Abstract
Adenosine-to-inosine (A-to-I) RNA editing is a post-transcriptional processing event involved in diversifying the transcriptome and is responsible for various biological processes. In this context, we developed a new method based on the highly selective cleavage activity of Endonuclease V against Inosine and the universal activity of sodium periodate against all RNAs to enrich the inosine-containing RNA and accurately identify the editing sites. We validated the reliability of our method in human brain in both Alu and non-Alu elements. The conserved sites of A-to-I editing in human cells (HEK293T, HeLa, HepG2, K562 and MCF-7) primarily occurs in the 3′UTR of the RNA, which are highly correlated with RNA binding and protein binding. Analysis of the editing sites between the human brain and mouse brain revealed that the editing of exons is more conserved than that in other regions. This method was applied to three neurological diseases (Alzheimer's, epilepsy and ageing) of mouse brain, reflecting that A-to-I editing sites significantly decreased in neuronal activity genes.
Graphical Abstract
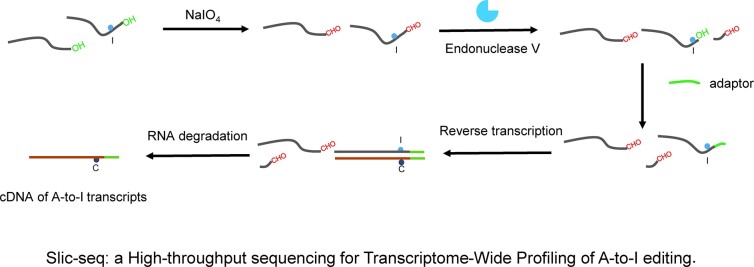
Graphical Abstract.
INTRODUCTION
A-to-I editing is one of the most abundant RNA modifications. It is catalyzed by the enzymes of the ADAR protein family, acting on dsRNA structures (1,2). The editing event alters the hydrogen bond pairing of nucleobases, and the editing site will be recorded as guanosine rather than the original adenosine. Moreover, A-to-I editing events are related to numerous critical biological processes, such as amino acid alterations (3), RNA splicing (3,4), nuclear retention (5), RNA interference (3,6,7) and innate cellular immunity (3,8,9). Then, the alteration of editing activity has been linked with multiple pathologies, including neurological disorders (10–12) and cancers (8,13,14). Nevertheless, further research must be conducted on the functions of most editing sites. Aiming to gain a comprehensive knowledge of these regulatory dynamics and biological roles, specifically their associations with different diseases, the key is robust discoveries and identification of A-to-I sites.
Several computational methods and tools have been developed to identify editing sites (15–17), yet most of these methods rely on multiple RNA-seq datasets and matched genomic DNA sequencing. In present computational methods, A-to-G fake positive signals possibly result from sequencing errors, SNPs, somatic mutations, unfavorable amplification of pseudogenes, PCR errors and spurious chemical alterations in RNA (18). Additionally, even though present transcriptome data have predicted millions of editing sites (19–21), low-expression transcripts and low-editing level sites may be ignored after rigorous bioinformatics screening from low coverage RNA-seq data. Therefore, extensive computational screening is necessary to predict low-editing rate A-to-I sites.
ICE-seq (22,23) improved the accuracy of discovering A-to-I RNA editing sites using acrylonitrile. Nonetheless, the method is limited in sensitivity due to the inability to enrich labeled inosine-containing transcripts. Subsequently, acrylamide derivatives (acrylamidofluorescein) (24) and acrylonitrile derivatives (25) have been explored. However, the derivative compounds are also reactive with pseudouridine and uridine, leading to off-target effects of this approach. Methods based on endonuclease activity (RNase T1 (26) and hEndoV (27)) to identify A-to-I editing sites have been developed. EndoVIPER-seq (28) uses Endonuclease V to enrich A-to-I sites in transcripts, improving the accuracy of recognition, however, the enrichment efficiency still has space for improvement. Accurate identification of A-to-I editing sites remains challenging.
In this paper, we developed a novel and effective biochemical method for transcriptome-wide identification of inosine based on Endonuclease V cleavage activity and high reactivity of sodium periodate to RNA 3′ terminal, which achieved the specific ligation of inosine-cleaved sequencing (Slic-seq). This robust and straightforward approach substantially enhances the detection and scope of A-to-I editing sites in cellular RNA while achieving the enrichment of inosine-containing RNAs.
MATERIALS AND METHODS
Cell culture
HEK293T, MCF-7, K562, HeLa and HepG2 were used in this study. HEK293T, MCF-7, HeLa and HepG2 cells were maintained in DMEM medium (Life Technologies) supplemented with 10% FBS and 1% penicillin/streptomycin. K562 was maintained in RPMI 1640 medium (Life Technologies) supplemented with 10% FBS and 1% penicillin/streptomycin. The cells were maintained at 37°C under a humidified atmosphere containing 5% CO2.
Mouse tissues
Mouse brains were isolated from adult C57BL/6 mice. Experimental protocols were approved by the IACUC of the Hubei Provincial Center for Disease Control and Prevention (Wuhan, China).
Procedure of poly(A)+ RNA isolation and fragmentation
Total RNA was isolated from cell lines or mouse tissues with TRIzol (Invitrogen) according to the manufacture's protocol. The mRNA was isolated by subjecting total RNA to oligo(dT) enrichment using Dynabeads Oligo(dT)25 (NEB) and gDNA was removed by TURBO DNase (Invitrogen). Purified polyadenylated RNA was fragmented using RNA fragmentation reagents (Ambion) at 70°C for 10 min. The human brain poly(A)+ RNA was purchased from Takara Bio Inc.
Validation of the effects of sodium periodate on RNA using LC–MS/MS analysis
After sodium periodate oxidation, 500 ng 40nt-poly(A) (Supplementary Table S1) RNA was digested by the combination of 2U nuclease P1 (Sigma, N8630) and 2U shrimp alkaline phosphatase (NEB, M0371S) in 50 μl water solution at 37°C for 2 h. Digested samples were filtered through 0.22-mm syringe filters before ultrahigh-performance LC–MS/MS analysis. The nucleosides were separated by an ultrahigh-performance liquid chromatographer (Shimadzu) equipped with a Shim-pack GIST C18 (100 mm × 2.1 mm i.d., 2.0 μm). Nucleosides were on-line analyzed using a triple quadrupole mass spectrometer after electrospray ionization with the following multiple reaction monitoring: m/z 268.1–136.1 (A), m/z 269.1–137.0 (I).
Validation of the effects of sodium periodate on RNA using NGS
100 ng model RNA-3 (Supplementary Table S1) was treated with sodium periodate. After purification, standard RNA seq libraries were prepared using NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (NEB) according to manufacturer's instructions. Sequencing was performed on Illumina HiSeq X Ten.
Blocking RNA 3′ end
Fragmented RNA was incubated with freshly prepared 50 mM NaIO4 (Sigma-Aldrich, S1878) at 25°C for 30 min in the dark.
Inosine-containing RNA cleavage
The RNA cleave assays were performed using Endonuclease V (NEB, M0305S) with standard reaction buffer (10 mM Tris–HCl pH 7.5, 0.5 mM MnCl2, 50 mM KCl, 1 mM dithiothreitol and 5% glycerol). The samples were incubated at 37°C for 60 min.
