Summary
Background
In individuals with malignancy or HIV-1 infection, antigen-specific cytotoxic T lymphocytes (CTLs) often display an exhausted phenotype with impaired capacity to eliminate the disease. Existing cell-based immunotherapy strategies are often limited by the requirement for adoptive transfer of CTLs. We have developed an immunotherapy technology in which potent CTL responses are generated in vivo by vaccination and redirected to eliminate target cells using a bispecific Redirector of Vaccine-induced Effector Responses (RoVER).
Methods
Following Yellow fever (YF) 17D vaccination of 51 healthy volunteers (NCT04083430), single-epitope YF-specific CTL responses were quantified by tetramer staining and multi-parameter flow cytometry. RoVER-mediated redirection of YF-specific CTLs to kill antigen-expressing Raji-Env cells, autologous CD19+ B cells or CD4+ T cells infected in vitro with a full-length HIV-1-eGFP was assessed in cell killing assays. Moreover, secreted IFN-γ, granzyme B, and TNF-α were analyzed by mesoscale multiplex assays.
Findings
YF-17D vaccination induced strong epitope-specific CTL responses in the study participants. In cell killing assays, RoVER-mediated redirection of YF-specific CTLs to autologous CD19+ B cells or HIV-1-infected CD4+ cells resulted in 58% and 53% killing at effector to target ratio 1:1, respectively.
Interpretation
We have developed an immunotherapy technology in which epitope-specific CTLs induced by vaccination can be redirected to kill antigen-expressing target cells by RoVER linking. The RoVER technology is highly specific and can be adapted to recognize various cell surface antigens. Importantly, this technology obviates the need for adoptive transfer of CTLs.
Funding
This work was funded by the Novo Nordisk Foundation (Hallas Møller NNF10OC0054577).
Keywords: Immunotherapy, Vaccine, Cytotoxic T lymphocytes, Redirection, Cancer, HIV-1 infection
Research in context.
Evidence before this study
Harnessing CD8+ T cells is a promising immunotherapeutic strategy and can be achieved by different means. Broad CD3-engaging bispecific T cell engagers (BITE) or Chimeric Antigen Receptor (CAR) methodologies have entered clinical utility. Bispecific molecules with greater selectivity for effector T cells has been proposed as a next generation therapeutic strategy.
Added value of this study
Here we show the possibility of inducing clonal expanded effector T cells by simple vaccination and hence provide the data to support the ability to redirect vaccine-generated T cells to eliminate cells expressing other antigens not comprised in the vaccine.
Implications of all the available evidence
This novel means of using in vivo vaccine-induced immunity to elimate cells expressing cancer- or HIV-antigens could lead to a new way of harnessing T cells for disease immunotherapy.
Introduction
Cytotoxic T lymphocytes (CTLs) are potent killers of malignant and virus-infected cells. Consequently, several existing immunotherapy strategies act by directing cytotoxic effector responses towards target cells as seen with chimeric antigen receptor (CAR) T cells and bispecific T cell engager (BiTE) constructs. In hematological malignancies, CAR T cells and BiTEs have improved treatment and outcomes.1, 2, 3, 4, 5, 6, 7, 8, 9 Thus, CAR T cells and BiTEs are now being explored to treat HIV-1 infection (clinical trials: NCT03617198, NCT04863066).10, 11, 12, 13, 14, 15 Both in malignant disease and during chronic HIV-1 infection, the immune system display an exhausted phenotype and is dysfunctional due to a chronic state of inflammation caused by sustained antigen exposure.16, 17, 18 Furthermore, the malignant and virus-infected cells evade immune-mediated clearance by escape mechanisms such as downregulation of Human Leukocyte Antigen class I (HLA-I) expression to prevent presentation of disease-associated antigens.19, 20, 21, 22, 23 Together, this suggests that the immune response is inefficient at eliminating a persistent malignancy as well as chronic HIV-1 infection, and thus emphasizes the need for assisting immunotherapies to induce effective immune-mediated killing. Despite the immense potential of the CAR T cell technology, the requirement for adoptive transfer of vast numbers of ex vivo genetically modified effector cells is an obvious obstacle limiting broader scale clinical application.4,5,24, 25, 26
A slightly different immunotherapy concept has been explored in pre-clinical cancer research achieving linkage of cancer target cells and CTLs by creating a chimeric molecule comprising an antibody or a single chain Fragment variable (scFv) specific for a tumor-associated antigen and a recombinant peptide-loaded HLA-I molecule directed against selected antigen-specific CTLs.27, 28, 29, 30, 31, 32, 33, 34, 35 Specific lysis of antigen-expressing cancer cells is thus achieved by redirecting CTLs specific for the peptide presented by the HLA-I domain. Several studies have shown that redirection of CTLs results in specific and potent killing of cancer cells in vitro27, 28, 29, 30, 31, 32, 33, 34, 35 as well as inhibition of tumor growth in vivo using tumor mouse models.28,29,32,34,35 Prominently, scFv-guided redirection of CTLs towards tumor cells by targeting cell-surface proteins is able to overcome the absence or low expression of HLA-I molecules.29,33,34 However, the clinical utility of this technology may be compromised by the inability to generate sufficient numbers of antigen-specific CTLs in vivo and the need for adoptive transfer of effector cells.
Here we describe an extension of the technology for redirection of an antigen-specific CTL response towards a target of choice. In this immunotherapy concept (Fig. 1a) a potent de novo CTL effector response is generated in vivo by simple vaccination followed by redirection of CTLs towards target cells by a recombinant RoVER (Redirector of Vaccine-induced Effector Responses). The RoVER is composed of two functionally distinct domains: 1) a scFv domain targeting specific cell surface antigens on target cells, and 2) an HLA class I molecule carrying an immunogenic vaccine epitope able to specifically engage and activate vaccine-induced effector T cells (Supplementary Fig. S1). We utilize the live-attenuated Yellow Fever (YF) 17D vaccine (Stamaril, Novartis), which induces remarkably strong CTL immune responses.36 This YF vaccine-induced generation of epitope-specific CTL responses is exploited in the adaptive immunotherapy technology to overcome the limitation of an inadequate and exhausted effector response.
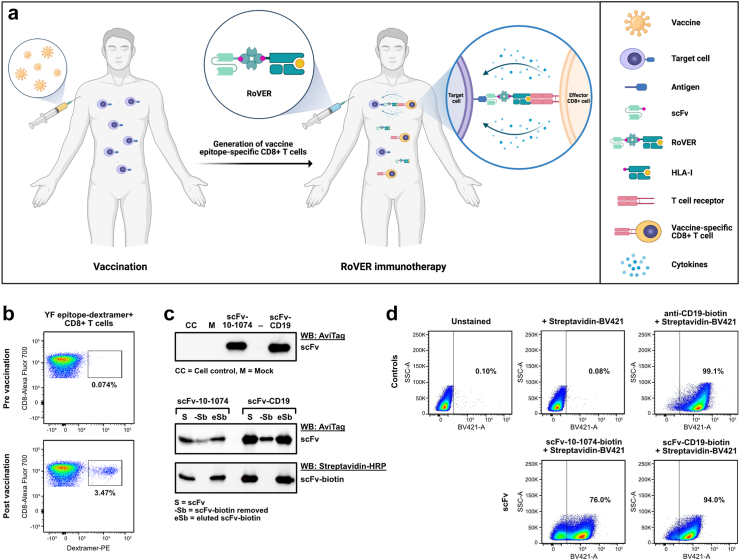
Fig. 1.
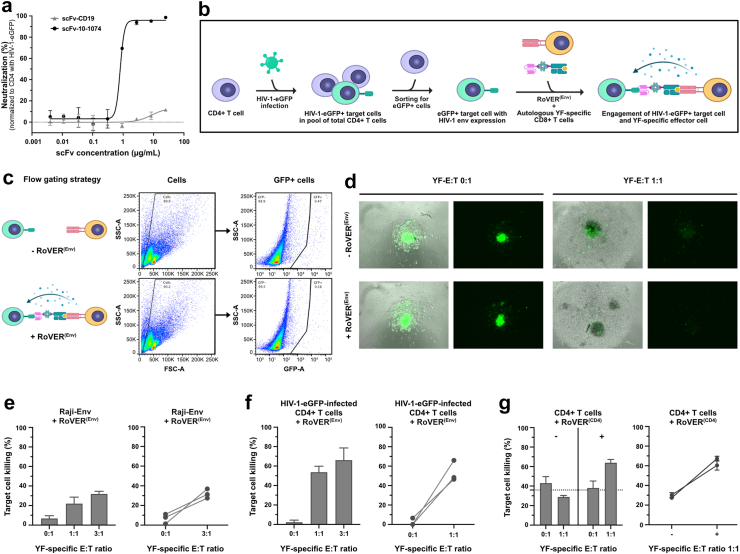
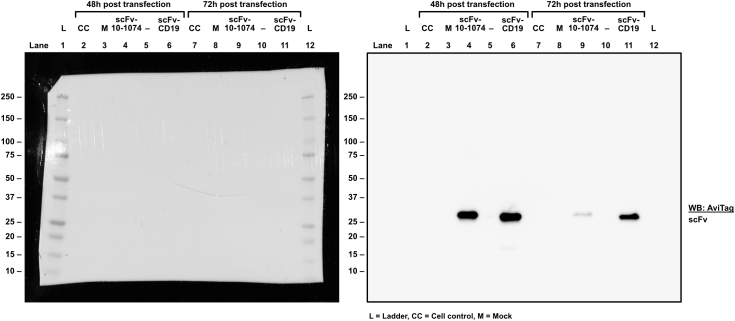
Functional assessment of RoVER components. a) Graphics of the RoVER technology. A vaccine induces potent vaccine epitope-specific CD8+ T cell responses which are redirected towards a cellular target of choice by administration of the bispecific RoVER. Graphics created with BioRender.com. b) Representative flow cytometry plots from dextramer staining of donor peripheral blood mononuclear cells (PBMCs) pre and post YF-17D vaccination (21 ± 3 days) showing vaccine-induced epitope-specific CD8+ T cell responses. c) Western blots with anti-AviTag or Streptavidin-HRP showing successful expression and biotinylation of scFv-10-1074 and scFv-CD19 proteins from transient transfection of mammalian cells with pcDNA3.1 (+) expression plasmids encoding the scFvs. d) Functional assessment of the target cell binding capacity of scFv-10-1074 and scFv-CD19 proteins by flow cytometry analyses using Raji-Env cells with stable expression of GFP, CD19 and HIV-1 envelope using BV421-conjugated streptavidin. Controls included unstained Raji-Env cells or cells stained with a biotin-conjugated anti-CD19 antibody and/or streptavidin-BV421.
