated to be ∼19.4 nM, which is similar to that found in a previous report (49). (E) Ratiometric imaging of MATase activity. As the AG-270 concentration increases, the green-to-red ratiometric signal decreases significantly in cells expressing the ratiometric SAM sensor. Cells expressing Pepper/RhoBAST control RNA showed no significant change in fluorescence ratio. Image acquisition time, 300 ms. Scale bar, 20 μm.
We next asked if the ratiometric SAM sensor can be used for SAM imaging in living cells. We expressed the ratiometric SAM sensor in HEK293T cells using the Tornado expression system (13). After incubating the HEK293T cells with HBC and TMR-DN fluorophores, we readily detected the green fluorescence of the Pepper-based SAM sensor and the red fluorescence of RhoBAST (Figure 5C). The ratiometric signal can be acquired by calculating the green-to-red fluorescence intensity (Figure 5C). Because RhoBAST fluorescence is not affected by SAM, the fluctuations of the ratiometric signal can therefore reflect the fluctuations of the cellular SAM level and MATase activity. These data showed that the ratiometric SAM sensor functions effectively in living cells.
We next asked if the ratiometric sensor can quantitate the inhibition efficacy of MATase inhibitors. We added AG-270 (49), a recently reported MATase inhibitor, to HEK293T cells expressing the ratiometric SAM sensor or a control RNA. After 24 h incubation, we assessed MATase activity with the ratiometric signal by measuring the green-to-red fluorescence ratio. The ratiometric signal in cells expressing the ratiometric SAM sensor was significantly reduced with increasing concentrations of AG-270, while the control group was not affected by AG-270 (Figure 5D, E). The decrease of the ratiometric signal can reflect the inhibition rate of MATase. By analyzing the ratiometric signal at various concentrations of AG-270, we plotted a dose–response curve and determined the IC50 value to be ∼19.4 nM (Figure 5D), which is consistent with the previous report (IC50 = 14 nM) (49). These results show that the ratiometric SAM sensor can accurately detect the efficacy of MATase inhibitors in living cells.
Modular generation for detection of other small molecules
To determine the versatility of the Pepper-based sensor, we incorporated other target-binding RNA sequences with Pepper and detected other small molecules. We generated the tetracycline sensor and the guanine sensor using Pepper (Figure 6A, E) (6,12,16,50). For the tetracycline sensor, an 83.7-fold fluorescence enhancement was observed in tetracycline sensors with the optimal transducer sequence after incubating with tetracycline (0.1 mM) (Figure 6B; Supplementary Figure S9A). Furthermore, the tetracycline sensor showed high selectivity with other antibiotics, including neomycin, tobramycin, gentamycin, ampicillin and kanamycin (Figure 6C). In addition, we found that doxycycline, a structural analog of tetracycline, was able to induce fluorescence of the tetracycline sensor, but this was significantly lower than tetracycline-induced fluorescence (Supplementary Figure S10). This is because doxycycline exhibits lower binding affinity (17.7 μM) toward the tetracycline aptamer compared with tetracycline (0.77 nM). For the guanine sensor, the optimal transducer gave a 5-fold signal-to-noise ratio and high specificity (Figure 6F, G; Supplementary Figure S9B). These data suggest that Pepper-based sensors can be applied to other small molecular metabolites and drugs.
Figure 6.
