Summary
Advanced sequencing technologies have expedited resolution of higher-level arthropod relationships. Yet, dark branches persist, principally among groups occurring in cryptic habitats. Among chelicerates, Solifugae (“camel spiders”) is the last order lacking a higher-level phylogeny and have thus been historically characterized as “neglected [arachnid] cousins”. Though renowned for aggression, remarkable running speed, and xeric adaptation, inferring solifuge relationships has been hindered by inaccessibility of diagnostic morphological characters, whereas molecular investigations have been limited to one of 12 recognized families. Our phylogenomic dataset via capture of ultraconserved elements sampling all extant families recovered a well-resolved phylogeny, with two distinct groups of New World taxa nested within a broader Paleotropical radiation. Divergence times using fossil calibrations inferred that Solifugae radiated by the Permian, and most families diverged prior to the Paleogene-Cretaceous extinction, likely driven by continental breakup. We establish Boreosolifugae new suborder uniting five Laurasian families, and Australosolifugae new suborder uniting seven Gondwanan families using morphological and biogeographic signal.
Subject areas: Zoology, Phylogenetics, Evolutionary biology, Phylogeny
Graphical abstract

Highlights
-
•
Ultraconserved elements unlock the first phylogeny of Solifugae (camel spiders)
-
•
A series of sensitivity analyses reconstruct robust evolutionary relationships
-
•
Ancestral areas of Solifugae suggest distributions resulted from Pangean breakup
-
•
New suborders for Laurasian (Boreosolifugae) and Gondwanan (Australosolifugae) taxa
Zoology; Phylogenetics; Evolutionary biology; Phylogeny
Introduction
Chelicerata is an ancient, monophyletic group of arthropods that is characterized by extensive diversity, high body plan disparity among their different orders, and inclusion of numerous charismatic taxa, such as spiders, scorpions, and horseshoe crabs. Despite their considerable diversity, nearly every group of chelicerate orders has benefitted from recent advances in molecular phylogenetics, genomics, and renewed interest in evolutionary dynamics. The past decade alone has witnessed the first molecular phylogenetic hypotheses for several orders, such as Scorpiones, Ricinulei (hooded tick-spiders), Palpigradi (microwhip scorpions), Thelyphonida (vinegaroons), Schizomida (short-tailed whip scorpions), and Amblypygi (whip spiders).1,2,3,4,5 More diverse chelicerate groups, such as Acari (mites), Araneae (spiders), Opiliones (harvestmen), and Pseudoscorpiones, have witnessed a surge of evolutionary inquiry in the past ten years.6,7,8,9,10,11,12,13,14,15,16,17 This decade is also notable for the completion of the first genomes for several chelicerate orders.18,19,20,21,22,23,24 These advances have revolutionized modern views of chelicerate phylogeny and evolution, prompting reevaluations of historical paradigms of habitat transition and evolution of complex characters.6,8,24,25,26,27,28
Solifugae, variously known as “camel spiders” or “sun spiders”, is a mesodiverse group (in comparison with other arachnid groups) that currently includes ca. 1,200 species classified in 12 families and 144 genera (Figure 1).29 Notorious for their fearsome appearance, large chelicerae (relative to body size), and high bite force,30 solifuges are one of seven chelicerate orders that are commonly referred to as “the neglected cousins” in the arachnological community, due to a dearth of systematic and phylogenetic studies.31 Various characteristics of solifuges distinguish them from other arachnids, such as a robust pair of two-segmented chelicerae, adhesive structures on the termini of the pedipalps for prey capture, the presence of malleoli (“racquet organs”, used for chemoreception and analogous to the pectines of scorpions) on the ventral surface of leg IV,32,33,34 and an extensive tracheal system.35 The last of these is understood to facilitate the rapid running speed of solifuges, as well as their inhabitation of some of the driest habitats on Earth. Historically, solifuges were thought to be closely related to pseudoscorpions36,37 a hypothesis that has since been contradicted by sperm ultrastructure,38 rare genomic changes, and the ensuing placement of pseudoscorpions as the sister group of scorpions,24 (but see39). Solifugae has more recently been recovered as part of a clade with acariform mites (Poecilophysidea), and possibly also palpigrades (Cephalosomata).27,40 A close relationship of these three orders is supported by the arrangement of the anterior body segments and the structure of the coxal glands, and is recovered by a subset of molecular analyses that have emphasized dense taxonomic sampling.27,40,41,42
Figure 1.
A sample of morphological diversity of Solifugae
(A) Adult female of Eremobates (Eremobatidae) from AZ, USA.
(B) Brooding female of Paragaleodes (Galeodidae) over a clutch of hatchlings, from Israel.
(C) Adult female of Mummucia (Mummuciidae) from Chile.
(D) Adult male of Procleobis patagonicus (Ammotrechidae) from Argentina.
(E) Adult female of unidentified Galeodidae (cf. Galeodes) from Israel.
(F) An unidentified Daesiidae from Israel.
(G) An unidentified Rhagodidae from Israel.
(H) An actively burrowing Hexisopodidae (cf. Hexisopus) from Namibia.
Photographs: G. Giribet (A); I. Armiach (B, E, F, and G); H. Iuri and A. Ojanguren-Affilastro (C and D); S. Aharon (H).
By contrast to their placement in higher-level chelicerate phylogeny, the internal relationships of Solifugae remain virtually unknown.31 A classification of the twelve extant families was proposed by Roewer43 based on overall similarity of characters that are, in turn, highly variable across genera and species; this classification largely remains in place.29 Beyond the incidence of poorly delimited higher-level taxa, a peculiarity of solifuge systematics is the concentration of diagnostic characters in the adult males of many lineages—in some groups, juveniles cannot be reliably assigned even to a specific family. As a result, few attempts have been made to infer phylogenetic relationships using either morphological or molecular datasets. Rhagodidae was suggested to be well-separated from the remaining solifuge families on the basis of its peculiar morphology, but this inference was not based on formal analyses and the polarity of these morphological characters is unknown.43 Investigations of specific character systems have documented broad diversity, such as in the flagellum of the male chelicera and the histology of sperm ultrastructure, but these data have not been applied toward the goal of formal inference of evolutionary relationships.44,45,46 Although molecular data offer considerable advantages over anatomical datasets for phylogenetic study of enigmatic groups like Solifugae, few works have addressed internal relationships within this order. For example, RADseq data supported the monophyly of genus Eremocosta and suggested post glacial colonizations in North American deserts.47 A recent analysis of Iranian solifuges based on one mitochondrial gene was able to sample seven families and the ensuing topology suggested the paraphyly of at least three families, but without significant nodal support for most interfamilial relationships.48 The only multilocus dataset applied to solifuge relationships examined the phylogeny of the North American family Eremobatidae and this four Sanger locus-based analysis uncovered extensive non-monophyly of constituent genera, together with limited nodal support for deep nodes.49
Given the presumably ancient diversification of Solifugae, as inferred from the appearance of crown group lineages by the Mesozoic and multiple fossil genera,29,50 bridging the knowledge gaps in solifuge higher-level phylogeny requires the application of phylogenomic datasets and tissue sampling from rare, often aging, tissue collections. We therefore amassed a set of 94 solifuge species drawn from natural history collections around the world and sequenced these for a chelicerate-specific suite of ultraconserved elements (UCEs), an approach notable for its demonstrable capability to accommodate even highly degraded tissues.51 Here, we provide a robustly resolved phylogenomic tree of Solifugae, in tandem with molecular dating efforts to provide a temporal context to solifuge diversification.
Results
We compiled a comprehensive dataset with high-quality ultraconserved profiles spanning all extant chelicerate orders, represented by 132 taxa. In this dataset, Solifugae was represented by 104 taxa, amounting to almost 10% of the global fauna,29 of which we generated UCE libraries for 93 terminals (89% of taxa) and in silico extracted UCEs from existing UCE libraries, transcriptomes or genomes for the remainder (Table S1). Selection of outgroups prioritized the sampling of basal splits, to facilitate node calibration in molecular dating. Locus count yields through paralogy filtering with liberal (65%) and stringent (80%) probe-to-library identity and coverage mapping thresholds and data completeness across 25% and 40% occupancy thresholds are listed in Table S1. In addition, to mitigate the impact of taxa with sparsely represented UCE loci (= high missing data), we generated a reduced matrix with higher total information content.
