Abstract
Lactobacillus and yeast obtained from fermented foods in North-East India were tested for safety and probiotic properties. All the lactobacilli and yeast tested negative for the catalase, indole, urease, phenylalanine, hemolysis, gelatin hydrolysis, and biogenic amine production tests, indicating that they are safe to use as probiotics in food supplements. Lactiplantibacillus plantarum KGL3A (accession no. MG722814) was capable of resisting the replicated gastric fluid (pH 2) till 2 h of exposure, whereas both KGL3A and Lacticaseibacillus rhamnosus K4E (accession no. KX950834.1) strains were able to resist pH 3 till 2 h of exposure with a reduction in overall viable cell count from 7.48 log CFU/mL to 1.09 log CFU/mL and 7.77 log CFU/mL to 0.83 log CFU/mL, respectively. In vitro gastric juice simulation conditions were tolerated by the yeast Saccharomyces cerevisiae WBS2A. The cell surface hydrophobicity (CSH) towards hydrocarbons (n-hexadecane) was seen highest in L. plantarum KGL3A (77.16± 0.84%) and Limosilactobacillus fermentum KGL4 accession no. MF951099 (72.60 ± 2.33%). The percentage auto-aggregation ranged from 8.70 to 25.53 after 2 h, which significantly increased to 10.50 to 26.94 during the fifth hour for cultures. Also, a higher percentage of co-aggregation was found for the culture L. rhamnosus K4E with S. typhi (34.18 ± 0.03%), E. coli (32.97 ± 0.02 %) and S. aureus (26.33 ± 0.06 %) and for the yeast S. cerevisiae WBS2A, a higher percentage of co-aggregation was found with Listeria monocytogenes (25.77 ± 0.22%). The antioxidant activity and proteolytic activity were found to be higher for Lactobacillus helveticus K14 and L. rhamnosus K4E. The proportion of decreased cholesterol was noticeably higher in KGL4 (29.65 ± 4.30%). β glucosidase activity was significantly higher in the L. fermentum KGL4 strain (0.359 ± 0.002), and α galactosidase activity was significantly higher in the L. rhamnosus K4E strain (0.415 ± 0.016). MTT assays suggested that KGL4 and WBS2A at a lower dose did not exhibit cytotoxicity.
Supplementary Information
The online version contains supplementary material available at 10.1007/s42770-023-01093-0.
Keywords: Lactobacillus, Yeast, Fermented foods, Safety, Anti-inflammatory, Probiotics
Introduction
Since ancient times, traditional fermented foods have been widely consumed and have played a significant role in our nutrition. It can be prepared at home or in a small business setting, utilizing relatively basic methods and tools [1]. Lactic acid bacteria (LAB) from several fermented products are highly regarded for their probiotic properties. Novel LAB strains with probiotic potential may be stored in fermented foods [2]. Probiotics have traditionally been believed to be best delivered by fermented foods [3]. The Lactobacillus genus is found in a variety of environments, along with fermented foods, leaf, and various body parts of the plant also. Normal human gut microbiota is primarily composed of lactobacilli, and some commensal species of these organisms have drawn attention for their favorable health impacts on the host [4]. In 2001, a definition of the probiotic was put out: “a preparation or product that contains enough viable, specified microorganisms to modify the microbiota in a compartment of the host and improve host health” [5]. Commercial uses of probiotic bacteria of human origin include L. rhamnosus GG, Lactobacillus acidophilus LA-1, and Lacticaseibacillus casei Shirota. LAB have been extensively studied in the last few decades for their ability to benefit human health, and it has been discovered that they build desirable gut microflora, thus being “generally regarded as safe” [6]. There are many uses for probiotics, one of which is the treatment of acute diarrhea [7]. Through fermentation and the probiotics' ability to reproduce in the gastrointestinal system, which releases lactase, probiotics are also used to reduce the amount of lactose in dairy products [8]. Studies have demonstrated that Lactiplantibacillus plantarum (KGL3A, accession no. MG722814) in fermented foods has antioxidant activity and antibacterial activity against certain pathogens such as S. typhimurium, E. faecalis, B. cereus, and E. coli. There have been numerous reports of L. plantarum strains producing the antibacterial compounds (bacteriocins) known as plantaricins. These substances have been demonstrated to be particularly potent against gastrointestinal infections and food-borne diseases while having widely varied activities and structures [9]. Probiotics have been proven in numerous studies to have negative impacts on the growth of harmful bacteria. The synthesis of inhibitory compounds like reuterin, bacteriocin, and hydroxyl radicals may be the cause of this adverse effect [10]. Currently, the yeast species Saccharomyces cerevisiae and Saccharomyces boulardii are employed as probiotics. It has been observed that supplementing S. cerevisiae as a live culture in animals improves their growth, health, and immunological response. However, specific considerations are needed when isolating and classifying yeasts from natural sources as probiotics. According to a recent study by Hati et al. [11], supplementing broilers with L. fermentum (accession no. MF951099) and S. cerevisiae (accession no. MG101828) accelerated growth, hematological characteristics, clinical biochemistry, and cecal and fecal microflora, and potential probiotic yeast and lactic acid bacteria maintain the gut health and development of broiler chicks during the 42 days study periods.
Study on the interactions of two kefir-isolated strains in an in vitro model revealed that various Lacticaseibacillus paracasei H9, (CGMCC NO.4780) traits improved the interaction with S. cerevisiae. This might be because of the polysaccharides in yeast cell walls and the proteins on bacterial cell surfaces, which are key components in the various probiotic capacities [12]. This beneficial relationship is typically linked to the yeast’s excretion of nutrients, namely peptides, amino acids, and vitamins [13]. This shows that yeast metabolites were crucial in improving L. rhamnosus survival. Fermented foods have well-known anti-inflammatory properties [14]. Probiotic fermented food is a factor in some of their beneficial properties [15]. Probiotics’ anti-inflammatory and immunomodulatory effects, which extend beyond the gut, have been linked to a variety of health advantages [16]. Consuming fermented foods containing probiotics can improve gut immunity and gut barrier integrity while maintaining gastrointestinal homeostasis [17] via several mechanisms, including the reduction of inflammation-promoting cytokines like IL-17F, IL-23 and Th17, antimicrobial peptide synthesis activation, and mucus secretion. In a study, L. plantarum Q7, L. plantarum F3-2 and L. plantarum YRL45 showed no negative effect or cytotoxicity on growth of RAW 364.7 and Caco-2 cells and also, suppressed the production of NO induced by LPS in RAW 264.7 cells and increased anti-inflammatory ability by inhibiting tumor necrosis factor-α (TNF-α) and Interleukin-1β (IL-1β) [18]. L. fermentum KGC1601 showed marked anti-inflammatory activity by regulating the expression of inflammatory cytokines. Overall, LPS treated media of culture L. fermentum KGC1601 decreased the expression of IL-1β, IL-6 and TNF-α [19]. According to research three indigenous yeast strains of S. cerevisiae namely TA4-10, LL1 and 4LBI-3 showed nontoxic effect on U937/PMA macrophage cell lines and showed potential anti-inflammatory activity by reducing the levels of reactive oxygen species superoxide anion radical and nitric oxide [20]. Studies have demonstrated that the polyphenolic substances in fermented foods promote the formation and metabolism of the microbiota and have the capacity to suppress inflammatory responses and prevent the production of inflammatory cytokines [21]. Given the mounting evidence for the essential function of fermented foods in illness prevention or health promotion [22], the current study’s goal is to assess the reduction of inflammation of Lactobacillus and yeast isolated from fermented foods of North-East India, as well as their safety and probiotic attributes for future application in functional fermented food developments with particular health aspects.
Materials and methods
Bacterial strains
Four LAB namely L. plantarum (KGL3A, accession no. MG722814), L. fermentum (KGL4, accession no. MF951099), L rhamnosus (K4E, accession no. KX950834.1), Lactobacillus helveticus (K14, accession no. KU644578.1) and one yeast strain namely S. cerevisiae (WBS2A, accession no. MG101828) isolated from traditional fermented foods of Garo Hills, Meghalaya, North Eastern part of India were considered for analyzing their safety aspects, probiotic attributes, techno-functional properties and in vitro cell culture study. Safety tests of all the strains were carried out namely Gram’s staining, negative staining, catalase test, indole test, urease test, phenylalanine test, hemolysis test, gelatin hydrolysis test, mucin degradation test, and biogenic amine production.
Safety aspects
Various safety aspects of Lactobacillus cultures and yeast were evaluated. Gram’s staining was carried out using the principles given by Smith and Hussey [23]. Catalase test [24] was performed in a test tube containing 5 mL of 24 h old growth of culture. Five drops of 10% H2O2 were added in the test tube and observed for the appearance of effervescence. The presence of effervescence in the test tube was considered positive result for the test. Indole test with slight modification [25] such as, tubes containing tryptophan broth (5ml) was prepared. 1 mL of active culture was inoculated to tryptophan broth. Tubes were incubated at 37 °C for 24 h. Escherichia coli ATCC 43888 was used as a positive control. After incubation, 2-3 drops of Kovac’s reagent (Himedia, India) were added. Formation of cherry red ring was considered as a positive indole reaction. Urease test [26] was performed in 5mL of Stuart’s Urea Broth inoculated with 24 h old pure culture. Proteus vulgaris ATCC6896 was used as positive control. The tubes were incubated at 37 °C for 24 h and further observed for a color change. Urease production was indicated by a bright pink color. Phenylalanine test [27] was performed in Phenylalanine agar (Sigma-Aldrich, USA) slants. Loop full of active culture was streaked on the slant and incubated for 37 °C for 24 h. P. vulgaris ATCC6896 was used as positive control. After incubation five to ten drops of 10.0% ferric chloride (LOBA-chemie, Mumbai, India) were dropped on the slant agar. When the test tube turned green within 1 to 5 min, it was regarded as a positive reaction. Hemolysis test [28] was followed with certain modifications such as, sheep blood agar (Himedia, India) plates were prepared by incorporating 7% sheep blood. Loop full of active culture was taken and streaked over the solidified sheep blood agar medium. Staphylococcus aureus MTCC 737 was used as positive control. The plates were incubated at 37 °C for 48 h. After incubation, this agar plates were examined for signs of β-hemolysis (Clear zone around colonies), α-hemolysis (green colored zones around the colonies) and γ-hemolysis (No clear zone). Gelatin hydrolysis test [27] was performed with slight modifications such as gelatin media for Lactobacillus strains was prepared by supplementing MRS broth with 12% gelatin (Himedia, India) and for control organism, nutrient gelatin was prepared and filled in tubes (10mL).1 mL of inoculum was added to the tubes and incubated at 37 °C for 7 days. S. aureus MTCC 737 was used as positive control. The gelatin tubes were removed daily from the incubator and placed at 4 °C to check for liquefaction. When a liquefaction reaction occurred at 4°C, it was regarded as a positive reaction. Mucin degradation test [27] was performed by inoculating 1mL of active culture each into 10 ml of (i) MRS basal medium containing 0.3% partially purified mucin (type III, Sigma-Aldrich, USA), (ii) MRS broth containing 1% glucose, (iii) MRS broth without glucose and (iv) MRS broth containing 1% glucose and 0.3% mucin. Tubes were incubated at 37 °C for 24 h. After incubation for 0, 8 and 24 h bacterial growth was assessed by measuring the absorbance at 600 nm. Biogenic amine production [29] was performed by streaking 24 h active culture previously grown in MRS broth supplemented with 1% L-ornithine (Sigma-Aldrich, USA) onto decarboxylase agar medium (Himedia, India) and incubated in anaerobic conditions for 4 days at 37 °C. A color change in medium (purple color) indicated an increase in pH and was considered as positive result.
Antibiotic disc assay [30] determined by Kirby-Bauer disk diffusion test. Standard antibiotic discs were procured from Himedia (India) which included Ampicillin (10 μg), Colistin (10 μg), Tetracycline (30 μg), Nalidixic acid (30 μg) Methicilin (5 μg), Rifampicin (5 μg), Erthromycin (15 μg) and Vancomycin (30 μg). MRS agar plates were prepared and 100μl of active culture was placed over the agar plates and spread over the solidified agar surface using a sterile cotton swab. After 10 min, the antibiotics disc was carefully placed on the agar by using a sterile forceps. The plates were incubated for 24 h at 37 °C. After incubation, the diameter of the inhibition zones was measured using Antibiotic zone scale (PW297, Himedia, India,).
Probiotic attributes
Several probiotic qualities, including bile salt tolerance, gastric juice endurance, intestinal juice endurance, cell hydrophobicity, auto-aggregation of cells, and cell co-aggregation activities of yeasts and lactic acid bacteria, have been tested.
