Abstract
Among the milk contaminating microorganisms, those which are able to form heat-resistant spores are concerning, especially for dairy companies that use ultra-high temperature (UHT) technology. These spores, throughout storage, can germinate and produce hydrolytic enzymes that compromise the quality of the final product. This study evaluated 184 UHT milk samples from different batches collected from seven Brazilian dairy companies with a possible microbial contamination problem. The bacteria were isolated, phenotypically characterized, clustered by REP-PCR, and identified through 16S rDNA sequencing. The presence of Bacillus sporothermodurans was verified using biochemical tests (Gram staining, catalase and oxidase test, glucose fermentation, esculin hydrolysis, nitrate reduction, and urease test). According to these tests, none of the isolates presented typical characteristics of B. sporothermodurans. In sequence, the isolates, that presented rod-shapes, were submitted to molecular analyses in order to determine the microbial biodiversity existing among them. The isolates obtained were grouped into 16 clusters, four of which were composed of only one individual. A phylogenetic tree was constructed using the sequences obtained from the 16S rDNA sequencing and some reference strains of species close to those found using BLAST search in the NCBI nucleotide database. Through this tree, it was possible to verify the division of the isolates into two large groups, the Bacillus subtilis and the Bacillus cereus groups. Furthermore, most isolates are phylogenetically closely related, which makes it even more difficult to identify them at the species level. In conclusion, it was possible to assess, in general, the groups of sporulated contaminants in Brazilian UHT milk produced in the regions evaluated. In addition, it was also possible to determine the biodiversity of spore-forming bacteria found in UHT milk samples, thus opening up a range of possible research topics regarding the effects of the presence of these microorganisms on milk quality.
Keywords: UHT milk, Spore-forming bacteria, Bacillus sp., Milk quality, Biodiversity
Introduction
Considering the availability of formal fluid milk, in Brazil, about 6977 billion liters (26.1%) were designated for the ultra-high temperature (UHT) milk sector in 2020, which was just below fluid milk used for cheese (33.8%) and powdered milk (26.6%) production. UHT milk consumption in Brazil has increased significantly and currently accounts for about 87% of the country’s fluid milk consumption [1]. This fact can be explained by the long shelf life of the product and its ease in storage, the practicality of the packaging, and the fact that the cold chain is eliminated during the internal logistics and distribution of the product.
Ultrapasteurization or UHT treatment consists of a heat treatment in continuous flow (130–150 °C/2–4 s), followed by cooling at 32 °C and aseptic packing in sterile and hermetically sealed packages [2]. This legislation establishes that UHT milk must not contain microorganisms capable of multiplying under normal conditions of storage and distribution [2]. This heat treatment process is able to eliminate all vegetative microorganism cells present in milk, as well as a large number of spores; except for heat resistant spores, such as is in the case of Geobacillus stearothermophilus, Bacillus subtilis, Bacillus megaterium, Bacillus coagulans, Bacillus licheniformis, and Bacillus cereus and Bacillus Sporothermodurans spores—bacterium that produce one of the most heat-resistant spores [3–6].
Thus, there is a great possibility that heat-resistant spores can germinate and, after germination, the bacteria can multiply during product storage and distribution, making it unfit for consumption and at odds with the standards of legislation [7]. In addition, after germination, the bacteria can multiply, achieve high counts, and produce hydrolytic enzymes that reduce product quality, mainly texture change and production of off flavors [3, 8, 9].
Based on this, the objective of this study is to detect the presence of spores and spore-forming bacteria in UHT milk samples from three Brazilian regions. Furthermore, to characterize the presence of B. sporothermodurans strains through phenotypic and genotypic analyses and, finally, determine the genetic biodiversity among the isolates through phylogenetic analysis.
Materials and methods
UHT milk samples
A total of 184 UHT milk samples from batches with possible microbial contamination and collected from seven Brazilian dairy companies (A, B, C, D, E, F, and G) were analyzed over 6 months (June to November).
Microbiological analyses of UHT milk samples and determination of spore-forming bacteria presence
Immediately upon receipt, the UHT milk samples were incubated for 7 days at 36 ± 1 °C. Subsequently, the samples were evaluated for the occurrence of changes in product characteristics (coagulation, flocculation, desorption, off-odor, or other). When evident change occurred, no further analyses were performed with the sample in question, and the result was reported as “altered product after incubation at 36 ± 1 °C for 7 days.”
Samples that did not display modification after incubation were ten-fold diluted in tubes containing 9 mL of 0.1% (w/v) peptone saline solution until 10−2 dilution. The dilutions were then pour plated onto brain heart infusion (BHI; Kasvi, São José dos Pinhais, PR, Brazil) agar and nutrient agar free of yeast extract (Kasvi), in duplicate. The plates were incubated at 30 ± 1 °C for 72 h. After incubation, the colonies that formed on the plates were counted and the results of aerobic mesophilic counts were expressed in CFU∙mL−1 [10].
To determine the presence of spore-forming bacteria, one aliquot of approximately 10 mL of each UHT milk sample was collected in sterile test tubes. Each of the aliquots was subjected to heat treatment (80 °C/10 min) [11] to eliminate all vegetative forms of the microorganisms present and to allow for the detection of spores.
After heat treatment, the aliquots were diluted (up to 10−2) in 0.1% (w/v) peptone saline solution and pour plated, in duplicate, using BHI agar (Kasvi) and nutrient agar free of yeast extract (Kasvi), and then followed by incubation at 30 ± 1 °C for 72 h. The colonies formed on the plates were counted, after incubation, and the results of spore-forming bacteria expressed in CFU∙mL−1.
Ten percent of the colonies were randomly selected from each BHI agar plate, referring to the count of aerobic mesophiles and germinated spores, followed by purification through the streak plate method on plate count agar (PCA; Kasvi). The pure cultures were stored at − 80 °C in BHI broth (Kasvi) supplemented with 30% (v/v) glycerol.