Methods
YF-17D vaccinated healthy donors and ethics
Healthy adult participants were recruited in the study (NCT04083430) and received vaccination with a single dose of 0.5 mL 17D live-attenuated YF vaccine (Stamaril, Sanofi Pasteur). Individuals self-reported any previous history of vaccination with YF-17D. Blood samples were taken (up to 100 days) prior to and at two timepoints (day 21 ± 3 and 100 ± 40) after vaccination. Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll density gradient centrifugation and cryopreserved in Heat Inactivated Foetal Bovine Serum (HI-FBS) with 10% DMSO. All participants provided informed consent under the approved protocol by the Danish Medicines Agency (2019-06-06-50), the Danish National Committee on Health Research Ethics (1-10-72-122-19) and the Danish Data Protection Agency (1-16-02-190-19).
Primary cells and cell lines
Peripheral blood mononuclear cells (PBMCs), primary B cells, CD4+, and CD8+ T cells obtained from YF-17D vaccinated study participants were cultured in RPMI 1640 medium supplemented with 10% HI-FBS, 1% Penicillin-Streptomycin (P/S) (cRPMI) and 100 IU/mL IL-2. Primary CD4+ T cells, CD8+ T cells and B cells were isolated from human PBMCs by negative selection using the CD4+ T Cell Isolation Kit (Miltenyi Biotec Cat# 130-096-533), CD8+ T Cell Isolation Kit (Miltenyi Biotec Cat# 130-096-495), and the B Cell Isolation Kit II (Miltenyi Biotec Cat# 130-091-151).
HEK293T cells obtained from the ATCC (ATCC Cat# CRL-3216™, RRID: CVCL_0063) were cultured in Dulbecco's Modified Eagle's Medium High Glucose (DMEM) supplemented with 10% HI-FBS and 1% P/S (cDMEM) at 37 °C in 5% CO2.
Expi293-F (Gibco Cat# A14527) and BirA-Expi293-F cells were cultured in Expi293 Expression Medium (Gibco Cat# A1435101) in sterile Erlenmeyer culture flasks at 37 °C in 8% CO2 with shaking. BirA-Expi293-F cells were generated by stabile transfection of Expi293-F cells with pDisplay-BirA-ER (Addgene Cat# 20856, RRID: Addgene_20856) plasmid expressing BirA enzyme. To select for BirA-expressing cells, 0.5 mg/mL G418 selection (Sigma–Aldrich Cat# G8168) was added to the culture medium, and 0.2 μg/mL biotin (Sigma–Aldrich Cat# B4639).
Raji-Env cells37 with stable expression of YU2 envelope and GFP under puromycin selection were cultured at 37 °C in 5% CO2 in cRPMI + 1 μg/mL puromycin (Gibco Cat# A1113803).
All cell lines were tested negative for mycoplasma.
HLA-typing of YF-specific CD8+ T cells
For all study participants, HLA class I (HLA-A, -B, -C) alleles were genotyped at the ASHI-accredited laboratory HistoGenetics (Ossining, NY) using sequencing-based typing (SBT).
Quantification of YF-specific CD8+ T cells
YF epitope-specific CD8+ T cells isolated from YF-17D vaccinated study participants were quantified by tetramer staining. To detect various HLA types, ten different biotin-conjugated HLA class I molecules carrying immunogenic YF peptides (ImmunAware Cat# 1001-03, 1002-09, 1016-04, 1020-04, 1048-05, 1058-02, 1072-02, 1088-02, 1105-02 or custom design) were analyzed: NS5286-295(KSEYMTSWFY)-A∗01:01, NS4B214-222 (LLWNGPMAV)-A∗02:01, NS34-12(VLWDIPTPK)-A∗03:01, NS324-32(IYGIFQSTF)-A∗24:02, NS5672-680 (RPIDDRFGL)-B∗07:02, NS373-81(SVKEDLVAY)-B∗15:01, NS2A4-13(HAVPFGLVSM)-B∗35:01, NS2B110-118(HPFALLLVL)-B∗35:01, E200-209 (TESWIVDRQW)-B∗44:02, NS1116-124(KTWGKNLVF)-B∗57:01. HLA class I monomers carrying a YF peptide were tetramerized using PE-conjugated streptavidin (BioLegend Cat# 405204). Tetramerization was obtained by three sequential additions of biotinylated YF peptide-HLA class I monomers to Streptavidin-PE in a 4:1 M ratio, followed by an incubation step at 4 °C for 15 min 1.5 × 106 PBMCs/flow tube were incubated for 30 min in the dark at room temperature (RT) in 40 μL tetramer solution prediluted in FACS buffer (PBS with 2% HI-FBS) to a final concentration of 30 nM. Subsequently, the cells were washed in PBS and stained with Live/Dead™ Fixable Near-IR Dead Cell Stain (Invitrogen Cat# L10119) and incubated for 5 min at RT in the dark. The PBMCs were further analyzed for T cell markers (CD3, CD8), extracellular T cell activation markers (CD38, HLA-DR) and CCR7 and CD45RA expression to define CD8+ T cell subsets using a panel of fluorochrome-conjugated antibodies (BD Biosciences Cat# 564001, RRID: AB_2744382, Cat# 565192, RRID: AB_2739104, Cat# 555488 RRID: AB_395879, Cat# 562444, RRID: AB_11151894, Cat# 748339, RRID: AB_2872758, BioLegend Cat# 353230, RRID: AB_2563630) in Brilliant Stain buffer (BD Biosciences Cat# 563794) for 20 min at RT in the dark. The cells were analyzed by flow cytometry (BD LSRFortessa™ X-20; BD Biosciences) using BD FACSDiva Software (BD Biosciences, RRID: SCR_001456).
Cloning of scFv
DNA sequences for scFv-CD19 or scFv-10-1074 were designed based on the CD19-targeting domain of blinatumomab (Blincyto, Amgen), or the broadly neutralizing antibody, 10-1074, targeting the V3-loop on the HIV-1 envelope.38 The scFv DNA sequences were ordered as gBlocks® Gene Fragments from Integrated DNA Technologies. For expression, the scFv DNA sequences were cloned into a pcDNA3.1 (+) backbone plasmid (Invitrogen Cat# V79020).
Production of biotinylated scFv proteins
Biotinylated scFv proteins were produced by BirA-Expi293F cells with expression plasmids encoding scFv-CD19 or scFv-10-1074 using PEI MAX® transfection reagent (Polysciences Cat# 24765) in a 1:4 ratio of DNA to PEI. Supernatants were collected on day 5 post transfection, and the scFv proteins were isolated on a HiTrap TALON crude column (Cytiva Cat# 29048565) capturing His-tagged protein and eluted in PBS supplemented with 10% glycerol. In brief, elution buffer (250 mM Imidazole, 50 mM NaH2PO4, 300 mM NaCl, pH 7.0) to a final concentration of 4% (v/v), Triton X-100 to a final concentration of 0.1% (v/v), and CaCl2 to at final concentration of 750 μM was added to the supernatant. The supernatant was incubated for 1 h at 4 °C under constant stirring and was subsequently centrifuged at 4000×g for 30 min and vacuum filtered using a cellulose acetate membrane with a pore size of 0.2 μM. The 6xHis tagged scFv proteins were captured on a 1 mL HiTrap TALON crude column (Cytiva Cat# 29048565). The column was washed with 10 column volumes wash buffer (50 mM NaH2PO4, 300 mM NaCl, pH 7.0) and the protein was eluted using 5 column volumes elution buffer. The eluted protein was dialyzed into PBS supplemented with 10% glycerol and sterile filtered using a cellulose acetate filter with a pore size of 0.2 μM attached to a syringe.
Western blot analysis of scFv proteins
Protein samples were mixed in a 3:1 ratio with 4x Laemmli Sample Buffer (Bio-Rad Cat# 1610747) supplemented with 5% 1 M dithiothreitol (DTT) and heated at 95 °C for 5 min. The denatured proteins were separated by SDS-PAGE on a Mini-PROTEAN® TGX Stain-Free™ Protein Gel 4–20% or a Criterion™ TGX Stain-Free™ Protein Gel 4–20% in 1x Tris/Glycine/SDS buffer and transferred to a PVDF membrane using a Trans-Blot Turbo Transfer System (Biorad). The membrane was blocked in Tris-buffered Saline (TBS) supplemented with 0.1% Tween®20 (TBS-T) and 3% Nonfat-dried Milk bovine powder (TBS-T-3%Milk) for 1 h at RT with rocking. Following blocking, the membrane was incubated with anti-Avi-tag (GenScript Cat# A00674, RRID: AB_915553) or horseradish peroxidase (HRP)-conjugated Direct-Blot™ anti-His tag (BioLegend Cat# 362614, RRID: AB_2734399) primary antibodies or streptavidin-HRP (Invitrogen™ Cat# S911) diluted 1:2000, 1:500 or 1:3333, respectively, in TBS-T-1%Milk overnight at 4 °C or at RT for 1 h with rocking. If applicable, the membrane was incubated with an HRP-conjugated anti-rabbit IgG (Genscript Cat# A01827) secondary antibody diluted 1:5000 in TBS-T-1%Milk for 1 h at RT with rocking. The membrane was visualized using Clarity™ Western ECL substrate (Biorad Cat# 1705060) and the ChemiDoc imaging system (Biorad).
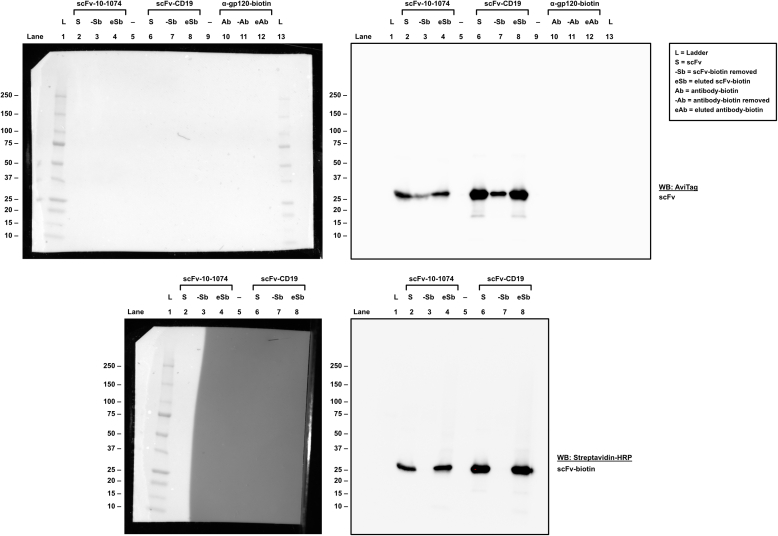
The degree of biotinylation of scFv proteins was assessed by Western blot analysis of scFv samples subjected to different conditions. In brief, samples containing biotinylated scFv proteins were compared to identical samples from which biotinylated proteins had been removed using Streptavidin Magnetic Beads (Thermo Scientific Cat# 88816). Subsequently, the samples were subjected to Western blot analysis as described. The degree of biotinylation was assessed by densiometric analyses using Image Lab software version 6.1.0 (Bio-Rad, RRID: SCR_014210).