Bile salt tolerance
The lactic acid bacteria and yeast were tested for bile salt tolerance in accordance with Patel et al. [31], with a few minor modifications. A 2% inoculation rate in de Man Rogosa and Sharpe (MRS, Himedia, India)/Yeast Malt (YM, Himedia, India) broth for 18 h was used to activate the cultures. After that, phosphate buffer saline was used to rinse the pellets twice (PBS), 10 min of centrifugation at 12,000 xg at 4 °C (Eppendorf Centrifuge, USA), and then suspended in PBS. To each tube containing 10 mL of MRS/YM broth (Himedia, India), these suspended cultures were added at a rate of 2% along with 0.5% bile and stirred. At intervals of 0, 2, and 4 h, 1 mL of sample was taken from each tube while they were all incubating at 37°C. The samples were homogenized in 10 mL of sterile water. The number of viable cells was determined and represented as CFU/mL after the appropriate dilutions of MRS/YM agar (Himedia, India) were placed on the plates. The plates were then kept at 37 °C and 25 °C for incubation, respectively, for 24–48 h.
Gastric and intestinal juice tolerance
The strains were cultured in MRS broth overnight at 37 °C, and cells were separated by centrifugation (Eppendorf Centrifuge, USA) at 17,700 xg for 15 min. Lactobacillus cells (adjusted to 108 CFU/mL) and yeast cells (adjusted to 105 CFU/mL) were inoculated in simulated gastric juice (NaCl: 0.73 g/L; KCl: 0.05 g/L; pepsin: 0.3 g/L) and pH was adjusted to 2.0 and 3.0. The cells were then incubated for 0, 2, and 4 h. Additionally, the survival rate was examined in terms of log CFU/mL following the isolates' exposure to synthetic digestive fluid (0.3% w/v bile salts and 0.1% w/v pancreatin, pH 8.0) for 0, 2, and 4 h of incubation. As a control, saline solution that is sterile (0.85% w/v NaCl), pH 7.0 was then set [32].
Cell surface hydrophobicity (CSH)
A biochemical measure for determining indirect adherence to the eukaryotic cells in the gut is hydrocarbon adhesion. The BATH (bacterial adhesion to hydrocarbons) method has been applied a little differently from Lee et al. [33]. The procedure for making the bacterial cell suspension in phosphate buffered saline (PBS) was the same as that detailed in the activity for bile salt tolerance. PBS was used to adjust the suspended cell concentration to OD600 0.5 ± 0.070 (A0). A mixture of 4.0 mL of n-hexadecane (Himedia, India) and 4.0 mL of bacterial suspension was vortexed rapidly for 2 min before being left undisturbed in a 37 °C incubator. The organic and aqueous phases were separated for 50 min at room temperature. The optical density (OD) was calculated after three mL of the aqueous phase were removed (A1). Using 4.0 mL PBS and 4.0 mL n-hexadecane, a blank was made in the same way as the test sample, and the OD value was recorded against it. After repeating the experiment, the average optical density value was calculated. The calculation was done using the equation below, the percentage hydrophobicity (%H):
Where,
Cell auto-aggregation
According to Kodaikkal [34], the cultures’ auto-aggregation experiment was conducted. The technique for making the bacterial suspension was the same as that for the bile salt tolerance test. Bacterial cell suspension (4 mL) was vortexed for 1 min to mix, and the auto-aggregation was monitored for 5 h at 37 °C. After removing 0.1 mL of the upper phase, the optical density was measured for 0, 2, and 5 h at 600 nm. The reading recorded at 0h is A0, followed by readings of A2 and A5. The following equation serves as the basis for measuring the percentage auto-aggregation (%Aa):
Where,
Cell co-aggregation
Co-aggregation was carried out with slight modifications to Kodaikkal [34] procedures. Bacterial suspension was prepared as per the procedure mentioned in the bile salt tolerance activity. Equal amounts of the pathogenic bacteria and LAB strains (2 mL each) were placed in test tubes, vortexed for 10 s, and incubated at 37 °C for 2 h. Control tubes containing 4 mL of each specific bacterial strain were also included in the experiment. Readings at 600 nm were taken spectrophotometrically after 2 h, and the findings were displayed as percent co-aggregation (% Co). The formula used to calculate the percentage of co-aggregation is as follows:
Bile salt hydrolysate activity (BSH)
Following Lee et al. [33], bile salt hydrolysate activity was carried out. 10 μL of active culture was spot inoculated onto MRS agar plates supplemented with 0.37g CaCl2 /L (Himedia, India) and 0.5% (w/v) sodium taurocholate (Himedia, India) and incubated anaerobically at 37 °C for 48h. BSH activity was considered positive for cultures showing zone of precipitation.
Techno-functional properties
Different lactic acid bacteria and yeast’s technologically useful characteristics were measured namely, antioxidative activity, proteolytic activity, cholesterol assimilation, α-galactosidase activity and β-glucosidase activity.
Antioxidant activity
The radical-scavenging capacity of various cultures was assessed based on a compound's ability to neutralize the stable ABTS (2, 2-Azino-bis, (3-ethylbenzothaizoline-6-sulfonic acid), Sigma-Aldrich, USA) radical. The antioxidative activity was carried out using the Das et al. [32] technique. The below equation was used to calculate the samples' capacity to neutralize free radicals:
Where,
ASample = the absorbance of sample
AControl = the absorbance of control sample
Proteolytic activity
The peptides generated by the yeast and Lactobacillus isolates in the skimmed milk medium have been measured using a spectrophotometer as the absorbance of free amino acids at 340 nm, according to the O-phthaldialdehyde (OPA) method of Donkor et al. [35].
Cholesterol assimilation
Cholesterol assimilation by the cultures was established by the technique given by Anandharaj et al. [36] with a few changes such as Lactobacillus and yeast cultures were inoculated (at 2% rate) in MRS broth (9 mL) and Yeast malt broth respectively, containing Bile salts (0.2% Sodium taurocholate (Himedia, India) and 0.3% Sodium thioglycolate (Himedia, India) and 50 μg/mL Cholesterol (Himedia, India). The O-phthalaldehyde (OPA) technique of Rudel and Morris (1973) was used to estimate % cholesterol assimilation by culture from media. The cholesterol assimilated by different strains was determined as follows:
Where,
C0: OD500 of MRS/ YM broth supernatant containing culture
C1: OD500 of MRS/ YM broth supernatant without culture
β-glucosidase and α-galactosidase Activity
α-Galactosidase and β-glucosidase activities were measured using the techniques described by Das et al. [32] and Otieno and Shah [37]. The underlying idea behind the enzyme assay is that when the enzyme galactosidase reacts with p-nitrophenyl-D-galactoside substrate, p-nitrophenol (pNP) into the medium as a result of a colorimetric reaction. By employing the Otieno and Shah [37] method to measure the rate of hydrolysis of p-nitrophenyl-D-glucopyranoside, α-glucosidase activity was identified. A UV-Vis spectrophotometer (Systronics, Ahmedabad) was used to spectrophotometrically detect the amount of emitted p-nitrophenol at 410 nm.
In vitro cell culture study
The following items were purchased from Hi-Media (India): MTT (03-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide), DMEM, Penicillin/Streptomycin (P/S) solution, and Fetal Bovine Serum (FBS). Sigma-Aldrich (USA) provided modified Griess reagent and LPS from E. coli. Elabscience, USA provided pro-inflammatory cytokine ELISA kits for IL-6, TNF-α and IL-1β. The NCCS (National Centre for Cell Science), Pune, India, supplied the murine macrophage cell lines (RAW 264.7). These cells were grown in 25-cm2 culture flasks of Dulbecco's Modified Eagle's Medium (DMEM) treated with 10% FBS and 0.1% penicillin-streptomycin antibiotic at 37 °C in a CO2 incubator (5% CO2). The flasks were passed every two days.
Cytotoxicity assay
Using the MTT assay, the cytotoxic effects of KGL4 and WBS2A on RAW 264.7 cells were studied [38]. In a 96-well microtitre plate, 2 x 105 RAW 264.7 cells were plated, cultured for 24 h, and then exposed to WBS2A and KGL4 at various doses (0.25, 0.5, 1, and 2 mg/mL). Each well received 10 μl of MTT (5 mg/mL in phosphate-buffered saline, pH 7.4), which was included following a 24-h incubation period at 37°C in an environment with 5% CO2 and 95% humidity. Incubation continued for another 4 h until a purple color appeared. After removing the supernatant, the formazan crystal had been dissolved in 100 μl of DMSO. Further, the absorbance was measured at 570 nm using a microplate reader (M200 PRO, Tecan Life Science). Based on the cell viability of treated and untreated samples, the percentage of viable cells was calculated.
Induction of NO production by LPS treatment on macrophages and its prevention by KGL4 and WBS2A
In a 48-well microplate, at a density of 1 x 106 /0.2 mL/well, RAW 264.7 cells were seeded and treated with 0.25 mg/mL each of KGL4 and WBS2A, either with or without the addition of 1 μg/mL of LPS from E. coli 026: B6. RAW cells that were given LPS alone (1 μg/mL) served as the positive control. After 16 h of incubation, 150 μl of supernatant was combined to 1% sulfanilamide (0.1% sulfuric acid) and 0.1% N-(1-naphthyl)-ethylenediamine dihydrochloride in 5% phosphoric acid [38]. After 30 min of incubation, the optical density (OD) of the solution was assessed at 540 nm in a microplate reader. The percentage of NO inhibition compared to control was estimated based on the amount of nitrite found in culture supernatants. The formula (OD of test/OD of positive control) × 100 was used to determine the percent NO generated.
In vitro pro-inflammatory cytokine analysis
The supernatants obtained under various interventions described above were used for determining IL-6, IL-1β and TNF-α levels, employing commercial products ELISA kits in accordance with the manufacturer’s institution, to detect pro-inflammatory cytokines (Elabscience, USA). At 450 nm, the absorbance/optical density (OD) generated were measured. A four-parameter logistic curve with standard concentration and OD values was plotted to determine the results.
Statistical analysis
Three experiments’ findings were given as mean standard error (SEM). Tests were run in triplicate. Using a one-way ANOVA, the analysis of variance was carried out, and the Duncan’s test with a 95% confidence level was used to investigate for any significant differences between the sample averages. Comparing various groups in a cell culture study, Tukey’s post hoc analysis was utilized after a one-way ANOVA. The data pertaining to the in vitro cell culture test were analyzed using GraphPad Prism 8.0 Software Inc., (La Jolla, CA, USA). The statistical significance level was set at P ≤ 0.05.
Results and discussion
Safety aspects
All the Lactobacillus cultures and yeast were discovered to be Gram positive. The catalase test was performed, and the positive control taken was S. aureus, which showed effervescence on adding 10% H2O2, whereas all the Lactobacillus and yeast lacked effervescence, showing negative for the catalase test. All the tested Lactobacillus and yeast were negative for indole test (positive control: E. coli ATCC 43888), Urease test (positive control: P. vulgaris ATCC 6896), and the phenylalanine test (positive control: P. vulgaris ATCC 6896). A hemolysis test was performed for all the Lactobacillus and yeast cultures. Red blood cells in the circulation are lysed by extracellular enzymes produced by specific bacterial species, which is referred to as hemolysis. These microbes produce hemolysins, which are extracellular enzyme, which emanate from the colonies and lyse red blood cells entirely or in part. As a positive control, S. aureus MTCC 737 was used, which showed hemolysis of blood cells. The gelatin hydrolysis test was negative for all the lactobacillus and yeast cultures, while the positive culture was S. aureus MTCC 737, which had produced gelatinases that liquefied gelatin (Supplementary material Fig S1). All the tested cultures were negative for biogenic amine production (Supplementary material Fig S1). In an antibiotic disc assay, L. plantarum KGL3A was found to be sensitive to tetracycline and erythromycin, while L. fermentum KGL4 was sensitive to tetracycline, rifampicin, and erythromycin.
Bobga et al. [39] isolated the L. fermentum strain PRI 29 (accession no. NR 113335.1) from Cameroonian fermented cow milk, which was found to be catalase negative and also lacked hemolytic activity. Damisa-Okorhi and Ataikiru [40] identified probiotic bacteria from fermented milk and milk products as L.fermentum MG-4, L. plantarum MN-5, Streptococcus thermophilus MC-2, Lactobacillus acidophilus MC-3, and Lactococcus lactis subspecies cremoris MG-1, whose biochemical testing showed that all of the LAB isolates were negative for indole and gelatine hydrolysis. Belicova et al. [41] studied L. plantarum from Slovak bryndza cheese has probiotic potential and safety characteristics and the study revealed that the majority of the examined L. plantarum isolates lack the capacity to produce these biogenic amines. Ji et al. [42] studied the functionality and security of Korean kimchi's lactic bacterial strains and found that all six tested lactic acid bacterial strains did not show any hemolytic activity, whose reliability was confirmed by a positive control, Bacillus cereus ATCC 27348.
It is commonly acknowledged that evaluating the safety of microorganisms used in food is important and has been one of the primary assurances needed by consumers for probiotic foods. All the Lactobacillus and yeast cultures assessed for safety aspects showed that the cultures are safe for consumption and/or use as a feed supplement.
Probiotic attributes
The resistance to bile salts, gastric juices, intestinal juices, hydrophobicity of the cell surface, cell auto-aggregation, and cell co-aggregation activities of lactic acid bacteria and yeasts were among the probiotic properties tested.