Phenotypic analyses of isolates
Each isolate from the UHT milk samples was submitted to biochemical tests, according to the methodology described by Normative Instruction no. 62 of 2003, with regards to phenotypic characterization to identify the presence of B. sporothermodurans. The following tests were performed: Gram staining, catalase and oxidase test, anaerobic growth, glucose fermentation, esculin hydrolysis, nitrate reduction, and urease test.
After performing the phenotypic tests, the isolates that showed bacilli morphology through Gram staining were cultivated in BHI broth (Kasvi) and subsequently subjected to molecular analysis.
Molecular biodiversity analyses
DNA extraction
A pellet of activated isolate culture was obtained by centrifugation at 10.000 × g for 10 min. The DNA of each isolate was extracted and purified using the DNA Purification Wizard® Genomic kit (Promega Corp., Madison, WI, USA). The quality and the concentration of the extracted DNA were measured on a NanoDrop™ Lite Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and the DNA concentration was standardized to 100 ng/μL.
Clustering of isolates by Rep-PCR
Rep-PCR analyses were performed according to the protocol described by Dal Bello et al. [12] using a single universal primer (GTG)5 (5′-GTGGTGGTGGTGGTG-3′). PCR reactions contained 12.5 μL of Go Taq Green Master Mix 2x (Promega), 1.0 μL of the primer (100 pMol), 100 ng DNA, and ultra-pure water (Promega) for a final volume of 25 μL. PCR amplification was carried out in a thermal cycler, and the cycle used was 95 °C for 5 min as initial step, 95 °C for 30 s, annealing at 40 °C for 30 s and 65 °C for 8 min 30 cycles, and a final extension step of 65 °C for 16 min [12].
The PCR products were visualized on 2% (w/v) agarose gel stained with GelRed (Biotium Inc., Hayward, CA, USA) and left for 2 h at a constant voltage of 75 V in 0.5 × TBE buffer, and developed using an LPIX transilluminator (Loccus Biotechnology, São Paulo, SP, Brazil). The banding profile was analyzed using BioNumerics software 6.6 (Applied Maths, Kortrijk, Belgium). The similarities between the profiles were calculated using the Pearson correlation. Dendrograms were constructed using the Unweighted Pair Group Method with Arithmetic (UPGMA).
16S rDNA sequencing
A representative isolate of each group generated by Rep-PCR analyses was randomly selected and subjected to 16S rDNA gene amplification using 8f (5″-CACGGATCCAGACTTTGATYMTGGCTCAG-3″) and 1512r (5″-GTGAAGCTTACGGYTAGCTTGTTACGACTT-3″) primers [13], which gives rise to fragments of approximately 1500 bp. The amplification reaction was done using 12.5 μL of Go Taq Green Master Mix 2x (Promega), 1.0 μL of each primer (0.1 µM), 100 ng DNA, and ultra-pure water (Promega) to a final volume of 25 μL. PCR amplification was carried out in a thermal cycler and the cycle used was 95 °C for 5 min as initial step, 94 °C for 20 s, annealing at 54 °C for 20 s and 68 °C for 2 min for 35 cycles, and a final extension step of 72 °C for 7 min. The products obtained from the PCR reaction were sequenced by Macrogen Inc. (South Korea). The obtained sequences were then aligned with 16S rRNA gene sequences present in the GenBank database (http://www.ncbi.nlm.nih.gov/genbank/) of the National Center for Biotechnology Information (NCBI) using BLAST software (Basic Local Alignment Search Tool) (http://www.ncbi.nlm.nih.gov/BLAST).
Phylogenetic analyses
A phylogenetic tree was constructed from the partial 16S rRNA sequences with MEGA 11 X version 11.0.10 [14] using MUSCLE [15] to align the sequences. Reference 16S rRNA gene sequences were retrieved from NCBI’s databases. The evolutionary history was inferred using the maximum likelihood method [16], with bootstrap analyses of 1000 replicates [17], and the Hasegawa-Kishino-Yano model [18] using a discrete gamma distribution (+ G) with 5 rate categories. The Interactive Tree Of Life (iTOL v.6.4) web-based tool was used to edit the phylogenetic tree (https://itol.embl.de/) [19].
Results
Microbiological quality of UHT milk samples
After incubation for 7 days at 36 ± 1 °C, only one UHT milk sample from company B presented gelation and was not considered for the following steps due to it already being considered unsatisfactory and of an unacceptable quality [20].
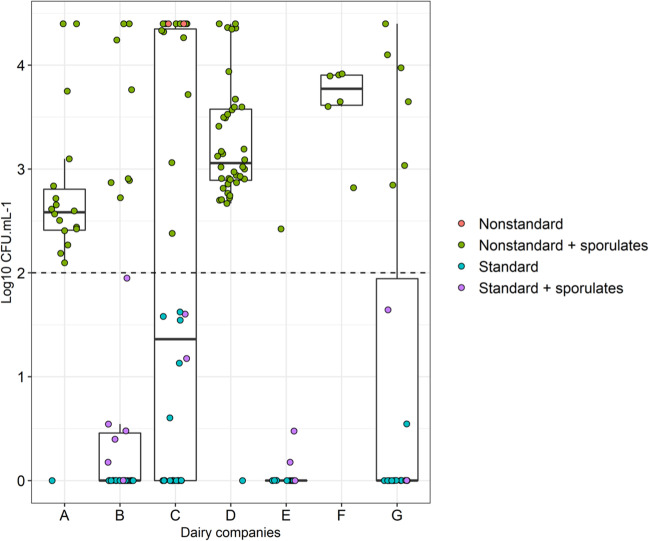
Regarding the microbiological quality, approximately 51% of the analyzed UHT milk samples presented viable aerobic mesophile counts above 100 CFU∙mL−1 (Fig. 1), which was the maximum limit allowed by current legislation at the time the samples were produced [2].
Fig. 1.
