Abstract
Staphylococcus aureus (SA) can thrive in a wide variety of hosts and environments, causing clinical infections and foodborne intoxications. In Brazil, SA is commonly isolated from traditional soft cheeses, especially those prepared from unpasteurized milk. In this research, the isolate S. aureus SABRC1 was evaluated for virulence traits under different conditions, including co-inoculation with Lactococcus lactis MC5 (isolated from “Fresh Minas Cheese”), which produces antibacterial peptides. Results from experiments with Caco-2 culture indicated S. aureus SABRC1 was able to adhere (42.83 ± 1.79%) and to invade (48.57 ± 0.41%) the intestinal cells. On the other hand, L. lactis MC5 presented anti-staphylococcal activity as indicated by agar assays, and it was also able to antagonize intestinal cell invasion by S. aureus. Moreover, Reverse Transcriptase-PCR experiments showed virulence genes of S. aureus SABRC1 (hla, icaA and sea) were differentially expressed under diverse culture conditions, which included Brain Heart Infusion modified or not by the addition of glucose, sodium chloride, milk or cheese. This suggests the virulence of S. aureus SABRC1 is influenced by compounds commonly found in daily diets, and not only by its genetic repertoire, adding a novel level of complexity for controlling infection by this pathogen.
Keywords: Cheese, Gene expression, Probiotic, Staphylococci, Virulence
Introduction
Staphylococcus aureus is an opportunistic and versatile Gram-positive pathogen commonly linked to superficial skin infections and self-limiting food poisoning, although life-threatening invasive diseases have also been reported [1, 2]. The ability of S. aureus to cause major infections relies on the coordinated action of its virulence factors for adhesion to cell surfaces, evasion from the immune system and secretion of substances that are harmful to host tissues, favoring cell invasion [3, 4].
Previous studies revealed populations of S. aureus are highly clonal, but the clonal complexes (CCs) are quite heterogeneous in relation to virulence parameters, such as cytotoxicity, blood survival and superantigen profile, as well as the ability to form biofilm [3, 5]. It has also been reported that some clonal strains are more likely to cause invasive disease than others, and thus, the analysis of virulence traits of diverse isolates of S. aureus is important to obtain a clear picture of its pathogenesis. Among the major CCs of S. aureus, the CC1 can be highlighted, as it is a pandemic S. aureus complex harboring isolates from food, animal and human sources [6]. Many S. aureus strains belonging to CC1 present virulence and immunomodulatory genes potentially linked to the occurrence of invasive infections, besides Multi Drug Resistance phenotypes, such as Methicillin Resistant S. aureus strains—MRSA [7, 8]. It is known the colonization of skin, nasal cavity and postoperative wounds are important risk factors for the establishment of staphylococcal infections. Moreover, the ability of S. aureus to colonize the human gastrointestinal tract (GIT), especially of hospitalized individuals, is also concerning if the oral route of infection is considered [9–11]. Although the persistence mechanisms of S. aureus in mammalian hosts has not been fully elucidated, it is known the bacterial ability to invade eukaryotic cells is a key factor for colonization of GIT [12, 13].
Based on this, finding strategies to prevent cell invasion by S. aureus is of utmost importance, and the use of beneficial microbes delivered by probiotic food or supplement can be an alternative. Probiotics are “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host” [14]. The majority of probiotic strains described so far belong to the group of Lactic Acid Bacteria (LAB), which may present the ability to diminish the adhesion of pathogens to eukaryotic cells, preventing infection [15].
LAB produce bioactive metabolites that can suppress pathogen growth, such as organic acids, hydrogen peroxide and bacteriocins [16]. Most bacteriocins are non-toxic antimicrobial peptides with GRAS status (Generally Recognized as Safe), which have been used for many years for food protection [16]. LAB and their bacteriocins also present potential for pharmaceutical applications alongside conventional antibiotics, since selected bacteriocins can present activity against multiresistant pathogens, including MRSA [17, 18].
For this study, virulence traits of S. aureus from CC1 were studied in combination with the bacteriocin producing cheese-isolated Lactococcus lactis MC5 strain.
Materials and methods
Bacterial strains and culture conditions
The bacterial strains used in this study were co-isolated from a “Fresh Minas cheese” (FMC) sample, obtained at an artisanal dairy producer located in Brazilian midwest: (i) S. aureus SABRC1, from CC1 (Sequence Type 3531), and (ii) Lactococcus lactis MC5. Both were maintained as frozen glycerol stocks at − 80 °C in Brain Heart Infusion (BHI) broth (Oxoid, UK). Working cultures were prepared in BHI medium (37 °C/24 h).
The isolation of S. aureus SABRC1 was previously described by Alves et al. [19]. For the isolation of the LAB with anti-staphylococcal activity, agar antimicrobial assays were performed according to Lewus and Montville [20] modified by De Martinis et al. [21]. In brief, L. lactis MC5 was screened out of ten isolates from FMC (data not shown). For that, LAB grown in MRS broth were spot plated (10 µL) on MRS agar and incubated for up to 72 h at 25 °C under anaerobiosis. Then, the plates were overlaid with semi-solid BHI agar (BHI plus 1% bacteriological agar) seeded with 105–106 CFU/mL of the target microorganism, and re-incubated anaerobically at 25 °C for 24 h, to exclude inhibition due to hydrogen peroxide. Antagonistic LAB were further assayed on Tryptic Soy agar with 0.6% of Yeast Extract (TSAYE) to exclude inhibition due to organic acids produced from glucose present in MRS agar. The peptidic nature of the inhibitory substances was checked through the enzymatic degradation assay [20, 21].
