Abstract
Trichosporon species are opportunistic pathogens, associated with a high mortality rate in immunocompromised patients. Oligonucleotide primers were used to amplify a 170-bp fragment of small-subunit ribosomal DNA of all species in the genus Trichosporon by PCR. The primers amplify DNAs of all species in the genus Trichosporon, including six causative agents of trichosporonosis. DNAs of other medically important yeasts, such as Candida albicans and Cryptococcus neoformans, are not amplified by this detection system.
Trichosporon Behrend is a medically important genus that includes the causative agents of deep-seated, mucosa-associated, and superficial infections. Since 1970, when Watson and Kallichurum (20) first reported disseminated infection (trichosporonosis) caused by Trichosporon species, there have been an increasing number of reports (10, 19) of this rare disease in immunocompromised hosts, such as patients who have undergone cytotoxic chemotherapy and recipients of organ transplants. The causative agent of trichosporonosis was believed to be Trichosporon cutaneum, but recent taxonomic research has indicated that six species, T. asahii, T. asteroides, T. cutaneum, T. inkin, T. mucoides, and T. ovoides, are associated with the infection (5, 7, 14, 15). Trichosporon species have four serotypes, I, II, III, and I-III, identified by using specific factor sera (8, 11). It has also been shown that the serotypes correlate well with the molecular phylogenetic tree (11, 12). Current microbiological diagnostic techniques used to identify Trichosporon species include production of arthroconidia, physiological characterization, and nuclear DNA-DNA hybridization experiments (5, 6, 13). However, a rapid detection system has not yet been developed.
In the present study, we describe a rapid PCR-based approach for detection of all species in the genus Trichosporon, including the causative agents of trichosporonosis.
The genera and species of 68 strains used in this study are listed in Table 1. They include all species of the genus Trichosporon and related medically important yeasts. DNA was prepared by the method of Makimura et al. (9). Briefly, one loop of yeast cells was suspended in lysing solution (100 mM Tris-HCl [pH 8.0], 30 mM EDTA, and 0.5% sodium dodecyl sulfate) and then heated at 100°C for 15 min. The solution was extracted with phenol-chloroform-isoamyl alcohol (25:24:1 [vol/vol]). DNA was precipitated with cold isopropanol. To select primers that would specifically amplify only Trichosporon species, the sequences of small-subunit (SSU) ribosomal DNA (rDNA) of pathogenic yeasts obtained from DNA sequence libraries (DDBJ, EMBL, and GenBank) were aligned. The primers, TRF (forward) (5′-AGAGGCCTACCATGGTATCA-3′) and TRR (reverse) (5′-TAAGACCCAATAGAGCCCTA-3′), were chosen to align with regions which were not conserved in other medically important yeasts. The PCR primers correspond to nucleotides 154 to 173 and 354 to 335 of Saccharomyces cerevisiae SSU rDNA. The oligonucleotide primers were obtained from Greiner Japan (Tokyo, Japan). Amplification reactions were performed in PCR buffer containing 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.0 mM MgCl2, 2.5 μM (each) dATP, dCTP, dGTP, and dTTP, 2 mM each oligonucleotide primer, and 0.5 U of Taq DNA polymerase (Takara, Shiga, Japan). The reaction mixtures were amplified in a Perkin-Elmer 9700 thermal cycler with the following program: 94°C for 3 min, followed by 30 cycles consisting of 94°C for 30 s, 56°C for 15 s, and 72°C for 15 s, with a final extension period at 72°C for 10 min. After thermal cycling, 5 μl of the amplified product was run on a 1.5% (wt/vol) agarose gel, stained with ethidium bromide, and visualized with UV light.
TABLE 1.