Functional assessment of scFvs
Primary CD4+ T cells infected with HIV-1-eGFP, primary B cells or Raji-Env cells were seeded in flow tubes at a density of 5.0 × 105 cells/tube. Cells were washed in FACS buffer and incubated with 9.25 μg/mL of scFv-10-1074-biotin or 9 μg/mL scFv-CD19-biotin at 4 °C for 1 h. After washing twice with FACS buffer, the cells were incubated with 10 μg/mL of BV421-conjugated Streptavidin (BioLegend Cat# 405226) in the dark at 4 °C for 20 min. Finally, the cells were washed twice in FACS buffer, fixed with 1% formaldehyde (Thermo Scientific Cat# 28908), and analyzed by flow cytometry (MACSQuant® Analyzer 16).
Preassembly of RoVER
The RoVER was preassembled according to the protocol for tetramerization of HLA-I monomers as described previously, however replacing half the HLA-I molecules with scFv or an antibody (biotin-conjugated CD4 antibody; Biolegend Cat# 344610, RRID: AB_2028490). Preassembly of RoVER was thus achieved by combining 1.627 mol scFv or HLA-I monomer to 1 mol Streptavidin. In in vitro cell killing assays, 1.8 × 10−5 μmol RoVER was used per 106 target cells.
In vitro cell killing assays
Target CD19+ or CD4+ cells isolated from PBMCs were stained with a 2.5 μM CellTrace™ Violet (Invitrogen Cat# C34557) according to manufacturer's protocol. In killing assays using blinatumomab, target cells were seeded in a round-bottom 96-well plate with 5.0 × 103 cells/well. 50 μL cRPMI + IL2 or 10 ng/mL blinatumomab was added immediately prior to adding CD8+ effector cells in different effector to target (E:T). For RoVER-mediated killing assays, the target cells were incubated with preassembled RoVER for 1 h at 4 °C. For peptide-mediated killing assays, the target cells were incubated with 10 μg/mL Antigen Peptide YFV Genome polyprotein HLA-A∗0201 (LLWNGPMAV) (JPT Peptide Technologies Cat# SP-MHCI-0063) for stimulation of NS4B-HLA-A∗02:01 specific CD8+ T cells or with 10 μg/mL Antigen Peptide Chicken Ovalbumin H-2 Kb (SIINFEKL) (JPT Peptide Technologies Cat# SP-MHCI-0016) as a control for 1 h at 4 °C. Target cells were seeded in a round-bottom 96-well plate at a density of 5.0 × 103 cells/well, and autologous CD8+ T cells were added in different YF-specific E:T) cell ratios. After 20 h incubation at 37 °C in 5% CO2, 100 μL supernatant from each well was collected and saved at −80 °C for cytokine analysis. To identify dead or dying cells, the cells in each well were stained with Live/Dead™ Fixable Near-IR Dead Cell Stain (LD-NIR) (Invitrogen Cat# L10119) for 5 min in the dark at RT. The samples were then analyzed by flow cytometry for dead (LD-NIR+) target cells (CellTrace™+) using a MACSQuant® Analyzer 16. We show the level of dead cells in the figures (including the background/negative condition) and report the adjusted percentage of killed cells by subtracting this background from that observed in the stimulated condition.
Detection of CD107α degranulation
Initially, RoVER-mediated killing was performed according to the protocol for In vitro cell killing assays as described above. Following co-culture of CD19+ and autologous CD8+ T cells in YF-E:T cell ratio 3:1, anti-CD107a (LAMP-1) antibody (Miltenyi, Cat# 130-119-869) was added along with BD GolgiStop (BD Biosciences Cat# 554724) in a final volume of 250 μL. Each E:T was tested in triplicates. After 3 h incubation at 37 °C in 5% CO2, the cells were stained with tetramer solution, Live/Dead™ Fixable Near-IR Dead Cell Stain and a panel of surface markers antibodies as described above in Quantification of YF-specific CD8+ T cells. The samples were analyzed by flow cytometry using a BD LSRFortessa™ X-20 and BD FACSDiva Software.
Cytokine release assays
Cytokines IFN-γ, TNF-α or Granzyme B in supernatant from killing assays were quantified by mesoscale multiplex assays: V-PLEX Custom Human Biomarkers based on Proinflammatory Panel 1 Human Kit (Meso Scale Discovery, Inc. Cat# K151A9H-1), U-PLEX Immuno-Oncology Group 1 (hu) (Meso Scale Discovery, Inc. Cat# K151AEL-1) or U-Plex Human Granzyme B assay (Meso Scale Discovery, Inc. Cat# K151H8K-1) according to the manufacturer's instructions. The samples were analyzed in duplicates on a MESO QuickPlex SQ 120 imager using DISCOVERY WORKBENCH® software (Meso Scale Discovery, RRID: SCR_019192).
HIV-1-eGFP virus production
A full-length HIV-1-eGFP reporter virus was generated using a pBR-HIV-1 M NL4-3 92TH14-12 plasmid39 kindly provided by the Institute of Molecular Virology, Ulm University Medical Center. The HIV-1-eGFP viruses were generated by transfection of HEK293T cells with the pBR-HIV-1 M NL4-3 92TH14-12 plasmid using PEI MAX® transfection reagent at a 4:1 M ratio of PEI:DNA. Viral supernatants were collected 72 h post transfection and filtered through a 0.45 μm Minisart® NML Syringe Filter. The viral supernatants were concentrated through Amicon Ultra-15 100K centrifugal filters and stored at −80 °C.
HIV-1 neutralization assay
CD4+ T cells were seeded in cRPMI + IL2 at a density of 3.0 × 104 cells/well in a round-bottom 96-well plate 48 h post activation. Each scFv was 3-fold serially diluted in cRPMI + IL2 with a starting concentration of 25 μg/mL and incubated with HIV-1-eGFP at 37 °C for 1 h. ScFv-virus mixture was transferred to the cells with a MOI of 0.1. Next, the plate was centrifuged at 1250×g at 30 °C for 2 h followed by incubation at 37 °C in 5% CO2. After 24 h, the cells were fixed with 1% formaldehyde, and analyzed by flow cytometry (MACSQuant® Analyzer 16) for GFP + cells.
HIV-1 env-expressing target cell killing assay
To assess RoVER-mediated killing of target cells expressing HIV-1 envelope, in vitro cell killing assays were performed using either a Raji cell line stably expressing YU2 envelope and GFP (Raji-Env), and autologous CD4+ T cells ex vivo infected with HIV-1-eGFP. Specifically, autologous CD4+ T cells were isolated from human PBMCs and activated for 48 h in cRPMI + IL2 supplemented with 1 μg/mL PHA at 37 °C in 5% CO2. Following activation, the PHA was removed and the cells were resuspended in HIV-1-eGFP diluted in cRPMI + IL2 for a MOI of 0.05 and spinoculated at 1250×g for 2 h at 30 °C. After 24 h incubation, the GFP-expressing HIV-1-infected cells were enriched using a MACSQuant® Tyto® Cell Sorter (Miltenyi). The HIV-1-eGFP-infected autologous CD4+ T cells or Raji-Env cells were seeded in a round-bottom 96 well plate with 2000–5000 GFP + target cells/well. In this HIV-1 Env-expressing target cell killing assay, a preassembled RoVER with a scFv-10-1074 targeting HIV-1 envelope was used. CD8+ T cells obtained from YF-17D vaccinated study participants were added in different YF-specific effector to GFP + target (YF-E:T) cell ratios and incubated at 37 °C in 5% CO2. After 20 h incubation, the HIV-1-eGFP-infected cells were fixated in 1% formaldehyde and analyzed for GFP + cells using a MACSQuant® Analyzer 16. Dead target cells were thus quantified by the disappearance of GFP + cells.
Statistics
Statistical analyses were performed using GraphPad Prism version 9.2.0 (GraphPad Software, RRID: SCR_002798), and the statistical analysis methods used as well as the specifics of data presentation are reported in the figure legends. Wilcoxon test was used to analyze paired data, whereas Mann–Whitney statistical test was used to analyze unpaired data. Statistical analyses were only performed on data with five or more data points. A p-value below 0.05 was considered statistically significant.
Role of funders
The funders were not involved in the conceptualization, study design, data collection, analysis or interpretation of data, and did not contribute to the writing or submission of this paper.
Results
Production and functional assessment of the RoVER components
To demonstrate RoVER-mediated redirection of a vaccine-induced effector response towards target cells, the RoVER was assembled from a biotinylated scFv protein (targeting either CD19 or HIV-1 envelope) and joined via streptavidin to a biotin-conjugated HLA-I molecule carrying a YF-vaccine epitope. Moreover, YF epitope-specific CD8+ T cells generated by YF-17D vaccination were detected by staining with HLA-I dextramers carrying YF epitopes (Fig. 1b). ScFvs were designed to contain a C-terminal AviTag for biotinylation and His-tag for purification purposes. Production and biotinylation of the scFv proteins were verified by Western blot analysis (Fig. 1c). Densiometric analysis showed that >90% of scFv-10-1074 and scFv-CD19 proteins were biotinylated, which was a prerequisite for successful assembly of the RoVER. Target cell binding capacity of the scFv-10-1074 and scFv-CD19 proteins were verified using a human B lymphoblastoid Raji-Env cell line expressing GFP, CD19 and HIV-1 envelope (Env) (Fig. 1d).37 As expected, flow cytometry analysis showed pronounced binding of the scFv-CD19 (94.0%) to the Raji-Env cell line, approaching the level of control anti-CD19 antibody binding (99.1%). Additionally, 76% of the HIV-1 Env expressing Raji-Env target cells bound scFv-10-1074 (Fig. 1d). Similar binding assays examining scFv-CD19 and scFv-10-1074 binding to primary CD19+ B cells or HIV-1-infected primary CD4+ T cells, respectively, further validated the scFv binding capacity (Supplementary Fig. S2). Thus, we show that efficient and high-level expression of selected biotinylated scFvs can be achieved and these scFvs enable binding with high specificity to desired target cells expressing either CD19 or HIV-1 envelope.