Boxplot containing an overview of the aerobic mesophilic and spore counts in the evaluated UHT milk samples, from each production company. The black dashed line indicates the limit of aerobic mesophiles allowed by legislation (100 CFU∙mL−1). Nonstandard + sporulates: UHT milk samples with total counts above the limit determined by legislation (100 CFU∙mL−1) and the presence of spore-forming bacteria; nonstandard: nonstandard samples that did not show counts of spore-forming bacteria; standard + sporulates: samples with total count within the limits determined by legislation and the presence of spore-forming bacteria; and standard: samples with total count within the standard determined by legislation without the presence of spore-forming bacteria
Most samples were found to be above the limit (100 CFU∙mL−1) established by legislation and are classified as nonstandard in relation to the aerobic mesophile and spore-forming bacteria counts after heat treatment. Only company C presented nonstandard samples that did not have any spore counts after heat treatment.
Dairy company “F” presented the worst results and the smallest variability, which is shown by the size of the rectangle in the boxplot. One hundred percent of the samples from dairy F were outside the standards established by the legislation, followed by companies “D” and “A,” with 97.5% and 94.4% of the samples above the limits, respectively (Fig. 1). Furthermore, 57.1% of 183 evaluated samples presented microbial counts after heat treatment (80 °C/10 min).
After enumeration of microorganisms in the UHT milk samples, a total of 890 isolates were obtained, of which 231 (25.9%) were isolated from heat-treated samples, therefore, spore-forming bacteria.
Phenotypic analyses of isolates
According to the Normative Instruction no. 62 of 2003, colonies of B. sporothermodurans present Gram-positive, rod-shaped morphologies, catalase, and oxidase positive reactions are not able to grow in anaerobiosis, hydrolyze esculin, do not ferment glucose, do not reduce nitrate, and do not produce urease [10]. Therefore, to be considered a strain of B. sporothermodurans, the isolates must exhibit all characteristics.
Although several isolates presented results consistent with those expected for B. sporothermodurans strains in some of the biochemical tests, none of them had all the necessary characteristics for such identification. Thus, according to the methodology used, none of the 890 isolates were identified as B. sporothermodurans.
Even though a negative result was obtained regarding the presence of B. sporothermodurans, 335 isolates were presented bacilli morphology and were subjected to molecular analyses to obtain more information regarding their taxonomy. This choice was made by the decision to keep the focus of the study on the possible representatives of the Bacillus genus, due to the ability of some species to form thermoresistant spores.
Molecular biodiversity analyses
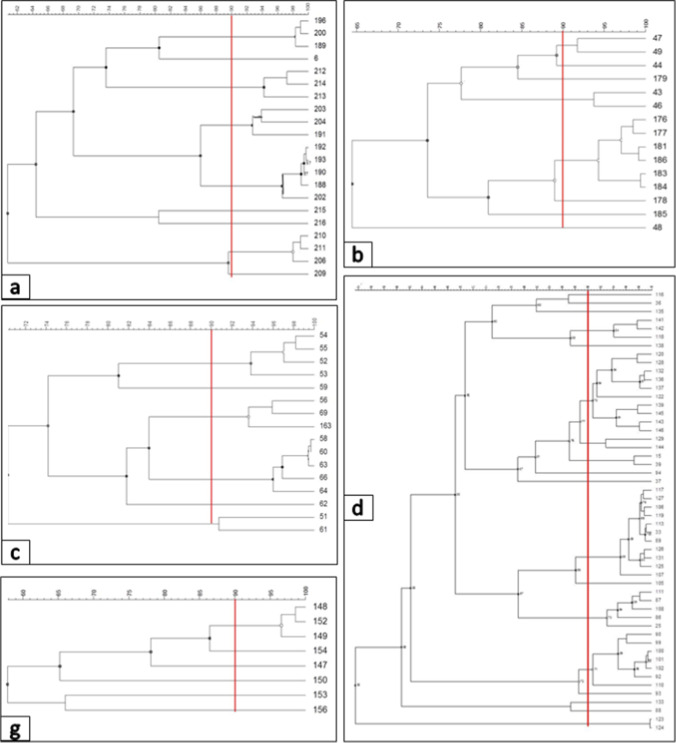
Rep-PCR analyses revealed a highly varied band profile, indicating a high diversity among the isolates. Even though the similarity between some isolates was very high, there was no case where it was 100%. Considering a level of 90% similarity, the dendrogram generated 25 clusters. Of the 25 clusters formed, 11 were formed by single strain, 11 clusters with 2 to 9 isolates, and 3 clusters with 12 to 17 isolates.
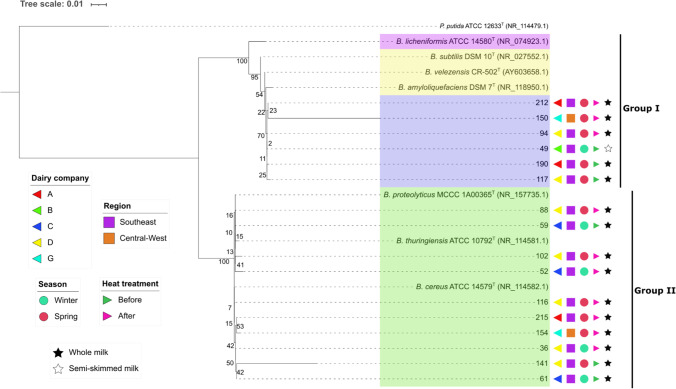
Regarding the geographic region, of the 25 groups obtained, 18 were composed only of isolates from the Southeast, 1 isolate from the Midwest, 5 isolates from the Southeast and Midwest regions, and 1 isolate from each of the three regions; and no group was formed, exclusively, by isolates from the Southern region (Fig. 3).
Fig. 3.
