Abstract
CRISPR-associated proteins such as Cas9 and Cas12a are programable RNA-guided nucleases that have emerged as powerful tools for genome manipulation and molecular diagnostics. However, these enzymes are prone to cleaving off-target sequences that contain mismatches between the RNA guide and DNA protospacer. In comparison to Cas9, Cas12a has demonstrated distinct sensitivity to protospacer-adjacent-motif (PAM) distal mismatches, and the molecular basis of Cas12a's enhanced target discrimination is of great interest. In this study, we investigated the mechanism of Cas12a target recognition using a combination of site-directed spin labeling, fluorescent spectroscopy, and enzyme kinetics. With a fully matched RNA guide, the data revealed an inherent equilibrium between a DNA unwound state and a DNA-paired duplex-like state. Experiments with off-target RNA guides and pre-nicked DNA substrates identified the PAM-distal DNA unwinding equilibrium as a mismatch sensing checkpoint prior to the first step of DNA cleavage. The finding sheds light on the distinct targeting mechanism of Cas12a and may better inform CRISPR based biotechnology developments.
Graphical Abstract
Graphical Abstract.
INTRODUCTION
Clustered-Regularly-Interspaced-Short-Palindromic-Repeats (CRISPR) and CRISPR-associated (Cas) proteins provide adaptive immunity for bacteria and archaea to combat invading viruses and other mobile genetic elements (1–4). CRISPR-Cas systems have been adapted and repurposed as biotechnology tools with applications in genome engineering, transcription regulation, molecular diagnostics, cellular therapeutics, and agriculture (5–8). Class II CRISPR systems that utilize a single effector nuclease, including Cas9 and Cas12a, have found significant utilization in research and commercial settings (9). While Cas9 is the most widely utilized enzyme for genome editing, Cas12a has been adopted for genome manipulation as well as molecular diagnostic applications (10,11).
Similar to Cas9, Cas12a uses an effector complex activated and programmed by CRISPR-encoded small RNA(s) (crRNA) to cleave double-stranded DNA (12,13). A specific target is identified by recognition of a short protospacer-adjacent-motif (PAM) within the DNA, followed by unwinding of a segment of the DNA designated as the protospacer to form the R-loop, in which the DNA target-strand (TS) is hybridized with the single-stranded guide of the crRNA, while the DNA non-target-strand (NTS) is unwound (Figure 1). The Watson-Crick base pairing between the RNA-guide and TS establishes a simple yet effective rule for targeting a desired genomic site and serves as the foundation for the Cas12a/Cas9-based technological evolution (14). However, it has also subjected these enzymes to undesired off-target effects, in which erroneous binding and/or cleavage occurs on off-target genomic sequences where imperfect complementarity between the RNA guide and protospacer exists (15–17). A mechanistic understanding of target discrimination is the key to combat the off-target effect, and remains an area under active investigation.
Figure 1.
Monitoring FnCas12a induced site-specific DNA unwinding via spectroscopic labels. The FnCas12a effector is shown in grey, with the colored line representing the on-target and off-target RNA guides that vary at positions +17 to +20. The DNA substrate duplex is shown with the PAM marked by an orange box; the protospacer (PAM +1 to +20) indicated by a box with coloring varied from the PAM-Proximal (blue) to the PAM-distal segment (purple); the R-loop Flank segment beyond the RNA/DNA hybrid marked by pink lines; and cleavage sites indicated by wedges. Chemical structures of the R5 nitroxide label (indicated by the colored arrows) and the 2-aminopurine label (indicated by pink circles) are shown on top, with the corresponding labeling positions marked on the DNA. Details on DNA and crRNA guide sequences can be found in Supplementary Table S1 and Table S2 respectfully.
Studies have revealed that the Cas12a-crRNA effector complex interrogates a DNA target via a series of coordinated conformational changes (18–21). Similar to Cas9, upon recognition of the PAM, Cas12a locally distorts and unwinds the base-pairs immediately adjacent to the PAM, establishing an R-Loop seed (22). Sequential unwinding of the DNA leads to formation of the full R-Loop, triggering allosteric activation and enabling DNA strand scission (18,20,21,23–25). However, Cas12a differs from Cas9 in key features of effector protein, crRNA, and target DNA (12,26). While Cas9 cleaves a DNA target using two nuclease domains in a parallel-sequential fashion (27–29), Cas12a causes double-stranded DNA breaks using a single RuvC nuclease domain following a sequential mechanism, with the NTS being cleaved first at a site located in the PAM-distal segment of the protospacer, around 14–18 nucleotides from the PAM (19). Following nicking of the NTS, the protein–RNA–DNA complex undergoes additional conformational changes, eventually enabling cleavage of the TS at a site located 1–2 nucleotides beyond the DNA/RNA hybrid within the R-loop flank (19,24,30,31). Consequently, Cas12a yields a mismatch tolerance pattern that is clearly different from that of Cas9 (32), with little to no tolerance for PAM-distal mismatches (33,34). However, while rapid progress has been made on understanding Cas12a structure (19–21) and targeting selection and discrimination (35–37), current understanding on Cas12a's target surveillance mechanism is not as comprehensive as that of Cas9 (38,39), and further in depth studies are needed on the structure-function relationship that governs Cas12a target selection.
Previous work has shown that in Cas9, DNA protospacer unwinding, which is directly connected with the allosteric activation of the protein, controls DNA cleavage activity and serves as a key determinant in sensing and discriminating DNA/guide mismatches (29,38,40,41). Given the differences between Cas12a and Cas9 (13), in particular the mismatch sensitivity patterns (32–34), the manner of Cas12a-indcued DNA unwinding and the dynamics of the R-loop is a critical element in deciphering its mechanism of DNA target interrogation. A large number of biophysical techniques, including single molecule fluorescence and force microscopies, have been applied to explore a variety of conformational dynamics of the Cas12a ternary complex along the pathway to DNA target cleavage (20,23,24,42–45), however, much remains to be learned about the R-loop conformational states of Cas12a and their roles in DNA target discrimination.
In this study, we used Francisella novicida Cas12a (FnCas12a) as a model system to examine Cas12a-induced DNA unwinding using a combination of site-directed spin labeling, 2-amino-purine (2AP) fluorescence, and nuclease cleavage activity measurements. The work leveraged a unique biophysical technique, site-directed spin labeling (SDSL), in which stable radicals (e.g. nitroxide spin labels) attached at specific sites of the parent molecule are utilized to obtain structural and dynamic information of the target system (46,47). In particular, SDSL in conjunction with a pulsed Electron Paramagnetic Resonance (EPR) technique, double electron-electron resonance (DEER) spectroscopy, allows for measurements of distances and distance distributions, which provide information on dynamic equilibria between conformational states in solution. Recently, we have applied SDSL-EPR to study Cas9 (41,48,49), including examining site-specific Cas9-induced DNA unwinding at a resolution of the individual nucleotides (41,49).
Results presented here reveal an inherent equilibrium between a DNA unwound state and a DNA-paired duplex-like state with an on-target Cas12a complex, in which the RNA guide fully matches the DNA protospacer. This DNA-paired/unwound equilibrium is tuned by the presence of PAM-distal RNA/DNA mismatches prior to the first step of NTS cleavage, but is no longer maintained with pre-nicked DNA substrates mimicking post-NTS cleavage states. Together, we propose a model for DNA target discrimination by Cas12a, where the DNA paired/unwound equilibrium serves as a checkpoint for licensing RuvC nuclease activity for the first step of cleavage of the NTS. The work advances our understanding on the role of DNA unwinding in directing Cas12a's nuclease activation to achieve target discrimination and sequential strand scission.
