Abstract
The Bacillus subtilis strain NDmed was isolated from an endoscope washer-disinfector in a medical environment. NDmed can form complex macrocolonies with highly wrinkled architectural structures on solid medium. In static liquid culture, it produces thick pellicles at the interface with air as well as remarkable highly protruding ‘‘beanstalk-like’’ submerged biofilm structures at the solid surface. Since these mucoid submerged structures are hyper-resistant to biocides, NDmed has the ability to protect pathogens embedded in mixed-species biofilms by sheltering them from the action of these agents. Additionally, this non-domesticated and highly biofilm forming strain has the propensity of being genetically manipulated. Due to all these properties, the NDmed strain becomes a valuable model for the study of B. subtilis biofilms. This review focuses on several studies performed with NDmed that have highlighted the sophisticated genetic dynamics at play during B. subtilis biofilm formation. Further studies in project using modern molecular tools of advanced technologies with this strain, will allow to deepen our knowledge on the emerging properties of multicellular bacterial life.
Keywords: Biofilms, Bacillus subilis, NDmed, CLSM, Pellicle, Colony
1. Introduction
Along with the constant environmental fluctuations, bacteria need to evolve adaptive strategies to survive, often by the formation of spatially structured assemblages encapsulated in a self-produced extracellular matrix called biofilms [1,2]. Microbial communities residing in these structured aggregates exhibit new emergent properties, such as resource capture by sorption, enzyme retention, social interactions, increased rate of genetic exchanges, enhanced tolerance and resistance to antimicrobials, and localized gradients due to the environmental micro-scale adaptations [3]. Such properties resulting in physiological diversification involve sophisticated gene regulation networks [4], whose study is important for the development of biotechnological applications using bacteria, as well as to better restrain bacterial pathogens in the medical field. A wide range of knowledge at the genetic level has been acquired from the highly tractable Gram-positive model organism Bacillus subtilis. In nature, B. subtilis is a soil-dwelling, non-pathogenic, motile bacterium promoting beneficial effects on plant growth by limiting the development of pathogenic species [5,6]. B. subtilis can also be found in animal and human gut microbiota, thanks to its capacity to sporulate and to form biofilms, both of which allow this species to pass the harsh gastric environment to reach and persist in the intestine [[7], [8], [9]]. B. subtilis has long been considered a GRAS (Generally Recognized As Safe) organism by the FDA (U.S. Food and Drug Administration) (e.g. FDA GRAS Notice GRN No. 562. http://wayback.archive-it.org/7993/20171031040136/https://www.fda.gov/downloads/Food/IngredientsPackagingLabeling/GRAS/NoticeInventory/UCM448213.pdf) [10,11], and is commercially available as a probiotic for human health, in the agricultural industry as a biocontrol agent, and in the food industry as a natto subspecies in traditional Japanese food from fermented soybeans [[12], [13], [14], [15]]. Due to its excellent protein secretion ability, it has been widely used as a cell factory to produce heterologous proteins [16]. Moreover, its capacity to form biofilms, associated with calcinogenic properties or synthesis of antimicrobial compounds find applications in the bio-remineralization of monumental stones of historical buildings or to protect ancient paintings from biodegradation [[17], [18], [19], [20]]. However, the formation of biofilms can be deleterious, generating problematic side effects in industrial pipeline clogging and biofouling, as well as hazards to human health by their persistence in medical environments and devices due to their resistance to biocides [21]. In this context, and besides being recognized as a non-pathogenic bacterium, B. subtilis can still be involved in post-surgery pathogenesis, leading to anastomotic leaks due to its high collagenolytic activity [22]. For all the above reasons, combined with the fact that B. subtilis is naturally competent, easy, and safe to be manipulated in the laboratory, it became the model for Gram-positive bacteria in physiological studies on the genetic regulations involved in general metabolism, or in specific biological processes such as sporulation [[23], [24], [25], [26]].
Differentiation of B. subtilis cells from motile to sessile ones has been observed to study the structured biofilm assemblages, particularly the development of complex macrocolonies on the air-solid interface and the formation of pellicles at the air-liquid interface. For instance, the wild-type strain NCIB3610 was able to form spatially organized wrinkled colonies and well-structured pellicles, contrary to the domesticated reference strain 168 that was only able to form smooth colonies and thin fragile pellicles [[27], [28], [29], [30], [31]]. A genetic comparison between the two strains made it possible to identify mutations in 168 responsible for its inability to form wrinkled and robust biofilms. These mutations were probably acquired during the mutagenic treatment of the “Marburg strain” in the late 1940s [30]. Besides, NCIB3610 possesses a large endogenous plasmid pBS32 which encodes a small protein ComI that inhibits transformation in this strain [32]. This explains the very low natural genetic competence ability of this natural isolate, which made it more difficult to manipulate for further genetic studies, contrary to 168, which lost this plasmid. Nevertheless, this did not preclude many genetic studies to be performed with NCIB3610, via SPP1 phage transduction [33], or using DK1042, a comIQ12L mutant NCIB3610 derivative strain [32]. These studies revealed various integrated regulatory pathways controlling biofilm formation, and unveiled several molecular mechanisms involved [34,35]. Besides, several other natural B. subtilis strains have been isolated more or less recently, presenting interesting biofilm phenotypes and being naturally competent, such as P9-B1 [36] or PS216 [37]. These strains have therefore also been used in many studies on different biofilm models, essentially macrocolony, floating pellicle, or even plant root colonization [[38], [39], [40]]. A submerged surface-associated biofilm model was developed with strain JH642 [41], but as being a close relative to the domesticated 168, this strain could not form robust wrinkled colonies [34], and the submerged biofilm formed remains thin and not highly structured. So, although this model was particularly relevant for the study of B. subtilis multicellular communities, practically no further studies was performed with it during the next 10 years, until using confocal laser scanning microscopy, we showed that several strains of B. subtilis from different origins are capable of forming such biofilms with complex structures on immersed surfaces [42,43].
In this review, we will present NDmed, another wild-type B. subtilis strain that we have been successfully using for several years in genetic studies on biofilms (Fig. 1). This strain can build highly structured biofilms in all described B. subtilis multicellular models (macro-colony, air/liquid pellicle and submerged), and is much more convenient for genetic manipulations than NCIB3610, which has greatly facilitated such studies.
Fig. 1.
Macro-colony of B. subtilis NDmed. Composite image of a colony of B. subtilis NDmed taken in digital photography (left part) and confocal scanning laser microscopy (right part); (diameter of the colony is approximately 2 cm). This artwork picture has been presented among 10 finalists at an artistic scientific photographs concourse organized by the French Embassy in Tokyo (Japan) in Dec. 2022.
2. NDmed, a hyper-biofilm forming B. subtilis strain
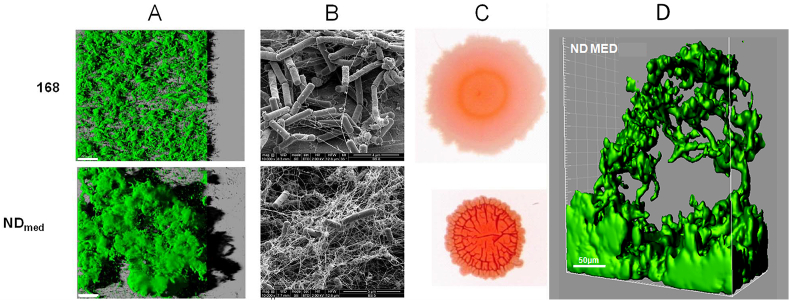
In a large number of ecological, industrial and hospital settings surface-associated microbial communities are the source of many problems, including public health issues such as nosocomial or foodborne infections [44,45]. For instance, some studies have reported the persistence of surface-associated bacteria on an endoscope even after cleaning and disinfecting procedures have taken place [46,47]. A developed biofilm provides bacteria with a protective environment and constitutes a survival strategy against stresses such as microbicide action, thus potentially leading to important healthcare issues. In the course of investigations aiming at unveiling resistance mechanisms behind such bacterial persistence and survival following biocide exposure in a medical environment, Martin et al. have isolated from an endoscope washer-disinfector a B. subtilis strain particularly resistant to high levels of disinfectants such as chlorine dioxide and hydrogen peroxide [21,48,49]. This now called NDmed strain (for non-domesticated strain isolated from medical environment) forms spatially architectural macro-colonies on solid agar medium and dramatically protruding ‘‘beanstalk-like’’ biofilm structures (with a height up to 300 μm) on submerged level, with the production of a notable high amount of exopolymeric substances (Fig. 2) [42,50]. Such complex three-dimensional structure of the NDmed biofilm appears to hinder the penetration and reactivity of oxidative agents, and thereby leads to hyper-resistance (Fig. 3).