Phylogenetic analysis by maximum likelihood method: Evolutionary history was inferred using the maximum likelihood method and Hasegawa-Kishino-Yano model [18]. The tree with the highest log likelihood (− 4572.85) is shown. Initial tree(s) for the heuristic search were obtained automatically by applying neighbor-join and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. A discrete gamma distribution was used to model evolutionary rate differences among sites (5 categories (+ G, parameter = 1.2803)). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Evolutionary analyses were conducted in MEGA11 [14]. Numbers at nodes are percentages of bootstrap values obtained by repeating analysis 1000 times to generate majority consensus tree, and the background colors identify the groups formed through these percentages (> 70%). Pseudomonas putida 12633 T was used as outgroup
Considering the clusters, there was a high diversity even within a single production line among isolates collected from a single company. Between the isolates of each company (from “A” to “G”), 9, 8, 6, 18, 1, 1, and 6 clusters were formed, respectively (Fig. 2). Except for companies “E” and “F,” which formed a cluster with only one isolate, the others would require a level between 50 and 70% of similarity for all the isolates to be grouped into a single cluster.
Fig. 2.
Dendrograms generated from REP-PCR print data of isolates from UHT milk samples of companies (a–d, g). Similarities between profiles were calculated using Pearson’s correlation. The dendrograms were constructed using the unweighted peer group method with arithmetic (UPGMA). The highlighted red line indicates the 90% similarity limit considered for clustering
Within each cluster of the 25 formed through Rep-PCR analyses, isolates were randomly selected for identification through 16S rDNA sequencing. Of the 25 strains sent for sequencing, 16 sequences were selected for construction of the phylogenetic tree due to the quality of the sequences obtained.
Considering a 95% similarity for the identification of the genus and 97% for the identification at the species level [21], the remaining 16 strains were identified as bacteria belonging to Bacillus genus. Although 16S rDNA sequencing was not possible to identify the strains to the species level, however, there were some species with which the sequences matched better. They were B. cereus, B. thuringienses, B. proteolyticus, B. subtilis, B. licheniformis, B. amyloliquefaciens, and B. velezensis.
Phylogenetic analyses
In the present study, the taxonomic relationship of the obtained isolates was inferred through phylogenetic analysis. The phylogenetic tree (Fig. 3) was constructed using the 16S rRNA gene sequences of the 16 selected strains, and 7 reference strains (belonging to the Bacillus groups that were close matches, according to BLAST), and Pseudomonas putida as an outgroup.
The numbers above and below tree branches correspond to the bootstrap support, which was performed with 1000 replications [17]. These numbers indicate the percentage of confidence of the formed clades. The higher the percentage, the more support given to the clades. In general, values above 70% are considered reliable [22].
When observing the phylogenetic tree, and considering the minimum value of 70% for the formation of clades, the sequences were grouped into two main clades. The first is composed of strains from the B. subtilis species group (B. subtilis, B. licheniformes, B. amyloliquefaciens, and B. velezensis) and the evaluated strains 212, 150, 94, 49, 190, and 117; and the second clade is composed of the B. cereus species group (B. cereus, B. proteolyticus, and B. thuringiensis) and the evaluated strains 88, 59, 102, 52, 116, 215, 154, 36, 141, and 61. The clade formed by individuals of the B. subtilis group was subdivided into three further clades, the first being composed of just B. licheniformis; the second of B. subtilis, B. velezensis, and B. amyloliquefaciens; and the last one formed by the 6 isolates evaluated in the study mentioned above. While the B. cereus clade was not subdivided, forming a single clade composed of B. proteolyticus, B. thuringiensis, B. cereus and the isolates 88, 59, 102, 52, 116, 215, 154, 36, 141, and 61. The formation of this clade reaffirms the phylogenetic proximity between these species, which had already been evidenced through the sequence analyses obtained in the 16S rDNA sequencing.
Discussion
Although there is no correlation between bacterial counts and spoilage potential, it is important to assess the quality of UHT milk by analyzing the contamination of the final product, as required by legislation. In this analysis, the average counts of contaminating mesophiles and spore-forming bacteria in the UHT milk samples ranged from 0 to 3.7 log CFU/mL, which represents a high variability of its microbiological contamination (Fig. 1). This high variability is confirmed in other studies that evaluated the quality of UHT milk in Brazil. Reported percentages of samples with viable aerobic mesophiles counts outside the limit allowed by legislation varied between 24 and 41.2% [23–25]. Pinto et al. [26] also evaluated samples of Brazilian UHT milk for the presence of spore-forming bacteria, and among 20 milk brands evaluated, 45% presented sporulated bacteria counts. In addition, 18.7% of the brands had sporulated counts greater than 100 CFU∙mL−1.
It is important to note that in 2018, MAPA (Ministério da Agricultura, Pecuária e Abastecimento) established a Manual of official methods for the analysis of food of animal origin (Normative Instruction No. 30/2018). In this manual, the microbiological quality of UHT milk is determined through the Commercial Sterility Test for Low Acid Foods—pH > 4.6—and the results are expressed as “positive” or “negative” for the presence of microorganisms [27].
Furthermore to the microbiological quality of the raw milk, the presence of contaminating microorganisms in UHT milk may be associated with failures in the production line, especially during packaging, insufficient heat treatment, and issues in piping sanitation procedures. All of which favors microbial adhesion and biofilm formation, as well as failures in packaging asepsis procedures [5, 6, 9, 28–30].
The presence of bacteria from the Bacillus genus, specifically, is more associated with raw milk with a high spore count and/or post-process contamination due to the presence of biofilms in the pipes [31–33]. The presence of Bacillus spp. in UHT milk, confirmed by this study, reaffirms the importance of the use of quality raw material and the lowest level of contamination possible, since some spore-forming microorganisms, besides producing heat resistant enzymes and forming biofilms, are considered pathogenic and represent a great risk to the health of the consumer, as is the case with B. cereus [34].