Specificity of Trichosporon primers
| Species and variety | No. of strains tested | PCR producta |
|---|---|---|
| Trichosporon speciesb | ||
| Serotype I | ||
| Trichosporon cutaneum | 4 | + |
| Trichosporon mucoides | 4 | + |
| Trichosporon jirovecii | 2 | + |
| Trichosporon moniliiforme | 1 | + |
| Serotype II | ||
| Trichosporon asahii var. asahii | 5 | + |
| Trichosporon asahii var. coremiformis | 2 | + |
| Trichosporon asahii var. faecalis | 2 | + |
| Trichosporon asteroides | 3 | + |
| Trichosporon aquatile | 2 | + |
| Trichosporon inkin | 2 | + |
| Trichosporon ovoides | 3 | + |
| Serotype III | ||
| Trichosporon brassicae | 1 | + |
| Trichosporon domesticum | 2 | + |
| Trichosporon montevideense | 1 | + |
| Serotype I-III | ||
| Trichosporon dulcitum | 1 | + |
| Trichosporon gracile | 1 | + |
| Trichosporon loubieri var. loubieri | 1 | + |
| Trichosporon loubieri var. laibachii | 1 | + |
| Trichosporon sporotrichoides | 1 | + |
| Nonreacting: Trichosporon pullulans | 3 | + |
| Other pathogenic yeasts | ||
| Candida albicans | 2 | − |
| Candida glabrata | 1 | − |
| Candida guilliermondii | 3 | − |
| Candida kefyr | 1 | − |
| Candida lusitaniae | 1 | − |
| Candida parapsilosis | 1 | − |
| Candida tropicalis | 1 | − |
| Cryptococcus neoformans var. neoformans | 5 | − |
| Cryptococcus neoformans var. gattii | 3 | − |
| Cryptococcus albidus var. albidus | 1 | − |
| Debaryomyces hansenii var. hansenii | 3 | − |
| Malassezia furfur | 3 | − |
| Malassezia pachydermatis | 1 | − |
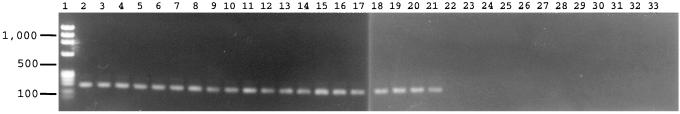
To detect Trichosporon species rapidly, we designed genus-specific primers for PCR, based on SSU rDNA sequences. The primers amplified the DNAs from all species in the genus Trichosporon, including clinical isolates, and produced a 170-bp fragment, as shown in Fig. 1. Other medically important yeasts (Table 1 and Fig. 1) were not amplified. At present, 17 species, including 5 varieties, are assigned to the genus Trichosporon (6, 13, 16). Partial sequence analysis of rRNA indicates that Trichosporon species are monophyletic, with the exception of T. pullulans (4, 6). Although the phylogenetic position of T. pullulans is remote from those of other Trichosporon species, the primers amplified the DNAs of all of the species in the genus, including T. pullulans. We took into consideration cross-reactivity with Cryptococcus neoformans DNA in the design of the genus-specific primers. The basidiomycetes are divided into three classes, the Urediniomycetes, Ustilaginomycetes, and Hymenomycetes. The Trichosporon species and C. neoformans are phylogenetically very closely related (17, 18) and are classified in the Hymenomycetes. Specific primers for Trichosporon species have been designed previously (3), but those primers amplified not only Trichosporon species, but also C. neoformans. Our primers amplified only Trichosporon species and did not amplify any strains of C. neoformans, including two varieties.
FIG. 1.
Agarose gel electrophoresis of PCR products of Trichosporon species and other medically important yeasts. Lane 1, molecular weight marker (HaeIII-digested φX174 DNA; Takara, Shiga, Japan); lane 2, T. cutaneum; lane 3, T. mucoides; lane 4, T. jirovecii; lane 5, T. moniliiforme; lane 6, T. asahii var. asahii; lane 7, T. asahii var. coremiformis; lane 8, T. asahii var. faecalis; lane 9, T. asteroides; lane 10, T. aquatile; lane 11, T. inkin; lane 12, T. ovoides; lane 13, T. brassicae; lane 14, T. domesticum; lane 15, T. montevideense; lane 16, T. dulcitum; lane 17, T. gracile; lane 18, T. loubieri var. loubieri; lane 19, T. loubieri var. laibachii; lane 20, T. sporotrichoides; lane 21, T. pullulans; lane 22, C. neoformans var. neoformans; lane 23, C. neoformans var. gattii; lane 24, Malassezia furfur; lane 25, Malassezia pachydermatis; lane 26, Candida albicans; lane 27, Candida glabrata; lane 28, Candida guilliermondii; lane 29, Candida kefyr; lane 30, Candida lusitaniae; lane 31, Candida parapsilosis; lane 32, Candida tropicalis; lane 33, Debaryomyces hansenii var. hansenii.