Phylogenomic analyses
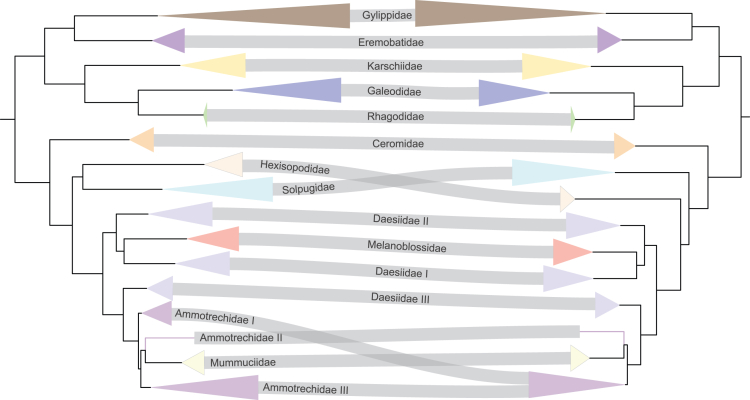
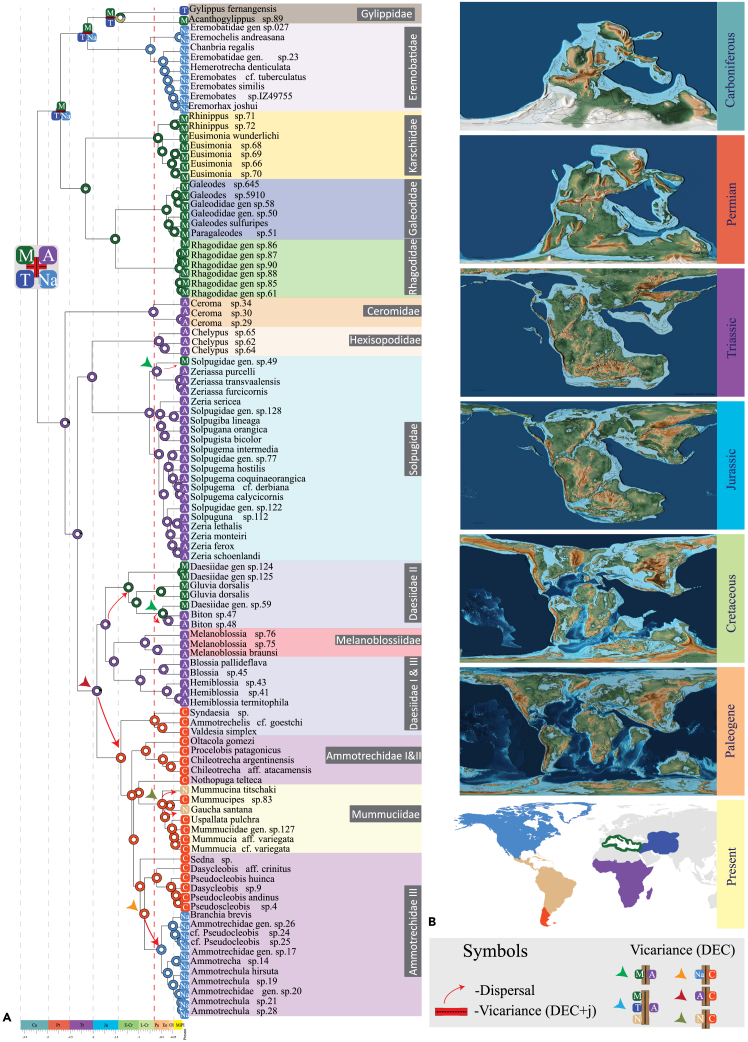
Maximum likelihood analyses based on 521 loci (25% occupancy; 65% probe-to-library identity threshold; GTR+F+I + G4 model) recovered a tree topology that divided solifuge families into two major clades, which we recognize as Boreosolifugae new suborder and Australosolifugae new suborder (Figure 6). Boreosolifugae was comprised of the Laurasian families Eremobatidae, Gylippidae, Karschiidae, Galeodidae, and Rhagodidae; these families are united by the presence of a sessile flagellum (or specialized long setae in the flagellar grove, in the case of Eremobatidae) in the chelicerae of adult males (see discussion). Australosolifugae was comprised of the Gondwanan families Ceromidae, Solpugidae, Hexisopodidae, Melanoblossiidae, Daesiidae, Mummuciidae and Ammotrechidae; these families are in turn diagnosed by the presence of a composite flagellum (see discussion). The automatic matrix reduction method (MARE) retained 267 loci and dropped the families Gylippidae, Karschiidae and Hexisopodidae, but retained the same order of interfamilial relationships and proposed suborders (Figure S1). All families except Ammotrechidae and Daesiidae were recovered as monophyletic with robust support (Figure 2). To rule out the possibility of systematic bias driven by compositional heterogeneity, GC-base proportions were mapped on the 25% occupancy phylogeny. Only two outgroup terminals, the parasitiform mite Galendroma occidentalis and Cryptocellus becki exhibited a high proportion of GC-content; excluding these taxa from the analysis did not affect the interfamilial relationships of Solifugae (Figure S2).
Figure 6.
Distribution of Solifugae families mapped on the 25% occupancy-based UCE phylogeny and world map indicating the deep split between Laurasian and Gondwanan taxa
Figure 2.
Phylogeny of Solifugae using ultraconserved elements
A maximum likelihood phylogeny of Chelicerata with all Solifugae family representatives using the 25% occupancy partitioned dataset of ultraconserved elements. Numbers at the nodes indicate percentage support values as Shimodaira-Hasegawa-like approximate likelihood ratio test and ultrafast bootstrap (not shown if > 95%).
The backbone tree topology of Solifugae was well-resolved and the basal split between the suborders was invariably recovered across analyses (Figures 2 and 3). Mummuciidae was almost always nested within Ammotrechidae, rendering the family paraphyletic (Figures 2 and 3). Melanoblossiidae was a sister group to the Daesiidae I clade, albeit with poor support (<95%). Partitioning by UCE loci with merging of similar partitions allowed did not affect any interfamilial relationship compared with phylogenies from their unpartitioned datasets across respective occupancies (Figures S3–S5).
Figure 3.
Comparison of Solifugae interfamilial relationships
Sankey comparison of interfamilial phylogenetic relationships of Solifugae compared between the 25% (left) and 40% (right) occupancy partitioned datasets at 65% contig to probe identity and coverage threshold.
Influence of stringent homology filtering
Matrices based on stringent filtering (80% identity and coverage probe-to-library match) discarded many loci, resulting in a more compact and denser dataset across different occupancies (Table S1). Some terminals were dropped at 40% occupancy because the stringency of this filter did not recover any UCE locus for those taxa. Nevertheless, at least one taxon representing each currently recognized family was present in each occupancy dataset, thereby not compromising the higher-level taxon sampling. The tree topologies recovered for matrices constructed under this threshold recovered similar relationships as previously reported for the intersuborders and interfamilial relationships. Among Solifugae, most interfamilial relationships were similar to that of the 25% occupancy phylogeny with 65% identity and coverage thresholds (Figures 2 and S6–S9). Gylippidae was recovered as paraphyletic in 25% occupancy matrix, but this is likely attributable to low locus representation (12 UCE loci for Gylippus ferganensis; Table S1), and was among the dropouts of 40% data matrix. Noticeable differences include, Ceromidae as a sister group of remaining Australosolifugae at 25% versus sister group of Solpugidae with 40% dataset. Ammotrechidae I and II formed a clade with both occupancies. Some outgroup taxa represented by very few loci were placed inside Solifugae at 40% occupancy, but with poor support. If these erroneous, poorly supported branches were to be pruned, solifuge interfamilial relationships continued to be robust even at this occupancy, which corresponds to a matrix of just 18 UCE loci (Figures S6–S9).
Divergence dating and biogeographic analyses
Divergence date optimizations calibrated with fossil age estimates in a least-squares (LSD2) and maximum likelihood (MCMCTree) frameworks, with and without ingroup fossils, recovered some non-overlapping age ranges (Figures 4 and S10–S13). Diversification of the crown group Solifugae was estimated within the Carboniferous (318–331 Ma) period. Boreosolifugae, and Australosolifugae diversified during the late diversified during the (257–278 Ma) Permian period. The MRCAs (most recent common ancestors) of most families including Gylippidae, Eremobatidae, Solpugidae, Daesiidae I, II, and Ammotrechidae I and III, originated during the Cretaceous period. The remaining families (Karschiidae, Galeodidae, Rhagodidae, Hexisopodidae, and Daesiidae III) originated post Cretaceous-Tertiary boundary.
Figure 4.
A time-calibrated phylogeny of Solifugae recovers Paleozoic radiation
(A) A maximum likelihood phylogeny of Chelicerata with all Solifugae families represented, time-calibrated using fossils reconstructed using the UCE dataset for occupancy 25%.
(B) Placement of fossils used as prior calibrations (marked with ‘star’ symbol) for the MCMCTree analysis.
Rhagodidae was estimated to have originated by late Miocene epoch, forming the most recently diverged family (Figure 4). LSD2 analysis recovered Carboniferous period as the origin for Crown Solifugae (Figures S7 and S13). However, for the suborders, LSD2 recovered strikingly recent divergence time estimates rendering them to have originated during the Triassic period. Exclusion of the two Solifugae fossils with LSD2 recovered more recent age range for all nodes (Figure S13).