Generation of Pepper-based sensors activated by diverse small molecules. (A) Sequences of the tetracycline sensor variants for biosensor optimization. The overall sequence and simplified structure of the tetracycline aptamer (blue) and transducer region (red dots boxed in black) are shown. (B) Optimization of the transducer for the tetracycline sensor. We generated tetracycline sensors with different transducers by in vitro transcription. The optimal transducer domain for the tetracycline sensor is transducer 4 (boxed in black), with an 83.7-fold increase in fluorescence signal upon incubation with tetracycline (0.1 mM). Ex = 485 nm, Em = 530 nm. The data are represented as the mean ± SD of three independent repeats. (C) The tetracycline sensor responds only to tetracycline and not to other antibiotics. To test the specificity of the tetracycline sensor, tobramycin, gentamicin, neomycin, kanamycin (aminoglycoside antibiotics) and ampicillin (β-lactam antibiotics) were selected. The results showed that the tetracycline sensor did not respond to other types of antibiotics, only tetracycline. Ex = 485 nm, Em = 530 nm. The data are represented as the mean ± SD of three independent repeats. (D) Using the tetracycline sensor to monitor the tetracycline level in living human cells. We expressed the tetracycline sensor in HEK293T cells. Cell culture medium was replaced with phenol red-free DMEM containing HBC (10 μM) in the presence or absence of tetracycline (0.1 mM) 1 h before imaging. In cells treated with tetracycline, enhanced fluorescence was readily observed. Image acquisition time, 500 ms. Scale bar, 20 μm. (E) Sequence, and design of the guanine sensor. The guanine aptamer (blue) is fused to Pepper (green) via a transducer domain (red dots boxed in black). The guanine sensors are designed so that Pepper folds only when guanine binds to the guanine aptamer. (F) Optimization of the transducer for the guanine sensor. The optimal transducer domain (transducer 4, boxed in black) was chosen because in the context of the sensor it displayed low background fluorescence, with a 5-fold increase in fluorescence signal upon incubation with guanine (0.1 mM). Ex = 485 nm, Em = 530 nm. The data are represented as the mean ± SD of three independent repeats. (G) The guanine sensor is only activated by guanine. We tested the selectivity of the guanine sensor with guanosine and adenine. Guanosine is a nucleotide synthesized from guanine, and adenine, like guanine, is a purine nucleobase. The results show that the guanine sensor can only be activated by guanine, but not by guanosine and adenine. Ex = 485 nm, Em = 530 nm. The data are represented as the mean ± SD of three independent repeats. (H) Monitoring the guanine level in living human cells with the guanine sensor. Cells expressing the guanine sensor were incubated in phenol red-free DMEM containing HBC (10 μM) in the presence or absence of guanine (0.1 mM) 1 h before imaging. The imaging results showed that the guanine sensor was able to detect the exogenous addition of guanine to living cells. Image acquisition time, 500 ms. Scale bar, 20 μm.
We next asked if the Pepper-based sensor can image tetracycline or guanine in living bacterial cells. We expressed the tetracycline sensor or the guanine sensor in BL21 E. coli cells incubated with HBC (2 μM). The E. coli cells expressing the sensor exhibit minimal fluorescence in the absence of the target. When incubated with a medium containing a tetracycline or guanine target (0.1 mM), cells expressing the tetracycline sensor or guanine target sensor showed significant fluorescence enhancement, respectively (Supplementary Figure S11A, B). These data suggest that the Pepper-based sensor can be designed to be a widely applicable small molecule imaging platform in bacteria.
To determine if these Pepper-based sensors could be used in living human cells, we expressed the circular tetracycline sensor or guanine sensor in HEK293T cells using the Tornado expression system. Upon incubation with a medium containing guanine or tetracycline, the cells exhibited significant fluorescence enhancement (Figure 6D, H; Supplementary Figure S4B, C, E, and S5B, C). Thus, the Pepper-based sensor can be designed for detection of various small molecule targets in living human cells.
Pepper-based sensors allow sensing of metal ions and proteins
We next asked if a Pepper-based sensor can detect metal ions. To test this, we designed a silver ions (Ag+) sensor using Pepper. Ag+ is harmful to human health; excess exposure to silver ions may lead to toxic symptoms including loss of organ function, growth retardation and mitochondrial damage through the promotion of oxidative stress (51). Therefore, imaging Ag+ helps to understand the mechanism of Ag+ damage to cells. However, there has been no report of imaging Ag+ in human cells with a genetically encoded fluorescent sensor. To detect Ag+ in living human cells, we designed a Pepper-based Ag+ sensor using cytosine–Ag+–cytosine (C–Ag+–C) metallo base pairs (52). We inserted C–C mismatches into Pepper stem–loop 1 (Figure 7A). In the absence of Ag+, the C–C mismatches resulted in an unstable stem–loop 1 structure, leading to low fluorescence. In the presence of Ag+, the C–Ag+–C metallo base pair was formed to stabilize the stem–loop 1 structure (53), leading to enhanced fluorescence (Figure 7A). We optimized the Ag+ sensor by adjusting the position and number of C–C mismatch insertions. The optimized sensor design achieves a 3.7-fold fluorescence enhancement (Figure 7B; Supplementary Figure S9C). Other metal ions failed to induce fluorescence in the Ag+ sensor (Figure 7C). These data demonstrate that a Pepper-based sensor can be designed for metal ion detection.