Slic-seq library preparation
The library was prepared following a previously reported procedure with slight changes (29). Fragmented RNA was first subjected to end repair by T4 PNK. For the treated sample, the RNA was treated with periodate followed by Endonuclease V cleavage.3′-adapter ligation (3′ adapter, /5rApp/NNNNNNTGGAATTCTCGGGTGCCAAGG/3ddC/) followed by the 3′ adapter removing by 5′-deadenylase and RecJf digestion. After purification, SuperScript III was used to perform RT (RT primer, ACACGACGCTCTTCCGATCT). After cDNA synthesis, 5′ adapter (5′ adapter, /5Phos/NNNNNNNNNNAGATCGGAAGAGCACACGTCTG/3ddC/) ligation was performed. After purification, qPCR was performed to evaluate the cycle numbers of each sample to avoid over-amplification. Library PCR amplification was performed using the NEB primers. The products were purified by NEBNext Sample Purification Beads or low melting point agarose gel and then used for sequencing.
RNA secondary structure prediction
The upstream and downstream 100bp sequences around candidate sites were extracted from transcriptome and subjected to RNA secondary structure prediction using RNA fold program from RNAStructure package (30).
Targeted amplicon sequencing
We first reverse transcribed the human brain poly(A)+ RNA (100 ng) using oligo(dT)18 primer and SuperScript III. After RNA removal with 0.1M NaOH, cDNA was purified using OCC Oligo Clean & Concentrator (Zymo Research). Regions flanking the targeting sites were selected for the design of primers, whose overhangs contained the paired Illumina adapter sequences. In addition, a 10-nt barcode was also added into each primer pair (Supplementary Table S2) to lower the detection limit from 10−3 to 10−5. The first round of PCR amplification was performed with NEBNext Q5 Master Mix (NEB, M0544L) using cDNA as an input template. After about ten cycles of amplification, the PCR products were purified with 1× AMPure XP beads and eluted with 0.1× TE buffer. Purified DNA samples were then subjected to the second round of amplification for roughly 15 cycles and assigned with different indexes followed by a purification with 0.8× AMPure XP beads and then used for sequencing.
Sequencing data processing
The 150 bp pair-end reads from Illumina Hiseq XTen system sequencing were first sent for adaptor and quality trimming using trim galore, and reads shorter than 30 nt after trimming were excluded. Note that a barcode of random hexamer (NNNNNNNNNN) was ligated to the fragments during library construction. These random barcodes serve to identify PCR duplicates from real different fragments with the identical sequences. Fastuniq was used to remove identical reads produced by PCR. Next, the random sequence of reads were removed and the clean reads were mapped to ribosomal RNAs, and the unmapped reads were mapped to the Genome (Gencode, mm10 for mouse and hg38 for human) using hisat2 with parameter ‘–mp 4,2 –rna-strandness RF –no-softclip –no-mixed –no-discordant’. Only the proper pair and primary alignments were persisted for the downstream pipelines.
Identification of putative inosine sites
A custom script was used to parse the pileup format into a tabular format summarizing the mutation at each position. Genes in the positive and negative strands of RNA were analyzed separately. To evaluate the distribution of the second base initiated by truncated reads, for all mismatch events, with at least 6 mismatches, and the coverage of truncation reads at least 10% were recorded. To reduce sequencing and alignment errors, we excluded three consecutive adjacent mutations of different types with a mutation rate of >20%. Since adjacent A-to-I editing sites might be lost after cleavage, we reduced the standard of site identification to at least 4 mismatches, and the coverage of truncation reads at least 5% within 3bp near the sites found under the previous standard. After the final filtration, A-G in the positive strand and T-C in the negative strand for the genome reference, with a base Phred quality score of ≥27 in the reads second position were candidates for A-to-I editing.
Annotation of inosine sites
The editing sites were annotated using ANNOVAR to find their location within host genes. Overlap genes containing A-to-I sites of human cell lines were subjected to Gene ontology enrichment analysis using DAVID online bioinformatics database (https://david.ncifcrf.gov/).
Genomic coordinates sites between human and mouse
The liftOver program (http://hgdownload.soe.ucsc.edu/admin/exe/linux.x86_64/liftOver) with default parameters was used to convert human putative inosine site location to mouse coordinates, and reconvert mouse putative inosine sites back to the human reference genome to verify the hits, only the loci that can be converted to each other are retained.
Targeted amplicon sequencing data analysis
We first grouped targeted amplicon sequencing FASTQ reads by the unique molecular identifier (UMI). First the adapter sequences of consensus reads were removed with cutadapt software (v.1.18). for reads in the same UMI group, we use fastuniq to removed PCR duplications. Then Cleaned reads were mapped to the reference index by bowtie2 with default parameters. Next, we filtered the mapped BAM files using the samtools view command (v.1.9) with parameters -f 3 -q 5. Finally, mutation rate statistics were performed using the pysam module.
Differential analysis of inosine
As enrichment fails to retain edit rate information, the number of truncated reads was utilized for evaluating editing discrepancies. Statistical analyses were performed using the R package (edgeR package), truncated read count data were first normalized with the trimmed mean of M-values normalization method. Only truncated reads were detected in all four samples were included in the differential analyses. edgeR package was then used to assess the statistical significance of observed differences in truncated read counts. After the statistical test, the P value was adjusted using the Benjamini and Hochberg method to control the false discovery rate (FDR). The editing sites were considered statistically different when their FDR was lower than 0.05 and the absolute log2-fold change was >1. For pathways analysis of differentially editing, GO Biological Process analyses in DAVID online tool.
RESULTS
High reliability of individual inosine sites identified by Slic-seq
Endonuclease V (Endo V) has been reported to be a highly active ribonuclease, particularly for inosine in RNA. It catalyzes the cleavage of the second RNA phosphodiester bone 3′ to the inosine, generating 3′-OH and 5′-P termini (31). Periodate is a regioselective oxidation agent capable of converting ortho-dihydroxy to dialdehyde. RNA 3′-terminals contain 2′,3′-hydroxyl groups of ribose, which can be efficiently oxidized to produce aldehyde groups (32,33). Using periodate oxidation followed by treatment with eEndo V, the inosine-containing RNAs produce 3′-OH. Thus, the inosine-containing RNAs will end in 3′-OH while the other RNAs will end in 3′-CHO. After adaptor ligation (only inosine-containing transcripts can be ligated), and reverse transcribed, inosine-containing RNA can be enriched for sequencing. We evaluated the oxidation reactivity of periodate and the feasibility of the strategy by polyacrylamide electrophoresis (Supplementary Figure 1A, B). LC–MS analysis showed that periodate has no detectable reactivity (<0.01%) in converting adenosine to inosine (Supplementary Figure 2). In addition, we used NGS to assess whether the model RNA-3 treated with sodium periodate would introduce mutations, and the results showed that the mutation rate at each position was maintained at background levels (Supplementary Figure 3A), and no A-to-G mutation could be observed after sodium periodate (Supplementary Figure 3B).