To infer ancestral areas and reconstruct the biogeographic history of Solifugae, we analyzed the dated tree topology in tandem with five broad geographic areas using RASP (Figure 5). Historical biogeographic area optimizations were assessed by the best fitting model using RASP on our 25% occupancy dataset. The model with highest AICc weight was the dispersal-extinction-cladogenesis with jump dispersal parameter model (DEC+j; 0.58). (see Table S5; Figure S14). The DEC+j model recovered a combination of Turanian, African tropics, Neotropical and Mediterranean regions as the area for the most recent common ancestor (MRCA) of Solifugae (Figure 5). The ancestral area for Boreosolifugae was a combination of Turanian, Neotropical and Mediterranean (i.e., fragments of Laurasia), whereas for Australosolifugae, the Afrotropics (i.e., a constituent of Gondwana) was recovered using both models (Figure 5). Most family distributions were limited to a single biogeographic region. One group of the polyphyletic Daesiidae (clade III) is distributed within Central Chile and the Patagonian (CCP) region and the other two groups have an African origin (Figure 5). At the crown node of the CCPian and Neotropical Ammotrechidae III clade, the DEC+j model optimized CCP as the ancestral area, which implied a dispersal event from CCP to the Neotropics. The exclusion of the jump parameter optimized a combination of CCP and Neotropical region as the ancestral area, thus implying a vicariance event dividing the descendant lineages (Figure 5). Seven dispersal events were implied by the DEC+j model (marked with red arrows in Figure 5). These dispersals included three events between the Mediterranean region and African tropics; one between Jurassic to Cretaceous, two events during Paleogene; two between Neotropical and the CCP regions during the Paleogene period; one dispersal event from CCP to the Nearctic region during the Paleogene; and one African tropics to CCP regions during the Jurassic period were implied by the DEC+j model. Overall, the DEC+j model favored a single area as origin where the descendant lineages occupying different regions recovered dispersal events. Alternative coding of areas by continents for extant taxa recovered Eurasia and Africa as the ancestral areas for Boreosolifugae and Australosolifugae, respectively. A further broader area coding by supercontinent recovered Laurasia as the ancestral area for Boreosolifugae and Gondwana for Australosolifugae (Figure S11).
Figure 5.
Historical biogeography of Solifugae supports Pangean diversification
(A) Biogeographic hypothesis obtained from the Dispersal-Extinction-Cladogenesis + jump (DEC+j) parameter model of RASP analyses on the fossil-calibrated MCMCTree analysis using the 25% occupancy dataset. The dotted line in red indicates the Cretaceous Triassic boundary.
(B) Graphics for Pangean breakup from Scotese, C.R., 2016. PALEOMAP Project, http://www.earthbyte.org/paleomap---paleoatlas---for---gplates/. Ancestral areas optimized for each family and higher-level nodes are marked at the respective MRCA nodes. Red arrows indicate dispersal implied by the DEC+j model whereas the colored arrowheads indicate alternative vicariance events implied by the exclusion of the jump parameter.
Discussion
The higher-level relationships of Solifugae
A recent exploration of long branch attraction effects in chelicerate phylogeny24 showed that taxonomic undersampling, and specifically, omitting the representation of basal nodes, exacerbated topological instability of fast-evolving arachnid orders like pseudoscorpions. Using a rare genomic change as a benchmark for phylogenetic accuracy, these analyses demonstrated that taxonomic sampling (especially of slowly evolving and basally branching groups) outperformed other approaches to mitigating long branch attraction, such as increasing matrix completeness, using coalescent-aware species tree approaches, filtering by evolutionary rate, and implementing site heterogeneous models in concatenated matrices. Ontano et al.24 reasoned that applying this remedy to other fast-evolving orders may greatly stabilize chelicerate phylogeny, given that at least four orders exhibit a propensity for long branch attraction artifacts in Chelicerata.26,42,52 This proposed remedy for long branch attraction is greatly potentiated by the availability of higher-level phylogenies for unstable groups, such as Acariformes11 and Palpigradi.2 However, representation of solifuges in phylogenomic works has historically been driven entirely by the availability of sequence-grade tissues, rather than by representation of phylogenetically significant exemplars, given that the internal phylogenetic structure of this order has never been fathomed.25,26,27,28,53,54 It is possible that this oversight may have underlain their known predilection for topological instability in some phylogenomic datasets.25,26,42
To redress this basic gap in our understanding of Solifugae, we generated the first higher-level phylogeny of the group, leveraging natural history collections worldwide to sample all extant families and reconcile a dated phylogeny against the fossil record. We obtained a robust backbone tree topology, with nearly all interfamilial nodes well-supported and recovered across an array of occupancy thresholds with support. Our results show that most solifuge families represent cohesive groups, with many families exhibiting high fidelity to specific biogeographic terranes. Curiously, we recovered the North American family Eremobatidae as the sister group of Gylippidae, which is restricted to the Old World (Central Asia and South Africa), with these two taxa in turn sister group to a large clade of Old World families (Galeodidae, Karschiidae, and Rhagodidae). This result was unanticipated because gylippids were previously considered part of Karschiidae,43 and because a North American sister group of this family implies a markedly disjunct distribution. However, this relationship is consistent with shared traits of cheliceral architecture and dentition in these two families.46
Paralleling this outcome, the other groups of New World solifuges were also recovered as nested within a clade of Old World taxa (Ceromidae, Solpugidae, Hexisopodidae, and Old World daesiids). Our analyses consistently recovered a clade formed by the South American family Mummuciidae, the New World family Ammotrechidae, and the South American subset of the paraphyletic group Daesiidae (the “Daesiidae III” clade). Within each, we additionally found close relationships between geographically proximate subtaxa. These results suggest a prominent phylogenetic signature in solifuge biogeographic distributions, and across varying depths of the phylogeny. This biogeographic signal, together with the stable relationships we recovered across analyses, thus serves as the namesake for the two suborders we establish herein for the Laurasian and Gondwanan solifuge clades. This definition of suborders by ancestral distribution is also upheld by alternative coding of biogeographic areas as continents or supercontinents.
Three relationships exhibited instability across analyses and invite further scrutiny. First, the fossorial group Hexisopodidae (commonly, “mole solifuges”), from southern Africa, exhibited some topological instability, being recovered as either the sister group of Solpugidae in a minority of analyses, or with a clade formed by Daesiidae + Mummuciidae + Ammotrechidae. Both placements are partially supported by available morphological data. Specifically, analyses of solifuge sperm ultrastructure previously showed that hexisopodids and solpugids share similarities in the fine structure of spermatozoa, such as a conical acrosomal vacuole that is located within the chromatin body;45 an older work had also suggested that hexisopodids may constitute a derived subtaxon of Solpugidae.55 But hexisopodids also exhibit digitiform protuberances of membranes and putative deposits of stored glycogen, which are similarly observed in Daesiidae and Ammotrechidae, in addition to Solpugidae.45 These data strongly accord with the recovery of these four families in a clade across our analyses, but we add the caveat that morphological data for solifuge sperm ultrastructure remain fragmentary; characteristics of the spermatozoa have been surveyed only in seven of the 12 families to date.44,45 In particular, data on the sperm ultrastructure of Mummuciidae, a member of this group, are not presently available.
The second relationship that exhibited instability across analyses was the placement of Mummuciidae. While mummuciids were typically recovered as nested within Ammotrechidae, we did recover this clade as the sister group of the ammotrechids in a minority of analyses (albeit without support). As with the previously discussed case, mummuciids were previously a subtaxon of Ammotrechidae, and thus a nested placement of this putative family would not be unprecedented.43 Given the complexity of ammotrechid subfamilies (a subset of which were sampled here), we submit that future systematic revisions of the South American solifuges must consider mummuciids as potentially derived members of Ammotrechidae.
Bird et al.46 defined three types of flagella: setiform, sessile and composite. The setiform flagellum has a uniform shape and maintains affinities with plumose setae. It is present in most Eremobatidae and Melanoblossinae. The sessile flagellum is clearly modified from its original setal form and its development does not appear to have involved a longitudinal invagination along the seta to form an alembic canal (as occurs in the composite flagellum). A sessile flagellum occurs in Galeodidae, Rhagodidae, and Karschiidae (with the possible exception of Karschia). The composite flagellum comprises three sections: a stalk, a base, and a shaft. It occurs in Ammotrechidae, Ceromidae, Daesiidae, Hexisopodidae, Mummuciidae and Solpugidae. The three sections are present in Ceromidae, Hexisopodidae, Solpugidae, and some Daesiidae, whereas the shaft is considered lost in Ammotrechidae, Mummuciidae, and most Daesiidae. There are two groups where the homology of the flagellum is not clear: Karschia and Gylippinae. For the genus Karschia, three sections are recognized resembling stalk, base, and shaft; however, there is no obvious homology with the composite flagellum, and the alembic canal seems to be absent. Bird et al.46 treated the karschiid flagellum as a sessile type. For the Gylippinae, Bird et al.46 considered the flagellum as composite, but without a stalk and base, because it is fused to the dorsal surface of the fixed finger, similarly to Solpugidae. However, Bird et al.46 also stated that the absence of stalk and base may suggest that the flagellum of Gylippinae is of the sessile type. In the context of our results, when mapping flagellum type only, it is equally parsimonious to consider the flagellum of Gylippinae as sessile or composite, but the loss of stalk and base implies more steps, and thus the condition of a sessile flagellum in Gylippinae becomes more parsimonious (Figures S15 and S16). Thus, Boreosolifugae is characterized by the presence of a sessile flagellum and the Australosolifugae by the presence of composite flagellum. The setiform flagellum is most likely a secondary condition that evolved independently in Eremobatidae and Melanoblossinae, as previously suggested by Bird et al.46 If the flagellum of Gylippinae were indeed of the composite type, this would suggest that the composite flagellum is the plesiomorphic condition, and that the sessile and setiform types are derived.