L. lactis MC5 was identified based on 16 s rRNA PCR amplification with the primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′- TACGGYTACCTTGTTACGACTT-3′), according to Devereux and Wilkinson [22]. Sequencing of the amplicon was done by Sanger method with 3500 Genetic Analyzer (ThermoFisher Scientific, USA), at the Multi User Laboratory of Nucleic Acids (FCFRP-USP, Ribeirão Preto, SP, Brazil). Data were analyzed with the program 4peaks (Nucleobytes Corporation Inc, USA) and compared to GenBank database using the BLASTn search program (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Epithelial cell lines
The human epithelial cell line Caco-2 (ATCC HTB-37) was obtained from BCRJ (Cell Bank of Rio de Janeiro, RJ, Brazil). The eukaryotic cells were grown in monolayers in 75-cm2 flasks with Dulbecco’s modified Eagle’s medium—DMEM (Sigma-Aldrich) containing 10% (v/v) fetal bovine serum (Wisent, Canada) added 1% (v/v) solution of antibiotics (100 U/mL penicillin and 100 g/mL streptomycin, Gibco), and incubated at 37 °C in humidified 5% CO2 atmosphere. The Caco-2 cells were subcultured by trypsinization, and to perform adhesion and invasion assays, they were grown as semi-confluent monolayers in 24-well plates, using DMEM without antibiotics for the assays with bacteria.
Bacterial adhesion assays in epithelial cells
These assays were carried out according to Moroni et al. [23] and Merghni et al. [24], with Caco-2 cells. Briefly, an exponential growth phase BHI broth culture of S. aureus SABRC1 was obtained at 37 °C, and the inoculum was prepared by centrifugation (4500 × g/15 min), double washing with sterilized Phosphate Buffered Saline (PBS, pH 7.0), and the suspend of the pellet in DMEM, to obtain a MOI (Multiplicity of Infection) of 100.
Confluent Caco-2 cells (ca. 2 × 105 cells per well) were inoculated with 250 µL of S. aureus SABRC1 suspension (ca. 107 CFU/mL) and incubated for 2 h at 37 °C under 5% CO2. After incubation, non-adhered bacterial cells were removed by triple washing of the monolayers of epithelial cells with PBS. Next, the eukaryotic cells were lysed by treatment with a solution of PBS added 0.1% (v/v) Triton X-100 (Sigma-Aldrich®), for 15 min at 37 °C under 5% CO2. Total bacteria (adherent plus invasive) were counted by seeding serial decimal dilutions of the cell lysate on BHI agar plates, incubated at 37 °C for 24 h. Adhesion rate (%) was calculated as the log of the total number of bacteria minus the log of the number of invasive bacteria (determined according to Sect. 2.4), divided by the log of the number of bacteria in the initial inoculum, multiplied by 100 [23, 24], using the equation:
The assays were performed at least as triplicates.
Bacterial invasion assays in epithelial cells
These assays were also done based on Moroni et al. [23] and Merghni et al. [24]. For that, confluent Caco-2 cell monolayers (ca. 2 × 105 cells per well) were obtained as explained in the previous section and inoculated with S. aureus SABRC1 (ca. 107 CFU/mL), followed by incubation for 2 h at 37 °C under 5% of CO2. The eukaryotic cells were washed three times with PBS to remove non-adherent bacteria, and next, DMEM plus 1 mg/mL gentamicin (Sigma-Aldrich) in PBS was added to kill adherent staphylococci only, since it does not penetrate the membrane of the epithelial cells. The antibiotic was removed from the Caco-2 cell monolayers by triple washing with PBS, and they were lysed with 0.1% Triton X-100 in PBS. Invasive bacteria were counted by seeding serial decimal dilutions of lysate on BHI agar plates, with incubation for 24 h at 37 °C. Invasion rate (%) was calculated considering the log number of invading cells, divided by log number of bacteria in the initial inoculum, multiplied by 100 [23, 24], using the equation:
Similar adhesion and invasion treatments of Caco-2 cells were carried out in the presence of L. lactis MC5 (107 CFU per well), and in the presence of semi-purified bacteriocin of L. lactis MC5, contained in Cell Free Supernatant (CFS). To obtain the CFS, the LAB was grown in MRS broth (37 °C, 24 h), the cells were pelleted by centrifugation, the supernatant was neutralized and filter sterilized (membrane 0.22 µm, GVWP, Millipore). The bacteriocin activity in the CFS was quantified by the critical dilution assay, which according to Mayr-Harting et al. [25] consists on the “deposition of uniform drops from each dilution on to the surface of a plate of nutrient medium that has been seeded with a uniform and standard inoculum of the sensitive indicator strain”. The results were expressed as Arbitrary Units (AUs) per mL, considering the end-point of inhibition for calculation [25]. In this research, twofold dilutions were done, drops of 10μL of each dilution were spotted, the nutrient medium used was BHI agar and sensitive indicator strain was S. aureus SBRAC 1.
