Abstract
Turkish White Cheese is a brined (or pickled) cheese with a salty, acidic flavor and a soft or semi-hard texture. It is the most produced and consumed type of cheese in Turkey. The purpose of this study was to determine the non-starter lactic acid bacteria and yeast microbiota of traditionally produced Turkish White Cheese and analyze the chemical properties and the aroma profile of the cheese. The results of the study identified 27 distinct strains belonging to 14 the non-starter lactic acid bacteria species and 49 different strains belonging to 11 yeast species. Lactobacillus plantarum was found to be the dominant species among the lactic acid bacteria, while Candida zeylanoides was the dominant yeast species in the White Cheese samples. In addition, Kluyveromyces lactis and Debaryomyces hansenii were prominent yeast species in cheese samples. Turkish White Cheese samples had different aromatic properties. The study is highly significant as it anaylzed both non-starter lactic acid bacteria and yeast microbiota of traditionally produced Turkish White Cheese through molecular methods. It also determined and analyzed a number of chemical and aromatic properties of White Cheese.
Keywords: White Cheese, Microbiota, Genotypic characterization, Aroma compounds
Introduction
White Cheese is the most produced and consumed type of cheese in Turkey and accounts for 60–80% of the total cheese production [1–3]. This cheese is produced in many countries around the world and is known by different names. For example, it is called as Feta in the Mediterranean region, Denmark, and Greece; Bjalo Salamureno Sirene in Bulgaria; Domiati in Egypt; Brinza in Israel; Teleme in Romania; and Queso Blanco in the USA [1, 4]
Although small dairy farms mostly produce White Cheese from raw milk and without the use of starter culture [1, 5] modern enterprises that make production using pasteurized milk and starter culture have emerged recently [6, 7]. However, the lack of industrial-scale starter culture production in Turkey has led cheese producers to be dependent on other countries. It has been observed that the imported starter cultures that are used fail to provide the characteristic taste, aroma, or textural qualities of industrial cheeses and also cause the cheese to ripen quickly and thus become unusable [6, 8, 9].
The industrial production of traditional dairy products is mainly possible through the correct determination of the microbiota of the traditional product. In order to identify and characterize microorganisms, classical methods with morphological, biochemical, and physiological characteristics and antibiotic susceptibility and serological typing of these microorganisms were initially used. However, with the development of molecular techniques that provide higher sensitivity identification at the strain level, a new phase has emerged. Each of these methods is aimed at classifying, describing, and characterizing microbial isolates at the level of genus, species, subspecies, and strain [10].
Dagdemir and Ozdemir [7] identified 24.43% of the microbiota as Enterococcus faecalis, 17.61% as Enterococcus faecium, and 19.88% as Lactobacillus fermentum in the identification study of lactic acid bacteria by analyzing fatty acid methyl esters in thirty White Cheese samples produced by the traditional method. Biochemical identification methods and Gram-positive ID test kits were applied to 77 strains isolated from seven White Cheese samples made from raw milk by Ertürkmen and Öner [11]. As a result of the analysis, 19 of the total isolates were Lactococcus lactis subsp. lactis, 4 Lactococcus lactis subsp. cremoris, 5 Enterococcus faecalis, 2 Enterococcus durans, 2 Enterococcus avium, 4 Pediococcus pentosaceus, 2 Enterococcus faecium, and 1 Enterococcus solitorius. Fourteen strains could not be identified with Gram-positive ID test kits. Karakuş et al. [12] identified a total of 348 bacterial strains isolated at the beginning and end of the 3-month ripening period from White Cheese produced without using starter culture in three different enterprises. The findings revealed that a great majority of the lactic flora at the beginning of ripening in cheeses is Lactococcus lactis subsp. lactis and Enterococcus species (E. faecalis and E. faecium) constitute the second-degree important group. Species belonging to the Lactobacillus and Leuconostoc genera of other lactic acid bacteria were also detected at low rates, and the identified species were Lactobacillus casei, L. plantarum, L. fermentum, L. brevis, Leuconostoc lactis, and L. mesenteroides subsp. dextranicum. While the lactococci ratio in the lactic flora decreased in the mature cheese samples, the Lactobacilli ratio increased. It was concluded that the dominant species in this group of bacteria were L. casei and L. plantarum.
Salum et al. [13] determined a total of 45 volatile components as a result of the volatile component analysis of the White Cheese samples obtained from various regions of Turkey and stated that the cheese samples had significant variations in terms of aroma properties.
The studies on White Cheese in Turkey have mostly focused on its physical and chemical properties. The studies on its microbiota and aromatic properties are insufficient. It is observed that Turkish White Cheese, produced traditionally in line with the literature, has different microbiological and sensory properties. The aim of this study is to contribute to the best possible characterization of White Cheese with the aim of identifying the most suitable starting material for industrial and standardized production.
Materials and methods
Production of White Cheese and sample collection
In the traditional production of Turkish White Cheese, pasteurized or unpasteurized milk can be used and no starter culture is added to the milk. First, the milk for cheese is strained; then, it is heated to fermentation temperatures in the fermentation vessel. Rennet is added to the extent that it will coagulate in 150 min. After the curd is formed, it is cut into pieces of 2 cm3, wrapped in gauze, and waited for 5–10 min to release the excess whey. The cheese curd is pressed with a 20-kg load on 100 kg of cheese until the whey stops dripping. After the weights are removed, the cheese mass is cut into 7 × 7 × 7 cm3 blocks and kept in brine (around 14%) for 2 h. Salted blocks are rested in cans in a row for 24 h. In this way, the boxes are filled with 17 kg of cheese in 4 days. Each layer is salted, and then, the cans are filled with a saline solution (around 10%) [1, 14].
The study utilized white Cheese samples that are produced by traditional methods and obtained from 10 different enterprises (n = 10) in 4 different provinces (Erzurum, Bayburt, Gümüşhane, Trabzon) of Turkey.
Chemical and microbiological analysis
Cheese samples were analyzed for dry matters, fat (Gerber method), ash, pH and salt (Mohr method), and titratable acidity (as lactic acid) in accordance with the methods described by Kurt et al. [15]. It was performed by calculating the ratio of salt and fat in dry matter. Free fatty acidity was determined according to ISO 660 [16].
Total aerobic mesophilic bacteria (TAMB) and yeast counts were reported according to Harrigan [17] and to Šuranská et al. [18] respectively, and lactococci and lactobacilli were counted according to Speck [19].
All chemical and microbiological analyses were carried out in parallel and the results were averaged.
LAB isolation
LAB isolation 10g cheese samples were homogenized in 90 ml of 2% sodium citrate solution using a stomacher. Serial dilutions (100–10−6) were run with PBS to a dilution factor of 10−6 and aliquots of these dilutions were plated to MRS (de Man, Rogosa and Sharpe, Merck, Germany) containing 0.1 g of cycloheximide per liter, BHI (Brain-heart infusion, Merck, Germany), and M17 agars (Merck, Germany) and incubated at 37 °C and 42 °C for 48 h. At the end of the incubation period, colonies with potential different morphologies were randomly taken from agar plates and grown under the same incubation conditions on both media and then tested for Gram stain, cell morphology, and catalase reaction. A total of 90 LAB isolates were further analyzed for their genotypic characterization. Stock solutions of LAB isolates were prepared in 20% (v/v) glycerol and stored at − 80 °C.
Isolation of yeast
For yeast isolation, aliquots of different dilutions were prepared as described above and spread on YPG (yeast extract-peptone-glucose, Oxoid, UK) agar plates. Plates were incubated at 25 °C for 3–5 days. After the growth, morphologically distinct colonies were selected and grown in Malt Extract Broth (Oxoid, UK) at 25 °C for 3 days [20]. Then, stock solutions of 80 yeast isolates were prepared with 20% (v/v) glycerol and stored at − 80 °C.
Genomic DNA isolation and genotypic characterization by RAPD-PCR and rep-PCR analysis
Genomic DNA isolation was performed through phenol chloroform method [21]. RAPD-PCR and rep-PCR analyses were carried out to perform a preliminary discrimination from NSLAB (non-starter lactic acid bacteria) and yeast species. Primer M13 (GAGGGTGGCGGTTCT) and primer (GTG)5 (5′-GTGGTGGTGGTGGTG-3′) were used for RAPD-PCR and rep-PCR mixes respectively; PCR and electrophoresis conditions were carried out in line with the method given in Yuvaşen et al. [22]. All primer sets were purchased commercially from Metabion (Martinsried, Germany).