YF-17D vaccination induces a potent response of yellow fever epitope-specific CD8+ T cells
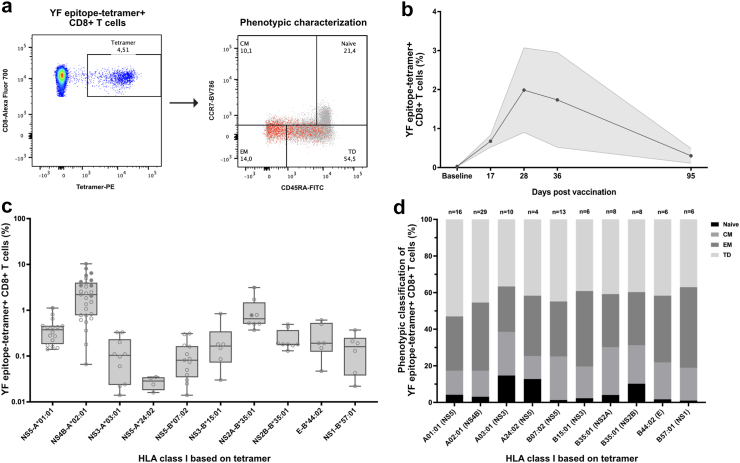
We performed vaccination of 51 healthy volunteers (Clinical trial: NCT04083430) with the live-attenuated yellow fever (YF) YF-17D vaccine (Stamaril, Novartis) to study vaccine-induced YF epitope-specific CD8+ T cell (YF CTL) responses (Fig. 2a and Supplementary Table S1).36,40, 41, 42, 43 We determined YF CTL peak responses in a small pilot longitudinal-sampling study of YF-17D vaccination of HLA-A∗02:01-positive healthy individuals (Fig. 2b). From previous publications and this pilot study, YF-17D vaccination was shown to generate a YF CTL response that peaks between day 14 and 30 following vaccination.36 Quantification of vaccine-induced YF CTLs showed that 79% (37/47 participants analyzed) had YF CTLs targeting more than one of the investigated epitopes (Fig. 2c and Supplementary Fig. S3). Furthermore, YF CTLs were detected for all the viral epitopes analyzed, thus demonstrating the broad specificity of the CTL responses induced by the YF-17D vaccine. However, the magnitude of CTL responses towards the different YF epitopes examined varied greatly among the analyzed donors regardless of HLA-I type and epitope specificity. The most potent YF CTL responses generated was observed for study participants with HLA-I types A∗01:01, A∗02:01, and B∗35:01 recognizing the HLA-I-restricted epitopes NS5286-295 (KSEYMTSWFY), NS4B214-222 (LLWNGPMAV), and NS2A4-13(HAVPFGLVSM), respectively. Quantification revealed that the most prevalent YF CTL response was observed for HLA-A∗02:01-positive study participants for which a median 2.20% (range 0.07–10.30%, n = 29) of all circulating CTLs were shown to be specific for the NS4B214-222 -restricted epitope. In comparison, NS5286-295 -specific CTL responses comprised 0.38% (median, range 0.14–1.12%, n = 16) of all circulating CTLs in HLA-A∗01:01-positive study participants, whereas 0.65% (range 0.37–3.13%, n = 8) of circulating CTLs were shown to be NS2A4-13-specific in HLA-B∗35:01-positive individuals.
Fig. 2.
Induction of potent response of yellow fever-specific CD8+ T cells by YF-17D vaccination. a) Flow cytometry gating strategy for quantification and phenotypic characterization of YF-epitope tetramer + CD8+ T cells in PBMCs obtained from YF-17D vaccinated study participants post vaccination (21 ± 3 days). The phenotypic characterization flow plot depicts the subset distribution of YF-epitope tetramer + CD8+ T cells (red) across the total CD8+ T cells (grey) based on surface staining for CD45RA and CCR7: Naïve (CD45RA+, CCR7+), central memory (CM) (CD45RA-, CCR7+), effector memory (EM) (CD45RA-, CCR7-), and terminal differentiated (TD) (CD45RA+, CCR7-). b) YF-17D vaccination pilot study showing the frequency of YF-epitope tetramer + CD8+ T cells in total CD8+ T cells from YF-17D vaccinated individuals at baseline pre-vaccination and at day 17, 28, 36 and 95 post vaccination (n = 2, data shown as mean ± SEM). c) The frequency of YF-epitope tetramer + CD8+ T cells in total CD8+ T cells from 47 vaccinated study participants obtained post vaccination (21 ± 3 days). Data is plotted according to the donor HLA class I. Data for each individual donor is depicted with circles, and filled circles denotes values for individuals of which PBMCs are used in vitro cell killing assays (refer to Fig. 3, Fig. 4, Fig. 5). Box-and-whisker plot of the net frequency of YF-epitope tetramer + CD8+ T cells according to the donor HLA class I. d) Phenotypic characterization of YF epitope tetramer + CD8+T cells from 47 YF-17D vaccinated study participants. Each bar represents the mean subset composition of YF-epitope tetramer + CD8+ T cells in individuals with the HLA class I denoted below the bar. The number of individuals in each analysis is denoted above the bars.
Further characterization of the vaccine-induced YF CTLs in terms of phenotypic distribution showed a similar pattern across the different HLA-I-types and epitope specificities with the terminal differentiated (TD) and effector memory (EM) subsets comprising the majority of YF CTLs (Fig. 2d and Supplementary Fig. S3). Specifically, NS5-A∗01:01-specific CTLs showed a phenotypic distribution of 54% TD and 30% EM (n = 16), NS4B-A∗02:01-specific CTLs comprised 46% TD and 37% EM (n = 29), and NS2A-B∗35:01-tetramer + CTLs showed a phenotypic distribution of 41% TD and 29% EM (n = 8). The dominating TD and EM CTL phenotypic subsets are characterized by high cytotoxic potential and the capacity for fast and potent effector functions upon antigen encounter.
In conclusion, YF-17D vaccination induced strong YF CTL responses which demonstrated broad specificity capable of targeting several YF viral epitopes. Regardless of HLA-type and epitope-specificity, the vaccine-induced YF-specific CTLs were predominantly of the TD and EM phenotypes. Based on the predominance of HLA-I-type A∗02:01 in the study population as well as the potent induction of vaccine epitope-specific CTL responses in individuals carrying the HLA-I alleles A∗01:01, A∗02:01, and B∗35:01 targeting the restricted epitopes NS5286-295, NS4B214-222, and NS2A4-13, these HLA-types and epitopes were selected for the functional assessment of the RoVER technology.
RoVER is able to redirect vaccine-induced effector cells to mediate killing of CD19+ target cells
To evaluate the RoVER technology, we initially tested the capacity of the RoVER to mediate state-of-the-art in vitro elimination of CD19+ B target cells by redirection of vaccine-induced YF CTLs. As control we used the FDA-approved BiTE, blinatumomab (Blincyto, Amgen) that is used to treat B cell leukemias by activation of CD3+ T cells to eliminate target B cells through CD19-CD3 linkage. In target cell killing assays, we were able to redirect the vaccine-induced YF epitope-specific effector response towards target CD19+ B cells using the RoVER(CD19), and achieved a dose-dependent effect of 36%, 41% and 54% (background adjusted) killing at YF-specific effector-to-target (YF-E:T) ratios of 1:1, 3:1 and 5:1, respectively (Fig. 3b). In contrast, no CD19+ B cell killing was observed without exposure to the RoVER(CD19) or upon exposure to scFv-CD19-streptavidin alone, thereby validating the specificity of the RoVER. This level of RoVER(CD19)-mediated target cell killing was comparable to killing assays using blinatumomab in which 52% (background adjusted) killing of CD19+ B target cells was achieved at effector-to-target (E:T) ratio 1:1 and 71% target killing at E:T 20:1 (Fig. 3c). The higher E:T ratios investigated for blinatumomab-mediated target cell killing was a consequence of the engagement of blinatumomab with all CD3+ T cells irrespective of antigen-specificity.
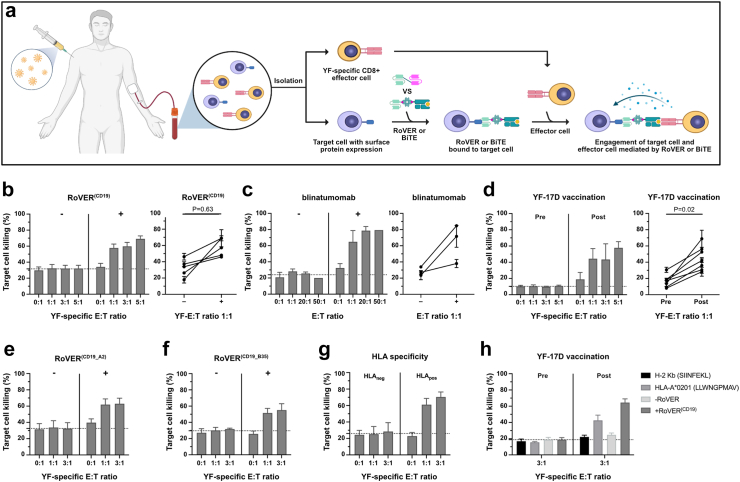
Fig. 3.
Specific killing of primary B target cells by autologous CD8+ T cells mediated by RoVER(CD19). a) Graphics of the workflow for in vitro target cell killing assays using the RoVER technology (Created with BioRender.com). b)In vitro cell killing assays investigating RoVER(CD19)-mediated killing of primary target CD19+ B cells using autologous CD8+ T cells obtained from YF-17D vaccinated study participants (n = 5). c)In vitro target cell killing of CD19+ B cells by autologous CD8+ T cells from healthy donor PBMCs using the FDA-approved BiTE, blinatumomab (n = 3). d) The dependence of RoVER(CD19)-mediated target cell killing for YF epitope-specific CD8+ T cells was investigated in vitro target cell killing assays using CD8+ T cells obtained pre and post YF-17D vaccination (21 ± 3 days) (bar graph: n = 3, XY-graph: n = 7). To investigate potential differences in RoVER-mediated killing capacity using two bispecific RoVER(CD19) containing different HLA class I-restricted YF epitopes together with autologous cells obtained from YF-17D vaccinated donors with matched HLA class I, we investigated in vitro target CD19+ B cell killing assays using a e) bispecific RoVER(CD19_A2) engaging CD19+ B target cells and NS4B-HLA-A∗02:01-specific CD8+ T effector cells (n = 3), or f) bispecific RoVER(CD19_B35) containing a NS2B-HLA-B∗35:01 domain (n = 2). g) HLA class I-specificity of RoVER(CD19)-mediated killing was tested in vitro cell killing assays using RoVER(CD19) containing a HLA class I domain matched to (HLApos) or different from (HLAneg) the donor HLA class I (n = 3). h)In vitro cell killing assays investigating peptide- and RoVER(CD19)-mediated killing of B cells using autologous CD8+ T cells obtained pre (n = 3) and post vaccination (21 ± 3 days) (n = 3, LLWNGPMAV; n = 4, SIINFEKL, −/+ RoVER). Data are the percentage of dead (Live/Dead stain) target cells (Celltrace+) out of total target cells shown as mean ± SEM for all donors (bar graph) or for individual donors (XY-graph) at the E:T or YF-E:T ratios indicated. p-values were calculated using Wilcoxon test. The dotted line is calculated based on the bars on the left side of each diagram and indicates the mean background target cell death.