Fig. 4.
Architecture of S. aureus AH478 and B. subtilis NDmed/S. aureus AH478 mixed biofilm. (A) 3D reconstruction of S. aureus AH478 biofilm. (B) 3D reconstruction of mixed species biofilm of B. subtilis NDmed (green)/S. aureus AH478 (red). Scale bars correspond to 20 μm (From Ref. [50]). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 2.
Comparison of architectures of biofilms formed by B. subtilis 168 and NDmed strains. (A) Aerial views of 168 and NDmed biofilm structure, with a virtual three-dimensional shadow projection on the right. Scale bars correspond to 50 μm. (B) Scanning Electron Microscopy images of 24-h biofilms. (C) Dye binding properties of 72 h macrocolonies grown on Congo red indicator medium. (D) Iso-surface representation of a particular ‘‘beanstalk-like’’ structure for NDmed (From Refs. [42,50]).
Fig. 3.
Peracetic acid (PAA) activity in B. subtilis biofilms. Visualization of the kinetics of membrane permeabilization (Chemchrome V6 fluorescence loss) in B. subtilis 168 and NDmed biofilms during PAA treatment (0.05%). Scale bars correspond to 20 μm (From Ref. [50]). Besides, when grown in mixed biofilm with Staphylococcus aureus, the B. subtilis NDmed strain demonstrated the ability to protect this pathogen from PAA action, thus enabling its persistence in the environment (Fig. 4) [50,51].
Whole genome sequencing of NDmed (4.06 Mb) revealed that this non-domesticated isolate is very close to the reference laboratory strain 168, with less than 100 single-nucleotide polymorphisms (SNPs) and less than 50 insertions/deletions (InDels) [52]. As in several other B. subtilis natural isolates, e.g. the biofilm-forming transformable strain PS216 [53], the SPβ prophage (134.4 kb) and the conjugative element ICEBs1 (20.5 kb) are missing, whereas a putative prophage (44.2 kb) is present immediately downstream of the glnA gene. It is noteworthy that among the up to now 708 sequenced genomes of B. subtilis strains from extremely various origins (https://www.ncbi.nlm.nih.gov/genome/browse/#!/prokaryotes/665/), one of the closest genome neighbors is that of strain PS216, displaying a gapped identity of 99.9823% with NDmed (https://www.ncbi.nlm.nih.gov/genome/neighbors/665?genome_assembly_id=205100). No plasmid was observed in NDmed, such as the one present in NCIB3610 which encodes the ComI transformation inhibitor [32]. In both strains 168 and its “ancestor” NCIB3610, the gene spsM (formerly ypqP) is disrupted by the SPβ prophage [[54], [55], [56], [57]]. This spsM gene is essential for adding polysaccharides to the spore envelope [58]. Thus, SPβ has to be excised during the sporulation process for the reconstitution of a functional spsM gene. This excision is restricted to the mother cell, whereas SPβ remains in the genome of the spore, and is transmitted to the next generation [59]. spsM encodes an UDP-GlcNAc 4,6-dehydratase involved in the first step of the biosynthesis of legionaminic acid from UDP-N-acetyl-α-d-glucosamine during sporulation. Legionaminic acid is a constituent of the crust, together with other carbohydrates and proteins, covering the spore surface. This outermost layer participates in the adhesion and spreading of spores into the environment [60]. In strains lacking SPβ, spsM is functional even during vegetative growth, and could therefore participate in synthesizing carbohydrates matrix components leading to the highly robust structured biofilms phenotype. Indeed, in PY79, a 168-derived laboratory strain cured of the SPβ prophage, the reestablishment of a functional spsM (ypqP) gene led to increased thickness and resistance to biocides of the associated submerged biofilms [61]. Likewise, deletion of spsM in the NDmed strain abolished its ability to protect S. aureus in a mixed submerged biofilm (Fig. 5), as well as the particularly remarkable submerged biofilm or macro-colony phenotype, which could be completely restored upon complementation by an ectopic wild-type copy of the gene (Fig. 6). Moreover, all the various B. subtilis strains containing a nondisrupted spsM gene that we have tested (NDmed, NDfood, PY79, BSn5, BSP1) formed denser submerged biofilms with more protruding structures than those formed by the strains whose spsM gene is disrupted by the SPβ prophage (168, NCIB 3610, ATCC 6051) [61]. It was therefore obvious that its product was an important determinant of B. subtilis surface biofilm architecture, through its involvement in the synthesis of matrix components participating to the protection against biocides [61]. On the other hand, on hosting the SPβ prophage in spsM, lysogenic strains acquire a bacteriocin gene cluster carried by this prophage, encoding sublancin, a lantibiotic with a broad spectrum of bactericidal activity [62]. This indicates the double importance of spsM, which depending on the environmental conditions and hosting or not SPβ, can play a defensive or offensive role, by synthesis of protective polysaccharides “shields” or antimicrobial “weapons”. Thus, the genome of NDmed provided some clues for a better understanding of B. subtilis social behavior in bacterial communities from natural environments. Although all the genes involved in biofilm formation found defective in strain 168 are wild-type in NDmed as in NCIB3610, some differences found between the latter strains (such as the SPβ prophage insertion) allowed to shed light on the specific biofilm phenotypes observed. However, as the biofilm morphology and matrix composition can be growth medium dependent, as shown with NCIB3610 strain for exopolymeric substances (EPS) composition [63], differences observed when comparing NDmed to NCIB3610 would vanish or turn around in case the growth medium or other environmental conditions were changed. Moreover NDmed has been proven easily transformable [50], which greatly facilitated our biofilms genetic studies.
Fig. 5.
Three-dimensional organization of B. subtilis NDmed and S. aureus mixed biofilms. Mixed biofilms of S. aureus mCherry (red) and B. subtilis GFP (green) strains were grown for 48 h. Representative 3D reconstruction images of S. aureus and B. subtilis NDmed Wild-Type (A) or spsM mutant (B) mixed biofilms are presented. The scale bars represent 50 μm (From Ref. [61]). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 6.
Visualization of the effect of ypqP (spsM) disruption on submerged-biofilm structure and complex colony morphology in B. subtilis NDmed. (A) Colonies of the NDmed Wild-Type, ypqP mutant, and ypqP-complemented strains were grown on TSB agar for 3 days. (B and C) Biofilms of the three strains were grown for 48 h and stained with SYTO9. For each strain, representative images of the adherent cells in contact with the surface (B) and the 3D reconstruction using IMARIS software (C) are presented. The scale bars represent 50 μm (From Ref. [61]).
3. NDmed, a versatile tool strain for genetic and structural B. subtilis biofilm studies
Studies of B. subtilis biofilms use different models corresponding to different types of multicellular communities encountered in nature. In aerial models, cells grow on the surface of a nutrient medium, at the interface with air, and thanks to their technical simplicity, these models allow to observe differences in the phenotypes between strains, without requiring complex tools. On a solid medium, the formation of macrocolonies with highly wrinkled architectural structures indicates a high capacity for extracellular matrix production. Wrinkles are formed by a lateral compressive force as a consequence of localized cell death, coupled with the stiffness provided by the extracellular matrix [64]. Beneath the wrinkles forms a remarkable network of well-defined channels providing the biofilm with an enhanced transport system to exchange water, nutrients, enzymes, and signals, to dispose of potentially toxic metabolites, allowing better metabolic cooperativity [65]. From a macrocolony, and in specific optimized conditions of medium, humidity and temperature, B. subtilis cells can swarm by organized collective movements, which include hyper-flagellated and highly motile cells, while proliferating and consuming nutrients [[66], [67], [68], [69], [70]]. This exploration behavior starting from a 3-D mother macrocolony structured biofilm to a monolayer multicellular communities can be seen as the formation of a 2-D developing biofilm. On a liquid medium, the formation of a thick pellicle depends on the synthesis of extracellular polymeric substances, essential for the complex 3D structure, as well as on amphiphilic properties of Bsla forming a hydrophobic layer at the interface with air. In the submerged model, cells grow on an inert solid surface (polystyrene) and at the interface between a liquid nutritive medium. Studies of these submerged biofilms require more customized laboratory tools, for observation and quantification of the 3D structure (thickness, roughness, and biovolume). An optimized framework for this consists in growth in microplates, combined with a confocal microscopy technique, allowing both spatial and temporal monitoring of the submerged biofilms down to a single-cell scale [71].