PCR amplification of the 16S rRNA region
After RAPD-PCR analysis, 16S rRNA gene PCR amplification was applied on colonies that were found to be genotypic. Universal primers AMP F (5′-GAGAGT TTGATYCTGGCTCAG-3′) and AMP R (3′-AAGGAGGTGATCCARCCGCA-5′) were used to amplify the c.1.5 kb 16S rRNA gene of LAB strains. PCR mixes were prepared with 1 μl DNA template from genomic DNA, 10 μl 5× PCR buffer for Taq polymerase, 1 μl of 20 mM primers AMP_F and AMP_R, 0.25 μl 5UTaq polymerase, and 0.4 μl dNTPs up to 50 μl of sterile H2O. PCR program consisted of 1 cycle denaturation at 95 °C for 2 min, 25 cycles: denaturation at 95 °C for 30 s, annealing at 55 °C for 20 s, polymerization at 72 °C for 30 s, and final polymerization at 72 °C for 5 min. All primer sets were purchased commercially from Metabion (Martinsried, Germany)
PCR amplification of the 26S rDNA region
After genomic DNA isolation, amplification of the D1/D2 region of the 26S rDNA gene was performed for yeast identification. Primers NL1 (5′-GCATATCAATAAGCGGAGGAAAAAG-3′) and NL4 (5′GGTCCGTGTTTCAAGACGG-3′) were used to perform this amplification. PCR mixes were prepared as described above using genomic DNAs of the corresponding yeast strains as templates and NL1 and NL4 as primer sets. The PCR process was conducted under specific conditions. It began with an initial denaturation step at 95 °C for a duration of 10 min. This was followed by 40 cycles, each consisting of a denaturation step at 94 °C for 1 min, an annealing step at 54 °C for 2 min, and finally an extension step at 72 °C for 7 min. These steps were performed to amplify the targeted 600-bp region. All primer sets were purchased commercially from Metabion (Martinsried, Germany)
After the PCR products were checked by gel electrophoresis, they were sequenced by sequencing service Medsantek (Istanbul, Turkey). Taxonomic identification of isolates was performed through the comparison of the obtained 16S rRNA gene sequences with the NCBI database using the BLAST algorithm with a similarity criterion of 98–100%.
Nucleotide access numbers and phylogenetic analyses
16S rRNA gene sequences of 27 lactic acid bacteria identified in this study were deposited in GenBank under accession number MT665007-MT669366 (27) and 26S rRNA gene sequences of 49 yeasts under accession number MT674912-MT808410.
Phylogenetic trees were constructed using neighbor-joining (NJ) method with 1000 bootstrap replicates. All phylogenetic analyses were performed using MEGA4.
Volatile-compound profiling of White Cheese by GC-MS analysis
The volatile compounds of 10 traditionally produced Turkish White Cheese were determined using GC-MS (GC-2010; Shimadzu Corporation, Kyoto, Japan) and headspace-SPME (solid phase microextraction) method. After 3 g of cheese sample was placed in a 20-ml SPME vial with silicone septum, 80 μl of internal standard (2-methyl-3-pentanone with a concentration of 20 ppm) was added. For the absorption of volatile compounds, SPME fiber (2 cm 50/30 mm Supelco, Bellefonte, USA) was immersed in a water bath at 45 °C for 30 min. The fiber was withdrawn and injected into the GC inlet and desorbed at 250 °C for 3 min. TR-WaxMS column (60 m length × 0.25 mm inner diameter × 0.25 μm film thickness, Thermo Fisher Scientific Bellefonte PA, USA) was used for analysis. The carrier gas was helium (1 ml/min) and mass range 35–350 m/z (mass/charge). The temperature program involved initially waiting at 40 °C for 10 min, followed by a gradual increase to 250 °C at a rate of 5 °C per minute, and finally maintaining this temperature for 10 min. Ion source temperature and transfer line were set as 260 °C (Eghbalian, 2017). The Wiley and NIST GC-MS library and external standard were used for the identification process. The retention index of peaks was calculated by using the C8-C20 alkane standard (Supelco, Bellefonte, PA, USA). The results were obtained as peak area of the volatile component/peak area of the internal standard.
Statistical analysis
The experimental data was analyzed through the SPSS statistical software program version 22 (SPSS Inc., Chicago, IL, USA), and Duncan’s multiple range tests were employed to determine differences between results.
Results and discussion
Chemical and microbiological analyses
Table 1 indicates the values of some microbiological and chemical analysis results of Turkish White Cheese. It was revealed that the pH values of the cheese samples were between 4.17 and 6.38, acidity 0.27–1.80, ash 3.96–12.27%, dry matter 42.20–58.1%, fat 13.0–26.5%, salt 2.63–8.03%, fat in dry matter 24.71–55.18%, salt in dry matter 4.99–16.72%, and free fatty acidity (FFA) 5.74–32.30% of oleic acid (Table 1). The number of Lactobacillus (or lactobacilli), Lactococcus (or lactococci), TMAB, and yeast was determined in the ranges of 5.57–8.49, 6.21–9.74, 5.88–10.52, and 4.49–8.49 log cfu/g, respectively. The statistical analysis results indicated that the cheese samples differed significantly (p < 0.01) from each other in terms of all microbiological and chemical properties given in Table 1.
Table 1.
Some microbiological and chemical analysis results of Turkish White Cheeses
| White Cheese samples | pH | Acidity (%) | Ash (%) | DM (%) | Fat (%) | Salt (%) | Fat in DM (%) | Salt in DM (%) | FFA (% oleic acid) | LB log cfu/g |
LC log cfu/g |
TAMB log cfu/g |
Yeast log cfu/g |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 5.24 ± 0.01e | 0.50 ± 0.05de | 9.78 ± 0.65cd | 55.70 ± 0.22b | 24.0 ± 0.71b | 6.20 ± 0.17b | 43.09 ± 0.10de | 11.13 ± 0.07f | 17.52 ± 0.22d | 7.43 ± 0.06b | 7.65 ± 0.19c | 8.23 ± 0.20bc | 7.81 ± 0.13c |
| 2 | 5.96 ± 0.00c | 0.27 ± 0.03g | 7.41 ± 0.40f | 42.20 ± 1.31h | 19.0 ± 0.00d | 5.20 ± 0.33c | 45.02 ± 0.24c | 12.32 ± 0.03e | 19.86 ± 0.08b | 7.18 ± 0.04cd | 7.75 ± 0.24c | 8.00 ± 0.17c | 8.35 ± 0.13ab |
| 3 | 6.09 ± 0.01b | 0.45 ± 0.22def | 8.10 ± 0.62e | 49.00 ± 0.10ef | 25.0 ± 0.00b | 4.56 ± 0.49d | 51.02 ± 0.00b | 9.30 ± 0.03h | 12.73 ± 0.07h | 6.66 ± 0.14e | 8.19 ± 0.16b | 7.94 ± 0.10c | 7.48 ± 0.08d |
| 4 | 6.38 ± 0.00a | 0.29 ± 0.00fg | 12.27 ± 0.03a | 48.02 ± 0.04f | 26.5 ± 0.71a | 8.03 ± 0.71a | 55.18 ± 0.81a | 16.72 ± 0.13a | 11.56 ± 0.08f | 6.04 ± 0.03g | 6.51 ± 0.14d | 6.57 ± 0.34d | 6.83 ± 0.11e |
| 5 | 5.98 ± 0.03c | 0.36 ± 0.00efg | 10.42 ± 0.14bc | 46.71 ± 0.75g | 20.0 ± 0.00d | 6.44 ± 0.16b | 42.81 ± 0.57e | 13.78 ± 0.07c | 13.76 ± 0.11e | 6.07 ± 0.01g | 6.36 ± 0.08d | 7.02 ± 0.03d | 7.63 ± 0.00cd |
| 6 | 4.97 ± 0.00h | 0.83 ± 0.00b | 11.14 ± 0.22b | 58.10 ± 0.00a | 17.5 ± 0.71e | 5.62 ± 0.00c | 30.12 ± 0.16g | 9.67 ± 0.00gh | 10.10 ± 0.03g | 6.48 ± 0.11f | 7.64 ± 0.24c | 5.88 ± 0.47e | 8.21 ± 0.07b |
| 7 | 4.17 ± 0.03ı | 1.80 ± 0.00a | 9.57 ± 0.31cd | 49.30 ± 0.21e | 13.5 ± 0.71f | 6.44 ± 0.16b | 27.38 ± 0.03h | 13.06 ± 0.11d | 19.02 ± 0.19c | 5.57 ± 0.11h | 6.21 ± 0.16d | 6.96 ± 0.14d | 4.49 ± 0.13g |
| 8 | 5.06 ± 0.00g | 0.86 ± 0.00b | 3.96 ± 0.01g | 52.61 ± 0.51c | 13.0 ± 0.71f | 2.63 ± 0.08e | 24.71 ± 0.03ı | 4.99 ± 0.06ı | 5.74 ± 0.11ı | 8.49 ± 0.18a | 9.74 ± 0.13a | 10.52 ± 0.47a | 8.49 ± 0.00a |
| 9 | 5.28 ± 0.00d | 0.58 ± 0.00cd | 9.13 ± 0.64de | 51.15 ± 0.07d | 20.0 ± 0.00d | 7.78 ± 0.08a | 39.10 ± 0.10f | 15.21 ± 0.00b | 32.30 ± 0.40a | 7.07 ± 0.07d | 8.53 ± 0.33b | 8.80 ± 0.16b | 6.15 ± 0.07f |
| 10 | 5.19 ± 0.01f | 0.67 ± 0.00c | 6.82 ± 0.08f | 51.32 ± 0.03d | 22.5 ± 0.71c | 5.12 ± 0.00c | 43.84 ± 0.00d | 9.97 ± 0.86g | 10.12 ± 0.16g | 7.32 ± 0.10bc | 8.57 ± 0.04b | 8.31 ± 0.10bc | 7.72 ± 0.03c |