Despite the high levels of contamination and the large number of isolates belonging to the Bacillus genus, it was not possible to detect B. sporothermodurans in the analyzed samples. In studies with Brazilian UHT milk samples, similar results were found regarding the absence of this bacterium [26, 35]. However, some researchers did find this bacterium in Brazilian UHT milk samples. Zacarchenco et al. [7] evaluated 100 samples of UHT milk from 6 Brazilian states and identified 24 isolates of B. sporothermodurans. Busatta et al. [29] and Pereira et al. [36] also identified the presence of this bacterium, respectively, in 54.5% and 60% of the brands evaluated. In a study on the presence of thermoresistant spore-forming bacteria in UHT milk from Thailand, Kmiha et al. [37] evaluated 41 samples that were taken at different stages during UHT milk manufacturing and the presence of B. sporothermodurans was identified only in raw milk samples. This difference between the results can be explained by the different techniques used to identify B. sporothermodurans. Among the studies carried out with Brazilian UHT milk samples, the presence of this species was confirmed based on the morphological characteristics and/or thermal resistance of the isolates, or on grouping isolates with a control strain belonging to B. sporothermodurans using RAPD typing technique. As presented throughout this article, species from the Bacillus genus are very close phylogenetically, which makes differentiation based only on the techniques used in these studies very difficult.
Through this study it was possible to compare molecular techniques and biochemical tests. Although they are simple and provide us with many interesting information about the metabolism and behavior of microorganisms, the biochemical tests are very laborious and require a lot of time and material to perform. In addition, the results are obtained, on average, after 72 h. Given the routine and flow of products from a dairy company, the identification of microorganisms through these techniques becomes completely unfeasible.
As shown in the results obtained through molecular techniques and phylogenetic analysis, the isolates belonging to the Bacillus genus, despite not having been identified at the species level, are phylogenetically close to some species of the genus, which is observed through the phylogenetic tree presented in Fig. 3.
The branches of the tree are associated with the transmission of genetic information from one generation to the next; in other words, the evolution of the descendants in relation to the node from which it is derived. The longer the branch, the greater the genetic change [38]. In general, except for isolates 141 and 150, the other individuals are evolutionarily very close to their ancestors, which indicates small genetic differences between them.
There was no pattern regarding the grouping of strains according to region, company, or season in which they were isolated. Furthermore, before and after heat treatment at 80 °C/10 min, the bacteria also subdivided between the main clades (group 1 and group 2). These results give us a general idea about the contaminants in Brazilian UHT milk. There is a high possibility that this is a common group of bacteria that do not depend on the region and season in which they were isolated.
Among the species associated with the isolates, the presence of B. cereus in food is very important from the point of view of food safety, since it is responsible for two food-borne diseases: the emetic and the diarrheal syndromes. The emetic syndrome is associated with the ingestion of a toxin previously produced in the food, whereas the diarrhea syndrome is the ingestion of the microorganism that in the intestine of the host produces an enterotoxin [3, 31, 39]. B. cereus strains were also found in other studies with Brazilian UHT milk samples [40–43]. In addition to UHT milk, this species has already been isolated from several dairy products such as raw milk, pasteurized milk, milk powder, ice cream, and fermented milk [31, 40, 41, 44–47].
Liu et al. [48] and Lorenzo et al. [3] identified B. proteolyticus and B. thuringiensis as a new species of the B. cereus group, respectively. B. proteolyticos strains are capable of producing proteinases that hydrolyze starch and casein [48]. In turn, B. thuringiensis, like B. cereus, is pathogenic and has been isolated in farm environments and in dairy products such as raw milk, cream, pasteurized milk, and cheese [45, 49]. These species were also associated with some sequences of the isolates evaluated in this study. This fact draws attention because, associated with the toxigenic potential of B. cereus, such bacteria have a high potential for deterioration. The strains associated with these species were mainly isolated from UHT milk samples from dairy companies in the Southeast.
Some isolates from the Southeast and Central-West UHT milk samples showed great similarity with B. subtilis. This species is related with the deterioration of various foods, which mainly include dairy products such as raw, pasteurized, and UHT milk [50], as it is capable of producing a variety of enzymes (proteolytic, aminolytic, lipolytic, and fibrolytic), a number of metabolites and volatile substances with antifungal activity, and bacteriocins [51–53]. Pinto et al. [26], in a study on the microbiological quality of Brazilian UHT milk, also reported the presence of B. subtilis corresponding to 68% of the 46 isolated strains.
B. licheniformis was associated with the strains isolated in the Southeast, and it is known for its ability to cause deterioration in dairy products due to the production of proteolytic and lipolytic enzymes that compromise the sensorial and functional properties of these products. In addition, it has the ability to form biofilms in processing plants and is considered by some researchers the predominant Bacillus species in dairy industries [31, 54–56]. Together with B. subtilis and B. cereus, these are the most predominant mesophilic aerobic spore-forming bacteria in raw milk [57].
B. velezensis and B. amyloliquefaciens are another species of the B. subtilis group [58–60] associated with evaluated isolate sequences. In recent studies they demonstrated the ability of B. velezensis strains to form biofilms and produce proteolytic and lipolytic enzymes that, consequently, compromise the quality of products in dairy industries [61, 62]. McKillip et al. [63] identified isolates of B. amyloliquefaciens in organic UHT milk and proved that it is a biofilm producer in amounts exceeding those of the B. cereus ATCC 14579 strain used as control.
It was possible to notice that most species of the Bacillus genus were genetically associated with the isolates found in the evaluated UHT milk samples and are producers of enzymes capable of hydrolyzing milk constituents and causing its destabilization. It is likely that one of these species was responsible for the gelation of the sample from company B. The gelation of UHT milk consists of the change in the physical state of the product from fluid to gel. This modification can occur due to the action of proteolytic enzymes produced by some bacteria. Kappa-casein is easily degraded by these proteases because it is located on the micelle surface. When it is hydrolyzed, a destabilization of the casein micelle can lead to milk coagulation [64]. The time required for gel formation over UHT milk storage is dependent on the extent of contamination of raw milk; however, each bacterium has its predilection to produce such enzymes, which leads us to conclude that the bacterial count is not always directly linked to the amount of enzyme produced [65].
Due to the typical characteristics of the Bacillus species, to which the isolates were phylogenetically close, it can be suggested that the problems associated with the batches to which the samples belonged to may be linked to the presence of biofilms in the processing line, as well as the use of raw milk with high microbiological and spore count for UHT milk production.
It is important that companies apply all the necessary tools for quality management and increasingly encourage their suppliers of raw material to always seek to produce high-quality milk. It is the only way possible to minimize the problems associated with microbiological contamination in UHT products and ensure the consumer has a high-quality product.