Figure 7.
Design of a Pepper-based sensor for ions and proteins in living cells. (A) Insertion of C–C mismatched base pairs into construct Pepper as an Ag+ sensor. We inserted a sequence containing C–C mismatches (blue) into stem–loop 1 as an Ag+-binding domain. In the absence of Ag+, the mismatches cause the stem–loop 1 structure to collapse. In the presence of Ag+, the C–C mismatches form C–Ag+–C metallo base pairs, thus stabilizing stem–loop 1. (B) Optimization of the transducer for the Ag+ sensor. We generated Ag+ sensors with different transducers by in vitro transcription. Sensors with different transducers (1 μM) were incubated with HBC (10 μM) in the presence or absence of Ag+ (0.1 mM) at 37°C for 1 h. The optimal stem for the Ag+ sensor has one C–C mismatch at the second base position only (boxed in black). Ex = 485 nm, Em = 530 nm. The data are represented as the mean ± SD of three independent repeats. (C) Selectivity of the Ag+ sensor. We incubated calcium (Ca2+), lithium (Li+), manganese (Mg2+) and sodium (Na+) ions with the Ag+ sensor, but none of these ions elicited a response from the Ag+ sensor. (D) Imaging the Ag+ ion in living human cells. We first expressed the Ag+ sensor in HEK293T cells. Cell culture medium was then replaced with phenol red-free DMEM containing HBC (10 μM) in the presence or absence of Ag+ (20 μM) 1 h before imaging. The imaging results showed that the Ag+ sensor was able to detect Ag+ in living human cells. Image acquisition time, 500 ms. Scale bar, 20 μm. (E) Secondary structure and modular design of the streptavidin sensors. The streptavidin aptamer (blue), transducer domain (red dots boxed in black) and Pepper domain (green) are depicted. The streptavidin sensors activate the fluorescence of HBC only in the presence of streptavidin. (F) Optimization of the transducer for the streptavidin sensor. We generated streptavidin sensors with different transducers by in vitro transcription. Sensors with different transducers (0.2 μM) were incubated with HBC (10 μM) in the presence or absence of streptavidin (2 μM) at 37°C for 1 h. The optimal transducer domain (transducer 6) showed a 7-fold increase in fluorescence upon streptavidin incubation. The optimal transducer is boxed in black. Ex = 485 nm, Em = 530 nm. The data are represented as the mean ± SD of three independent repeats. (G) Selectivity of the streptavidin sensor. The fluorescence was measured in the presence of 2 μM streptavidin or competing proteins. The data are represented as the mean ± SD of three independent repeats. (H) Imaging streptavidin in individual cells with the streptavidin sensor. To observe streptavidin in living cells, BL21 E. coli cells expressing either the streptavidin sensor alone (left) or streptavidin and the streptavidin sensor (right) were incubated with HBC and imaged. The data show that only cells co-expressing the streptavidin sensor and streptavidin exhibit strong fluorescence. Images were acquired with the microscope. Image acquisition time, 500 ms. Scale bar, 5 μm.
We next expressed the Ag+ sensor in HEK293T cells. Cell fluorescence was measured by confocal microscopy or flow cytometry after incubation with Ag+ (20 μM) for 1 h. Ag+-dependent cell fluorescence was clearly observed (Figure 7D; Supplementary Figures S4D, E and S5D). In addition, we observed Ag+-dependent cell fluorescence in living E. coli cells (Supplementary Figure S11C). These data demonstrate that an Ag+ sensor is for the first time imaged in mammalian cells using a genetically encoded fluorescent sensor, and the Pepper-based sensor is a comprehensive imaging platform for this metal ion in living cells.