B. subtilis NDmed was phenotypically compared to NCIB3610 and 168 strains in four multicellular models. In this context, NDmed could form highly structured macrocolonies, pellicles, as well as submerged biofilms and was able to swarm efficiently on semi-solid medium [72]. Moreover, several NDmed-derived mutants defective in genes previously described as triggering biofilm formation in other strains were also compared through this multiculturing approach (Fig. 7). This global view over different biofilm models currently used in genetic studies on both motility and biofilm formation highlighted the value of NDmed as an undomesticated, naturally competent B. subtilis isolate to point out the involvement of several genes in the formation of different structural biofilms.
Fig. 7.
Comparative phenotype for B. subtilis strains and NDmed mutants on different multicellular culture assays. Macrocolonies were grown on 1.5% agar TSA for 6 days at 30 °C. For swarming, 0.7% agar B-medium plates were inoculated on the middle and incubated for 24 h at 30 °C. Pellicles were obtained after 24 h of culture at 30 °C of bacteria in TSB in a 24-well plate. Macrocolony, swarming, and pellicle images are representative of the majority of the phenotype from at least three replicates for each strain revealing the effect of mutations on the biofilm formation. In a microplate system, immersed biofilms are labeled by SYTO 9 after 24 h of incubation at 30 °C. The shadow on the right represents the vertical projection of the submerged biofilm (scale bars represent 40 μm) (From Ref. [72]).
4. Dynamics and structural determinants of B. subtilis NDmed submerged biofilms
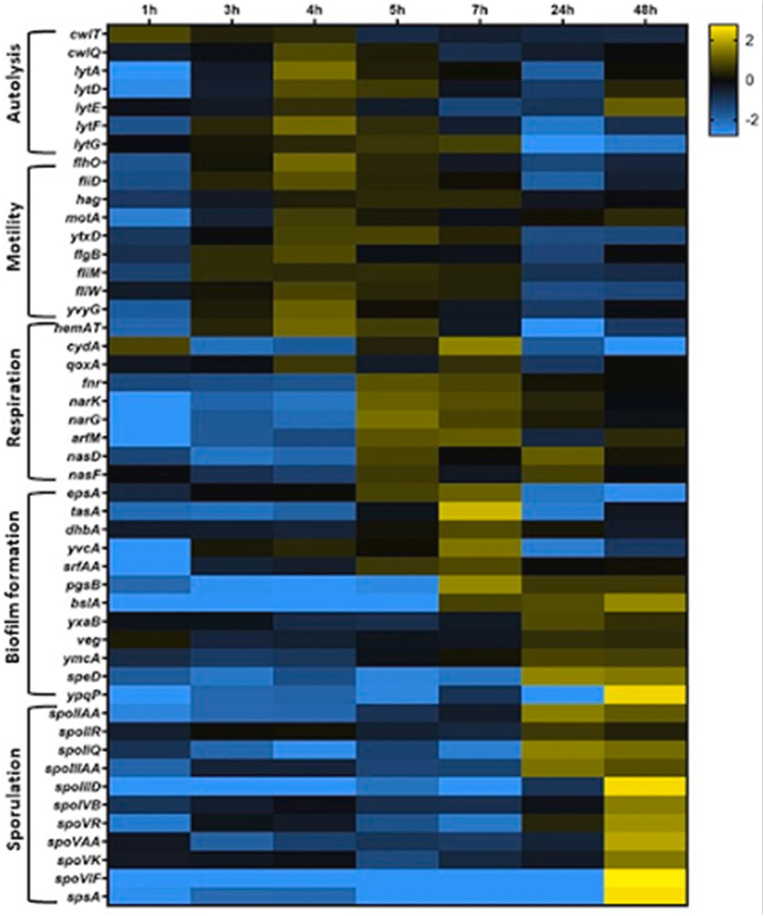
B. subtilis submerged biofilms can be a good model representative of some Bacilli natural habitats such as soil and plant roots surface [5,42,73]. NDmed has proven to be a good tool for studying the formation dynamics of such submerged biofilms. Various photonic and electronic microscopic techniques allowed us to analyze the three-dimensional biofilm architecture with this strain (Fig. 8) [43]. The kinetics of bacterial colonization on the surface could be followed by time-lapse confocal laser scanning microscopy, which revealed an unexpected biphasic submerged biofilm development of NDmed. Measurements of oxygen concentration and reporting the expression of genes involved in motility, matrix synthesis and anaerobiosis allowed to decipher the phenomenon: cells first adhere to the surface, forming elongated chains, which are suddenly fragmented, releasing free motile cells. This switching coincides with an oxygen depletion, which precedes the formation of the pellicle at the liquid-air interface. Residual bacteria still associated with the solid surface start then to express matrix genes under anaerobic metabolism to build the typical biofilm protruding structures (Fig. 9). The same behavior was also observed for all B. subtilis strains tested, notably 168 and NCIB3610, but seems to be very particular to this species, as it was not observed with close relative but different Bacilli (B. cereus, B. licheniformis and B. amyloliquefaciens) [74]. A transcriptome analysis by tiling arrays over a temporal scale confirmed these microscopic observations. During the first hours the genes encoding basic functions essential for cellular growth are expressed at a constant rate. Upon oxygen depletion, when none of the aerobic respiratory genes is expressed, genes required for autolysis and motility start to be upregulated, leading to elongated sessile chains fragmenting into motile cells. Shortly after, upregulation of anaerobic respiration genes can be observed, followed by expression of biofilm matrix genes, the time when the biofilms (submerged and pellicle) are in the process of formation and stabilization of complex architecture. Finally, genes related to sporulation are strongly upregulated in the old biofilm (Fig. 10) [74].
Fig. 8.
3D architecture of B. subtilis NDmed biofilm. (A) Three-dimensional reconstruction of biofilm from Confocal Laser Scanning Microscopy (CLSM) stack images. (C) Field Emission Scanning Electron Microscopy (FESEM) micrograph of biofilm. (B and D) Environmental Scanning Electron Microscopy (ESEM) micrographs of biofilm at pressure in a microscope chamber of 4 and 5 Torr, respectively (From Ref. [43]).
Fig. 9.
The biphasic process of submerged biofilm formation by B. subtilis NDmed. Left panel: (A) 4D-CLSM of B. subtilis NDmed GFP on submerged surfaces. Imaris Easy 3D reconstructions (top) and sections views as an XZ projection (bottom) at specific time points of a representative experiment of three independent experiments. The shadow on the right represents a vertical (YZ) projection of the submerged biofilm (scale bars represent 20 μm). (B) Space-time kymograph generated with BiofilmQ from 4D-CLSM series showing the brutal apparition of free cells in all the wells 3 h after biofilm initiation and the late initiation of submerged biofilm after 7 h. dz represents the distance to the surface in μm and Ich1 the GFP fluorescence intensity in relative arbitrary units. Representative of n = 3 independent biofilms. (C) Individual cell length coordinately and brutally drops during chain fragmentation 2–3 h after biofilm initiation. Chains fragmentation is correlated with an increased number of detected individual objects in the medium. Mean cell length ± SD calculated from n = 3 experiments. Right panel: Space-time kymographs for reporters (D) hag (motility), (E) tapA (matrix), (F) fnr (anaerobiosis) transcription during submerged biofilm formation of B. subtilis NDmed. Representative of n = 3 independent biofilms for each reporter. Kymographs were constructed with BiofilmQ visualization toolbox from 4D-CLSM image sequences with fluorescent transcriptional fusions (NDmed547 [amyE::Phag-gfp sacA::PtapA-mKate2] and GM3361 [Pfnr-gfpmut3]). dz represents the distance to the surface in μm and Ich1 the fluorescent reporter intensity in relative arbitrary units. (G) graph representing the oxygen concentration measured in two wells with a microelectrode showing a sharp decrease of oxygen concentration that drops from around 185 ppm at t = 0 below the probe detection limit after less than 5 h (From Ref. [74]).