Next, we wanted to test the dependence of RoVER-mediated killing on vaccine-induced YF CTLs. We compared the killing of CD19+ B cells by autologous CD8+ T cells obtained pre and post YF-17D vaccination. RoVER(CD19)-mediated CD19+ B cell killing of 33% was obtained at YF-E:T ratio 1:1 upon exposure to CTLs obtained from study participants post vaccination and thus after induction of a YF-specific effector response. The target cell killing increased with increasing numbers of effector cells reaching 45% target cell killing at YF-E:T 5:1. In contrast, no increase in target cell killing was observed compared to background using autologous CD8+ T cells obtained pre vaccination (Fig. 3d).
Collectively, we showed that the RoVER(CD19) was able to both recruit and activate YF-specific CD8+ T cells leading to target CD19+ B cell killing in a range comparable to the FDA-approved BiTE, blinatumomab. Further, RoVER(CD19)-mediated target cell killing was dependent on both the presence of the bispecific RoVER(CD19) and vaccine-induced YF CTLs.
RoVER-mediated target killing can be achieved using various HLA-I molecules and vaccine epitopes
To investigate the broader utility of the RoVER(CD19) technology with various HLA-I types, in vitro target cell killing assays were performed using RoVER(CD19) comprising different HLA-I domains together with vaccine-induced YF CTLs from study participants with a variety of HLA-haplotypes.
Target cell killing could be achieved using RoVER(CD19) comprising either NS4B-HLA-A∗02:01 or NS2B-HLA-B∗35:01 with specific CD19+ B cell killing of 41% and 30%, respectively, at YF-E:T ratio 1:1 (Fig. 3e and f). To confirm that target cell killing was HLA-I dependent, in vitro target cell killing assays were performed using RoVER(CD19) containing either HLA-I domains matching (HLApos) or not-matching (HLAneg) the HLA-I haplotype of the donor (Fig. 3g). In this setup, donors with mixed HLA-I haplotype combinations (HLA-A2+/HLA-B35-, and HLA-A1+/HLA-A2-) were used to account for potential differences in immunogenicity of the presented YF peptides. For HLA-A2+/HLA-B35- individuals, a RoVER(CD19_A2) comprising a NS4B-HLA-A∗02:01 domain was used as a matching RoVERpos, whereas a RoVER(CD19_B35) comprising a NS2B-B∗35:01 was used as an unspecific RoVERneg. Likewise, for HLA-A1+/HLA-A2- individuals, a RoVER (CD19_A1) comprising a NS5-A∗01:01 domain was used as RoVERpos and a RoVER (CD19_A2) as RoVERneg. No target cell killing was observed using a bispecific RoVER with an unmatched HLA-I domain (RoVERneg), whereas a marked increase in background adjusted target cell killing of 47% at YF-E:T 1:1 and 57% at YF-E:T 3:1 were achieved using a matching bispecific RoVER (RoVERpos) (Fig. 3g).
In conclusion, RoVER-mediated target cell killing was HLA-I dependent and similar levels of in vitro killing could be achieved using RoVERs containing different HLA-I molecules and vaccine epitopes.
Subsequently, we wanted to evaluate the effect of RoVER on the killing capacity of YF-specific CD8+ T cells. We performed parallel RoVER(CD19)- and peptide-mediated target cell killing of CD19+ B cells by autologous CD8+ T cells obtained pre and post YF-17D vaccination. The level of target cell killing obtained upon exposure to RoVER(CD19) and LLWNGPMAV using CTLs obtained post vaccination was 60% and 35%, respectively at YF-E:T ratio 3:1. In contrast, no CD19+ B cell killing was observed without exposure to RoVER(CD19) or upon exposure to irrelevant control peptide SIINFEKL (Fig. 3h). This validates specific stimulation of NS4B-HLA-A∗02:01 specific CD8+ T cells following exposure of RoVER(CD19) and LLWNGPMAV. Additionally, the data show that RoVER-mediated target cell killing does not negatively affect the killing capacity of the effector cells compared to YF peptide pulsed cells.
Degranulation and cytokine release in target cell killing assays
The speicificty of RoVER-mediated target cell killing was further evaluated by flow cytometric detection of degranulation. Quantifcation of CD107α surface expressing YF CTLs showed degranulated YF-specific CD8+ T cells following RoVER-mediated killing of CD19+ B cells by autologous CD8+ T cells obtained post vaccination. Stimulation without RoVER and with CD8+ T cells obtained pre vaccination did not result in CD107α surface expression. Hence, CD107α degranulation was specific to both vaccine-induced YF CTLs and the presence of RoVER(CD19) (Fig. 4a and Supplementary Fig. S5).
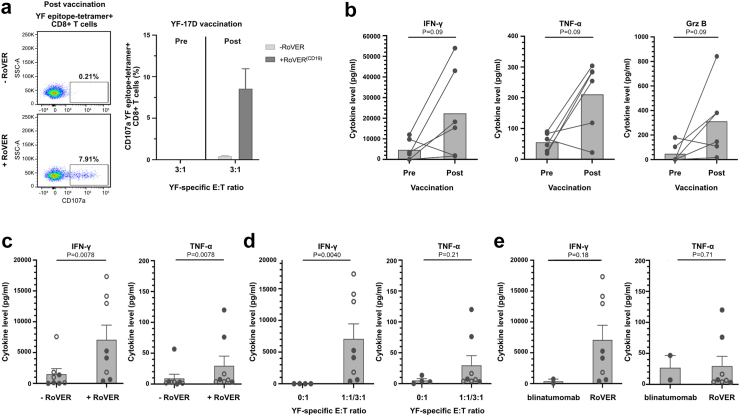
Fig. 4.
Degranulation and cytokine release from in vitro target killing assays using blinatumomab or RoVER. Detetction of CD107α degranulation and Mesoscale multiplex analyses of supernatants from in vitro cell killing assays. a) Representative flow cytometry plots of CD107α surface expression following in vitro cell killing assay showing RoVER-induced degranulating YF epitope-specific CD8+ T cells (left). Percentage of degranulating YF epitope-specific CD8+ T cells (n = 4) (right). b) Analysis of IFN-γ, TNF-α or Grz B in supernatant from RoVER-mediated killing of target cells using PBMCs obtained pre and post vaccination (21 ± 3 days) (n = 6, 100,000 PBMCs/well) (). IFN-γ or TNF-α cytokine release from RoVER-mediated target cell killing assays c) with or without exposure to RoVER (n = 8) or d) at YF-E:T 0:1 or 1:1 (n = 4, solid circles) and 3:1 (n = 4, open circles) using YF epitope-specific CD8+ T cell obtained from study participants post YF-17D vaccination (21 ± 3 days) (n = 8). e) Comparison of IFN-y and TNF-a release from in vitro target cell killing using blinatumomab (n = 2) or RoVER (n = 8). Data are mean ± SEM. p-values were calculated using Wilcoxon test when comparing paired data and Mann–Whitney statistical test when comparing unpaired data.
The induction of an effector response was further assessed by the level of cytokines released from RoVER-mediated killing of target cells. Assessing a pool of PBMCs obtained from study participants post-vaccination, we observed pronounced induction of the cytokines Interferon-gamma (IFN-γ), Tumor Necrosis Factor-alpha (TNF-α), and Granzyme B (Grz B) upon RoVER(CD19)-mediated target cell killing with a fold change of 3.8, 2.8 and 5.7, respectively, compared to using PBMCs obtained pre-vaccination (Fig. 4b).
Further investigations into cytokine release upon RoVER-mediated killing of target cells showed that induction of IFN-γ and TNF-α secretion was dependent on exposure to the RoVER(CD19) (Fig. 4c) as wells as vaccine-induced CD8+ T cells (Fig. 4d). This was evident from pronounced increases in cytokine secretion upon RoVER(CD19)-mediated killing of B cells with a fold change of 5.8 IFN-γ and 2.2 TNF-α at YF-E:T ratio 1:1 compared to conditions without exposure to RoVER(CD19) (Fig. 4c), and fold changes of 665.0 IFN-γ and 9.0 TNF-α compared to conditions without the presence of CD8+ T effector cells (Fig. 4d). Moreover, the cytokine response induced upon RoVER-mediated target cell killing was in a comparable range to cytokine release upon blinatumomab-mediated target cell killing with fold changes of 5.7 IFN-γ and 0.92 TNF-α at E:T ratio 1:1 (Fig. 4e). The level of cytokines released from peptide-mediated killing of target cells was also assessed to further compare the effector response induced by RoVER (CD19) and YF peptide pulsed cells. In agrrement with the findings in Fig. 3h, the induction of IFN-γ, TNF-α, and Grz B from RoVER-mediated killing of target cells was in a range comparable to the release from LLWNGPMAV pulsed killing (Supplementary Fig. S4).
In summary, RoVER-mediated target cell killing was associated with CD107α surface upregulation and pronounced release of the cytokines IFN-γ, TNF-α and Grz B, which is in accordance with the induction of a potent effector response and cytolysis of target cells.
The RoVER technology is adaptable
Next, we wanted to demonstrate that the RoVER technology can be adapted to various targets by altering the scFv domain of the RoVER. Therefore, we replaced the scFv-CD19 with the HIV-1 envelope-specific scFv-10-1074.
To assess the functional efficacy of the scFv-10-1074 used in the assembly of the RoVER(Env), we show the capacity of the scFv-10-1074 to specifically bind HIV-1 Env-expressing cells (Fig. 1d and Supplementary Fig. S2) and neutralize cell-free HIV-1 (Fig. 5a). The scFv-10-1074 was able to neutralize full-length HIV-1-eGFP and prevent infection of primary CD4+ T cells showing an IC50 of 0.8 μg/mL. This scFv-10-1074 neutralization of HIV-1-eGFP was dose-dependent and envelope-specific as a scFv-CD19 was unable to neutralize the virus (Fig. 5a).
Fig. 5.
Specific killing of HIV-1 envelope-expressing target cells using the RoVER technology. a) Neutralization data for scFv-10-1074 and scFv-CD19 showing specific binding of scFv-10-1074 to HIV-1 envelope (n = 3). Normalized to HIV-1-eGFP-infected CD4+ T cell control without scFv. b) Graphics of the workflow for killing of HIV-1-infected CD4+ target cells by autologous CD8+ T cells using RoVER(Env) (Created with BioRender.com). c) Flow cytometry gating strategy for HIV-1 target cell killing assays. d) Fluorescence microscopy images of killing assay wells for the autologous setup with HIV-1-eGFP-infected CD4+ cells at different YF-E:T ratios with or without exposure to RoVER(Env). HIV-1-eGFP-infected target cells are identified by eGFP expression (green). RoVER(Env)-mediated killing of e) Raji-Env cells expressing HIV-1 envelope and GFP (n = 3) or f) autologous CD4+ T cells obtained from YF-17D vaccinated study participants and ex vivo infected with HIV-1-eGFP (n = 3). g) RoVER(CD4)-mediated killing of autologous CD4+ T cells (n = 2). All HIV-1 killing assay data shows the disappearance of HIV-1 envelope-expressing target cells (GFP+) normalized to the number of target cells in control wells without exposure to RoVER(Env). CD4 killing data shows the percentage of dead (Live/Dead stain) target cells (Celltrace+). The killing data are mean ± SEM for all donors (bar graph) or for individual donors (XY-graph) at indicated YF-E:T ratios. The dotted line is calculated based on the bars on the left side of each diagram and indicates the mean background target cell death.
