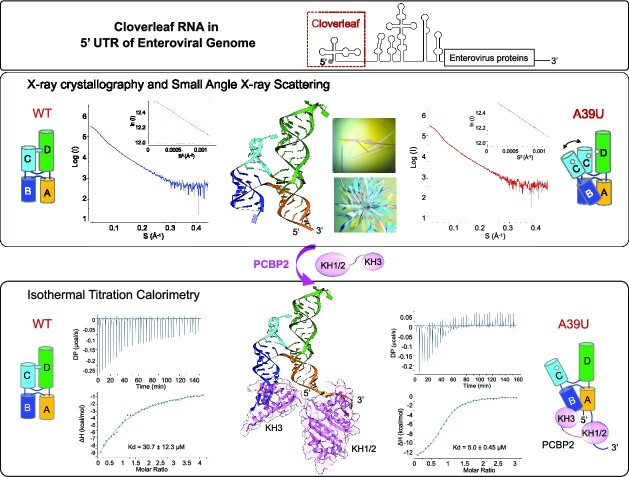
Abstract
The genomes of positive-strand RNA viruses serve as a template for both protein translation and genome replication. In enteroviruses, a cloverleaf RNA structure at the 5′ end of the genome functions as a switch to transition from viral translation to replication by interacting with host poly(C)-binding protein 2 (PCBP2) and the viral 3CDpro protein. We determined the structures of cloverleaf RNA from coxsackievirus and poliovirus. Cloverleaf RNA folds into an H-type four-way junction and is stabilized by a unique adenosine-cytidine-uridine (A•C-U) base triple involving the conserved pyrimidine mismatch region. The two PCBP2 binding sites are spatially proximal and are located on the opposite end from the 3CDpro binding site on cloverleaf. We determined that the A•C-U base triple restricts the flexibility of the cloverleaf stem–loops resulting in partial occlusion of the PCBP2 binding site, and elimination of the A•C-U base triple increases the binding affinity of PCBP2 to the cloverleaf RNA. Based on the cloverleaf structures and biophysical assays, we propose a new mechanistic model by which enteroviruses use the cloverleaf structure as a molecular switch to transition from viral protein translation to genome replication.
Graphical Abstract
Graphical Abstract.
INTRODUCTION
In positive-strand RNA viruses, the viral genome is the template for both viral protein translation and genome replication. Viral polymerases are unable to synthesize RNA from a positive-sense RNA genome that is actively being translated (1). Thus, viruses require a mechanism to switch from genome translation to replication. Enteroviruses and flaviviruses, two of the most studied positive-strand RNA viruses, use a structured region of their RNA genome as an RNA promoter to modulate the transition between translation and replication (2,3). Enteroviruses, small non-enveloped RNA viruses of the family Picornaviridae, cause a wide array of diseases in humans. Well known enterovirus pathogens include poliovirus (PV) that causes poliomyelitis, and coxsackieviruses and echoviruses that cause diseases with symptoms ranging from respiratory illness and aseptic meningitis to acute myocarditis or myelitis (4). Enteroviruses encapsidate a ∼7.5 kb, positive-sense RNA genome covalently linked to a peptide primer VPg (virion protein genome linked). The genome consists of a 5′ untranslated region (UTR), a single open reading frame (ORF) and a 3′-UTR terminating in a poly(A) tail. The 5′ UTR is approximately 750 nt long and contains six conserved RNA domains, I–VI that are involved in the regulation of genome replication and polyprotein translation (Figure 1A). Domain I forms a four-way junction structure, called cloverleaf, which functions as the promoter for negative-strand RNA synthesis by the viral polymerase 3Dpol (2,5). The cloverleaf structure also acts as a switch to direct the transition from viral translation to genome replication and serves as the assembly site of the viral replication complex (6–9). The other domains, II–VI, form an internal ribosome entry site (IRES) that recruits the ribosome for initiation of viral polyprotein translation (10).
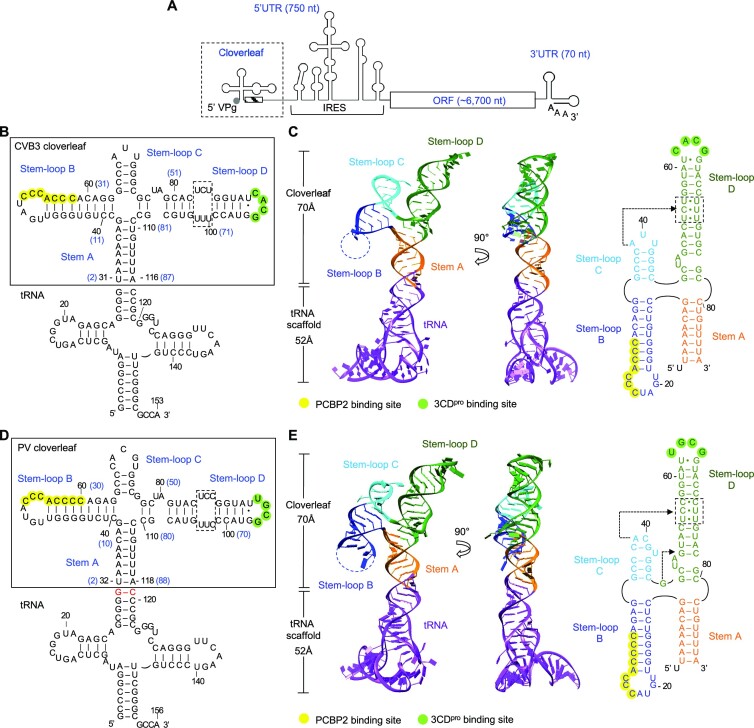
Figure 1.
Enterovirus cloverleaf RNA forms an H-type four-way junction. (A) Schematic representation of an enterovirus genome. The genome contains one open reading frame (ORF), flanked by a highly structured 5′ untranslated region (UTR) and a 3′ UTR with a poly(A) tail. The 5′ UTR contains the cloverleaf and internal ribosome entry site (IRES) structures and is covalently linked to VPg (viral protein genome-linked). The cloverleaf structure is located at the 5′ terminus of the 5′ UTR immediately followed by the 3′ poly-rC region (hatched box). (B) Design of tRNA-fused coxsackievirus B3 (CVB3) cloverleaf. CVB3 cloverleaf (nucleotides 2–87) is inserted into the anticodon loop of human tRNALys scaffold sequence to create tRNA-CLCVB3. The nucleotide numbers in cloverleaf are also shown on the secondary structure in parenthesis in blue. Cloverleaf consists of stem A and stem–loops B, C and D. The PCBP2 binding site in stem–loop B is colored in yellow, and the 3CDpro binding site is colored in green. The pyrimidine mismatch region in stem–loop D is boxed. (C) Crystal structure of CVB3 cloverleaf fused with a tRNA-scaffold. Cloverleaf RNA is colored by domain: stem A, orange; stem–loop B, blue; stem–loop C, cyan; and stem–loop D, green. The tRNA scaffold is colored purple. The missing nucleotides in stem–loop B are indicated by a dotted line. The secondary structure of CVB3 cloverleaf derived from the crystal structure is shown on the right. The PCBP2 binding site, the 3CDpro binding site and the pyrimidine mismatch region are highlighted as in (B). The nucleotides involved in the A•C-U base triple are indicated by an arrow. (D) Design of tRNA-fused poliovirus (PV) cloverleaf. The PV cloverleaf sequence (nucleotides 2–88) is inserted into the anticodon loop of human tRNALys. Additional G-C base pair was introduced between the tRNA scaffold and PV cloverleaf sequence to facilitate crystallization (colored in red). Functional regions in tRNA-CLPV are indicated as in (B). (E) Crystal structure of PV cloverleaf fused with a tRNA-scaffold. The tRNA-CLPV construct is colored by domain as in (C). The secondary structure derived from the crystal structure is shown on the right. PV cloverleaf has two base triples A•C-U and G•G-C, and their locations are indicated with arrows.
Enteroviruses control the relative levels of viral protein translation and genome replication by modulating interactions of cellular and viral proteins with the 5′ UTR of the genome. At the early stages of viral infection, the cellular RNA-binding protein, poly(C)-binding protein 2 (PCBP2) binds the cloverleaf RNA and IRES, and increases rate and quantity of viral protein translation (11). Upon translation, virally encoded 3CDpro, the precursor protein for 3C protease (3Cpro) and 3D polymerase (3Dpol) forms a ternary complex with cloverleaf RNA and PCBP2, which then represses viral translation and promotes genomic RNA synthesis (6,12,13). Interestingly, the affinity of PCBP2 for cloverleaf increases ∼100-fold in the presence of 3CDpro (12), suggesting that 3CDpro stabilizes the interaction between PCBP2 and cloverleaf. This differential interaction of PCBP2 with cloverleaf RNA in the presence of 3CDpro directs the switch from viral genome translation to RNA replication (negative strand RNA synthesis). However, the molecular mechanism by which the cloverleaf mediates the switch is not well understood. Here, we report the crystal structures of cloverleaf RNA from coxsackievirus B3 (CVB3) and PV. The ‘H’-shaped structure of cloverleaf RNA is stabilized by a unique adenosine-cytidine-uridine (A•C-U) base triple between stem–loops C and D. Disruption of the A•C-U base triple enhances PCBP2 binding to cloverleaf, suggesting a new model for the translation-to-replication switch.
MATERIALS AND METHODS
Construction, expression and purification of CVB3 and PV tRNA-cloverleaf (tRNA-CL)
The tRNA-CL fusion constructs for CVB3 and PV were designed based on the tRNA scaffold approach (14,15). The CVB3 cloverleaf sequence (nt 2–87 of AY752944.2) or PV cloverleaf sequence (nt 2–88 of V01149.1) was inserted into the anticodon loop of human tRNALys to generate tRNA-CLCVB3 (153 nt) and tRNA-CLPV (156 nt), respectively (Figures 1B and 1D). Secondary structure prediction using the RNAfold program (16) indicated that the tRNA-CL RNA maintains the same predicted folds for the individual tRNA and cloverleaf.