We next asked if Pepper can generate sensors of proteins. To test this idea, we fused a streptavidin-binding aptamer to Pepper via different transducer stems with different thermodynamic stabilities (Figure 7E) (9). We next tested the fluorescence responsiveness of each RNA against streptavidin. The optimal sensor exhibited a 7.2-fold fluorescence increase following the addition of streptavidin (2 μM), and shows high selectivity (Figure 7F, G; Supplementary Figure S9D).
We next imaged protein expression levels in living cells using a Pepper-based sensor. To test this, we expressed the streptavidin sensor alone or co-expressed it with streptavidin target in BL21 E. coli cells. Cells expressing the sensor alone exhibit low background fluorescence. In contrast, cells co-expressing the sensor and streptavidin showed significantly enhanced fluorescence (Figure 7H). These data demonstrate that Pepper-based sensors provide a generalizable approach for generating bright and stable sensors for proteins.
DISCUSSION
Here we converted Pepper, an RG4-free, bright, stable and multiple-spectral fluorogenic RNA, into an RNA-based sensor for measuring the abundance and signaling of intracellular molecules. Pepper-based sensors show high brightness and stability (25). Importantly, Pepper does not harbor the RG4 motif to trigger the cellular RNA unfolding machinery, which enables it to be developed as various robust sensors in living cells. Crucially, Pepper exhibits other key merits for sensing study, including high brightness, high stability, high folding, high binding affinity and high pH tolerance, allowing Pepper to be constructed as robust RNA-based sensors in living cells.
We designed sensors for metabolite detection in vitro and in vivo. We first rationally designed a Pepper-based sensor for SAM, a small molecular metabolite that is a methyl donor for nearly all cellular methylation reactions. The Pepper-based SAM sensor only exhibits fluorescence when SAM is present. We showed that the sensor works both in vitro and in vivo with high activation and high brightness.
Intracellular physiological SAM concentrations vary in different cell types. Therefore, the ideal SAM sensor allows high-range detection of SAM. We found thqt the previously designed Broccoli-based SAM sensor exhibits an artificial fluorescence decrease on SAM, when the SAM concentration gradually increases above 100 μM. In contrast, the Pepper-based SAM sensor exhibits the corresponding response to SAM at a physiological concentration, and is less prone to inhibition by high concentrations of SAM. Therefore, a Pepper-based sensor is more suitable for detecting physiologically relevant SAM concentrations compared with a Broccoli-based sensor.
With a Pepper-based sensor, we revealed SAM signal function over time in living human cells, and tracked the biosynthesis pathway of SAM with cell-to-cell variation. Furthermore, we monitored the activity of MATase and determined the inhibition of MATase inhibitors in living individual human cells. Additionally, our SAM sensor detected the gene expression of MATase and verified the agonist drug to accumulate SAM in cancer cells, which has not been reported by other fluorogenic RNA-based or fluorescent protein-based sensors.
We constructed a ratiometric RNA-based sensor to determine the IC50 of MATase inhibitors in live cells. To the best of our knowledge, this is the first time that the IC50 of a drug has been identified by an RNA-based sensor directly in live cells. Therefore, we expect that RNA-based sensors can be used to identify or screen drugs in living human cells.
Furthermore, we demonstrated the versatility of the sensor. We designed Pepper-based sensors for various other targets, including metabolites, synthetic compounds, proteins and metal ions in living cells, demonstrating the universality of Pepper-based sensors. One key process is that this work is the first time that Ag+ has been observed with a genetically encoded sensor in living mammalian cells. The monitoring of intracellular levels of various heavy metal ions is of great significance. However, heavy metal ions cause denaturation of the protein (54), which may limit the application of fluorescent proteins to sense heavy metal ions. Our study demonstrated that RNA has the potential to bind metal ions selectively, and we designed an RNA-based sensor for Ag+ in living human cells for the first time, introducing a new approach for ionomics study.