| P | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** |
DM dry matter, FFA free fatty acid, LC Lactococcus, LB Lactobacillus, TAMB total aerobic mesophilic bacteria
**Significant at p < 0.01 probability level
It is stated in the Turkish Standards for White Cheese (TS 591) that pH value of classical White Cheese must be at least 4.5, titratable acidity (in terms of lactic acid) 3%, dry matter content at least 40%, the fat rate at least 45% (m/m) in dry matter, and salt at most 10% in dry matter [23]. While all cheese samples were in accordance with the TS 591 in terms of acidity and dry matter, it was observed that 1 sample in terms of pH, 7 samples in terms of fat in dry matter, and 6 samples in terms of salt content in dry matter were not in accordance with the standards. In their study investigating the chemical composition of 38 Turkish White Cheese, Turantaş et al. [24] determined the average dry matter and salt ratios as 41.82 and 3.56, respectively. It was observed that these values are lower than the values of dry matter and salt obtained in this study. It is believed that the high values of dry matter obtained in this study possibly result from the high salt content and the use of milk from different species of animals in the production of cheese, the absence of a standard method in the production of cheese, or different ripening/age periods of cheese. Öner et al. [14] stated that while the dry matter and pH values of Turkish White Cheese decrease during the ripening period, the salt content increases. Bakirci et al. [25] on the other hand determined that the ash and salt ratios in the same type of cheese increased during the storage period and stated that this could be explained by the moisture loss of the cheese samples during storage. The conversion of triglycerides into free fatty acids (FFA) by microorganisms and natural milk lipase has an important role in the development of different flavoring substances in cheese. Short- and medium-chain free fatty acids are especially used as precursors in the formation of aromatic products such as ethyl ketones, esters, and thioesters [26–28]. The FFA values of the cheese samples were determined between 5.74 and 32.29 (samples 8 and 9). There are a limited number of studies on the lipolysis level of White Cheese [8]. Çelik et al. [29] determined the acidity level of cheese in order to determine the level of lipolysis and calculated the result as 0.93–3.86 mg KOH/100 g fat.
It was observed that the number of lactococci was higher than that of lactobacilli in all cheese samples. A number of previous reports stated that lactococci were the dominant microbiota in the early stages of ripening in raw milk cheeses [14, 30, 31].
The TAMB number in cheese samples was determined as 5.88–9.52 log cfu/g. There are many factors that affect the total number of aerobic mesophyll viable bacteria in food. The fact that the milk used in cheese making is not pasteurized, hygienic rules are not followed, and the cheese is served fresh without ripening has an effect on the microbiological quality of the cheese [32].
NSLAB microbiota of Turkish White Cheese
As a result of genotypic characterization of 90 NSLAB bacteria isolated from Turkish White Cheeses, 27 different strains belonging to 14 LAB species were detected (Table 2).
Table 2.
Distribution of identified NSLAB isolates in White Cheese samples (+ and - represent the presence of each species within the corresponding sample).
| White Cheese samples | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| Aerococcus urinaeequi | - | - | + | - | - | - | - | - | - | - |
| Enterobacter cloacae | - | - | + | - | - | - | - | - | - | - |
| Enterococcus faecalis | - | - | - | - | - | - | + | - | + | - |
| Enterococcus faecium | + | + | - | - | - | - | - | - | - | + |
| Enterococcus italicus | - | - | - | - | + | - | - | - | - | - |
| Levilactobacillus brevis | - | - | - | - | + | - | - | - | - | - |
| Lacticaseibacillus paracasei | - | - | - | - | + | - | - | + | - | - |
| Lactiplantibacillus plantarum | - | - | + | + | - | + | - | + | + | - |
| Lactococcus garvieae subsp. bovis | - | + | - | - | - | - | - | - | - | - |
| Lactococcus lactis | + | - | - | - | + | - | - | - | - | - |
| Lactococcus lactis subsp. cremoris | - | - | - | - | + | - | - | - | - | - |
| Leuconostoc mesenteroides subsp. jonggajibkimchii | - | - | + | - | + | - | - | - | - | - |
| Staphylococcus sciuri | - | - | - | - | - | + | - | - | - | - |
| Streptococcus lutetiensis | - | - | + | - | - | - | - | - | - | - |
| Streptococcus thermophilus | - | - | - | - | - | - | - | - | + | - |
| Weissella thailandensis | - | - | - | - | - | - | + | - | - | - |
While 8 LAB species: Aerococcus urinaeequi (isolated from cheese sample 3), Enterobacter cloacae (isolated from cheese sample 3), Enterococcus italicus (isolated from cheese sample 5), Levilactobacillus brevis ( formerly Lactobacillus brevis; isolated from cheese sample 5), Lactococcus garvieae (isolated from cheese sample 2), Streptococcus lutetiensis (isolated from cheese sample 3), Streptococcus thermophilus (isolated from cheese sample 9), and Weissella thailandensis (isolated from cheese sample 7), were detected in only one sample, Enterococcus faecalis (isolated from cheese samples 7 and 9), Lacticaseibacillus paracasei (formerly Lactobacillus paracasei; isolated from cheese samples 5 and 8), Lactococcus lactis (isolated from cheese samples 1 and 5), and Leuconostoc mesenteroides (isolated from cheese samples 3 and 5) were detected in two samples each; Enterococcus faecium were detected in 3 (isolated from cheese samples 1, 2, and 10) and Lactiplantibacillus plantarum (formerly Lactobacillus plantarum) were detected in 5 samples (isolated from cheese samples 3, 4, 6, 8, and 9). Apart from these, 2 Staphylococcus sciuri strains were detected as non-LAB of the microbiota of one cheese sample (sample 6).
Percentage representation of LAB species identified in Turkish White Cheese samples is given in Fig. 1. Among 14 LAB species detected in 10 cheese samples, Lactiplantibacillus plantarum was found to be the most abundant with a rate of 22%. Then, the rest follows respectively as Enterococcus faecium (15%), Lactococcus lactis (11%), Enterococcus faecalis, Lacticaseibacillus paracasei, Leuconostoc mesenteroides (7%), Aerococcus urinaeequi, Enterobacter cloacae, Enterococcus italicus, Levilactobacillus brevis, Lactococcus garvieae, Streptococcus lutetiensis, Streptococcus thermophilus, and Weissella thailandensis (4%).
Fig. 1.
Proportional (%) distribution of NSLAB in Turkish White Cheese
The analysis of Fig. 2, which shows the phylogenetic relationship of different NSLAB strains based on the alignment of 16S rRNA genes with MEGA4, indicates that Lactococcus lactis (3 strain), Lactococcus garvieae (1), Streptococcus lutetiensis (1), Streptococcus thermophilus (1), Weissella thailandensis (1), Enterococcus italicus (1), Enterococcus faecalis (2), Enterococcus faecium (4) cluster together, Lacticaseibacillus paracasei (2), Leuconostoc mesenteroides (2), Aerococcus urinaeequi (1), Levilactobacillus brevis (1), and Lactiplantibacillus plantarum (6) are clustered into the same group, and Enterobacter cloacae (1) is also separated from other isolates and forms a different group.
Fig. 2.