DNA encoding the designed tRNA-CLCVB3 was synthesized at Epoch Biosciences (Houston, TX), and inserted into pBluescript II SK + vector under the control of E. coli lpp promoter and rrnC terminator. The tRNA-CLPV was generated from tRNA-CLCVB3 by sequential site-directed mutagenesis replacing the sequence of CVB3 cloverleaf RNA with PV cloverleaf. Additionally, a G-C base pair was inserted between the tRNA scaffold and the PV cloverleaf sequence to facilitate crystallization (Figure 1D). The tRNA-CLPV containing the A39U mutation was generated by site-directed mutagenesis of tRNA-CLPV.
The plasmids containing either tRNA-CLCVB3 or tRNA-CLPV were freshly transformed into E. coli BL21 cells, and the cells were grown overnight in 2x YT medium at 37°C. The cells were harvested and resuspended in 15 ml of 10 mM Tris-HCl buffer (pH 7.4) containing 10 mM magnesium acetate. Saturated phenol (10–15 ml) was added to the cell suspension to extract RNA, and the solution was centrifuged at 20,000 g for 30 min at 20°C to remove cell debris. The resulting aqueous phase was mixed with 0.1 volumes of 5 M NaCl and 2 volumes of absolute ethanol, and the RNA pelleted by centrifugation at 20,000 g for 30 min at 4°C. The RNA pellet was dissolved in buffer A (40 mM sodium phosphate or Tris–Cl, pH 7.0), and loaded on HiLoad 16/60 Q-HP anion-exchange column (GE Healthcare, Buckinghamshire, UK) equilibrated with buffer A. The tRNA-CL constructs were eluted with 0.5–0.6 M NaCl gradient in buffer A. The RNA fractions were analyzed by electrophoresis on 8% urea-polyacrylamide gel. The pooled samples were buffer-exchanged to 20 mM Tris–HCl, pH 7.4, containing 100 mM NaCl, and concentrated to 4 mg/ml (tRNA-CLPV) and 30 mg/ml (tRNA-CLCVB3) for crystallization.
Expression and purification of PCBP2 proteins
DNA plasmid encoding human PCBP2 protein was purchased from Addgene (Watertown, MA). The full-length PCBP2 clone was used to subclone KH1–KH2 domain (KH1/2 constituting amino acids 1–168) and the KH3 domain (amino acids 283-358) with an N-terminal hexa-histidine tag followed by a TEV protease cleavage site. Both KH1/2 and KH3 proteins were expressed in Escherichia coli BL21(DE3) cells. Cells were grown in LB supplemented with 30 μg/ml of kanamycin at 37°C to an O.D.600 of ∼0.8, and protein expression was induced by the addition of 1 mM IPTG with growth continued overnight at 18°C. For protein purification, the cell pellet from a 2 L culture was resuspended in 30 ml of lysis buffer (40 mM HEPES, pH 7.4, 200 mM NaCl, 5% (v/v) glycerol, 5 mM β-mercaptoethanol and cOmplete™ EDTA-free protease inhibitor cocktail (Roche)) and lysed by sonication. Protein in the soluble fraction of the lysate was loaded onto TALON™ (Clontech) metal-affinity chromatography resin pre-equilibrated in lysis buffer (without the protease inhibitor cocktail). Bound proteins were eluted using a gradient of 5–150 mM imidazole in elution buffer (40 mM HEPES, pH 7.4, 200 mM NaCl, 5% (v/v) glycerol and 5 mM β-mercaptoethanol). KH1/2 was further purified using size exclusion chromatography on HiLoad Superdex 16/60 S200 prep-grade column (Cytiva) equilibrated with elution buffer. Purity was assessed using SDS-PAGE.
Crystallization, X-ray data collection and structure determination
The tRNA-CLCVB3 was crystallized by the hanging-drop vapor diffusion method at 20°C by mixing the RNA with an equal volume of reservoir solution containing 100 mM Bis-Tris, pH 6.5 and 48% polypropylene glycol (PPG)-P400. Crystals grew to full size within a week. For data collection, crystals from the drop were directly flash-frozen in liquid nitrogen. PPG-P400 served as a cryoprotectant. Diffraction data to 2.9 Å resolution were collected at 100 K with a wavelength of 0.9785 Å at the Advanced Photon Source beamline 21-ID-D (Argonne National Laboratory, Chicago). The data set was processed using HKL2000 (17). The crystal belonged to space group P21 with unit cell dimensions of 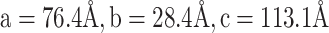 and
and 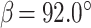 , and contained one molecule in the asymmetric unit with solvent content of 66%. The initial solutions were found by molecular replacement with tRNALys (PDB code 7LYF) and short dsRNA helix (PDB code 6HU6) as search models using the program PHASER in the PHENIX suite (18). Manual model building was carried out with Coot and iterative refinement was performed with phenix.refine (18,19). Reflections with I/σ(I) > 0.7 were included during refinement. The final tRNA-CLCVB3 structure contains entire tRNA-CL except nt 44 to 58 (corresponding to nt 15 to 29 of cloverleaf) and nt 152–153 (two nucleotides at the 3′ end of tRNA). The R and Rfree factors of the final model are 29.0 and 30.8%, respectively (Table 1).
, and contained one molecule in the asymmetric unit with solvent content of 66%. The initial solutions were found by molecular replacement with tRNALys (PDB code 7LYF) and short dsRNA helix (PDB code 6HU6) as search models using the program PHASER in the PHENIX suite (18). Manual model building was carried out with Coot and iterative refinement was performed with phenix.refine (18,19). Reflections with I/σ(I) > 0.7 were included during refinement. The final tRNA-CLCVB3 structure contains entire tRNA-CL except nt 44 to 58 (corresponding to nt 15 to 29 of cloverleaf) and nt 152–153 (two nucleotides at the 3′ end of tRNA). The R and Rfree factors of the final model are 29.0 and 30.8%, respectively (Table 1).
Table 1.
X-ray crystallographic data collection and refinement statistics
| tRNA-CLCVB3 | tRNA-CLPV | |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 0.9785 | 1.542 |
| No. of reflections | 9250 | 8743 |
| Space group | P21 | C2 |
| Unit cell dimensions (Å) |
a = 76.4, b = 28.4, c = 113.1, 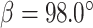
|
a = 150.0, b = 28.0, c = 111.3, 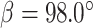
|
| Resolution (Å) | 44.7–2.9 (2.97–2.92)* | 47.5–3.1 (3.15–3.1)* |
| Completeness (%) | 83.3 (76.3) | 99.5 (99.8) |
| Redundancy | 2.5 (2.1) | 5.4 (4.9) |
| I/σI | 5.74 (1.1) | 6.5 (1.1) |
| R p.i.m | 0.094 (0.401) | 0.127 (0.635) |
| Refinement | ||
| No of reflections | 7020 | 8743 |
| Resolution range (Å) | 44.7–2.9 | 47.5–3.1 |
| R work/Rfree | 29.0/30.8 | 27.5/28.9 |
| No. of atoms | ||
| Nucleotide | 2896 | 3109 |
| r.m.s deviations | ||
| Bond angles (°) | 0.69 | 0.56 |
| Bond length (Å) | 0.002 | 0.007 |
| B-factors (Å2) | ||
| Nucleotide | 105 | 114 |
*Values in parentheses are for the highest resolution shell.
The tRNA-CLPV was crystallized by the hanging-drop vapor diffusion method at 20°C by mixing the RNA with an equal volume of reservoir solution containing 47% PPG-P400 and 100 mM Imidazole, pH 7.4. Before sealing the well, 10% (v/v) methanol was added to the reservoir solution, but not the drop. The crystals grew to full size in ∼2 weeks. For X-ray data collection, the crystals were flash-frozen in nitrogen stream. X-ray diffraction data to 3.1 Å resolution were collected at 100 K at a wavelength of 1.542 Å using the in-house Rigaku FRE++DW superbright X-ray source, which is coupled to a Rigaku R-AXIS IV++ image-plate detector. The data set was processed with HKL3000 (17) and the crystal belonged to space group C2 with unit cell dimensions of 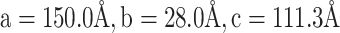 and
and 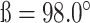 . The asymmetric unit contained one molecule with a solvent content of 66%. The structure was determined by molecular replacement with the individual tRNA and cloverleaf of tRNA-CLCVB3 as search models using the program PHASER in the PHENIX suite (18). Manual model building and iterative refinement were carried out with Coot and phenix.refine (18,19). The final model contained one tRNA-CLPV molecule with the R and Rfree factors of 27.5 and 28.9%, respectively (Table 1). The tRNA-CLPV model contains the entire tRNA-CLPV sequence except nt 47–55 (corresponding to nt 17 to 25 of cloverleaf) and nt 156 (the 3′ end of tRNA).
. The asymmetric unit contained one molecule with a solvent content of 66%. The structure was determined by molecular replacement with the individual tRNA and cloverleaf of tRNA-CLCVB3 as search models using the program PHASER in the PHENIX suite (18). Manual model building and iterative refinement were carried out with Coot and phenix.refine (18,19). The final model contained one tRNA-CLPV molecule with the R and Rfree factors of 27.5 and 28.9%, respectively (Table 1). The tRNA-CLPV model contains the entire tRNA-CLPV sequence except nt 47–55 (corresponding to nt 17 to 25 of cloverleaf) and nt 156 (the 3′ end of tRNA).
Bioinformatic and structural analysis of enterovirus cloverleaf RNA
A total of 162 enterovirus sequences were downloaded from Rfam (RF00386) (20). The cloverleaf sequences containing ambiguous nucleotides were removed, and porcine enterovirus sequences that have shorter 5′ UTR were manually adjusted. The final aligned sequences of 153 cloverleaf RNAs were used for R-scape analysis to determine conservation and co-variance (21). The r.m.s.d. of the CVB3 and PV cloverleaf structures were calculated using Chimera (22).