Recent taxonomic studies have indicated that trichosporonosis is caused by six species; T. asahii, T. asteroides, T. cutaneum, T. inkin, T. mucoides, and T. ovoides (5, 7, 14, 15). T. domesticum and T. pullulans are not major causative agents of trichosporonosis and were rarely isolated from patients. Moreover, it has been suggested that the major causative agents of trichosporonosis differ in the type of infection they cause. T. asahii is involved in deep-seated infection. T. asteroides and T. cutaneum are associated with superficial infection. T. ovoides is involved in capital white piedra, and T. inkin is involved in white piedra of the genital area. The majority of trichosporonosis cases reported involving deep-seated infections have been associated with an underlying neoplastic disease, particularly hematologic malignancies (10, 17). This infection is life threatening, with a high mortality rate, and the prognosis for patients with deep-seated trichosporonosis is very poor. Early diagnosis and treatment is therefore of paramount importance to trichosporonosis patients. It is clinically significant that all the pathogenic species are detected by a single test by using our PCR detection system. We have not yet performed direct detection of Trichosporon DNA from trichosporonosis patients’ specimens, such as blood and urine specimens, but we are in process of developing the methodology.
Moreover, Trichosporon species are also involved in summer-type hypersensitivity pneumonitis (1, 2). We did not perform specificity testing against nonpathogenic yeast using this PCR method, but it may be possible to distinguish Trichosporon species from environmental isolates.
In conclusion, a method for the rapid detection of Trichosporon species, including all causative agents of trichosporonosis, was established. The method presented in this study can specifically and rapidly detect the DNAs of Trichosporon species, and we anticipate that it will be a useful microbiological tool for the diagnosis of trichosporonosis.
REFERENCES
- 1.Ando M, Arima K, Yoneda R, Tamura M. Japanese summer-type hypersensitivity pneumonitis. Am Rev Respir Dis. 1991;144:765–769. doi: 10.1164/ajrccm/144.4.765. [DOI] [PubMed] [Google Scholar]
- 2.Ando M, Sakata T, Yoshida K, Yamasaki H, Araki S, Onoue K, Shinoda T. Serotype-related antigen of Trichosporon cutaneum in the induction of summer-type hypersensitivity pneumonitis: correlation between serotype of inhalation challenge-positive antigen and that of the isolates from patients’ homes. J Allergy Clin Immunol. 1990;85:36–44. doi: 10.1016/0091-6749(90)90218-s. [DOI] [PubMed] [Google Scholar]
- 3.Fell J W. rDNA targeted oligonucleotide primers for the identification of pathogenic yeasts in a polymerase chain reaction. J Ind Microbiol. 1995;14:475–477. doi: 10.1007/BF01573961. [DOI] [PubMed] [Google Scholar]
- 4.Guého E, Improvisi L, Christen R, de Hoog G S. Phylogenetic relationships of Cryptococcus neoformans and some related basidiomycetous yeasts determined from partial large subunit rRNA sequences. Antonie Leeuwenhoek. 1993;63:175–189. doi: 10.1007/BF00872392. [DOI] [PubMed] [Google Scholar]
- 5.Guého E, Improvisi L, de Hoog G S, Dupont B. Trichosporon on humans: a practical account. Mycoses. 1994;37:3–10. doi: 10.1111/j.1439-0507.1994.tb00277.x. [DOI] [PubMed] [Google Scholar]