Fig. 10.
Temporal tiling array transcriptome of Bacillus subtilis NDmed colonizing microplate wells. All the biomass from the wells was collected for the transcriptome analysis 1, 3, 4, 5, 7, 24, and 48 h after inoculation. A log2 fold change (log2FC) of expression was calculated for the genes from the ratio of expression over the average of expression across all temporal samples. The heatmap displays data for 48 genes selected from Subtiwiki categories, as representatives for the different functional categories [75]. The yellow and the blue represent respectively an upregulation or a downregulation of a gene compared to its average expression over the time course, with a scale adjusted to a log2FC of ±2.8 (From Ref. [74]). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
5. Spatio-temporal heterogeneity of gene expression in B. subtilis surface-associated multicellular assemblages
Bacterial cells in multicellular communities (macrocolony, pellicle, submerged biofilm, swarming cells) are not only spatially localized in microenvironmental settings different from each other, but also subjected to different chemical gradients within each model [4]. For example, in aerial biofilms, the permeability of oxygen in the biomass decreases gradually from the outer top layer to the inside bottom layers, whereas the nutrient gradient is the opposite, with higher concentrations near the surface (nutrient agar or liquid surface). On the other hand, in the submerged biofilms the oxygen and the nutrient gradients are parallel, with gradually decreasing concentrations through the biomass from the top to the bottom inert surface. These chemical gradients generate within each biofilm model local microenvironments associated with physiologically heterogeneous bacterial subpopulations that differ both spatially and temporally and not necessarily bringing into play the same genetic elements nor at the same level. Multicellular communities developing throughout different environmental culturing conditions can present some similarities, but can also display considerable differences at the structural, chemical, and gene expression heterogeneity levels [72,76].
A spatio-temporal correlation could take place between the phenotype and the patterns of gene expression, which can lead to subpopulations with different functions in coordination with time. Indeed, it has been shown that B. subtilis biofilm growth is highly regulated and organized into discrete ontogenetic stages, analogous to those of eukaryotic embryos, recapitulating phylogeny at the gene expression level [77]. Thus, various types of B. subtilis cells are present at the same time in a biofilm, such as motile cells, surfactant producers, matrix producers and sporulating ones [78]. These subpopulations with distributed different tasks are important for the growth and migration of cells seeking nutrients [[79], [80], [81], [82]]. A whole transcriptional analysis of the differently localized heterogeneous compartments of these different biofilm models allowed us to further understand the core of the transcriptional network taking place between them during NDmed biofilms development. To unveil the spatial transcriptional heterogeneity between the different communities, various spatio-physiological populations selected from different spatially organized B. subtilis NDmed communities were analyzed by RNA-seq, which led to a global characterisation of genes specifically expressed in each compartmental population [83]. Following this mesoscale analysis, the patterns of expression of several selected genes were reported by fluorescent transcriptional reporter fusions at a single-cell scale with time-lapse confocal laser scanning microscopy (CLSM) (Fig. 11A). This also permitted to unveil spectacular mosaic expression patterns of genes involved in antagonist functions within a biofilm, such as motility vs matrix synthesis (Fig. 11B). Especially, a particular attention on expression of oppositely regulated genes of the carbon central metabolism allowed to identify in a same biofilm bacterium under either glycolytic or gluconeogenic regimes, coexisting as spatially segregated populations. Altogether, this study gave novel insights into the development and dispersal of B. subtilis NDmed surface-associated communities [83].
Fig. 11.
CLSM of NDmed 547 reporting in green the expression of hag (motility) and in red the expression of tapA (matrix synthesis). (A) 4D-CLSM of the biphasic submerged biofilm formation process. The scale bars represent 50 μm. (B) CLSM visualization of the wells colonization after 24 h, both on the surface (with a zoom on submerged biofilm on the bottom right with a scale bar of 30 μm) and at the liquid-air interface (with a zoom on a floating pellicle on the up right with a scale bar of 30 μm) (From Ref. [74]). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
6. Future contributions of advanced technologies in the study of NDmed biofilms
Our exploration of the mechanisms underlying biofilm formation and architecture in the NDmed strain already provided a huge amount of data regarding the spatiotemporal expression of genes. This genetically tractable strain is now attracting considerable interest as a model biofilm-forming Bacillus to expand our knowledge of the gene regulatory network behind this developmental switch using advanced genetic tools.
The recent progress of the CRISPR-Cas technology, in combination with phage derived lambda-red recombineering system has improved genome editing and genetic engineering in a wide range of bacteria. The CRISPR/cas9 derived from Streptococcus pyogenes, has been already proved useful in assisting genome editing in both domesticated and undomesticated Bacilli [[84], [85], [86]]. The high level of genetic identity of the NDmed strain with the laboratory B. subtilis model strain 168 makes it possible to take advantage, by simple transformation, of the comprehensive collection of BKE/BKK single mutants targeting each of the non-essential genes of this bacterium [87]. Combined with CRISPR methodology, this invaluable collection, available from the Bacillus Genetic Stock Center (BGSC, USA) could be also leveraged to perform CRISPR-assisted targeted genetic engineering in other B. subtilis strains [88].
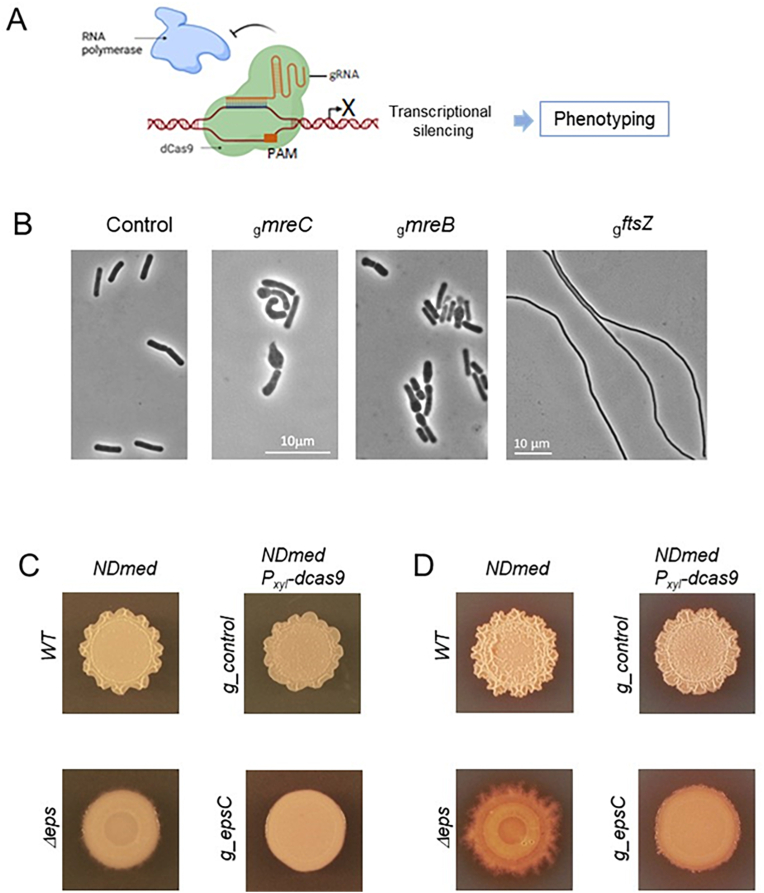
The CRISPR-dCas9 gene silencing system is also a very effective loss-of-function tool to study the relationships between genotype and phenotype without requiring the alteration of genes. Using a catalytically inactive Cas9 protein (dCas9) and single gene-targeting guide RNAs (sgRNAs), CRISPR interference (CRISPRi) has emerged as a powerful genetic methodology to dissect the functions of genes in various bacterial species [[89], [90], [91], [92], [93]]. Such an approach has been used to investigate 258 essential gene functions in B. subtilis 168 [94]. This CRISPRi system, composed of chromosomally inserted modules expressing a xylose-inducible dcas9 gene and single gRNAs, is easily transferable into other B. subtilis strains with highly genomic similarities such as NDmed. Indeed, the Pxyl-based CRISPRi system is functional in NDmed and can be successfully used to block cytokinesis by targeting essential genes involved in cell division and elongation (Fig. 12). CRISPR-mediated knockdown of ftsZ encoding the FtsZ protein involved in the formation of the Z-ring required for the constriction of the septum during division, triggers extensive elongation of cells only a few hours after induction, similar to a B. subtilis ftsZ mutant [95] (Fig. 12B). The downregulation of expression of mreB or mreC, involved in controlling cell morphogenesis generates expected bulged and shapeless cells consequential to a defect in cell wall synthesis [96] (Fig. 12B). This approach is also powerful for studying the function of genes involved in the formation and development of multicellular communities. As illustrated in Fig. 12CD, silencing of epsC and downstream genes of the eps operon, responsible for the synthesis of exopolysaccharides, leads to a smooth biofilm-deficient phenotype of macrocolony, similar to that of a ΔepsA-O strain. Compared to the intricate tri-dimensional structure exhibited by a wild-type strain or a strain expressing the dcas9 together with a neutral non-targeting gRNA, this observation shows that the CRISPRi technology can be successfully applied to long-term phenotypic studies and is relevant to investigate bacterial responses during the transitional switch to biofilm formation. Another interesting aspect of this approach is not only its ability to target multiple genes, but also to probe non-coding elements of the bacterial genome. In all living organisms, non-coding RNAs (ncRNAs) are playing an important role in many biological processes by affecting the translation or the stability of mRNA [97]. Some ncRNAs were found involved in biofilm formation in various bacteria [98,99]. However, their potential regulatory role during biofilm development in Bacillus remains largely unexplored.