Small-angle X-ray scattering (SAXS)
SAXS experiments were performed using a Rigaku FR-E++ X-ray source (λ = 1.542 Å) and BioSAXS-1000 camera (Woodlands, TX, USA). Samples for SAXS analysis were freshly prepared from purified tRNA-CLPV wild-type and A39U mutant in a buffer of 10 mM Tris pH 7.4 and 100 mM NaCl. Scattering intensities, I(q) for the RNA samples and buffer were recorded as a function of scattering vector q (q = 4πsin θ/λ, where 2θ is the scattering angle and λ is the X-ray wavelength). SAXS data were collected for several concentrations of wild-type (0.39, 0.95, 1.85 and 3.3 mg/ml) and A39U mutant (0.33, 0.85, 1.6 and 3.1 mg/ml) to evaluate the effects of concentration on the scattering curves. For each sample and buffer measurement, SAXS data were collected in 1-h X-ray exposures to assess radiation damage, and the 1-h frames with no significant changes from the first frame were averaged using SAXSLab (Rigaku) to produce 1D curves. The buffer scattering contributions were subtracted from the sample scattering curve using the SAXNS web server (https://xray.utmb.edu/saxns-es.html). Data analysis was performed using PRIMUS from the program ATSAS suite 2.7.1, as previously reported (23,24). Experimental SAXS data from different RNA concentrations were first analyzed for sample aggregation using the Guinier plot, where the forward scattering intensity I(0) and the radius of gyration RG were plotted using the Guinier approximation: I(q) ≈ I(0) exp[(−q2RG2)/3], with the limit qRG < 1.3. The RNAs with high concentrations (>1.6 mg/ml) displayed a mild effect of inter-particle repulsion and thus were removed from further analyses. The pairwise-distance distribution function P(r) was calculated from the entire scattering patterns from WT (0.95 mg/ml) and A39U (0.85 mg/ml) via indirect Fourier inversion of the scattering intensity I(q). The RG and maximum particle diameter (Dmax) were also determined from P(r) function for each RNA sample. The SAXS data extrapolated to zero concentration also have similar RG and Dmax values (25). The RG and Dmax values of tRNA-CLPV were calculated from the crystal structure using the program CRYSOL (23) for comparison to the experimentally determined values.
Isothermal titration calorimetry binding studies
For ITC measurements, PCBP2 proteins and RNA samples were dialyzed overnight at 4°C in ITC buffer containing 20 mM HEPES, pH 7.4, 200 mM NaCl, 2 mM MgCl2 and 5% (v/v) glycerol. PCBP2 protein (KH1/2 and KH3 domains) concentrations were measured using Bradford assays. RNA controls, stem–loop B (5′ GCUCUGGGGUUGUACCCACCCCAGAGC 3′) and 3′ poly-rC (5′ACUCCCUUCCCGUA 3′) were synthesized (IDT, Coralville, IA). RNA concentrations were measured using NanoDrop™ 1000 spectrophotometer (ThermoFisher). ITC experiments were performed using MicroCal PEAQ-ITC (Malvern, UK). PCBP2 protein (300 μl) was loaded in the cell in concentrations ranging from 37–70 μM, and RNA (45 μl) was loaded in the syringe in concentrations ranging from 320–570 μM. Each run was performed at 4°C and consisted of 25–30 injections of 1.25 or 1.75 μl each. ITC measurements for each combination of PCBP2 proteins (KH1/2 and KH3) and RNAs (stem–loop B, 3′poly-rC, tRNA-CLPV and tRNA-CLPV(A39U)) were repeated at least twice. Data was analyzed using MicroCal PEAQ ITC analysis software.
Modeling of cloverleaf RNA with PCBP2 and 3CDpro
The cloverleaf RNA and PCBP2 complex was modeled using the Nova-2 KH3 domain structure in complex with a 20 nt RNA hairpin (PDB accession code 1EC6) (26). The nova-2 RNA sequence that interacts with the KH3 domain (11AUCAC15) corresponds to 22UCCCA26 and 22ACCCA26 in CVB3 and PV cloverleaf, respectively (see Figure 5B). The cloverleaf stem–loop B and Nova-2 RNA hairpin were overlaid by the stem to generate the cloverleaf and PCBP2 KH3 domain complex model. The cloverleaf and 3CDpro interaction was investigated using the PV 3CDpro structure (PDB accession code 2IJD) (27). Interacting residues in 3Cpro and stem–loop D of cloverleaf have previously been identified (28,29). NMR studies of stem–loop D in the presence of 3Cpro suggest that the apical tetraloop and 53C, 61U, 66G and 69C nucleotides contribute to 3Cpro interaction (28). Further, mutational studies in 3Cpro identified that R84, D85, I86, T154, G155, K156 and R176 are involved in the stem–loop D binding (29). Thus, 3CDpro could be manually modelled by placing these 3Cpro residues near the apical loop of stem–loop D and the 3Dpol domain oriented along the dsRNA helix of stem–loop D in cloverleaf (see Figure 5D), since 3Dpol was also reported to contribute to cloverleaf RNA interaction (30,31).
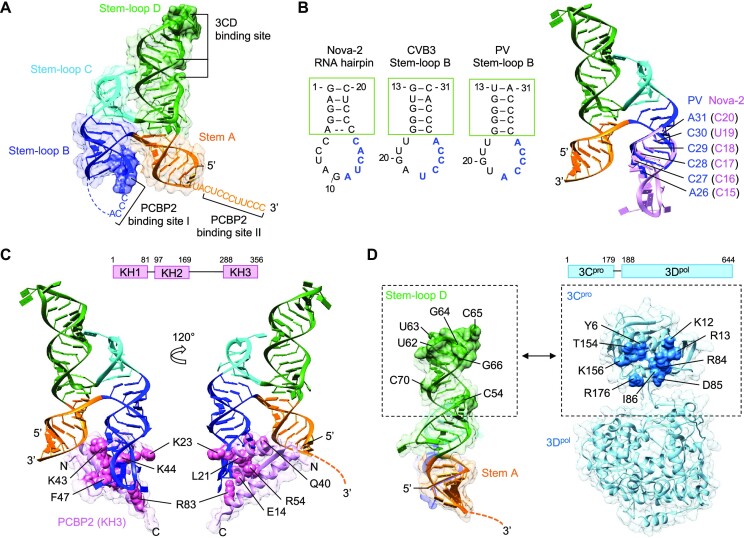
Figure 5.
Model of the ternary complex containing cloverleaf RNA, PCBP2, and 3CDpro. (A) Locations of PCBP2 and 3CDpro binding sites in cloverleaf RNA. Cloverleaf RNA is shown as ribbon diagram in a transparent surface and colored as in Figure 1. The nucleotides indicated in PCBP2 binding in stem–loop B (blue surface) and 3CDpro binding in stem–loop D (green surface) are depicted with darker colors in PV cloverleaf. The 3′ poly-rC site adjacent to stem A (PCBP2 binding site II) is also indicated. (B, C) Model of the cloverleaf-PCBP2 complex. The cloverleaf-PCBP2 (KH3 domain) model was generated using the Nova-2 KH3 domain structure complexed with a 20 nt RNA hairpin (PDB code 1EC6). The nova-2 RNA sequence that interacts with the KH3 domain (11AUCAC15, blue) corresponds to 22UCCCA26 and 22ACCCA26 in CVB3 and PV cloverleaf, respectively (B). The cloverleaf stem–loop B and nova-2 RNA hairpins were overlaid by the stem (green box) to generate a complete cloverleaf structure. Superposed cloverleaf and Nova-2 nucleotides are shown in blue and pink, respectively. For clarity, the PCBP2 KH3 domain is omitted. The cloverleaf-PCBP2 (KH3 domain) complex (C). The KH3 domain residues that directly interact with RNA is shown in pink spheres and labeled. The schematic of the PCBP2 domains is shown on top. (D) Cloverleaf and 3CDpro interaction. Stem-loop D nucleotides and the 3Cpro residues implicated in the cloverleaf and 3Cpro interaction are labeled and shown as green surface on the cloverleaf and blue spheres on the 3CDpro structure (PDB code 2IJD), respectively. 3Dpol also contributes to cloverleaf interaction, but its interaction site on cloverleaf is not known. The schematic of the 3CDpro domains is shown on top.
RESULTS
Enterovirus cloverleaf RNA forms an H-type four-way RNA junction
Cloverleaf RNAs from different enteroviruses share a high degree of sequence identity and show similar predicted secondary structures, comprising of a stem A and three stem–loops B, C and D (Figures 1B, D and S1). The structures of CVB3 and PV cloverleaf RNAs were determined using the tRNA-scaffold approach as previously described (14,15). Briefly, the anticodon loop of human tRNALys was replaced with the CVB3 (nt 2–87) or PV cloverleaf sequence (nt 2–88) to generate the chimeric tRNA-cloverleaf constructs, tRNA-CLCVB3 (153 nt) and tRNA-CLPV (156 nt) (Figures 1B and D). In these chimeras, stem A of cloverleaf is continuous with the anticodon stem of tRNALys. The tRNA-CL constructs enabled large-scale recombinant expression and purification of stable cloverleaf RNA from E. coli. The tRNA-scaffold was also necessary to obtain crystals, since the tRNA moiety was involved in crystal contacts in both RNA structures. The structures of CVB3 and PV tRNA-CL were determined to 2.9 and 3.1 Å resolution, respectively (Table 1). Entire cloverleaf could be modeled except the apical loop in stem–loop B, 15 nt in CVB3 and 8 nt in PV cloverleaf (Figures 1C, E and S2).