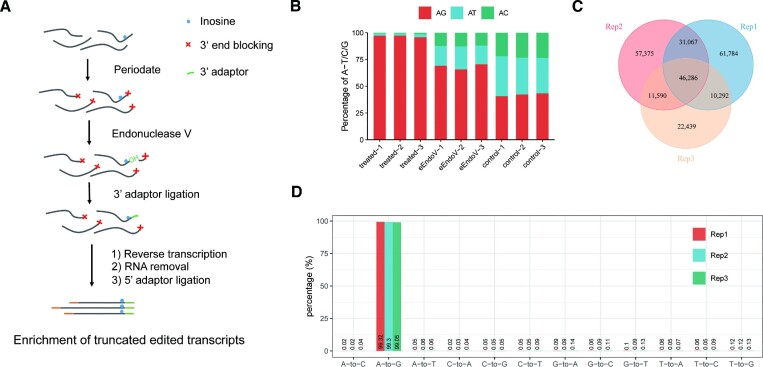
Briefly, the method is mainly composed of these steps: (1) 3′ end blocking, (2) inosine-containing RNA cleavage, (3) 3′ adaptor ligation, (4) reverse transcribe and ligate the adaptor to the 5′ end, followed by PCR amplification. The principle of our method is outlined (Figure 1A). Next, we tested Slic-seq on human RNAs to optimize the reaction conditions and examine its sensitivity and specificity. In each experiment, we constructed three different RNA libraries, comprising a treated sample, an input sample, and an eEndo V treated control sample without 3′ end blocking. In the treated sample, the A to G mismatch was significant increase compared with the two control groups. The A to G mutation (>95%) dominates in the second base (Figure 1B), while all mutation types were equally distributed in the control sample. To improve the sequencing quality of the second base, we increased six random sequences on the 3′ adaptor.
Figure 1.
Identification and validation of A-to-I RNA editing sites using Slic-seq. (A) Workflow of Slic-seq; (B) the A to G/T/C mutation proportions at second base of reads in the treated samples (periodate treated followed eEndo V treated), eEndo V treated samples and periodate treated samples; (C) the overlapping sites among three replicates in the HEK293T identified by Slic-seq; (D) the putative A-to-I RNA editing sites (∼99% being the A-to-G type among 12 types of mutations).
We were subsequently inspired to generate a pipeline to identify editing sites based on mutation and truncation. Since adjacent A-to-I editing sites might be lost after cleavage, we searched for potential sites a second time within a 3-bp window of the inosine identified in the first search (Materials and Methods). We found typical sites that matched the characteristics of our method from the IGV (Supplementary Figure 4A, B). In the three biological replicates, the proportions of A-to-G mismatches at all sites were 99.32%, 99.3% and 99.05%, respectively (Figure 1D). Assuming that the error rates for all 12 mismatch types were equal, the average false discovery rate was (0.78%/11)/99.22% = 0.07%. Notably, inclusion of specific genomic data of samples was unnecessary for our method and access to RNA-seq data was sufficient (External Databases S1).
Subsequently, we observed a high correlation at both truncated reads and FPKM between Slic-seq replicates (Supplementary Figure 5 and 6), indicating the high reproducibility of Slic-seq. The selected procedure executed three biological replicates and maintained recurring candidate sites found in two replicates at a minimum. A total of 99235 high-confidence inosine sites was detected (Figure 1C, External Databases S2). Unedited RNAs were discarded, and information about editing levels was obtained from additional RNA-seq (External Databases S3).
Characteristics of the Slic-seq
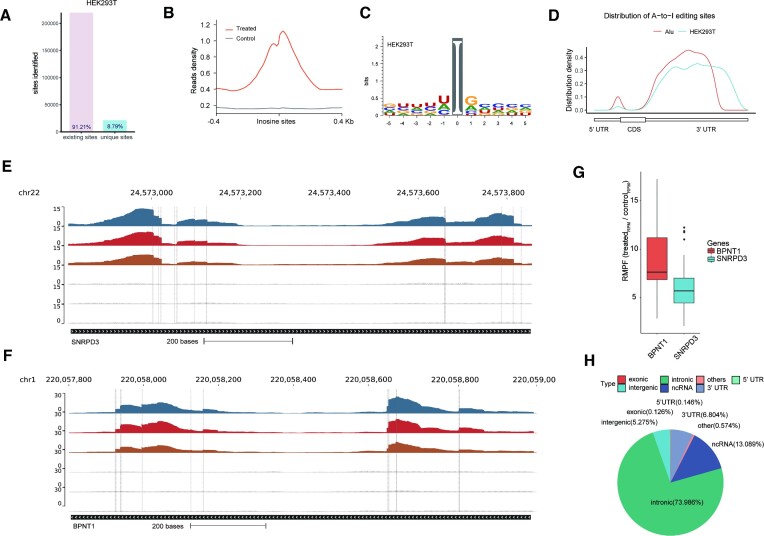
Combining the sites identified from three replicates, a total of 240 833 sites of HEK293T were identified, including 219 655 known or predicted editing sites (91.21%) deposited in the REDIportal (19) database (Figure 2A). There remains a number of novel editing sites that are absent in the database. In addition, no saturation was seen with Slic-seq at ∼1 × 108 uniquely mapped reads (Supplementary Figure 7A), consistent with the fact that a large number of editing sites exist in the human transcriptome. The newly discovered sites are mainly located on introns and intergenic regions (Supplementary Figure 7B). We then assessed expression levels of edited-site transcripts for new and known sites from RNA-seq. Low-expressed transcripts (FPKM < 1) contain more newly discovered editing sites (Supplementary Figure 7C). We compared the editing rates of new sites and known sites identified by Slic-seq in the corresponding RNA-seq and found that the newly identified sites are generally sites with lower editing rates. The proportion of the new sites with editing rates lower than 0.1 is significantly higher than that for the known sites (Supplementary Figure 7D). To confirm whether the newly discovered sites are located on dsRNA, we extracted the ±100 nt sequences around the newly identified CDS sites, then performed RNA secondary structure prediction (34) to assess these sequences and showed editing sites in dsRNA (Supplementary Figure 8).
Figure 2.
Slic-seq demonstrates high accurate features of A-to-I editing sites. (A) The proportion of A-to-I editing sites identified by Slic-seq from HEK293T in (existing sites) or not in (unique sites) REDIportal; (B) density plots showing the distribution of distance between the center of inosine peaks/control; (C) sequence motif obtained by HEK293T surrounding the editing sites presented by Slic-seq; (D) distribution of A-to-I editing sites and Alu peaks. A-to-I editing sites identified in HEK293T are tend to be located in 3′ UTRs, which had a similar distribution with Alu distribution; (E, F) representative views of the inosine sites on mRNA (SNRPD3 and BPNT1) by Slic-seq. Confident editing sites are highlighted with gray lines; (G) RPMF (fold of Reads Per Million) of mRNA (SNRPD3 and BPNT1) signals between treated samples and control samples. Center line, median; box limits, upper and lower quartiles; whiskers, 1.5 × interquartile range; points, outliers; (H) pie chart presenting the fraction of inosine sites of HEK293T in non-overlapping transcript segments.