In addition to Pepper, other fluorogenic RNA aptamers without an RG4 motif have been reported, including Squash (43), RhoBAST (47) and Riboglow (32), but most aptamers have not been designed as RNA-based sensors. Recently reported Squash/Broccoli were designed as RNA-based ratiometric sensors (43). Although Squash alone does not contain an RG4 motif, Squash incorporates RG4-containing Broccoli for ratiometric sensing, potentially preventing its application.
Additionally, Pepper-based sensors show multicolor modulation, exhibiting the high flexibility of RNA-based sensors. Such flexibility is especially useful when used for ratiometric biosensors, orthogonal sensors or for creating stable cell lines or transgenic animals.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Professor Fangqing Zhao and Zhongxuan Zhang for their comments and suggestions.
Contributor Information
Zhenyin Chen, Beijing Institute of Life Sciences, Chinese Academy of Sciences, Beijing 100101, China; Department of Pulmonary and Critical Care Medicine, Department of Inflammation and Clinical Allergology, the Affiliated Hospital of Southwest Medical University, Luzhou, Sichuan 646000, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Wei Chen, Beijing Institute of Life Sciences, Chinese Academy of Sciences, Beijing 100101, China; Institute of Cytology and Genetics, the Hengyang Key Laboratory of Cellular Stress Biology, Hengyang Medical School, University of South China, Hengyang, Hunan 421001, China.
Zhayila Reheman, Beijing Institute of Life Sciences, Chinese Academy of Sciences, Beijing 100101, China; School of Life Science, Hebei University, Baoding, Hebei 071000, China.
Haodong Jiang, Beijing Institute of Life Sciences, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Jiahui Wu, Department of Chemistry, University of Massachusetts, Amherst, MA01003, USA.
Xing Li, Beijing Institute of Life Sciences, Chinese Academy of Sciences, Beijing 100101, China; Department of Pulmonary and Critical Care Medicine, Department of Inflammation and Clinical Allergology, the Affiliated Hospital of Southwest Medical University, Luzhou, Sichuan 646000, China; Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, Tianjin 300308, China; State Key Laboratory of Integrated Management of Pest Insects and Rodents, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China.
DATA AVAILABILITY
All data are available in the main text or the supplementary data.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Science Foundation of China [32271515 and 32311530120 to X. Li]; Tianjin Synthetic Biotechnology Innovation Capacity Improvement Project [TSBICIP-CXRC-038 to X. Li]; the University of Massachusetts [start-up grant to J. Wu]; and Beijing Institutes of Life Science, Chinese Academy of Sciences [Talent research start-up fund to X. Li].
Conflict of interest statement. X. Li and Z. Chen are authors of a Chinese patent application on MATase activity detection and MATase inhibitor identification, which are related to the technology described in this manuscript. The remaining authors declare no competing interests.
REFERENCES
- 1. Palmer A.E., Qin Y., Park J.G., McCombs J.E.. Design and application of genetically encoded biosensors. Trends Biotechnol. 2011; 29:144–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Romoser V.A., Hinkle P.M., Persechini A.. Detection in living cells of Ca2+-dependent changes in the fluorescence emission of an indicator composed of two green fluorescent protein variants linked by a calmodulin-binding sequence. A new class of fluorescent indicators. J. Biol. Chem. 1997; 272:13270–13274. [DOI] [PubMed] [Google Scholar]