The cloverleaf RNAs of both CVB3 and PV form an ‘H’ shaped molecule ∼70 Å long and ∼45 Å wide with its four arms stacked into two coaxial dsRNA helices (Figures 1C and E). The stem A and stem–loop D form a contiguous, coaxial dsRNA helix (one arm of the H) and the stem–loops B and C form the second coaxial dsRNA helix (the other arm of the H). The two larger stem–loops B and D are on diagonally opposite ends of the H junction, antiparallel to each other. The structures belong to the H-family of 4-way RNA junctions, one of the nine known 4-way junction families (32). In CVB3 cloverleaf, both sets of stacked helices are flush, with no mismatched bases, resulting in a stable contiguous double helical RNA structure (Figure 1C). In PV cloverleaf, stem–loops B and C are stacked similar to CVB3 cloverleaf, whereas the stem A and stem–loop D stack is mismatch-mediated by a single base G46 insertion at the junction (Figure 1E). G46 in PV cloverleaf interacts with the 51G-C78 base pair and form a G•G-C base triple, providing additional stability (see Figure 2D and G). The secondary structures of CVB3 and PV cloverleaf derived from the crystal structures are identical to the prediction except a nucleotide at the four-way junction in the PV cloverleaf, where C81 base pairs with G9, instead of the predicted G46 (see Figure 1D, E).
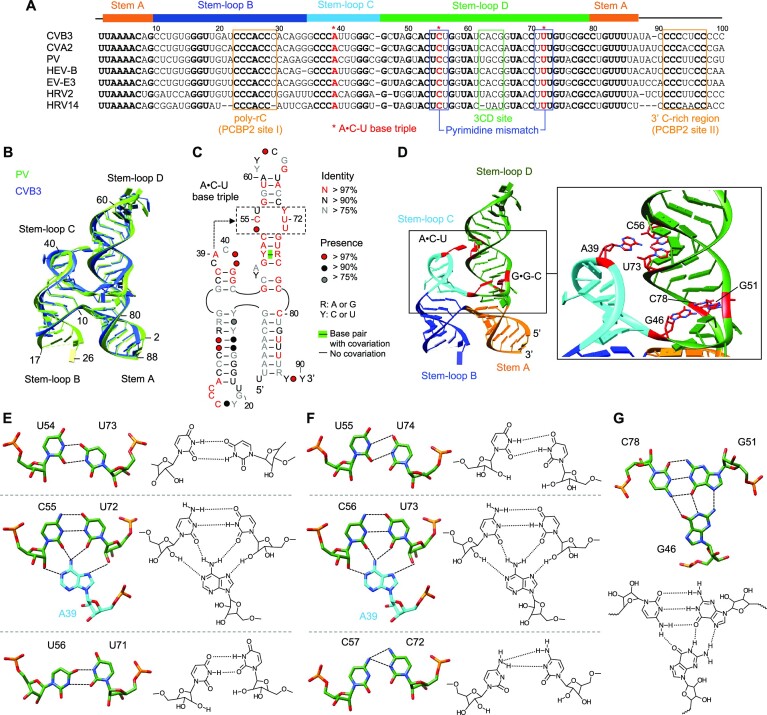
Figure 2.
Cloverleaf RNA is stabilized by the A•C-U base triple. (A) Sequence alignment of enterovirus cloverleaf RNA. The cloverleaf sequences from coxsackievirus B3 and A2 (CVB3, AY752944.2; CVA, NC_038306.1), poliovirus (PV, V01149.1), human enterovirus (HEV-B, NC_001472.1), echovirus (EV-E3, AB647326.1) and human rhinovirus 2 and 14 (HRV2, AB647326.1; HRV14, K02121.1) are aligned. Conserved nucleotides are shown in bold. The overall sequence identity among the seven RNAs is 55%. Structural assignment of the cloverleaf domains (stem A and stem–loops B, C and D) is shown at the top of the alignment. The positions of the PCBP2 binding sites (I and II), 3CDpro binding site, pyrimidine mismatch and the A•C-U base triple are indicated. (B) Superposition of CVB3 and PV cloverleaf structures. The CVB3 (blue) and PV (green) cloverleafs can be superimposed with an r.m.s.d. of 1.99 Å over 64 atom pairs (C3’). The 5′ and 3′ ends of nucleotides are numbered for PV cloverleaf. (C) Conservation of enterovirus cloverleaf. Nucleotide conservation was calculated by R-scape from a total of 153 enterovirus sequences and nucleotides are colored based on the conservation score, red, >97% conservation; black, > 90% conservation; and grey, >75% conservation. Closed circles represent the occupancy of nucleotide positions in the sequences. The pyrimidine mismatch region (boxed) and the location of the A•C-U base triple are indicated. (D) Tertiary interactions in cloverleaf. The positions of the A•C-U and G•G-C base triples in the structure of PV cloverleaf are shown in red. A close-up view is shown on the right. e. f. The A•C-U base triple in CVB3 (E) and PV (F) cloverleaf RNAs. The three base pairs in the pyrimidine mismatch are shown with the chemical structures. A39 base (stem–loop C) is inserted into the central C-U base pair within the pyrimidine mismatch region (stem–loop D) to form the A•C-U base triple. Hydrogen bonds are shown with dotted lines. (G) The G•G-C base triple in PV cloverleaf. The unpaired nucleotide G46 located at the base of stem–loop D interacts with G51-C78 base pair in stem–loop D. The chemical structures with hydrogen bonds are shown below.
Cloverleaf RNA is stabilized by a conserved A•C-U base triple
Enterovirus cloverleaf is highly conserved, and CVB3 and PV cloverleaf RNAs share a 57% sequence identity (Figure 2A). The overall structures of CVB3 and PV cloverleaf RNAs are thus similar, and the two structures are superimposable with an r.m.s.d. of 1.99 Å over 64 C3’ atom pairs (Figure 2B). The biggest difference between the two structures lies in the proximal portion of stem–loop D, due to an additional nucleotide G46 in PV cloverleaf. Sequence alignment of 153 enterovirus cloverleaf RNAs were further used to identify conserved nucleotides and base pair covariance by R-scape (Figure 2C) (21). The highly conserved nucleotides (> 97% conservation) were found in the poly-rC sequence in stem–loop B as well as stem–loops C and D.
In the crystal structures, stem–loop C forms a conserved GC-rich dsRNA stem with the apical loop containing a highly conserved adenosine A39 (Figure 2C). Stem-loop D also forms a GC-rich stem containing a three-pyrimidine mismatch (54UCU56 and 71UUU73 in CVB3 cloverleaf and 55UCC57 and 72UUC74 in PV cloverleaf). In both structures, the pyrimidine mismatch forms three non-Watson-Crick base pairs, U-U, C-U and U-U (or C-C) with two hydrogen bonds per base pair (Figures 2D-F). Further, the structures show a new tertiary interaction between stem–loops C and D, where the conserved A39 of stem–loop C is inserted into the minor groove of the pyrimidine mismatch, interacting with the central C-U base pair. The A39 base forms two hydrogen bonds with each nucleotide of the C-U base pair via its Watson-Crick and Hoogsteen edges (Figures 2E and F). This novel A•C-U base triple, observed in both CVB3 (A39•C55-U71) and PV (A39•C56-U72) cloverleaf structures, differs from the more common A-minor motif, wherein the sugar edge (N1-C2-N3 edge) of A is inserted into the minor groove of a dsRNA helix (33,34). Formation of the A•C-U base triple requires narrowing the width of the minor groove from a typical ∼11 Å (C1’-C1’ distance) to ∼8.5 Å at the central C-U base pair in the pyrimidine mismatch, indicating that the pyrimidine mismatch functions as a structural funnel to reduce the width of the minor groove (Figures 2E and F). The existence of the A•C-U base triple is also consistent with SHAPE analysis, in which A39 and pyrimidine mismatch nucleotides (54UCU56 and 71UUU73 in CVB3) show low reactivity (Figure S3) (35). These extensive interactions in the base triple are unique to the cloverleaf RNA structure and have not been reported for any RNA structures in the RNA base triple database (http://rna.bgsu.edu/triples/triples.php) (36).
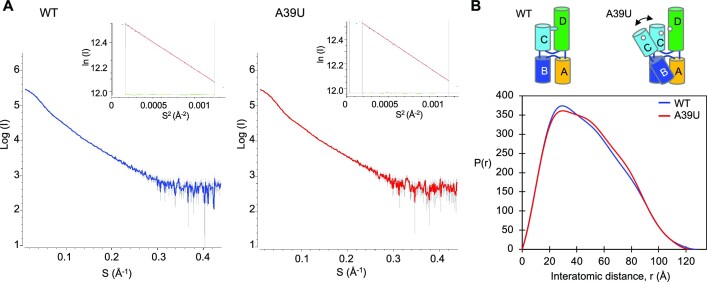
The A•C-U base triple would stabilize the overall ‘H’ conformation of cloverleaf, and likely restrict hinge-like motions between the two arms, i.e. the stacked helix of stem–loops B and C, relative to the stacked helix of stem A and stem–loop D. To test the effect of the A•C-U base triple on cloverleaf RNA dynamics, we used small-angle X-ray scattering (SAXS) and measured the scattering profiles of wild-type and A39U mutant PV cloverleaf RNAs in solution (Figure 3A). The A39U mutant cannot form the A•C-U base triple, and thus stem–loop C would move in relation to stem–loop D, broadening the spatial distribution of the ensemble of cloverleaf structures. Such a change would be visible in the pairwise distance distribution function P(r), which represents the distribution of distances between pairs of atoms within the RNA sample. The P(r) function for the wild-type PV cloverleaf displays a skewed distribution with a peak in the 30–40 Å range, corresponding to the atomic distances between two parallel helices, and the maximum distance of 128 Å, consistent with the elongated shape of tRNA-CLPV. In comparison, the P(r) function of the A39U mutant shows a reduced maximum in the 30–40 Å interatomic range and an increased shoulder peak in the 50–80 Å range, suggesting that the loss of the restricting tertiary interaction (i.e. A•C-U base triple) leads to subsequent increase in the relative motion between the two helices (Figure 3B). The results thus indicate that the two stacked helices in the ‘H’ shaped cloverleaf are stabilized by the A•C-U base triple.