Solifuge diversification reflects ancient vicariance across Pangaea
The chronological sequence of Pangean fragmentation is well-documented, and numerous cases of biotic distributions and divergence time estimates have been shown to retain the signature of supercontinental breakup.56,57,58,59 Our biogeographic analysis revealed the general pattern that the distribution of most solifuge clades is delimited by single biogeographic regions. Ancestral area optimizations in the internal nodes and the distribution of extant species (e.g., near-absence on oceanic and Darwinian islands) suggest that solifuges are habitat specialists adapted to specific environments, being largely confined to xeric habitats. In previous works, the origin of Solifugae has been estimated to span the late Cambrian or Ordovician, albeit with limited sampling of ingroup taxa.3,10,13,15,28 Like many arachnid orders, the crown group of Solifugae dates to the Carboniferous or earlier, as reflected by its fragmentary fossil record.29,50 Many solifuge families and genera are relatively young, with diversification of many genera occurring after the Cretaceous-Paleogene extinction event. While this may reflect an artifact of limited sampling, one possibility for the observation of relatively young ages of genus-level clades is that the recent aridification and the opening of comparatively young deserts may have opened new ecological niches for Solifugae, driving the diversification of this arid-adapted arachnid group by the mid-Cenozoic.49,60,61 Their absence on dry continental landmasses such as Australia may be attributable to the recent timing of aridification of the Sahul Shelf, as well as the mesic and cooler past conditions of the Australian interior.62 However, assessing this hypothesis using parametric tests requires extensive sampling of extant diversity, in tandem with consideration of variance in the ages of deserts worldwide. Targeted approaches like UCE sequencing offer the promise of renewed utility of natural history collections and revitalized prospects for hypothesis testing with poorly studied taxa.
We emphasize that various subtaxa were not included in this study, which is aimed specifically at higher-level relationships between families. Given the fidelity we observed between solifuges and the terranes that they inhabit, we anticipate that future sampling efforts will identify additional cases of non-monophyletic taxa from regions not represented in this study (e.g., Dinorhax; the putative Daesiidae of southern Asia), which will require intensive rounds of systematic revision under a phylogenomics lens. Such genomics-driven revisionary efforts have demonstrated marked success and efficiency in modernizing the classification and evolutionary analysis of comparably diverse groups, such as scorpions and harvestmen.4,12,63,64,65
Of immediate interest in this regard are the putative “Gylippidae” of southern Africa (the genera Bdellophaga, Lipophaga, and Trichotoma, which collectively form the subfamily Lipophaginae), whose distribution represents a biogeographic anomaly if they were indeed nested within Boreosolifugae (Figure 6). As an initial step to testing this placement, we inferred the relationships of this group by assembling a Sanger dataset. Sequence data for four Sanger loci (one of these a nuclear ribosomal gene) were previously generated for a single species of Trichotoma, as an outgroup exemplar in an investigation of Eremobatidae.49 Given that nuclear ribosomal bycatch is commonly present in both transcriptomes and UCE assemblies, we were able to generate a 22-taxon alignment sampling the major clades of Solifugae for the 421-bp region of 28S available for Trichotoma michaelseni. As shown in Figure S17, the gene tree of 28S recovered the basal split between Boreosolifugae and Australosolifugae, but placed Trichotoma within Australosolifugae, with support (as the sister group of Ceromidae). These results reinforce a basal split of solifuges that is consistent with subdivision into Laurasian and Gondwanan clades, and tentatively suggest that Lipophaginae should be elevated to family status. Extrapolating from these biogeographic trends, and based upon morphological correspondences of the chelicera,46 we strongly suspect that the southeast Asian genus Dinorhax (putatively a melanoblossid) is a member of Boreosolifugae.
The phylogenetic tree topopology presented herein bookends a ca. 20-year gap of available higher-level phylogenies for the orders of Chelicerata.31 The relationships inferred herein are anticipated to aid taxonomic revisionary efforts, and revitalize morphological comparative studies and biogeographic efforts within this understudied group.
Limitations of the study
We note that taxon sampling in this study is limited to available and sequence-grade exemplars of extant families; we were unable to obtain material for some unusual taxa, such as Bdellophaga, Lipophaga, and Dinorhax to test the current placement of this group. Sampling of these taxa is anticipated to test the reconstruction of the biogeographic history of the two Solifugae suborders defined herein. Future investigations of solifuge evolutionary history must emphasize the role of global perturbations of the past and the downstream effects of climatic cycles on diversification and range expansion of these cryptic, long-neglected arthropods.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Data matrices were deposited on Mendeley | This study | https://doi.org/10.17632/r6cmspfr25.1 |
| Raw reads are available from the NCBI Sequence Read Archive | This study | BioProject PRJNA1000606 |
| Experimental models: Organisms/strains | ||
| Solifugae species | This study | Table S2 |
| Software and algorithms | ||
| PHYLUCE pipeline | Faircloth66 | https://phyluce.readthedocs.io/en/latest/ |
| IQ-TREE2 | Nguyen et al.67 | http://www.iqtree.org/ |
| BBMap | Bushnell68 | https://github.com/BioInfoTools/BBMap |
| LSD2 | To et al.69 | https://github.com/tothuhien/lsd2 |
| MCMCTREE | part of PAML v.4.8; Yang70; dos Reis and Yang71 | https://github.com/abacus-gene/paml |
| RASP 4.0 | Yu et al.72 | https://github.com/sculab/RASP |
| MUSCLE v.3.2.1 | Edgar73 | https://www.ebi.ac.uk/Tools/msa/muscle/ |
| MARE v0.1.2-rc | Meyer et al.74 | https://bonn.leibniz-lib.de/en/research/research-centres-and-groups/mare |
| Other | ||
| Spider2Kv1 probe set | Kulkarni et al.14 | https://doi.org/10.1111/1755-0998.13099 |
Resource availability
Lead contact
Further information and requests for resources can be directed to and will be fulfilled by the lead contact, Dr. Siddharth Kulkarni (sskulkarni24@wisc.edu).
Materials availability
This study did not generate new unique reagents.
Method details
Species sampling
Specimens sequenced for this study were collected from field sites as part of our recent collecting campaigns, as well as contributed by collections of the Museo Argentino de Ciencias Naturales “Bernardino Rivadavia”, Buenos Aires, Argentina (MACN); The National Natural History Collections, The Hebrew University, Jerusalem, Israel (NNHC); the Denver Museum of Nature & Science, Colorado, United States (DMNS); the National Collection of Arachnida, Agricultural Research Council, South Africa (ARC-PCP), and the Zoological Museum Hamburg (ZMH). Collecting permits for study taxa were issues to different laboratories over several years; permitting data are available upon request from the authors. For de novo sequencing of UCEs, sampled exemplars of each of the 12 described extant families, as follows: 22 Ammotrechidae, three Ceromidae, 14 Daesiidae, two Eremobatidae, three Galeodidae, two Gylippidae, three Hexisopodidae, seven Karschiidae, three Melanoblossiidae, seven Mummuciidae, six Rhagodidae, and 21 Solpugidae. To this dataset, we added UCE loci from four published solifuge transcriptomes and one de novo transcriptome, comprising two Eremobatidae and two Galeodidae and from four UCE assemblies comprising three Eremobatidae and one Daesiidae.
Outgroup sampling was influenced by previous works that have inferred various possible placements of Solifugae in the chelicerate Tree of Life, such as a sister group to Acariformes and Palpigradi, or part of a clade with Riniculei, Opiliones, and Xiphosura.25,26,27,40,42 Given this instability across studies, we sampled 2–3 terminals of every extant chelicerate order, prioritizing the sampling of basal splits in each outgroup order. Outgroup datasets were drawn from published transcriptomes and from our previous UCE assemblies that were captured with the same probe set (detailed below). All tree topologies were rooted with Pycnogonida. Accession data for all specimens in this study are provided in Table S2.