MATERIALS AND METHODS
Cas12a protein expression and purification
Catalytically-active FnCas12a and Catalytically-inactive dFnCas12a were expressed in Escherichia coli and purified following reported protocols (50,51). Briefly, a plasmid encoding the desired Cas12a protein with a His-MBP tag at the N-terminus was transformed into Rosetta (DE3) Competent E. coli (Novagen) by heat shock. A single colony from the transformation was inoculated into Lysogeny Broth with appropriate antibiotic (50 ug/ml Kanamycin) and incubated at 37°C overnight. Then, the small-scale culture was added to terrific Broth with 50 ug/ml antibiotic and incubated at 37°C until the optical density at 600 nm reached 0.5–0.6. The temperature was then reduced to 18°C, and incubation continued for 20 minutes. Overexpression was then induced by adding 200 uM IPTG and shaking at 18°C for 16–20 h. The cells were harvested via centrifugation and resuspended in cold lysis buffer [20 mM Tris pH 8.0, 500 mM NaCl, 10 mM imidazole], with protease inhibitor (Roche). The cells were lysed at 4°C via sonication and then subjected to ultracentrifugation at 4°C to separate cellular debris. The supernatant underwent immobilized metal affinity chromatography and the pooled fractions containing His-MBP-Cas12a underwent TEV protease digestion to cleave the His-MBP tag. The crude FnCas12a was then subjected to heparin chromatography (HiTrap Heparin Hp column Cytivia). The pooled fractions were further purified by size exclusion chromatography (Cytivia S200 Increase) with 20 mM Tris pH 8.0, 500mM KCl, and 1mM DTT as the running buffer, before being concentrated and stored at –80°C in storage buffer [20 mM Tris pH 8.0, 500 mM KCl, 1 mM DTT and 20% glycerol]. Concentrations of proteins were determined according to absorbance at 280 nM with an extinction coefficient of 144 000 M−1 cm−1.
DNA substrate preparation
All DNA oligonucleotides, including those with specific 2AP or phosphorothioate modifications, were synthesized via solid-phase chemical synthesis and obtained commercially (Integrated DNA Technologies, Coralville, Iowa). Complete information of DNA substrate sequences is described in Supporting Information Table S1. DNA concentrations were determined by measuring the absorbance at 260 nm and using the extinction coefficient values provided by the vendor.
Formation of the target DNA duplex was carried out via mixing a 1:1 molar ratio of the desired target and non-target strand in annealing buffer (50 mM Tris, pH 7.5, 100 mM NaCl) and incubating overnight at room temperature. The annealed duplexes were purified by size exclusion chromatography (SEC) and buffer-exchanged into 1× CRISPR reaction buffer (20 mM Tris, pH 7.5; 150 mM KCl, 5% glycerol and 5 mM MgCl2) and stored at –20°C until measurement.
crRNA preparation
The crRNA’s were prepared by T7 in vitro transcription using a T7 top-stranded primer and a single-stranded DNA oligonucleotide template (see Supplementary Table S2 for detailed sequences). Transcription reactions were carried out following reported procedures (44). The crude transcription products were purified via denaturing polyacrylamide gel electrophoresis and redissolved in ME buffer (10 mM MOPS, pH 6.5 and 1mM EDTA) and stored at -20°C. RNA concentrations were determined based on the absorbance at 260 nm, with the molar extinction coefficients estimated by ε = number of nucleotide × 10 000 M−1 cm−1. crRNA sequences are listed in Supplementary Table S3.
Preparation of dFnCas12a ternary complexes for spectroscopy measurements
Ternary complexes were assembled in 1× CRISPR reaction buffer with the dFnCas12a/crRNA//DNA molar ratio being 1:1.2:1.1. To assemble the complex, first the crRNA was refolded via heating at 95°C for 1min in ME buffer and then cooled at room temperature for 5 min. The crRNA was adjusted to an appropriate amount of salt to match the 1× CRISPR reaction buffer and then mixed with the dFnCas12a protein. The dFnCas12a/crRNA mixture was incubated at 37°C for 10 minutes to form the effector complex, and then the appropriate amount of DNA duplex was added. The dFnCas12a/crRNA/DNA mixture was incubated at 37°C for an additional 10 minutes before purification using SEC in 1× reaction buffer. SEC fractions containing ternary complexes with 2AP modified DNA were pooled and used directly for 2AP fluorescence measurements. Ternary complexes containing spin-labeled DNA were further processed as described below.
2-Amino-purine (2AP) fluorescence measurements
2AP measurements were conducted using a Molecular Devices SpectraMax® iD5 plate reader. 100uL of purified 2AP-modified duplex DNA or dFnCas12a ternary complex was transferred to sample wells in a black Corning® 384 well flat bottom polystyrene plate (Mfr No. 35766). Utilizing the onboard monochromator, the excitation was fixed at 315 nm with the fluorescence emission maxima collected at 368nm. In parallel, absorbance measurements on the corresponding samples were obtained on a DU800 UV−Vis spectrometer (Beckman Coulter, Fullerton, CA). Following a prior report (41), the background-corrected emission at 368 nm (F368, obtained by subtracting the corresponding buffer emission) was normalized by the 260 nm absorbance (A260) of the same sample to obtain:
 |
(1) |
where ϵ is the extinction coefficient of the particular sample, ϵ2AP is the extinction coefficient of 2AP at 315 nm, φ is the quantum yield of 2AP, and K is a constant independent of the sample concentration. Furthermore, one can normalize the 2AP quantum yield of a given sample to that of the corresponding free duplex as:
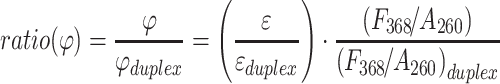 |
(2) |
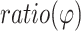 , which specifically reports on the 2AP quantum yield while minimizing the impact of local DNA sequence variations, was used to access DNA unwinding.
, which specifically reports on the 2AP quantum yield while minimizing the impact of local DNA sequence variations, was used to access DNA unwinding.
Site-directed spin labelling
Site-specific labeling with the R5 nitroxide label was conducted following a previously described protocol (52,53). Briefly, crude phosphorothioate-modified DNA oligonucleotide was mixed with an acetonitrile/water (50%/50% by volume) solution containing a R5 precursor [3-Iodomethyl-1-oxyl-2,2,5,5-tetramethylpyrroline (Toronto Research Chemicals, cat. no. I709500)] (60–100 mM) and 100 mM MES (pH 5.8). The mixture was incubated at room temperature for 12–24 h. The labeled oligonucleotide was then purified into sterile deionized water via application of a desalting spin column (Thermo Scientific Zeba 7K MWCO, cat. No. 89883). Note that follow-up SEC purification of the DNA duplex or the ternary complex served as an additional step to remove unattached nitroxides. Labeling efficiency was assessed by a previously reported spin counting procedure (54) and was generally >90%. The R5-labled oligonucleotide was stored at –20°C until DNA substrate preparation.
Continuous wave EPR measurements
Spin-labeled samples (10–50uM) were transferred to round glass capillary tubes (0.6 mm ID, 0.8 mm OD; Vitrocom, Inc.) for measurement. The continuous-wave EPR (cw-EPR) spectra were measured utilizing a Bruker EMX spectrometer housing an ER4119HS cavity. For spectral acquisition, the incident microwave power was 2 mW, and the field modulation was 2 G at a frequency of 100 kHz. Each spectrum was acquired with 512 points, corresponding to a spectral range of 100 G, then corrected for background baseline, and intensity normalized following reported protocols (54).
Double electron-electron resonance (DEER) spectroscopy
Doubly R5-labeled samples were buffer exchanged to 1× CRISPR reaction buffer dissolved in deuterium oxide via a 100 kDa spin filter concentrator, then combined with the appropriate volume of deuterated glycerol to produce DEER samples in 30% (v/v) glycerol. Each DEER sample was approximately 30 ul, with the sample concentration estimated to be 60–80 uM. The samples were loaded into quartz EPR capillary tubes and flash frozen with liquid nitrogen immediately before measurement.
DEER measurements were carried out at 50 K on a Bruker ELEXSYS E580 X-band spectrometer with an ER4118-MS3-EN resonator. A dead-time free four-pulse scheme was used (55), with the pump pulse frequency set at the center of the nitroxide spectrum and the observer frequency being approximately 70 MHz lower. The observer p pulse was 32 ns. The pump π pulse, optimized using a nutation experiment, was set between 8 and 16 ns. The delay between the observer π/2 and π pulses was set to 420 ns to minimize deuterium ESEEM. The video bandwidth was fixed at 20 MHz. The shot repetition time was set between 1000 and 1400 μs. Accumulation time in each measurement was approximately 20–24 h, with 100 shots per point.