Figure 3.
Small-angle X-ray scattering (SAXS) data analysis. (A) Scattering curves of PV wild-type and A39U mutant cloverleaf. Experimental scattering patterns for PV wild-type cloverleaf (0.95 mg/ml, blue) and A39U mutant cloverleaf (0.89 mg/ml, red) are shown. In the Guinier plot (inset), the Guinier region (qRG < 1.3) is delimited by the gray vertical lines. The red solid lines are the linear regression fit in the Guinier region, and the residuals are colored green. The radius of gyration (RG) determined from the Guinier plot is 36.2 and 36.9 Å for wild-type and A39U cloverleaf, respectively. (B) Pairwise distance distribution function P(r) analysis of PV wild-type and A39U mutant cloverleaf. The pairwise distance distribution function P(r) calculated from the SAXS curves shows that the A39U mutant displays an increased population in the 50–80 Å interatomic distance range compared to the wild-type, consistent with a conformational change within cloverleaf. The radius of gyration (RG) and maximal distance (Dmax) determined from the pairwise distance distribution function are 38.0 and 128 Å for wild-type and 38.2 and 124 Å for A39U mutant. In comparison, the RG and Dmax values calculated from the crystal structure are 38 and 130 Å, respectively.
Binding interaction of PCBP2 domains to cloverleaf RNA is modulated by the A•C-U base triple
Enterovirus cloverleaf RNA has two poly-rC sites, one within stem–loop B (PCBP2-binding site I) and the second in the C-rich region adjacent to the 3′ end of the cloverleaf structure (3′poly-rC, PCBP2-binding site II) (Figures 1A, 2A and S1). Mutations in either stem–loop B (C24 to A) or 3′poly-rC (C91/93/97/99 to A) of the PV cloverleaf resulted in loss of PCBP2 binding, indicating that both sites are critical for PCBP2 interaction and viral replication (8,31,37,38). PCBP2, a member of the hetero nuclear ribonucleoprotein (hn-RNP) family of proteins, consists of three K-homology (KH) RNA-binding domains, KH1-3, with a 120 aa-long linker between the KH2 and KH3 domains (see Figure 5C). Each KH domain recognizes ∼4 nt long single stranded C-rich sequence, and the individual KH1 and KH3 domains have been shown to interact with cloverleaf RNA (39). However, the binding affinities of individual KH domains are lower than that of full-length PCBP2 (39). For example, the binding affinity of KH1 domain to PV cloverleaf is ∼10 fold lower than that of full-length PCBP2 (40,41). Further, mutations in KH2 and KH3 domains significantly decrease PV cloverleaf interaction, suggesting that the entire PCBP2 likely contributes to binding (40,41).
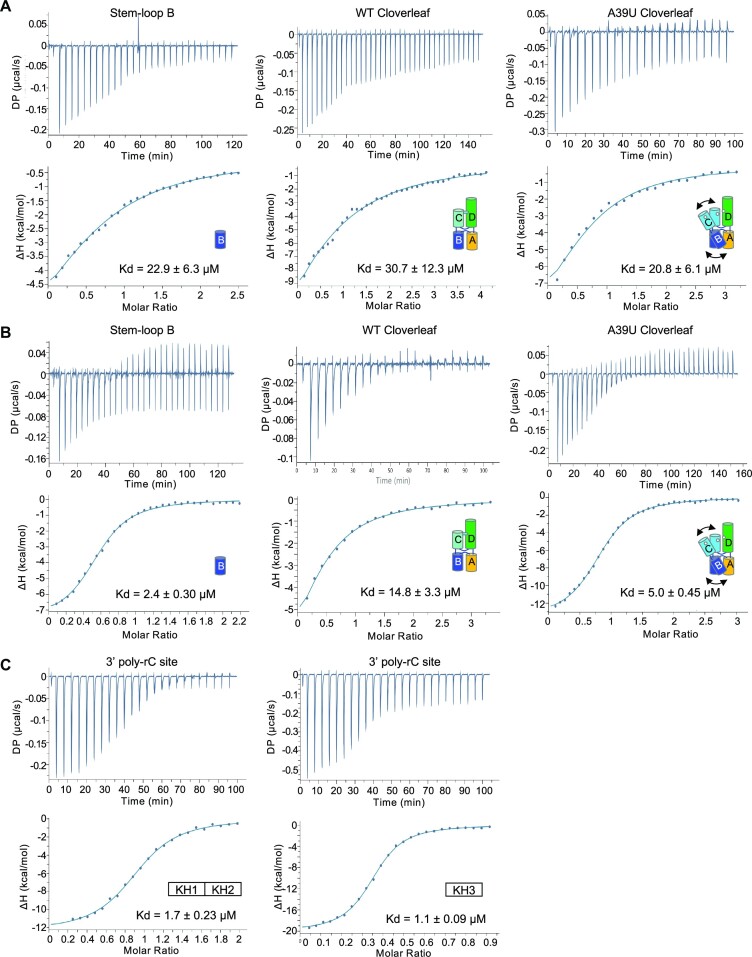
The two PCBP2 binding sites in cloverleaf are structurally distinct. The poly-rC site in stem–loop B is restricted in a stem–loop and tucked against stem A, while the 3′ poly-rC site is single stranded and flexible. Thus, the KH domains of PCBP2 may recognize the two poly-rC sites differently. Since cloverleaf has been shown to interact with KH1 and KH3 domains, we generated KH1–KH2 domain (KH1/2) and KH3 domain of PCBP2 separately and measured their interactions with PV cloverleaf RNAs. The KH1/2 domain was chosen because the KH1 and KH2 domains likely function as a single RNA binding unit in the context of the full-length PCBP2 protein. The KH1–KH2 domains of PCBP2 form a stable heterodimer in solution that buries a large hydrophobic surface (42). The RNA interaction sites are on the opposite side of the dimer interface, and binding of nucleic acid does not disrupt the KH1–KH2 domain interactions (42). In contrast, the KH3 domain exists as a monomer in solution and does not form a homo or heterodimer with itself or other KH domains (42). The KH1/2 and KH3 domain were then used to measure their interactions with synthetic PV stem–loop B (5′ GCUCUGGGGUUGUACCCACCCCAGAGC 3′) and the 3′poly-rC site (5′ ACUCCCUUCCCGUA 3′) using isothermal titration calorimetry (ITC). The KH1/2 domain binds stem–loop B and 3′poly-rC site with binding affinity (Kd) of 22.9 ± 6.3 and 1.7 ± 0.23 μM, respectively (Figure 4A and C). Thus, the KH1/2 domain preferentially binds the linear 3′poly-rC site with ∼14-fold higher affinity than stem–loop B. The KH1/2 domain binds both stem–loop B and 3′poly-rC site in a 1:1 molar ratio, indicating that a single KH1/2 interacts with either PCBP2-binding site (Figures 4A and B). In contrast, the KH3 domain binds stem–loop B and 3′poly-rC site with similar Kd values of 2.4 ± 0.30 and 1.1 ± 0.09 μM, respectively (Figure 4B and C). The KH3 domain binds stem–loop B with 1:1 stoichiometry, while it binds 3′poly-rC with 2:1 (protein/RNA) stoichiometry (Figure 4C). This is consistent with the previous observations based on ITC and structural data that multiple KH3 domains (but not the KH1/2 domain) can interact with tandem poly C regions in ssRNA (43,44).
Figure 4.
Isothermal calorimetry of PCBP2 domains with poly-rC containing RNAs. (A) Interaction of PCBP2 KH1/2 domain with PV cloverleaf RNA. Isolated stem–loop B, WT cloverleaf (tRNA-CLPV) and A39U mutant cloverleaf (tRNA-CLA39U) were titrated into PCBP2 KH1/2 domain. Thermograms and fitted binding curves are shown. Data were fit with single binding-site model. Titrations were repeated at least twice. (B) Interaction of PCBP2 KH3 domain with PV cloverleaf RNA. Isolated stem–loop B, WT cloverleaf and A39U mutant cloverleaf were titrated into PCBP2 KH3 domain. The data were fit with single binding-site model. Note KH3 domain binds A39U mutant cloverleaf with higher affinity than WT cloverleaf, suggesting that stem–loop B site is not accessible in WT cloverleaf. (C) Interaction of PCBP2 domains with PV 3′poly-rC site. The linear ssRNA containing the 3′poly-rC sequence was titrated into either KH1/2 or KH3 domains. The binding affinities to the linear ssRNA are similar for both PCBP2 domains. Note that the KH3 domain binds 3′poly-rC with 2:1 (protein/RNA) stoichiometry. Summary of parameters are listed in Table S1.
We next asked whether the PCBP2 domains can recognize stem–loop B in the context of the full-length PV cloverleaf. The KH1/2 domain binds the wild-type cloverleaf with Kd of 30.7 ± 12.3 μM, similar to that of isolated stem–loop B (Figure 4A). Surprisingly, the KH3 domain binds wild-type cloverleaf with Kd of 14.8 ± 3.3 μM, 5-fold lower affinity than that of isolated stem–loop B (2.9 μM) (Figure 4B). The result suggests that the KH3 domain cannot access the binding site of stem–loop B in the full-length cloverleaf. Since the cloverleaf structure is conformationally restricted by the A•C-U base triple between stem–loops C and D, we measured PCBP2 domain interactions with A39U mutant cloverleaf, which increases the flexibility between the stem–loops. The KH1/2 domain binds A39U cloverleaf with Kd of 20.8 ± 6.1 μM, a small increase from 31 μM for wild-type cloverleaf. However, the KH3 domain binds the A39U cloverleaf with 5.0 ± 0.45 μM affinity, a 3-fold increase from the wild-type cloverleaf. The Kd value is similar to that of the stem–loop B control, suggesting that the poly-rC site in stem–loop B is again accessible in the A39U mutant cloverleaf. Hence, the A•C-U base triple of cloverleaf restricts the binding of PCBP2 domains and consequently, relaxing the A•C-U base triple increases the PCBP2 binding affinity. Taken together, these data lead to a model where KH3 binds the tip of stem–loop B and KH1/2 binds to the 3′ poly-rC site. The modulation of PCBP2 binding via the A•C-U base triple could be a mechanism for enhanced PCBP2 interaction with cloverleaf RNA in the presence of 3CDpro (see Discussion).