Dendrogram showing multiple sequence alignment of 16S rRNA gene sequences of strains isolated from Turkish White Cheese. Pairwise phylogenetic distances were calculated based on 1400 nt of 16S rRNA gene
Lactiplantibacillus plantarum was the dominant species in Turkish White Cheese (22%). It was detected in 5 cheese samples. Tzanetakis and Litopoulou-Tzanetaki [33], Manolopoulou et al. [30], and Rantsiou et al. [34] determined that L. plantarum was the dominant microbiota during ripening in traditionally produced Feta cheese. Karakuş et al. [12] stated that although Lactococcus species decreased during ripening, Lactobacillus species such as L. casei and L. plantarum increased and formed an important part of the lactic microbiota in mature types of cheese. As a result, they reported that L. casei and L. plantarum should be considered in the formulation of the starter culture suitable for White Cheese production. However, L. casei was not detected in the current study. Durlu Özkaya [35] stated that L. plantarum is among the predominantly detected species in the microbiota of White Cheese, and L. lactis subsp. lactis, L. plantarum, and E. faecium can be included in the starter culture combination.
Akbulut et al. [36] collected 30 White Cheese samples from different parts of Turkey and identified 41 isolates from 17 species in his study, where they identified lactic acid bacteria and determined 1 of them as L. plantarum. Importantly, although Lentilactobacillus buchneri was shown as the dominant LAB in White Cheese samples in their report, this species was not detected in the study.
Enterococcus faecium (15%) and Enterococcus faecalis (7%) were other NSLAB strains found in traditional White Cheese samples. E. faecium was the second most abundant NSLAB species. It was identified in 3 cheese samples. However, E. faecalis was detected in 2 samples. Durlu Özkaya [35] determined that E. faecium was the most common microbiota found in White Cheese samples. Dagdemir and Ozdemir [7] found that E. faecalis (24.71%), E. faecium (17.97%), and Lb. fermentum (19.66%) strains were included at a high rate and stated that in addition to E. faecalis strains, E. faecium strains, which were predominantly found in cheese, could be used as an auxiliary culture in the production of White Cheese. Lb. fermentum was not detected in our study.
Karakuş et al. [12] concluded that Enterococcus species (E. faecalis and E. faecium) constitute the second most important group in their study in which they examined lactic acid bacteria during the ripening process of White Cheese and stated that E. faecalis was found in higher numbers in fresh cheeses and E. faecium in mature cheese. In addition, it was stated that enterococci, which were numerically important in the microbiota of cheese, might contribute to the production and maturation of White Cheese but the fact that this group of bacteria was an indicator of fecal contamination in food microbiology and also caused food poisoning due to toxic metabolites formed by some strains prevented the adoption of recommendations to use them as starter cultures [37].
In general, the presence of enterococci in milk and cheese is explained by the contamination of farm animal feces, water sources, used milk equipment, and milk storage tanks [38]. E. faecalis and E. faecium are species frequently encountered in cheese, and they constitute the dominant microbiota in cheese due to their ability to survive at pasteurization temperatures, their resistance to high salt concentrations, and their ability to grow at low temperatures [39].
Lactococcus lactis (11%) was the third most dominant NSLAB species in traditionally produced White Cheese samples. It was detected in 2 samples. Although this strain was not detected by Akbulut et al. [36], Durlu Özkaya [35] isolated 13 L. lactis subsp. lactis strains of a total 85 lactic acid bacteria from 30 White Cheese samples collected from various provinces of Turkey. Karakuş et al. [12] isolated a total of 348 lactic acid bacteria at the beginning and end of the 3-month maturation period from White Cheese produced without using starter culture and obtained from three different enterprises, and defined 55 of these bacteria as L. lactis subsp. lactis, 9 of which as L. lactis subsp. cremoris and 2 of which as L. lactis subsp. lactis biovar. diacetylactis. Dagdemir and Ozdemir [7] defined 2 of 3 strains of Lactococcus genus from 178 lactic acid bacteria isolated from commercial types of White Cheese as L. lactis subsp. lactis, 1 of which as L. lactis subsp. cremoris.
Lacticaseibacillus paracasei and Leuconostoc mesenteroides (7%) were other NSLAB species detected in cheese samples. Akbulut et al. [36] detected four of the 41 isolates as L. paracasei, but not Leuc. mesenteroides. In the study of Durlu Özkaya [35], neither species was detected. Karakuş et al. [12] detected 7 out of 348 strains as Leuc. mesenteroides but detected no L. paracasei in any strains.
Aerococcus urinaeequi, Enterobacter cloacae, Enterococcus italicus, Levilactobacillus brevis, Lactococcus garvieae, Streptococcus lutetiensis, Streptococcus thermophilus, and Weissella thailandensis (4%) were the other NSLAB species detected in cheese samples. The presence of L. brevis was also revealed by Karakuş et al. [12], Dagdemir and Ozdemir [7], and Akbulut et al. [36] and the presence of S. thermophilus by Karakuş et al. [12] in traditionally produced White Cheese samples.
As non-LAB species, Staph. sciuri was presented in a cheese sample, albeit at very low levels, and previous studies demonstrated the presence of this species in White Cheese [36, 40]. Staph. sciuri can be contaminated with cheese made from raw milk, factory tools and equipment, or brine bath [41]. Although the possibilities of using this species as a starter culture have been investigated by Bockelmann et al. [41], it has been described as an opportunistic pathogen in certain clinical situations by Irlinger [42].
Yeast microbiota of Turkish White Cheese
Since yeasts are both spoilage organisms and contribute to the characteristic taste of cheese, there has been an increasing interest in the yeast microbiota of cheese [22, 43, 44]. Presumably, the presence of some yeast strains has a beneficial effect on cheese quality at limited concentrations [45].
As a result of genotypic characterization of 80 yeast isolates collected from Turkish White Cheese samples, 49 different isolates belonging to 11 different yeast species were identified (Table 3). The study revealed the rich yeast microbiota of traditionally produced Turkish White Cheese samples.
Table 3.
Distribution of identified yeast isolates in White Cheese samples (+ and - represent the presence of each species within the corresponding sample)
| White Cheese samples | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| Candida inconspicua | - | - | - | - | - | - | - | + | - | - |
| Candida zeylanoides | - | + | + | - | + | - | + | + | - | + |
| Debaryomyces hansenii | - | + | - | + | + | + | + | - | + | + |
| Debaryomyces mycophilus | - | - | - | + | - | - | - | - | - | - |
| Kluyveromyces lactis | + | - | + | + | + | - | - | + | + | + |
| Kluyveromyces marxianus | - | - | - | - | - | - | - | + | + | - |
| Meyerozyma guilliermondii | - | - | + | - | - | - | - | - | - | - |
| Pichia fermentans | - | - | - | - | - | - | - | + | + | + |
| Rhodotorula mucilaginosa | - | - | - | - | - | - | - | + | - | - |
| Rhodotorula sp | - | - | - | - | - | + | - | - | - | - |
| Torulaspora delbrueckii | - | - | - | - | - | - | - | - | + | - |
Kluyveromyces lactis and Debaryomyces hansenii were found in 7 cheese samples; Candida zeylanoides in 6; Pichia fermentans in 3; and Kluyveromyces marxianus in 2; Candida inconspicua, Debaryomyces mycophilus, Meyerozyma guilliermondii, Rhodotorula mucilaginosa, Rhodotorula sp., and Torulaspora delbrueckii were detected in 1 sample each (Table 3).
In Fig. 3, which shows the percentage of yeast species identified in Turkish White Cheese samples, Candida zeylanoides (27%) was found to be the dominant yeast species of the microbiota in the cheese samples. It was followed by K. lactis (23%) and D. hansenii (18%). K. marxianus, P. fermentans 8%, C. inconspicua, M. guilliermondii 4%, Debaryomyces mycophilus, Rhodotorula mucilaginosa, Rhodotorula sp., and Torulaspora delbrueckii were the yeast species found in 2% of White Cheese samples.
Fig. 3.