- 6.Guého E, Smith M T, de Hoog G S, Grand G B, Christen R, Batenburg-van der Vegte W H. Contributions to a revision of the genus Trichosporon. Antonie Leeuwenhoek. 1992;61:289–316. doi: 10.1007/BF00713938. [DOI] [PubMed] [Google Scholar]
- 7.Herbrecht R, Koening H, Waller K, Liu L, Guého E. Trichosporon infections: clinical manifestations and treatment. J Mycol Med. 1993;3:129–136. [Google Scholar]
- 8.Ikeda R, Yokota M, Shinoda T. Serological characterization of Trichosporon cutaneum and related species. Microbiol Immunol. 1996;40:813–819. doi: 10.1111/j.1348-0421.1996.tb01146.x. [DOI] [PubMed] [Google Scholar]
- 9.Makimura K, Murayama Y S, Yamaguchi H. Detection of a wide range of medically important fungal species by polymerase chain reaction (PCR) J Med Microbiol. 1994;40:358–364. doi: 10.1099/00222615-40-5-358. [DOI] [PubMed] [Google Scholar]
- 10.Nahass G T, Rosenberg S P, Leonardi C L, Penneys N S. Disseminated infection with Trichosporon beigelii. Arch Dermatol. 1993;129:1020–1023. doi: 10.1001/archderm.129.8.1020. [DOI] [PubMed] [Google Scholar]
- 11.Nishiura Y, Nakagawa-Yoshida K, Suga M, Shinoda T, Guého E, Ando M. Assignment and serotyping of Trichosporon species: the causative agents of summer-type hypersensitivity pneumonitis. J Med Vet Mycol. 1997;35:45–52. doi: 10.1080/02681219780000861. [DOI] [PubMed] [Google Scholar]
- 12.Sugita T, Makimura K, Nishikawa A, Uchida K, Yamaguchi H, Shinoda T. Partial sequences of large subunit ribosomal DNA of a new yeast species, Trichosporon domesticum, and related species. Microbiol Immunol. 1997;41:571–573. doi: 10.1111/j.1348-0421.1997.tb01893.x. [DOI] [PubMed] [Google Scholar]
- 13.Sugita T, Nishikawa A, Shinoda T. Reclassification of Trichosporon cutaneum by DNA relatedness by using the spectrophotometric method and the chemiluminometric method. J Gen Appl Microbiol. 1994;40:397–408. [Google Scholar]
- 14.Sugita T, Nishikawa A, Shinoda T, Kume H. Taxonomic position of deep-seated, mucosa-associated, and superficial isolates of Trichosporon cutaneum from trichosporonosis patients. J Clin Microbiol. 1995;33:1368–1370. doi: 10.1128/jcm.33.5.1368-1370.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sugita T, Nishikawa A, Shinoda T, Kusunoki T. Taxonomic studies on clinical isolates from superficial trichosporonosis patients by DNA relatedness. Jpn J Med Mycol. 1996;37:107–110. [Google Scholar]
- 16.Sugita T, Nishikawa A, Shinoda T, Yoshida K, Ando M. A new species, Trichosporon domesticum, isolated from the house of a summer-type hypersensitivity pneumonitis patient in Japan. J Gen Appl Microbiol. 1995;41:429–436. [Google Scholar]
- 17.Swann E C, Taylor J W. Phylogenetic diversity of yeast-producing basidiomycetes. Mycol Res. 1995;99:1205–1210. [Google Scholar]
- 18.Swann, E. C., and J. W. Taylor. 1995. Phylogenetic perspectives on basidiomycete systematics: evidence from the 18S rRNA gene. Can. J. Bot. 73(Suppl. 1):S862–S868.
- 19.Walsh T J. Trichosporonosis. Infect Dis Clin N Am. 1989;3:43–52. [PubMed] [Google Scholar]
- 20.Watson K C, Kallichurum S. Brain abscess due to Trichosporon cutaneum. J Med Microbiol. 1970;3:191–193. doi: 10.1099/00222615-3-1-191. [DOI] [PubMed] [Google Scholar]