- 3. Pendin D., Greotti E., Lefkimmiatis K., Pozzan T.. Exploring cells with targeted biosensors. J. Gen. Physiol. 2017; 149:1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sun Z., Wu R., Zhao B., Zeinert R., Chien P., You M.. Live-cell imaging of guanosine tetra- and pentaphosphate (p)ppGpp with RNA-based fluorescent sensors. Angew. Chem. Int. Ed. Engl. 2021; 60:24070–24074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Miyawaki A. Development of probes for cellular functions using fluorescent proteins and fluorescence resonance energy transfer. Annu. Rev. Biochem. 2011; 80:357–373. [DOI] [PubMed] [Google Scholar]
- 6. Paige J.S., Nguyen-Duc T., Song W., Jaffrey S.R.. Fluorescence imaging of cellular metabolites with RNA. Science. 2012; 335:1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Paige J.S., Wu K.Y., Jaffrey S.R.. RNA mimics of green fluorescent protein. Science. 2011; 333:642–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Filonov G.S., Moon J.D., Svensen N., Jaffrey S.R.. Broccoli: rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J. Am. Chem. Soc. 2014; 136:16299–16308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Song W., Strack R.L., Jaffrey S.R.. Imaging bacterial protein expression using genetically encoded RNA sensors. Nat. Methods. 2013; 10:873–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Strack R.L., Jaffrey S.R.. New approaches for sensing metabolites and proteins in live cells using RNA. Curr. Opin. Chem. Biol. 2013; 17:651–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Strack R.L., Song W., Jaffrey S.R.. Using Spinach-based sensors for fluorescence imaging of intracellular metabolites and proteins in living bacteria. Nat. Protoc. 2014; 9:146–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. You M., Litke J.L., Jaffrey S.R.. Imaging metabolite dynamics in living cells using a Spinach-based riboswitch. Proc. Natl Acad. Sci. USA. 2015; 112:E2756–E2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Litke J.L., Jaffrey S.R.. Highly efficient expression of circular RNA aptamers in cells using autocatalytic transcripts. Nat. Biotechnol. 2019; 37:667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. You M., Litke J.L., Wu R., Jaffrey S.R.. Detection of low-abundance metabolites in live cells using an RNA integrator. Cell Chem. Biol. 2019; 26:471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moon J.D., Wu J., Dey S.K., Litke J.L., Li X., Kim H., Jaffrey S.R.. Naturally occurring three-way junctions can be repurposed as genetically encoded RNA-based sensors. Cell Chem. Biol. 2021; 28:1569–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu R., Karunanayake Mudiyanselage A., Shafiei F., Zhao B., Bagheri Y., Yu Q., McAuliffe K., Ren K., You M.. Genetically encoded ratiometric RNA-based sensors for quantitative imaging of small molecules in living cells. Angew. Chem. Int. Ed. Engl. 2019; 58:18271–18275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yu Q., Ren K., You M.. Genetically encoded RNA nanodevices for cellular imaging and regulation. Nanoscale. 2021; 13:7988–8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chakraborty K., Veetil A.T., Jaffrey S.R., Krishnan Y.. Nucleic acid-based nanodevices in biological imaging. Annu. Rev. Biochem. 2016; 85:349–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mukkayyan N., Poon R., Sander P.N., Lai L.Y., Zubair-Nizami Z., Hammond M.C., Chatterjee S.S.. In vivo detection of cyclic-di-AMP in Staphylococcus aureus. ACS Omega. 2022; 7:32749–32753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Su Y., Hickey S.F., Keyser S.G., Hammond M.C.. In vitro and in vivo enzyme activity screening via RNA-based fluorescent biosensors for S-adenosyl-l-homocysteine (SAH). J. Am. Chem. Soc. 2016; 138:7040–7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guo J.U., Bartel D.P.. RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. Science. 2016; 353:aaf5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen M.C., Tippana R., Demeshkina N.A., Murat P., Balasubramanian S., Myong S., Ferre-D’Amare A.R.. Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36. Nature. 