Distance distributions from the DEER data were obtained using Consensus Deer analysis, an analysis package accompanied in the DEERAnalysis software kit (56,57). Modeling of inter-R5 distance distributions was performed using ALLNOX (58,59). The input structure for the free DNA was a linear B-form duplex built with the P3 sequence using Avogadro (60). The input structure for the ternary complex was a FnCas12a crystal structure (PDB 6i1k) showing unwound DNA (19). Modeling used previously reported parameters (59), except that the torsion angles were varied with a step size of 60°. Modeling yielded the ensemble of pairwise distances between the nitrogen atoms of the pyrroline of the allowed R5 conformers (N-N), from which the average distance was computed (58,59).
DNA cleavage assay
DNA substrate duplexes containing terminal fluorescent labels were purified by SEC into 1× CRISPR buffer. For a given reaction, a FnCas12a/crRNA effector complex (200 nM) was assembled with a protein/crRNA molar ratio of 1:1.25 and incubated in 1× CRISPR reaction buffer for 10 min at 37°C. The desired DNA substrate (25 nM) was then added. The reaction was allowed to proceed at 20°C for the desired time, before being stopped via addition of denaturing gel loading buffer (95% formamide, 20 mM EDTA, 0.025% SDS) with the volume ratio between the dye and the sample being 1:4. The cleavage products and the precursor were resolved via a denaturing polyacrylamide gel. The resulting gels were imaged using a ChemiDoc MP system (Bio-Rad) using the appropriate fluorescein imaging settings. The band intensities of the DNA species and background were quantified using ImageJ (1.47). The fraction of precursor for each timepoint was calculated as:
 |
(3) |
with Ip being the background corrected uncleaved precursor band intensity, and Ic being the background corrected intensity of the cleaved product. Cleavage experiments were performed in triplicate, and the average frac(pre) was plotted for each timepoint with the standard deviation shown as error bars. When appropriate, the time dependence of average frac(pre) was fit to a single-exponential decay using an in-house program written in Matlab:
 |
(4) |
with kobs representing the rate constant, and a representing the active fraction of the precursor.
RESULTS
A DNA duplex-like paired state coexists with the unwound state in the on-target ternary complex prior to DNA cleavage
To evaluate Cas12a mediated DNA unwinding using SDSL, we constructed a model DNA substrate duplex termed P3, and attached R5 nitroxide labels onto desired specific backbone positions (Figure 1; Supplementary Table S1). The labeled duplex was assembled into a ternary complex with a catalytically-inactive FnCas12a/crRNA effector (dFnCas12a). The dFnCas12a ternary complex was purified (Supplementary Figure S1A) then subjected to DEER measurements. Control measurements showed only minor impact on DNA cleavage rates due to spin-labeling, indicating the R5 labels did not significantly perturb the native folding and function of the Cas12a ternary complex (Supplementary Figure S2).
DEER measurements were first carried out with a pair of R5 labels attached at backbone positions directly across the duplex at the PAM + 10 protospacer positions (Figure 2A and B), which are located in the middle of the protospacer (Figure 1). With the free (unbound) DNA duplex, the observed dipolar evolution trace measurement showed oscillations with fast decay and yielded a distance distribution showing a single population centered at 26Å (Figure 2A). This is consistent with previously reported studies of R5-labled DNA duplexes (52). With an on-target ternary complex, where the DNA was bound to a dFnCas12a effector containing an RNA guide that fully matches the DNA protospacer, the measured dipolar evolution showed oscillations containing features of both fast and slow decay (Figure 2B). The resulting distance distribution showed two well-defined major populations, one centered at 28 Å and another centered at 41 Å (Figure 2B). Modeling using a reported dFnCas12a on-target complex crystal structure (PDB 6i1k), which shows unwinding of the DNA protospacer (19) resulted in a predicted average inter-R5 distance of 43.5Å at PAM + 10 (Figure 2B, Model). Considering the 2–3 Å range of variation in average inter-R5 distances in the modelling (58,59), the predicted PAM + 10 distance on the 6i1k structure showing DNA unwinding matches very well with the measured long-distance population (Figure 2B), leading to the conclusion that the observed long distance represents the DNA unwound state.
Figure 2.
A DNA duplex-like paired state coexists with the unwound state in the on-target ternary complex prior to DNA cleavage. Distance measurements are shown for the PAM + 10 (panels (A) and (B)) and the PAM + 17 + 26 (panels (C) and (D)) datasets obtained with intact NTS strands. For each data set, shown on the left is the DEER dipolar evolution trace; middle is the resulting distance distribution with the error contour represented by the shaded band; and right is the rotamer modeling obtained with ALLNOX. To aid comparison, the distance distributions include a black dashed line to mark the maximum distance measured from the corresponding free DNA duplex, and a red dashed line to mark the maxima of the long-distance populations measurand on the complex samples, which represent the DNA in an unwound state. Additional details can be found in Supplementary Figures S3, S4, and Table S4.
It is interesting that the DEER data reveal a significant 28 Å-population at PAM + 10 within the on-target complex (Figure 2B). This distance is within 2 Å of the inter-R5 distance value predicted using a uniform B-form duplex model (Figure 2A, Model) as well as that measured with the corresponding free DNA (Figure 2A). It indicates that the corresponding population undergoes very little, if any, strand separation (i.e. unwinding), and therefore is assigned as a ‘duplex-like’ state with the DNA remaining paired at the PAM + 10 position. Note that control measurements confirmed that the large majority of the R5-labeled DNA was bound within the dFnCas12a complex (Supplementary Figure S1B). Given the significant presence of the 28Å population (Figure 2B), it is impossible that it arose from DNA dissociated from the ternary complex. In addition, R5 labels attached at the PAM + 10 sites, which are located at the middle of the protospacer, likely incur a fair number of contacts to Cas12a and crRNA within the ternary complex, as indicated by cw-EPR measurements (Supplementary Figure S5). This may contribute to the difference between the observed 28Å distance within the complex vs. that measured with the free DNA (26Å, Figure 2A). As such, the PAM + 10 DEER data (Figure 2B) reveal that the bound DNA exists in equilibrium between a duplex-like state and an unwound state.
Although control measurements showed only minor impacts on DNA cleavage rates due to R5 labeling (Supplementary Figure S2), one cannot completely exclude the possibility that R5 labels presented at PAM + 10 sites bias DNA unwinding to some degree. To further examine the possible simultaneous presence of the duplex-like and unwound states in the on-target ternary complex, we prepared a P3 construct with R5 attached at the TS PAM + 17 and TS PAM + 26 positions (Figure 1, Supplementary Table S1), and measured this PAM + 17 + 26 distance to examine the target-strand conformation (Figure 2C and D). DEER measurements with the free duplex revealed a population with a distance distribution centered at 31 Å (Figure 2C), which agrees quite well with the model-predicted distance (33.2Å, Figure 2C, model). When bound to the on-target dFnCas12a ternary complex, the DEER measured distance profile showed a clear bimodal distribution (Figure 2D). A major population was centered at 41 Å. This matches the predicted value of 40.1 Å obtained using the 6i1k crystal structure (Figure 2D, Model), which shows DNA unwinding at the PAM + 17 position and kinking of the R-loop flank segment beyond the 20-base-pair RNA/DNA hybrid. This population therefore represents the unwound DNA. Importantly, the other major population is centered at 31 Å, which matches exactly with the distance measured in the free linear duplex (Figure 2C), and therefore was assigned as the duplex-like state representing the paired DNA. As such, the data reveal that the PAM-distal segment of the DNA target strand exists in equilibrium between an unwound state and a duplex-like paired state. We note that the PAM + 10 data set seems to show a higher percentage of the duplex-like population than that observed in the PAM + 17 + 26 data set (Figure 2B and D), which may reflect site-specific variations of R5’s impact on the bound DNA conformations. None the less, the clear presence of the bimodal distribution observed with the PAM + 17 + 26 data set unambiguously support the conclusion that within the on-target complex, an equilibrium of the unwound and duplex-like paired states exists at the protospacer segment distal from the PAM.