For RoVER(Env)-mediated killing of HIV-1 Env-expressing cells, both a Raji-Env cell line37 expressing HIV-1 Env and GFP, and autologous CD4+ cells ex vivo infected with a full-length HIV-1-eGFP reporter virus were used as target cells. RoVER(Env)-mediated killing of HIV-1-infected or HIV-1 Env-expressing cells could thus be assessed by the disappearance of GFP + cells (Fig. 5b–d). Using the RoVER(Env), we were able to redirect the vaccine-induced YF CTL effector response to target Raji-Env cells achieving 22% and 32% killing at YF-E:T ratios 1:1 and 3:1, respectively (Fig. 5e). Importantly, when infecting autologous CD4+ T cells with full-length HIV-1-eGFP, RoVER-linked vaccine-induced CD8+ T cells were able to mediate specific killing of 53% at YF-E:T ratio 1:1 and 65% target killing at YF-E:T ratio 3:1 (Fig. 5f).
As a proof of RoVER adaptability, we also assembled a RoVER(CD4) using a biotin-conjugated anti-CD4 antibody to enable assessment of RoVER-mediated CD4+ target cell killing. As for CD19+ and HIV-1 Env + target cell killing, RoVER(CD4) showed equal capacity to eliminate CD4+ T cells achieving 64% killing at YF-E:T ratio 1:1 (Fig. 5g).
Collectively, the RoVER technology can be adapted to redirect effector responses towards distinct cell surface antigens of interest including CD19+ B cells, CD4+ T cells and HIV-1 Env-expressing cells by changing the scFv domain of the bispecific RoVER.
Discussion
Here we present an adaptive immunotherapy concept for redirection of vaccine-induced effector responses towards specified cellular targets through our bispecific RoVER. We provide data to show the ability to redirect vaccine-induced CD8+ T cells with different TCR specificities and further highlight the capacity of the technology to enable targeting cells of choice.
Adoptive CAR T cell therapy rely on the capacity to instill a new antigen specificity in autologous or allogeneic CD8+ T cells through genetic engineering of the cells to express a synthetic chimeric antigen receptor. This treatment require leukapheresis followed by ex vivo genetic manipulation and expansion of the cells prior to reinfusion of the genetically modified T cells. Usually, around 108 CAR T cells are reinfused into the patient.44 Here we adopt a different approach by exploring the capacity of the immune system for in vivo clonal expansion following simple vaccination. We use the YF-17D vaccine which has been described to contain several immunodominant epitopes that drive highly expanded CD8+ T cell specificities.36,40, 41, 42, 43,45 One such immunodominant epitope is the YF NS4B214-222 response observed in HLA-A∗02:01 individuals. We show that on average 2.2% (and up to 10.3%) of total circulating CD8+ T cells are specific for this single epitope following vaccination. With an estimated total body load of approximately 4 × 1011 CD8+ T cells in an adult person46 and assuming a blood sample represents the distribution in primary and secondary immune tissues, the YF NS4B214-222 epitope-specific effector cells would comprise approximately 1010 CD8+ T cells in an HLA-A∗02:01 vaccinated individual. Furthermore, the vast majority (>80%) of these NS4B214-222 epitope-specific CD8+ T cells display an effector memory or terminal differentiated effector phenotype that is committed to immune killing. Thus, by using the chimeric RoVER with a recombinant HLA-I displaying the NS4B214-222 epitope described here, such combination strategy provides ample effector cells to mediate target cell killing in vivo. Thus, the requirement for adoptive transfer of vast numbers of CTLs is obviated in the RoVER technology by exploiting the in vivo generation of potent CTL immune responses induced by simple vaccination.
To investigate the personalized tissue type-specificity of the RoVER technology, we explored whether the RoVER technology was applicable to various HLA-I types and YF epitopes. Using RoVER with either A∗01:01-restricted NS5286-295, A∗02:01-restricted NS4B214-222, and B∗35:01-restricted NS2A4-13, we demonstrated that RoVER-mediated target cell killing was specific for the HLA-I type of the donor. Importantly, identical levels of target cell killing could be achieved using three different HLA-I-matched YF epitopes indicating similar cell-intrinsic killing capacities independent of vaccine epitope and HLA-I background. This is important for the utility of the RoVER technology as it needs to be adapted dependent on the individual HLA-I types.
HIV-1 persistence despite effective antiretroviral therapy (ART) is a major barrier to achieve viral eradication. A major focus of HIV-1 cure research is to harness immunotherapy strategies and CD8+ T cells in particular to mediate HIV-1-infected cell killing. Clearance of HIV-1 infected cells is often limited by a dysfunctional immune effector response. The RoVER technology generates a de novo vaccine-induced effector response towards a vaccine-epitope unrelated to HIV-1 and thus not per se limited by exhaustion. ScFv-guided redirection of this potent and functional de novo vaccine-induced effector response towards target cells offers a potential mechanism to overcome immune exhaustion.
A requirement for effective treatment of HIV-1 with the RoVER technology is the expression of cell surface antigens specifically targeted by the scFv domain of the RoVER. HIV-1 envelope surface expression easily discriminates infected cells from healthy cells although levels of HIV-1 Env expression is dependent on cell activity and viral replication. Further, resistant viral strains and the high mutation rate of HIV-1 may also compromise the binding capacity of the RoVER scFv domain and thus challenge the effectivity of the RoVER technology. Consequently, by designing the RoVER against target cells using a scFv domain targeting multiple clades of HIV-1, or by using combinations of different scFv domains targeting distinct HIV-1 epitopes, we may reduce the risk of immune escape from the RoVER technology. Intriguingly, although HIV-1-infected cells only have low surface expression of HIV-1 envelope,47,48 we observed potent RoVER-mediated killing of autologous CD4+ T cells ex vivo infected with a full-length HIV-1-eGFP.
An important challenge of immunotherapeutic strategies using bispecific molecules that engage CD3 on the entire lymphocyte population (such as blinatumomab) is the risk of excessive immune activation4,5,24, 25, 26 sometimes leading to the development of cytokine release syndrome which can be fatal and thus limits the clinical application of these technologies. This excessive immune activation is often a consequence of broad engagement of the entire T cell compartment irrespective of lineage commitment or phenotype. Since the RoVER specifically engages and activates only vaccine epitope-specific CTLs and not the entire CD3+ T cell compartment, this technology may avoid excessive immune activation. Moreover, treatment with the RoVER technology, in contrast to CAR T cell therapy, can be stopped at any time in the event of associated toxicities, since this would simply leave the patient with a natural expanded T lymphocyte memory pool that will generate natural immune memory. In contrast, the technology may have limitation in terms of both potency and effector cell availability in vivo and as the technology relies on epitope-specific immunity a risk of immune waining may also diminish the long-term utility.
The RoVER technology is dependent on preconditioning with a vaccination to generate an expansion of vaccine epitope-specific effector cells. Vaccination with the live-attenuated YF-17D as used here is not recommended in individuals with impaired immunity.49 HIV-1-infected individuals with a CD4+ T cell count above 200/mm3 and on suppressive antiretroviral treatment are not advised against vaccination with the live-attenuated YF-17D vaccine.49 Here, YF-vaccination was used as proof-of-principle to generate a CD8+ T cell response that can be redirected towards a different antigen via the RoVER technology. Importantly, there are also several inactivated YF vaccines being developed as potential alernatives to the live-attenuated YF-17D vaccine50, 51, 52, 53 for use in vulnerable patient populations. Moreover, in antigen-experienced people, the potency of recall effector responses induced upon re-vaccination needs to be addressed. Lastly, this technology needs to be evaluated in appropriate animal models to explore efficacy. However, the choice of model system will be important as the technology and all the in vitro data presented here rely on a haplotype match. Therefore, the translation to animal models need to take immunodominat epitope induction as well as haplotype-mathed RoVER molecule into consideration.
Our RoVER technology offers a simple mechanism for induction and redirection of a CTL effector response. By simple vaccination, this immunotherapy strategy is able to generate vast numbers of CTLs in vivo and thus obviates the need for adoptive transfer by exploiting the patient's own immune system. Importantly, while several existing bispecific immunotherapies induce broad T cell activation to eliminate target cells, the RoVER specifically engage only vaccine epitope-specific CTLs and thereby may reduce the risk of excessive immune activation and the development of cytokine release syndrome. Moreover, by scFv-guided redirection of vaccine-induced CTLs towards target cells, the RoVER technology may be able to overcome the obstacle of HLA-I downregulation often observed in chronic HIV-1 infection and during tumor progression. Besides relying on a safe and well-tested intervention (vaccination) as a simple means to generate vast numbers of effector CD8+ T cells in vivo, another promise of this technology is the adaptability allowed by altering the scFv-binding domain of the RoVER thus targeting a cell-surface antigen of choice.
Contributors
Conceptualization, MT, CVK, MHS; Methodology, MT, MHS, CVK, EFI, JDG, AH, RH; Investigation, CVK, EFI, MHS, IM, AH, JDG; Resources, OSS, MT, LJØ; RH; Writing manuscript, CVK, MT, EFI; Visualization, CVK, EFI, MHS, JDG; Supervision, MT, MHS, OSS; Project Administration, MT; Funding Acquisition, MT. All authors have read, edited, and approved the final manuscript.
Data sharing statement
The published article includes all data generated and analysed for this study. Further information and requests for resources, reagents or primary data are available on request from the lead contact, Martin Tolstrup (marttols@rm.dk).
Declaration of interests
The authors declare no competing interests. CVK, JDG, MHS and MT are listed as inventors on a patent application filed by Aarhus University.
Acknowledgements
We would like to show our great appreciation to the study participants who devoted their time for research and kindly donated their biological material for this study. We would like to thank Institute of Molecular Virology, Ulm University Medical Center for providing HIV-1-eGFP plasmids and Pasteur Institute for providing the Raji-Env cells used in this study. This study was funded by the Novo Nordisk Foundation (Hallas Møller NNF10OC0054577 to MT).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104785.