Ultraconserved element sequencing and phylogenomic analyses
For newly sequenced specimens, 1–4 legs or tissue teased from a single chelicera from single specimens were used for DNA extractions using the DNeasy Blood and Tissue kit and the QIAamp DNA Mini kit (Qiagen Inc., Valencia, CA). Libraries were prepared and enriched following protocols outlined by Kulkarni et al.14 and Miranda et al.75 All pools were enriched with the Spider2Kv1 probe set14 following the myBaits protocol 4.01 (Arbor Biosciences). Sequencing was performed on an Illumina NovoSeq platform. Assembly, alignment, trimming and concatenation of data were performed using the PHYLUCE pipeline (publicly available at https://phyluce.readthedocs.io/en/latest/). UCE contigs were extracted using the Spider2Kv1 probe set14 to target 2,021 UCE loci (locus recovery listed in Table S1). To augment the UCE dataset with RNASeq datasets, we followed the assembly, sanitation, reading frame detection, and UCE retrieval pipeline outlined by Kulkarni et al.76 Homology screening was performed by employing liberal (65%) and stringent (80%) probe-to-library identity and coverage mapping thresholds, following recent implementations.77
Phylogenomic analyses
The assembly, alignment, trimming and concatenation of data were done using the PHYLUCE pipeline (publicly available at https://phyluce.readthedocs.io/en/latest/). We generated a more complete matrix subset to minimize missing data by (1) filtering partitions by occupancies 25% and 40%, and (2) the automatic matrix reduction criterion using MARE v0.1.2-rc74 with default parameters. Phylogenetic analyses were performed on the unpartitioned nucleotide data using IQ-TREE67 version 2. Model selection was allowed for each unpartitioned dataset using the TEST function and an independent partitioned analysis using MFP+MERGE function to accommodate a best-fit partition by merging across prescribed locus partitions.78,79 Nodal support was estimated via 2,000 ultrafast bootstrap replicates79 with 10,000 iterations. To reduce the risk of overestimating branch support with ultrafast bootstrap due to model violations, we appended the command -bnni. With this command, the ultrafast bootstrap optimizes each bootstrap tree using a hill-climbing nearest neighbor interchange (NNI) search based on the corresponding bootstrap alignment.79 We also trialed a multispecies coalescent analysis with Accurate Species TRee Algorithm (ASTRAL)80,81 using gene trees, with input tree topologies reconstructed using IQ-TREE. However, the genes trees and the species tree were largely discordant. This phenomenon has been noted previously and is understood to result from the short lengths of typical UCE locus alignments,82,83,84 and thus we did not proceed with this analysis.
GC-content
GC content can bias the phylogenetic inferences reconstructed using genome scale data.85 To address this, we computed GC content for each taxon in the concatenated alignment using a custom script paired with BBMap (https://sourceforge.net/projects/bbmap/).
Phylogenomic dating
As no molecular phylogeny of the order exists and the solifuge fossil record is sparse, the timeframe of solifuge diversification is effectively unknown. To provide a temporal context to the divergences we inferred, we performed divergence time estimation using two approaches: a least-squares approach method (LSD2)69 and a Bayesian inference approach (MCMCTree; part of PAML v.4.870,71). As LSD2 requires at least one fixed date, we used an absolute calibration of 314.6 Ma for the crown Orthosterni fossil, Compsoscorpius buthiformis. We optimized the fossil information-based calibrations on the tree topology inferred from the 25% occupancy data set for the basis for divergence time estimation, implementing uniform node age priors to accommodate the scarcity of terrestrial chelicerate fossils. The root age was set to a range of 550–600 Mya, following Wolfe et al.86 The crown age of Solifugae was constrained using a minimum age bound of 305 Ma, based on the Carboniferous fossil Prosolpuga carbonaria (Petrunkevitch, 1913). The stem age of Ceromidae was constrained using a minimum age bound of 115 Ma, based on the Cretaceous fossil Cratosolpuga wunderlichi (Selden and Shear 2016). The recently described Burmese amber fossil Cushingia ellenbergeri was not included for calibration, as characters of this species potentially accord with a melanoblossid, a gylippid, or a rhagodid identity, which precludes its use for calibrating a specific node.87,88 Outgroup nodes were calibrated using the oldest unambiguous fossils representing those clades. Justifications and references for node calibrations are provided in Table S6.
Ancestral area reconstruction
We reconstructed ancestral areas on internal nodes of the dated preferred tree and the combined tree using the package BioGeoBEARS89 implemented in RASP 4.0.72 Each of the terminals was assigned to one of the following biogeographic regions: Turanian, African tropics, Neotropical, Nearctic, Central Chile-Patagonia and Mediterranean. We chose this scheme of area coding based on the distribution of the extant solifuges and following commonly used area definitions from the literature. Some taxa such as ammotrechids and eremobatids are distributed in the United States and Mexico. For these taxa, we coded the area as Nearctic following the delimitation of this region by Escalante et al.90 Additionally, to assess the influence of area coding, we alternatively coded areas by continents (corresponding to their geological plate); and more coarsely, as either Laurasia or Gondwana, based on the geological origin of those areas.
Assessment of Trichotoma placement
To infer the placement of the southern African “Gylippidae”, we obtained a sequence of 28S rRNA from GenBank for Trichotoma michaelseni (accession no. KT276815.1). Using BLASTn, we retrieved the corresponding sequence of 28S rRNA from libraries of Acanthogylippus sp. (Gylippidae), Ammotrechula sp. 28, Biton sp. 47, Blossia sp. 45, Ceroma sp. 29, Ceroma sp. 30, Chanbria regalis, Chelypus sp. 64, Dasycleobis sp. 9, Eremochelis andreasana, Eusimonia wunderlichi, Galeodes sp., Gluvia dorsalis, Rhagodidae sp. 86, Rhagodidae sp. 87, Rhinippus sp. 71, Uspallata pulchra, Zeria monteiri, and Zeriassa purcelli. The tree topology was rooted with exemplars of Ricinulei (Ricinoides feae; JX951360.1) and Opiliones (Troglosiro longifossa; DQ518045.1). Alignment was performed using MUSCLE v.3.2.173 (Edgar 2004) and the tree topology was inferred using maximum likelihood in IQ-TREE, with automated model-fitted and 1000 ultrafast bootstrap resampling replicates (Figure S17).
Acknowledgments
We are grateful to Nuria Macias (Departamento de Zoología de la Universidad de La Laguna) for providing a specimen of Eusimonia wunderlichi from the Canary Islands, and to Marc Domènech and Miquel Arnedo (University of Barcelona) for providing a specimen of Gluvia dorsalis from Spain. Photographs of live specimens were kindly provided by Igor Armiach Steinpress and Shlomi Aharon. Sequencing was performed at the Biotechnology Center of the University of Wisconsin-Madison. We thank Gustavo Hormiga for access to the Colonial One High Performance Computing Facility at The George Washington University to conduct some of the analyses. This project was supported by Binational Science Foundation grant no. BSF-2019216 to E.G.R. and P.P.S.; and National Science Foundation grant no. DEB-1754587 to P.E.C. S.S.K., G.G., and P.P.S. were additionally supported by National Science Foundation NSF grant no. IOS-2016141 to P.P.S.
Author contributions
Conceptualization, S.K., P.E.C., A.A.O.A., E.G.R., and P.S.; Methodology, S.K., H.S., H.I., and P.S.; Resources, E.L.G., H.I., R.J., J.A.B., G.G., M.R.G., D.H., R.L., A.A.O.A., C.E.S.L., G.S.D.M., P.E.C., E.G.R., and P.S. Writing – Original Draft, S.K. and P.S.; Writing – Review and Editing, H.S., E.L.G., H.I., R.J., J.A.B., G.G., M.R.G., D.H., R.L., A.A.O.A., C.E.S.L., G.S.D.M., P.E.C., E.G.R., and P.S., Supervision, P.S.; Project Administration, P.S.; Funding Acquisition, E.G.R. and P.S.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: August 19, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107684.
Supplemental information
Geographical data, collection date and collector are provided for the material sequenced in this study.