The concentrations (v/v) of CFS tested were 10%, 25%, 50%, 75% and 100%, by replacing DMEM during the steps of incubation with S. aureus SABRC1 for adhesion and invasion assays.
Microscopy of bacterial adhesion and invasion tests in epithelial cells
Confluent Caco-2cells were cultured as described previously, but using sterilized discs suitable for confocal laser scanning microscopy—CLSM (SPL Life Sciences, Kasvi, USA), with incubation for 48 h at 37 °C in 5% CO2. The adhered cells were washed three times with PBS. S. aureus SABRC1 and L. lactis MC5 were grown in test tubes containing 4 mL of BHI and incubated at 37 °C for 24 h. Bacteria were centrifuged at 4500 g for 15 min, the supernatants were removed and PBS was added. Bacterial suspensions were added to the culture plates of Caco-2 cells and incubated for 2 h at 37 °C in 5% CO2 to allow for invasion and adhesion.
The discs appropriate for CLSM with the Caco-2 and bacterial cells were stained with Live/Dead Cell viability assay kit (Thermo Fisher, USA), as instructed by the manufacturer, and observed by CLSM at the Multiuser Laboratory of the Faculty of Medicine of Ribeirão Preto—USP (Leica TCS-SP5 AOBS, 63 × objective). The argon laser (488 nm) was used to observe cells stained with Syto 9 (green fluorescence—viable, intact membrane), and the helium laser (543 nm) was used for the cells stained with propidium iodide (red fluorescence—non viable, damaged membrane). The images were processed with the LAS software version: 2.6.0 builds 7266 (Leica).
Expression of hla, icaA and sea virulence genes in Staphylococcus aureus SABRC1
S. aureus SABRC1 was cultured overnight at 37 °C in different culture media to compare the expression of the virulence genes hla, icaA and sea. The media evaluated were: (i) BHI broth (control); (ii) BHI broth + Glucose (1%); (iii) BHI broth + Glucose (1%) + NaCl (4%) [26]; (iv) milk culture media [27]; (v) milk powder medium [28] and (vi) “Fresh Minas Cheese” medium [29, 30].
Total RNA was extracted using the PureLink® RNA Mini kit, as instructed by the manufacturer (Thermo Fisher), and the synthesis of complementary DNA (cDNA) was performed with primers according to the literature for alfa toxin—hla [31], intercellular adhesin—icaA [32], enterotoxin—sea [31] (Table 1). The housekeeping gene 16S rRNA was used as reference for calculation of differential gene expression [33]. The runs were performed in a 7500 Fast Real Time PCR System (Applied Biosystems, USA), and the relative quantification of mRNA levels in S. aureus was evaluated under different culture conditions, by comparison with cultures obtained in BHI broth (control), with the ∆∆Ct comparative method described by Livak and Schmittgen [34].
Table 1.
Sequences of gene primers used to study virulence in a Staphylococcus aureus isolate from Fresh Minas cheese
| Gene | Primer | Sequence | Source |
|---|---|---|---|
| Alfa toxin: hla |
hla Fwr hla Rev |
GGGGACCATATGATAGAGATT TGTAGCGAAGTCTGGTGAAA |
[31] |
| Intercelular adesin: icaA |
icaA Fwr icaA Rev |
AGTTGTCGACGTTGGCTA CCAAAGACCTCCCAATGT |
[32] |
| Enterotoxina: sea |
SEA Fwr SEA Rev |
CGA AGG TTC TGT AGA CGA AGG TTC TGT AGA |
[31] |
| 16S rRNA* |
16S Fwd 16S Rev |
CGGTCCAGACTCCTACGGGAGGCAGCA GCGTGGACTACCAGGGTATCTAATCCT |
[33] |
*Gene 16S rRNA used as housekeeping gene
Statistical analysis
The t-Student’s test (unpaired data) was used for data treatment of the experiments of adhesion, invasion and gene expression. Unless otherwise stated, data were obtained from at least three independent experiments using at least intra assays duplicates. Statistical analyses were done in Prism Statistics, Version 6 (GraphPad Prism, USA).
Results
The CFS of L. lactis MC5A contained antimicrobial substances that were destroyed by proteases, indicating it was a bacteriocin-like substance—BLIS (data not shown). The agar dilution assay showed the BLIS produced by L. lactis MC5A had an activity of 800 AU/mL against S. aureus SBRAC1.
With regard to Caco-2 cell culture assays, S. aureus SABRC1 presented high ability for adhesion (42.83 ± 1.79%), which was not affected by co-culture with L. lactis MC5 (Table 2). However, significant differences (p < 0.01) in the adhesion capacity of S. aureus were observed in the presence of CFS of L. lactis MC5 at higher concentrations (50%, 75% and 90%), as shown in Table 3.
Table 2.
Adhesion to intestinal cells (Caco-2) by the cheese-isolate Staphylococcus aureus SABRC1 alone or co-inoculated with Lactococcus lactis MC5
| Condition* | Adhesion (%) |
|---|---|
| S. aureus | 42.83 ± 1.79 |
| S. aureus + L. lactis | 41.69 ± 1.47a |
*Baird-Parker agar was used to count staphylococci selectively
aNo significant difference in comparison with control (p > 0.01)
Table 3.