The PAM-distal DNA paired/unwound equilibrium serves as a checkpoint to license the first-step NTS cleavage
Next, we examined how DNA unwinding is impacted by mismatches between the DNA target and the RNA guide. We focused on mismatches at the PAM-distal positions, as there is a clear difference in mismatch sensitivity at this segment between Cas12a and Cas9 (32,33). A P3 off-target crRNA (Supplementary Table S3) was synthesized to assemble an off-target ternary complex with dFnCas12a and the P3 DNA, with the PAM + 17 to + 20 positions (i.e. the most PAM-distal portion of the RNA/DNA hybrid) all containing RNA/DNA mismatches. DEER measurements were carried out for this P3 off-target ternary complex with the R5-labeled PAM + 17 + 26 DNA. The resulting distance distribution showed only one population with the maxima located at 31Å (Figure 3A) that is nearly identical to that measured from the free duplex (Figure 3B). Furthermore, comparing to the P3 on-target complex, the off-target complex clearly lost the 41Å population that represents the unwound DNA (Figure 3B). The DEER data therefore revealed that the four mismatches disrupt the DNA unwinding equilibrium – the DNA unwound state is lost, and the DNA remains in the duplex-like paired state in the ternary complex.
Figure 3.
The PAM-distal DNA unwinding equilibrium serves as a checkpoint for the first step of NTS cleavage. (A) Distance measurements on the PAM + 17 + 26 intact duplex bound to an off-target effector that contains 4 consecutive protospacer mismatches at positions PAM + 17–20. (B) Comparison of the PAM + 17 + 26 distance distributions between the free duplex and the off-target complex (left) and the on-target and off-target complexes (right). To aid comparison, the distance distributions include a black dashed line to mark the maxima measured from the corresponding DNA duplex, and a red dashed line to mark the maxima of the long-distance (unwound DNA) population measurand on the complex samples. Additional details can be found in Supplementary Figures S3, S4, and Table S4. (C) Normalized fluorescence quantum yield (ratio(ϕ)) obtained with the 2AP incorporated at the NTS PAM + 18 position of the P3.2 construct. Additional details can be found in Supplementary Table S5. (D) Representative gels showing time course measurements of NTS (left) and TS (right) cleavage by FnCas12a.
Further investigations on PAM-distal DNA unwinding were carried out using a 2AP fluorescence assay developed in our prior Cas9 study (41). A slightly modified DNA substrate, P3.2–2AP (Supplementary Table S1), was generated to substitute a 2AP at the NTS PAM + 18 position. P3.2–2AP was assembled into either an on-target complex (i.e. RNA/DNA match from PAM + 1 to PAM + 20) or an off-target complex (i.e. mismatches at PAM + 17 to PAM + 20). Following our prior work (41),  (eq.2), which reports on the 2AP quantum yield and serves as a sensor of the state of stacking of the DNA base, was obtained to access DNA unwinding. For the on-target complex, the measured
(eq.2), which reports on the 2AP quantum yield and serves as a sensor of the state of stacking of the DNA base, was obtained to access DNA unwinding. For the on-target complex, the measured  value was 2.87 ± 0.47, which is clearly higher than that obtained for the free duplex (1.00 ± 0.10) (Figure 3C, Supplementary Table S5). This is consistent with the expected DNA unwinding at PAM + 18, as unwinding would reduce DNA base stacking, thus increasing 2AP quantum yield. For the off-target complex, the
value was 2.87 ± 0.47, which is clearly higher than that obtained for the free duplex (1.00 ± 0.10) (Figure 3C, Supplementary Table S5). This is consistent with the expected DNA unwinding at PAM + 18, as unwinding would reduce DNA base stacking, thus increasing 2AP quantum yield. For the off-target complex, the 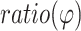 value was 1.47 ± 0.27, which increases only slightly compared to that of the duplex (Figure 3C, Supplementary Table S5). This indicates little DNA unwinding. As such, the 2AP measurements, which were conducted at room temperature under the same buffer condition used for assessing DNA cleavage activities, strongly support the conclusions drawn from the DEER data, that the PAM-distal mismatches push the system back to the duplex-like paired state, while the DNA unwound state is not populated.
value was 1.47 ± 0.27, which increases only slightly compared to that of the duplex (Figure 3C, Supplementary Table S5). This indicates little DNA unwinding. As such, the 2AP measurements, which were conducted at room temperature under the same buffer condition used for assessing DNA cleavage activities, strongly support the conclusions drawn from the DEER data, that the PAM-distal mismatches push the system back to the duplex-like paired state, while the DNA unwound state is not populated.
We further carried out in vitro DNA cleavage measurements using catalytically-active FnCas12a to examine the functional consequences of perturbing the equilibrium in Cas12a induced DNA unwinding. Under saturating enzyme concentrations, a complex containing the on-target RNA cleaved the NTS to completion within 120 minutes, while a complex containing the off-target RNA resulted in no observable products during the same time period (Figure 3D), indicating a loss of the ability to cleave NTS. Given that with Cas12a, NTS cleavage is required for TS cleavage, the off-target complex showed no cleavage product for the TS, while the on-target complex cleaved the TS to completion as expected (Figure 3D). Furthermore, control experiments confirmed that the DNA substrate was completely bound by either the on-target or the off-target complex (Figure S6).
Taken together, with four RNA/DNA mismatches at the PAM-distal segment, pairings between the DNA TS and NTS dominate, and the system is trapped in the duplex-like DNA paired state. This results in a complete loss of the DNA unwound state, preventing the first-step cleavage of the NTS and subsequently the cleavage of the TS. As such, the DNA paired/unwound equilibrium acts as checkpoint, with occupancy of the unwound state serves as a license to allow progression to DNA strand scission.
Nicking the non-target-strand alters DNA conformation at the target-strand cleavage site
Prior studies have shown that following DNA unwinding and R-loop formation, FnCas12a first nicks the NTS at a site located close to PAM + 18 and has trimming activity on the NTS up to PAM + 15 (19,30) (Figure 1). The activated RuvC nuclease then continues to cleave TS at/near PAM + 22, which is located beyond the 20-base-pair RNA/DNA hybrid (Figure 1), to generate a double-stranded DNA break (19,30). It is believed that conformational changes of DNA must occur at the target-strand cleavage site after NTS nicking but prior to TS cleavage, however, details remain largely unknown (24,30,45). In the SDSL and 2AP measurements described above, dFnCas12a and P3 duplexes containing an intact NTS and TS (Figures 2 and 3) were used to investigate DNA unwinding prior to the first catalytic step, i.e. NTS nicking by Cas12a. To further investigate DNA conformational changes at the target-strand cleavage site in solution beyond the first catalytic step, we developed P3-nicked DNA constructs, in which the intact TS oligonucleotide (i.e. P3a, Supplementary Table S1) is hybridized with two oligonucleotides to generate a fully paired duplex with a nick at/near the NTS cleavage site (Supplementary Table S1). The nicked substrates were reconstituted with dFnCas12a and various crRNAs to mimic post NTS-nicking ternary complexes that are progressing towards TS cleavage.
SDSL and 2AP measurements were first conducted to investigate DNA conformation at the TS cleavage site. A pair of R5 nitroxides were attached at cross-strand positions at PAM + 22, and the labeled DNAs were bound with the fully-matched crRNA effectors to assemble on-target ternary complexes (Figure 4). With the P3 construct containing an intact NTS, the on-target complex gave a dipolar evolution trace showing oscillations with a fast decay, and the resulting distance distribution showed a dominant population centered at 26 Å (Figure 4A, complex). A minor peak was also observed with the center being at 20Å (Figure 4A, complex), although given its large error contour it is not clear whether this represents an artifact or a true minor population. The major 26Å population agrees well with the predicted value of 28.2 Å obtained using the 6i1k structure (Figure 4A, complex model), which is representative of an intact NTS in an on-target complex. Furthermore, PAM + 22 measurement on the free intact DNA yielded a distance distribution with one population centered at 26 Å (Figure 4A, duplex), and cw-EPR measurements indicated that the DNA local environment at NTS + 22 is similar to that of a free duplex without incurring contacts to the Cas12a effector (Supplementary Figure S5C). Together, SDSL indicated that prior to NTS nicking, in solution the target-strand cleavage site (i.e. PAM + 22) remains at the duplex-like DNA paired state. In addition, with 2AP incorporated at NTS PAM + 23, which is directly adjacent to the target-strand cleavage site, the complex gave a  value of 0.95 ± 0.04 (Figure 4C), which is comparable to the free duplex (Figure 4C, intact) and indicates the base remains well stacked. Overall, prior to NTS nicking, while the protospacer segment exists in equilibrium between the unwound and duplex-like paired states, the target-strand cleavage site, which is at the R-Loop flank downstream of the protospacer, remains in a duplex-like state.
value of 0.95 ± 0.04 (Figure 4C), which is comparable to the free duplex (Figure 4C, intact) and indicates the base remains well stacked. Overall, prior to NTS nicking, while the protospacer segment exists in equilibrium between the unwound and duplex-like paired states, the target-strand cleavage site, which is at the R-Loop flank downstream of the protospacer, remains in a duplex-like state.