Bile salt tolerance
Surviving under gastro-intestinal tract conditions is an essential property in probiotics. The microorganism’s viability decreases after passing through the harsh conditions of an acidic stomach and must pass through the bile juices in order to reach the intestine. Therefore, probiotic bacteria's ability to endure bile conditions is crucial. Bile salts render live cells inactive by rupturing the cell membrane. The typical bile salt concentration in the human colon is 0.3% (w/v), which is also the concentration used to calculate the growth lag time of LAB strains. Bile salt concentration varies with digesting time [43]. There is disagreement on the precise concentration that the chosen strains should be tolerant of. At intervals of 0, 2, and 4 h, the cultures were evaluated for 0.5% bile salt tolerance. All the lactic acid bacteria strains were unable to tolerate 0.5% bile salt at 2 and 4 h of incubation. In contrast, the yeast strain S. cerevisiae WBS2A was capable of withstanding 0.5% bile during 2 and 4 h of incubation. While the total number of cells decreased after 4 h of incubation from 7.37 log CFU/mL to 6.72 log CFU/mL, the viable cell counts modestly decreased with an increase in incubation time (Table 1). Han et al. [44] found that the lag times of the bile-tolerant strains Levilactobacillus brevis R4, L. curvatus, L. curvatus R5, and Lactiplantibacillus pentosus were 1.58, 3.50, 3.08, and 2.17 h, respectively. While it was discovered that the bile-sensitive bacteria Pediococcus pentosaceus R1, L. acidophilus, and L. fermentum R6 had lag times of over 9 h. The probiotic potential of traditional fermented Bambangan's (Mangifera pajang) isolated lactic acid bacteria in Malaysia was evaluated by Seah et al. [45]. L. rhamnosus strain 0504 had a viable cell count of 0.26 ± 0.01 log CFU/h, while L. plantarum strains varied from 0.41 to 0.68 log CFU/h. Probiotic properties of strains of L. fermentum isolated from cheese from Tulum were examined by Tulumoglu et al. [46]. LP1, LP2, LP5, LP6, and LP7 strains of L. fermentum did not survive 0.5% bile salt after 4 h of exposure. Syal and Vohra [47] screened twenty yeasts for their probiotic potential that were isolated from traditional Indian fermented foods. All the yeasts were able to survive 0.5% bile, where the survival percentage ranged from 93.00 ± 0.83 % to 100.00 ± 0.39 %. The present findings support the Syal and Vohra [47] study, where the yeasts are tolerant to 0.5% bile exposure.
Table 1.
Survival rate of lactobacilli and yeast isolates during exposure to bile salt
| Organism (O) |
Treatment (T) |
Time (H) | T mean | ||
|---|---|---|---|---|---|
| 0 h | 2 h | 4 h | |||
| KGL3A | 0.5% bile | 7.47b | 0.00c | 0.00c | 5.32 |
| Control | 7.49b | 8.47a | 8.49a | ||
| KGL4 | 0.5% bile | 7.41b | 0.00c | 0.00c | 5.29 |
| Control | 7.43b | 8.42a | 8.47a | ||
| K4E | 0.5% bile | 3.76a | 0.00b | 0.00b | 2.59 |
| Control | 3.78a | 3.95a | 4.06a | ||
| K14 | 0.5% bile | 3.68a | 0.00b | 0.00b | 2.55 |
| Control | 3.63a | 3.90a | 4.08a | ||
| WBS2A | 0.5% bile | 7.37b | 7.07bc | 6.72c | 7.52 |
| Control | 7.37b | 8.22a | 8.36a | ||
| Source | SEm | CD (0.05) | CV% | ||
| O | 0.08 | 0.23 | 7.32 | ||
| T | 5.08 | 0.14 | |||
| O*T | 0.11 | 0.32 | |||
| H | 6.22 | 0.18 | |||
| O*H | 0.14 | 0.39 | |||
| T*H | 8.79 | 0.18 | |||
| O*T*H | 0.20 | 0.56 | |||
*Values with different superscripts differ significantly (p≤0.05), log CFU/mL, n=3
Furthermore, in the study by Seah et al. [45], the number of L. rhamnosus and L. plantarum strains that survived was very small, and L. fermentum strain R6 of Han et al. [44] was found sensitive to 0.5% bile concentrations, which supports our findings that lactic acid bacteria strains are unable to survive exposure to bile salts. Chang et al. [48] discovered the probiotic qualities of lactic acid bacteria isolated from kimchi. Among the various lactobacillus strains tested for bile salt tolerance, L. plantarum strains LA89402, LA89409, and LA89911 were unable to survive at 0.5% bile salt. Similar observations were seen in our study, where the L. plantarum strain KGL3A did not survive at 0.5% bile exposure.
Gastric and intestinal juice tolerance
The upper gastrointestinal tract environment, where gastric acidity ranges from pH 2.5–3.5, protects well against the entrance of foreign microorganisms, and is where LAB strains must first adapt to survive in the intestinal system. According to the results shown in Table 2 and Table 3, four lactic acid bacterial strains could not survive after being exposed for 4 h to artificial gastric juice with 0.3% pepsin. However, L. plantarum KGL3A was able to resist pH 2 until the second hour of exposure, whereas both L. plantarum KGL3A and L. rhamnosus K4E strains were able to resist pH 3 until the second hour of exposure with a reduction in overall viable cell count from 7.48 log CFU/mL to 1.09 log CFU/mL and 7.77 log CFU/mL to 0.83 log CFU/mL, respectively. Yeast strain WBS2A was able to endure pH values of 2 and 3 in artificial gastric juice. The total viable cell counts for the WBS2A strain exposed to low pH 2 were found to be decreasing with increasing exposure time, from 6.79 log CFU/mL up to 6.45 log CFU/mL at 4 h after the initial exposure (0.34 log CFU/mL reduction). Additionally, the WBS2A strain was able to survive gastric juice with a pH of 3 despite a minor decline in viable cell count from 6.63 log CFU/mL after initial contact to 6.45 log CFU/mL at 4 h (a decline of 0.18 log CFU/mL). Nigerian native yeasts isolated from fermented food products were examined for their in vitro probiotic activities by Adesokan et al. [49]. Both pH 2 and pH 3 were tolerated by the S. cerevisiae PAW02 strain of digestive fluid. The viable cell count was determined to be 8.31 ± 0.05 log CFU/mL at zero hours at pH 2, rising to 9.50 ± 0.05 log CFU/mL at 4 h. In contrast, the viable cell count at pH 3 decreased from 8.86 ± 0.05 log CFU/mL at zero hour to 7.48 ± 0.04 log CFU/mL after 1 h. The lactic acid bacteria's probiotic properties in Indonesia's naturally fermented milk, specifically dangke and dadih, were described by Jatmiko et al. [50]. Of the 20 strains screened for acid tolerance, only 5% of the strains demonstrated greater tolerance to more acidic conditions (pH 2-4), namely L. plantarum SL2.7. S. cerevisiae HM535662, which was isolated from the traditional fermented cuisine “Bhaturu” of the Western Himalayas, was examined in vitro by Sourabh et al. [51]. The S. cerevisiae (HM535662) strain was able to withstand pH values for simulated gastric juice at 2 and 3. After 240 min of exposure, viability was reduced at pH 2 at a greater rate (2.71 log CFU/mL to 4.12 log CFU/mL) than at pH 3 (0.88 log CFU/mL to 3.06 log CFU/mL), suggesting this isolate’s inherent tolerance. Muna and Adel [52] evaluated the probiotic potential of camel's milk Lactobacillus strains. L. rhamnosus strains M6 and M19 and L. plantarum strains M7 and M11 could not resist the pH 3 gastric juice after 3 h of incubation. The results of Adesokan et al. [49] and Sourabh et al. [51] are supported by S. cerevisiae ability to survive in stomach fluids at pH 2 and pH 3, as well as the fact that our yeast strain S. cerevisiae WBS2A survived the gastric juices comparatively well.
Table 2.
In vitro gastric juice tolerance ability of lactobacilli and yeast at pH 2
| Organism (O) |
Treatment (T) |
Time (H) | T mean | ||
|---|---|---|---|---|---|
| 0 h | 2 h | 4 h | |||
| KGL3A | pH 2 | 7.23a | 3.53b | 0.00c | 5.47 |
| Control | 7.35a | 7.36a | 7.37a | ||
| KGL4 | pH 2 | 4.95b | 0.00c | 0.00c | 3.50 |
| Control | 5.00a | 5.29a | 5.79a | ||
| K4E | pH 2 | 7.09a | 0.00b | 0.00b | 4.80 |
| Control | 7.21a | 7.25a | 7.26a | ||
| K14 | pH 2 | 7.13a | 0.00b | 0.00b | 4.83 |
| Control | 7.26a | 7.27a | 7.31a | ||
| WBS2A | pH 2 | 6.79a | 6.58a | 6.45a | 6.78 |
| Control | 6.90a | 6.94a | 7.03a | ||
| Source | SEm | CD (0.05) | CV% | ||
| O | 0.17 | 0.47 | 13.91 | ||
| T | 0.11 | 0.30 | |||
| O*T | 0.24 | 0.67 | |||
| H | 0.13 | 0.36 | |||
| O*H | 0.29 | 0.82 | |||
| T*H | 0.18 | 0.36 | |||
| O*T*H | 0.41 | 1.15 | |||
*Values with different superscripts differ significantly (p≤0.05), log CFU/mL, n=3
Table 3.
In vitro gastric juice tolerance ability of lactobacilli and yeast at pH 3
| Organism (O) |
Treatment (T) |
Time (H) | T mean | ||
|---|---|---|---|---|---|
| 0 h | 2 h | 4 h | |||
| KGL3A | pH 3 | 7.48b | 1.09c | 0.00d | 5.21 |
| Control | 7.39a | 7.57a | 7.71a | ||
| KGL4 | pH 3 | 6.21a | 0.00b | 0.00b | 4.25 |
| Control | 6.23a | 6.31a | 6.78a | ||
| K4E | pH 3 | 7.77a | 0.83b | 0.00b | 5.35 |
| Control | 7.81a | 7.83a | 7.84a | ||
| K14 | pH 3 | 7.84a | 0.00b | 0.00b | 5.22 |
| Control | 7.81a | 7.83a | 7.86a | ||
| WBS2A | pH 3 | 6.63a | 6.58a | 6.45a | 6.87 |
| Control | 6.75a | 7.38a | 7.42a | ||
| Source | SEm | CD (0.05) | CV% | ||
| O | 0.15 | 0.42 | 11.72 | ||
| T | 9.40 | 0.27 | |||
| O*T | 0.21 | 0.59 | |||
| H | 0.12 | 0.33 | |||
| O*H | 0.26 | 0.73 | |||
| T*H | 0.16 | 0.33 | |||
| O*T*H | 0.36 | 1.03 | |||
*Values with different superscripts differ significantly (p≤0.05), log CFU/mL, n=3
All four lactic acid bacteria strains and one yeast strain were able to tolerate intestinal juice (pH 8) during the course of incubation. The KGL3A strain showed better survival in intestinal juice as compared to other bacterial strains, whereas the viable log cell count was found to be 7.24 log CFU/mL during initial exposure, which was slightly reduced after 4 h of exposure to 7.12 log CFU/mL. The WBS2A yeast strain also survived in intestinal juice with a slight reduction in viable cell count, which was 6.61 log CFU/mL previously and at 4 h of incubation reached 6.26 log CFU/mL (Table 4). Abudoleh et al. [53] examined the probiotic properties of microorganisms isolated from indigenous pickled and fermented foods from Jordan. L. fermentum strain G2 tolerated simulated intestinal juice; after 4 h of incubation, a growth of 6.2 ± 0.16 log CFU/mL was seen. L. fermentum strain J2 grew at a rate of 6.5 ± 0.31 log CFU/mL. Adesokan et al. [49] showed that all the tested yeast cultures survived the intestinal juice pH8, where S. cerevisiae PAW02 showed higher survivability with a viable cell count of 8.90 ± 0.05 log CFU/mL at zero hours, which fell to 8.63 ± 0.05 log CFU/mL over the course of the 4-h incubation. Zhai et al. [54] investigated the potential for lactic acid bacteria to protect against cadmium toxicity. The likelihood that different L. plantarum strains will survive in simulated intestinal juice ranged from 87.56 ± 0.80 % to 92.98 ± 0.22 %. L. rhamnosus strain CCFM311 had a survival rate of 85.59 ± 0.25 %. Survival of L. fermentum strain KGL4 in simulated intestinal juice was found to be better as compared to the L. fermentum strains used by Abudoleh et al. [53]. The previous study of our work carried out by Mishra et al. [55] observed that the two lactobacillus isolates, namely KGL4 and KGL3A, showed the highest resistance in simulated intestinal fluid following a 4 h incubation with corresponding cell counts of 6.65 log CFU/mL and 6.42 log CFU/mL. In our present study we have observed a higher resistance towards simulated gastric juice by KGL3A with corresponding cell counts of 6.65 log CFU/mL and 6.42 log CFU/mL.