Proportional (%) distribution of yeasts in Turkish White Cheese
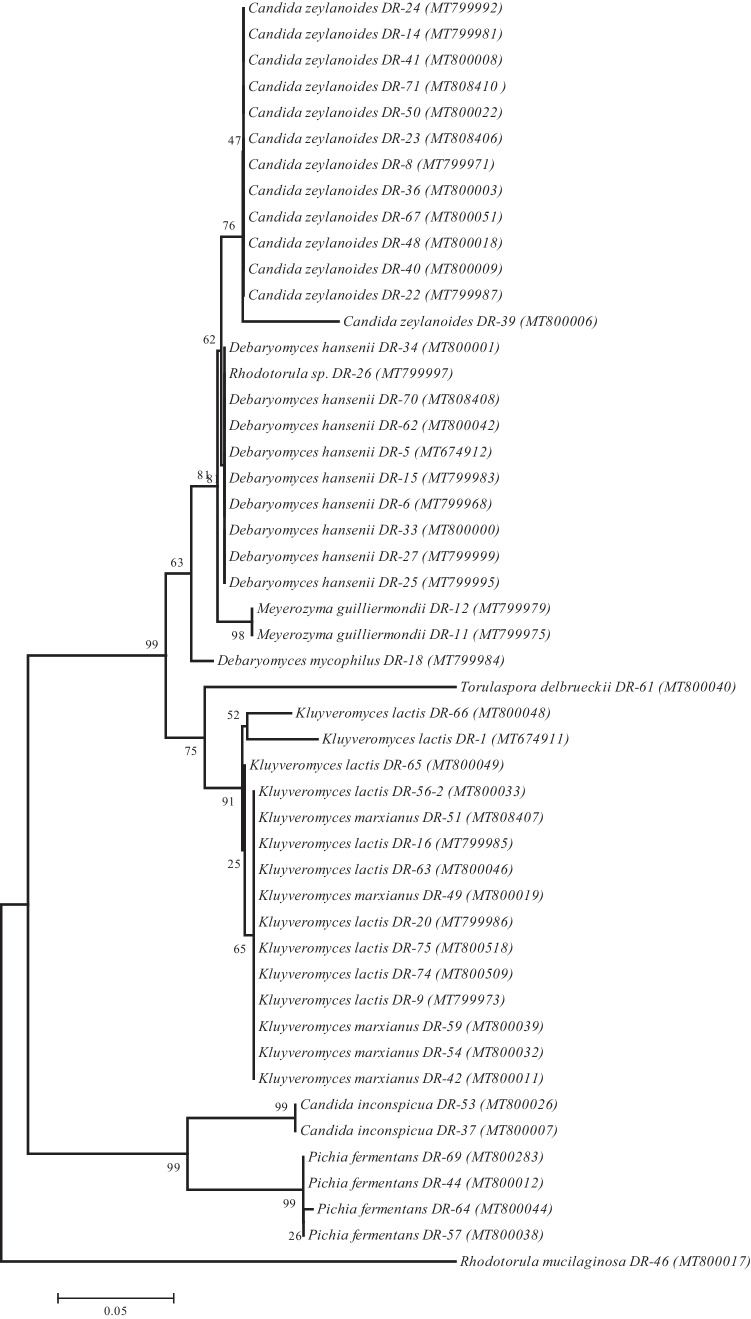
Figure 4, which shows the phylogenetic relationship of the yeast strains isolated from Turkish White Cheese, indicates that C. zeylanoides, D. hansenii, Rhodotorula sp., M. guilliermondii, D. mycophilus, Torulaspora delbrueckii, K. lactis, and K. marxianus are classified in one group and C. inconspicua and P. fermentans in another one, and Rhodotorula mucilaginosa is separated from other isolates and forms a different group.
Fig. 4.
Dendrogram showing multiple sequence alignment of D1/D2 domain of the 26S rDNA region of yeast strains isolated in this study. Pairwise phylogenetic distances were calculated based on 600 nt of this region
It was observed that Candida zeylanoides (27%) formed the dominant yeast microbiota in cheese samples. It was detected in 6 cheese samples. In their study on the molecular characterization of yeasts isolated from traditional Turkish cheeses, Ozmen Togay et al. [44] suggested that the dominant microbiota was D. hansenii and T. delbrueckii, and the two isolates isolated from White Cheese were C. zeylanoides and T. delbrueckii. In addition to C. zeylanoides, T. delbrueckii and D. hansenii were among the yeast species detected in the study. T. delbrueckii is among the species identified as the dominant microbiota in Feta cheeses by Westall and Filtenborg [45], and the author stated that this strongly fermented species causes the boxes to swell.
Kluyveromyces lactis (23%, in 7 samples), Debaryomyces hansenii (18%, in 7 samples), Pichia fermentans (8%; in 3 samples), and Kluyveromyces marxianus (8%, in 2 samples) were other yeast species detected in White Cheese samples. Tornadijo et al. [46] stated that K. lactis constituted 28% of the yeasts isolated from 1-week-old Amada cheese produced from raw goat milk, and the importance of K. lactis and K. marxianus in dairy products is due to their ability to ferment lactose. In another study examining the yeast and mold microbiota of different kinds of cheese, it was determined that D. hansenii was the dominant microbiota. The prevalence of D. hansenii in cheese is probably due to several physiological characteristics. This organism has the capacity to grow at high salt concentrations and low pH, use lactate as the main carbon source, produce proteolytic and lipolytic enzymes that can metabolize milk proteins and fat, and grow at low temperature and low water activity [47, 48]. Westall and Filtenborg [45] and Ozmen Togay et al. [44] reported that K. lactis and D. hansenii with similar properties can contribute to the taste and texture of cheese. Goerges et al. [49] reported that K. marxianus isolated from cheese has strong antilisterial activity. Hatoum et al. [50] determined that some D. hansenii and P. fermentans strains obtained from raw milk and cheese products exhibited a significant inhibitory effect against Listeria ivanovii and stated that these isolates have the potential to be used as biological preservatives.
Candida inconspicua, Meyerozyma guilliermondii 4%, Debaryomyces mycophilus, Rhodotorula mucilaginosa, Rhodotorula sp., and Torulaspora delbrueckii 2% were the yeast species found in White Cheese samples. Laurenčík et al. [51] stated that C. inconspicua was among the species occasionally found in the microbiota of traditional Slovak sheep cheese. The presence of R. mucilaginosa and M. guilliermondii in traditional types of Turkish cheese was revealed by Ozmen Togay et al. [44]. There seems no study in the literature in which D. mycophilus was detected in cheese samples. It was isolated from the intestines of woodlice species by Thanh et al. [52].
Aroma profile of traditionally produced Turkish White Cheese
Aroma substances detected by GC-MS in cheese samples are presented in Table 4, and results are given as volatile compound peak area/internal standard peak area (arbitrary unit). Aroma compounds detected in cheese samples were classified as carboxylic acids, esters, ketones, alcohols, monoterpenes, aldehydes, and other compounds.
Table 4.
Aroma compounds detected in traditionally produced Turkish White Cheese
| White Cheese Samples | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| Carboxylic acids | ||||||||||
| Acetic acid | 17.60 | 2.48 | ||||||||
| Butanoic acid | 30.26 | 37.17 | 34.13 | 33.33 | 43.28 | 32.24 | 21.87 | 18.20 | 34.81 | 30.42 |
| Hexanoic acid | 29.23 | 33.67 | 32.82 | 35.43 | 32.93 | 20.82 | 26.96 | 15.54 | 36.10 | 28.63 |
| Octanoic acid | 6.83 | 9.27 | 5.53 | 7.55 | 4.62 | 5.70 | 17.8 | 3.9 | 4.05 | |
| 3-Methyl butanoic acid | 2.02 | 2.76 | 2.32 | 4.13 | 3.35 | |||||
| Benzene carboxylic acid | 3.3 | |||||||||
| Esters | ||||||||||
| Butanoic acid, 3-methyl-, butyl ester | 2.39 | |||||||||
| Butanoic acid, ethyl ester | 3.58 | 1.35 | 4.00 | |||||||
| Decanoic acid, ethyl ester | 1.55 | 2.29 | 0.84 | |||||||
| Hexanoic acid, ethyl ester | 10.3 | 2.16 | 7.12 | 3.19 | 1.93 | 4.17 | 6 | 7.7 | 8.71 | 7.01 |
| Hexanoic acid, methyl ester | 2.64 | 2.59 | ||||||||
| Octanoic acid, 3-methylbutyl ester | 1.26 | |||||||||
| Octanoic acid, ethyl ester | 5.96 | 1.72 | 2.18 | 2.79 | 5.11 | 4.37 | 5 | 2.25 | ||
| Octanoic acid, methyl ester | 1.19 | |||||||||
| Ketones | ||||||||||
| 1-Phenylethanone (acetophenone) | 2.32 | 3.23 | 2.05 | 7.15 | 5.93 | 2.43 | 1.97 | 2.28 | ||
| 2,3-Butandione | 1.06 | 0.82 | 2.71 | 2.55 | 0.68 | 2.14 | ||||
| 2-Butanone | 0.15 | 1.82 | 2.27 | |||||||
| 2-Heptanone | 4.98 | 2.7 | 3.59 | 2.06 | 5.08 | 1.93 | 2.18 | 2.95 | ||
| 3-Hydroxy-2-butanone | 1.91 | 2.34 | 1.06 | 2.33 | 0.53 | |||||
| Acetone | 1.43 | 1.47 | ||||||||
| Alcohols | ||||||||||
| 1-Heptanol | 4.29 | |||||||||
| 2-Butanol | 0.65 | 0.72 | 1.53 | 1.10 | ||||||
| 2-Ethylhexanol | 1.11 | 0.97 | 2.72 | 2.43 | 0.63 | 0.40 | ||||
| 2-Propanol | 1.02 | 0.82 | 1.41 | |||||||
| 3-Methyl-1-butanol | 0.59 | |||||||||
| Benzeneethanol | 2.30 | 1.8 | ||||||||
| Ethanol | 3.78 | 2.99 | 3.82 | 4.14 | 3.58 | 10.43 | 3.11 | 4.78 | ||
| Monoterpenes | ||||||||||
| dl-Limonene | 1.53 | 3.52 | 5.29 | 2.08 | 0.77 | 2.23 | ||||
| l-Limonene | 1.83 | |||||||||
| Aldehydes | ||||||||||
| 3-Methylbutanal | 6.03 | |||||||||
| Other compounds | ||||||||||
| Hexane | 2.34 | 5.27 | ||||||||
| 2-Methylbutane | 2.06 | |||||||||
| Ammonium acetate | 1.9 | |||||||||
| Oxirane, 2-3-methyl | 1.69 | |||||||||
| Thiirane (methoxymethyl)- | 1.01 | |||||||||
Although carboxylic acids, which are mainly products of carbohydrate and lipid metabolism [53], were the dominant volatile aroma compound in all cheese samples, significant changes were observed in cheese samples. This value ranged from 59.13 (sample 6) to 88.55 (sample 5). Similar results have been obtained by many researchers [13, 54, 55]. Salum et al. [13] reported that as a result of aroma analysis of 9 White Cheese samples obtained from different regions of Turkey, the ratio of acid compounds was between 72.9 and 92.1%. Marangoz and Bostan [55] determined that acid compounds ranged from 80.65 to 75.46% in odor-problematic and control group cheese. In both studies, it was stated that acids were the dominant flavoring agents. Gürkan [56] found the dominant flavoring agents to be esters and alcohols, while Sahingil et al. [57] determined that the most common aroma were acids, ketones, and alcohols.