Conclusion
Through the results obtained, it can be concluded that spore-forming bacteria of the Bacillus genus comprise an important group of contaminants in UHT milk in Brazil. Although there was a possibility that the evaluated UHT milk samples belong to batches with problems, the presence of isolates phylogenetically close to species capable of causing food-borne diseases, such as B. cereus, gives us future perspectives to search for faster detection and identification methods for these contaminants. In addition, more effective conservation methods, compatible with the reality of the dairy industries, against this class of microorganisms must also be evaluated. At the same time, one should always seek to raise awareness among producers about the importance of raw material quality.
Acknowledgements
The authors are thankful to Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, Brasília, DF, Brazil, Financial code 001), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brasília, DF, Brazil), and Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG, Belo Horizonte, MG, Brazil).
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Responsible Editor: Luis Augusto Nero
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.ABLV (Associação Brasileira de Leite Longa Vida) (2020) Relatório Anual da Administração – 2020
- 2.BRASIL. Ministério da Agricultura Pecuária e Abastecimento (1997) Portaria no 370, de 04 de setembro de 1997. Regulamento Técnico para Fixação de Identidade e Qualidade do Leite U.H.T (U.A.T). Diário Oficial da República Federativa do Brasil 19700
- 3.Lorenzo JM, Munekata PE, Dominguez R et al (2018) Main groups of microorganisms of relevance for food safety and stability: general aspects and overall description. In: Innovative technologies for food preservation: Inactivation of spoilage and pathogenic microorganisms. Elsevier, pp 53–107
- 4.Pettersson B, Lembke F, Hammer P, et al. Bacillus sporothemodurans, a new species producing heat-resistant endospores highly. Int J Syst Bacteriol. 1996;46:759–764. doi: 10.1099/00207713-46-3-759. [DOI] [PubMed] [Google Scholar]
- 5.Menezes MF, Simeoni CP, Bortoluzzi D, et al. Microbiota e conservação do leite. Revista Eletrônica em Gestão, Educação e Tecnologia Ambiental. 2014;18:76–89. doi: 10.5902/2236117013033. [DOI] [Google Scholar]
- 6.Techer C, Baron F, Jan S (2014) Spoilage of animals products | Microbial milk spoilage. In: Encyclopedia of Food Microbiology, 2nd edn. pp 446–452. 10.1016/B978-0-12-384730-0.00443-2
- 7.Zacarchenco PB, de Freitas Leitão MF, Destro MT, Andrigheto C. Ocorrência de Bacillus sporothermodurans em leite UAT/UHT brasileiro e a influência do tratamento térmico. Ciênc Tecnol Aliment. 2000;20(363):368. doi: 10.1590/S0101-20612000000300014. [DOI] [Google Scholar]
- 8.Zhang D, Li S, Palmer J et al (2020) The relationship between numbers of Pseudomonas bacteria in milk used to manufacture UHT milk and the effect on product quality. Int Dairy J 105:104687. 10.1016/j.idairyj.2020.104687
- 9.Elzhraa F, Al-Ashmawy M, El-Sherbini M, Abdelkhalek A. Evaluation of physicochemical properties and microbiological quality of UHT milk regularly introduced to resident patients in Mansoura University hospitals. Veterinarija ir Zootechnika. 2021;79:9–16. [Google Scholar]
- 10.BRASIL. Ministério da Agricultura Pecuária e Abastecimento (2003) Instrução Normativa no 62, de 26 de agosto de 2003. Oficializa os Métodos Analíticos Oficiais para Análises Microbiológicas para Controle de Produtos de Origem Animal e Água. Diário Oficial da República Federativa do Brasil. Brasília, DF, 14, sessão 1
- 11.Doll EV, Scherer S, Wenning M (2017) Spoilage of microfiltered and pasteurized extended shelf life milk is mainly induced by psychrotolerant spore-forming bacteria that often originate from recontamination. Front Microbiol 8:135. 10.3389/fmicb.2017.00135 [DOI] [PMC free article] [PubMed]
- 12.Dal Bello B, Rantsiou K, Bellio A, et al. Microbial ecology of artisanal products from North West of Italy and antimicrobial activity of the autochthonous populations. LWT. 2010;43:1151–1159. doi: 10.1016/j.lwt.2010.03.008. [DOI] [Google Scholar]
- 13.Felske A, Rheims H, Wokerink A, et al. Ribosome analysis reveals prominent activity of an uncultured member of the class Actinobacteria in grassland soils. Microbiology (N Y) 1997;143:2983–2989. doi: 10.1099/00221287-143-9-2983. [DOI] [PubMed] [Google Scholar]
- 14.Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38:3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 17.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution (N Y) 1985;39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 18.Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 19.Letunic I, Bork P. Interactive Tree of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:w293–w296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.BRASIL. Ministério da Agricultura Pecuária e Abastecimento (2019) Instrução Normativa n° 60, de 23 de dezembro de 2019. Listas de padrões microbiológicos para alimentos prontos para oferta ao consumidor. Diário Oficial da República Federativa do Brasil. Brasília, DF, 133