Table 4.
Survival rate of lactobacilli and yeast during exposure to intestinal juice
| Organism (O) |
Treatment (T) |
Time (H) | T mean | ||
|---|---|---|---|---|---|
| 0 h | 2 h | 4 h | |||
| KGL3A | 0.1% pancreatin | 7.24a | 7.19a | 7.12a | 7.00 |
| Control | 7.22a | 7.67a | 5.55b | ||
| KGL4 | 0.1% pancreatin | 6.83abc | 6.62abcd | 6.41bcd | 6.67 |
| Control | 6.10d | 6.91ab | 7.17a | ||
| K4E | 0.1% pancreatin | 7.05a | 6.91abc | 6.17d | 6.66 |
| Control | 6.02b | 6.86abc | 6.95ab | ||
| K14 | 0.1% pancreatin | 6.91abc | 6.83abcd | 6.15e | 6.72 |
| Control | 6.29de | 7.06ab | 7.10a | ||
| WBS2A | 0.1% pancreatin | 6.61abcd | 6.35d | 6.26d | 6.52 |
| Control | 6.69abc | 6.82ab | 7.17a | ||
| Source | SEm | CD (0.05) | CV% | ||
| O | 8.27 | 0.23 | 5.21 | ||
| T | 5.23 | NS | |||
| O*T | 0.12 | 0.33 | |||
| H | 6.41 | 0.18 | |||
| O*H | 0.14 | 0.41 | |||
| T*H | 9.06 | 0.18 | |||
| O*T*H | 0.20 | 0.57 | |||
*Values with different superscripts differ significantly (p≤0.05), log CFU/mL, n=3
Cell surface hydrophobicity (CSH)
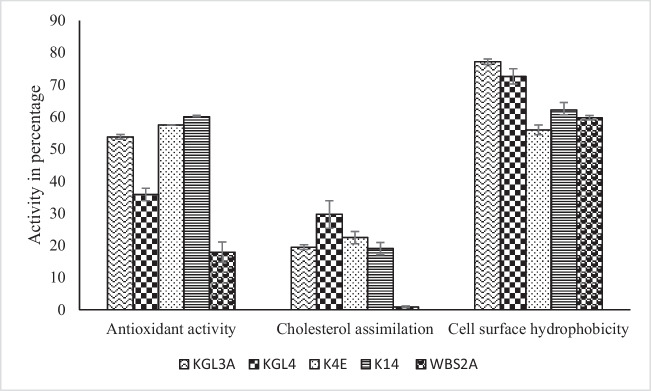
An effective probiotic’s next obstacle is adhering to small intestinal cells after surviving the upper gastrointestinal transit. Microorganism adhesion and proliferation on intestinal epithelial cells are believed to be significantly influenced by cell surface hydrophobicity [55]. n-hexadecane was used to measure hydrophobicity because, as compared to other hydrocarbons, it has been shown to provide more accurate results without requiring cell lysis for the assessment of probiotics’ ability to adhere to surfaces. Microorganisms’ hydrophobicity to hydrocarbons on their cell surfaces (n-hexadecane) was determined (Fig. 1). Individual cultures’ percentage CSH values to n-hexadecane ranged from 55.91 to 77.16%. L. plantarum KGL3A (77.16± 0.84 %) was found to be highly hydrophobic to n-hexadecane and was significantly at par with L. fermentum KGL4 (72.60 ± 2.33 %), followed by L. helveticus K14 (62.17 ± 2.28 %), WBS2A (59.76 ± 0.67 %), and L. rhamnosus K4E (55.91 ± 1.51 %). S. cerevisiae DABRP5 was isolated from the batter in a bollo, a typical Goan fermented dish, and Pereira et al. [56] evaluated its probiotic potential. S. cerevisiae strain DABRP5 (accession no. MT712864) showed the highest cell surface hydrophobicity of 69.80 ± 0.87 % towards n-hexadecane, whereas S. cerevisiae strain DABRP12 (accession no. MT712866) showed the least activity of 45.09 ± 0.74 %. 11 different strains of L. fermentum isolated from fermented dairy products and newborn feces were evaluated by Panicker et al. [57] for their in vitro probiotic characteristics. The hydrophobicity of the surfaces of cultured cells, L. fermentum MTCC-8711, was found to be 29.99 ± 2.97 %, whereas the hydrophobicity of the culture L. rhamnosus GG was found to be 32.14 ± 3.11% towards n-hexadecane. Devi et al. [58] used comparative analysis to assess the probiotic qualities of lactic acid bacteria. The hydrophobicity % to n-hexadecane of L. plantarum LP was found to be 45.3 ± 0.06 %, and that of L. fermentum F14 was found to be 48.6 ± 0.06 %. Deng et al. [59] examined the cell surface characteristics of five yeast strains that degrade polycyclic aromatic compounds. Among the yeast strains tested, S. cerevisiae had the lowest cell surface hydrophobicity to hexadecane at 0.7 ± 0.1 %. The cell surface hydrophobicity of the S. cerevisae strain that was utilised in our investigation is greater than the Deng et al. [59] studies, which showed 0.7 ± 0.1% towards n-hexadecane. The cell surface hydrophobicity of L. fermentum, L. plantarum, and L. rhamnosus strains used in the study was higher as compared to Panicker et al. [57] and Devi et al. [58] studies. Mishra et al. [55] found that the cell surface hydrophobicity of lactic acid bacteria ranged from 47.44 ± 0.64 % to 68.30 ± 0.78%. Whereas, in present study, we have observed a better CSH activity, ranging from 55.91 to 77.16 %.
Fig. 1.
Antioxidant activity, cholesterol assimilation, and cell surface hydrophobicity of selected cultures
Cell auto-aggregation
Adhesion has been more strongly correlated with autoaggregation than hydrophobicity [60]; therefore, a substantial quantity of autoaggregation capacity may be responsible for this indigenous isolate’s adhesion trait. Microorganism adhesion can be substantially correlated with isolates with superior autoaggregation capacity in addition to good hydrophobicity values. Despite the fact that these two qualities are distinct from one another, they continue to be connected to a particular microbe’s ability to adhere. The auto-aggregation study for cultures lasted 5 h. It was discovered that the rate of aggregation was growing over time. After 2 h, the percentage auto-aggregation ranged from 8.70 to 25.53, which significantly increased to 10.50 to 26.94 during the fifth hour. L. rhamnosus K4E and L. helveticus K14 dominated the experiment's maximum aggregation at all times (Table 5). The Indian fermented foods include yeasts with probiotic potential was assessed by Sunita et al. [61]. The yeast strain MH425 had the lowest auto-aggregation percentage of 3.2 % and the highest auto-aggregation percentage of 37.1 %. Reuben et al. [62] carried out a study to characterize and evaluate lactic acid bacteria for potential probiotic properties in indigenous raw milk. The auto-aggregation percentage of L. plantarum strain C16 was 38.5 ± 13.44 % and that of L. fermentum strain G9 was 41.5 ± 6.39 %. In order to study the probiotic qualities and bioactive properties of the lactic acid bacteria, Divisekera et al. [63] isolated the bacteria from finger millet flour that had undergone fermentation. The auto-aggregation percentage for L. plantarum R17 (MF405176.1) increased from 5.56 ± 0.10 at 2 h to 60.44 ± 1.71 at 5 h of incubation, whereas for L. fermentum RV02 (MF033346.1) it increased from 16.09 ± 0.48 at 2 h to 46.25 ± 0.55 at 5 h of incubation. The increase in auto-aggregation percentage was 54.88 % for L. plantarum R17 and 30.16 % for L. fermentum RV02. Garcia-Cayuela et al. [64] investigated the bond properties of dairy L. plantarum strains with a phenotype of aggregation. Auto-aggregation ability of various L. plantarum strains ranged from 6.31 ± 1.13 % to 22.13 ± 0.89 % and L. rhamnosus strain GR-1 showed 6.70 ± 0.14 %. The L. rhamnosus culture used in our study showed better auto-aggregation as compared to Garcia-Cayuela et al. [64]. The previous investigation by Mishra et al. [65] revealed that the lactic acid bacteria's cell auto-aggregation ranged from 30.69 to 81.32% which was higher than the one reported in the current study.
Table 5.
Auto-aggregation ability of cultures at different time intervals
| Culture | Incubation time (h) | |
|---|---|---|
| 2 h | 5 h | |
| KGL3A | 10.00 ± 0.058cd | 10.50 ± 0.349e |
| KGL4 | 10.96 ± 0.201bc | 15.85 ± 0.124cd |
| K4E | 10.28 ± 0.916c | 17.10 ± 0.490b |
| K14 | 8.70 ± 0.131d | 14.77 ± 0.280d |
| WBS2A | 25.53 ± 0.105a | 26.94 ± 0.709a |
*Values with different superscripts differ significantly (p≤0.05), auto-aggregation (%) mean ± SEM, n=3
Cell co-aggregation
An important characteristic of Lactobacillus is coaggregation, which suggests a possible capacity for both competing with pathogens and preventing their colonization through antagonistic interactions. The co-aggregation of cultures was determined against E. coli MTCC 1687, Salmonella typhimurium ATCC 14028, S. aureus MTCC 737, and Listeria monocytogenes MTCC 657. The percentage co-aggregation of lactobacillus cultures and yeast with L. monocytogenes, S. aureus, S. typhi, and E. coli is shown in Table 6. A higher percentage of co-aggregation was found for culture K4E with S. typhi (34.18 ± 0.03%), E. coli (32.97 ± 0.02 %) and S. aureus (26.33 ± 0.06 %). A significantly higher percentage of co-aggregation of WBS2A was found with L. monocytogenes (25.77 ± 0.22 %). In order to test potential probiotic Lactobacillus spp. strains against Salmonella strains, Fadare et al. [66] used garlic extract as a synbiotic antibacterial agent. L. plantarum strains AM3, NG13, DB3, and DS11 recorded co-aggregation values of 34.1%, 21.2%, 24.4%, and 19.8%, respectively, against S. typhi. Lactic acid bacteria isolated from fermented foods sources have probiotic effects were researched by Bindu and Lakshmidevi [67]. L. fermentum strain Cu3-PM8 (MCC4233) showed the least E. coli co-aggregation at 09.97 ± 0.04 % whereas L. plantarum strain Cu2-PM7 (MCC4246) showed 14.92 ± 0.06 % co-aggregation with E. coli and 17.06 ± 0.07% with L. monocytogenes at 2 h. L. fermentum strain IB-PM15 showed the least co-aggregation of 15.02 ± 0.05% with S. aureus. Lee et al. [68] evaluated the impact of the kimchi-derived probiotic L. plantarum KU200656 on microorganisms and biofilms. The co-aggregation assay of L. plantarum against various pathogens was carried out, and the co-aggregation percentages were 21.45 ± 4.35 %, 21.35 ± 2.36 %, 22.52 ± 3.62 %, and 24.81 ± 3.26 % counter to S. typhimurium, S. aureus, L. monocytogenes, and E. coli, respectively. Lactic acid bacteria that were obtained from traditional fermented Thai foods were studied for their probiotic potential by Suwannaphan [69]. Among all the isolates tested for co-aggregation with pathogenic bacteria, L. fermentum strain K9 showed a lower level of aggregation with E. coli (5.56 ± 0.74 %) and S. aureus (10.15 ± 2.07 %). L. fermentum strain K4 exhibited the least amount of aggregation with S. typhimurium (10.74 ± 1.95 %). The cultures in the study demonstrated greater co-aggregation activity of L. plantarum and L. fermentum with E. coli, S. aureus and L. monocytogenes than Bindu and Lakshmidevi [67] and L. plantarum co-aggregation activity with L. monocytogenes, S. typhimurium, and E. coli than Lee et al. [68].
Table 6.
Co-aggregation ability of cultures to various pathogens
| Culture | L. monocytogenes | S. typhi | E. coli | S. aureus |
|---|---|---|---|---|
| KGL3A | 23.98 ± 0.117b | 24.08 ± 0.192c | 24.03 ± 0.088c | 19.35 ± 0.161b |
| KGL4 | 22.33 ± 0.060c | 22.63 ± 0.130d | 22.25 ± 0.153e | 16.57 ± 0.142d |
| K4E | 22.05 ± 0.087c | 34.18 ± 0.033a | 32.97 ± 0.017a | 26.33 ± 0.060a |
| K14 | 22.27 ± 0.117c | 23.67 ± 0.033c | 23.03 ± 0.101d | 11.05 ± 0.050e |
| WBS2A | 25.77 ± 0.219a | 26.48 ± 0.520b | 26.12 ± 0.117b | 18.15 ± 0.104c |
*Values with different superscripts differ significantly (p≤0.05), cell co-aggregation ability (%), mean ± SEM, n=3
Bile salt hydrolysate activity (BSH)
BSH activity was observed only in the case of the culture WBS2A (Supplementary material Fig S2). Lactobacillus cultures did not show prominent bile salt hydrolysate activity. Inhibitory activity of bile salt hydrolase and lipase in reconstituted skim milk fermented with lactic acid bacteria were investigated by Gil-Rodriguez and Beresford [70]. L. brevis strains nos. 36 and 38, L. curvatus strain nos. 73, L. paracasei strain nos. 9 and 87, and L. rhamnosus did not show BSH activity, and the cultures were tested without prior exposure to bile. Hernandez-Gomez [71] evaluated BSH activity of L. plantarum DGIA1 against all bile acid conjugates namely sodium glycocholate, sodium glycodeoxycholate, sodium taurocholate, and sodium taurodeoxycholate. Sharma et al [72] isolated probiotic lactic acid bacteria from camel and identified probiotic potentials. L. plantarum 15 showed BSH activity against sodium deoxycholate and sodium taurodeoxycholate.