Adhesion of Staphylococcus aureusSABRC1 to intestinal cells (Caco-2), in the presence of Cell Free Supernatant (CFS) of Lactococcus lactis MC5
| Conditions | Adhesion (%) |
|---|---|
| Control (DMEM) | 43.94 ± 0.10 |
| 10% CFS | 41.80 ± 2.94a |
| 25% CFS | 43.21 ± 1.14a |
| 50% CFS | 37.82 ± 1.52b |
| 75% CFS | 28.78 ± 1.28b |
| 90% CFS | 33.39 ± 0.39b |
aNo significant difference in the comparison with the control (p > 0.1)
bSignificant difference in comparison with control (p < 0.1)
Concerning the ability of S. aureus SABRC1 to invade Caco-2 cells (Table 4), the rate was 48.57 ± 0.41% for single culture, but when staphylococci were co-inoculated with L. lactis MC5, the invasiveness decreased to 31.20 ± 0.17% (p<0.05). Figure 1 shows Caco-2 cells invaded by S. aureus SABRC1, stained with fluorescent dyes and observed by CLSM. Moreover, it was observed the capacity of S. aureus SABRC1 to invade Caco-2 cells was significantly reduced (p<0.01) in the presence of CFS from L. lactis MC5, to the point that in the concentrations of 75% and 90% CFS, staphylococci were not invasive (Table 5).
Table 4.
Invasion of intestinal cells (Caco-2) by the cheese-isolate Staphylococcus aureus SABRC1 alone or co-inoculated with Lactococcus lactis MC5
| Condition* | Invasion (%) |
|---|---|
| S. aureus | 48.57 ± 0.41 |
| S. aureus + L. lactis | 31.20 ± 0.17a |
*Baird-Parker agar was used to count staphylococci
aSignificant difference in comparison with control (p < 0.05)
Fig. 1.

Microphotography illustrating results from invasion assays of intestinal cells (Caco-2) by Staphylococcus aureus SABRC1, stained with Live/Dead viability assay kit (Thermo Fisher, USA) and observed with a confocal laser scanning microscope (Leica TCS-SP5 AOBS, software LAS-AF)
Table 5.
Invasion of intestinal cells (Caco-2) by Staphylococcus aureus SABRC1 in the presence or in the absence of Cell Free Supernatant (CFS) of Lactococcus lactis MC5
| Condition | Adhesion (%) |
|---|---|
| Control (DMEM) | 48.85 ± 1.45 |
| CFS 10% | 39.01 ± 1.19a |
| CFS 25% | 33.43 ± 1.08a |
| CFS 50% | 23.14 ± 1.47a |
| CFS 75% | ND* |
| CFS 90% | ND* |
aSignificant differences in comparison with the control (p < 0.01)
*Not determined (bacterial count below the quantification limit of 10 CFU per plate)
Results of quantitative RT-PCR experiments revealed the influence of common diet components on the expression of the virulence genes hla, icaA and sea in S. aureus SABRC1. With regard to hla (Fig. 2) there was a significant decrease of expression in the conditions BHI + Glucose (p < 0.0001), BHI + Glucose + NaCl (p < 0.0001), Milk (p < 0.0001), Milk Powdered (p < 0.01) and “Fresh Minas Cheese” medium (p < 0.01), compared to BHI only.
Fig. 2.

Expression of hla gene in Staphylococcus aureus SABRC1, measured by quantitative Reverse Transcriptase PCR assay, based on Livak and Schmittgen (2001).(i) Brain Heart Infusion broth—BHI (control); (ii) BHI + 1% Glucose; (iii) BHI + 1% Conditions tested for bacterial growth: Glucose + 4% NaCl; (iv) Milk medium; (v) Milk Powder medium; (vi) “Fresh Minas Cheese” medium. Each determination was done in triplicate, and statistics (t test) is shown as: **p < 0.01, ***p < 0.0001, ****p < 0.0001
Although no significant difference was observed for the expression of icaA in S. aureus SABRC1 cultured in “Fresh Minas Cheese” medium in comparison with the BHI control (Fig. 3), this gene was downregulated in BHI + Glucose (p < 0.0001), BHI + Glucose + NaCl (p < 0.0001), Milk (p < 0.0001) and Powdered Milk (p < 0.05).
Fig. 3.