We found much stronger signals in Slic-seq when compared to RNA-seq, and the inflection point was the exact position of an inosine site (Figure 2B). In this study, we identified base preferences adjacent to every site by expanding the context sequences. While the +1 position preferred G > A > C > U, the –1 position preferred U > A > C >> G (Figure 2C). The preference sequence was similar to the binding sequence of ADARs (35), implying minimal Slic-seq sequence bias. The distribution of inosine within transcriptomes revealed strong enrichment in the downstream 3′UTRs (Figure 2D). As anticipated, these A-to-I sites overlapped with the Alu repeat elements well, where A-to-I editing is prevalent.
The SNRPD3 and BPNT1 mRNAs, were known editing sites and consistently identified in three biological replicates of Slic-seq. The inosine-containing RNA have been enriched and the base-resolution editing sites were highlighted at the second base of the reads (Figure 2E and F). The neighbor editing sites continued to have high detection accuracy for sites that are very close to each other. Both SNRPD3 and BPNT1 had 12 confident editing sites. Compared to the control, the BPNT1 editing region was enriched ∼8-fold and SNRPD3 was enriched ∼6-fold (Figure 2G). The dsRNAs formed by inverted repeats in introns or UTRs are typical targets of ADAR (9). As expected, most inosine (67.17%) was found on intronic transcripts, while others were within the 3′ UTR (19.42%) and noncoding sequences (9.53%) in HEK293T cells (Figure 2H).
A-to-I editing sites identified by Slic-seq are more reliable
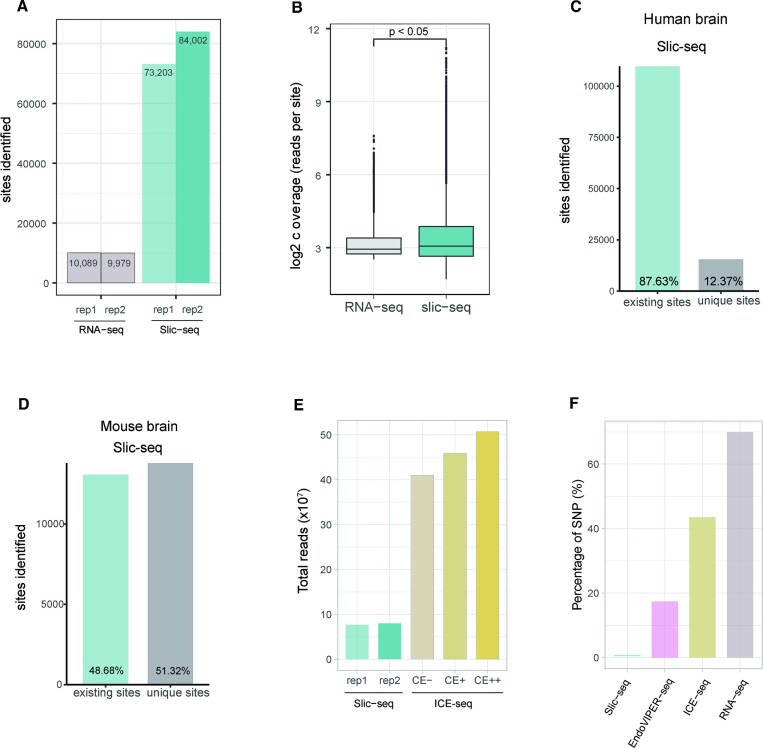
We then performed RNA-seq and Slic-seq on the same human brain mRNA samples. We employed the REDITools (16) package-associated filtering steps to identify A-to-I editing sites from RNA-seq. At same sequencing volumes, the number of identified A-to-I editing sites was significantly higher in Slic-seq, which achieved 7.8-fold identified A-to-I editing sites compared to RNA-seq (Figure 3A). Similarly, by comparing the coverage of the editing sites in RNA-seq and Slic-seq, we observed a significantly higher coverage of A-to-I editing sites from Slic-seq. All these findings suggest that Slic-seq remarkably improved the sensitivity in detecting A-to-I editing than the standard RNA-seq (Figure 3B).
Figure 3.
Comparison of A-to-I editing sites identified by Slic-seq and other high-throughput sequencing methods. (A) Number of identified sites from RNA-seq and Slic-seq at the same sequencing depth; (B) Box and Whisker plot shows that read coverages at A-to-I editing sites are significantly increased by Slic-seq (two-sided t-tests, P < 0.05); (C) the proportion of A-to-I editing sites identified by Slic-seq from human brain in (existing sites) or not in (unique sites) REDIportal; (D) the proportion of A-to-I editing sites identified by Slic-seq from mouse brain in (existing sites) or not in (unique sites) REDIportal; (E) fractions of all sequenced read pairs (Slic-seq and ICE-seq) before trimming and mapping; (F) SNPs’ fractions of A-to-G candidate sites for HEK293T in RNA-seq, EndoVIPER-seq and Slic-seq.
Slic-seq identified 125 146 A-to-I editing sites in human brain mRNA overlapped most with the database (109 667 sites overlap, accounting for 87.63%) (Figure 3C), and it also showed a similar ratio in HEK293T cells. Compared with EndoVIPER-seq (28) results in human brain mRNA, which also uses Endonuclease V to enrich A-to-I transcripts, Slic-seq performed higher overlap with databases (Figure 3C and Supplementary Figure 9A), and Slic-seq has a slightly higher proportion of common sites between replicates than EndoVIPER-seq (Supplementary Figure 9B, C). Additionally, we compared mouse brain A-to-I editing with Nascent RNA-seq data (36). This ratio (48.68%) was much lower than annotations in the Slic-seq and human databases, suggesting that mouse edited annotations are still insufficient (Figure 3D).
To further evaluate the reliability and performance of Slic-seq, we compared Slic-seq results with published ICE-seq (23) data in human brain RNA. Our two replicates used 76770037 and 79676223 pairs of reads, respectively. In the ICE-seq, a single biological replicate contained three samples (CE-, CE+, CE++) 409 843 384; 459 198 334 and 507 338 206 pairs of reads, respectively (Figure 3E). Slic-seq detected approximately four times as many editing sites, using <25% sequencing depth of ICE-seq.
Most existing methods have difficulty in identifying A-to-I editing sites in non-Alu regions, which have relatively fewer sites and substantial mutations (37). We then assessed the the A-to-T/C/G fractions in Alu elements, non-Alu repetitive elements and non-repetitive elements across different genic regions (intron, exon, 5′UTR, 3′UTR). For human brain mRNA, both RNA-seq and Slic-seq showed great accuracy in all Alu regions, meanwhile Slic-seq can significantly improve the detection accuracy in non-Alu repetitive and non-repetitive regions (Supplementary Figure 10A-B) and showed good stability in different replicates of different samples (HEK293T, human brain tissue, mouse brain tissue). (Supplementary Figure 10A and 10C, D).
We then used targeted amplicon sequencing to interrogate new editing sites in CDS of human brain. We selected 34 editing sites detected simultaneously by two replicates. Within the 27 sites that were successfully amplified, we validated 26 unique editing sites by Slic-seq, with the editing ratio from 0.19% to 24.61% (Supplementary Figure11).