2018; 558:465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hou Q., Jaffrey S.R.. Synthetic biology tools to promote the folding and function of RNA aptamers in mammalian cells. RNA Biol. 2023; 20:198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Trachman R.J. 3rd, Autour A., Jeng S.C.Y., Abdolahzadeh A., Andreoni A., Cojocaru R., Garipov R., Dolgosheina E.V., Knutson J.R., Ryckelynck M.et al.. Structure and functional reselection of the Mango-III fluorogenic RNA aptamer. Nat. Chem. Biol. 2019; 15:472–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen X., Zhang D., Su N., Bao B., Xie X., Zuo F., Yang L., Wang H., Jiang L., Lin Q.et al.. Visualizing RNA dynamics in live cells with bright and stable fluorescent RNAs. Nat. Biotechnol. 2019; 37:1287–1293. [DOI] [PubMed] [Google Scholar]
- 26. Huang K., Chen X., Li C., Song Q., Li H., Zhu L., Yang Y., Ren A.. Structure-based investigation of fluorogenic Pepper aptamer. Nat. Chem. Biol. 2021; 17:1289–1295. [DOI] [PubMed] [Google Scholar]
- 27. Rees H.C., Gogacz W., Li N.S., Koirala D., Piccirilli J.A.. Structural basis for fluorescence activation by Pepper RNA. ACS Chem. Biol. 2022; 17:1866–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen W., Zhao X., Yang N., Li X.. Single mRNA Imaging with fluorogenic RNA aptamers and small-molecule fluorophores. Angew. Chem. Int. Ed. Engl. 2023; 62:e202209813. [DOI] [PubMed] [Google Scholar]
- 29. Baugh C., Grate D., Wilson C.. 2.8 Å crystal structure of the malachite green aptamer. J. Mol. Biol. 2000; 301:117–128. [DOI] [PubMed] [Google Scholar]
- 30. Shelke S.A., Shao Y., Laski A., Koirala D., Weissman B.P., Fuller J.R., Tan X., Constantin T.P., Waggoner A.S., Bruchez M.P.et al.. Structural basis for activation of fluorogenic dyes by an RNA aptamer lacking a G-quadruplex motif. Nat. Commun. 2018; 9:4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tan X., Constantin T.P., Sloane K.L., Waggoner A.S., Bruchez M.P., Armitage B.A.. Fluoromodules consisting of a promiscuous RNA aptamer and red or blue fluorogenic cyanine dyes: selection, characterization, and bioimaging. J. Am. Chem. Soc. 2017; 139:9001–9009. [DOI] [PubMed] [Google Scholar]
- 32. Braselmann E., Wierzba A.J., Polaski J.T., Chrominski M., Holmes Z.E., Hung S.T., Batan D., Wheeler J.R., Parker R., Jimenez R.et al.. A multicolor riboswitch-based platform for imaging of RNA in live mammalian cells. Nat. Chem. Biol. 2018; 14:964–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sunbul M., Jaschke A.. SRB-2: a promiscuous rainbow aptamer for live-cell RNA imaging. Nucleic Acids Res. 2018; 46:e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Song W., Filonov G.S., Kim H., Hirsch M., Li X., Moon J.D., Jaffrey S.R.. Imaging RNA polymerase III transcription using a photostable RNA-fluorophore complex. Nat. Chem. Biol. 2017; 13:1187–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li X., Kim H., Litke J.L., Wu J., Jaffrey S.R.. Fluorophore-promoted RNA folding and photostability enables imaging of single Broccoli-tagged mRNAs in live mammalian cells. Angew. Chem. Int. Ed. Engl. 2020; 59:4511–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Livak K.J., Schmittgen T.D.. Analysis of relative gene expression data using real-time quantitative PCR and the 2(–ΔΔC(T)) method. Methods. 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 37. Sun Z., Nguyen T., McAuliffe K., You M.. Intracellular imaging with genetically encoded RNA-based molecular sensors. Nanomaterials (Basel). 2019; 9:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim H., Jaffrey S.R.. A fluorogenic RNA-based sensor activated by metabolite-induced RNA dimerization. Cell Chem. Biol. 2019; 26:1725–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li X., Mo L., Litke J.L., Dey S.K., Suter S.R., Jaffrey S.R.. Imaging intracellular S-adenosyl methionine dynamics in live mammalian cells with a genetically encoded red fluorescent RNA-based sensor. J. Am. Chem. Soc. 2020; 142:14117–14124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lemke E.A., Schultz C.. Principles for designing fluorescent sensors and reporters. Nat. Chem. Biol. 2011; 7:480–483. [DOI] [PubMed] [Google Scholar]