Fig. 12.
Gene silencing by CRISPRi in B. subtilis NDmed. (A) Schematic view of CRISPRi-mediated silencing of gene expression. (B) Phase contrast images of NDmed_Pxyl-dcas9 cells expressing gRNAs targeting the mreB, mreC or ftsZ genes. Cells were cultivated in the presence of xylose1% for 5 h prior to observation. Control cells do not contain targeting gRNA sequences. Scale bars represent 10 μm. (C and D) Biofilm macrocolony assay. NDmed_Pxyl-dcas9 cells expressing gRNAs targeting the epsC gene or a negative control guide were inoculated at the center of a MSgg agar plate containing 1% xylose and grown at 30 °C for 40 h (C) or 60 h (D). The macrocolony phenotype resulting from the CRISPRi-mediated gene silencing of epsC was compared to those of the NDmed Wild-Type and ΔepsA-O mutant. The macrocolony images are representative of three replicates.
In combination with NGS sequencing technologies, CRISPRi pool (or CRISPi-seq) is now used successfully to perform large-scale functional genetic screening using genome-wide libraries of gRNAs. These screens allow to quickly identify genes or genetic elements whose repression confers an advantage or a disadvantage in a particular physiological condition [100]. CRISPRi pools enable the interrogation of the fitness of genes upon exposure to biological stressors. This approach can be now timely used to investigate genes and regulatory pathways affecting biofilm formation when subjected to chemical or physical challenges such as biocides or extreme environments such as altered gravity. Based on the RNAseq data generated in our previous transcriptome studies, we have already constructed in NDmed a biofilm-oriented library of guide RNAs targeting a subset of genes upregulated during the early stage of biofilm formation.
We project to use NDmed as a model strain for studying microbial biofilms in microgravity and hypergravity conditions. Microgravity corresponds to conditions encountered in the International Space Station (ISS), in which the establishment and development of biofilms on many different hardware surfaces can lead to significant problems [101,102]. Thus understanding the particularities in the mechanisms involved in such conditions is a real challenge toward the limitation of these problems susceptible to arise beyond the ISS in long spaceship journeys and in extraterrestrial human base settlements with lower gravity (Moon, Mars …).
To conclude, the B. subtilis strain NDmed possesses a remarkable ability to form highly structured biofilms with different morphologies such as complex macrocolonies, thick pellicles, and beanstalk-like submerged biofilm structures. It is also hyper-resistant to biocides and can protect pathogens in mixed-species biofilms. Along with its ease of genetic manipulation, NDmed stands out as a valuable bacterial model for biofilm studies using modern molecular and microscopic techniques.
CRediT authorship contribution statement
Yasmine Dergham: Conceptualization, Writing – original draft, Writing – review & editing. Dominique Le Coq: Conceptualization, Writing – original draft, Writing – review & editing. Arnaud Bridier: Writing – review & editing. Pilar Sanchez-Vizuete: Writing – review & editing. Hadi Jbara: Writing – original draft, Writing – review & editing. Julien Deschamps: Writing – review & editing. Kassem Hamze: Writing – review & editing, Funding acquisition. Ken-ichi Yoshida: Writing – review & editing. Marie-Françoise Noirot-Gros: Conceptualization, Writing – original draft. Romain Briandet: Conceptualization, Writing – review & editing, Funding acquisition, Project administration.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the MICA department and the Micalis Institute of INRAE. Yasmine Dergham was the recipient of funding from the Union of Southern Suburbs Municipalities of Beirut, INRAE, Campus France PHC CEDRE 42280 PF and Fondation AgroParisTech (France). Hadi Jbara is the recipient of funding from the European Space Agency (ESA) OSIP IDEA: I-2021-03383 and INRAE. Pilar Sanchez-Vizuete was the recipient of a PhD grant from the Région Ile-de-France (DIM ASTREA). Arnaud Bridier was the recipient of a PhD grant from the Medicen foundation. We thank Adrien Forge for technical contribution in CRISPR-mediated phenotyping. This work is performed under the umbrella of the European Space Agency Topical Team: Biofilms from an interdisciplinary perspective.
Data availability
No data was used for the research described in the article.
References
- 1.Flemming H.-C., Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 2.Flemming H.-C., Wuertz S. Bacteria and archaea on Earth and their abundance in biofilms. Nat Rev Microbiol. 2019;17:247–260. doi: 10.1038/s41579-019-0158-9. [DOI] [PubMed] [Google Scholar]
- 3.Flemming H.-C., Wingender J., Szewzyk U., Steinberg P., Rice S.A., Kjelleberg S. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol. 2016;14:563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 4.Stewart P., Franklin M. Physiological heterogeneity in biofilms. Nat Rev Microbiol. 2008;6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y., Yan F., Chai Y., Liu H., Kolter R., Losick R., et al. Biocontrol of tomato wilt disease by Bacillus subtilis isolates from natural environments depends on conserved genes mediating biofilm formation. Environ Microbiol. 2013;15:848–864. doi: 10.1111/j.1462-2920.2012.02860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng Y., Zhu Y., Wang P., Zhu L., Zheng J., Li R., et al. Complete genome sequence of Bacillus subtilis BSn5, an endophytic bacterium of Amorphophallus konjac with antimicrobial activity for the plant pathogen Erwinia carotovora subsp. carotovora. J Bacteriol. 2011;193 doi: 10.1128/JB.00129-11. 2070–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbosa T.M., Serra C.R., La Ragione R.M., Woodward M.J., Henriques A.O. Screening for Bacillus isolates in the broiler gastrointestinal tract. Appl Environ Microbiol. 2005;71:968–978. doi: 10.1128/AEM.71.2.968-978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tam N.M.K., Uyen N.Q., Hong H.A., Duc L.H., Hoa T.T., Serra C.R., Henriques A.O., Cutting S.M. The intestinal life cycle of Bacillus subtilis and close relatives. J Bacteriol. 2006;188:2692–2700. doi: 10.1128/JB.188.7.2692-2700.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong H.A., Khaneja R., Tam N.M.K., Cazzato A., Tan S., Urdaci M., et al. Bacillus subtilis isolated from the human gastrointestinal tract. Res Microbiol. 2009;160:134–143. doi: 10.1016/j.resmic.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 10.de Boer Sietske A., Diderichsen B. On the safety of Bacillus subtilis and B. amyloliquefaciens: a review. Appl Microbiol Biotechnol. 1991;36:1–4. doi: 10.1007/BF00164689. [DOI] [PubMed] [Google Scholar]
- 11.Sewalt V., Shanahan D., Gregg L., La Marta J., Carrillo R. The generally recognized as safe (GRAS) process for industrial microbial enzymes. Ind Biotechnol. 2016;12:295–302. doi: 10.1089/ind.2016.0011. [DOI] [Google Scholar]
- 12.Earl A.M., Losick R., Kolter R. Ecology and genomics of Bacillus subtilis. Trends Microbiol. 2008;16:269–275. doi: 10.1016/j.tim.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujita M., Nomura K., Hong K., Ito Y., Asada A., Nishimuro S. Purification and characterization of a strong fibrinolytic enzyme (nattokinase) in the vegetable cheese natto, a popular soybean fermented food in Japan. Biochem Biophys Res Commun. 1993;197:1340–1347. doi: 10.1006/bbrc.1993.2624. [DOI] [PubMed] [Google Scholar]