Almost all methods need to remove SNPs when identifying confident editing sites. However, Slic-seq achieved high accuracy without SNP removal. We then selected A-to-G candidate sites at EndoVIPER-seq, ICE-seq and RNA-seq, which had been performed according to the respective pipeline without SNP removal. By comparing the candidate sites and identified sites by Slic-seq with SNP database, the proportion of SNPs was calculated in Slic-seq, EndoVIPER-seq, ICE-seq and RNA-seq. In the RNA-seq data, ∼69% A-to-G candidate sites are overlapped with SNP database. In ICE-seq, the processed samples screened out some SNPs, but ∼43% of the candidate sites were SNPs. In EndoVIPER-seq, there remained ∼17% SNPs in the candidate inosines sites. In Slic-seq, SNPs account for only ∼0.03%. This finding indicates that Slic-seq can effectively shield the interference of the A-to-G mutation caused by non-inosine (Figure 3F). Additionally, we examined sites overlapping with the SNP database in Slic-seq and found that most of these sites were enriched and had distinct truncation features (Supplementary Figure 12). In line with the properties of Slic-seq, these sites should not be discarded as SNPs. This confirms the potentiality of our method to be applied in other species with considerable somatic mutations and unknown SNPs. Together, these results indicate that Slic-seq has greater sensitivity and higher reliability for detection of A-to-I editing sites.
In the human brain, all the three methods showed similar motif trends, with a depletion of guanosine immediately upstream of the edited site, and some enrichment of G immediately downstream (Supplementary Figure 13). Additionally, we found that the editing motif was similar in different regions of different samples (HEK293T, human brain tissue, mouse brain tissue) (Supplementary Figure 14).
Slic-seq analysis of the different cell lines and brain tissues
RNA editing in cell lines has been reported to markedly lower than that in tissue samples (38). The RNA editing landscape in cell lines has not yet been well characterized. We further identified the editing sites of four cell lines, including HeLa, HepG2, K562 and MCF-7 (External Databases S1–S3). A good correlation was observed from the scatterplot of FPKM values and truncation values, consistent with HEK293T (Supplementary Figure 5 and 6). The clustering of all significantly enriched sequences recapitulated the HEK293T editing consensus sequence to a great extent (Supplementary Figure 15). Additionally, four cell lines showed a similar distribution with the HEK293T and two brain tissues editing increased in the CDS region (Supplementary Figure 16).
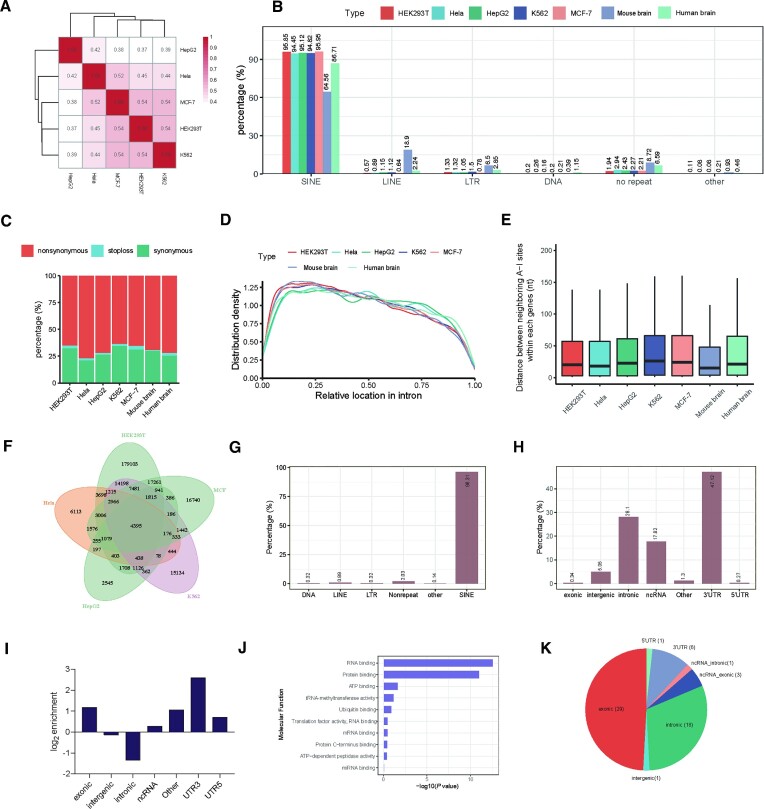
The correlation between different cells (Figure 4A) reflected the conservation of A-to-I editing sites (37,39). The editing sites of the human mRNA are primarily linked with repeat elements. Most of the editing sites were deposited in short interspersed nuclear elements (SINEs), and some were distributed in long terminal repeats (LTRs), long interspersed nuclear elements (LINEs), and nonrepeat regions (Figure 4B). Among different cells, the transcriptomic distribution displayed similar patterns.
Figure 4.
Transcriptome-wide distribution of inosine revealed by Slic-seq in multiple cell. (A) Correlation of inosine sites between different cells. A spearman correlation analysis was performed based on the coverage of treated samples; (B) the repeat class distribution of editing sites corresponding to inosine locus for different cell lines; (C) the percentages of different codon mutation types; (D) The relative position of inosine sites within introns; (E) distance between two neighboring inosine sites within each gene. Center line, median; box limits, upper and lower quartiles; whiskers, 1.5× interquartile range; (F) conserved editing sites in different cells; (G) the percentages of editing sites associated with a particular repeat; (H) the fractions of A-to-I editing sites in different transcript segments; (I) relative enrichment of A-to-I editing sites across transcript segments; (J) gene ontology (GO) analysis suggesting that conserved editing sites in different cells are significantly enriched in RNA binding and protein binding; (K) conserved sites in human and mouse brain.
Compared with noncoding sites, editing sites in coding regions (recoding sites) were much less prevalent. Additionally, alterations in amino acid assignment resulting from A-to-I editing in CDS may potentially modulate protein function. Then, we focused on the coding region of editing sites. The synonymous and nonsynonymous mutations were comparable, accounting for approximately 25% and 70% across different cell lines, respectively. In addition, some editing sites in the CDS resulted in translation termination (Figure 4C). We calculated the relative distances of inosine sites to intron-exon boundaries in each intron (Figure 4D). The findings showed an even distribution of inosine along the intron regions, with a slight depletion toward the 3′ end. Using the inosine positions identified by Slic-seq, we calculated the distances between each two adjacent inosine sites within each gene. Our results verified that two adjacent inosine sites tended to be clustered (40) and the distance is generally < 50nt between neighboring inosine sites (Figure 4E). The distance was slightly shorter than that between tandem ADAR proteins, which bind dsRNA substrates in the cellular context (41). From the perspective of several types of cells and two brain tissues, editing sites prefer to be within the intronic transcripts, 3′UTR regions and non-coding sequences (Supplementary Figure 17).