- 21.Guinebretière MH, Auger S, Galleron N, et al. Bacillus cytotoxicus sp. nov. is a novel thermotolerant species of the Bacillus cereus group occasionally associated with food poisoning. Int J Syst Evol Microbiol. 2013;63:31–40. doi: 10.1099/ijs.0.030627-0. [DOI] [PubMed] [Google Scholar]
- 22.Hall BG. Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol. 2013;30:1229–1235. doi: 10.1093/molbev/mst012. [DOI] [PubMed] [Google Scholar]
- 23.Coelho PS, Silva N, Brescia MV, Siqueira AP. Avaliação da qualidade microbiológica do leite UAT integral comercializado em Belo Horizonte. Arq Bras Med Vet Zootec. 2001;53:256–262. doi: 10.1590/S0102-09352001000200021. [DOI] [Google Scholar]
- 24.Bersot LDS, Galvão JA, Raymundo NKL, et al. Evaluation of microbiological and physical-chemical quality of UHT milk produced in the Parana State – Brazil. Semin Cienc Agrar. 2010;31(645):652. [Google Scholar]
- 25.Souza LV, da S Meloni VA, de Souza Batista C, et al. Avaliação da qualidade microbiológica e físico-química de leite UHT integral processado em indústrias do estado de Minas Gerais, Brasil. Revista Brasileira de Agropecuária Sustentável (RBAS) 2014;4(6):15. [Google Scholar]
- 26.Pinto CLO, Souza LV, Meloni VAS, et al. Microbiological quality of Brazilian UHT milk: Identification and spoilage potential of spore-forming bacteria. Int J Dairy Technol. 2018;71:20–26. doi: 10.1111/1471-0307.12339. [DOI] [Google Scholar]
- 27.BRASIL. Ministério da Agricultura Pecuária e Abastecimento (2018) Instrução Normativa no 30, de 26 de junho de 2018. Oficializa os métodos constantes do Manual de Métodos Oficiais para Análise de Alimentos de Origem Animal. Diário Oficial da República Federativa do Brasil 9
- 28.Chavan RS, Chavan SR, Khedkar CD, Jana AH. UHT milk processing and effect of plasmin activity on shelf life: a review. Compr Rev Food Sci Food Saf. 2011;10:251–268. doi: 10.1111/j.1541-4337.2011.00157.x. [DOI] [Google Scholar]
- 29.Pereira JR, Tamanini R, Rios EA, et al. Microbiota mesófila aeróbia contaminante do leite UHT. Revista do Instituto de Laticínios Cândido Tostes. 2013;68:25–31. doi: 10.5935/2238-6416.20130039. [DOI] [Google Scholar]
- 30.Costa EA, Carioca LJ, Freitas VV, et al. Avaliação da eficiência de sanitizantes sobre bactérias esporuladas isoladas de leite UHT integral. Revista do Instituto de Laticínios Cândido Tostes. 2017;71:01. doi: 10.14295/2238-6416.v71i1.442. [DOI] [Google Scholar]
- 31.Gopal N, Hill C, Ross PR et al (2015) The prevalence and control of Bacillus and related spore-forming bacteria in the dairy industry. Front Microbiol 6:1418. 10.3389/fmicb.2015.01418 [DOI] [PMC free article] [PubMed]
- 32.Velázquez-Ordoñez V, Valladares-Carranza B, Tenorio-Borroto E et al (2019) Microbial contamination in milk quality and health risk of the consumers of raw milk and dairy products. In: Nutrition in Health and Disease-Our Challenges Now and Forthcoming Time, vol 11. pp 181–205
- 33.Ledina T, Djordjevic J, Bulajic S (2021) Spore-forming bacteria in the dairy chain. In: IOP Conference Series: Earth and Environmental Science. IOP Publishing Ltd, p 012051
- 34.Montanhini MTM, Colombo M, Nero LA, Bersot LS. Short communication: presence of neutral metallopeptidase (npr) gene and proteolytic activity of Bacillus cereus isolated from dairy products. J Dairy Sci. 2013;96:5641–5643. doi: 10.3168/jds.2013-6886. [DOI] [PubMed] [Google Scholar]
- 35.Rezer APDS (2010) Avaliação da qualidade microbiológica e físico-química do leite UHT integral comercializado no Rio Grande do Sul. Universidade Federal de Santa Maria, p 73
- 36.Busatta B, Valdruga C, Cansian E, Luis R. Ocorrência de Bacillus sporothermodurans em leite UAT integral e desnatado. Food Sci Technol. 2005;25:408–411. doi: 10.1590/S0101-20612005000300003. [DOI] [Google Scholar]
- 37.Kmiha S, Aouadhi C, Klibi A, et al. Seasonal and regional occurrence of heat-resistant spore-forming bacteria in the course of ultra-high temperature milk production in Tunisia. J Dairy Sci. 2017;100:6090–6099. doi: 10.3168/jds.2016-11616. [DOI] [PubMed] [Google Scholar]
- 38.Baldauf SL. Phylogeny for the faint of heart: a tutorial. Trends Genet. 2003;19:345–351. doi: 10.1016/S0168-9525(03)00112-4. [DOI] [PubMed] [Google Scholar]
- 39.Senesi S, Ghelardi E. Production, secretion and biological activity of Bacillus cereus enterotoxins. Toxins (Basel) 2010;2:1690–1703. doi: 10.3390/toxins2071690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vidal-Martins AMC, Rossi OD, Jr, Rezende-Lago NC. Microrganismos heterotróficos mesófilos e bactérias do grupo do Bacillus cereus em leite integral submetido a ultra alta temperatura. Arq Bras Med Vet Zootec. 2005;57:396–400. doi: 10.1590/S0102-09352005000300019. [DOI] [Google Scholar]
- 41.Rezende-Lago NCM, Rossi OD, Jr, Vidal-Martins AMC, Amaral LA. Occurrence of Bacillus cereus in Whole milk and enterotoxigenic potential of the isolated strains. Arq Bras Med Vet Zootec. 2007;59:1563–1569. doi: 10.1590/S0102-09352007000600032. [DOI] [Google Scholar]
- 42.Montanhini MTM, Pinto JPDAN, Bersot LDS. Ocorrência de Bacillus cereus em leite comercializado nos estados do Paraná, Santa Catarina e São Paulo. UNOPAR Cientifíca - Ciências biológicas e da Saúde. 2012;14:155–158. doi: 10.17921/2447-8938.2012v14n3p%p. [DOI] [Google Scholar]
- 43.de Rezende NCM, Rossi OD, doAmaral LA. Ocorrência de bactérias do grupo do Bacillus cereus em leite UHT integral (Uitra-High-Temperature) Revista Brasileira de Ciência Veterinária. 2000;7:162–166. doi: 10.4322/rbcv.2015.205. [DOI] [Google Scholar]