Data and code availability
-
•
Raw reads data have been deposited at NCBI Sequence Read Archive and are publicly available as of the date of publication. Original data matrices have been deposited on Mendeley. DOIs are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Murienne J., Benavides L.R., Prendini L., Hormiga G., Giribet G. Forest refugia in Western and Central Africa as ‘museums’ of Mesozoic biodiversity. Biol. Lett. 2013;9:20120932. doi: 10.1098/rsbl.2012.0932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giribet G., McIntyre E., Christian E., Espinasa L., Ferreira R.L., Francke Ó.F., Harvey M.S., Isaia M., Kováč Ĺ., McCutchen L., et al. The first phylogenetic analysis of Palpigradi (Arachnida)—the most enigmatic arthropod order. Invertebr. Systemat. 2014;28:350. doi: 10.1071/is13057. [DOI] [Google Scholar]
- 3.Fernández R., Giribet G. Unnoticed in the Tropics: Phylogenomic Resolution of the Poorly Known Arachnid Order Ricinulei (Arachnida) R. Soc. Open Sci. 2015;2:150065. doi: 10.1098/rsos.150065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma P.P., Fernández R., Esposito L.A., González-Santillán E., Monod L. Phylogenomic resolution of scorpions reveals multilevel discordance with morphological phylogenetic signal. Proc. Biol. Sci. 2015;282:20142953. doi: 10.1098/rspb.2014.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clouse R.M., Branstetter M.G., Buenavente P., Crowley L.M., Czekanski-Moir J., General D.E.M., Giribet G., Harvey M.S., Janies D.A., Mohagan A.B., et al. First global molecular phylogeny and biogeographical analysis of two arachnid orders (Schizomida and Uropygi) supports a tropical Pangean origin and mid-Cretaceous diversification. J. Biogeogr. 2017;44:2660–2672. doi: 10.1111/jbi.13076. [DOI] [Google Scholar]
- 6.Fernández R., Hormiga G., Giribet G. Phylogenomic Analysis of Spiders Reveals Nonmonophyly of Orb Weavers. Curr. Biol. 2014;24:1772–1777. doi: 10.1016/j.cub.2014.06.035. [DOI] [PubMed] [Google Scholar]
- 7.Hedin M., Starrett J., Akhter S., Schönhofer A.L., Shultz J.W. Phylogenomic Resolution of Paleozoic Divergences in Harvestmen (Arachnida, Opiliones) via Analysis of Next-Generation Transcriptome Data. PLoS One. 2012;7:e42888. doi: 10.1371/journal.pone.0042888.t004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bond J.E., Garrison N.L., Hamilton C.A., Godwin R.L., Hedin M., Agnarsson I. Phylogenomics Resolves a Spider Backbone Phylogeny and Rejects a Prevailing Paradigm for Orb Web Evolution. Curr. Biol. 2014;24:1765–1771. doi: 10.1016/j.cub.2014.06.034. [DOI] [PubMed] [Google Scholar]
- 9.Garrison N.L., Rodriguez J., Agnarsson I., Coddington J.A., Griswold C.E., Hamilton C.A., Hedin M., Kocot K.M., Ledford J.M., Bond J.E. Spider phylogenomics: untangling the Spider Tree of Life. PeerJ. 2016;4:e1719. doi: 10.7717/peerj.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernández R., Sharma P.P., Tourinho A.L., Giribet G. The Opiliones tree of life: shedding light on harvestmen relationships through transcriptomics. Proc. Biol. Sci. 2017;284:20162340–20162410. doi: 10.1098/rspb.2016.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klimov P.B., OConnor B.M., Chetverikov P.E., Bolton S.J., Pepato A.R., Mortazavi A.L., Tolstikov A.V., Bauchan G.R., Ochoa R. Comprehensive phylogeny of acariform mites (Acariformes) provides insights on the origin of the four-legged mites (Eriophyoidea), a long branch. Mol. Phylogenet. Evol. 2018;119:105–117. doi: 10.1016/j.ympev.2017.10.017. [DOI] [PubMed] [Google Scholar]
- 12.Santibáñez-López C.E., González-Santillán E., Monod L., Sharma P.P. Phylogenomics facilitates stable scorpion systematics_ Reassessing the relationships of Vaejovidae and a new higher-level classification of Scorpiones (Arachnida) Mol. Phylogenet. Evol. 2019;135:22–30. doi: 10.1016/j.ympev.2019.02.021. [DOI] [PubMed] [Google Scholar]
- 13.Benavides L.R., Cosgrove J.G., Harvey M.S., Giribet G. Phylogenomic interrogation resolves the backbone of the Pseudoscorpiones tree of life. Mol. Phylogenet. Evol. 2019;139:106509. doi: 10.1016/j.ympev.2019.05.023. [DOI] [PubMed] [Google Scholar]
- 14.Kulkarni S., Wood H., Lloyd M., Hormiga G. Spider-specific probe set for ultraconserved elements offers new perspectives on the evolutionary history of spiders (Arachnida, Araneae) Mol. Ecol. Resour. 2020;20:185–203. doi: 10.1111/1755-0998.13099. [DOI] [PubMed] [Google Scholar]
- 15.Ballesteros J.A., Setton E.V.W., Santibáñez-López C.E., Arango C.P., Brenneis G., Brix S., Corbett K.F., Cano-Sánchez E., Dandouch M., Dilly G.F., et al. Phylogenomic Resolution of Sea Spider Diversification through Integration of Multiple Data Classes. Mol. Biol. Evol. 2021;38:686–701. doi: 10.1093/molbev/msaa228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kallal R.J., Kulkarni S.S., Dimitrov D., Benavides L.R., Arnedo M.A., Giribet G., Hormiga G. Converging on the orb: denser taxon sampling elucidates spider phylogeny and new analytical methods support repeated evolution of the orb web. Cladistics. 2021;37:298–316. doi: 10.1111/cla.12439. [DOI] [PubMed] [Google Scholar]
- 17.Santibáñez-López C.E., Aharon S., Ballesteros J.A., Gainett G., Baker C.M., González-Santillán E., Harvey M.S., Hassan M.K., Abu Almaaty A.H., Aldeyarbi S.M., et al. Phylogenomics of Scorpions Reveal Contemporaneous Diversification of Scorpion Mammalian Predators and Mammal-Active Sodium Channel Toxins. Syst. Biol. 2022;71:1281–1289. doi: 10.1093/sysbio/syac021. [DOI] [PubMed] [Google Scholar]
- 18.Sanggaard K.W., Bechsgaard J.S., Fang X., Duan J., Dyrlund T.F., Gupta V., Jiang X., Cheng L., Fan D., Feng Y., et al. Spider genomes provide insight into composition and evolution of venom and silk. Nat. Commun. 2014;5:3765–3811. doi: 10.1038/ncomms4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gulia-Nuss M., Nuss A.B., Meyer J.M., Sonenshine D.E., Roe R.M., Waterhouse R.M., Sattelle D.B., de la Fuente J., Ribeiro J.M., Megy K., et al. Genomic insights into the Ixodes scapularis tick vector of Lyme disease. Nat. Commun. 2016;7:10507. doi: 10.1038/ncomms10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoy M.A., Waterhouse R.M., Wu K., Estep A.S., Ioannidis P., Palmer W.J., Pomerantz A.F., Simão F.A., Thomas J., Jiggins F.M., et al. Genome Sequencing of the Phytoseiid Predatory Mite Metaseiulus occidentalis Reveals Completely Atomized Hox Genes and Superdynamic Intron Evolution. Genome Biol. Evol. 2016;8:1762–1775. doi: 10.1093/gbe/evw048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kenny N.J., Chan K.W., Nong W., Qu Z., Maeso I., Yip H.Y., Chan T.F., Kwan H.S., Holland P.W.H., Chu K.H., Hui J.H.L. Ancestral whole-genome duplication in the marine chelicerate horseshoe crabs. Heredity. 2016;116:190–199. doi: 10.1038/hdy.2015.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwager E.E., Sharma P.P., Clarke T., Leite D.J., Wierschin T., Pechmann M., Akiyama-Oda Y., Esposito L., Bechsgaard J., Bilde T., et al. The house spider genome reveals an ancient whole-genome duplication during arachnid evolution. BMC Biol. 2017;15:62. doi: 10.1186/s12915-017-0399-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gainett G., González V.L., Ballesteros J.A., Setton E.V.W., Baker C.M., Barolo Gargiulo L., Santibáñez-López C.E., Coddington J.A., Sharma P.P. The genome of a daddy-long-legs (Opiliones) illuminates the evolution of arachnid appendages. Proc. Biol. Sci. 2021;288:20211168. doi: 10.1098/rspb.2021.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ontano A.Z., Gainett G., Aharon S., Ballesteros J.A., Benavides L.R., Corbett K.F., Gavish-Regev E., Harvey M.S., Monsma S., Santibáñez-López C.E., et al. Taxonomic Sampling and Rare Genomic Changes Overcome Long-Branch Attraction in the Phylogenetic Placement of Pseudoscorpions. Mol. Biol. Evol. 2021;38:2446–2467. doi: 10.1093/molbev/msab038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma P.P., Kaluziak S.T., Pérez-Porro A.R., González V.L., Hormiga G., Wheeler W.C., Giribet G. Phylogenomic Interrogation of Arachnida Reveals Systemic Conflicts in Phylogenetic Signal. Mol. Biol. Evol. 2014;31:2963–2984. doi: 10.1093/molbev/msu235. [DOI] [PubMed] [Google Scholar]