Appendix A. Supplementary data
Figure S1. Molecular mechanism of RoVER. Graphics showing the molecular mechanism of RoVER. The biotinylated (4) scFv domain (3) of the RoVER binds specific cell surface antigens (2) on target cells (1) and is joined via streptavidin molecule (5) to a biotin-conjugated (4) HLA-I domain (6) carrying a highly immunogenic vaccine-epitope (7) for specific engagement and activation of vaccine epitope-specific CD8+ T cells (9) through T cell receptor (8) recognition. The graphics is created with BioRender.com.
Figure S2. scFv binding capacity to primary CD19+ B cells or HIV-1-eGFP-infected CD4+ T cells. a) Flow cytometry analysis of scFv-CD19 and scFv-10-1074 binding to CD19+ B cells or HIV-1-eGFP-infected (GFP+) CD4+ T cells. Flow cytometry analysis of scFv-CD19 and scFv-10-1074 binding capacity to CD19+ B cells and HIV-1-eGFP-infected CD4+ T cells, respectively. Controls included unstained cells and cells stained with Streptavidin-BV421. ScFv-10-1074 binding capacity to HIV-1-eGFP-infected cells is shown for gated GFP+ cells. b) Flow cytometry analysis of scFv-10-1074 binding specificity to HIV-1-eGFP-infected or uninfected CD4+ T cells. Flow cytometry analysis of scFv-10-1074 binding capacity to HIV-1-eGFP-infected or uninfected CD4+ T cells. Controls included unstained cells and cells stained with Streptavidin-BV421. ScFv-10-1074 binding capacity is shown for total CD4+ T cells including uninfected (GFP-) cells.
Supplementary table 1. Study participant demographics. Table showing demographic characteristics of the clinical study participants (NCT04083430) receiving YF-17D vaccination from which PBMCs were obtained for functional assays investigating the RoVER technology.
Figure S3. Flow gating strategy for quantification of tetramer+ CD8+ T cells and phenotypic characterization. Flow cytometry gating strategy for quantification of tetramer+ CD8+ T cells in PBMCs isolated from YF-17D vaccinated study participants. Quantification was achieved by tetramer staining and multicolor flow cytometry using a live/dead stain along with fluorophore-conjugated antibodies against CD3 and CD8 T cell markers and using various HLA-I tetramers carrying immunogenic YF peptides. The CD8+ T cells were further characterized in respect to phenotype based on the expression of the surface markers CD45RA and CCR7 into the distinct cellular subsets: Naïve (N) (CCR7+, CD45RA+), Central memory (CM) (CCR7+, CD45RA-), Effector memory (EM) (CCR7-, CD45RA-) and Terminally differentiated (TD) (CCR7-, CD45RA+) CD8+ T cells. Furthermore, back gating of the tetramer+ CD8+ T cells allowed for quantification of the distribution of tetramer+ CD8+ T cells (red) across the phenotypic subsets of total CD8+ T cells (grey).
Figure S4. Cytokine release from in vitro target killing assays using peptides or RoVER. Mesoscale multiplex analyses of supernatants from in vitro cell killing assays. Analysis of IFN-γ, TNF-α or Grz B in supernatant from peptide- or RoVER-mediated killing of B cells by autologous CD8+ T cells obtained pre (n = 3) and post vaccination (21 ± 3 days) (n = 3, LLWNGPMAV; n = 4, SIINFEKL, −/+ RoVER). Data are mean ± SEM.
Figure S5. Flow gating strategy for detection of CD107α degranulation. Flow cytometry gating strategy for quantification of CD107α+ tetramer+ CD8+ T cells in in vitro cell killing assays. Representative analysis of CD107α surface expression following RoVER-mediated killing of B cells cells by autologous CD8+ T cells obtained post vaccination (21 ± 3 days). Quantification was achieved by tetramer staining and multicolor flow cytometry using a live/dead stain along with fluorophore-conjugated antibodies against CD3 and CD8 T cell markers and using various HLA-I tetramers carrying immunogenic YF peptides. A threshold of 3 events in the CD107α gate was set to discriminate the positive signal from the background.
S6_ Supplemental Western blots.
S_6Supplemental Western blots_Fig 1C_Full blots.
References
- 1.Kalos M., Levine B.L., Porter D.L., et al. T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci Transl Med. 2011;3(95) doi: 10.1126/scitranslmed.3002842. 95ra73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brentjens R.J., Davila M.L., Riviere I., et al. CD19-Targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci Transl Med. 2013;5(177):177ra38. doi: 10.1126/scitranslmed.3005930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neelapu S.S., Locke F.L., Bartlett N.L., et al. Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N Engl J Med. 2017;377(26):2531–2544. doi: 10.1056/NEJMoa1707447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maude S.L., Laetsch T.W., Buechner J., et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N Engl J Med. 2018;378(5):439–448. doi: 10.1056/NEJMoa1709866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuster S.J., Svoboda J., Chong E.A., et al. Chimeric antigen receptor T cells in refractory B-cell lymphomas. N Engl J Med. 2017;377(26):2545–2554. doi: 10.1056/NEJMoa1708566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abramson J.S., Irwin K.E., Frigault M.J., et al. Successful anti-CD19 CAR T-cell therapy in HIV-infected patients with refractory high-grade B-cell lymphoma. Cancer. 2019;125(21):3692–3698. doi: 10.1002/cncr.32411. [DOI] [PubMed] [Google Scholar]
- 7.Bargou R., Leo E., Zugmaier G., et al. Tumor regression in cancer patients by very low doses of a T cell-engaging antibody. Science. 2008;321(5891):974–977. doi: 10.1126/science.1158545. [DOI] [PubMed] [Google Scholar]
- 8.Viardot A., Goebeler M.-E., Hess G., et al. Phase 2 study of the bispecific T-cell engager (BiTE) antibody blinatumomab in relapsed/refractory diffuse large B-cell lymphoma. Blood. 2016;127(11):1410–1416. doi: 10.1182/blood-2015-06-651380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dufner V., Sayehli C.M., Chatterjee M., et al. Long-term outcome of patients with relapsed/refractory B-cell non-Hodgkin lymphoma treated with blinatumomab. Blood Adv. 2019;3(16):2491–2498. doi: 10.1182/bloodadvances.2019000025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hale M., Mesojednik T., Romano Ibarra G.S., et al. Engineering HIV-resistant, anti-HIV chimeric antigen receptor T cells. Mol Ther. 2017;25(3):570–579. doi: 10.1016/j.ymthe.2016.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scholler J., Brady T.L., Binder-Scholl G., et al. Decade-long safety and function of retroviral-modified chimeric antigen receptor T cells. Sci Transl Med. 2012;4(132):132ra53. doi: 10.1126/scitranslmed.3003761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deeks S.G., Wagner B., Anton P.A., et al. A phase II randomized study of HIV-specific T-cell gene therapy in subjects with undetectable plasma viremia on combination antiretroviral therapy. Mol Ther. 2002;5(6):788–797. doi: 10.1006/mthe.2002.0611. [DOI] [PubMed] [Google Scholar]
- 13.Mitsuyasu R.T., Anton P.A., Deeks S.G., et al. Prolonged survival and tissue trafficking following adoptive transfer of CD4ζ gene-modified autologous CD4+ and CD8+ T cells in human immunodeficiency virus–infected subjects. Blood. 2000;96(3):785–793. [PubMed] [Google Scholar]
- 14.Anthony-Gonda K., Bardhi A., Ray A., et al. Multispecific anti-HIV duoCAR-T cells display broad in vitro antiviral activity and potent in vivo elimination of HIV-infected cells in a humanized mouse model. Sci Transl Med. 2019;11(504) doi: 10.1126/scitranslmed.aav5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leibman R.S., Richardson M.W., Ellebrecht C.T., et al. Supraphysiologic control over HIV-1 replication mediated by CD8 T cells expressing a re-engineered CD4-based chimeric antigen receptor. PLoS Pathog. 2017;13(10) doi: 10.1371/journal.ppat.1006613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wherry E.J. T cell exhaustion. Nat Immunol. 2011;12(6):492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- 17.Wherry E.J., Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol. 2015;15(8):486–499. doi: 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fenwick C., Joo V., Jacquier P., et al. T-cell exhaustion in HIV infection. Immunol Rev. 2019;292(1):149–163. doi: 10.1111/imr.12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwartz O., Maréchal V., Gall S.L., Lemonnier F., Heard J.-M. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV–1 Nef protein. Nat Med. 1996;2(3):338–342. doi: 10.1038/nm0396-338. [DOI] [PubMed] [Google Scholar]
- 20.Collins K.L., Chen B.K., Kalams S.A., Walker B.D., Baltimore D. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature. 1998;391(6665):397–401. doi: 10.1038/34929. [DOI] [PubMed] [Google Scholar]
- 21.Dhatchinamoorthy K., Colbert J.D., Rock K.L. Cancer immune evasion through loss of MHC class I antigen presentation. Front Immunol. 2021;12(469) doi: 10.3389/fimmu.2021.636568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang C.-C., Pirozzi G., Wen S.-H., et al. Multiple structural and epigenetic defects in the human leukocyte antigen class I antigen presentation pathway in a recurrent metastatic melanoma following immunotherapy. J Biol Chem. 2015;290(44):26562–26575. doi: 10.1074/jbc.M115.676130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maeurer M.J., Gollin S.M., Storkus W.J., et al. Tumor escape from immune recognition: loss of HLA-A2 melanoma cell surface expression is associated with a complex rearrangement of the short arm of chromosome 6. Clin Cancer Res. 1996;2(4):641–652. [PubMed] [Google Scholar]
- 24.Sterner R.C., Sterner R.M. CAR-T cell therapy: current limitations and potential strategies. Blood Cancer J. 2021;11(4):69. doi: 10.1038/s41408-021-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park J.H., Rivière I., Gonen M., et al. Long-term follow-up of CD19 CAR therapy in acute lymphoblastic leukemia. N Engl J Med. 2018;378(5):449–459. doi: 10.1056/NEJMoa1709919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teachey D.T., Rheingold S.R., Maude S.L., et al. Cytokine release syndrome after blinatumomab treatment related to abnormal macrophage activation and ameliorated with cytokine-directed therapy. Blood. 2013;121(26):5154–5157. doi: 10.1182/blood-2013-02-485623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogg G.S., Dunbar P.R., Cerundolo V., McMichael A.J., Lemoine N.R., Savage P. Sensitization of tumour cells to lysis by virus-specific CTL using antibody-targeted MHC class I/peptide complexes. Br J Cancer. 2000;82(5):1058–1062. doi: 10.1054/bjoc.1999.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmittnaegel M., Levitsky V., Hoffmann E., et al. Committing cytomegalovirus-specific CD8 T cells to eliminate tumor cells by bifunctional major histocompatibility class I antibody fusion molecules. Cancer Immunol Res. 2015;3(7):764–776. doi: 10.1158/2326-6066.CIR-15-0037. [DOI] [PubMed] [Google Scholar]