Technofunctional properties
Antioxidant activity
Higher antioxidant activity was found in culture K14 activity (60.00 ± 0.48 %) which was found to be at par with culture K4E (57.50 ± 0.00%). Lower antioxidant activity was obtained for culture WBS2A (17.92 ± 3.13 %) (Fig. 1). Yang et al. [73] investigated the immune system benefits and probiotic properties of L. plantarum 200655 isolates. The findings of radical scavenging (%) by ABTS revealed different scavenging activities were displayed by different LAB strains, with higher activity for L. plantarum 200655 (38.13%), followed by L. plantarum KCTC 3108 (35.03%) and L. rhamnosus GG (24.76%). Kathiriya et al. [74] assessed Lactic Acid Bacteria in vitro probiotic capacity, wherein higher antioxidant activity was obtained for the culture L. rhamnosus NS6 (9.50±0.88 %ABTS activity), followed by S. thermophilus MD2 (2.45±0.37 % ABTS activity) and S. thermophilus MD8 (1.88±0.16 % ABTS activity). P. pentosaceus R1 (42.4%), L. plantarum (40.1%), and L. sake (38.5%) were three lactic acid bacteria with the highest ABTS+ scavenging rates. In milk whey, antioxidant activity was produced by Virtanen et al. [75] during lactic acid bacteria fermentation. There was shown to be more radical scavenging activity in Leuconostoc mesenteroides ssp. cremoris B53 (53 ± 5.10 %), whereas cultured L. rhamnosus ATCC 7469 showed 29 ± 0.42 % activity, and L. helveticus E showed 28 ± 3.18 % activity. The antioxidant activity of L. plantarum, L. fermentum, and L. rhamnosus was found to contrast with our studies by Yang et al. [73], Kathiriya et al. [74], and Virtanen et al. [75].
Proteolytic activity
Significantly higher proteolytic activity was given by culture K4E (7.29 ± 0.15 mg/mL), which was also on par with all the remaining Lactobacillus cultures, namely K14 (7.28 ± 0.18 mg/mL), KGL4 (7.24 ± 0.10 mg/mL), and KGL3A (7.00 ± 0.11 mg/mL). However, lower proteolytic activity was given by yeast culture WBS2A (Table 7). When sheep milk is fermented, antimicrobial, antimicrobial peptide, antioxidative, and anti-inflammatory properties as well as the generation of ultra-filtered antioxidative are examined by Ashokbhai et al. [10]. From 6.10 mg/mL (12 h incubation, 1.5% inoculation rate) to 10.40 mg/mL (48 h incubation, 2.5% inoculation rate), L. fermentum proteolytic activity was measured. Dineshbhai et al. [76] investigated the possibility of biofunctionalities of Saccharomyces and Lactobacillus, as well as bioactive peptides released from fermented whey protein. S. cerevisiae and L. fermentum cultures were reported to have proteolytic activities of 7.24 mg/mL and 8.59 mg/mL, respectively, with a 2.5% inoculation rate after 48 h of incubation. According to Shukla et al. [77], when culture was administered at a rate of 2% v/v, total proteolytic activity in L. plantarum ranged from 7.04 mg/mL at 12 h to 9.32 mg/mL at 48 h. Patel et al. [78] investigated ultrafiltration peptide fractions from fermented camel milk as potential sources of antioxidant peptides for their antioxidative and anti-inflammatory effects. The proteolytic activity obtained during camel milk fermentation after 24 h was found to be 7.47 ± 0.11 mg/mL. The proteolytic activity of L. fermentum in our study was found to be similar to that of Dineshbhai et al. [76].
Table 7.
Proteolytic activity, β-glucosidase activity, and α-galactosidase activity of cultures
| Culture | Proteolytic activity | β-Glucosidase activity of cultures after 24 h | α-Galactosidase activity of cultures after 24 h |
|---|---|---|---|
| KGL3A | 7.00 ± 0.11a | 0.134 ± 0.004e | 0.258 ± 0.007c |
| KGL4 | 7.24 ± 0.10a | 0.359 ± 0.002a | 0.339 ± 0.014b |
| K4E | 7.29 ± 0.15a | 0.300 ± 0.002d | 0.415 ± 0.016a |
| K14 | 7.28 ± 0.18a | 0.330 ± 0.001c | 0.354 ± 0.007b |
| WBS2A | 5.68 ± 0.05b | 0.348 ± 0.001b | 0.326 ± 0.003b |
*Values with different superscripts differ significantly (p≤0.05), proteolytic activity (mg/ml), β-glucosidase activity (OD420nm), α-galactosidase activity (OD420nm), mean ± SEM, n=3
Cholesterol assimilation
Bacterially growing cells may reduce cholesterol through assimilation and/ or integration into the cellular of the probiotic microorganism, inhibiting the body’s ability to reabsorb cholesterol. In KGL4, the proportion of cholesterol decreased was noticeably higher (29.65 ± 4.30 %) as compared to K4E (22.47 ± 1.92 %), KGL3A (19.42 ± 0.75 %), K14 (19.02 ± 1.90 %), and WBS2A (0.86 ± 0.25 %), as shown in Fig. 1. Castorena-Alba et al. [79] evaluated the reference strains and probiotic bacteria derived from food for their ability to assimilate cholesterol, acid, and bile. The highest percentage of absorption of cholesterol was observed in strains B. lactis (47.39%), L. fermentum (49.34%), and L. acidophilus (54.26%). In terms of assimilation of cholesterol, L. rhamnosus (13.21%) and L. pentosus (4.31%) strains showed the lowest percentages. Angmo et al. [80] looked at the probiotic characteristics of lactic acid bacteria from fermented Ladakhi foods and beverages. Although isolate 11 showed higher cholesterol assimilation at 19.75%, the assimilation of cholesterol by several lactic acid bacteria ranged from 1.14% to 19%. A potential probiotic bacterium with cholesterol-lowering capabilities called L. fermentum SM-7, was characterized by a Pan et al. [81] study. L. fermentum strain SM-2 showed the lowest cholesterol lowering ability of 15.2 ± 2.0 % whereas the highest activity was seen in L. fermentum strain SM-7 at 66.8 ± 5.0 %. Compared to research conducted by Castorena-Alba et al. [79], L. rhamnosus had a higher cholesterol assimilation activity.
β-Glucosidase and α-galactosidase activity
Probiotic bacteria produce β-glucosidase enzymes, which aid in the removal of the glycoside moiety from glycosylated flavonoids such as those found in soybean products. This hydrolysis makes flavonoids more absorbable through the digestive tract, which is required for their beneficial effects on human health [82]. This enzyme also acts on lactose and hydrolyses it into readily digestible galactose and glucose, which helps to reducing the symptoms of lactose intolerance in people [83]. β-glucosidase activity was significantly higher in the KGL4 strain (0.359 ± 0.002), followed by WBS2A (0.348 ± 0.001), K14 (0.330 ± 0.001), K4E (0.300 ± 0.002) and KGL3A (0.134 ± 0.004) as shown in Table 7. Hati et al [84] observed that at different incubation temperatures and various levels of skim milk powder additions in yoghurt culture (1% of L. bulgaricus and S. thermophilus) to soymilk improved overall β-Glucosidase activity with highest activity (4.97 U/ml). According to Jang et al. [85], the L. plantarum Ln1, a probiotic isolated from kimchi was tested for its antioxidant properties. β-glucosidase was produced by L. plantarum KCTC 3108 and L. plantarum Ln1, with yields of 1.49 and 7.04 mU/mL, respectively. Zhu et al. [86] optimized conditions for lactic acid fermentation in fermented tofu whey drinks that contain isoflavone-rich aglycones. At 20 h, L. rhamnosus GG (LGG) has 6.71± 0.32 mU/mL of β-glucosidase activity. Hati et al [87] also studied Lactobacillus cultures for their β-glucosidase activity during fermentation in soymilk. Highest activity was observed in L. rhamnosus C6 as 1.66 U/mL whereas lowest activity was noted by L. rhamnosus NCDC24 (0.54 U/mL).
A digestive enzyme called α-galactosidase converts the complex sugars in beans into simpler ones, making them easier for people to digest. Since, the α-galactosidase enzyme is absent in human digestive tract the digestion of sugars is difficult. So, the presence of this enzyme activity in probiotic bacteria is essential for their use as a food supplement. The α-galactosidase activity of the K4E strain was significantly higher (0.415 ± 0.016). Lower α-galactosidase activity was detected in KGL3A (0.258 ± 0.007). Apart from it the α-galactosidase activity of KGL4 (0.339 ± 0.014), K14 (0.354 ± 0.007) and WBS2A (0.326 ± 0.003) were at par with each other (Table 7). The potential for bean products was investigated by Liu et al. [88] in relation to the thermostability of probiotics and their 𝛼-galactosidases. L. rhamnosus strain 910 gave a α-galactosidase activity of 0.21 U/mL. Keat-hui et al. [89] investigated the bioactivity and growth properties of probiotics during preservation in a media based on tofu. L. fermentum strain FTD 13 gave highest α-galactosidase activity of 1.904 ± 0.036 U/mg of protein. Mandal and Bagchi [83] examined native lactobacillus isolates to find those with the most health-promoting qualities. L. fermentum strain FA 5 isolated from fermented soybean seeds showed α-galactosidase activity of 9.627 ± 0.131U/mg of protein, L. helveticus strain FA 7 isolated from fermented rice gave 8.150 ± 0.007 U/mg of protein, L. plantarum strain GRI-2 isolated from human gut showed 6.011 ± 0.178 U/mg of protein.
Anti-inflammatory activity of KGL4 and WBS2A cultures in the RAW macrophage cell line
Numerous disorders, including inflammatory bowel disease, non-alcoholic fatty liver disease, diabetes, obesity, metabolic syndrome, and chronic kidney disease, are brought on by inflammation [90]. The development of chronic low-grade inflammation and metabolic disorders is brought on by microbial imbalances within the body (gut dysbiosis), which increase the number of gut-derived lipopolysaccharides and interact with macrophages and intestinal epithelial cells. This is accomplished by activating a number of signalling pathways, including nuclear factor-kB and MAPK. Probiotics can alter the composition of the gut microbiota, according to numerous in vivo and in vitro studies or the cell-wall-related components that are involved in the inflammatory response [91].
Dineshbhai et al. [76] investigated potential for biofunctionalities of Saccharomyces and Lactobacillus in the production of bioactive peptides from fermented whey protein. KGL4 (48 h, 37 °C,) and WBS2A (48 h, 25 °C) yielded highest concentrations of peptides, 7.24 mg/mL and 8.59 mg/mL, respectively. Chopada et al. [92] isolated and characterized the novel ACE inhibitory and antioxidant peptides from co-fermented whey protein concentrate made by the co-growth of L. paracasei and S. cerevisiae. Maximum proteolytic activity was seen at 37 °C for 48 h for M11 (6.50 mg/mL) and at 25 °C for WBS2A (8.59 mg/mL). Hati et al. [11] discovered a favorable result when adding L. fermentum (KGL4) and S. cerevisiae (WBS2A) to broiler diets. The KGL4- fed group had improved hematological features, increased body weight gain, decreased FCR, and increase triglycerides and HDL content. From the work published earlier, cultures KGL4 and WBS2A were found promising and were selected for in vitro anti-inflammatory studies in the RAW 264.7 macrophage study.