To identify functional and vital editing sites, evolutionary conservation at each site should be considered. Afterward, we evaluated the shared editing sites from different cells. The 4395 editing sites overlapped with a number of inosine-containing genes in five human cell line experiments (Figure 4F). These sites were mainly located in SINE element (Figure 4G), and the remaining sites were scattered in 3′UTRs (47.5%), introns (28.1%) and ncRNAs (17.32%) (Figure 4H). Approximately 4-fold enrichment over the expected distribution was calculated, and 3′UTR segments were most enriched in editing sites (Figure 4I). Moreover, Gene Ontology (GO) analysis of the conserved editing sites revealed that the predominant functions were RNA binding and protein binding (Figure 4J).
We then compared editing sites between human and mouse brains and found 59 conserved sites (External Databases S4), 11 of which were newly identified and 29 of which were deposited on exons (Figure 4K). We selected 2 newly identified sites on exons above. These sites, located on genes namely POUSF3 and FBXL17, are predicted to be located in dsRNA regions in both species (Supplementary Figure 18). Due to their low coverage in RNA-seq, we are unable to obtain reliable editing rates of the sites. Partially as the identified conserved sites are, most of them are enriched in exons, verifying the conservation of exons across species and the nonrandom distribution of editing sites. Recent studies have focused on the coding sites and found that highly edited sites are evolutionarily conserved in non-primate mammals (42).
Analysis of inosine sites in neurological diseases
A-to-I editing has now been identified as a reliable, differential biomarker in a number of neurological disorders. Numerous studies have covered the A-to-I editing of Alzheimer's disease (43) and epilepsy (44–47), focusing on limited editing sites lacking whole transcript-wide assays.
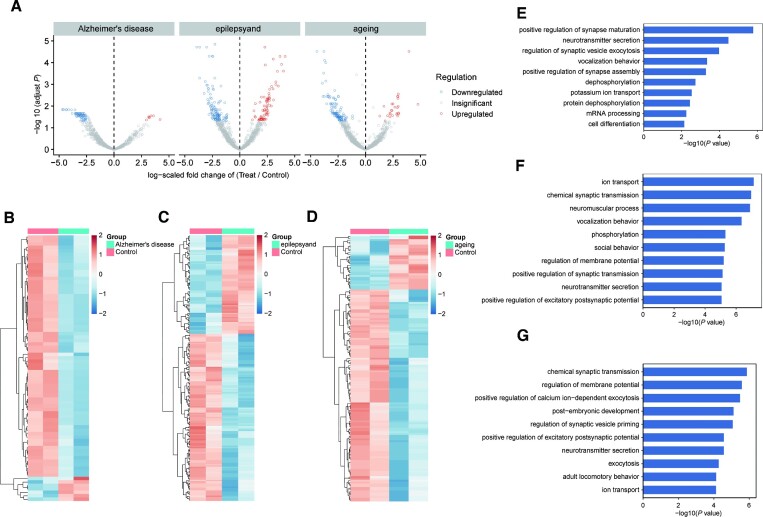
Accurate detection of differential editing, especially those with low editing rates, is even more challenging (9). We next examined the ability of Slic-seq to be applied to detect editing differences. We performed Slic-seq on mouse brain tissue RNA and screened 1005, 1364 and 1570 reliable sites in Alzheimer, epilepsy and ageing mouse brain RNAs respectively for analysis of differential editing sites (External Databases S5), which were both detected in the models of disease and normal mouse brain in all replicates (Figure 5A). The differential A-to-I editing was based on the difference in the number of edited reads between disease samples with normal samples. In Alzheimer's disease, we found 77 significantly differential edited sites (|log2FC| > 1 and FDR < 0.05), 70 of which were reduced in cases compared with the controls (Figure 5A and B). In epilepsy, 202 editing sites presented significant differential editing levels (|log2FC| > 1 and FDR < 0.05), 127 of which exhibited decreased editing levels (Figure 5A and C). In ageing, 148 sites showed statistically significant differential editing levels (|log2FC| > 1 and FDR < 0.05). On results similar with Alzheimer's disease and epilepsy, 118 of which were edited down (Figure 5A and D). Among the differential editing sites, we analyzed the expression differences of the genes in the corresponding RNA-seq, most genes with differential RNA editing sites did not display differential gene expression (|log2FC| > 1 and FDR < 0.05) (Supplementary Figure 19 and External Databases S5).
Figure 5.
Differential editing sites identified in three neurological diseases of mouse brain tissue. (A) Volcano plots showing A-to-I editing differentially between WT and treated models. Editing level significantly upregulated (red) or downregulated (blue) were identified in brain tissue. Editing sites detected in each sample of the control groups and the experimental groups were selected. Editing sites number, Alzheimer's disease: n = 1005, epilepsy: n = 1346; ageing: n = 1570; (B–D) heat map of the significant differential editing sites. (B) 77 differential editing sites in Alzheimer's disease, (C) 202 differential editing sites in epilepsy; (D) 148 differential editing sites in ageing were identified using the edgeR package (FDR < 0.05; log2(fold change) > 1); (E, F) GO analysis of enriched terms of genes with downregulated inosine abundance. GO enrichment analysis of genes with editing sites significantly downregulated or upregulated in (E) Alzheimer's disease, (F) epilepsy and (G) ageing. Significance of enrichment was determined by –log10(adjusted P value). The P value was provided by DAVID online tool and calculated with Fisher's exact test.
These significantly different sites were consistently reduced in cases compared with controls. With GO analysis, we observed significant enrichment for processes in various synaptic activity and neurotransmitter transport functions (Figure 5E–G). Differential RNA editing showed a high relevance to neuronal activity between the case and control mice. We further investigated whether the A-to-I editing differences in transcripts were impacted by differentially expressed genes. Here, differences in A-to-I editing sites displayed a low correlation with differentially expressed genes (DEGs) (r = 0.34 in Alzheimer's disease, r = 0.22 in epilepsy and r = 0.22 in ageing), implying RNA editing as a possible post-transcriptional mechanism for the regulation of gene expression (Supplementary Figure 20).
Functional enrichment analysis was performed for genes significantly (|log2FC| > 1 and FDR < 0.05) differentially expressed between cases and controls. We also observed minimal overlap between GO terms enriched among editing sites and those enriched among DEGs (Supplementary Figure 21). These results suggest that differential gene expression alone does not account for the observed differences in editing sites between cases and controls. Compared with differences in gene expression, differences in A-to-I editing showed a stronger correlation with neural activity. These results suggest that in neurodegenerative diseases, decreased A-to-I editing in genes involved in synaptic formation and in their activity may be responsible for reduced neural activity.
DISCUSSION
Identification of individual editing sites remains difficult due to the limitations of RNA-seq experiments, which require large amounts of input RNA material and high sequencing depth. Determining the rare or tissue/developmental specific sites is difficult due to the small sample size. Editing sites tend to be located in Alu dsRNAs such secondary structure may impact the incorporation into standard RNA-seq libraries.
In this study, we developed Slic-seq as a new method for the enrichment of inosine-containing transcripts from cellular RNA. The eEndo V is equally active on both single- and double-stranded RNAs (31), thus facilitating the identification of editing sites in different regions. Integrating enrichment, truncation and its own mutation signals, Slic-seq greatly improves the detection range and accuracy in both repetitive and non-repetitive sequences. Notably, the method is almost SNP-independent, revealing its potential to be applied to other species and further study of editing functions.