Figure 4.
Non-target strand nicking alters DNA conformation at the target-strand cleavage site. (A) Distance measurements of intact DNA labeled across the duplex at the target strand cleavage site (PAM + 22). Shown on the left is the DEER dipolar evolution trace; middle is the resulting distance distribution with the error contour represented by the shaded band and the maximum distance measured from the free intact DNA duplex marked by a dotted line; and right is the rotamer modeling obtained with ALLNOX. (B) Distance measurements of the pre-nicked P3-n15 duplexes labeled at PAM + 22, with dipolar evolution traces shown on the left and the resulting distance distribution shown on the right. (C) Fluorescence quantum yield measurements of 2AP incorporated at NTS PAM + 23 for the intact P3 and pre-nicked P3-n15 constructs. Additional details on DEER measurements can be found in Supplementary Figures S3, S4, and additional detail on 2AP measurements can be found in Supplementary Table S5.
When the P3 construct with a NTS nick at PAM + 15 (P3-n15, Supplementary Table S1) was assembled into the on-target complex, the PAM + 22 DEER data differed clearly from that of the intact P3 (Figure 4). The free P3-n15 construct behaved as a typical duplex: the dipolar evolution trace showed oscillations, and the resulting distance distribution showed one major population centered at 26Å (Figure 4B, duplex). DEER signal, which require the simultaneous presence of R5 at the TS and the nicked NTS, was also observed from bound P3-n15 (Figure 4B, complex), indicating the nicked strand remained associated with the ternary complex. However, oscillations were not observed in the dipolar evolution trace of the bound P3-n15 (Figure 4B, complex), and the resulting distance distribution showed a broad profile with no definable populations (Figure 4B, complex). This highly heterogeneous PAM + 22 distance profile likely reflects a number of aspects of conformational changes that occur upon NTS nicking. The DNA segment may move into multiple new positions with respect to the protein and/or RNA, resulting in heterogeneous contacts between the R5 labels and the parent complex that alter inter-R5 distances. This is supported by the observed changes in the cw-EPR spectra between the nicked and intact PAM + 22 NTS complex (Supplementary Figure S5C). More importantly, with these movements, the DNA segment likely experiences distortion/unwinding. This is supported by 2AP fluorescence data. With a 2AP substituted at NTS PAM + 23, the P3-n15 construct gave a  value of 4.80 ± 0.88 (Figure 4C, nicked). This is substantially higher than that of the free nicked duplex (Figure 4C, nicked), and indicates significant unstacking of the base at the TS cleavage site. Overall, with the NTS nicked, the SDSL and 2AP data reveal alterations of DNA conformation at the TS cleavage site in solution. This likely reflects the series of complex transformations that occur as the system transitions toward target-strand cleavage, although discerning details about these transitions are beyond the capability of the spectroscopic tools used in this work.
value of 4.80 ± 0.88 (Figure 4C, nicked). This is substantially higher than that of the free nicked duplex (Figure 4C, nicked), and indicates significant unstacking of the base at the TS cleavage site. Overall, with the NTS nicked, the SDSL and 2AP data reveal alterations of DNA conformation at the TS cleavage site in solution. This likely reflects the series of complex transformations that occur as the system transitions toward target-strand cleavage, although discerning details about these transitions are beyond the capability of the spectroscopic tools used in this work.
The PAM-distal DNA paired/unwound equilibrium checkpoint functions prior to nicking of the non-target strand
To further examine PAM-distal DNA unwinding and its role in target discrimination after the first-step of NTS nicking, we measured the PAM + 17 + 26 distances using a P3 construct with a nick at NTS 18 (P3-n18, Supplementary Table S1). With the free duplex, the distance distribution obtained for the nicked duplex is centered at the same value as that of the intact duplex, and it shows a slightly larger width that is consistent with an increasing flexibility due to nicking (Figure 5A, duplex). However, with the on-target complex, the dipolar oscillation trace with P3-n18 showed decay with no oscillation, and the resulting distance distribution was very broad (Figure 5A, on-target complex). Different from that of the intact construct (Figure 2D), the nicked on-target complex distance distribution showed no discernible discrete populations (Figure 5A, on-target complex). The profile showed highest occupancy at 35 Å, which is slightly longer that the 31Å maxima of the free nicked duplex (Figure 5A). A fair amount of occupancy was also represented at the 41Å segment that was identified as the unwound/bent conformation (Figure 5A on-target complex). The data indicate that alteration of DNA conformation upon nicking the NTS is not only confined to the TS cleavage site (i.e. PAM + 22 & PAM + 23, Figure 4), but also to the R-Loop flank downstream of the protospacer. The system no longer occupies, in an equilibrium fashion, the discrete unwound state and the duplex-like paired state (Figure 5B, on-target complex), but instead samples a larger number of conformations. Considering that the complex was constituted with dFnCas12a, which is unable to execute DNA strand scission, it is likely that the system samples continuously through multiple states along the reaction pathway, resulting in the observed highly heterogeneous distance distribution profile (Figure 5A, on-target complex).
Figure 5.
The PAM-distal DNA unwinding equilibrium is lost upon non-target-strand nicking. (A) Distance measurements obtained with the pre-nicked P3-n18 constructs labeled at PAM + 17 + 26. Shown on the left is the DEER dipolar evolution trace; and on the right is the resulting distance distribution with the error contour represented by the shaded band. For comparison, on the distance distributions a black dotted line marks the maxima of the free nicked DNA duplex, and a red dotted line marks the maxima of the unwound DNA measured from the intact PAM + 17 + 26 sample (see Figure 2D). Additional details can be found in Supplementary Figures S3 and Table S4. (B) Comparisons of distance distributions obtained for the NTS pre-nicked and intact constructs. (C) Representative gels showing TS cleavage by Cas12a on the pre-nicked P3-n18 substrates, (D) Fitting analysis to obtain TS cleavage rate constants. Data shown in Figure 5C are used to obtain values for the NTS pre-nicked substrate. Data shown in Figure 3D are used to obtain values for the NTS intact substrate with the on-target complex, while values for the off-target complex is not obtained due to the lack of reaction progression. Note that FnCas12a exhibits imprecise NTS cleavage and trimming activity between positions NTS + 15 to NTS + 18 (19,30). While P3-n18 and P3-n15 (Figure 4) place the nick at different NTS sites, they both mimic the post-NTS-cleavage DNA, and serve as the proper constructs to study progression towards TS cleavage by Cas12a.
When the P3-n18 construct was assembled into the off-target complex, the measured PAM + 17 + 26 distance distribution profile was again very broad and without discernible discrete populations (Figure 5A, off-target complex). The maximum of the off-target complex differs from that of the on-target complex (Figure 5A), indicating that the RNA/DNA mismatches do impact the DNA conformation at the R-loop flank segment. Furthermore, compared to the intact off-target complex (Figure 5B, off-target complex), the nicked off-target complex showed clearly higher relative occupancy at the long-distance region that includes the 41 Å segment that is identified as the unwound/bent conformation. This indicates that with NTS pre-nicked, the off-target complex is able to sample through the conformational states required for progressing toward TS cleavage. Indeed, an active FnCas12a with the off-target RNA cleaved the TS of the nicked P3-n18 substrate to completion (Figure 5C), which is drastically different from that of the intact substrate (Figure 5D). Furthermore, off-target cleavage with the nicked P3-n18 was 6.5 times slower than that of on-target cleavage (Figure 5D). This indicates conformation at the R-loop flank segment remains different between the on-target and off-target complexes, and is consistent with the difference in the observed PAM + 17 + 26 distance distribution profiles (Figure 5A).