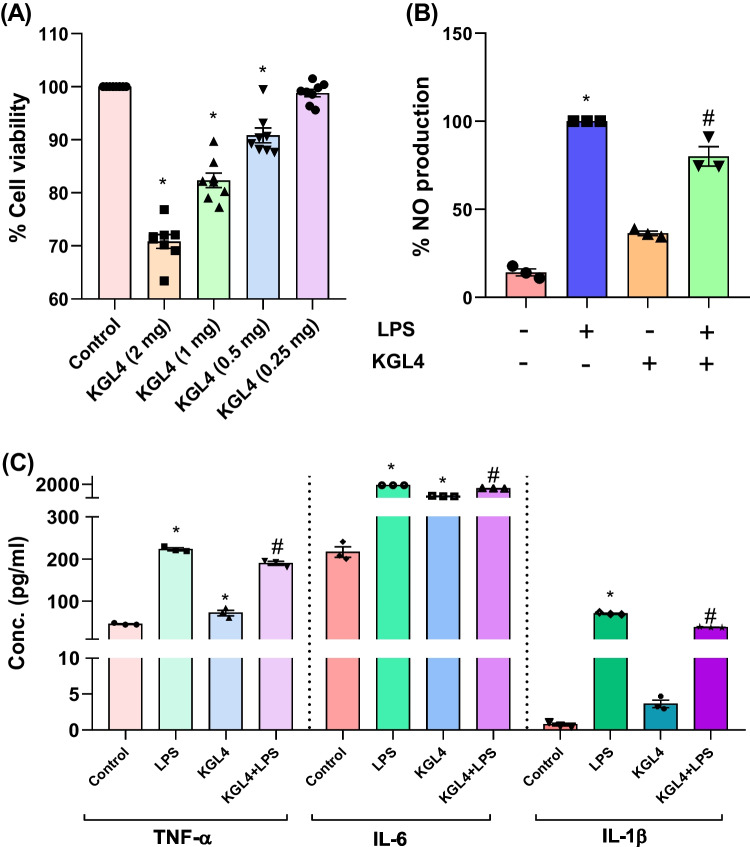
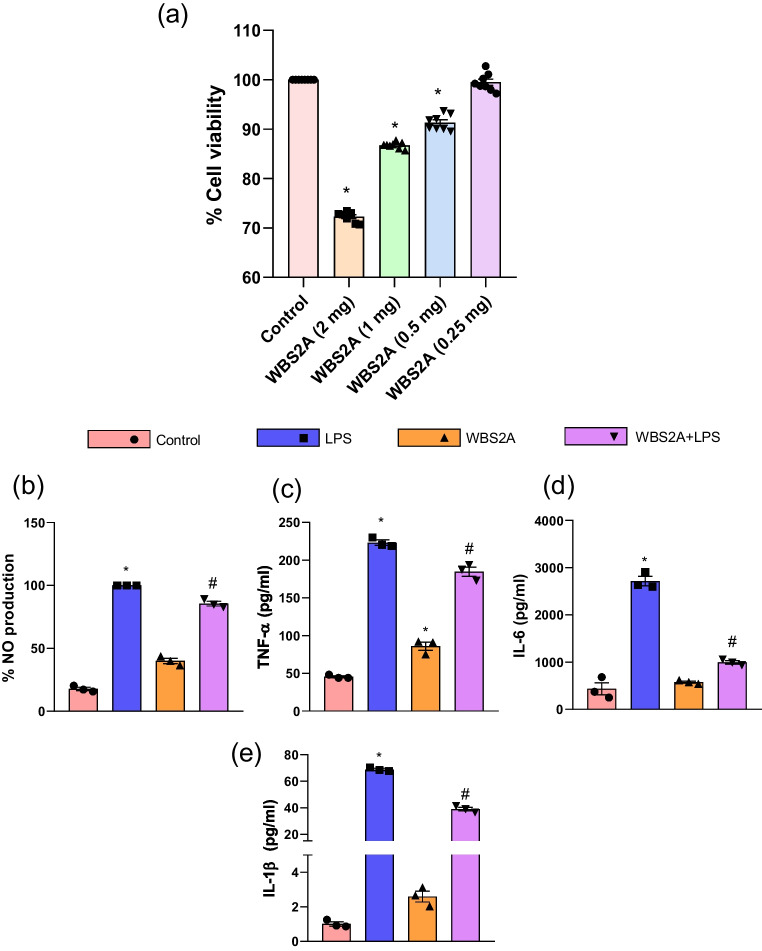
Lower dose of KGL4 and WBS2A did not induce cytotoxicity
The cell viability after exposing KGL4 and WBS2A on the RAW 264.7 macrophages at different concentration (0.25, 0.5, 1, and 2 mg/mL) was illustrated in the figure (Figs. 2 and 3a). Cells exposed to KGL4 and WBS2A did not exhibit cytotoxicity at 0.25 mg/mL (Figs. 2 and 3a) relative to the control cells. In case of KGL4 and WBS2A, treatment at 0.25 mg/mL gave highest viability whereas treatment at 0.5, 1 and 2 mg/mL exhibited loss in cell viability. With this observation, we have chosen 0.25 mg/mL for in vitro testing for the inhibition of cytokines and mediators that promote inflammation, as well as TNF-α, NO, IL-6, and IL-1β.
Fig. 2.
Effect of the KGL4 on A cell viability; B nitric oxide productions; C TNF-α; D IL-6; and E IL-1β measured in the supernatants of LPS-stimulated RAW 264.7 macrophages
Fig. 3.
Effect of the WBS2A on A cell viability; B NO production; C TNF-α; D IL-6; and E IL-1β production in the supernatants of LPS-stimulated RAW 264.7 cells
Low doses of KGL4 and WBS2A prevented LPS-induced NO and pro-inflammatory cytokine production by the RAW 264.7
The effect of KGL4 and WBS2A on LPS-induced inflammation in macrophages were investigated by assessing NO response which is one of the essential inflammatory parameters. It was found that LPS stimulation resulted in a remarkable elevation of nitric oxide response ten times higher than control (Fig. 2b). This NO production by LPS-activated cells appeared to be reduced by KGL4 and WBS2A at 0.25 mg/mL suggesting a promising anti-inflammatory activity (Fig. 2b).
Cytokine analysis in the supernatants of RAW 264.7 cells
The generation of TNF-α, IL-6, and IL-1β by RAW 264.7 macrophages stimulated with LPS for 16 h was investigated. As shown in figure, the TNF-α, IL-6, and IL-1β levels were significantly elevated following stimulation of the cells with LPS which is further reduced in KGL4 and WBS2A treatment group (Figs. 2c and 3c–e). KGL4 and WBS2A were tested in vitro, and the results showed considerable suppression of TNF-α, IL-6, and IL-1β in LPS-stimulated RAW 264.7 macrophages (Figs. 2c and 3c–e). The amount of pro-inflammatory indicators produced was decreased by treatment with KGL4 and WBS2A at 0.25 mg/mL.
MTT assays suggested that KGL4 and WBS2A at a lower dose do not exhibit cytotoxicity. The most effective inducer of an inflammatory response is NO, which cultivates TLR-4 on the surface of macrophages to activate the nuclear transcription factor NF-κB [93]. Furthermore, the binding causes the production of pro-inflammatory mediators for instance TNF-α, NO, IL-6, and IL-1β among others [94]. LPS promotes inflammatory conditions by causing the RAW 264.7 cells to cause inflammatory mediators such TNF-α, IL-1β, and IL-6 [95]. Our findings concur with those of Michels et al. [96], Xiaoqing et al. [97], and Song et al. [98], who analyzed the anti-inflammatory effect of bacterial strains and their fermented peptides in RAW 264.7 cells. We conclude that KGL4 and WBS2A may act like potent anti-inflammatory peptides by suppressing pro-inflammatory cytokine production.
Conclusion
Lactobacilli and yeast were initially isolated from traditional foods (fermented rice beverage, wanti and fermented fish) of Meghalaya, India. The strains possessed promising probiotics properties and were identified to be safe for use as food supplement. L. plantarum KGL3A survived in gastric juice for 2 h and also showed highest cell surface hydrophobicity towards n-hexadecane. Whereas, the antioxidant activity and proteolytic activity were found maximum for L. helveticus K14 and L. rhamnosus K4E. L. fermentum KGL4 produced a substantially greater percentage reduction in cholesterol contents. Moreover, L. fermentum KGL4 and S. cerevisiae WBS2A also exhibited strong anti-inflammatory activity on LPS-induced inflammation in RAW 264.7 cells. Therefore, these data confirm that WBS2A and KGL4 may be a potent product for the alleviation of inflammation and related comorbidities. Further investigation might lead to more insights into their potential health benefits through in vivo studies with scientifically proven health benefits to improve quality of human life.
Supplementary information
(DOCX 1432 kb)
Author contribution
Subrota Hati designed the work; supervision; writing of the manuscript and editing; interpreted statistical analysis; involved in funding acquisition.
Krupali Ramanuj carried out wet lab experiment and statistical analysis for safety aspects, probiotic attributes and techno-functional properties of Lactic acid bacteria and yeast.
Bethsheba Basaiawmoit and Krupali Ramanuj were major contributors in writing the manuscript.
Ruchika Maurya, Kanthi Kiran Kondepudi and Mahendra Bishnoi performed in vitro cell culture assay and its statistical analysis, writing of the manuscript and editing.
Sreeja V contributed in editing the manuscript.
B. K. Mishra contributed in editing the manuscript.
Funding
Funding was provided by the Department of Biotechnology, Ministry of Science and Technology [Grant BT/PR41738/NER/95/1857/2021], Government of India.
Data availability
Raw data not published in supplementary materials and are available on reasonable request from the corresponding author.
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Aidoo KE, Nout NJR, Sarkar PK. Occurrence and function of yeasts in Asian indigenous fermented foods. FEMS Yeast Res. 2006;6(1):30–39. doi: 10.1111/j.1567-1364.2005.00015.x. [DOI] [PubMed] [Google Scholar]
- 2.Taked S, Yamasaki K, Takeshita M, Kikuchi Y, Tsend-Ayush C. The investigation of probiotic potential of lactic acid bacteria isolated from traditional Mongolian dairy products. Anim Sci J. 2011;82:571–579. doi: 10.1111/j.1740-0929.2011.00874.x. [DOI] [PubMed] [Google Scholar]
- 3.Kumari A, Angmo K, Bhalla TC. Probiotic attributes of indigenous Lactobacillus spp. isolated from traditional fermented foods and beverages of north-western Himalayas using in vitro screening and principal component analysis. J Food Sci Technol. 2016;53(5):2463–2475. doi: 10.1007/s13197-016-2231-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aruoma OI, Somanah J, Bourdon E, Rondeau P, Bahorun T. Diabetes as a risk factor to cancer: functional role of fermented papaya preparation as phytonutraceutical adjunct in the treatment of diabetes and cancer. Mutat Res Fundam Mol Mech Mutagen. 2014;768:60–68. doi: 10.1016/j.mrfmmm.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 5.Schrezenmeir J, de Vrese M. Probiotics, prebiotics, and synbiotics—approaching a definition. Am J Clin Nutr. 2001;73(2):361s–364s. doi: 10.1093/ajcn/73.2.361s. [DOI] [PubMed] [Google Scholar]
- 6.Fijan S. Microorganisms with claimed probiotic properties: an overview of recent literature. Int J Environ Res Public Health. 2014;11(5):4745–4767. doi: 10.3390/ijerph110504745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanders ME, Guarner F, Guerrant R, Holt PR, Quigley EM, Sartor RB, Sherman PM, Mayer EA. An update on the use and investigation of probiotics in health and disease. Gut. 2013;62(5):787–796. doi: 10.1136/gutjnl-2012-302504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rolfe RD. The role of probiotic cultures in the control of gastrointestinal health. J Nutr. 2000;130(2):396S–402S. doi: 10.1093/jn/130.2.396S. [DOI] [PubMed] [Google Scholar]
- 9.Ashokbhai JK, Basaiawmoit B, Das S, Sakure A, Maurya R, Bishnoi M, Kondepudi KK, Padhi S, Rai AK, Liu Z, Hati S. Antioxidative, antimicrobial and anti-inflammatory activities and release of ultra-filtered antioxidative and antimicrobial peptides during fermentation of sheep milk: in-vitro, in-silico and molecular interaction studies. Food Biosci. 2022;47:101666. doi: 10.1016/j.fbio.2022.101666. [DOI] [Google Scholar]
- 10.Kolida S, Saulnier DM, Gibson GR. Gastrointestinal microflora: probiotics. Adv Appl Microbiol. 2006;59:187–219. doi: 10.1016/s0065-2164(06)59007-0. [DOI] [PubMed] [Google Scholar]
- 11.Hati S, Ramanuj K, Basaiawmoit B, Koringa P, Desai M, Ghodasara DJ, Joshi KV, Pathan M, Bhagora NJ, Savaliya FP, Mishra BK (2022) Significance of Limosilactobacillus fermentum and Saccharomyces cerevisiae on the growth performance, haematological traits, serum biochemistry, faecal and caeca microbiota of broiler chickens. J Am Nutr Assoc:1–20. 10.1080/27697061.2022.2149634 [DOI] [PubMed]
- 12.Xie N, Zhou T, Li B. Kefir yeasts enhance probiotic potentials of Lactobacillus paracasei H9: the positive effects of coaggregation between the two strains. Food Res Int. 2012;45(1):394–401. doi: 10.1016/j.foodres.2011.10.045. [DOI] [Google Scholar]
- 13.Liu SQ, Tsao M. Enhancing stability of lactic acid bacteria and probiotics by Williopsis saturnus var. saturnus in fermented milks. Nutr Food Sci. 2010;40(3):314–322. doi: 10.1108/00346651011044014. [DOI] [Google Scholar]
- 14.Zulkawi N, Ng KH, Zamberi R, Yeap SK, Satharasinghe D, Jaganath IB, Jamaluddin AB, Tan SW, Ho WY, Alitheen NB. In vitro characterization and in vivo toxicity, antioxidant and immunomodulatory effect of fermented foods; Xeniji™. BMC Complement Altern Med. 2017;17:344. doi: 10.1186/s12906-017-1845-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bell V, Ferrao J, Pimentel L, Pintado M, Fernandes T. One health, fermented foods, and gut microbiota. Foods. 2018;7:195. doi: 10.3390/foods7120195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shahbazi R, Yasavoli-Sharahi H, Alsadi N, Ismail N, Matar C. Probiotics in treatment of viral respiratory infections and neuroinflammatory disorders. Molecules. 2020;25(21):4891. doi: 10.3390/molecules25214891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng Y, Huang Y, Wang Y, Wang P, Song H, Wang F. Antibiotics induced intestinal tight junction barrier dysfunction is associated with microbiota dysbiosis, activated NLRP3 inflammasome and autophagy. PLoS One. 2019;14:e0218384. doi: 10.1371/journal.pone.0218384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bu Y, Liu Y, Liu Y, Wang S, Liu Q, Hao H, Yi H. Screening and probiotic potential evaluation of bacteriocin-producing Lactiplantibacillus plantarum in vitro. Foods. 2022;11(11):1575. doi: 10.3390/foods11111575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim H, Lee YS, Yu HY, Kwon M, Kim KK, In G, Hong SK, Kim SK. Anti-inflammatory effects of Limosilactobacillus fermentum KGC1601 isolated from panax ginseng and its probiotic characteristics. Foods. 2022;11(12):1707. doi: 10.3390/foods11121707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siesto G, Pietrafesa R, Infantino V, Thanh C, Pappalardo I, Romano P, Capece A. In vitro study of probiotic, antioxidant and anti-inflammatory activities among indigenous Saccharomyces cerevisiae strains. Foods. 2022;11(9):1342. doi: 10.3390/foods11091342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng Y, Zhang M, Mujumdar AS, Gao Z. Recent research process of fermented plant extract: A review. Trends Food Sci Technol. 2017;65:40–48. doi: 10.1016/j.tifs.2017.04.006. [DOI] [Google Scholar]