- 44.Wong H-C, Chang M-H, Fan J-Y. Incidence and characterization of Bacillus cereus isolates contaminating dairy products. Appl Environ Microbiol. 1988;54:699–702. doi: 10.1128/aem.54.3.699-702.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bartoszewicz M, Hansen BM, Swiecicka I. The members of the Bacillus cereus group are commonly present contaminants of fresh and heat-treated milk. Food Microbiol. 2008;25:588–596. doi: 10.1016/j.fm.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 46.Zhou G, Liu H, He J, et al. The occurrence of Bacillus cereus, B. thuringiensis and B. mycoides in Chinese pasteurized full fat milk. Int J Food Microbiol. 2008;121:195–200. doi: 10.1016/j.ijfoodmicro.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 47.Zhou G, Zheng D, Dou L, et al. Occurrence of psychrotolerant Bacillus cereus group strains in ice creams. Int J Food Microbiol. 2010;137:143–146. doi: 10.1016/j.ijfoodmicro.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 48.Liu Y, Du J, Lai Q, et al. Proposal of nine novel species of the Bacillus cereus group. Int J Syst Evol Microbiol. 2017;67:2499–2508. doi: 10.1099/ijsem.0.001821. [DOI] [PubMed] [Google Scholar]
- 49.Molva C, Sudagidan M, Okuklu B. Extracellular enzyme production and enterotoxigenic gene profiles of Bacillus cereus and Bacillus thuringiensis strains isolated from cheese in Turkey. Food Control. 2009;20:829–834. doi: 10.1016/j.foodcont.2008.10.016. [DOI] [Google Scholar]
- 50.Heyndrickx M, Scheldeman P (2002) Bacilli associated with spoilage in dairy products and other food. In: Applications and Systematics of Bacillus and Relatives. pp 64–82. 10.1002/9780470696743.ch6
- 51.Schallmey M, Singh A, Ward OP. Developments in the use of Bacillus species for industrial production. Can J Microbiol. 2004;50:1–17. doi: 10.1139/w03-076. [DOI] [PubMed] [Google Scholar]
- 52.Kai M, Effmert U, Berg G, Piechulla B. Volatiles of bacterial antagonists inhibit mycelial growth of the plant pathogen Rhizoctonia solani. Arch Microbiol. 2007;187:351–360. doi: 10.1007/s00203-006-0199-0. [DOI] [PubMed] [Google Scholar]
- 53.Chen H, Xiao X, Wang J, et al. Antagonistic effects of volatiles generated by Bacillus subtilis on spore germination and hyphal growth of the plant pathogen, Botrytis cinerea. Biotechnol Lett. 2008;30:919–923. doi: 10.1007/s10529-007-9626-9. [DOI] [PubMed] [Google Scholar]
- 54.De Jonghe V, Coorevits A, De Block J, et al. Toxinogenic and spoilage potential of aerobic spore-formers isolated from raw milk. Int J Food Microbiol. 2010;136:318–325. doi: 10.1016/j.ijfoodmicro.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 55.Reginensi SM, González MJ, Olivera JA, et al. RAPD-based screening for spore-forming bacterial populations in Uruguayan commercial powdered milk. Int J Food Microbiol. 2011;148:36–41. doi: 10.1016/j.ijfoodmicro.2011.04.020. [DOI] [PubMed] [Google Scholar]
- 56.Dhakal R, Chauhan K, Seale RB, et al. Genotyping of dairy Bacillus licheniformis isolates by high resolution melt analysis of multiple variable number tandem repeat loci. Food Microbiol. 2013;34:344–351. doi: 10.1016/j.fm.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 57.Westhoff DC, Dougherty SL. Characterization of Bacillus species iIsolated from spoiled ultrahigh temperature processed milk. J Dairy Sci. 1981;64:572–580. doi: 10.3168/jds.S0022-0302(81)82614-8. [DOI] [Google Scholar]
- 58.Ruiz-García C, Béjar V, Martínez-Checa F, et al. Bacillus velezensis sp. nov., a surfactant-producing bacterium isolated from the river Vélez in Málaga, southern Spain. Int J Syst Evol Microbiol. 2005;55:191–195. doi: 10.1099/ijs.0.63310-0. [DOI] [PubMed] [Google Scholar]
- 59.Wang LT, Lee FL, Tai CJ, Kuo HP. Bacillus velezensis is a later heterotypic synonym of Bacillus amyloliquefaciens. Int J Syst Evol Microbiol. 2008;58:671–675. doi: 10.1099/ijs.0.65191-0. [DOI] [PubMed] [Google Scholar]
- 60.Jeyaram K, Romi W, Singh TA, et al. Distinct differentiation of closely related species of Bacillus subtilis group with industrial importance. J Microbiol Methods. 2011;87:161–164. doi: 10.1016/j.mimet.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 61.Elegbeleye JA, Buys EM. Molecular characterization and biofilm formation potential of Bacillus subtilis and Bacillus velezensis in extended shelf-life milk processing line. J Dairy Sci. 2020;103:4991–5002. doi: 10.3168/jds.2019-17919. [DOI] [PubMed] [Google Scholar]
- 62.Elegbeleye JA, Buys EM (2022) Potential spoilage of extended shelf-life (ESL) milk by Bacillus subtilis and Bacillus velezensis. LWT 153:112487. 10.1016/j.lwt.2021.112487
- 63.McKillip JL, Grutsch A, Wagner ER, Klug C. Bacillus amyloliquefaciens from UHT organic milk produces biofilm and demonstrates virulence potential. J Anim Sci. 2016;94:283–283. doi: 10.2527/jam2016-0597. [DOI] [Google Scholar]
- 64.Bulgari O, Caroli AM. Protein profile of ultra high temperature (UHT) milk: occurence of para-kapa-casein in the sediment. Scienza e Tecnica Lattiero-Casearia. 2017;68:53–58. [Google Scholar]
- 65.Deeth HC (2019) The effect of UHT processing and storage on milk proteins. In: Milk Proteins: From Expression To Food. Elsevier, pp 385–421. 10.1016/B978-0-12-815251-5.00010-4