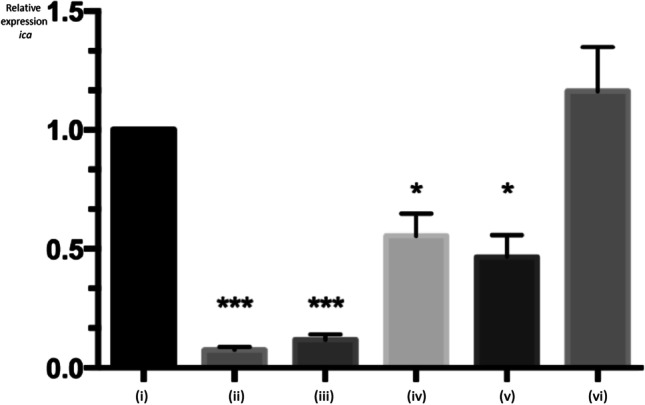
Expression of icaA gene in Staphylococcus aureus SABRC1, measured by quantitative Reverse Transcriptase PCR assay, based on Livak and Schmittgen (2001). Conditions tested for bacterial growth: (i) Brain Heart Infusion broth—BHI (control); (ii) BHI + 1% Glucose; (iii) BHI + 1% Conditions tested for bacterial growth: Glucose + 4% NaCl; (iv) Milk medium; (v) Milk Powder medium; (vi) “Fresh Minas Cheese” medium. Each determination was done in triplicate, and statistics (t test) is shown as: *p < 0.05, ***p < 0.0001
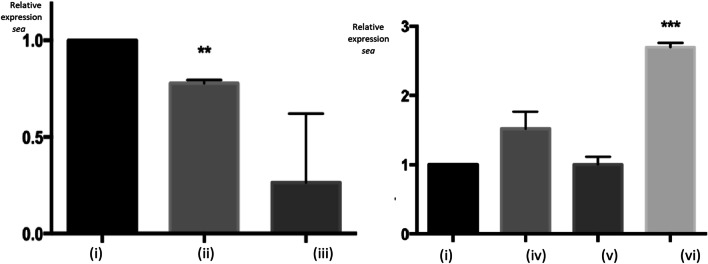
The expression of the gene sea was also studied for S. aureus SABRC1, and the results are shown in Fig. 4. There were no statistically significant differences in the cultures obtained in BHI + Glucose + NaCl (p > 0.01), Milk (p > 0.0001) and Milk Powdered (p > 0.0001). However, it was interesting to note the downregulation of sea in BHI + Glucose (p < 0.01), in contrast with its upregulation in “Fresh Minas Cheese” medium (p < 0.0001), compared to culture in regular BHI.
Fig. 4.
Expression of sea gene in Staphylococcus aureus SABRC1, measured by quantitative Reverse Transcriptase PCR assay, based on Livak and Schmittgen (2001). Conditions tested for bacterial growth—panel A:(i) Brain Heart Infusion broth—BHI (control); (ii) BHI + 1% Glucose; (iii) BHI + 1% Glucose + 4% NaCl. Conditions tested for bacterial growth—panel B: (i) Brain Heart Infusion broth—BHI (control); (iv) Milk medium; (v) Milk Powder medium; (vi) “Fresh Minas Cheese” medium. Each determination was done in triplicate, and statistics (t test) is shown as: **p < 0.01 and ***p < 0.0001
Discussion
The intestinal carriage of S. aureus is not very common; nonetheless, it is worrying because it may contribute to the bacterial spread into the environment and to increase the risk of infections, as well as it may represent a reservoir of antibiotic resistance genes [35, 36]. Thus, strategies to avoid infections by S. aureus are very important, especially those alternatives to the prescription of antibiotics. In this sense, it is an innovative approach to use beneficial microbes to counteract the adhesion and invasion of intestinal cells by staphylococci.
In this study, the results of adhesion assays (Table 2) showed S. aureus SABRC1 adhered to Caco-2 cells, and the adhesion rate was not affected by the presence of L. lactis MC5 (Table 3). However, the adhesion of S. aureus SABRC1 to Caco-2 cells was diminished in presence of CFS of L. lactis MC5 (Table 4). According to recent literature [37], L. lactis may antagonize pathogens by the production of antimicrobial components, competition for nutrients and for binding sites, besides the modulation of host immune response. One possible explanation for the decreased adhesion rate observed for S. aureus SABRC1 to Caco-2 cells in the presence of CFS from L. lactis MC5 is the direct action of BLIS on the bacterial cells.
It was also observed in this research L. lactis MC5 was able to inhibit the invasion of Caco-2 cells by S. aureus (Table 4), similar to the effect of CFS from the LAB, mainly at higher concentrations (Table 5). According to the literature [38, 39], the virulence factors of S. aureus include several cell wall-anchored proteins (CWA), and the majority of strains expresses two CWA fibronectin-binding adhesins that promote cell invasion. Interestingly, it has also been reported the bacteriocin nisin produced by L. lactis may modulate the expression of fibronectin-binding proteins involved in cell invasion [40], which may partially explain the decreased ability of S. aureus SABRC1 to invade Caco-2 cells in the presence of L. lactis MC5 or its CFS.
Moreover, the virulence of S. aureus can be directly influenced by culture conditions, with variable expression of genes such as hla, icaA and sea [41, 42]. In this study, the expression of virulence genes was evaluated using several culture media, and the results are shown in Figs. 2, 3 and 4. The genes hla, icaA and sea were downregulated in S. aureus SABRC1 grown in BHI + Glucose or BHI + Glucose + NaCl, in comparison with regular BHI. For S. aureus and other Gram-positive pathogens, it has been reported glucose can inhibit toxin production [43–45], which is partially in agreement with the results from this research. However, sea gene was upregulated in “Fresh Minas Cheese” medium (Fig. 4), corroborating results from Ferreira et al. [46] who reported sea was detected among the enterotoxin-related genes in S. aureus from artisanal cheeses, which are products usually linked to foodborne outbreaks [47].