With the P3-n18 construct, both on-target and off-target complexes gave broad PAM + 17 + 26 distance distribution profiles showing no discrete populations (Figure 5A), indicating that the system is sampling a number of conformations instead of occupying the discrete DNA paired and DNA unwound states. In parallel, the nicked construct gave a much smaller off-target rate reduction in TS cleavage (Figure 5D). Together, this indicates that the equilibrium between the duplex-like paired state and the DNA unwound state serves as a checkpoint to discriminate against RNA/DNA mismatches prior to NTS nicking, but is no longer effective after the first-step DNA strand scission.
DISCUSSION
Based on the data presented above, we propose a model in which an equilibrium between a duplex-like paired state and a DNA unwound state serves as a check-point to interrogate PAM-distal RNA/DNA mismatches, which licenses Cas12a for the first-step of NTS cleavage (Figure 6). As the Cas12a effector engages an intact DNA duplex, PAM-recognition followed by R-loop formation at the PAM adjacent 5–8 bp segment achieves binding of the DNA to the ternary complex. For an on-target DNA, propagation of DNA unwinding and R-loop formation from the mid-protospacer to the PAM-distal segment establishes the fully unwound conformation, which has been previously observed in crystal structures (19) and is captured by DEER measurements in solution (Figure 2). Interestingly, the DEER data revealed another conformation, with the bound DNA showing distance distributions matching that of a paired duplex (Figure 2B and 2D). We note such a duplex-like paired DNA conformation at PAM-mid to distal segment is also observed during R-loop formation in newly reported cryo-EM structures (21). Importantly, in this study, the observed DEER data, which were obtained on SEC-purified samples that must have reached equilibrium, clearly show that both the duplex-like (paired) and the unwound states are present simultaneously (Figure 2B and 2D), indicating that they co-exist in solution at equilibrium (Figures 2 and 6A). This conclusion is consistent with several prior studies showing that the Cas12a R-loop is reversible and can sample multiple states in a dynamic equilibrium(24,35,42–45). We also note that DEER and 2AP data obtained on FnCas12a here show that with intact NTS, the TS cleavage site (i.e. PAM + 22) remains at the duplex-like paired state with little DNA distortion (Figure 4A and C). While this is consistent with the reported FnCas12a crystal structure (19) (Figure 4A), it contrasts with other reports with LbCas12a (24,30). This may reflect variations in R-loop dynamics and mismatch discriminations among different Cas12a orthologs reported in a number of prior work (30,33,37,43).
Figure 6.
Proposed mechanism of Cas12a DNA target discrimination. (A) FnCas12a-induced unwinding of an intact DNA target adopts an inherent equilibrium between a duplex-like paired state and a DNA unwound state. This paired/unwound equilibrium serves as a checkpoint for the first-step of NTS cleavage, as on-target RNA/DNA pairing promotes PAM-distal DNA unwinding and licenses the target for the first-step NTS cleavage; while off-target mismatches trap the target at the duplex-like paired state and inhibit NTS cleavage. (B) First-step nicking of NTS alters DNA conformations at the TS cleavage site, allowing TS scission. The DNA paired/unwound equilibrium is no longer maintained to discriminate against RNA/DNA mismatches.
The presence of a duplex-like paired DNA state within the ternary complex in solution has significant functional implications. Cas12a possesses one nuclease domain belonging to the RuvC family, which acts as a single-stranded nuclease (10,12,13,19). As such, occupancy of the unwound state, in which NTS is exposed as a single-strand, is a pre-requisite for NTS nicking, while occupancy at the duplex-like paired state would prevent NTS cleavage (nicking). Data presented here clearly show that PAM-distal RNA/DNA mismatches (Figure 3) tunes the equilibrium between the duplex-like state and the unwound state, thereby regulating occupancy of the unwound state and controlling NTS nicking. The large shift of the equilibrium towards the paired state (Figure 6A) with the PAM-distal RNA/DNA mismatch can be rationalized based on the weakening of the RNA/DNA hybrid interaction, which in turn favors the DNA TS/NTS pairing. As such, the paired-unwound equilibrium is utilized by Cas12a to interrogate the proper complementarity between the RNA guide and the DNA target. It serves as a checkpoint that licenses the first step of NTS nicking, and dictates the entire Cas12a reaction pathway.
Data presented further indicate that the paired/unwound mismatch sensing equilibrium is maintained only prior to the first step of NTS nicking (Figure 5). A number of studies have revealed DNA conformational changes post NTS-nicking in Cas12a (24,30,31,45). Consistent with these reports, data obtained here using pre-nicked DNA constructs show that DNA conformational changes occur prior to the second step of TS cleavage across both the R-loop flank (Figure 4) and PAM-distal region of the TS (Figure 5). The DEER measurements revealed that post-NTS cleavage, the PAM-Distal segment of the DNA samples a number of conformations (Figure 4B and Figure 5) as opposed to occupying discrete unwound or duplex-like paired states (Figure 2). Furthermore, the data indicate that with the pre-nicked construct, the off-target complex likely maintains a certain ability to sample through the conformational states to achieve TS cleavage (Figure 5A), thus drastically reducing discrimination (Figure 5C and D). Overall, this leads to our proposed model that the paired/unwound equilibrium is a checkpoint placed prior to the first-step of the cleavage reaction pathway (Figure 6).
The proposed paired/unwound equilibrium checkpoint in Cas12a is quite distinct from that of Cas9 and accounts for the observed differences in the mismatch sensitivity patterns (32,33). Work has shown that Cas9 possess an ‘excess’ unwinding ability prior to strand scission: It completely unwinds the protospacer with an on-target full-length RNA guide (41,49); and supports a certain degree of unwinding either in the presence of PAM-distal RNA/DNA mismatches (38,39) or with truncated guides that do not allow PAM-distal RNA/DNA pairing (41). Such ‘excess’ unwinding allows the ternary complex to transition into the catalytically active conformation, leading to promiscuous DNA cleavage. This accounts for the observed high frequency occurrence of PAM-distal mismatches in the off-target pattern (32). On the other hand, with the FnCas12a studied here, the on-target complex does not excessively tilt the system towards unwinding, as a paired duplex-like state is observed to coexist with the fully unwound state (Figure 2B and D). Such an equilibrium enables the native Cas12a to place a more stringent requirement for matching between the RNA guide and the DNA, as each and all of the RNA/DNA pairings are required to maintain occupancy of the unwound state. Indeed, this conclusion is supported by a complete collapse of the unwound state in the off-target complex, which prevents NTS cleavage (Figure 3). Coupled with the sequential cleavage model requiring nicking NTS first prior to TS cleavage, Cas12a processes an exclusive ability to distinguish PAM-distal mismatches, resulting in very low PAM-distal off-targets that sets it apart from Cas9 (33–35).
Expanding on prior work in Cas9 (41,48,49), data presented here showcase the unique ability of SDSL in deriving insight on the structure-dynamics-function relationship governing CRISPR target discrimination. The ability of SDSL and DEER to reveal ensemble population distributions in solution (61) enables the experimental observation of the Cas12a paired/unwound equilibrium that senses PAM-distal RNA/DNA mismatches (Figures 2 and 3). The R5 label has a smaller size and a short linker (Figure 1), and therefore are much more intimately coupled to the DNA as compared to most fluorophores. This is critical for the experimental identification of a duplex-like state within the Cas12a bound DNAs (Figures 2–4). While this study focuses on DNA conformations, work is on-going to apply SDSL to connect conformational changes in CRISPR protein domains to DNA unwinding and target discrimination.
In conclusion, this work harnesses the utility of SDSL to assess conformational equilibrium in solution and combines it with 2AP fluorescence spectroscopy and enzyme kinetics to characterize the mechanism of Cas12a target DNA unwinding and specificity. The data reveals an inherent DNA unwinding equilibrium that serves as a check-point for sensing PAM-distal mismatches prior to first-step cleavage of NTS. This sets a foundation for future exploration on enhancing specificity in Cas12a-based applications via manipulation of this unwinding equilibrium.