- 22.Wilburn J, Ryan E (2017) Fermented foods in health promotion and disease prevention: An overview. In Fermented Foods in Health and Disease Prevention :3–19. 10.1016/B978-0-12-802309-9.00001-7
- 23.Smith AC, Hussey MA. Gram stain protocols. American Society for Microbiology; 2005. p. 14. [Google Scholar]
- 24.Reiner K (2010) Catalase test protocol. American Society of Microbiology in 2016. https://asm.org/getattachment/72a871fc-ba92-4128-a194-6f1bab5c3ab7/CatalaseTest-Protocol.pdf
- 25.Harley JP. Laboratory exercises in microbiology. 6. New York, NY: McGraw Hill; 2005. [Google Scholar]
- 26.Brink B. Urease test protocol. Washington, DC: American Society for Microbiology; 2010. [Google Scholar]
- 27.Mi-Sun K, Yeu J-E, Hong S-P. Safety evaluation of oral care probiotics Weissella cibaria CMU and CMS1 by phenotypic and genotypic analysis. Int J Mol Sci. 2019;20(11):2693. doi: 10.3390/ijms20112693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bennani S, Mchiouer K, Rokni Y, Meziane M. Characterization and identification of lactic acid bacteria isolated from Moroccan raw cow’s milk. J Mater Environ Sci. 2017;8:4934–4944. [Google Scholar]
- 29.Pisano MB, Viale S, Conti S, Fadda ME, Deplano M, Melis MP, Deiana M, Cosentino S. Preliminary evaluation of probiotic properties of Lactobacillus strains isolated from Sardinian dairy products. Biomed Res Int. 2014;2014:286390. doi: 10.1155/2014/286390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang L, Zhuo-Yang Z, Ke D, Jian-Ping Y, Xiao-Kui G. Antibiotic resistance of probiotic strains of lactic acid bacteria isolated from marketed foods and drugs. Biomed Environ Sci. 2009;22(5):401–412. doi: 10.1016/S0895-3988(10)60018-9. [DOI] [PubMed] [Google Scholar]
- 31.Patel A, Prajapati JB, Holst O, Ljungh A. Determining probiotic potential of exopolysaccharide producing lactic acid bacteria isolated from vegetables and traditional Indian fermented food products. Food Biosci. 2014;5:27–33. doi: 10.1016/j.fbio.2013.10.002. [DOI] [Google Scholar]
- 32.Das S, Mishra BK, Hati S. Techno-functional characterization of indigenous Lactobacillus isolates from the traditional fermented foods of Meghalaya, India. Curr Res Food Sci. 2020;3:9–18. doi: 10.1016/j.crfs.2020.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee H, Yoon H, Ji Y. Functional properties of Lactobacillus strains isolated from kimchi. Int J Food Microbiol. 2011;145:155–161. doi: 10.1016/j.ijfoodmicro.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 34.Kodaikkal V (2008) Adhesion characteristics of probiotic lactobacilli in gastrointestinal tract. M.Sc. Thesis submitted to Anand Agricultural University, Anand.
- 35.Donkor ON, Henriksson A, Vasiljevic T, Shah NP. Proteolytic activity of dairy lactic acid bacteria and probiotics as determinant of growth and in vitro angiotensin-converting enzyme inhibitory activity in fermented milk. Lait. 2007;87(1):21–38. doi: 10.1051/lait:2006023. [DOI] [Google Scholar]
- 36.Anandharaj M, Sivasankari B, Santhanakaruppu R, Manimaran M, Rani RP, Sivakumar S. Determining the probiotic potential of cholesterol-reducing Lactobacillus and Weissella strains isolated from gherkins (fermented cucumber) and south Indian fermented koozh. Res Microbiol. 2015;166(5):428–439. doi: 10.1016/j.resmic.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 37.Otieno DO, Shah NP. Endogenous β-glucosidase and β-galactosidase activities from selected probiotic micro-organisms and their role in isoflavone biotransformation in soymilk. J Appl Microbiol. 2007;103(4):910–917. doi: 10.1111/j.1365-2672.2007.03438.x. [DOI] [PubMed] [Google Scholar]
- 38.Khare P, Maurya R, Bhatia R, Mangal P, Singh J, Podili K, Bishnoi M, Kondepudi KK. Polyphenol rich extracts of finger millet and kodo millet ameliorate high fat diet-induced metabolic alterations. Food Funct. 2020;11(11):9833–9847. doi: 10.1039/D0FO01643H. [DOI] [PubMed] [Google Scholar]
- 39.Bobga PT, Fossi BT, Taiwe GS, Nkanpira KT, Yolande NE, Ngwa FA, Tatsinkou LLT, Wanyu BY, Ndip LM. Evaluation of the anti-diabetic potential of probiotic Lactobacillus fermentum (PRI 29) isolated from cameroonian fermented cow milk in alloxan induced diabetes type-1 mice model. Saudi J Pathol Microbiol. 2022;7(10):381–393. doi: 10.36348/sjpm.2022.v07i10.001. [DOI] [Google Scholar]
- 40.Damisa-Okorhi FB, Ataikiru TL (2015) Preliminary study on antimicrobial activity of friendly bacteria isolated from dairy products. 10.15739/ibspr.009
- 41.Belicova A, Mikulasova M, Dusinsky R (2013) Probiotic potential and safety properties of Lactobacillus plantarum from Slovak Bryndza cheese. Biomed Res Int. 10.1155/2013/760298 [DOI] [PMC free article] [PubMed]
- 42.Ji Y, Kim H, Park H, Lee J, Lee H, Shin H, Kim B, Franz CM, Holzapfel WH. Functionality and safety of lactic bacterial strains from Korean kimchi. Food Control. 2013;31(2):467–473. doi: 10.1016/j.foodcont.2012.10.034. [DOI] [Google Scholar]
- 43.Guo CF, Zhang LW, Li JY, Zhang YC, Xue CH, Yi HX, Han X. Screening of bile salt hydrolase-active lactic acid bacteria for potential cholesterol-lowering probiotic use. Adv Mater Res. 2012;345:139–146. doi: 10.4028/www.scientific.net/AMR.345.139. [DOI] [Google Scholar]
- 44.Han Q, Kong B, Chen Q, Sun F, Zhang H. In vitro comparison of probiotic properties of lactic acid bacteria isolated from Harbin dry sausages and selected probiotics. J Funct Foods. 2017;32:391–400. doi: 10.1016/j.jff.2017.03.020. [DOI] [Google Scholar]
- 45.Ng S, Koon SS, Padam BS, Chye FY. Evaluation of probiotic potential of lactic acid bacteria isolated from traditional Malaysian fermented Bambangan (Mangifera pajang) CYTA J Food. 2015;13(4):563–572. doi: 10.1080/19476337.2015.1020342. [DOI] [Google Scholar]
- 46.Tulumoglu S, Kaya HI, Simsek O. Probiotic characteristics of Lactobacillus fermentum strains isolated from tulum cheese. Anaerobe. 2014;30:120–125. doi: 10.1016/j.anaerobe.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 47.Syal P, Vohra A. Probiotic potential of yeasts isolated from traditional Indian fermented foods. Int J Microbiol Res. 2013;5(2):390. doi: 10.9735/0975-5276.5.2.390-398. [DOI] [Google Scholar]
- 48.Chang JH, Shim YY, Cha SK, Chee KM. Probiotic characteristics of lactic acid bacteria isolated from kimchi. J Appl Microbiol. 2010;109(1):220–230. doi: 10.1111/j.1365-2672.2009.04648.x. [DOI] [PubMed] [Google Scholar]
- 49.Adesokan IA, Sanni AI, Kanwar SS. In vitro evaluation of probiotic properties of indigenous yeasts isolated from Nigeria fermented food products. Asian Food Sci J. 2021;20(5):75–85. doi: 10.9734/AFSJ/2021/v20i530300. [DOI] [Google Scholar]
- 50.Jatmiko YD, Howarth GS, Barton MD. AIP conference proceedings. 2017. Assessment of probiotic properties of lactic acid bacteria isolated from Indonesian naturally fermented milk; p. 050008. [Google Scholar]
- 51.Sourabh A, Kanwar SS, Sharma OP. In vitro characterization of Saccharomyces cerevisiae HM535662 obtained from an indigenous fermented food “Bhaturu” of Western Himalayas. Afr J Biotechnol. 2012;11(52):11447–11454. doi: 10.5897/AJB11.3295. [DOI] [Google Scholar]
- 52.Muna MA, Adel MM. Isolation of Lactobacillus strains with probiotic potential from camel’s milk. Afr J Microbiol Res. 2014;8(15):1645–1655. doi: 10.5897/AJMR2013.6598. [DOI] [Google Scholar]
- 53.Abudoleh SM, Hamdan SO, Mahasneh AM, Al-Khani ZM, Talhouni AA. Isolation and characterization of potential probiotic bacteria from Jordanian traditional pickled and fermented foods. Acta Pol Pharm. 2021;78(4):515–520. doi: 10.32383/appdr/141300. [DOI] [Google Scholar]
- 54.Zhai Q, Yin R, Yu L, Wang G, Tian F, Yu R, Zhao J, Liu X, Chen YQ, Zhang H, Chen W. Screening of lactic acid bacteria with potential protective effects against cadmium toxicity. Food Control. 2015;54:23–30. doi: 10.1016/j.foodcont.2015.01.037. [DOI] [Google Scholar]
- 55.Mishra BK, Das S, Nandy SK, Patel M, Hati S (2022) Genomic and probiotic attributes of Lactobacillus strains from rice-based fermented foods of North Eastern India. J Food Sci Technol. 10.1007/s13197-022-05633-8 [DOI] [PMC free article] [PubMed]
- 56.Pereira RP, Jadhav R, Baghela A, Barretto DA. In vitro assessment of probiotic potential of Saccharomyces cerevisiae DABRP5 isolated from bollo batter, a traditional Goan fermented food. Probiotics Antimicrob Proteins. 2021;13(3):796–808. doi: 10.1007/s12602-020-09734-8. [DOI] [PubMed] [Google Scholar]
- 57.Panicker AS, Ali SA, Anand S, Panjagari NR, Kumar S, Mohanty AK, Behare PV. Evaluation of some in vitro probiotic properties of Lactobacillus fermentum Strains. J Food Sci Technol. 2018;55(7):2801–2807. doi: 10.1007/s13197-018-3197-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Devi SM, Archer AC, Halami PM. Screening, characterization and in vitro evaluation of probiotic properties among lactic acid bacteria through comparative analysis. Probiotics Antimicrob Proteins. 2015;7(3):181–192. doi: 10.1007/s12602-015-9195-5. [DOI] [PubMed] [Google Scholar]
- 59.Deng Y, Zhang Y, Hesham AEL, Liu R, Yang M. Cell surface properties of five polycyclic aromatic compound-degrading yeast strains. Appl Microbiol Biotechnol. 2010;86(6):1933–1939. doi: 10.1007/s00253-010-2477-7. [DOI] [PubMed] [Google Scholar]
- 60.Del Re B, Sgorbati B, Miglioli M, Palenzona D. Adhesion, autoaggregation and hydrophobicity of 13 strains of Bifidobacterium longum. Lett Appl Microbiol. 2000;31:438–442. doi: 10.1046/j.1365-2672.2000.00845.x. [DOI] [PubMed] [Google Scholar]
- 61.Sunita A, Aparajita S, Aayushi R, Arti N. Evaluation of the probiotic potential of yeasts isolated from Indian fermented food items. Res. J Biotechnol. 2021;16:10. doi: 10.25303/1610rjbt3342. [DOI] [Google Scholar]