- 29.Noy R., Haus-Cohen M., Oved K., Voloshin T., Reiter Y. Recruitment of oligoclonal viral-specific T cells to kill human tumor cells using single-chain antibody-peptide-HLA fusion molecules. Mol Cancer Ther. 2015;14(6):1327–1335. doi: 10.1158/1535-7163.MCT-14-0467. [DOI] [PubMed] [Google Scholar]
- 30.Robert B., Guillaume P., Luescher I., Romero P., Mach J.-P. Antibody-conjugated MHC class I tetramers can target tumor cells for specific lysis by T lymphocytes. Eur J Immunol. 2000;30(11):3165–3170. doi: 10.1002/1521-4141(200011)30:11<3165::AID-IMMU3165>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 31.Lev A., Novak H., Segal D., Reiter Y. Recruitment of CTL activity by tumor-specific antibody-mediated targeting of single-chain class I MHC-peptide complexes. J Immunol. 2002;169(6):2988–2996. doi: 10.4049/jimmunol.169.6.2988. [DOI] [PubMed] [Google Scholar]
- 32.Schütz C., Varela J.C., Perica K., Haupt C., Oelke M., Schneck J.P. Antigen-specific T cell Redirectors: a nanoparticle based approach for redirecting T cells. Oncotarget. 2016;7(42):68503–68512. doi: 10.18632/oncotarget.11785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mous R., Savage P., Remmerswaal E.B.M., Van Lier R.A.W., Eldering E., Van Oers M.H.J. Redirection of CMV-specific CTL towards B-CLL via CD20-targeted HLA/CMV complexes. Leukemia. 2006;20(6):1096–1102. doi: 10.1038/sj.leu.2404185. [DOI] [PubMed] [Google Scholar]
- 34.Novak H., Noy R., Oved K., Segal D., Wels W.S., Reiter Y. Selective antibody-mediated targeting of class I MHC to EGFR-expressing tumor cells induces potent antitumor CTL activity in vitro and in vivo. Int J Cancer. 2007;120(2):329–336. doi: 10.1002/ijc.22168. [DOI] [PubMed] [Google Scholar]
- 35.Fischer C., Munks M.W., Hill A.B., et al. Vaccine-induced CD8 T cells are redirected with peptide-MHC class I-IgG antibody fusion proteins to eliminate tumor cells in vivo. MAbs. 2020;12(1) doi: 10.1080/19420862.2020.1834818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akondy R.S., Monson N.D., Miller J.D., et al. The yellow fever virus vaccine induces a broad and polyfunctional human memory CD8+ T cell response. J Immunol. 2009;183(12):7919–7930. doi: 10.4049/jimmunol.0803903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dufloo J., Guivel-Benhassine F., Buchrieser J., et al. Anti- HIV -1 antibodies trigger non-lytic complement deposition on infected cells. EMBO Rep. 2020;21(2) doi: 10.15252/embr.201949351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mouquet H., Scharf L., Euler Z., et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci U S A. 2012;109(47):E3268–E3277. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Münch J., Rajan D., Schindler M., et al. Nef-mediated enhancement of virion infectivity and stimulation of viral replication are fundamental properties of primate lentiviruses. J Virol. 2007;81(24):13852–13864. doi: 10.1128/JVI.00904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barrett A.D., Teuwen D.E. Yellow fever vaccine—how does it work and why do rare cases of serious adverse events take place? Curr Opin Immunol. 2009;21(3):308–313. doi: 10.1016/j.coi.2009.05.018. [DOI] [PubMed] [Google Scholar]
- 41.Monath T.P., Vasconcelos P.F.C. Yellow fever. J Clin Virol. 2015;64:160–173. doi: 10.1016/j.jcv.2014.08.030. [DOI] [PubMed] [Google Scholar]
- 42.Collins N.D., Barrett A.D.T. Live attenuated yellow fever 17D vaccine: a legacy vaccine still controlling outbreaks in modern day. Curr Infect Dis Rep. 2017;19(3):14. doi: 10.1007/s11908-017-0566-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pulendran B., Oh J.Z., Nakaya H.I., Ravindran R., Kazmin D.A. Immunity to viruses: learning from successful human vaccines. Immunol Rev. 2013;255(1):243–255. doi: 10.1111/imr.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hartmann J., Schüßler-Lenz M., Bondanza A., Buchholz C.J. Clinical development of CAR T cells—challenges and opportunities in translating innovative treatment concepts. EMBO Mol Med. 2017;9(9):1183–1197. doi: 10.15252/emmm.201607485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kongsgaard M., Bassi M.R., Rasmussen M., et al. Adaptive immune responses to booster vaccination against yellow fever virus are much reduced compared to those after primary vaccination. Sci Rep. 2017;7(1) doi: 10.1038/s41598-017-00798-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jenkins M.K., Chu H.H., McLachlan J.B., Moon J.J. On the composition of the preimmune repertoire of T cells specific for peptide–major histocompatibility complex ligands. Annu Rev Immunol. 2010;28(1):275–294. doi: 10.1146/annurev-immunol-030409-101253. [DOI] [PubMed] [Google Scholar]
- 47.Zhu P., Chertova E., Bess J., et al. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc Natl Acad Sci U S A. 2003;100(26):15812–15817. doi: 10.1073/pnas.2634931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chertova E., Bess J.W., Crise B.J., et al. Envelope glycoprotein incorporation, not shedding of surface envelope glycoprotein (gp120/SU), is the primary determinant of SU content of purified human immunodeficiency virus type 1 and simian immunodeficiency virus. J Virol. 2002;76(11):5315–5325. doi: 10.1128/JVI.76.11.5315-5325.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jonker E.F.F., Visser L.G., Roukens A.H. Advances and controversies in yellow fever vaccination. Ther Adv Vaccines. 2013;1(4):144–152. doi: 10.1177/2051013613498954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Monath T.P., Lee C.K., Julander J.G., et al. Inactivated yellow fever 17D vaccine: development and nonclinical safety, immunogenicity and protective activity. Vaccine. 2010;28(22):3827–3840. doi: 10.1016/j.vaccine.2010.03.023. [DOI] [PubMed] [Google Scholar]
- 51.Gaspar L.P., Mendes Y.S., Yamamura A.M.Y., et al. Pressure-inactivated yellow fever 17DD virus: implications for vaccine development. J Virol Methods. 2008;150(1-2):57–62. doi: 10.1016/j.jviromet.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 52.Schäfer B., Holzer G.W., Joachimsthaler A., et al. Pre-clinical efficacy and safety of experimental vaccines based on non-replicating vaccinia vectors against yellow fever. PLoS One. 2011;6(9) doi: 10.1371/journal.pone.0024505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Monath T.P., Fowler E., Johnson C.T., et al. An inactivated cell-culture vaccine against yellow fever. N Engl J Med. 2011;364(14):1326–1333. doi: 10.1056/NEJMoa1009303. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Molecular mechanism of RoVER. Graphics showing the molecular mechanism of RoVER. The biotinylated (4) scFv domain (3) of the RoVER binds specific cell surface antigens (2) on target cells (1) and is joined via streptavidin molecule (5) to a biotin-conjugated (4) HLA-I domain (6) carrying a highly immunogenic vaccine-epitope (7) for specific engagement and activation of vaccine epitope-specific CD8+ T cells (9) through T cell receptor (8) recognition. The graphics is created with BioRender.com.
Figure S2. scFv binding capacity to primary CD19+ B cells or HIV-1-eGFP-infected CD4+ T cells. a) Flow cytometry analysis of scFv-CD19 and scFv-10-1074 binding to CD19+ B cells or HIV-1-eGFP-infected (GFP+) CD4+ T cells. Flow cytometry analysis of scFv-CD19 and scFv-10-1074 binding capacity to CD19+ B cells and HIV-1-eGFP-infected CD4+ T cells, respectively. Controls included unstained cells and cells stained with Streptavidin-BV421. ScFv-10-1074 binding capacity to HIV-1-eGFP-infected cells is shown for gated GFP+ cells. b) Flow cytometry analysis of scFv-10-1074 binding specificity to HIV-1-eGFP-infected or uninfected CD4+ T cells. Flow cytometry analysis of scFv-10-1074 binding capacity to HIV-1-eGFP-infected or uninfected CD4+ T cells. Controls included unstained cells and cells stained with Streptavidin-BV421. ScFv-10-1074 binding capacity is shown for total CD4+ T cells including uninfected (GFP-) cells.
Supplementary table 1. Study participant demographics. Table showing demographic characteristics of the clinical study participants (NCT04083430) receiving YF-17D vaccination from which PBMCs were obtained for functional assays investigating the RoVER technology.
Figure S3. Flow gating strategy for quantification of tetramer+ CD8+ T cells and phenotypic characterization. Flow cytometry gating strategy for quantification of tetramer+ CD8+ T cells in PBMCs isolated from YF-17D vaccinated study participants. Quantification was achieved by tetramer staining and multicolor flow cytometry using a live/dead stain along with fluorophore-conjugated antibodies against CD3 and CD8 T cell markers and using various HLA-I tetramers carrying immunogenic YF peptides. The CD8+ T cells were further characterized in respect to phenotype based on the expression of the surface markers CD45RA and CCR7 into the distinct cellular subsets: Naïve (N) (CCR7+, CD45RA+), Central memory (CM) (CCR7+, CD45RA-), Effector memory (EM) (CCR7-, CD45RA-) and Terminally differentiated (TD) (CCR7-, CD45RA+) CD8+ T cells. Furthermore, back gating of the tetramer+ CD8+ T cells allowed for quantification of the distribution of tetramer+ CD8+ T cells (red) across the phenotypic subsets of total CD8+ T cells (grey).
Figure S4. Cytokine release from in vitro target killing assays using peptides or RoVER. Mesoscale multiplex analyses of supernatants from in vitro cell killing assays. Analysis of IFN-γ, TNF-α or Grz B in supernatant from peptide- or RoVER-mediated killing of B cells by autologous CD8+ T cells obtained pre (n = 3) and post vaccination (21 ± 3 days) (n = 3, LLWNGPMAV; n = 4, SIINFEKL, −/+ RoVER). Data are mean ± SEM.
Figure S5. Flow gating strategy for detection of CD107α degranulation. Flow cytometry gating strategy for quantification of CD107α+ tetramer+ CD8+ T cells in in vitro cell killing assays. Representative analysis of CD107α surface expression following RoVER-mediated killing of B cells cells by autologous CD8+ T cells obtained post vaccination (21 ± 3 days). Quantification was achieved by tetramer staining and multicolor flow cytometry using a live/dead stain along with fluorophore-conjugated antibodies against CD3 and CD8 T cell markers and using various HLA-I tetramers carrying immunogenic YF peptides. A threshold of 3 events in the CD107α gate was set to discriminate the positive signal from the background.