- 26.Ballesteros J.A., Sharma P.P. A Critical Appraisal of the Placement of Xiphosura (Chelicerata) with Account of Known Sources of Phylogenetic Error. Syst. Biol. 2019;68:896–917. doi: 10.1093/sysbio/syz011. [DOI] [PubMed] [Google Scholar]
- 27.Ballesteros J.A., Santibáñez-López C.E., Baker C.M., Benavides L.R., Cunha T.J., Gainett G., Ontano A.Z., Setton E.V.W., Arango C.P., Gavish-Regev E., et al. Comprehensive Species Sampling and Sophisticated Algorithmic Approaches Refute the Monophyly of Arachnida. Mol. Biol. Evol. 2022;39:msac021. doi: 10.1093/molbev/msac021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ban X.C., Shao Z.K., Wu L.J., Sun J.T., Xue X.F. Highly diversified mitochondrial genomes provide new evidence for interordinal relationships in the Arachnida. Cladistics. 2022;38:452–464. doi: 10.1111/cla.12504. [DOI] [PubMed] [Google Scholar]
- 29.World Solifugae Catalog. https://wac.nmbe.ch/order/solifugae/6
- 30.van der Meijden A., Langer F., Boistel R., Vagovic P., Heethoff M. Functional morphology and bite performance of raptorial chelicerae of camel spiders (Solifugae) J. Exp. Biol. 2012;215:3411–3418. doi: 10.1242/jeb.072926. [DOI] [PubMed] [Google Scholar]
- 31.Harvey M.S. The neglected cousins: what do we know about the smaller arachnid orders? J. Arachnol. 2002;30:357–372. [Google Scholar]
- 32.Brownell P.H., Farley R.D. The organization of the malleolar sensory system in the solpugid, Chanbria sp. Tissue Cell. 1974;6:471–485. doi: 10.1016/0040-8166(74)90039-1. [DOI] [PubMed] [Google Scholar]
- 33.Cushing P.E., Brookhart J.O., Kleebe H.-J., Zito G., Payne P. The suctorial organ of the Solifugae (Arachnida, Solifugae) Arthropod Struct. Dev. 2005;34:397–406. doi: 10.1016/j.asd.2005.02.002. [DOI] [Google Scholar]
- 34.Willemart R.H., Santer R.D., Spence A.J., Hebets E.A. A sticky situation: solifugids (Arachnida, Solifugae) use adhesive organs on their pedipalps for prey capture. J. Ethol. 2011;29:177–180. doi: 10.1007/s10164-010-0222-4. [DOI] [Google Scholar]
- 35.Franz-Guess S., Klußmann-Fricke B.J., Wirkner C.S., Prendini L., Starck J.M. Morphology of the tracheal system of camel spiders (Chelicerata: Solifugae) based on micro-CT and 3D-reconstruction in exemplar species from three families. Arthropod Struct. Dev. 2016;45:440–451. doi: 10.1016/j.asd.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 36.Weygoldt P., Paulus H.F. Untersuchungen zur Morphologie, Taxonomie und Phylogenie der Chelicerata1 II. Cladogramme und die Entfaltung der Chelicerata. J. Zool. Syst. Evol. Res. 2009;17:177–200. doi: 10.1111/j.1439-0469.1979.tb00699.x. [DOI] [Google Scholar]
- 37.Shultz J.W. A phylogenetic analysis of the arachnid orders based on morphological characters. Zool. J. Linn. Soc. 2007;150:221–265. doi: 10.1111/j.1096-3642.2007.00284.x. [DOI] [Google Scholar]
- 38.Alberti G., Peretti A.V. Fine structure of male genital system and sperm in solifugae does not support a sister-group relationship with pseudoscorpiones (Arachnida) J. Arachnol. 2002;30:268–274. doi: 10.1636/0161-8202(2002)030[0268:FSOMGS]2.0.CO;2. [DOI] [Google Scholar]
- 39.Michalski H., Harms D., Runge J., Wirkner C.S. Evolutionary morphology of coxal musculature in Pseudoscorpiones (Arachnida) Arthropod Struct. Dev. 2022;69:101165. doi: 10.1016/j.asd.2022.101165. [DOI] [PubMed] [Google Scholar]
- 40.Pepato A.R., da Rocha C.E.F., Dunlop J.A. Phylogenetic position of the acariform mites: sensitivity to homology assessment under total evidence. BMC Evol. Biol. 2010;10:235. doi: 10.1186/1471-2148-10-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dunlop J.A., Krüger J., Alberti G. The sejugal furrow in camel spiders and acariform mites. Arachnol. Mitteil. 2012;43:8–15. doi: 10.5431/aramit4303. [DOI] [Google Scholar]
- 42.Ballesteros J.A., Santibáñez López C.E., Kováč Ľ., Gavish-Regev E., Sharma P.P. Ordered phylogenomic subsampling enables diagnosis of systematic errors in the placement of the enigmatic arachnid order Palpigradi. Proc. Biol. Sci. 2019;286:20192426. doi: 10.1098/rspb.2019.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roewer C., Carl F. Bronn’s Klassen und Ordnungen des Tierreichs. 5: Arthropoda. IV: Arachnoidea. 5(IV)(4)(4–5) Akademische Verlagsgesellschaft M.B.H.; 1934. Solifugae, palpigradi; pp. 481–723. [Google Scholar]
- 44.Klann A.E., Bird T., Peretti A.V., Gromov A.V., Alberti G. Ultrastructure of spermatozoa of solifuges (Arachnida, Solifugae): Possible characters for their phylogeny? Tissue Cell. 2009;41:91–103. doi: 10.1016/j.tice.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 45.Klann A.E., Bird T.L., Talarico G. Ultrastructural characterization of Hexisopus psammophilus (Arachnida: Solifugae: Hexisopodidae) spermatozoa in comparison to other solifuge spermatozoal traits. J. Arachnol. 2011;39:280–286. doi: 10.1636/CB10-94.1. [DOI] [Google Scholar]
- 46.Bird T., Wharton R., Prendini L. Cheliceral Morphology in Solifugae (Arachnida): Primary Homology, Terminology, and Character Survey. Bull. Am. Mus. Nat. Hist. 2015;394:1–355. [Google Scholar]
- 47.Santibáñez-López C.E., Cushing P.E., Powell A.M., Graham M.R. Diversification and post-glacial range expansion of giant North American camel spiders in genus Eremocosta (Solifugae: Eremobatidae) Sci. Rep. 2021;11:22093. doi: 10.1038/s41598-021-01555-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maddahi H., Khazanehdari M., Aliabadian M., Kami H.G., Mirshamsi A., Mirshamsi O. Mitochondrial DNA phylogeny of camel spiders (Arachnida: Solifugae) from Iran. Mitochondrial DNA Part A. 2017;28:909–919. doi: 10.1080/24701394.2016.1209194. [DOI] [PubMed] [Google Scholar]
- 49.Cushing P.E., Graham M.R., Prendini L., Brookhart J.O. A multilocus molecular phylogeny of the endemic North American camel spider family Eremobatidae (Arachnida: Solifugae) Mol. Phylogenet. Evol. 2015;92:280–293. doi: 10.1016/j.ympev.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 50.Dunlop J.A. Geological history and phylogeny of Chelicerata. Arthropod Struct. Dev. 2010;39:124–142. doi: 10.1016/j.asd.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 51.Derkarabetian S., Benavides L.R., Giribet G. Sequence capture phylogenomics of historical ethanol-preserved museum specimens: Unlocking the rest of the vault. Mol. Ecol. Resour. 2019;19:1531–1544. doi: 10.1111/1755-0998.13072. [DOI] [PubMed] [Google Scholar]
- 52.Ontano A.Z., Steiner H.G., Sharma P.P. How many long branch orders occur in Chelicerata? Opposing effects of Palpigradi and Opilioacariformes on phylogenetic stability. Mol. Phylogenet. Evol. 2022;168:107378. doi: 10.1016/j.ympev.2021.107378. [DOI] [PubMed] [Google Scholar]
- 53.Borner J., Rehm P., Schill R.O., Ebersberger I., Burmester T. A transcriptome approach to ecdysozoan phylogeny. Mol. Phylogenet. Evol. 2014;80:79–87. doi: 10.1016/j.ympev.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 54.Arribas P., Andújar C., Moraza M.L., Linard B., Emerson B.C., Vogler A.P. Mitochondrial Metagenomics Reveals the Ancient Origin and Phylodiversity of Soil Mites and Provides a Phylogeny of the Acari. Mol. Biol. Evol. 2020;37:683–694. doi: 10.1093/molbev/msz255. [DOI] [PubMed] [Google Scholar]
- 55.Hewitt J. Descriptions of new South African Araneae and Solifugae. Ann. Transvaal Mus. 1919;6:63–111. [Google Scholar]
- 56.Sanmartín I., Ronquist F. Southern Hemisphere Biogeography Inferred by Event-Based Models: Plant versus Animal Patterns. Syst. Biol. 2004;53:216–243. doi: 10.1080/10635150490423430. [DOI] [PubMed] [Google Scholar]