- 14.Bais H.P., Fall R., Vivanco J.M. Biocontrol of Bacillus subtilis against infection of arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol. 2004;134:307–319. doi: 10.1104/pp.103.028712. http://www.plantphysiol.org/cgi/doi/10.1104/pp.103.028712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marzorati M., Van den Abbeele P., Bubeck S., Bayne T., Krishnan K., Young A., Mehta D., DeSouza A. Bacillus subtilis HU58 and Bacillus coagulans SC208 probiotics reduced the effects of antibiotic-induced gut microbiome dysbiosis in an M-SHIME® model. Microorganisms. 2020;8:1–15. doi: 10.3390/microorganisms8071028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Dijl J., Hecker M. Bacillus subtilis: from soil bacterium to super-secreting cell factory. Microb Cell Factories. 2013;12:3. doi: 10.1186/1475-2859-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tiano P., Biagiotti L., Mastromei G. Bacterial bio-mediated calcite precipitation for monumental stones conservation: methods of evaluation. J Microbiol Methods. 1999;36:139–145. doi: 10.1016/s0167-7012(99)00019-6. [DOI] [PubMed] [Google Scholar]
- 18.Gadd G.M. Metals, minerals and microbes: geomicrobiology and bioremediation. Microbiology. 2010;156:609–643. doi: 10.1099/mic.0.037143-0. [DOI] [PubMed] [Google Scholar]
- 19.Caselli E., Pancaldi S., Baldisserotto C., Petrucci F., Impallaria A., Volpe L., et al. Characterization of biodegradation in a 17th century easel painting and potential for a biological approach. PLoS One. 2018;13(12) doi: 10.1371/journal.pone.0207630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han Z., Wang J., Zhao H., Tucker M.E., Zhao Y., Wu G., Zhou J., Yin J., Zhang H., Zhang X., Yan H. Mechanism of biomineralization induced by Bacillus subtilis J2 and characteristics of the biominerals. Minerals. 2019;9(4):218. doi: 10.3390/min9040218. [DOI] [Google Scholar]
- 21.Martin D.J.H., Denyer S.P., McDonnell G., Maillard J.-Y. Resistance and cross-resistance to oxidising agents of bacterial isolates from endoscope washer disinfectors. J Hosp Infect. 2008;69:377–383. doi: 10.1016/j.jhin.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 22.Jasper B., van Praagh J.B., Luo J.N., Zaborina O., Alverdy J.C. Involvement of the commensal organism Bacillus subtilis in the pathogenesis of anastomotic leak. Surg Infect. 2020;21:865–870. doi: 10.1089/sur.2019.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi K., Ehrlich S.D., Albertini A., Amati G., Andersen K.K., Arnaud M., et al. Essential Bacillus subtilis genes. Proc Natl Acad Sci USA. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piggot P.J., Hilbert D.W. Sporulation of Bacillus subtilis. Curr Opin Microbiol. 2004;7:579–586. doi: 10.1016/j.mib.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 25.Nicolas P., Mäder U., Dervyn E., Rochat T., Leduc A., Pigeonneau N., et al. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science. 2012;335:1103–1106. doi: 10.1126/science.1206848. [DOI] [PubMed] [Google Scholar]
- 26.Buescher J.M., Liebermeister W., Jules M., Uhr M., Muntel J., Botella E., et al. Global network reorganization during dynamic adaptations of Bacillus subtilis metabolism. Science. 2012;335:1099–1103. doi: 10.1126/science.1206871. [DOI] [PubMed] [Google Scholar]
- 27.Toole G.O., Kaplan H.B., Kolter R. Biofilm Formation as microbial development. Annu Rev Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 28.Branda S.S., González-Pastor J.E., Ben-Yehuda S., Losick R., Kolter R. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci U S A. 2001;98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Branda S.S., Chu F., Kearns D.B., Losick R., Kolter R. A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol. 2006;59:1229–1238. doi: 10.1111/j.1365-2958.2005.05020.x. [DOI] [PubMed] [Google Scholar]
- 30.McLoon A.L., Guttenplan S.B., Kearns D.B., Kolter R., Losick R. Tracing the domestication of a biofilm-forming bacterium. J Bacteriol. 2011;193:2027–2034. doi: 10.1128/JB.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cairns L.S., Hobley L., Stanley-Wall N.R. Biofilm formation by Bacillus subtilis: new insights into regulatory strategies and assembly mechanisms. Mol Microbiol. 2014;93:587–598. doi: 10.1111/mmi.12697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konkol M.A., Blair K.M., Kearns D.B. Plasmid-encoded comI inhibits competence in the ancestral 3610 strain of Bacillus subtilis. J Bacteriol. 2013;195:4085–4093. doi: 10.1128/JB.00696-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Branda S.S., González-Pastor J.E., Dervyn E., Ehrlich S.D., Losick R., Kolter R. Genes involved in formation of structured multicellular communities by Bacillus subtilis. J Bacteriol. 2004;186:3970–3979. doi: 10.1128/JB.186.12.3970-3979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vlamakis H., Chai Y., Beauregard P., Losick R., Kolter R. Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol. 2013;11:157–168. doi: 10.1038/nrmicro2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arnaouteli S., Bamford N.C., Stanley-Wall N.R., Kovács A.T. Bacillus subtilis biofilm formation and social interactions. Nat Rev Microbiol. 2021;19:600–614. doi: 10.1038/s41579-021-00540-9. [DOI] [PubMed] [Google Scholar]
- 36.Thérien M., Kiesewalter H.T., Auria E., Charron-Lamoureux V., Wibowo M., Maróti G., Kovács A.T., Beauregard P.B. Surfactin production is not essential for pellicle and root-associated biofilm development of Bacillus subtilis. Biofilms. 2020;2 doi: 10.1016/j.bioflm.2020.100021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stefanic P., Mandic-Mulec I. Social interactions and distribution of Bacillus subtilis pherotypes at microscale. J Bacteriol. 2009;191:1756–1764. doi: 10.1128/JB.01290-08. https://doi:10.1128/JB.01290-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spacapan M., Danevcic T., Mandic-Mulec I. ComX-induced exoproteases degrade ComX in Bacillus subtilis PS-216. Front Microbiol. 2018;9:105. doi: 10.3389/fmicb.2018.00105. https://doi:10.3389/fmicb.2018.00105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spacapan M., Danevčič T., Štefanic P., Mandic-Mulec I. Quorum sensing in Bacillus subtilis slows down biofilm formation by enabling sporulation bet hedging. bioRxiv. 2019 doi: 10.1101/768671. preprint. [DOI] [Google Scholar]
- 40.Krajnc M., Stefanic P., Kostanjšek R., Mandic-Mulec I., Dogsa I., Stopar D. Systems view of Bacillus subtilis pellicle development. npj Biofilms Microbiomes. 2022;8:25. doi: 10.1038/s41522-022-00293-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamon M.A., Lazazzera B.A. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol Microbiol. 2001;42:1199–1209. doi: 10.1046/j.1365-2958.2001.02709.x. [DOI] [PubMed] [Google Scholar]
- 42.Bridier A., Le Coq D., Dubois-Brissonnet F., Thomas V., Aymerich S., Briandet R. The spatial architecture of Bacillus subtilis biofilms deciphered using a surface-associated model and in situ imaging. PLoS One. 2011;6 doi: 10.1371/journal.pone.0016177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bridier A., Meylheuc T., Briandet R. Realistic representation of Bacillus subtilis biofilms architecture using combined microscopy (CLSM, ESEM and FESEM) Micron. 2013;48:65–69. doi: 10.1016/j.micron.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 44.Hall-Stoodley L., Stoodley P. Evolving concepts in biofilm infections. Cell Microbiol. 2009;11:1034–1043. doi: 10.1111/j.1462-5822.2009.01323.x. [DOI] [PubMed] [Google Scholar]
- 45.Bridier A., Sanchez-Vizuete P., Guilbaud M., Piard J.C., Naïtali M., Briandet R. Biofilm-associated persistence of food-borne pathogens. Food Microbiol. 2015;45:167–178. doi: 10.1016/j.fm.2014.04.015. [DOI] [PubMed] [Google Scholar]
- 46.Deva A.K., Vickery K., Zou J., West R.H., Selby W., Benn R.A.V., Harris J.P., Cossart Y.E. Detection of persistent vegetative bacteria and amplified viral nucleic acid from in-use testing of gastrointestinal endoscopes. J Hosp Infect. 1998;39:149–157. doi: 10.1016/S0195-6701(98)90329-2. [DOI] [PubMed] [Google Scholar]