Lactobacilli species such as L. paracasei convert amino acids into keto and hydroxy acids, and these compounds are then transformed into carboxylic acids by some strains of Lactococcus, such as Lactococcus lactis subsp. cremoris. Therefore, such cooperation between L. lactis and some strains of Lactobacilli can promote aroma development in cheese [58]. Lactococcus lactis subsp. cremoris was detected only in sample 5 and the amount of carboxylic acid in sample 5 may be related to the coexistence of these two strains.
The most common carboxylic acids are butanoic acid, hexanoic acid, and octanoic acids respectively. The study results regarding carboxylic acid types and rates align with Sahingil et al. [57] and Yaşa [54]. Butanoic acid and hexanoic acid were detected in all cheese samples. Butanoic acid is one of the most important odor compounds in cheeses and its density in raw milk cheese is much higher than in pasteurized milk cheeses [59]. Sahingil et al. [57] stated that butanoic acid, hexanoic acid, and acetic acid were the most acid compounds in White Cheese samples and stated that acid compounds in cheeses increased as maturation progressed. Octanoic acid is among the acid compounds detected by these researchers.
Acetic acid was detected in only 2 samples (7 and 9). Enterococcus faecalis was detected only in these two samples, and acetic acid production may be related to this species. 3-Methyl butanoic acid and benzene carboxylic acid were the other carboxylic acids detected in cheese samples. 3-Methyl butanoic acid was detected in 5 samples, and it can be said that it is one of the characteristic aroma compounds of Turkish White Cheese. Benzene carboxylic acid was detected in only one sample. Although benzene carboxylic acid or benzoic acid is widely used as a food preservative in the food industry, it is also produced naturally in cheese in different amounts depending on the incubation temperature, starter type, and incubation time [60, 61].
Esters were the second major flavoring agents in the White Cheese samples. Cheese samples had a rich variety of esters. Eight different ester compounds were detected. Hexanoic acid ethyl ester of these compounds was detected in each sample in varying ratios of 1.93–10.3.
Ester compounds were detected in varying ratios between 1.93 and 19.07%. Ester formation in cheese results from direct esterification of alcohols and carboxylic acids or alcoholysis [62]. Marangoz and Bostan [55] detected ester compounds varying between 8.3 and 9.0% in White Cheese samples. They stated that the most frequently defined subgroups of these esters were ethyl esters, propyl esters, and methyl esters. Hexanoic acid ethyl ester was detected in all cheese samples (1.93–10.30%). Marangoz and Bostan [55] reported that the predominant ethyl esters were ethyl hexanoate, which had a strong fruity, pineapple-like aroma. Octanoic acid ethyl ester was also one of the prominent aromatic ester compounds in the study.
Ketone compounds (1-phenylethanone, 2,3-butandione, 2-butanone, 2-heptanone, 3-hydroxy-2-butanone, and Acetone) were detected in cheese samples, the ratios of which varied between 2.49 and 14.05%. 2-Heptanone and 1-phenylethanone were detected in 8 samples each, and it is possible to note that they constitute important aromatic ketone compounds of Turkish White Cheese.
Another group of volatile aroma compounds detected in cheese samples was alcohol. Six different alcohol compounds were detected and they constituted 6.12% of the total volatile components. The most prominent among these was ethanol, which was detected in 8 samples at rates ranging from 2.99 to 10.43%. A number of studies have stated that the most common volatile compound in similar cheeses is ethanol [63, 64]. No ethanol detected in two samples. The reason for this is probably related to the fact that these types of cheese are sold without packages. 1-Heptanol (CAS), 2-butanol, 2-ethylhexanol, 2-propanol, 3-methyl-1-butanol, and benzeneethanol were other alcohol compounds.
Aldehydes were available at low levels compared to other major aroma groups and 3-methylbutanal was the only aldehyde compound detected in cheese samples. It was detected at the rate of 6.03% in only 1 sample (sample number 6). 3-Methylbutanal and 1-heptanol, one of the alcohol compounds, were detected only in sample 7. Staphylococcus sciuri and Rhodotorula species were detected only in this sample, and these species or the complex microbiota including these species may be related to the production of these aroma substances [56, 65].
dl-Limonene and l-limonene were monoterpenes detected in cheese samples. Monoterpene compound was detected in 6 cheese samples. dl-Limonene was more common. Aminifar et al. [66] detected limonene compounds in Lighvan cheese and stated that its ratio increased during storage. A number of studies have detected these compounds in milk and cheese and reported that they may be of plant origin [67].
In addition, 5 different other compounds were identified. The rate of other compounds detected in cheese samples was 1.43%.
Conclusion
This study revealed a number of chemical properties, NSLAB, and yeast microbiota and aroma profile of traditionally produced Turkish White Cheese. It was determined that traditionally produced Turkish White Cheese types have different chemical properties and cheese samples with different microbiota have different aroma profiles. These findings may contribute to the development of local standard starter cultures for the production of White Cheese or to the enrichment of existing cultures.
Author contribution
EM and NY contributed to the analysis, EM contributed to the writing of the first draft and the creation of tables and figures, and ED contributed to the revision of the article. All authors approved the manuscript.
Funding
The authors would like to thank Bayburt University for the internal fund.
Data availability
All data generated or analyzed during this study are included in this published article.