Taken altogether, the results from this in vitro study showed S. aureus is able to adhere and to invade epithelial cells, but the presence of LAB and/or BLIS may interfere with the infection process. Moreover, it was demonstrated the composition of the culture media greatly influences the expression of virulence genes in S. aureus highlighting the need to take these into account for the correct assessment of the virulent potential of S. aureus.
Acknowledgements
MGP especially acknowledges CAPES for a post-doctoral fellowship (PNPD). ECPDM is a research fellow of the National Council for Scientific and Technological Development (CNPq)—Brazil and FCNA received a Master fellowship from CNPq.
Author contribution
All authors contributed to the development of the study. Material preparation, data collection and analysis were performed by FCNA and MGP. VFA collaborated by discussing experiments and drafting the manuscript. ECPDM coordinated a financial grant for this project, designed and supervised the microbiological tests. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
The authors are grateful to São Paulo Research Foundation—Brazil for a research grant (FAPESP 2012/50507–1), to the Multiuser Laboratory of Confocal Microscopy—LMMC (FAPESP 2004/08868–0) and also to Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brazil (CAPES; finance code 001).
Data availability
The data that support the findings of this study are available from the corresponding author upon request.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cheung GYC, Bae JS, Otto M. Pathogenicity and virulence of Staphylococcus aureus. Virulence. 2021;12(1):547–569. doi: 10.1080/21505594.2021.1878688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silva-Santana G, Cabral-Oliveira GG, Oliveira DR, Nogueira BA, Pereira-Ribeiro PMA, Mattos-Guaraldi AL. Staphylococcus aureus biofilms: an opportunistic pathogen with multidrug resistance. Rev Med Microbiol. 2021;32(1):12–21. doi: 10.1097/MRM.0000000000000223. [DOI] [Google Scholar]
- 3.Pérez-Montarelo D, Viedma E, Murcia M, Muñoz-Gallego I, Larrosa N, Brañas P, Fernández-Hidalgo N, Gavaldà J, Almirante B, Chaves F. Pathogenic characteristics of Staphylococcus aureus endovascular infection isolates from different clonal complexes. Front Microbiol. 2017;19(8):917. doi: 10.3389/fmicb.2017.00917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pivard M, Moreau K, Vandenesch F. Staphylococcus aureus arsenal to conquer the lower respiratory tract. mSphere. 2021;6(3):e00059-21. doi: 10.1128/mSphere.00059-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.King J, Kulhankova K, Christopher S, Bao V, Salgado-Pabón W. Phenotypes and virulence among Staphylococcus aureus USA100, USA200, USA300, USA400, and USA600 clonal lineages. Cell Host Microbe. 2016;1(3):e00071-16. doi: 10.1128/mSphere.00071-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dittman KK, Chaul LT, Lee S, Oxaran V, Corassin CH, Oliveira CAF, De Martinis ECP, Alves VF, Gram L. Staphylococcus aureus in some Brazilian dairy industries: changes of contamination and diversity. Front Microbiol. 2017;24(8):2049. doi: 10.3389/fmicb.2017.020497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alba P, Feltrin F, Cordaro G, Porrero MC, Kraushaar B, Argudín MA, Nykäsenoja S, Monaco M, Stegger M, Aarestrup FM, Butaye P, Franco A, Battisti A. Livestock-associated methicillin resistant and methicillin susceptible Staphylococcus aureus Sequence Type (CC)1 in European farmed animals: high genetic relatedness of isolates from Italian cattle herds and humans. PLoS One. 2015;10(8):e0137143. doi: 10.1371/journal.pone.0137143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ilczyszyn WM, Sabat AJ, Akkerboom V, Szkarlat A, Klepacka J, Sowa-Sierant I, Wasik B, Kosecka-Strojek M, Buda A, Miedzobrodzki J, Friedrich AW. Clonal structure and characterization of Staphylococcus aureus strains from invasive infections in paediatric patients from south Poland: association between age, spa types, clonal complexes, and genetic markers. PLoS One. 2016;11(3):e0151937. doi: 10.1371/journal.pone.0151937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Acton DS, Plat-Sinnige MJ, van Wamel W, de Groot N, van Belkum A. Intestinal carriage of Staphylococcus aureus: how does its frequency compare with that of nasal carriage and what is its clinical impact? Eur J Clin Microbiol Infect Dis. 2009;28(2):115–127. doi: 10.1007/s10096-008-0602-7. [DOI] [PubMed] [Google Scholar]