- 47.Machado A.P., Pimenta A.T.M., Contijo P.P., Geocze S., Fischman O. Microbiologic profile of flexible endoscope disinfection in two Brazilian hospitals. Arq Gastroenterol. 2006;43:255–258. doi: 10.1590/s0004-28032006000400002. [DOI] [PubMed] [Google Scholar]
- 48.Martin Deborah J.H. Cardiff University; Cardiff (UK): 2009. Understanding microbial survival in, and the development of resistance to, high-level disinfection. PhD thesis. [Google Scholar]
- 49.Martin D.J.H., Wesgate R.L., Denyer S.P., McDonnell G., Maillard J.Y. Bacillus subtilis vegetative isolate surviving chlorine dioxide exposure: an elusive mechanism of resistance. J Appl Microbiol. 2015;119:1541–1551. doi: 10.1111/jam.12963. [DOI] [PubMed] [Google Scholar]
- 50.Bridier A., Sanchez-Vizuete MdP., Le Coq D., Aymerich S., Meylheuc T., Maillard J.Y., Thomas V., Dubois-Brissonnet F., Briandet R. Biofilms of a Bacillus subtilis hospital isolate protect Staphylococcus aureus from biocide action. PLoS One. 2012;7 doi: 10.1371/journal.pone.0044506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanchez-Vizuete P., Orgaz B., Aymerich S., Le Coq D., Briandet R. Pathogens protection against the action of disinfectants in multispecies biofilms. Front Microbiol. 2015;6:705. doi: 10.3389/fmicb.2015.00705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanchez-Vizuete P., Tanaka K., Bridier A., Shirae Y., Yoshida K., Bouchez T., Aymerich S., Briandet R., Le Coq D. Genome sequences of two nondomesticated Bacillus subtilis strains able to form thick biofilms on submerged surfaces. Genome Announc. 2014;2 doi: 10.1128/genomeA.00946-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Durrett R., Miras M., Mirouze N., Narechania A., Mandic-Mulec I., Dubnau D. Genome sequence of the Bacillus subtilis biofilm-forming transformable strain PS216. Genome Announc. 2013;1(3) doi: 10.1128/genomeA.00288-13. https://doi:10.1128/genomeA.00288-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kunst F., Ogasawara N., Moszer I., Albertini A.M., Alloni G., Azevedo V., et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 55.Nye T.M., Schroeder J.W., Kearns D.B., Simmons L.A. Complete genome sequence of undomesticated Bacillus subtilis strain NCIB 3610. Genome Announc. 2017;5 doi: 10.1128/genomeA.00364-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brito P.H., Chevreux B., Serra C.R., Schyns G., Henriques A.O., Pereira-Leal J.B. Genetic competence drives genome diversity in Bacillus subtilis. Genome Biol Evol. 2018;10:108–124. doi: 10.1093/gbe/evx270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kohm K., Floccari V.A., Lutz V.T., Nordmann B., Mittelstädt C., Poehlein A., Dragoš A., Commichau F.M., Hertel R. The Bacillus phage SPβ and its relatives: a temperate phage model system reveals new strains, species, prophage integration loci, conserved proteins and lysogeny management components. Environ Microbiol. 2022;24:2098–2118. doi: 10.1111/1462-2920.15964. [DOI] [PubMed] [Google Scholar]
- 58.Abe K., Kawano Y., Iwamoto K., Arai K., Maruyama Y., Eichenberger P., et al. Developmentally-regulated excision of the SPβ prophage reconstitutes a gene required for spore envelope maturation in Bacillus subtilis. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abe K., Takamatsu T., Sato T. Mechanism of bacterial gene rearrangement: SprA-catalyzed precise DNA recombination and its directionality control by SprB ensure the gene rearrangement and stable expression of spsM during sporulation in Bacillus subtilis. Nucleic Acids Res. 2017;45:6669–6683. doi: 10.1093/nar/gkx466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dubois T., Krzewinski F., Yamakawa N., Lemy C., Hamiot A., Brunet L., Lacoste A.S., Knirel Y., Guerardel Y., Faille C. The sps genes encode an original legionaminic acid pathway required for crust assembly in Bacillus subtilis. mBio. 2020;11 doi: 10.1128/mBio.01153-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sanchez-Vizuete P., Le Coq D., Bridier A., Herry J.-M., Aymerich S., Briandet R. Identification of ypqP as a New Bacillus subtilis biofilm determinant that mediates the protection of Staphylococcus aureus against antimicrobial agents in mixed-species communities. Appl Environ Microbiol. 2015;81:109–118. doi: 10.1128/AEM.02473-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dragoš A., Andersen A.J.C., Lozano-Andrade C.N., Kempen P.J., Kovacs A.T., Strube M.L. Phages carry interbacterial weapons encoded by biosynthetic gene clusters. Curr Biol. 2021;31 doi: 10.1016/j.cub.2021.05.046. 3479-3489.e5. [DOI] [PubMed] [Google Scholar]
- 63.Dogsa I., Brloznik M., Stopar D., Mandic-Mulec I. Exopolymer diversity and the role of levan in Bacillus subtilis biofilms. PLoS One. 2013;8(4) doi: 10.1371/journal.pone.0062044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Asally M., Kittisopikul M., Rué P., Du Y., Hu Z., Çağatay T., Robinson A.B., Lu H., Garcia-Ojalvo J., Süel G.M. Localized cell death focuses mechanical forces during 3D patterning in a biofilm. Proc Natl Acad Sci USA. 2012;109:18891–18896. doi: 10.1073/pnas.1212429109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilking J.N., Zaburdaev V., De Volder M., Losick R., Brenner M.P., Weitz D.A. Liquid transport facilitated by channels in Bacillus subtilis biofilms. Proc Natl Acad Sci USA. 2013;110:848–852. doi: 10.1073/pnas.1216376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hamze K., Autret S., Hinc K., Laalami S., Julkowska D., Briandet R., Renault M., Absalon C., Holland I.B., Putzer H., Séror S.J. Single-cell analysis in situ in a Bacillus subtilis swarming community identifies distinct spatially separated subpopulations differentially expressing hag (flagellin), including specialized swarmers. Microbiology. 2011;157:2456–2469. doi: 10.1099/mic.0.047159-0. [DOI] [PubMed] [Google Scholar]
- 67.Julkowska D., Obuchowski M., Holland I.B., Séror S.J. Branched swarming patterns on a synthetic medium formed by wild-type Bacillus subtilis strain 3610: detection of different cellular morphologies and constellations of cells as the complex architecture develops. Microbiology. 2004;150:1839–1849. doi: 10.1099/mic.0.27061-0. [DOI] [PubMed] [Google Scholar]
- 68.Julkowska D., Obuchowski M., Holland I.B., Séror S.J. Comparative analysis of the development of swarming communities of Bacillus subtilis 168 and a natural wild type: critical effects of surfactin and the composition of the medium. J Bacteriol. 2005;187:65–76. doi: 10.1128/JB.187.1.65–76.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hamouche L., Laalami S., Daerr A., Song S., Holland B.I., Séror S.J., Hamze K., Putzer H. Bacillus subtilis swarmer cells lead the swarm, multiply, and generate a trail of Quiescent descendants. mBio. 2017;8:1–14. doi: 10.1128/mBio.02102-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Debois D., Hamze K., Guérineau V., Le Caër J.P., Holland I.B., Lopes P., Ouazzani J., Séror S.J., Brunelle A., Laprévote O. In situ localisation and quantification of surfactins in a Bacillus subtilis swarming community by imaging mass spectrometry. Proteomics. 2008;8:3682–3691. doi: 10.1002/pmic.200701025. [DOI] [PubMed] [Google Scholar]