- 57.Giribet G., Sharma P.P., Benavides L.R., Boyer S.L., Clouse R.M., De Bivort B.L., Schwendinger P.J., Dimitrov D., Kawauchi G.Y., Murienne J., Schwendinger P.J. Evolutionary and biogeographical history of an ancient and global group of arachnids (Arachnida: Opiliones: Cyphophthalmi) with a new taxonomic arrangement. Biol. J. Linn. Soc. 2012;105:92–130. [Google Scholar]
- 58.Mao K., Milne R.I., Zhang L., Peng Y., Liu J., Thomas P., Mill R.R., Renner S.S. Distribution of living Cupressaceae reflects the breakup of Pangea. Proc. Natl. Acad. Sci. USA. 2012;109:7793–7798. doi: 10.1073/pnas.1114319109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murienne J., Daniels S.R., Buckley T.R., Mayer G., Giribet G. A living fossil tale of Pangaean biogeography. Proc. Biol. Sci. 2014;281:20132648. doi: 10.1073/pnas.1102473108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Z., Ramstein G., Schuster M., Li C., Contoux C., Yan Q. Aridification of the Sahara desert caused by Tethys Sea shrinkage during the Late Miocene. Nature. 2014;513:401–404. doi: 10.1038/nature13705. [DOI] [PubMed] [Google Scholar]
- 61.Zheng H., Wei X., Tada R., Clift P.D., Wang B., Jourdan F., Wang P., He M. Late Oligocene–early Miocene birth of the Taklimakan Desert. Proc. Natl. Acad. Sci. USA. 2015;112:7662–7667. doi: 10.1073/pnas.1424487112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Byrne M., Steane D.A., Joseph L., Yeates D.K., Jordan G.J., Crayn D., Aplin K., Cantrill D.J., Cook L.G., Crisp M.D., et al. Decline of a biome: evolution, contraction, fragmentation, extinction and invasion of the Australian mesic zone biota. J. Biogeogr. 2011;38:1635–1656. doi: 10.1111/j.1365-2699.2011.02535.x. [DOI] [Google Scholar]
- 63.Derkarabetian S., Starrett J., Tsurusaki N., Ubick D., Castillo S., Hedin M. A stable phylogenomic classification of Travunioidea (Arachnida, Opiliones, Laniatores) based on sequence capture of ultraconserved elements. ZooKeys. 2018;760:1–36. doi: 10.3897/zookeys.760.24937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Benavides L.R., Hormiga G., Giribet G. Phylogeny, evolution and systematic revision of the mite harvestman family Neogoveidae (Opiliones Cyphophthalmi) Invertebr. Systemat. 2019;1509:1–81. doi: 10.1071/is18018. [DOI] [Google Scholar]
- 65.Santibáñez-López C.E., Ojanguren-Affilastro A.A., Sharma P.P. Another one bites the dust: taxonomic sampling of a key genus in phylogenomic datasets reveals more non-monophyletic groups in traditional scorpion classification. Invertebr. Systemat. 2020;34:133–211. doi: 10.1071/is19033. [DOI] [Google Scholar]
- 66.Faircloth B.C. PHYLUCE is a software package for the analysis of conserved genomic loci. Bioinformatics. 2016;32:786–788. doi: 10.1093/bioinformatics/btv646. [DOI] [PubMed] [Google Scholar]
- 67.Nguyen L.-T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bushnell B. BBMap : A Fast, Accurate, Splice-Aware Aligner. No. LBNL-7065E. Lawrence Berkeley National Lab; 2014. [Google Scholar]
- 69.To T.-H., Jung M., Lycett S., Gascuel O. Fast Dating Using Least-Squares Criteria and Algorithms. Syst. Biol. 2016;65:82–97. doi: 10.1093/sysbio/syv068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 71.dos Reis M., Yang Z. In: Evolutionary Genomics: Statistical and Computational Methods Methods in Molecular Biology. Anisimova M., editor. Springer; 2019. Bayesian Molecular Clock Dating Using Genome-Scale Datasets; pp. 309–330. [DOI] [PubMed] [Google Scholar]
- 72.Yu Y., Blair C., He X. RASP 4: Ancestral State Reconstruction Tool for Multiple Genes and Characters. Mol. Biol. Evol. 2020;37:604–606. doi: 10.1093/molbev/msz257. [DOI] [PubMed] [Google Scholar]
- 73.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Meyer B., Meusemann K., Misof B. 2011. MARE V. 0.1.2-rc: MAtrix REduction—a tool to select optimized data subsets from supermatrices for phylogenetic inference. http://mare.zfmk.de. [Google Scholar]
- 75.De Miranda G.S., Kulkarni S.S., Tagliatela J., Baker C.M., Giupponi A.P.L., Labarque F.M., Gavish-Regev E., Rix M.G., Carvalho L.S., Fusari L.M., et al. 2022. The Rediscovery of a Relict Unlocks the First Global Phylogeny of Whip Spiders (Amblypygi) (Genomics) [DOI] [PubMed] [Google Scholar]
- 76.Kulkarni S., Kallal R.J., Wood H., Dimitrov D., Giribet G., Hormiga G. Interrogating genomic-scale data to resolve recalcitrant nodes in the Spider Tree of Life. Mol. Biol. Evol. 2021;38:891–903. doi: 10.1093/molbev/msaa251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bossert S., Danforth B.N. On the universality of target-enrichment baits for phylogenomic research. Methods Ecol. Evol. 2018;9:1453–1460. doi: 10.1111/2041-210X.12988. [DOI] [Google Scholar]
- 78.Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., von Haeseler A., Jermiin L.S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hoang D.T., Chernomor O., Von Haeseler A., Minh B.Q., Vinh L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mirarab S., Warnow T. ASTRAL-II: coalescent-based species tree estimation with many hundreds of taxa and thousands of genes. Bioinformatics. 2015;31:i44–i52. doi: 10.1093/bioinformatics/btv234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mirarab S., Reaz R., Bayzid M.S., Zimmermann T., Swenson M.S., Warnow T. ASTRAL: genome-scale coalescent-based species tree estimation. Bioinformatics. 2014;30:i541–i548. doi: 10.1093/bioinformatics/btu462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Meiklejohn K.A., Faircloth B.C., Glenn T.C., Kimball R.T., Braun E.L. Analysis of a Rapid Evolutionary Radiation Using Ultraconserved Elements: Evidence for a Bias in Some Multispecies Coalescent Methods. Syst. Biol. 2016;65:612–627. doi: 10.1093/sysbio/syw014. [DOI] [PubMed] [Google Scholar]
- 83.Baca S.M., Alexander A., Gustafson G.T., Short A.E.Z. Ultraconserved elements show utility in phylogenetic inference of Adephaga (Coleoptera) and suggest paraphyly of ‘Hydradephaga’: Phylogeny of Adephaga inferred with UCEs. Syst. Entomol. 2017;42:786–795. doi: 10.1111/syen.12244. [DOI] [Google Scholar]
- 84.Bossert S., Murray E.A., Pauly A., Chernyshov K., Brady S.G., Danforth B.N. Gene Tree Estimation Error with Ultraconserved Elements: An Empirical Study on Pseudapis Bees. Syst. Biol. 2021;70:803–821. doi: 10.1093/sysbio/syaa097. [DOI] [PubMed] [Google Scholar]
- 85.Benjamini Y., Speed T.P. Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Res. 2012;40:e72. doi: 10.1093/nar/gks001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wolfe J.M., Daley A.C., Legg D.A., Edgecombe G.D. Fossil calibrations for the arthropod Tree of Life. Earth Sci. Rev. 2016;160:43–110. doi: 10.1016/j.earscirev.2016.06.008. [DOI] [Google Scholar]
- 87.Dunlop J.A., Bird T.L., Brookhart J.O., Bechly G. A camel spider from Cretaceous Burmese amber. Cretac. Res. 2015;56:265–273. doi: 10.1016/j.cretres.2015.05.003. [DOI] [Google Scholar]
- 88.Bartel C., Dunlop J.A., Bird T.L. The Second Camel Spider (Arachnida, Solifugae) from Burmese Amber. Arachnology. 2016;17:161–164. doi: 10.13156/arac.2006.17.3.161. [DOI] [Google Scholar]
- 89.Matzke N.J. 2018. Nmatzke/BioGeoBEARS: BioGeoBEARS: BioGeography with Bayesian (And Likelihood) Evolutionary Analysis with R Scripts. [DOI] [Google Scholar]
- 90.Escalante T., Rodríguez-Tapia G., Szumik C., Morrone J.J., Rivas M. Delimitation of the Nearctic region according to mammalian distributional patterns. J. Mammal. 2010;91:1381–1388. doi: 10.1644/10-MAMM-A-136.1. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Geographical data, collection date and collector are provided for the material sequenced in this study.
Data Availability Statement
-
•
Raw reads data have been deposited at NCBI Sequence Read Archive and are publicly available as of the date of publication. Original data matrices have been deposited on Mendeley. DOIs are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.