Declarations
Consent for publication
Written informed consent for publication was obtained from all participants.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hayaloglu AA, Guven M, Fox PF. Microbiological, biochemical and technological properties of Turkish White cheese ‘Beyaz Peynir’. Int Dairy J. 2002;12:635–648. doi: 10.1016/S0958-6946(02)00055-9. [DOI] [Google Scholar]
- 2.Alichanidis E, Polychroniadou A. Characteristics of major traditional regional cheese varieties of East-Mediterranean countries: a review. Dairy Sci Technol. 2008;88:495–510. doi: 10.1051/dst:2008023. [DOI] [Google Scholar]
- 3.Üçüncü M. Süt ve Mamulleri Teknolojisi. İzmir: Meta Basım; 2013. [Google Scholar]
- 4.Bintsis T, Papademas P. Microbiological quality of white-brined cheeses: a review. Int J Dairy Technol. 2002;55(3):113–120. doi: 10.1046/j.1471-0307.2002.00054.x. [DOI] [Google Scholar]
- 5.Karakuş M, Alperden I. Effect of starter composed of various species of lactic bacteria on quality and ripening of Turkish white pickled cheese. LWT - Food Sci Technol. 1995;28(4):404–409. doi: 10.1016/0023-6438(95)90024-1. [DOI] [Google Scholar]
- 6.Kılıç S. Türkiye sütçülüğünde saf kültür kullanımı ve karşılaşılan sorunlar. Akad Gida. 2005;3(4):5–10. [Google Scholar]
- 7.Dagdemir E, Ozdemir S. Technological characterization of the natural lactic acid bacteria of artisanal Turkish White Pickled cheese. Int J Dairy Technol. 2008;61(2):133–140. doi: 10.1111/j.1471-0307.2008.00394.x. [DOI] [Google Scholar]
- 8.Çelik Ș, Uysal Ș. Composition, quality, microflora and ripening of Beyaz cheese. Atatürk Univ J Agric Fac. 2009;40(1):141–151. [Google Scholar]
- 9.Ersoy M. Characterization of some technological properties of Lactococcus strains isolated from traditionally made cheeses. Bursa, Turkey: Health Sciences Institute, Uludağ University; 2009. [Google Scholar]
- 10.Franco-Duarte R, Černáková L, Kadam SS, Kaushik K, Salehi B, Bevilacqua A, et al. Advances in chemical and biological methods to identify microorganisms—from past to present. Microorganisms. 2019;7(5):130. doi: 10.3390/microorganisms7050130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ertürkmen P, Öner Z. Determination of lactic acid bacteria properties as starter cultures isolated from the beyaz cheese by biochemical methods. Süleyman Demirel Univ Fen Bilim Enst Dergisi. 2015;19(3):9–16. [Google Scholar]
- 12.Karakuş M, Borcaklı M, Alperden I (1992) Lactic acid bacteria of white pickled cheese in the ripening process. Gıda17(6):363–369. https://dergipark.org.tr/tr/pub/gida/issue/6962/92814
- 13.Salum P, Govce G, Kendirci P, Bas D, Erbay Z. Composition, proteolysis, lipolysis, volatile compound profile and sensory characteristics of ripened white cheeses manufactured in different geographical regions of Turkey. Int Dairy J. 2018;87:26–36. doi: 10.1016/j.idairyj.2018.07.011. [DOI] [Google Scholar]
- 14.Öner Z, Karahan AG, Aloğlu H. Changes in the microbiological and chemical characteristics of an artisanal Turkish white cheese during ripening. LWT - Food Sci Technol. 2006;39(5):449–454. doi: 10.1016/j.lwt.2005.03.015. [DOI] [Google Scholar]
- 15.Kurt A, Cakmakci S, Caglar A (1996) A guide book of analysis methods of milk and milk products. Atatürk Univ Agric Fac Pub (18) Erzurum/Turkey:1–238
- 16.ISO (2009) ISO 660: Animal and vegetable fats and oils: determination of acid value and acidity.
- 17.Harrigan WF. Laboratory methods in food microbiology. San Diego, USA: Academic Press; 1998. [Google Scholar]
- 18.Šuranská H, Raspor P, Uroić K, Golić N, Kos B, Mihajlović S, Begović J, Šušković J, Topisirović L, Čadež N. Characterisation of the yeast and mould biota in traditional white pickled cheeses by culture-dependent and independent molecular techniques. Folia Microbiol. 2016;61:455–463. doi: 10.1007/s12223-016-0455-x. [DOI] [PubMed] [Google Scholar]
- 19.Speck ML. Compendium of methods for the microbiological examination of foods. Washington: American Public Health Association; 1984. [Google Scholar]
- 20.Gulitz A, Stadie J, Wenning M, Ehrmann MA, Vogel RF. The microbial diversity of water kefir. Int J Food Microbiol. 2011;151:284–288. doi: 10.1016/j.ijfoodmicro.2011.09.016. [DOI] [PubMed] [Google Scholar]
- 21.İspirli H. Master’s Thesis, Graduate School of Natural and Applied Sciences, Department of Food Engineering. Türkiye.: Bayburt University, Bayburt; 2016. Isolation of lactic acid bacteria (lab) from Erzincan Tulum Cheese, determination by molecular methods and genetically determining of their exopolysaccharides (eps) production potential. [Google Scholar]
- 22.Yuvaşen A, Macit E, Dertli E. Microbial species playing roles for the production of traditional Kasar cheese during pre-maturation period. LWT - Food Sci Technol. 2018;91:406–413. doi: 10.1016/j.lwt.2018.01.075. [DOI] [Google Scholar]
- 23.TS (2006). Beyaz Peynir. Turk Standartlari Enstitusu. The Institute of Turkish Standards, TS 591, Ankara.
- 24.Turantaş F, Ünlütürk A, Göktan D. Microbiological and compositional status of Turkish white cheese. Int J Food Microbiol. 1989;8(1):19–24. doi: 10.1016/0168-1605(89)90076-7. [DOI] [PubMed] [Google Scholar]
- 25.Bakirci I, Kavaz A, Macit E. Effect of different brine concentrations and ripening period on some quality properties of Turkish white pickled cheese. Afr J Biotechnol. 2011;10(56):11925–11931. [Google Scholar]
- 26.McSweeney PL, Sousa MJ. Biochemical pathways for the production of flavour compounds in cheeses during ripening: a review. Lait. 2000;80(3):293–324. doi: 10.1051/lait:2000127. [DOI] [Google Scholar]
- 27.Collins YF, McSweeney PL, Wilkinson MG. Lipolysis and free fatty acid catabolism in cheese: a review of current knowledge. Int Dairy J. 2003;13(11):841–866. doi: 10.1016/S0958-6946(03)00109-2. [DOI] [Google Scholar]
- 28.Fernández-García E, Carbonell M, Calzada J, Nuñez M. Seasonal variation of the free fatty acids contents of Spanish ovine milk cheeses protected by a designation of origin: a comparative study. Int Dairy J. 2006;16(3):252–261. doi: 10.1016/j.idairyj.2005.02.010. [DOI] [Google Scholar]
- 29.Çelik Ş, Bakırcı İ, Özdemir S. Effects of high heat treatment of milk and brine concentration on the quality of Turkish white cheese. Milchwissenschaft. 2005;60(2):147–151. [Google Scholar]
- 30.Manolopoulou E, Sarantinopoulos P, Zoidou E, Aktypis A, Moschopoulou E, Kandarakis IG, Anifantakis EM. Evolution of microbial populations during traditional Feta cheese manufacture and ripening. Int J Food Microbiol. 2003;82(2):153–161. doi: 10.1016/S0168-1605(02)00258-1. [DOI] [PubMed] [Google Scholar]
- 31.Milani E, Shahidi F, Mortazavi SA, Vakili SAR, Ghoddusi HB. Microbiological, biochemical and rheological changes throughout ripening of Kurdish Cheese. J Food Prot. 2014;34(2):168–175. doi: 10.1111/jfs.12110. [DOI] [Google Scholar]
- 32.Yoon Y, Lee S, Choi KH. Microbial benefits and risks of raw milk cheese. Food Control. 2016;63:201–215. doi: 10.1016/j.foodcont.2015.11.013. [DOI] [Google Scholar]
- 33.Tzanetakis N, Litopoulou-Tzanetaki E. Changes in numbers and kinds of lactic acid bacteria in Feta and Teleme, two Greek cheeses from ewes’ milk. J Dairy Sci. 1992;75(6):1389–1393. doi: 10.3168/jds.S0022-0302(92)77891-6. [DOI] [Google Scholar]
- 34.Rantsiou K, Urso R, Dolci P, Comi G, Cocolin L. Microflora of Feta cheese from four Greek manufacturers. Int J Food Microbiol. 2008;126(1-2):36–42. doi: 10.1016/j.ijfoodmicro.2008.04.031. [DOI] [PubMed] [Google Scholar]
- 35.Durlu Özkaya F. Doctoral Thesis. Graduate School of Natural and Applied Sciences Department of Food Engineering. Ankara University, Ankara, Turkey; 2001. A comparative study on proteolytic activity, bacteriocin efficiency and biogenic amine formation of some lactococcus, enterococcus and lactobacillus strains isolated from white pickled cheese. [Google Scholar]
- 36.Akbulut S, Baltaci MO, Adiguzel G, Adiguzel A. Identification and potential biotechnological characterization of lactic acid bacteria isolated from white cheese samples. J Pure Appl Microbiol. 2022;16(4):2912–2922. doi: 10.22207/JPAM.16.4.66. [DOI] [Google Scholar]