- 10.Misawa Y, Kelley KA, Wang X, Wang L, Park WB, Birtel J, Saslowsky D, Lee JC. Staphylococcus aureus colonization of the mouse gastrointestinal tract is modulated by wall teichoic acid, capsule, and surface proteins. PLoS Pathog. 2015;11(7):e1005061. doi: 10.1371/journal.ppat.1005061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jayashree S, Karthikeyan R, Nithyalakshmi S, Ranjani J, Gunasekaran P, Rajendhran J. Anti-adhesion property of the potential probiotic strain Lactobacillus fermentum 8711 against methicillin-resistant Staphylococcus aureus (MRSA) Front Microbiol. 2018;8(9):411. doi: 10.3389/fmicb.2018.00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouchard DS, Rault L, Berkova N, Le Loir Y, Even S. Inhibition of Staphylococcus aureus invasion into bovine mammary epithelial cells by contact with live Lactobacillus casei. Appl Environ Microbiol. 2013;79(3):877–885. doi: 10.1128/AEM.03323-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim S, Lee J, Lee S, Ha J, Lee J, Choi Y, Oh H, Yoon Y, Kyoung-Hee C. Invasion of intestinal cells by Staphylococcus aureus is mediated by pyruvate formate lyase protein (PFLB) J Pure Appl Microbiol. 2019;13(2):647–652 . doi: 10.22207/JPAM.13.2.01. [DOI] [Google Scholar]
- 14.Hill C, Guarner F, Reid G, Gibson G, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, Calder PC, Sanders ME. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11(8):506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 15.Nowak A, Motyl I (2017) In vitro anti-adherence effect of probiotic Lactobacillus strains on human enteropathogens. Biotech Food Sci. 81(2): 103–112. http://212.51.210.149/handle/11652/1620
- 16.Pato U, Riftyan E, Ayu DF, Jonnaidi NN, Wahyuni MS, Feruni JA, Abdel-Wahhab, Antibacterial efficacy of lactic acid bacteria and bacteriocin isolated from Dadih’s against Staphylococcus aureus. Food Sci Technol. 2022;42:e27121. doi: 10.1590/fst.27121. [DOI] [Google Scholar]
- 17.Newstead LL, Varjonen K, Nuttall T, Paterson GK. Staphylococcal-produced bacteriocins and antimicrobial peptides: their potential as alternative treatments for Staphylococcus aureus infections. Antibiotics (Basel) 2020;9(2):40. doi: 10.3390/antibiotics9020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yaacob SN, Wahab RA, Misson M, Sabullah MK, Huyop F, Zin NM. Lactic acid bacteria and their bacteriocins: new potential weapons in the fight against methicillin-resistant Staphylococcus aureus. Future Microbiol. 2022;17:683–699. doi: 10.2217/fmb-2021-0256. [DOI] [PubMed] [Google Scholar]
- 19.Alves VF, Nino-Arias FC, Pitondo-Silva A, Frazilio DA, Gonçalves LO, Toubas LC, Torres IMS, Oxaran V, Dittmann KK, De Martinis ECP. Molecular characterisation of Staphylococcus aureus from some artisanal Brazilian dairies. Int Dairy J. 2018;85(10):247–253. doi: 10.1016/j.idairyj.2018.06.008. [DOI] [Google Scholar]
- 20.Lewus CB, Montville TJ. Detection of bacteriocins produced by lactic acid bacteria. J Microbiol Meth. 1991;13(2):145–150. doi: 10.1016/0167-7012(91)90014-H. [DOI] [Google Scholar]
- 21.De Martinis ECP, Públio MRP, Santarosa PR, Freitas FZ. Antilisterial activity of lactic acid bacteria isolated from vacuum-packaged Brazilian meat and meat products. Braz J Microbiol. 2001;32:32–37. doi: 10.1590/S1517-83822001000100008. [DOI] [Google Scholar]
- 22.Devereux R, Wilkinson SS. Amplification of ribosomal RNA sequences. In: Akkermans ADL, editor. Molecular microbial ecology manual. Dordrecht: Kluwer Academic; 2004. pp. 509–522. [Google Scholar]
- 23.Moroni O, Kheadr E, Boutin Y, Lacroix C, Fliss I. Inactivation of adhesion and invasion of food-borne Listeria monocytogenes by bacteriocin-producing Bifidobacterium strains of human origin. Appl Environ Microbiol. 2006;72:11. doi: 10.1128/AEM.00928-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merghni A, Nejma MB, Helali I, Hentati H, Bongiovanni A, Lafont F, Aouni M, Mastouri M. Assessment of adhesion, invasion and cytotoxicity potential of oral Staphylococcus aureus strains. Microb Pathog. 2015;86:1–9. doi: 10.1016/j.micpath.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 25.Mayr-Harting A, Hedges AJ, Berkeley RCW. Methods in microbiology. London: Academic Press; 1972. Methods for studying bacteriocins; pp. 315–342. [Google Scholar]
- 26.Ping J, Abercrombie N, Jeffrey K, Leung P. An improved medium for growing Staphylococcus aureus biofilm. J Microbiol Methods. 2012;90(2):115–118. doi: 10.1016/j.mimet.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 27.Snel GGM, Malvisi M, Pilla R, Piccinini R. Evaluation of biofilm formation using milk in a flow cell model and microarray characterization of Staphylococcus aureus strains from bovine mastites. Vet Microbiol. 2014;174(3–4):489–495. doi: 10.1016/j.vetmic.2014.09.020. [DOI] [PubMed] [Google Scholar]
- 28.Ikeda T, Yamaguchi K, Makino S. Quantitative analysis of Staphylococcus aureus in skimmed milk powder by real time PCR. J Vet Med Sci. 2005;67(10):1037–1041. doi: 10.1292/jvms.67.1037. [DOI] [PubMed] [Google Scholar]