By applying Slic-seq to multiple cells, we evaluated the conservation of A-to-I editing sites. These sites, enriched in 3′UTR, may affect the RNA binding and protein binding. Additionally, we analyzed the differential editing sites between normal mouse and three neurological models and found that the A-to-I editing were decreased in neuronal activity-related genes. Such enrichment strategies can be used for high-precision studies of differential editing levels.
We anticipate that the approach will substantially augment our knowledge of A-to-I RNA editing, particularly its global regulation and dynamics, and unveil additional information crucial to advance our understanding of disease progression and biological processes.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Supercomputing Center of Wuhan University for providing the supercomputing system.
Contributor Information
Qi Wei, College of Chemistry and Molecular Sciences, Key Laboratory of Biomedical Polymers-Ministry of Education, Wuhan University, Wuhan, Hubei 430072, PR China.
Shaoqing Han, College of Chemistry and Molecular Sciences, Key Laboratory of Biomedical Polymers-Ministry of Education, Wuhan University, Wuhan, Hubei 430072, PR China.
Kexin Yuan, College of Chemistry and Molecular Sciences, Key Laboratory of Biomedical Polymers-Ministry of Education, Wuhan University, Wuhan, Hubei 430072, PR China.
Zhiyong He, College of Chemistry and Molecular Sciences, Key Laboratory of Biomedical Polymers-Ministry of Education, Wuhan University, Wuhan, Hubei 430072, PR China.
Yuqi Chen, College of Chemistry and Molecular Sciences, Key Laboratory of Biomedical Polymers-Ministry of Education, Wuhan University, Wuhan, Hubei 430072, PR China.
Xin Xi, College of Chemistry and Molecular Sciences, Key Laboratory of Biomedical Polymers-Ministry of Education, Wuhan University, Wuhan, Hubei 430072, PR China.
Jingyu Han, College of Chemistry and Molecular Sciences, Key Laboratory of Biomedical Polymers-Ministry of Education, Wuhan University, Wuhan, Hubei 430072, PR China.
Shen Yan, College of Chemistry and Molecular Sciences, Key Laboratory of Biomedical Polymers-Ministry of Education, Wuhan University, Wuhan, Hubei 430072, PR China.
Yingying Chen, College of Chemistry and Molecular Sciences, Key Laboratory of Biomedical Polymers-Ministry of Education, Wuhan University, Wuhan, Hubei 430072, PR China.
Bifeng Yuan, School of Public Health, Wuhan University, Wuhan, HuBei 430071, PR China.
Xiaocheng Weng, College of Chemistry and Molecular Sciences, Key Laboratory of Biomedical Polymers-Ministry of Education, Wuhan University, Wuhan, Hubei 430072, PR China.
Xiang Zhou, College of Chemistry and Molecular Sciences, Key Laboratory of Biomedical Polymers-Ministry of Education, Wuhan University, Wuhan, Hubei 430072, PR China; Department of Hematology, Zhongnan Hospital, Wuhan University, Wuhan, Hubei 430071, PR China; Taikang Center for Life and Medical Sciences, Wuhan University, Wuhan, Hubei 430072, PR China.
DATA AVAILABILITY
Code for the analyses described in this paper is available in the GitHub repository (https://github.com/sqhan-whu/A-I_stop, permanent doi: 10.5281/zenodo.8084456). All data sets have been deposited in the Gene Expression Omnibus under accession number GSE169710. Other data and materials are available from the authors upon reasonable request.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [21 721 005, 91 753 201]; National Key R&D Program of China [2022YFA 1303500]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. The authors declare no competing financial interest.
REFERENCES
- 1. Bass B.L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002; 71:817–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Roundtree I.A., Evans M.E., Pan T., He C.. Dynamic RNA mod-ifications in gene expression regulation. Cell. 2017; 169:1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nishikura K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 2016; 17:83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rueter S., Dawson T., Emeson R.. Regulation of alterna-tive splicing by RNA editing. Nature. 1999; 399:75–80. [DOI] [PubMed] [Google Scholar]
- 5. Chen L., DeCerbo J.N., Carmichael G.G.. Alu element-mediated gene silencing. EMBO J. 2008; 27:1694–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kawahara Y., Zinshteyn B., Sethupathy P., Iizasa H., Hatzigeorgiou A.G., Nishikura K.. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007; 315:1137–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ota H., Sakurai M., Gupta R., Valente L., Wulff B.E., Ariyoshi K., Iizasa H., Davuluri R.V., Nishikura K.. ADAR1 forms a complex with dicer to promote microRNA processing and RNA-induced gene silencing. Cell. 2013; 153:575–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ishizuka J.J., Manguso R.T., Cheruiyot C.K., Bi K., Panda A., Iracheta-Vellve A., Miller B.C., Du P.P., Yates K.B., Dubrot J.et al.. Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature. 2019; 565:43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eisenberg E., Levanon E.Y.. A-to-I RNA editing—immune protector and transcriptome diversifier. Nat. Rev. Genet. 2018; 19:473–490. [DOI] [PubMed] [Google Scholar]
- 10. Maas S., Kawahara Y., Tamburro K.M., Nishikura K.. A-to-I RNA editing and human disease. RNA Biol. 2006; 3:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tran S.S., Jun H.I., Bahn J.H., Azghadi A., Ramaswami G., Van Nostrand E.L., Nguyen T.B., Hsiao Y.E., Lee C., Pratt G.A.et al.. Widespread RNA editing dysregulation in brains from autistic individuals. Nat. Neurosci. 2019; 22:25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Breen M.S., Dobbyn A., Li Q., Roussos P., Hoffman G.E., Stahl E., Chess A., Sklar P., Li J.B., Devlin B.et al.. Global landscape and genetic regulation of RNA editing in cortical samples from individuals with schizophrenia. Nat. Neurosci. 2019; 22:1402–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Han L., Diao L., Yu S., Xu X., Li J., Zhang R., Yang Y., Werner H.M.J., Eterovic A.K., Yuan Y.et al.. The genomic landscape and clinical relevance of A-to-I RNA editing in human cancers. Cancer Cell. 2015; 28:515–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tan M.H., Li Q., Shanmugam R., Piskol R., Kohler J., Young A.N., Liu K.I., Zhang R., Ramaswami G., Ariyoshi K.et al.. Dynamic landscape and regulation of RNA editing in mammals. Nature. 2017; 550:249–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Oakes E., Vadlamani P., Hundley H.A.. Methods for the detection of adenosine-to-inosine editing events in cellular RNA. In: Shi Y. (eds) mRNA processing. Methods Mol. Biol. 2017; 1648:103–127. [DOI] [PubMed] [Google Scholar]
- 16. Picardi E., Pesole G.. REDItools: high-throughput RNA editing detection made easy. Bioinformatics. 2013; 29:1813–1814. [DOI] [PubMed] [Google Scholar]
- 17. John D., Weirick T., Dimmeler S., Uchida S.. RNAEditor: easy detection of RNA editing events and the introduction of editing islands. Brief. Bioinform. 2016; 18:993–1001. [DOI] [PubMed] [Google Scholar]