- 62.Reuben RC, Roy PC, Sarkar SL, Alam ARU, Jahid IK. Characterization and evaluation of lactic acid bacteria from indigenous raw milk for potential probiotic properties. J Dairy Sci. 2020;103(2):1223–1237. doi: 10.3168/jds.2019-17092. [DOI] [PubMed] [Google Scholar]
- 63.Divisekera DM, Samarasekera JK, Hettiarachchi C, Gooneratne J, Choudhary MI, Gopalakrishnan S, Wahab AT. Lactic acid bacteria isolated from fermented flour of finger millet, its probiotic attributes and bioactive properties. Ann Microbiol. 2019;69(2):79–92. doi: 10.1007/s13213-018-1399-y. [DOI] [Google Scholar]
- 64.Garcia-Cayuela T, Korany AM, Bustos I, Gomez de Cadinanos LP, Requena T, Pelaez C, Martinez-Cuesta M. Adhesion abilities of dairy Lactobacillus plantarum strains showing an aggregation phenotype. Food Res Int. 2014;57:44–50. doi: 10.1016/j.foodres.2014.01.010. [DOI] [Google Scholar]
- 65.Mishra BK, Hati S, Das S, Kumari R. Evaluation of probiotic potentials of Lactobacillus isolated from traditional fermented foods of Garo Hills, Meghalaya, India. Rev Med Microbiol. 2018;29(3):120–128. doi: 10.1097/MRM.0000000000000139. [DOI] [Google Scholar]
- 66.Fadare OS, Singh V, Enabulele OI, Shittu OH, Pradhan D. In vitro evaluation of the synbiotic effect of probiotic Lactobacillus strains and garlic extract against Salmonella species. LWT. 2022;153:112439. doi: 10.1016/j.lwt.2021.112439. [DOI] [Google Scholar]
- 67.Bindu A, Lakshmidevi N. Identification and in vitro evaluation of probiotic attributes of lactic acid bacteria isolated from fermented food sources. Arch Microbiol. 2021;203(2):579–595. doi: 10.1007/s00203-020-02037-0. [DOI] [PubMed] [Google Scholar]
- 68.Lee JE, Lee NK, Paik HD. Antimicrobial and anti-biofilm effects of probiotic Lactobacillus plantarum KU200656 isolated from kimchi. Food Sci Biotechnol. 2021;30(1):97–106. doi: 10.1007/s10068-020-00837-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Suwannaphan S. Isolation, identification and potential probiotic characterization of lactic acid bacteria from Thai traditional fermented food. AIMS Microbiol. 2021;7(4):431. doi: 10.3934/microbiol.2021026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gil-Rodriguez AM, Beresford T. Bile salt hydrolase and lipase inhibitory activity in reconstituted skim milk fermented with lactic acid bacteria. J Funct Foods. 2021;77:104342. doi: 10.1016/j.jff.2020.104342. [DOI] [Google Scholar]
- 71.Hernandez-Gomez JG, Lopez-Bonilla A, Trejo-Tapia G, Avila-Reyes SV, Jimenez-Aparicio AR, Hernandez-Sanchez H. In vitro bile salt hydrolase (BSH) activity screening of different probiotic microorganisms. Foods. 2021;10(3):674. doi: 10.3390/foods10030674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sharma A, Lavania M, Singh R, Lal B. Identification and probiotic potential of lactic acid bacteria from camel milk. Saudi J Biol Sci. 2021;28(3):1622–1632. doi: 10.1016/j.sjbs.2020.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang SJ, Lee JE, Lim SM, Kim YJ, Lee NK, Paik HD. Antioxidant and immune-enhancing effects of probiotic Lactobacillus plantarum 200655 isolated from kimchi. Food Sci Biotechnol. 2019;28(2):491–499. doi: 10.1007/s10068-018-0473-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kathiriya MR, Hati S, Prajapati JB, Vekariya YV. Assessment of in vitro probiotic potential of lactic acid bacteria. Res Rev J Dairy Sci Tech. 2018;5(1):17–30. doi: 10.37591/rrjodst.v5i1.487. [DOI] [Google Scholar]
- 75.Virtanen T, Pihlanto A, Akkanen S, Korhonen H. Development of antioxidant activity in milk whey during fermentation with lactic acid bacteria. J Appl Microbiol. 2007;102(1):106–115. doi: 10.1111/j.1365-2672.2006.03072.x. [DOI] [PubMed] [Google Scholar]
- 76.Dineshbhai CK, Basaiawmoit B, Sakure A, Maurya R, Bishnoi M, Kondepudi KK, Patil GB, Mankad M, Liu Z, Hati S. Exploring the potential of Lactobacillus and Saccharomyces for biofunctionalities and the release of bioactive peptides from whey protein fermentate. Food Biosci. 2022;48:101758. doi: 10.1016/j.fbio.2022.101758. [DOI] [Google Scholar]
- 77.Shukla P, Sakure A, Pipaliya R, Basaiawmoit B, Maurya R, Bishnoi M, Kondepudi KK, Hati S. Exploring the potential of Lacticaseibacillus paracasei M11 on antidiabetic, anti-inflammatory, and ACE inhibitory effects of fermented dromedary camel milk (Camelus dromedaries) and the release of antidiabetic and anti-hypertensive peptides. J Food Biochem. 2022;46:e14449. doi: 10.1111/jfbc.14449. [DOI] [PubMed] [Google Scholar]
- 78.Patel D, Sakure A, Lodha D, Basaiawmoit B, Maurya R, Das S, Bishnoi M, Kondepudi KK, Hati S (2021) Significance of Lactobacillus fermentum on antioxidative and anti-inflammatory activities and ultrafiltration peptide fractions as potential sources of antioxidative peptides from fermented camel milk (Indian breed). J Am Coll Nutr:1–10. 10.1080/07315724.2021.1983485 [DOI] [PubMed]
- 79.Castorena-Alba MM, Vazquez-Rodriguez JA, Lopez-Cabanillas Lomeli M, Gonzalez-Martinez BE. Cholesterol assimilation, acid and bile survival of probiotic bacteria isolated from food and reference strains. CyTA J Food. 2018;16(1):36–41. doi: 10.1080/19476337.2017.1335347. [DOI] [Google Scholar]
- 80.Angmo K, Kumari A, Bhalla TC. Probiotic characterization of lactic acid bacteria isolated from fermented foods and beverage of Ladakh. LWT-Food Sci Technol. 2016;66:428–435. doi: 10.1016/j.lwt.2015.10.057. [DOI] [Google Scholar]
- 81.Pan DD, Zeng XQ, Yan YT. Characterisation of Lactobacillus fermentum SM-7 isolated from koumiss, a potential probiotic bacterium with cholesterol-lowering effects. J Sci Food Agric. 2011;91(3):512–518. doi: 10.1002/jsfa.4214. [DOI] [PubMed] [Google Scholar]
- 82.Strahsburger E, de Lacey AML, Marotti I, DiGioia D, Biavati B, Dinelli G. In vivo assay to identify bacteria with β-glucosidase activity. Electron J Biotechnol. 2017;30:83–87. doi: 10.1016/j.ejbt.2017.08.010. [DOI] [Google Scholar]
- 83.Mandal H, Bagchi T. In vitro screening of indigenous Lactobacillus isolates for selecting organisms with better health-promoting attributes. Appl Biochem Biotechnol. 2018;185(4):1060–1074. doi: 10.1007/s12010-018-2709-3. [DOI] [PubMed] [Google Scholar]
- 84.Hati S, Ningtyas DW, Khanuja JK, Prakash S. β-Glucosidase from almonds and yoghurt cultures in the biotransformation of isoflavones in soy milk. Food Biosci. 2020;34:100542. doi: 10.1016/j.fbio.2020.100542. [DOI] [Google Scholar]
- 85.Jang HJ, Song MW, Lee NK, Paik HD. Antioxidant effects of live and heat-killed probiotic Lactobacillus plantarum Ln1 isolated from kimchi. J Food Sci Technol. 2018;55(8):3174–3180. doi: 10.1007/s13197-018-3245-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhu Y, Wang Z, Zhang L. Optimization of lactic acid fermentation conditions for fermented tofu whey beverage with high-isoflavone aglycones. LWT. 2019;111:211–217. doi: 10.1016/j.lwt.2019.05.021. [DOI] [Google Scholar]
- 87.Hati S, Vij S, Singh BP, Mandal S. β-Glucosidase activity and bioconversion of isoflavones during fermentation of soymilk. J Sci Food Agric. 2015;95(1):216–220. doi: 10.1002/jsfa.6743. [DOI] [PubMed] [Google Scholar]
- 88.Liu X, Champagne CP, Lee BH, Boye JI, Casgrain M (2014) Thermostability of probiotics and their α-galactosidases and the potential for bean products. Biotechnol Res Int 2014. 10.1155/2014/472723 [DOI] [PMC free article] [PubMed]
- 89.Keat-hui N, Lye HS, Easa AM, Liong MT. Growth characteristics and bioactivity of probiotics in tofu-based medium during storage. Ann Microbiol. 2008;58(3):477–487. doi: 10.1007/BF03175546. [DOI] [Google Scholar]
- 90.Furman D, Campisi J, Verdin E, Carrera-Bastos P, Targ S, Franceschi C, Ferrucci L, Gilroy DW, Fasano A, Miller GW, Miller AH. Chronic inflammation in the etiology of disease across the life span. Nat Med. 2019;25(12):1822–1832. doi: 10.1038/s41591-019-0675-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cristofori F, Dargenio VN, Dargenio C, Miniello VL, Barone M, Francavilla R. Anti-inflammatory and immunomodulatory effects of probiotics in gut inflammation: a door to the body. Front Immunol. 2021;12:178. doi: 10.3389/fimmu.2021.578386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chopada K, Basaiawmoit B, Sakure A, Maurya R, Bishnoi M, Kondepudi KK, Solanki D, Singh BP, Padhi S, Rai AK, Liu Z (2022) Purification and characterization of novel antihypertensive and antioxidative peptides from whey protein fermentate: in vitro, in silico, and molecular interactions studies. J Am Nutr Assoc:1–20. 10.1080/27697061.2022.2110966 [DOI] [PubMed]
- 93.Jia J, Sun Y, Hu Z, Li Y, Ruan X. Propofol inhibits the release of interleukin-6, 8 and tumor necrosis factor-α correlating with high-mobility group box 1 expression in lipopolysaccharides-stimulated RAW 264.7 cells. BMC Anesthesiol. 2017;17(1):1–9. doi: 10.1186/s12871-017-0441-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hou Y, Yu H, Liu X, Li G, Pan J, Zheng C, Yu W. Gingipain of Porphyromonas gingivalis manipulates M1 macrophage polarization through C5a pathway. In Vitro Cell Dev Biol Anim. 2017;53(7):593–603. doi: 10.1007/s11626-017-0164-z. [DOI] [PubMed] [Google Scholar]
- 95.Khanna S, Bishnoi M, Kondepudi KK, Shukla G. Isolation, characterization and anti-inflammatory mechanism of probiotics in lipopolysaccharide-stimulated RAW 264.7 macrophages. World J Microbiol Biotechnol. 2020;36(5):1–11. doi: 10.1007/s11274-020-02852-z. [DOI] [PubMed] [Google Scholar]
- 96.Michels M, Jesus GFA, Voytena APL. Immunomodulatory effect of Bifidobacterium, Lactobacillus, and Streptococcus strains of paraprobiotics in lipopolysaccharide-stimulated inflammatory responses in RAW-264.7 Macrophages. Curr Microbiol. 2022;79(1):1–14. doi: 10.1007/s00284-021-02708-1. [DOI] [PubMed] [Google Scholar]
- 97.Xiaoqing Xu, Yu Qiao, Qing Peng, Bo Shi, Vermont P, Dia (2022) Antioxidant and immunomodulatory properties of partially purified exopolysaccharide from Lactobacillus Casei isolated from Chinese Northeast Sauerkraut. Immunol Investig 51(4): 748-765. 10.1080/08820139.2020.1869777 [DOI] [PubMed]
- 98.Song D, Lee HB, Kim GB, Kang SS. Whey fermented by Enterococcus faecalis M157 exhibits antiinflammatory and antibiofilm activities against oral pathogenic bacteria. J Dairy Sci. 2022;105(3):1900–1912. doi: 10.3168/jds.2021-21233. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 1432 kb)
Data Availability Statement
Raw data not published in supplementary materials and are available on reasonable request from the corresponding author.