Two poly-rC sites in cloverleaf RNA are spatially proximal, suggesting cooperative binding of PCBP2 domains
The ‘H’ conformation of the cloverleaf RNA would place the two PCBP2 binding sites, stem–loop B and 3′poly-rC site spatially proximal to one another (Figure 5A), suggesting that the two poly-rC sites may function together to interact with KH domains within a single PCBP2 molecule. The poly-rC site in stem–loop B consists of two consecutive CCC sequences, i.e. 23CCCACCC29 in CVB3 or 23CCCACCCC30 in PV (Figure 2A). The first half of the sequence (23CCC25) is in the apical loop, while the second half (28CCC30) is in the stem portion of stem–loop B that stacks against stem A (Figure 5A). Since KH domains bind ssRNA, PCBP2 would likely interact with the poly-rC site in the apical loop (23CCC25) of stem–loop B. Further, it has been shown that when 28CCC30 in PV was replaced with UUU (maintaining the U-G base pairs), the mutant virus was still able to replicate (45). Thus, the second half of poly-rC site in the stem may be required to maintain the structural integrity of stem–loop B.
PCBP2 KH domains show a common binding mode for a C-rich RNA/DNA, which allows for modeling of the cloverleaf RNA and PCBP2 KH3 domain interaction. Single-stranded RNA/DNA binds in an extended conformation across one side of the KH domain in a cleft formed by two helices and a strand (46). To visualize PCBP2 and cloverleaf interaction, we modeled the KH3 domain of PCBP2 on the apical loop of stem–loop B using the structure of a homologous Nova-2 KH3 domain complexed with a 20 nt hairpin (PDB accession code 1EC6) (26). The 20 nt hairpin in the Nova-2 KH3 domain complex has a similar stem sequence and loop size to stem–loop B of cloverleaf (Figure 5B). The cloverleaf and KH3 complex was thus modeled by superposition of the Nova-2 RNA and stem–loop B of cloverleaf (Figure 5C). In the superposed structure, the KH3 domain of PCBP2 is positioned towards stem A of the cloverleaf with minor clashes (Figure 5C). Factors contributing to the relaxation of the H conformation of the cloverleaf would relieve the steric clashes between the PCBP2 KH3 domain and stem A, facilitating increased access to the site. Thus, the superposition is in agreement with the lower binding affinity of KH3 domain with full-length cloverleaf than the isolated stem–loop B (Figure 4B). Furthermore, this arrangement of KH3 minimally affects the 3′poly-rC site, and there is sufficient space for a second KH domain (KH1/2) to bind the 3′poly-rC site (Figure 5C). Thus, it is possible that one PCBP2 molecule binds across the two poly-rC sites simultaneously facilitated by the long linker between KH2 and KH3 domains, rather than two distinct PCBP2 proteins individually interacting with each of the two sites. This cooperative binding of the KH domains across both poly-rC sites would increase both specificity and affinity of the protein–RNA interaction compared to individual KH domain interactions of multiple PCBP2 molecules (see Figure 6).
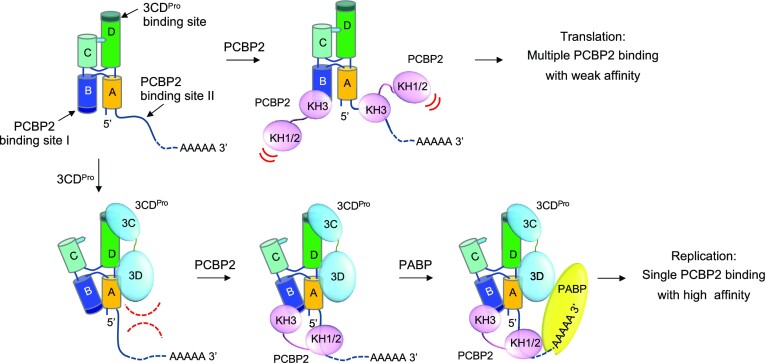
Figure 6.
Model of 3CDpro-enhanced PCBP2 interaction with cloverleaf. Enterovirus genome contains the 5′ cloverleaf and the 3′ poly-A tail. In the absence of 3CDpro, multiple PCBP2 molecules can bind the two poly-rC sites of cloverleaf with low affinity. Following viral protein translation, 3CDpro binds stem–loop D and ‘unlocks’ the A•C-U base triple. This allows a hinge-like motion between the two arms of the H configuration, and exposes the PCBP2-binding site on stem–loop B. Additionally, 3CDpro binding to stem–loop D may restrict the flexibility of the 3′ C-rich region (PCBP2-binding site II). This orients the 3′ C-rich region such that a single PCBP2 molecule can bind to both poly-rC sites simultaneously, increasing the binding affinity of PCBP2 to cloverleaf. The resulting cloverleaf-PCBP2-3CDpro ternary complex then facilitates genome circularization by recruiting the poly(A) binding protein associated with the 3′ poly(A) tail, and subsequent initiation of viral RNA synthesis.
Viral protein 3CDpro binding site on the cloverleaf is distal to the PCBP2 binding sites
The enteroviral 3CDpro protein binds cloverleaf RNA primarily via the apical loop of stem–loop D (30,31,47). In the cloverleaf RNA structures, stem–loops B and D are antiparallel to each other, and their apical loops (i.e. the PCBP2 and 3CDpro binding sites) are separated by > 80 Å (Figure 5A). Thus, 3CDpro does not likely interact with PCBP2 directly, consistent with a previous report (41). Both size and sequence of the apical loop are critical for virus-specific 3CDpro interaction. For example, CVB3 3CDpro binds stem–loop D of CVB3, PV or HRV-2 (with CACG, UGCG or UACG in the apical loop, respectively), but is unable to bind that of HRV-14 (with UAU in the apical loop) (Figures 1B, D and S1) (47–49). Deletion of C64 in the apical loop of stem–loop D (62CACG65) in CVB3 leads to significant decrease in negative strand RNA synthesis, suggesting that 3CDpro may specifically recognize C64 (37). The apical loops of stem–loop D in CVB3 (CACG) and PV (UGCG) are structurally similar to the UNCG tetraloop motif (N = any base) that is commonly found in RNA-protein interaction sites (50). The two terminal bases, 62C and 65G in the CVB3 tetraloop or 63U and 66G in the PV tetraloop, form hydrogen bonds stabilizing the loop, while the critical C64 in CVB3 and the equivalent C65 in PV are solvent exposed (Figures 5D and S4c).
3CDpro is a large molecule (∼100Å in length), and the major cloverleaf binding site is in 3Cpro. NMR studies of CVB3 stem–loop D in the presence of 3Cpro suggest that the apical tetraloop, C53, 61U-G66 base pair and C69 (corresponding to C54, 62U-G67 base pair and C70 in PV sequence) contribute to 3Cpro interaction (Figures 1C and 5D) (28). In addition, mutational studies in 3Cpro identified 82KFRDI86 and 153TGK155 residues (PV sequence) as the major cloverleaf-binding site in 3CDpro (Figure 5D) (29). Thus, the 3Cpro domain could be placed near the apical loop of stem–loop D away from stem–loop C in the cloverleaf and 3CDpro complex (see Figure 6). The 3Dpol domain of 3CDpro also interacts with cloverleaf RNA, since 3CDpro binds cloverleaf with ∼10-fold higher affinity than the 3Cpro domain alone (30,31). Although it is not clear how 3CDpro is oriented on cloverleaf, 3CDpro could reach the bottom of stem A that is coaxially stacked with stem–loop D if it is oriented along the dsRNA helix of stem–loop D (Figure 5D).
DISCUSSION
Proposed model of 3CDpro-enhanced PCBP2 interaction with cloverleaf
Following protein translation, the viral 3CDpro and cellular PCBP2 proteins bind cloverleaf RNA to form a ternary complex at the 5′ end of the viral genome, which then promotes negative-strand RNA synthesis from the 3′ end. In the ternary complex, the presence of 3CDpro increases the binding affinity of PCBP2 for the cloverleaf structure by ∼100-fold (from 95 nM to 1 nM) (12). However, the mechanism by which 3CDpro stabilizes the interaction between cloverleaf and PCBP2 is unknown. In one model, stem–loops B and D are proposed to interact directly, such that 3CDpro binding to stem–loop D induces a conformational change in stem–loop B, to which PCBP2 binds with higher affinity (51). However, the crystal structures of CVB3 and PV cloverleaf RNAs show that the stem–loops B and D are spatially distant, located on the opposite ends of the H junction structure (Figures 1 and 5A). Thus, direct interaction between stem–loops B and D cannot explain the cooperativity of 3CDpro and PCBP2 binding to cloverleaf. Consistent with the structures, mutations in cloverleaf RNA that disrupt PCBP2 binding do not have any effect on its interaction with 3CDpro, further confirming that the PCBP2 and 3CDpro binding sites are spatially separated (52). In another model, PCBP2 interacts with 3CDpro directly when 3CDpro binds stem–loop D, and this direct interaction between 3CDpro and PCBP2 would lead to an increased affinity to cloverleaf. However, it also seems unlikely that 3CDpro binding to stem–loop D creates an additional binding site for PCBP2, since the direct interaction between PCBP2 and 3CDpro has not been observed (41).