- 18. Pinto Y., Levanon E.Y.. Computational approaches for de-tection and quantification of A-to-I RNA-editing. Methods. 2019; 156:25–31. [DOI] [PubMed] [Google Scholar]
- 19. Picardi E., D’Erchia A.M., Giudice C.L., Pesole G.. REDIportal: a comprehensive database of A-to-I RNA editing events in humans. Nucleic Acids Res. 2016; 45:D750–D757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ramaswami G., Li J.B.. RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 2013; 42:D109–D113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kiran A., Baranov P.V.. DARNED: a DAtabase of RNa EDiting in humans. Bioinformatics. 2010; 26:1772–1776. [DOI] [PubMed] [Google Scholar]
- 22. Sakurai M., Yano T., Kawabata H., Ueda H., Suzuki T.. Inosine cyanoeth-ylation identifies A-to-I RNA editing sites in the human transcriptome. Nat. Chem. Biol. 2010; 6:733–740. [DOI] [PubMed] [Google Scholar]
- 23. Sakurai M., Ueda H., Yano T., Okada S., Terajima H., Mitsuyama T., Toyoda A., Fujiyama A., Kawabata H., Suzuki T.. A biochemical land-scape of A-to-I RNA editing in the human brain transcriptome. Genome Res. 2014; 24:522–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Knutson S.D., Ayele T.M., Heemstra J.M.. Chemical label-ing and affinity capture of inosine-containing RNAs using acrylamidofluorescein. Bioconjugate Chem. 2018; 29:2899–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li Y., Gohl M., Ke K., Vanderwal C.D., Spitale R.C.. Identification of adenosine-to-inosine RNA editing with acrylonitrile reagents. Org. Lett. 2019; 21:7948–7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cattenoz P.B., Taft R.J., Westhof E., Mattick J.S.. Transcriptome-wide identification of A >I RNA editing sites by inosine specific cleavage. RNA. 2013; 19:257–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen J.J., You X.J., Li L., Xie N.B., Ding J.H., Yuan B.F., Feng Y.Q.. Single-base resolution detection of adeno-sine-to-inosine RNA editing by endonuclease-mediated sequencing. Anal. Chem. 2022; 94:8740–8747. [DOI] [PubMed] [Google Scholar]
- 28. Knutson S.D., Arthur R.A., Johnston H.R., Heemstra J.M.. Selective enrichment of A-to-I edited transcripts from cellular RNA using endonuclease V. J. Am. Chem. Soc. 2020; 142:5241–5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou H., Rauch S., Dai Q., Cui X., Zhang Z., Nachtergaele S., Sepich C., He C., Dickinson B.C.. Evolution of a reverse transcriptase to map N1-methyladenosine in human messenger RNA. Nat. Methods. 2019; 16:1281–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reuter J.S., Mathews D.H.. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinf. 2010; 11:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vik E.S., Nawaz M.S., Strøm Andersen P., Fladeby C., Bjørås M., Dalhus B., Alseth I.. Endonuclease V cleaves at inosines in RNA. Nat. Commun. 2013; 4:2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Honda S., Loher P., Shigematsu M., Palazzo J.P., Suzuki R., Imoto I., Rigoutsos I., Kirino Y.. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E3816–E3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Evans M.E., Clark W.C., Zheng G., Pan T.. Determination of tRNA aminoacylation levels by high-throughput sequencing. Nucleic Acids Res. 2017; 45:e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lorenz R., Bernhart S.H., Zu Siederdissen H., Tafer H., Flamm C., Stadler P.F., Hofacker I.L.. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Eggington J., Greene T., Bass B.. Predicting sites of ADAR editing in double-stranded RNA. Predicting sites of ADAR editing in double-stranded RNA. Nat. Commun. 2011; 2:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Licht K., Kapoor U., Amman F., Picardi E., Martin D., Bajad P., Jantsch M.F.. A high resolution A-to-I editing map in the mouse identifies editing events controlled by pre-mRNA splicing. Genome Res. 2019; 29:1453–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ramaswami G., Zhang R., Piskol R., Keegan L.P., Deng P., O’Connell M.A., Li J.B.. Identifying RNA editing sites using RNA sequencing data alone. Nat. Methods. 2013; 10:128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schaffer A.A., Kopel E., Hendel A., Picardi E., Levanon E.Y., Eisenberg E.. The cell line A-to-I RNA editing catalogue. Nucleic Acids Res. 2020; 48:5849–5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang Q., Xiao X.. Genome sequence–independent identification of RNA editing sites. Nat. Methods. 2015; 12:347–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Levanon E.Y., Eisenberg E., Yelin R., Nemzer S., Hallegger M., Shemesh R., Fligelman Z.Y., Shoshan A., Pollock S.R., Sztybel D.et al.. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004; 22:1001–1005. [DOI] [PubMed] [Google Scholar]
- 41. Song Y., Yang W., Fu Q., Wu L., Zhao X., Zhang Y., Zhang R.. irCLASH reveals RNA sub-strates recognized by human ADARs. Nat. Struct. Mol. Biol. 2020; 27:351–362. [DOI] [PubMed] [Google Scholar]
- 42. Gabay O., Shoshan Y., Kopel E., Ben-Zvi U., Mann T.D., Bressler N., Cohen-Fultheim R., Schaffer A.A., Roth S.H., Tzur Z.et al.. Landscape of aden-sine-to-inosine RNA recoding across human tissues. Nat. Commun. 2022; 13:1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Khermesh K., D’Erchia A.M., Barak M., Annese A., Wachtel C., Levanon E.Y., Picardi E., Eisenberg E.. Reduced levels of protein recoding by A-to-I RNA editing in Alzheimer's disease. RNA. 2016; 22:290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brusa R., Zimmermann F., Koh D.S., Feldmeyer D., Gass P., Seeburg P.H., Sprengel R.. Early-onset epilepsy and postnatal lethality associated with an editing-deficient GluR-B allele in mice. Science. 1995; 270:1677–1680. [DOI] [PubMed] [Google Scholar]
- 45. Srivastava P.K., Bagnati M., Delahaye-Duriez A., Ko J.H., Rotival M., Langley S.R., Shkura K., Mazzuferi M., Danis B., van Eyll J.et al.. Genome-wide analysis of differential RNA editing in epilepsy. Genome Res. 2017; 27:440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Johannesen K.M., Gardella E., Linnankivi T., Courage C., de Saint Martin A., Lehesjoki A.E., Mignot C., Afenjar A., Lesca G., Abi-Warde M.T.et al.. Defining the phenotypic spectrum of SLC6A1 mutations. Epilepsia. 2018; 59:389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang J., Poliquin S., Mermer F., Eissman J., Delpire E., Wang J., Shen W., Cai K., Li B.M., Li Z.Y.et al.. Endoplasmic reticulum retention and degradation of a mutation in SLC6A1 associated with epilepsy and autism. Mol Brain. 2020; 13:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Code for the analyses described in this paper is available in the GitHub repository (https://github.com/sqhan-whu/A-I_stop, permanent doi: 10.5281/zenodo.8084456). All data sets have been deposited in the Gene Expression Omnibus under accession number GSE169710. Other data and materials are available from the authors upon reasonable request.