- 71.Bridier A., Dubois-Brissonnet F., Boubetra A., Thomas V., Briandet R. The biofilm architecture of sixty opportunistic pathogens deciphered using a high throughput CLSM method. J Microbiol Methods. 2010;82:64–70. doi: 10.1016/j.mimet.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 72.Dergham Y., Sanchez-Vizuete P., Le Coq D., Deschamps J., Bridier A., Hamze K., et al. Comparison of the genetic features involved in Bacillus subtilis biofilm formation using multi-culturing approaches. Microorganisms. 2021;9:633. doi: 10.3390/microorganisms9030633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pandin C., Le Coq D., Canette A., Aymerich S., Briandet R. Should the biofilm mode of life be taken into consideration for microbial biocontrol agents? Microb Biotechnol. 2017;10:719–734. doi: 10.1111/1751-7915.12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sanchez-Vizuete P., Dergham Y., Bridier A., Deschamps J., Dervyn E., Hamze K., Aymerich S., Le Coq D., Briandet R. The coordinated population redistribution between Bacillus subtilis submerged biofilm and liquid-air pellicle. Biofilm. 2021;4 doi: 10.1016/j.bioflm.2021.100065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pedreira T., Elfmann C., Stülke J. The current state of SubtiWiki, the database for the model organism Bacillus subtilis. Nucleic Acids Res. 2022;50:D875–D882. doi: 10.1093/nar/gkab943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bridier A., Briandet R. Microbial biofilms: structural plasticity and emerging properties. Microorganisms. 2022;10:138. doi: 10.3390/microorganisms10010138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Futo M., Opašić L., Koska S., Čorak N., Široki T., Ravikumar V., Thorsell A., Lenuzzi M., Domagoj K., et al. Embryo-like features in developing Bacillus subtilis biofilms. Mol Biol Evol. 2021;38:31–47. doi: 10.1093/molbev/msaa217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Qin Y., Angelini L.L., Chai Y. Bacillus subtilis cell differentiation, biofilm formation and environmental prevalence. Microorganisms. 2022;10:1108. doi: 10.3390/microorganisms10061108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lopez D., Vlamakis H., Kolter R. Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol Rev. 2009;33:152–163. doi: 10.1111/j.1574-6976.2008.00148.x. [DOI] [PubMed] [Google Scholar]
- 80.López D., Kolter R. Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis. FEMS Microbiol Rev. 2010;34:134–149. doi: 10.1111/j.1574-6976.2009.00199.x. [DOI] [PubMed] [Google Scholar]
- 81.van Gestel J., Vlamakis H., Kolter R. From cell differentiation to cell collectives: Bacillus subtilis uses division of labor to migrate. PLoS Biol. 2015;13:1–29. doi: 10.1371/journal.pbio.1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jeckel H., Matthey N., Dresher K. Common concepts for bacterial collectives. Elife. 2019;8:8–10. doi: 10.7554/eLife.47019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dergham Y., Le Coq D., Nicolas P., Deschamps J., Huillet E., Sanchez-Vizuete P., Hamze K., Briandet R. Multi-scale transcriptome unveils spatial organisation and temporal dynamics of Bacillus subtilis biofilms. bioRxiv. 2023 doi: 10.1101/2023.01.06.522868. preprint. [DOI] [Google Scholar]
- 84.Altenbuchner J. Editing of the Bacillus subtilis genome by the CRISPR-Cas9 system. Appl Environ Microbiol. 2016;82:5421–5427. doi: 10.1128/AEM.01453-16. https://doi:10.1128/AEM.01453-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang K., Duan X., Wu J. Multigene disruption in undomesticated Bacillus subtilis ATCC 6051a using the CRISPR/Cas9 system. Sci Rep. 2016;6 doi: 10.1038/srep27943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang Y., Wang D., Wang X., Tao H., Feng E., Zhu L., Pan C., Wang B., Liu C., Liu X., Wang H. Highly efficient genome engineering in Bacillus anthracis and Bacillus cereus using the CRISPR/Cas9 system. Front Microbiol. 2019;10:1932. doi: 10.3389/fmicb.2019.01932. https://doi:10.3389/fmicb.2019.01932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Koo B.-M., Kritikos G., Farelli J.D., Todor H., Tong K., Kimsey H., Wapinski I., et al. Construction and analysis of two genome-scale deletion libraries for Bacillus subtilis. Cell Syst. 2017;4:291–305. doi: 10.1016/j.cels.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sachla A.J., Alfonso A.J., Helmann J.D. A simplified method for CRISPR-Cas9 engineering of Bacillus subtilis. Microbiol Spectr. 2021;9 doi: 10.1128/Spectrum.00754-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Qi L., Larson M., Gilbert L., Doudna J., Weissman J., Arkin A., Lim W. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Choudhary E., Thakur P., Pareek M., Agarwal N. Gene silencing by CRISPR interference in mycobacteria. Nat Commun. 2015;6:6267. doi: 10.1038/ncomms7267. [DOI] [PubMed] [Google Scholar]
- 91.Rock J.M., Hopkins F.F., Chavez A., Diallo M., Chase M.R., Gerrick E.R., et al. Programmable transcriptional repression in mycobacteria using an orthogonal CRISPR interference platform. Nat Microbiol. 2017;2 doi: 10.1038/nmicrobiol.2016.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Noirot-Gros M.F., Forrester S., Malato G., Larsen P., Noirot P. CRISPR interference to interrogate genes that control biofilm formation in Pseudomonas fluorescens. Sci Rep. 2019;9 doi: 10.1038/s41598-019-52400-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Guzzo M., Castro L.K., Reisch C.R., Guo M.S., Laub M.T. A CRISPR interference system for efficient and rapid gene knockdown in Caulobacter crescentus. mBio. 2020;11 doi: 10.1128/mBio.02415-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Peters J.M., Colavin A., Shi H., Czarny T.L., Larson M.H., Wong S., Hawkins J.S., Lu C.H.S., Koo B.M., Marta E., et al. A comprehensive, CRISPR-based functional analysis of essential genes in bacteria. Cell. 2016;165:1493–1506. doi: 10.1016/j.cell.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Erickson H.P., Anderson D.E., Osawa FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol Mol Biol Rev. 2010;74:504–528. doi: 10.1128/MMBR.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Carballido-López R., Formstone A. Shape determination in Bacillus subtilis. Curr Opin Microbiol. 2007;10:611–616. doi: 10.1016/j.mib.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 97.Repoila F., Darfeuille F. Small regulatory non-coding RNAs in bacteria: physiology and mechanistic aspects. Biol Cell. 2009;101:117–131. doi: 10.1042/BC20070137. [DOI] [PubMed] [Google Scholar]
- 98.Mitra A., Mukhopadhyay S. Regulation of biofilm formation by non-coding RNA in prokaryotes. Curr Res Pharmacol Drug Discov. 2022;4 doi: 10.1016/j.crphar.2022.100151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huber M., Lippegaus A., Melamed S., Siemers M., Wucher B.R., Hoyos M., Nadell C., Storz G., Papenfort K. An RNA sponge controls quorum sensing dynamics and biofilm formation in Vibrio cholerae. Nat Commun. 2022;13:7585. doi: 10.1038/s41467-022-35261-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Geissler A.S., Fehler A.O., Poulsen L.D., González-Tortuero E., Kallehauge T.B., Alkan F., Anthon C., Seemann S.E., Rasmussen M.D., Breüner A., Hjort C., Vinther J., Gorodkin J. CRISPRi screen for enhancing heterologous α-amylase yield in Bacillus subtilis. J Ind Microbiol Biotechnol. 2023:50. doi: 10.1093/jimb/kuac028. kuac028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vélez Justiniano Y.-A., Goeres D.M., Sandvik E.L., Kjellerup B.V., Sysoeva T.A., Harris J.S., et al. Mitigation and use of biofilms in space for the benefit of human space exploration. Biofilms. 2023;5 doi: 10.1016/j.bioflm.2022.100102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Marra D., Karapantsios T., Caserta S., Secchi E., Holynska M., Labarthe S., Polizzi B., Ortega S., Kostoglou M., Lasseur C., Karapanagiotis I., Lecuyer S., Bridier A., Noirot-Gros M.-F., Briandet R. Migration of surface-associated microbial communities in spaceflight habitats. Biofilm. 2023;5 doi: 10.1016/j.bioflm.2023.100109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.