Based on the arrangement of the two PCBP2 binding sites in relation to the 3CDpro binding site on the cloverleaf, and the increased binding affinity observed for PCBP2 KH3 domain to the A39U cloverleaf, we propose a new model, in which 3CDpro interaction with stem–loop D induces global conformational changes in cloverleaf (Figure 6). First, 3CDpro interaction with stem–loop D allows hinge-like motions between the two arms of ‘H’ junction by ‘unlocking’ the A•C-U base triple between stem–loops C and D. This in turn exposes stem–loop B to bind PCBP2 with increased affinity. Additionally, the large 3CDpro interaction with stem–loop D and likely with stem A may restrict the conformational flexibility in the 3′poly-rC site, orienting it towards stem–loop B (PCBP2-binding site I) (Figure 6). This new arrangement of the two poly-rC sites would be conducive to a single PCBP2 binding to both sites and explain the increased binding affinity for PCBP2 in the presence of 3CDpro (Figure 6). The ternary complex of cloverleaf, PCBP2, and 3CDpro then interacts with poly(A)-binding protein bound at the 3′ poly-A tail, promoting the circularization of the viral genome (53). 3CDpro is then cleaved to 3Cpro and 3Dpol, and the viral polymerase 3Dpol bound to the cloverleaf RNA is now positioned to initiate RNA synthesis from the 3′ terminus of the circularized genome.
Implications for clinically isolated enterovirus replication
Enteroviruses with 5′ terminal deletions within cloverleaf (15–50 nt) have been isolated in patients with myocarditis and dilated cardiomyopathy along with small amount of intact viral genome (54,55). These viruses replicate slowly and establish persistent infection in the heart muscle, leading to viral myocarditis. Perhaps the slow viral replication rate helps the virus avoid host immune surveillance (54). In replication assays, viral RNAs with 15–36 nt deletions at the 5′ terminus replicate in vitro without any helper virus, while viral RNAs with deletions of 37–50 nt at the 5′ terminus do not replicate and require a small amount of intact viral genome (as helper virus) (56,57). It is surprising that the 5′ terminally deleted viruses can replicate, considering the essential role of cloverleaf RNA in viral replication. The 36 nt deletion in the cloverleaf RNA destroys double-stranded stem A and removes entire stem–loop B but maintains tertiary interaction of stem–loops C and D (Figure 1). Our structures suggest that stem–loop B and the 3′ C-rich region are individually capable of interacting with KH domains in a single PCBP2. Thus, even in the absence of stem–loop B, PCBP2 likely binds to the 3′ C-rich region of the truncated cloverleaf, thus maintaining a ternary complex with 3CDpro. Such a ternary complex would preserve a productive arrangement of cloverleaf, PCBP2 and 3CDpro similar to that of the full-length cloverleaf, supporting viral replication.
While our manuscript was under review, a crystal structure of CVB3 cloverleaf RNA was published (58). The structure was determined using a Fab fragment as a protein scaffold for crystallization and thus the cloverleaf sequence was modified to include the Fab-binding RNA sequence (5′-GAAACAC-3′) in stem–loop B and a G65 to C substitution in stem–loop D. Despite Fab-binding sequence being in stem–loop B, the heavy and light chains of Fab molecules make extensive lattice contacts with all four RNA stems in the crystal. Nevertheless, the overall structure of the Fab-bound cloverleaf is similar to our WT CVB3 structure including the conserved A•C-U base triple. The two cloverleaf structures could be superposed with an r.m.s.d. of 3.4 Å over 71 common backbone atom pairs (Figure S4a). The largest differences are found in Fab interaction sites or the G65C mutation site (Figure S4a). First, while our cloverleaf shows coaxially stacked stem A and stem–loop D, the helical axis of stem A in the Fab-bound structure is tilted by ∼15° relative to that of stem–loop D and interacts with Fab. Second, the Fab-binding sequence in stem–loop B is stabilized by the Fab interaction, while our structure lacks density for the apical loop of stem–loop B, indicating the highly dynamic nature of the loop. Third, the unpaired dinucleotide bulge in stem–loop D (48UA49) shows different conformations (Figure S4b). In our structure, both nucleotides are flipped out of the helix, allowing uninterrupted base stacking in the helix. In the Fab-bound structure, A49 is flipped out of the helix and interacts with Fab, while U48 forms hydrogen bonds with the G50-C77 base pair. This creates a kink that disrupts the continuous stacking (Figure S4b). Finally, the apical tetraloop in stem–loop D, the 3CDpro binding site, shows different orientations. In our structure, the tetraloop (62CACG65) forms an UNCG-type configuration, where the two penultimate bases (C62 and G65) are involved in hydrogen bonding (Figure S4c). The tetraloop is tilted away from the helical axis of stem D and projects the apical loop for 3CDpro interaction. In comparison, the Fab-bound structure contains a G65C mutation in the 62CACC65 tetraloop and shows a GNRA-type tetraloop configuration (58) (Figure S4c). The tetraloop is roughly parallel to the helical axis of stem D and more proximal to stem C (Figure S4a). Previous binding studies with 3Cpro suggested that the local RNA structure of the loop, determined by the length and sequence, may be critical for its recognition by 3Cpro or the 3CDpro protein (47). Indeed, the G65C mutant cloverleaf binds 3Cpro with 13-fold lower affinity than WT cloverleaf (58). Thus, a single nucleotide change in the tetraloop can perturb the native conformation and disfavor the biologically active structure for 3Cpro interaction.
Supplementary Material
ACKNOWLEDGEMENTS
This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). The authors acknowledge the Sealy Center for Structural Biology and Molecular Biophysics at the University of Texas Medical Branch at Galveston for providing research resources. We thank Dr Krishna Rajarathnam for helpful advice on ITC.
Contributor Information
Keerthi Gottipati, Department of Biochemistry and Molecular Biology, Sealy Center for Structural Biology and Molecular Biophysics, University of Texas Medical Branch, 301 University Boulevard, Galveston, TX 77555, USA; Department of Molecular and Cellular Biochemistry, Indiana University, 212 S. Hawthorne Drive, Bloomington, IN 47405, USA.
Sean C McNeme, Department of Biochemistry and Molecular Biology, Sealy Center for Structural Biology and Molecular Biophysics, University of Texas Medical Branch, 301 University Boulevard, Galveston, TX 77555, USA.
Jerricho Tipo, Department of Pharmacology and Toxicology, Sealy Center for Structural Biology and Molecular Biophysics, University of Texas Medical Branch, 301 University Boulevard, Galveston, TX 77555, USA.
Mark A White, Department of Biochemistry and Molecular Biology, Sealy Center for Structural Biology and Molecular Biophysics, University of Texas Medical Branch, 301 University Boulevard, Galveston, TX 77555, USA.
Kyung H Choi, Department of Biochemistry and Molecular Biology, Sealy Center for Structural Biology and Molecular Biophysics, University of Texas Medical Branch, 301 University Boulevard, Galveston, TX 77555, USA; Department of Molecular and Cellular Biochemistry, Indiana University, 212 S. Hawthorne Drive, Bloomington, IN 47405, USA.
DATA AVAILABILITY
All data are available in the main text or the supplementary materials. The poliovirus and coxsackievirus B3 cloverleaf structures were deposited to PDB with accession numbers 8S95 and 8SP9, respectively.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH [R01AI 187856, R21AI 157336 to KHC]; Jeane B. Kempner and McLaughlin predoctoral fellowship (to S.M.).
Conflict of interest statement. None declared.