Supplementary Material
ACKNOWLEDGEMENTS
We thank X. Zhang, I. Schuster, D. Wu, and Y. Liu for comments on the manuscript.
Author contributions: J.S. and P.Z.Q. designed the studies and wrote the manuscript; J.S. and A.A. prepared protein and RNA reagents; J.S. and K.G.L. carried out the enzyme kinetics measurements; J.S. carried out spin-labeling measurements and modelling; J.S. and W.J. carried out 2AP measurements.
Notes
Present address: Wei Jiang, Sanofi Institute for Biomedical Research, 100 Chongwen Rd, Suzhou Industrial park, Suzhou 215001, China.
Contributor Information
Jaideep Singh, Department of Chemistry, University of Southern California, Los Angeles, CA 90089, USA.
Kevin G Liu, Department of Chemistry, University of Southern California, Los Angeles, CA 90089, USA.
Aleique Allen, Department of Chemistry, University of Southern California, Los Angeles, CA 90089, USA.
Wei Jiang, Department of Chemistry, University of Southern California, Los Angeles, CA 90089, USA.
Peter Z Qin, Department of Chemistry, University of Southern California, Los Angeles, CA 90089, USA.
DATA AVAILABILITY
All relevant data are included in the main text or the supplemental information.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institution of General Medical Sciences [R01GM124413, R35GM145341, RR028992 to P.Z.Q., T32-GM118289 to J.S.]; Anton B. Burg Foundation; J.S. is a recipient of the Alfred E. Mann Innovation in Chemistry Doctoral Fellowship. Funding for open access charge: NIGMS [R35GM145341].
Conflict of interest statement. None declared.
REFERENCES
- 1. Sontheimer E.J., Barrangou R.. The Bacterial Origins of the CRISPR Genome-Editing Revolution. Human Gene. Therapy. 2015; 26:413–424. [DOI] [PubMed] [Google Scholar]
- 2. Hille F., Charpentier E.. CRISPR-Cas: biology, mechanisms and relevance. Philos. Trans. Roy. Soc. B Biol. Sci. 2016; 371:20150496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Marraffini L.A. CRISPR-Cas immunity in prokaryotes. Nature. 2015; 526:55–61. [DOI] [PubMed] [Google Scholar]
- 4. Van Der Oost J., Westra E.R., Jackson R.N., Wiedenheft B.. Unravelling the structural and mechanistic basis of CRISPR–Cas systems. Nat. Rev. Microbiol. 2014; 12:479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McCarty N.S., Graham A.E., Studená L., Ledesma-Amaro R.. Multiplexed CRISPR technologies for gene editing and transcriptional regulation. Nat. Commun. 2020; 11:1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kaminski M.M., Abudayyeh O.O., Gootenberg J.S., Zhang F., Collins J.J.. CRISPR-based diagnostics. Nat. Biomed. Eng. 2021; 5:643–656. [DOI] [PubMed] [Google Scholar]
- 7. Chavez M., Chen X., Finn P.B., Qi L.S.. Advances in CRISPR therapeutics. Nat. Rev. Nephrol. 2023; 19:9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhu H., Li C., Gao C.. Applications of CRISPR–Cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol. 2020; 21:661–677. [DOI] [PubMed] [Google Scholar]
- 9. Makarova K.S., Wolf Y.I., Iranzo J., Shmakov S.A., Alkhnbashi O.S., Brouns S.J., Charpentier E., Cheng D., Haft D.H., Horvath P.. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020; 18:67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., Doudna J.A.. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018; 360:436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li S.-Y., Cheng Q.-X., Wang J.-M., Li X.-Y., Zhang Z.-L., Gao S., Cao R.-B., Zhao G.-P., Wang J.. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 2018; 4:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., Van Der Oost J., Regev A.et al.. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015; 163:759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Swarts D.C., Jinek M.. Cas9 versus Cas12a/Cpf1: structure–function comparisons and implications for genome editing. Wiley Interdiscipl. Rev. RNA. 2018; 9:e1481. [DOI] [PubMed] [Google Scholar]
- 14. Shivram H., Cress B.F., Knott G.J., Doudna J.A.. Controlling and enhancing CRISPR systems. Nat. Chem. Biol. 2021; 17:10–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cho S.W., Kim S., Kim Y., Kweon J., Kim H.S., Bae S., Kim J.-S.. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014; 24:132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Manghwar H., Li B., Ding X., Hussain A., Lindsey K., Zhang X., Jin S.. CRISPR/Cas systems in genome editing: methodologies and tools for sgRNA design, off-target evaluation, and strategies to mitigate off-target effects. Adv. Sci. 2020; 7:1902312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Naeem M., Majeed S., Hoque M.Z., Ahmad I.. Latest developed strategies to minimize the off-target effects in CRISPR-Cas-mediated genome editing. Cells. 2020; 9:1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Swarts D.C., van der Oost J., Jinek M.. Structural basis for guide RNA processing and seed-dependent DNA targeting by CRISPR-Cas12a. Mol. Cell. 2017; 66:221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Swarts D.C., Jinek M.. Mechanistic insights into the cis-and trans-acting DNase activities of Cas12a. Mol. Cell. 2019; 73:589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stella S., Mesa P., Thomsen J., Paul B., Alcón P., Jensen S.B., Saligram B., Moses M.E., Hatzakis N.S., Montoya G.. Conformational activation promotes CRISPR-Cas12a catalysis and resetting of the endonuclease activity. Cell. 2018; 175:1856–1871. [DOI] [PubMed] [Google Scholar]
- 21. Strohkendl I., Moy C., Nguyen A., Russell R., Taylor D.W.. Structural basis of Cas12a R-loop propagation on pathway to DNA cleavage. 2023; bioRxiv doi:22 June 2023, preprint: not peer reviewed 10.1101/2023.03.13.532460. [DOI]
- 22. Yamano T., Zetsche B., Ishitani R., Zhang F., Nishimasu H., Nureki O.. Structural basis for the canonical and non-canonical PAM recognition by CRISPR-Cpf1. Mol. Cell. 2017; 67:633–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wörle E., Newman A., D’Silva J., Burgio G., Grohmann D. Allosteric activation of CRISPR-Cas12a requires the concerted movement of the bridge helix and helix 1 of the RuvC II domain. Nucleic Acids Res. 2022; 50:10153–10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Naqvi M.M., Lee L., Montaguth O.E.T., Diffin F.M., Szczelkun M.D.. CRISPR–Cas12a-mediated DNA clamping triggers target-strand cleavage. Nat. Chem. Biol. 2022; 18:1014–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Saha A., Arantes P.R., Hsu R.V., Narkhede Y.B., Jinek M., Palermo G.. Molecular dynamics reveals a DNA-Induced dynamic switch triggering activation of CRISPR-Cas12a. J. Chem. Inform. Model. 2020; 60:6427–6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Babu K., Kathiresan V., Kumari P., Newsom S., Parameshwaran H.P., Chen X., Liu J., Qin P.Z., Rajan R.. Coordinated actions of Cas9 HNH and RuvC nuclease domains are regulated by the bridge helix and the target DNA sequence. Biochemistry. 2021; 60:3783–3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Raper A.T., Stephenson A.A., Suo Z.. Functional insights revealed by the kinetic mechanism of CRISPR/Cas9. J. Am. Chem. Soc. 2018; 140:2971–2984. [DOI] [PubMed] [Google Scholar]
- 29. Gong S., Yu H.H., Johnson K.A., Taylor D.W.. DNA unwinding is the primary determinant of CRISPR-Cas9 activity. Cell Rep. 2018; 22:359–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cofsky J.C., Karandur D., Huang C.J., Witte I.P., Kuriyan J., Doudna J.A.. CRISPR-Cas12a exploits R-loop asymmetry to form double-strand breaks. Elife. 2020; 9:e55143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Saha A., Arantes P.R., Schmitz M., Chanez C., Jinek M., Palermo G.. An alpha-helical lid guides the target DNA toward catalysis in CRISPR-Cas12a. 2022; bioRxiv doi:05 September 2022, preprint: not peer reviewed 10.1101/2022.09.05.506663. [DOI] [PMC free article] [PubMed]