- 37.Terzić-Vidojević A, Veljović K, Begović J, Filipić B, Popović D, Tolinački M, Miljković M, Kojić M, Golić N. Diversity and antibiotic susceptibility of autochthonous dairy enterococci isolates: are they safe candidates for autochthonous starter cultures? Front Microbiol. 2015;6:954. doi: 10.3389/fmicb.2015.00954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gelsomino R, Vancanneyt M, Cogan TM, Condon S, Swings J. Source of enterococci in a farmhouse raw-milk cheese. Appl Environ Microbiol. 2002;68(7):3560–3565. doi: 10.1128/AEM.68.7.3560-3565.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giraffa G. Enterococci from foods. FEMS Microbio Rev. 2002;26(2):163–171. doi: 10.1111/j.1574-6976.2002.tb00608.x. [DOI] [PubMed] [Google Scholar]
- 40.Pangallo D, Šaková N, Koreňová J, Puškárová A, Kraková L, Valík L, Kuchta T. Microbial diversity and dynamics during the production of May bryndza cheese. Int J Food Microbiol. 2014;170:38–43. doi: 10.1016/j.ijfoodmicro.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 41.Bockelmann W, Hoppe-Seyler T, Krusch U, Hoffmann W, Heller KJ. The microflora of Tilsit cheese. Part 2. Development of a surface smear starter culture. Food/Nahrung. 1997;41(4):213–218. doi: 10.1002/food.19970410406. [DOI] [Google Scholar]
- 42.Irlinger F. Safety assessment of dairy microorganisms: coagulase-negative staphylococci. Int J Food Microbiol. 2008;126(3):302–310. doi: 10.1016/j.ijfoodmicro.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 43.Geronikou A, Srimahaeak T, Rantsiou K, Triantafillidis G, Larsen N, Jespersen L. Occurrence of yeasts in white-brined cheeses: methodologies for identification, spoilage potential and good manufacturing practices. Front Microbiol. 2020;11:582778. doi: 10.3389/fmicb.2020.582778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ozmen Togay S, Capece A, Siesto G, Aksu H, Sandikci Altunatmaz S, Yilmaz Aksu F, Karagul Yuceer Y. Molecular characterization of yeasts isolated from traditional Turkish cheeses. Food Sci Technol. 2020;40:871–876. doi: 10.1590/fst.24319. [DOI] [Google Scholar]
- 45.Westall S, Filtenborg O. Yeast occurrence in Danish feta cheese. Food Microbiol. 1998;15(2):215–222. doi: 10.1006/fmic.1997.0161. [DOI] [Google Scholar]
- 46.Tornadijo ME, Fresno JM, Sarmiento RM, Carballo J. Study of the yeasts during the ripening process of Armada cheeses from raw goat’s milk. Lait. 1998;78(6):647–659. doi: 10.1051/lait:1998657. [DOI] [Google Scholar]
- 47.Fadda ME, Mossa V, Pisano MB, Deplano M, Cosentino S. Occurrence and characterization of yeasts isolated from artisanal Fiore Sardo cheese. Int J Food Microbiol. 2004;95(1):51–59. doi: 10.1016/j.ijfoodmicro.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 48.Banjara N, Suhr MJ, Hallen-Adams HE. Diversity of yeast and mold species from a variety of cheese types. Curr Microbiol. 2015;70(6):792–800. doi: 10.1007/s00284-015-0790-1. [DOI] [PubMed] [Google Scholar]
- 49.Goerges S, Aigner U, Silakowski B, Scherer S. Inhibition of Listeria monocytogenes by food-borne yeasts. Appl Environ Microbiol. 2006;72(1):313–318. doi: 10.1128/AEM.72.1.313-318.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hatoum R, Labrie S, Fliss I. Identification and partial characterization of antilisterial compounds produced by dairy yeasts. Probiotics Antimicrob. 2013;5(1):8–17. doi: 10.1007/s12602-012-9109-8. [DOI] [PubMed] [Google Scholar]
- 51.Laurenčík M, Sulo P, Sláviková E, Piecková E, Seman M, Ebringer L. The diversity of eukaryotic microbiota in the traditional Slovak sheep cheese—Bryndza. Int J Food Microbiol. 2008;127(1-2):176–179. doi: 10.1016/j.ijfoodmicro.2008.06.016. [DOI] [PubMed] [Google Scholar]
- 52.Thanh VN, Van Dyk MS, Wingfield MJ. Debaryomyces mycophilus sp. nov., a siderophore-dependent yeast isolated from woodlice. FEMS Yeast Res. 2002;2(3):415–427. doi: 10.1016/S1567-1356(02)00132-0. [DOI] [PubMed] [Google Scholar]
- 53.Dertli E, Çon AH. Microbial diversity of traditional kefir grains and their role on kefir aroma. LWT - Food Sci Technol. 2017;85:151–157. doi: 10.1016/j.lwt.2017.07.017. [DOI] [Google Scholar]
- 54.Yaşa M. Master’s Thesis. Graduate School of Natural and Applied Sciences of Food Engineering. Konya, Turkey: Necmettin Erbakan University; 2018. Determination of fatty acids, volatile aroma compounds and textural properties of white pickled cheese made from thermosonicated milk. [Google Scholar]
- 55.Marangoz B, Bostan K. Identification of off-odor compounds in Turkish white cheese with putrid defects by SPME-GC/MS. Acta Vet Eurasia. 2020;46(3):125–132. doi: 10.5152/actavet.2020.20017. [DOI] [Google Scholar]
- 56.Gürkan H. Graduate School of Natural and Applied Sciences Department of Food Engineering. Malatya, Turkey: Inönü University; 2019. Determination of bioactive peptide production performance with volatile aroma compounds and contribution to proteolysis level in white cheese of some yeasts. [Google Scholar]
- 57.Sahingil D, Hayaloglu AA, Simsek O, Ozer B. Changes in volatile composition, proteolysis and textural and sensory properties of white-brined cheese: effects of ripening temperature and adjunct culture. Dairy Sci Technol. 2014;94:603–623. doi: 10.1007/s13594-014-0185-2. [DOI] [Google Scholar]
- 58.Kieronczyk A, Skeie S, Langsrud T, Yvon M. Cooperation between Lactococcus lactis and nonstarter lactobacilli in the formation of cheese aroma from amino acids. Appl Environ Microbiol. 2003;69(2):734–739. doi: 10.1128/AEM.69.2.734-739.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cornu A, Rabiau N, Kondjoyan N, Verdier-Metz I, Pradel P, Tournayre P, Berdague JL, Martin B. Odour-active compound profiles in Cantal-type cheese: effect of cow diet, milk pasteurization and cheese ripening. Int Dairy J. 2009;19(10):588–594. doi: 10.1016/j.idairyj.2009.04.008. [DOI] [Google Scholar]
- 60.Del Olmo A, Calzada J, Nuñez M. Benzoic acid and its derivatives as naturally occurring compounds in foods and as additives: uses, exposure, and controversy. Crit Rev Food Sci Nutr. 2017;57(14):3084–3103. doi: 10.1080/10408398.2015.1087964. [DOI] [PubMed] [Google Scholar]
- 61.Park SY, Yoo MY, Paik HD, Lim SD. Production of benzoic acid as a natural compound in fermented skim milk using commercial cheese starter. J Dairy Sci. 2017;100(6):4269–4275. doi: 10.3168/jds.2016-12399. [DOI] [PubMed] [Google Scholar]
- 62.Hayaloglu AA, Cakmakci S, Brechany EY, Deegan KC, McSweeney PLH. Microbiology, biochemistry, and volatile composition of Tulum cheese ripened in goat’s skin or plastic bags. J Dairy Sci. 2007;90(3):1102–1121. doi: 10.3168/jds.S0022-0302(07)71597-7. [DOI] [PubMed] [Google Scholar]
- 63.Kondyli E, Katsiari MC, Masouras T, Voutsinas LP. Free fatty acids and volatile compounds of low-fat Feta-type cheese made with a commercial adjunct culture. Food Chem. 2002;79(2):199–205. doi: 10.1016/S0308-8146(02)00132-2. [DOI] [Google Scholar]
- 64.Tosun I. Master’s Thesis. Graduate School of Natural and Applied Sciences, Departmant of Food Engineering. Bursa, Turkey: Uludağ University; 2009. Beyaz peynirin uçucu flavor bileşikleri üzerine, starter kültür ve olgunlaştırmanın etkisi. [Google Scholar]
- 65.Gatzias IS, Karabagias IK, Kontominas MG, Badeka AV. Geographical differentiation of feta cheese from northern Greece based on physicochemical parameters, volatile compounds and fatty acids. LWT - Food Sci Technol. 2020;131:109615. doi: 10.1016/j.lwt.2020.109615. [DOI] [Google Scholar]
- 66.Aminifar M, Hamedi M, Emam-Djomeh Z, Mehdinia A. Investigation on proteolysis and formation of volatile compounds of Lighvan cheese during ripening. J Food Sci Technol. 2014;51(10):2454–2462. doi: 10.1007/s13197-012-0755-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bozoudi D, Claps S, Abraham EM, Parissi ZM, Litopoulou-Tzanetaki E. Volatile organic compounds of mountainous plant species and the produced milk as affected by altitude in Greece: a preliminary study. Int J Dairy Technol. 2019;72(1):159–164. doi: 10.1111/1471-0307.12573. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.