- 29.Furtado MM, Neto JPML. Manual técnico para a produção industrial de queijos. Dipermar, São Paulo: Tecnologia de queijos; 1994. pp. 1–118. [Google Scholar]
- 30.Silva FT (2005) Queijo Minas Frescal. In: Coleção Agroindústria Familiar—Empresa Brasileira de Pesquisa Agropecuária—Informação Tecnológica. Embrapa, Brasília - DF, pp. 13–30. https://ainfo.cnptia.embrapa.br/digital/bitstream/item/11884/2/00076200.pdf. Acessed 12 July 2022
- 31.Oogai Y, Matsuo M, Hashimoto M, Kato F, Sugai M, Komatsuzawa H. Expression of virulence factors in Staphylococcus aureus grown in serum. Appl Environ Microbiol. 2011;77(22):8097–8105. doi: 10.1128/AEM.05316-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar G, Negi Y, Gaur A, Khanna D. Detection of virulence genes in Staphylococcus aureus isolated from paper currency. Int J Infect Dis. 2009;13(6):e450–e455. doi: 10.1016/j.ijid.2009.02.020. [DOI] [PubMed] [Google Scholar]
- 33.Eleaume H, Jabbouri S. Comparison of two standarsation methods in real-time quantitative RT-PCR to follow Staphylococcus aureus genes expression during in vitro growth. J Microbiol Meth. 2004;59(3):363–370. doi: 10.1016/j.mimet.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Fukuta Y, Cunningham CA, Harris PL, Wagener MM, Muder RR. Identifying the risk factors for hospital-acquired methicillin-resistant Staphylococcus aureus (MRSA) infection among patients colonized with MRSA on admission. Infect Control Hosp Epidemiol. 2012;33(12):1219–1225. doi: 10.1086/668420. [DOI] [PubMed] [Google Scholar]
- 36.Rolain J. Food and human gut as reservoirs of transferable antibiotic resistance encoding genes. Front Microbiol. 2013;24(4):173. doi: 10.3389/fmicb.2013.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vieco-Saiz N, Belguesmia Y, Raspoet R, Auclair E, Gancel F, Kempf I, Drider D. Benefits and inputs from lactic acid bacteria and their bacteriocins as alternatives to antibiotic growth promoters during food-animal production. Front Microbiol. 2019;11(10):57. doi: 10.3389/fmicb.2019.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schaffner F, Ray A, Dontenwill M. Integrin α5β1, the fibronectinreceptor, as a pertinent therapeutic target in solid tumors. Cancers (Basel) 2013;5(1):27–47. doi: 10.3390/cancers5010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Speziale P, Pietrocola G. The multivalent role of fibronectin-binding proteins A and B (FnBPA and FnBPB) of Staphylococcus aureus in host infections. Front Microbiol. 2020;26(11):2054. doi: 10.3389/fmicb.2020.02054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edwards AM, Potts JR, Josefsson E, Massey RC. Staphylococcus aureus host cell invasion and virulence in sepsis is facilitated by the multiple repeats within FnBPA. Plos Pathog. 2010;6(6):e-1000964. doi: 10.1371/journal.ppat.1000964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.André MCDPB, Campos L, Borges A, Kipnis F, Pimenta A, Serafini B. Comparison of Staphylococcus aureus isolates from food handlers, raw bovine milk and Minas Frescal cheese by antibiogram and pulsed-field gel electrophoresis following SmaI digestion. Food Control. 2008;19(2):200–207. doi: 10.1016/j.foodcont.2007.03.010. [DOI] [Google Scholar]
- 42.Bar-Gal G, KahilaBlum SE, Hadas L, Ehricht R, Monecke S, Leitner G. Host-specificity of Staphylococcus aureus causing intramammary infections in dairy animals assessed by genotyping and virulence genes. Vet Microbiol. 2015;176(1–2):143–54. doi: 10.1016/j.vetmic.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 43.Smith J, Bencivengo M, Buchanan R, Kunsch C. Enterotoxin A production in Staphylococcus aureus: inhibition by glucose. Arch Microbiol. 1986;144(2):131–136. doi: 10.1007/BF00414722. [DOI] [PubMed] [Google Scholar]
- 44.Méndez MB, Goñi A, Ramirez W, Grau R. Sugar inhibits the production of the toxins that trigger clostridial gas gangrene. Microb Pathog. 2012;52(1):85–91. doi: 10.1016/j.micpath.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 45.Yang SC, Lin CH, Sung CT, Fang JY. Antibacterial activities of bacteriocins: application in foods and pharmaceuticals. Front Microbiol. 2014;26(5):241. doi: 10.3389/fmicb.2014.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferreira GB, Oliveira ACS, Marson JM, Terra APS. Research of Staphylococcus aureus in “Minas Frescal” Cheese marketed in the region of Triângulo Mineiro. Rev Baiana Saúde Pública. 2010;34(3):575–589. doi: 10.22278/2318-2660.2010.v34.n3.a57. [DOI] [Google Scholar]
- 47.Argudín M, Mendoza M, Rodicio M. Food poisoning and Staphylococcus aureus enterotoxins. Toxins. 2010;2:1751–1773. doi: 10.3390/toxins2071751. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request.