- 32. Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F.. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016; 351:84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kleinstiver B.P., Tsai S.Q., Prew M.S., Nguyen N.T., Welch M.M., Lopez J.M., McCaw Z.R., Aryee M.J., Joung J.K.. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat. Biotechnol. 2016; 34:869–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kleinstiver B.P., Sousa A.A., Walton R.T., Tak Y.E., Hsu J.Y., Clement K., Welch M.M., Horng J.E., Malagon-Lopez J., Scarfò I.. Engineered CRISPR–Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat. Biotechnol. 2019; 37:276–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Strohkendl I., Saifuddin F.A., Rybarski J.R., Finkelstein I.J., Russell R.. Kinetic basis for DNA target specificity of CRISPR-Cas12a. Mol. Cell. 2018; 71:816–824.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jones S.K., Hawkins J.A., Johnson N.V., Jung C., Hu K., Rybarski J.R., Chen J.S., Doudna J.A., Press W.H., Finkelstein I.J.. Massively parallel kinetic profiling of natural and engineered CRISPR nucleases. Nat. Biotechnol. 2021; 39:84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Murugan K., Seetharam A.S., Severin A.J., Sashital D.G.. CRISPR-Cas12a has widespread off-target and dsDNA-nicking effects. J. Biol. Chem. 2020; 295:5538–5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bravo J.P.K., Liu M.-S., Hibshman G.N., Dangerfield T.L., Jung K., McCool R.S., Johnson K.A., Taylor D.W.. Structural basis for mismatch surveillance by CRISPR–Cas9. Nature. 2022; 603:343–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pacesa M., Lin C.-H., Cléry A., Saha A., Arantes P.R., Bargsten K., Irby M.J., Allain F.H.-T., Palermo G., Cameron P.et al.. Structural basis for Cas9 off-target activity. Cell. 2022; 185:4067–4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Okafor I.C., Singh D., Wang Y., Jung M., Wang H., Mallon J., Bailey S., Lee J.K., Ha T.. Single molecule analysis of effects of non-canonical guide RNAs and specificity-enhancing mutations on Cas9-induced DNA unwinding. Nucleic Acids Res. 2019; 47:11880–11888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li Y., Liu Y., Singh J., Tangprasertchai N.S., Trivedi R., Fang Y., Qin P.Z.. Site-specific labeling reveals Cas9 induces partial unwinding without RNA/DNA pairing in sequences distal to the PAM. CRISPR J. 2022; 5:341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jeon Y., Choi Y.H., Jang Y., Yu J., Goo J., Lee G., Jeong Y.K., Lee S.H., Kim I.-S., Kim J.-S.et al.. Direct observation of DNA target searching and cleavage by CRISPR-Cas12a. Nat. Commun. 2018; 9:2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Singh D., Mallon J., Poddar A., Wang Y., Tippana R., Yang O., Bailey S., Ha T.. Real-time observation of DNA target interrogation and product release by the RNA-guided endonuclease CRISPR Cpf1 (Cas12a). Proc. Natl. Acad. Sci. U.S.A>. 2018; 115:5444–5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang L., Sun R., Yang M., Peng S., Cheng Y., Chen C.. Conformational dynamics and cleavage sites of Cas12a are modulated by complementarity between crRNA and DNA. Iscience. 2019; 19:492–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Son H., Park J., Hwang I., Jung Y., Bae S., Lee S.. Mg2+-dependent conformational rearrangements of CRISPR-Cas12a R-loop complex are mandatory for complete double-stranded DNA cleavage. Proc. Natl. Acad. Sci. U.S.A>. 2021; 118:e2113747118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sowa G.Z., Qin P.Z.. Conn P.M. Site-directed Spin Labeling Studies on Nucleic Acid Structure and Dynamics. Progress in Nucleic Acid Research and Molecular Biology. 2008; 82:Academic Press; 147–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ding Y., Nguyen P., Tangprasertchai N.S., Reyes C.V., Zhang X., Qin P.Z.. Chechik V., Murphy D.M., Gilbert B.. Nucleic acid structure and dynamics: perspectives from site-directed spin labeling. Electron Paramagnetic Resonance. 2015; 24:The Royal Society of Chemistry; 122. [Google Scholar]
- 48. Vazquez Reyes C., Tangprasertchai N.S., Yogesha S.D., Nguyen R.H., Zhang X., Rajan R., Qin P.Z.. Nucleic acid-dependent conformational changes in CRISPR–Cas9 revealed by site-directed spin labeling. Cell Biochem. Biophys. 2017; 75:203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tangprasertchai N.S., Di Felice R., Zhang X., Slaymaker I.M., Vazquez Reyes C., Jiang W., Rohs R., Qin P.Z.. CRISPR–Cas9 mediated DNA unwinding detected using site-directed spin labeling. ACS Chem. Biol. 2017; 12:1489–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jiang W., Singh J., Allen A., Li Y., Kathiresan V., Qureshi O., Tangprasertchai N., Zhang X., Parameshwaran H.P., Rajan R.. CRISPR-Cas12a nucleases bind flexible DNA duplexes without RNA/DNA complementarity. ACS Omega. 2019; 4:17140–17147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mohanraju P., Van Der Oost J., Jinek M., Swarts D.C. Heterologous expression and purification of the CRISPR-Cas12a/Cpf1 protein. Bio-protocol. 2018; 8:e2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cai Q., Kusnetzow A.K., Hubbell W.L., Haworth I.S., Gacho G.P.C., Van Eps N., Hideg K., Chambers E.J., Qin P.Z.. Site-directed spin labeling measurements of nanometer distances in nucleic acids using a sequence-independent nitroxide probe. Nucleic Acids Res. 2006; 34:4722–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Qin P.Z., Haworth I.S., Cai Q., Kusnetzow A.K., Grant G.P.G., Price E.A., Sowa G.Z., Popova A., Herreros B., He H.. Measuring nanometer distances in nucleic acids using a sequence-independent nitroxide probe. Nature Protocols. 2007; 2:2354–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang X., Cekan P., Sigurdsson S.T., Qin P.Z.. Chapter 15 - Studying RNA Using Site-Directed Spin-Labeling and Continuous-Wave Electron Paramagnetic Resonance Spectroscopy. Methods in Enzymology. 2009; 469:Academic Press; 303–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pannier M., Veit S., Godt A., Jeschke G., Spiess H.W.. Dead-time free measurement of dipole–dipole interactions between electron spins. J. Magn. Reson. 2011; 213:316–325. [DOI] [PubMed] [Google Scholar]
- 56. Fábregas Ibáñez L., Jeschke G., Stoll S.. DeerLab: a comprehensive software package for analyzing dipolar electron paramagnetic resonance spectroscopy data. Magn. Reson. 2020; 1:209–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Worswick S.G., Spencer J.A., Jeschke G., Kuprov I.. Deep neural network processing of DEER data. Science Advances. 2018; 4:eaat5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Beasley K.N., Sutch B.T., Hatmal M.M., Langen R., Qin P.Z., Haworth I.S.. Qin P.Z., Warncke K.. Chapter Twenty-One - Computer Modeling of Spin Labels: NASNOX, PRONOX, and ALLNOX. Methods in Enzymology, Electron Paramagnetic Resonance Investigations of Biological Systems by Using Spin Labels, Spin Probes, and Intrinsic Metal Ions. 2015; 563:Part A. Academic Press; 569–593. [DOI] [PubMed] [Google Scholar]
- 59. Ding Y., Kathiresan V., Zhang X., Haworth I.S., Qin P.Z.. Experimental validation of the ALLNOX program for studying protein–nucleic acid complexes. J. Phys. Chem. A. 2019; 123:3592–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hanwell M.D., Curtis D.E., Lonie D.C., Vandermeersch T., Zurek E., Hutchison G.R.. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J. Cheminformatics. 2012; 4:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. del Alamo D., DeSousa L., Nair R.M., Rahman S., Meiler J., Mchaourab H.S.. Integrated AlphaFold2 and DEER investigation of the conformational dynamics of a pH-dependent APC antiporter. Proc. Natl. Acad. Sci. U.S.A. 2022; 119:e2206129119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are included in the main text or the supplemental information.