- 41. Lu C., Smith A.M., Fuchs R.T., Ding F., Rajashankar K., Henkin T.M., Ke A.. Crystal structures of the SAM-III/S(MK) riboswitch reveal the SAM-dependent translation inhibition mechanism. Nat. Struct. Mol. Biol. 2008; 15:1076–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pendleton K.E., Chen B., Liu K., Hunter O.V., Xie Y., Tu B.P., Conrad N.K.. The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017; 169:824–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dey S.K., Filonov G.S., Olarerin-George A.O., Jackson B.T., Finley L.W.S., Jaffrey S.R.. Repurposing an adenine riboswitch into a fluorogenic imaging and sensing tag. Nat. Chem. Biol. 2022; 18:180–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lu S.C S-Adenosylmethionine. Int. J. Biochem. Cell Biol. 2000; 32:391–395. [DOI] [PubMed] [Google Scholar]
- 45. Lombardini J.B., Talalay P.. Formation, functions and regulatory importance of S-adenosyl-L-methionine. Adv. Enzyme Regul. 1970; 9:349–384. [DOI] [PubMed] [Google Scholar]
- 46. Huang J., Barr E., Rudnick D.A.. Characterization of the regulation and function of zinc-dependent histone deacetylases during rodent liver regeneration. Hepatology. 2013; 57:1742–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sunbul M., Lackner J., Martin A., Englert D., Hacene B., Grun F., Nienhaus K., Nienhaus G.U., Jaschke A.. Super-resolution RNA imaging using a rhodamine-binding aptamer with fast exchange kinetics. Nat. Biotechnol. 2021; 39:686–690. [DOI] [PubMed] [Google Scholar]
- 48. Filonov G.S., Jaffrey S.R.. RNA imaging with dimeric Broccoli in live bacterial and mammalian cells. Curr. Protoc. Chem. Biol. 2016; 8:1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Konteatis Z., Travins J., Gross S., Marjon K., Barnett A., Mandley E., Nicolay B., Nagaraja R., Chen Y., Sun Y.et al.. Discovery of AG-270, a first-in-class oral MAT2A inhibitor for the treatment of tumors with homozygous MTAP deletion. J. Med. Chem. 2021; 64:4430–4449. [DOI] [PubMed] [Google Scholar]
- 50. Suess B., Hanson S., Berens C., Fink B., Schroeder R., Hillen W.. Conditional gene expression by controlling translation with tetracycline-binding aptamers. Nucleic Acids Res. 2003; 31:1853–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jiao Y., Gao Y., Meng Y., Lu W., Liu Y., Han H., Shuang S., Li L., Dong C.. One-step synthesis of label-free ratiometric fluorescence carbon dots for the detection of silver ions and glutathione and cellular imaging applications. ACS Appl. Mater. Interfaces. 2019; 11:16822–16829. [DOI] [PubMed] [Google Scholar]
- 52. Kondo J., Tada Y., Dairaku T., Saneyoshi H., Okamoto I., Tanaka Y., Ono A.. High-resolution crystal structure of a silver(I)–RNA hybrid duplex vontaining Watson–Crick-like C–Silver(I)–C metallo-base pairs. Angew. Chem. Int. Ed. Engl. 2015; 54:13323–13326. [DOI] [PubMed] [Google Scholar]
- 53. Yu Q., Shi J., Mudiyanselage A., Wu R., Zhao B., Zhou M., You M.. Genetically encoded RNA-based sensors for intracellular imaging of silver ions. Chem. Commun. (Camb.). 2019; 55:707–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tamas M.J., Sharma S.K., Ibstedt S., Jacobson T., Christen P.. Heavy metals and metalloids as a cause for protein misfolding and aggregation. Biomolecules. 2014; 4:252–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Davis J.H., Szostak J.W.. Isolation of high-affinity GTP aptamers from partially structured RNA libraries. Proc. Natl Acad. Sci. USA. 2002; 99:11616–11621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zeng Z., Huang Z.Z., Chen C., Yang H., Mao Z., Lu S.C.. Cloning and functional characterization of the 5'-flanking region of human methionine adenosyltransferase 1A gene. Biochem. J. 2000; 346:475–482. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or the supplementary data.