REFERENCES
- 1. Barton D.J., Morasco B.J., Flanegan J.B. Translating ribosomes inhibit poliovirus negative-strand RNA synthesis. J. Virol. 1999; 73:10104–10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ogram S.A., Flanegan J.B.. Non-template functions of viral RNA in picornavirus replication. Curr. Opin. Virol. 2011; 1:339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Choi K.H. The role of the stem-loop A RNA promoter in flavivirus replication. Viruses. 2021; 13:1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baggen J., Thibaut H.J., Strating J., van Kuppeveld F.J.M.. The life cycle of non-polio enteroviruses and how to target it. Nat. Rev. Microbiol. 2018; 16:368–381. [DOI] [PubMed] [Google Scholar]
- 5. Barton D.J., O’Donnell B.J., Flanegan J.B.. 5' cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 2001; 20:1439–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gamarnik A.V., Andino R.. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998; 12:2293–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pascal S.M., Garimella R., Warden M.S., Ponniah K.. Structural biology of the enterovirus replication-linked 5'-cloverleaf RNA and associated virus proteins. Microbiol. Mol. Biol. Rev. 2020; 84:e0062-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Toyoda H., Franco D., Fujita K., Paul A.V., Wimmer E.. Replication of poliovirus requires binding of the poly(rC) binding protein to the cloverleaf as well as to the adjacent C-rich spacer sequence between the cloverleaf and the internal ribosomal entry site. J. Virol. 2007; 81:10017–10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Beckham S.A., Matak M.Y., Belousoff M.J., Venugopal H., Shah N., Vankadari N., Elmlund H., Nguyen J.H.C., Semler B.L., Wilce M.C.J.et al.. Structure of the PCBP2/stem–loop IV complex underlying translation initiation mediated by the poliovirus type I IRES. Nucleic Acids Res. 2020; 48:8006–8021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Daijogo S., Semler B.L.. Mechanistic intersections between picornavirus translation and RNA replication. Adv. Virus Res. 2011; 80:1–24. [DOI] [PubMed] [Google Scholar]
- 11. Blyn L.B., Towner J.S., Semler B.L., Ehrenfeld E.. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 1997; 71:6243–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gamarnik A.V., Andino R.. Interactions of viral protein 3CD and poly(rC) binding protein with the 5' untranslated region of the poliovirus genome. J. Virol. 2000; 74:2219–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Parsley T.B., Towner J.S., Blyn L.B., Ehrenfeld E., Semler B.L.. Poly (rC) binding protein 2 forms a ternary complex with the 5'-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA. 1997; 3:1124–1134. [PMC free article] [PubMed] [Google Scholar]
- 14. Ponchon L., Beauvais G., Nonin-Lecomte S., Dardel F.. A generic protocol for the expression and purification of recombinant RNA in Escherichia coli using a tRNA scaffold. Nat. Protoc. 2009; 4:947–959. [DOI] [PubMed] [Google Scholar]
- 15. Lee E., Bujalowski P.J., Teramoto T., Gottipati K., Scott S.D., Padmanabhan R., Choi K.H.. Structures of flavivirus RNA promoters suggest two binding modes with NS5 polymerase. Nat. Commun. 2021; 12:2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lorenz R., Bernhart S.H., Höner Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L.. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Otwinowski Z., Minor W.. Carter C.W.J., Sweet R.M.. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997; 276:NY: Academic Press; 307–326. [DOI] [PubMed] [Google Scholar]
- 18. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W.et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D, Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta. Crystallogr. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 20. Kalvari I., Nawrocki E.P., Ontiveros-Palacios N., Argasinska J., Lamkiewicz K., Marz M., Griffiths-Jones S., Toffano-Nioche C., Gautheret D., Weinberg Z.et al.. Rfam 14: expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Res. 2021; 49:D192–D200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rivas E., Clements J., Eddy S.R.. A statistical test for conserved RNA structure shows lack of evidence for structure in lncRNAs. Nat. Methods. 2017; 14:45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 23. Franke D., Petoukhov M.V., Konarev P.V., Panjkovich A., Tuukkanen A., Mertens H.D.T., Kikhney A.G., Hajizadeh N.R., Franklin J.M., Jeffries C.M.et al.. ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Crystallogr. 2017; 50:1212–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bussetta C., Choi K.H.. Dengue virus nonstructural protein 5 adopts multiple conformations in solution. Biochemistry. 2012; 51:5921–5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jacques D.A., Trewhella J.. Small-angle scattering for structural biology–expanding the frontier while avoiding the pitfalls. Protein Sci. 2010; 19:642–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lewis H.A., Musunuru K., Jensen K.B., Edo C., Chen H., Darnell R.B., Burley S.K.. Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell. 2000; 100:323–332. [DOI] [PubMed] [Google Scholar]
- 27. Marcotte L.L., Wass A.B., Gohara D.W., Pathak H.B., Arnold J.J., Filman D.J., Cameron C.E., Hogle J.M.. Crystal structure of poliovirus 3CD protein: virally encoded protease and precursor to the RNA-dependent RNA polymerase. J. Virol. 2007; 81:3583–3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ohlenschlager O., Wohnert J., Bucci E., Seitz S., Hafner S., Ramachandran R., Zell R., Gorlach M.. The structure of the stemloop D subdomain of coxsackievirus B3 cloverleaf RNA and its interaction with the proteinase 3C. Structure. 2004; 12:237–248. [DOI] [PubMed] [Google Scholar]
- 29. Walker P.A., Leong L.E., Porter A.G.. Sequence and structural determinants of the interaction between the 5'-noncoding region of picornavirus RNA and rhinovirus protease 3C. J. Biol. Chem. 1995; 270:14510–14516. [DOI] [PubMed] [Google Scholar]
- 30. Andino R., Rieckhof G.E., Trono D., Baltimore D. Substitutions in the protease (3Cpro) gene of poliovirus can suppress a mutation in the 5' noncoding region. J. Virol. 1990; 64:607–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Andino R., Rieckhof G.E., Achacoso P.L., Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5'-end of viral RNA. EMBO J. 1993; 12:3587–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Laing C., Schlick T.. Analysis of four-way junctions in RNA structures. J. Mol. Biol. 2009; 390:547–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nissen P., Ippolito J.A., Ban N., Moore P.B., Steitz T.A.. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:4899–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Torabi S.F., Vaidya A.T., Tycowski K.T., DeGregorio S.J., Wang J., Shu M.D., Steitz T.A., Steitz J.A.. RNA stabilization by a poly(A) tail 3'-end binding pocket and other modes of poly(A)-RNA interaction. Science. 2021; 371:eabe6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mahmud B., Horn C.M., Tapprich W.E.. Structure of the 5' untranslated region of enteroviral genomic RNA. J. Virol. 2019; 93:e01288-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Petrov A.I., Zirbel C.L., Leontis N.B.. WebFR3D–a server for finding, aligning and analyzing recurrent RNA 3D motifs. Nucleic Acids Res. 2011; 39:W50–W55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sharma N., Ogram S.A., Morasco B.J., Spear A., Chapman N.M., Flanegan J.B.. Functional role of the 5' terminal cloverleaf in Coxsackievirus RNA replication. Virology. 2009; 393:238–249. [DOI] [PubMed] [Google Scholar]
- 38. Zell R., Ihle Y., Seitz S., Gundel U., Wutzler P., Gorlach M.. Poly(rC)-binding protein 2 interacts with the oligo(rC) tract of coxsackievirus B3. Biochem. Biophys. Res. Commun. 2008; 366:917–921. [DOI] [PubMed] [Google Scholar]
- 39. Zell R., Ihle Y., Effenberger M., Seitz S., Wutzler P., Gorlach M.. Interaction of poly(rC)-binding protein 2 domains KH1 and KH3 with coxsackievirus RNA. Biochem. Biophys. Res. Commun. 2008; 377:500–503. [DOI] [PubMed] [Google Scholar]
- 40. Walter B.L., Parsley T.B., Ehrenfeld E., Semler B.L.. Distinct poly(rC) binding protein KH domain determinants for poliovirus translation initiation and viral RNA replication. J. Virol. 2002; 76:12008–12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Silvera D., Gamarnik A.V., Andino R.. The N-terminal K homology domain of the poly(rC)-binding protein is a major determinant for binding to the poliovirus 5'-untranslated region and acts as an inhibitor of viral translation. J. Biol. Chem. 1999; 274:38163–38170. [DOI] [PubMed] [Google Scholar]
- 42. Du Z., Fenn S., Tjhen R., James T.L.. Structure of a construct of a human poly(C)-binding protein containing the first and second KH domains reveals insights into its regulatory mechanisms. J. Biol. Chem. 2008; 283:28757–28766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Teplova M., Malinina L., Darnell J.C., Song J., Lu M., Abagyan R., Musunuru K., Teplov A., Burley S.K., Darnell R.B.et al.. Protein-RNA and protein-protein recognition by dual KH1/2 domains of the neuronal splicing factor Nova-1. Structure. 2011; 19:930–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Backe P.H., Messias A.C., Ravelli R.B., Sattler M., Cusack S.. X-ray crystallographic and NMR studies of the third KH domain of hnRNP K in complex with single-stranded nucleic acids. Structure. 2005; 13:1055–1067. [DOI] [PubMed] [Google Scholar]
- 45. Vogt D.A., Andino R.. An RNA element at the 5'-end of the poliovirus genome functions as a general promoter for RNA synthesis. PLoS Pathog. 2010; 6:e1000936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Valverde R., Edwards L., Regan L.. Structure and function of KH domains. FEBS J. 2008; 275:2712–2726. [DOI] [PubMed] [Google Scholar]
- 47. Zell R., Sidigi K., Bucci E., Stelzner A., Gorlach M.. Determinants of the recognition of enteroviral cloverleaf RNA by coxsackievirus B3 proteinase 3C. RNA. 2002; 8:188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rieder E., Xiang W., Paul A., Wimmer E.. Analysis of the cloverleaf element in a human rhinovirus type 14/poliovirus chimera: correlation of subdomain D structure, ternary protein complex formation and virus replication. J. Gen. Virol. 2003; 84:2203–2216. [DOI] [PubMed] [Google Scholar]
- 49. Cornell C.T., Semler B.L.. Subdomain specific functions of the RNA polymerase region of poliovirus 3CD polypeptide. Virology. 2002; 298:200–213. [DOI] [PubMed] [Google Scholar]
- 50. Thapar R., Denmon A.P., Nikonowicz E.P.. Recognition modes of RNA tetraloops and tetraloop-like motifs by RNA-binding proteins. Wiley Interdiscip. Rev. RNA. 2014; 5:49–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Warden M.S., Cai K., Cornilescu G., Burke J.E., Ponniah K., Butcher S.E., Pascal S.M.. Conformational flexibility in the enterovirus RNA replication platform. RNA. 2019; 25:376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Blair W.S., Parsley T.B., Bogerd H.P., Towner J.S., Semler B.L., Cullen B.R.. Utilization of a mammalian cell-based RNA binding assay to characterize the RNA binding properties of picornavirus 3C proteinases. RNA. 1998; 4:215–225. [PMC free article] [PubMed] [Google Scholar]
- 53. Herold J., Andino R.. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell. 2001; 7:581–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chapman N.M., Kim K.S., Drescher K.M., Oka K., Tracy S.. 5' terminal deletions in the genome of a coxsackievirus B2 strain occurred naturally in human heart. Virology. 2008; 375:480–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bouin A., Nguyen Y., Wehbe M., Renois F., Fornes P., Bani-Sadr F., Metz D., Andreoletti L.. Major persistent 5' terminally deleted coxsackievirus B3 populations in human endomyocardial tissues. Emerg. Infect. Dis. 2016; 22:1488–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Glenet M., N’Guyen Y., Mirand A., Henquell C., Lebreil A.L., Berri F., Bani-Sadr F., Lina B., Schuffenecker I., Andreoletti L.et al.. Major 5'terminally deleted enterovirus populations modulate type I IFN response in acute myocarditis patients and in human cultured cardiomyocytes. Sci. Rep. 2020; 10:11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kim K.S., Tracy S., Tapprich W., Bailey J., Lee C.K., Kim K., Barry W.H., Chapman N.M.. 5'-Terminal deletions occur in coxsackievirus B3 during replication in murine hearts and cardiac myocyte cultures and correlate with encapsidation of negative-strand viral RNA. J. Virol. 2005; 79:7024–7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Das N.K., Hollmann N.M., Vogt J., Sevdalis S.E., Banna H.A., Ojha M., Koirala D. Crystal structure of a highly conserved enteroviral 5' cloverleaf RNA replication element. Nat. Commun. 2023; 14:1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or the supplementary materials. The poliovirus and coxsackievirus B3 cloverleaf structures were deposited to PDB with accession numbers 8S95 and 8SP9, respectively.